Abstract
Convenient methods for the preparation of gene delivery platforms based on branched low molecular weight polyethylenimine (PEI) were described. Firstly, PEI lipids, with a low molecular weight PEI headgroup and hexadecyl chain tail group, were prepared through a highly efficient ring-opening reaction of glycidyl hexadecyl ether (EpoxyC16) by amine from PEI. Then, the PEI lipids were used as a component of cationic liposomes and as a surfactant for the preparation of poly(D,L-lactide-co-glycolide) (PLGA) nanoparticle (NP) via solvent extraction/evaporation method. As potential effective gene delivery platforms, their preparation, size, size distribution, toxicities, plasmid DNA loading, in vitro transfection and intracellular trafficking were studied. Both facile platforms showed less toxicity and higher transfection efficacy when compared to high molecular weight PEI in vitro, and may have further versatile applications in the gene delivery field.
Keywords: Gene Delivery, Low Molecular Weight Polyethylenimine, Liposome, Core–Shell Nanoparticle
INTRODUCTION
Current gene therapy delivery systems lack desired convenience, efficiency and safety. While viral-based systems appear to be the most effective way to transfer genes to cells, safety concerns and long-term oncologic effects have prompted extensive research on non-viral delivery systems. Starting with a calcium carbonate-based delivery technique, several non-viral delivery techniques have shown comparable transfection efficacies to those of viral-based gene delivery systems. Among these non-viral delivery techniques, cationic polymers (polylysine,1–5 chitosan,6–11 PAMAM dendrimer,12,13 synthetic cationic polymer or lipid14–20 and PEI based systems21–28) have attracted increasing attention. PEI is a readily commercially available polycation containing a high density of primary, secondary and tertiary amines. Before recent investigation into its use as a gene delivery polymer,29 PEI was previously used as a chelating and flocculation agent.
Taking advantage of its proton sponge effect and ability to protect DNA from degradation by enzymes, PEI shows relatively high transfection efficacy in vitro and in vivo. However, both the transfection efficacy and toxicity of PEI increase with its molecular weight.26,30–35 An effective transfection by PEI largely relies on the effective condensation of DNA by the high cationic density from high molecular PEI. However, higher cationic density also leads to higher cytotoxicity because of reduced backbone biodegradability. For example, 800 Da PEI has lower cytotoxicity but has almost no transfection efficacy; and 25 kDa PEI is effective in gene transfection but has high cytotoxicity. Modification of PEI with various chemical moieties has been studied to both decrease toxic effects and increase gene delivery efficacy.36–44
Various strategies have been reported to chemically modify its polymeric backbone. These modifications increase biodegradability by reducing the positive charge of PEI. In these reports, low molecular weight PEI was cross-linked with a series of diacrylate,45–47 and higher molecular weight PEI analogs with biodegradable backbones were complexed with DNA or siRNA. Hydrolysis of their ester bonds occur under physiological conditions within the cell after transfection, and convert the cross-linked high molecular weight PEI into a low-toxicity. Using these methods, higher transfection efficacies and lower toxicities were achieved as compared to 25 kDa PEI. Similarly, degradable PEIs with acid-labile imine linkers were synthesized by low molecular weight PEI and glutadialdehyde.34 These PEIs rapidly degraded into nontoxic, low molecular weight PEI in acidic endosome. Disulfide bonds have also been introduced to the backbone using cross-linking reagents. After being complexed with DNA, higher molecular weight PEI analogs can be cleaved into low molecular weight pieces, which reduces toxicity and potentially allows favorable access for transcription machinery.
To optimize delivery, highly specific intracellular stimuli were designed to trigger PEI systems. Two studies reported successful methods, which were derived from stimuli-triggered degradability of disulfide polyester cross-linked PEI48,49 and pH-sensitive polycations.50 Cationic lipids are another widely used type of non-viral vectors for gene delivery, and are especially attractive because they can easily be synthesized and used for systematic elucidation of structure-activity relationships by verifying their structures. The polar hydrophilic headgroup of cationic lipids enable the condensation of nucleic acids and further govern transfection efficacy. They can be quaternary ammonium ions, amines, amino acids or peptides, guanidinium ions, heterocyclic headgroups, and other sophisticatedly designed functional headgroups.19,20,51,52 Very recently, low molecular weight PEI was modified by amphiphilic Tween 85 via esterification reaction and purification. The result of lipid-like Tween 85-modified PEI2K could condense DNA efficiently and significantly increase the cellular uptake of complexes with higher transfection efficacy.53
In this study, we adopted the highly efficient ring-opening reaction of epoxides by amine groups for the modification of low molecular weight PEI with long alkyl chains, and obtained amphiphilic PEI lipids with different amounts of hexadecyl tail groups. The lipids were further used for the formulation of cationic liposomes by simple sonication method, and also as a surfactant in the PLGA NP via the solvent extraction/evaporation method54,55 (Fig. 1). The properties of these two types of gene delivery platforms were systematically studied. Through the physical self-assembly of low molecular weight PEI on the surface of NP, higher transfection efficacy and lowered toxicity in both facile gene delivery platforms were realized as compared to high molecular weight PEI 25 kDa. Although PEI based NP as a gene delivery system has been reported before,53 PEI lipid from highly efficient chemistry, and later as a component of liposome or surfactant for PLGA NPs, has never been reported as an effective gene delivery or co-delivery system.
Figure 1.
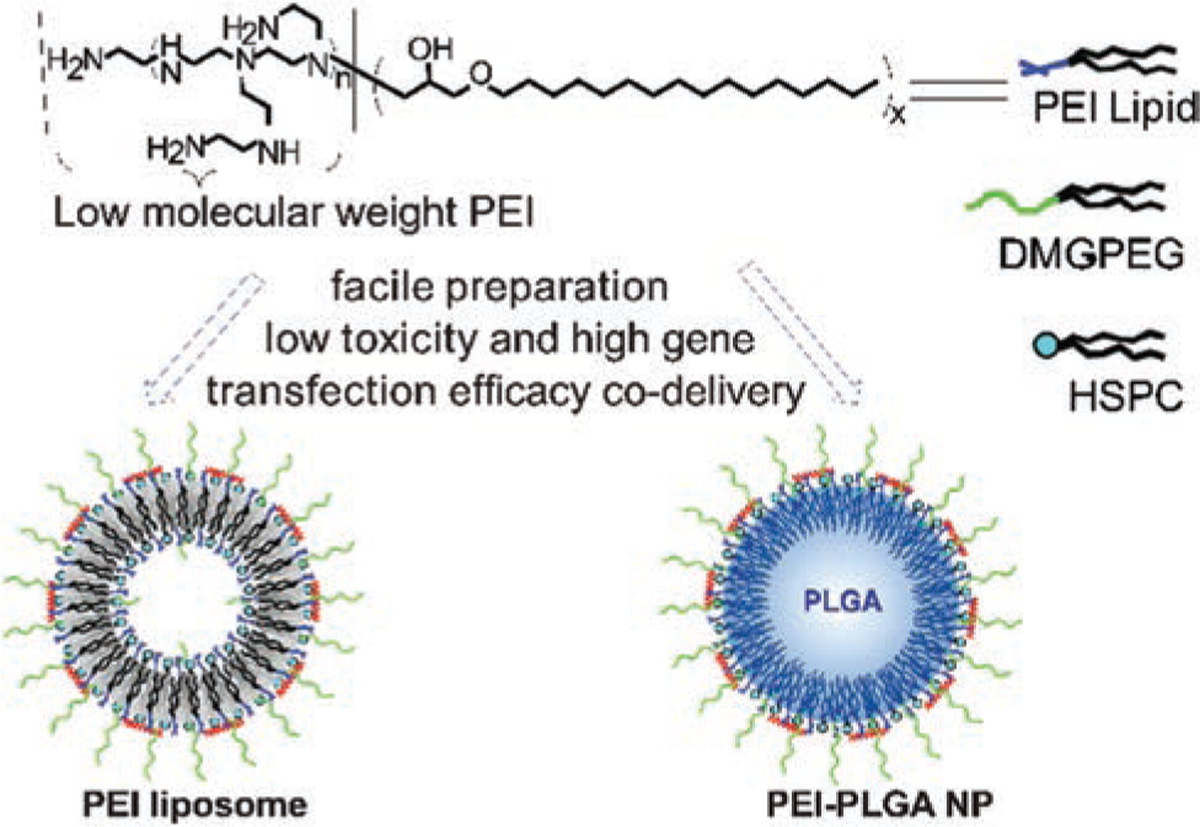
Scheme of facile gene delivery platforms with low molecular weight PEI by highly efficient chemistry.
