Abstract
Spontaneous clearance of acute hepatitis C virus (HCV) infection is associated with single nucleotide polymorphisms (SNPs) on the MHC class II. We fine-mapped the MHC region in European (n = 1,600; 594 HCV clearance/1,006 HCV persistence) and African (n = 1,869; 340 HCV clearance/1,529 HCV persistence) ancestry individuals and evaluated HCV peptide binding affinity of classical alleles. In both populations, HLA-DQβ1Leu26 (p valueMeta = 1.24 × 10−14) located in pocket 4 was negatively associated with HCV spontaneous clearance and HLA-DQβ1Pro55 (p valueMeta = 8.23 × 10−11) located in the peptide binding region was positively associated, independently of HLA-DQβ1Leu26. These two amino acids are not in linkage disequilibrium (r2 < 0.1) and explain the SNPs and classical allele associations represented by rs2647011, rs9274711, HLA-DQB1∗03:01, and HLA-DRB1∗01:01. Additionally, HCV persistence classical alleles tagged by HLA-DQβ1Leu26 had fewer HCV binding epitopes and lower predicted binding affinities compared to clearance alleles (geometric mean of combined IC50 nM of persistence versus clearance; 2,321 nM versus 761.7 nM, p value = 1.35 × 10−38). In summary, MHC class II fine-mapping revealed key amino acids in HLA-DQβ1 explaining allelic and SNP associations with HCV outcomes. This mechanistic advance in understanding of natural recovery and immunogenetics of HCV might set the stage for much needed enhancement and design of vaccine to promote spontaneous clearance of HCV infection.
Keywords: hepatitis C virus, fine-mapping, HLA-DQβ1, HCV clearance, trans-ancestral, host genetics, MHC, GWAS, HLA imputation, infection
Introduction
An estimated 71.1 million people worldwide have chronic hepatitis C viral (HCV) infection,1,2 which can result in liver cirrhosis, hepatocellular cancer, and liver failure.3 Not all acute infections become chronic since approximately 30% of infected individuals spontaneously clear HCV infection. Spontaneous clearance is associated with sex, race, and host genetics.4,5 Host genetic variants in the Interferon Lamba 3 and Interferon Lambda 4 (IFNL4, IFNL3) genes, the MHC region, and G protein-coupled receptor 158 (GPR158) gene5, 6, 7, 8, 9 are all associated with HCV spontaneous clearance. Interestingly, the association in the MHC class II region which spans 305.2 kb across the HLA-DQB1/HLA-DQA1 and HLA-DQA2 genes is consistent across populations of different genetic ancestries and demographic histories.9, 10, 11, 12, 13, 14, 15, 16 Specifically, the main genetic signals are localized 48.5 kb upstream of HLA-DQA2 and 33.3 kb upstream of the HLA-DQB1 gene;5,6 however, the causal variants for this association remain elusive.
The tight linkage disequilibrium (LD) across disease-associated MHC haplotype and the highly polymorphic nature of associated variants makes determining the contribution of specific classical HLA alleles and amino acid residues to the spontaneous clearance of HCV infection challenging. To identify putative causal amino acid positions and residues associated with HCV spontaneous clearance and their potential impact on peptide presentation by HLA molecules, we investigated the HLA region in individuals of African and European ancestry with HCV spontaneous clearance and persistence. We identified key amino acids that were present in both European and African ancestry individuals, suggesting they are not specific to ancestry populations and may be targeted across diverse groups. We also predicted the peptide binding affinity of HLA class II molecules identified as relevant for HCV clearance or persistence.
Material and methods
Samples
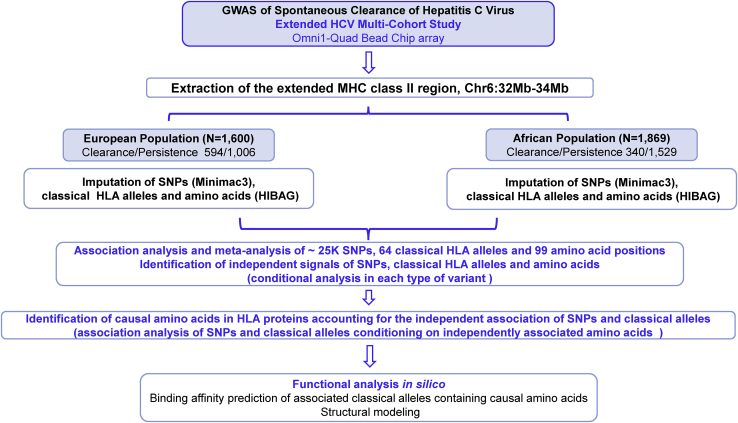
The study design is presented in Figure 1. The study included 3,469 individuals participating in the Extended HCV Genetics Consortium.5,6,17 This is a multi-site international consortium including multiple studies from Europe and United States in which HCV infection outcomes were ascertained. They include ALIVE (AIDS Link to the Intravenous Experience),18 BBAASH (Baltimore Before and After Acute Study of Hepatitis),19 BAHSTION (Boston Acute Hepatitis C Virus Study: Transmission, Immunity and Outcomes Network),13 Cramp and colleagues’ study,20 HGDS (Hemophilia Growth and Development Study),21 Mangia and colleagues’ study,22 MHCS (Multicenter Hemophilia Cohort Study) and MHCS-II,23 REVELL (Correlates of Resolved Versus Low-Level Viremic Hepatitis C Infection in Blood Donors) study,24 the Swan Project,25 the Toulouse, France cohort,15 WIHS (Women’s Interagency HIV Study),26 the United Kingdom Drug Use cohort,27 and the Urban Health Study (UHS)28,29 as described in detail in elsewhere.5,6,17 These studies were selected because they had well-established hepatitis C virus (HCV) outcomes, as previously described,5,6,17 available DNA, and IRB approval for genetic testing. Information about HIV infection status was also obtained in the included individuals since it is a determinant of HCV clearance. All subjects gave written informed consent for this study, approved by each Institutional Review Board.13 The complete sample corresponded to 1,600 individuals of European ancestry (594 with HCV clearance/1,006 with HCV persistence) and 1,869 of African ancestry (340 with clearance/1,529 with persistence). Detailed information about the study cohorts is available in supplemental material and methods and distribution of the analyzed individuals by ancestry group, sex, and HIV infection status is presented in Table 1.
Figure 1.
Outline of the trans-ancestral fine mapping strategy used to identify causal variants in the MHC region associated with HCV spontaneous clearance in European and African ancestry populations
Table 1.
Demographic characteristics of the analyzed studies by genetically determined ancestry groups
| Genetically determined ancestry group | N | HCV infection persistence:clearance | (+) HIV infection (%) | Female sex (%) |
|---|---|---|---|---|
| African ancestry | 1,869 | 1,529:340 | 38.6 | 33.3 |
| European ancestry | 1,600 | 1,006:594 | 16.1 | 32.1 |
| Total | 3,469 | 2,535:934 | 28.2 | 32.8 |
Genotyping, imputation of SNPs, classical HLA alleles, and amino acids
Our initial dataset consisted of genotypes obtained using the Illumina Omni1-Quad BeadChip array (Illumina)5,6 and processed using standard genome-wide association study protocols for quality control6 (dbGaP accession numbers dbGaP:phs000454.v1.p1 and dbGaP:phs000248.v1.p1). We extracted genotyped markers from a 2 Mb segment of the MHC corresponding to the extended class II region30 delimited by chr6:32,000,000–34,000,000 (coordinates based on the Genome Reference Consortium Human Build 37, GRCh37/hg19). SNP imputation was done using Minimac331 software through the publicly available Michigan Imputation Server.6 As part of the quality control, we excluded SNPs with imputation r2 < 0.3 and minor allele frequency (MAF) < 1%.
In each ancestry group, we used the method implemented in HIBAG (HLA Imputation using attribute BAGging)32 to impute classical HLA alleles from genotyped/imputed SNPs for HLA-DPB1, HLA-DQA1, HLA-DQB1, and HLA-DRB1 loci producing genotypes at two field-resolution for each individual. This method takes advantage of the extended haplotype structure within the MHC and makes predictions of classical HLA alleles by averaging HLA-type posterior probabilities over an ensemble of classifiers built on bootstrap samples. Classifiers are trained from large databases of individuals with known HLA and SNP genotypes within the MHC region.32 In order to capture the appropriate genetic background, we used a paired ancestry group of classifiers for each target population. For European ancestry we used pre-built classifiers computed from individuals of European ancestry from the HLARES database32 included with the software, and for individuals of African ancestry we built classifiers based on the HLA types and genotypes from the “Consortium on Asthma among African-ancestry Populations in the Americas” (CAAPA, dbGaP Study Accession: dbGaP:phs001123.v1.p1)33,34 (supplemental material and methods). For the imputation of classical HLA class II alleles and amino acids, we used markers that intersected between our target populations and those included in the classifiers for each ancestry group, corresponding to 3,569 SNPs for European ancestry and 991 for African ancestry.
