Abstract
Coronavirus disease 2019 (COVID-19) is a contagious disease caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) that can be transmitted through human interaction. In this paper, we present a Piecewise Susceptible–Exposed–Infectious–Unreported–Removed model for infectious diseases and discuss qualitatively and quantitatively. The parameters are explored by mathematical and statistical methods. Numerical simulations of these models are performed on COVID-19 US data and Python is used in the visualization of results. Outbreak factor is generated by piecewise model to explore the future trend of the US pandemic. Several error metrics are given to discuss the accuracy of the models. The main achievement of this paper is to propose the piecewise model and find the relationship between spread of pandemic and mitigation measures to control it by observing the results of numerical simulations. Performance analysis of piecewise model is presented based on COVID-19 data obtained by ‘worldmeter’.
Keywords: Dynamic parameters, SEIUR, Outbreak factor, COVID-19, Unreported population
1. Introduction
Since the end of 2019, the COVID-19 pandemic [1], [2], caused by the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), is raging around the world [16]. In December 2019, the first cases of COVID-19 were detected in Wuhan, Hubei Province, China. As of March 11, 2021, more than 118 million confirmed cases have been reported in 219 countries and regions, of which more than 2.621 million died and 66.926 million recovered. At present, the number of confirmed cases is still rising rapidly [3].
Hamer, Ross, and others have done a lot of work in the establishment of mathematical models of infectious diseases until 1927 [7], [10]. Kermack and McKendrick studied the SIR compartmental model during the Black Death in London [7], [14], and the SIS model was established in 1932 [15], [19]. Based on the study of these models, the threshold theory in the dynamics of infectious diseases was proposed [17]. The SIR model of Kermack and McKendrick is the most classic and basic model among infectious disease models and has made a foundational contribution to the study of infectious disease dynamics. The SIR model divides the total population into the following three populations: susceptible , which represents the people who are not infected but are likely to be infected by this type of disease; infectious , which represents the people who have been infected as patients and has the capability of transmitting the disease; and removed , which stands for the people who have been removed from the infected, see also [12], [13] and references therein. Nowadays, the SEIR model [8] is widely used to analyze infectious diseases, see also [9], [20] and references therein. In this model, individuals experience a long incubation duration (the “exposed” category), such that the individual is infected but not yet symptomatic. For example, chicken pox and even vector-borne diseases such as dengue hemorrhagic fever have a long incubation duration where the individual cannot yet transmit the pathogen to others.
The SEIR model is as follows;
| (1) |
| (2) |
| (3) |
| (4) |
The meaning of each parameter and variable is shown below:
: Total population.
: The number of susceptible individuals at time . They are not infected at this point.
: The number of asymptomatic individuals exposed to the virus at time . They are in the incubation period and capable of transmitting the disease but have not exhibited any symptoms.
: The number of symptomatic infected individuals at time . This group has tested positive and corresponds to the Worldmeter data for active cases. [5]
: The number of individuals who have been removed from the infected at time due to either death or recovery.
: Transmission rate.
: Removed rate.
: Incubation rate.
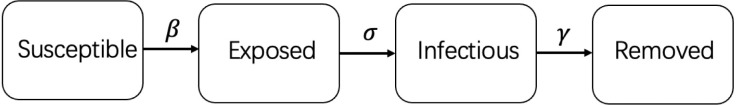
The SEIR diagram below shows how individuals move through each compartment in the model (see Fig. 1):
Fig. 1.
SEIR model.
The SEIR model can be applied to most infectious diseases, but there are some limitations when applied to the COVID-19 data. Due to the characteristics of COVID-19, exposed individuals also have the capability of transmitting the disease. The traditional SEIR model assumes that the members of the compartment are infected but not infectious during the incubation period[8]. The SEIR model also ignores the group of unreported cases. Due to the lack of medical resources and variability in the testing policy, there is no guarantee that all infected people are tested and reported during a wide-spread pandemic. This group of people is likely to become hidden transmitters in the population. The SEIR model with constant parameters cannot be used for long-term simulation. This is because the transmission rate and removed rate must be changed over a long period of time.
In this paper, we propose the piecewise SEIUR model and the use of a type of least-squares method to estimate the parameters, which overcomes the main difficulty in the SEIR model.
2. SEIUR model
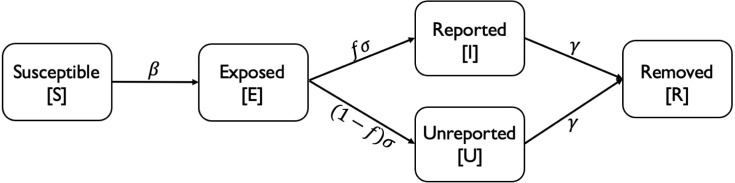
We expect to address the two weaknesses of the SEIR model. First, to overcome the second weakness, the SEIUR model divides the infection equation (3) from SEIR model into two new parts: an unreported symptomatic infection equation (13) and a reported symptomatic infection equation (12). Compartment represents individuals who are infectious and have been tested (and thus reported). The new compartment represents people who are symptomatic and infectious but have not been tested for various reasons. Let be the proportion from to , and be the proportion from to . To handle the first weakness, the infectious population in Eqs. (10), (11) is replaced by the sum of exposed population and unreported infectious cases , because the individuals in groups and have the ability to spread the virus to others during the infection. On the contrary, people in group were required to be quarantined at home or in hospital and could not infect others.
