Abstract
Two isolates of Streptococcus pneumoniae having different optochin susceptibilities were recovered from a blood sample of a 2-year-old boy with community-acquired pneumonia. The two isolates were documented to belong to a single clone on the basis of the isolates' identical serotype (23F), antibiograms by the E-test, random amplified polymorphic DNA patterns generated by arbitrarily primed PCR, pulsed-field gel electrophoresis, and restriction fragment length polymorphism of the penicillin-binding protein genes pbp2b and pbp2x.
CASE REPORT
A 2-year-3-month-old boy was admitted to our ward with the chief problem of cough and rhinorrhea for 2 weeks and high fever with generalized tonic-clonic convulsion on the day of admission. He had a history of febrile convulsion. Physical findings were unremarkable except for an injected throat. His activity was fair without any meningeal sign. A chest radiograph showed infiltration over bilateral lung fields. Laboratory testing revealed a leukocyte count of 37,650/mm3 with 88% neutrophils and a markedly elevated C-reactive protein level (14.2 mg/dl, normal level being <0.5 mg/dl). Two sets of blood cultures yielded Streptococcus pneumoniae, with an inhibition zone around the oxacillin disk (1-μg disk) (BBL Microbiology Systems, Cockeysville, Md.) of 8 mm. Cefazolin (100 mg/kg of body weight/day) was given for 2 days, and the fever subsided before the blood culture result was available. Since the patient's general condition was healthy, cefazolin was given for a further 3 days, treatment was then shifted to oral cephalexin (25 mg/kg/day) for 1 week, and the patient was discharged in stable condition. The child did well in the following 6 months.
Microbiological investigation.
Two sets of blood cultures both grew gram-positive cocci in BACTEC 6A aerobic bottles (Becton Dickinson, Sparks, Md.). After subculture, the organisms grew well on Trypticase soy agar supplemented with 5% sheep blood agar plates (BBL Microbiology Systems) at 37°C in ambient air. The colonies were alpha-hemolytic and nonmucoid. The optochin sensitivity test for the organisms was performed on Trypticase soy agar supplemented with 5% sheep blood agar according to a previous description (18). An inhibition zone of 15 mm in diameter around the 6-mm optochin disk was identified, and several colonies (isolate A) also grew within the inhibition zone (7 to 9 mm from the center of the disk). When the colonies grown inside (isolate A) and outside (isolate B) the inhibition zone, respectively, were inoculated onto two plates of Trypticase soy agar supplemented with 5% sheep blood and were incubated for 24 h, no inhibition zone around the optochin disk was found for isolate A and a complete inhibition zone of 15 mm around the disk (no scattered colonies within the zone) was demonstrated for isolate B. Isolates A and B were positive by the bile solubility test, and their biochemical profiles generated by the API 32 Strep System (bioMeriuex Vitek, Marcy l'Etoile, France) were in accordance with the identification of S. pneumoniae.
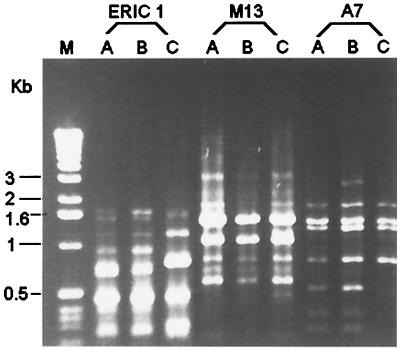
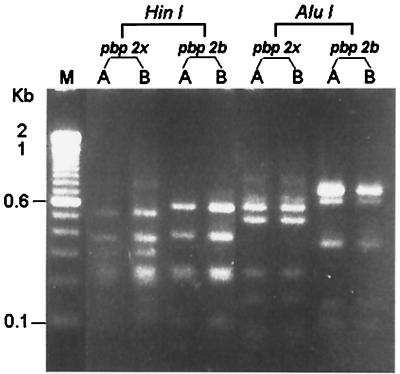
The two isolates of S. pneumoniae (isolates A and B) both belonged to serotype 23F as determined by the capsular swelling test (quellung reaction) by specific antisera as previously described (8). For both isolates, penicillin and cefotaxime MICs were 2 and 1 μg/ml, respectively, determined by means of the E-test (PDM Epsilometer; AB Biodisk, Solna, Sweden). MICs of other agents for these two isolates were also identical: erythromycin, ≥256 μg/ml; rifampin, 1 μg/ml; clindamycin, ≥256 μg/ml; and vancomycin, 0.5 μg/ml. Molecular typing of the two isolates by random amplified polymorphic DNA (RAPD) analysis of chromosomal DNA generated by arbitrarily primed PCR and pulsed-field gel electrophoresis and restriction fragment length polymorphism (RFLP) profiles of penicillin-binding protein genes (pbp2b and pbp2x) after digestion with restriction enzymes HinfI and AluI (Gibco BRL, Gaithersburg, Md.) were performed in accordance with previous descriptions (7, 12, 19). For RAPD analysis, three primers were used: ERICI (5′-GTGAATCCCCAGGAGCTTACAT-3′), M13 (5′-GAGGGTGGCGGTTCT-3′), and OPA-7 (5′-GAAACGGGTG-3′). For molecular typing studies, one S. pneumoniae isolate recovered from another patient was included as a control strain. The two isolates had identical RAPD patterns (Fig. 1), pulsotypes, and RFLP profiles of pbp2b and pbp2x (Fig. 2).
