Abstract
The human retrovirus human T-cell leukemia virus type I (HTLV-1) infects human T cells by vertical transmission from mother to child through breast milk or horizontal transmission through blood transfusion or sexual contact. Approximately 5% of infected individuals develop adult T-cell leukemia/lymphoma (ATL) with a poor prognosis, while 95% of infected individuals remain asymptomatic for the rest of their lives, during which time the infected cells maintain a stable immortalized latent state in the body. It is not known why such a long latent state is maintained. We hypothesize that the role of functional proteins of HTLV-1 during early infection influences the phenotype of infected cells in latency. In eukaryotic cells, a mRNA quality control mechanism called nonsense-mediated mRNA decay (NMD) functions not only to eliminate abnormal mRNAs with nonsense codons but also to target virus-derived RNAs. We have reported that HTLV-1 genomic RNA is a potential target of NMD, and that Rex suppresses NMD and stabilizes viral RNA against it. In this study, we aimed to elucidate the molecular mechanism of NMD suppression by Rex using various Rex mutant proteins. We found that region X (aa20–57) of Rex, the function of which has not been clarified, is required for NMD repression. We showed that Rex binds to Upf1, which is the host key regulator to detect abnormal mRNA and initiate NMD, through this region. Rex also interacts with SMG5 and SMG7, which play essential roles for the completion of the NMD pathway. Moreover, Rex selectively binds to Upf3B, which is involved in the normal NMD complex, and replaces it with a less active form, Upf3A, to reduce NMD activity. These results revealed that Rex invades the NMD cascade from its initiation to completion and suppresses host NMD activity to protect the viral genomic mRNA.
Keywords: HTLV-1 rex, NMD inhibition, viral RNA, Upf1, Upf3, SMG5/7
1. Introduction
In the period 1980–1981, human T-cell leukemia virus type I (HTLV-1) was discovered as a human retrovirus in Japan and the USA, and was later shown to be the causative agent of adult T-cell leukemia/lymphoma (ATL) [1,2,3,4,5]. HTLV-1 mainly targets CD4+ T cells and causes ATL in approximately 5% of infected individuals after an incubation period of 50–60 years [6]. In contrast, 95% of infected individuals remain asymptomatic throughout their lives, and infected cells maintain a stable immortalized latent state in the body for decades [7]. We hypothesize that the function of HTLV-1 viral accessory proteins from infection to transition to latency influences the phenotype of infected cells during latent infection.
HTLV-1 is a sense-stranded RNA virus belonging to the genus Deltaretrovirus in the family Retroviridae. After infection, the virus undergoes reverse transcription and is permanently integrated into the human genomic DNA of the host cell as a 9 kb provirus with a long terminal repeat (LTR) at both ends. The provirus first transcribes a fully (twice) spliced Tax/Rex mRNA. Tax strongly activates the LTR, promotes transcription of viral genes, and disrupts various gene expression, signal transduction, cell cycle regulation, and DNA damage responses in the host cell. It is thought to be involved in the immortalization and transformation of HTLV-1-infected T cells [7,8,9,10,11,12]. Rex binds specifically to the Rex recognition element (RxRE) in the 3′-UTR of HTLV-1 mRNA and mediates nuclear export of viral mRNA in a CRM1-dependent manner [13,14]. In particular, Rex facilitates the extranuclear trafficking of unstable non-spliced Gag/Pro/Pol mRNA and incompletely spliced Env mRNA to promote the production of viral particles [15,16,17,18,19]. This is accompanied by a decrease in the level of Tax/Rex mRNA expression and a shift to a latent infection state. Rex is therefore thought to regulate the timing of HTLV-1 replication and latency [20,21,22].
Rex is a 27 kDa phosphoprotein consisting of 189 amino acids and has seven phosphorylation sites: Thr-22, Ser-36, Thr-37, Ser-70, Ser-97, Ser-106 and Thr-174, of which Ser-97 and Thr-174 are thought to be important in the transport of viral mRNA [23]. Rex has several domains, which are nuclear/nucleolus localization signals (NLS) for importin-β-mediated nuclear translocation and arginine-rich motifs (ARM) for RNA binding via the RxRE (aa1–19); the nuclear export signal (NES) (aa79–99); the multimerization domains (MD) (aa57–66 and aa106–124), and the stability domain (SD) (aa170–189) [19,24]. Rex shuttles between the nucleus and the cytoplasm, transporting viral mRNA-encoding structural proteins to the site of translation and regulating virus particle formation [21,22].
On the other hand, eukaryotic cells are universally equipped with nonsense-mediated mRNA decay (NMD), which targets and degrades aberrant mRNAs with premature termination codons (PTCs) that exist more than 50–55 nt upstream of the exon junction complex (EJC). When the ribosome stops at the stop codon on the mRNA, the translation termination factors, eRF1 and eRF3, and the central regulators of NMD, Upf1 and SMG1, are recruited to the ribosome, forming the SURF complex. When the stop codon is located upstream of the EJC, the SURF complex and EJC form a decay inducing complex (DECID), which is recognized as an aberrant mRNA with PTCs. When DECID is formed, Upf1 is phosphorylated by SMG1, initiating the NMD pathway. Phosphorylated Upf1 and the aberrant mRNA are transferred to the processing body (p-body), where Upf1 dephosphorylation by SMG5/7 or SMG6 dissociates Upf1 from the mRNA and recruits nuclease, completing the degradation of the aberrant mRNA. Recently, it has been shown that the levels of normal mRNAs containing upstream open reading frame (uORF), programmed ribosomal frameshift (PRF) signal, and a long 3′-untranslated region (UTR) of more than 1000 nt are fine-tuned by NMD; thus, NMD is an essential mechanism for maintaining cellular homeostasis through the control of intracellular mRNA levels and quality [25,26,27].
It has been widely reported that viral RNA is a target of NMD, and RNA viruses have evolved their own strategies to protect viral RNA from NMD [28,29,30,31]. HTLV-1 encodes more than 10 different viral proteins within its 9 kbp proviral genome. This is made possible by multiple alternative splicing sites, sub-optimal AUGs, -1 ribosomal frameshift signal (-1RFS) and other RNA signals in the viral genome. We have previously shown that unspliced HTLV-1 mRNA is degraded as a target of NMD in human cells, while Rex has function to suppress NMD, stabilizes unspliced HTLV-1 mRNA and promotes translation of viral structural proteins [32]. It has also been reported that Tax inhibits NMD via interaction with Upf1 and eIF3E/INT6 [33,34]. Recently, Prochasson et al. elegantly reviewed the relationship between HTLV-1 and host NMD [35]. However, the offense and defense between HTLV-1 and NMD are complex and it remains unclear how HTLV-1 evades this powerful host mRNA quality control mechanism. We hypothesize that, in addition to Tax-mediated NMD repression, Rex also represses NMD during HTLV-1 mRNA trafficking, but its molecular mechanism remains unclear. In the present study, we aimed to identify the “NMD repressor domain” of Rex and to elucidate the mechanism of NMD repression by Rex by examining the interaction between Rex and host NMD regulatory proteins.
2. Materials and Methods
2.1. Cell Culture
CEM, Molt-4, Jurkat (T-ALL patient-derived T-cell lines), MT-2 (HTLV-1-immortalized T-cell lines), HeLa (cervical cancer-derived epithelial cell line), HEK293T (human embryonic kidney-derived cell line containing the SV40 T-antigen), and HEK293FT were used in the present study. All cell lines were obtained and maintained as previously reported [36]. About the origin of the cell lines, MT-2 was provided from Gunma University, Japan. Authentication was conducted in our laboratory using the proviral integration-site sequencing technique [37]. We also confirmed production of infectious HTLV-1 viral particles and expression of Gag-Tax fusion protein. Jurkat and HEK293FT were obtained from Riken Cell Bank, Japan. CEM, Molt-4, HeLa, and HEK293T cells were provided by the Japanese Foundation for Cancer Research (JFCR).
2.2. Construction of Protein Expression Plasmids in Mammalian Cells
2.2.1. Construction of WT-Rex and Other Protein Expression Plasmids
The cDNAs were amplified by PCR with Platinum Taq High Fidelity (Invitrogen, Thermo Fisher Scientific, Inc., Waltham, MA, USA using the original plasmid containing the cDNAs encoding each protein as a template. The primers with appropriate restriction enzyme sites were used for PCRs. The amplicon after PCR was separated by agarose-gel electrophoresis, TA-ligated into pGEM-T-Easy vector (Promega, Corp., Madison, WI, USA), amplified in DH-5α E. coli strain, and purified by a miniprep kit (SIGMA-Aldrich, Merck KGaA, Darmstadt, Germany). After cleavage with restriction enzymes, the target cDNA was extracted by electrophoresis and ligated with the final vector. The sequence was confirmed before used in experiments. The list of template plasmids, primers (all are shown in 5′→ 3′) and final vectors used for plasmid preparation is shown below (Table 1).
Table 1.
Primers for constructions of protein expression plasmids.
| Plasmid Name | Template | Primers | Subcloning to |
|---|---|---|---|
| His-CRM1 | HeLa cDNA library | hCRM1(BamHI)-For: GGATCCATGCCAGCAATTATG hCRM1(XhoI)-Rev: CTCGAGTTAATCACACATTTC |
pCDNA3.1C /his (Invitrogen, Thermo Fisher Scientific, Inc., Waltham, MA, USA) |
| GST-UPF1 | UPF1 fragment was exerted from UPF1-pCDNA3.1/his C (Invitrogen, Thermo Fisher Scientific, Inc., Waltham, MA, USA) [32] and subcloned in pMEG-2 at EcoRI and XbaI. | ||
| EYFP-UPF1 | pEYFP-C1 (Clontech, Takara Bio Inc., Japan) | EYFP(HindIII)-For: CCCAAGCTTATCATGGTGAGCAAGG EYFP(BamHI)-Rev: CGGGGATCCCTTGTACAGCTCGTC |
EYFP cDNA was subcloned in UPF1-pCDNA3.1/his C (Invitrogen, Thermo Fisher Scientific, Inc., Waltham, MA, USA) [32] at HindIII and BamHI. |
| ECFP-Rex | Rex-pME-FLAG [32] | P27Rex(HindIII)-For: CCCAAGCTTCTATGCCCAAGACCCGTCGG P27Rex(BamHI)-Rev: GCGGATCCCGGTCACATGGGGCAGGAG |
pECFP-C1 (Clontech, Takara Bio Inc., Shiga, Japan) |
| DCP2-mCherry | DCP2 fragment was exerted from pMAG1-MN1(MBL Co., LTD, Japan)-DCP2 plasmid (Kind gift from Dr. Tadanori Yamochi) and subcloned to pmCherry-C1 (Clontech, Takara Bio Inc., Shiga, Japan) at BamH1. | ||
| GST-UPF2 | pMEG-2 [38] | GST(NotI)-For: GCGGCCGCCAATGTCCCCTATACTAGG GST+Stop(NheI)-Rev: GCTAGCTTATTTTGGAGGATGGTC |
GST cDNA was subcloned to Rent2-GFP-pCDNA3.1 TOPO (gifted from Dr. Hal Dietz (Plasmid #17709; http://n2t.net/addgene:17709 (accessed on 20 December 2021); RRID: Addgene_17709, Addgene, Watertown, MA, USA) [39]. |
| HA-UPF3A | UPF3A fragment was exerted from UPF3A-puc57 (Synthesized by GENEWIZ, South Plainfield, NJ, USA) and subcloned in pCDNA3-HA (Invitrogen, Thermo Fisher Scientific, Inc., Waltham, MA, USA) at NotI and XbaI. | ||
| His-UPF3B | UPF3B fragment was exerted from UPF3B-puc57 (Synthesized by GENEWIZ, South Plainfield, NJ, USA) and subcloned in pCDNA3.1C/his (Invitrogen, Thermo Fisher Scientific, Inc., Waltham, MA, USA) at BamH1 and XhoI. | ||
| GST-Rex | Rex-pME-FLAG | P27Rex(EcoRI)-For: GAATTCATGCCCAAGACCCG P27Rex(NotI)-Rev: GCGGCCGCTCACATGGGGCAGGAG |
pMEG-2 [38] |
| Rex-pRxPuro | Rex-pME-FLAG | P27Rex(EcoRI)-For: GAATTCTCGCCACCATGCCCAAGACCCG P27Rex(NotI)-Rev: GCGGCCGCTCACATGGGGCAGGAG |
pRx-Puro [40] |
2.2.2. Construction of Mutant Protein Expression Plasmids
For the construction of expression plasmids of Rex mutants, Upf1 mutants and Rex(-)-pFL-MT-2, we used a primer set containing the desired deletion/substitution and introduced the mutations using the Prime STAR Mutagenesis Basal Kit (Takara Bio Inc., Shiga, Japan). The point mutants in the RNA-binding motif of Rex were adapted from those constructed by Rimsky et al. [41]. M1 and M2 were mutated in the ARM, and M3–5 were mutated in the aa20–56 region, respectively. M1 and M2 were shown to lose RNA-binding ability, and their localization was observed in the cytoplasm and in the nucleus/cytoplasm, whereas the RNA-binding activity of M3–5 was comparable to that of wild-type Rex. The Rex phosphorylation mutants are based on the comprehensive identification of phosphorylation sites in Rex by Kesic et al. [23]. The hyperphosphorylated mutant (G495R/G497E) [42] and the ATPase-deficient mutant (R843C) [43] of Upf1 were constructed based on previous reports. The template plasmids used to construct the mutant expression plasmids and the mutagenesis primers (all are shown in 5′→ 3′) are shown below (Table 2). In the present study, a HTLV-1 infectious clone, pFL-MT-2 [44], was used to establish the site of HTLV-1 genomic expression. To construct the Rex(-)-pFL-MT-2, the HTLV-1 infectious plasmid without Rex expression, the mutagenesis primers shown in Table 2 were used.
