Abstract
Simple PCR and sequencing assays that utilize a single pair of degenerate primers were used to characterize a 438-bp-long DNA fragment internal (sodAint) to the sodA gene encoding the manganese-dependent superoxide dismutase in 19 enterococcal type strains (Enterococcus avium, Enterococcus casseliflavus, Enterococcus cecorum, Enterococcus columbae, Enterococcus dispar, Enterococcus durans, Enterococcus faecalis, Enterococcus faecium, Enterococcus flavescens, Enterococcus gallinarum, Enterococcus hirae, Enterococcus malodoratus, Enterococcus mundtii, Enterococcus pseudoavium, Enterococcus raffinosus, Enterococcus saccharolyticus, Enterococcus seriolicida, Enterococcus solitarius, and Enterococcus sulfureus). Sequence analysis of the sodAint fragments enabled reliable identification of 18 enterococcal species, including E. casseliflavus-E. flavescens and E. gallinarum. The sodAint fragments of E. casseliflavus and E. flavescens were almost identical (99.5% sequence identity), which suggests that they should be associated in a single species. Our results confirm that the sodA gene constitutes a more discriminative target sequence than 16S rRNA gene in differentiating closely related bacterial species.
Enterococci, although not highly virulent microorganisms, have emerged worldwide in the last decade as one of the leading causes of nosocomial bacteremia, surgical wound infections, and urinary tract infections (9, 10, 13, 24). This evolution is mainly due to the appearance of multiresistant strains of enterococci that are resistant to most antibiotics used in treatment (e.g., ampicillin, aminoglycosides, and glycopeptides). Most human enterococcal infections (≥90%) are caused by Enterococcus faecalis and Enterococcus faecium; however, the incidence of other species, such as Enterococcus casseliflavus and Enterococcus gallinarum, could be underestimated because of bacterial misidentification. In clinical laboratories, accurate identification of enterococcal species is required to carry out a proper epidemiologic surveillance and may help in the management of infected patients in case of relapse. This is usually done by testing tolerance to bile esculine and tellurite, growth in 6.5% NaCl broth, and specific carbohydrate utilization (2, 6); by characterizing bacterial motility and pigment production (1); and by using commercial biochemical test systems, such as the API 20STREP or Rapid ID 32 Strep systems. However, these phenotypic methods are often not reliable and the automated systems, such as the Vitek and MicroScan systems, do not properly identify enterococci other than E. faecalis and E. faecium in the absence of additional tests (11). Consequently, several genotypic methods based on the analysis of PCR products derived from selected target DNA have been developed for the species identification of enterococci (3, 14, 22). This includes the determination of the 16S ribosomal DNA (rDNA) sequence (18), a strategy which is now greatly facilitated by the use of universal 16S PCR primers associated with the development of simplified, partially automated, and cost-effective sequencing technologies. However, the interpretation of these data may be complicated by the fact that divergent 16S rDNA sequences may exist within a single organism (23) or, alternatively, that closely related species may have identical 16S rDNA sequences (8), as recently shown in the genera Enterococcus for E. casseliflavus and E. gallinarum (18). To solve this problem, it is possible to use alternative monocopy target sequences which exhibit a higher divergence than that of the 16S rDNA. The sodA gene of the gram-positive cocci which encodes the manganese-dependent superoxide dismutase fulfills these criteria and we recently reported that sequencing of the sodA PCR product with the use of a single pair of degenerate primers constitutes a valuable approach to the genotypic identification of the 29 streptococcal species (20). In this work, the same universal primers (19) were used to construct a sodA database of 19 enterococcal species, including E. casseliflavus and E. gallinarum. We show the usefulness of this library for a rapid sequence-based identification method of enterococcal isolates.
