Abstract
Lycium species (goji), belonging to Solanaceae, are widely spread in the arid to semiarid environments of Eurasia, Africa, North and South America, among which most species have affinal drug and diet functions, resulting in their potential to be a superior healthy food. However, compared with other crop species, scientific research on breeding Lycium species lags behind. This review systematically introduces the present germplasm resources, cytological examination and molecular-assisted breeding progress in Lycium species. Introduction of the distribution of Lycium species around the world could facilitate germplasm collection for breeding. Karyotypes of different species could provide a feasibility analysis of fertility between species. The introduction of mapping technology has discussed strategies for quantitative trait locus (QTL) mapping in Lycium species according to different kinds of traits. Moreover, to extend the number of traits and standardize the protocols of trait detection, we also provide 1,145 potential traits (275 agronomic and 870 metabolic) in different organs based on different reference studies on Lycium, tomato and other Solanaceae species. Finally, perspectives on goji breeding research are discussed and concluded. This review will provide breeders with new insights into breeding Lycium species.
Keywords: Lycium species, molecular breeding, strategy of breeding, standardization, trait detection
Introduction
Goji, the general name of the plants in the genus Lycium, are members of the Solanaceae family found in arid to semiarid regions of Eurasia, Africa, and North and South America. Worldwide, 31 species of Lycium have been reported as medicines and/or foods, such as L. ciliatum (medicine for digestive inflammation), L. cinereum (fruit as food and medicine for rheumatism and headache; root as medicine for perfume, kidney disease, and anodyne), L. pallidum (fruit as food; root as medicine for chickenpox and toothache), L. richii (fruit as food), L. intricatum (seed and fruit as medicine for helminthiasis and eye diseases, respectively), among which L. chinense and L. barbarum have been widely regarded as superfoods with affinal drug and dietary properties in recent years (Yao et al., 2018b). Goji (wolfberry), the fruit of Lycium plants, contains trace elements such as zinc, iron, calcium, and phosphorus as well as metabolic components such as flavonoids, carotenoids, and polysaccharides in red fleshy fruits with affinal drug and dietary functions (Yin and Dang, 2008; Wang C.C. et al., 2010; Wang J. et al., 2010; Qiu et al., 2014). Antioxidative compounds, polysaccharides (Dahech et al., 2013), and hydroxycinnamic acid amides (HCCAs) provide potential anti-inflammatory effects (Wang S. et al., 2017). Lycium barbarum polysaccharides (LBPs) have neuroprotective (Xing et al., 2016), liver-protective (Liu et al., 2015), anti-radiation (Duan et al., 2015), anti-fatigue (Reeve et al., 2010), antitumor (Peifei et al., 2015) and anti-aging properties (Ye et al., 2015), and they scavenge free radicals (Mocan et al., 2014), boost immunity (Su et al., 2014), reduce ischemia/reperfusion injury (Lu and Zhao, 2010), protect against cardiac poisoning (Xin et al., 2007, 2011), and improve reproductive function (Qian and Yu, 2016). Goji (Lycium Linn) is a significant medicinal plant, part of which goji fruit (gouqizi) and root (digupi) are two of the most important Chinese medicinal materials for diabetes prevention and treatment. Goji leaf tea, on the other hand, has a hypoglycemic effect on diabetes (Sun et al., 2018).
China is, in fact, the world’s largest producer and exporter. Goji production areas in China are estimated at 88,000 ha, covering the entire northwest to central China, including Xinjiang, Ningxia, Gansu, Qinghai, Shanxi, Inner Mongolia and Hubei (Chen et al., 2018). The yield of dry goji berries in China reached 410,608 t in 2017, while the yield in the Ningxia region, which is considered the largest goji planting area, is expected to be approximately 108,473 t (State Forestry Administration of China [SFAC], 2018). As domestic and worldwide market demands grow, the manufacturing capacity of Ningxia Province cannot keep pace (Yao et al., 2018b). Thus, it is necessary to increase goji berry production in more favorable cultivation areas (Cao and Wu, 2015; Gong et al., 2019). Furthermore, all of the cultivars used in the goji industry are for dry fruit cultivation in Ningxia Province, China, and are rarely used for other purposes. The goji industry’s constraint is a lack of cultivars for other uses (Nan et al., 2017). As a result, new cultivars with high production yield and for other uses in the goji industry must be developed.
Starting from the genetic origin of Lycium fruit, continuously researching and developing improved new varieties on the basis of the original varieties of Lycium species and improving the breeding technology system of good varieties of Lycium are effective methods to overcoming this bottleneck (Shi et al., 2016). However, there are few reports on the molecular markers and regulatory genes related to edible traits such as the fruit size, shape and hardness of Lycium species. With the development of the social economy, consumer demand for wolfberry has increased and includes products for medicinal uses, fresh food, processing, fruits, and vegetables. As such, traditional wolfberry varieties and industrial models have been unable to meet the constantly rich market demand; thus, wolfberry breeding work will be toward selection research on special types of wolfberry varieties to breed and the specificity to meet diverse usage requirements in industry (Nan et al., 2017).
In terms of breeding methods, the same problems that exist in the selection of special cultivars and ordinary cultivars of traditional Lycium breeding methods, especially individual plant selection methods, have been emphasized, while biological technology breeding methods have been neglected. With the development of biotechnology, molecular marker-assisted selection (MAS) breeding has been widely used in crop breeding. For example, researchers cloned for the first time a high-yield gene that increases the grain number per panicle after mapping QTLs using populations constructed by Koshihikari (cultivars with tall plants and low yields) and Habataki (cultivars with short plants and high yields). With this gene, a new type of super rice with high yield and lodging resistance was developed by MAS (Ashikari et al., 2005). The same MAS procedure was also successfully executed in disease resistance traits in tomato, taste and virus resistance selection in pepper, and bacterial wilt resistance in eggplant (Foolad and Panthee, 2012; Hanson et al., 2016; Khapte et al., 2018; Moodley et al., 2019). In the breeding of special varieties of Lycium species, we should strengthen the organic combination of traditional breeding technology and molecular marker-assisted breeding. It cannot only improve the selection efficiency but also greatly shorten the breeding time (fewer years) to implementing early assisted selection with molecular marker-based traditional breeding as the main method (Nan et al., 2017). The present attention of practical breeding is to develop an increasing number of markers using biotechnology tools to enable molecular-assisted selection to accelerate the delivery of improved varieties to agriculturalists (Rubiales et al., 2021).
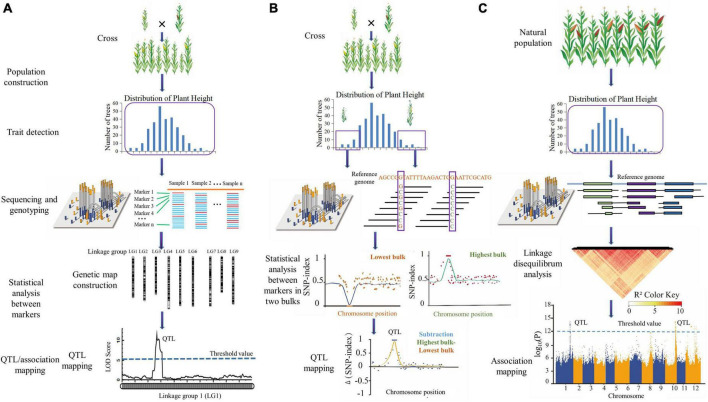
As shown in Figure 1, the procedure of MAS combined with next-generation sequencing (NGS) breeding mainly includes genetic population construction, morphological trait detection, population sequencing and genotyping, statistical analysis between markers, and QTL/association mapping.
FIGURE 1.
The procedure of MAS combining next-generation sequencing (NGS). (A) Linkage mapping; (B) bulk segregation analysis (BSA); (C) association mapping. Genetic population construction, morphological trait detection, population sequencing and genotyping, statistical analysis between markers, and QTL/association mapping are common procedures in MAS combining NGS.
Thus, in this study, we summarize all factors in molecular MAS, including germplasm resources, compatibility between species with karyotypes, candidate traits and their protocols for detection, and the progress of molecular MAS in Lycium. Moreover, we also discuss research strategies for different traits and prospective directions of selective breeding in Lycium species.
Germplasm Resources of Goji Berry
By 2001, approximately 80 species of Lycium had been found and named from South America to North America, Australia to the Pacific Islands and Eurasia to the South (Fukuda et al., 2001; Shi et al., 2012), whereas in 2018, the number exceeded 97 (Yao et al., 2018a). Lycium species are found in a variety of habitats, primarily in temperate and subtemperate climates (Fukuda et al., 2001; Shi et al., 2012). According to statistics, there are 14 species of goji in Eurasia, 26 in South Africa, and 25 in the southern part of North America. Up to 32 species are widely distributed in South America, whereas there are only three in Australia.
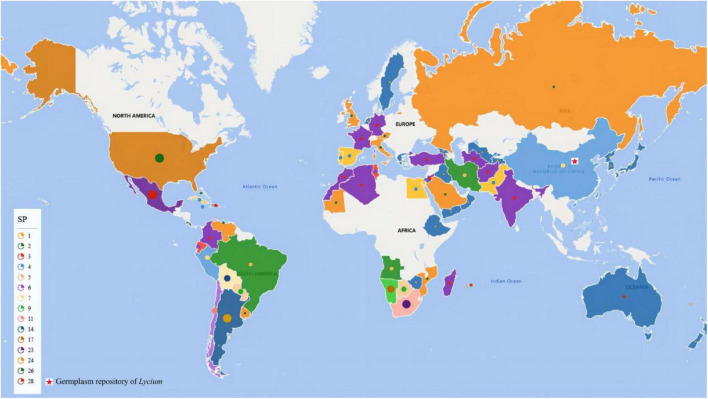
To date, as shown in Figure 2, the Lycium resources of 86 countries/regions have been reported, among which Mexico and the US have the most abundant Lycium species (Editorial Committee of Chinese Flora, Chinese Academy of Sciences, 1978; Yao et al., 2018a). Goji is primarily found in the northern parts of China (Yao et al., 2018a). There are seven species and three varieties of Lycium in China including L. cylindricum, L. yunnanense, L. dasystemum along with mutants (L. dasystemum var. rubricaulium), L. truncatum, L. ruthenicum (Figure 3A), L. barbarum (Figures 3D,E) along with mutants (L. barbarum L. var. auranticarpum K. F. Ching) (Figure 3F), and L. chinense Mill. (Figures 3B,C) along with mutants [L. chinense Mill. var. potaninii (Pojark.) A. M. Lu] (Editorial Committee of Chinese Flora, Chinese Academy of Sciences, 1978; Dong et al., 2008; Yao et al., 2018a), among which five species have medical functions and four can be regarded as food (Editorial Committee of Chinese Flora, Chinese Academy of Sciences, 1978; Li Y. C. et al., 2001; Azadi et al., 2007; Olatunji et al., 2018; Wang et al., 2018; Li et al., 2019a; Ye and Jiang, 2020).
FIGURE 2.
Distribution of Lycium species worldwide. SP, species number. Dots with different colors and sizes indicate species numbers in the country marked by them. The red pentacle indicates the location of the germplasm repository of Lycium.
FIGURE 3.
Some collections of Lycium species in the resources. (A) L. ruthenicum; (B,C) L. chinense; (D,E) L. barbarum; (F) L. barbarum variant; (G,H,K) L. ruthenicum variant; (I) offspring between L. barbarum and L. chinense; and (J,L) offspring cross between L. barbarum and L. ruthenicum.
However, among the Lycium species, according to the Chinese Pharmacopeia, only L. barbarum is recorded as a medicine (Zhang et al., 2001). Several cultivars of L. barbarum have been reported, including the Ningqi series (from Ningqi No. 1 to No. 10), Chaiqi series (Chaiqi No. 1, No. 2), Qingqi series (No. 1, No. 2), Jingqi series (from Jingqi No. 1 to No. 6) (Qin and Dai, 2017), and Zhongkelvchuan No. 1 (Yang et al., 2015).
In addition to new species, intercross plants have also been reported, such as L. ciliaturn × L. cestroides (Bernardello et al., 1995), L. barbarum × L. chinense (Rehman et al., 2020; Figure 3I), and L. barbarum × L. yunnanense (Zhao et al., 2021). In the germplasm repository of the Chinese Academy of Sciences (E 106.04968°, 38.4398563°), offspring of L. barbarum and L. ruthenicum (Figures 3J,L) have also been created. Intercross-demonstration offspring can be used to map QTLs with interspecies crosses.
To provide a reference for germplasm collection in genetic breeding, we summarized the resource diversity in all countries reported (Figure 2 and Supplementary Table 1). These resources with multiple trait variations will provide multiple materials for genetic research on traits related to fruits (color, size, and metabolism), leaves (length, width, and thickness), resistances and so on.
Currently, the Lycium germplasm repository at the Goji Engineering Technology Center mainly focuses on Ningxia Province, and it comprises seven species and three varieties distributed naturally in China as well as germplasm imported from the United States, South Korea, Malaysia, and other countries, totaling 60 varieties (lines) and more than 2,000 intermediate materials (1,500 red fruits, 120 yellow fruits, 300 black fruits and 150 other fruits) (NingXia Academy of Agricultural and Forestry Sciences, 2015). The second germplasm resource is in the Chinese Academy of Sciences, which has collected 35 core Lycium species resources with different characteristics from all over the world and obtained approximately 10,000 hybrid progenies through intraspecific and interspecific hybridization, providing abundant germplasm resources for breeding and parent plants with rich phenotypes for the construction of NAM (nesting association mapping) populations of Lycium species.
Karyotyping
Cytological examination is a necessary procedure in molecular breeding (Xu, 2010), for validation of both intercrosses and intracrosses in relation to fertility and for avoiding individuals containing translocations and polyploid species containing monosomes or partial chromosomes from being used as mapping parents (Xu, 2010). The cross between tetraploid and diploid L. barbarum showed strong cross affinity or backcross affinity; however, the progeny were highly sterile, and the seed satiation rate was very low, making the breeding of triploid seedless plants difficult (Li J. et al., 2001), so it was not conducive to the construction of hybrid populations.
The karyotypes of all of the studied taxa can reveal differences among the species, including size, major visible chromosomal rearrangements, cryptic structural changes, paracentric inversions or reciprocal translocations of segments of similar length (Stiefkens and Bernardello, 2002). A karyotypic analysis showed that L. barbarum from Ningxia had 12 pairs of chromosomes (2n = 24) (Chen J. et al., 2013). In the karyotype study, 57 kinds of species, varieties, and hybrid plant were used out of a total of 97 species. Most of the Lycium species are diploid with 12 pairs of chromosomes (Laura et al., 2010). Some others are teraploid, octaploid, decaploid, and even hendecaploid (Bernardello et al., 1995; Stiefkens and Bernardello, 2005; Chen J. et al., 2013; Stiefkens et al., 2020; Table 1).
TABLE 1.