MATERIALS AND METHODS
Materials
Low molecular branched PEI (Mn 600) (PEI800), high molecular branched PEI (Mn 10000) (PEI25K), glycidyl hexadecyl ether, and 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) were obtained from Sigma. Hydrogenated soybean phosphatidylcholine (HSPC, MW 762) was purchased from Avanti Polar Lipids. 1,2-Dimyristoyl-sn-glycerol, methoxypolyethylene Glycol (DMG-PEG, 2000) was purchased from NOF America. Poly(D,L-lactide-co-glycolide) (50/50) with terminal carboxylate groups (PLGA; inherent viscosity, 0.55–0.75 dl/g in hexafluoroisopropanol; MW 44 kDa) was obtained from Absorbable Polymers International. YOYO-1 and Lyso-Tracker Red were purchased from Thermo Fisher Scientific. All other reagents were from commercial resources and were used as received.
Full length plasmid DNA IL-4 (pIL-14) encoded with GFP (pIL-4/GFP) was amplified using Escherichia coli DH5α and purified using QIAprep Spin Miniprep Kit (QIAGEN). The concentration of DNA was determined by measuring UV absorbance at 260 nm and 280 nm, and the ratio of absorbance at 260 nm and 280 nm was greater than 1.8. Hela cells were obtained from the American Type Culture Collection (ATCC) and cultured in DMEM containing 10% fetal bovine serum (FBS), 100 Unit/mL penicillin G sodium, and 100 µg/mL streptomycin sulfate (complete DMEM medium). Cells were maintained at 37 °C in a humidified and 5% CO2 incubator.
SEC and FT-IR
The tetrahydrofuran (THF) size exclusion chromatography (SEC) was equipped with Polymer Standards Services (PSS) columns (guard, 105, 103, and 102 Å SDV columns) at 35 °C with THF flow rate = 1.0 mL/min, a differential refractive index (RI) detector (Wyatt Technology) using PSS WinGPC 7.5 software. FT-IR spectra were obtained with Jasco FT/IR-6300.
Synthesis of PEI Lipid
EpoxyC16 (MW = 298) and branched PEI800 with fixed molar ratio of 12/1, 8/1 and 4/1 were mixed in a glass vial. The vial was sealed and stirred at 40 °C for 1 day. Products as white solids were obtained after cooling down to room temperature. The products were characterized by FT-IR and SEC and used in following experiments without further purification.
Preparation of PEI Liposome
The formation of PEI8C16 liposome was used as an example. PEI lipid (20 mg), HSPC (10 mg), and DMG-PEG (2 mg) were mixed and dissolved in 1 mL of DCM/ethanol (2/1, v/v). The solvent was slowly evaporated, and a thin film was finally formed. After drying in a vacuum for 24 hrs, the lipid film was hydrated at 40 °C with 2.5 mL double distilled water to obtain large multilamellar vesicle (LMV) suspensions. The LMV suspension was broken down to small unilamellar vesicle (LUV) suspensions by sonication with a probe type sonicator at 40 °C. After about 10 minutes, a slightly hazy transparent suspension was obtained, and the size remained unchanged even after a longer time of sonication. After cooling down, the suspension was adjusted to 2.5 mL with double distilled water to obtain a liposome stock suspension and stored at 4 °C.
Preparation of PEI-PLGA NPs
PEI/PLGA NPs were prepared by a modified solvent extraction/evaporation method.55,56 Briefly, PLGA (15 mg), PEI8C16 (25 mg), and DMG-PEG (2 mg) were dissolved in 0.5 mL of DCM. The solution was combined with 2.5 mL of DD water in a glass vial. The mixture was stirred and sonicated with a probe type sonicator for 5 minutes to obtain a fine emulsion. The emulsion was further stirred overnight at 4 °C in the vial without a cap to yield a slightly hazy transparent suspension. The suspension was washed using Amicon Ultra-4 centrifugal filter (Millipore, Billerica, MA) with a molecular weight cut-off of 100 kDa, and adjusted to 2.5 mL with double distilled water to obtain a stock PEI-PLGA NPs suspension, which was stored at 4 °C.
Preparation of pDNA Loaded Liposomes and PEI-PLGA NPs
N/P ratios of pIL-4/GFP loaded liposome and PEI-PLGA NPs were the ratios of moles of the amine groups of chitosan to those of the phosphate groups of pIL-4/GFP. For the calculation of N/P ratios, 330 Da was used as the average mass per charge for pIL-4/GFP and 43 Da was used as the mass per charge PEI.57 The nanoformulation (0.2 µg/µL) was mixed with pIL-4/GFP or YOYO-1 labeled pIL-4 solution at different N/P ratios in PBS (10 µL) and incubated at room temperature for 30 min.
Gel Retardation Assay
Liposomes or NPs were mixed with 0.2 µg pIL-4/GFP with different N/P ratios,57 and the effect of liposome on the condensation of pIL-4/GFP was investigated using electrophoresis on 1% agarose gel with Tris-acetate (TAE) running buffer at 100 V for 30 min. DNA was visualized with ethidium bromide (0.2 mg/mL).
Zetasizer
The size, size distribution, and zeta potential were acquired by a Zetasizer Nano ZS (Malvern). For each sample, the liposome or NPs were tested three times at 25 °C in double distilled water at a concentration 1 mg/mL.
Scanning Electron Microscope (SEM)
Changes in the morphologies of the obtained NPs were observed by using a SEM (JEOL JSM-6010LA, Tokyo, Japan). In order to avoid the breaking down of the liposomes and NPs, we placed 50 µl of liposomes and NPs onto the cover glass and added one drop of 2% of uranyl acetate (Cat. No. 22400, Electron Microscopy Sciences Hattfield, PA) for 15 min. Then the slide was washed out with DI, and the sample was kept in the vacuum desiccator to dry out for 24 h. The dried samples were coated with gold/Palladium using Gold/Palladium Target 57 mm diameter × 0.1 mm thick (Gold/Palladium Target: Electron Microscopy Sciences Cat. No. 91017PD) with research grade argon gas (EMS Quorum EMS 150R ES sputter coater Laughton, East Sussex, United Kingdom) to observe SEM, which were operated at 15 kV with top view.
Cytotoxicity Assay
Hela cells were seeded in 96 well plates at a density of 1 × 104 cells per well with 200 µL of culture medium for 24 h. Then, PEI Liposomes and PEI 25 K at concentration of 5, 7.5, 10, 15, 20 and 25 ng/mL were added to cultures. No treated cultures served as control. After 24 h treatment, the medium was removed and 0.1 mL of MTT solution (5 mg/mL) was added to each well to react for 45 minutes at 37 °C. The supernatant was removed and 0.1 mL DMSO was added to dissolve the MTT formazan crystals. The plates were gently shaken for 10 min to let the purple formazan crystals dissolve. Absorbance at 490 nm was measured and the results were normalized by controls. Each experiment was done in triplicate.
Transfection of pIL-4/GFP by PEI Liposomes and PEI-PLGA NPs
Hela cells and mouse bone marrow derived macrophages (BMMs) were seeded into 48-well plates and cultured overnight in complete DMEM with 10% FBS (37 °C, 5% CO2) until 70% confluence was reached before in vitro DNA transfection assays. For transfection, pIL-4/GFP liposomes and PEI-PLGA NPs were prepared at different N/P ratio containing 0.4 µg or 0.8 µg of pIL-4/GFP and incubated for 30 min at room temperature, then added into each well of 0.2 mL medium. After 6 h, the media were replaced with fresh, FBS-containing media (0.5 mL/well). The cells were further cultured for 48 h. PEI25K/pIL-4/GFP complexes, at N/P ratio of 10 and 0.8 µg DNA, were used as positive control.22 The GFP expressing cells were visualized by Nikon Ti fluorescence microscopy (Nikon Instruments Inc., Melville, NY).
Flow Cytometry
The cells were fixed with 2% paraformaldehyde to determine the GFP positive cell percentage and GFP fluorescence intensity by Gallios™ Flow Cytometer. Total of 5 × 105 cells were quantitatively analyzed by flow cytometry and analyzed with Flowjo software.
Enzyme-Linked Immunosorbent Assay (ELISA)
The human and mouse specific anti-IL-4 antibodies were used to detect IL-4 protein levels in culture medium at day 5 post transfection by ELISA assay according to the manufacturer’s protocol (Abcam, Cambridge, MA, USA). Each well was repeated three times.
Intracellular Tracking pIL-4
Hela cells were seeded on 10 mm2 glass coverslips, placed in 24-well plates and incubated for 24 h. pIL-4 was labeled with green fluorescence (YOYO-1) at a ratio of 10/1; the nucleic acids base pair to dye molecules. The resulting YOYO-1 labeled pIL-4 was then loaded into PEI nanoformulations at the N/P ratio of 20/1. YOYO-1 labeled nanoformulations were added to the cells at a DNA concentration of 1 µg/mL and incubated with the cells for 1, 2 and 24 hr. After being stained with Lysotracker-Red according to the manufacturer’s protocol, the cells were immediately fixed with 4% paraformaldehyde. Nuclei were stained with DAPI. Intracellular localization was visualized using Nikon Ti fluorescence microscopy.
Statistical Analyses
One-way analysis of variance was used to compare each of the outcomes. If the overall F-test was statistically significant, then pairwise comparisons were conducted with adjustments made for multiple comparisons, using Tukey’s method. P values less than 0.05 were considered statistically significant.