The establishment of the SEIUR model is based on the following assumptions:
-
•
Keep first three assumptions same as SEIR model
-
•
Exposed and Unreported infected individuals are capable of transmitting the disease
-
•
Reported infectious individuals do not have capability of transmitting the disease
-
•
Unreported infected individuals cannot become the reported infected individuals
The SEIUR model is given by;
| (5) |
| (6) |
| (7) |
| (8) |
| (9) |
Keep , , , , , and same as SEIR Model.
: The number of reported infected individuals at time who are symptomatic and have been tested.
: The number of individuals infected with the virus but have not been tested at time and thus not reported.
: The proportion of Exposed individuals that become reported infected individuals (Reported fraction) (see Fig. 2).
Fig. 2.
SEIUR model.
The transition diagram is shown as:
No matter how the SEIR model changes, the core parameters are always and . Therefore, the estimation of and is particularly vital for both SEIR model and SEIUR model.
2.1. Piecewise SEIUR model
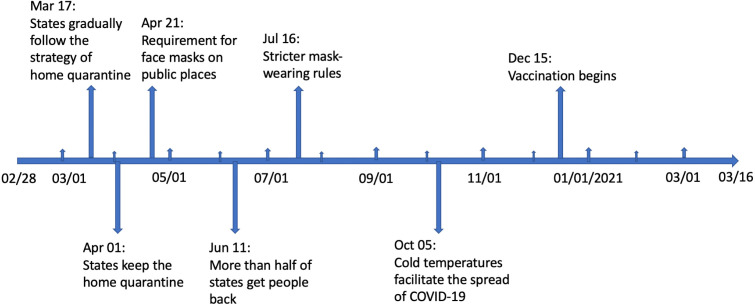
The SEIR and SEIUR models have a fixed value for and for the entire time period. Since the values of and are affected by many factors, such as government epidemic prevention measures, quarantine, and vaccination, it is unreasonable to set and as constants. In addition, another popular method is to use a function to fit and respectively. This approach makes the values of and continuous, and the changes are too frequent. Therefore, this paper combines these two ideas, assuming that the values of and are constant in a period of time, and when entering the next period, the values of and are updated. The selection of update points is based on the time when the government implemented the relevant strategies or policies, and the phase between two consecutive update points is defined as the “period”. The strategies release timeline is shown in Fig. 3.
Fig. 3.
Strategies [18] release timeline.
Using Fig. 3, we separate the whole time period from 2/28/2020 to 3/16/2021 into eight different parts:
-
1.
From 2/28/2020 to 3/16/2020
-
2.
From 3/17/2020 to 3/31/2020
-
3.
From 4/1/2020 to 4/20/2020
-
4.
From 4/21/2020 to 6/10/2020
-
5.
From 6/11/2020 to 7/15/2020
-
6.
From 7/16/2020 to 10/4/2020
-
7.
From 10/5/2020 to 1/15/2021
-
8.
From 1/15/2021 to 3/16/2021
For each period , we have an independent SEIUR model, and corresponding specific parameters and . The piecewise SEIUR model is shown below:
| (10) |
| (11) |
| (12) |
| (13) |
| (14) |
: Transmission rate during the period .
: Removed rate during the period .
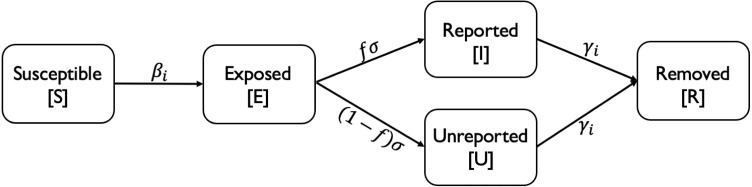
Other notations have the same meaning as before and the transition diagram for period is shown in Fig. 4.
Fig. 4.
SEIUR model for period .
In an epidemic, we can obviously judge whether the epidemic is breaking out or disappearing through the change rate of the number of infected people. In the piecewise SEIUR model, E, I and U represent the number of infections. The difference is that those in E are in the incubation period, while those in I and U have been infected.
Thus, we can conclude that the epidemic is still in outbreak when the change rate of the summation of , and is greater than , that is,
which implies
By the SEIUR model
It follows that
Since , , are all positive, we have
When the rate of change of , we have that the virus is disappearing.
Using the previous approach, we have that
When the rate of change of , we have that the virus is under control and may coexist with humans for a long time.
Therefore, we have
By the mathematical analysis above, in each period , the formula of the outbreak factor at time generated by the piecewise SEIUR model is defined by
And we define to represent the average value of outbreak factor in period .
3. Learning of the piecewise SEIUR model
3.1. Data
The data used in this paper is from the reference website ‘Worldmeter’ [5]. This website provides a variety of real-time statistical data. The website belongs to Dadax, an independent digital media company in the United States. This website records all pandemic data in the United States from February 15, 2020 to the present, and the data is updated daily.
The data is comprised of three different contents. The first is active cases starting from 2/15/2020 to 3/16/2021. The second and third contents are in same period and include the total cases and total deaths respectively. The detail of the contents is shown in Table 1:
Table 1.
Data content of total cases, active cases and total deaths.
| Data content | Data description | Data period | Data size |
|---|---|---|---|
| Total cases | Total number of infections | 2/15/2020 to 3/16/2021 | (1, 396) |
| (including deaths and recoveries) | |||
| Active cases | Current number of infections | 2/15/2020 to 3/16/2021 | (1, 396) |
| (excluding deaths and recoveries) | |||
| Total deaths | Total number of deaths | 2/15/2020 to 3/16/2021 | (1, 396) |
We shift the initial data point from 2/15/2020 to 2/28/2020. Although the United States announced the first case of the coronavirus on 1/21/2020, the lack of effective detection methods and insufficient understanding of the coronavirus have led to the low accuracy of the data in the early stage of the pandemic. Thus, we discarded the data from the early period of the pandemic and set 2/28/2020 as the new initial data point.