FIG. 1.
RAPD patterns generated by arbitrarily primed PCR by three primers, ERICI, M13, and OPA-7. Lane M, molecular markers; lanes C, control strain; lanes A and B, isolates A and B, respectively.
FIG. 2.
RFLP patterns of the pbp2b and pbp2x genes generated with the restriction enzymes HinfI and AluI. See the legend to Fig. 1 for lane definitions.
Discussion.
S. pneumoniae is a major cause of community-acquired pneumonia, otitis media, paranasal sinusitis, bacteremia, and meningitis (16). The emergence of drug-resistant strains of S. pneumoniae has complicated treatment of these common infections (3, 4, 6, 8, 10, 11). Pneumococci are identified in the Clinical Microbiology Laboratory of the National Taiwan University Hospital by the following three reactions: alpha-hemolysis on sheep blood agar, catalase negativity, and solubility in bile salts or susceptibility to ethylhydrocupreine (optochin) (18). In recent years, a number of isolates have been found to be optochin resistant, which has led cautious microbiologists to rely more on the use of bile solubility for definitive identification (9, 15, 17). In fact, it has been reported that up to 5% of S. pneumoniae strains may be optochin indeterminate or resistant (1, 15). Furthermore, the bile solubility test is not always specific for S. pneumoniae (5). The use of these standard tests can result in ambiguous phenotypes for certain organisms, leading to identification difficulties for routine microbiology laboratories (2, 14).
To our knowledge, bacteremia due to a single clone of S. pneumoniae which simultaneously possessed two isolates with different optochin susceptibilities has never been documented previously. By use of the antibiotyping and molecular typing methods, this report described an episode of bacteremia caused by a single clone of serotype 23F and penicillin-resistant S. pneumoniae that obviously possessed two subpopulations with different optochin susceptibilities.
In summary, we describe a case with invasive infection caused by two isolates of multidrug-resistant S. pneumoniae exhibiting different optochin susceptibilities which were later documented to belong to a single clone. Clinical microbiologists need not only to be aware of a continued increase in antimicrobial drug resistance in clinical isolates of S. pneumoniae but also to understand the potential difficulties of identification of this organism by conventional methods, particularly the optochin susceptibility test.
