Abstract
Planktonic microbial communities mediate many vital biogeochemical processes in wetland ecosystems, yet compared to other aquatic ecosystems, like oceans, lakes, rivers or estuaries, they remain relatively underexplored. Our study site, the Florida Everglades (USA)—a vast iconic wetland consisting of a slow-moving system of shallow rivers connecting freshwater marshes with coastal mangrove forests and seagrass meadows—is a highly threatened model ecosystem for studying salinity and nutrient gradients, as well as the effects of sea level rise and saltwater intrusion. This study provides the first high-resolution phylogenetic profiles of planktonic bacterial and eukaryotic microbial communities (using 16S and 18S rRNA gene amplicons) together with nutrient concentrations and environmental parameters at 14 sites along two transects covering two distinctly different drainages: the peat-based Shark River Slough (SRS) and marl-based Taylor Slough/Panhandle (TS/Ph). Both bacterial as well as eukaryotic community structures varied significantly along the salinity gradient. Although freshwater communities were relatively similar in both transects, bacterioplankton community composition at the ecotone (where freshwater and marine water mix) differed significantly. The most abundant taxa in the freshwater marshes include heterotrophic Polynucleobacter sp. and potentially phagotrophic cryptomonads of the genus Chilomonas, both of which could be key players in the transfer of detritus-based biomass to higher trophic levels.
Keywords: Florida Everglades, coastal microbiology, bacterioplankton, eukaryotic microbial communities, 16S rRNA amplicon sequencing, 18S rRNA amplicon sequencing
1. Introduction
Coastal environments, which are among the most diverse and productive habitats in the world, provide ecosystem services worth trillions of dollars annually [1,2]. Over the last century, freshwater diversion and the conversion of wetlands to agricultural land has reduced the area of global coastal ecosystems by more than two-fold [3]. These developments have made the ecological well-being of coastal ecosystems a pressing global issue, and especially conservation of unique and iconic sites that are inscribed on the UNESCO World Heritage List and considered in ‘critical’ status, like the Everglades National Park (ENP), are in the focus of local and national environmental policies. Consequently, the Comprehensive Everglades Restoration Plan, with an approximate 50-year construction schedule, was initiated in the 1990s to restore the quantity, timing and distribution of the pre-drainage water flow through the Everglades that was changed profoundly due to urban development, agriculture and extensive drainage [4,5].
Coastal environments are heavily affected by a heterogeneous variety of terrestrial, freshwater and marine aquatic ecosystems and therefore represent highly complex and dynamic habitats. This complexity challenges informed management decision-making that relies on sufficient knowledge on the state of the system and its responses to external environmental pressures. In the Everglades specifically, shifts in fresh and marine water supply influence total phosphorous concentrations, salinity, and inundation, which in turn affect the net carbon balance in the ecosystem along the freshwater-marine gradient [6]. Most of the current ENP landscape is less than 1.5 m above the mean sea level [7], which makes it highly susceptible to a projected sea level rise of up to 2 m by 2100 [8,9]. Flower et al. [10] estimated that a 0.5 m rise in sea level would lead to seawater and mangroves encroaching 15 km into ENP by 2060. In comparison, paleoecological records demonstrate that in the past 3000 years, the gradual replacement of freshwater marsh by mangrove forests has shifted the ecotone only 20 km inland [11].
While there is considerable effort in using algae as bioindicators for the ‘health’ of aquatic ecosystems worldwide [12], and in the Everglades [13,14,15], prokaryotic communities have largely been overlooked by environmental monitoring programs [16], despite being essential for the functioning of all aquatic ecosystems and food webs [17,18]. Prokaryotic communities respond rapidly to environmental changes and therefore provide valuable bioindicators, which can predict short- to medium term regime shifts of an entire ecosystem [19,20]. Especially the use of high-throughput sequencing has revolutionized studies on microbial communities and has made effective monitoring of microbial communities feasible [19]. In the Everglades, to date only sediment bacterial communities have been analyzed using low-throughput molecular methods. These studies indicated differences in community composition between ecosystem types along the salinity gradient from freshwater to marine habitats. The salinity and total phosphorous in the water column were identified as the main factors explaining these shifts [21].
There are two main major water drainages in ENP: Shark River Slough (SRS), a large long-hydroperiod and peat-based drainage that receives freshwater from the Tamiami Canal and flows directly into the Gulf of Mexico (GoM; Figure 1), and Taylor Slough Panhandle (TS/Ph), a smaller short-hydroperiod and marl-based drainage, located in the eastern boundary of the ENP that is indirectly connected to the GoM through vast seagrass meadows in Florida Bay (FB). Here, we use high-throughput amplicon sequencing of 16S and 18S rRNA genes to determine the composition of bacterial and eukaryotic microbial communities in surface waters along the SRS and TS/Ph transects. Our main questions were: (1) who are the key members of planktonic microbial communities in the Everglades, and (2) how do environmental variables shape these communities along the transects from freshwater to marine habitats? This analysis of microbial communities in the Everglades provides crucial information about an essential part of its food web and serves as a reference to study the impact of environmental change in this unique ecosystem.
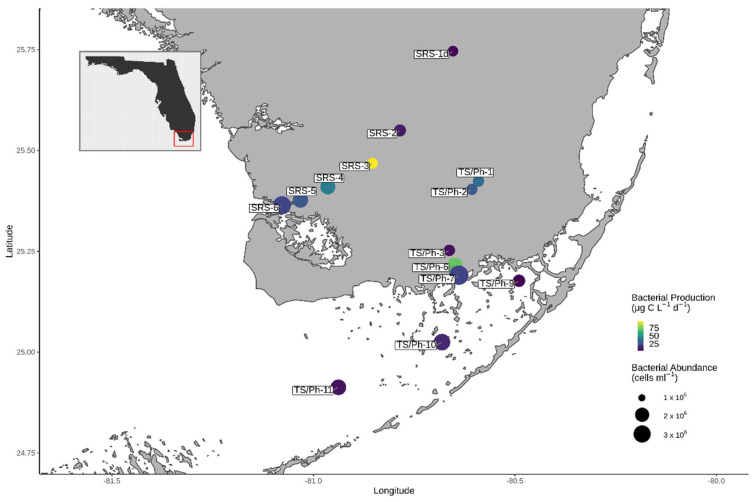
Figure 1.
Map of the study area and sampling sites in Everglades National Park, Florida (USA), covering Shark River Slough (SRS 1d-5), Taylor Slough (TS/Ph 1–7) and Florida Bay (TS/Ph 9–11). The size and color of site markers indicate bacterial abundances and bacterial production, respectively.
2. Materials and Methods
2.1. Sample Collection
Two liters of surface water samples were obtained from 14 FCE LTER core sampling sites during the subtropical wet season in August of 2017 (Table 1; Figure 1; https://fce-lter.fiu.edu/research/sites/, accessed on 14 December 2021). Samples were kept on ice until filtration and were processed within 12 h. To determine bacterial abundances (BA), 9 mL subsamples were separated for flow cytometry and fixed with paraformaldehyde (final concentration 1%; pH = 7.4), incubated at room temperature (RT) for 60 min, and stored at −20 °C until analyses. The microbial communities were separated during filtration into two size fractions using 5 µm (>5 µm fraction) and 0.22 µm (0.22 µm–5 µm fraction) nitrocellulose membranes (MF-Millipore, Darmstadt, Germany). The physicochemical background data and bacterial secondary production values were obtained via standard FCE-LTER protocols (https://fcelter.fiu.edu/data/protocols/index.html, accessed on 14 December 2021). Stations 1–3 in SRS and stations TS/Ph 1–3 are in freshwater marshes. The remaining three stations in SRS (4–6) and stations 6 and 7 of TS/Ph are considered ecotones. Stations TS/Ph 9–11 are situated in Florida Bay. Details on the sites, including coordinates, can be found at: https://fce-lter.fiu.edu/research/sites/index.php (accessed on 114 December 2021).
Table 1.
Station locations, bacterial abundances (BA) and bacterial productivities (BP) and selected environmental data (Chlorophyll a—Chl a; total nitrogen—TN; total phosphorous—TP).
| Station | BA (cells mL−1) |
BP (μg C L−1 d−1) |
Chl a (μg L−1) |
Salinity (PSU) | TN (μM) | TP (μM) | DOC (μM) |
|---|---|---|---|---|---|---|---|
| TS/Ph-1 | 1,206,442 | 36.53 | 2.89 | 0.2 | 33.67 | 0.6 | 851.67 |
| TS/Ph-2 | 1,200,550 | 29.99 | 0.11 | 0.2 * | 43.70 * | NA | 746.67 |
| TS/Ph-3 | 1,233,549 | 6.54 | 2.71 | 0.2 | 66.23 | 0.53 | 893.33 |
| TS/Ph-6 | 2,586,481 | 71.87 | 1.16 | 6.1 * | 75.40 * | 0.4 * | 1254.17 |
| TS/Ph-7 | 3,884,709 | 22.58 | 1.56 | 14.8 * | 68.84 * | 0.41* | 1085.83 |
| TS/Ph-9 | 1,493,439 | 3.98 | 0.07 | 39.50 | 45.39 | 0.23 | 460.42 ** |
| TS/Ph-10 | 2,811,578 | 13.82 | 0.12 | 41.40 | 53.62 | 0.23 | 631.67 ** |
| TS/Ph-11 | 2,455,006 | 7.58 | 0.06 | 39.49 | 23.25 | 0.30 | 270 ** |
| SRS-1d | 1,048,253 | 5.11 | 2.49 | 0.2 | 70.29 | 0.54 | 1268.33 |
| SRS-2 | 1,390,989 | 9.51 | 1.63 | 0.1 | 55.93 | 0.37 | 1148.08 |
| SRS3 | 1,127,995 | 95.80 | 1.25 | 0.2 | 53.25 | 1.00 | 1243.33 |
| SRS4 | 2,272,470 | 42.85 | 1.79 | 0.4 | 63.76 | 0.71 | 1157.33 |
| SRS5 | 2,381,611 | 29.66 | 1.61 | 2.1 | 51.76 | 0.63 | 1066.67 |
| SRS6 | 3,272,441 | 22.64 | 1.3 | 19.3 | 37.94 | 0.70 | 929.25 |
* Measurement taken 1–3 days apart from collection of water sample for microbial analyses. ** TOC (μM), DOC data not available. NA = not analyzed.
