Abstract
The genus Enterobacter is a member of the ESKAPE group, which contains the major resistant bacterial pathogens. Enterobacter cloacae complex (ECC) has emerged as a clinically significant cause of a wide variety of nosocomial infections. Carbapenem-nonsusceptible Enterobacter cloacae complex (CnsECC) has become an emerging threat to public health but there is still a lack of comprehensive molecular and clinical epidemiological analysis. A total of 157 CnsECC isolates were recovered during October 2011 to August 2017. hsp60 gene sequencing and pulsed-field gel electrophoresis (PFGE) were applied to discriminate the species, genetic clusters and clonal relatedness. All the isolates were subjected to polymerase chain reaction (PCR) analysis for carbapenemase, AmpC-type β-lactamase, and extended spectrum β-lactamase (ESBL) genes. Clinical data were collected on all patients for comparing clinical risks and outcomes between patients with carbapenemase-producing (CP)-CnsECC compared with non-CP-CnsECC infection. The most commonly identified species was E. hormaechei subsp. hoffmannii (47.1%), followed by E. hormaechei subsp. steigerwaltii (24.8%). Different species of CnsECC isolates showed heterogeneity in resistance patterns to piperacillin/tazobactam, cefepime and levofloxacin. In the present study, we observed that E. hormaechei subsp. hoffmannii was characterized with higher cefepime and levofloxacin resistance rate but lower piperacillin/tazobactam resistance rate relative to other species of CnsECC. CP-CnsECC comprised 41.1% (65 isolates) and all of these isolates carried IMP-8. In this study, 98% of patients had antimicrobial therapy prior to culture, with a total of 57/150 (38%) patients being exposed to carbapenems. Chronic pulmonary disease (OR: 2.51, 95% CI: 1.25–5.06), received ventilator support (OR: 5.54, 95% CI: 2.25–12.03), steroid exposure (OR: 3.88, 95% CI: 1.91–7.88) and carbapenems exposure (OR: 2.17, 95% CI: 1.10–4.25) were considered risk factors associated with CP-CnsECC infection. The results suggest that CP-CnsECC are associated with poorer outcomes including in-hospital mortality, 30-day mortality and 100-day mortality. Our study provides insights into the epidemic potential of IMP-8-producing E. cloacae for healthcare-associated infections and underscores the importance of understanding underlying resistance mechanisms of CnsECC to direct antibiotic treatment decisions.
Keywords: Enterobacter cloacae complex, carbapenem, carbapenemase-producing Enterobacteriaceae, molecular epidemiology
1. Introduction
The Enterobacter cloacae complex (ECC) is a group of Enterobacteriaceae widely distributed in nature [1,2]. Currently, ECC has emerged as a clinically significant cause of a wide variety of nosocomial infections, such as pneumonia, urinary tract infections, intraabdominal infection, and bacteremia [3,4,5]. Although ECC is composed of multiple species, including E. cloacae, E. hormaechei, E. asburiae, E. kobei, E. ludwigii, E. nimipressuralis and E. mori [6], the routine identification of bacteria in clinical microbiology laboratories is not able to differentiate the species in the ECC by using commercialized systems [7,8]. Molecular and genomic techniques are needed to identify the ECC species precisely. Sequencing of the heat shock protein 60 (hsp60) gene, which can discriminate of this complex into 13 genetic clusters (C-I to CXIII) appears to be a valuable tool to identify of ECC species [3]. Furthermore, multilocus sequence analysis (MLSA) of housekeeping genes was also strongly validated by whole genome sequence (WGS) for phylogenetic classification of Enterobacter species [9]. Despite the fact that misidentification of ECC species has little impact on antibiotic therapy previously, as the species of the ECC have the similar antibiotic resistance profiles, these molecular approaches still have been used recently for precise identification of the species in the ECC for understanding the epidemiology, pathogenesis, and microbiological features [10].
Because of the extended overuse of antibiotics, multidrug resistant (MDR) ECC strains have emerged and spread globally. The genus Enterobacter is a member of the ESKAPE group, which are described as the major resistant bacterial pathogens and leading cause of nosocomial infections throughout the world [11]. The undesirable antibiotic resistance, especially carbapenem-resistant Enterobacteriaceae (CRE), is a major public health crisis because these agents are regarded as one of the last effective therapies available for treating serious infections caused by extended-spectrumβ-lactamase (ESBL)- or AmpC-producing Enterobacteriaceae [12]. Infections caused by CRE can lead to severe clinical outcomes and high hospitalization cost [13]. Recent surveillance studies have displayed that Enterobacter spp. are often the second or third most common CRE [14,15,16,17]. Therefore, Carbapenem-nonsusceptible Enterobacter cloacae complex (CnsECC) may become an emerging threat to public health.
In Taiwan, Lai et al. reported that E. cloacae has the highest rates of carbapenem-insusceptibility among Enterobacteriaceae species from patients admitted to intensive care units (ICUs) in 2016 [18]. The results from Study for Monitoring Antimicrobial Resistance Trends in Taiwan (SMART) between 2016 and 2018 found that isolates of Enterobacter species showed higher rates of nonsusceptibility to ertapenem than E. coli or K. pneumoniae isolates [4]. The production of carbapenemases is the most important causes of carbapenem resistance in CRE [17]. The most common genes encoding carbapenemases are the Klebsiella pneumoniae carbapenemases (KPCs), imipenemase metallo-β-lactamases (IMPs), Verona integron encoded metallo-β-lactamases (VIMs), New Delhi metallo-β-lactamase (NDMs), and oxacillin (OXA) 48-like enzymes [19]. Global surveillance showed diversity of genes encoding carbapenemases in CnsECC [7,17]. In Taiwan, IMP type carbapenemase is highly prevalent in CnsECC [20,21,22,23,24]. IMP type carbapenemases were first discovered in Japan during the 1990s and have up to 18 varieties [19]. IMP-8 was the most common carbapenemase in E. cloacae from Taiwan [23]. However, comprehensive data regarding the species diversity, microbiological features, resistance plasmid characteristics and clinical relevance of CnsECC remains limited. This study was conducted to understand the epidemiology of species, clonal relatedness, carbapenemase-producing genes, and clinical features of CnsECC in Taiwan.