RESULTS
Synthesis of PEI Lipid
Lipids with PEI headgroups51 and hexadecyl chains tail groups were synthesized by epoxy-amine chemistry58 between the epoxide group from EpoxyC16 and the amine group from low molecular weight PEI.59–61 One mol of branched PEI800 has approximately 18 mol of amine groups, in which 4.5 mol (25%) are tertiary amines, 9 mol (50%) are secondary amines and 4.5 mol (25%) are primary amines according to the information provided by the supplier. Both secondary amines and primary amines can react with epoxide groups. One secondary amine group can react with one equivalent epoxide group, and one primary amine group is capable of engaging in two ring opening reactions. Theoretically, one mole of PEI800 can react with 18 mol of EpoxyC16. In our study, the molar ratios at 12 to 1, 8 to 1, and 4 to 1 between EpoxyC16 and PEI800 were designed and studied. The reactions under three fold ratios went smoothly without any solvent at 40 °C (Fig. 2(A)). After a 24-hour reaction, we labeled the products as PEI4C16, PEI8C16, and PEI12C16. They were characterized by FT-IR and SEC. By comparing the FT-IR spectra of all three products with the starting material EpoxyC16 (Fig. 2(B)), we can see that the characteristic peaks of epoxide group at 850 cm−1 and 910 cm−1 fully disappeared in the spectra of products. FT-IR spectra indicated that EpoxyC16 fully reacted with PEI800, and no EpoxyC16 was left in the unpurified products. In order to characterize the molecular weight and molecular weight distribution of the obtained products by THF SEC, their acetylation derivatives were prepared. From SEC elution curves (Fig. 2(C)) of acetylation derivatives of PEI800, PEI4C16, PEI8C16 and PEI12C16 (labeled respectively as Ac PEI800, Ac PEI4C16, Ac PEI4C16 and Ac PEI4C16), three obtained PEI lipids had different molecular weights and molecular weight distributions as compared to the original PEI800, indicating the complete reaction of PEI800 with EpoxyC16. Thus, three PEI lipid headgroups with different amounts of hexadecyl tail groups were obtained with convenience and high purity by the ring-opening reaction of epoxide by PEI800.
Figure 2.
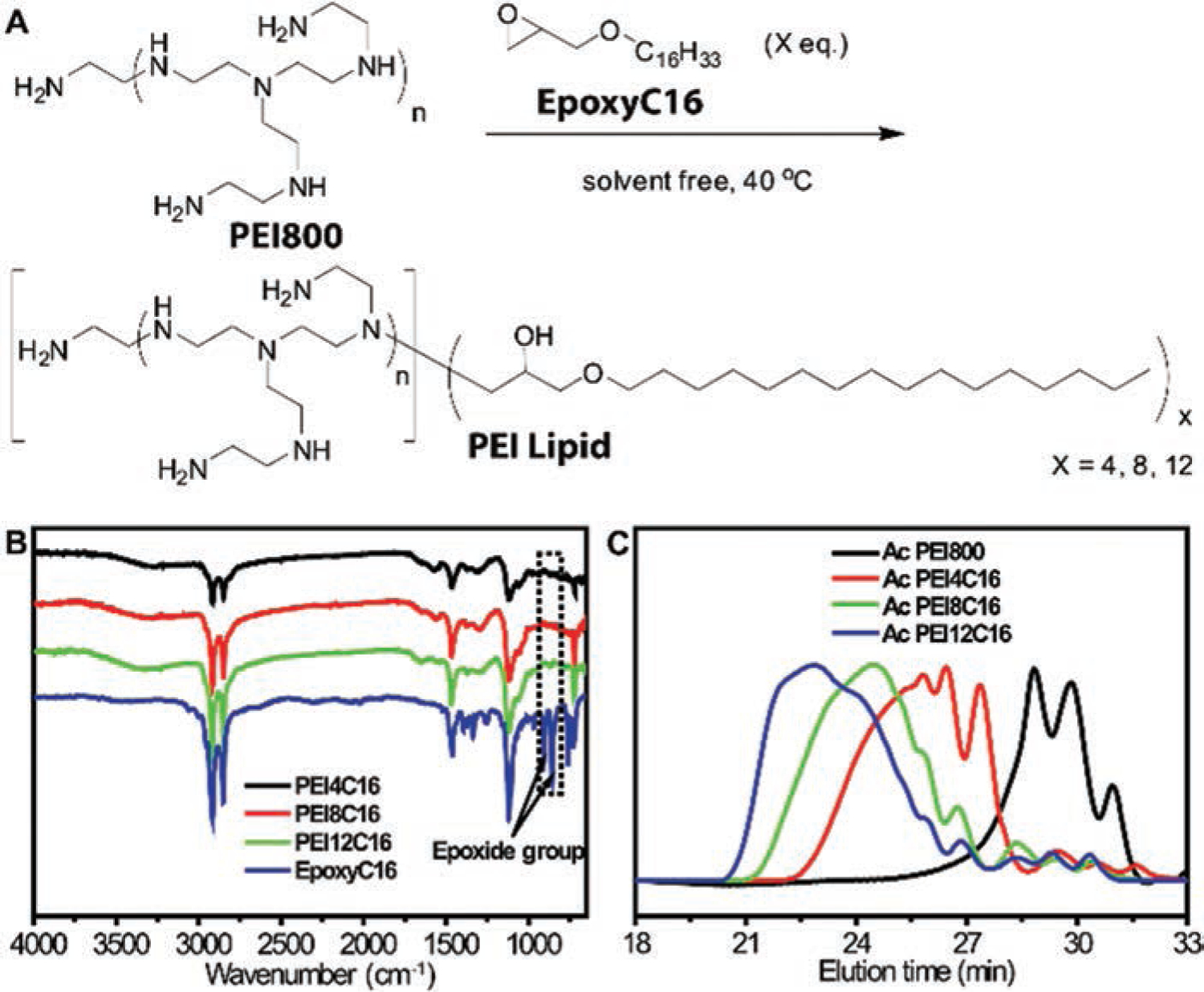
(A) Synthesis of PEI lipid by highly efficient reaction of EpoxyC16 with PEI800. (B) FT-IR spectra of EpoxyC16, PEI4C16, PEI8C16 and PEI12C16. (C) THF SEC elution curves of Ac PEI800, Ac PEI4C16, Ac PEI4C16 and Ac PEI4C16.
Preparation and Characterization of PEI Liposomes and PEI-PLGA NP
Liposomes were made from PEI lipid, HSPC and DMG-PEG, a polyethylene glycol modified lipid (Fig. 1). DMG-PEG, which will provide the PEG corona for the formed liposome, was used to increase the stability of PEI liposome in medium or with in vivo circulation.62 For this study, liposomes were simply prepared without extrusion by hydration and sonication, known as the microfluid method.63,64 After a series of tentative transfection experiments, we found a higher mass percentage of PEI headgroups in the liposome composition was very important to the final transfection efficacy. The transfection efficacy of PEI liposomes was low (fewer than 0.5% GFP positive cell) when the mass percentage of PEI headgroups of the liposome composition was less than 8.3%. By increasing the ratio of PEI lipids in the liposome composition, the mass percentage of PEI headgroups was successfully elevated to 18.6%, 15.7% and 8.8% in the liposomes made from PEI4C16, PEI8C16 and PEI12C16 respectively. The size and size distribution of prepared liposomes, as characterized by Zetasizer, were listed in Table I. All nanoformulations showed narrow size distribution (Fig. 3(A)). However, these three PEI lipids were not capable of forming particles smaller than 400 nm in size when the mass percentage of PEI headgroups were further increased.
Table I.
Preparation and characterization of PEI liposome and PEI-PLGA nanoparticles.
| Formulation (PEI lipid:HSPC:DMG-PEG:PLGA) (mg) |
PEI mass content (%) |
Size (nm) | PDI | Zeta potential (mV) | |
|---|---|---|---|---|---|
| PEI12C16 LIPO | 27.5:27.5:2:0 | 8.8 | 148.5 ± 1.36 | 0.13 | +43.13 ± 0.80 |
| PEI8C16 LIPO | 20:10:2:0 | 15.7 | 115.10 ± 1.56 | 0.25 | +37.93 ± 0.76 |
| PEI4C16 LIPO | 12.5:12.5:2:0 | 18.6 | 75.24 ± 1.92 | 0.20 | +32.0 ± 1.63 |
| PEI-PLGA NP | 25:0:2:15 | 15.0 | 146.36 ± 1.89 | 0.18 | +41.1 ± 1.05 |
Note: LIPO (liposome).
Figure 3.

The particle size and size distribution were analyzed by DLS and graphed in (A). SEM was used to characterize PEI liposomes (B) and PEI-PLGA NP (C). The particles appear to be smooth in surface and spherical in shape. In contrast to PEI liposomes, the bright color indicates the hollow core of PEI-PLGA NP in SEM image.
