Abstract
Leptospira is a highly diverse genus comprising many species and serogroups in Brazil as well as all over the world. However, a study by arbitrarily primed PCR of 44 leptospiral strains isolated from humans during three different outbreaks in Brazilian urban centers reveals that 43 of 44 isolates exhibit very similar fingerprints. Analysis of these isolates indicates that they belong to a clonal subpopulation of Leptospira interrogans sensu stricto.
The large diversity within Leptospira interrogans sensu lato is demonstrated by the 223 serovars presently recognized according to serological criteria (6). The microscopic agglutination and cross-agglutinin absorption tests have been used for identification of leptospires at the serovar level. Serologically related serovars are functionally clustered within 25 serogroups. Serogrouping is a simple and reliable test which requires only 25 serum samples. In contrast, serotyping at the serovar level requires complete collections of both reference strains (up to 223) and corresponding sera. Cross-adsorption, a complex and partially subjective procedure, is then performed.
Since the mid-1980s, 10 Leptospira species isolated from humans or animals have been distinguished on the basis of DNA-DNA hybridization studies, namely, L. interrogans sensu stricto (hereafter called L. interrogans), L. kirshneri, L. weilii, L. noguchii, L. borgpetersenii, L. santarosai L. meyeri, L. inadai, L. fainei, and L. alexanderi (1, 8, 10, 12). It should be emphasized that serogroups are unequally distributed into species. For example, serogroup Pomona comprises serovars kunning, mozdok, and tsaratsovo, which belong to the species L. kirshneri, while the seven remaining serovars from this serogroup belong to three other species. Arbitrarily primed PCR (AP-PCR) allows the direct generation of highly discriminant fingerprints and is probably the simplest DNA-based subtyping method described to date. It has successfully been applied to the characterization of Leptospira, showing species assignments that were in agreement with species assignments based on DNA-DNA homology (9).
Our objective was to evaluate an AP-PCR method for the rapid comparison of a large number of leptospiral isolates from different sources, sites, and years in Brazil and for the establishment of a possible clonal connection between isolates from a common-source outbreak.
The 44 Leptospira Brazilian isolates from human sources (Table 1) were provided by the Instituto Oswaldo Cruz (Rio de Janeiro, Brazil), the Center for Zoonosis Control (São Paulo, Brazil), and the University of São Paulo (São Paulo, Brazil). The isolates originated from three outbreaks of leptospirosis which had taken place in Brazil, in the states of São Paulo in 1994 and 1995 and Rio de Janeiro in 1996. Strains (Table 2) from the Collection of the World Health Organization Collaborating Center for Leptospirosis, Institut Pasteur, Paris, France, were used as reference strains.
TABLE 1.
Identification of the Leptospira serogroups and species from isolates involved in human epidemic outbreaks among humans in Brazil
| Isolate(s) | Isolation time, location | Serogroup | Species |
|---|---|---|---|
| SP-03, SP-04, SP-17, SP-22, SP-25, SP-42, SP-44, SP-59, SP-67, SP-69, SP-73, SP-74, SP-75, SP-80, SP-86, SP-104 | 1994, Sao Paulo | Icterohaemorrhagiae | L. interrogans |
| SP-58 | 1994, Sao Paulo | Pomona | L. kirshneri |
| SP-10, SP-14, SP-18, SP-20, SP-36, SP-49, SP-50, SP-52, SP-61, SP-69, SP-70, SP-102, SP-103, SP-149, SP-150, SP-151, SP-163, SP-204, SP-212, SP-217 | 1995, Sao Paulo | Icterohaemorrhagiae | L. interrogans |
| SP-13, SP-28, SP-68, SP-167 | 1995, Sao Paulo | Icterohaemorrhagiaea | L. interrogans |
| SP-53 | 1994, Sao Paulo | Icterohaemorrhagiaea | L. interrogans |
| 1010, 1015 | 1996, Rio de Janeiro | Icterohaemorrhagiae | L. interrogans |
This strain should have been identified as belonging to serogroup canicola.
TABLE 2.
Representative Leptospira strains from the Pasteur Institute collection
| Serovar | Strain | Serogroup | Species |
|---|---|---|---|
| copenhageni | Winjberg | Icterohaemorrhagiae | L. interrogans |
| canicola | Hond Utrecht | Canicola | L. interrogans |
| canicola | chiffon | Canicola | L. interrogans |
| pomona | Pomona | Pomona | L. interrogans |
| grippotyphosa | Moskva V | Grippotyphosa | L. kirshneri |
| kunming | K5 | Pomona | L. kirshneri |
| mozdok | 5621 | Pomona | L. kirshneri |
| tsaratsovo | B 81/7 | Pomona | L. kirshneri |
The identification at the serogroup level (Table 1) was carried out, in Brazil, by previously described procedures (4). All SP (São Paulo) isolates belonged to the Icterohaemorrhagiae serogroup but with some uncertainty regarding SP-18, SP-20, SP-53, and SP-167. Isolates 1010 and 1015 also belonged to the Icterohaemorrhagiae serogroup. Isolate SP-58 was from the Pomona serogroup.