Cytological characteristics of Lycium species.
| Taxon | Code | Chromosome number | Karyotype | Tlb | c | r | A1 | A2 | St | R | References | |
| 1 | L. elongatum | Diploid | 24 | 11 m* + 1 sm | 25.01 | 2.08 | 1.22 | 0.16 | 0.12 | 1 A | − | Stiefkens and Bernardello, 1996 |
| 2 | L. afrum | Haploid | 12 | − | − | − | − | − | − | − | − | Minne et al., 1994 |
| 3 | L. ameghinoi | Diploid | 24 | 11 m* + 1 sm | 20.2 | 1.7 | 1.23 | 0.17 | 0.16 | 2A | 1.7 | Stiefkens and Bernardello, 2005 |
| 4 | L. americanum | Diploid | 24 | 11 m* + 1 sm | 22.74 | 1.89 | 1.22 | 0.16 | 0.12 | 1 A | − | Stiefkens and Bernardello, 1996 |
| 5 | L. amoenum | Diploid | 24 | 10 m* + 2 sm | 26.76 | 2.23 | 1.29 | 0.2 | 0.1 | 1A | 1.41 | Laura et al., 2010 |
| 6 | L. arenicolum | Triploid | 36 | − | − | − | − | − | − | − | − | Minne et al., 1994 |
| 7 | L. barbarum | Diploid | 24 | 12 m | 49.99 | 4.17 | 1.23 | − | − | − | 1.51 | Chen J. et al., 2013 |
| 8 | L. bosciifolium | Diploid | 24 | 11 m* + 1 sm | 25.04 | 2.09 | 1.25 | 0.24 | 0.12 | 1A | 1.49 | Laura et al., 2010 |
| 9 | L. cestroides | Diploid | 24 | 11 m* + 1 sm | 25.78 | 2.14 | 1.22 | 0.17 | 0.12 | 1 A | − | Stiefkens and Bernardello, 1996 |
| Diploid | 24 | 11 mb + 1 sm | 19.19 | 1.6 | 1.21 | 0.15 | 0.14 | − | − | Bernardello et al., 1995 | ||
| 10 | L. chanar | Diploid | 24 | 11 m* + 1 sm | 22.68 | 1.89 | 1.17 | 0.11 | 0.14 | 2A | − | Stiefkens and Bernardello, 2002 |
| 11 | L. chinense | Diploid | 24 | 12 m | 50 | 4.17 | 1.31 | − | − | − | 1.93 | Chen J. et al., 2013 |
| 12 | L. chilense var. chilense | Diploid/Tetraploid | 24/48 | 11 m* + 1 sm | 20 | 1.7 | 1.14 | 0.1 | 0.13 | 1A | 1.52 | Stiefkens and Bernardello, 2005 |
| 13 | L. chilense var. confertifolium | Tetraploid | 48 | 22 m + 2 sm | 22.61 | 1.88 | − | − | − | − | − | Stiefkens et al., 2020 |
| 14 | L. chilense var. descolei | Diploid/Tetraploid | 24/48 | 11 m* + 1 sm | 23.9 | 2 | 1.19 | 0.14 | 0.12 | 1A | 1.46 | Stiefkens and Bernardello, 2005 |
| 15 | L. chilense var. Descolei | Tetraploid | 48 | 22 m + 2 sm | 26.32 | 2.19 | − | − | − | − | − | Stiefkens et al., 2020 |
| 16 | L. chilense var. filifolium | Diploid/Tetraploid | 24/48 | 11 m* + 1 sm | 30 | 2.5 | 1.2 | 0.15 | 0.12 | 1A | 1.51 | Stiefkens and Bernardello, 2005 |
| 17 | L. ciliatum | Diploid | 24 | 11 m* + 1 sm | 21.2 | 1.7 | 1.21 | 0.15 | 0.11 | 2A | 1.4 | Stiefkens and Bernardello, 2005 |
| Tetraploid | 48 | 22 m + 2 sm | 22.03 | 1.84 | − | − | − | − | − | Stiefkens and Bernardello, 2002 | ||
| 18 | L. ciliatum | Diploid | 24 | 11 mb + 1 sm | 20.57 | 1.71 | 1.24 | 0.17 | 0.11 | − | − | Bernardello et al., 1995 |
| 19 | L. cuneatum | Diploid | 24 | 11 m* + 1 sm | 20.34 | 1.69 | 1.18 | 0.12 | 0.14 | 2A | − | Stiefkens and Bernardello, 2002 |
| 20 | L. depressum | Diploid | 24 | 11 m + 1 sm | 56.48 | 4.71 | 1.44 | − | − | 2A | − | Sheidai et al., 1999 |
| Diploid | 24 | 12 m | 64.08 | 5.34 | 1.3 | − | − | 1A | − | Sheidai et al., 1999 | ||
| Diploid | 24 | 12 m | 67.18 | 5.60 | 1.83 | − | − | 1A | − | Sheidai et al., 1999 | ||
| 21 | L. ferocissimum | Diploid | 24 | 10 m* + 2 sm | 22.16 | 2.02 | 1.3 | 0.21 | 0.08 | 1A | 1.29 | Laura et al., 2010 |
| 22 | L. fremontii | Octaploid | 96 | − | − | − | − | − | − | − | − | Chen J. et al., 2013 |
| 23 | L. fuscum | Diploid | 24 | 11 m* + 1 sm | 22.7 | 1.89 | 1.22 | 0.16 | 0.14 | 2A | − | Stiefkens and Bernardello, 2002 |
| 24 | L. gilliesianum | Diploid | 24 | 11 m* + 1 sm | 20.66 | 1.72 | 1.25 | 0.18 | 0.17 | 2A | − | Stiefkens and Bernardello, 2002 |
| 25 | L. horridum | Diploid | 24 | − | − | − | − | − | − | − | − | Minne et al., 1994 |
| 26 | L. humile | Diploid | 24 | 11 m + 1 sm | 21.45 | 1.79 | 1.34 | 0.16 | 0.11 | - | 1.71 | Stiefkens et al., 2020 |
| 27 | L. infaustum | Diploid | 24 | 11 m* + 1 sm | 21.52 | 1.79 | 1.25 | 0.18 | 0.13 | 1 A | - | Stiefkens and Bernardello, 1996 |
| 28 | L. kopetdaghi | Diploid | 24 | 12 m | 50.91 | 4.24 | 1.38 | − | − | 1A | − | Sheidai et al., 1999 |
| 29 | L. makranicum | Diploid | 24 | 12 m | 51.36 | 4.28 | 1.32 | − | − | 1A | − | Sheidai et al., 1999 |
| 30 | L. minutifolium | Diploid | 24 | 11 m* + 1 sm | 24.93 | 2.07 | 1.21 | 0.16 | 0.08 | 1A | − | Stiefkens and Bernardello, 2002 |
| 31 | L. morongii | Diploid | 24 | 11 m* + 1 sm | 23.23 | 1.94 | 1.2 | 0.14 | 0.12 | 1A | − | Stiefkens and Bernardello, 2002 |
| 32 | L. nodosum | Diploid | 24 | 11 m* + 1 sm | 20.03 | 1.67 | 1.27 | 0.19 | 0.13 | 2A | − | Stiefkens and Bernardello, 2002 |
| 33 | L. oxycarpum | Diploid | 24 | 10 m* + 2 sm | 23.22 | 1.93 | 1.3 | 0.2 | 0.08 | 1A | 1.3 | Laura et al., 2010 |
| 34 | L. pallidum | Diploid | 24 | 10 m + 2 sm | 49.99 | 4.17 | 1.22 | − | − | − | 1.55 | Chen J. et al., 2013 |
| 35 | L. rachidocladum | Diploid | 24 | 11 m* + 1 sm | 28.01 | 2.33 | 1.28 | 0.20 | 0.16 | 1 A | − | Stiefkens and Bernardello, 1996 |
| 36 | L. repens | Hendecaploid | 132 | − | 22.68 | 1.89 | − | − | − | − | − | Stiefkens et al., 2020 |
| 37-1 | L. ruthenicum | Diploid | 24 | 9 m + 3 sm | 70.87 | 5.91 | 1.5 | − | − | 2A | − | Sheidai et al., 1999 |
| 37-2 | L. ruthenicum | Diploid | 24 | 9 m + 3 sm | 66.84 | 5.57 | 1.52 | − | − | 2A | − | Sheidai et al., 1999 |
| 37-3 | L. ruthenicum | Diploid | 24 | 12 m | 50.18 | 4.18 | 1.25 | − | − | − | 1.53 | Chen J. et al., 2013 |
| 38 | L. shawii | Diploid | 24 | 11 m + 1 sm | 40.92 | 3.41 | 1.35 | − | − | 2A | − | Sheidai et al., 1999 |
| 39 | L. stenophyllum | Diploid | 24 | 11 m* + 1 sm | 22.9 | 1.9 | 1.17 | 0.12 | 0.13 | 1A | − | Stiefkens and Bernardello, 2002 |
| 40 | L. tenue | Diploid | 24 | 10 m* + 2 sm | 21.65 | 1.8 | 1.28 | 0.18 | 0.1 | 2A | 1.43 | Laura et al., 2010 |
| 41 | L. tenuispinosum | Diploid | 24 | 11 m* + 1 sm | − | − | − | − | − | − | − | Stiefkens and Bernardello, 1996 |
| 42 | L. tenuispinosum var. friesii | Diploid | 24 | 11 m* + 1 sm | 25.29 | 2.10 | 1.21 | 0.15 | 0.12 | 1 A | − | Stiefkens and Bernardello, 1996 |
| 43 | L. tenuispinosum var. tenuispinosum | Diploid | 24 | 11 m* + 1 sm | 23.63 | 1.96 | 1.21 | 0.16 | 0.1 | 1 A | − | Stiefkens and Bernardello, 1996 |
| 44 | L. tenuispinosum var. calysinum | Diploid | 24 | 11 m* + 1 sm | 21.52 | 1.79 | 1.21 | 0.15 | 0.12 | 1 A | − | Stiefkens and Bernardello, 1996 |
| 45 | L. tetrandrum | Triploid | 36 | − | − | − | − | − | − | − | − | Minne et al., 1994 |
| 46 | L. villosum | Diploid | 24 | − | − | − | − | − | − | − | − | Minne et al., 1994 |
| 47 | L. vimineum | Diploid | 24 | 11 m* + 1 sm | 20.01 | 1.67 | 1.24 | 0.17 | 0.13 | 2A | - | Stiefkens and Bernardello, 2002 |
| 48 | L. australe | Diploid | 24 | 10 m + 2 sm | 31.43 | 2.62 | 1.25 | 0.16 | 0.12 | − | 1.61 | Stiefkens et al., 2020 |
| 49 | L. berlandieri | Diploid | 24 | 10 m + 2 sm | 38.7 | 3.23 | 1.36 | 0.22 | 0.12 | − | 1.55 | Stiefkens et al., 2020 |
| 50 | L. californicum | Diploid | 24 | 11 m + 1 sm | 29.8 | 2.48 | 1.19 | 0.14 | 0.1 | − | 1.46 | Stiefkens et al., 2020 |
| Tetraploid | 48 | 22 m + 2 sm | 39.65 | 3.3 | − | − | − | − | − | Stiefkens et al., 2020 | ||
| 51 | L. exsertum | Tetraploid | 48 | 20 m + 4 sm | 40.26 | 3.35 | − | − | − | − | − | Stiefkens et al., 2020 |
| 52 | L. fremontii | Decaploid | 120 | – | 29.24 | 2.44 | − | − | − | − | − | Stiefkens et al., 2020 |
| 53 | L. intricatum | Diploid | 24 | 10 m + 2 sm | 33.59 | 2.8 | 1.24 | 0.21 | 0.16 | - | 1.81 | Stiefkens et al., 2020 |
| 54 | L. pallidum | Diploid | 24 | 10 m + 2 sm | 29.98 | 2.5 | 1.37 | 0.25 | 0.11 | - | 1.49 | Stiefkens et al., 2020 |
| 55 | L. parishii | Diploid | 24 | 11 m + 1 sm | 30.67 | 2.55 | 1.15 | 0.12 | 0.08 | - | 1.36 | Stiefkens et al., 2020 |
| 56 | Lycium sp. | Tetraploid | 48 | 20 m + 4 sm | 24.52 | 2.04 | − | − | − | − | − | Stiefkens et al., 2020 |
| 57 | Hybrid (L. ciliaturn × L. cestroides) | Diploid | 24 | 11 mb + 1 sm | 22.23 | 1.85 | 1.22 | 0.16 | 0.13 | − | − | Bernardello et al., 1995 |
Tl, mean total haploid chromosome length; C, mean chromosome length; r, mean arm ratio. Mean asymmetry indices: A1, intrachromosomic; A2, interchromosomic; St, Stebbins’ (1971), category of asymmetry; R, ratio between largest and smallest chromosomes in complement. Lengths in μm. m, metacentric chromosome; sm, submetacentric chromosome. An asterisk indicates that the first chromosome pair has a satellite on the short arm.
Traits for Quantitative Trait Locus Mapping in Lycium
Although Lycium species are found all over the world, only L. barbarum is frequently planted as fruit trees and medicinal plants, particularly in China (Shi et al., 2012). Thus, the majority of breeding selection research has been conducted in China. Although the Chinese government has recognized nearly 10 top varieties of Lycium, only “Ningqi No. 1,” “Ningqi Cai No. 1,” “Mengqi No. 1,” “Ningqi No. 4,” “Ningqi No. 7,” and “Zhongkelvchuan No. 1” have been actively promoted and implemented in production (An et al., 2009; Chen et al., 2018), and the number of new cultivars is still growing. However, the majority of goji fruits on the market are dried fruits in medical use, and fruit juice and fresh fruits are rarely promoted to the market (Huang et al., 2013). As a result, single usage (only dry fruits for medical use) cannot adjust to the demands of industrial product diversification (An et al., 2009). The usage needs to be extended to food, tea, juice, fruit, and chemical extraction uses, such as goji leaf tea, wolfberry tea, wolfberry juice, and wolfberry wine.
Breeders have been grappling with how to standardize the evaluation of germplasm resources and new varieties in recent years, as goji plants have become more widely planted and the demands for germplasm resources have grown (Zhang and Sun, 2011). The State Forestry Administration and the Ministry of Agriculture are currently the main authorities in China for examining and approving new varieties. In anticipation of the acceptance of new Lycium variants, the two departments have developed two sets of “Guidelines for the Conduct of Tests for Distinctness, Uniformity, and Stability of Lycium species” (DUS) (LY/T2099-2013 and NY/T2528-2013). The number of agricultural qualities covered by the two DUS guidelines is limited, and the breeding requirements cannot be fully met. The DUS testing guidelines for new plant varieties need to be further improved (Zhang and Sun, 2011). Tomato, eggplant, potato, and goji are all members of the Solanaceae family, which is one of the most significant families in agriculture. It possesses a highly conserved genomic sequence and a phenotypic trait set that is extremely diverse. Therefore, comparative genomics across different Solanaceae species can frequently lead to major findings (Bombarely et al., 2010). Currently, international studies on other Solanaceae species, such as tomato and potato, are quite in depth regardless of the variety of features, gene research, or other themes. Plants have been investigated for more than 1,000 trait descriptions worldwide,1 of which 387 are for the Solanaceae family (solgenomics.net). The Sol Genetic Network (SGN)2 is a professional portal that provides genomic and phenotypic data on Solanaceae and allied species. It compiles information on the genomic and phenotypic characteristics of tomato, potato, pepper, eggplant, tobacco, and other species (Bombarely et al., 2010). In contrast to other crops, research on the resource base of Lycium species is still in its early stages, and systematic and in-depth research must be strengthened (An et al., 2009). We combine data from SGN, DUS, “Normative description and data standard of L. barbarum germplasm resources” (Shi et al., 2012), and other published papers to provide additional traits and their standardized detection methods and QTL mapping for breeders to facilitate standardization of trait research.
Whole-Plant Traits
Many genetic traits, such as ground diameter, crown width, plant height, branch hardness, and natural plant type, are dominated by whole-plant attributes, which can be utilized as the primary reference indices for Lycium species breeding selection (Yuan et al., 2013). Trunk diameter (TD), for example, is related to the yield, resistance and growth rate of the plant. Unlike the branch angle or tree height, the trunk diameter is unaffected by trimming and can properly depict the plant’s growth state. It is a crucial index for precisely reflecting the plant’s growth status, such as the amount of growth (Miller and Scorza, 2010; Wang et al., 2015b). Wang et al. (2015b) mapped four QTL loci influencing the growth of cherry trunk using a high-density genetic map of Prunus avium and discovered that the QTL loci controlling the growth of cherry trunk varied with distinct developmental stages (Wang et al., 2015b). These findings suggested that the QTLs governing trunk growth were stage-specific (Liu et al., 2008). To make production management easier, cultivars with a large branching angle, small tree body, and large trunk diameter have been produced (Wang et al., 2015b). Thus, trunk diameter is extremely important in production.
Whole-plant traits, such as plant height, trunk diameter, and internode length, are regulated by genes that can be detected by QTL mapping in Lycium and neighboring species (Frary et al., 2003; Barchi et al., 2009; Gong et al., 2019). Thus, mapping of QTLs and gene mining of plant-related traits are useful.
To standardize the detection of whole-plant-related traits, we summarized thirty kinds of traits and their references (Supplementary Table 2).
Resistance Traits
Plants of the Lycium genus attract a variety of pests due to their lush stems and leaves as well as their sweet fruit juice. According to research and investigation, there are approximately 38 pests in L. barbarum in Ningxia, including seven major pests and four diseases, the majority of which are unique to goji plants. If prevention and control are not strengthened in a timely manner, they frequently cause serious goji production problems, even failure, and severely affect yield (Cao and He, 2013). Planting resistant cultivars and spraying fungicides are two possible approaches to controlling pests (Villegas-Fernández et al., 2021). However, continuous use of pesticides can result in loss of biodiversity, environmental contamination, and a slew of ecological issues. Furthermore, widespread contamination of the environment and accumulation in the food chain can pose a risk of teratogenic, genotoxic, oncogenic, and carcinogenic effects on humans (Singh et al., 2017). Thus, stress-resistant cultivar breeding is required.
The first strategy to breeding cultivars with continuous resistance is to select plants with more than resistance genes with the help of robust molecular markers (Rubiales et al., 2021). Research on resistance to biotic stress on neighboring species in Solanaceae, including Capsicum (pepper), Solanum (potato), and Lycopersicon (tomato), showed that these resistances were regulated by resistance genes (R genes). Moreover, homologous genes in the R family (Pto, N, Sw-5, I2, and Prf) in pepper were discovered in syntenous sites in genomes of other solanaceous species and occasionally also discovered to extra sites neighboring phenotypically defined R genes in Solanaceae (Grube et al., 2000). Because resistance is regulated by resistance genes, QTL mapping can be conducted for resistance traits. QTL mapping can be used to further understand the genetic basis of various traits, including resistance to biotic stress (Iordãchescu et al., 2020).
The obtainability of a variety of resistant sources and of a dependable screening technique are crucial to execute a resistance selection project. Study on the molecular and inherited foundation of resistance can improve breeding efficiency (Rispail et al., 2020). To date, there are still no reports about genetic breeding in resistance research on Lycium. To facilitate genetic research on resistance in Lycium species and genetic resource conservation and evaluation, standardized methods to evaluate resistance to 11 kinds of insects/disease and references were summarized (Supplementary Table 3).