For association analyses, we excluded classical HLA genotypes imputed at two-field resolution with a posterior probability lower than 0.535,36 and alleles with MAF lower than 1%. Details of HLA imputation performance of classical alleles for the different HLA genes are presented in Table S1.
Translation of classical HLA alleles to amino acid residues in the antigen binding domain of the HLA proteins was performed based on the classical HLA alleles imputed by HIBAG and the peptide sequence of alleles available in the HLA database using the HIBAG software.32 For association analyses with HCV spontaneous clearance, we included all amino acid polymorphic positions with residues with a frequency greater than 1% (Figure 1).
Statistics
We performed association analyses with each type of variant (SNPs, classical HLA alleles, and amino acid residues in the proteins) and conditional analyses to identify independent SNPs, classical HLA alleles, and amino acid signals within each type of variant. Finally, we used conditional analyses across the variant types to identify which amino acids explained the independent associations (Figure 1).
Association analyses and meta-analysis of each type of variant
For each type of variant that met quality control thresholds, we tested for association with HCV spontaneous clearance using an additive logistic regression model adjusting for HIV infection status and the first two principal components using PLINK 2.0 alpha version37,38 for SNPs, and HIBAG32 for HLA classical allele and amino acid residue in the proteins (supplemental material and methods). Population-specific PCs were calculated using the smartpca program in EIGENSOFT32,39 separately in the European and the African ancestry populations (excluding 1000 Genomes reference populations) using 16,142 genomic independent markers. In each ancestry group, individuals grouped in a tight cluster, and PC1 and PC2 explained most of the variance in the data (Figure S1). We extended the analysis to consider HIV status and applied the same regression model and meta-analysis to those variants in individuals with and without HIV independently in each population. In the European ancestry population, we tested associations with 22,259 genotyped/imputed SNPs; 54 classical alleles in the genes HLA-DRB1, HLA-DQA1, HLA-DQB1, and HLA-DPB1 and 205 residues (and combinations of residues) in 99 amino acid positions in these proteins. For the African ancestry population, we evaluated associations with 25,565 SNPs, 33 classical HLA alleles in the genes HLA-DRB1 and HLA-DQB1, and 155 residues and their combinations. Classical HLA alleles and amino acids in HLA-DQA1 and HLA-DPB1 were not imputed in the African ancestry population since data for those genes was not available in the reference panel used for imputation (Table S1).
For each type of variant, we analyzed each ancestry group separately and then performed a fixed effect inverse-variance weighted meta-analysis, using METAL.40 A locus-specific p value threshold of significance for the meta analysis of this chromosome region would be adequate (p valueMeta 2.3 × 10−6), but we chose to use a more conservative population-specific genome-wide p value threshold of significance (p valueMeta) of 2.05 × 10−7 for the SNPs in the meta-analysis,6 and a p valueMeta of 7.8 × 10−4 (64 classical HLA alleles tested in the meta-analysis) and 5.0 × 10−4 (99 amino acid positions tested in the meta-analysis) was used for classical HLA alleles and amino acids, respectively.
Identification of independent signals for each type of variants
We performed iterative conditional analyses using additive logistic models in each type of variant and meta-analyzed the results from the two populations, in each iteration step. In brief, we first identified the most strongly associated variant in the meta-analysis (lowest p valueMeta) and performed forward iterative conditional logistic regression to identify other independent signals. The significance threshold for the conditional analysis was set at a p valueMeta < 5 × 10−6 for SNPs and a p valueMeta < 1.0 × 10−3 for classical HLA alleles and amino acid residues given the number of tests for each type of variant. If another variant of the same type was significantly associated in the conditional analysis, we introduced this variant in the model as a covariate to evaluate the effect of the remaining variants, until no variant was significant.
Identification of amino acids accounting for independent signals
We performed conditional analyses with customized scripts in R41 and PLINK 2.0 alpha version37,38 to determine the amino acid residues accounting for the independently associated SNPs and classical HLA alleles by using iterative additive logistic regression models until the association of the top SNPs (or classical HLA alleles) were diminished and no other SNP (or classical HLA allele) was significantly associated in the region (Figure 2) at the conditional p valueMeta threshold. In the conditional models, we included every associated amino acid as a covariate as well as HIV infection status and two principal components.42 We prioritized the amino acids with the strongest associations and those identified as independently associated and their combinations. For each analysis we meta-analyzed the results from the two populations in each step.
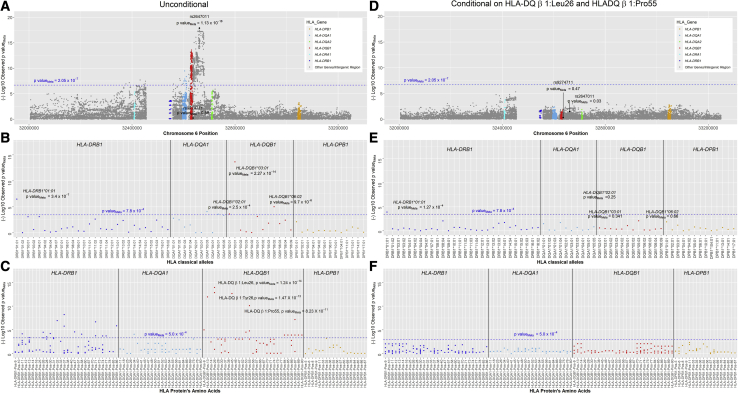
Figure 2.
Results of the fixed-effect meta-analyses in 932 individuals with HCV spontaneous clearance and 2,532 persons with persistent HCV infection from African and European ancestry populations
(A) Association results of the unconditional analysis of SNPs.
(B) Association results of the unconditional analysis of HLA classical alleles.
(C) Association results of the unconditional analysis of amino acid residues.
(D–F) Results of the same variants after conditioning on HLADRβ1Leu26 and HLADRβ1Pro55. Each point corresponds to the p valueMeta in each type of variant. The blue line and the p valueMeta colored in blue represent the threshold level of fixed effects meta-analysis significance corrected by multiple comparisons.
Epitope prediction of associated classical HLA class II alleles containing causal amino acids
We selected the classical class II HLA alleles positively (clearance) and negatively (persistence) associated with HCV spontaneous clearance that represented the associated amino acids of interest. To investigate the capabilities of associated alleles to bind epitopes derived from the entire HCV polyprotein sequence (supplemental material and methods), we then performed T cell epitope prediction analyses. Using tools available in IEDB-Analysis Resources (see web resources),43 we analyzed the HCV polyprotein sequence (GenBank:AFE48416.1) in combination with the associated clearance and persistence HLA alleles. Predictions were performed by applying the NetMHCIIpan 3.2 algorithm44 and extracting all the predicted IC50 values for each possible 15-mer spanning the entire HCV proteome (supplemental material and methods). IC50 values in nano-Molar (nM) represent the concentration of the test peptide, which will displace 50% of a standard peptide from the HLA molecule in question. The lower the predicted IC50 values are, the stronger the binding peptide-HLA allele combination is. To be considered an epitope, a cutoff of binding of ≤1000 nM has been applied, as previously reported.45 The geometric mean ratio (described in supplemental material and methods) has been calculated only in peptides predicted to be epitopes in at least one of the classical HLA class II alleles analyzed in this study. The higher the geometric mean ratio the stronger is the binding affinity in clearance alleles versus persistence ones.
We aimed to address the global prediction patterns of these alleles using a PCA on the entire set of predicted peptides per allele after logarithmic transformation using the “prcomp” function in R.41 We then performed a more in-depth analysis, considering separately the predicted binding distribution of each specific class II allele where each dot represents the predicted value of the specific peptide derived from the HCV proteome. Combined clearance IC50 values (all clearance alleles) versus combined persistence (all persistent alleles) were compared using Mann-Whitney U test. Statistical analyses and graphs were performed using Prism 8 (GraphPad Software). A p value < 0.05 was considered as significant in this analysis.
Location of amino acid residues on HLA proteins
To delineate the physical location of the causal amino acids in the tertiary structure of the HLA proteins, we used X-ray structures from the Protein Data Bank (PDB).46 This provides structural visualization of HLA-DQα1/DQβ1 heterodimer localizing the amino acids that account for the signal in MHC class II region. The accession code for the selected structure was PDB: 1jk847 for HLA-DQα1/DQβ1 and was visualized using PyMOL v. 2.3.2 (The PyMOL Molecular Graphics System, Version 2.0 Schrödinger, LLC).
Study approval
Each individual study cohort obtained consent for genetic testing from their governing Institutional Review Board (IRB) and the Johns Hopkins School of Medicine Institutional Review Board approved the overall analysis and written informed consent was received from participants prior to inclusion in the study.
