Abstract
Objectives
It has been suggested that smoking affects the oral microbiome, but its effects on sites other than the subgingival microbiome remain unclear. This study investigated the composition of the salivary and tongue bacterial communities of smokers and nonsmokers in periodontally healthy adults.
Methods
The study population included 50 healthy adults. The bacterial composition of resting saliva and the tongue coating was identified through barcoded pyrosequencing analysis of the 16S rRNA gene. The Brinkman index (BI) was used to calculate lifetime exposure to smoking. The richness and diversity of the microbiome were evaluated using the t‐test. Differences in the proportions of bacterial genera between smokers and nonsmokers were evaluated using the Mann–Whitney U test. The quantitative relationship between the proportions of genera and the BI was evaluated using Pearson's correlation analysis.
Results
The richness and diversity of the oral microbiome differed significantly between saliva and the tongue but not between smokers and nonsmokers. The saliva samples from smokers were enriched with the genera Treponema and Selenomonas. The tongue samples from smokers were enriched with the genera Dialister and Atopobium. The genus Cardiobacterium in saliva, and the genus Granulicatella on the tongue, were negatively correlated with BI values. On the other hand, the genera Treponema, Oribacterium, Dialister, Filifactor, Veillonella, and Selenomonas in saliva and Dialister, Bifidobacterium, Megasphaera, Mitsuokella, and Cryptobacterium on the tongue were positively correlated with BI values.
Conclusions
The saliva and tongue microbial profiles of smokers and nonsmokers differed in periodontally healthy adults. The genera associated with periodontitis and oral malodor accounted for high proportions in saliva and on the tongue of smokers without periodontitis and were positively correlated with lifetime exposure to smoking. The tongue might be a reservoir of pathogens associated with oral disease in smokers.
Keywords: 16S rRNA gene sequencing, cigarette smoking, oral microbiome, saliva, tongue
1. INTRODUCTION
Periodontal disease is one of the most common chronic diseases and the cause of tooth loss among adults (Frencken et al., 2017). Tobacco smoking is recognized as the most important environmental risk factor for periodontal disease. A recent systematic review has reported that tobacco smoking increases the risk of periodontal disease by 85% (Leite et al., 2018). Smokers have deeper probing depths, greater attachment loss, more bone resorption, and fewer teeth than nonsmokers (Johnson & Hill, 2004). When implants are used, being a smoker significantly affects the failure rate, the risk of postoperative infection, and marginal bone loss (Chrcanovic et al., 2015). Cigarette smoking has a variety of effects on host‐pathogen interactions in the oral cavity, such as reduction of cell‐mediated and humoral immune responses, promotion of infection with microbial pathogens, interference with antimicrobial therapies, and strengthening of antimicrobial resistance (Barbour et al., 1997; Bateson, 1993; Epstein et al., 1993; Feldman & Anderson, 2013). Furthermore, with the development of analytical technology, many studies have been conducted on the effects of cigarette smoking on the oral microbiome. The microbiome of gingival crevicular fluid has been compared between smokers and nonsmokers in healthy individuals, patients with chronic periodontal disease, and in patients with peri‐implantitis (Mason et al., 2015; Moon et al., 2015; Tsigarida et al., 2015). Those studies have reported differences in the gingival crevicular microbiome between smokers and nonsmokers. A study that investigated the changes in microbial composition associated with stopping smoking reported that the subgingival microbial community was recolonized by a greater number of health‐associated species following nonsurgical periodontal therapy and cessation of smoking (Delima et al., 2010).
Cigarette smoke affects not only the gingival sulcus but also bacteria on the tongue, buccal mucosa, and plaque. However, the effects of smoking on sites other than the gingival crevicular fluid have rarely been investigated, except in studies conducted on oral wash samples (Wu et al., 2016) and buccal mucosa (Yu et al., 2017). Investigating the effect of smoking on the microbial ecosystems in various sites of the oral cavity is important for developing an oral health strategy. The tongue occupies a large area in the oral cavity and has a different microbial community than the periodontal pockets and dental plaque (Simón‐Soro et al., 2013). The tongue coating is an important cause of oral malodor (Scully et al., 1997), and differences in tongue microbiomes with and without oral malodor have been reported (Bernardi et al., 2020). However, the effect of cigarette smoking on the tongue microbiome has not been investigated.
Therefore, this study investigated the differences in the microbial composition of the tongue directly exposed to cigarette smoke in smokers with that of nonsmokers. As the tongue microbiome is affected by periodontal disease (Tanaka et al., 2004), healthy young people without periodontal disease were targeted. In addition, the study subjects did not have periodontal pockets; therefore, resting saliva was sampled to investigate the microbiota existing around the gingival sulcus.