Phenology Traits
As domestic and worldwide market demand grows, the manufacturing capacity of Ningxia Province cannot keep pace (Yao et al., 2018b). Thus, it is necessary to increase goji berry output in other favorable agricultural areas (Barchi et al., 2009; Gong et al., 2019). The study of phenology aids in the development of Lycium species variants that are appropriate for various climate conditions. Higher latitudes and altitudes and shorter growing seasons than the current optimal areas limit the number of cultivars growers choose in outdoor production (Kevany et al., 2008). Qinghai Province, for example, is a plateau-climate producing area (Yao et al., 2018b). The harvest season for goji fruit is from July to November each year. However, in September in Qinghai, the temperature drops below 10°C, drastically shortening the harvest season and resulting in lower yield (Zheng et al., 2018). Growing early maturing cultivars is one way to address climate-related constraints. Growers benefit from these varieties as well because the earliest fruit to market in a season might command a higher price (Kevany et al., 2008). Thus, the ability to have fruits mature early is a very desirable and important feature (Kevany et al., 2008).
Early ripening occurs when the ethylene receptor LeETR4 is suppressed, but fruit size, yield, and flavor-related chemical composition remain mostly intact. Moreover, gibberellins (GAs) were also found to have negative effects on fruit ripening. Restriction of GAs can activate ripening regulator genes such as CNR, RIN, and NOR, mediating the biosynthesis of ethylene. Further study showed that the SlGA2ox1 gene regulated GA catabolism, resulting in reductions in GA levels in fruit tissues and leading to early ripening in tomato (Li et al., 2019b). Biotechnology could provide important tools for the creation of early ripening cultivars as our understanding of the molecular control of fruit ripening grows (Kevany et al., 2008). Thus, to apply MAS in phenology trait QTL mapping and standardize the detection method, we summarized 25 phenology traits in Supplementary Table 4.
Fruit Traits
The fruit size of L. barbarum impacts the price of dry goji fruits and is one of the most important indicators of goji fruit grading (Ma et al., 2017). The higher the wolfberry grade and the larger the fruit size, the higher the price. Therefore, in regard to goji cultivation, breeders are more likely to choose types with large fruit sizes and high yields. Fruit weight, fruit length, and fruit breadth are subdivided into indices for analyzing goji fruit size (An et al., 2007). Meanwhile, crop yield is considered positively correlated with fruit weight, fruit length, and fruit width, showing that superior farming varieties with large fruit length, width, and weight cannot only improve fruit quality but also increase yield (Henareh et al., 2015). Artificial domestication can boost the productivity and quality of numerous crops. When compared with predomestication, the size and weight of tomato fruits grew dramatically with favorable features following long-term artificial domestication (Tanksley, 2004).
Fruit weight in most tomato species is controlled by a major QTL (fw2.2) (Alpert et al., 1995). In Lycium species, QTLs for fruit traits were first detected by Zhao et al. (2019) with an interspecies genetic map, and 41 stable QTLs were detected (Zhao et al., 2019). QTL mapping for fruit traits was executed by Rehman et al. (2020), with two stable QTLs (Rehman et al., 2020). Genetics are a key factor for fruit traits, and it is feasible to map QTLs with a combination of genetic mapping data and morphological data. Standardized techniques were used to determine the 129 fruit-related traits (Supplementary Table 5).
Leaf Traits
Leaf traits of Lycium include leaf length, leaf width, leaf thickness, petiole length, leaf color et al. (State Forestry Administration of China [SFAC], 2013). The distance between the upper and lower surfaces of the blade is measured as the leaf thickness (LT) (Coneva et al., 2017). Leaf thickness is an important leaf shape parameter for crop plant type improvement and serves as a reference index in the breeding of high-yield crop varieties (Chen D. et al., 2015). Leaf thickness and crop yield have been discovered to have a substantial positive association in rice studies. Plants with thick leaves produce more fruit (Liu C. et al., 2014). As a result, genetic improvement of crop leaf thickness is critical for increasing crop output (Chen D. et al., 2015). Meanwhile, research has discovered that thicker leaves are better for plant growth in arid environments (Coneva et al., 2017).
Photosynthesis is a leaf feature that describes the process by which plants receive light energy and transform it into chemical energy, and it is the foundation of plant organic matter synthesis (Richards, 2000). It is critical to a plant’s ability to synthesize its own chemicals (Long et al., 2006; Zhu et al., 2012). Photosynthetic features include the photosynthetic rate (Pn), stomatal conductance, intercellular carbon dioxide, the transpiration rate, the limiting value of the stoma, water usage efficiency, the chlorophyll content, and a variety of other variables (Qu et al., 2017). Research on other plants shows that polygenes play a major role in photosynthesis (Yang et al., 2007; Kumar et al., 2012; Czyczylo-Mysza et al., 2013). Moreover, Gong et al. (2019) detected 29 QTLs for photosynthetic traits, among which eight are stable QTLs (Gong et al., 2019), indicating that photosynthesis in Lycium species is regulated by genes and is applicable to mapping QTLs in Lycium species.
Thus, we provide 33 kinds of leaf trait descriptions and their references for standardized detection (Supplementary Table 6).
Flower Traits
Flowering transition, meristem identity choice, floral organ initiation, and floral organ morphogenesis are the four major steps of tomato flower development (Liu J. et al., 2014). (1) Plants respond to the external environment and their own signals during the flowering transition stage, transitioning from vegetative to reproductive growth. This process is controlled by a set of genes associated to flowering time (Liu J. et al., 2014). In tomato, two flowering control routes have been identified: photoperiod and autonomic. The photoperiod pathway is regulated by uniflora (UF) and compound inflorescence (S) gene, whereas the autonomous flowering pathway is regulated by single flower truss (SFT) gene and jointless (J) gene (Dielen et al., 2004; Molinero-Rosales et al., 2004; Liu J. et al., 2014). Flowering time-related genes S and J are positioned downstream of UF, which is a crucial gene in tomato that regulates flowering time (Quinet et al., 2006). Florigen signals are stimulated by SFT protein. The overexpression of SFT gene could stimulate flowering in tomatoes (Lifschitz and Eshed, 2006). (2) Plants respond to signals from diverse flowering time regulatory pathways to activate meristem characteristic genes and define meristem attributes during the meristem identity choice stage. After flowering induction, the vegetative meristem becomes the inflorescence meristem (IM) (Liu J. et al., 2014). Lateral organs replace leaves on the inflorescence meristem, which is followed by the flower meristem (FM) (Chandler, 2012). In tomato, the IM motifs SFT, UF, J, and Macrocalyx (MC) have been identified (Liu J. et al., 2014). (3) Meristem signature genes activate floral organ signature genes in various areas during the floral organ initiation stage. Meristem identity genes are critical genes that control the process of IM formation FM and determine the transition from flowering to floral organogenesis. Falsiflora (FA) and anantha (AN) were the FM-regulated genes in tomato (Liu J. et al., 2014). (4) Floral organ characteristic genes activate downstream organ morphogenesis genes during the floral organ morphogenesis stage, determining the precise cell types and tissues that make up each organ (Jack, 2004; Liu J. et al., 2014). The ABCE model is used to describe the mechanism of floral organ development (Krizek and Fletcher, 2005). The ABC genes are initially activated by floral meristem identification genes including leafy (LFY), apetala 1 (AP1), AP2, and unusual floral organs (UFO) gene, but later expression patterns are adjusted through interactions between the ABC genes. Four activities are present in adjacent whorls of the flower: A, B, C, and E. These four activities are thought to work together with different combinations to determine the identity of the flower’s four organs: sepals, petals, stamens, and carpels (Jack, 2004; Liu J. et al., 2014). In the whole steps, Auxin is also the key factor in determination of the discrimination and quantity of floral organs. The disorder of auxin signaling, transportation or biosynthesis always results in failure of floral discrimination (Cheng and Zhao, 2007).
The relative location of the pistil and stamen has a significant impact on plant fertility. The pistil is more exposed than the stamen in many plant species, and it is the main feature of cross-pollination (Chen and Tanksley, 2004). In hybrid systems of crops, particularly in the application of hybrid rice, the stigma exertion rate plays a vital function (Zhou et al., 2017). The stigma and the top of the stamen tube are almost equal in most wild tomatoes, indicating cross-pollination, whereas in domesticated tomatoes, the stigma and the top of the stamen tube being almost equal indicates self-pollination (Ranc et al., 2008).
The relative location of the pistil and stamen has a significant impact on plant fertility (Chen and Tanksley, 2004). Cross-pollination occurs in the majority of tomato species with exposed stigmas. On the other hand, most tomato species with similar lengths of pistils and stamens or with pistils shorter than the stamen display autocompatibility (Chen and Tanksley, 2004). The pistil of the sterile male varietal Ningqi No. 5 was substantially higher than the stamen among cross-pollinated L. barbarum varieties (Chen C. et al., 2017). Thus, the relative location of the pistil and stamen could be an indicator for identifying selfing fertility in Lycium plants.
In this section, we describe 27 flower-related traits and their detection references for standardizing the detection method.
Seed Traits
The seed is the primary site for endogenous hormone synthesis and accumulation, which is critical for fruit development. Previous studies in several fruits have revealed a strong link between fruit seed number and fruit size (Zheng et al., 2015). The incidence of the preharvest decline of fruits is associated with the loss of hormones from the seeds of almost ripe apples (Luckwill, 1948). Seed weight, length, and width have a substantial effect on fruit size development in L. barbarum according to previous studies. Seed number, on the other hand, has a greater impact on fruit size development than seed weight (Zheng et al., 2015; Wang J. et al., 2017). For the eight evaluated peach rootstocks, the fresh mass of fruits exhibits a strong positive association with the fresh mass of seeds (Souza et al., 2016). The fresh mass of seeds has a strong association with the germination speed index and mean germination time (Souza et al., 2016). Therefore, it can also serve as a predictor of plant reproductivity. Fruit yield was found to be positively associated with average seed yield and fruit weight per plant in a tomato study. The seed vigor index has the greatest direct effect on seed yield and is regarded as a useful measure for predicting seed lot performance in the field (Sharma, 2012).
According to research, there is a link between fruit development and seed development (Zheng et al., 2012). It reveals that the initial rapid growth phase of the fruit is also the first rapid growth period of the seed, but the seed’s growth ratio is faster than the fruit’s, and the seed’s endosperm grows rapidly during this period. Seeds had slow development characteristics when fruits entered the slow growth stage, and the rate of rise in seed length and width was much lower than the rate of growth of seeds in the initial fast growth stage, during which seeds mostly underwent embryo differentiation. Fruit volume and weight increased fast during the second rapid development stage, although seed length and width increased very slightly. In this stage, only the embryo grew larger (Zheng et al., 2012). Goji seed research shows a similar trend to tomato seed development (Wm Hilhorst et al., 1998). In the early stages, a significant increase in ABA content is invariably followed by an increase in seed weight, however, in the later stages, the ABA content falls and the seed enters the sluggish development stage (Wm Hilhorst et al., 1998).
In seed development stage, a variety of metabolites and jasmonic acid (JA) accumulate in certain organs and tissues. Jasmonate-inducible genes, are expressed in tandem with the accumulation, including developmentally regulated genes and defense genes (Wasternack et al., 2013). In seed formation, coordinated expression of embryo and endosperm is necessary, which is aided by both sporophytic and gametophytic genes from parents (Chaudhury et al., 2001). In the early stages of seed formation, the SlNAC3 gene (No apical meristem, NAM; ATAF1/ATAF2, and Cup-shaped cotyledon, CUC genes in tomato, Solanaceae) in tomato regulates embryo development, while factor for inversion stimulation (FIS) regulates endosperm development (Chaudhury et al., 2001; Han et al., 2014).
Although the chromosome ploidy of parents has an effect on seed fertility and the seed satiation rate (Li J. et al., 2001), in the case of good parental compatibility, the seed traits are mainly regulated by genetics. Research on Arabidopsis show that seed weight, size as well as the accumulation of seed oil and protein are regulated by APETALA2 (AP2) (Jofuku et al., 2005). Research on overexpression and mutation of da1-1 allele demonstrated that genes in this family play a key role as controller of organ and seed size in vegetation (Li et al., 2008). Thus, the feasibility of QTLs for seed traits was validated.
Here, eight kinds of traits and their standardized reference are provided (Supplementary Table 8).
Metabolites
The plant metabolome has a wide range of functions and is frequently viewed as a link between the genome and the phenome (Chen et al., 2016). It can make the link between genes and agronomic features easier. We found additional candidate genes potentially responsible for the variation in traits such as grain color and size using parallel mGWAS (metabolic GWAS) and pGWAS (phenotype GWAS) and provided evidence of a metabotype-phenotype linkage (Chen et al., 2016). Thus, research on mQTLs and mGWAS is crucial. With the advancement of liquid chromatography–mass spectrometry (LC–MS) technology, a large number of metabolites can be detected on a large scale with a targeted or untargeted metabolic profile, relative content, or absolute content (Chen W. et al., 2013; Chen et al., 2016). Khan et al. (2012) discovered 418 metabolites in apple peel and 254 in apple pulp using untargeted LC–MS metabolic profiling, of which 50% in the peel and 44% in the pulp had their mQTLs successfully identified (Khan et al., 2012). Chen et al. (2016) discovered 587 metabolites in cereal using a targeted metabolic profile, of which 331 (56.4%) showed at least one significant relationship, indicating a link between metabotype and phenotype (Chen et al., 2016). Shi created a broad-targeted metabolome approach for detecting 612 compounds in goji fruits in 2019 (Shi et al., 2019). Thus, LC–MS may be used to detect a large number of metabolites for mQTLs (or mGWASs). As Supplementary Table 9 shows, 870 metabolic traits and their reference detection methods have been described (Supplementary Table 9).
In addition, although the metabolic traits include the basic nutritional and medical functions of goji, according to International Standard of wolfberry (ISO 23193:2020), American Herbal Pharmacopoeia, British Pharmacopoeia Commission, European Pharmacopoeia, The standards for wolfberry in China (GB/T 18672-2014), Chinese Pharmacopoeia, Korean Pharmacopoeia, and Vietnam Pharmacopoeia, polysaccharide, total sugar, protein, zeaxanthin, zeaxanthin dipalmitate, betaine, and ash contents are regarded as quality indicators in wolfberry, and directly determine the level of quality (Korea Food and Drug Administration, 2014; Chinese Pharmacopoeia Commission, 2015; European Directorate for the Quality of Medicines, 2016; Japanese Pharmacopoeia Editorial Committee, 2016; British Pharmacopoeia Commission, 2017; Yao et al., 2018a; The American Herbal Pharmacopoeia, 2019). Thus, they can be key traits in breeding.
Postharvest Traits
The fruit processing and storage procedures for L. barbarum are determined by its postharvest quality. For example, Ningqi No. 1 and Ningqi No. 7 (elite L. barbarum cultivars) have excellent yield and disease resistance and are suitable for the production of dried fruits, but they are not suitable for the production of fruit juice due to their bitter and heavy medicinal flavor. Wolfberry with yellow fruits is ideal for making fruit juice because it has no unpleasant tastes, is bright in color, and leaves little residue after juicing (Wang et al., 2016). By examining the changes in the decay index and physiological indices of fresh L. barbarum fruit after harvest, Ge et al. (2008) discovered the ideal storage temperature for fresh fruits after harvest (Ge et al., 2008). Thus, mapping QTLs for postharvest traits can help to accelerate cultivar selection for the goji industry for different uses. In tomato, fruit firmness was validated to be regulated by GA2-oxidase, which is encoded by the FIS1 gene (Li et al., 2020). In Lycium species, Rehman et al. (2020) also mapped one QTL (qFF10–1) for fruit firmness traits (Rehman et al., 2020), confirming that fruit firmness is a gene-regulated trait and that mapping QTLs by linkage mapping is feasible. Twenty-three kinds of postharvest traits are described in this section for further research (Supplementary Table 10).
Progress of Marker-Assisted Selection Technology in Lycium
Genetic Marker Development
Molecular markers are based on nucleotide sequence changes, directly reflecting DNA sequence variations. Because molecular markers are not affected by environmental changes, development, differentiation, or cellular defense, compared with traditional phenotypic markers, they are more stable and observable (Agarwal et al., 2008).
The ideal genetic markers have the following characteristics: high genetic polymorphism, high reproducibility (the results can be repeated in different laboratories), codominance (the heterozygosity and homozygosity can pass an experimental test), low price (low cost of marker development and genotype identification), simple operation (automated operation, procedure, and sequencing), neutral selection (no multiple gene effects), markers uniformly distributed throughout the genome, and clear differentiation of alleles (Xu, 2010). Traditional molecular marker techniques include restriction fragment length polymorphism (RFLP), random amplified polymorphic DNA (RAPD), amplified fragment length polymorphism (AFLP), simple sequence repeat (SSR), and NGS-based single nucleotide polymorphism (SNP) (Blenda et al., 2012).
In 2000, molecular markers such as RAPD were first applied in Lycium species for the validation of the genetic diversity of L. barbarum (Cheng et al., 2000), while RAPD was also validated for its feasibility to distinguish close species of Lycium (Zhang et al., 2001). To date, simple sequence repeat (SSR), conserved ortholog set II (COSII), characterized amplified region (SCAR), amplified fragment length polymorphism (AFLP), restriction site−associated DNA sequencing (RAD-seq), internal transcribed spacer (ITS), sequence-related amplified polymorphism (SRAP), intersimple sequence repeat (ISSR), and random amplified, microsatellite polymorphism (RAMP)-PCR markers have been applied in genetic diversity, population structure, morphological variation, phylogenetic inference, traceability, and cultivar/species identification and discrimination (Zhang et al., 2001, 2018; Yin et al., 2005; Sze et al., 2008; Chung et al., 2009; Kwon et al., 2009; Levin et al., 2009; Zhao et al., 2010; Balasubramani et al., 2011; Liu et al., 2012, 2020b,c; Tripathi, 2013; Xin et al., 2013; Chen and Zhong, 2014; Wang et al., 2015a; Chen C. et al., 2017; Chen J. et al., 2017; McCulloch et al., 2020; Jung et al., 2021). SSR markers can be used for the detection of genetic polymorphisms of species, calculation of genetic relationships (genetic distance) between varieties, identification of cultivars, and even construction of genetic maps and QTL mapping with sufficient SSR markers (Chen H. et al., 2015; Essid et al., 2015; Ibrahim et al., 2016; Thammina et al., 2017; Portis et al., 2018b). One hundred fifteen SSR and 12 ILP markers were applied to construct a genetic map with the F1 population (Hu, 2015). However, because of the limited number of markers and huge average genetic distance between two adjacent markers, QTL mapping was not suitable (Hu, 2015). To address this problem, Chen C. et al. (2017) developed 22,537 EST-SSR markers using transcriptomes of different fruit ripening stages.