Results
Unconditional and conditional association analyses and meta-analyses
Unconditional and conditional association analysis with SNPs
There were 1,219 SNPs significantly associated with HCV spontaneous clearance in a 305.3 kb genomic segment of the MHC class II region (chr6:32,376,360–32,681,631) harboring the genes HLA-DRA1, HLA-DRB1, HLA-DQA1, and HLA-DQB1 (Figure 2A, Table S2). The lead SNP (rs2647011, C>A, p valueMeta = 1.13 × 10−18) is in the intergenic region between HLA-DQB1 and HLA-DQA2 (Figure 2A). This SNP, located 1,333 bp from rs2647006, which was detected as the top signal in a previous trans-ethnic GWAS6 and both SNPs are in LD in European (r2 = 0.96) and African ancestry populations (r2 = 0.97).
In the analysis conditioning on rs2647011, we detected a second independent association with rs9274711 (p valueMeta conditional on rs2647011 = 4.18 × 10−6, African population OR T allele = 0.71 [95% CI:0.59–0.86], p value = 4.1 × 10−4, European population OR = 0.72 [95% CI:0.58–0.89], p value = 2.7 × 10−3) (Figure S2B). This variant is not associated in the individual SNP analysis (p valueMeta unconditional = 0.58, African population OR T allele = 0.99 [95% CI: 0.83–1.16], p value = 0.56, European population OR T allele = 0.94 [95% CI:0.77–1.15], p value = 0.83), as shown in Figure S2A and Figure 2. Further analyses determined that allele frequency of the C allele of rs2647011 is 0.37 and 0.39, in European and African ancestry, respectively. Similarly, the frequency of the T allele of rs9274711 is 0.26 and 0.22 those populations, respectively. Both SNPs are not in linkage disequilibrium (r2 = 0.19 and r2 = 0.17 in European and African ancestry populations, respectively). Moreover, a formal test of SNP×SNP interaction was not significant in any of the populations (p value rs2647011 × rs9274711 = 0.12 and p value = 0.86 in Europeans and African ancestry populations, respectively). Interestingly, rs2647011A allele (favorable for HCV clearance) is frequently in the same haplotype as rs9274711 T allele (non favorable for HCV clearance, Haplotype Frequency AT = 0.25 and 0.21 for European and African ancestry, respectively). The complementary alleles form a more frequent haplotype CA = 0.37 and 0.39 for European and African ancestry, respectively. Thus, it is plausible that the independent effect of the rs9274711T allele is only present when we condition on rs2647011. When including these two variants in the association analysis, no other SNP was significantly associated in the MHC Class II region in the meta-analysis (Figure S2C).
Unconditional and conditional association analysis with classical HLA alleles
SNP data were then used to impute HLA alleles. The strongest signal of association was HLA-DQB1∗03:01 (p valueMeta = 2.27 × 10−14), while associations with lower statistical significance were found with HLA-DRB1∗01:01 (p valueMeta = 3.4 × 10−7), HLA-DQB1∗06:02 (p valueMeta = 9.7 × 10−6), HLA-DQB1∗05:01 (p valueMeta = 7.3 × 10−4), HLA-DQB1∗02:01 (p valueMeta = 2.5 × 10−4), HLA-DQA1∗03:03 (p valueMeta = 8.5 × 10−5), HLA-DQA1∗05:01 (p valueMeta = 2.6 × 10−4), and HLA-DQA1∗05:05 (p valueMeta = 4.4 × 10−4) (Figure 2B, Table S3).
In the conditional analysis of classical HLA alleles, HLA-DQB1∗03:01 and HLA-DRB1∗01:01 (p valueMeta conditional on HLA-DQB1∗03:01 = 5.61 × 10−9) were independently associated with HCV spontaneous clearance. Including those two alleles in the model eliminated all residual association of classical HLA alleles (Figure S3).
Unconditional and conditional association analysis with amino acid residues
We then identified specific amino acid residues in the HLA-DQβ1 and HLA-DRβ1 proteins associated with HCV spontaneous clearance with high significance (Figure 2C, Table S4). In HLA-DQβ1, the strongest association was HLA-DQβ1-26; among the three possible residues (Leu, Tyr, and Gly), the HLA-DQβ1Leu26 was associated with 35%–45% lower likelihood of clearance (p valueMeta = 1.24 × 10−14), whereas HLA-DQβ1Tyr26 was associated with 64%–81% higher likelihood of HCV spontaneous clearance (p valueMeta = 1.47 × 10−13); this finding was consistent across ancestry groups (Tables 2 and S4).
Table 2.
Results of the unconditional association analysis and fixed effect meta-analysis of the two amino acids in HLA-DQβ1 protein that explain the association in the MHC class II region, HLA-DQβ1Leu26, and HLA DQβ1Pro55 and alternatives HLA-DQβ1Tyr26 and HLA DQβ1Ala71 in the European and African ancestry populations
| Amino acid position | Effect allele/non effect allele |
European ancestry population (N = 1,600) |
African ancestry population (N = 1,869) |
p valueMeta | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
|
Frequency effect allele |
OR (95% CI) | p value |
Frequency effect allele |
OR (95% CI) | p value | |||||
| Clearance (n = 594) | Persistence (n = 1,006) | Clearance (n = 340) | Persistence (n = 1,529) | |||||||
| 26∗ | L/G,Y | 0.45 | 0.55 | 0.64 (0.54–0.75) | 8.63 × 10−8 | 0.35 | 0.49 | 0.55 (0.45–0.68) | 2.61 × 10−8 | 1.25 × 10−14 |
| 26 | L,Y/G | 0.77 | 0.78 | 0.92 (0.76–1.11) | 3.96 × 10−1 | 0.67 | 0.7 | 0.86 (0.7–1.07) | 1.89 × 10−1 | 1.29 × 10−1 |
| 26∗ | Y/G,L | 0.31 | 0.23 | 1.64 (1.36–1.98) | 1.62 × 10−7 | 0.32 | 0.21 | 1.81 (1.45–2.27) | 1.81 × 10−7 | 1.47 × 10−13 |
| 55 | L/R,P | 0.09 | 0.14 | 0.59 (0.45–0.79) | 3.04 × 10−4 | 0.07 | 0.08 | 0.82 (0.56–1.21) | 3.20 × 10−1 | 9.00 × 10−4 |
| 55∗ | L,P/R | 0.56 | 0.52 | 1.25 (1.07–1.48) | 6.10 × 10−3 | 0.43 | 0.36 | 1.42 (1.16–1.74) | 7.57 × 10−4 | 1.69 × 10−5 |
| 55∗ | P/R,L | 0.47 | 0.38 | 1.51 (1.28–1.77) | 8.79 × 10−7 | 0.36 | 0.27 | 1.6 (1.29–1.99) | 2.11 × 10−5 | 8.23 × 10−11 |
| 71 | D,K,T/A | 0.83 | 0.86 | 0.76 (0.61–0.95) | 1.73 × 10−2 | 0.76 | 0.8 | 0.81 (0.64–1.02) | 7.67 × 10−2 | 3.23 × 10−3 |
| 71 | K/A,D,T | 0.09 | 0.14 | 0.59 (0.44–0.79) | 3.43 × 10−4 | 0.07 | 0.08 | 0.82 (0.56–1.2) | 3.17 × 10−1 | 1.00 × 10−3 |
| 71 | T/A,D,K | 0.67 | 0.64 | 1.06 (0.88–1.27) | 5.49 × 10−1 | 0.6 | 0.61 | 0.9 (0.73–1.12) | 3.47 × 10−1 | 8.33 × 10−1 |
| 71 | D/A,K,T | 0.03 | 0.03 | 0.92 (0.58–1.46) | 7.21 × 10−1 | 0.09 | 0.08 | 1 (0.7–1.43) | 9.98 × 10−1 | 7.96 × 10−1 |
∗Asterisk indicates the amino acids significantly associated in the fixed-effect unconditional meta-analyses. Single letter and three letter amino acid code: A, Ala; C, Cys; D, Asp; E, Glu; F, Phe; G, Gly; H, His; I, Ile; K, Lys; L, Leu; M, Met; N, Asn; P, Pro; Q, Gln; R, Arg; T, Thr; S, Ser; Y, Tyr; V, Val; W, Trp.
The conditional analysis of amino acids also identified two independent signals, corresponding to the lead amino acid residue HLA-DQβ1Leu26 and HLA-DQβ1Pro55 (p valueMeta unconditional = 8.23 × 10−11, p valueMeta conditional on HLA-DQβ1Leu26 = 2.8 × 10−4). After including these two amino acid residues in the model, no other amino acids in the MHC class II region were significantly associated (Figures 2F and S4).