The main characteristics of the bacterial strains used in this study, including the type strains, are listed in Table 1 and 2. Rapid extraction of bacterial genomic DNA was carried out by using the InstaGene Matrix (Bio-Rad, Hercules, Calif.) on cells collected from 2 ml of an overnight culture. The sodA degenerate primers d1 (5′-CCITAYICITAYGAYGCIYTIGARCC-3′) and d2 (5′-ARRTARTAIGCRTGYTCCCAIACRTC-3′) were used to amplify an internal fragment, designated sodAint, representing approximately 85% of their sodA genes. PCRs were performed on a Gene Amp System 9600 instrument (Perkin-Elmer Cetus, Roissy, France) in a final volume of 50 μl containing 250 ng of DNA as template, 0.5 μM each primer, 200 μM each deoxynucleoside triphosphate, and 1 U of AmpliTaq Gold DNA polymerase (Perkin-Elmer) in a 1× amplification buffer (10 mM Tris-HCl [pH 8.3], 50 mM KCl, 1.5 mM MgCl2). The PCR mixtures were denatured (3 min at 95°C) and then subjected to 30 cycles of amplification (60 s of annealing at 37°C, 60 s of elongation at 72°C, and 30 s of denaturation at 95°C) and 7 min at 72°C for the last elongation cycle. A single DNA fragment corresponding to the expected 480-bp amplification product, sodAint, was observed in all cases following agarose gel electrophoresis and ethidium bromide staining (data not shown). PCR products were purified on an S-400 Sephadex column (Pharmacia, Uppsala, Sweden) and directly sequenced on both strands with the oligonucleotides d1 and d2 by using the ABI-PRISM Big Dye terminator sequencing kit on a Genetic ABI-PRISM 310 Sequencer Analyzer (Perkin-Elmer). The cycle sequencing protocol was optimized as follows: the sequencing mixtures were subjected to 40 cycles of amplification consisting of 10 s of denaturation at 96°C, 5 s of annealing at 40°C, and 4 min of elongation at 60°C.
TABLE 1.
Enterococcal type strains used in this study
| Straina | Other designationb | sodAint accession no. |
|---|---|---|
| E. avium CIP 103019 T | ATCC 14025 | AJ387906 |
| E. casseliflavus CIP 103018 T | ATCC 25788 | AJ387907 |
| E. cecorum CIP 103676 T | ATCC 43198 | AJ387908 |
| E. columbae CIP 103675 T | ATCC 51263 | AJ387909 |
| E. dispar CIP 103646 T | ATCC 51266 | AJ387910 |
| E. durans CIP 55.125 T | ATCC 19432 | AJ387911 |
| E. faecalis CIP 103015 T | ATCC 19433 | AJ387912 |
| E. faecium CIP 103014 T | ATCC 19434 | AJ387913 |
| E. flavescens CIP 103525 T | ATCC 49996 | AJ387914 |
| E. gallinarum CIP 103013 T | ATCC 49573 | AJ387915 |
| E. hirae CIP 53.48 T | ATCC 8043 | AJ387916 |
| E. malodoratus CIP 103012 T | ATCC 43197 | AJ387917 |
| E. mundtii CIP 103010 T | ATCC 43186 | AJ387918 |
| E. pseudoavium CIP 103647 T | ATCC 49372 | AJ387919 |
| E. saccharolyticus CIP 103246 T | ATCC 43076 | AJ387920 |
| E. solitarius CIP 103330 T | NCTC 12193 | AJ387921 |
| E. raffinosus CIP 103329 T | ATCC 49427 | AJ387922 |
| E. seriolicida CIP 104369 T | ATCC 49156 | AJ387923 |
| E. sulfureus CIP 104373 T | DSM 6905 | AJ387924 |
| L. garvieae CIP 102507 T | DSM 20684 | AJ387925 |
CIP, Collection de l'Institut Pasteur.
ATCC, American Type Culture Collection; DSM, Deutsche Sammlung von Mikrooganismen und Zelkulturen; NCTC, National Collection of Type Cultures.
TABLE 2.