Linkage Mapping
Linkage mapping is based on the frequency of recombination between two markers, which can be used to determine their genetic distance on chromosomes. The greater the genetic distance between two markers on a chromosome, the higher their exchange frequency (Xu, 2010). A genetic map depicts the linear order of DNA markers on a chromosome based on their relative positions (Madhusudhana, 2015). The linkage mapping procedure mainly includes constructing mapping population(s), trait data collection, sequencing and genotyping with polymorphic markers, genetic map construction, and QTL mapping (marker-trait associations between genotypic and trait phenotypic data Madhusudhana, 2015; Figure 1A). Linkage mapping can be applied under QTL mapping for multiple traits at the same time (Meuwissen and Goddard, 2004). The linkage mapping (genetic map) population is always constructed with a cross of two parents (Yu et al., 2008). There are several advantages of linkage mapping: (1) The complexity of the population structure is reduced due to parental confirmation, which improves accuracy and reduces false-positives; and (2) the mapping population is constructed by combining the offspring of two parents. Even if the features are regulated by rare genes, the offspring contains a significant proportion of unusual genes. As a result, linkage mapping may be able to locate rare genes (Mackay and Powell, 2007; Yu et al., 2008).
Linkage mapping has been successfully applied in Lycium species (Table 2). The intraspecies genetic map consisted of 23,967 markers with a size of 964.03 cM (Gong et al., 2019). According to the genome published by Cao, the size of the genome in L. barbarum is 1.67 Gb. Thus, we determined that the average DNA content between two neighboring markers (genome size/marker number) was 69,679 bp. However, the two interspecies genetic maps had higher genetic distances (1702.45 and 1649.03 cM) and lower marker numbers (6,733 and 3,495 cM) (Zhao et al., 2019; Rehman et al., 2020), and the average DNA content between two neighboring markers was 248,032 and 477,825 bp. Obviously, intraspecies genetic maps have higher resolution than intraspecies genetic maps. However, all three maps still have higher resolution than most other species, such as maize (130,000 bp) and grape (676,000 and 606,000 bp in two parent maps) (Chen et al., 2014; Chen J. et al., 2015), which were successfully mapped to QTLs. Thus, mapping QTLs with linkage mapping is applicable to Lycium species.
TABLE 2.
Genetic map construction and QTL mapping in Lycium species.
| Population type | Parents | Population size | Chr | Length (cM) | No. of markers | Marker interval (cM) | Max interval (cM) | Traits | QTL | References |
| Intraspecies | Lb × Lb | 305 | 12 | 964.03 | 23,967 | 0.04 | 1.98 | 6 | 32 | Gong et al., 2019 |
| Interspecies | Lb × Lc | 302 | 12 | 1702.45 | 6,733 | 0.31 | 11.7 | 7 | 55 | Zhao et al., 2019 |
| Interspecies | Lc × Lb | 305 | 12 | 1649.03 | 3,495 | 0.47 | 16.99 | 11 | 117 | Rehman et al., 2020 |
Lb, L. barbarum; Lc, L. chinense; Chr, chromosome.
Other Marker-Assisted Selection Techniques With Potential Value
Bulk Segregation Analysis
Bulk segregant analysis (BSA) is a QTL mapping technique for locating genomic regions with genetic loci that impact a trait of interest (Michelmore et al., 1991; Magwene et al., 2011). Individuals are assayed for the focal characteristic in a segregating population derived from a genetic cross, and two pools (bulks) of segregants are established by choosing individuals from the phenotypic distribution tails (other sampling designs can also be used as discussed below). Individual genotyping or the production of pooled DNA samples from which allele frequencies are obtained are used to estimate genotype frequencies for the two bulks. In genomic regions with no loci affecting the trait, allele frequencies should be roughly comparable between the two bulks. Allele frequencies should differ between bulks in regions of the genome that include causative loci. High marker density and accurate allele frequency estimation inside bulks are the most effective features of BSA (Ehrenreich et al., 2009; Magwene et al., 2011; Lu et al., 2021).
Although utilization of BSA to map QTLs in Lycium species has not yet been reported, BSA, with its cost-effectiveness and efficiency, has been widely used to map QTLs in woody, herbaceous species, including apple and watermelon (Dong et al., 2018; Liu et al., 2021), as well as neighboring species in Solanaceae, such as fruit color in pepper, tomato, and eggplant (Lee et al., 2020; Lu et al., 2021; Tang et al., 2021). Thus, it has the potential for application in Lycium species.
The BSA procedure is performed by following key points: establishing a segregating population, collecting trait data and performing bulk selection. Individuals with high and low values for the trait of interest are phenotyped and selected. These individuals’ DNAs are pooled into high and low bulks, which are then sequenced and SNP-called, obviating the requirements to design markers ahead of time and to statistically analyze linkage distances among markers of the two bulks. SNPs discovered in reads derived from regions not connected to the trait of interest should be approximately 50% of the reads in bulks selected from populations (Figure 1B). Depending on the bulk, SNPs in reads aligning to genomic regions directly associated with the trait should be over- or underrepresented. Under QTL mapping, QTL identification can be accomplished by comparing relative allele depths or SNP indices (defined as the number of reads containing an SNP divided by the total sequencing depth at that SNP) between the bulks (Mansfeld and Grumet, 2018). BSA is a cost-effective and efficient approach to fine-mapping genes in unsequenced genomes by targeting SNPs to specific genomic intervals (Trick et al., 2012).
Association Mapping
Genome-wide association study (GWAS) is an analytical method based on linkage disequilibrium of genes or loci among biological populations, combining genotype and phenotypic data from mapping populations and analyzing the relationship between detection markers or loci and traits using statistical methods (Yu and Buckler, 2006). The procedure of association mapping comprises population selection, morphological data collection, NGS and genotyping, linkage disequilibrium analysis, and association mapping between markers and traits (Zhu et al., 2008; Figure 1C).
GWAS mapping technique is regarded as a substitute for linkage mapping for research on association between genotype and phenotype. Based on recombination to reorganize the genome, both techniques are for evaluating associations between genotype and phenotype. Linkage disequilibrium (LD), the term to evaluate the non-random connotation between markers, was the basis of GWAS (Barchi et al., 2019). GWAS offers three advantages over linkage analysis: (1) High precision and resolution. Linkage disequilibrium (LD) declines quickly after multiple hybridization, recombination, and mutation information from associated populations, and localization precision is considerably increased with high resolution. (2) Without the constraints of the parental population, multiple gene types can be discovered on the same locus. (3) Time efficiency is achieved by using natural populations as a source of material, which reduces the time required to produce genetic populations (Yu and Buckler, 2006; Madhusudhana, 2015; Hu et al., 2018). To date, there is still no report about the utilization of association mapping in Lycium species.
However, utilization in neighboring species in Solanaceae, such as QTLs for leaf-, plant-, and fruit-related traits in eggplant (Portis et al., 2015), virus resistance in pepper (Tamisier et al., 2020), and multiple root- and fruit quality-related traits in tomato (Tamisier et al., 2020), demonstrates potential utilization in the breeding of Lycium species.
Mapping Population and Strategies of High-Density Genetic Map Construction in Lycium
Classification of Mapping Populations
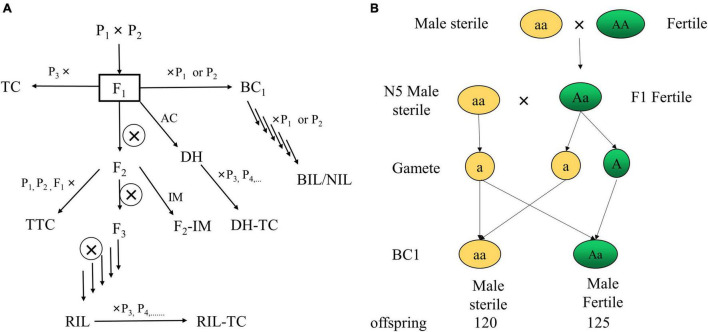
The mapping population can be classified into anther culture (AC), backcross population (BC), backcross inbred line (BIL), double haploid (DH), intermating (IM), near-isogenic line (NIL), recombinant inbred line (RIL), testcross (TC), and triple testcross (TTC) (Xu, 2010; Figure 4A).
FIGURE 4.
Classification of mapping populations (A) (Xu, 2010) and strategy of population construction through sterile male QTL mapping (B). Pn, parent; F1, filial generation 1; AC, mapping population can be classified into anther culture; BC, backcross population; BIL, backcross inbred line; DH, double haploid; IM, intermating; NIL, near-isogenic line; RIL, recombinant inbred line; TC, testcross; TTC, triple testcross; N5, a sterile male cultivar of L. barbarum.
Mapping With F1 Population
Self-incompatibility is common among woody plants. After a lengthy period of evolution, Lycium species, like other woody plants, are genetically polymorphic and extremely heterozygous (Zhao et al., 2010). Goji are woody plants with a long development cycle, high genomic heterozygosity, and difficulty acquiring homozygous genes, making the genetic map of woody plants more difficult to generate than that of herbaceous species (Song et al., 2012; Guo et al., 2015; Zhang et al., 2015). However, the high heterozygosity of woody plants satisfies the requirements for genetic map construction and QTL mapping using the F1 population. Thus, all genetic maps of woody plants and some highly heterozygous perennial herbs, such as Prunus avium, Juglans regia, Camellia sinensis, Paeonia Sect., Prunus mume, and Cynara cardunculus, are currently based on F1 populations (Cai et al., 2015; Ma et al., 2015; Wang et al., 2015b; Zhang et al., 2015; Zhu et al., 2015; Portis et al., 2018a). To date, three genetic maps of Lycium species have been constructed using the F1 population. The first high-density genetic map was constructed with an intraspecies cross population of L. barbarum (Gong et al., 2019), while the other two were L. barbarum and L. chinense interspecies cross populations (Zhao et al., 2019; Rehman et al., 2020).
Mapping With Other Population
Although the F1 population is suitable for genetic map development, as shown in Figure 4B, several features, particularly qualitative features governed by recessive alleles, show no distribution in F1 offspring, according to our research. Thus, selfing, sib mating and back-crossing to construct F2 or BC1 populations are necessary. For example, Ningqi No. 5 (N5) is a L. barbarum (Chen C. et al., 2017) sterile male cultivar, and we crossed N5 with Zhongkeluchuan No. 1 to examine sterile male features. The F1 offspring, on the other hand, are entirely reproductive males. As a result, we backcrossed with N5 and obtained 120 sterile males and 125 fertile plants. Thus, we obtained an F2 population with male sterility segregation (sterile males: fertile plants = 1:1) that can be used for QTL mapping, and we discovered that male sterility is regulated by a single recessive gene. The segregations derived from diploid parents can be classified into eight segregations (ab × cd, ef × eg, hk × hk, lm × ll, nn × np, aa × bb, ab × cc and cc × ab), and only homozygous segregation pattern (aa × bb) is suitable for constructing the genetic map in BC1 and F2 populations (Fernando, 1982; Yu et al., 2013; Zhang et al., 2013). According to Zhao et al. (2019) and Rehman et al. (2020), the number of genotypes (aa × bb) in the parent (L. barbarum × L. chinense) is 17,513 (17.02%) and 24,329 (59.9%), respectively, indicating that the F2 and BC1 populations can be used to generate genetic maps and map QTLs in Lycium species. Furthermore, using the newly published (Cao et al., 2021) L. barbarum genome by Cao et al. (2021) as a reference genome, we can map QTLs using a more cost-effective method, such as BSA.
Multiparental Population
When investigating natural populations, genome-wide association analysis offers numerous advantages, but it also has many drawbacks: (1) the population structure has an impact on the accuracy of GWAS analysis. Locus identification is prone to false-positives when the population structure is complicated. (2) Rare genes in populations are challenging to find using GWAS. Due to their low proportion and wide distribution in the population (linkage mapping), rare genes are easily filtered out of analyses (Lander and Schork, 1994). Linkage map analysis, on the other hand, has the following drawbacks: (1) due to the low frequency of marker recombination, the coverage of markers in broad chromosome regions is low (Yu et al., 2008; Hu et al., 2018); (2) a mapped population cannot be used for numerous traits because the population is only derived from the cross of two parents and the number of variant characteristics is limited compared with association mapping (Yu et al., 2008); and (3) it takes a long time to build the population because it is built by crossing (Madhusudhana, 2015).
However, in terms of prior knowledge, cross-validation, and statistical power, linkage analysis and association mapping are complementary to one another (Wilson et al., 2004). The genetic population of association mapping, on the other hand, is a natural population, whereas linkage mapping is invariably a biparental population. Yu et al. (2008) suggested the nested association mapping (NAM) technique to integrate the benefits of both linkage and association mapping strategies (Madhusudhana, 2015). The Buckler lab at Cornell University introduced nested association mapping (NAM) for the first time in 2008. Large populations with many small populations can be created by hybridizing multiple parents with a common parent (Buckler et al., 2009). The inbred line B73 was chosen as the common parent in the maize NAM population construction, and it was crossed with 25 other inbred lines with a wide genetic variety and then self-crossed to the F5 generation using the single-grain transfer method. Finally, 25 RIL populations were acquired. There were 200 recombinant inbred lines (RILs) in each population (Madhusudhana, 2015). The use of the NAM population for QTL mapping has two obvious benefits: (1) because the NAM population has distinct parents, both association and linkage analyses can be performed at the same time, and mapping approaches are more diversified and versatile; (2) the population structure can be simplified, and the false-positives generated by the complicated population structure can be minimized because there is a common parent and a clear population structure (Madhusudhana, 2015). Minor changes are required to apply the NAM population to Lycium. Unlike herbaceous plants that have a brief lifespan, goji species are shrubs that live for decades (Cao and He, 2013) and can be bred by cuttage (Huang et al., 2015), which is convenient for germplasm resource conservation. As a result, there is no need to create a recombination inbred line. Furthermore, because goji species, like other shrub plants, are heterozygous (Zhao et al., 2010), they are excellent for mapping with F1 populations. Conclusively, NAM populations can be created using F1 populations crossed by a common parent with distinct cultivars/lines. After modest modifications, other multiparental populations, such as multiparental advanced generation inter-crosses (MAGIC) (Pascual et al., 2015), random-open-parent association mapping (ROAM) (Glowinski and Flint-Garcia, 2018), and complete-diallel plus unbalanced breeding-derived inter-crosses (CUBIC) (Liu et al., 2020a), can be used in Lycium breeding.
Perspectives of Lycium Species Breeding
Mechanized Harvest
In China, the level of mechanization of Chinese medicinal materials is rather low; production is primarily through manpower, and the instruments are simple (Junfa et al., 2009), particularly for goji berries. Currently, the harvesting of goji is mainly reliant on manpower, resulting in low picking efficiency and high picking costs (Zheng et al., 2018). Furthermore, with the rapid migration of rural workers to urban areas, agricultural labor costs are rising. With a comprehensive understanding of farmers’ labor (Junfa et al., 2009) costs, it is critical to build harvesting machinery systems (Zheng et al., 2018). A one-time picking technique, which requires crop growth periods and harvest times to be coordinated, can be utilized to cut labor costs and use mechanization (Zheng et al., 2018). However, due to the complicated growth features of goji fruit, mechanical harvesting is challenging due to the presence of ripe fruits, immature fruits, and blossoms on the same branch at the same time (Zheng et al., 2018). As a result, cultivating new varieties that are compact, tolerant to dense planting, and suitable for mechanized operation and have synchronized fruit ripening and compact plant architecture, has become the primary goal of agricultural genetic improvement (Guo et al., 2019). In coffee fruits, synchronized flowering and fruiting and a reduction in fruit removal force were shown to be ideal for mechanical harvesting (Tongumpai, 1993). Thus, we need to determine which characteristic makes fruit ripening more coordinated. The fruit ripening period of plants with a flower terminal is comparatively concentrated, according to tomato research, which is helpful to mechanical harvesting (Li et al., 2018; Guo et al., 2019). Flower production and limited growth at the apex of the branch can promote uniform fruit ripening, providing theoretical support and practical direction for plant type breeding suitable for mechanical harvesting operations (Guo et al., 2019). For many years, crop physiology, culture, and breeding have focused on improving flower terminal characteristics. Centroradialis (CEN) gene expression increases in the inflorescence, and the apex stops growing and blossoms, indicating apical capping (Bradley et al., 1996). The flower terminal of the tomato was found to be controlled by a gene, and gene editing might be used to domesticate it (Pnueli et al., 1998; Li et al., 2018).