For the independently associated SNPs, classical alleles, and amino acids, an analysis stratified by HIV infection status demonstrated no difference in associations between those with and without HIV (Table S5). We also ran the conditional analysis with HLA-DQβ1Tyr26 and identified two residual signals given by HLA-DQβ1Ala71 (p valueMeta = 9.94 × 10−8) and by HLA-DRβ1Glu28 (p valueMeta = 1.64 × 10−6). After conditioning on HLA-DQβ1Tyr26 and HLA-DQβ1Ala71, we observed no additional amino acids associated in the MHC class II region.
Amino acid residues explain SNP and classical HLA allele associations with HCV clearance
After analyzing models including SNPs and classical HLA alleles and conditioning on the associated amino acid residues and combination of residues (Tables S6 and S7), we found that HLA-DQβ1Leu26 and HLA-DQβ1Pro55 reduced the independent SNP association of rs2647011 (p valueMeta = 0.03) and rs9274711 (p valueMeta = 0.47) (Figure 2D), indicating that the amino acids likely explain the association of rs2647011. Similarly, conditioning on those two key amino acids diminished the association of HLA-DQB1∗03:01 (p valueMeta = 0.341) and reduced the association with HLA-DRB1∗01:01 (p valueMeta = 1.27 × 10−4) (Figure 2E) further underscoring the role of those amino acids explaining the strong association in this region. Conditioning on HLA-DQβ1Tyr26 and HLA-DQβ1Ala71 also reduced the classical HLA associations, but to a lesser extent (rs2647011 = 3.98 × 10−3; rs9274711 = 0.03; HLA-DQB1∗03:01 = 0.68, and HLA-DRB1∗01:01 = 1.86 × 10−3) (Table S6).
In summary, two amino acids in HLA-DQβ1 protein, HLA-DQβ1Leu26 and HLA DQβ1Pro55 (or alternatively HLA-DQβ1Tyr26 and HLA DQβ1Ala71), explain the SNP and classical allele associations with HCV spontaneous clearance in the MHC class II region.
Epitope predictions of associated classical HLA class II alleles containing causal amino acids
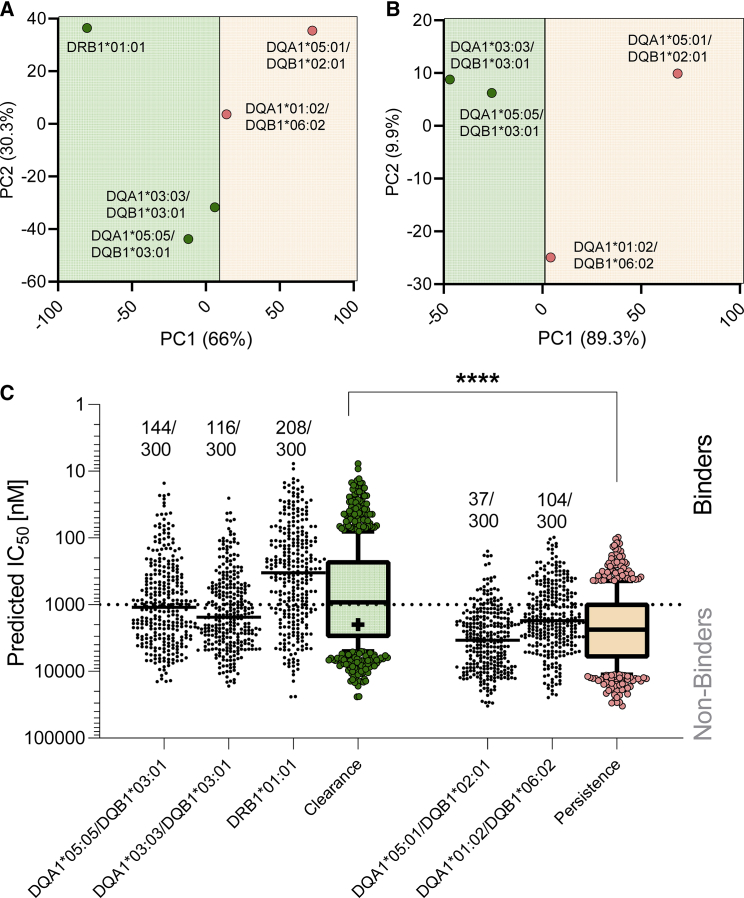
HLA class II binding predictions were performed with classical HLA alleles for clearance (DRB1∗01:01, DQA1∗03:03/DQB1∗03:01, DQA1∗05:05/DQB1∗03:01) and for persistence (DQA1∗01:02/DQB1∗06:02 and DQA1∗05:01/DQB1∗02:01). HLA-DQB1 clearance/persistence groups of associated alleles were stratified by the putative causal amino acid in position 26 of HLA-DQβ1, as described above and shown in Table 3. Clearance alleles were separated from the persistence alleles by two principal components derived from the binding prediction, chiefly PCA1 (66% variance, Figure 3A). The separation was augmented when analyzing only the alleles of the HLA-DQ locus (Figure 3B). Significantly higher frequencies of predicted epitopes were observed in the amino acid sequences of clearance compared to persistence alleles, both in terms of the fractions predicted to bind with an IC50 value of ≤1000 nM and the geometric means of the combined IC50 values of the clearance allele group (geometric mean: 761.7 nM) versus the persistent group (2,321 nM, p value = 1.35 × 10−38 by Mann-Whitney U test) (Figure 3C, Table S8). The observation was maintained when considering only the HLA-DQ locus which showed higher frequencies of predicted epitopes (260 versus 141) and binding affinity compared to persistence alleles (geometric mean: 1,107 nM versus 2,321 nM, p value = 4.07 × 10−18).
Table 3.
Amino acid positions and residues associated with HCV spontaneous clearance, significantly associated HLA class II alleles containing them and location of the amino acids in the pockets of each protein
| HLA classical allele associated with HCV clearance/persistence& | Amino acid residues associated with HCV clearance/persistence and location in pockets of HLA-DQβ1 and HLA-DRβ1 proteins | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Pocket in the protein | P6 | P4 | P4 | P1 | P1 | P1 | P1 | P1 | ||||
| HLA-DQB1 | 9 | 13 | 26 | 45 | 53 | 55 | 84 | 85 | 86 | 87 | 89 | 90 |
| DQB1∗03:01 | Y | A∗ | Y∗ | E∗ | L | P+ | Q | L | E | L | T | T |
| DQB1∗06:02& | F∗& | G& | L∗+& | G& | Q& | R∗& | E& | V& | A& | F∗& | G& | I& |
| DQB1∗02:01& | Y | G& | L∗+& | G∗& | L | L∗ | Q | L | E | L | T | T |
| P6 | P4 | P4 | P6 | P1 | P7 | P9 | P4 | P4 | P1 | |||
| HLA-DRB1 | 11 | 13 | 26 | 28 | 30 | 31 | 32 | 47 | 58 | 71 | 74 | 86 |
| DRB1∗01:01 | L | F | L | E | C | I | Y | Y | A& | R | A | G |
Amino acids in alleles are coded in a single-letter code. All amino acids in the table reached significance level in the meta-analysis and were associated with clearance except for those with the & symbol which were associated with persistence. Asterisks (∗) indicate those that accounted the association of the HLA allele and those with + symbol accounted for the observed association with SNP in MHC class II region. Single letter and three letter amino acid code: A, Ala; C, Cys; E, Glu; F, Phe; G, Gly; I, Ile; L, Leu; P, Pro; Q, Gln; R, Arg; S, Ser; Y, Tyr. Abbreviations: P: Pocket.
Figure 3.
T cell prediction analysis of HLA class II alleles associated with HCV spontaneous clearance
Predicted IC50 values of all the possible 15-mer peptides encompassing the entire HCV proteome in combination with HLA class II clearance (HLA-DRB1∗01:01, HLA-DQA1∗03:03/HLA-DQB1∗03:01, HLA-DQA1∗05:05/HLA-DQB1∗03:01) and persistent alleles (HLA-DQA1∗01:02/HLA-DQB1∗06:02 and HLA-DQA1∗05:01/HLA-DQB1∗02:01).
(A) Principal component analysis (PCA) of the entire prediction distribution based on clearance (green) or persistence alleles (red).
(B) PCA based on HLA class II alleles related to the HLA-DQ locus only.
(C) Predicted IC50 values for each peptide-HLA classical allele combination and for the combined clearance (green) or persistence alleles (red). Epitopes were defined as peptides with a predicted IC50 ≤1,000 nM (above dotted line, denoted as “binders”) and the numbers of predicted epitopes are shown as a fraction of those considered for each of the HLA class II classical alleles in analysis. Combined data in (C) are expressed as whiskers 10–90 percentile, the plus sign representing the mean. ∗∗∗∗p value < 0.0001 by Mann-Whitney U test.
Overall, classical HLA class II alleles clustered based on their predicted binding pattern capabilities. Alleles positively associated with HCV spontaneous clearance had more predicted HLA binders, potentially recognized by T cells.