REFERENCES
- 1.Austrian R. Random gleanings from a life with the pneumococcus. J Infect Dis. 1975;131:474–484. doi: 10.1093/infdis/131.4.474. [DOI] [PubMed] [Google Scholar]
- 2.Borek P A, Dressel D C, Hussong J, Peterson L R. Evolving clinical problems with Streptococcus pneumoniae: increasing resistance to antimicrobial agents, and failure of traditional optochin identification in Chicago, Illinois, between 1993 and 1996. Diagn Microbiol Infect Dis. 1997;29:209–214. doi: 10.1016/s0732-8893(97)00141-7. [DOI] [PubMed] [Google Scholar]
- 3.Bradley J S, Kaplan S L, Klugman K P, Leggiadro R J. Consensus: management of infections in children caused by Streptococcus pneumoniae with decreased susceptibility to penicillin. Pediatr Infect Dis J. 1995;14:1037–1041. [PubMed] [Google Scholar]
- 4.Caputo G M, Appelbaum P C, Liu H H. Infections due to penicillin-resistant pneumococci. Arch Intern Med. 1993;153:1301–1310. [PubMed] [Google Scholar]
- 5.Cowan S T, Steel K C. Manual for the identification of medical bacteria. London, England: Cambridge University Press; 1974. p. 29. [Google Scholar]
- 6.Friedland I R, McCracken G H. Management of infections caused by antibiotic resistant Streptococcus pneumoniae. N Engl J Med. 1994;331:377–382. doi: 10.1056/NEJM199408113310607. [DOI] [PubMed] [Google Scholar]
- 7.Hsueh P-R, Teng L-J, Lee L-N, Yang P-C, Ho S-W, Luh K-T. Dissemination of high-level penicillin-, extended-spectrum cephalosporin-, and erythromycin-resistant Streptococcus pneumoniae clones in Taiwan. J Clin Microbiol. 1999;37:221–224. doi: 10.1128/jcm.37.1.221-224.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hsueh P-R, Teng L-J, Lee L-N, Yang P-C, Ho S-W, Luh K-T. Extremely high incidence of macrolide and trimethoprim-sulfamethoxazole resistance among clinical isolates of Streptococcus pneumoniae in Taiwan. J Clin Microbiol. 1999;37:897–901. doi: 10.1128/jcm.37.4.897-901.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kontiainen S, Sivonen A. Optochin resistance in Streptococcus pneumoniae strains isolated from blood and middle ear fluid. Eur J Clin Microbiol. 1987;6:422–424. doi: 10.1007/BF02013101. [DOI] [PubMed] [Google Scholar]
- 10.Lee H J, Park J Y, Jang S H, Kim J H, Kim E C, Choi K W. High incidence of resistance to multiple antimicrobials in clinical isolates of Streptococcus pneumoniae from a university hospital in Korea. Clin Infect Dis. 1995;20:826–835. doi: 10.1093/clinids/20.4.826. [DOI] [PubMed] [Google Scholar]
- 11.Leggiadro R J, Davis Y, Tenover F C. Outpatient drug-resistant pneumococcal bacteremia. Pediatr Infect Dis J. 1994;13:1144–1146. doi: 10.1097/00006454-199412000-00014. [DOI] [PubMed] [Google Scholar]
- 12.McDougal L K, Facklam R, Reeves M, Hunter S, Swenson J M, Hill B C, Tenover F C. Analysis of multiply antimicrobial-resistant isolates of Streptococcal pneumoniae from the United States. Antimicrob Agents Chemother. 1992;36:2176–2184. doi: 10.1128/aac.36.10.2176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Moissenet D, Valcin M, Marchand V, Garabedian E, Geslin P, Garbarg-Chenon A, Vu-Thien H. Molecular epidemiology of Streptococcus pneumoniae with decreased susceptibility to penicillin in a Paris children's hospital. J Clin Microbiol. 1997;35:298–301. doi: 10.1128/jcm.35.1.298-301.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mundy L S, Janoff E N, Schwebke K E, Shanholtzer C J, Willard K E. Ambiguity in the identification of Streptococcus pneumoniae: optochin, bile solubility, quellung, and the Accu Probe DNA probe tests. Am J Clin Pathol. 1998;109:55–61. doi: 10.1093/ajcp/109.1.55. [DOI] [PubMed] [Google Scholar]
- 15.Munoz R, Fenoll A, Vicioso D, Casal J. Optochin-resistant variants of Streptococcus pneumoniae. Diagn Microbiol Infect Dis. 1990;13:63–66. doi: 10.1016/0732-8893(90)90056-2. [DOI] [PubMed] [Google Scholar]
- 16.Musher D M. Infections caused by Streptococcus pneumoniae: clinical spectrum, pathogenesis, immunology, and treatment. Clin Infect Dis. 1992;14:801–809. doi: 10.1093/clinids/14.4.801. [DOI] [PubMed] [Google Scholar]
- 17.Phillips G, Barker R, Brogan O. Optochin resistant Streptococcus pneumoniae. Lancet. 1988;ii:281. doi: 10.1016/s0140-6736(88)92573-1. [DOI] [PubMed] [Google Scholar]
- 18.Ruoff K L. Streptococcus. In: Murray P R, Baron E J, Pfaller M A, Tenover F C, Yolken R H, editors. Manual of clinical microbiology. 6th ed. Washington, D.C.: ASM Press; 1995. pp. 299–314. [Google Scholar]
- 19.Smith A M, Klugman K P, Coffey T J, Spratt B G. Genetic diversity of penicillin-binding protein 2B and 2X genes from Streptococcus pneumoniae in South Africa. Antimicrob Agents Chemother. 1993;37:1938–1944. doi: 10.1128/aac.37.9.1938. [DOI] [PMC free article] [PubMed] [Google Scholar]