2. MATERIALS AND METHODS
2.1. Study population
The study population comprised 50 healthy volunteers (39 men and 11 women; mean age 25.6 ± 2.1; range 23–31 years). Dental and health checkups were conducted before collecting samples. Periodontal status was assessed using the community periodontal index probe. Participants scored 0 for both bleeding on probing and probing pocket depth based on the criteria of the WHO (World Health Organization, 2013). None of the dental or health checkups detected any problems in the participants that required treatment. No participant had taken antibiotics within the prior 3 months. The study was approved by, and conducted under the supervision of, the Ethics Committee for Clinical Research of Fukuoka Gakuen (Approval No. 249). All participants understood the purpose and content of the study and provided written informed consent to participate.
The cigarette‐smoking status of the participants was determined using a self‐completed questionnaire. None of the study subjects used electronic cigarettes or smokeless cigarettes. Smoking status was defined in the questionnaire as “smoker”, an individual who had smoked ≥100 cigarettes in total after starting smoking, and “nonsmoker”, an individual who had either never smoked or had smoked <100 cigarettes in total after starting smoking (Hanioka et al., 2007). The Brinkman index (BI), which is defined as (number of cigarettes per day) × (number of years for which a person smoked) (Brinkman & Coates Jr., 1963), was used to calculate lifetime exposure to smoking.
2.2. Sampling
Participants were asked to collect 3 ml of resting saliva in a disposable tube at 3:30 pm at least 2.5 h after smoking, eating, or brushing their teeth. The 1‐ml whole saliva samples were pelleted through centrifugation and stored at −30°C until use. Subsequently, tongue samples were collected using the MS Tongue Cleaner (Morita, Osaka, Japan), suspended in 10 ml phosphate‐buffered saline, pelleted by centrifugation, and stored at −30°C until use.
2.3. 16S rRNA gene sequencing analysis
DNA was extracted as described previously (Takeshita et al., 2010). Three of the saliva samples did not have sufficient bacterial DNA; therefore, 47 saliva and 50 tongue samples were investigated in this study. The V3–V4 regions of the 16S rRNA gene were amplified and sequenced on a 454 Life Sciences Genome Sequencer FLX instrument (Roche, Basel, Basel, Switzerland) from Takara Bio Inc. (Otsu, Japan).
Sequences were excluded from the analysis if they were shorter than 240 bases, and were subsequently removed if they did not include the correct primer sequence. The remaining sequences were assigned to each subject by examining the six‐base barcode sequence. UCHIME v6.1.544 (Edgar et al., 2011) was used to remove supposed chimeric sequences, and sequences with 80% of their nucleotides with fragment quality scores below 20. The remaining sequences were assigned to operational taxonomic units using CD‐HIT with a threshold of 98% pairwise identity (Li & Godzik, 2006). Rarefaction curves calculated using QIME2 (Bolyen et al., 2019) indicated that a sufficient number of reads was obtained for 16S rRNA analyses. Each sequence was compared to 1647 sequences of the 16S rRNA gene from oral bacteria deposited in HOMD (Chen et al., 2010) (HOMD 16S rRNA RefSeq Extended Version 1.1) using the BLAST algorithm, with a similarity score of 98.5% and a minimum coverage of 97% assigned to the best BLAST hit.
2.4. Statistical analysis
The richness and diversity of the microbiome were assessed by the number of species and the Shannon–Weiner Index, respectively. The effects of smoking on sex, age, and the richness and diversity of the microbiome were evaluated using the t‐test. The Mann–Whitney U‐test was used to compare the proportions of bacterial genera between smokers and nonsmokers. Pearson's correlation analysis was used to assess the relationships between the proportion of bacterial genera and BI values. R software (version 4.0.3) (The R project homepage, 2021) was used for all statistical analyses. The level of significance was set at p < 0.05.
3. RESULTS
3.1. Study population and samples
Eighteen participants (16 men and two women; mean age, 26.8 ± 2.4 years) were smokers and 32 (23 men and nine women; mean age, 25.0 ± 1.6 years) were nonsmokers. No association between sex and smoking status was found (p = 0.163). Smokers were older than nonsmokers (p = 0.006). BI values in smokers ranged from 35 to 450, and the average value (±SD) was 162.8 ± 120.4. The BI value of one patient was >400, and that patient was considered a heavy smoker. Three saliva samples–from two smokers and one nonsmoker–and two tongue samples–from one smoker and one nonsmoker–could not be analyzed because an insufficient amount of DNA was extracted from the samples.
3.2. Richness and diversity of the microbiome
In total, 99 bacterial genera and 228 species were detected in the saliva and tongue samples. The average number (± SD) of species in the saliva was 64.0 ± 19.5, and the number on the tongue was 50.0 ± 10.7 (Figure 1a p = 0.000). By contrast, the average number of species in the saliva was 68.5 ± 2.12 in smokers and 61.5 ± 17.8 in nonsmokers, while the tongue hosted 49.6 ± 10.9 species in smokers and 50.2 ± 10.8 in nonsmokers (Figure 1a p < 0.05). Subsequently, the difference in the overall phylogenetic community between smokers and nonsmokers was assessed using the Shannon–Wiener Index. A distinct overall bacterial community composition was observed in saliva (3.42 ± 0.53) and on the tongue (3.69 ± 0.44) (Figure 1b p = 0.013). No significant differences in the diversity of the bacterial communities between smokers and nonsmokers were observed in saliva or on the tongue (Figure 1b).