Location of amino acid residues accounting for the association on MHC class II
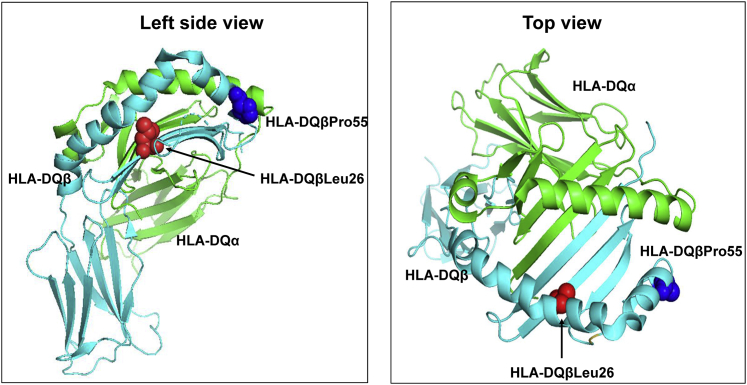
Amino acid in position 26 of the HLA-DQβ1 chain (HLA-DQβ26) is located in the floor of the putative peptide binding groove in the second hypervariable region as part of the peptide-binding pocket P4 (Table 3, Figure 4) and can harbor a hydrophobic Leucine versus a hydrophilic polar non-charged tyrosine (and in a very small frequency glycine). Likewise, HLA-DQβ55 forms part of a peptide-binding region in the HLA-DQ molecule, and the change from arginine to proline implies a physio-chemical shift from hydrophilic to hydrophobic, but we cannot verify the effect of these changes on the affinity of the HLA molecule.
Figure 4.
3D ribbon models for HLA-DQ protein
Molecular structure of the extracellular region of HLA-DQ molecule displaying the peptide binding groove along with the associated amino acid residues that totally accounted for the observed signal in the MHC class II region in European and African ancestry groups. The Protein Data Bank: 1jk8 was used to generate the 3D structures HLA-DQ using PyMOL (The PyMOL Molecular Graphics System, v.2.0 Schrödinger, LLC). Abbreviations: Pro, proline; Leu, leucine.
Discussion
This study maps the association of HLA class II with HCV spontaneous clearance to two amino acid positions in the peptide binding regions of HLA-DQβ. The finding corresponds with HCV peptide class II binding predictions and is sustained in persons of both African and European ancestry. Greater understanding of how some individuals spontaneously clear HCV infection might be useful for designing urgently needed vaccines that promote spontaneous clearance of HCV infection.48, 49, 50
The results of our study confirm the association of HCV spontaneous clearance with classical HLA alleles and SNPs that we and others have already reported. For example, Duggal et al. described the association of HLA-DQB1∗03:01 with HCV spontaneous clearance in a European ancestry population.5,9 Similarly, this allele has being identified in other studies with individuals of European ancestry,9,51, 52, 53 two ethnically distinct Chinese populations,54 and in families from Egypt.55 While those studies provided unmistakable evidence of an association with the HLA locus, no prior study has been able to identify the causal variants. In this study, we identify two single amino acid residues in the peptide binding groove of the protein HLA-DQβ1 (Leu26 and Pro55) that explain the association of both individual SNPs and classical HLA alleles with HCV clearance.
The amino acid residues within the peptide-binding site of the β-chain of HLA-DQβ1 molecule play an important role in the selectivity of peptide binding. For example, the presence of Leu26 in pocket 4 in the HLA-DQβ1 chain that we found to be associated with HCV clearance also has been identified in traits such as the anticentromere autoantibody response in a small group of European individuals with systemic sclerosis,56 as well as autoantibody responses such as anti-ro (SS-A) response in patients of European and African ancestry with systemic lupus erythematosus (SLE) or Sjögren’s syndrome,57 and anti-ribosomal P antibodies in another large multiethnic cohort of patients with SLE including individuals of European and Asian ancestry.58 Similarly, an association with rs1130380, the SNP that causes the amino acid substitution Arg55Pro in the HLA-DQβ1 was previously detected in a genome-wide association study for chronic HCV infection and healthy controls in a Japanese cohort.59 However, the specific association analysis of the amino acid residue and HCV clearance was not implemented in the Japanese cohort as we did in our analysis.
Both codons we identified as associated with HCV clearance form part of a peptide-binding groove in the HLA-DQ molecule. This finding suggests that HLA-DQ binding may be critical to the net effectiveness of HCV immune responses. This inference is supported by our formal, independent HLA binding analysis. Indeed, the HLA-DQ haplotypes linked to clearance (HLA-DQA1∗03:03/HLA-DQB1∗03:01 and HLA- DQA1∗05:05/HLA- DQB1∗03:01) bound a greater number of predicted HCV peptides and showed binding with higher affinity compared to those linked to persistence (HLA-DQA1∗01:02/HLA-DQB1∗06:02 and HLA-DQA1∗05:01/HLA-DQB1∗02:01). Additionally, this finding agrees with Cramp et al. who demonstrated more robust CD4+ T cell responses in those who were HLA-DQB1∗03:01 positive, as well as a strong association with spontaneous resolution of HCV infection.60 Likewise, Kovacs et al. demonstrated that HLA-DQB1∗03:01 was associated with increased CD8+ T cell activation.61
Even though several amino acids forming classical HLA alleles were associated with HCV spontaneous clearance, their association was highly dependent on HLA-DQβ1 Leu26 and Pro55 as well as the association of the classical HLA alleles itself. The direction of the effect of the association of each residue in position 26 of HLA-DQβ1 is highly concordant with the associated classical allele containing them. Leu26 tags the two classical HLA alleles negatively associated with clearance (HLA-DQB1∗06:02 and HLA-DQB1∗02:01) and Tyr 26 is contained in the classical HLA allele that favors HCV spontaneous clearance, HLA-DQB1∗03:01. Furthermore, HLA-DQβ26 is the main amino acid position that is different between the clearance and persistence alleles including in our epitope binding assay. Consequently, it is reasonable to understand the association considering this differential affinity for HCV peptides. Although additional work would be needed to prove the inference, there are no experimental models of HCV clearance. Nonetheless, ex vivo studies that directly demonstrate differential binding would confirm this inference.
We present consistent fine-mapping results across ancestral populations. This study was limited by the lack of HCV genotypes, which is an additional factor associated with HCV spontaneous clearance, and lack of imputation of the HLA-DQA alleles and binding affinity assays in the African populations restricting the generalization of the associations. As with other complex diseases, the penetrance of these HLA alleles is not 100%, other genes also play a role62,63 and HCV outcome may be mediated or interact with age and initial HCV viral load.
We used HIBAG,32 a robust imputation program that references large databases with high genetic diversity and has demonstrated sucess in multi-ethnic benchmarking studies.33,64, 65, 66 Moreover, we confirmed that the allelic frequency of the HIBAG imputed alleles is in concordance with that from other populations reported in large databases such as the Allele Net Frequency Database67 and Immuno Polymorphism Database,68 which supports the accuracy of the imputation. Additionally, we performed a sensitivity analysis in a subsample of 31 individuals by estimating the HLA classical alleles using HLA-HD, an HLA imputation method based on next generation sequencing data (NGS).69 We observed high imputation accuracy for HLA-DRB1 (97.6%), HLA-DQA1 (100%), reaching 98% for HLA-DQB1 and HLA-DPB1 genes (Table S9). The observed that differences in 2 out of 3 discordant alleles were at the second field of resolution (e.g., DRB1∗13:01, DRB1∗13:02) and likely reflect differences in the reference sequences used and their ability to capture less frequent alleles. None of the three discordant alleles resulted in changes to the associated top amino acids. We also included HIV infection status in the model to account for any effect of HIV infection in HCV clearance.
In summary, fine mapping of the MHC region associated with the HCV spontaneous clearance revealed the key amino acids in the HLA-DQβ1 protein that explain the association in persons of both European and African ancestry. The mechanistic advance is that we have mapped an association of a large genomic region to two amino acids in HLA-DQβ1 and specifically to their binding with HCV peptides. This advance in understanding of natural recovery might set the stage for much needed enhancement of HCV vaccine efforts which are urgently needed as new HCV infections in the United States have tripled in the past 10 years.
Acknowlegments
Funding provided by National Institutes of Health R01 DA013324, AI0148049, DA033541, DA04334, R01 DA12568, U01DA036297, U01AI131314, U19AI066345, R01 HL076902, R01 DA16159, R01 DA21550, UL1 RR024996, R01HD41224, R01DA038632, R01DA026141, AI082630, DA033541, AI082630, AI066345, AI091649, and U19AI088791; AIDS Research through the Center for Inherited Diseases at Johns Hopkins University and National Institute of Allergy and Infectious Diseases.. Data in this manuscript were collected by MACS and WIHS, now the MACS/WIHS Combined Cohort Study (MWCCS), which is supported by the National Institutes of Health. N.V. has received funding from the European Union’s Horizon 2020 research and innovation program under the Marie Skłodowska-Curie grant agreement No. 846520. A.S. has been funded by NIH NIAID contract Nr. 75N93019C00001. R.T.C. was supported by the MGH Research Scholars Program. This project has been funded in whole or in part with federal funds from the Frederick National Laboratory for Cancer Research, under Contract No. HHSN261200800001E. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government.