The obtained PEI lipid, with its amphiphilic nature, is a functional surfactant. Next, we designed a core–shell NP using PEI lipids as a surfactant, with PLGA as the core, to develop a core–shell nanoparticle (PEI-PLGA NP) by solvent extraction/evaporation method.55,56 By comparison of three PEI liposomes, PEI8C16 liposomes had the highest transfection efficacy. We have chosen PEI8C16 as the surfactant to formulate core–shell NP. It can be envisaged that the obtained PEI-PLGA NP has a hydrophobic PLGA core, with C16 chains inserted within hydrophilic cationic PEI shells. PEI-PLGA NP has the diameter and zeta potential of 146.36 nm and +41.1 mV respectively (Fig. 3(A)). The positive zeta potential value was in agreement with the structure of the obtained PEI-PLGA NP, with its cationic PEI shell. SEM was used to analyze the morphology of PEI liposomes and PEI-PLGA NP. The SEM images exhibit well-dispersed PEI liposomes (Fig. 3(B)) and PEI-PLGA NP (Fig. 3(C)), which were smooth in surface and spherical in shape. The bright color showing the cores of PEI-PLGA NP is an indication of a hollow core. The results clearly show that the shape and size of PEI liposome and PEI-PLGA NPs were under 150 nm. These measurements correspond with the zetasizer measurement. In contrast to PEI liposome, PEI-PLGA NPs is a firmer shape because of the PLGA core and PEI shell structure. In addition, the shape and size of PEI liposome and PEI-PLGA NP were uniform for months, indicating both chemical and physical stability.
Plasmid DNA (pIL-4/GFP) Encapsulation
The pIL-4/GFP were successfully loaded into liposome due to the electrostatic interaction between the ionizable amino groups located on the liposome surface, and the polyanionic nucleic acids of the pIL-4/GFP. We investigated the effect of pIL-4/GFP encapsulation on size and zeta potential of the PEI4C16 liposome by loading pIL-4/GFP with different nanoformulation (nano)/pIL-4/GFP ratios based on calculations of nitrogen to phosphate (N/P ratio).57 Gel retardation assay was performed to evaluate the ability of PEI4C16 liposome to condense pIL-4/GFP at N/P ratios of 1/0, 20/1, 10/1, 5/1, 2/1, and 1/1 (Fig. 4(A)). The results showed that pIL-4/GFP was totally loaded onto the PEI4C16 liposomes when the N/P ratio was greater than 5/1. When N/P ratio was reduced from 5/1 to 1/1, the unencapsulated pIL-4/GFP was detected with the appearance of a free pIL-4/GFP band. Three liposomes of PEI12C16, PEI8C16 and PEI4C16 had similar pIL-4/GFP loading ability (data not shown). The variations in size and zeta potential of PEI4C16 liposome were also detected for these N/P ratios. By increasing pIL-4/GFP, the sizes were enlarged (Fig. 4(B), red line), while the zeta potential (Fig. 4(B), blue line) declined.
Figure 4.

The effect of pIL-4/GFP encapsulation on size and zeta potential was determined by agarose gel electrophoresis and zetasizer assays. Results of agarose gel reaction for pIL-4/GFP in PEI4C16 liposome showed in the lanes at different N/P ratios (A). The size and zeta potential of the modified PEI4C16 liposome at different N/P ratios showed in (B). The increase in size and decrease in zeta potential were associated to the elevation of pIL-4/GFP loading. For pIL-4/GFP concentration, N/P ratio at 1/1 includes 0.2 µg plasmid.
Cytotoxicity
Lowering the cytotoxicity of the gene delivery platform is one of the major goals for current gene delivery research.12,19,26,30–35,45,46,65 In this study, MTT assay was performed to assess the viability of Hela cells following PEI liposome and PEI-PLGA NP uptake. PEI NP showed significantly lower cytotoxicity after 24 hrs treated to cultures at all experimental concentrations (Fig. 5). In contrast to PEI25K, PEI4C16 and PEI8C16 liposomes exhibited notable lower cytotoxicity at concentrations under 20 ug/ml (Fig. 5). The reduced cytotoxicity of PEI liposome and PEI-PLGA NP was conferred by the low molecular weight of PEI head groups, and will greatly benefit the clinical applications of therapeutic delivery. Among these three PEI liposomes, the toxicity increased as the amount of hexadecyl tail groups increased. We deduced that PEI liposomes with a higher density of tail groups had greater ability to damage the cell bilayer membrane when they interacted with cells. This phenomenon is in accordance with the mechanism of action for membrane disruptive effects of cationic lipids proposed by Sean.19
Figure 5.
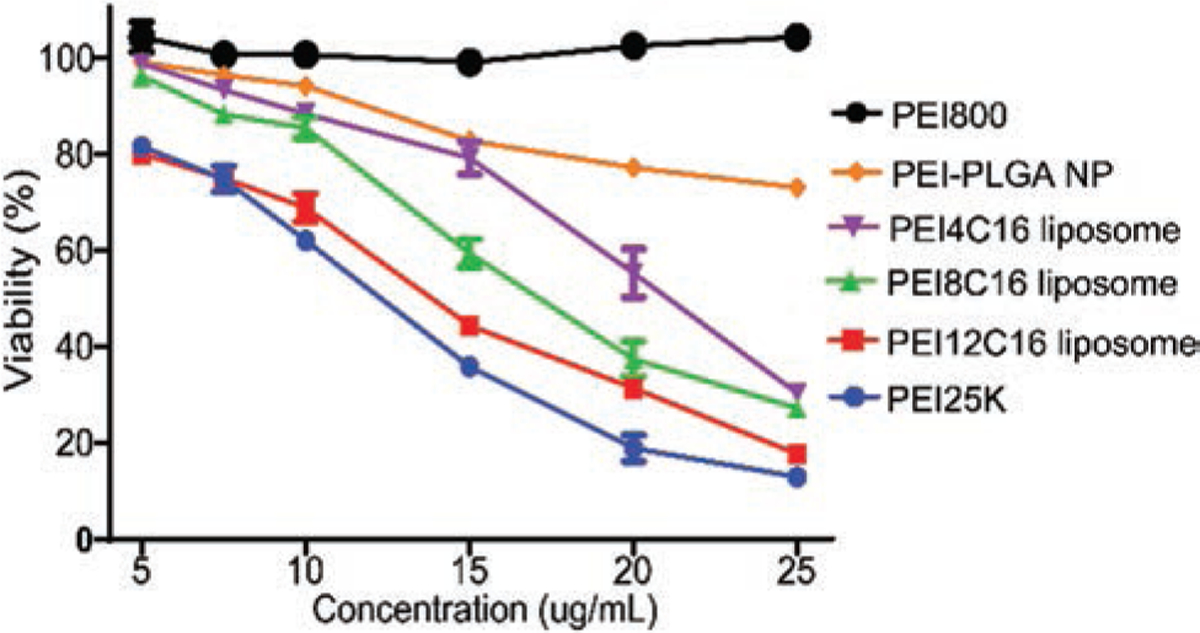
Viability of Hela cells after incubation for 24 hrs with three types of PEI liposome, PEI-PLGA NP, PEI800 and PEI25K at different concentrations. The commercial PEI25K served as control. The concentrations were converted to PEI concentrations. Data are expressed as mean ± SD (n = 3) (P < 0.01).
Cell Uptake and DNA Release
The efficacy of a gene delivery system depends on uptake, the condensation and release of the gene, and its ability to escape from the endosome. Double labeling of endosomes by LysoTracker (red) and pIL-4/GFP by YOYO-1 (green), the microscopy images were further used to observe the intracellular distribution of YOYO-1-pIL-4/GFP loaded PEI nanocarriers. When the cells were incubated with YOYO-1-pIL-4/GFP-PEI liposomes for 1 hr, green spots of YOYO-1-pIL-4/GFP were observed in the cytoplasm, indicating the quick uptake by the cultures. The co-localization indicated the accordance of endosomal (Fig. 6, red) entrapping of YOYO-1-pIL-4/GFP-PEI liposomes (Fig. 6, green) via the endocytosis uptake mechanism. After 2 hrs incubation, YOYO-1-pIL-4/GFP was separated from endosomes (Fig. 6, arrow head). Importantly, when the incubation time was further increased to 24 hrs, the separated YOYO-1-pIL-4/GFP entered and accumulated in nuclei (Fig. 6, arrow). This phenomenon indicates that successful separation of DNA from endosomes and subsequent integration into the host’s genome were the key steps for an effective gene transfection.16,66,67
Figure 6.