2.2. Flow Cytometry
Samples for flow cytometry were incubated with SYBR Green I nucleic acid stain for 30 min at RT. Flow cytometry analyses were performed on a Guava easyCyte HT (Luminex, Austin, TX, USA) at a flow rate of 0.24 μL s−1. Cell populations were discriminated via green fluorescence (532 nm) and side scatter channels using a blue laser (488 nm). High nucleic acid and low nucleic acid content bacterial cell counts were pooled together to obtain total bacterial abundances (BA, [22]). Samples were analyzed using Guava’s InCyte software (Luminex, Austin, TX, USA).
2.3. Molecular Methods
DNA extractions were carried out with the Qiagen PowerWater Kit following the manufacturer’s recommended protocol (Qiagen, Hilden, Germany). DNA sequence data was generated using Illumina paired-end sequencing (151 bp × 12 bp × 151 bp MiSeq run) at the Environmental Sample Preparation and Sequencing Facility at Argonne National Laboratory (Lemont, IL, USA). DNA extracts were used as templates for the amplification of the V4 hypervariable region of the 16S rRNA gene (515F-806R primer pair, [23]) and V9 hypervariable region of the 18S rRNA gene (1389F-EukB primer pair, [24]). In addition, primers contained sequencer adapter sequences and the reverse amplification primer also contained a twelve base barcode sequence for multiplexing. Each 25-µL PCR reaction contained 9.5 µL of MO BIO PCR Water (Certified DNA-Free), 12.5 µL of QuantaBio’s AccuStart II PCR ToughMix (2× concentration, 1× final), 1 µL Forward Primer (5 µM concentration, 200 pM final), 1 µL Golay barcode tagged Reverse Primer (5 µM concentration, 200 pM final), and 1 µL of template DNA. The PCR conditions to amplify the 16S rRNA gene were as follows: 94 °C for 3 min to denature the DNA, with 35 cycles at 94 °C for 45 s, 50 °C for 60 s, and 72 °C for 90 s; final extension of 10 min at 72 °C to ensure complete amplification. The PCR conditions to amplify the 18S rRNA gene were as follows: 94 °C for 3 min to denature the DNA, with 35 cycles at 94 °C for 45 s, 57 °C for 60 s, and 72 °C for 90 s; final extension of 10 min at 72 °C. Amplicons were then quantified using PicoGreen (Invitrogen, Waltham, MA, USA) and a plate reader (Infinite 200 PRO, Tecan). Once quantified, volumes of each of the products were pooled into a single tube so that each amplicon is represented in equimolar amounts. This pool was then cleaned up using AMPure XP Beads (Beckman Coulter, Brea, CA, USA), and afterwards quantified using a fluorometer (Qubit, Invitrogen). After quantification, the molarity of the pool was determined and diluted to 2 nM, denatured, and diluted to a final concentration of 6.75 pM with a 10% PhiX spike for sequencing on an Illumina MiSeq.
2.4. Bioinformatics
The QIIME 2 microbiome analysis package [25], was used for sequence analyses. Quality filtering, chimera identification and merging of paired-end reads was carried out using the DADA2 plugin [26], as implemented in QIIME2. SILVA release 132 (Ref NR 99) taxonomy and q2-feature-classifier were used for classification of 16S rRNA gene sequences [27,28]. Data filtering and statistical analyses were carried out with R version 3.2.0 (R Core Team 2014). Vegan package was used to carry out permutational multivariate analysis of variance using distance matrices (vegdist and adonis functions, with 999 permutations) and perform detrended correspondence analyses (decorana function) with environmental fitting (envfit function) [29]. Sequence variants (SVs) classified as chloroplasts or mitochondria were discarded from the dataset. Demultiplexed raw data was submitted to the Sequence Read Archive under accession number PRJNA525456.
2.5. Preprocessing of the Dataset
A total of 15,698 sequence variants (SVs) of microbial eukaryotes and bacteria were obtained from the analyses of 14 water samples (Table 2). Amplicon sequencing of the 16S rRNA gene (V4 hypervariable region) was used to analyze bacterioplankton community composition (BCC) within the 0.22–5.0 µm size fraction. After quality control and the removal of mitochondrial and chloroplast ribosomal rRNA sequences, a total of 1,207,635 partial 16S rRNA gene sequences were utilized in this study (average of 83,890 sequences per sample, Table 2). All samples were rarefied to 50,000 sequences for statistical analyses. A total of 4755 prokaryotic sequence variants (SVs) were observed, which could be assigned to 562 genus-level taxa (prokaryotic genera, PG, as defined by classification level 6 in QIIME2), out of which 75 contributed more than 0.1% of the sequences in the entire dataset.
Table 2.
Total number of analyzed sequences, observed sequence variants (SV) and genera-level taxa (Qiime2 classifier, level 6), as well as Shannon diversity indices (H’) for 16S rRNA (bac.) and 18S rRNA (euk.) gene data.
| Station | Sequences (bac.) | SV (bac.) |
Genus-Level Taxa (bac.) | H’ (bac.) |
Sequences (euk.) | SV (euk.) |
Genus-Level Taxa (euk.) | H’ (euk.) |
|---|---|---|---|---|---|---|---|---|
| TS/Ph-1 | 80125 | 281 | 151 | 6.36 | 173981 | 1654 | 213 | 8.41 |
| TS/Ph-2 | 70198 | 1302 | 165 | 4.58 | 269629 | 1302 | 148 | 7.02 |
| TS/Ph-3 | 90170 | 1227 | 145 | 4.19 | 198954 | 1227 | 135 | 7.02 |
| TS/Ph-6 | 68729 | 752 | 87 | 4.75 | 305749 | 752 | 115 | 6.02 |
| TS/Ph-7 | 50720 | 1008 | 75 | 4.61 | 208940 | 1008 | 154 | 6.71 |
| TS/Ph-9 | 56338 | 488 | 50 | 4.51 | 77764 | 488 | 103 | 4.83 |
| TS/Ph-10 | 53636 | 1084 | 64 | 3.85 | 102902 | 281 | 52 | 5.50 |
| TS/Ph-11 | 170185 | 1654 | 128 | 5.82 | 135226 | 1084 | 146 | 6.81 |
| SRS-1d | 56789 | 1174 | 104 | 4.66 | 377682 | 1174 | 130 | 5.34 |
| SRS-2 | 58820 | 1029 | 86 | 4.42 | 183907 | 1029 | 141 | 6.42 |
| SRS-3 | 76305 | 1358 | 168 | 5.81 | 273752 | 1358 | 172 | 6.20 |
| SRS-4 | 97050 | 490 | 139 | 5.88 | 389092 | 490 | 95 | 6.65 |
| SRS-5 | 149420 | 1748 | 188 | 5.71 | 315255 | 1748 | 203 | 6.60 |
| SRS-6 | 129150 | 1252 | 133 | 4.44 | 188495 | 1252 | 188 | 6.77 |
The composition of eukaryotic communities was determined via 18S rRNA gene amplicon sequencing (>5 µm size fraction). After discarding all sequences classified as Bacteria and Metazoa, a total of 3,201,328 sequences (average 218,606 per sample, Table 2) was used for further analyses. A relatively large fraction of eukaryotic SVs (10,943) remained classified only as Eukaryota (11.0%). To improve these classifications, eukaryotic sequence variants were clustered into operational taxonomic units (OTUs, with 95% sequence similarity threshold), which lowered the fraction of unclassified eukaryotes to 5.4%. There were 55 OTUs (out of a total of 5020) that contributed to at least 1% of sequences in at least one of the samples.
