Abstract
Background:
Coronavirus disease 2019 (COVID-19), caused by novel severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), is a global epidemic with a high mortality rate. In this study, our goal was to identify the function and associated targets of SARS-CoV-2 from circulating monocytes in the blood and peripheral blood mononuclear cell (PBMC) dataset of patients with COVID-19.
Methods:
The Gene Expression Omnibus database (GSE164805 and GSE180594) was used to identify differentially expressed genes (DEGs). Gene ontology function analysis and Kyoto Encyclopedia of Genes and Genomes pathway enrichment analyses of the DEGs were performed using the DAVID database.
Results:
Gene ontology analysis of DEG revealed that GSE164805 and GSE180594 were involved in the regulation of cell migration, upregulation of cell proliferation, and in the activation of the mitogen-activated protein kinase signaling pathway. Kyoto Encyclopedia of Genes and Genomes analysis of GSE164805 revealed that the DEGs were enriched in peroxisome, melanogenesis, and actin regulation. Peroxisome genes were highly expressed in patients with mild and severe COVID-19. Bioinformatics analysis to compare GSE180594 and public data for the single-cell atlas of the peripheral immune response in patients with COVID-19 showed that interferon-associated genes were highly increased in acute COVID-19 PBMC and in CD14+ and CD16+ monocytes from patients with COVID-19.
Conclusions:
We comprehensively analyzed the blood cell gene expression profile data of patients with COVID-19 using bioinformatics methods to preliminary understand the functions and associated targets of DEGs in the blood cells of these patients. Thus, our data provide targets for potential therapies against COVID-19.
Keywords: Severe acute respiratory syndrome coronavirus 2, immune response, interferon
Introduction
Coronaviruses are enveloped viruses with a large plus-strand RNA genome 1 that is capped and polyadenylated. Coronaviruses were identified in 2002 as pathogenic agents during the severe acute respiratory syndrome (SARS) outbreak that occurred in Guangdong province in China. 2 There is evidence that the SARS-CoV virus originated from a nonhuman host, such as bats, can affect humans. Several pneumonia cases of unknown origin, named as SARS coronavirus 2 (SARS-CoV-2), first occurred in early December of 2019 and then worldwide in 2020.3,4 Coronavirus disease 2019 (COVID-19) is the acute respiratory infectious disease that occurs following infection with novel SARS-CoV-2. 4 Several studies have shown that SARS-CoV-2 is similar to SARS-CoV and MERS-CoV viruses in genomic structure and pathogenesis, but it is more transmissible than SARS-CoV and Middle East respiratory syndrome coronavirus (MERS-CoV) viruses.5-8
A considerable proportion of individuals who have recovered from COVID-19 suffer from persistent and prolonged symptoms, a condition known as long-COVID or postacute COVID-19 syndrome9-11 and defined as persistent symptoms for more than 6 months. Those symptoms include residual inflammation (convalescent phase), immune dysregulation, asthenia, dyspnea, and organ damage as well as nonspecific effects following hospitalization or prolonged ventilation (postintensive care syndrome), and social isolation.10-12
There is evidence that acute respiratory distress syndrome caused by SARS-CoV infection results in a cytokine storm that is readily followed by the immune system “attacking” the body. 7 Cytokine storms cause acute respiratory distress syndrome, multiple organ failure, and death in severe cases of SARS-CoV-2 infection. 13 The release of immune effector cells with abundant proinflammatory cytokines (interferon [IFN]α, IFNγ, interleukin [IL]-18, IL-33, IL-1β, IL-6, IL-12, tumor necrosis factor [TNF]α, transforming growth factor β) and chemokines (C-C motif chemokine ligand [CCL]2, CCL3, CCL5, C-X-C motif chemokine ligand [CXCL]10, CXCL8, and CXCL9) results in an abnormal systemic inflammatory response.4,7
Although a few therapeutic agents for COVID-19 have been improved, there are no effective drugs for uncontrollable infection. 14 In this study, to identify the functional effect and targets of COVID-19 in blood cells, datasets and bioinformatics approaches were conducted. Our results identified several genes associated with peroxisome and IFN response, thus providing potential biomarkers and targets for potential therapies against COVID-19.
Materials and Methods
Datasets
Differentially expressed genes in the transcription profile were identified in the National center for biotechnology information (NCBI) Gene expression omnibus (GEO) archive 15 and analyzed using the GEO2R tool. 16 Gene expression data for COVID-19 were analyzed using the GSE180594 and GSE164805 datasets. Differentially expressed genes were characterized for each sample (P < .05) to identify enriched biological processes. The GSE180594 dataset was prepared from circulating monocytes from fresh blood samples in patients with confirmed COVID-19 at emergency room arrival, before receiving any treatment (acute COVID-19), in individuals who overcame COVID-19 6 months ago (post-COVID-19), and in healthy controls (HCs). The GSE164805 dataset contains information obtained using Peripheral blood mononuclear Cells (PBMCs) from patients with severe and mild COVID-19 along with 5 HCs.