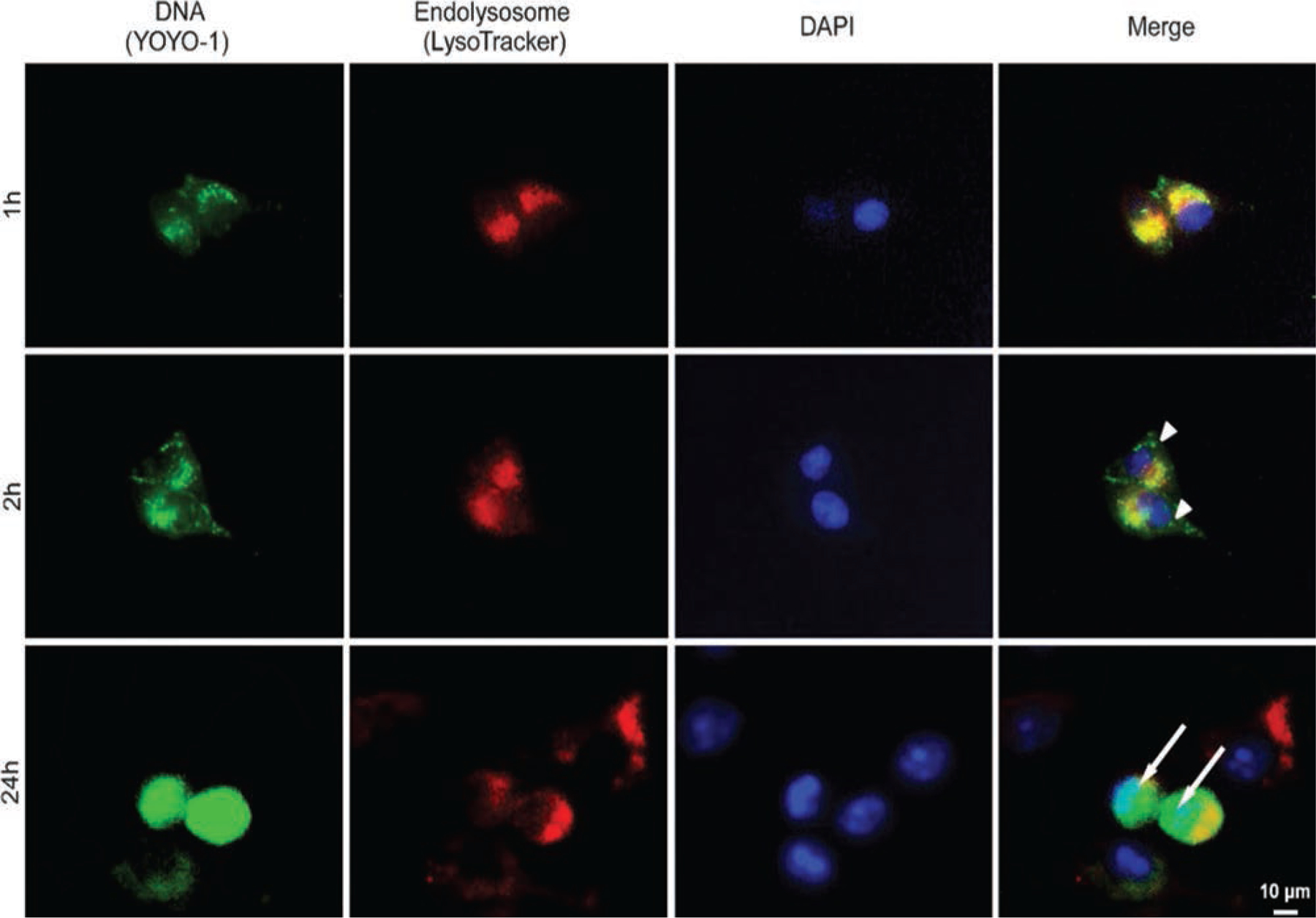
The pIL-4/GFP was labeled with YOYO-1 (green) before loading into PEI liposomes. Hela cells were treated with YOYO-1-pIL-4/GFP loaded PEI liposomes for 1, 2 and 24 hrs. The lysosomes were labeled with LysoTracker (red), and the nuclei were stained with DAPI (blue). The co-localization of YOYO-1-pIL-4/GFP (green) and lysosomes (red) was determined by florescence microscopy at 1 h. With 2 hrs of incubation, the YOYO-1 labeled pIL-4/GFP was released from lysosomes presenting in cytoplasm (arrow head). After 24 hrs, the amount of YOYO-1-pIL-4/GFP escaped from lysosomes and entered nuclei (arrow).
Gene Transfer Efficiency
The pIL-4/GFP containing full-length GFP-IL4 DNA was loaded into PEI4C16 liposome, PEI8C16 liposome and PEI-PLGA NP for optimizing transfection efficacy. The commercially available PEI25K22 was used as control. Hela cells were transfected at different N/P ratio with the pIL-4/GFP concentration of 2 µg/mL or 4 µg/mL. With a series of tentative transfection experiments, the GFP expression was analyzed by fluorescence microscopy (Fig. 7(A)) and flow cytometry (Fig. 7(B)). PEI8C16 and PEI-PLGA NP provided greater transfection efficacy. The transfection efficacy could not be achieved when pIL-4/GFP concentrations were increased at the same N/P ratio, which may result from elevated toxicity. At the pIL-4/GFP concentration of 2 µg/mL, PEI4C16 liposome treated cultures exhibited fewer GFP positive cells. By taking advantage of the less toxic PEI8C16 liposome, the highest percentage of GFP positive cells was at the N/P ratio of 15/1 (Fig. 7(C)). Quantitative analysis of GFP protein production was measured by flow cytometry.12 PEI-PLGA NP produced greater GFP fluorescence intensity in comparison to PEI-liposomes and PEI125K (Fig. 7(D)). To better identify and characterize functional pIL-4 delivery, mouse BMMs were transfected with human pIL-4 (hIL-4) and mouse pIL-4 (mIL-4). The culture medium was collected to perform ELISA test. The human and mouse IL-4 protein levels were significantly increased at day 5 post-transfection compared to the non-transfected controls (Fig. 7(E)). In transfected cultures with human pIL-4 obtained 103.08 ± 4.82 pg/mL of human IL4 protein. Mouse BMMs transfected with mouse pIL-4 increased IL-4 protein production to 505.1 ± 11.86 pg/mL in comparison to 20.5 ± 3.38 pg/mL seen in the controls. Both human and mouse pIL-4 were efficiently transfected into cells in which human IL-4 expression were markedly higher than control.
Figure 7.
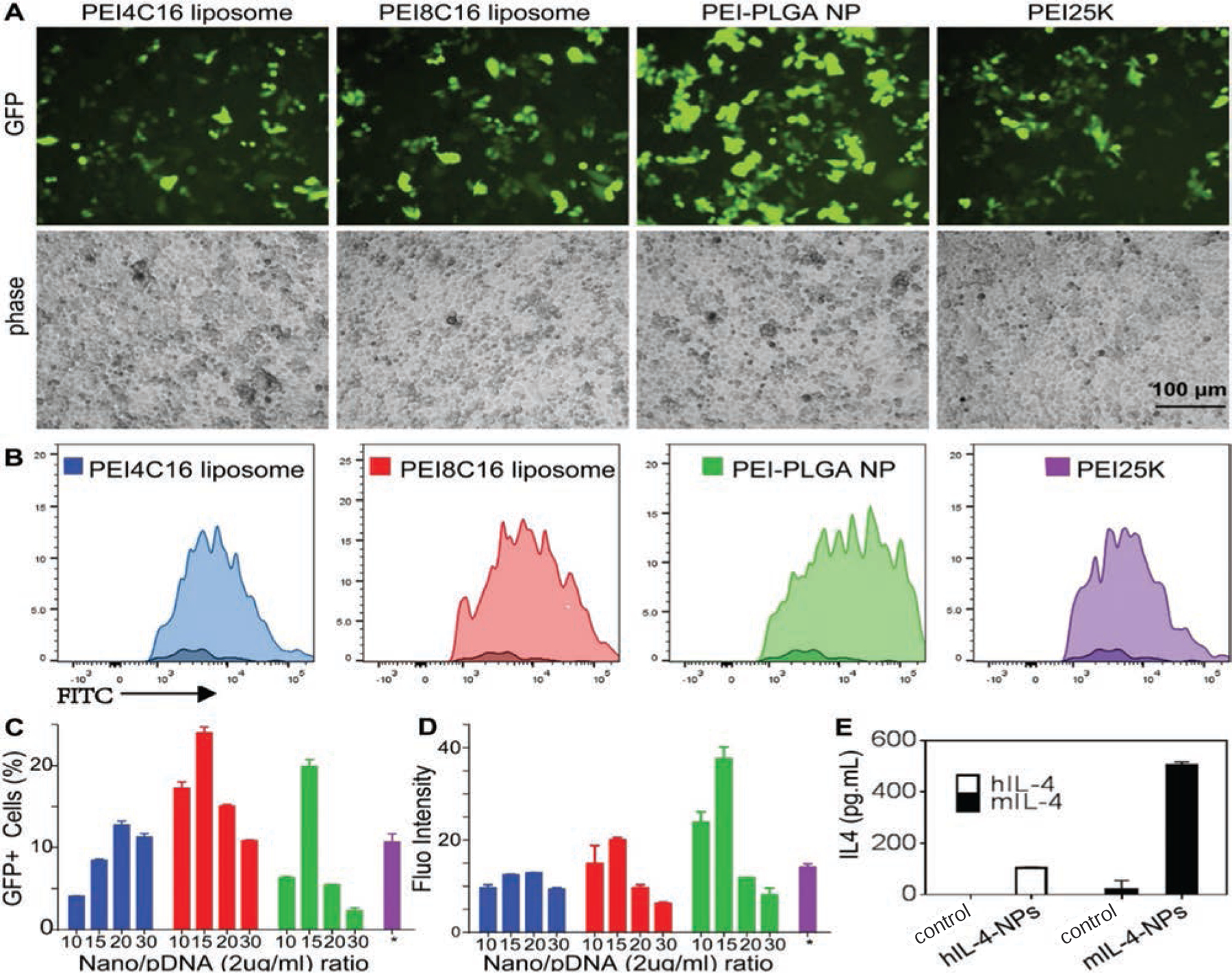
Fluorescence microscopy and flow cytometry assays were used to determine the transfection of pIL-4/GFP loaded PEI4C16 liposome, PEI8C16 liposome, PEI-PLGA NP and PEI25K under their best transfection conditions. The transfection experiments were performed with pIL-4/GFP concentration at 4 µg/mL in PEI4C16 liposome, and 2 µg/mL in PEI8C16 liposome and PEI-PLGA NP. The transfection by PEI25K was at its optimized N/P ratio of 10/1 (4 µg/mL). The images of GFP expression were observed at various N/P ratios (A). Transfection efficiency of pIL-4/GFP in cells was determined by flow cytometry (B). Quantitative analysis of fluorescence microscopy images was used to calculate the percentage of GFP positive cells (C). The levels of GFP expression were measured by flow cytometry to determine the fluorescence intensity (D). To identify the expression of exogenous IL4, mouse bone marrow derived macrophages (BMMs) were transfected with human pIL-4 (hIL-4) and mouse pIL-4 (mIL-4) (E). The culture medium were collected to perform ELISA test. The levels of human and mouse IL-4 protein in BMMs culture medium were significantly increased at day 5 post-transfection as compared to the control groups (E). Data are expressed as mean ± SD (n = 3) (P < 0.01).