3. Results
3.1. General Overview of the Datasets
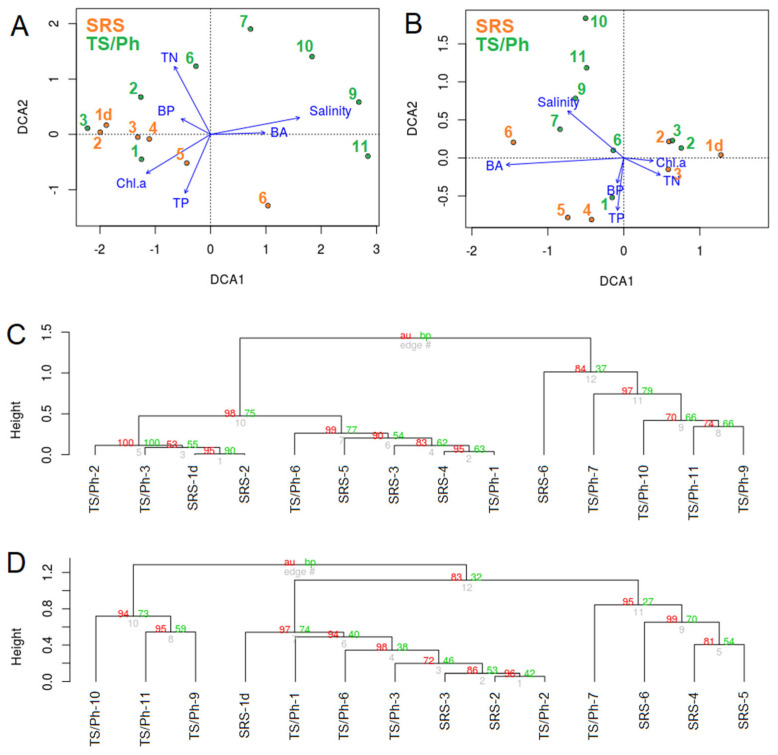
In both transects, bacterial and microbial eukaryotic communities were clearly different among freshwater, ecotone, and marine wetlands. Freshwater communities of both transects were similar to each other, while microbial communities in the more saline samples (ecotone, mangroves), and in Florida Bay, were distinctly different (Figure 2). The bacterial community composition (BCC) was most significantly correlated with salinity (R2 = 0.92; p < 0.001), concentrations of total nitrogen (TN; R2 = 0.66; p < 0.01), and chlorophyll a (Chl a, R2 = 0.63; p < 0.01; Figure 2). Permutational multivariate analysis using distance matrices was used to quantify the effects of environmental factors, transects (SRS, TS/Ph and FB) and ecosystem type (freshwater marshes, ecotone and marine). The ecosystem type better explained variability in BCC (R2 = 0.44; p < 0.001) than salinity (R2 = 0.10; p < 0.05), which was the only environmental variable with significant effect (Table S1). When the ecosystem types in different transects were split into separate groups, even more variability in BCC was explained (R2 = 0.58; p < 0.001).
Figure 2.
Detrended correspondence analyses of the prokaryotic (A) and eukaryotic (B) communities fitted with environmental vectors: bacterial abundance (BA), bacterial production (BP), total nitrogen (TN), total phosphorous (TP) and chlorophyll a (Chl a) (Table 1). Cluster dendrograms of the prokaryotic (C) and eukaryotic (D) communities based on sequence variant level composition (cluster method: average; distance: correlation; 1000 bootstrap replications), supplemented with approximately unbiased p-values (au, red), bootstrap probability values (bp, green), and edge numbers (grey).
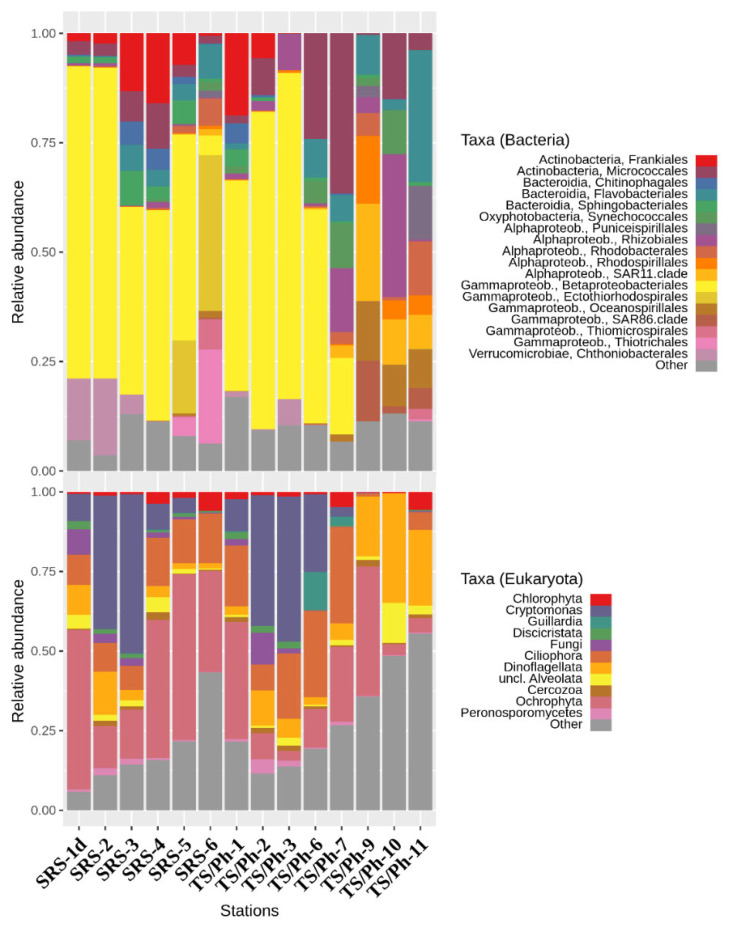
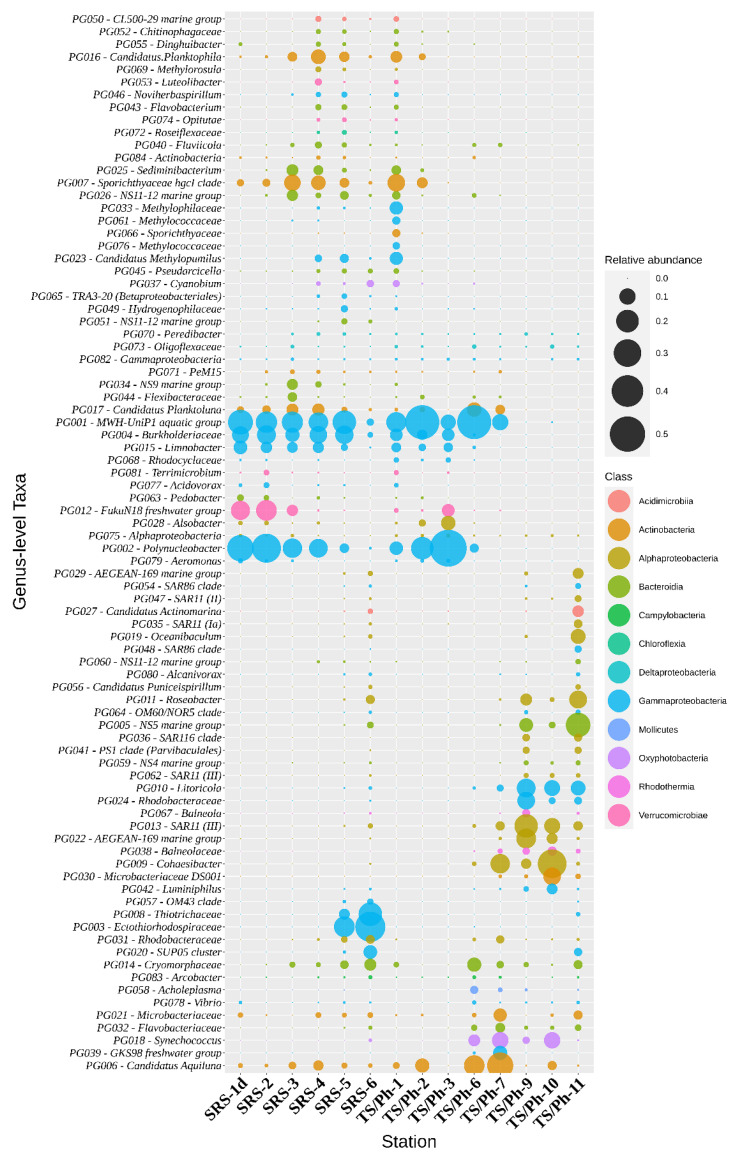
At a higher taxonomic level, BCC followed patterns documented for other estuarine and wetland ecosystems with a strong salinity gradient [30,31], with Betaproteobacteria and Actinobacteria dominating low salinity environments, and Alpha- and Gammaproteobacteria dominating coastal marine environments. Relative abundances of the Bacteroidetes phylum stayed relatively constant in both fresh and saltwater environments (Figure 3). Although the dataset contained 101 different class-level bacterial taxa, 94.4% of sequences could be assigned to just ten higher taxa: Betaproteobacteria (39.2%), Alphaproteobacteria (14.4%), Actinobacteria (13.6%), Gammaproteobacteria (10.3%), Flavobacteriia (5.7%), Sphingobacteriia (3.2%), Spartobacteria (3.2%), Cyanobacteria (2.3%), Proteobacteria Incertae Sedis (1.9%) and Acidimicrobiia (0.6%) (Figure 3). In total, 562 genus-level taxa were identified, out of which 75 contributed to more than 0.1% of all the sequences and can therefore be considered ‘common’ in the ecosystem (Figure 4).
Figure 3.
Bacterioplankton community composition on order-level taxa (top) and eukaryotic microbial community composition represented by variable higher taxa (bottom). In both cases, only taxa that contributed to more than 1% of the respective sequence datasets were included.
Figure 4.
Relative abundance of the bacterial tax comprising more than 0.1% of the bacterial sequences in each sample. Taxa are classified to genera (prokaryotic genera, PG, QIIME2 classifier level 6) or the lowest rank possible according to the classifier. The size of the bubbles represents their relative abundance, and the color indicates the class.