Table 2.
Primers for mutagenesis.
| Plasmid Name | Template | Primers for Mutagenesis |
|---|---|---|
| ΔARM-Rex | Rex-pME-FLAG | ΔARM-Rex-Top: CCTCGAGTGGCCCACTTCCCAGGGT ΔARM-Rex-Bottom: GTGGGCCACTCGAGGAATTCCTTGTC |
| Δaa20-56-Rex | Rex-pME-FLAG | Δaa20-56-Rex-Top: AACACCACCCGCCTACATCGTCACG Δaa20-56-Rex-Botom: TAGGCGGGTGGTGTTGGTGGTCTTTT |
| ΔN-MD-Rex | Rex-pME-FLAG | ΔN-MD-Rex-Top: TGTTCGGCCTGTCCAGAGCATCAGA ΔN-MD-Rex-Bottom: TGGACAGGCCGAACATAGTCCCCCAG |
| ΔNES-Rex | Rex-pME-FLAG | ΔNES-Rex-Top: CCCCATCGAGAGAACCTCTAAGACCC ΔNES-Rex-Bottom: GTTCTCTCGATGGGGTCCCAGGTGA |
| ΔC-MD-Rex | Rex-pME-FLAG | ΔC-MD-Rex-Top: AAGACCCTCCAGGCCATGCGCAAAT ΔC-MD-Rex-Bottom: GGCCTGGAGGGTCTTAGAGGTTCTCT |
| ΔARM/NES-Rex | ΔNES-Rex | ΔARM-Rex-Top: CCTCGAGTGGCCCACTTCCCAGGGT ΔARM-Rex-Bottom: GTGGGCCACTCGAGGAATTCCTTGTC |
| ΔN/C-MD-Rex | ΔC-MD-Rex | ΔN-MD-Rex-Top: TGTTCGGCCTGTCCAGAGCATCAGA ΔN-MD-Rex-Bottom: TGGACAGGCCGAACATAGTCCCCCAG |
| Δaa125-139-Rex | Rex-pME-FLAG | Rex(Δaa125-139)-Top: TCCTTCCAACCCACCCTTGGGCAGC Rex(Δaa125-139)-Bottom: GGTGGGTTGGAAGGAGGGTGGAATGT |
| Δaa125-149-Rex | Rex-pME-FLAG | Rex(Δaa125-149)-Top: TCCTTCCTGTCTTTTCCAGACCCCG Rex(Δaa125-149)-Bottom: AAAAGACAGGAAGGAGGGTGGAATGT |
| Δaa125-159-Rex | Rex-pME-FLAG | Rex(Δaa125-159)-Top: TCCTTCCAAAACCTGTACACCCTCT Rex(Δaa125-159)-Bottom: CAGGTTTTGGAAGGAGGGTGGAATGT |
| Δaa125-169-Rex | Rex-pME-FLAG | Rex(Δaa125-169) -Top: CTCCTTCCTTGTCTGCATGTACCTCT Rex(Δaa125-169)-Bottom: CAGACAAGGAAGGAGGGTGGAATGT |
| Δaa170-189-Rex | Rex-pME-FLAG | Rex(Δaa170-189)-Top: AGGCTCCGTGAACTAGTCTAGAGAAA Rex(Δaa170-189)-Bottom: TAGTTCACGGAGCCTCCCCAGAGGG |
| Δaa160-189-Rex | Rex-pME-FLAG | Rex(Δaa160-189)-Top: CCGGCCCCTGAACTAGTCTAGAGAAA Rex(Δaa160-189)-Bottom: TAGTTCAGGGGCCGGAGTCCGGGGT |
| Δaa150-189-Rex | Rex-pME-FLAG | Rex(Δaa150-189)-Top: CCCAACCCTGAACTAGTCTAGAGAAA Rex(Δaa150-189)-Bottom: TAGTTCAGGGTTGGGAGGTGCTGCC |
| Δaa150-159-Rex | Rex-pME-FLAG | Rex(Δaa150-159)-Top: CCAACCCAAAACCTGTACACCCTCT Rex(Δaa150-159)-Bottom: CAGGTTTTGGGTTGGGAGGTGCTGCC |
| Δaa160-169-Rex | Rex-pME-FLAG | Rex(Δaa160-169)-Top: CGGCCCCTTGTCTGCATGTACCTCT Rex(Δaa160-169)-Bottom: GCAGACAAGGGGCCGGAGTCCGGGGT |
| Δaa150-169-Rex | Rex-pME-FLAG | Rex(Δaa150-169)-Top: CCAACCCTTGTCTGCATGTACCTCT Rex(Δaa150-169)-Bottom: GCAGACAAGGGTTGGGAGGTGCTGCC |
| Rex-M1 (RRR5/6/7DL) | Rex-pME-FLAG | Rex(M1) -Top: AGACCGATCTGCCCCGCCGATCCCAAAG Rex(M1)-Bottom: GGGGCAGATCGGTCTTGGGCATCTCGAG |
| Rex-M2 (KR14/15DL) | Rex-pME-FLAG | Rex(M2)-Top: AAAGAGATCTACCACCAACACCATGGCC Rex(M2)-Bottom: GTGGTAGATCTCTTTGGGATCGGCGGGG |
| Rex-M3 (FF30/31DL) | Rex-pME-FLAG | Rex(M3)-Top: GAGTCGATCTTTCGGATACCCAGTCTAC Rex(M3)-Bottom: CCGAAAGATCGACTCTGTCCAAACCCTG |
| Rex-M4 (DTQ33/34/35DL) | Rex-pME-FLAG | Rex(M4)-Top: TTTCGGATCTGTCTACGTGTTTGGAGAC Rex(M4)-Bottom: TAGACAGATCCGAAAAGAAGACTCTGTC |
| Rex-M5 (YK43/44DL) | Rex-pME-FLAG | Rex(M5)-Top: CTGTGGATCTGGCGACTGGTGCCCCATC Rex(M5)-Bottom: TCGCCAGATCCACAGTCTCCAAACACGT |
| Rex (T22A) | Rex-pME-FLAG | Rex (T22A)-Top: CCAACACCATGGCCCGCTTCCCAGGGTTTGG Rex (T22A)-Bottom: CCAAACCCTGGGAAGCGGGCCATGGTGTTGG |
| Rex (S36A) | Rex-pME-FLAG | Rex (S36A)-Top: TTTTCGGATACCCAGGCTACGTGTTTGGAGA Rex (S36A)-Bottom: TCTCCAAACACGTAGCCTGGGTATCCGAAAA |
| Rex (T37A) | Rex-pME-FLAG | Rex (T37A)-Top: TCGGATACCCAGTCTGCGTGTTTGGAGACTG Rex (T37A)-Bottom: CAGTCTCCAAACACGCAGACTGGGTATCCGA |
| Rex (S70A) | Rex-pME-FLAG | Rex (S70A)-Top: TGGCCACCTGTCCAGGCCATCAGATCACCTGG Rex (S70A)-Bottom: CCAGGTGATCTGATGGCCTGGACAGGTGGCCA |
| Rex (S97A) | Rex-pME-FLAG | Rex (S97A)-Top: CTCGACTCCCCTCCTGCCCCACCCAGAGAAC Rex (S97A)-Bottom: GTTCTCTGGGTGGGGCAGGAGGGGAGTCGAG |
| Rex (S106A) | Rex-pME-FLAG | Rex (S106A)-Top: GAACCTCTAAGACCCGCAAGGTCCTTACCCC Rex (S106A)-Bottom: GGGGTAAGGACCTTGCGGGTCTTAGAGGTTC |
| Rex (T174A) | Rex-pME-FLAG | Rex (T174A)-Top: CCGTTGTCTGCATGTGCCTCTACCAGCTTTC Rex (T174A)-Bottom: GAAAGCTGGTAGAGGCACATGCAGACAACGG |
| GST- UPF1(G495R/G497E) | GST-UPF1 | hUPF1(G495R/G497E)-Top: AGGGCCCGCCAAGAACGGAGAAGACGGTGAC hUPF1(G495R/G497E)-Bottom: TCACCGTCTTCTCCGTTCTTGGCGGGCCCT |
| GST- UPF1(R843C) | GST-UPF1 | hUPF1(R843C)-Top: TCCTGTCCTGTGTGTGTGCCAACGAGCACCA hUPF1(R843C)-Bottom: TGGTGCTCGTTGGCACACACACAGGACAGGA |
| Rex(-)-pFL-MT2 | pFL-MT2 [44] | p27Rex(ATG-to-CTG)-Top: CCTCAAGCGAGCTGCCTGCCCAAGACCCGTC p27Rex(ATG-to-CTG)-Bottom: GACGGGTCTTGGGCAGGCAGCTCGCTTGAGG |
2.3. Establishment of Stable Rex-Expressing T-Cell Lines by Retroviral Expression
In order to establish T-cell lines that express Rex stably, we employed a retroviral expression system using pRx-Puro vector for Rex-expressing recombinant retrovirus preparation. After 24 h, HEK293FT cells were seeded on a 6 cm dish at 5 × 105 cells/mL, 5 mL/dish, and 5 μg of gag/pol and 5 μg of env vector, 10 μg of pRx-Puro (Mock) or 10 μg of pRx-Puro-Rex were co-transfected by the calcium phosphate method. The culture medium was changed after 4 h and the supernatant h was filtered after 48 by a filter with a pore size of 0.45 μm and used as a virus solution. An amount of 1.0 × 106 cells/1 mL/well of CEM or Molt4 were seeded on 6-well plates and 1 mL of the Mock or Rex-expressing retrovirus solution and 8 μg/mL of polybrene were added to infect the cells. After 48 h, puromycin selection (0.5 μg/mL) was started and the cells were used for experiments after 12 days.
2.4. Measurement of Cellular NMD Activity
2.4.1. The β-Globin Luciferase Reporter Assay System
In this study, a highly sensitive and quantitative β-globin (WT/PTC) dual luciferase reporter system was used to measure the activity of NMD [32]. pCDNA6-RSV-Renilla-Luc-β-globin (WT) (Renilla-WT) is expressed independent of NMD activity in the cell, whereas pCDNA6-RSV-Firefly-Luc-β-globin (PTC) (Firefly-PTC), in which PTC is introduced into the second exon, is expressed dependent on NMD activity because its mRNA is targeted by NMD. NMD activity is therefore calculated as the ratio between Renilla-luciferase activity and Firefly-luciferase activity (Renilla-WT/Firefly-PTC). At 24 h after HeLa cells were seeded at 2 × 104 cells/well, 6 wells each for one condition (i.e., n = 6) in 48-well plates, 100 ng each of Renilla-WT and Firefly-PTC, and 200 ng of various effector plasmids for a well were co-transfected by the calcium phosphate method. Then, the luciferase assay was performed after 24 h. Rex/Mock Molt-4 cells or Rex/Mock CEM cells were seeded at 2.5 × 105 cells/well in 48-well plates, 6 wells each for one condition (i.e., n = 6), and co-transfected with 100 ng of Renilla-WT and 300 ng of Firefly-PTC for a well with Lipofectamine 2000 (Invitrogen, Thermo Fisher Scientific, Inc., Waltham, MA, USA). The luciferase assay was performed after 24 h. To examine the effect of the HTLV-1 infectious clone (pFL-MT2) on NMD activity, firstly sHeLa cells stably expressing Renilla-WT and Firefly-PTC were established. sHeLa cells were seeded at 2.0 × 104 cells/well on 48-well plates well, 6 wells each for one condition (i.e., n = 6). After 24 h, 400 ng of pFL-MT2 or Rex(-)-pFL-MT-2 for a well was transfected using Lipofectamine 2000 (Invitrogen, Thermo Fisher Scientific, Inc., Waltham, MA, USA), and 48 h later, Luciferase assay was performed. The Luciferase assay was conducted using a dual-Luciferase reporter system (Promega, Corp., Madison, WI, USA) to measure the activity of Renilla-luciferase and Firefly-luciferase using a Centro LB 960 luminometer (Berthold Technologies GmbH & Co KG, Bad Wildbad, Germany).