DISCUSSION
In this study, we synthesized PEI lipids with low molecular weight PEI as the headgroup51 and hexadecyl chain as the tail group by highly efficient chemical reactions. The ring-opening reaction of epoxides by amine groups was carried out under mild conditions without any solvent. This process is highly efficient and specific, and yields product with negligible impurities. Compared to a similar modification process, no further purification is needed.53 In our study, under the molar ratios of 12 to 1, 8 to 1, and 4 to 1 of EpoxyC16 to PEI800, PEI800 was found to be fully modified by reaction with EpoxyC16, and produced three PEI headgroup lipids.
The obtained PEI lipids, with their hydrophobic C16 alkyl chains and hydrophilic cationic PEI head, are suitable components for preparing liposomes and a proper surfactant for the preparation of PLGA nanoparticle by a solvent extraction/evaporation method.55,56 PEI liposome and PEI-PLGA NP developed by the low molecular weight PEI lipids, were capable of effectively delivering full-length pIL-4/GFP into the cells for gene transfection. The positive zeta potential value was in agreement with the structure of the obtained PEI liposome and PEI-PLGA NP with cationic PEI shell. All four nanoformulations made from PEIs successfully transferred plasmid DNA into Hela cells. The key for efficient plasmid DNA transfection is formulating a higher mass percentage of PEI headgroups to liposome composition. We successfully increased the mass percentage of three PEI lipids to 18.6%, 15.7%, and 8.8% to produce the liposomes. In addition, the unique features of PEI-PLGA NP as a co-delivery system68,69 for the packaging of hydrophobic molecules inside of PLGA core and to load genes onto PEI shell, is certainly realized.25,70
PEI liposomes and PEI-PLGA NP are effective in gene delivery due to the proton sponge effect of PEI, which promotes intracellular release and escape from the endosomes.71,72 The positive potential obtained from PEI liposomes and PEI-PLGA NP was found to promote cellular uptake. By labeling DNA with YOYO-1 (green), we were able to visualize the uptake, release, and intracellular distribution.73 Double fluorescence labeling of pIL-4/GFP and lysosome effectively detected the delivery efficacy by visualizing the subcellular localization of pIL-4/GFP (green) and trafficking the escape from lysosomes (red). We found that the quick uptake of PEI liposomes and PEI-PLGA NP led to an increase of intracellular pIL-4/GFP. The release of DNA from lysosomes is a critical step for pIL-4/GFP transfection. These NPs, with negative surface charge, were capable of effectively loading pIL-4/GFP.
In vitro, all nanoformulations formed with low molecular weight PEI lipids showed lower toxicity and higher transfection efficacy than high molecular weight PEI control. The reduced cytotoxicity of nanocarriers from modified low molecular weight PEI has great potential for gene delivery and clinical applications. These two delivery platforms may have versatile applications in gene therapy due to readily available raw materials, the facile, efficient and clean preparation, and the potential of small therapeutic molecules co-delivery. We are currently carrying out a further study on improving chemical structures and formulation with the aim of enhancing gene delivery and co-delivery of small therapeutic molecules, with specific targeting and appropriate stimuli response.
Acknowledgments:
This study was supported by NIH 1R01GM114851–01A1, the Seed Grant of Texas Tech University Health Sciences Center El Paso and the international program of excellent lecturers of Shandong province education department, Shandong, China. We gratefully thank Beverley Court, Michelle Castro, and Gloria Chavez for their outstanding administrative support.
REFERENCES
- 1.Midoux P and Monsigny M, Efficient gene transfer by histidylated polylysine pIL-4/GFP complexes. Bioconjugate Chem 10, 406 (1999). [DOI] [PubMed] [Google Scholar]
- 2.Putnam D, Gentry CA, Pack DW, and Langer R, Polymer-based gene delivery with low cytotoxicity by a unique balance of side-chain termini. Proc. Natl. Acad. Sci. USA 98, 1200 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zauner W, Ogris M, and Wagner E, Polylysine-based transfection systems utilizing receptor-mediated delivery. Advanced Drug Delivery Reviews 30, 97 (1998). [DOI] [PubMed] [Google Scholar]
- 4.Tao Y, Han J, and Dou H, Surface modification of paclitaxel-loaded polymeric nanoparticles: Evaluation of in vitro cellular behavior and in vivo pharmacokinetic. Polymer 53, 5078 (2012). [Google Scholar]
- 5.Tao YH, Han JF, and Dou HY, Brain-targeting gene delivery using a rabies virus glycoprotein peptide modulated hollow liposome: Bio-behavioral study. J. Mater. Chem 22, 11808 (2012). [Google Scholar]
- 6.Tao Y, Han J, and Dou H, Paclitaxel-loaded tocopheryl succinate-conjugated chitosan oligosaccharide nanoparticles for synergistic chemotherapy. J. Mater. Chem 22, 8930 (2012). [Google Scholar]
- 7.Termsarasab U, Cho H-J, Kim DH, Chong S, Chung S-J, Shim C-K, Moon HT, and Kim D-D, Chitosan oligosaccharide-arachidic acid-based nanoparticles for anti-cancer drug delivery. Int. J. Pharm 441, 373 (2013). [DOI] [PubMed] [Google Scholar]
- 8.Termsarasab U, Yoon I-S, Park J-H, Moon HT, Cho H-J, and Kim D-D, Polyethylene glycol-modified arachidyl chitosan-based nanoparticles for prolonged blood circulation of doxorubicin. Int. J. Pharm 464, 127 (2014). [DOI] [PubMed] [Google Scholar]
- 9.Lu H, Dai Y, Lv L, and Zhao H, Chitosan-graft-polyethylenimine/DNA nanoparticles as novel non-viral gene delivery vectors targeting osteoarthritis. PLoS One 9, e84703 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fang JK, Chen L, Lu XG, Cao D, Guo LL, Zhang YS, Li LB, Zhang LF, Kuang YT, and Wang SL, Optimization of transforming growth factor-beta1 siRNA loaded chitosan-tripolyphosphate nanoparticles for the treatment of colorectal cancer hepatic metastasis in a mouse model. J. Biomed. Nanotechnol 12, 1489 (2016). [DOI] [PubMed] [Google Scholar]
- 11.Duan S, Song M, He J, Zhou N, Zhou S, Zhao J, Fang Y, Yi P, Huang X, Luo G, Lai C, Yu X, Zhang Z, Xie Y, Zhao Y, and Lu X, Folate-modified chitosan nanoparticles coated interferon-inducible protein-10 gene enhance cytotoxic T lymphocytes’ responses to hepatocellular carcinoma. J. Biomed. Nanotechnol 12, 700 (2016). [DOI] [PubMed] [Google Scholar]
- 12.Chang H, Wang H, Shao N, Wang M, Wang X, and Cheng Y, Surface-engineered dendrimers with a diaminododecane core achieve efficient gene transfection and low cytotoxicity. Bioconjugate Chem 25, 342 (2014). [DOI] [PubMed] [Google Scholar]
- 13.Tang MX, Redemann CT, and Szoka FC Jr., In vitro gene delivery by degraded polyamidoamine dendrimers. Bioconjug. Chem 7, 703 (1996). [DOI] [PubMed] [Google Scholar]