In addition to the above three sets of data, this paper also needs the total removed cases and total recovered cases. These two sets of data can be obtained by simple calculations between total cases, active cases, and total deaths. The formulas is followed:
In previous sections, we clearly explained the reasons for constructing the piecewise model and how we segmented the data into distinct periods.
Table 2 shows the details about each time period.
Table 2.
Split data according to strategies [18].
| Period # | Time period | Data size | Related policy |
|---|---|---|---|
| Period 1 | 2/28/2020 to 3/16/2020 | (1, 18) | No control measures implemented |
| Period 2 | 3/17/2020 to 3/31/2020 | (1, 15) | States gradually follow the strategy |
| of home quarantine | |||
| Period 3 | 4/1/2020 to 4/20/2020 | (1, 20) | States keep the home quarantine |
| Period 4 | 4/21/2020 to 6/10/2020 | (1, 51) | Requirement for face masks on public places |
| Period 5 | 6/11/2020 to 7/15/2020 | (1, 35) | More than half of states get people back |
| to work | |||
| Period 6 | 7/16/2020 to 10/4/2020 | (1, 81) | Stricter mask-wearing rules |
| Period 7 | 10/5/2020 to 12/14/2020 | (1, 71) | Cold temperatures facilitate the spread |
| of COVID-19 | |||
| Period 8 | 12/15/2020 to 3/16/2021 | (1, 92) | Vaccination begins |
According to the piecewise SEIUR model, in each period , there is a corresponding SEIUR model. In addition to showing the accuracy of SEIUR model in each period , this paper also illustrates the performance of this model in the short future term. In other words, we obtain the SEIUR model in period , and use it to estimate the data at the beginning of period . For that reason, the training data for SEIUR model in period is all data from itself, and the test data consists of the first seven days from period . For the last period, since the next period data is not available, we divide the period itself into training data and test data with test data being the last seven days.
3.2. Reported rate estimation and algorithms
Our first goal is to estimate an appropriate value of the proportion . Recalling the definition explained in the previous section, stands for the proportion of that become . Let represent the exposed individuals in time , represent the removed rate, is the incubation rate, represent the daily COVID-19 tests per thousand people at time , stand for the daily positive rate (the share of COVID-19 tests that are positive) at time , and is the total population. And we assume that all individuals in are newly infected and the effect of new exposed cases from to on the results is ignored, then the proportion at time is given by
| (15) |
In fact, the reported rate at time can be approximately equal to the proportion of that would be converted into reported cases. Consequently, to estimate , is served as the denominator, and the numerator should be the number of newly reported cases from time to the time that all is converted into reported cases or unreported cases. By the definition, we have that represents the daily positive tests at time . stands for the average number of days it takes for a newly infected individual (in ) to move to the next compartment ( or ). Hence, can be used to estimate the number of newly reported cases from time to the time that all is converted into reported cases or unreported cases. The result is the formula (15).
Since the number of exposed people cannot be directly counted, it is almost impossible to find relevant statistical data on . Here we use the SEIR model to generate historical data of . is assumed in previous section. is obtained by the least square method. is the total population of United States. For and , we use data from ‘Our World in Data’ [11], which provide the daily COVID-19 tests per thousand people and the daily positive rate. Now, we can generate the proportion at any day .
For simplicity, we define as the constant proportion rate for all data and use it in the numerical simulation.
By the formula (15) and the provided data, we estimate
We can easily obtain the numerical solution of the SEIUR model and piecewise SEIUR model. The algorithms are shown as Algorithms 1 and 2.
| Algorithm 1 Determining the numerical solution of SEIUR model |
| at all time |
| Input: |
| The initial value of variables: , , , and ; |
| The optimal parameters: , , and ; |
| Total population of the United States: ; |
| The number of iterations or days: ; |
| Output |
| , , , and at all time |
| Procedure |
| For in to |
| Return, , , and |
| Algorithm 2 Determining the numerical solution of piecewise SEIUR model |
| at time for |
| where is the start point of period and |
| Input: |
| The initial value of variables: , , , and ; |
| The optimal parameters for each period : , , and ; |
| Total population of the United States: ; |
| The number of iterations or days: ; |
| Output |
| , , and at all time |
| Procedure |
| For in to |
| For in to |
| Return, , , and |
In the SEIUR model, there are four unknown parameters to be estimated: , , , and . We have estimated the proportion of to as 0.6, that is, . The incubation rate of the COVID-19 has been estimated between 2 to 11 days ( to percentile) and the mean incubation period as 6.4 days (95% CI: 5.6–7.7) [6]. Thus, we assume . Least square method is employed to find the optimal parameters of and . The algorithm is shown as Algorithm 3.
| Algorithm 3 Parameter Estimation for SEIUR model |
| Input |
| Sequence ; |
| Sequence ; |
| Parameters: , ; |
| Numerical Solution (Algorithm 1) for at day , denoted as ; |
| Real Data [5] for at day , denoted as |
| The number of iterations or days: ; |
| Output |
| Optimal Parameters: , |
| Procedure |
| For in to |
| For in to |
| Find and corresponding indices and |
| Return and |
In the piecewise SEIUR model, we also have and . Parameter estimation of and for each period is shown in Algorithm 4.