Gene ontology and pathway enrichment analysis of differentially expressed genes
The 250 differentially expressed genes (DEGs) from GSE180594 and GSE164805, identified using the GEO2R tool, were analyzed using the DAVID v6.8 (https://david.ncifcrf.gov/) online database. Differentially expressed genes were selected with a value of P < .05. Gene ontology and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analyses were performed for signaling pathway enrichment analysis, with a value of P < .05. For gene ontology (GO) and KEGG enrichment analysis, the background species were selected as Homo sapiens in DAVID.
Blood atlas
Single-cell data for peroxiredoxin 1 (PRDX1), carnitine O-acetyltransferase (CRAT), and peroxisomal membrane protein 4 (PXMP4) were obtained from the Human Protein Atlas ver20.0 (Available from http://www.proteinatlas.org/, accessed November 3, 2021).
Data for the single-cell atlas of the peripheral immune response in patients with severe COVID-19 were obtained from Wilk data 17 which was obtained using single-cell RNA sequencing to profile PBMCs from 7 patients hospitalized for COVID-19 and 6 HCs.
The online tool Venn Diagram (http://bioinformatics.psb.ugent.be/webtools/Venn/) was used to sketch a Venn diagram to identify genes common to GSE180594, GSE164805, CD14+ monocytes, or CD16+ monocytes in patients with severe COVID-19 based on Wilk data. 17
Protein-protein interaction network analysis
After identified genes common to GSE180594 and CD14+ monocytes or CD16+ monocytes in patients with severe COVID-19 using Wilk data, a protein-protein interaction network was drawn using the STRING database (https://string-db.org/) to evaluate the interactions between genes.
Results
Gene ontology analysis
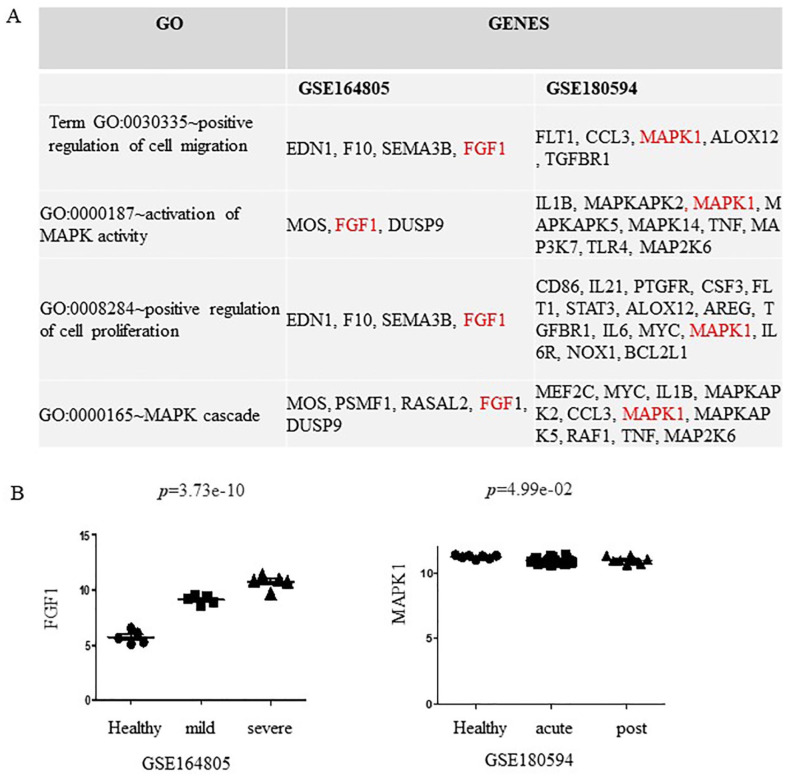
Gene ontology and KEGG analyses were performed to determine the functions of the DEGs from GSE180594 and GSE 164805 using the DAVID tool. Common biological processes from GSE180594 and GSE164805 are shown in Figure 1A. Significant enrichment was found in GO:0030335: positive regulation of cell migration, GO:0000187: activation of mitogen-activated protein kinase (MAPK) activity, GO:0008284: positive regulation of cell proliferation, and GO:0000165: MAPK cascade. Fibroblast growth factor 1 (FGF1) from GSE164805 and MAPK1 from GSE180594 are involved in these 4 signaling pathways. Fibroblast growth factor 1 is highly expressed in patients with mild and severe COVID-19 compared with HCs (Figure 1A). Mitogen-activated protein kinase 1 was slightly downregulated in patients with acute and post-COVID-19 compared with HCs (Figure 1B).