2. Results
2.1. Species Identification of CnsECC Isolates
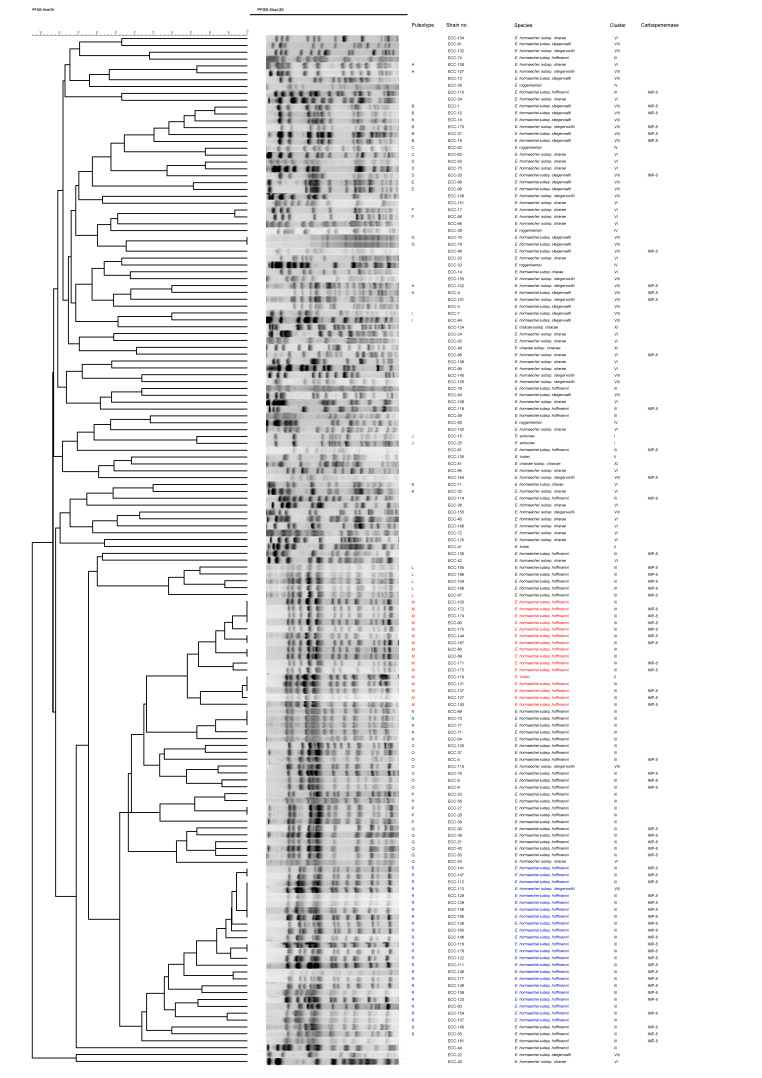
A total of 157 isolates were collected with initial reports as CnsECC from the Kaohsiung Medical University Hospital from October 2011 to August 2017. To identify species of the ECCs, we used hsp60 gene sequencing to classify the CnsECCs into seven species (clusters) (Table 1). The most commonly identified species was E. hormaechei subsp. hoffmannii (cluster III) (74/157, 47.1%), followed by E. hormaechei subsp. steigerwaltii (cluster VIII) (39/157, 24.8%), E. hormaechei subsp. oharae (cluster VI) (30/157, 19.1%), E. roggenkampii (cluster IV) (6/157, 3.8%), E. kobei (cluster II) (3/157, 1.9%), E. cloacae subsp. cloacae (cluster XI) (3/157, 1.9%) and E. asburiae (cluster I) (2/157, 1.3%). For further discrimination, pulsed-field gel electrophoresis (PFGE) analysis was performed. Among the 157 CnsECC isolates, 150 isolates were successfully obtained the PFGE banding results (Figure 1). In PFGE analysis, 96 of 150 isolates (64%) can be clustered into 19 pulsotypes (A-S) with >75% similarity in PFGE banding patterns and the two predominant patterns were Pulsotype R (23 isolates) and Pulsotype M (16 isolates). In these two pulsotypes, 22 and 15 isolates belonged to E. hormaechei subsp. hoffmannii, respectively.
Table 1.
Species identification by hsp60 gene sequencing and PFGE pulsotypes of CnsECC isolates.
| Species | Cluster | No. of Isolates (%) | PFGE Pulsotype (n) |
|---|---|---|---|
| Enterobacter asburiae | I | 2 (1.3) | J (2) |
| Enterobacter kobei | II | 3 (1.9) | M (1) |
| Enterobacter hormaechei subsp. hoffmannii | III | 74 (47.1) | L (5) M (15) N (5) O (6) P (5) Q (5) R (22) S (2) |
| Enterobacter roggenkampii | IV | 6 (3.8) | C (1) |
| Enterobacter hormaechei subsp. oharae | VI | 30 (19.1) | A (1) C (1) D (2) F (2) K (2) Q (1) |
| Enterobacter hormaechei subsp. steigerwaltii | VIII | 39 (24.8) | A (1) B (6) D (1) E (2) G (2) H (2) I (2) O (1) R (1) |
| Enterobacter cloacae subsp. cloacae | XI | 3 (1.9) | |
| Total | 157 | 96 |
Figure 1.
Dendrogram of pulsed-field gel electrophoresis (PFGE) cluster analysis of 150 carbapenem-nonsusceptible Enterobacter cloacae complex isolates. The two predominant patterns Pulsotype R (23 isolates) and Pulsotype M (16 isolates) are marked with blue and red color.
2.2. Antimicrobial Susceptibility of CnsECC Isolates
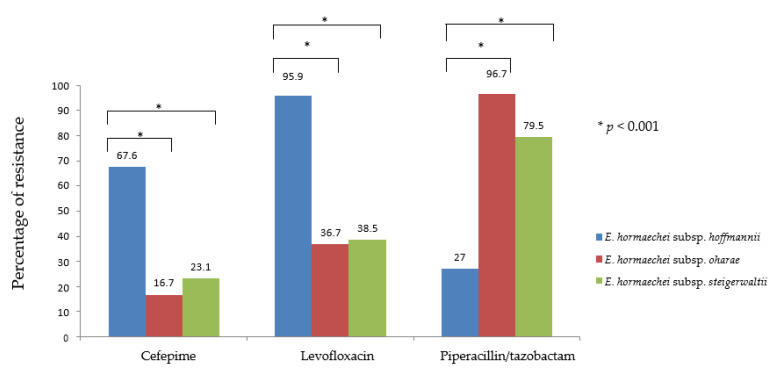
All of the 157 CnsECC isolates were resistant to multiple antimicrobial agents, such as cefazolin, cefmetazole, and ampicillin. Most of the isolates were resistant to ampicillin-sulbactam, ceftazidime, sulfamethoxazole-trimethoprim and levofloxacin. Almost all of the isolates remain susceptible to amikacin (99.4%). The antimicrobial susceptibility testing is summarized in Table 2. Notably, the prevalence of resistance was significantly different among three predominant species (E. hormaechei subsp. hoffmannii, E. hormaechei subsp. oharae and E. hormaechei subsp. steigerwaltii) in cefepime, levofloxacin and piperacillin/tazobactam (Table 3 and Figure 2). The cefepime resistance was higher in E. hormaechei subsp. Hoffmannii (67.6%) than in E. hormaechei subsp. oharae and E. hormaechei subsp. steigerwaltii (16.7% and 23.1%) respectively (p < 0.001). The overall resistant rate to levofloxacin was 65.6% and was significantly higher in E. hormaechei subsp. hoffmannii (95.9%) and lower in E. hormaechei subsp. oharae (36.7%) and E. hormaechei subsp. steigerwaltii (38.5%) (p < 0.001). In contrast, resistance to piperacillin/tazobactam was 58.0% overall, but was significantly lower in E. hormaechei subsp. hoffmannii (27.0%) and higher in E. hormaechei subsp. oharae (96.7%) and E. hormaechei subsp. steigerwaltii (79.5%) (p < 0.001) (Table 3 and Figure 2).
Table 2.
