Abstract
Mink lung cells were more sensitive than the commonly used MDCK or pRhMK cells for rapid detection of influenza virus A from clinical specimens. Mixed Mv1Lu and A549 cells in a single shell vial were synergistic for detection of influenza virus A and were as sensitive as individual cells for detection of other respiratory viruses.
Isolation and identification of respiratory viruses typically require the use of multiple cell lines, including primary monkey kidney (PMK) cells, as well as a battery of continuous cell lines, most often MRC-5, HEp-2, A549, WI-38, MDCK, and/or HEK.
This report describes the use of mink lung (Mv1Lu) cells, a continuous cell line, for rapid detection of influenza viruses. It also introduces a mixture of two continuous cell lines (Mv1Lu and A549) that broaden the susceptibility profile to include all of the respiratory viruses.
Clinical specimens of nasopharyngeal aspirates were collected by insertion of a 5-French nasogastric feeding tube into the nasopharynx and aspiration with a syringe. The aspirates were expelled into viral transport medium (M4-RT; Micro Test, Inc., Liburn, Ga.) and then delivered to the laboratory. The specimens were vigorously vortexed for 10 s, followed by centrifugation at 1,400 × g for 5 min to pellet the epithelial cells. The cell pellet was washed twice with phosphate-buffered saline and tested by indirect immunofluorescent assay (IFA). The supernatant was used for shell vial culture. A processed specimen of 0.2 ml was inoculated into each shell vial of MDCK cells, A549 cells, HEp-2 cells, Mv1Lu cells, a mixture of Mv1Lu and A549 cells (Diagnostic Hybrids, Inc., Athens, Ohio), and pRhMK cells (Viromed, Minneapolis, Minn.). The vials were recapped and centrifuged at 700 × g for 45 to 60 min at 24°C. After overnight incubation at 36°C in a CO2 incubator, the coverslips of cells were fixed, removed from the shell vial, and immunostained with respiratory IFA kits (Intracel, Inc., division of Bartels Immunodiagnostics, Bellevue, Wash.).
Twenty specimens that originally tested positive by IFA were inoculated into shell vials in OPTI-MEM without serum during overnight incubation. After incubation, the cells were fixed and then stained with anti-influenza virus A monoclonal antibody, and the number of positive cells was counted (data not shown). Cells that stained positive were identified in 20 of 20 specimens in Mv1Lu and pRhMK cells, while MDCK cells detected only 16 of 20 positive specimens. Although pRhMK cells detected all positive specimens, the number of positive cells was generally much lower than with Mv1Lu cells (data not shown). MDCK cells were substantially less sensitive than Mv1Lu and pRhMK cells.
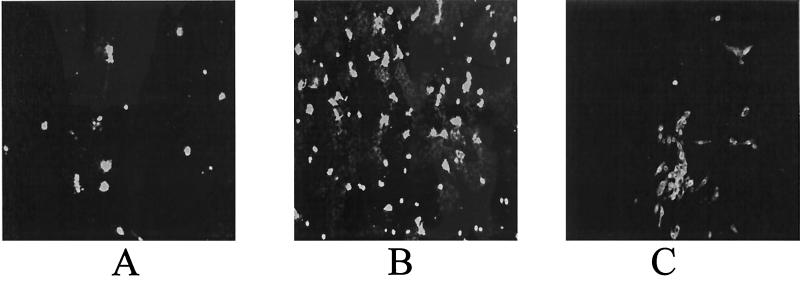
A549 reportedly has been highly sensitive for adenovirus detection (2, 5, 6) and to be equally sensitive as Hep-2 for rapid detection of respiratory syncytial virus (RSV) (3). Thus, A549 cells were mixed with Mv1Lu cells in a single vial. The mixed cells were compared with individual vials of Mv1Lu and pRhMK cells for rapid detection of influenza virus A. Twenty IFA-tested positive frozen specimens were inoculated into each vial and detected as described above. Mv1Lu and mixed A549-Mv1Lu cells detected 20 of 20 specimens, and pRhMK cells detected 16 of 20 specimens (Table 1). Interestingly, mixed A549-Mv1Lu cells showed more positive cells than Mv1Lu cells alone in 17 of 20 specimens. The average numbers of positive cells from 20 clinical specimens were 827 for Mv1Lu (range, 1 to 3,500), 1,819 for A549-Mv1Lu (range, 1 to 11,000), and 527 for pRhMK (range, 0 to 2,336). A549 is only moderately sensitive for influenza virus A detection (1), and in the mixed cells, most of the positive cells were the Mv1Lu cells and cells at the junction of the A549 and Mv1Lu cells. It is very unlikely, but some strains of influenza virus A might grow extremely well in A549 cells. The reason for the synergistic effect is unclear. Representative samples of positive fluorescent-staining cells of a clinical specimen from three different cell types are shown in Fig. 1. Typically, the Mv1Lu and mixed A549-Mv1Lu cells showed evenly scattered individual positive cells over the entire coverslip, while pRhMK cells showed small foci of positive cells. A typical focus in pRhMK cells was shown in Fig. 1, but such foci only appeared every several fields.
TABLE 1.