Figure 1.
Gene ontology analysis of differentially expressed genes (DEGs) in patients with COVID-19. (A) Gene ontology analysis of DEGs from GSE164805 and GSE180594. (B) FGF1 is upregulated in patients with mild and severe COVID-19 (left). MAPK1 is slightly downregulated in patients with acute and COVID-19 (right). COVID-19 indicates coronavirus disease 2019; FGF1, fibroblast growth factor 1; GO, gene ontology; MAPK1, mitogen-activated protein kinase 1.
Based on KEGG analysis, the enriched pathways of DEGs from GSE164805 were related to Hsa04810: regulation of actin cytoskeleton, Hsa04916: melanogenesis, and hsa04146: peroxisome (Figure 2A). The top 10 enriched pathways of DEGs from GSE180594 based on the P value were hsa05323: rheumatoid arthritis, hsa04668: TNF signaling pathway, hsa05164: influenza A, hsa04060: cytokine-cytokine receptor interaction, has05321: inflammatory bowel disease, hsa05145: toxoplasmosis, hsa04010: MAPK signaling pathway, hsa05133: pertussis, has05142: Chagas disease (American trypanosomiasis), and hsa04620: Toll-like receptor signaling pathway (Figure 2B).
Figure 2.
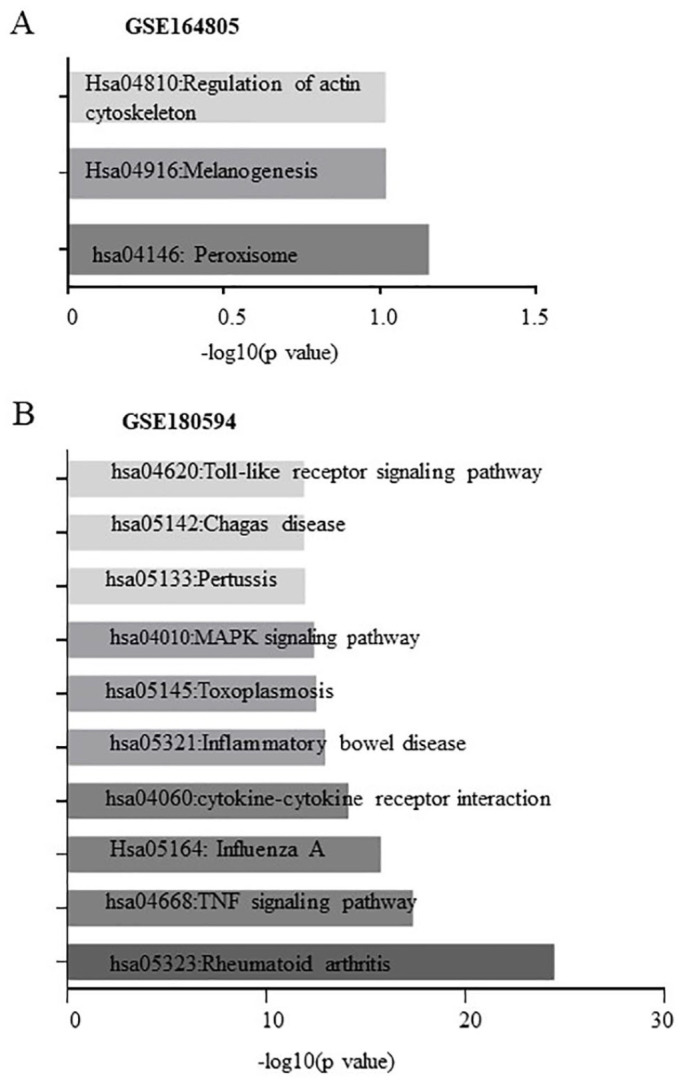
Kyoto Encyclopedia of Genes and Genomes pathways enriched by differentially expressed genes (DEGs) from (A) GSE164805 and (B) GSE180594.