- 14.Zou L, Tao Y, Payne G, Do L, Thomas T, Rodriguez J, and Dou H, Targeted delivery of nano-PTX to the brain tumor-associated macrophages. Oncotarget 8, 6564 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yan Y, Xue L, Miller JB, Zhou K, Kos P, Elkassih S, Liu L, Nagai, Xiong H, and Siegwart DJ, One-pot synthesis of functional poly(amino ester sulfide)s and utility in delivering pIL-4/GFP and siRNA. Polymer 72, 271 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang Z, Yin L, Xu Y, Tong R, Lu Y, Ren J, and Cheng J, Facile functionalization of polyesters through thiol-yne chemistry for the design of degradable, cell-penetrating and gene delivery dual-functional agents. Biomacromolecules 13, 3456 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen J, Liu Q, Xiao J, and Du J, EpCAM-antibody-labeled non-cytotoxic polymer vesicles for cancer stem cells-targeted delivery of anticancer drug and siRNA. Biomacromolecules 16, 1695 (2015). [DOI] [PubMed] [Google Scholar]
- 18.Felgner PL, Gadek TR, Holm M, Roman R, Chan HW, Wenz M, Northrop JP, Ringold GM, and Danielsen M, Lipofection: A highly efficient, lipid-mediated DNA-transfection procedure. Proceedings of the National Academy of Sciences 84, 7413 (1987). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Semple SC, Akinc A, Chen J, Sandhu AP, Mui BL, Cho CK, Sah DW, Stebbing D, Crosley EJ, Yaworski E, Hafez IM, Dorkin JR, Qin J, Lam K, Rajeev KG, Wong KF, Jeffs LB, Nechev L, Eisenhardt ML, Jayaraman M, Kazem M, Maier MA, Srinivasulu M, Weinstein MJ, Chen Q, Alvarez R, Barros SA, De S, Klimuk SK, Borland T, Kosovrasti V, Cantley WL, Tam YK, Manoharan M, Ciufolini MA, Tracy MA, de Fougerolles A, MacLachlan I, Cullis PR, Madden TD, and Hope MJ, Rational design of cationic lipids for siRNA delivery. Nat. Biotechnol 28, 172 (2010). [DOI] [PubMed] [Google Scholar]
- 20.Whitehead KA, Dorkin JR, Vegas AJ, Chang PH, Veiseh O, Matthews J, Fenton OS, Zhang Y, Olejnik KT, Yesilyurt V,Chen D, Barros S, Klebanov B, Novobrantseva T, Langer R, and Anderson DG, Degradable lipid nanoparticles with predictable in vivo siRNA delivery activity. Nat. Commun 5:4227 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Godbey WT, Wu KK, and Mikos AG, Poly(ethylenimine) and its role in gene delivery. J. Controlled Release 60, 149 (1999). [DOI] [PubMed] [Google Scholar]
- 22.Boussif O, Lezoualc’h F, Zanta MA, Mergny MD, Scherman D, Demeneix B, and Behr JP, A versatile vector for gene and oligonucleotide transfer into cells in culture and in vivo: Polyethylenimine. Proc. Natl. Acad. Sci. USA 92, 7297 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Coll J-L, Chollet P, Brambilla E, Desplanques D, Behr J-P, and Favrot M, In vivo delivery to tumors of DNA complexed with linear polyethylenimine. Human Gene Therapy 10, 1659 (1999). [DOI] [PubMed] [Google Scholar]
- 24.Wightman L, Kircheis R, Rössler V, Carotta S, Ruzicka R, Kursa M, and Wagner E, Different behavior of branched and linear polyethylenimine for gene delivery in vitro and in vivo. The Journal of Gene Medicine 3, 362 (2001). [DOI] [PubMed] [Google Scholar]
- 25.Mimi H, Ho KM, Siu YS, Wu A, and Li P, Polyethyleneimine-based core–shell nanogels: A promising siRNA carrier for argininosuccinate synthetase mRNA knockdown in Hela cells. J. Controlled Release 158, 123 (2012). [DOI] [PubMed] [Google Scholar]
- 26.Yao H, Ng S-MS, Tang G-P, and Lin MC, Development of a novel low toxicity and high efficiency PEI-based nanopolymer for gene delivery in vitro and in vivo. Molecular Therapy 17, S64 (2009). [Google Scholar]
- 27.Lungwitz U, Breunig M, Blunk T, and Göpferich A, Polyethylenimine-based non-viral gene delivery systems. European Journal of Pharmaceutics and Biopharmaceutics 60, 247 (2005). [DOI] [PubMed] [Google Scholar]
- 28.Yamada H, Loretz B, and Lehr C-M, Design of starch-graft-PEI polymers: An effective and biodegradable gene delivery platform. Biomacromolecules 15, 1753 (2014). [DOI] [PubMed] [Google Scholar]
- 29.Jager M, Schubert S, Ochrimenko S, Fischer D, and Schubert US, Branched and linear poly(ethylene imine)-based conjugates: Synthetic modification, characterization, and application. Chem. Soc. Rev 41, 4755 (2012). [DOI] [PubMed] [Google Scholar]
- 30.Moghimi SM, Symonds P, Murray JC, Hunter AC, Debska G, and Szewczyk A, A two-stage poly(ethylenimine)-mediated cytotoxicity: Implications for gene transfer/therapy. Mol. Ther 11, 990 (2005). [DOI] [PubMed] [Google Scholar]
- 31.Zintchenko A, Philipp A, Dehshahri A, and Wagner E, Simple modifications of branched PEI lead to highly efficient siRNA carriers with low toxicity. Bioconjugate Chem 19, 1448 (2008). [DOI] [PubMed] [Google Scholar]
- 32.Fischer D, Li Y, Ahlemeyer B, Krieglstein J, and Kissel T, In vitro cytotoxicity testing of polycations: Influence of polymer structure on cell viability and hemolysis. Biomaterials 24, 1121 (2003). [DOI] [PubMed] [Google Scholar]
- 33.Gao J-Q, Zhao Q-Q, Lv T-F, Shuai W-P, Zhou J, Tang G-P, Liang W-Q, Tabata Y, and Hu Y-L, Gene-carried chitosan-linked-PEI induced high gene transfection efficiency with low toxicity and significant tumor-suppressive activity. Int. J. Pharm 387, 286 (2010). [DOI] [PubMed] [Google Scholar]
- 34.Kim YH, Park JH, Lee M, Kim Y-H, Park TG, and Kim SW, Polyethylenimine with acid-labile linkages as a biodegradable gene carrier. J. Controlled Release 103, 209 (2005). [DOI] [PubMed] [Google Scholar]
- 35.Beyerle A, Irmler M, Beckers J, Kissel T, and Stoeger T, Toxicity pathway focused gene expression profiling of PEI-based polymers for pulmonary applications. Molecular Pharmaceutics 7, 727 (2010). [DOI] [PubMed] [Google Scholar]
- 36.Boeckle S, Fahrmeir J, Roedl W, Ogris M, and Wagner E, Melittin analogs with high lytic activity at endosomal pH enhance transfection with purified targeted PEI polyplexes. J. Control. Release 112, 240 (2006). [DOI] [PubMed] [Google Scholar]
- 37.Cheng W, Yang C, Hedrick JL, Williams DF, Yang YY, and Ashton-Rickardt PG, Delivery of a granzyme B inhibitor gene using carbamate-mannose modified PEI protects against cytotoxic lymphocyte killing. Biomaterials 34, 3697 (2013). [DOI] [PubMed] [Google Scholar]
- 38.Huang FW, Wang HY, Li C, Wang HF, Sun YX, Feng J, Zhang XZ, and Zhuo RX, PEGylated PEI-based biodegradable polymers as non-viral gene vectors. Acta Biomater 6, 4285 (2010). [DOI] [PubMed] [Google Scholar]
- 39.Kleemann E, Neu M, Jekel N, Fink L, Schmehl T, Gessler T, Seeger W, and Kissel T, Nano-carriers for DNA delivery to the lung based upon a TAT-derived peptide covalently coupled to PEG-PEI. J. Control. Release 109, 299 (2005). [DOI] [PubMed] [Google Scholar]
- 40.Li B, Zhou F, Huang K, Wang Y, Mei S, Zhou Y, and Jing T, Environmentally friendly chitosan/PEI-grafted magnetic gelatin for the highly effective removal of heavy metals from drinking water. Sci. Rep 7, 43082 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Muthiah M, Che HL, Kalash S, Jo J, Choi SY, Kim WJ, Cho CS, Lee JY, and Park IK, Formulation of glutathione responsive anti-proliferative nanoparticles from thiolated Akt1 siRNA and disulfide-crosslinked PEI for efficient anti-cancer gene therapy. Colloids Surf. B Biointerfaces 126, 322 (2015). [DOI] [PubMed] [Google Scholar]
- 42.Rudolph C, Sieverling N, Schillinger U, Lesina E, Plank C, Thunemann AF, Schonberger H, and Rosenecker J, Thyroid hormone (T3)-modification of polyethyleneglycol (PEG)-polyethyleneimine (PEI) graft copolymers for improved gene delivery to hepatocytes. Biomaterials 28, 1900 (2007). [DOI] [PubMed] [Google Scholar]