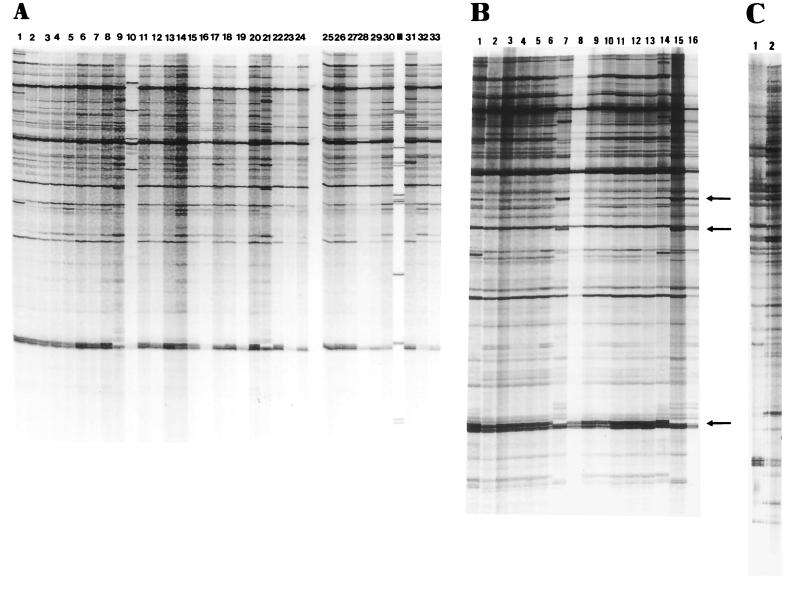
Each isolate was grown to the exponential phase of growth in EMJH medium (3, 5) at 30°C with shaking. Approximately 5 × 108 cells of each strain were used for DNA extraction, which was performed with the Dynabeads kit (Dynal) according to the instructions of the supplier. DNA was eluted from the Dynabeads complex in 40 μl of TE (10 mM Tris-HCl, 1 mM EDTA [pH 8]) and was kept at −20°C. Five microliters was used for each AP-PCR experiment. Fingerprints were generated as described previously (9) by using the KG primer (5′-CAC ACG CAC ACG GAA GAA-3′; purchased from Eurogentec). We compared the AP-PCR patterns of the isolates tested in this study with those of strains of the Icterohaemorrhagiae and Pomona serogroups from the collection, as identified above, which belonged to diverse pathogenic species. The fingerprints of 38 isolates were similar to those of a copenhageni serovar strain from the collection (Fig. 1A, lanes 2 to 8, 11 to 16, 18 to 20, 22 to 30, 32 to 33 compared to lane 1, and Fig. 1B, lanes 2 to 6 and 8 to 13 compared to lanes 1 and 18). The fingerprints of five isolates (SP-28, SP-53, SP-13, SP-167, and SP-68, Fig. 1A, lanes 9, 17, 21, and 31, and Fig. 1B, lane 7, respectively) showed DNA profiles closest to that of the canicola serovar reference strain (Fig. 1B, lanes 20 and 21).
FIG. 1.
AP-PCR fingerprints of genomic DNAs of the Leptospira isolates described in Table 1 and Table 2. An autoradiograph of a 4% acrylamide–50% urea gel is shown. (A) Lane 1, serovar copenhageni strain Winjberg; lanes 2 to 33, SP-03, SP-04, SP-10, SP-22, SP-25, SP-42, SP-44, SP-53, SP-58, SP-59, SP-67, SP-69, SP-73, SP-80, SP-104, SP-13, SP-14, SP-18, SP-20, SP-28, SP-36, SP-49, SP-50, SP-61, SP-70, SP-102, SP-149, SP-150, SP-163, SP-167, SP-212, and SP-217, respectively; lane M, marker. (B) Lanes 1 and 16, serovar copenhageni strain Winjberg; lanes 2 to 13; SP-74, SP-75, SP-86, SP-17, SP-52, SP-68, SP-69, SP-103, SP-151, SP-204, 1010, and 1015, respectively; lanes 14 and 15, serovar canicola strains chiffon and Hond Utrecht, respectively. The arrows correspond to major differences between the profiles of serovars canicola and copenhageni. (C) Lane 1, SP-58; lane 2, serovar grippotyphosa strain Moskva V.
While the fingerprints of the reference strains of serovars copenhageni and canicola have a great deal of similarity, there are recognizable differences in major bands (in Fig. 1B, the two upper arrows indicate bands present in the profile for serovar canicola and absent from the profile for serovar copenhageni, while the lower arrow indicates a band present in the profile for serovar copenhageni and absent from the profile for serovar canicola). The 38 isolates serotyped to the Icterohaemorrhagiae serogroup and the 5 strains serotyped to the Canicola serogroup were found to be similar to serovar copenhageni strains and serovar canicola strains from collections, respectively. However, small differences in two or three minor bands could be found. Since serovars canicola and copenhageni belong to the L. interrogans species, it can be concluded that with one exception (strain SP-58 [Fig. 1A, lane 10]) all of the 44 human isolates are L. interrogans. Isolate SP-58 showed a profile similar to that of serovar grippotyphosa and was identified as L. kirshneri (Fig. 1C, lane 1 compared to lane 2).