Peroxisomes genes are associated with COVID-19
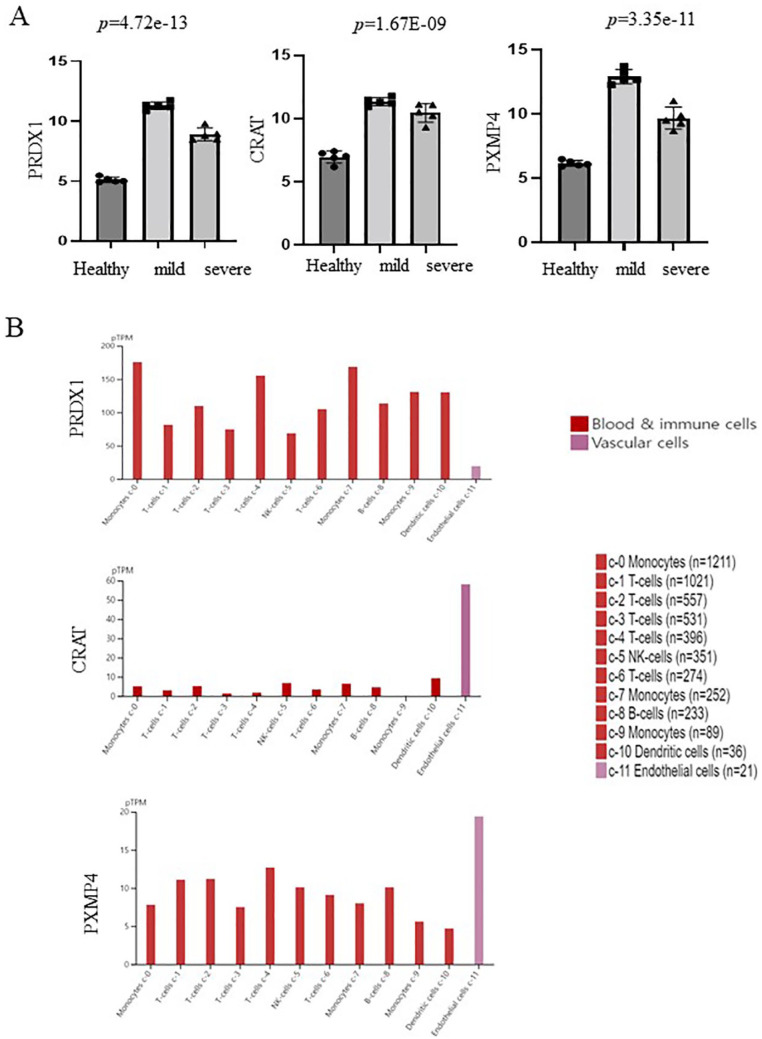
PRDX1, CRAT, and PXMP4 from GSE164805 are involved in peroxisomes, which have not been well studied in COVID-19. Therefore, we evaluated the levels of these genes in patients with mild and severe COVID-19 based on GSE164805. As shown in Figure 3A, PRDX1, CRAT, and PXMP4 were highly increased in patients with mild and severe COVID-19.
Figure 3.
Peroxisome-related genes (PRDX1, CRAT, and PXMP4) associated with COVID-19. (A) Upregulation of peroxisome-related genes (PRDX1, CRAT, and PXMP4) in patients with mild and severe COVID-19. (B) Single-cell distribution of peroxisome-related genes (PRDX1, CRAT, and PXMP4) in PBMC. Single-cell data from the blood for PRDX1, CRAT, and PXMP4 were obtained from the Human Protein Atlas ver20.0 (available from http://www.proteinatlas.org/, accessed on November 3, 2021). COVID-19 indicates coronavirus disease 2019; CRAT, carnitine O-acetyltransferase; PRDX1, peroxiredoxin 1; PXMP4, peroxisomal membrane protein 4.
To determine the role of PRDX1, CRAT, and PXMP4 in peripheral immune cells, single-cell data for the blood were obtained from the human protein atlas. As shown in Figure 3B, CRAT and PXMP4 were highly expressed in endothelial cells, whereas PRDX1 was expressed in all immune cells such as monocytes, T cells, dendritic cells, and B cells but not endothelial cells.