Figure 1.
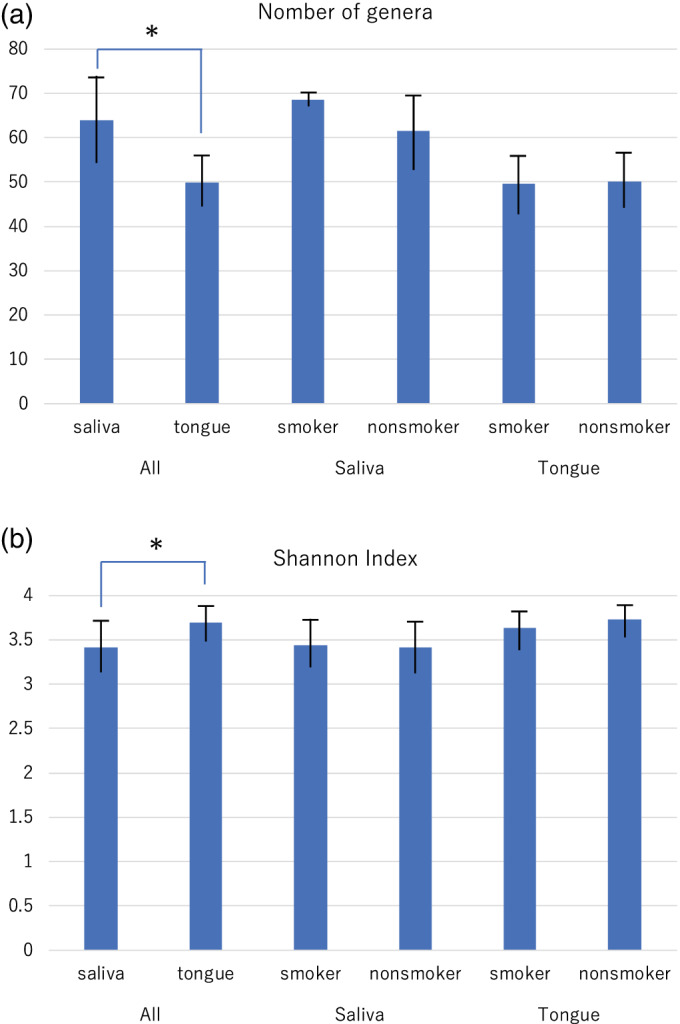
Comparison of the number of bacterial genera (a) and the Shannon index (b) Between the saliva and tongue, and between smokers and nonsmokers in the saliva, and on the tongue. *p < 0.05 between saliva and tongue according to t‐test
3.3. Comparison of the proportions of bacterial genera in smokers and nonsmokers
The proportions of bacterial genera were compared between smokers and nonsmokers (Figure 2). The genera Streptococcus, Prevotella, Neisseria, and Actinomyces comprised the highest proportions in the saliva microbiota, whereas the genera Streptococcus, Prevotella, Actinomyces, and Veillonella comprised high proportions in the tongue microbiota. Compared to those from nonsmokers, the saliva samples from smokers were significantly enriched in the genera Treponema (p = 0.030) and Selenomonas (p = 0.037), and depleted in Capnocytophaga (p = 0.014) and Cardiobacterium (p = 0.010) (Figure 3a). By contrast, the tongue samples from the smokers were enriched in the genera Dialister (p = 0.003) and Atopobium (p = 0.007), and depleted in Haemophilus (p = 0.044), Gemella (p = 0.008), Peptostreptococcus (p = 0.040), Granulicatella (p = 0.022), Catonella (p = 0.049), and Peptostreptococcaceae (p = 0.027) (Figure 3b).
Figure 2.