| Algorithm 4 Parameter Estimation for piecewise SEIUR model |
| Input |
| The number of periods: ; |
| Sequence ; |
| Sequence ; |
| Parameters: , ; |
| Numerical Solution (Algorithm 2) for at day , denoted as ; |
| Real Data [5] for at day , denoted as ; |
| The number of iterations or days: ; |
| Note that: ; |
| where is the start point of period and ; |
| Output |
| Optimal Parameters: , for all in to |
| Procedure |
| For in to |
| For in to |
| For in to |
| (Mean Square Error) |
| Find and corresponding indices and |
| Return and |
4. Simulation results
4.1. Parameter estimation results
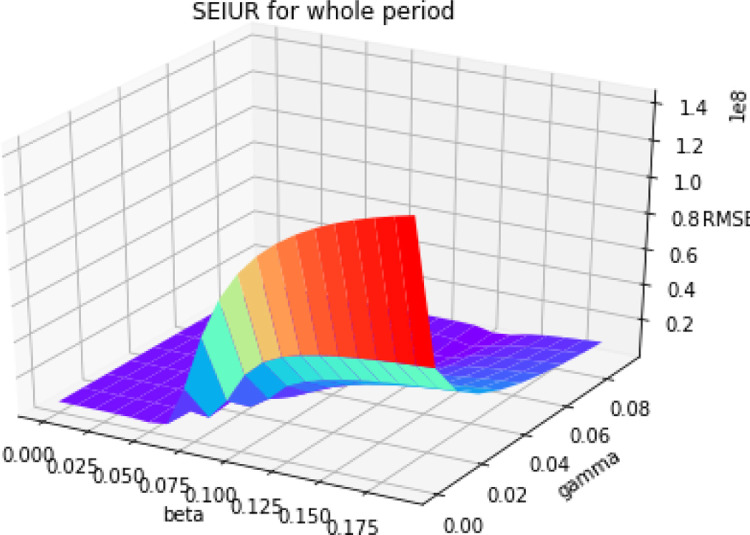
Based on the parameter estimation Algorithm 3, we estimate the parameters and for SEIUR model in this section. Fig. 5 is the 3D error graph for different values of and .
Fig. 5.
3D error graph with different and for SEIUR model.
We know that the optimal parameters can be derived from the lowest error point. For the SEIUR model, the optimal parameters are and
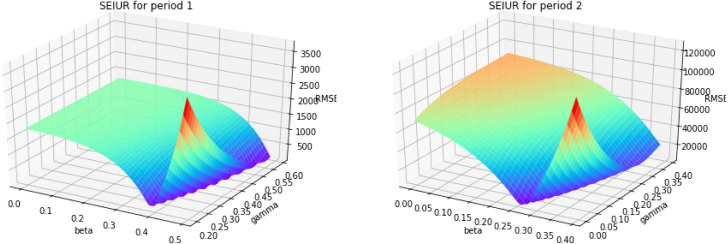
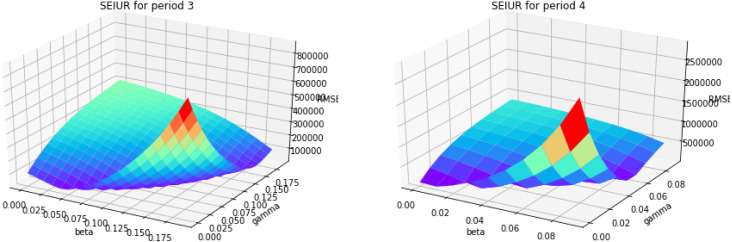
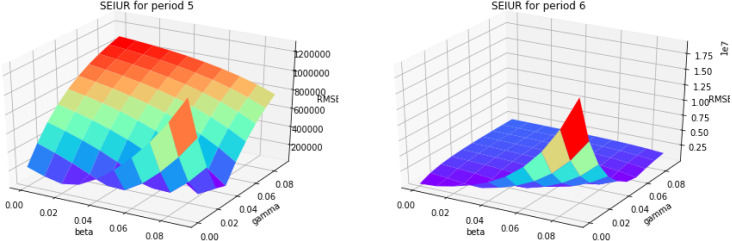
Based on the parameter estimation Algorithm 4, we estimate the parameters and for piecewise SEIUR model in each period . Fig. A.18, Fig. A.19, Fig. A.20, Fig. A.21 are the 3D error graphs showing the optimal values of parameters for each period.
Fig. A.18.
3D error graph for different and for periods 1 and 2.
Fig. A.19.
3D error graph for different and for periods 3 and 4.
Fig. A.20.
3D error graph for different and for periods 5 and 6.
Fig. A.21.
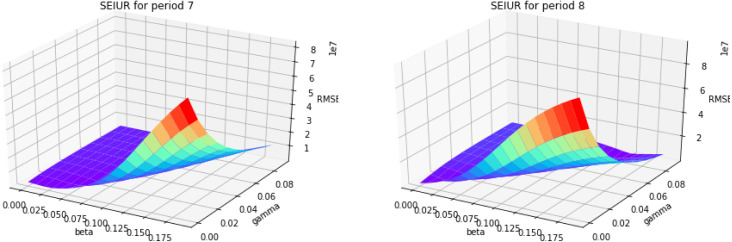
3D error graph for different and for periods 7 and 8.
Applying the same method from the SEIUR model to the piecewise SEIUR model, we can find the and related to the lowest error for each period . This allows us to generate the optimal parameters for all periods in Table 3.