Antimicrobial susceptibility of CnsECC isolates a.
| Antimicrobial Agents | R | I/SDD b | S | MIC (μg/mL) | |||||
|---|---|---|---|---|---|---|---|---|---|
| n | % | n | % | n | % | MIC50 | MIC90 | Range | |
| Ampicillin | 157 | 100.0 | 0 | 0 | 0 | 0 | - | - | - |
| Amikacin | 1 | 0.6 | 0 | 0 | 156 | 99.4 | ≤2 | 16 | ≤2–≥64 |
| Ceftazidime | 148 | 94.3 | 2 | 1.3 | 7 | 4.5 | ≥64 | ≥64 | 2–≥64 |
| Cefmetazole | 157 | 100.0 | 0 | 0 | 0 | 0 | ≥64 | ≥64 | ≥64 |
| Cefazolin | 157 | 100.0 | 0 | 0 | 0 | 0 | ≥64 | ≥64 | ≥64 |
| Ertapenem | 156 | 99.4 | 1 | 0.6 | 0 | 0 | 4 | ≥8 | 1–≥8 |
| Cefepime | 65 | 41.4 | 33 | 21.0 | 59 | 37.6 | 8 | ≥64 | ≤1–≥64 |
| Gentamicin | 38 | 24.2 | 7 | 4.5 | 112 | 71.3 | 2 | ≥16 | ≤1–≥16 |
| Levofloxacin | 103 | 65.6 | 12 | 7.6 | 42 | 26.8 | ≥8 | ≥8 | ≤0.12–≥8 |
| Meropenem | 31 | 19.7 | 5 | 3.2 | 121 | 77.1 | 0.5 | 8 | ≤0.25–≥16 |
| Ampicillin/sulbactam | 156 | 99.4 | 0 | 0 | 1 | 0.6 | - | - | - |
| Sulfamethoxazol/trimethoprim | 113 | 72.0 | 0 | 0 | 44 | 28.0 | ≥16/304 | ≥16/304 | ≤1/19–≥16/304 |
| Tigecycline | 81 | 51.6 | 10 | 6.4 | 66 | 42.0 | ≥8 | ≥8 | ≤0.5–≥128 |
| Piperacillin/tazobactam | 91 | 58.0 | 37 | 23.6 | 29 | 18.5 | ≥128/4 | ≥128/4 | ≤4/4–≥128/4 |
a Abbreviations: R, resistant; I, intermediate; S, susceptible; SDD, susceptible-dose dependent. b SDD for cefepime.
Table 3.
Antimicrobial resistance in each species of CnsECC isolates.
| Species (n) Cluster | AM | AN | CAZ | CMZ | CZ | ETP | FEP | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| n | % | n | % | n | % | n | % | n | % | n | % | n | % | ||
| E. asburiae (2) | I | 2 | 100 | 0 | 0.0 | 2 | 100 | 2 | 100 | 2 | 100 | 2 | 100 | 0 | 0.0 |
| E. kobei (3) | II | 3 | 100 | 0 | 0.0 | 3 | 100 | 3 | 100 | 3 | 100 | 3 | 100 | 1 | 33.3 |
| E. hormaechei subsp. hoffmannii (74) | III | 74 | 100 | 0 | 0.0 | 71 | 95.9 | 74 | 100 | 74 | 100 | 74 | 100 | 50 | 67.6 |
| E. roggenkampii (6) | IV | 6 | 100 | 0 | 0.0 | 4 | 66.7 | 6 | 100 | 6 | 100 | 6 | 100 | 0 | 0.0 |
| E. hormaechei subsp. oharae (30) | VI | 30 | 100 | 1 | 3.3 | 28 | 93.3 | 30 | 100 | 30 | 100 | 30 | 100 | 5 | 16.7 |
| E. hormaechei subsp. steigerwaltii (39) | VIII | 39 | 100 | 0 | 0.0 | 38 | 97.4 | 39 | 100 | 39 | 100 | 38 | 97.4 | 9 | 23.1 |
| E. cloacae subsp. cloacae (3) | XI | 3 | 100 | 0 | 0.0 | 2 | 66.7 | 3 | 100 | 3 | 100 | 3 | 100 | 0 | 0.0 |
| Total (157) | 157 | 100 | 1 | 0.6 | 148 | 94.3 | 157 | 100 | 157 | 100.0 | 156 | 99.4 | 65 | 41.4 | |
| Species (n) Cluster | GM | LEV | MEM | SAM | SXT | TGC | TZP | ||||||||
| n | % | n | % | n | % | n | % | n | % | n | % | n | % | ||
| E. asburiae (2) | I | 0 | 0.0 | 2 | 100 | 0 | 0.0 | 2 | 100 | 0 | 0.0 | 0 | 0.0 | 2 | 100 |
| E. kobei (3) | II | 0 | 0.0 | 2 | 66.7 | 0 | 0.0 | 3 | 100 | 2 | 66.7 | 1 | 33.3 | 3 | 100 |
| E. hormaechei subsp. hoffmannii (74) | III | 18 | 24.3 | 71 | 95.9 | 24 | 32.4 | 74 | 100 | 72 | 97.3 | 52 | 70.3 | 20 | 27.0 |
| E. roggenkampii (6) | IV | 0 | 0.0 | 1 | 16.7 | 0 | 0.0 | 6 | 100 | 1 | 16.7 | 0 | 0.0 | 4 | 66.7 |
| E. hormaechei subsp. oharae (30) | VI | 8 | 26.7 | 11 | 36.7 | 2 | 6.7 | 30 | 100 | 15 | 50.0 | 12 | 40.0 | 29 | 96.7 |
| E. hormaechei subsp. steigerwaltii (39) | VIII | 12 | 30.8 | 15 | 38.5 | 5 | 12.8 | 38 | 97.4 | 22 | 56.4 | 16 | 41.0 | 31 | 79.5 |
| E. cloacae subsp. cloacae (3) | XI | 0 | 0.0 | 1 | 33.3 | 0 | 0.0 | 3 | 100 | 1 | 33.3 | 0 | 0.0 | 2 | 66.7 |
| Total (157) | 38 | 24.2 | 103 | 65.6 | 31 | 19.7 | 156 | 99.4 | 113 | 72.0 | 81 | 51.6 | 91 | 58.0 | |
AM, ampicillin; AN, amikacin; CAZ, ceftazidime; CMZ, cefmetazole; CZ, cefazolin; ETP, ertapenem; FEP, cefepime; GM, gentamicin; LEV, levofloxacin; MEM, meropenem; SAM, ampicillin/sulbactam; SXT, sulfamethoxazol/trimethoprim; TGC, tigecycline; TZP, piperacillin/tazobactam.
Figure 2.
The statistical analysis among three predominant species (E. hormaechei subsp. hoffmannii, E. hormaechei subsp. oharae and E. hormaechei subsp. steigerwaltii) in cefepime, levofloxacin and piperacillin/tazobactam.
2.3. Detection of Genes Encoding Carbapenemases, AmpC and ESBLs in CnsECC Isolates
The 157 CnsECC isolates were examined for the presence of genes encoding the Carbapenemase (blacarbapenemase), ESBL (blaESBL) and AmpC (blaAmpC), which is demonstrated in Table 4. Carbapenemase-producing Enterobacteriaceae comprised 41.4% (65 isolates) of the 157 CnsECC isolates and all of them were positive for IMP-8. Among 65 IMP-8-positive CnsECC isolates, 81.5% (53 isolates) belonged to E. hormaechei subsp. hoffmannii (cluster III), 16.9% (11 isolates) were E. hormaechei subsp. steigerwaltii (cluster VIII) and 1.5% (1 isolate) was E. hormaechei subsp. oharae (cluster VI). There were 33 (21%) isolates carried ESBL genes and the predominant blaESBL was SHV-12 (31 isolated, 94%). While 59 (37.6%) isolates were blaAmpC-positive, and genes encoding ACT, CMY, DHA-1 and MIR accounted for 78% (46 isolates), 5% (2 isolates), 8% (5 isolates) and 10% (6 isolates) of the blaAmpC-positive CnsECC isolates. It is worth nothing that coexistence of ESBL and AmpC, and the carriage of multiple ESBL and/or AmpC was common, especially in E. hormaechei subsp. hoffmannii (cluster III). Cocarriage of IMP-8 and SHV-12 was detected in eight isolates and cocarriage of IMP-8 and ACT was detected in 24 isolates, respectively. Besides, coexistence of IMP-8, SHV-12 and ACT was noted in four isolates of CnsECC (Table 4).