Declaration of interests
J.C.P. has received research grant support to her institution from Gilead Sciences, Merck, and Abbvie and has served on an advisory board for Gilead Sciences and Theratechnolgies. S.H.M. have received speaker fees from Gilead Sciences not related to this work. A.H.K. serves on the Data Monitoring Committee for Kintor Pharmaceutical.
Published: January 27, 2022
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.ajhg.2022.01.001.
Data and code availability
The datasets used in this study are available at dbGaP accession numbers dbGaP:phs000454.v1.p1 (https://www.ncbi.nlm.nih.gov/projects/gap/cgi-bin/study.cgi?study_id=phs000454.v1.p1) and dbGaP: phs000248.v1.p1 (https://www.ncbi.nlm.nih.gov/projects/gap/cgi-bin/study.cgi?study_id=phs000248.v1.p1). The reference panel derived from the “Consortium on Asthma among African-ancestry Populations in the Americas” is avalaible at dbGaP accession number dbGaP:phs001123.v1.p1 (https://www.ncbi.nlm.nih.gov/projects/gap/cgi-bin/study.cgi?study_id=phs001123.v1.p1). The HCV polyprotein sequence is available at GenBank: AFE48416.1 (https://www.ncbi.nlm.nih.gov/protein/AFE48416.1). Molecular structure used to generate HLA-DQ 3D structures is available at PDB:1jk8 (https://www.rcsb.org/structure/1jk8).
Web resources
HLA database used for imputation, http://hla.alleles.org/alleles/p_groups.html
IEDB-Analysis Resources, http://tools.iedb.org/main
PyMOL Molecular Graphics System program, https://pymol.org/2
RCSB Protein Data Bank, http://www.rcsb.org/pdb/home/home.do
Supplemental information
References
- 1.Anonymous Global Hepatitis Report 2017 . 2017. Geneva: World Health Organization.https://www.who.int/publications/i/item/global-hepatitis-report-2017 [Google Scholar]
- 2.Polaris Observatory HCV Collaborators Global prevalence and genotype distribution of hepatitis C virus infection in 2015: a modelling study. Lancet Gastroenterol. Hepatol. 2017;2:161–176. doi: 10.1016/S2468-1253(16)30181-9. [DOI] [PubMed] [Google Scholar]
- 3.Lavanchy D. The global burden of hepatitis C. Liver Int. 2009;29(Suppl 1):74–81. doi: 10.1111/j.1478-3231.2008.01934.x. [DOI] [PubMed] [Google Scholar]
- 4.Thomas D.L., Astemborski J., Rai R.M., Anania F.A., Schaeffer M., Galai N., Nolt K., Nelson K.E., Strathdee S.A., Johnson L., et al. The natural history of hepatitis C virus infection: host, viral, and environmental factors. JAMA. 2000;284:450–456. doi: 10.1001/jama.284.4.450. [DOI] [PubMed] [Google Scholar]
- 5.Duggal P., Thio C.L., Wojcik G.L., Goedert J.J., Mangia A., Latanich R., Kim A.Y., Lauer G.M., Chung R.T., Peters M.G., et al. Genome-wide association study of spontaneous resolution of hepatitis C virus infection: data from multiple cohorts. Ann. Intern. Med. 2013;158:235–245. doi: 10.7326/0003-4819-158-4-201302190-00003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vergara C., Thio C.L., Johnson E., Kral A.H., O’Brien T.R., Goedert J.J., Mangia A., Piazzolla V., Mehta S.H., Kirk G.D., et al. Multi-Ancestry Genome-Wide Association Study of Spontaneous Clearance of Hepatitis C Virus. Gastroenterology. 2019;156:1496–1507.e7. doi: 10.1053/j.gastro.2018.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Prokunina-Olsson L., Muchmore B., Tang W., Pfeiffer R.M., Park H., Dickensheets H., Hergott D., Porter-Gill P., Mumy A., Kohaar I., et al. A variant upstream of IFNL3 (IL28B) creating a new interferon gene IFNL4 is associated with impaired clearance of hepatitis C virus. Nat. Genet. 2013;45:164–171. doi: 10.1038/ng.2521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Aka P.V., Kuniholm M.H., Pfeiffer R.M., Wang A.S., Tang W., Chen S., Astemborski J., Plankey M., Villacres M.C., Peters M.G., et al. Association of the IFNL4-ΔG Allele With Impaired Spontaneous Clearance of Hepatitis C Virus. J. Infect. Dis. 2014;209:350–354. doi: 10.1093/infdis/jit433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Huang H., Duggal P., Thio C.L., Latanich R., Goedert J.J., Mangia A., Cox A.L., Kirk G.D., Mehta S., Aneja J., et al. Fine-mapping of genetic loci driving spontaneous clearance of hepatitis C virus infection. Sci. Rep. 2017;7:15843. doi: 10.1038/s41598-017-16011-2. 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.McKiernan S.M., Hagan R., Curry M., McDonald G.S., Kelly A., Nolan N., Walsh A., Hegarty J., Lawlor E., Kelleher D. Distinct MHC class I and II alleles are associated with hepatitis C viral clearance, originating from a single source. Hepatology. 2004;40:108–114. doi: 10.1002/hep.20261. [DOI] [PubMed] [Google Scholar]
- 11.Thio C.L., Gao X., Goedert J.J., Vlahov D., Nelson K.E., Hilgartner M.W., O’Brien S.J., Karacki P., Astemborski J., Carrington M., Thomas D.L. HLA-Cw∗04 and hepatitis C virus persistence. J. Virol. 2002;76:4792–4797. doi: 10.1128/JVI.76.10.4792-4797.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hraber P., Kuiken C., Yusim K. Evidence for human leukocyte antigen heterozygote advantage against hepatitis C virus infection. Hepatology. 2007;46:1713–1721. doi: 10.1002/hep.21889. [DOI] [PubMed] [Google Scholar]
- 13.Kim A.Y., Kuntzen T., Timm J., Nolan B.E., Baca M.A., Reyor L.L., Berical A.C., Feller A.J., Johnson K.L., Schulze zur Wiesch J., et al. Spontaneous control of HCV is associated with expression of HLA-B 57 and preservation of targeted epitopes. Gastroenterology. 2011;140:686–696.e1. doi: 10.1053/j.gastro.2010.09.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kuniholm M.H., Kovacs A., Gao X., Xue X., Marti D., Thio C.L., Peters M.G., Terrault N.A., Greenblatt R.M., Goedert J.J., et al. Specific human leukocyte antigen class I and II alleles associated with hepatitis C virus viremia. Hepatology. 2010;51:1514–1522. doi: 10.1002/hep.23515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Alric L., Fort M., Izopet J., Vinel J.P., Charlet J.P., Selves J., Puel J., Pascal J.P., Duffaut M., Abbal M. Genes of the major histocompatibility complex class II influence the outcome of hepatitis C virus infection. Gastroenterology. 1997;113:1675–1681. doi: 10.1053/gast.1997.v113.pm9352872. [DOI] [PubMed] [Google Scholar]
- 16.Fanning L.J., Levis J., Kenny-Walsh E., Wynne F., Whelton M., Shanahan F. Viral clearance in hepatitis C (1b) infection: relationship with human leukocyte antigen class II in a homogeneous population. Hepatology. 2000;31:1334–1337. doi: 10.1053/jhep.2000.7437. [DOI] [PubMed] [Google Scholar]
- 17.Wojcik G.L., Thio C.L., Kao W.H., Latanich R., Goedert J.J., Mehta S.H., Kirk G.D., Peters M.G., Cox A.L., Kim A.Y., et al. Admixture analysis of spontaneous hepatitis C virus clearance in individuals of African descent. Genes Immun. 2014;15:241–246. doi: 10.1038/gene.2014.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vlahov D., Muñoz A., Anthony J.C., Cohn S., Celentano D.D., Nelson K.E. Association of drug injection patterns with antibody to human immunodeficiency virus type 1 among intravenous drug users in Baltimore, Maryland. Am. J. Epidemiol. 1990;132:847–856. doi: 10.1093/oxfordjournals.aje.a115727. [DOI] [PubMed] [Google Scholar]
- 19.Cox A.L., Netski D.M., Mosbruger T., Sherman S.G., Strathdee S., Ompad D., Vlahov D., Chien D., Shyamala V., Ray S.C., Thomas D.L. Prospective evaluation of community-acquired acute-phase hepatitis C virus infection. Clin. Infect. Dis. 2005;40:951–958. doi: 10.1086/428578. [DOI] [PubMed] [Google Scholar]
- 20.Cramp M.E., Carucci P., Underhill J., Naoumov N.V., Williams R., Donaldson P.T. Association between HLA class II genotype and spontaneous clearance of hepatitis C viraemia. J. Hepatol. 1998;29:207–213. doi: 10.1016/s0168-8278(98)80005-6. [DOI] [PubMed] [Google Scholar]