2.4.2. Measurement of NMD Activity Adapted for Flow Cytometry
β-globin(WT) and β-globin(PTC) cDNA fragments were subcloned into pmCherry-C1 or pEGFP-C1 (both from Clontech, Takara Bio, Inc., Shiga, Japan), respectively. These two reporter plasmids and pME-FLAG-Rex or pME-FLAG (5 μg each) were introduced into Jurkat (5 × 105 cells) by electroporation (250 V, 1050 F, 720 Ω). After 24 h, EGFP-expressing cells and mCherry-expressing cells were detected by a flow cytometer (FACS Calibur, BD Biosciences, Franklin Lakes, NJ, USA) and the ratio of mCherry (WT)/EGFP (PTC) was calculated as relative NMD activity.
2.5. Inhibition of Rex Phosphorylation by H-7 (Protein Kinase C Inhibitor)
H-7 is a protein kinase C inhibitor and has been reported to inhibit the phosphorylation of Rex [45,46]. HeLa cells were seeded in 48 wells at 2 × 104 cells/250 L/well on 48-well plates After 24 h, the cells were co-transfected with 100 ng each of Renilla-WT and Firefly-PTC, and 200 ng of pME-FLAG (Mock) or pME-FLAG-Rex by Lipofectoamine2000 (Invitrogen, Thermo Fisher Scientific, Inc., Waltham, MA, USA). After 24 h, H-7 was added to a final concentration of 50 μM (20 h treatment). Similarly, H-7 was added at 40 h (4 h treatment), 42 h (2 h treatment), and at 44 h, and Luc assay was performed simultaneously with H-7-free (0 h treatment) samples.
2.6. Interaction between Rex and Host NMD Regulatory Complex Proteins
2.6.1. Identification of NMD Regulatory Proteins Interacting with Rex
HEK293FT cells were seeded at a concentration of 5 × 105 cells/mL and 10 mL/10 cm culture dish one day before transfection. At 24 h after seeding, 10 μg pMEG-2 or Rex-pMEG-2 plasmid was transfected by polyethylenimine (PEI). After 48 h, the cells were collected and whole-cell lysate was prepared in TBS buffer containing 1% NP-40. GST-Rex was then collected using Glutathione Sepharose 4G (Cytiva, Marlborough, MA, USA) and the coprecipitated NMD complex proteins were detected by Western blotting.
2.6.2. Interaction between Upf2 and Upf3A or Upf3B with/without Rex
HEK293FT cells were seeded at a concentration of 5 × 105 cells/mL and 10 mL/10 cm culture dish one day before transfection. At 24 h after seeding, 10 μg each of GST-Upf2, HA-UP3A, His-Upf3B with/without SRα-Rex plasmid was transfected by PEI. For the negative control, cells transfected with pMEG-2 (GST only) instead of GST-Upf2 were also prepared. After 48 h, the cells were collected and whole-cell lysate was prepared in TBS buffer containing 1% NP-40. After that, GST-Upf2 was collected using Glutathione Sepharose 4G (Cytiva, Marlborough, MA, USA), and the coprecipitated Upf3A and Upf3B were detected by Western blotting.
2.6.3. Subcellular Interaction between Rex and Upf1 by FRET
In the present study, we employed fluorescence resonance energy transfer (FRET) analysis to identify the subcellular sites of Rex and Upf1 interaction. An energy transfer from donor (ECFP) to acceptor (EYFP) occurs when the distance between ECFP and EYFP is less than 10 nm, in which the two molecules are highly likely to be physically interacting.
We first prepared the expression plasmids of ECFP-Rex and EYFP-Upf1. HeLa cells were seeded at 1 × 105 cells/mL, 200 μL/well, on a 4-well culture glass slide. After 24 h, both ECFP-Rex and EYFP-Upf1 expression plasmids were transfected to HeLa cells using Lipofectamine2000 (Invitrogen, Thermo Fisher Scientific, Inc., Waltham, MA, USA). After a further 24 h, cells were fixed in 4% paraformaldehyde, and sealed in the mounting medium containing DAPI in 80% glycerol. To eliminate the FRET signal derived from non-specific interactions other than Rex and Upf1, we prepared a series of negative-control HeLa cells expressing ECFP and EYFP; ECFP and EYFP-Upf1; ECFP-Rex and EYFP. To confirm that our FRET system indeed detects high efficiency FRET, we also prepared positive-control HeLa cells expressing tandemly connected ECFP + EYFP.
For high-efficiency FRET detection, we employed the acceptor-photobleaching using LSM710 confocal microscope (Carl Zeiss, AG., Oberkochen, Germany). Briefly, EYFP molecules in regions of interest (ROIs) of nuclei, cytoplasm, and p-bodies were photobleached by the laser of 541 nm at 100% power and 100-times iterations. The intensity of ECFP before and after photobleaching was measured every 1 s for 10 s, respectively, i.e., 10 measurements each before and after photobleaching. Calculation of FRET efficiency is as follows. FRET efficiency (%) = [1-(average of pre-photobleaching intensity of ECFP)/(average of post-photobleaching intensity of ECFP)] ×100. Higher FRET efficiency means closer positioning of ECFP and EYFP. Data were collected from 6 to 10 ROIs in one image (n = 6–10).
2.7. Western Blotting
Sample cells were suspended in RIPA buffer (10 mM Tris-HCl (pH 7.4), 1% NP-40, 0.1% sodium deoxycholate, 0.1% SDS, 0.15 M NaCl, 1 mM EDTA (pH 8.0)) with protein inhibitor cocktail (Nacalai Tesque, Inc., Kyoto, Japan) and PMSF. Protein assay reagent (Bio-Rad Laboratories, Inc., Hercules, CA, USA) was used to determine the protein concentration. The whole-cell lysate was diluted to 1 mg protein/mL in sample buffer and boiled for 5 min to obtain a sample. Proteins were then separated by SDS-PAGE using a 12.5% acrylamide gel and transferred to a PVDF membrane (MerckMillipore, Merck KGaA, Darmstadt, Germany) using a wet transfer device. The transferred membrane was immersed in blocking solution (5% skim milk in 0.1% Tween-20 containing Tris-buffered saline, TBST) and blocked for 1 h. After blocking, the membrane was immersed in the primary antibody solution 2000-fold diluted in blocking solution at 4 °C for 20 h. The membrane was then washed 3 times for 15 min in TBST. The membrane was then incubated with the secondary antibody solution diluted 2000-fold in blocking solution for 2 h at room temperature. Finally, the membrane was washed with TBST for 5 min x 3 times and the bands were detected in NBT/BCIP (Promega, Corp., Madison, WI, USA) solution. The primary and secondary antibodies used in this experiment were as follows.
Primary antibodies: Upf1(#9435S, Cell Signaling Technology, Inc. Danvers, MA, USA), Upf2 (#ab-28712-200, Abcam, Plc. Cambridge, UK), Upf3A (#H00065110-M06, Abnova, Corp.), Upf3B (#A303-688A, Bethyl Laboratories, Inc., Montgomery, TX, USA), SMG1 (#9149S, Cell Signaling Technology, Inc.), SMG5 (#ab-129107, Abcam, Plc.), SMG6 (#sc-50984-R, Santa Cruz Biotechnology, Inc., Dallas, TX, USA), SMG7 (#sc-134857, Santa Cruz Biotechnology, Inc., Dallas, TX, USA), β-Actin (#sc-69879, Santa Cruz Biotechnology, Inc., Dallas, TX, USA), FLAG (#F1804, SIGMA-Aldrich, Merck KgaA, Darmstadt, Germany), GST (#Cytiva 27-4577-01, SIGMA-Aldrich, Merck KgaA, Darmstadt, Germany), and monoclonal antibodies against HTLV-1 proteins (Tax, Rex, Gag(p53), and Env(gp24)) were generated by Yuetsu Tanaka at the University of Ryukyus.
Secondary antibodies: Alkaline phosphatase (AP)-conjugated anti-mouse IgG (#S3721), AP-conjugated anti-rabbit IgG (#S3731), AP-conjugated anti-rat IgG (#S383A), AP-conjugated anti-goat IgG (#V-1151) (all from Promega, Corp., Madison, WI, USA).
2.8. Statistical Analysis
Throughout the present study, two-tailed paired Student’s t-tests were performed to test the statistical difference between the experimental groups. Asterisks in the figures indicate a significant difference between the tested groups (*, p < 0.05; **, p < 0.01; and ***, p < 0.001, n > 3).
3. Results
3.1. NMD Inhibition by Rex at the Site of HTLV-1 Viral Genome Expression
3.1.1. Rex Inhibits NMD in Human T Cells
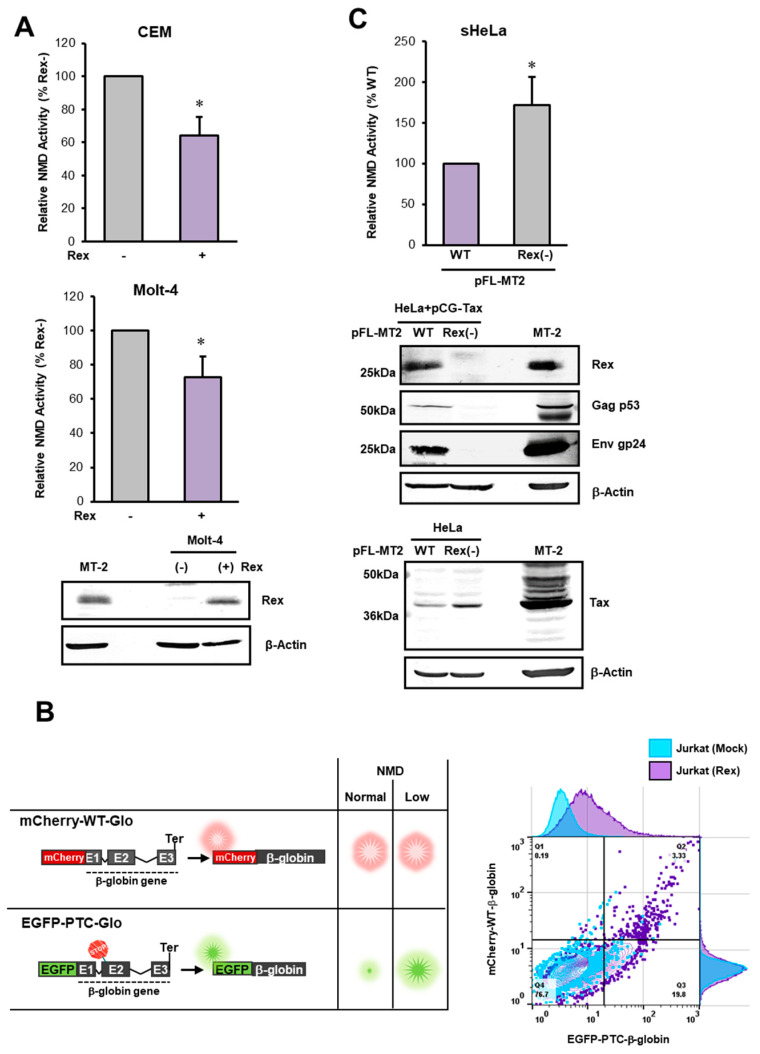
We have previously reported that Rex inhibits NMD in HeLa cells [32], but not in human T cells, the site of HTLV-1 infection. In this experiment, we stably overexpressed Rex in CEM and Molt-4 T-cell lines using a retroviral expression system and analyzed the activity of NMD using the β-globin NMD reporter system. The results showed that NMD was significantly suppressed in Rex-expressing cells compared to control cells in both T cell lines (Figure 1A). The NMD activity in Jurkat cells with or without Rex was also analyzed by flow cytometry. The expression of EGFP-β-globin (PTC) was significantly increased in the presence in Rex, indicating that β-globin (PTC) mRNA was stabilized by the reduction in NMD activity (Figure 1B). These results indicate that host NMD is suppressed by Rex in T cells, which are the target of HTLV-1 infection.
Figure 1.