Like the BCC, the composition of the eukaryotic microbial communities (EMC) also varied along the physicochemical gradients (Figure 2). Permutational multivariate analysis demonstrated that in the case of the EMC the ecosystem type (R2 = 0.27; p < 0.001; Table S2) also outperformed salinity (R2 = 0.11; p < 0.05) and Chl a (R2 = 0.09; p < 0.05) which there were only two environmental variables with a significant effect. Differences between the transects were not significant. However, discriminating ecosystem types between transects constrained more variability than ecosystem types alone (R2 = 0.41; p < 0.005).
Most sequences that could be assigned to eukaryotic taxa included members of Ochrophyta (27.3%), Cryptophyta (15.0%), Ciliophora (14.3%), Dinoflagellata (13.2%), Chlorophyta (2.3%) and Fungi (2.3%) (Figure 3). Clear shifts in community composition were evident even for higher taxonomic levels in both BCC and EMC. Therefore freshwater, ecotone and marine estuary ecosystems are discussed separately below for a more comprehensive overview.
3.2. Freshwater Marsh Communities
Stations 1–3 in SRS and stations TS/Ph-1–2 are in freshwater marshes. TS/Ph-3 is heavily impacted by water flow (tides and freshwater input), had a very low salinity at the time of sampling (Table 1), and was thus clustered with the other freshwater sites. The lowest bacterial cell abundances were found in freshwater stations compared to the other stations, in both transects (in TS/Ph-1–3 around 1.2 × 106 cells L−1 and 1.0 × 106 up to 1.4 × 106 cells L−1 in SRS 1d–3; Figure 1). Bacterial secondary production (BP), a proxy for the integration of DOM into bacterial biomass, did not exhibit a clear trend and varied between the freshwater stations (Figure 1). In TS/Ph, BP rates decreased about five times from TS/Ph-1 (36.5 μg-C L−1 d−1) to TS/Ph-3, while the highest values for the entire dataset were observed in SRS3 (95.8 μg-C L−1 d−1) (Figure 1).
Most of the 16S rRNA gene sequences in the freshwater samples were assigned to the orders Betaproteobacteriales (63.4%), Chthoniobacterales (Verrucomicrobiae, 7.3%), Frankiales (Actinobacteria, 7.0%), Rhizobiales (Alphaproteobacteria, 4.4%) and Micrococcales (Actinobacteria, 3.9%) (Figure 3). At the genus level (Qiime2 classifier, level 6), core taxa (mean relative abundance over 1% in all marsh sites) were Polynucleobacter (26.1%), MWH-UniP1 aquatic group (21.7%, Burkholderiales), FukuN18 freshwater group (7.0%, Verrucomicrobia), hgcI clade (5.0%, Sporichthyaceae), Limnobacter (4.0%, Burkholderiaceae), Candidatus Aquilina (2.0%, Microbacteriaceae), Alsobacter (2.0%, Rhizobiales), NS11-12 marine group (1.9%, Sphingobacteriales), Sediminibacterium (1.4%, Chitinophagaceae), Candidatus Planktoluna (1.7%, Microbacteriaceae), unclassified Methylophilaceae (1.1%), and Candidatus Methylopumilus (1.1%, Methylophilaceae) (Figure 4).
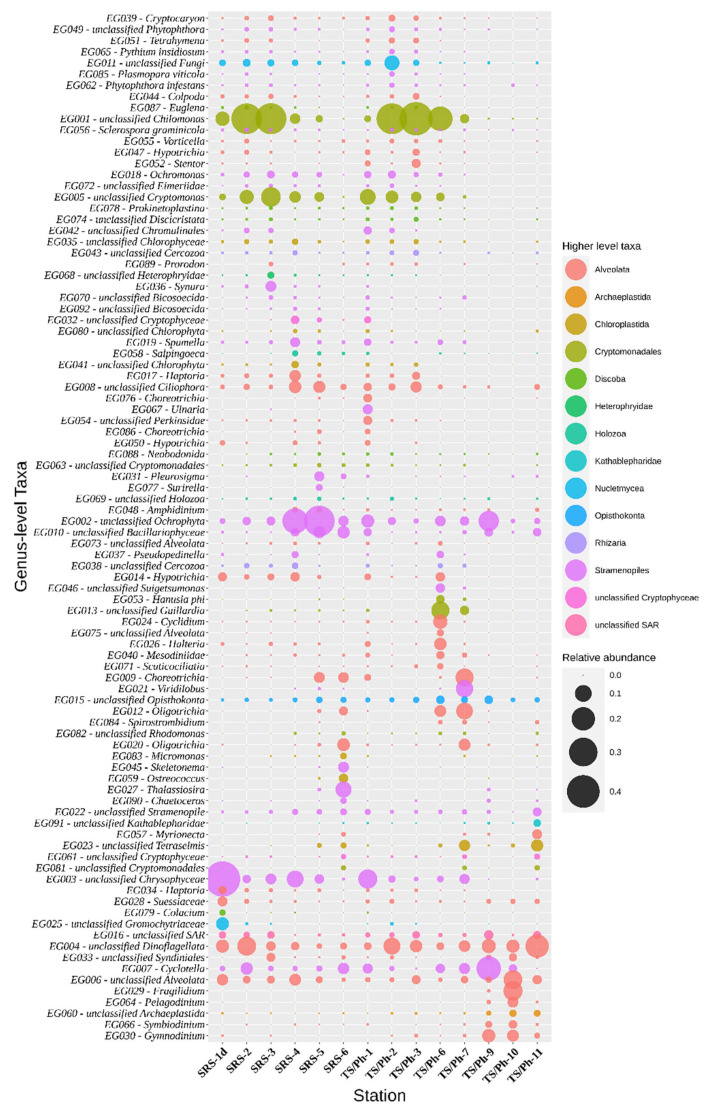
The freshwater EMC were dominated by Chryptophyta/Cryptomonads (on average 29.3%), followed by Ochrophyta (21.5%), Ciliophora (12.4%), Dinoflagellata (6.3%), Fungi (4.6%) and Chlorophyta (1.2%) (Figure 3). In both transects, cryptomonads dominated the P-limited freshwater areas, yet their relative abundance decreased towards the mangrove ecotone, and they were nearly absent in marine samples. The most abundant OTUs were assigned to Chilomonas (OTU1, average 26.2%), and Cryptomonas (OTU8, 2.7% and OTU14, 2.4%) (Figure 5).
Figure 5.
Relative abundance of eukaryotic taxa comprising more than 0.1% of the bacterial sequences in each sample. Operational taxonomic units (OTU) are classified to the lowest rank possible according to the classifier. The size of the bubbles represents their relative abundance, and the color indicates the higher-level classification, as depicted on the right. Percentages in parentheses are sequence similarity values to the listed taxon.
3.3. Ecotone
Stations 4–5 in SRS are situated within mangrove forests and TS/Ph-6 and TS/Ph-7 are located in an estuarine ecotone with mangrove islands [32]. The highest bacterial abundances in our datasets were found in ecotone stations bordering marine estuaries, reaching 3.9 × 106 cells L−1 in TS/Ph-7 (Table 2). In the TS/Ph transect, BP peaked in TS/Ph-6 (71.9 μg C L−1 d−1) and decreased nearly 20-fold towards the estuary. A similar trend was observed in SRS, where rates dropped from 42.9 to 22.6 μg C L−1 d−1 within the ecotone (Figure 1).
The major differences in the BCC of the ecotones compared to the freshwater stations were decreases of the relative abundances of Betaproteobacteria (to 33.4%), Sphingobacteriia (to 3.6%) and Spartobacteria (to 0.1%), and concomitant increase in the relative abundances of Actinobacteria (to 20.6%), Gammaproteobacteria (to 13.6%), Alphaproteobacteria (to 7.7%), Proteobacteria Incertae Sedis (to 5.2%) and Cyanobacteria (to 3.6%). In parallel, increases in the relative abundances of eukaryotic taxa were found among Ochrophyta (to 34.6%) and Ciliophora (to 21.7%). Significant differences were also detected among both the BMC and EMC of the two distinct ecotones. Members of Micrococcales (Actinobacteria) became the second most abundant prokaryotic group in the TS/Ph ecotone (30.4%), while contributing only to 4.9% in waters of the SRS ecotone. Similarly, Synechococcales and Rhizobiales were found to be more abundant in the ecotone of Taylor Slough (on average 7.7% and 7.6%, respectively), while they both contributed to less than 1% in the Shark River Slough ecotone. A large fraction of BCC at the SRS ecotone were identified as Ectothiorhodospirales (17.4%), Thiotrichales (8.6%) and Frankiales (7.9%), while these three orders were absent from the TS/Ph- ecotone. The relative abundance of Flavobacterales were similar between the ecotones, contributing 5.1% at TS/Ph and 7.4% at SRS.
Within EMCs, the relative abundance of Ochrophyta increased to over 40% at the SRS ecotone (59.8% in SRS5) and more gradually from TS/Ph-6 to TS/Ph-7 (13.0–27.4%). The fraction of ciliates (Ciliophora) also increased in the ecotone to an average of 17.3% at SRS and 32.4% at TS/Ph. The average relative abundance of cryptomonads decreased within ecotones of both transects (Figure 3). Compared to TS/Ph, EMC at SRS contained a larger fraction of diatoms, mainly OTU3 (Urosolenia; 21.3%), OTU17 (Thalassiosira; 4.5%) and OTU16 (Pleurosigma; 3.5%). Heterotrophic protists were more prominent in TS/Ph, especially at TS/Ph-7 (OTU7, Pelagostrobilidium, 14.0%; OTU10, Strombidium, 11.9%; and raphidophyte OTU18, Viridilobus, 12.9%).