- 21.Hilgartner M.W., Donfield S.M., Willoughby A., Contant C.F., Jr., Evatt B.L., Gomperts E.D., Hoots W.K., Jason J., Loveland K.A., McKinlay S.M., et al. Hemophilia growth and development study. Design, methods, and entry data. Am. J. Pediatr. Hematol. Oncol. 1993;15:208–218. doi: 10.1097/00043426-199305000-00009. [DOI] [PubMed] [Google Scholar]
- 22.Mangia A., Gentile R., Cascavilla I., Margaglione M., Villani M.R., Stella F., Modola G., Agostiano V., Gaudiano C., Andriulli A. HLA class II favors clearance of HCV infection and progression of the chronic liver damage. J. Hepatol. 1999;30:984–989. doi: 10.1016/s0168-8278(99)80250-5. [DOI] [PubMed] [Google Scholar]
- 23.Goedert J.J., Chen B.E., Preiss L., Aledort L.M., Rosenberg P.S. Reconstruction of the hepatitis C virus epidemic in the US hemophilia population, 1940-1990. Am. J. Epidemiol. 2007;165:1443–1453. doi: 10.1093/aje/kwm030. [DOI] [PubMed] [Google Scholar]
- 24.Tobler L.H., Bahrami S.H., Kaidarova Z., Pitina L., Winkelman V.K., Vanderpool S.K., Guiltinan A.M., Cooper S., Busch M.P., Murphy E.L. A case-control study of factors associated with resolution of hepatitis C viremia in former blood donors (CME) Transfusion. 2010;50:1513–1523. doi: 10.1111/j.1537-2995.2010.02634.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Edlin B.R., Shu M.A., Winkelstein E., Des Jarlais D.C., Busch M.P., Rehermann B., O’Brien T.R., Talal A.H., Tobler L.H., Zeremski M., Beeder A.B. More rare birds, and the occasional swan. Gastroenterology. 2009;136:2412–2414. doi: 10.1053/j.gastro.2009.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kuniholm M.H., Gao X., Xue X., Kovacs A., Marti D., Thio C.L., Peters M.G., Greenblatt R.M., Goedert J.J., Cohen M.H., et al. The relation of HLA genotype to hepatitis C viral load and markers of liver fibrosis in HIV-infected and HIV-uninfected women. J. Infect. Dis. 2011;203:1807–1814. doi: 10.1093/infdis/jir192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Khakoo S.I., Thio C.L., Martin M.P., Brooks C.R., Gao X., Astemborski J., Cheng J., Goedert J.J., Vlahov D., Hilgartner M., et al. HLA and NK cell inhibitory receptor genes in resolving hepatitis C virus infection. Science. 2004;305:872–874. doi: 10.1126/science.1097670. [DOI] [PubMed] [Google Scholar]
- 28.Kral A.H., Bluthenthal R.N., Lorvick J., Gee L., Bacchetti P., Edlin B.R. Sexual transmission of HIV-1 among injection drug users in San Francisco, USA: risk-factor analysis. Lancet. 2001;357:1397–1401. doi: 10.1016/S0140-6736(00)04562-1. [DOI] [PubMed] [Google Scholar]
- 29.Kral A.H., Lorvick J., Gee L., Bacchetti P., Rawal B., Busch M., Edlin B.R. Trends in human immunodeficiency virus seroincidence among street-recruited injection drug users in San Francisco, 1987-1998. Am. J. Epidemiol. 2003;157:915–922. doi: 10.1093/aje/kwg070. [DOI] [PubMed] [Google Scholar]
- 30.Horton R., Wilming L., Rand V., Lovering R.C., Bruford E.A., Khodiyar V.K., Lush M.J., Povey S., Talbot C.C., Jr., Wright M.W., et al. Gene map of the extended human MHC. Nat. Rev. Genet. 2004;5:889–899. doi: 10.1038/nrg1489. [DOI] [PubMed] [Google Scholar]
- 31.Das S., Forer L., Schönherr S., Sidore C., Locke A.E., Kwong A., Vrieze S.I., Chew E.Y., Levy S., McGue M., et al. Next-generation genotype imputation service and methods. Nat. Genet. 2016;48:1284–1287. doi: 10.1038/ng.3656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zheng X., Shen J., Cox C., Wakefield J.C., Ehm M.G., Nelson M.R., Weir B.S. HIBAG--HLA genotype imputation with attribute bagging. Pharmacogenomics J. 2014;14:192–200. doi: 10.1038/tpj.2013.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vince N., Limou S., Daya M., Morii W., Rafaels N., Geffard E., Douillard V., Walencik A., Boorgula M.P., Chavan S., et al. CAAPA Association of HLA-DRB1∗09:01 with tIgE levels among African-ancestry individuals with asthma. J. Allergy Clin. Immunol. 2020;146:147–155. doi: 10.1016/j.jaci.2020.01.011. [DOI] [PubMed] [Google Scholar]
- 34.Daya M., Rafaels N., Brunetti T.M., Chavan S., Levin A.M., Shetty A., Gignoux C.R., Boorgula M.P., Wojcik G., Campbell M., et al. CAAPA Association study in African-admixed populations across the Americas recapitulates asthma risk loci in non-African populations. Nat. Commun. 2019;10:880–887. doi: 10.1038/s41467-019-08469-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Levin A.M., Adrianto I., Datta I., Iannuzzi M.C., Trudeau S., McKeigue P., Montgomery C.G., Rybicki B.A. Performance of HLA allele prediction methods in African Americans for class II genes HLA-DRB1, -DQB1, and -DPB1. BMC Genet. 2014;15:72. doi: 10.1186/1471-2156-15-72. 72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Khor S.S., Yang W., Kawashima M., Kamitsuji S., Zheng X., Nishida N., Sawai H., Toyoda H., Miyagawa T., Honda M., et al. High-accuracy imputation for HLA class I and II genes based on high-resolution SNP data of population-specific references. Pharmacogenomics J. 2015;15:530–537. doi: 10.1038/tpj.2015.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chang C.C., Chow C.C., Tellier L.C., Vattikuti S., Purcell S.M., Lee J.J. Second-generation PLINK: rising to the challenge of larger and richer datasets. Gigascience. 2015;4:7–8. doi: 10.1186/s13742-015-0047-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Purcell S., Neale B., Todd-Brown K., Thomas L., Ferreira M.A., Bender D., Maller J., Sklar P., de Bakker P.I., Daly M.J., Sham P.C. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Price A.L., Patterson N.J., Plenge R.M., Weinblatt M.E., Shadick N.A., Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat. Genet. 2006;38:904–909. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- 40.Willer C.J., Li Y., Abecasis G.R. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics. 2010;26:2190–2191. doi: 10.1093/bioinformatics/btq340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.R Core Team . 2013. A Language and Environment for Statistical Computing. R Foundation for Statistical Computing.Vienna, Austria.http://www.r-project.org/ [Google Scholar]
- 42.Tian C., Hromatka B.S., Kiefer A.K., Eriksson N., Noble S.M., Tung J.Y., Hinds D.A. Genome-wide association and HLA region fine-mapping studies identify susceptibility loci for multiple common infections. Nat. Commun. 2017;8:599. doi: 10.1038/s41467-017-00257-5. 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dhanda S.K., Mahajan S., Paul S., Yan Z., Kim H., Jespersen M.C., Jurtz V., Andreatta M., Greenbaum J.A., Marcatili P., et al. IEDB-AR: immune epitope database-analysis resource in 2019. Nucleic Acids Res. 2019;47(W1):W502–W506. doi: 10.1093/nar/gkz452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jensen K.K., Andreatta M., Marcatili P., Buus S., Greenbaum J.A., Yan Z., Sette A., Peters B., Nielsen M. Improved methods for predicting peptide binding affinity to MHC class II molecules. Immunology. 2018;154:394–406. doi: 10.1111/imm.12889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Peters B., Nielsen M., Sette A. T Cell Epitope Predictions. Annu. Rev. Immunol. 2020;38:123–145. doi: 10.1146/annurev-immunol-082119-124838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Berman H.M., Westbrook J., Feng Z., Gilliland G., Bhat T.N., Weissig H., Shindyalov I.N., Bourne P.E. The Protein Data Bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lee K.H., Wucherpfennig K.W., Wiley D.C. Structure of a human insulin peptide-HLA-DQ8 complex and susceptibility to type 1 diabetes. Nat. Immunol. 2001;2:501–507. doi: 10.1038/88694. [DOI] [PubMed] [Google Scholar]