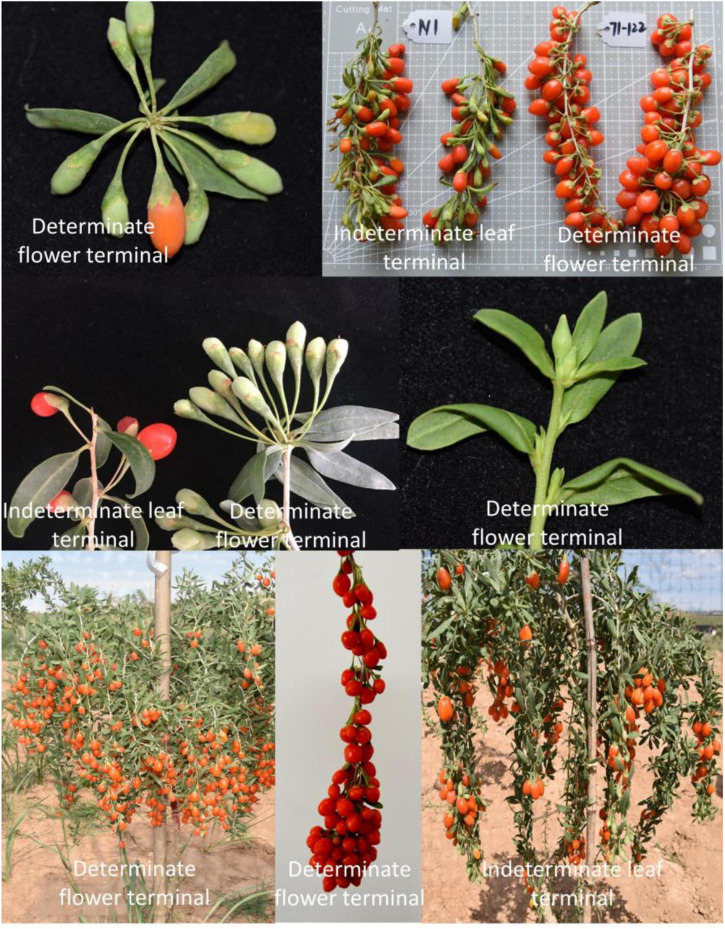
The M1 strain developed in the early stages of our research group was used as the female parent, and Zhongkeluchuan1 was used as the male parent. A mutant plant with the flower terminal attribute was established in the established genetic population (population size, 425 plants). The branch tip stops growing when the new branches reach 40 cm, and the apical meristem develops into a cluster of flowers (Figure 5). The ripening rate of the fruits on the same branch is very consistent. The discovery of this germplasm will aid in the future identification of candidate genes for controlling flower terminal characteristics in L. barbarum. The directional breeding of congruent varieties of L. barbarum, as well as the functional verification of genes and the establishment of molecular markers, provides a solid theoretical and practical foundation.
FIGURE 5.
Comparison of cultivars with indeterminate leaf terminal and determinate flower terminal.
Thus, to fulfill this aim, we could select approximately 300 offspring and their parents as genetic populations to construct a high-density genetic map with linkage mapping (SLAF-seq or ddRAD-seq) technology according to previous studies (Figure 1A, steps 1, 3, and 4) (Gong et al., 2019; Zhao et al., 2019; Rehman et al., 2020). For flower terminal trait detection, traits No. 8 (flower terminal) and No. 9 (flower terminal rate) in Supplementary Table 7 can be reference methods (Figure 1A, step 2). Finally, as Figure 1A and Step 5 show, QTLs for flower terminals could be mapped in a genetic map with the combination of sequencing and phenotypic data. In addition, BSA-seq could also be used as an alternative, as shown in Figure 1B.
The detected QTLs could be developed as markers for determinate terminal trait selection in Lycium species, which will accelerate the selection of flower terminal cultivars. Moreover, the publication of the genome of L. barbarum (Cao et al., 2021) enables the mining of candidate genes for further study, such as candidate gene validation, rapid breeding of cultivars with determinate flower terminal using transgenic technology, and studies on the mechanism of terminals flower development.
Cultivars for Fruits
The goji products on the market right now are mostly dried fruits; however, the drying process causes losses of proteins, carotenoids, fatty acids, and other bioactive elements (Reeve et al., 2010; Hu et al., 2011). Fresh fruits, with their complete nutritional value and delicious flavor, can meet consumer demands for functional fruits. Therefore, fresh wolfberry, as a new functional fruit, has large market potential (Huang et al., 2013). The goji industry should be built on the basis of medicinal value and expanded on the basis of editable value. Thus, we should move beyond the research category of therapeutic impact and broaden the area of fresh food research to broaden the application and industrial scope (Qin and Dai, 2017). Fresh fruits, as opposed to dried fruits, must have a stunning appearance, a lengthy fruit, thick flesh, hard skin, a sweet and good taste, and other favorable traits. However, most modern cultivars have plump pulp, thin skin, and a high moisture content. They are easily damaged while being picked and during transportation, resulting in cracked peels and eventually moldy fruits (Li et al., 2010). Therefore, fruit color, size (especially length), pulp thickness, peel stiffness, and sensory taste should be prioritized in Lycium fruit breeding.
Obviously, there are many phenotypic requirements in fruit cultivars. To facilitate the breeding of fruit cultivars, we need a population with segregations of all traits required. However, it is not easy to construct a biparental population with so many distinguishing variations. Thus, the NAM population, constructed by crossing multiple parents and one common parent (Figure 6), with sharp segregations of multiple kinds of traits, could be a suitable option. NAM populations plus both association mapping and linkage mapping technologies could map QTLs for multiple fruit use related traits, with both high resolution and precision. Moreover, besides the contribution to QTL to facilitate breeding of fruit use cultivar (same to QTL for flower terminal trait), NAM population could cause more plant with more significant phenotypic variations, such as fruit color, fruit shape, leaf shape, plant shape, and et al. (Figures 3G–J), which facilitate the preservation of germplasm resources for ornamental horticulture and other potential uses.
FIGURE 6.
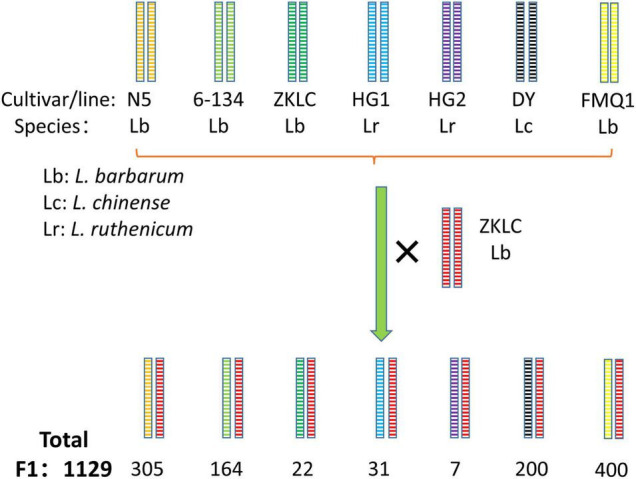
Construction of NAM population in Lycium. cultivar/line of L. barbarum: N5, 6-134, ZKLC, FMQ1; cultivar/line of L. chinense: DY; cultivar/line of L. ruthenicum: HG1, HG2; × : cross.
Improving Light Use Efficiency
Global food crop production has doubled in the last two decades and continues to rise (Evans, 1996). According to Richard’s research, the increases in grain yield in recent decades have two characteristics: (1) photosynthesis theory can be used to boost agricultural yields; (2) photosynthesis-related genetic theories have not been used to boost crop yields (Richards, 2000). Photosynthesis is genetically controlled and plays a crucial role in plant development, biomass buildup, and crop yield (Qu et al., 2017). Therefore, photosynthesis has not been completely utilized in modern agricultural production despite having a great development potential for increasing crop yields (Long et al., 2015). Improving the photosynthetic rate is thought to be one of the most essential ways to boost crop yield (Makino, 2011). There are a variety of techniques to boost the net photosynthetic rate, including increasing light gathering capacity, light energy conversion efficiency, CO2 fixation ability, and CO2 conversion efficiency in leaves (Ort et al., 2015). However, there have been few findings on genes that control the photosynthetic biosynthesis pathway (Osipova et al., 2016). The genes that affect the photosynthetic rate and water use efficiency are being studied to improve these qualities (Zheng et al., 2011).
There are sharp differences in photosynthesis between cultivars and species in Lycium, such as L. cylindricum (net photosynthetic rate, Pn, 9.40 μmol CO2 s–1 m–2), L. yunnanense (14.18 μmol CO2 s–1 m–2), L. dasystemum (19.46 μmol CO2 s–1 m–2), L. ruthenicum (7.29 μmol CO2 s–1 m–2), L. barbarum L. var. auranticarpum K. F. Ching (8.69 μmol CO2 s–1 m–2), L. barbarum “Ningqi-5” (9.20 μmol CO2 s–1 m–2) and L. barbarum L. “Zhongkelüchuan-1” (18.19 μmol CO2 s–1 m–2) (Cao and Wu, 2015; Gong et al., 2019), providing multiple resources for research on photosynthetic traits. Moreover, Gong et al. (2019) detected 29 QTLs for photosynthetic traits, including Pn, water use efficiency (WUE), limiting value of the stoma (Ls), transpiration rate (Trmmol), intracellular carbon dioxide (Ci), and stomatal conductance (Cond) (Gong et al., 2019), which will greatly shorten the breeding procedure. Existing photosynthetic QTLs could be developed as markers for photosynthetic trait selection in Lycium species, which will accelerate the selection of flower terminal cultivars. The other steps, including candidate gene mining, validation, rapid breeding of cultivars with highly efficient photosynthesis and study of the mechanism of photosynthesis, are the same as the procedure for research on flower terminal traits mentioned above.
Role of Marker-Assisted Selection in the Cultivation and Conservation of Germplasm Resources and Research on Multiple Traits
MAS with NGS consists of germplasm resource collection, population construction, trait detection, sequencing and genotyping, genetic statistical analysis, and QTL mapping (Figure 1). Thus, research on multiple traits with MAS could facilitate not only the breeding of elite cultivars but also cultivation and conservation of new germplasm resources.
Worldwide, there are 97 species with different variations and functions in Lycium (Yao et al., 2018a), and they provide a variety of germplasm resources for cultivation and breeding in goji plants. Here, we summarized the distribution of all species in Lycium, species richness in all countries worldwide, location of existing germplasm repositorys, and functions of different species according to existing published studies (Figure 2 and Supplementary Table 1), simplifying and facilitating the collection and conservation of germplasm resources.
Most of the population in MAS was constructed by crossing. Different ploidies of chromosomes in parents may cause highly sterile progeny and a low rate of seed satiation, resulting in difficult breeding of seedless plants (Li J. et al., 2001). Validation of karyotype analysis in parents is necessary (Xu, 2010). To select intra- and interspecies parents for crossing and evaluate their fertilities, Table 2 could be referenced. Table 2 summarizes the karyotype analysis of 50 species, eight varieties, and one hybrid plant, among which 42 species had the diploid type. Crosses can occasionally create plants with new variations, such as fruit color (Figure 3) and flower terminals (Figure 5), which will facilitate the cultivation of new germplasms.
Compared with other crops, studies on the resource base of Lycium species, including systematic and in-depth research, are still lagging behind (An et al., 2009). Conversely, studies on other Solanaceae species, such as tomato and potato, are quite in depth, regardless of gene research, the variety of features, or other themes. Thus, the conclusion in our research on 1,144 traits (274 agronomic and 870 metabolic) and their standardized detection based on tomato, Lycium, and other Solanaceae species in the prospective trait supplying section (Supplementary Tables 2–10) will facilitate the morphological evaluation, conservation and breeding of Lycium germplasm.
QTLs derived from sequencing data and phenotypic data have two kinds of usages. One is for marker development, which could accelerate the selection for elite cultivars with specific trait selection. The present emphasis on practical breeding is incorporating biotechnological methods for the development of a variety of robust markers to execute MAS, in order to speed up the breeding of elite cultivars and delivery of them to the producer (Rubiales et al., 2021), such as the breeding of elite cultivars in grain, tomato, pepper, and eggplant (Foolad and Panthee, 2012; Hanson et al., 2016; Khapte et al., 2018; Moodley et al., 2019). The other is for genetic and mechanistic research of the traits, for example, rice grain. A gene (Gn1a) encoding cytokinin oxidase/dehydrogenase was detected after mapping QTLs for grain yield. The enzyme was found to restrict the synthesis of the phytohormone cytokinin. After a series of validations, the mechanism was successfully demonstrated as gene-cytokinin oxidase/dehydrogenase-phytohormone cytokinin-grain yield (Ashikari et al., 2005).
Finally, after reviewing many studies, we came to a conclusion on the future breeding research direction of Lycium species. This review will help to guide future Lycium breeding studies.
Author Contributions
HG: conceptualization, methodology, software, writing—review and editing, and visualization. FR, SZ, ZL, and TY: validation and formal analysis. BA: investigation. YM: resources. DW: data curation. JH: writing—original draft preparation. YW: supervision, project administration, and funding acquisition. All authors contributed to the article and approved the submitted version.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We thank Qin Ken from NingXia Academy of Agricultural and Forestry Sciences for providing Pictures.
Footnotes
Funding
This research was funded by the National Key Research and Development Program Project (Grant No. 2019YFC1711100), the Sub-Project of Chinese Academy of Sciences Pilot Project (Grant No. XDA24030502), the Guangdong Provincial Special Fund for Modern Agriculture Industry Technology Innovation Teams, China (Grant No. 2020KJ148), and the National Natural Science Foundation of China (Grant No. 31770334).