Rex inhibits NMD in T cells. (A). NMD activity was measured in CEM or Molt-4 cells constantly expressing Rex. The results showed a significant decrease in NMD activity in Rex-expressing cells of both cell lines (n = 6, mean ± SD, * p < 0.05). The image below shows Rex expression in Rex(+/-)-Molt-4 cells as detected by Western blotting. The whole-cell lysate of HTLV-1-infected immortalized cell line MT-2 is used as the positive control in Western blotting. (B). NMD-activity reporters (mCherry-WT (red) and EGFP-PTC (green)) were introduced into Jurkat, and the expression levels of mCherry and EGFP in the presence and absence of Rex were detected by flow cytometry (left figure). The results showed that the percentage of cells expressing EGFP increased in Rex-expressing cells compared to Mock cells (right panel), indicating that EGFP-β-globin (PTC) mRNA is stabilized in the presence of Rex. (C). Wild-type HTLV-1 infectious clone, pFL-MT2, or Rex-deficient clone, Rex(-)-pFL-MT2, were transfected into sHeLa cells stably expressing Renilla-WT and Firefly-PTC and evaluated for NMD activity. The results showed that NMD suppression was abolished in Rex (-) clone-transfected cells (top graph). In the cells with Rex(-)-pFL-MT2, the expression of Tax was upregulated and the expression of viral structural proteins such as Gag-p53 and Env-gp24 was markedly reduced (bottom image) (n = 6, mean ± SD, * p < 0.05).
3.1.2. Effect of Rex on NMD Activity at the Site of HTLV-1 Viral Genome Expression
To clarify the effect of Rex on NMD in a more realistic HTLV-1 expression setting, a wild-type HTLV-1-infected clone, pFL-MT2 [44], and a Rex-deficient clone Rex(-)-pFL-MT-2 were transfected into sHeLa cells stably expressing the NMD reporter, and evaluated the NMD activity. The results showed that NMD was repressed in pFL-MT2 transfected cells, whereas NMD repression was abolished in Rex(-)-pFL-MT-2 transfected cells (Figure 1C). In Rex(-)-pFL-MT-2 transfected cells, the expression of Gag-p53 and Env-gp24 was decreased and that of Tax was increased in association with the loss of Rex expression (Figure 1C). These results indicate that the presence or absence of Rex directly affects host NMD activity at the site of HTLV-1 genome expression.
3.2. Identification of Rex Domains Important for NMD Suppression
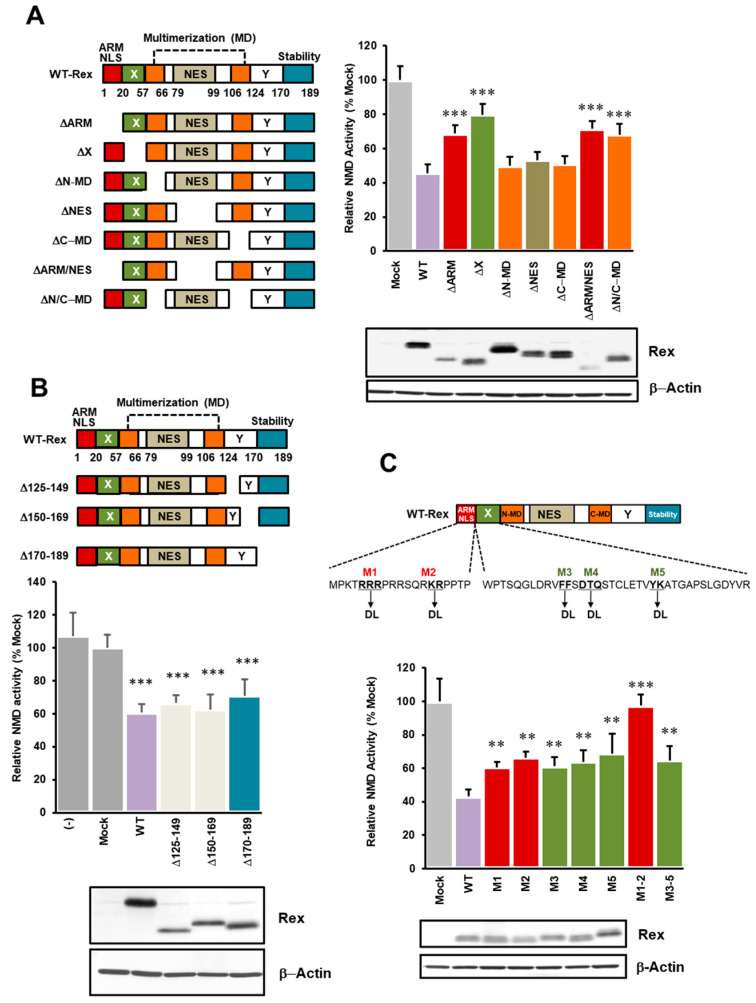
In this study, we compared the inhibitory effects of various deletion mutants of Rex on NMD and attempted to identify the Rex domain(s) that play an important role in NMD suppression. The results showed that the NMD repressive function of four Rex deletion mutants, ΔARM, ΔX (aa20–56), ΔARM/NES and ΔN/C-MD, was significantly reduced compared to wild-type (WT) Rex (Figure 2A). The C-terminal deletion mutants, Δaa125–149, Δaa150–169 and Δaa170–189, did not differ significantly in their ability to suppress NMD compared to WT-Rex (Figure 2B). Although it has been reported that this region contains a stability domain [24], the protein expression levels of these three mutants were similar to WT-Rex in the present study. Figure S1 shows that deletion of aa150–159 in the Y region resulted in almost no protein expression (mutant 3 and 4 compared with 2). Deletion of only the stability domain (aa170–189) (mutant 5) had no significant effect on protein expression, whereas deletion of aa160–169 with the stability domain markedly reduced protein expression (mutant 6 and 7 compared with 5, 9, and 10). These data indicate that part of the Y region plays an important for Rex stabilization. It was confirmed that there is no significant difference in the NMD inhibitory effect between mutant 10 and WT-Rex (Figure S1C). These results indicate that the ARM, an X region of unknown function (aa20–56), and at least one of the two multimerization domains are important for the function of Rex in NMD suppression.
Figure 2.
N-terminal region of Rex plays important roles in NMD inhibition. (A). We generated seven domain-deficient mutants of Rex (left panel) and compared the degree of NMD repression with WT-Rex using the NMD activity reporter assay system in HeLa cells. As shown in the graph on the right, NMD repression was significantly reduced when the ARM region, X region or both multimerization domains were deleted. Western blotting below shows the expression levels of each Rex mutant (n = 6, mean ± SD, *** p < 0.001, compared with WT-Rex). (B). For the C-terminal side, where the stabilizing domain of Rex is located, we also generated three types of deletion mutants (upper panel) and compared the degree of NMD repression with WT-Rex using the NMD activity reporter assay system in HeLa cells. As shown in the lower graph, there was no significant change in NMD repression in any of the mutants compared to WT-Rex. Western blotting below shows the expression levels of each Rex mutant (n = 6, mean ± SD, *** p < 0.001, compared with Mock). (C). We generated five point mutants in the ARM and X regions of Rex (upper panel) and compared the degree of NMD repression with WT-Rex using the NMD activity reporter assay system in HeLa cells. As shown in the lower graph, the degree of NMD suppression was significantly reduced in all mutants compared to WT-Rex. The reduction in NMD repression was particularly pronounced for both M1 and M2 mutants. Western blotting below shows the expression levels of each Rex mutant (n = 6, mean ± SD, ** p < 0.01; *** p < 0.001, compared with WT-Rex).
3.3. ARM and X Region of Rex Play Important Roles in NMD Inhibition
Next, we used point mutants in the ARM and X regions (aa20–56) to elaborate on their relationship to the NMD repression mechanism: M1 (RRR5-7DL) and M2 (KR14-15DL) were mutated in the ARM region, M3 (FF30-31DL), M4 (DTQ33-35DL) and M5 (YK43-44DL) in the X region (Figure 2C). Comparison of the NMD repression of these mutants with that of WT-Rex showed that NMD activity was significantly restored in all mutants, confirming the importance of the ARM and X regions in NMD repression. The most significant recovery of NMD activity was observed when M1 and M2 of the ARM region were mutated simultaneously. Rimsky et al. [41] mentioned that M1 and M2 mutants lack the biological function of Rex to nuclear-export HTLV-1 Env mRNA and thus express Env protein, while M3, M4, and M5 maintained the Rex function. For Env expression, Rex needs to be transported to the nucleus and to bind to the RxRE of Env mRNA. The authors also showed that M1 localizes only in the cytoplasm, while M2 evenly localizes both in the nucleus and the cytoplasm. Our results show that M2, which can shuttle between the nucleus and the cytoplasm also lack the NMD suppressive activity. Therefore, the RNA-binding function of Rex may be related to its NMD-suppressive function.
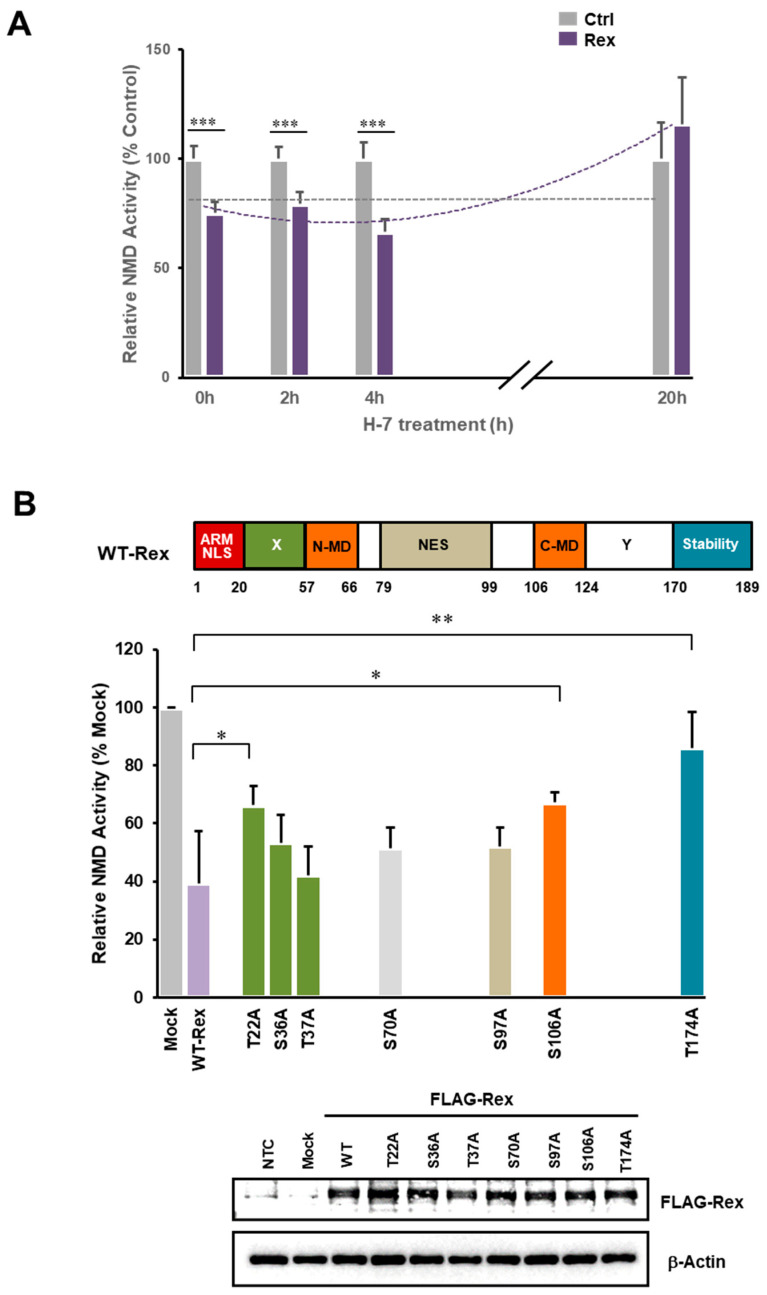
3.4. Relationship between Rex Phosphorylation State and NMD Inhibition
It is known that the activity of Rex is regulated by the phosphorylation status at multiple sites [23]. In the present study, H-7 (protein kinase C inhibitor) treatment abolished NMD inhibition (Figure 3A). Since H-7 treatment reduces the overall Rex phosphorylation to one-tenth of non-treated control [45], Figure 3A suggested that phosphorylation is also important to regulate the Rex activity in NMD inhibition. We then generated Rex phosphorylation mutants in which serine or threonine was replaced by alanine at seven phosphorylation sites [23], and compared the NMD-suppressive activity of these mutants with that of WT-Rex. We found that T22A, S106A and T174A mutants showed significantly reduced NMD repressive function (Figure 3B). These phosphorylation sites are located in the X region, the C-terminal multimerization domain and the stability domain, respectively, suggesting that phosphorylation in these regions may be important for NMD suppression.