- 48.Thomas D.L. Global Elimination of Chronic Hepatitis. N. Engl. J. Med. 2019;380:2041–2050. doi: 10.1056/NEJMra1810477. [DOI] [PubMed] [Google Scholar]
- 49.Bailey J.R., Barnes E., Cox A.L. Approaches, Progress, and Challenges to Hepatitis C Vaccine Development. Gastroenterology. 2019;156:418–430. doi: 10.1053/j.gastro.2018.08.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cox A.L. Challenges and Promise of a Hepatitis C Virus Vaccine. Cold Spring Harb. Perspect. Med. 2020;10:a036947. doi: 10.1101/cshperspect.a036947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Minton E.J., Smillie D., Neal K.R., Irving W.L., Underwood J.C., James V., Members of the Trent Hepatitis C Virus Study Group Association between MHC class II alleles and clearance of circulating hepatitis C virus. J. Infect. Dis. 1998;178:39–44. doi: 10.1086/515599. [DOI] [PubMed] [Google Scholar]
- 52.Thio C.L., Thomas D.L., Goedert J.J., Vlahov D., Nelson K.E., Hilgartner M.W., O’Brien S.J., Karacki P., Marti D., Astemborski J., Carrington M. Racial differences in HLA class II associations with hepatitis C virus outcomes. J. Infect. Dis. 2001;184:16–21. doi: 10.1086/321005. [DOI] [PubMed] [Google Scholar]
- 53.Harris R.A., Sugimoto K., Kaplan D.E., Ikeda F., Kamoun M., Chang K.M. Human leukocyte antigen class II associations with hepatitis C virus clearance and virus-specific CD4 T cell response among Caucasians and African Americans. Hepatology. 2008;48:70–79. doi: 10.1002/hep.22287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Huang J., Xu R., Wang M., Liao Q., Huang K., Shan Z., You Q., Li C., Rong X., Fu Y. Association of HLA-DQB1∗03:01 and DRB1∗11:01 with spontaneous clearance of hepatitis C virus in Chinese Li ethnicity, an ethnic group genetically distinct from Chinese Han ethnicity and infected with unique HCV subtype. J. Med. Virol. 2019;91:1830–1836. doi: 10.1002/jmv.25531. [DOI] [PubMed] [Google Scholar]
- 55.El-Bendary M., Neamatallah M., Esmat G., Kamel E., Elalfy H., Besheer T., Eldeib D., Eladl A.H., El-Setouhy M., El-Gilany A.H., El-Waseef A. Associations of human leucocyte antigen class II-DQB1 alleles with hepatitis C virus infection in Egyptian population: a multicentre family-based study. J. Viral Hepat. 2016;23:961–970. doi: 10.1111/jvh.12573. [DOI] [PubMed] [Google Scholar]
- 56.Reveille J.D., Owerbach D., Goldstein R., Moreda R., Isern R.A., Arnett F.C. Association of polar amino acids at position 26 of the HLA-DQB1 first domain with the anticentromere autoantibody response in systemic sclerosis (scleroderma) J. Clin. Invest. 1992;89:1208–1213. doi: 10.1172/JCI115704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Reveille J.D., Macleod M.J., Whittington K., Arnett F.C. Specific amino acid residues in the second hypervariable region of HLA-DQA1 and DQB1 chain genes promote the Ro (SS-A)/La (SS-B) autoantibody responses. J. Immunol. 1991;146:3871–3876. [PubMed] [Google Scholar]
- 58.Arnett F.C., Reveille J.D., Moutsopoulos H.M., Georgescu L., Elkon K.B. Ribosomal P autoantibodies in systemic lupus erythematosus. Frequencies in different ethnic groups and clinical and immunogenetic associations. Arthritis Rheum. 1996;39:1833–1839. doi: 10.1002/art.1780391109. [DOI] [PubMed] [Google Scholar]
- 59.Miki D., Ochi H., Takahashi A., Hayes C.N., Urabe Y., Abe H., Kawaoka T., Tsuge M., Hiraga N., Imamura M., et al. HLA-DQB1∗03 confers susceptibility to chronic hepatitis C in Japanese: a genome-wide association study. PLoS ONE. 2013;8:e84226. doi: 10.1371/journal.pone.0084226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Cramp M.E., Rossol S., Carucci P., Williams R., Naoumov N.V., Donaldson P.T. The influence of the HLA class II allele DQB1∗0301 on HCV specific T helper cell responses. J. Hepatol. 1998;28:94. [Google Scholar]
- 61.Kovacs A.A.Z., Kono N., Wang C.H., Wang D., Frederick T., Operskalski E., Tien P.C., French A.L., Minkoff H., Kassaye S., et al. Association of HLA Genotype With T-Cell Activation in Human Immunodeficiency Virus (HIV) and HIV/Hepatitis C Virus-Coinfected Women. J. Infect. Dis. 2020;221:1156–1166. doi: 10.1093/infdis/jiz589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Thomas D.L., Thio C.L., Martin M.P., Qi Y., Ge D., O’Huigin C., Kidd J., Kidd K., Khakoo S.I., Alexander G., et al. Genetic variation in IL28B and spontaneous clearance of hepatitis C virus. Nature. 2009;461:798–801. doi: 10.1038/nature08463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Vergara C., Duggal P., Thio C.L., Valencia A., O’Brien T.R., Latanich R., Timp W., Johnson E.O., Kral A.H., Mangia A., et al. Multi-ancestry fine mapping of interferon lambda and the outcome of acute hepatitis C virus infection. Genes Immun. 2020;21:348–359. doi: 10.1038/s41435-020-00115-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Adebamowo S.N., Adeyemo A.A., ACCME Research Group as part of the H3Africa Consortium Classical HLA alleles are associated with prevalent and persistent cervical high-risk HPV infection in African women. Hum. Immunol. 2019;80:723–730. doi: 10.1016/j.humimm.2019.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Shen J., Mehrotra D.V., Dorr M.B., Zeng Z., Li J., Xu X., Nickle D., Holzinger E.R., Chhibber A., Wilcox M.H., et al. Genetic Association Reveals Protection against Recurrence of Clostridium difficile Infection with Bezlotoxumab Treatment. MSphere. 2020;5:e00232-20. doi: 10.1128/mSphere.00232-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Degenhardt F., Wendorff M., Wittig M., Ellinghaus E., Datta L.W., Schembri J., Ng S.C., Rosati E., Hübenthal M., Ellinghaus D., et al. Construction and benchmarking of a multi-ethnic reference panel for the imputation of HLA class I and II alleles. Hum. Mol. Genet. 2019;28:2078–2092. doi: 10.1093/hmg/ddy443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Gonzalez-Galarza F.F., McCabe A., Santos E.J.M.D., Jones J., Takeshita L., Ortega-Rivera N.D., Cid-Pavon G.M.D., Ramsbottom K., Ghattaoraya G., Alfirevic A., et al. Allele frequency net database (AFND) 2020 update: gold-standard data classification, open access genotype data and new query tools. Nucleic Acids Res. 2020;48(D1):D783–D788. doi: 10.1093/nar/gkz1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Robinson J., Barker D.J., Georgiou X., Cooper M.A., Flicek P., Marsh S.G.E. IPD-IMGT/HLA Database. Nucleic Acids Res. 2020;48(D1):D948–D955. doi: 10.1093/nar/gkz950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kawaguchi S., Higasa K., Shimizu M., Yamada R., Matsuda F. HLA-HD: An accurate HLA typing algorithm for next-generation sequencing data. Hum. Mutat. 2017;38:788–797. doi: 10.1002/humu.23230. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets used in this study are available at dbGaP accession numbers dbGaP:phs000454.v1.p1 (https://www.ncbi.nlm.nih.gov/projects/gap/cgi-bin/study.cgi?study_id=phs000454.v1.p1) and dbGaP: phs000248.v1.p1 (https://www.ncbi.nlm.nih.gov/projects/gap/cgi-bin/study.cgi?study_id=phs000248.v1.p1). The reference panel derived from the “Consortium on Asthma among African-ancestry Populations in the Americas” is avalaible at dbGaP accession number dbGaP:phs001123.v1.p1 (https://www.ncbi.nlm.nih.gov/projects/gap/cgi-bin/study.cgi?study_id=phs001123.v1.p1). The HCV polyprotein sequence is available at GenBank: AFE48416.1 (https://www.ncbi.nlm.nih.gov/protein/AFE48416.1). Molecular structure used to generate HLA-DQ 3D structures is available at PDB:1jk8 (https://www.rcsb.org/structure/1jk8).