Identification of various enterococcal strains by sequencing the sodAint fragment
| Strain | Relevant characteristic(s)a | Bacterial speciesb | Accession no. |
|---|---|---|---|
| NEM1616 | E. faecalis; vanA | E. faecalis (99.5) | AJ387927 |
| NEM1617 | E. faecalis; vanA | E. faecalis (98.6) | AJ387928 |
| NEM1618 | Enterococcus sp. | E. durans (99.3) | AJ387929 |
| NEM1619 | E. hirae | E. durans (99.5) | AJ387930 |
| NEM1620 | Enterococcus sp. | E. durans (99.1) | AJ387931 |
| NEM1621 | E. hirae | E. hirae (99.8) | AJ387932 |
| NEM1622 | Enterococcus sp. | E. hirae (99.5) | AJ387933 |
| NEM1623 | E. casseliflavus | E. casseliflavus (99.1) | AJ387934 |
| NEM1624 | E. faecium; vanB | E. faecium (99.5) | AJ387935 |
| NEM1625 | E. faecium; vanA | E. faecium (100) | AJ387936 |
| NEM1626 | E. faecium; vanB | E. faecium (99.8) | AJ387937 |
| NEM1627 | E. faecium; multiply resistant strain | E. faecium (99.8) | AJ387938 |
| NEM1628 | E. faecium; multiply resistant strain | E. faecium (99.8) | AJ387939 |
| NEM1629 | Enterococcus sp. | E. gallinarum (98.6) | AJ387940 |
| NEM1630 | E. avium | E. avium (100) | AJ387941 |
Bacterial strains were all clinical isolates from our collection which were identified by using conventional microbiological tests and the ID 32 Strep System (API-bioMérieux). Presence of vanA (NEM1616, NEM1617, and NEM1625) and vanB (NEM1624 and NEM1626) was determined by PCR with specific primers (3).
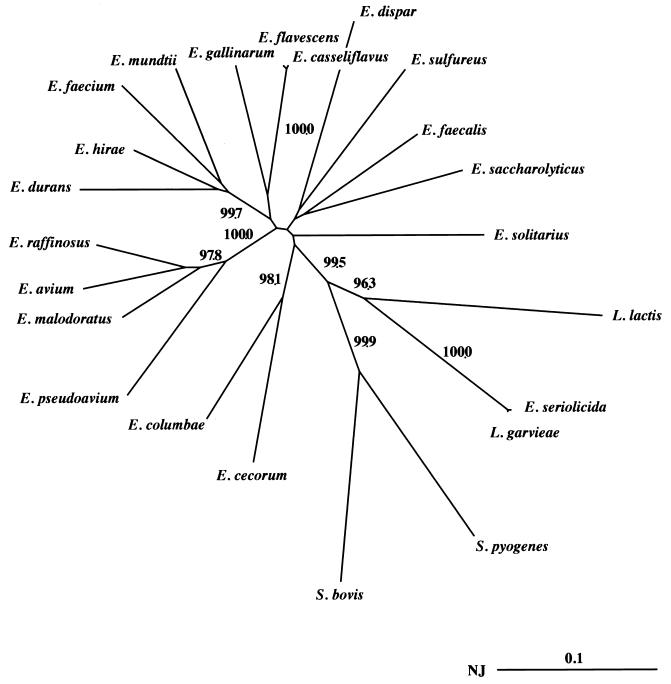
The species identification was based on the phylogenic position of the sodAint fragment of the strain studied relative to those of the type strains, as shown in Fig. 1. Numbers in parentheses indicate the percentage of identity of the sodAint fragment with that of the corresponding type strains.