Table 4.
β-lactamase genes of CnsECC isolates.
| β-Lactamase Genes (n) | ||||||
|---|---|---|---|---|---|---|
| Species (n) | Cluster | Carbapenemase | bla ESBL | bla AmpC | Co-Existence of blaESBL, blaAmpC, or Carbapenemase Genes | Other β-Lactamase Genes |
| E. asburiae (2) | I | ACT (2) | ||||
| E. kobei (3) | II | |||||
| E.hormaechei subsp. hoffmannii (74) | III | IMP-8 (53) | SHV-12 (18) CTX-M9 (1) |
CMY (2) ACT (43) DHA (1) |
IMP-8 + SHV-12 + ACT (4) IMP-8 + SHV-12 (2) IMP-8 + CMY-2 + ACT (1) IMP-8 + CMY-2 (1) IMP-8 + ACT (23) SHV-12 + CTX-M9 + ACT (1) SHV-12 + ACT (9) |
TEM-1 (11) OXA-1 (1) |
| E. roggenkampii (6) | IV | MIR (6) | ||||
| E. hormaechei subsp. oharae (30) | VI | IMP-8 (1) | SHV-12 (6) | ACT (1) DHA-1 (3) |
SHV-12 + DHA-1 (3) | TEM-1 (13) OXA-1 (1) |
| E. hormaechei subsp. steigerwaltii (39) | VIII | IMP-8 (11) | SHV-12 (7) CTX-M14 (1) |
DHA-1 (1) | IMP-8 + SHV-12 (6) | TEM-1 (13) OXA-1 (2) |
| E. cloacae subsp. cloacae (3) | XI | TEM-1 (2) | ||||
| Total (157) | 65 (41.4%) | 33 (21.0%) | 59 (37.6%) | 50 (31.8%) | 43 (27.4%) | |
2.4. Carbapenem MIC Distribution of CnsECC Isolates with and without IMP-8
IMP-8 is the only carbapenemase-encoding gene detected in this study. To evaluate possible effects of IMP-8 on the carbapenems MIC, we conducted antimicrobial susceptibility testing of ertapenem and meropenem. As shown in Table 5, regardless of harboring a carbapenemase-encoding gene, almost all (99.4%) the CnsECC strains were resistant to ertapenem (MIC50 = 4 μg/ mL). In contrast, in 65 IMP-8-positive strains, 35 (53.8%) remained susceptible to meropenem and among the 92 non-carbapenemase producing CnsECC (non-CP-CnsECC) strains, most of them (86 isolates, 93.5%) were susceptible to meropenem.
Table 5.
Carbapenem MIC distribution among CnsECC isolates with or without carbapenemase IMP-8.
| IMP-8/Carbapenem | MIC (μg/mL) | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| ≤0.25 | 0.5 | 1 | 2 | 4 | 8 | ≥8 | ≥16 | MIC50 | MIC90 | Range | |
| IMP-8 positive (n = 65) | |||||||||||
| Ertapenem | 1 | 2 | 36 | 26 | 4 | ≥8 | 1–≥8 | ||||
| Meropenem | 8 | 10 | 17 | 4 | 9 | 4 | 13 | 1 | ≥16 | ≤0.25–≥16 | |
| IMP-8 negative (n = 92) | |||||||||||
| Ertapenem | 31 | 51 | 10 | 4 | ≥8 | 2–≥8 | |||||
| Meropenem | 68 | 6 | 12 | 1 | 3 | 1 | 1 | ≤0.25 | 1 | ≤0.25–≥16 | |
Gray-shaded cells indicate resistance.
2.5. Clinical Features and Data Analysis
Between October 2011 and August 2017, a total of 150 patients with nonduplicate CnsECC isolates were identified. Baseline characteristics and prior health care exposure of the patients with CnsECC are shown in Table 6. Their median age was 68 years, 62 (41.3%) were female, and 64 (42.7%) were carbapenemase-producing Enterobacter cloacae complex (CP-ECC) carriers. Almost all individuals had underlying comorbid conditions; the majority of patients had underlying cardiovascular disease (n = 105, 70%), followed by chronic kidney disease (n = 94, 62.7%), chronic pulmonary disease (n = 92, 61.3%), diabetes mellitus (n = 65, 43.3%), chronic liver disease (n = 62, 41.3%), cerebrovascular disease (n = 49, 32.7%), hematological malignancies (n = 48, 32.0%), and solid tumor (n = 46, 30.7%). Most of the patients with CnsECC had catheter devices indwelling (n = 138, 92%), ventilator support (n = 93, 62%) and antibiotics exposure (n = 147, 98%) within 3 months prior to isolation of CnsECC. The average length of days from admission to positive CnsECC culture was 26 days (IQR: 5.25–33). In the statistics analysis for CP-ECC acquisition compared with CnsECC without carbapenemase production (non-CP-ECC), significantly greater chronic pulmonary disease (OR: 2.51; 95% CI: 1.25–5.06), ventilator support (OR: 5.54; 95% CI: 2.55–12.03) and carbapenem exposure (OR: 2.17; 95% CI: 1.10–4.25) were associated with the CP-ECC group.
Table 6.
Baseline characteristics and health care exposure of CnsECC.
| Variables | CnsECC n = 150 |
Non-CP-EnsCC n = 86 |
CP-EnsCC n = 64 |
OR (95% CI) |
p |
|---|---|---|---|---|---|
| Sex, female | 62 | 36 | 26 | 0.95 (0.49–1.83) | 0.88 |
| Age (years), median (range) | 68 (4–95) | 70.5 (4–95) | 63 (34–94) | 0.64 | |
| Cardiovascular disease | 105 (70%) | 62 | 43 | 0.79 (0.39–1.60) | 0.52 |
| Cerebrovascular disease | 49 (32.7%) | 26 | 23 | 1.29 (0.65–2.57) | 0.46 |
| Diabetes mellitus | 65 (43.3%) | 35 | 30 | 1.29 (0.67–2.47) | 0.45 |
| Chronic pulmonary disease | 92 (61.3%) | 45 | 47 | 2.51 (1.25–5.06) | 0.01 * |
| Chronic liver disease | 62 (41.3) | 40 | 22 | 0.60 (0.31–1.17) | 0.14 |
| Chronic kidney disease | 94 (62.7) | 55 | 39 | 0.88 (0.45–1.71) | 0.71 |
| Solid tumor | 46 (30.7) | 28 | 18 | 0.81 (0.40–1.65) | 0.56 |
| Hematological malignancies | 48 (32.0) | 27 | 21 | 1.05 (0.65–1.67) | 0.85 |
| Health care exposure (within 3 months) | |||||
| Indwelling devices | 138 (92) | 78 | 60 | 1.54 (0.44–5.39) | 0.50 |
| Hemodialysis | 26 (17.3) | 14 | 12 | 1.18 (0.51–2.78) | 0.69 |
| Ventilator support | 93 (62) | 40 | 53 | 5.54 (2.55–12.03) | <0.001 * |
| Surgery | 63 (42) | 36 | 27 | 1.01 (0.53–1.96) | 0.97 |
| Chemotherapy | 12 (8) | 6 | 6 | 1.38 (0.42–4.49) | 0.59 |
| Steroid exposure | 53 (35.8) | 19 | 34 | 3.88 (1.91–7.88) | <0.001 * |
| Antibiotics exposure | 147 (98) | 83 | 64 | 5.41 (0.27–106.6) | 0.27 |
| Carbapenems exposure | 57 (38) | 25 | 31 | 2.17 (1.10–4.25) | 0.02 * |
Abbreviations: CnsECC, Carbapenem-nonsusceptible Enterobacter cloacae complex; CP, carbapenemase-producing. * Statistical significance, p < 0.05.