The 43 strains represent a cluster of isolates that are quite similar, even though they were isolated from human infections that occurred in different places and at different times during so-called epidemic periods. It should be noted that a very large set of serovars belonging to several serogroups and species does exist in wildlife in Brazil (2, 7, 11). In addition, the 10 Brazilian serovars described in the Leptospira collection (1) correspond to eight serogroups and four species. The present data confirm the predominance of a clonal subpopulation of L. interrogans as the cause of human leptospirosis in the large urban centers of Brazil. Similar strains have been isolated (data not shown) from dogs and rodents, which are the main reservoirs and shedders of leptospires in urban areas.
AP-PCR fingerprinting analysis is a rapid method for use in epidemiological studies. It is possible to compare the profiles of a large number of strains and reach meaningful and epidemiologically relevant conclusions concerning the geographic distributions of Leptospira populations.
Acknowledgments
We thank Antonio José Alves and Leticia Bastos for technical assistance in the cultivation of leptospires. We thank Danielle Margarita, Elisabeth Bellenger, Edith Fournié-Amazouz,and Natacha Sertour for help during the course of this study.
This work was supported by grants from the National Council for Research and Technological Development (fellowship to M. M. Pereira) and the National Foundation for Health/Brazilian Ministry of Health.
REFERENCES
- 1.Brenner D J, Kaufman A F, Sulzer K R, Steigerwalt A G, Rogers F C, Weyant R S. Further determination of DNA relatedness between serogroups and serovars in the family Leptospiraceae with a proposal for Leptospira alexanderi sp. nov. and four new Leptospira genomospecies. Int J Syst Bacteriol. 1999;49:839–858. doi: 10.1099/00207713-49-2-839. [DOI] [PubMed] [Google Scholar]
- 2.Cordeiro F, Sulzer C R, Ramos A A. Leptospira interrogans em diversas espécies de animais silvestres na regiâo sudeste do Brasil. Pesq Vet Bras. 1981;1:19–29. [Google Scholar]
- 3.Ellinghausen H C, McCullough W G. Nutrition of Leptospira pomona and growth of 13 other serotypes: fractionation of oleic albumin complex and medium bovine albumin and polysorbate 80. Am J Vet Res. 1965;26:45–51. [PubMed] [Google Scholar]
- 4.Faine S. Guidelines for the control of leptospirosis. Publication no. 67. Geneva, Switzerland: World Health Organization; 1982. [Google Scholar]
- 5.Johnson R C, Harris V G. Differentiation of pathogenic and saprophytic Leptospires. I. Growth at low temperatures. J Bacteriol. 1967;94:27–31. doi: 10.1128/jb.94.1.27-31.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kmety E, Dikken H. Classification of the species Leptospira interrogans and history of its serovars. Groningen, The Netherlands: University Press; 1993. [Google Scholar]
- 7.Pereira M M, Korver H, Mazzonelli J M, Andrade J, Moraes G. Search for leptospires and specific antibodies in wild animals trapped in a periurban area of Rio de Janeiro, Brazil. In: Kobayashi Y, editor. Leptospirosis, Proceedings of the Leptospirosis Research Conference, 1990. 1991. Matsuyama, Japan. [Google Scholar]
- 8.Perolat P, Chappel R J, Adler B, Baranton G, Bulach D M, Billinghurst M L, Letocart M, Merien F, Serrano M S. Leptospira fainei sp. nov. isolated from pigs in Australia. Int J Syst Bacteriol. 1998;48:851–858. doi: 10.1099/00207713-48-3-851. [DOI] [PubMed] [Google Scholar]
- 9.Ralph D, McClelland M, Welsh J, Baranton G, Perolat P. Leptospira species categorized by arbitrarily primed polymerase chain reaction (PCR) and by mapped restriction polymorphisms in PCR-amplified rRNA genes. J Bacteriol. 1993;175:973–981. doi: 10.1128/jb.175.4.973-981.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ramadass P, Jarvis B D W, Corner R J, Penny D, Marshall R B. Genetic characterization of pathogenic Leptospira species by DNA hybridization. Int J Syst Bacteriol. 1992;42:215–219. doi: 10.1099/00207713-42-2-215. [DOI] [PubMed] [Google Scholar]
- 11.Santa Rosa C A, Sulszer C R, Yanaguita R M, Silva A S. Leptospirosis in wildlife in Brazil. Isolation of serovars canicola, pyrogenes and grippotyphosa. Int J Zoonoses. 1980;7:40–43. [PubMed] [Google Scholar]
- 12.Yasuda P H, Steigerwalt A G, Sulzer K R, Kaufmann A F, Rogers F, Brenner D J. Deoxyribonucleic acid relatedness between serogroups and serovars in the family Leptospiraceae with proposals for seven new Leptospira species. Int J Syst Bacteriol. 1987;3:407–415. doi: 10.1099/00207713-49-2-839. [DOI] [PubMed] [Google Scholar]