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.802936/full#supplementary-material
References
- Agarwal M., Shrivastava N., Padh H. (2008). Advances in molecular marker techniques and their applications in plant sciences. Plant Cell Rep. 27 617–631. 10.1007/s00299-008-0507-z [DOI] [PubMed] [Google Scholar]
- Alpert K. B., Grandillo S., Tanksley S. D. (1995). Fw 2.2: a major qtl controlling fruit weight is common to both red-and green-fruited tomato species. Theor. Appl. Genet. 91 994–1000. 10.1007/BF00223911 [DOI] [PubMed] [Google Scholar]
- An W., Zhang H., He J., Li X., Fan Y. (2009). Research progress in goji breeding. N. Hortic. 5 125–128. [Google Scholar]
- An W., Zhao J., Shi Z., Jiao E. (2007). Evaluation criteria of some fruit quantitative characteristics of wolfberry (Lycium) genetic r esources. J. Fruit Sci. 24 172–175. [Google Scholar]
- Ashikari M., Sakakibara H., Lin S., Yamamoto T., Takashi T., Nishimura A., et al. (2005). Cytokinin oxidase regulates rice grain production. Science 309 741–745. 10.1126/science.1113373 [DOI] [PubMed] [Google Scholar]
- Azadi N., Nazeri V., Namaki Shoushtari A. A. H., Kazem Pour Ousalou S. (2007). Lycium dasystemum pojark. (Solanaceae), a new record from iran. Iranian J. Bot. 13 109–111. [Google Scholar]
- Balasubramani S. P., Goraya G. S., Venkatasubramanian P. (2011). Development of its sequence-based markers to distinguish berberis aristata dc. From b. Lycium royle and b. Asiatica roxb. 3 Biotech 1 11–19. 10.1007/s13205-010-0001-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barchi L., Lefebvre V., Sage-Palloix A., Lanteri S., Palloix A. (2009). Qtl analysis of plant development and fruit traits in pepper and performance of selective phenotyping. Theor. Appl. Genet. 118 1157–1171. 10.1007/s00122-009-0970-0 [DOI] [PubMed] [Google Scholar]
- Barchi L., Portis E., Toppino L., Rotino G. L. (2019). Molecular Mapping, Qtl Identification, and Gwa Analysis. Cham: Springer International Publishing, 41–54. [Google Scholar]
- Bernardello L., Rodriguez I., Stiefkens L., Galetto L. (1995). The hybrid nature of Lycium ciliatum × cestroides (Solanaceae): experimental, anatomical, and cytological evidence. Can. J. Bot. 73 1995–2005. 10.1139/b95-214 [DOI] [Google Scholar]
- Blenda A., Fang D. D., Rami J., Garsmeur O., Luo F., Lacape J. M. (2012). A high density consensus genetic map of tetraploid cotton that integrates multiple component maps through molecular marker redundancy check. PLoS One 7:e0045739. 10.1371/journal.pone.0045739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bombarely A., Menda N., Tecle I. Y., Buels R. M., Strickler S., Fischer-York T., et al. (2010). The sol genomics network (solgenomics.net): growing tomatoes using perl. Nucleic Acids Res. 39 D1149–D1155. 10.1093/nar/gkq866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradley D., Carpenter R., Copsey L., Vincent C., Rothstein S., Coen E. (1996). Control of inflorescence architecture in antirrhinum. Nature 379 791–797. 10.1038/379791a0 [DOI] [PubMed] [Google Scholar]
- British Pharmacopoeia Commission (2017). British Pharmacopoeia. London: British Pharmacopoeia Commission, 91–92. [Google Scholar]
- Buckler E. S., Holland J. B., Bradbury P. J., Acharya C. B., Brown P. J., Browne C., et al. (2009). The genetic architecture of maize flowering time. Science 325 714–718. 10.1126/science.1174276 [DOI] [PubMed] [Google Scholar]
- Cai C., Cheng F., Wu J., Zhong Y., Liu G. (2015). The first high-density genetic map construction in tree peony (Paeonia sect. Moutan) using genotyping by specific-locus amplified fragment sequencing. PLos One 10:e0128584. 10.1371/journal.pone.0128584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Y., He J. (2013). Cultivation of Lycium. Shenyang: Yangguang Press. [Google Scholar]
- Cao Y., Li Y., Fan Y., Li Z., Yoshida K., Wang J. Y., et al. (2021). Wolfberry genomes and the evolution of Lycium (solanaceae). Commun. Biol. 4:671. 10.1038/s42003-021-02152-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Y., Wu P. (2015). Wolfberry Germplasm Resources in China. Beijing: China Forestry Publishing House. [Google Scholar]
- Chandler J. W. (2012). Floral meristem initiation and emergence in plants. Cell. Mol. Life Sci. 69 3807–3818. 10.1007/s00018-012-0999-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhury A. M., Koltunow A., Payne T., Luo M., Tucker M. R., Dennis E. S., et al. (2001). Control of early seed development. Annu. Rev. Cell Dev. Biol. 17 677–699. 10.1146/annurev.cellbio.17.1.677 [DOI] [PubMed] [Google Scholar]
- Chen C., Xu M., Wang C., Qiao G., Wang W., Tan Z., et al. (2017). Characterization of the Lycium barbarum fruit transcriptome and development of est-ssr markers. PLoS One 12:e187738. 10.1371/journal.pone.0187738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen D., Zhou X., Li L., Liu C., Chen Y. (2015). Research progress on rice (Oryza sativa. L) leaf thickness. J. Agric. 5 22–25. [Google Scholar]
- Chen H., Song Y., Li L., Khan M. A., Li X., Korban S. S., et al. (2015). Construction of a high-density simple sequence repeat consensus genetic map for pear (Pyrus spp.). Plant Mol. Biol. Rep. 33 316–325. 10.1007/s11105-014-0745-x [DOI] [Google Scholar]
- Chen H., Zhong Y. (2014). Microsatellite markers for Lycium ruthenicum (Solananeae). Mol. Biol. Rep. 41 5545–5548. 10.1007/s11033-014-3442-9 [DOI] [PubMed] [Google Scholar]
- Chen J., Chao C. T., Wei X. (2018). Gojiberry Breeding: Current Status and Future Prospects. London: IntechOpen. [Google Scholar]
- Chen J., Liu X., Zhu L., Wang Y. (2013). Nuclear genome size estimation and karyotype analysis of Lycium species (Solanaceae). Sci. Hortic. 151 46–50. 10.1016/j.scienta.2012.12.004 [DOI] [Google Scholar]
- Chen J., Wang N., Fang L., Liang Z., Li S. H., Wu B. H. (2015). Construction of a high-density genetic map and QTLs mapping for sugars and acids in grape berries. BMC Plant Biol. 15:28. 10.1186/s12870-015-0428-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J., Zhang D., Zhang C., Xu M., Yin W. (2017). Physiological characterization, transcriptomic profiling, and microsatellite marker mining of Lycium ruthenicum. J. Zhejiang Univ. Sci. B 18 1002–1021. 10.1631/jzus.B1700135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen K. Y., Tanksley S. D. (2004). High-resolution mapping and functional analysis of se2.1: a major stigma exsertion quantitative trait locus associated with the evolution from allogamy to autogamy in the genus Lycopersicon. Genetics 168 1563–1573. 10.1534/genetics.103.022558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W., Gong L., Guo Z., Wang W., Zhang H., Liu X., et al. (2013). A novel integrated method for large-scale detection, identification, and quantification of widely targeted metabolites: application in the study of rice metabolomics. Mol. Plant 6 1769–1780. 10.1093/mp/sst080 [DOI] [PubMed] [Google Scholar]
- Chen W., Wang W., Peng M., Gong L., Gao Y., Wan J., et al. (2016). Comparative and parallel genome-wide association studies for metabolic and agronomic traits in cereals. Nat. Commun. 7:12767. 10.1038/ncomms12767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z., Wang B., Dong X., Liu H., Ren L., Chen J., et al. (2014). An ultra-high density bin-map for rapid qtl mapping for tassel and ear architecture in a large f2 maize population. BMC Genomics 15:433. 10.1186/1471-2164-15-433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng K. T., Chang H. C., Huang H., Lin C. T. (2000). Rapd analysis of Lycium barbarum medicine in Taiwan market. Bot. Bull. Acad. Sin. 41 11–14. 10.7016/BBAS.200001.0011 [DOI] [Google Scholar]
- Cheng Y., Zhao Y. (2007). A role for auxin in flower development. J. Integr. Plant Biol. 49 99–104. 10.1111/j.1744-7909.2006.00412.x [DOI] [Google Scholar]
- Chinese Pharmacopoeia Commission (2015). Chinese Pharmacopoeia 1. Beijing: China Medical Science Press, 124–249. [Google Scholar]
- Chung J., Lee G., Bang K., Park C., Park Y. (2009). Cultivar discrimination of Korean and Chinese boxthorn (Lycium chinense mill. and Lycium barbarum L.) using SSR markers. Korean J. Med. Crop Sci. 17 445–451. [Google Scholar]
- Coneva V., Frank M. H., Balaguer M. A. D. L., Li M., Sozzani R., Chitwood D. H., et al. (2017). Genetic architecture and molecular networks underlying leaf thickness in desert-adapted tomato Solanum pennellii. Plant Physiol. 175 376–391. 10.1104/pp.17.00790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czyczylo-Mysza I., Tyrka M., Marcinska I., Skrzypek E., Karbarz M., Dziurka M., et al. (2013). Quantitative trait loci for leaf chlorophyll fluorescence parameters, chlorophyll and carotenoid contents in relation to biomass and yield in bread wheat and their chromosome deletion bin assignments. Mol. Breed. 32 189–210. 10.1007/s11032-013-9862-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahech I., Farah W., Trigui M., Ben Hssouna A., Belghith H., Belghith K. S., et al. (2013). Antioxidant and antimicrobial activities of Lycium shawii fruits extract. Int. J. Biol. Macromol. 60 328–333. 10.1016/j.ijbiomac.2013.05.020 [DOI] [PubMed] [Google Scholar]
- Dielen V., Quinet M., Chao J., Batoko H., Havelange A., Kinet J. M. (2004). Uniflora, a pivotal gene that regulates floral transition and meristem identity in tomato (Lycopersicon esculentum). New Phytol. 161 393–400. 10.1046/j.1469-8137.2003.00937.x [DOI] [PubMed] [Google Scholar]
- Dong J., Yang J., Wang Y. (2008). Species resources of Lycium in china and research progress domestic and overseas. China J. Chin. Mater. Med. 5 2020–2027. [PubMed] [Google Scholar]
- Dong W., Wu D., Li G., Wu D., Wang Z. (2018). Next-generation sequencing from bulked segregant analysis identifies a dwarfism gene in watermelon. Sci. Rep. U.K. 8:2908. 10.1038/s41598-018-21293-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan Y., Yao X., Wang C., Chen X., Zhang J., Li X. (2015). Protective effects of Lycium ruthenicum murr on x-radiation injured mice. Nat. Product Res. Dev. 27 148–152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Editorial Committee of Chinese Flora, Chinese Academy of Sciences (1978). Flora of China. Beijing: Science press. [Google Scholar]
- Ehrenreich I. M., Gerke J. P., Kruglyak L. (2009). Genetic dissection of complex traits in yeast: insights from studies of gene expression and other phenotypes in the byxrm cross. Cold Spring Harb. Symp. Quant. Biol. 74 145–153. 10.1101/sqb.2009.74.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Essid A., Aljane F., Ferchichi A., Ignacio Hormaza J. (2015). Analysis of genetic diversity of tunisian caprifig (Ficus carica L.) accessions using simple sequence repeat (SSR) markers. Hereditas 152:1. 10.1186/s41065-015-0002-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- European Directorate for the Quality of Medicines (2016). European Pharmacopoeia. Strasbourg: European Directorate for the Quality of Medicines, 1263–1264. [Google Scholar]
- Evans L. T. (1996). Crop Evolution, Adaptation and Yield. New York: Cambridge university press. [Google Scholar]
- Fernando C. (1982). New combinations and new variety of Lycium (Solanaceae) from north America. Bot. Sci. 43 5–8. 10.17129/botsci.1263 [DOI] [Google Scholar]
- Foolad M. R., Panthee D. R. (2012). Marker-assisted selection in tomato breeding. Crit. Rev. Plant Sci. 31 93–123. 10.1080/07352689.2011.616057 [DOI] [Google Scholar]
- Frary A., Doganlar S., Daunay M. C., Tanksley S. D. (2003). QTL analysis of morphological traits in eggplant and implications for conservation of gene function during evolution of Solanaceous species. Theor. Appl. Genet. 107 359–370. 10.1007/s00122-003-1257-5 [DOI] [PubMed] [Google Scholar]
- Fukuda T., Yokoyama J., Ohashi H. (2001). Phylogeny and biogeography of the genus Lycium (Solanaceae): inferences from chloroplast DNA sequences. Mol. Phylogenet. Evol. 19 246–258. 10.1006/mpev.2001.0921 [DOI] [PubMed] [Google Scholar]
- Ge Y., Cao Y., Xu X., Zhao J., Zhang B. (2008). Study on quality change of fresh fruit of Lycium barbarum after harvest. N. Hortic. 7 227–229. [Google Scholar]
- Glowinski A., Flint-Garcia S. (2018). “Germplasm resources for mapping quantitative traits in maize,” in The Maize Genome, eds Bennetzen J., Flint-Garcia S., Hirsch C., Tuberosa R. (Cham: Springer International Publishing; ), 143–159. 10.1007/978-3-319-97427-9_10 [DOI] [Google Scholar]
- Gong H., Rehman F., Yang T., Li Z., Zeng S., Pan L., et al. (2019). Construction of the first high-density genetic map and QTL mapping for photosynthetic traits in Lycium barbarum l. Mol. Breed. 39:106. 10.1007/s11032-019-1000-9 [DOI] [Google Scholar]
- Grube R. C., Radwanski E. R., Jahn M. (2000). Comparative genetics of disease resistance within the Solanaceae. Genetics 155 873–887. 10.1093/genetics/155.2.873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo F., Yu H., Tang Z., Jiang X., Wang L., Wang X., et al. (2015). Construction of a SNP-based high-density genetic map for pummelo using rad sequencing. Tree Genet. Genomes 11:18. 10.1007/s11295-014-0831-0 [DOI] [Google Scholar]
- Guo P., Ren G., Li Z., Zeng S., De T. (2019). Progress of determinate growth genes and gene editing breeding. Chinese J. Trop. Crops 40 2016–2021. 10.3969/j.issn.1000-2561.2019.10.014 [DOI] [Google Scholar]
- Han Q. Q., Song Y. Z., Zhang J. Y., Liu L. F. (2014). Studies on the role of the slnac3 gene in regulating seed development in tomato (Solanum lycopersicum). J. Hortic. Sci. Biotech. 89 423–429. 10.1080/14620316.2014.11513101 [DOI] [Google Scholar]
- Hanson P., Lu S., Wang J., Chen W., Kenyon L., Tan C. W., et al. (2016). Conventional and molecular marker-assisted selection and pyramiding of genes for multiple disease resistance in tomato. Sci. Hortic. Amsterdam 201 346–354. 10.1016/j.scienta.2016.02.020 [DOI] [Google Scholar]
- Henareh M., Dursun A., Mandoulakani B. A. (2015). Genetic diversity in tomato landraces collected from turkey and Iran revealed by morphological characters. Acta Sci. Pol. Hortorum Cultus 14 87–96. [Google Scholar]
- Hu J., Guo C., Wang B., Ye J., Liu M., Wu Z., et al. (2018). Genetic properties of a nested association mapping population constructed with semi-winter and spring oilseed rapes. Front. Plant Sci. 9:1740. 10.3389/fpls.2018.01740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu W. (2015). Development and Utility of Est-Ssr and Est-Snp Markers for Four Medicinal Plants. Huairou: University of Chinese Academy of Sciences. [Google Scholar]
- Hu Y. F., Hu H. Y., Jiang C. J., Zhu Y. H., Wang J. M. (2011). The influence of different product process on the effect ingredient content of Ningxia Lycium barbarum. Adv. Mat. Res. 20 2083–2086. [Google Scholar]
- Huang T., Liu J., Qin K., Dai G., Zhang B. (2013). Study on quality requirements for fresh wolfberry. Ningxia J. Agric. For. Sci. Technol. 54 151–154. [Google Scholar]
- Huang W., Liao S., Lv H., Khaldun A. B. M., Wang Y. (2015). Characterization of the growth and fruit quality of tomato grafted on a woody medicinal plant, Lycium chinense. Sci. Hortic. Amsterdam 197 447–453. 10.1016/j.scienta.2015.10.005 [DOI] [Google Scholar]
- Ibrahim S. D., Adawy S. S., Atia M. A. M., Alsamman A. M., Mokhtar M. M. (2016). Genetic diversity, variety identification and gene detection in some Egyptian grape varieties by SSR and SCOT markers. Plant Omics 9 311–318. 10.21475/poj.09.05.16.pne125 [DOI] [Google Scholar]
- Iordãchescu M., Udrişte A. A., Bãdulescu L. (2020). Use of molecular markers in improving resistance to biotic stress in Solanaceae-a review. Sci. Pap. Ser. B Hortic. 64 405–412. [Google Scholar]
- Jack T. (2004). Molecular and genetic mechanisms of floral control. Plant Cell 16 S1–S17. 10.1105/tpc.017038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Japanese Pharmacopoeia Editorial Committee (2016). Japanese Pharmacopoeia (17th). Tokyo: Ministry of Health, Labour and Welfare, 1909–1910. [Google Scholar]
- Jofuku K. D., Omidyar P. K., Gee Z., Okamuro J. K. (2005). Control of seed mass and seed yield by the floral homeotic gene apetala2. Proc. Natl. Acad. Sci. U.S.A. 102 3117–3122. 10.1073/pnas.0409893102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Junfa W., Xu M., Liuxuan M., Chuanhua Y. (2009). The current states and technology study of harvesting equipments of rhizome traditional Chinese medicinal materials. J. Agric. Mech. Res. 12 242–246. [Google Scholar]
- Jung W., Chung I., Kim S., Chi H., Yu C. Y., Ghimire B. K., et al. (2021). Direct shoot organogenesis from Lycium chinense miller leaf explants and assessment of genetic stability using ISSR markers. Agronomy 11:503. 10.3390/agronomy11030503 [DOI] [Google Scholar]