Figure 3.
Phosphorylation status of Rex is related to its NMD inhibitory function. (A). Time course changes in the ability of Rex to inhibit NMD after treatment with the PKC inhibitor H-7 were examined using the NMD activity reporter assay system in HeLa cells. The results showed that Rex significantly inhibited NMD activity up to 4 h after H-7 treatment compared with the control but lost its ability to inhibit NMD after 20 h (n = 6, mean ± SD, *** p < 0.001). (B). Each mutant of seven phosphorylation sites of Rex was prepared and the NMD inhibitory ability was compared with that of WT-Rex using the NMD activity reporter assay system in HeLa cells. The results showed that the phosphorylation-deficient mutants at T22 in the X region, S106 in the C-terminal multimerization domain and T174 in the stability domain showed significantly reduced NMD repression. Western blotting below shows the expression levels of each Rex mutant (n = 6, mean ± SD, * p < 0.05; ** p < 0.01).
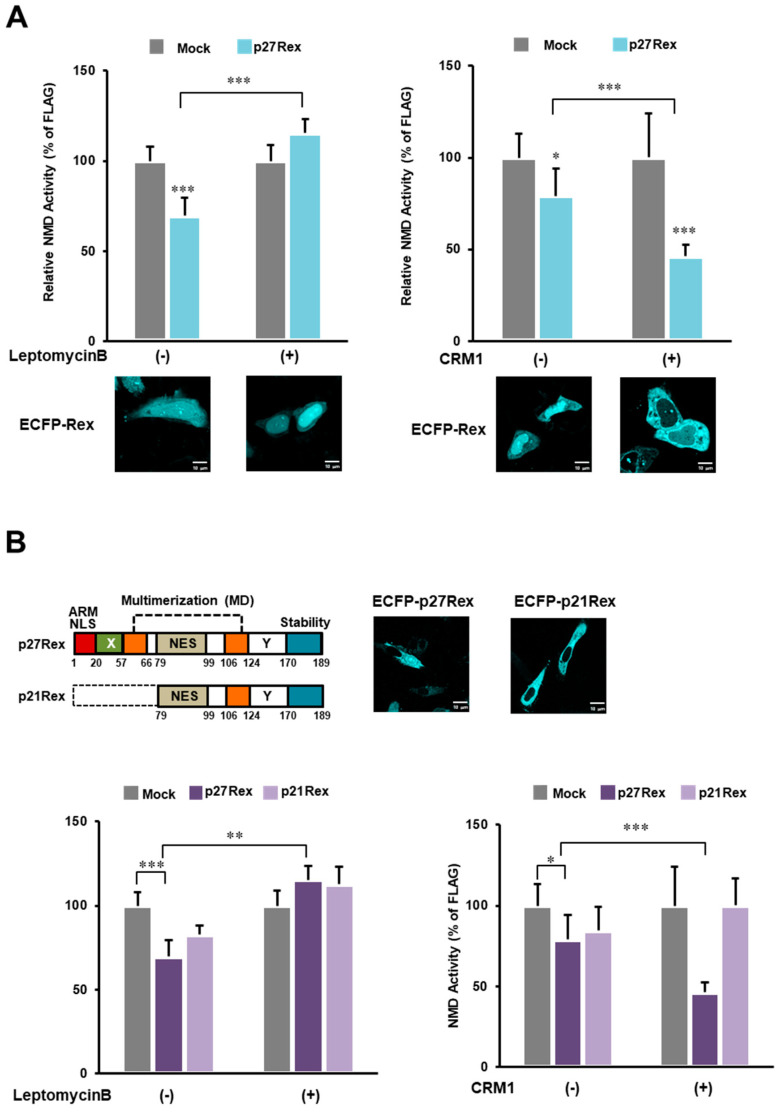
3.5. Verification of CRM1 Dependence of NMD Suppression by Rex
It is known that Rex binds to the cellular nuclear export protein CRM1 and is transported to the cytoplasm [13,14]. In this study, we investigated how CRM1 inhibition by Leptomycin B treatment or CRM1 overexpression affects the ability of Rex to inhibit NMD. Leptomycin B treatment abolished the ability of Rex to inhibit NMD, whereas overexpression of CRM1 enhanced the ability of Rex to inhibit NMD (Figure 4A). This suggests that Rex exerts its NMD repressive function in the cytoplasm. This was also confirmed by experiments using p21Rex, a Rex mutant lacking N-terminal aa1–78 due to translation starting at the second ATG of the Rex ORF [47,48]. P21Rex lacks the NLS/ARM and therefore localizes to the cytoplasm. Because of its cytoplasmic localization, NMD activity in p21Rex-expressing cells was not affected by Leptomycin B treatment or CRM1 overexpression compared to p27(WT)Rex (Figure 4B). Thus, it appears that CRM1-dependent transport of Rex into the cytoplasm is required for Rex-mediated NMD repression.
Figure 4.
Rex needs to be nuclear-exported to inhibit NMD. (A). Treatment with Leptomycin B, an inhibitor of the nuclear export protein CRM1, significantly reduced the ability of Rex to inhibit NMD (left panel, n = 6, mean ± SD, *** p < 0.001). In contrast, overexpression of CRM1 significantly enhanced the ability of Rex to inhibit NMD (right panel, n = 6, mean ± SD, * p < 0.05; *** p < 0.001). The confocal laser microscopy images below show the subcellular localization of ECFP-Rex in HeLa cells subjected to each treatment. The scale bar indicates 10 μm (B). The data of the p21Rex mutant, which lacks NLS and thus localizes only in the cytoplasm (upper panel, The scale bar indicates 10 μm.), are added to the graphs of WT(p27)-Rex to compare the NMD-suppressive ability of WT(p27)-Rex with that of p21Rex under Leptomycin B treatment or CRM1 overexpression. Upon Leptomycin B treatment, only WT-Rex showed a significant decrease in NMD repression (left panel, n = 6, mean ± SD, ** p < 0.01; *** p < 0.001). When CRM1 was overexpressed, WT-Rex showed a significant enhancement of NMD repression, whereas p21Rex showed no change in NMD repression (right panel, n = 6, mean ± SD, * p < 0.05; *** p < 0.001).
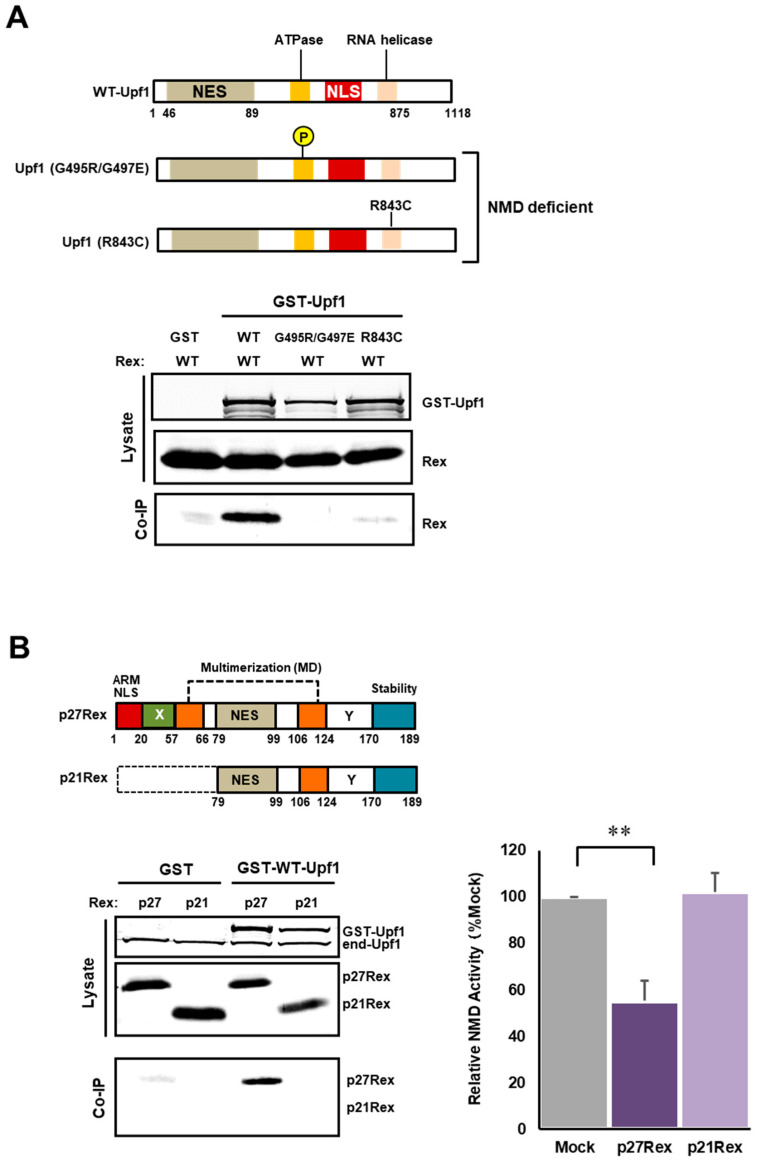
3.6. Physiological Interaction between Rex and Upf1
Upf1 is a central regulator of NMD; phosphorylation and dephosphorylation of Upf1 are required for the activation and completion of the NMD pathway [49,50]. In addition, the ATPase activity of Upf1 is required for the completion of NMD and the disassembly of mRNPs [51]. We therefore generated NMD-deficient Upf1 mutants, a hyper-phosphorylation mutant Upf1 (G495R/G497E) [42] and an ATPase-deficient mutant Upf1 (R843C) [43], and examined their interaction with Rex. We found that Rex interacted only with WT-Upf1, but not with the NMD-activation-deficient Upf1 mutants (Figure 5A). Additionally, when the interaction of p27Rex and p21Rex with WT-Upf1 was compared, an interaction was observed only between p27Rex and WT-Upf1 (Figure 5B). As p21 Rex does not show NMD inhibition (Figure 5B, right panel graph), this suggests that the interaction of Rex with Upf1 via aa1–78, which contains the ARM and X regions, is required for NMD repression by Rex.
Figure 5.
Only WT(p27)-Rex interacts with WT-Upf1. (A). Hyperphosphorylated and RNA helicase-deficient Upf1 mutants were generated (upper panel) and their interaction with WT-Rex was examined by GST pulldown assay in GST-Upf1 and FLAG-Rex-expressing HEK293FT cells. The results showed that Rex interacted only with WT-Upf1 and not with the NMD-deficient Upf1 mutants (lower panel). (B). The interaction of WT-Rex and p21Rex with WT-Upf1 was examined by GST pulldown assay in GST-Upf1 and FLAG-Rex-expressing HEK293FT cells. The results showed that only WT-Rex interacted with Upf1 (bottom left), which was correlated with NMD inhibitory capacity measured by the NMD activity reporter assay system in HeLa cells (bottom right) (n = 6, mean ± SD, ** p < 0.01).
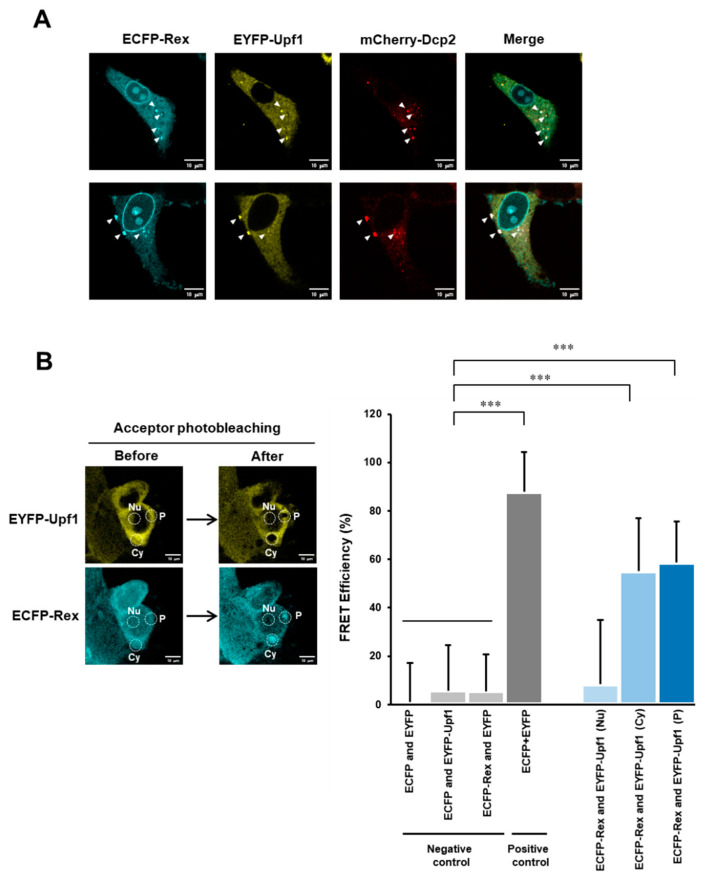
3.7. Subcellular Interaction between Rex and Upf1
When ECFP-Rex, EYFP-Upf1 and mCherry-Dcp2 were co-expressed in HeLa cells, Rex and Upf1 were shown to co-localize in the cytoplasm and the p-body (Figure 6A). We then calculated the FRET efficiency of ECFP-Rex and EYFP-Upf1 in these cells in the cytoplasm, the p-body and the nucleus. A significant increase in FRET efficiency was observed in the cytoplasm and the p-body (Figure 6B). These results indicate that Rex and Upf1 interact with each other in the cytoplasm and the p-body.