The nucleotide sequences of the sodAint fragments from the type strains of Enterococcus avium, E. casseliflavus, Enterococcus cecorum, Enterococcus columbae, Enterococcus dispar, Enterococcus durans, E. faecalis, E. faecium, Enterococcus flavescens, E. gallinarum, Enterococcus hirae, Enterococcus malodoratus, Enterococcus mundtii, Enterococcus pseudoavium, Enterococcus raffinosus, Enterococcus saccharolyticus, Enterococcus seriolicida, Enterococcus solitarius, Enterococcus sulfureus, and Lactococcus garvieae were determined (Table 1). We assumed that the PCR products sequenced were actual sodAint fragments, since the corresponding deduced polypeptides all contained the amino acids characteristic of the manganese-dependent superoxide dismutase (16, 17) at the expected positions (data not shown). Multiple alignment of these sodAint DNA sequences plus those of L. garvieae (Table 1), Lactococcus lactis (19), Streptococcus bovis (20), and Streptococcus pyogenes (20) was carried out with the Clustal X program (12), and an unrooted phylogenetic tree was constructed by the neighbor-joining (NJ) method (21). The sequences of the degenerate oligonucleotides d1 and d2 and alignment gaps were not taken into consideration for calculations. The reliability of the tree nodes was evaluated by calculating the percentage of 1,000 bootstrap resamplings that support each topological element. Only the nodes having a bootstrap value greater than 95% are shown in Fig. 1, since this critical value could be used to define the monophyly of a clade of related organisms (7). This analysis revealed that, as expected, the members of the genus Enterococcus, with the exception of E. seriolicida, were clustered within a clade supported by 99.5% of the bootstrap replicates. The sodAint sequences of E. seriolicida and of L. garvieae were almost identical (99.5% sequence identity) and were clustered with that of L. lactis within a clade supported by 96.3% of the bootstrap confidence limit (Table 3 and Fig. 1). These results are consistent with the redesignation of E. seriolicida as L. garvieae (4). The phylogenetic tree representing the enterococcal sodAint sequences (Fig. 1) has the same topology as the NJ tree constructed from the analysis of their 16S rDNA sequences (18). It is worth noting that the sodAint sequences of the E. casseliflavus and E. gallinarum type strains displayed 16.9% sequence divergence, a value similar to the 19.7% sequence divergence observed between the ddl genes encoding the d-Ala–d-Ala ligases in these species (5). These results do not support the suggestion that E. casseliflavus and E. gallinarum comprise a single species (18). By contrast, the fact that the 16S rDNA sequence (18) and the ddl (15), vanC (3), and sodAint (Table 3) genes of the E. casseliflavus and E. flavescens type strains were almost identical (99.9, 99.5, 96, and 98% sequence identity, respectively) suggests that they should be associated in a single species.
FIG. 1.
Phylogenetic unrooted tree showing relationships among the sodAint fragments from various enterococcal type strains. The tree was established from an analysis of the sequences listed in Table 1 by using the NJ method. The sodAint sequences of L. lactis, L. garvieae, S. bovis, and S. pyogenes type strains were included in this work. The value on each branch is the estimated confidence limit (expressed as a percentage) for the position of the branch as determined by bootstrap analysis. Only bootstrap values greater than 95%, which were considered significant, are indicated. The scale bar (NJ distance) represents a 10% difference in nucleotide sequences.
TABLE 3.