- 43.Siegman S, Truong NF, and Segura T, Encapsulation of PEGy-lated low-molecular-weight PEI polyplexes in hyaluronic acid hydrogels reduces aggregation. Acta Biomater 28, 45 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhou Y, Yu F, Zhang F, Chen G, Wang K, Sun M, Li J, and Oupicky D, Cyclam-modified PEI for combined VEGF siRNA silencing and CXCR4 inhibition to treat metastatic breast cancer. Biomacromolecules 19, 392 (2018). [DOI] [PubMed] [Google Scholar]
- 45.Dong W, Jin G-H, Li S-F, Sun Q-M, Ma D-Y, and Hua Z-C, Cross-linked polyethylenimine as potential DNA vector for gene delivery with high efficiency and low cytotoxicity. Acta Biochimica et Biophysica Sinica 38, 780 (2006). [DOI] [PubMed] [Google Scholar]
- 46.Forrest ML, Koerber JT, and Pack DW, A degradable polyethylenimine derivative with low toxicity for highly efficient gene delivery. Bioconjugate Chem 14, 934 (2003). [DOI] [PubMed] [Google Scholar]
- 47.Xun M-M, Xiao Y-P, Zhang J, Liu Y-H, Peng Q, Guo Q, Wu W-X, Xu Y, and Yu X-Q, Low molecular weight PEI-based polycationic gene vectors via Michael addition polymerization with improved serum-tolerance. Polymer 65, 45 (2015). [Google Scholar]
- 48.Wang F, Shen Y, Zhang W, Li M, Wang Y, Zhou D, and Guo S, Efficient, dual-stimuli responsive cytosolic gene delivery using a RGD modified disulfide-linked polyethylenimine functionalized gold nanorod. J. Controlled Release 196, 37 (2014). [DOI] [PubMed] [Google Scholar]
- 49.Petersen H, Merdan T, Kunath K, Fischer D, and Kissel T, Poly(ethylenimine-co-l-lactamide-co-succinamide): A biodegradable polyethylenimine derivative with an advantageous pH-dependent hydrolytic degradation for gene delivery. Bioconjugate Chem 13, 812 (2002). [DOI] [PubMed] [Google Scholar]
- 50.Wei H, Volpatti LR, Sellers DL, Maris DO, Andrews IW, Hemphill S, Chan LW, Chu DSH, Horner PJ, and Pun SH, Dual Responsive, stabilized nanoparticles for efficient in vivo plasmid delivery. Angew. Chem. Int. Ed 52, 5377 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhi D, Zhang S, Cui S, Zhao Y, Wang Y, and Zhao D, The headgroup evolution of cationic lipids for gene delivery. Bioconjugate Chem 24, 487 (2013). [DOI] [PubMed] [Google Scholar]
- 52.Gehin C, Montenegro J, Bang EK, Cajaraville A, Takayama S, Hirose H, Futaki S, Matile S, and Riezman H, Dynamic amphiphile libraries to screen for the “fragrant” delivery of siRNA into Hela cells and human primary fibroblasts. J. Am. Chem. Soc 135, 9295 (2013). [DOI] [PubMed] [Google Scholar]
- 53.Xiao J, Duan X, Yin Q, Chen L, Zhang Z, and Li Y, Low molecular weight polyethylenimine-graft-tween 85 for effective gene delivery: Synthesis and in vitro characteristics. Bioconjugate Chem 23, 222 (2012). [DOI] [PubMed] [Google Scholar]
- 54.Chen YC, Zhu XD, Zhang XJ, Liu B, and Huang L, Nanoparticles modified with tumor-targeting scFv deliver siRNA and miRNA for cancer therapy. Molecular Therapy 18, 1650 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Freitas S, Merkle HP, and Gander B, Microencapsulation by solvent extraction/evaporation: Reviewing the state of the art of micro-sphere preparation process technology. J. Control. Release 102, 313 (2005). [DOI] [PubMed] [Google Scholar]
- 56.Liu XY, Ruan LM, Mao WW, Wang JQ, Shen YQ, and Sui MH, Preparation of RGD-modified long circulating liposome loading matrine, and its in vitro anti-cancer effects. International Journal of Medical Sciences 7, 197 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhao QQ, Chen JL, Lv TF, He CX, Tang GP, Liang WQ, Tabata Y, and Gao JQ, N /P ratio significantly influences the transfection efficiency and cytotoxicity of a polyethylenimine/chitosan/DNA complex. Biol. Pharm. Bull 32, 706 (2009). [DOI] [PubMed] [Google Scholar]
- 58.Kang T, Amir RJ, Khan A, Ohshimizu K, Hunt JN, Sivanandan K, Montanez MI, Malkoch M, Ueda M, and Hawker CJ, Facile access to internally functionalized dendrimers through efficient and orthogonal click reactions. Chem. Commun. (Camb) 46, 1556 (2010). [DOI] [PubMed] [Google Scholar]
- 59.Wan D, Wang G, Pu H, and Jin M, Can nonspecific host-guest interaction lead to highly specific encapsulation by a supramolecular nanocapsule? Macromolecules 42, 6448 (2009). [Google Scholar]
- 60.Wan D, Pu H, Jin M, Pan H, and Chang Z, Enhancing the unimolecularity and control for guest release of a macromolecular nanocapsule via core engineering. Reactive and Functional Polymers 70, 916 (2010). [Google Scholar]
- 61.Zou L, Zhu W, Chen Y, and Xi F, Modification of side chain terminals of PEGylated molecular bottle brushes—A toolbar of molecular nanoobjects. Polymer 54, 481 (2013). [Google Scholar]
- 62.Kim JY, Kim JK, Park JS, Byun Y, and Kim CK, The use of PEGylated liposomes to prolong circulation lifetimes of tissue plasminogen activator. Biomaterials 30, 5751 (2009). [DOI] [PubMed] [Google Scholar]
- 63.Allen TM and Cullis PR, Liposomal drug delivery systems: From concept to clinical applications. Advanced Drug Delivery Reviews 65, 36 (2013). [DOI] [PubMed] [Google Scholar]
- 64.Vemuri S and Rhodes CT, Preparation and characterization of liposomes as therapeutic delivery systems: A review. Pharmaceutica Acta Helvetiae 70, 95 (1995). [DOI] [PubMed] [Google Scholar]
- 65.Hunter AC, Molecular hurdles in polyfectin design and mechanistic background to polycation induced cytotoxicity. Advanced Drug Delivery Reviews 58, 1523 (2006). [DOI] [PubMed] [Google Scholar]
- 66.Varkouhi AK, Scholte M, Storm G, and Haisma HJ, Endosomal escape pathways for delivery of biologicals. J. Controlled Release 151, 220 (2011). [DOI] [PubMed] [Google Scholar]
- 67.Gabrielson NP and Cheng J, Multiplexed supramolecular self-assembly for non-viral gene delivery. Biomaterials 31, 9117 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sun T-M, Du J-Z, Yao Y-D, Mao C-Q, Dou S, Huang S-Y, Zhang P-Z, Leong KW, Song E-W, and Wang J, Simultaneous delivery of siRNA and paclitaxel via a “two-in-one” micelleplex promotes synergistic tumor suppression. ACS Nano 5, 1483 (2011). [DOI] [PubMed] [Google Scholar]
- 69.Wang Y, Gao S, Ye W-H, Yoon HS, and Yang Y-Y, Co-delivery of drugs and DNA from cationic core–shell nanoparticles self-assembled from a biodegradable copolymer. Nat. Mater 5, 791 (2006). [DOI] [PubMed] [Google Scholar]
- 70.Zhu J, Tang A, Law LP, Feng M, Ho KM, Lee DKL, Harris FW, and Li P, Amphiphilic core–shell nanoparticles with poly(ethylenimine) shells as potential gene delivery carriers. Biocon-jugate Chem 16, 139 (2005). [DOI] [PubMed] [Google Scholar]
- 71.Fichter KM, Ingle NP, McLendon PM, and Reineke TM, Polymeric nucleic acid vehicles exploit active interorganelle trafficking mechanisms. ACS Nano 7, 347 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Yue Y, Jin F, Deng R, Cai J, Dai Z, Lin MC, Kung HF, Mattebjerg MA, Andresen TL, and Wu C, Revisit complexation between DNA and polyethylenimine-effect of length of free polycationic chains on gene transfection. J. Control. Release 152, 143 (2011). [DOI] [PubMed] [Google Scholar]
- 73.Zhang J, Fu Y, and Lakowicz JR, Fluorescence images of DNA-bound YOYO between coupled silver particles. Langmuir 23, 11734 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]