Table 3.
Optimal model parameters by period.
| Period | ||
|---|---|---|
| 1 | 0.44 | 0.36 |
| 2 | 0.26 | 0.10 |
| 3 | 0.10 | 0.04 |
| 4 | 0.03 | 0.01 |
| 5 | 0.08 | 0.03 |
| 6 | 0.06 | 0.03 |
| 7 | 0.11 | 0.06 |
| 8 | 0.10 | 0.05 |
4.2. Performance
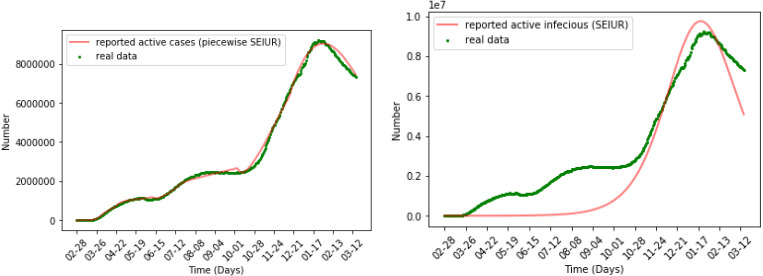
Fig. 6 shows the numerical simulation results of the SEIUR model on all data. We find that when simulating all of the data at once, the SEIUR model can successfully simulate the trend of data, but it cannot accurately reflect the true value of the data at each point. We have a more intuitive reflection in the error analysis in the next section.
Fig. 6.
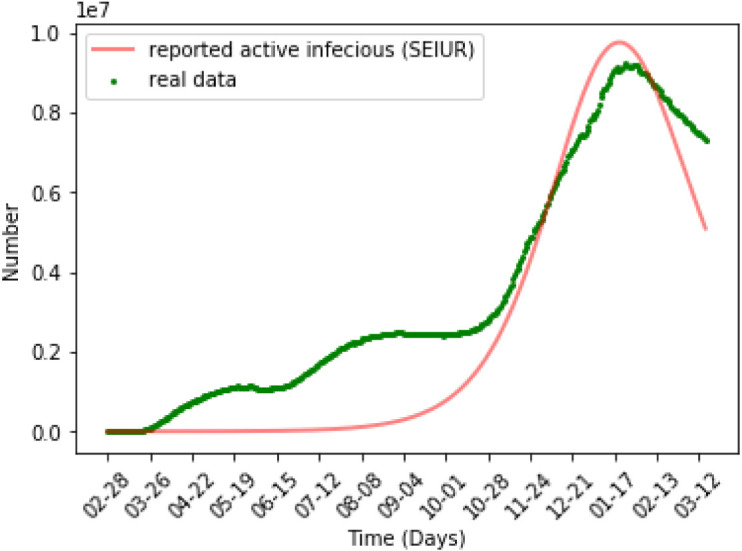
Numerical simulation for SEIUR.
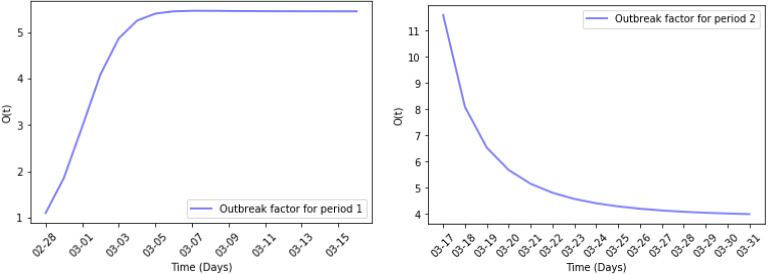
Fig. 7 shows the estimated reported active cases and the real data in periods 1 and 2 where the blue dots are training data and the green dots are test data. Fig. 8 shows the daily outbreak factor in periods 1 and 2. In period 1, no control measures were implemented which led to an exponential outbreak, and we calculate that in period 1. This is much greater than 1. This is consistent with what we proved in previous section: when is much greater than 1, the pandemic is in a major outbreak stage. Observing the simulation results on the test data, the SEIUR model in the first period can still simulate the data at the beginning of the second period very well. This also means that at the beginning of the second period the home quarantine strategy has not yet contributed to a significant impact, and the pandemic was still breaking out. The of the entire second period also confirmed this point. In this period, , which is slightly lower than that of the period 1 but still far greater than 1. This indicates that in the middle and late stages of period 2, the home quarantine strategy has achieved initial results, but the epidemic was still in outbreak. The test data of period 2 showed that at the beginning of the period 3, the true value of the test data is significantly smaller than the results obtained by SEIUR model generated from parameters estimated in period 2. This implies that the home quarantine strategy implemented in the period 2 achieved a significant effect in the period 3.
Fig. 7.
Numerical simulation in periods 1 and 2. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Fig. 8.
Outbreak factor in periods 1 and 2.