Figure 6.
Rex interacts with Upf1 in the cytoplasm and in the p-body. (A). Confocal laser microscopy images of ECFP-Rex, EYFP-Upf1, and mCherry-Dcp2 in HeLa cells, where Dcp2 is a p-body marker. The white arrowhead indicates the site of the p-body. The scale bar indicates 10 μm. (B). The strength of the interaction between Rex and Upf1 in the cytoplasm, the p-body and the nucleus was analyzed by FRET. The left panel shows an example of images before and after acceptor photobleaching by confocal laser microscopy in HEK293FT cells co-expressing ECFP-Rex and EYFP-Upf1. The dotted white circles indicate regions where photobleaching was applied (Cy = cytoplasm, P = p-body, and Nu = nucleus, The scale bar indicates 10 μm.). The graph on the right shows the FRET efficiency (%). Significantly higher FRET efficiency was detected in the positive control (tandem ECFP + EYFP) than in the negative controls (ECFP and EYFP, ECFP and EYFP-Upf1, ECFP-Rex and EYFP), demonstrating that FRET efficiency is correctly measured in this method. The FRET efficiency between ECFP-Rex and EYFP-Upf1 was significantly higher in the cytoplasm and the p-body, indicating that Rex and Upf1 interact at these intracellular sites (n = 6–10, mean ± SD, *** p < 0.001).
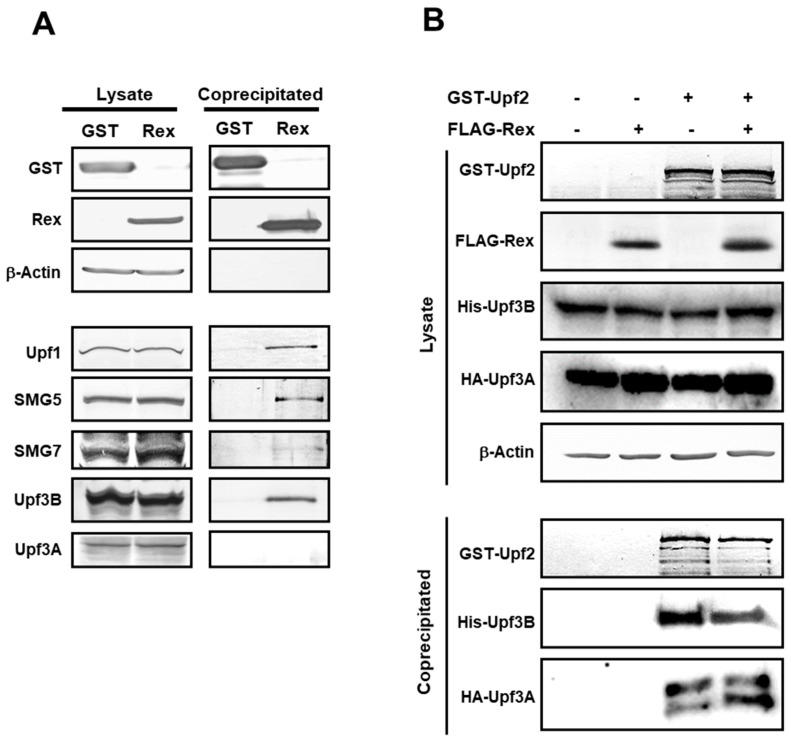
3.8. Interaction between Rex and NMD Regulatory Complex Proteins
Finally, the interaction of Rex with NMD regulatory proteins was investigated by coimmunoprecipitation assay with GST-Rex. The results showed that Rex interacted with SMG5 and Upf3B as well as Upf1. It also interacted weakly with SMG7 (Figure 7A). We noted that Rex binds to Upf3B but not to Upf3A. The normal NMD complex contains Upf3B, which mediates the interaction between EJC and Upf1 via Upf2 when the NMD pathway is activated. On the other hand, it is known that when Upf3A is included in the NMD complex, NMD activity is reduced [52]. Therefore, we hypothesized that Rex selectively binds to Upf3B and inhibits its entry into the NMD complex, thereby suppressing NMD. We examined how the amounts of His-Upf3B and HA-Upf3A interacting with GST-Upf2 changed in the presence and absence of Rex by GST-Upf2 coimmunoprecipitation assay. As such, in the presence of Rex, the amount of Upf3B interacting with Upf2 decreased, whereas the amount of Upf3A increased (Figure 7B).
Figure 7.
Rex interacts with Upf1 and SMG5/7 and contributes to the substitution of Upf3B and Upf3A. (A). NMD regulatory complex proteins interacting with Rex in HEK293FT cells were investigated by GST-Rex coimmunoprecipitation assay. The results showed that Rex interacts not only with Upf1 but also with SMG5, SMG7 and Upf3B. In contrast, Rex did not interact with Upf3A, the isoform of Upf3B. (B). Amount of Upf3B which binds to Upf2 in the NMD complex in the presence or absence of Rex detected by coimmunoprecipitation assay of GST-Upf2 with His-Upf3B and HA-Upf3A in HEK293FT cells. The amount of Upf3B interacting with Upf2 was reduced in the presence of Rex, and instead the interaction with Upf3A, the less active form of Upf3, was increased.
4. Discussion
Host cell NMD is a threat to the virus because many mRNAs derived from viral genomes have structures and RNA signals that are not found in eukaryotic cells and can be degraded via NMD as foreign substances. Previously, we have reported that Rex, an RNA-binding protein of HTLV-1, suppresses host NMD activity and stabilizes HTLV-1 genomic RNA [32]. However, the mechanism by which Rex achieves such a function has not been elucidated. In the present study, we “dissected” Rex to identify the important domain(s) of Rex for NMD repression by using various domain-deleted mutants of Rex. We also showed that Rex intervenes in the steps of NMD from the trigger to completion and may influence the function of multiple host factors that regulate NMD activity.
4.1. NMD Inhibition by Rex at the Mimiced Site of HTLV-1 Infection
In the present study, analysis of NMD activity in a stable Rex-expressing T cell line showed that Rex suppressed NMD in T cells, the host cells of HTLV-1 infection (Figure 1A,B). When the HTLV-1 infectious clone, pFL-MT2, was used to simulate a more realistic HTLV-1 infection, the activity of NMD was significantly increased in cells transfected with the Rex(-) clone compared with those transfected with the Rex(+) clone (Figure 1C). Because Rex represses Tax expression, Tax expression was increased in Rex(-)-pFL-MT-2 transfected cells compared with pFL-MT2 cells (Figure 1C, lower Western blotting image). It has been reported that Tax also represses NMD [33,34]. In our previous study, Tax was also found to inhibit NMD [32]. However, in the present study, NMD activity was significantly increased in cells transfected with Rex(-)-pFL-MT-2, even though Tax expression was upregulated. This suggests that Rex is mainly responsible for the NMD repressive function at the site where all HTLV-1 accessory proteins are expressed.
4.2. Identification of Rex Domains Important for NMD Suppression
The current study shows that ARM, the X region (aa20–56), whose function was previously unknown, or at least one of the two multimerization domains are important for the ability of Rex to suppress NMD (Figure 2A,C). In particular, loss of the X region was associated with a significant loss of NMD-suppressive activity, suggesting that the X region plays a central role in Rex-mediated NMD suppression (Figure 2A). Simultaneous loss of the two multimerization domains resulted in decreased NMD repression (Figure 2A), suggesting that multimerization is essential for Rex-mediated NMD repression. On the other hand, the Y region (aa124–169) and the stabilizing region (aa170–189) did not affect the expression level of Rex or its ability to suppress NMD (Figure 2B). As shown in Figure S1, deletion of aa150–159 in the Y region, as well as deletion of aa160–169 with the stability domain, markedly reduced protein. Therefore, the Y region may contain a motif which is important for stabilization of Rex protein.
4.3. Rex Phosphorylation State and NMD Inhibition
It is known that phosphorylation and multimerization of Rex are essential for the nuclear export and stabilization of unspliced and partially-spliced mRNAs of HTLV-1 [45,46]. We hypothesized that Rex phosphorylation is also required for NMD repression. We first inhibited the entire phosphorylation of Rex by H-7 and found that Rex-mediated NMD repression was abolished after 20 h of H-7 treatment (Figure 3A). This indicates that Rex phosphorylation is required for Rex-mediated NMD repression. Next, we used a series of Rex phosphorylation mutants to identify the phosphorylation sites of Rex that are important for NMD suppression. Of the seven phosphorylation sites in Rex, phosphorylation at T22, S106 and S174 was shown to be important for NMD inhibition (Figure 3B). T22 is located in the X region, S106 in the C-MD and S174 in the stability domain. The importance of the X region and MDs is consistent with the results using the domain-deficient mutant Rex proteins (Figure 2). Phosphorylation of S174 has also been reported to be essential for the activation of Rex [23]. The relationship between phosphorylation of T22 in the X region and its activation, which is important for NMD repression, needs to be further investigated.
4.4. Verification of CRM1 Dependence of NMD Suppression by Rex
To test the importance of nuclear export of Rex itself in NMD inhibition, we compared the ability of Rex to repress NMD after treatment with the CRM1 inhibitor Leptomycin B or overexpression of CRM1. We found that CRM1 inhibition reduced the ability of Rex to inhibit NMD, whereas overexpression of CRM1 enhanced the ability of Rex to inhibit NMD (Figure 4A). Since p21Rex, which lacks NLS and localizes only to the cytoplasm, had no such effect (Figure 4B), it is likely that CRM1-dependent transport of Rex to the cytoplasm is required for NMD repression. Since NMD functions in the cytoplasm in concert with translation [25], it is reasonable that NMD repression by Rex occurs in the cytoplasm.
In Figure 2A, however, ΔNES-Rex demonstrated the same NMD inhibitory activity as WT-Rex. Figure S2 shows that ΔNES-Rex is mainly in the nucleus but also in the cytoplasm at a lower level in HeLa cells. Therefore, we speculate that ΔNES-Rex in the cytoplasm exhibits NMD inhibitory activity, at least by ectopic overexpression.
4.5. Examination of Physical Interaction between Rex and NMD Factors
Finally, we investigated which interactions between Rex and NMD regulatory proteins are important for Rex-mediated NMD repression. We first examined the interaction between Rex and Upf1, the key regulator of NMD. The NMD cascade begins with phosphorylation of Upf1 by SMG1 and is completed by dephosphorylation by SMG5/7 [49,50]. Therefore, phosphorylation and dephosphorylation of Upf1 are essential for the execution of the NMD pathway. It has also been reported that the helicase activity of Upf1 is essential for the completion of NMD [51]. Previously, it has been reported that hyperphosphorylated Upf1 (G495R/G497E) and ATPase helicase-deficient Upf1 (R843C) are unable to activate NMD [42,43]. In the present study, we investigated the interaction between Rex and these Upf1 mutants by GST-Upf1 coimmunoprecipitation assay. The results showed that Rex interacted only with WT-Upf1 and hardly bound to mutant Upf1 (Figure 5A). It is possible that the helicase-deficient Upf1 mutant is not part of the active NMD complex and therefore has no opportunity to interact with Rex. It was also recently reported that hyper-phosphorylated Upf1 also plays an important role in the protein quality control machinery that transports C-terminal-deficient peptides, by-products of NMD target mRNAs, to the aggresome [53]. By not interacting with hyper-phosphorylated Upf1, Rex may avoid being transported to aggresome by Upf1.
When the interaction of WT-Upf1 with p27Rex or p21Rex was compared, only p27Rex interacted with Upf1 (Figure 5B). This result suggests that Rex interacts with Upf1 via the ARM/NLS and X regions of the N-terminal region, and that the interaction of p27Rex with Upf1 via the N-terminal region is important in NMD repression, as p21Rex did not show NMD repression (Figure 5B, right side graph). This result is in agreement with the results, which showed that the ARM and X regions of Rex are important for NMD repression (Figure 2 and Figure 3).