Identity matrix based on pairwise comparisons of sodAint fragments of enterococcal type strains
| Strain no. | Speciesa | % Identity with:
|
|||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 | 16 | 17 | 18 | 19 | ||
| 1 | E. avium | 74.0 | 67.4 | 71.9 | 70.3 | 70.3 | 73.7 | 69.9 | 74.2 | 75.1 | 75.6 | 71.0 | 80.1 | 72.6 | 74.0 | 85.4 | 87.9 | 60.7 | 67.1 |
| 2 | E. casseliflavus | 66.4 | 70.8 | 72.6 | 72.4 | 77.9 | 72.4 | 99.5 | 83.1 | 78.5 | 77.4 | 71.5 | 73.5 | 74.0 | 76.7 | 76.7 | 66.4 | 75.6 | |
| 3 | E. cecorum | 78.8 | 72.4 | 66.0 | 68.7 | 66.2 | 66.9 | 67.4 | 70.3 | 64.2 | 65.5 | 71.7 | 65.8 | 67.6 | 66.0 | 62.8 | 68.7 | ||
| 4 | E. colombae | 69.4 | 68.9 | 71.7 | 69.2 | 70.8 | 72.6 | 73.1 | 68.7 | 69.6 | 72.1 | 72.8 | 73.3 | 71.9 | 67.1 | 69.2 | |||
| 5 | E. dispar | 70.3 | 77.4 | 68.7 | 72.8 | 72.1 | 73.5 | 72.8 | 70.5 | 71.0 | 69.4 | 72.1 | 70.3 | 62.6 | 74.9 | ||||
| 6 | E. durans | 73.1 | 81.3 | 72.4 | 76.3 | 84.9 | 80.1 | 69.6 | 72.8 | 70.5 | 71.5 | 73.3 | 62.1 | 73.7 | |||||
| 7 | E. faecalis | 72.6 | 78.3 | 77.6 | 77.9 | 72.4 | 71.2 | 78.8 | 73.5 | 77.9 | 75.1 | 67.1 | 76.5 | ||||||
| 8 | E. faecium | 72.4 | 77.2 | 83.1 | 81.7 | 67.4 | 72.6 | 69.4 | 71.9 | 71.9 | 62.1 | 72.1 | |||||||
| 9 | E. flavescens | 83.1 | 78.3 | 77.2 | 72.1 | 72.8 | 74.4 | 77.2 | 76.9 | 65.8 | 74.9 | ||||||||
| 10 | E. gallinarum | 80.8 | 78.5 | 73.3 | 76.0 | 73.5 | 77.4 | 75.1 | 66.2 | 76.7 | |||||||||
| 11 | E. hirae | 83.6 | 71.5 | 73.3 | 73.5 | 77.4 | 76.7 | 63.5 | 75.3 | ||||||||||
| 12 | E. mundtii | 70.8 | 69.4 | 69.6 | 71.9 | 73.5 | 63.9 | 74.4 | |||||||||||
| 13 | E. pseudoavium | 70.5 | 69.2 | 81.7 | 80.4 | 62.8 | 65.3 | ||||||||||||
| 14 | E. saccharolyticus | 72.4 | 75.1 | 71.0 | 62.1 | 76.7 | |||||||||||||
| 15 | E. solitarius | 74.2 | 72.8 | 64.8 | 72.1 | ||||||||||||||
| 16 | E. malodoratus | 87.9 | 63.5 | 70.5 | |||||||||||||||
| 17 | E. raffinosus | 64.6 | 67.6 | ||||||||||||||||
| 18 | E. seriolicida | 61.6 | |||||||||||||||||
| 19 | E. sulfureus | ||||||||||||||||||
The main characteristics of the strains are listed in Table 1.
The phylogenetic tree shown in Fig. 1 revealed the presence of two major clusters within the enterococcal species which we have designated the faecium group (E. faecium, E. durans, E. hirae, and E. mundtii) and the avium group (E. avium, E. malodoratus, E. pseudoavium, and E. raffinosus). Within each group, the 16S rDNA sequences exhibited more than 99% sequence identity (18), whereas the highest percentage of similarity found between two sodAint sequences was 87.9% (Table 3). These results confirm that the gene sodA constitutes a more discriminative target sequence than 16S RNA in differentiating closely related bacterial species.
Fifteen enterococcal isolates were identified by using conventional microbiological tests, Rapid ID 32 Strep, and the sodAint systems (Table 2). In all cases, the sodAint sequences of the isolates displayed less than 1.5% divergence from the corresponding type strain. For 10 strains (NEM1616, NEM1617, NEM1621, NEM1623, NEM1624, NEM1625, NEM1626, NEM1627, NEM1628, and NEM1630), the two methods gave the same results. Four isolates (NEM1618, NEM1620, NEM1622, and NEM1629) were identified at the species level with the sodAint system but not with conventional microbiological tests or the Rapid ID 32 Strep system. The remaining isolate, NEM1619, was identified with the Rapid ID 32 Strep system as E. hirae but was identified with the sodAint system as E. durans (Table 2). The reliability of the molecular identification of NEM1164 was based on the fact that its sodAint fragment displays 99.5 and 85% sequence identity with the type strains of E. durans and E. hirae, respectively.