3.4. Florida Bay
In FB, the BCC was dominated by Flavobacteriales (13.8%), SAR11 clade (13.4%), Rhizobiales (12.3%), Oceanospirillales (10.7%), Rhodospirillales (8.1%), SAR86 clade (6.8%), Micrococcales (6.4%), Rhodobacterales (6.1%), Puniceispirillales (4.9%) and Synechococcales (4.0%). While organic carbon concentrations were very low in the FB sites, the highest concentration of TOC was found at the central part of the bay, TS/Ph-10 (Table 1), accompanied by the highest bacterial cell counts and BP rates within FB (2.8 × 106 cells L−1; 13.8 μg C L−1 d−1), surpassing the corresponding values of TS/Ph-9 (1.5 × 106 cells L−1; 4.0 μg C L−1 d−1). Ochrophyta (average 28.5%, mostly diatoms), Dinoflagellata (16.2%), Ciliophora (8.8%) and unclassified Opisthokonta (3.5%) were the most abundant eukaryotic microbial taxa in these waters. The northeastern part of the bay (TS/Ph-9) had higher inorganic nutrient concentrations (Table 1) and autotrophic diatoms dominated the EMC; Cyclotella (OTU4, 27.4%) and Chaetoceros (OTU11, 17.8%) were the most abundant genus-level taxa.
The BCC at this location was dominated by Cohaesibacter (32.6%) that presumably can decompose casein, cellulose, xanthine and hypoxanthine [33]. In addition, other coastal heterotrophs, like Microbacteriaceae DS001 (11.9%) and Litoricola (9.5%) and unicellular picocyanobacteria of the genus Synechococcus (10.1%), which have been reported to form blooms in FB [34], were abundant.
The accompanying EMC were dominated by dinoflagellates (Figure 3): Fragilidium (OTU9, 22.3%) Pyrophacus (OTU12, 15.1%), Prorocentrum (OTU2, 14.2%) and Pelagodinium (OTU20, 9.1%). Members of Fragilidium are mixotrophic dinoflagellates and also known to prey upon red-tide and toxic dinoflagellates, including Prorocentrum species [35].
4. Discussion
4.1. Freshwater Marsh Communities
Long-term monitoring data indicated that abrupt and sustained increases in TP and DOC from marine storm surges and severe low-temperature events increase bacterioplankton productivity for extended periods, and that these responses are more pronounced in SRS than in the TS/Ph transect [36]. Despite these striking differences in BP, the composition of the freshwater microbial communities was not significantly different between the two transects.
The abundance of Betaproteobacteria in numerous freshwater habitats is heavily affected by salinity [37,38,39,40]. A similar trend is apparent in this study as well. The most abundant Betaproteobacteria were composed of two genus-level taxa: Polynucleobacter (PG002, 26.0%) and MWH-UniP1 aquatic group (PG001, on average 21.6%). Both taxa belong to the order Burkholderiales, together with less abundant PG016 (unclassified) and PG050 (Limnobacter), which made up 3.9% and 0.4% of all bacterial sequences in the freshwater marshes, respectively.
Polynucleobacter-related sequences were divided between two free-living species P. cosmopolitanus and P. asymbioticus, which are ubiquitous and frequently abundant members of freshwater bacterioplankton [41,42,43] in habitats that vary in chemical and climatic conditions [44,45,46]. As 16S rRNA data does not provide sufficient resolution for the identification of Polynucleobacter species [47], the composition and ecological function of these highly abundant organisms in the marshes of the Everglades warrants further detailed studies. Cultured strains of P. asymbioticus originate mainly from humic-rich habitats, where they can utilize products of photodegradation of humic substances [42,48]. The high relative abundance of Polynucleobacter in the Everglades watershed is likely explained by the prevailing high concentrations of humic substances, which compose about 50% of the DOC in this environment and are in part mineralized by solar radiation [49].
Verrucomicrobia, which were mostly represented by the order Chtioniobacterales in the class Spartobacteria, had a relative abundance of 7.3% in the ENP freshwater marshes (Figure 3). These bacteria are present in a large variety of terrestrial and aquatic ecosystems and are also a dominant group in many humic lakes, composing up to 19% of the respective BCCs [50,51,52]. Most Verrucomicrobia are specialized in the degradation of algal polymers, specifically polysaccharides, such as cellulose and chitin [53]. Therefore, their occurrence is usually correlated with the biomass dynamics of phytoplankton, including Chrysophyceae [54], a group that was also present in the freshwater stations in ENP. There were many similarities between the BCC of the ENP transects and that of the Brazos and Mississippi Rivers that also flow into GoM, including high relative abundances of Limnohabitans, Polynucleobacter, acI clade, LD28 and others [19,55,56].
Cryptophyta have mostly been considered autotrophic, but exceptions were reported for cryptophytes in ice-covered lakes in Antarctica, where mixotrophic behavior for survival under light-limited conditions was observed during winter [57,58]. Williams and Trexler [59] demonstrated the importance of ‘detrital’ carbon flow in the Everglades, indicating that the microbial loop provides a major route of energy flow to higher trophic levels. The most abundant group of cryptophytes in the marshes, Chilomonas, are heterotrophs that can feed on detritus in the form of particulate organic matter [60] and could therefore be a key taxon in the carbon cycle of the Everglades.
Fungi are considered important heterotrophic degraders in periphytic communities [15,61] and could contribute to same processes in the water column. The highest relative abundances of Fungi were observed in the freshwater marshes (8.3% in SRS1d and 10.6% at TS/Ph-2). The most abundant fungal OTU (OTU29) could only be classified as a member of the division Glomeromycota, which includes all species involved in arbuscular mycorrhizal symbioses. We cannot exclude the possibility that these sequences come from spores rather than metabolically active cells.
4.2. Ecotone
Ecotones are transitions between ecosystems that are characterized by steep environmental gradients [62]. They are critical in regulating the transport of DOM and nutrients into coastal waters [63,64,65]. The hydrographic changes in the Everglades over the last 100 years have had the most impact on its ecotones [66].
Mangrove plant roots excrete organic compounds and release oxygen to the rhizosphere, thus changing the chemical characteristics in the sediment area around the roots [67,68,69]. Nevertheless, mangrove sediments are primarily anaerobic with a thin aerobic sediment layer on top [69,70], thus providing chemical characteristics which encourage anoxygenic photolithotrophic and chemolithotrophic sulfur-oxidizing bacteria, which in this study were also found in the water column of the SRS ecotone (e.g., Ectothiorhodospirales, Thiotrichaceae and SUP05 cluster). Our results indicate that anaerobic processes in the rhizosphere have a high impact on microbial communities in overlaying water column.
Diatoms showed increased diversity and relative abundance towards the ecotone in our dataset, most notably along the SRS transect (Figure 5). Concomitant with the increase of relative abundance in diatoms, an increase in the abundances of bacterial taxa that have been shown to be associated with diatom blooms, like Flavobacteriales and Rhodobacterales [71] was also observed (Figure 4). Representatives of Flavobacteriales are well-known degraders and consumers of high-molecular-mass organic matter [53,72,73], and Rhodobacterales are known to utilize exopolymer particles [74] and could therefore play an important role in the degradation of these compounds in the ecotones in the Everglades.
4.3. Florida Bay
FB is a large and shallow estuary, with an average depth of only 1.5 m [75]. In our dataset, the samples from the FB are represented by stations TS/Ph-9, TS/Ph-10 and TS/Ph-11 (Figure 1). The bay receives freshwater runoff from the Everglades marsh mainly through the C-111 Canal and Taylor Slough. Its west side opens to the GoM, the main source of phosphorous for FB, resulting in a phosphorous gradient between the eastern and western parts of FB [76]. Drainage canals are a major source of contamination to the local reef environments, and the nutrients that leak into the estuaries in South Florida lead to occasional/regular algal blooms. Florida Bay waters have been divided into six segments based on their biogeochemical characteristics [77,78]. Our data are in line with some of these general features, as the northern part of the bay (TS/Ph-9) has higher nutrient concentrations, and the central part of the bay (TS/Ph-10) has higher Chl a level (Table 1). The highest bacterial cell counts and BP within the FB were also observed in TS/Ph-10 (2.8 × 106 cells L−1; 13.8 μg-C L−1 d−1), surpassing the corresponding values of TS/Ph-9 (1.5 × 106 cells L−1; 4.0 μg-C L−1 d−1).
The highly variable abundance of certain taxa within the FB sites indicates that a higher spatiotemporal resolution is needed to accurately describe the composition of microbial plankton communities in these habitats. The northeastern part of the bay (TS/Ph-9) had higher inorganic nutrient concentrations (Table 1) and autotrophic diatoms dominated the EMC; Cyclotella (OTU4, 27.4%) and Chaetoceros (OTU11, 17.8%) were the most abundant genus-level taxa. The accompanying BCC contained a large fraction of SAR11 clade (22.2%) and Rhodospirillales (15.5%) that specialize in the active uptake of low-molecular weight monomers during diatom dominated phytoplankton blooms [71,79]. Flavobacteriales made up 9% of BCC at TS/Ph-9 and might be key players in the initial degradation of organic matter derived from the observed algae [53,72,73]. The closest station to the Gulf of Mexico, TS/Ph-11, exhibited a high abundance of the NS5 marine group (Flavobacteriaceae, 23.7%), Roseobacter (12.2%) and Oceanibaculum (SAR116 clade, 8.3%), which are also abundant in northern parts of the Gulf of Mexico [19,80].