2.6. Culture Sources and Outcomes of Patients with CnsECC Infection
The distributions of the culture source and outcome of CnsECC cases are shown in Table 7. A total of 150 consecutive nonduplicate CnsECC isolates were originated from different anatomical sites: blood (n = 15, 10.0%), sputum (n = 49, 32.7%), urine (n = 51, 34.0%), pus (n = 21, 14.0%), body fluid (n = 7, 4.6%), catheter (n = 2, 1.3%), catheter (n = 1, 3.4%), bronchoalveolar lavage (n = 3, 2.0%), and swab (n = 2, 1.3%). The incidence was no different in each culture source between CP-ECC and non-CP-ECC. The average length of stay for all CnsECC patients was 37.5 days (IQR, 20–64). There was no difference in length of hospitalization, between CP-ECC and non-CP-ECC. Patients with CP-ECC infection were more likely to have a higher in-hospital mortality (OR: 2.6; 95% CI: 1.25–5.40), 30-day mortality (OR: 3.01; 95% CI: 1.34–6.72) and 100-day mortality (OR: 2.84; 95% CI: 1.34–6.03) compared with non-CP-ECC.
Table 7.
Culture source and clinical outcome among CnsECC.
| Variables | CnsECC n = 150 |
Non-CP-EnsCC n = 86 |
CP-EnsCC n = 64 |
OR (95% CI) |
p |
|---|---|---|---|---|---|
| Culture source | 0.10 | ||||
| Blood | 15 (10.0%) | 9 | 6 | 0.89 (0.30–2.63) | 0.83 |
| Sputum | 49 (32.7%) | 23 | 26 | 1.03 (0.52–2.04) | 0.93 |
| Urine | 51 (34.0%) | 29 | 22 | 1.87 (0.94–3.74) | 0.07 |
| Abscess | 21 (14.0%) | 15 | 6 | ||
| Body fluid | 7 (4.6%) | 6 | 1 | ||
| Catheter | 2 (1.3%) | 2 | 0 | ||
| BAL | 3 (2.0%) | 0 | 3 | ||
| Swab | 2 (1.3%) | 2 | 0 | ||
| Outcomes | |||||
| Length of hospitalization, (median days, IQR) | 37.5 (20–64) | 29.5 (17–57) | 45.5 (28–64) | 0.48 | |
| In-hospital mortality | 42 (28) | 17 | 25 | 2.60 (1.25–5.40) | 0.01 * |
| 30-day mortality | 33 (22) | 12 | 21 | 3.01 (1.34–6.72) | 0.007 * |
| 100-day mortality | 39 (26) | 15 | 24 | 2.84 (1.34–6.03) | 0.007 * |
Abbreviations: CnsECC, carbapenem-nonsusceptible Enterobacter cloacae complex; CP, carbapenemase-producing; IQR, interquartile range. * Statistical significance, p < 0.05.
3. Discussion
An increasing incidence of CnsECC poses a great threat to public health [7,25]. There, it is urgent to characterize the clinical molecular epidemiology of CnsECC infection. In this study, we demonstrated that the most common species of CnsECC in southern Taiwan was E. hormaechei subsp. hoffmannii (cluster III) (47.1%), which is similar to previous study for ECC isolates co-resistant to carbapenem and colistin in southeast China [3]. The second common species of CnsECC in this study was E. hormaechei subsp. steigerwaltii (cluster VIII) (24.8%), which was the predominant strain in northeast China (38.9%) [17] and Japan (33.3%) [26].
It is well known that ECC has an intrinsic resistance to ampicillin, amoxicillin and first- and second-generation cephalosporins owing to low expression of chromosomal ampC gene encoding an inducible AmpC β-lactamase under a basal condition [7,11,27]. Expectedly, most of the CnsECC isolates in the present study showed resistant to ampicillin, ampicillin/sulbactam, and first- and second-generation cephalosporins. It is noteworthy that the different species of CnsECC isolates showed heterogeneity in resistance patterns to cefepime, levofloxacin and piperacillin/tazobactam. Our data demonstrated E. hormaechei subsp. hoffmannii (cluster III) was characterized with higher cefepime and levofloxacin resistance rate but lower piperacillin/tazobactam resistance rate relative to other species of CnsECC. A similar pattern of results was also revealed in a recent study, which reported different antibiotics resistant pattern between two identical genetic clusters among carbapenem-resistant ECC isolates [17]. These observations indicate multiple drug-resistant mechanisms may participate in CnsECC isolates and warrant further investigation.
The resistant mechanisms of CRE are the presence of either carbapenemase or ESBLs and/or AmpC enzyme as well as losing outer membrane porin proteins [23,28,29]. Our study showed the predominant mechanism of CnsECC isolates were carbapenemase production (41.4%) and AmpC overproduction (37.3%). This result is compatible with previous studies demonstrating carbapenemase production and overproduction of AmpC are the main carbapenem resistance mechanisms in CRE isolates [28,29,30]. The prevalence of CP-CnsECC among clinical CnsECC isolates was around 41.4% (65/157 isolates) and IMP-8 was the only carbapenemase detected in the CP-ECC isolates (65/65 isolates, 100%) in this study. IMP-encoding ECC have been reported globally, including in China (IMP-1 and IMP-34), Thailand (IMP-14), Japan (IMP-1 and IMP-11), Australia (IMP-4) and Korea (IMP-4) [31]. In Taiwan, IMP-8-encoding CnsECC has been reported since the early 2000s [24]. In a large surveillance report for carbapenem-nonsusceptible Enterobacteriaceae during 2010–2012 in Taiwan observed 96.3% of CP-CnsECC had IMP-8 which agrees with our findings [23].
In the present study, we defined CnsECC as ECC isolates with nonsusceptibility to ertapenem. Among the 157 CnsECC isolates, we observed that isolates tests showing susceptibility to meropenem were common (77.1%, 121/157). Similar results have been found by other studied [23,24,32]. Meanwhile, we found almost all non-CP-CnsECC were susceptible to meropenem (93.5%, 86/92) and lower meropenem susceptibility rates in CP-CnsECC isolates (53.8%, 35/65). This finding is also correlated with previous studies for carbapenem-nonsusceptible Klebsiella pneumoniae (CnsKP), which showed CP-CnsKP was more nonsusceptible to meropenem than non-CP-CnsKP [33]. Consequently, non-CP-CnsECC might be effectively treated by meropenem in current circumstances. Aside from the carbapenem, several novel antibiotics, such as ceftazidime-avibactam, ceftozolane-tazobactam, cefepime-zidebactam, meropenem-vaborbactam, cefiderocol and eravacycline have been developed to treat multidrug-resistant organisms but many of them have limited activity against strains bearing class B metallo-lactamases (MBLs) such as IMP. Therefore, using novel rapid molecular/ lateral flow infectious disease diagnostic tools such as Xpert® Carba-R test (Cepheid, Sunnyvale, CA, USA), Biofire FilmArray panels (Biomerieux, Nürtingen, Germany) or NG-Test Carba 5 (NG Biotech, Guipry, France) to identify the type of carbapenemase present in the carbapenem nonsusceptible isolate is important for optimizing antibiotics therapy [34,35].