In conclusion, we have determined the sodAint sequences of the type strains of E. avium, E. casseliflavus, E. flavescens, E. cecorum, E. columbae, E. dispar, E. durans, E. faecalis, E. faecium, E. gallinarum, E. hirae, E. malodoratus, E. mundtii, E. pseudoavium, E. raffinosus, E. saccharolyticus, E. seriolicida, E. solitarius, and E. sulfureus and demonstrated the usefulness of this database for the species identification of enterococcal isolates. The identification method presented in this study is not accessible to routine clinical microbiology laboratories, but it may become the “gold standard” technique in reference and large research hospital laboratories for epidemiologic purposes and/or identifying problematic strains.
Nucleotide sequence accession number. Representative nucleotide sequences have been submitted to the EMBL database and have been given accession no. AJ387906 to AJ387925 and AJ387927 to AJ387941.
Acknowledgments
We thank C. Bizet for the gift of enterococcal type strains (CIP), O. Gaillot for the gift of clinical isolates, O. Gaillot and S. Nair for a critical reading of the manuscript, and P. Berche for his interest in this work.
This work was supported by the Institut Pasteur and by the University of Paris V.
REFERENCES
- 1.Cartwright C P, Stock F, Fahle G A, Gill V J. Comparison of pigment production and motility tests with PCR for reliable identification of intrinsically vancomycin-resistant enterococci. J Clin Microbiol. 1995;33:1931–1933. doi: 10.1128/jcm.33.7.1931-1933.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Devriese L A, Pot B, Kersters K, Lauwers S, Haesebrouck F. Acidification of methyl-alpha-d-glucopyranoside: a useful test to differentiate Enterococcus casseliflavus and Enterococcus gallinarum from Enterococcus faecium species group and from Enterococcus faecalis. J Clin Microbiol. 1996;34:2607–2608. doi: 10.1128/jcm.34.10.2607-2608.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dutka-Malen S, Evers S, Courvalin P. Detection of glycopeptide resistance genotypes and identification to the species level of clinically relevant enterococci by PCR. J Clin Microbiol. 1995;33:24–27. doi: 10.1128/jcm.33.1.24-27.1995. . (Erratum, 33:1434.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Eldar A, Ghittino C, Asanta L, Bozzetta E, Goria M, Prearo M, Bercovier H. Enterococcus seriolicida is a junior synonym of Lactococcus garvieae, a causative agent of septicemia and meningoencephalitis in fish. Curr Microbiol. 1996;32:85–88. doi: 10.1007/s002849900015. [DOI] [PubMed] [Google Scholar]
- 5.Evers S, Casadewall B, Charles M, Dutka-Malen S, Galimand M, Courvalin P. Evolution of structure and substrate specificity in d-alanine:d-alanine ligases and related enzymes. J Mol Evol. 1996;42:706–712. doi: 10.1007/BF02338803. [DOI] [PubMed] [Google Scholar]
- 6.Facklam R R, Collins M D. Identification of Enterococcus species isolated from human infections by a conventional test scheme. J Clin Microbiol. 1989;27:731–734. doi: 10.1128/jcm.27.4.731-734.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Felsenstein J. Confidence limits on phylogeny and approach using the bootstrap. Evolution. 1985;39:783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- 8.Fox G E, Wisotzkey J D, Jurtshuk P., Jr How close is close: 16S rRNA sequence identity may not be sufficient to guarantee species identity. Int J Syst Bacteriol. 1992;42:166–170. doi: 10.1099/00207713-42-1-166. [DOI] [PubMed] [Google Scholar]
- 9.Hunt C P. The emergence of enterococci as a cause of nosocomial infection. Br J Biomed Sci. 1998;55:149–156. [PubMed] [Google Scholar]