5. Conclusions
Pelagic microbial communities differed significantly in habitats along the salinity gradient in the Florida Coastal Everglades. In the freshwater marshes, detrital ‘brown’ carbon flow is essential for the food web and, accordingly, the most abundant organisms were heterotrophs that are presumably capable of degradation of complex organic carbon. In these habitats, solar radiation generates dissolved organic matter via photo-dissolution of flocculent, detrital material and terrestrial humic-like components that can contribute up to 70% of the chromophoric DOM [81]. Polynucleobacter, the most abundant prokaryotic group that we detected in the marsh samples, have been shown to utilize products of photodegradation humic substances—a capability that might explain their abundance in this ecosystem. Potentially phagotrophic cryptomonads of the genus Chilomonas were identified as predominant eukaryotic microorganisms, indicating that they could hold a key position in this ecosystem by transferring detritus-based biomass to higher trophic levels. In wetland ecotones, oxygen production and excretion of organic matter by mangrove roots, as well as the increased concentration of sulfate from marine waters, create niches for aerobic as well as anaerobic microbial communities, even in the water column. We identified photolithotrophic and chemolithotrophic sulphur-oxidizing bacteria (Ectothiorhodospirales and Thiotrichales) as predominant members of BCC at the SRS ecotone. Similarly, marine microbial communities at the three sampling sites in Florida Bay were heterogeneous, indicating the presence of (micro)niches and the need of higher resolution of sampling sites. Further data is necessary to pinpoint these trends and to analyze the potential seasonality of microbial communities in these systems. Nevertheless, the datasets presented here will provide valuable baseline information for environmental monitoring in these habitats.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/microorganisms10020215/s1, Table S1. Quantitative effects of environmental parameters, ecosystem type (freshwater marshes, ecotone, marine) and transect (SRS, TS/Ph, FB+) on the bacterial community variation by permutational multivariate analysis of variance (using ‘adonis’ function of vegan package) based on weighted UniFrac distance. R2 values present the proportion of variation constrained by factors. Table S2. Quantitative effects of environmental parameters, ecosystem type (freshwater marshes, ecotone, marine) and transect (SRS, TS/Ph, FB+) on the eukaryotic microbial community variation by permutational multivariate analysis of variance (using ‘adonis’ function of vegan package) based on weighted UniFrac distance. R2 values present the proportion of variation constrained by factors.
Author Contributions
Conceptualization, U.S., P.L., E.E.G. and J.S.K.; formal analysis, P.L. and S.S.; resources, K.U., R.T. and S.S.; writing—original draft preparation, P.L.; writing—review and editing, P.L., E.E.G., J.S.K. and U.S.; project administration, E.E.G., J.S.K. and U.S.; funding acquisition, E.E.G., J.S.K. and U.S. All authors have read and agreed to the published version of the manuscript.
Funding
This material is based upon work supported by the National Science Foundation through the Florida Coastal Everglades Long Term Ecological Research program under Cooperative Agreements #DEB-2025954, #DEB-1832229, and #DEB-1237517.Additional support was provided by USDA/NIFA [HATCH project FLA-FTL-005631 to US]. EEG is supported by the George M. Barley Jr. Endowment from the Institute of Environment at Florida International University, where this is contribution #1398.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Demultiplexed raw data was submitted to the Sequence Read Archive under accession number PRJNA525456.
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Costanza R., d’Arge R., de Groot R., Farber S., Grasso M., Hannon B., Limburg K., Naeem S., ONeill R., Paruelo J., et al. The value of the world’s ecosystem services and natural capital. Nature. 1997;387:253–260. doi: 10.1038/387253a0. [DOI] [Google Scholar]
- 2.de Groot R., Brander L., van der Ploeg S., Costanza R., Bernard F., Braat L., Christie M., Crossman N., Ghermandi A., Hein L., et al. Global estimates of the value of ecosystems and their services in monetary units. Ecosyst. Serv. 2012;1:50–61. doi: 10.1016/j.ecoser.2012.07.005. [DOI] [Google Scholar]
- 3.Davidson N.C. How much wetland has the world lost? Long-term and recent trends in global wetland area. Mar. Freshw. Res. 2014;65:934–941. doi: 10.1071/MF14173. [DOI] [Google Scholar]
- 4.Sklar F., McVoy C., Van Zee R., Gawlik D., Swift D., Park W., Fitz C., Wu Y., Rudnick D., Fontaine T., et al. Everglades Interim Report. SFWMD; West Palm Beach, FL, USA: 1999. Hydrologic needs: The effects of altered hydrology on the everglades. [Google Scholar]
- 5.McVoy C., Park Said W., Obeysekera J., VanArman J.A., Dreschel T.W. Landscapes and Hydrology of the Predrainage Everglades. University Press of Florida; Gainesville, FL, USA: 2011. [Google Scholar]
- 6.Troxler T., Gaiser E., Barr J., Fuentes J., Jaffe R., Childers D., Collado-Vides L., Rivera-Monroy V., Castaneda-Moya E., Anderson W., et al. Integrated carbon budget models for the everglades terrestrial-coastal-oceanic gradient current status and needs for inter-site comparisons. Oceanography. 2013;26:98–107. doi: 10.5670/oceanog.2013.51. [DOI] [Google Scholar]
- 7.Titus J., Richman C. Maps of lands vulnerable to sea level rise: Modeled elevations along the US Atlantic and Gulf coasts. Clim. Res. 2001;18:205–228. doi: 10.3354/cr018205. [DOI] [Google Scholar]
- 8.Church J., Clark P., Cazenave A., Gregory J., Jevrejeva S., Levermann A., Merrifield M., Milne G., Nerem R., Nunn P., et al. Sea-level rise by 2100. Science. 2013;342:1445. doi: 10.1126/science.342.6165.1445-a. [DOI] [PubMed] [Google Scholar]
- 9.Haigh I., Wahl T., Rohling E., Price R., Pattiaratchi C., Calafat F., Dangendorf S. Timescales for detecting a significant acceleration in sea level rise. Nat. Commun. 2014;5:3635. doi: 10.1038/ncomms4635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Flower H., Rains M., Fitz C. Visioning the future: Scenarios modeling of the florida coastal everglades. Environ. Manag. 2017;60:989–1009. doi: 10.1007/s00267-017-0916-2. [DOI] [PubMed] [Google Scholar]
- 11.Yao Q., Liu K. Dynamics of marsh-mangrove ecotone since the mid-holocene: A palynological study of mangrove encroachment and sea level rise in the shark river estuary, Florida. PLoS ONE. 2017;12:e0173670. doi: 10.1371/journal.pone.0173670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stevenson J. Ecological assessments with algae: A review and synthesis. J. Phycol. 2014;50:437–461. doi: 10.1111/jpy.12189. [DOI] [PubMed] [Google Scholar]
- 13.Lane C., Brown M. Diatoms as indicators of isolated herbaceous wetland condition in Florida, USA. Ecol. Indic. 2007;7:521–540. doi: 10.1016/j.ecolind.2006.06.001. [DOI] [Google Scholar]
- 14.Gaiser E. Periphyton as an indicator of restoration in the Florida everglades. Ecol. Indic. 2009;9:S37–S45. doi: 10.1016/j.ecolind.2008.08.004. [DOI] [Google Scholar]
- 15.Gaiser E., McCormick P., Hagerthey S., Gottlieb A. Landscape patterns of periphyton in the Florida Everglades. Crit. Rev. Environ. Sci. Technol. 2011;41:92–120. doi: 10.1080/10643389.2010.531192. [DOI] [Google Scholar]
- 16.Caruso G., Azzaro M., Caroppo C., Decembrini F., Monticelli L., Leonardi M., Maimone G., Zaccone R., La Ferla R. Microbial community and its potential as descriptor of environmental status. ICES J. Mar. Sci. 2016;73:2174–2177. doi: 10.1093/icesjms/fsw101. [DOI] [Google Scholar]
- 17.Azam F., Malfatti F. Microbial structuring of marine ecosystems. Nat. Rev. Microbiol. 2007;5:782–791. doi: 10.1038/nrmicro1747. [DOI] [PubMed] [Google Scholar]
- 18.Gasol J., Pinhassi J., Alonso-Saez L., Ducklow H., Herndl G., Koblizek M., Labrenz M., Luo Y., Moran X., Reinthaler T., et al. Towards a better understanding of microbial carbon flux in the sea. Aquat. Microb. Ecol. 2008;53:21–38. doi: 10.3354/ame01230. [DOI] [Google Scholar]
- 19.Henson M., Hanssen J., Spooner G., Fleming P., Pukonen M., Stahr F., Thrash J. Nutrient dynamics and stream order influence microbial community patterns along a 2914 kilometer transect of the Mississippi river. Limnol. Oceanogr. 2018;63:1837–1855. doi: 10.1002/lno.10811. [DOI] [Google Scholar]
- 20.Markussen T., Happel E., Teikari J., Huchaiah V., Alneberg J., Andersson A., Sivonen K., Riemann L., Middelboe M., Kisand V. Coupling biogeochemical process rates and metagenomic blueprints of coastal bacterial assemblages in the context of environmental change. Environ. Microbiol. 2018;20:3083–3099. doi: 10.1111/1462-2920.14371. [DOI] [PubMed] [Google Scholar]
- 21.Ikenaga M., Guevara R., Dean A.L., Pisani C., Boyer J.N. Changes in community structure of sediment bacteria along the florida coastal everglades marsh-mangrove-seagrass salinity gradient. Microb. Ecol. 2010;59:284–295. doi: 10.1007/s00248-009-9572-2. [DOI] [PubMed] [Google Scholar]
- 22.Lebaron P., Servais P., Agogue H., Courties C., Joux F. Does the high nucleic acid content of individual bacterial cells allow us to discriminate between active cells and inactive cells in aquatic systems? Appl. Environ. Microbiol. 2001;67:1775–1782. doi: 10.1128/AEM.67.4.1775-1782.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Caporaso J., Lauber C., Walters W., Berg-Lyons D., Huntley J., Fierer N., Owens S., Betley J., Fraser L., Bauer M., et al. Ultra-high-throughput microbial community analysis on the illumina Hiseq and Miseq platforms. ISME J. 2012;6:1621–1624. doi: 10.1038/ismej.2012.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Amaral-Zettler L., McCliment E., Ducklow H., Huse S. A method for studying protistan diversity using massively parallel sequencing of v9 hypervariable regions of small-subunit ribosomal RNA genes. PLoS ONE. 2009;4:e6372. doi: 10.1371/annotation/50c43133-0df5-4b8b-8975-8cc37d4f2f26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bolyen E., Rideout J., Dillon M., Bokulich N., Abnet C., Al-Ghalith G., Alexander H., Alm E., Arumugam M., Asnicar F., et al. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 2019;37:852–857. doi: 10.1038/s41587-019-0209-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Callahan B., McMurdie P., Rosen M., Han A., Johnson A., Holmes S. DADA2: High-resolution sample inference from illumina amplicon data. Nat. Methods. 2016;13:581–583. doi: 10.1038/nmeth.3869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Quast C., Pruesse E., Yilmaz P., Gerken J., Schweer T., Yarza P., Peplies J., Glockner F. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 2013;41:D590–D596. doi: 10.1093/nar/gks1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bokulich N., Kaehler B., Rideout J., Dillon M., Bolyen E., Knight R., Huttley G., Caporaso J. Optimizing taxonomic classification of marker-gene amplicon sequences with QIIME 2′s q2-feature-classifier plugin. Microbiome. 2018;6:90. doi: 10.1186/s40168-018-0470-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Oksanen J., Blanchet F.G., Friendly M., Kindt R., Legendre P., McGlinn D., Minchin P.R., O’Hara R., Simpson G., Solymos P. Vegan: Community Ecology Package. R Package Version 2.5-6. 2019. 2020. [(accessed on 14 December 2021)]. Available online: https://cran.r-project.org/web/packages/vegan/vegan.pdf.