Comparison of positive clinical specimens identified in different cells by shell vial
| Virus | No. of specimens positive
|
||||
|---|---|---|---|---|---|
| A549-Mv1Lu | A549 | Mv1Lu | HEp-2 | pRhMK | |
| Influenza virus A | 20 | NTa | 20 | NT | 16 |
| Influenza virus B | 27 | 27 | 27 | NT | 27 |
| Adenovirus | 16 | 16 | NT | NT | NT |
| RSV | 12 | 12 | 12 | 13 | NT |
| Parainfluenza virus 1 | 5 | 5 | 5 | NT | 5 |
| Parainfluenza virus 2 | 2 | 2 | 2 | NT | 2 |
| Parainfluenza virus 3 | 12 | 12 | 12 | NT | 12 |
NT, not tested.
FIG. 1.
IFA staining of influenza virus A-infected cells from a clinical specimen. (A) Mv1Lu cells. (B) Mixed A549-Mv1Lu cells. (C) pRhMK cells. Magnification, ×100.
To assess whether mixed A549-Mv1Lu cells can support the rapid detection of other respiratory viruses as individual cells in one vial, specimens containing other respiratory viruses were evaluated as described above. Twenty-seven influenza virus B, 5 parainfluenza virus type 1, 2 parainfluenza virus type 2, and 12 parainfluenza virus type 3 IFA-tested positive specimens were evaluated in A549, Mv1Lu, A549-Mv1Lu, and pRhMK cells. Eighteen previously identified adenovirus-positive (by IFA) clinical specimens were compared in A549 and mixed A549-Mv1Lu cells. Sixteen IFA-tested respiratory syncytial virus (RSV)-positive specimens were also compared in A549, mixed A549-Mv1Lu, Mv1Lu, and Hep-2 cells. The results are summarized in Table 1. Surprisingly, influenza virus B and each of three parainfluenza virus types were detected in all of the four cell preparations. For adenovirus, both cell preparations missed the same two specimens, indicating that A549 in mixed cells is as sensitive as A549 alone for detection of adenovirus, whereas adenovirus was only detected in A549 cells, but not in Mv1Lu cells. For RSV, the Hep-2 cells detected one additional positive specimen that had only one positive cell. Two IFA-positive adenovirus specimens and three IFA-positive RSV specimens were missed by this overnight culture detection method. This may due to the fact that infected cells in the samples were removed for the IFA test and/or the fact that a longer incubation period may be required to increase the rate of detection for these two viruses. Taken together, these data indicate that mixed A549-Mv1Lu cells are comparable with the sensitive individual cells for detection of multiple respiratory viruses.
Huang and Turchek reported the high sensitivity of Mv1Lu cells for rapid culture detection of influenza viruses in a shell vial format (Y. T. Huang, Letter, Pan Am. Soc. Clin. Virol. 25:1–4, 1998; Y. T. Huang and B. M. Turchek, Fourteenth Annu. Clin. Virol. Symp., abstr. M15, 1998). Recently, Schultz-Cherry et al. compared Mv1Lu cells to MDCK cells and determined that different subtypes of influenza viruses A and B were able to replicate on Mv1Lu cells, and virus titers produced on both cell lines were comparable, with the exception that Mv1Lu cells supported the replication of more subtypes of influenza virus A than MDCK cells (4). They suggest that Mv1Lu cells may be useful for culturing influenza viruses for vaccine production and virus isolation.
In this report, we demonstrated that Mv1Lu is highly sensitive for rapid detection of influenza virus A. The combination of A549 and Mv1Lu cells in a single vial not only broadens the virus susceptibility to include other respiratory viruses, but also enhances the number of positive cells for detection of influenza virus A. Further examination of mixed A549-Mv1Lu cells with other respiratory viruses is warranted. Use of mixed Mv1Lu and A549 cells in a rapid shell vial format has the potential to replace the need for pRhMK cell culture for detection of respiratory viruses.
Acknowledgments
This study was supported by a research fund from Pathology Associates of University Hospitals of Cleveland, Inc., Cleveland, Ohio.
We thank Tandalea Harney for preparation of the manuscript.
REFERENCES
- 1.Brumback B G, Wade C D. Simultaneous rapid culture for four respiratory viruses in the same cell monolayer using a differential multicolored fluorescent confirmatory stain. J Clin Microbiol. 1996;34:798–801. doi: 10.1128/jcm.34.4.798-801.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mahafzah A M, Landry M L. Evaluation of immunofluorescent reagents, centrifugation, and conventional cultures for the diagnosis of adenovirus infection. Diagn Microbiol Infect Dis. 1989;12:407–411. doi: 10.1016/0732-8893(89)90111-9. [DOI] [PubMed] [Google Scholar]
- 3.Rabalais G P, Stout G G, Ladd K L, Cost K M. Rapid diagnosis of respiratory viral infections by using a shell vial assay and monoclonal antibody pool. J Clin Microbiol. 1992;30:1505–1508. doi: 10.1128/jcm.30.6.1505-1508.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schultz-Cherry S, Dybdahl-Sissoko N, McGregor M, Hinshaw V S. Mink lung epithelial cells: unique cell line that supports influenza A and B virus replication. J Clin Microbiol. 1998;36:3718–1720. doi: 10.1128/jcm.36.12.3718-3720.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Smith C D, Craft D W, Shiromoto R S, Yan P O. Alternative cell line for virus isolation. J Clin Microbiol. 1986;24:265–268. doi: 10.1128/jcm.24.2.265-268.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Woods G L, Young A. Use of A-549 cells in a clinical virology laboratory. J Clin Microbiol. 1988;26:1026–1028. doi: 10.1128/jcm.26.5.1026-1028.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]