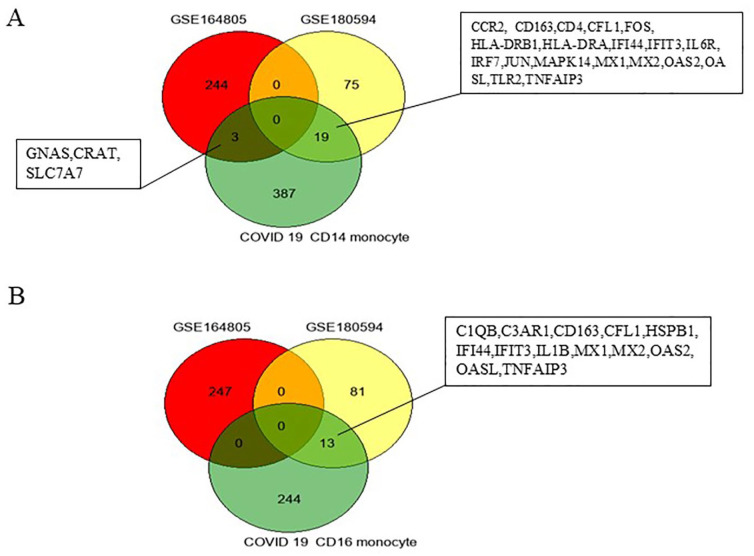
Next, to identify the DEGs from GSE180594 and GSE164805 in peripheral immune cells from patients with COVID-19, Wilk data, 17 which were obtained using single-cell RNA sequencing to profile PBMCs from 7 patients hospitalized for COVID-19, were analyzed. The Venn diagrams in Figure 4A indicate that 19 genes (CCR2, CD163, CD4, CFL1, FOS, HLA-DRB1, HLA-DRA, IFI44, IFIT3, IL6R, IRF7, JUN, MAPK14, MX1, MX2, OAS2, OASL, TLR2, and TNFAIP3) overlapped genes between GSE180594 and CD4+ monocytes in peripheral immune cells from COVID-19. Three genes (GNAS, CRAT, and SLC7A7) overlapped between GSE180594 and CD4+ monocytes in peripheral immune cells from COVID-19 (Figure 4A).
Figure 4.
Venn diagram showing overlap between differentially expressed genes (DEGs) from GSE164805, GSE180594, and single-cell atlas of the peripheral immune response in patients with severe COVID-19 based on Wilk data. 17 (A) Intersection of DEGs from GEO2R was identified from GSE164805, GSE180594, and genes expressed in CD14+ monocytes of patients with COVID-19 (Wilk data). (B) Intersection of GSE164805, GSE180594, and genes expressed in CD16+ monocytes of patients with COVID-19 (Wilk data). COVID-19 indicates coronavirus disease 2019
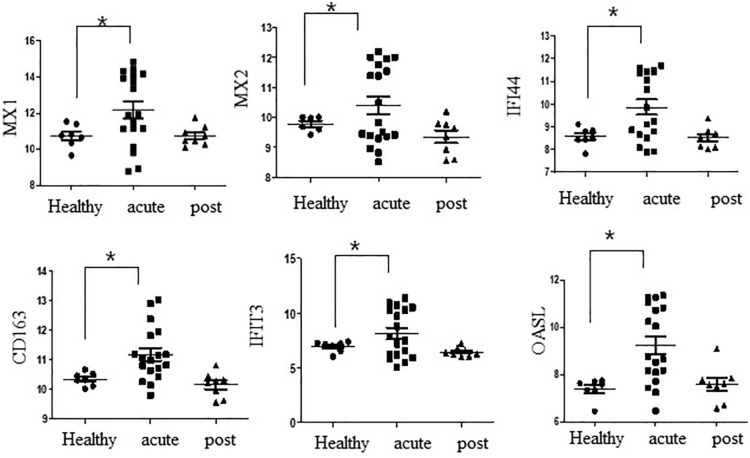
Thirteen genes (C1QB, C3AR1, CD163, CFL1, HSPB1, IFI44, IFIT3, IL1B, MX1, MX2, OAS2, OASL, and TNFAIP3) overlapped between the GSE180594 and CD16+ monocytes in peripheral immune cells from COVID-19 (Figure 4B). MX1, MX2, IFI44, CD163, IFIT3, and OASL were highly expressed in acute COVID-19, whereas these genes were not changed in post-COVID-19 individuals based on GSE180594 (Figure 5).
Figure 5.
Relative expression of MX1, MX2, IFI44, CD163, IFIT3, and OASL in healthy controls and patients with acute and post-COVID-19. COVID-19 indicates coronavirus disease 2019.
*P < .05 by unpaired t test.
Furthermore, to understand the roles of overlapping genes between GSE180594 and CD4+ monocytes in COVID-19 pathophysiology, 19 overlapping genes between GSE180594 and CD4+ monocytes were evaluated using STRING. Interferon-induced protein 44 (IFI44), 2′-5′-oligoadenylate synthetase-like (OASL), 2′-5′-oligoadenylate synthetase 2 (OAS2), IFIT3, myxovirus resistance genes (MX)1, MX2, and IFN regulatory factor 7 (IRF7) were mainly associated with CD4, Toll-like receptor 2 (TLR2), JUN, FOX, major histocompatibility complex, class II, DR alpha (HLA-DRA), major histocompatibility complex, class II, DR beta 1 (HLA-DRB1), and MAPK14 (Figure 6A). As shown in Figure 6B, in CD16+ monocytes in COVID-19, overlapping genes (OAS, IFI44, MX2, MX1, OASL, IFIT3, TNFAIP3, IL1B, HSPB1, CFL1, CD163, C3AR1, and C1QB) between GSE180594 and CD16+ monocytes in COVID-19 were mainly interacted.