- 30.Herlemann D.P., Woelk J., Labrenz M., Jürgens K. Diversity and abundance of “Pelagibacterales” (SAR11) in the Baltic Sea salinity gradient. Syst. Appl. Microbiol. 2014;37:601–604. doi: 10.1016/j.syapm.2014.09.002. [DOI] [PubMed] [Google Scholar]
- 31.Fortunato C.S., Eiler A., Herfort L., Needoba J.A., Peterson T.D., Crump B.C. Determining indicator taxa across spatial and seasonal gradients in the columbia river coastal margin. ISME J. 2013;7:1899–1911. doi: 10.1038/ismej.2013.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ewe S., Gaiser E., Childers D., Iwaniec D., Rivera-Monroy V., Twilley R. Spatial and temporal patterns of aboveground net primary productivity (anpp) along two freshwater-estuarine transects in the florida coastal everglades. Hydrobiologia. 2006;569:459–474. doi: 10.1007/s10750-006-0149-5. [DOI] [Google Scholar]
- 33.Hwang C., Cho B. Cohaesibacter gelatinilyticus gen. Nov., sp nov., a marine bacterium that forms a distinct branch in the order Rhizobiales, and proposal of Cohaesibacteraceae fam. Nov. Int. J. Syst. Evol. Microbiol. 2008;58:267–277. doi: 10.1099/ijs.0.65016-0. [DOI] [PubMed] [Google Scholar]
- 34.Boyer J., Dailey S., Gibson P., Rogers M., Mir-Gonzalez D. The role of dissolved organic matter bioavailability in promoting phytoplankton blooms in Florida bay. Hydrobiologia. 2006;569:71–85. doi: 10.1007/s10750-006-0123-2. [DOI] [Google Scholar]
- 35.Jeong H. The ecological roles of heterotrophic dinoflagellates in marine planktonic community. J. Eukaryot. Microbiol. 1999;46:390–396. doi: 10.1111/j.1550-7408.1999.tb04618.x. [DOI] [Google Scholar]
- 36.Kominoski J., Gaiser E., Castaneda-Moya E., Davis S., Dessu S., Julian P., Lee D., Marazzi L., Rivera-Monroy V., Sola A., et al. Disturbance legacies increase and synchronize nutrient concentrations and bacterial productivity in coastal ecosystems. Ecology. 2020;101:e02988. doi: 10.1002/ecy.2988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zwart G., van Hannen E.J., Kamst-van Agterveld M.P., Van der Gucht K., Lindström E.S., Van Wichelen J., Lauridsen T., Crump B.C., Han S.K., Declerck S. Rapid screening for freshwater bacterial groups by using reverse line blot hybridization. Appl. Environ. Microbiol. 2003;69:5875–5883. doi: 10.1128/AEM.69.10.5875-5883.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zwisler W., Selje N., Simon M. Seasonal patterns of the bacterioplankton community composition in a large mesotrophic lake. Aquat. Microb. Ecol. 2003;31:211–225. doi: 10.3354/ame031211. [DOI] [Google Scholar]
- 39.Buck U., Grossart H., Amann R., Pernthaler J. Substrate incorporation patterns of bacterioplankton populations in stratified and mixed waters of a Humic Lake. Environ. Microbiol. 2009;11:1854–1865. doi: 10.1111/j.1462-2920.2009.01910.x. [DOI] [PubMed] [Google Scholar]
- 40.Wu Q., Zwart G., Schauer M., Kamst-van Agterveld M., Hahn M. Bacterioplankton community composition along a salinity gradient of sixteen high-mountain lakes located on the Tibetan plateau, China. Appl. Environ. Microbiol. 2006;72:5478–5485. doi: 10.1128/AEM.00767-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Percent S.F., Frischer M.E., Vescio P.A., Duffy E.B., Milano V., McLellan M., Stevens B.M., Boylen C.W., Nierzwicki-Bauer S.A. Bacterial community structure of acid-impacted lakes: What controls diversity? Appl. Environ. Microbiol. 2008;74:1856–1868. doi: 10.1128/AEM.01719-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jezberova J., Jezbera J., Brandt U., Lindstrom E., Langenheder S., Hahn M. Ubiquity of Polynucleobacter necessarius ssp. asymbioticus in lentic freshwater habitats of a heterogenous 2000 km2 area. Environ. Microbiol. 2010;12:3302. doi: 10.1111/j.1462-2920.2009.02106.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Newton R., Jones S., Eiler A., McMahon K., Bertilsson S. A guide to the natural history of freshwater lake bacteria. Microbiol. Mol. Biol. Rev. 2011;75:14–49. doi: 10.1128/MMBR.00028-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wu Q., Hahn M. High predictability of the seasonal dynamics of a species-like Polynucleobacter population in a freshwater lake. Environ. Microbiol. 2006;8:1660–1666. doi: 10.1111/j.1462-2920.2006.01049.x. [DOI] [PubMed] [Google Scholar]
- 45.Ghai R., Rodriguez-Valera F., McMahon K.D., Toyama D., Rinke R., Cristina Souza de Oliveira T., Wagner Garcia J., Pellon de Miranda F., Henrique-Silva F. Metagenomics of the water column in the pristine upper course of the amazon river. PLoS ONE. 2011;6:e23785. doi: 10.1371/journal.pone.0023785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hahn M., Koll U., Jezberova J., Camacho A. Global phylogeography of pelagic polynucleobacter bacteria: Restricted geographic distribution of subgroups, isolation by distance and influence of climate. Environ. Microbiol. 2015;17:829–840. doi: 10.1111/1462-2920.12532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hahn M., Scheuerl T., Jezberova J., Koll U., Jezbera J., Simek K., Vannini C., Petroni G., Wu Q. The passive yet successful way of planktonic life: Genomic and experimental analysis of the ecology of a free-living Polynucleobacter population. PLoS ONE. 2012;7:e32772. doi: 10.1371/journal.pone.0032772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hoetzinger M., Schmidt J., Jezberova J., Koll U., Hahn M. Microdiversification of a pelagic Polynucleobacter species is mainly driven by acquisition of genomic islands from a partially interspecific gene pool. Appl. Environ. Microbiol. 2017;83:e02266-16. doi: 10.1128/AEM.02266-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Qualls R., Richardson C. Factors controlling concentration, export, and decomposition of dissolved organic nutrients in the everglades of Florida. Biogeochemistry. 2003;62:197–229. doi: 10.1023/A:1021150503664. [DOI] [Google Scholar]
- 50.Kolmonen E., Sivonen K., Rapala J., Haukka K. Diversity of cyanobacteria and heterotrophic bacteria in cyanobacterial blooms in lake joutikas, finland. Aquat. Microb. Ecol. 2004;36:201–211. doi: 10.3354/ame036201. [DOI] [Google Scholar]
- 51.Kolmonen E., Haukka K., Rantala-Ylinen A., Rajaniemi-Wacklin P., Lepisto L., Sivonen K. Bacterioplankton community composition in 67 finnish lakes differs according to trophic status. Aquat. Microb. Ecol. 2011;62:241–250. doi: 10.3354/ame01461. [DOI] [Google Scholar]
- 52.Arnds J., Knittel K., Buck U., Winkel M., Amann R. Development of a 16S rRNA-targeted probe set for Verrucomicrobia and its application for fluorescence in situ hybridization in a humic lake. Syst. Appl. Microbiol. 2010;33:139–148. doi: 10.1016/j.syapm.2009.12.005. [DOI] [PubMed] [Google Scholar]
- 53.Hugerth L., Larsson J., Alneberg J., Lindh M., Legrand C., Pinhassi J., Andersson A. Metagenome-assembled genomes uncover a global brackish microbiome. Genome Biol. 2015;16:279. doi: 10.1186/s13059-015-0834-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Parveen B., Mary I., Vellet A., Ravet V., Debroas D. Temporal dynamics and phylogenetic diversity of free-living and particle-associated verrucomicrobia communities in relation to environmental variables in a mesotrophic lake. FEMS Microbiol. Ecol. 2013;83:189–201. doi: 10.1111/j.1574-6941.2012.01469.x. [DOI] [PubMed] [Google Scholar]