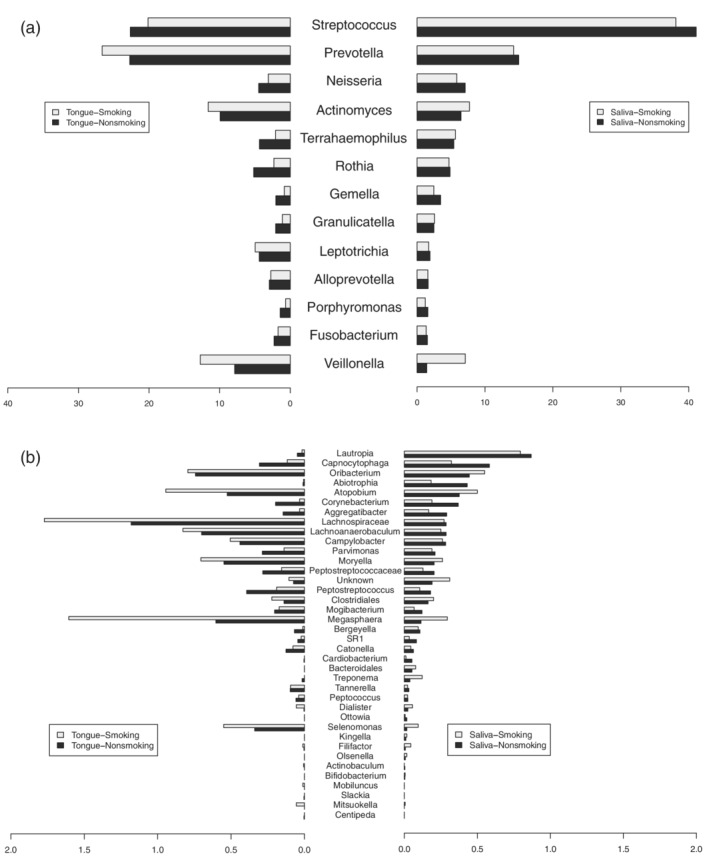
Comparison of the proportion of bacterial genera between smokers and nonsmokers. Genera with high proportions (a) and genera with low proportions (b). The left side is the tongue microbiome and the right side is the saliva microbiome. Light‐gray bars show the proportions of genera in smokers, and black bars show the proportions of genera in nonsmokers
Figure 3.
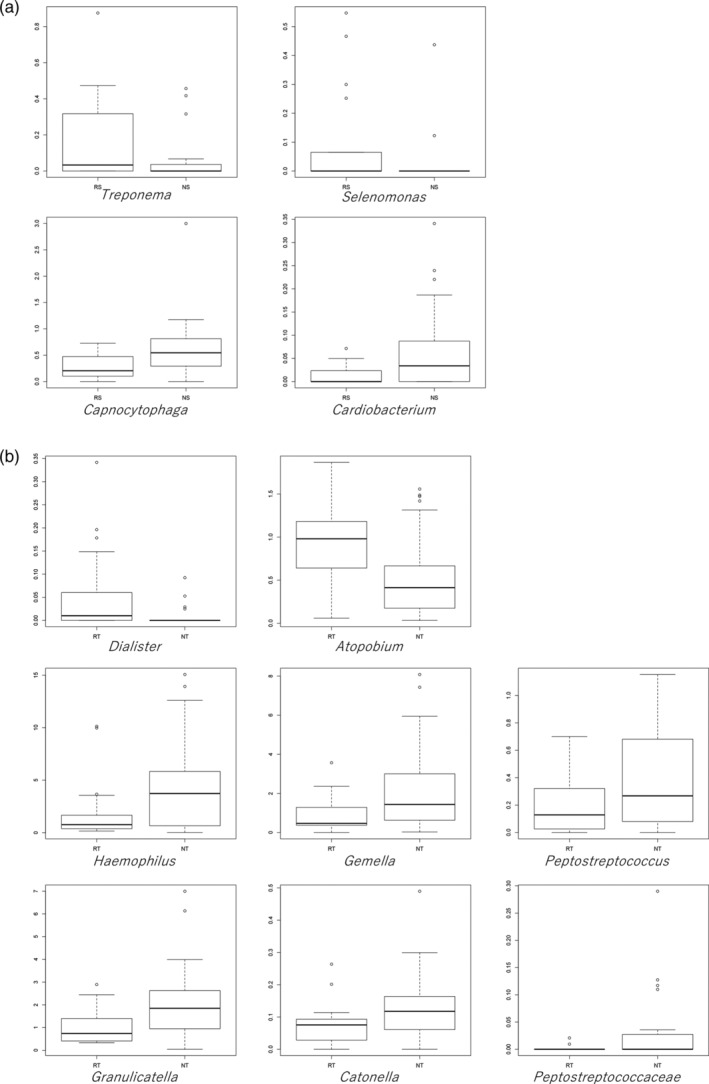
Bacterial genera in saliva (a) and on the tongue (b) that differed significantly between smokers and nonsmokers
3.4. Bacterial genera related to lifetime exposure to smoking
Table 1 shows the bacterial genera correlated with BI values at α < 0.05. The genus Bifidobacterium was positively correlated with BI values in the tongue samples (r = 0.680). The genus Dialister was positively correlated with BI values in the saliva and tongue samples. The genera Cardiobacterium and Granulicatella were negatively correlated with BI values in saliva and the tongue, respectively. The genera Treponema and Selenomonas, which were predominant in the saliva of smokers compared with nonsmokers, were positively correlated with BI values in the saliva samples.
Table 1.