In this study, 98% of patients had antimicrobial therapy prior to culture, with a total of 57/150 (38%) patients being exposed to carbapenems. Chronic pulmonary disease and carbapenems exposure were considered risk factors associated with CP-CnsECC. Besides, we observed CP-CnsECC associated with poorer outcomes including in-hospital mortality, 30-day mortality and 100-day mortality. A previous study demonstrated CP-CRE may be more virulent than non-CP-CRE and is associated with poorer outcomes [36]. However, several studies showed there was no difference in clinical outcomes [25,26,37]. The heterogeneity could be related to various carbapenemase-producing genes in CRE were predominant in different species of Enterobacteriaceae or geographic regions and it highlights the need to understand the local epidemiology to tailor prevention efforts in individual regions. This is the first study to make comparison of clinical characteristics and patient outcomes between carbapenemase-producing and non-carbapenemase-producing CnsECC isolates in Taiwan.
This study has several limitations. Due to the lack of available medical information for ascertainment of infections, our data likely overestimated the proportion of lower respiratory and urinary tract infections. Furthermore, we did not have a control group of carbapenem-susceptible ECC. Therefore, we were not able to conduct risk factor analysis on CnsECC. There is also the possibility that novel carbapenemases or rarer enzymes that were not performed. Finally, as in any observational study, information on clinical characteristics and outcomes could not be completely acquired and our analysis of clinical outcome was subject to confounding biases.
4. Materials and Methods
4.1. Bacterial Isolates
A total of 157 ertapenem-nonsusceptible Enterobacter cloacae complex isolates (minimum inhibitory concentrations [MICs] of ertapenem >1 μg/mL) were collected from Kaohsiung Medical University Hospital during October 2011 to August 2017 and defined as CnsECC. The identification of ECC and testing of ertapenem susceptibility were performed as routine clinical microbiology laboratory procedures. The collection and testing of clinical specimens were approved by the Review Board Committee of KMUH.
4.2. Antimicrobial Susceptibility Testing
Antimicrobial susceptibility was tested by the broth dilution method according to the guidelines of the Clinical and Laboratory Standards Institute (CLSI) [38]. All isolates were tested for minimal inhibitory concentrations (MICs) of β-lactam agents, including penicillins (ampicillin, ampicillin/sulbactam, piperacillin-tazobactam), cephalosporins (cefazolin, cefmetazole, ceftazidime and cefepime), carbapenems (ertapenem and meropenem); and non-β-lactams, including fluoroquinolone (levofloxacin), aminoglycosides (gentamicin and amikacin), trimethoprim-sulfamethoxazole and tigecycline.
4.3. Amplification and Sequencing of the hsp60 Gene
Polymerase chain reaction (PCR) analysis for partial sequencing of the hsp60 gene was performed by a protocol described previously [39]. A 341-bp fragment of the hsp60 gene was amplified and sequencing. The sequences of a 272-bp fragment of the hsp60 gene obtained for 157 strains were analyzed by the nucleotide BLAST program searches against the NCBI database and each isolate was assigned to its respective species, subspecies, and cluster according to taxonomic studies published previously [6,39].
4.4. Pulsed Field Gel Electrophoresis
Pulsed-field gel electrophoresis (PFGE) XbaI (New England Biolabs, Beverly, MA, USA)-digested genomic DNA was conducted to delineate the genetic relatedness of the isolates using procedures described previously [40]. PFGE patterns were interpreted in accordance with the criteria of Tenover et al. [41]. Restriction fragments were analyzed using GelCompar II software 6.5 (Applied Maths, Austin, TX, USA), and dendrograms of the patterns were constructed using the unweighted pair group method with arithmetic mean (UPGMA) based on the Dice similarity index.
4.5. Detection of Genes Encoding Carbapenemases, AmpC, and ESBLs
All verified isolates were subjected to polymerase chain reaction (PCR) detection of genes encoding carbapenemases (IMP, KPC, OXA, NDM, VIM, BIC, IMI, SME, AIM, DIM, GIM, SPM, SIM, and GES) [42], AmpC genes (CMY, DHA, and ACT) [43] and ESBL genes (CTX-M, SHV, and TEM) [44].
4.6. Clinical Data Collection and Statistical Analyses
This was a single-center, retrospective, observational study of patients with positive cultures of CnsECC from October 2011 to August 2017 at KUMH. Patient information was retrospectively retrieved via electronic medical records. The parameters included demographic data, comorbidities, healthcare exposures (such as indwelling devices, hemodialysis, mechanical ventilation, and surgeries), exposure to antimicrobials within 3 months prior to isolation of CnsECC, and the clinical outcomes. Clinical outcomes were assessed by either in-hospital mortality or patient survival. The Student t-test was used for continuous variables. The chi-square test or Fisher exact test was used to compare categorical variables. For categorical variables with more than two categories, we used ordinary logistic regression to perform multiple comparisons. Statistical significance was set at p < 0.05. All statistical analyses were performed using MedCalc statistical software version 20.013 (MedCalc Software Corporation, Ostend, Belgium).
5. Conclusions
In conclusion, nearly all the CnsECC isolates in the present study were resistant to ampicillin and first- and second-generation cephalosporins but the majority remained susceptible to amikacin and meropenem. Different species of CnsECC isolates showed heterogeneity in resistance patterns to cefepime, levofloxacin and piperacillin/tazobactam. The most common species of CnsECC was E. hormaechei subsp. Hoffmannii, which was characterized with higher cefepime and levofloxacin resistance rate and with higher prevalence of CP-CnsECC. IMP-8 was the only carbapenemases detected among the CnsECC isolates in this study. Analysis on clinical data revealed patient with CP-CnsECC infection had poorer clinical outcomes. This study highlights the need to understand the local molecular epidemiology of carbapenem-nonsusceptible ECC and shows that using novel rapid infectious disease diagnostic tools to identify the type of carbapenemase is important for optimizing antibiotics therapy against CnsECC.
Author Contributions
Conceptualization, P.-L.L. and C.-Y.C.; data curation, C.-J.C., S.-H.J. and C.-Y.C.; investigation, S.-H.J., H.-L.F. and P.-H.H.; data collection, P.-L.L. and C.-Y.C.; supervision, P.-L.L. and C.-Y.C.; writing—original draft, C.-J.C.; review and editing, P.-L.L. and C.-Y.C. All authors have read and agreed to the published version of the manuscript.
Funding
This study was financially supported by the Ministry of Science and Technology of Taiwan (MOST 107-2314-B-037-081) and Kaohsiung Medical University Hospital (KMUH107-7R22).
Institutional Review Board Statement
The collection and testing of clinical specimens were approved by the Review Board Committee of KMUH (KMUHIRB-E(II)-20180101).