- 55.Staley C., Unno T., Gould T., Jarvis B., Phillips J., Cotner J., Sadowsky M. Application of illumina next-generation sequencing to characterize the bacterial community of the upper Mississippi river. J. Appl. Microbiol. 2013;115:1147–1158. doi: 10.1111/jam.12323. [DOI] [PubMed] [Google Scholar]
- 56.Becker J., Rodibaugh K., Hahn D., Nowlin W. Bacterial community composition and carbon metabolism in a subtropical riverscape. Hydrobiologia. 2017;792:209–226. doi: 10.1007/s10750-016-3058-2. [DOI] [Google Scholar]
- 57.Marshall W., Laybourn-Parry J. The balance between photosynthesis and grazing in antarctic mixotrophic cryptophytes during summer. Freshw. Biol. 2002;47:2060–2070. doi: 10.1046/j.1365-2427.2002.00950.x. [DOI] [Google Scholar]
- 58.Laybourn-Parry J., Marshall W., Marchant H. Flagellate nutritional versatility as a key to survival in two contrasting antarctic saline lakes. Freshw. Biol. 2005;50:830–838. doi: 10.1111/j.1365-2427.2005.01369.x. [DOI] [Google Scholar]
- 59.Williams A., Trexler J. A preliminary analysis of the correlation of food-web characteristics with hydrology and nutrient gradients in the southern everglades. Hydrobiologia. 2006;569:493–504. doi: 10.1007/s10750-006-0151-y. [DOI] [Google Scholar]
- 60.Scherwass A., Fischer Y., Arndt H. Detritus as a potential food source for protozoans: Utilization of fine particulate plant detritus by a heterotrophic flagellate, Chilomonas paramecium, and a ciliate, Tetrahymena pyriformis. Aquat. Ecol. 2005;39:439–445. doi: 10.1007/s10452-005-9012-4. [DOI] [Google Scholar]
- 61.Kuehn K., Francoeur S., Findlay R., Neely R. Priming in the microbial landscape: Periphytic algal stimulation of litter-associated microbial decomposers. Ecology. 2014;95:749–762. doi: 10.1890/13-0430.1. [DOI] [PubMed] [Google Scholar]
- 62.Kark S., van Rensburg B. Ecotones: Marginal or central areas of transition? Isr. J. Ecol. Evol. 2006;52:29–53. doi: 10.1560/IJEE.52.1.29. [DOI] [Google Scholar]
- 63.Chen R., Twilley R. Patterns of mangrove forest structure and soil nutrient dynamics along the shark River estuary, Florida. Estuaries. 1999;22:955–970. doi: 10.2307/1353075. [DOI] [Google Scholar]
- 64.Davis S., Cable J., Childers D., Coronado-Molina C., Day J., Hittle C., Madden C., Reyes E., Rudnick D., Sklar F. Importance of storm events in controlling ecosystem structure and function in a Florida gulf coast estuary. J. Coast. Res. 2004;20:1198–1208. doi: 10.2112/03-0072R.1. [DOI] [Google Scholar]
- 65.Sutula M., Perez B., Reyes E., Childers D., Davis S., Day J., Rudnick D., Sklar F. Factors affecting spatial and temporal variability in material exchange between the southern everglades wetlands and Florida bay (USA) Estuar. Coast. Shelf Sci. 2003;57:757–781. doi: 10.1016/S0272-7714(02)00403-1. [DOI] [Google Scholar]
- 66.Ross M., Meeder J., Sah J., Ruiz P., Telesnicki G. The southeast saline everglades revisited: 50 years of coastal vegetation change. J. Veg. Sci. 2000;11:101–112. doi: 10.2307/3236781. [DOI] [Google Scholar]
- 67.Andersen F., Kristensen E. Oxygen microgradients in the rhizosphere of the mangrove Avicennia marina. Mar. Ecol. Prog. Ser. 1988;44:201–204. doi: 10.3354/meps044201. [DOI] [Google Scholar]
- 68.Alongi D., Clough B., Robertson A. Nutrient-use efficiency in arid-zone forests of the mangroves Rhizophora stylosa and Avicennia marina. Aquat. Bot. 2005;82:121–131. doi: 10.1016/j.aquabot.2005.04.005. [DOI] [Google Scholar]
- 69.Reef R., Feller I., Lovelock C. Nutrition of mangroves. Tree Physiol. 2010;30:1148–1160. doi: 10.1093/treephys/tpq048. [DOI] [PubMed] [Google Scholar]
- 70.Holguin G., Vazquez P., Bashan Y. The role of sediment microorganisms in the productivity, conservation, and rehabilitation of mangrove ecosystems: An overview. Biol. Fertil. Soils. 2001;33:265–278. doi: 10.1007/s003740000319. [DOI] [Google Scholar]
- 71.Teeling H., Fuchs B., Becher D., Klockow C., Gardebrecht A., Bennke C., Kassabgy M., Huang S., Mann A., Waldmann J., et al. Substrate-controlled succession of marine bacterioplankton populations induced by a phytoplankton bloom. Science. 2012;336:608–611. doi: 10.1126/science.1218344. [DOI] [PubMed] [Google Scholar]
- 72.Cottrell M., Kirchman D. Natural assemblages of marine proteobacteria and members of the cytophaga-flavobacter cluster consuming low- and high-molecular-weight dissolved organic matter. Appl. Environ. Microbiol. 2000;66:1692–1697. doi: 10.1128/AEM.66.4.1692-1697.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kirchman D. The ecology of cytophaga-flavobacteria in aquatic environments. FEMS Microbiol. Ecol. 2002;39:91–100. doi: 10.1016/S0168-6496(01)00206-9. [DOI] [PubMed] [Google Scholar]
- 74.Taylor J., Cottingham S., Billinge J., Cunliffe M. Seasonal microbial community dynamics correlate with phytoplankton-derived polysaccharides in surface coastal waters. ISME J. 2014;8:245–248. doi: 10.1038/ismej.2013.178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Hall A., Wells R., Sweeney J., Townsend F., Balmer B., Hohn A., Rhinehart H. Annual, seasonal and individual variation in hematology and clinical blood chemistry profiles in bottlenose dolphins (Tursiops Truncatus) from Sarasota Bay, Florida. Comp. Biochem. Physiol. Part A Mol. Integr. Physiol. 2007;148:266–277. doi: 10.1016/j.cbpa.2007.04.017. [DOI] [PubMed] [Google Scholar]
- 76.Fourqurean J., Zieman J., Powell G. Phosphorus limitation of primary production in Florida Bay—Evidence from C-N-P ratios of the dominant seagrass Thalassia testudinum. Limnol. Oceanogr. 1992;37:162–171. doi: 10.4319/lo.1992.37.1.0162. [DOI] [Google Scholar]
- 77.Briceno H., Boyer J. Climatic controls on phytoplankton biomass in a sub-tropical estuary, Florida Bay, USA. Estuaries Coasts. 2010;33:541–553. doi: 10.1007/s12237-009-9189-1. [DOI] [Google Scholar]
- 78.Briceño H.O., Boyer J.N., Castro J., Harlem P. Biogeochemical classification of south Florida’s estuarine and coastal waters. Mar. Pollut. Bull. 2013;75:187–204. doi: 10.1016/j.marpolbul.2013.07.034. [DOI] [PubMed] [Google Scholar]
- 79.Klindworth A., Mann A.J., Huang S., Wichels A., Quast C., Waldmann J., Teeling H., Glöckner F.O. Diversity and activity of marine bacterioplankton during a diatom bloom in the North Sea assessed by total RNA and pyrotag sequencing. Mar. Genom. 2014;18:185–192. doi: 10.1016/j.margen.2014.08.007. [DOI] [PubMed] [Google Scholar]
- 80.King G., Smith C., Tolar B., Hollibaugh J. Analysis of compostion and structure of coastal to mesopelagic bacterioplankton communities in the northern Gulf of Mexico. Front. Microbiol. 2013;3:438. doi: 10.3389/fmicb.2012.00438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Pisani O., Yamashita Y., Jaffe R. Photo-dissolution of flocculent, detrital material in aquatic environments: Contributions to the dissolved organic matter pool. Water Res. 2011;45:3836–3844. doi: 10.1016/j.watres.2011.04.035. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Demultiplexed raw data was submitted to the Sequence Read Archive under accession number PRJNA525456.