Pearson's correlation coefficients between the relative abundances of the oral bacterial genera and Brinkman index values (α < 0.05)
| Saliva (n = 47) | Tongue (n = 48) | ||
|---|---|---|---|
| Genus | r | Genus | r |
| Treponema | 0.366 | Granulicatella | −0.282 |
| Cardiobacterium | −0.292 | Dialister | 0.416 |
| Oribacterium | 0.324 | Bifidobacterium | 0.680 |
| Dialister | 0.300 | Megasphaera | 0.295 |
| Filifactor | 0.373 | Mitsuokella | 0.522 |
| Veillonella | 0.336 | Cryptobacterium | 0.437 |
| Selenomonas | 0.369 | ||
Note: r, Pearson's correlation coefficient. Genera with three or fewer detected samples were omitted.
4. DISCUSSION
This is the first study to report the effects of smoking on the microbiome of resting saliva and the tongue. The bacterial diversities of the different oral micro‐niches are dependent on location (Simón‐Soro et al., 2013). The effect of smoking on the microbiome is also expected to differ from site to site. Many studies have examined the effects of smoking on the microbiome in gingival crevicular fluid (Mason et al., 2015; Moon et al., 2015; Tsigarida et al., 2015; Delima et al., 2010), and a few studies have examined the microbiome in mouth‐rinse water and buccal mucosa (Wu et al., 2016; Yu et al., 2017; Morris et al., 2013). Studies investigating microbiomes based on mouth‐rinse samples and bronchoscopic alveolar lavage samples have reported differences in the oral microbiomes of smokers, although the lung microbiomes did not differ significanlty (Morris et al., 2013). 16S rRNA sequencing of supra‐ and subgingival dental plaque, saliva, soft oral tissue, and nasal swab samples has revealed lower alpha diversity in smokers than in nonsmokers in the buccal mucosa, whereas samples from other sites did not differ significantly in microbial diversity or composition (Yu et al., 2017). These findings indicate that the oral microbiome is potentially susceptible to smoking. The current study found that microbial diversity differed significantly between resting saliva and the tongue coating, but there was no significant difference in the microbial diversity of saliva or on the tongue between smokers and nonsmokers, and some predominant genera in smokers were found at the genus level. Other studies have reported significant differences in the microbial diversity of subgingival plaque (Mason et al., 2015) and oral wash samples (Wu et al., 2016) between smokers and nonsmokers. The participants in the current study were young and had healthy periodontal tissues; therefore, no differences were observed. However, it is noteworthy that there were generic‐level microbial differences between smokers and nonsmokers, even though the subjects had no illness or symptoms. In particular, periodontopathic bacteria and the organisms relative to oral malodor increase were found in higher proportions in smokers.
The tongue is the most important anatomical structure in the oral cavity due to its location and functions (Roldán et al., 2003). Oral microorganisms existing on the tongue dorsum have easy access to nutrients, including saliva, epithelium, and food debris (Roldán et al., 2003). The tongue coating is an important source of volatile sulfur compounds, the main component of oral malodor (Scully et al., 1997). It has also been suggested to function as a reservoir for periodontopathic pathogens (Tanaka et al., 2004). Tongue morphology is reported to be negatively affected by smoking (Konstantinidis et al., 2010). Hence, it was strongly predicted that the tongue microbiota would be affected and changed by smoking. Major species on the tongue coating were Streptococcus, Prevotella, Actinomyces, and Veillonella in the present study, which is similar to previous reports that investigated the bacterial composition of the tongue dorsum (Aas et al., 2005; Washio et al., 2005). Washio et al. (Washio et al., 2005) identified differences in the numbers of hydrogen‐sulfide‐producing bacteria, including Prevotella, Actinomyces, and Veillonella, between subjects with and without oral malodor, while the bacterial community of the tongue had similar compositions in the two groups. Notably, the proportions of these species tended to be higher in smokers than in nonsmokers in this study, although the difference was not significant (Figure 2). Furthermore, the proportions of Atopobium and Dialister species, which have been reported as oral malodor‐related species in previous reports, were significantly higher in the tongue samples from smokers than in those from nonsmokers (Figure 3) (Kazor et al., 2003; Takeshita et al., 2012). Our previous study investigated species in the hydrogen‐sulfide‐dominant group and the methyl‐mercaptan‐dominant group in subjects with oral malodor, and the proportions of Atopobium and Dialister species were higher in the methyl‐mercaptan‐dominant group than the no‐odor group (Takeshita et al., 2012). The levels of these species increase in the subgingival plaque of patients with chronic periodontal disease (Kumar et al., 2005).
The proportions of the genera Treponema and Selenomonas in resting saliva were significantly higher in smokers than in nonsmokers (Figures 2 and 3). Furthermore, those organisms were positively correlated with BI values (Table 1). Resting saliva has been reported to have a microbial composition that differs from that of other sites in the oral cavity (Simón‐Soro et al., 2013), indicating that it represents some bacteria that do not colonize the teeth, gingival sulcus, or tongue. The current study detected a significant difference in microbial diversity between resting saliva and the tongue (Figure 1). The genera Treponema and Selenomonas are motile bacilli related to aggressive periodontitis and oral malodor. Both are potent hydrogen sulfide producers in the presence of L‐cysteine (Persson et al., 1990). However, Selenomonas species were significantly more predominant in the methyl‐mercaptan‐dominant group than the no‐odor group in our previous study (Takeshita et al., 2012). The increases in these organisms in resting saliva imply inflammation of the gingival crevice.