- Kevany B. M., Taylor M. G., Klee H. J. (2008). Fruit-specific suppression of the ethylene receptor leetr4 results in early-ripening tomato fruit. Plant Biotechnol. J. 6 295–300. 10.1111/j.1467-7652.2007.00319.x [DOI] [PubMed] [Google Scholar]
- Khan S. A., Chibon P. Y., de Vos R. C., Schipper B. A., Walraven E., Beekwilder J., et al. (2012). Genetic analysis of metabolites in apple fruits indicates an mQTL hotspot for phenolic compounds on linkage group 16. J. Exp. Bot. 63 2895–2908. 10.1093/jxb/err464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khapte P., Singh T. H., Lakshmana D. C. (2018). Screening of elite eggplant (Solanum melongena) genotypes for bacterial wilt (Ralstonia solanacearum) in field conditions and their genetic association by using ssr markers. Indian J. Agr. Sci. 88 1502–1511. [Google Scholar]
- Korea Food and Drug Administration (2014). Korean Pharmacopoeia (11th). Seoul: Shinil Publishing Company, 1862–1863. [Google Scholar]
- Krizek B. A., Fletcher J. C. (2005). Molecular mechanisms of flower development: an armchair guide. Nat. Rev. Genet. 6 688–698. 10.1038/nrg1675 [DOI] [PubMed] [Google Scholar]
- Kumar S., Sehgal S. K., Kumar U., Prasad P. V. V., Joshi A. K., Gill B. (2012). Genomic characterization of drought tolerance-related traits in spring wheat. Euphytica 186 265–276. 10.1007/s10681-012-0675-3 [DOI] [Google Scholar]
- Kwon S., Lee G., Lee S., Park Y., Gwag J., Kim T. S., et al. (2009). Isolation and characterization of 21 microsatellite loci in Lycium chinense and cross-amplification in Lycium barbarum. Conserv. Genet. 10 1557–1560. 10.1007/s10592-008-9792-x [DOI] [Google Scholar]
- Lander E. S., Schork N. (1994). genetic dissection of complex traits. Science 5181 2037–2048. [DOI] [PubMed] [Google Scholar]
- Laura S., Peñas M. L. L., Bernardello G., Levin R. A., Miller J. S. (2010). Karyotypes and fluorescent chromosome banding patterns in southern African Lycium (solanaceae). Caryologia 63 50–61. 10.1080/00087114.2010.10589708 [DOI] [Google Scholar]
- Lee S. B., Kim J. E., Kim H. T., Lee G. M., Kim B. S., Lee J. M. (2020). Genetic mapping of the c1 locus by GBS-based BSA-seq revealed pseudo-response regulator 2 as a candidate gene controlling pepper fruit color. Theor. Appl. Genet. 133 1897–1910. 10.1007/s00122-020-03565-5 [DOI] [PubMed] [Google Scholar]
- Levin R. A., Whelan A., Miller J. S. (2009). The utility of nuclear conserved ortholog set II (COSII) genomic regions for species-level phylogenetic inference in Lycium (Solanaceae). Mol. Phylogenet. Evol. 53 881–890. 10.1016/j.ympev.2009.08.016 [DOI] [PubMed] [Google Scholar]
- Li H., Huang M., Luo Q., Hong X., Ramakrishna S., So K. F. (2019a). Lycium barbarum (wolfberry) increases retinal ganglion cell survival and affects both microglia/macrophage polarization and autophagy after rat partial optic nerve transection. Cell Transplant. 28 607–618. 10.1177/0963689719835181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Wu H., Qi Q., Li H., Li Z., Chen S., et al. (2019b). Gibberellins play a role in regulating tomato fruit ripening. Plant Cell Physiol. 60 1619–1629. 10.1093/pcp/pcz069 [DOI] [PubMed] [Google Scholar]
- Li J., Wang J., Wang L., Huang Z. (2001). New variety breeding of seedless Ningxia wolfberry. Acta Bot. Boreali Occidentalia Sin. 21 446–450. [Google Scholar]
- Li R., Sun S., Wang H., Wang K., Yu H., Zhou Z., et al. (2020). Fis1 encodes a ga2-oxidase that regulates fruit firmness in tomato. Nat. Commun. 11:5844. 10.1038/s41467-020-19705-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li T., Yang X., Yu Y., Si X., Zhai X. (2018). Domestication of wild tomato is accelerated by genome editing. Nat. Biotechnol. 36 1160–1163. 10.1038/nbt.4273 [DOI] [PubMed] [Google Scholar]
- Li X., He J., Cao Y. (2010). The sales problems and solutions of fresh wolfberry fruit. N. Hortic. 6 175–176. [Google Scholar]
- Li Y., Zheng L., Corke F., Smith C., Bevan M. W. (2008). Control of final seed and organ size by the da1 gene family in Arabidopsis thaliana. Gene Dev. 22 1331–1336. 10.1101/gad.463608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y. C., Wu P. E., Zhou J. (2001). Standards Collection of Materia Medica. Chengdu: Science and Technology Press. [Google Scholar]
- Lifschitz E., Eshed Y. (2006). Universal florigenic signals triggered by ft homologues regulate growth and flowering cycles in perennial day-neutral tomato. J. Exp. Bot. 57 3405–3414. 10.1093/jxb/erl106 [DOI] [PubMed] [Google Scholar]
- Liu C., Zhou X., Chen D., Li L., Li J., Chen Y. D. (2014). Natural variation of leaf thickness and its association to yield traits in Indica rice. J. Integr. Agr. 13 316–325. 10.1016/S2095-3119(13)60498-0 [DOI] [Google Scholar]
- Liu D., Sun D., Liang J., Dou J., Yang S., Zhu H., et al. (2021). Characterization and bulk segregant analysis of ‘moon and star’ appearance in watermelon. Sci. Hortic. Amsterdam 285:110140. 10.1016/j.scienta.2021.110140 [DOI] [Google Scholar]
- Liu G. F., Yang J., Xu H. M., Hayat Y., Zhu J. (2008). Genetic analysis of grain yield conditioned on its component traits in rice (Oryza sativa L.). Aust. J. Agr. Res. 59 189–195. 10.1071/AR07163 [DOI] [Google Scholar]
- Liu H., Wang X., Xiao Y., Luo J., Qiao F., Yang W., et al. (2020a). Cubic: an atlas of genetic architecture promises directed maize improvement. Genome Biol. 21:20. 10.1186/s13059-020-1930-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J., Liang Y., Qin L., Wang L. (2014). Research progress on the molecular biology of floral development in tomato (Solanum lycopersicum L.). J. Agric. Biotechnol. 22 351–361. [Google Scholar]
- Liu X., Cheng J., Mei Z., Wei C., Khan M. A., Peng J., et al. (2020b). Scar marker for identification and discrimination of specific medicinal Lycium chinense miller from Lycium species from RAMP-PCR RAPD fragments. 3 Biotech 10:334. 10.1007/s13205-020-02325-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X., Du J., Khan M. A., Cheng J., Wei C., Mei Z., et al. (2020c). Analysis of genetic diversity and similarities between different Lycium varieties based on ISSR analysis and RAMP-PCR markers. World Acad. Sci. J. 2 83–90. 10.3892/wasj.2020.39 [DOI] [Google Scholar]
- Liu Y., Cao L., Du J., Jia R., Wang J. (2015). Protective effects of Lycium barbarum polysaccharides against carbon tetrachloride-induced hepatotoxicity in precision-cut liver slices in vitro and in vivo in common carp (Cyprinus carpio L.). Comp. Biochem. Phys. C 169 65–72. 10.1016/j.cbpc.2014.12.005 [DOI] [PubMed] [Google Scholar]
- Liu Z., Shu Q., Wang L., Yu M., Hu Y., Zhang H., et al. (2012). Genetic diversity of the endangered and medically important Lycium ruthenicum Murr. Revealed by sequence-related amplified polymorphism (SRAP) markers. Biochem. Syst. Ecol. 45 86–97. 10.1016/j.bse.2012.07.017 [DOI] [Google Scholar]
- Long S. P., Marshall-Colon A., Zhu X. (2015). Meeting the global food demand of the future by engineering crop photosynthesis and yield potential. Cell 161 56–66. 10.1016/j.cell.2015.03.019 [DOI] [PubMed] [Google Scholar]
- Long S. P., Zhu X. G., Naidu S. L., Ort D. R. (2006). Can improvement in photosynthesis increase crop yields? Plant Cell Environ. 29 315–330. 10.1111/j.1365-3040.2005.01493.x [DOI] [PubMed] [Google Scholar]
- Lu S., Zhao P. (2010). Chemical characterization of Lycium barbarum polysaccharides and their reducing myocardial injury in ischemia/reperfusion of rat heart. Int. J. Biol. Macromol. 47 681–684. 10.1016/j.ijbiomac.2010.08.016 [DOI] [PubMed] [Google Scholar]
- Lu Y., Luo S., Li N., Li Q., Du W., Zhang W., et al. (2021). Candidate gene, smcpr1, encoding cpr1 related to plant height of the eggplant dwarf mutant dwf. Horticulturae 7:196. 10.3390/horticulturae7070196 [DOI] [Google Scholar]
- Luckwill L. C. (1948). The hormone content of the seed in relation to endosperm development and fruit drop in the apple. J. Hortic. Sci. 24 32–44. 10.1080/00221589.1948.11513679 [DOI] [Google Scholar]
- Ma J., Huang L., Ma C., Jin J., Li C., Wang R. K., et al. (2015). Large-scale snp discovery and genotyping for constructing a high-density genetic map of tea plant using specific-locus amplified fragment sequencing (SLAF-seq). PLoS One 10:e0128798. 10.1371/journal.pone.0128798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma J., Zhang D., He J., Kang C., Yi W. (2017). Mechanical design of automatic conveying and grading device for wolfberry. J. Agric. Mech. Res. 39 108–116. 10.3969/j.issn.1003-188X.2017.02.023 [DOI] [Google Scholar]
- Mackay I., Powell W. (2007). Methods for linkage disequilibrium mapping in crops. Trends Plant Sci. 12 57–63. 10.1016/j.tplants.2006.12.001 [DOI] [PubMed] [Google Scholar]
- Madhusudhana R. (2015). “Linkage mapping,” in Sorghum Molecular Breeding, eds Madhusudhana R., Rajendrakumar P., Patil J. V. (New Delhi: Springer India; ), 47–70. [Google Scholar]
- Magwene P. M., Willis J. H., Kelly J. K. (2011). The statistics of bulk segregant analysis using next generation sequencing. PLoS Comput. Biol. 7:e1002255. 10.1371/journal.pcbi.1002255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makino A. (2011). Photosynthesis, grain yield, and nitrogen utilization in rice and wheat. Plant Physiol. 155 125–129. 10.1104/pp.110.165076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansfeld B. N., Grumet R. (2018). Qtlseqr: an r package for bulk segregant analysis with next-generation sequencing. Plant Genome 11:180006. 10.3835/plantgenome2018.01.0006 [DOI] [PubMed] [Google Scholar]
- McCulloch G. A., Mauda E. V., Chari L. D., Martin G. D., Gurdasani K., Morin L., et al. (2020). Genetic diversity and morphological variation in African boxthorn (Lycium ferocissimum) – characterising the target weed for biological control. Biol. Control 143:104206. 10.1016/j.biocontrol.2020.104206 [DOI] [Google Scholar]
- Meuwissen T. H., Goddard M. E. (2004). Mapping multiple qtl using linkage disequilibrium and linkage analysis information and multitrait data. Genet. Sel. Evol. 36 261–279. 10.1186/1297-9686-36-3-261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michelmore R. W., Paran I., Kesseli R. V. (1991). Identification of markers linked to disease-resistance genes by bulked segregant analysis: a rapid method to detect markers in specific genomic regions by using segregating populations. Proc. Natl. Acad. Sci. U.S.A. 88 9828–9832. 10.1073/pnas.88.21.9828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller S. S., Scorza R. (2010). Response of two novel peach tree growth habits to in-row tree spacing, training system, and pruning: effect on growth and pruning. J. Am. Pomol. Soc. 64 199–217. [Google Scholar]
- Minne L., Spies J. J., Venter H. J. T., Venter A. M. O. F. (1994). Breeding systems in some representatives of the genus Lycium (Solanaceae). Bothalia 24 107–110. 10.4102/abc.v24i1.759 [DOI] [Google Scholar]
- Mocan A., Vlase L., Vodnar D. C., Bischin C., Hanganu D., Gheldiu A. M., et al. (2014). Polyphenolic content, antioxidant and antimicrobial activities of Lycium barbarum l. And Lycium chinense Mill. Leaves. Molecules 19 10056–10073. 10.3390/molecules190710056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molinero-Rosales N., Latorre A., Jamilena M., Lozano R. (2004). Single flower truss regulates the transition and maintenance of flowering in tomato. Planta 218 427–434. 10.1007/s00425-003-1109-1 [DOI] [PubMed] [Google Scholar]
- Moodley V., Naidoo R., Gubba A., Mafongoya P. L. (2019). Development of potato virus y (pvy) resistant pepper (Capsicum annuum L.) lines using marker-assisted selection (MAS). Physiol. Mol. Plant P 105 96–101. 10.1016/j.pmpp.2018.12.002 [DOI] [Google Scholar]
- Nan X., Shao Q., Wang J., Chang H., Wang H. (2017). Prospect of Special Wolfberry Varieties Breeding. Yinchuan: Ningxia Forestry News, 48–52. [Google Scholar]
- NingXia Academy of Agricultural and Forestry Sciences (2015). The only Lycium germplasm resources garden in the word were built in Ningxia. Ningxia J. Agric. For. Sci. Technol. 56:58. [Google Scholar]
- Olatunji O. J., Chen H., Zhou Y. (2018). Lycium chinense leaves extract ameliorates diabetic nephropathy by suppressing hyperglycemia mediated renal oxidative stress and inflammation. Biomed. Pharmacother. 102 1145–1151. 10.1016/j.biopha.2018.03.037 [DOI] [PubMed] [Google Scholar]
- Ort D. R., Merchant S. S., Alric J., Barkan A., Blankenship R. E., Bock R., et al. (2015). Redesigning photosynthesis to sustainably meet global food and bioenergy demand. Proc. Natl. Acad. Sci. U.S.A. 112 8529–8536. 10.1073/pnas.1424031112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osipova S., Permyakov A., Permyakova M., Pshenichnikova T., Verkhoturov V., Rudikovsky A., et al. (2016). Regions of the bread wheat d genome associated with variation in key photosynthesis traits and shoot biomass under both well watered and water deficient conditions. J. Appl. Genet. 57 151–163. 10.1007/s13353-015-0315-4 [DOI] [PubMed] [Google Scholar]
- Pascual L., Desplat N., Huang B. E., Desgroux A., Bruguier L., Bouchet J. P., et al. (2015). Potential of a tomato magic population to decipher the genetic control of quantitative traits and detect causal variants in the resequencing era. Plant Biotechnol. J. 13 565–577. 10.1111/pbi.12282 [DOI] [PubMed] [Google Scholar]
- Peifei L., Bingxiu X., Huilin C. (2015). Lycium Barbarum and Tumors in the Gastrointestinal Tract. Dordrecht: Springer. [Google Scholar]
- Pnueli L., Carmel-Goren L., Hareven D., Gutfinger T., Alvarez J. (1998). The self-pruning gene of tomato regulates vegetative to reproductive switching of sympodial meristems and is the ortholog of cen and tfl1. Development 125 1979–1989. 10.1242/dev.125.11.1979 [DOI] [PubMed] [Google Scholar]
- Portis E., Acquadro A., Tirone M., Pesce G. R., Mauromicale G., Lanteri S. (2018a). Mapping the genomic regions encoding biomass-related traits in Cynara cardunculus L. Mol. Breed. 38:64. 10.1007/s11032-018-0826-x [DOI] [Google Scholar]
- Portis E., Cericola F., Barchi L., Toppino L., Acciarri N., Pulcini L., et al. (2015). Association mapping for fruit, plant and leaf morphology traits in eggplant. PLoS One 10:e135200. 10.1371/journal.pone.0135200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Portis E., Lanteri S., Barchi L., Portis F., Valente L. (2018b). Comprehensive characterization of simple sequence repeats in eggplant (Solanum melongena L.) genome and construction of a web resource. Front. Plant Sci. 9:401. 10.3389/fpls.2018.00401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian L., Yu S. (2016). Protective effect of polysaccharides from Lycium barbarum on spermatogenesis of mice with impaired reproduction system induced by cyclophosphamide. Am. J. Reprod. Immunol. 76 383–385. 10.1111/aji.12558 [DOI] [PubMed] [Google Scholar]
- Qin K., Dai G. (2017). Progress and prospect of wolfberry strains breeding. Ningxia J. Agric. Forest 58 25–28. [Google Scholar]
- Qiu S., Chen J., Chen X., Fan Q., Zhang C., Wang D., et al. (2014). Optimization of selenylation conditions for Lycium barbarum polysaccharide based on antioxidant activity. Carbohyd. Polym. 103 148–153. 10.1016/j.carbpol.2013.12.032 [DOI] [PubMed] [Google Scholar]
- Qu M., Zheng G., Hamdani S., Essemine J., Song Q., Wang H., et al. (2017). Leaf photosynthetic parameters related to biomass accumulation in a global rice diversity survey. Plant Physiol. 175 248–258. 10.1104/pp.17.00332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinet M., Dielen V., Batoko H., Boutry M., Havelange A. (2006). Genetic interactions in the control of flowering time and reproductive structure development in tomato (Solanum lycopersicum). New Phytol. 170 701–710. 10.1111/j.1469-8137.2006.01717.x [DOI] [PubMed] [Google Scholar]
- Ranc N., Muños S., Santoni S., Causse M. (2008). A clarified position for Solanum lycopersicum var. Cerasiformein the evolutionary history of tomatoes (Solanaceae). BMC Plant Biol. 8:130. 10.1186/1471-2229-8-130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reeve V. E., Allanson M., Arun S. J., Domanski D., Painter N. (2010). Mice drinking goji berry juice (Lycium barbarum) are protected from uv radiation-induced skin damage via antioxidant pathways. Photoch. Photobio. Sci. 9:601. 10.1039/b9pp00177h [DOI] [PubMed] [Google Scholar]
- Rehman F., Gong H., Li Z., Zeng S., Yang T., Ai P., et al. (2020). Identification of fruit size associated quantitative trait loci featuring SLAF based high-density linkage map of goji berry (Lycium spp.). BMC Plant Biol. 20:474. 10.1186/s12870-020-02567-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards R. A. (2000). Selectable traits to increase crop photosynthesis and yield of grain crops. J. Exp. Bot. 51 447–458. 10.1093/jexbot/51.suppl_1.447 [DOI] [PubMed] [Google Scholar]