Figure 6.
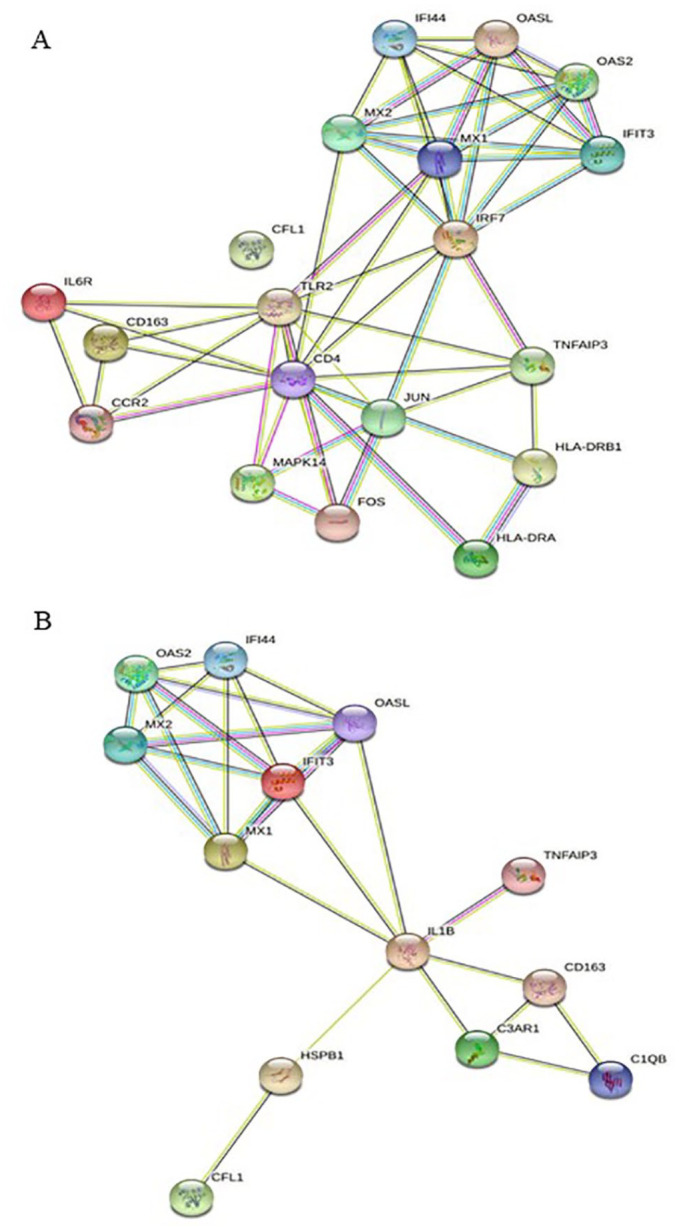
Protein-protein interactions of genes from monocyte in patients with COVID-19 analyzed using STRING. (A) Protein-protein interaction of genes from GSE180594 and CD14+ monocytes in patients with severe COVID-19 based on Wilk data. (B) Protein-protein interaction of genes from GSE180594 and CD16+ monocytes in patients with severe COVID-19 based on Wilk data. COVID-19 indicates coronavirus disease 2019.
Discussion
We integrated 2 publicly available GEO datasets and used multiple bioinformatics tools to identify functional targets in patients with COVID-19. Gene ontology analysis revealed positive regulation of cell migration, activation of MAPK activity, positive regulation of cell proliferation, and MAPK cascade. It is well known that the MAPK pathway is activated during viral infections 18 and regulates apoptosis, 19 immune response. 20 In addition, human SARS-CoV-1 infection activates the p38 MAPK pathway and enhances phosphorylation of its downstream-regulated proteins, particularly kinases. 21 In this study, FGF1 from GSE164805 and MAPK1 from GSE180594 were found to be involved in these 4 signaling pathways. Fibroblast growth factor 1 is highly expressed in patients with mild and severe COVID-19 compared with HCs. Mitogen-activated protein kinase 1 was slightly downregulated in patients with acute and post-COVID-19 compared with HCs. There is evidence that FGF1, an acidic FGF, is a cellular growth factor and strong angiogenic factor that controls the development of new blood vessels. 22 Fibroblast growth factor 1 and MAPK1 may be effective therapeutic agents for COVID-19.