Most genera that were positively correlated with BI values were strictly anaerobic and have been reported to be periodontitis‐ and oral‐malodor‐associated microorganisms (Table 1). The quantitative relationship between these genera and tobacco exposure is supported by previous studies, in which 12 months of smoking cessation reduced the proportions of Treponema and Dialister in subgingival plaques (Delima et al., 2010). Bifidobacterium was positively correlated with the amount of smoke on the tongue. A recent investigation using mouth‐rinse samples reported that the genus Bifidobacterium is enriched among current‐smokers compared with never‐smokers (Yang et al., 2019). The genera Bifidobacterium, Megasphaera, and Mitsuokella are adapted to low‐pH conditions (Russell, 1991; Levine et al., 2012). It is unknown why smokers have an increased number of bacteria adapted to low‐pH conditions, but it may indirectly explain the involvement of smoking and secondhand smoke in dental caries (Hanioka et al., 2011; Jiang et al., 2019).
This was a cross‐sectional study; thus, the relationship between differences in the microbiome and future onset of periodontal disease cannot be clarified. In addition, if oral malodor could be evaluated, the current relationship between differences in the microbiome and oral malodor could have been clarified; however, oral malodor was not evaluated in this study. It would be necessary to ask the subjects to quit smoking for 12 h or more to accurately determine oral malodor because otherwise it would be affected by the smell of cigarettes.
In conclusion, our findings indicate that the microbial profiles of smokers and nonsmokers in the saliva and on the tongue differed at the generic level in healthy Japanese adults. Because of the characteristics of the genera that were common to smokers and that correlated with smoking exposure, smokers may be at risk for oral malodor and future periodontitis, even if they have a clinically normal oral cavity.
CONFLICT OF INTEREST
The author declares there is no potential conflict of interest.
AUTHOR CONTRIBUTIONS
Nao Suzuki designed the study, collected and analyzed the data, and wrote the manuscript. Yoshio Nakano analyzed and interpreted the data, and wrote the Materials and methods and Results sections of the manuscript. Masahiro Yoneda and Takao Hirofuji were mainly involved in writing the Discussion section. Takashi Hanioka mainly wrote the Introduction and Discussion. All authors approved the final manuscript and take responsibility for its contents.
ACKNOWLEDGMENTS
Textcheck (www.textcheck.com) provided English language editing. The study was supported in part by JSPS KAKENHI Grant Number 19K10433, and by the Oral Medicine Research Center of Fukuoka Dental College.
Suzuki, N. , Nakano, Y. , Yoneda, M. , Hirofuji, T. , & Hanioka, T. (2022). The effects of cigarette smoking on the salivary and tongue microbiome. Clinical and Experimental Dental Research, 8, 449–456. 10.1002/cre2.489
Funding information JSPS KAKENHI, Grant/Award Number: 19K10433; the Oral Medicine Research Center of Fukuoka Dental College
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are available from the corresponding author, N. S., upon reasonable request.
REFERENCES
- Aas, J. A. , Paster, B. J. , Stokes, L. N. , Olsen, I. , & Dewhirst, F. E. (2005). Defining the normal bacterial flora of the oral cavity. Journal of Clinical Microbiology, 43, 5721–5732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbour, S. E. , Nakashima, K. , Zhang, J. B. , Tangada, S. , Hahn, C. L. , Schenkein, H. A. , & Tew, J. G. (1997). Tobacco and smoking: Environmental factors that modify the host response (immune system) and have an impact on periodontal health. Critical Reviews in Oral Biology and Medicine, 8, 437–460. [DOI] [PubMed] [Google Scholar]
- Bateson, M. C. (1993). Cigarette smoking and helicobacter pylori infection. Postgraduate Medical Journal, 69, 41–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernardi, S. , Karygianni, L. , Filippi, A. , Anderson, A. C. , Zürcher, A. , Hellwig, E. , Vach, K. , Macchiarelli, G. , & Al‐Ahmad, A. (2020). Combining culture and culture‐independent methods reveals new microbial composition of halitosis patients' tongue biofilm. Microbiology, 9, e958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolyen, E. , Rideout, J. R. , Dillon, M. R. , Bokulich, N. A. , Abnet, C. C. , al‐Ghalith, G. A. , Alexander, H. , Alm, E. J. , Arumugam, M. , Asnicar, F. , Bai, Y. , Bisanz, J. E. , Bittinger, K. , Brejnrod, A. , Brislawn, C. J. , Brown, C. T. , Callahan, B. J. , Caraballo‐Rodríguez, A. M. , Chase, J. , … Caporaso, J. G. (2019). Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nature Biotechnology, 37, 852–857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brinkman, G. L. , & Coates, E. O., Jr. (1963). The effect of bronchitis, smoking, and occupation on ventilation. The American Review of Respiratory Disease, 87, 684–693. [DOI] [PubMed] [Google Scholar]