- Rispail N., Prats E., Rubiales D. (2020). “Medicago truncatula as a model to study powdery mildew resistance,” in The Model Legume Medicago Truncatula, ed. de Bruijn F. J. (Hoboken: John Wiley & Sons; ), 390–397. 10.1093/pcp/pcab021 [DOI] [Google Scholar]
- Rubiales D., Fondevilla S., Fernández-Aparicio M. (2021). Development of pea breeding lines with resistance to Orobanche crenata derived from pea landraces and wild Pisum spp. Agronomy 11:36. 10.3390/agronomy11010036 [DOI] [Google Scholar]
- Sharma B. (2012). Correlation and path coefficient analysis for quantitative and qualitative traits for fruit yield and seed yield in tomato genotypes. Indian J. Hortic. 69 540–544. [Google Scholar]
- Sheidai M., Narengi Z., Khatamsaz M. (1999). Karyotype and seed protein analyses of Lycium (Solanaceae) in Iran. Edinburgh J. Bot. 56 253–264. 10.1017/S0960428600001116 [DOI] [Google Scholar]
- Shi Y., Wei C., Chen Z., Hou Y. (2016). Research progress on postharvest physiology and storage technology of fresh fruit of Lycium barbarum L. Storage Process 16 102–106. 10.3969/j.issn.1009-6221.2016.03.021 [DOI] [Google Scholar]
- Shi Z., Men H., Du H. (2012). Description Specification and Data Standard of Lycium Germplasm Resources. Beijing: China Forestry Publishing House. [Google Scholar]
- Shi Z., Wei F., Wan R., Li Y., Wang Y. (2019). Impact of nitrogen fertilizer levels on metabolite profiling of the Lycium barbarum L. Fruit. Molecules 24:3879. 10.3390/molecules24213879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh N. S., Sharma R., Parween T., Patanjali P. K. (2017). Pesticide Contamination and Human Health Risk Factor. Cham: Springer International Publishing, 49–68. [Google Scholar]
- Song W., Li Y., Zhao Y., Liu Y., Niu Y., Pang R., et al. (2012). Construction of a high-density microsatellite genetic linkage map and mapping of sexual and growth-related traits in half-smooth tongue sole (Cynoglossus semilaevis). PLoS One 7:e0052097. 10.1371/journal.pone.0052097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Souza A. D. G., Smiderle O. J., Spinelli V. M., Souza R. O. D., Bianchi V. J. (2016). Correlation of biometrical characteristics of fruit and seed with twinning and vigor of prunus persica rootstocks. J. Seed. Sci. 38 322–328. 10.1590/2317-1545v38n4164650 [DOI] [Google Scholar]
- State Forestry Administration of China [SFAC] (2013). Guideline for the Conduct of Tests for Distinctness, Uniformity and Stability — Lycium. LY/T 2099-2013. Beijing: Standards Press of China. [Google Scholar]
- State Forestry Administration of China [SFAC] (2018). China Forestry Statistics Yearbook-2017. Beijing: China Forestry Publishing. [Google Scholar]
- Stebbins G. L. (1971). Chromosomal Evolution in Higher Plants. London: Edward Arnold. [Google Scholar]
- Stiefkens L., Bernardello G. (2002). Karyotypic studies in Lycium section mesocope (Solanaceae) from South America. Caryologia 55 199–206. 10.1080/00087114.2002.10589278 [DOI] [Google Scholar]
- Stiefkens L., Bernardello G. (2005). Karyotype studies in Lycium sections schistocalyx and sclerocarpellum (Solanaceae). Edinburgh J. Bot. 62 53–67. 10.1017/S0960428606000023 [DOI] [Google Scholar]
- Stiefkens L., Bernardello L. (1996). Karyotypic studies in South American Lycium (Solanaceae). Cytologia 61 395–402. 10.1508/cytologia.61.395 [DOI] [Google Scholar]
- Stiefkens L., Las Peñas M. L., Levin R. A., Miller J. S., Bernardello G. (2020). Chromosome evolution in the cosmopolitan genus Lycium (solanaceae). Taxon 69 124–141. 10.1002/tax.12205 [DOI] [Google Scholar]
- Su C., Duan X., Liang L., Feng-Wang, Zheng J., Fu X. Y., et al. (2014). Lycium barbarum polysaccharides as an adjuvant for recombinant vaccine through enhancement of humoral immunity by activating tfh cells. Vet. Immunol. Immunopathol. 158 98–104. 10.1016/j.vetimm.2013.05.006 [DOI] [PubMed] [Google Scholar]
- Sun Y., Lei Y., Wu Z., Yang C. (2018). The Intellectual Property Protection of Traditional Chinese Medicine Lycium for the Prevention and Treatment of Diabetes. Chengdu: Sichuan Province, China. [Google Scholar]
- Sze S. C., Song J., Wong R. N., Feng Y., Ng T., Tong Y., et al. (2008). Application of scar (sequence characterized amplified region) analysis to authenticate Lycium barbarum (wolfberry) and its adulterants. Biotechnol. Appl. Biochem. 51:15. 10.1042/BA20070096 [DOI] [PubMed] [Google Scholar]
- Tamisier L., Szadkowski M., Nemouchi G., Lefebvre V., Szadkowski E. (2020). Genome-wide association mapping of QTLs implied in potato virus y population sizes in pepper: evidence for widespread resistance QTL pyramiding. Mol. Plant Pathol. 21 3–16. 10.1111/mpp.12874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Y., Ren J., Liu C., Jiang J., Yang H. (2021). Genetic characteristics and QTL analysis of the soluble sugar content in ripe tomato fruits. Sci. Hortic. Amsterdam 276:109785. 10.1016/j.scienta.2020.109785 [DOI] [Google Scholar]
- Tanksley S. D. (2004). The genetic, developmental, and molecular bases of fruit size and shape variation in tomato. Plant Cell 16 S181–S189. 10.1105/tpc.018119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thammina C. S., Olsen R. T., Kramer M., Pooler M. R. (2017). Genetic relationships of boxwood (Buxus L.) Accessions based on genic simple sequence repeat markers. Genet. Resour. Crop Evol. 64 1281–1293. 10.1007/s10722-016-0436-6 [DOI] [Google Scholar]
- The American Herbal Pharmacopoeia (2019). Lycium (Goji) Berry. Scotts Valley, CA: The American Herbal Pharmacopoeia. [Google Scholar]
- Tongumpai P. (1993). Strategies for Machine Harvesting of Mature Coffee (Coffea Arabica l.) Fruits. Dissertation. Corvallis, OR: Oregon State University. [Google Scholar]
- Trick M., Adamski N. M., Mugford S. G., Jiang C. C., Febrer M., Uauy C. (2012). Combining SNP discovery from next-generation sequencing data with bulked segregant analysis (BSA) to fine-map genes in polyploid wheat. BMC Plant Biol. 12:14. 10.1186/1471-2229-12-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tripathi V. (2013). Assessment of genetic diversity in berberis Lycium royle complex using RAPD markers. J. Cell Biol. Genet. 3 1–13. 10.5897/JCBG11.003 [DOI] [Google Scholar]
- Villegas-Fernández ÁM., Amarna A. A., Moral J., Rubiales D. (2021). Crop diversification to control powdery mildew in pea. Agronomy 11:690. 10.3390/agronomy11040690 [DOI] [Google Scholar]
- Wang C. C., Chang S. C., Inbaraj B. S., Chen B. H. (2010). Isolation of carotenoids, flavonoids and polysaccharides from Lycium barbarum L. and evaluation of antioxidant activity. Food Chem. 120 184–192. 10.1016/j.foodchem.2009.10.005 [DOI] [Google Scholar]
- Wang H., Li J., Tao W., Zhang X., Gao X., Yong J., et al. (2018). Lycium ruthenicum studies: molecular biology, phytochemistry and pharmacology. Food Chem. 240 759–766. 10.1016/j.foodchem.2017.08.026 [DOI] [PubMed] [Google Scholar]
- Wang J., Chen W., Chen J., Zhou X., Xu D., Li J. H., et al. (2015a). Population genetic diversity of wild Lycium ruthenicum in qaidam inferred from AFLP markers. Chinese J. Plant Ecol. 39 1003–1011. 10.17521/cjpe.2015.0097 [DOI] [Google Scholar]
- Wang J., Hu Y., Wang D., Zhang F., Zhao X., Abula S., et al. (2010). Lycium barbarum polysaccharide inhibits the infectivity of newcastle disease virus to chicken embryo fibroblast. Int. J. Biol. Macromol. 46 212–216. 10.1016/j.ijbiomac.2009.11.011 [DOI] [PubMed] [Google Scholar]
- Wang J., Wang Q., Liu F., Li Y., Jiang J. (2017). The correlation between seed characters and fruit characters of Lycium ruthenicum Murr. Seed 36 80–83. [Google Scholar]
- Wang J., Zhang K., Zhang X., Yan G., Zhou Y., Feng L., et al. (2015b). Construction of commercial sweet cherry linkage maps and QTL analysis for trunk diameter. PLoS One 10:e0141261. 10.1371/journal.pone.0141261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S., Suh J. H., Zheng X., Wang Y., Ho C. (2017). Identification and quantification of potential anti-inflammatory hydroxycinnamic acid amides from wolfberry. J. Agric. Food Chem. 65 364–372. 10.1021/acs.jafc.6b05136 [DOI] [PubMed] [Google Scholar]
- Wang Y., Guo S., An W., Liu L., Yin Y. (2016). Study on the characters and major constituents in the fruits of five wolfberry varieties. J. For. Environ. 36 367–372. [Google Scholar]
- Wasternack C., Forner S., Strnad M., Hause B. (2013). Jasmonates in flower and seed development. Biochimie 95 79–85. 10.1016/j.biochi.2012.06.005 [DOI] [PubMed] [Google Scholar]
- Wilson L. M., Whitt S. R., Ibaénez A. M., Rocheford T. R., Goodman M. M., Buckler E. S., IV (2004). Dissection of maize kernel composition and starch production by candidate gene association. Plant Cell 16 2719–2733. 10.1105/tpc.104.025700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wm Hilhorst H., Pc Groot S., J Bino R. (1998). The tomato seed as a model system to study seed development and germination. Acta Bot. Neerlandica 47 169–183. [Google Scholar]
- Xin T., Yao H., Gao H., Zhou X., Ma X. (2013). Super food Lycium barbarum (Solanaceae) traceability via an internal transcribed spacer 2 barcode. Food Res. Int. 54 1699–1704. 10.1016/j.foodres.2013.10.007 [DOI] [Google Scholar]
- Xin Y., Wan L., Peng J., Guo C. (2011). Alleviation of the acute doxorubicin-induced cardiotoxicity by Lycium barbarum polysaccharides through the suppression of oxidative stress. Food Chem. Toxicol. 49 259–264. 10.1016/j.fct.2010.10.028 [DOI] [PubMed] [Google Scholar]
- Xin Y., Zhou G., Deng Z., Chen Y., Wu Y., Xu P. S., et al. (2007). Protective effect of Lycium barbarum on doxorubicin-induced cardiotoxicity. Phytother. Res. 21 1020–1024. 10.1002/ptr.2186 [DOI] [PubMed] [Google Scholar]
- Xing X., Liu F., Xiao J., So K. F. (2016). Neuro-protective mechanisms of Lycium barbarum. Neuromol. Med. 18 253–263. 10.1007/s12017-016-8393-y [DOI] [PubMed] [Google Scholar]
- Xu Y. (2010). Molecular Plant Breeding. Cambridge, MA: CABI North American Office. [Google Scholar]
- Yang D., Jing R., Chang X., Li W. (2007). Quantitative trait loci mapping for chlorophyll fluorescence and associated traits in wheat (Triticum aestivum). J. Integr. Plant Biol. 49 646–654. 10.1111/j.1672-9072.2007.00443.x [DOI] [Google Scholar]
- Yang T., Dong J., Yue J., Wang Y. (2015). A new wolfberry cultivar‘zhongke lüchuan 1’. Acta Hortic. Sin. 42 2557–2558. 10.16420/j.issn.0513-353x.2014-1003 [DOI] [Google Scholar]
- Yao R., Heinrich M., Zou Y., Reich E., Zhang X., Chen Y., et al. (2018b). Quality variation of goji (fruits of Lycium spp.) In china: a comparative morphological and metabolomic analysis. Front. Pharmacol. 9:151. 10.3389/fphar.2018.00151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao R., Heinrich M., Weckerle C. S. (2018a). The genus Lycium as food and medicine: a botanical, ethnobotanical and historical review. J. Ethnopharmacol. 212 50–66. 10.1016/j.jep.2017.10.010 [DOI] [PubMed] [Google Scholar]
- Ye M., Moon J., Yang J., Lim H. H., Hong S. B., Shim I., et al. (2015). The standardized Lycium chinense fruit extract protects against alzheimer’s disease in 3xtg-ad mice. J. Ethnopharmacol. 172 85–90. 10.1016/j.jep.2015.06.026 [DOI] [PubMed] [Google Scholar]
- Ye X., Jiang Y. (2020). Phytochemicals in Goji Berries. Boca Raton, FL: CRC Press. [Google Scholar]
- Yin G., Dang Y. (2008). Optimization of extraction technology of the Lycium barbarum polysaccharides by box-behnken statistical design. Carbohyd. Polym. 74 603–610. 10.1016/j.carbpol.2008.04.025 [DOI] [Google Scholar]
- Yin X. L., Fang K. T., Liang Y. Z., Wong R. N., Ha A. W. (2005). Assessing phylogenetic relationships of Lycium samples using rapd and entropy theory. Acta Pharmacol. Sin. 26 1217–1224. 10.1111/j.1745-7254.2005.00197.x [DOI] [PubMed] [Google Scholar]
- Yu J., Buckler E. S. (2006). Genetic association mapping and genome organization of maize. Curr. Opin. Biotech. 17 155–160. 10.1016/j.copbio.2006.02.003 [DOI] [PubMed] [Google Scholar]
- Yu J., Holland J. B., McMullen M. D., Buckler E. S. (2008). Genetic design and statistical power of nested association mapping in maize. Genetics 178 539–551. 10.1534/genetics.107.074245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J., Zhang K., Li S., Yu S., Zhai H. (2013). Mapping quantitative trait loci for lint yield and fiber quality across environments in a Gossypium hirsutum × Gossypium barbadense backcross inbred line population. Theor. Appl Genet. 126 275–287. 10.1007/s00122-012-1980-x [DOI] [PubMed] [Google Scholar]
- Yuan H., An W., Li L., Cao Y., Weihua L. (2013). The investigation and cluster analysis of main morphological characters for germplasm of Chinese wolfberry. J. Plant Genet. Resour. 14 627–633. [Google Scholar]
- Zhang D., Xia T., Dang S., Fan G., Wang Z. (2018). Investigation of Chinese wolfberry (Lycium spp.) germplasm by restriction site-associated DNA sequencing (rad-seq). Biochem. Genet. 56 575–585. 10.1007/s10528-018-9861-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., Zhang Q., Cheng T., Yang W., Pan H., Zhong J., et al. (2015). High-density genetic map construction and identification of a locus controlling weeping trait in an ornamental woody plant (Prunus mume sieb. Et zucc). DNA Res. 22 183–191. 10.1093/dnares/dsv003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang K. Y., Leung H. W., Yeung H. W., Wong R. N. (2001). Differentiation of Lycium barbarum from its related Lycium species using random amplified polymorphic dna. Planta Med. 67 379–381. 10.1055/s-2001-14310 [DOI] [PubMed] [Google Scholar]
- Zhang X., Sun Z. (2011). Development status of new plant variety protection and DUS testing. For. Res. 24 247–252. [Google Scholar]
- Zhang Y., Wang L., Xin H., Li D., Ma C. (2013). Construction of a high-density genetic map for sesame based on large scale marker development by specific length amplified fragment (SLAF) sequencing. BMC Plant Biol. 13:141. 10.1186/1471-2229-13-141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J., Li H., Xu Y., Yin Y., Huang T., Zhang B., et al. (2021). A consensus and saturated genetic map provides insight into genome anchoring, synteny of Solanaceae and leaf- and fruit-related QTLs in wolfberry (Lycium linn.). BMC Plant Biol. 21:350. 10.1186/s12870-021-03115-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J., Xu Y., Li H., Yin Y., An W., Li Y., et al. (2019). A snp-based high-density genetic map of leaf and fruit related quantitative trait loci in wolfberry (Lycium linn.). Front. Plant Sci. 10:977. 10.3389/fpls.2019.00977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao W., Chung J., Cho Y., Rha W., Lee G., Ma K. H., et al. (2010). Molecular genetic diversity and population structure in Lycium accessions using SSR markers. CR Biol. 333 793–800. 10.1016/j.crvi.2010.10.002 [DOI] [PubMed] [Google Scholar]
- Zheng G., Su X., Ma Y., Qi G., Yang J. (2015). Effect of seed characters on fruit size of Lycium barbarum. N. Hortic. 6 134–137. [Google Scholar]
- Zheng G., Zhang L., Wang J., Hu Z. (2012). The morphology development of fruit and seed in Lycium barbarum. Guangxi Zhiwu Guihaia 32 810–815. [Google Scholar]
- Zheng T. C., Zhang X. K., Yin G. H., Wang L. N., Han Y. L., Chen L., et al. (2011). Genetic gains in grain yield, net photosynthesis and stomatal conductance achieved in Henan province of China between 1981 and 2008. Field Crops Res. 122 225–233. 10.1016/j.fcr.2011.03.015 [DOI] [Google Scholar]
- Zheng Z., Wang R., Zhang Y., Zhao Z., Gao L. (2018). “Present situation and development of harvesting mechanization of chinese medicinal materials,” in Proceedings of the 2018 ASABE Annual International Meeting, (St. Joseph, MI: ASABE; ). [Google Scholar]
- Zhou H., Li P., Xie W., Hussain S., Li Y., Xia D., et al. (2017). Genome-wide association analyses reveal the genetic basis of stigma exsertion in rice. Mol. Plant 10 634–644. 10.1016/j.molp.2017.01.001 [DOI] [PubMed] [Google Scholar]
- Zhu C., Gore M., Buckler E. S., Yu J. (2008). Status and prospects of association mapping in plants. Plant Genome 1 5–20. 10.3835/plantgenome2008.02.0089 [DOI] [Google Scholar]
- Zhu X., Song Q., Ort D. R. (2012). Elements of a dynamic systems model of canopy photosynthesis. Curr. Opin. Plant Biol. 15 237–244. 10.1016/j.pbi.2012.01.010 [DOI] [PubMed] [Google Scholar]
- Zhu Y., Yin Y., Yang K., Li J., Sang Y., Huang L., et al. (2015). Construction of a high-density genetic map using specific length amplified fragment markers and identification of a quantitative trait locus for anthracnose resistance in walnut (Juglans regia L.). BMC Genom. 16:614. 10.1186/s12864-015-1822-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.