In addition, we analyzed in detail the intracellular interaction between Rex and Upf1. First, we co-expressed ECFP-Rex, EYFP-Upf1 and mCherry-Dcp2 in HeLa cells and investigated the subcellular localization of each protein. The results showed that Rex and Upf1 co-localized in the cytoplasm and the p-body as indicated by Dcp2 (Figure 6A). In addition, when the interaction between Rex and Upf1 was examined in the cell by FRET, the FRET efficiency of Rex and Upf1 was significantly increased in the cytoplasm and the p-body (Figure 6B). The p-body is a site of mRNA processing and mRNA degradation by the NMD pathway [54,55,56,57]. It has also been reported that viral mRNAs and proteins accumulate in the p-body [58]. Recently, it was also reported that p-bodies store translationally repressed mRNAs and regulate the efficiency of mRNA translation [59]. The initiation of the Upf1-centred NMD pathway occurs in the cytoplasm and is completed in the p-body. Therefore, Rex may interact with Upf1 from initiation to termination of NMD to regulate its activity.
Our GST-Rex immuno-coprecipitation assay shows that Rex interacts with Upf1, SMG5, Upf3B, and slightly with SMG7 (Figure 7A). SMG5 and SMG7 are responsible for dephosphorylation of Upf1, which is essential for completion of NMD [49,50]. Thus, Rex may inhibit NMD completion by suppressing the function of these proteins. Our results also showed that Rex interacted with Upf3B but did not interact with Upf3A (Figure 7A). The normal NMD complex mainly contains Upf3B interacting with Upf2. On the other hand, it has been reported that NMD activity decreases when Upf3A is included [52]. We investigated the possibility that Rex alters the amounts of Upf3B and Upf3A in the NMD complex. Our results demonstrated that, in the presence of Rex, the amount of Upf3B bound to Upf2 decreased and the amount of Upf3A increased (Figure 7B). These results suggest that Rex may contribute to the replacement of Upf3B and Upf3A in the NMD complex. We believe that Rex selectively binds to Upf3B, thereby preventing Upf3B from entering the NMD complex and allowing Upf3A to enter instead, thereby suppressing NMD activity.
4.6. Summary
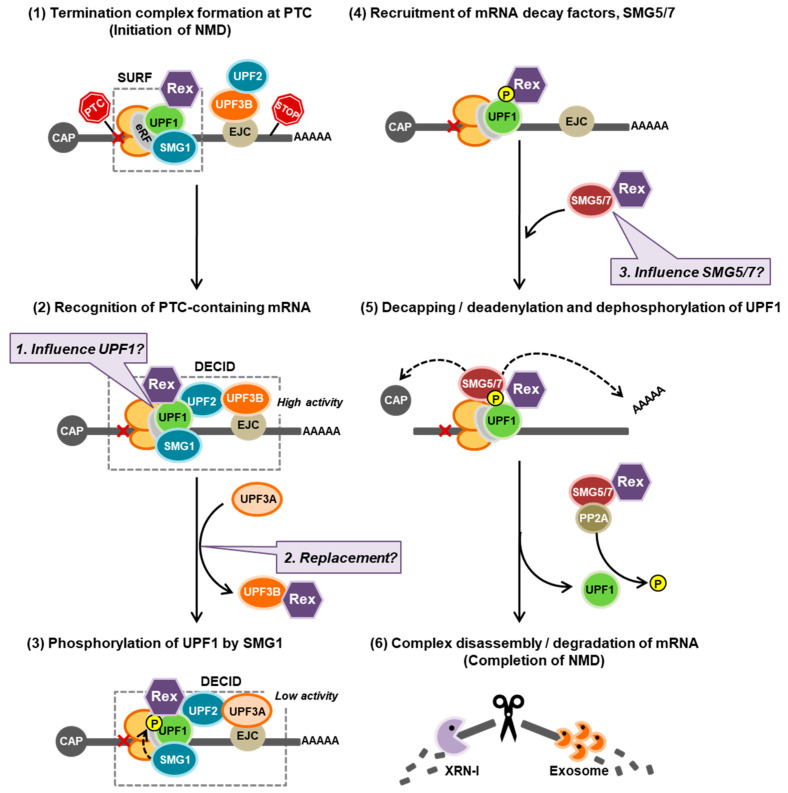
In the current study, we found that the ARM region and the X region, whose function was previously unknown, at the N-terminus of Rex play important roles in NMD repression. This may be because Rex interacts with the NMD key regulator Upf1 via this region. Furthermore, the interaction between Rex and Upf1 was observed in the cytoplasm and p-bodies, where the NMD cascade functions. Indeed, Rex interacts not only with Upf1, but also with SMG5 and SMG7, which are essential for Upf1 dephosphorylation and NMD completion, suggesting that Rex may intervene in the NMD regulatory complex from initiation to termination of NMD. Finally, we found that Rex binds selectively to Upf3B, one of the two isoforms of Upf3, and that in the presence of Rex, the amount of Upf3B in the NMD complex is reduced and the intervention of Upf3A is promoted; since the intervention of Upf3A reduces NMD activity [52], it is possible that Rex may suppress NMD by swapping Upf3B and Upf3A in the complex. Thus, Rex may enter the NMD complex from the beginning to the end of NMD and may influence the cellular NMD activity (Figure 8). Such interaction between Rex and NMD regulatory proteins may occur on the HTLV-1 mRNA, because the HTLV-1 genomic mRNA that Rex transports out of the nucleus contains multiple RNA signals that activate NMD.
Figure 8.
Possible model of Rex-NMD complex interaction and new questions. NMD progresses in a cascade as shown in (1–6). (1): Our results suggest that Rex may inhibit NMD by intervening from the beginning to the end of this cascade by interacting with the NMD key regulator Upf1 and may have some effect on its activity (Baloon-1). (2): Rex interacts only with Upf3B, suggesting that Rex substitutes Upf3A for Upf3B in the NMD complex, resulting in the inclusion of less active Upf3A in the NMD complex, thereby suppressing NMD activity (Baloon-2). (4,5): The mRNA decay factors, the SMG5/7 complex, are recruited to phosphorylated Upf1. The SMG5/7 complex recruits the decapping complex (DCP2/DCP1a) and the deadenylation complex CCR4–NOT to enhance decapping and deadenylation of the NMD target mRNA. Additionally, the SMG5/7 complex recruits protein phosphatase 2A (PP2A) for dephosphorylation of Upf1. Since dephosphorylation of Upf1 is essential for NMD completion, Rex may influence the dephosphorylation pathway of Upf1 via SMG5/7 interaction, and thus may influence NMD completion (Baloon-3).
Protecting viral mRNA from host NMD is an essential function in viral replication. In this study, we proposed a new possibility that Rex may protect HTLV-1 genomic mRNA, a particularly vulnerable NMD target, from degradation by interacting with various NMD regulators. Rex may also regulate the timing of the initial active viral replication phase and the subsequent stable latent infection phase not only by the already-known function to transport viral mRNAs encoding structural proteins from the nucleus, but also by controlling the activity of NMD that targets these viral mRNAs.
Acknowledgments
We appreciate Hiroo Hoshino in Gunma University, Japan, for his generous gift of MT-2 cells. We thank Hisatoshi Shida in Hokkaido University, Japan, very much for his kind gift of the Rex expressing plasmid, SRα-Rex.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/v14020344/s1, Figure S1: Y-region contains a motif important for stabilization of Rex; Figure S2: Subcellular localization of FLAG-Rex in HeLa cells.
Author Contributions
K.N., N.K. and M.H. performed experiments; Y.T. produced and provided monoclonal antibodies against HTLV-1 proteins; T.O. produced and provided HTLV-1 infectious clone pFL-MT2; K.N., K.U. and T.W. designed the research and wrote this paper. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by Grants-in-Aid for Scientific Research from the Ministry of Education, Culture, Sports, Science, and Technology of Japan, to KN (15K06827) and to KN (19K07573).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable (No public data was used in this study. Data in this study are not available in public.)
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Poiesz B.J., Ruscetti F.W., Gazdar A.F., Bunn P.A., Minna J.D., Gallo R.C. Detection and isolation of type C retrovirus particles from fresh and cultured lymphocytes of a patient with cutaneous T-cell lymphoma. Proc. Natl. Acad. Sci. USA. 1980;77:7415–7419. doi: 10.1073/pnas.77.12.7415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Poiesz B.J., Ruscetti F.W., Reitz M.S., Kalyanaraman V.S., Gallo R.C. Discovery of new type C retrovirus (HTLV-1) Nature. 1981;294:268–271. doi: 10.1038/294268a0. [DOI] [PubMed] [Google Scholar]
- 3.Yoshida M., Miyoshi I., Yorio H. Isolation and characterization of retrovirus from cell lines of human adult T-cell leukemia and its implications in the disease. Proc. Natl. Acad. Sci. USA. 1982;79:2031–2035. doi: 10.1073/pnas.79.6.2031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Takatsuki K. Discovery of adult T-cell leukemia. Retrovirology. 2005;3:16. doi: 10.1186/1742-4690-2-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gallo R.C. The discovery of the first human retrovirus: HTLV-1 and HTLV-2. Retrovirology. 2005;7:17. doi: 10.1186/1742-4690-2-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Iwanaga M., Watanabe T., Utsunomiya A., Okayama A., Uchimaru K., Koh K.-R., Ogata M., Kikuchi H., Sagara Y., Uozumi K., et al. Human T-cell leukemia virus type I (HTLV-1) proviral load and disease progression in asymptomatic HTLV-1 carriers: A nationwide prospective study in Japan. Blood. 2010;116:1211–1219. doi: 10.1182/blood-2009-12-257410. [DOI] [PubMed] [Google Scholar]
- 7.Hirons A., Khoury G., Purcell D.F.J. Human T-cell lymphotropic virus type-1: A lifelong persistent infection, yet never truly silent. Lancet Infect. Dis. 2021;21:e2–e10. doi: 10.1016/S1473-3099(20)30328-5. [DOI] [PubMed] [Google Scholar]
- 8.Johnson J.M., Harrod R., Franchini G. Molecular biology and pathogenesis of the human T-cell leukaemia/lymphotropic virus Type-1 (HTLV-1) Int. J. Exp. Pathol. 2001;82:135–147. doi: 10.1046/j.1365-2613.2001.00191.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Franchini G., Fukumoto R., Fullen J.R. T-cell control by human T-cell leukemia/lymphoma virus type 1. Int. J. Hematol. 2003;78:280–296. doi: 10.1007/BF02983552. [DOI] [PubMed] [Google Scholar]
- 10.Nicot C., Harrod R.L., Ciminale V., Franchini G. Human T-cell leukemia/lymphoma virus type 1 nonstructural genes and their functions. Oncogene. 2005;24:6026–6034. doi: 10.1038/sj.onc.1208977. [DOI] [PubMed] [Google Scholar]
- 11.Kashanchi F., Brady J.N. Transcriptional and post-transcriptional gene regulation of HTLV-1. Oncogene. 2005;24:5938–5951. doi: 10.1038/sj.onc.1208973. [DOI] [PubMed] [Google Scholar]
- 12.Kannian P., Green P.L. Human T Lymphotropic Virus Type 1 (HTLV-1): Molecular Biology and Oncogenesis. Viruses. 2010;2:2037–2077. doi: 10.3390/v2092037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hakata Y., Umemoto T., Matsushita S., Shida H. Involvement of human CRM1 (exportin 1) in the export and multimerization of the Rex protein of human T-cell leukemia virus type 1. J. Virol. 1998;72:6602–6607. doi: 10.1128/JVI.72.8.6602-6607.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hakata Y., Yamada M., Shida H. Rat CRM1 Is Responsible for the Poor Activity of Human T-Cell Leukemia Virus Type 1 Rex Protein in Rat Cells. J. Virol. 2001;75:11515–11525. doi: 10.1128/JVI.75.23.11515-11525.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Inoue J., Seiki M., Yoshida M. The second pX product p27 chi-III of HTLV-1 is required for gag gene expression. FEBS Lett. 1986;209:187–190. doi: 10.1016/0014-5793(86)81108-5. [DOI] [PubMed] [Google Scholar]
- 16.Hidaka M., Inoue J., Yoshida M., Seiki M. Post-transcriptional regulator (rex) of HTLV-1 initiates expression of viral structural proteins but suppresses expression of regulatory proteins. EMBO J. 1988;7:519–523. doi: 10.1002/j.1460-2075.1988.tb02840.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gröne M., Koch C., Grassmann R. The HTLV-1 Rex protein induces nuclear accumulation of unspliced viral RNA by avoiding intron excision and degradation. Virology. 1996;218:316–325. doi: 10.1006/viro.1996.0200. [DOI] [PubMed] [Google Scholar]