- Chen, T. , Yu, W. H. , Izard, J. , Baranova, O. V. , Lakshmanan, A. & Dewhirst, F. E. (2010). The human oral microbiome database: A web accessible resource for investigating oral microbe taxonomic and genomic information. Database (Oxford), 2010, baq013. [DOI] [PMC free article] [PubMed]
- Chrcanovic, B. R. , Albrektsson, T. , & Wennerberg, A. (2015). Smoking and dental implants: A systematic review and meta‐analysis. Journal of Dentistry, 43, 487–498. [DOI] [PubMed] [Google Scholar]
- Delima, S. L. , McBride, R. K. , Preshaw, P. M. , Heasman, P. A. , & Kumar, P. S. (2010). Response of subgingival bacteria to smoking cessation. Journal of Clinical Microbiology, 48, 2344–2349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar, R. C. , Haas, B. J. , Clemente, J. C. , Quince, C. , & Knight, R. (2011). UCHIME improves sensitivity and speed of chimera detection. Bioinformatics, 27, 2194–2200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epstein, J. B. , Freilich, M. M. , & Le, N. D. (1993). Risk factors for oropharyngeal candidiasis in patients who receive radiation therapy for malignant conditions of the head and neck. Oral Surgery, Oral Medicine, and Oral Pathology, 76, 169–174. [DOI] [PubMed] [Google Scholar]
- Feldman, C. , & Anderson, R. (2013). Cigarette smoking and mechanisms of susceptibility to infections of the respiratory tract and other organ systems. The Journal of Infection, 67, 169–184. [DOI] [PubMed] [Google Scholar]
- Frencken, J. E. , Sharma, P. , Stenhouse, L. , Green, D. , Laverty, D. , & Dietrich, T. (2017). Global epidemiology of dental caries and severe periodontitis – A comprehensive review. Journal of Clinical Periodontology, 18, S94–S105. [DOI] [PubMed] [Google Scholar]
- Hanioka, T. , Ojima, M. , Tanaka, K. , & Yamamoto, M. (2011). Does secondhand smoke affect the development of dental caries in children? A systematic review. International Journal of Environmental Research and Public Health, 8, 1503–1519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanioka, T. , Ojima, M. , Tanaka, K. , & Aoyama, H. (2007). Relationship between smoking status and tooth loss: Findings from national databases in Japan. Journal of Epidemiology, 17, 125–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The R project homepage, https://www.r-project.org, 2021.
- Jiang, X. , Jiang, X. , Wang, Y. , & Huang, R. (2019). Correlation between tobacco smoking and dental caries: A systematic review and meta‐analysis. Tobacco Induced Diseases, 17, 34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson, G. K. , & Hill, M. (2004). Cigarette smoking and the periodontal patient. Journal of Periodontology, 75, 196–209. [DOI] [PubMed] [Google Scholar]
- Kazor, C. E. , Mitchell, P. M. , Lee, A. M. , Stokes, L. N. , Loesche, W. J. , Dewhirst, F. E. , & Paster, B. J. (2003). Diversity of bacterial populations on the tongue dorsa of patients with halitosis and healthy patients. Journal of Clinical Microbiology, 41, 558–563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konstantinidis, I. , Chatziavramidis, A. , Printza, A. , Metaxas, S. , & Constantinidis, J. (2010). Effects of smoking on taste: Assessment with contact endoscopy and taste strips. The Laryngoscope, 120, 1958–1963. [DOI] [PubMed] [Google Scholar]
- Kumar, P. S. , Griffen, A. L. , Moeschberger, M. L. , & Leys, E. J. (2005). Identification of candidate periodontal pathogens and beneficial species by quantitative 16S clonal analysis. Journal of Clinical Microbiology, 43, 3944–3955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leite, F. R. M. , Nasciment, G. G. , & Scheutz, F. (2018). Effect of smoking on periodontitis: A systematic review and meta‐regression. American Journal of Preventive Medicine, 54, 831–841. [DOI] [PubMed] [Google Scholar]
- Levine, U. Y. , Bearson, S. M. , & Stanton, T. B. (2012). Mitsuokella jalaludinii inhibits growth of salmonella enteria serover Typhimurium . Veterinary Microbiology, 159, 115–112. [DOI] [PubMed] [Google Scholar]
- Li, W. , & Godzik, A. (2006). Cd‐hit: A fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics, 22, 1658–1659. [DOI] [PubMed] [Google Scholar]