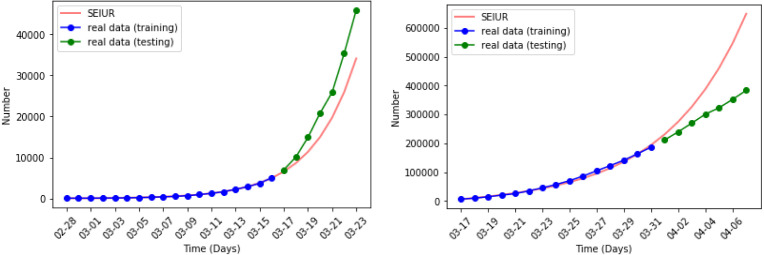
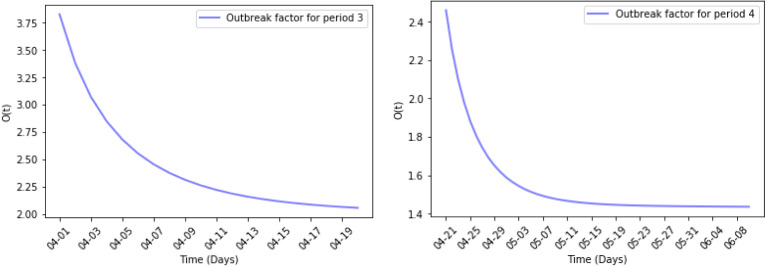
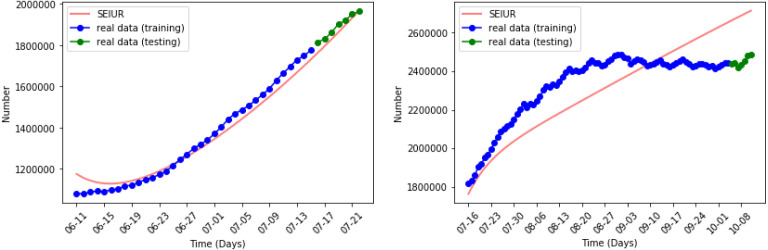
Fig. 9, Fig. 10 show the results of the numerical simulations and the outbreak factor in periods 3 and 4. in period 3, which has a 52% decrease compared to period 2. This confirms the analysis made in the discussion of period 2. The home quarantine strategy has slowed the spread of the epidemic. In period 4, mask wearing further reduced the spread of pandemic. The figure about period 4 illustrates the growth rate of the active cases has decreased compared to period 3 and reached the first turning point at the end of May. Quantitatively, in period 4, which is a decrease of 34% compared to period 3.
Fig. 9.
Numerical simulation in periods 3 and 4.
Fig. 10.
Outbreak factor in periods 3 and 4.
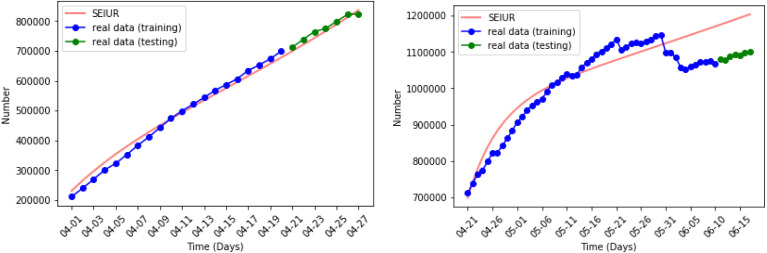
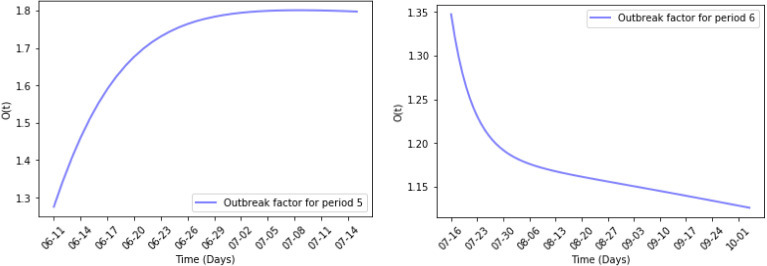
Fig. 11, Fig. 12 show the results of numerical simulations and outbreak factors in periods 5 and 6. In period 5, many states are relaxing their restrictions, with more than half set to be partially reopened. This led to the pandemic rebounding rapidly after reaching the first turning point and active cases starting to rise again. The average outbreak factor also rose from 1.5362 to 1.7176. In period 6, stricter mask-wearing rules slowed the spread of pandemic, and the second turning point was reach at the end of August with .
Fig. 11.
Numerical simulation in periods 5 and 6.
Fig. 12.
Outbreak factor in periods 5 and 6.
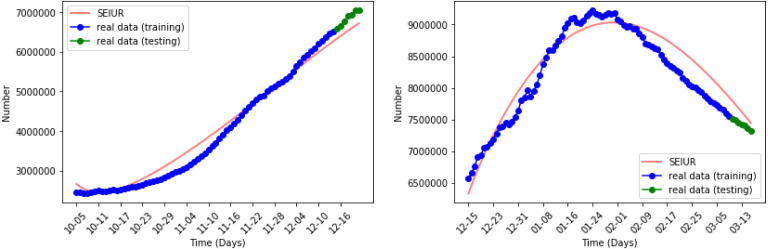
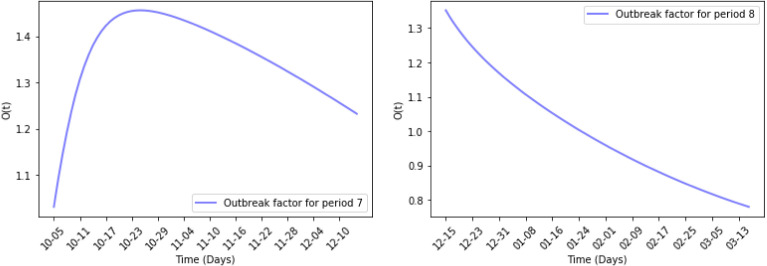
Fig. 13, Fig. 14 show the results of numerical simulations and outbreak factors in periods 7 and 8. Winter brings shorter days and lower temperatures to the United Stated which facilitate the spread of COVID-19. Period 7 shows this process. With the advent of winter, active cases began to increase again after reaching the second turning point, and rose back to 1.3414. Period 8 is the latest period so far, and vaccinations began on the first day of this period. The graph in period 8 shows that the vaccination quickly controlled the spread of the pandemic, and the third turning point appeared at the end of January 2021. So far, the reported active cases have continued to decline. In period 8, which is less than 1. If this continues, the epidemic is likely to be completely controlled in the next few months.