A recent study reported that peroxisomes are associated with COVID-19, in which the small viral protein encoded by ORF14 binds to the critical peroxisome biogenesis factor PEX14. 23 Peroxisomes play a role in metabolizing lipids and reactive oxygen species 24 and are involved in proinflammatory and immune pathways. 25 Our data showed that peroxisome-related genes such as CRAT, PRDX1, and PXMP4 were highly expressed in patients with mild and severe COVID-19. Thus, our data suggest that the peroxisome is important in COVID-19.
In this study, MX1, IFI44, CD163, IFIT3, and OASL were highly expressed in acute COVID-19, although these genes were not changed in post-COVID-19 (Figure 5). In addition, these genes were expressed in COVID-19 CD4+ and CD16+ monocytes. Previous studies showed that MX1 and MX2 expression was significantly higher in patients with COVID-19 than in a non-COVID-19 group. MX1 is an IFN-inducible protein associated with viral infections such as influenza and viral encephalitis 26 and is triggered by SARS-CoV-2. 27 MX2 is also essential for type I IFN-induced postentry inhibition of HIV-1 infection. 28 Overexpression of IFI44 is necessary for restricting respiratory syncytial virus infection at an early time postinfection. 29 OASL, an IFN-stimulating gene, plays a role in antiviral defense mechanisms 30 and in the inflammatory response to SARS-CoV-2. 31 CD163, a scavenger receptor, plays a role as of inflammation and a marker of anti-inflammatory (M2) macrophages. 32 After SARS-CoV-2 infection, immune cells are activated and secrete a large number of inflammatory cytokines, resulting in a cytokine storm that can result in death.13,33 Analyzing bioinformatics data from peripheral blood samples revealed that MX1, IFI44, IFIT3, and OASL are associated with IFN responses. Determining the changes in IFN response genes in the human blood after SARS-CoV-2 infection can improve our understanding of the mechanism, diagnosis, and treatment of COVID-19.
Conclusions
We comprehensively analyzed the blood cell gene expression profile data of patients with COVID-19 using bioinformatics methods to preliminary understand the functions and associated targets of DEGs in the blood cells of these patients. Our results suggest that positive regulation of cell migration, activation of MAPK activity, positive regulation of cell proliferation, and MAPK cascade are associated with COVID-19. Peroxisome-associated genes such as CRAT, PRDX1, and PXMP4 were highly expressed in patients with mild and severe COVID 19. In addition, MX1, IFI44, CD163, IFIT3, and OASL are highly expressed in patients with acute COVID 19 and CD14+ and CD16+ monocytes in patients with COVID-19. Thus, these genes may be useful as diagnostic markers and therapeutic targets for COVID-19.
Footnotes
Author Contributions: EJS performed the writing, protocol, and draft of the article.
Declaration of Conflicting Interests: The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This was supported by the National Research Foundation of Korea funded by the Ministry of Education (NRF-2021R1I1A1A01052609).
Consent for Publication: The authors gave their consent for their data to be used in the article
ORCID iD: Eun Jung Sohn  https://orcid.org/0000-0001-6095-5106
https://orcid.org/0000-0001-6095-5106
References
- 1. Fehr AR, Perlman S. Coronaviruses: an overview of their replication and pathogenesis. Methods Mol Biol. 2015;1282:1-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Peiris JS, Yuen KY, Osterhaus AD, Stohr K. The severe acute respiratory syndrome. N Engl J Med. 2003;349:2431-2441. [DOI] [PubMed] [Google Scholar]
- 3. Li J, Gong X, Wang Z, et al. Clinical features of familial clustering in patients infected with 2019 novel coronavirus in Wuhan, China. Virus Res. 2020;286:198043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Huang CL, Wang YM, Li XW, et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395:497-506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Liu Y, Gayle AA, Wilder-Smith A, Rocklov J. The reproductive number of COVID-19 is higher compared to SARS coronavirus. J Travel Med. 2020;27:taaa021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Lauer SA, Grantz KH, Bi Q, et al. The Incubation Period of Coronavirus Disease 2019 (COVID-19) from publicly reported confirmed cases: Estimation and Application. Ann Intern Med. 2020;172:577-582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Coperchini F, Chiovato L, Croce L, Magri F, Rotondi M. The cytokine storm in COVID-19: An overview of the involvement of the chemokine/chemokine-receptor system. Cytokine Growth Factor Rev. 2020;53:25-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Li X, Geng M, Peng Y, Meng L, Lu S. Molecular immune pathogenesis and diagnosis of COVID-19. J Pharm Anal. 2020;10:102-108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Sudre CH, Murray B, Varsavsky T, et al. Attributes and predictors of long COVID. Nat Med. 2021;27:626-631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Oronsky B, Larson C, Hammond TC, et al. A review of persistent post-COVID syndrome (PPCS) [published online ahead of print February 20, 2021]. Clin Rev Allergy Immunol. doi: 10.1007/s12016-021-08848-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Utrero-Rico A, Gonzalez-Cuadrado C, Chivite-Lacaba M, et al. Alterations in circulating monocytes predict COVID-19 severity and include chromatin modifications still detectable six months after recovery. Biomedicines. 