- 10.Huycke M M, Sahm D F, Gilmore M S. Multiple-drug resistant enterococci: the nature of the problem and an agenda for the future. Emerg Infect Dis. 1998;4:239–249. doi: 10.3201/eid0402.980211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Iwen P C, Kelly D M, Linder J, Hinrichs S H. Revised approach for identification and detection of ampicillin and vancomycin resistance in Enterococcus species by using MicroScan panels. J Clin Microbiol. 1996;34:1779–1783. doi: 10.1128/jcm.34.7.1779-1783.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jeanmougin F, Thompson J D, Gouy M, Higgins D G, Gibson T J. Multiple sequence alignment with Clustal X. Trends Biochem Sci. 1998;23:403–405. doi: 10.1016/s0968-0004(98)01285-7. [DOI] [PubMed] [Google Scholar]
- 13.Moellering R C., Jr Vancomycin-resistant enterococci. Clin Infect Dis. 1998;26:1196–1199. doi: 10.1086/520283. [DOI] [PubMed] [Google Scholar]
- 14.Monstein H J, Quednau M, Samuelsson A, Ahrne S, Isaksson B, Jonasson J. Division of the genus Enterococcus into species groups using PCR-based molecular typing methods. Microbiology. 1998;144:1171–1179. doi: 10.1099/00221287-144-5-1171. [DOI] [PubMed] [Google Scholar]
- 15.Navarro F, Courvalin P. Analysis of genes encoding d-alanine–d-alanine ligase-related enzymes in Enterococcus casseliflavus and Enterococcus flavescens. Antimicrob Agents Chemother. 1994;38:1788–1793. doi: 10.1128/aac.38.8.1788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Parker M W, Balke C C F. Crystal structure of manganese superoxide dismutase from Bacillus stearothermophilus at 2.4 A resolution. J Mol Biol. 1988;199:649–661. doi: 10.1016/0022-2836(88)90308-7. [DOI] [PubMed] [Google Scholar]
- 17.Parker M W, Blake C C F. Iron- and manganese-containing superoxide dismutases can be distinguished by analysis of their primary structures. FEBS Lett. 1988;229:377–382. doi: 10.1016/0014-5793(88)81160-8. [DOI] [PubMed] [Google Scholar]
- 18.Patel R, Piper K E, Rouse M S, Steckelberg J M, Uhl J R, Kohner P, Hopkins M K, Cockerill III F R, Kline B C. Determination of 16S rRNA sequences of enterococci and application to species identification of nonmotile Enterococcus gallinarum isolates. J Clin Microbiol. 1998;36:3399–3407. doi: 10.1128/jcm.36.11.3399-3407.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Poyart C, Berche P, Trieu-Cuot P. Characterization of superoxide dismutase genes from Gram-positive bacteria by polymerase chain reaction using degenerate primers. FEMS Microbiol Lett. 1995;131:41–45. doi: 10.1016/0378-1097(95)00232-t. [DOI] [PubMed] [Google Scholar]
- 20.Poyart C, Quesne G, Coulon S, Berche P, Trieu-Cuot P. Identification of streptococci to species level by sequencing the gene encoding the manganese-dependent superoxide dismutase. J Clin Microbiol. 1998;36:41–47. doi: 10.1128/jcm.36.1.41-47.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 22.Tyrrell G J, Bethune R N, Willey B, Low D E. Species identification of enterococci via intergenic ribosomal PCR. J Clin Microbiol. 1997;35:1054–1060. doi: 10.1128/jcm.35.5.1054-1060.1997. . (Erratum, 35:2434.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ueda K, Seki T, Kudo T, Yoshida T, Kataoka M. Two distinct mechanisms cause heterogeneity of 16S rRNA. J Bacteriol. 1999;181:78–82. doi: 10.1128/jb.181.1.78-82.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Woodford N. Glycopeptide-resistant enterococci: a decade of experience. J Med Microbiol. 1998;47:849–862. doi: 10.1099/00222615-47-10-849. [DOI] [PubMed] [Google Scholar]