Fig. 13.
Numerical simulation in periods 7 and 8.
Fig. 14.
Outbreak factor in periods 7 and 8.
Fig. 15 shows the numerical simulation results of the piecewise SEIUR model and SEIUR model, where the result of piecewise SEIUR is obtained by combining the graphs of eight periods. It can be seen from the figure that the piecewise SEIUR model performs better than the SEIUR model. This also means that it is reasonable to divide a long periods of time into different sub-period according to policy dates, and then use the SEIUR model to simulate each period respectively.
Fig. 15.
Numerical simulation for piecewise SEIUR and SEIUR.
4.3. Error metrics
After performing the numerical simulation of the model, error analysis is an effective way to assess the accuracy of the model. There are many error metrics, such as mean absolute error (MAE), mean absolute percentage error (MAPE), symmetric mean absolute percentage error (SMAPE), mean squared error (MSE), root mean squared error (RMSE) and score. In this section we discuss several representative error metrics for error analysis.
The Mean Absolute Percentage Error (MAPE) [4] is one of the most commonly used to measure model accuracy. It is the average of the relative error. It is given by the following formula:
The Mean Absolute Error (MAE) [4] is a popular error metric to measure model accuracy. As the name implies, it is the average of the absolute error. The formula is shown below:
The Mean Squared Error (MSE) [4] is an error metric to measure model accuracy. Since MSE is a continuous function, it is often used together with the least square method and gradient descent method. It is defined as the average squared error. The formula is shown below:
The score [4] function computes the coefficient of determination, usually denoted as . It represents the proportion of variance (of ) that has been explained by the independent variables in the model. It provides an indication of goodness of fit and therefore a measure of how well unseen samples are likely to be predicted by the model through the proportion of explained variance.
If is the predicted value of the th sample and is the corresponding true value for total samples, the estimated is defined as:
From the results shown in Fig. 16, it can be seen that the piecewise SEIUR model has a small MAPE in each period and over the entire data. Specifically, the MAPE from period 3 to period 8 is lower than 5%, the MAPE in all periods is lower than 10%, and the MAPE in the entire period is only 4%. In contrast, the MAPE of SIEIUR model exceeds 50%.
Fig. 16.
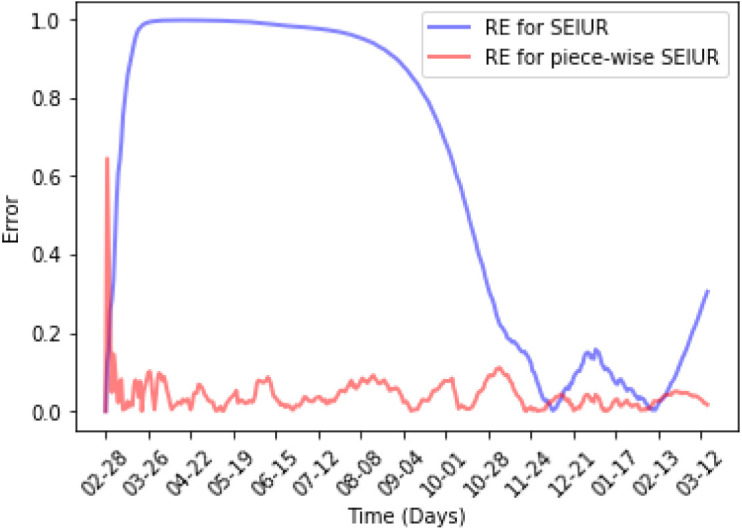
Relative error for Piecewise SEIUR and SEIUR.
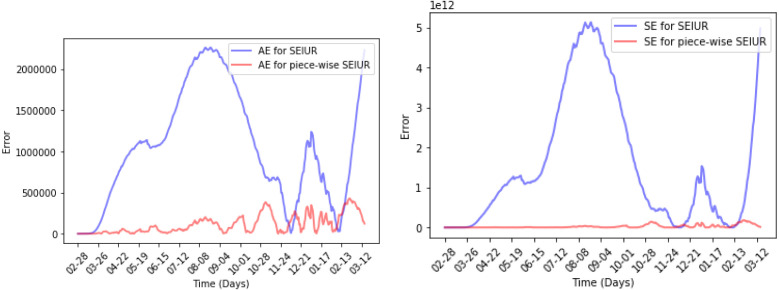
MAE and MSE are shown in Fig. 17 and produce the same results. No matter which error metric is used, the piecewise SEIUR model has a better performance than the SEIUR model. Quantitatively, the error of piecewise SEIUR model under the MAE metric is only one-tenth of that of the SEIUR, and under the MSE metric it is only one-sixth.
Fig. 17.
Absolute error (AE) and Square error (SE) for Piecewise SEIUR and SEIUR.
The results from score also illustrate that piecewise SEIUR model has an almost perfect fitting. The score of piecewise SEIUR model is approximately equal to 1 (see Table 4).
Table 4.