- Mason, M. R. , Preshaw, P. M. , Nagaraja, H. N. , Dabdoub, S. M. , Rahman, A. , & Kumar, P. S. (2015). The subgingival microbiome of clinically healthy current and never smokers. The ISME Journal, 9, 268–272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon, J. H. , Lee, J. H. , & Lee, J. Y. (2015). Subgingival microbiome in smokers and non‐smokers in Korean chronic periodontitis patients. Molecular Oral Microbiology, 30, 227–241. [DOI] [PubMed] [Google Scholar]
- Morris, A. , Back, J. M. , Schloss, P. D. , Campbell, T. B. , Crothers, K. , Curtis, J. L. , Flores, S. C. , Fontenot, A. P. , Ghedin, E. , Huang, L. , Jablonski, K. , Kleerup, E. , Lynch, S. V. , Sodergren, E. , Twigg, H. , Young, V. B. , Bassis, C. M. , Venkataraman, A. , Schmidt, T. M. , & Weinstock, G. M. (2013). Comparison of the respiratory microbiome in healthy nonsmokers and smokers. American Journal of Respiratory and Critical Care Medicine, 187, 1067–1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Persson, S. , Edlund, M. B. , Claesson, R. , & Carlsson, J. (1990). The formation of hydrogen sulfide and methyl mercaptan by oral bacteria. Oral Microbiology and Immunology, 5, 195–201. [DOI] [PubMed] [Google Scholar]
- Petersen, P. E. , Baez, R. J. , & World Health Organization. (2013). Oral health surveys: basic methods, 5th ed. World Health Organization. [Google Scholar]
- Roldán, S. , Herrera, D. , & Sanz, M. (2003). Biofilms and the tongue: Therapeutical approaches for the control of halitosis. Clinical Oral Investigations, 7, 189–197. [DOI] [PubMed] [Google Scholar]
- Russell, J. B. (1991). Intercellular pH of acid‐tolerant ruminal bacteria. Applied and Environmental Microbiology, 57, 3383–3384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scully, C. , El‐Maaytah, M. , Porter, S. R. , & Greenman, J. (1997). Breath odor: Etiopathogenesis, assessment and management. European Journal of Oral Sciences, 105, 287–293. [DOI] [PubMed] [Google Scholar]
- Simón‐Soro, A. , Tomás, I. , Cabrera‐Rubio, R. , Catalan, M. D. , Nyvad, B. , & Mira, A. (2013). Microbial geography of the oral cavity. Journal of Dental Research, 92, 616–621. [DOI] [PubMed] [Google Scholar]
- Takeshita, T. , Suzuki, N. , Nakano, Y. , Shimazaki, Y. , Yoneda, M. , Hirofuji, T. , & Yamashita, Y. (2010). Relationship between oral malodor and the global composition of indigenous bacterial populations in saliva. Applied and Environmental Microbiology, 76, 2806–2814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeshita, T. , Suzuki, N. , Nakano, Y. , Yasui, M. , Yoneda, M. , Shimazaki, Y. , Hirofuji, T. , & Yamashita, Y. (2012). Discrimination of oral microbiota associated with high hydrogen sulfide and methyl mercaptan production. Scientific Reports, 2, 215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka, M. , Yamamoto, Y. , Kuboniwa, M. , Nonaka, A. , Nishida, N. , Maeda, K. , Kataoka, K. , Nagata, H. , & Shizukuishi, S. (2004). Contribution of periodontal pathogens on tongue dorsa analyzed with real‐time PCR to oral malodor. Microbes and Infection, 6, 1078–1083. [DOI] [PubMed] [Google Scholar]
- Tsigarida, A. A. , Dabdoub, S. M. , Nagaraja, H. N. , & Kumar, P. S. (2015). The influence of smoking on the peri‐implant microbiome. Journal of Dental Research, 94, 1202–1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Washio, J. , Sato, T. , Koseki, T. , & Takahashi, N. (2005). Hydrogen sulfide‐producing bacteria in tongue biofilm and their relationship with oral malodor. Journal of Medical Microbiology, 54(Pt9), 889–895. [DOI] [PubMed] [Google Scholar]
- Wu, J. , Peters, B. A. , Dominianni, C. , Zhang, Y. , Pei, Z. , Yang, L. , Ma, Y. , Purdue, M. P. , Jacobs, E. J. , Gapstur, S. M. , Li, H. , Alekseyenko, A. V. , Hayes, R. B. , & Ahn, J. (2016). Cigarette smoking and the oral microbiome in a large study of American adults. The ISME Journal, 10, 2435–2446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, Y. , Zheng, W. , Cai, Q. Y. , Shrubsole, M. J. , Pei, Z. , Brucker, R. , Steinwandel, M. D. , Bordenstein, S. R. , Li, Z. , Blot, W. J. , Shu, X. O. , & Long, J. (2019). Cigarette smoking and oral microbiota in low‐income and African‐American populations. Journal of Epidemiology and Community Health, 73, 1108–1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, G. , Phillips, S. , Gail, M. H. , Goedert, J. J. , Humphrys, M. S. , Ravel, J. , Ren, Y. , & Caporaso, N. E. (2017). The effect of cigarette smoking on the oral and nasal microbiota. Microbiome, 5, 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data that support the findings of this study are available from the corresponding author, N. S., upon reasonable request.