Informed Consent Statement
Informed consent was waived because of the retrospective nature of the study and the analysis used anonymous clinical data.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Mezzatesta M.L., Gona F., Stefani S. Enterobacter cloacae complex: Clinical impact and emerging antibiotic resistance. Future Microbiol. 2012;7:887–902. doi: 10.2217/fmb.12.61. [DOI] [PubMed] [Google Scholar]
- 2.Pavlovic M., Konrad R., Iwobi A.N., Sing A., Busch U., Huber I. A dual approach employing MALDI-TOF MS and real-time PCR for fast species identification within the Enterobacter cloacae complex. FEMS Microbiol. Lett. 2012;328:46–53. doi: 10.1111/j.1574-6968.2011.02479.x. [DOI] [PubMed] [Google Scholar]
- 3.Liu S., Fang R., Zhang Y., Chen L., Huang N., Yu K., Zhou C., Cao J., Zhou T. Characterization of resistance mechanisms of Enterobacter cloacae Complex co-resistant to carbapenem and colistin. BMC Microbiol. 2021;21:208. doi: 10.1186/s12866-021-02250-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jean S.S., Lee Y.L., Liu P.Y., Lu M.C., Ko W.C., Hsueh P.R. Multicenter surveillance of antimicrobial susceptibilities and resistance mechanisms among Enterobacterales species and non-fermenting Gram-negative bacteria from different infection sources in Taiwan from 2016 to 2018. J. Microbiol. Immunol. Infect. 2021 doi: 10.1016/j.jmii.2021.07.015. [DOI] [PubMed] [Google Scholar]
- 5.Potter R.F., D’Souza A.W., Dantas G. The rapid spread of carbapenem-resistant Enterobacteriaceae. Drug Resist. Update. 2016;29:30–46. doi: 10.1016/j.drup.2016.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Davin-Regli A., Lavigne J.P., Pages J.M. Enterobacter spp.: Update on Taxonomy, Clinical Aspects, and Emerging Antimicrobial Resistance. Clin. Microbiol. Rev. 2019;32:e00002-19. doi: 10.1128/CMR.00002-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Annavajhala M.K., Gomez-Simmonds A., Uhlemann A.C. Multidrug-Resistant Enterobacter cloacae Complex Emerging as a Global, Diversifying Threat. Front. Microbiol. 2019;10:44. doi: 10.3389/fmicb.2019.00044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Godmer A., Benzerara Y., Normand A.C., Veziris N., Gallah S., Eckert C., Morand P., Piarroux R., Aubry A. Revisiting Species Identification within the Enterobacter cloacae Complex by Matrix-Assisted Laser Desorption Ionization-Time of Flight Mass Spectrometry. Microbiol. Spectr. 2021;9:e00661-21. doi: 10.1128/Spectrum.00661-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Singh N.K., Bezdan D., Checinska Sielaff A., Wheeler K., Mason C.E., Venkateswaran K. Multi-drug resistant Enterobacter bugandensis species isolated from the International Space Station and comparative genomic analyses with human pathogenic strains. BMC Microbiol. 2018;18:175. doi: 10.1186/s12866-018-1325-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wu W., Feng Y., Zong Z. Precise Species Identification for Enterobacter: A Genome Sequence-Based Study with Reporting of Two Novel Species, Enterobacter quasiroggenkampii sp. nov. and Enterobacter quasimori sp. nov. mSystems. 2020;5:e00527-20. doi: 10.1128/mSystems.00527-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.De Oliveira D.M.P., Forde B.M., Kidd T.J., Harris P.N.A., Schembri M.A., Beatson S.A., Paterson D.L., Walker M.J. Antimicrobial resistance in eskape pathogens. Clin. Microbiol. Rev. 2020;33:e00181-19. doi: 10.1128/CMR.00181-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Peirano G., Matsumura Y., Adams M.D., Bradford P., Motyl M., Chen L., Kreiswirth B.N., Pitout J.D.D. Genomic Epidemiology of Global Carbapenemase-Producing Enterobacter spp., 2008–2014. Emerg. Infect. Dis. 2018;24:1010–1019. doi: 10.3201/eid2406.171648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Davin-Regli A., Pages J.M. Enterobacter aerogenes and Enterobacter cloacae; versatile bacterial pathogens confronting antibiotic treatment. Front. Microbiol. 2015;6:392. doi: 10.3389/fmicb.2015.00392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wilson B.M., El Chakhtoura N.G., Patel S., Saade E., Donskey C.J., Bonomo R.A., Perez F. Carbapenem-Resistant Enterobacter cloacae in Patients from the US Veterans Health Administration, 2006–2015. Emerg. Infect. Dis. 2017;23:878–880. doi: 10.3201/eid2305.162034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Aoki K., Harada S., Yahara K., Ishii Y., Motooka D., Nakamura S., Akeda Y., Iida T., Tomono K., Iwata S., et al. Molecular Characterization of IMP-1-Producing Enterobacter cloacae Complex Isolates in Tokyo. Antimicrob. Agents Chemother. 2018;62:e02091-17. doi: 10.1128/AAC.02091-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jean S.S., Lee N.Y., Tang H.J., Lu M.C., Ko W.C., Hsueh P.R. Carbapenem-Resistant Enterobacteriaceae Infections: Taiwan Aspects. Front. Microbiol. 2018;9:2888. doi: 10.3389/fmicb.2018.02888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen J., Tian S., Nian H., Wang R., Li F., Jiang N., Chu Y. Carbapenem-resistant Enterobacter cloacae complex in a tertiary Hospital in Northeast China, 2010-2019. BMC. Infect. Dis. 2021;21:611. doi: 10.1186/s12879-021-06250-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lai C.C., Chen Y.S., Lee N.Y., Tang H.J., Lee S.S., Lin C.F., Lu P.L., Wu J.J., Ko W.C., Lee W.S., et al. Susceptibility rates of clinically important bacteria collected from intensive care units against colistin, carbapenems, and other comparative agents: Results from Surveillance of Multicenter Antimicrobial Resistance in Taiwan (SMART) Infect. Drug Resist. 2019;12:627–640. doi: 10.2147/IDR.S194482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Suay-García B., Pérez-Gracia M.T. Present and Future of Carbapenem-resistant Enterobacteriaceae (CRE) Infections. Antibioticcs. 2019;8:122. doi: 10.3390/antibiotics8030122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chang C.L., Su L.H., Lu C.M., Tai F.T., Huang Y.C., Chang K.K. Outbreak of ertapenem-resistant Enterobacter cloacae urinary tract infections due to a contaminated ureteroscope. J. Hosp. Infect. 2013;85:118–124. doi: 10.1016/j.jhin.2013.06.010. [DOI] [PubMed] [Google Scholar]
- 21.Huang L.F., Lee C.T., Su L.H., Chang C.L. A Snapshot of Co-Resistance to Carbapenems and Tigecycline in Clinical Isolates of Enterobacter cloacae. Microb. Drug Resist. 2017;23:1–7. doi: 10.1089/mdr.2015.0311. [DOI] [PubMed] [Google Scholar]
- 22.Jean S.S., Hsueh P.R., Group S.A.-P. Distribution of ESBLs, AmpC β-lactamases and carbapenemases among Enterobacteriaceae isolates causing intra-abdominal and urinary tract infections in the Asia-Pacific region during 2008–2014: Results from the Study for Monitoring Antimicrobial Resistance Trends (SMART) J. Antimicrob. Chemother. 2017;72:166–171. doi: 10.1093/jac/dkw398. [DOI] [PubMed] [Google Scholar]