- 18.Ye J., Silverman L., Lairmore M.D., Green P.L., Dc W. HTLV-1 Rex is required for viral spread and persistence in vivo but is dispensable for cellular immortalization in vitro. Blood. 2003;102:3963–3969. doi: 10.1182/blood-2003-05-1490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Baydoun H.H., Bellon M., Nicot C. HTLV-1 Yin and Yang: Rex and p30 Master Regulators of Viral mRNA Trafficking. AIDS Rev. 2008;10:195–204. doi: 10.1016/j.bbi.2008.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rende F., Cavallari I., Corradin A., Silic-Benussi M., Toulza F., Toffolo G.M., Tanaka Y., Jacobson S., Taylor G.P., D’Agostino D.M., et al. Kinetics and intracellular compartmentalization of HTLV-1 gene expression: Nuclear retention of HBZ mRNAs. Blood. 2011;117:4855–4859. doi: 10.1182/blood-2010-11-316463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nakano K., Watanabe T. HTLV-1 Rex: The courier of viral messages making use of the host vehicle. Front. Microbiol. 2012;3:330. doi: 10.3389/fmicb.2012.00330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nakano K., Watanabe T. HTLV-1 Rex tunes the cellular environment favorable for viral replication. Viruses. 2016;8:58. doi: 10.3390/v8030058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kesic M., Doueiri R., Ward M., Semmes O.J., Green P.L. Phosphorylation regulates human T-cell leukemia virus type 1 Rex function. Retrovirology. 2009;6:105. doi: 10.1186/1742-4690-6-105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xie L., Kesic M., Yamamoto B., Li M., Younis I., Lairmore M.D., Green P.L. Human T-cell leukemia virus type 2 Rex carboxy terminus is an inhibitory/stability domain that regulates Rex functional activity and viral replication. J. Virol. 2009;83:5232–5243. doi: 10.1128/JVI.02271-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Karousis E.D., Mühlemann O. Nonsense-mediated mRNA decay begins where translation ends. Cold Spring Harb. Perspect. Biol. 2019;11:a032862. doi: 10.1101/cshperspect.a032862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fiorini F., Bagchi D., Le Hir H., Croquette V. Human Upf1 is a highly processive RNA helicase and translocase with RNP remodelling activities. Nat. Commun. 2015;6:7581. doi: 10.1038/ncomms8581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kurosaki T., Popp M.W., Maquat L.E. Quality and quantity control of gene expression by nonsense-mediated mRNA decay. Nat. Rev. Mol. Cell Biol. 2019;20:406–420. doi: 10.1038/s41580-019-0126-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Popp M.W.L., Cho H., Maquat L.E. Viral subversion of nonsense-mediated mRNA decay. Rna. 2020;26:1509–1518. doi: 10.1261/rna.076687.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Quek B.L., Beemon K. Retroviral strategy to stabilize viral RNA. Curr. Opin. Microbiol. 2014;18:78–82. doi: 10.1016/j.mib.2014.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Moon S.L., Barnhart M.D., Wilusz J. Inhibition and avoidance of mRNA degradation by RNA viruses. Curr. Opin. Microbiol. 2012;15:500–505. doi: 10.1016/j.mib.2012.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dickson A.M., Wilusz J. Strategies for viral RNA stability: Live long and prosper. Trends Genet. 2011;27:286–293. doi: 10.1016/j.tig.2011.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nakano K., Ando T., Yamagishi M., Yokoyama K., Ishida T., Ohsugi T., Tanaka Y., Brighty D.W., Watanabe T. Viral interference with host mRNA surveillance, the nonsense-mediated mRNA decay (NMD) pathway, through a new function of HTLV-1 Rex: Implications for retroviral replication. Microbes Infect. 2013;15:491–505. doi: 10.1016/j.micinf.2013.03.006. [DOI] [PubMed] [Google Scholar]
- 33.Fiorini F., Robin J.P., Kanaan J., Borowiak M., Croquette V., Le Hir H., Jalinot P., Mocquet V. HTLV-1 Tax plugs and freezes Upf1 helicase leading to nonsense-mediated mRNA decay inhibition. Nat. Commun. 2018;9:431. doi: 10.1038/s41467-017-02793-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mocquet V., Neusiedler J., Rende F., Cluet D., Robin J., Terme J., Dodon M.D., Morris C., Ciminale V., Jalinot P. The Human T-Lymphotropic Virus Type 1 Tax Protein Inhibits Nonsense-Mediated mRNA Decay by Interacting with INT6/EIF3E and Upf1. J. Virol. 2012;86:7530–7543. doi: 10.1128/JVI.07021-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Prochasson L., Jalinot P., Mocquet V. The complex relationship between HTLV-1 and nonsense-mediated mRNA decay (NMD) Pathogens. 2020;9:287. doi: 10.3390/pathogens9040287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nakano K., Uchimaru K., Utsunomiya A., Yamaguchi K., Watanabe T. Dysregulation of c-Myb pathway by aberrant expression of proto-oncogene MYB provides the basis for malignancy in adult T-cell leukemia/lymphoma cells. Clin. Cancer Res. 2016;22:5915–5928. doi: 10.1158/1078-0432.CCR-15-1739. [DOI] [PubMed] [Google Scholar]
- 37.Firouzi S., López Y., Suzuki Y., Nakai K., Sugano S., Yamochi T., Watanabe T. Development and validation of a new high-throughput method to investigate the clonality of HTLV-1-infected cells based on provirus integration sites. Genome Med. 2014;6:46. doi: 10.1186/gm568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ishida T., Tojo T., Aoki T., Kobayashi N., Ohishi T., Watanabe T., Yamamoto T., Inoue J.I. TRAF5, a novel tumor necrosis factor receptor-associated factor family protein, mediates CD40 signaling. Proc. Natl. Acad. Sci. USA. 1996;93:9437–9442. doi: 10.1073/pnas.93.18.9437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mendell J.T., Medghalchi S.M., Lake R.G., Noensie E.N., Dietz H.C. Novel Upf2p Orthologues Suggest a Functional Link between Translation Initiation and Nonsense Surveillance Complexes. Mol. Cell. Biol. 2000;20:8944–8957. doi: 10.1128/MCB.20.23.8944-8957.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ishida T., Mizushima S.I., Azuma S., Kobayashi N., Tojo T., Suzuki K., Aizawa S., Watanabe T., Mosialos G., Kieff E., et al. Identification of TRAF6, a novel tumor necrosis factor receptor-associated factor protein that mediates signaling from an amino-terminal domain of the CD40 cytoplasmic region. J. Biol. Chem. 1996;271:28745–28748. doi: 10.1074/jbc.271.46.28745. [DOI] [PubMed] [Google Scholar]
- 41.Rimsky L., Dodon M.D., Dixon E.P., Greene W.C. Trans-dominant inactivation of HTL V-I and HIV-1 gene expression by mutation of the HTLV-1 Rex transactivator. Nature. 1989;341:453–456. doi: 10.1038/341453a0. [DOI] [PubMed] [Google Scholar]
- 42.Isken O., Kim Y.K., Hosoda N., Mayeur G.L., Hershey J.W.B., Maquat L.E. Upf1 phosphorylation triggers translational repression during nonsense-mediated mRNA decay. Cell. 2008;133:314–327. doi: 10.1016/j.cell.2008.02.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kaygun H., Marzluff W.F. Regulated degradation of replication-dependent histone mRNAs requires both ATR and Upf1. Nat. Struct. Mol. Biol. 2005;12:794–800. doi: 10.1038/nsmb972. [DOI] [PubMed] [Google Scholar]
- 44.Ohsugi T., Kumasaka T., Urano T. Construction of a full-length human T cell leukemia virus type I genome from MT-2 cells containing multiple defective proviruses using overlapping polymerase chain reaction. Anal. Biochem. 2004;329:281–288. doi: 10.1016/j.ab.2004.02.036. [DOI] [PubMed] [Google Scholar]
- 45.Adachi Y., Nosaki T., Hatanaka M. Protein kinase inhibitor H-7 blocks accumulation of unspliced mRNA of Human T-cell leukemia virus type I (HTLV-I) Biochem. Biophys. Res. Commun. 1990;169:469–475. doi: 10.1016/0006-291X(90)90355-Q. [DOI] [PubMed] [Google Scholar]
- 46.Adachi Y., Copeland T.D., Takahashi C., Nosaka T., Ahmed A., Oroszlan S., Hatanaka M. Phosphorylation of the Rex protein of human T-cell leukemia virus type I. J. Biol. Chem. 1992;267:21977–21981. doi: 10.1016/S0021-9258(19)36709-2. [DOI] [PubMed] [Google Scholar]
- 47.Orita S., Kobayashi H., Aono Y., Saiga A., Maeda M., Igarashi H. p21X mRNA is expressed as a singly spliced pX transcript from defective provirus genomes having a partial delection of the pol-env region in human T-cell leukemia virus type 1-infected cells. Nucleic Acids Res. 1993;21:3799–3807. doi: 10.1093/nar/21.16.3799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Saiga A., Aono Y., Imai J., Kinoshita K., Orita S., Igarashi H. Presence of antibodies to p21X and/or p27rex proteins in sera from human T-cell leukemia virus type I-infected individuals. J. Virol. Methods. 1996;57:157–168. doi: 10.1016/0166-0934(95)01979-0. [DOI] [PubMed] [Google Scholar]
- 49.Ohnishi T., Yamashita A., Kashima I., Schell T., Anders K.R., Grimson A., Hachiya T., Hentze M.W., Anderson P., Ohno S. Phosphorylation of hUpf1 induces formation of mRNA surveillance complexes containing hSMG-5 and hSMG-7. Mol. Cell. 2003;12:1187–1200. doi: 10.1016/S1097-2765(03)00443-X. [DOI] [PubMed] [Google Scholar]
- 50.Unterholzner L., Izaurralde E. SMG7 acts as a molecular link between mRNA surveillance and mRNA decay. Mol. Cell. 2004;16:587–596. doi: 10.1016/j.molcel.2004.10.013. [DOI] [PubMed] [Google Scholar]
- 51.Franks T.M., Singh G., Lykke-Andersen J. Upf1 ATPase-Dependent mRNP Disassembly Is Required for Completion of Nonsense- Mediated mRNA Decay. Cell. 2010;143:938–950. doi: 10.1016/j.cell.2010.11.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chan W.-K., Bhalla A.D., Le Hir H., Nguyen L.S., Huang L., Gécz J., Wilkinson M.F. A Upf3-mediated regulatory switch that maintains RNA surveillance. Nat. Struct. Mol. Biol. 2009;16:747–753. doi: 10.1038/nsmb.1612. [DOI] [PubMed] [Google Scholar]
- 53.Park Y., Park J., Hwang H.J., Kim B., Jeong K., Chang J., Lee J.B., Kim Y.K. Nonsense-mediated mRNA decay factor Upf1 promotes aggresome formation. Nat. Commun. 2020;11:3106. doi: 10.1038/s41467-020-16939-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Luo Y., Na Z., Slavoff S.A. P-Bodies: Composition, Properties, and Functions. Biochemistry. 2018;57:2424–2431. doi: 10.1021/acs.biochem.7b01162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Brogna S., Ramanathan P., Wen J. Upf1 P-body localization. Biochem. Soc. Trans. 2008;36:698–700. doi: 10.1042/BST0360698. [DOI] [PubMed] [Google Scholar]
- 56.Eulalio A., Behm-Ansmant I., Izaurralde E. P bodies: At the crossroads of post-transcriptional pathways. Nat. Rev. Mol. Cell Biol. 2007;8:9–22. doi: 10.1038/nrm2080. [DOI] [PubMed] [Google Scholar]
- 57.Parker R., Sheth U. P bodies and the control of mRNA translation and degradation. Mol. Cell. 2007;25:635–646. doi: 10.1016/j.molcel.2007.02.011. [DOI] [PubMed] [Google Scholar]
- 58.Beckham C.J., Parker R. P bodies, stress granules, and viral life cycles. Cell Host Microbe. 2008;3:206–212. doi: 10.1016/j.chom.2008.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hubstenberger A., Courel M., Bénard M., Souquere S., Ernoult-Lange M., Chouaib R., Yi Z., Morlot J.B., Munier A., Fradet M., et al. P-Body Purification Reveals the Condensation of Repressed mRNA Regulons. Mol. Cell. 2017;68:144–157.e5. doi: 10.1016/j.molcel.2017.09.003. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Not applicable (No public data was used in this study. Data in this study are not available in public.)