Error table for SEIUR and Piecewise SEIUR models.
| Error metrics |
||||
|---|---|---|---|---|
| Models name | MAPE | MAE | MSE | |
| Piecewise SEIUR model | 4.083% | 0.9972 | ||
| SEIUR model | 59.526% | 0.8239 | ||
5. Conclusion
In this paper, we have presented the SEIUR model and the piecewise SEIUR model. These two models are tested on COVID-19 data in the United States to demonstrate their performance. We estimated piecewise parameters and for each period of the model. These two models were applied to COVID-19 data in the United States to demonstrate their performance. The piecewise SEIUR model is seen to produce higher simulation accuracy than the SEIUR model. The MAPE of the piecewise SEIUR is only 4%. The error of piecewise SEIUR model under MAE and MSE metrics is far less than that of the SEIUR model. The score of the piecewise SEIUR model is 0.9972 which is close to 1. The outbreak factor generated by the piecewise SEIUR model can be applied to highlight the impact of the epidemic prevention strategies in each period. This also provides a mathematical tool for future research on the impact of different strategies on epidemics.
Appendix. Error graphs for the seiur model
References
- 1.Covid-19 Pandemic, Wikipedia, Wikimedia Foundation, https://en.wikipedia.org/wiki/COVID-19_pandemic.
- 2.Coronavirus Disease (Covid-19) - Events as They Happen, World Health Organization. World Health Organization, https://www.who.int/emergencies/diseases/novel-coronavirus-2019/events-as-they-happen.
- 3.Coronavirus Cases: Worldometer. https://www.worldometers.info/coronavirus/.
- 4.3.3. Metrics and Scoring: Quantifying the Quality of Predictions, Scikit.
- 5.United States COVID Cases, Worldometer.
- 6.Backer Jantien A., Klinkenberg Don, Wallinga Jacco. Incubation period of 2019 novel coronavirus (2019-nCoV) infections among travellers from wuhan, China, 20–28 2020. Eurosurveillance. 2020;25(5) doi: 10.2807/1560-7917.ES.2020.25.5.2000062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brauer Fred. Mathematical epidemiology: Past, present, and future. Infect. Dis. Model. 2017;2(2):113–127. doi: 10.1016/j.idm.2017.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Carcione José M., Santos Juan E., Bagaini Claudio, Ba Jing. A simulation of a COVID-19 epidemic based on a deterministic SEIR model. Front. Public Health. 2020;8:230. doi: 10.3389/fpubh.2020.00230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hamdy Youssef, Alghamdi Najat, Ezzat Magdy A., El-Bary Alaa A., Shawky Ahmed M. Study on the SEIQR model and applying the epidemiological rates of COVID-19 epidemic spread in Saudi Arabia. Infect. Dis. Model. 2021;6:678–692. doi: 10.1016/j.idm.2021.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hammer W.N. Epidemic disease in england-the evidence of variability and the persistence of type. Lancet. 1906;II:733–739. [Google Scholar]
- 11.Hannah Ritchie, Mathieu Edouard, Rodés-Guirao Lucas, Appel Cameron, Giattino Charlie, Ortiz-Ospina Esteban, Hasell Joe, Macdonald Bobbie, Beltekian Diana, Roser Max. 2020. Coronavirus (COVID-19) testing - statistics and research. Our world in data. https://ourworldindata.org/coronavirus-testing. [Google Scholar]
- 12.Ilyin Sergey O. A recursive model of the spread of COVID-19: Modelling study. JMIR Public Health Surveill. 2021;7(4) doi: 10.2196/21468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jie Long, Khaliq A.Q.M, Furati K.M. Identification and prediction of time-varying parameters of COVID-19 model: a data-driven deep learning approach. Int. J. Comput. Math. 2021;98:1617–1632. [Google Scholar]
- 14.Kermack ., Ogilvy William, McKendrick Anderson G. A contribution to the mathematical theory of epidemics. Proc. R. Soc. Lond. Ser. A. 1927;115(772):700–721. [Google Scholar]
- 15.Kermack ., Ogilvy William, McKendrick Anderson G. Contributions to the mathematical theory of epidemics, ii.—The problem of endemicity. Proc. R. Soc. Lond. Ser. A. 1932;138(834):55–83. [Google Scholar]
- 16.Mohan B.S, Nambiar V. Covid-19: an insight into SARS-CoV-2 pandemic originated at wuhan city in hubei province of China. J. Infect. Dis. Epidemiol. 2020;6(4):146. [Google Scholar]
- 17.Siettos Constantinos I., Russo Lucia. Mathematical modeling of infectious disease dynamics. Virulence. 2013;4(4):295–306. doi: 10.4161/viru.24041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.The New York Times . The New York Times. The New York Times; 2020. See Reopening Plans and Mask MandAtes for All 50 States. https://www.nytimes.com/interactive/2020/us/states-reopen-map-coronavirus.html. [Google Scholar]
- 19.Yang Junyuan, Chen Yuming, Xu Fei. Effect of infection age on an SIS epidemic model on complex networks. J. Math. Biol. 2016;73(5):1227–1249. doi: 10.1007/s00285-016-0991-7. [DOI] [PubMed] [Google Scholar]
- 20.Youssef Hamdy M., Alghamdi Najat A., Ezzat Magdy A., El-Bary Alaa A., Shawky Ahmed M. A new dynamical modeling SEIR with global analysis applied to the real data of spreading COVID-19 in Saudi Arabia. Math. Biosci. Eng. 2020;17:7018–7044. doi: 10.3934/mbe.2020362. [DOI] [PubMed] [Google Scholar]