- 23.Wang J.T., Wu U.I., Lauderdale T.L., Chen M.C., Li S.Y., Hsu L.Y., Chang S.C. Carbapenem-nonsusceptible Enterobacteriaceae in Taiwan. PLoS ONE. 2015;10:e0121668. doi: 10.1371/journal.pone.0121668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yang F.C., Yan J.J., Hung K.H., Wu J.J. Characterization of ertapenem-resistant Enterobacter cloacae in a Taiwanese university hospital. J. Clin. Microbiol. 2012;50:223–226. doi: 10.1128/JCM.01263-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Marimuthu K., Venkatachalam I., Khong W.X., Koh T.H., Cherng B.P.Z., Van La M., De P.P., Krishnan P.U., Tan T.Y., Choon R.F.K., et al. Clinical and Molecular Epidemiology of Carbapenem-Resistant Enterobacteriaceae Among Adult Inpatients in Singapore. Clin. Infect. Dis. 2017;64:S68–S75. doi: 10.1093/cid/cix113. [DOI] [PubMed] [Google Scholar]
- 26.Tetsuka N., Hirabayashi A., Matsumoto A., Oka K., Hara Y., Morioka H., Iguchi M., Tomita Y., Suzuki M., Shibayama K., et al. Molecular epidemiological analysis and risk factors for acquisition of carbapenemase-producing Enterobacter cloacae complex in a Japanese university hospital. Antimicrob. Resist. Infect. Control. 2019;8:126. doi: 10.1186/s13756-019-0578-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ito A., Nishikawa T., Ota M., Ito-Horiyama T., Ishibashi N., Sato T., Tsuji M., Yamano Y. Stability and low induction propensity of cefiderocol against chromosomal AmpC β-lactamases of Pseudomonas aeruginosa and Enterobacter cloacae. J. Antimicrob. Chemother. 2018;73:3049–3052. doi: 10.1093/jac/dky317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Satlin M.J., Chen L., Patel G., Gomez-Simmonds A., Weston G., Kim A.C., Seo S.K., Rosenthal M.E., Sperber S.J., Jenkins S.G., et al. Multicenter Clinical and Molecular Epidemiological Analysis of Bacteremia Due to Carbapenem-Resistant Enterobacteriaceae (CRE) in the CRE Epicenter of the United States. Antimicrob. Agents Chemother. 2017;61:e02349-16. doi: 10.1128/AAC.02349-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Alizadeh N., Ahangarzadeh Rezaee M., Samadi Kafil H., Hasani A., Soroush Barhaghi M.H., Milani M., Yeganeh Sefidan F., Memar M.Y., Lalehzadeh A., Ghotaslou R. Evaluation of Resistance Mechanisms in Carbapenem-Resistant Enterobacteriaceae. Infect. Drug Resist. 2020;13:1377–1385. doi: 10.2147/IDR.S244357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu S., Huang N., Zhou C., Lin Y., Zhang Y., Wang L., Zheng X., Zhou T., Wang Z. Molecular mechanisms and epidemiology of carbapenem-resistant Enterobacter cloacae complex isolated from chinese patients during 2004–2018. Infect. Drug Resist. 2021;14:3647–3658. doi: 10.2147/IDR.S327595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lee J.H., Bae I.K., Lee C.H., Jeong S. Molecular Characteristics of First IMP-4-Producing Enterobacter cloacae Sequence Type 74 and 194 in Korea. Front. Microbiol. 2017;8:2343. doi: 10.3389/fmicb.2017.02343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Endimiani A., Perez F., Bajaksouzian S., Windau A.R., Good C.E., Choudhary Y., Hujer A.M., Bethel C.R., Bonomo R.A., Jacobs M.R. Evaluation of updated interpretative criteria for categorizing Klebsiella pneumoniae with reduced carbapenem susceptibility. J. Clin. Microbiol. 2010;48:4417–4425. doi: 10.1128/JCM.02458-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chiu S.K., Ma L., Chan M.C., Lin Y.T., Fung C.P., Wu T.L., Chuang Y.C., Lu P.L., Wang J.T., Lin J.C., et al. Carbapenem Nonsusceptible Klebsiella pneumoniae in Taiwan: Dissemination and Increasing Resistance of Carbapenemase Producers During 2012–2015. Sci. Rep. 2018;8:8468. doi: 10.1038/s41598-018-26691-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tenover F.C. Using Molecular Diagnostics to Develop Therapeutic Strategies for Carbapenem-Resistant Gram-Negative Infections. Front. Cell. Infect. Microbiol. 2021;11:715821. doi: 10.3389/fcimb.2021.715821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Messacar K., Parker S.K., Todd J.K., Dominguez S.R. Implementation of Rapid Molecular Infectious Disease Diagnostics: The Role of Diagnostic and Antimicrobial Stewardship. J. Clin. Microbiol. 2017;55:715–723. doi: 10.1128/JCM.02264-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tamma P.D., Goodman K.E., Harris A.D., Tekle T., Roberts A., Taiwo A., Simner P.J. Comparing the Outcomes of Patients with Carbapenemase-Producing and Non-Carbapenemase-Producing Carbapenem-Resistant Enterobacteriaceae Bacteremia. Clin. Infect. Dis. 2017;64:257–264. doi: 10.1093/cid/ciw741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lin Q., Wu M., Yu H., Jia X., Zou H., Ma D., Niu S., Huang S. Clinical and Microbiological Characterization of Carbapenem-Resistant Enterobacteriales: A Prospective Cohort Study. Front. Pharmacol. 2021;12:716324. doi: 10.3389/fphar.2021.716324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.CLSI . Performance Standards for Antimicrobial Susceptibility Testing. 31st ed. Clinical and Laboratory Standards Institute; Malvern, PA, USA: 2021. CLSI Supplement M100. [Google Scholar]
- 39.Hoffmann H., Roggenkamp A. Population genetics of the nomenspecies Enterobacter cloacae. Appl. Environ. Microbiol. 2003;69:5306–5318. doi: 10.1128/AEM.69.9.5306-5318.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.D’Agata E.M., Gerrits M.M., Tang Y.W., Samore M., Kusters J.G. Comparison of pulsed-field gel electrophoresis and amplified fragment-length polymorphism for epidemiological investigations of common nosocomial pathogens. Infect. Control Hosp. Epidemiol. 2001;22:550–554. doi: 10.1086/501950. [DOI] [PubMed] [Google Scholar]
- 41.Tenover F.C., Arbeit R.D., Goering R.V., Mickelsen P.A., Murray B.E., Persing D.H., Swaminathan B. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: Criteria for bacterial strain typing. J. Clin. Microbiol. 1995;33:2233–2239. doi: 10.1128/jcm.33.9.2233-2239.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chiu S.K., Wu T.L., Chuang Y.C., Lin J.C., Fung C.P., Lu P.L., Wang J.T., Wang L.S., Siu L.K., Yeh K.M. National surveillance study on carbapenem non-susceptible Klebsiella pneumoniae in Taiwan: The emergence and rapid dissemination of KPC-2 carbapenemase. PLoS ONE. 2013;8:e69428. doi: 10.1371/journal.pone.0069428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Alvarez M., Tran J.H., Chow N., Jacoby G.A. Epidemiology of conjugative plasmid-mediated AmpC β-lactamases in the United States. Antimicrob. Agents Chemother. 2004;48:533–537. doi: 10.1128/AAC.48.2.533-537.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Eckert C., Gautier V., Saladin-Allard M., Hidri N., Verdet C., Ould-Hocine Z., Barnaud G., Delisle F., Rossier A., Lambert T., et al. Dissemination of CTX-M-type β-lactamases among clinical isolates of Enterobacteriaceae in Paris, France. Antimicrob. Agents Chemother. 2004;48:1249–1255. doi: 10.1128/AAC.48.4.1249-1255.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.