2021;9:1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Nalbandian A, Sehgal K, Gupta A, et al. Post-acute COVID-19 syndrome. Nat Med. 2021;27:601-615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Xu Z, Shi L, Wang Y, et al. Pathological findings of COVID-19 associated with acute respiratory distress syndrome. Lancet Respir Med. 2020;8:420-422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Li G, Ruan S, Zhao X, Liu Q, Dou Y, Mao F. Transcriptomic signatures and repurposing drugs for COVID-19 patients: findings of bioinformatics analyses. Comput Struct Biotechnol J. 2021;19:1-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Barrett T, Troup DB, Wilhite SE, et al. NCBI GEO: archive for high-throughput functional genomic data. Nucleic Acids Res. 2009;37:D885-D890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Barrett T, Wilhite SE, Ledoux P, et al. NCBI GEO: archive for functional genomics data sets —update. Nucleic Acids Res. 2013;41:D991-D995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Wilk AJ, Rustagi A, Zhao NQ, et al. A single-cell atlas of the peripheral immune response in patients with severe COVID-19. Nat Med. 2020;26:1070-1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kumar R, Khandelwal N, Thachamvally R, et al. Role of MAPK/MNK1 signaling in virus replication. Virus Res. 2018;253:48-61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Bian J, Wang K, Kong X, et al. Caspase- and p38-MAPK-dependent induction of apoptosis in A549 lung cancer cells by Newcastle disease virus. Arch Virol. 2011;156:1335-1344. [DOI] [PubMed] [Google Scholar]
- 20. Gaur P, Munjhal A, Lal SK. Influenza virus and cell signaling pathways. Med Sci Monit. 2011;17:RA148-RA154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Mizutani T, Fukushi S, Saijo M, Kurane I, Morikawa S. Phosphorylation of p38 MAPK and its downstream targets in SARS coronavirus-infected cells. Biochem Biophys Res Commun. 2004;319:1228-1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Marwa BA, Raguema N, Zitouni H, et al. FGF1 and FGF2 mutations in preeclampsia and related features. Placenta. 2016;43:81-85. [DOI] [PubMed] [Google Scholar]
- 23. Knoblach B, Ishida R, Hobman TC, Rachubinski RA. Peroxisomes exhibit compromised structure and matrix protein content in SARS-CoV-2-infected cells. Mol Biol Cell. 2021;32:1273-1282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Smith JJ, Aitchison JD. Peroxisomes take shape. Nat Rev Mol Cell Biol. 2013;14:803-817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Dixit E, Boulant S, Zhang Y, et al. Peroxisomes are signaling platforms for antiviral innate immunity. Cell. 2010;141:668-681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ciancanelli MJ, Abel L, Zhang SY, Casanova JL. Host genetics of severe influenza: from mouse Mx1 to human IRF7. Curr Opin Immunol. 2016;38:109-120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Bizzotto J, Sanchis P, Abbate M, et al. SARS-CoV-2 infection boosts MX1 antiviral effector in COVID-19 patients. Iscience. 2020;23:101585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Dicks MDJ, Betancor G, Jimenez-Guardeño JM, et al. Multiple components of the nuclear pore complex interact with the amino-terminus of MX2 to facilitate HIV-1 restriction. PLoS Pathog. 2018;14:e1007408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Busse DC, Habgood-Coote D, Clare S, et al. Interferon-induced Protein 44 and interferon-induced Protein 44-like restrict replication of respiratory syncytial virus. J Virol. 2020;94:18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Leisching G, Ali A, Cole V, Baker B. 2′-5′-Oligoadenylate synthetase-like protein inhibits intracellular M. Mol Immunol. 2020;118:73-78. [DOI] [PubMed] [Google Scholar]
- 31. Shaath H, Vishnubalaji R, Elkord E, Alajez NM. Single-cell transcriptome analysis highlights a role for neutrophils and inflammatory macrophages in the pathogenesis of severe COVID-19. Cells. 2020;9:2374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Etzerodt A, Moestrup SK. CD163 and inflammation: biological, diagnostic, and therapeutic aspects. Antioxid Redox Signal. 2013;18:2352-2363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Gao X, Liu Y, Zou S, et al. Genome-wide screening of SARS-CoV-2 infection-related genes based on the blood leukocytes sequencing data set of patients with COVID-19. J Med Virol. 2021;93:5544-5554. [DOI] [PMC free article] [PubMed] [Google Scholar]