Abstract
Purpose:
Congenital anomalies of the kidneys and urinary tract (CAKUT) constitute the leading cause of chronic kidney disease in children. In total, 174 monogenic causes of isolated or syndromic CAKUT are known. However, syndromic features may be overlooked when the initial clinical diagnosis of CAKUT is made. We hypothesized that the yield of a molecular genetic diagnosis by exome sequencing (ES) can be increased by applying reverse phenotyping, by re-examining the case for signs/symptoms of the suspected clinical syndrome that results from the genetic variant detected by ES.
Methods:
We conducted ES in an international cohort of 731 unrelated families with CAKUT. We evaluated ES data for variants in 174 genes, in which variants are known to cause isolated or syndromic CAKUT. In cases in which ES suggested a previously unreported syndromic phenotype, we conducted reverse phenotyping.
Results:
In 83 of 731 (11.4%) families, we detected a likely CAKUT-causing genetic variant consistent with an isolated or syndromic CAKUT phenotype. In 19 of these 83 families (22.9%), reverse phenotyping yielded syndromic clinical findings, thereby strengthening the genotype–phenotype correlation.
Conclusion:
We conclude that employing reverse phenotyping in the evaluation of syndromic CAKUT genes by ES provides an important tool to facilitate molecular genetic diagnostics in CAKUT.
Keywords: Congenital anomalies of the kidneys and urinary tract (CAKUT), Exome sequencing, Monogenic disease causation, Renal developmental gene
Introduction
Congenital anomalies of the kidneys and urinary tract (CAKUT) comprise a broad spectrum of birth defects that are caused by impaired embryonic development of the genitourinary tract.1–3 They represent the most frequent birth defect and the leading cause of chronic kidney disease (CKD) in children and young adults (49%) and frequently lead to end-stage renal disease.4,5 It has been hypothesized that CAKUT may occur because of germline variants in single genes that constitute a Mendelian disease on the basis of the following arguments: (1) familial occurrence; (2) presence of CAKUT as part of multiorgan syndromes, the fact that specific genes govern renal development; (3) existence of monogenic mouse models with CAKUT; and (4) the congenital nature of CAKUT.6–8 Owing to the rapid advances in high-throughput DNA sequencing within the past years, 174 genes have been identified to cause CAKUT in humans, if mutated.9 These genes can be categorized on the basis of their resulting phenotypes: 23 genes cause isolated CAKUT, ie, without extrarenal symptoms (Supplemental Table 1). These genes are closely linked to dysregulated renal and urinary tract morphogenesis, eg, ACE, AGT, AGTR1, and REN, which cause renal tubular dysgenesis10 or the genes that converge in the SLIT2-ROBO2 pathway.11 In contrast, 16 genes are known to cause syndromic CAKUT, here defined as the presence of extrarenal manifestations in ≥50% of cases with pathogenic variants in these 16 genes (Supplemental Table 2). Pathogenic variants in these genes cause an impairment of renal and urinary tract development, concomitantly disturbing development of other organ systems. Many of them, such as PAX2,12 are transcription factors that mediate developmental processes in a variety of organ systems. A number of these monogenic human CAKUT genes have been shown to interact in certain developmental pathways (reviewed in van der Ven et al8). In addition to these 23 forms of isolated CAKUT (ie, restricted to the kidneys and urinary tract; Supplemental Table 1) and the 16 forms of syndromic CAKUT in humans (Supplemental Table 2), 135 multiorgan syndromes caused by single-gene alterations in 135 distinct monogenic genes are known to facultatively exhibit CAKUT features in <50% of cases (Supplemental Table 3). In this latter group of genes causing syndromes with facultative CAKUT, the pathogenetic mechanisms are often not fully unraveled. However, these genes can still be considered candidate genes for CAKUT.
The broad interindividual phenotypic variability has been previously considered a general feature of CAKUT and has been explained to be predominantly due to both the complex underlying mechanisms in urinary tract development and variable expressivity, which is especially apparent within the high number of autosomal dominant CAKUT traits (reviewed in van der Ven et al8).
Finally, in some instances, CAKUT can be phenocopied by variants in other genes that mimic CAKUT phenotypes, such as small kidneys or hydronephrosis. This applies particularly to renal cystic ciliopathies13 (Supplemental Table 4).
Targeted gene panel sequencing, and later on, exome sequencing (ES) allowed the identification of pathogenic variants in these monogenic genes in cohorts of patients with CAKUT, leading to a detection rate of 6% to 20% in pediatric cohorts.6,14–22 In 104 pediatric renal transplant recipients, ES revealed a monogenic cause of CAKUT as causative of CKD in 11% of the total cohort and in 18% of the 55 patients with CAKUT.23 Furthermore, in several studies on adult patients with CKD, including those with CKD of unknown cause, a pathogenic variant in a gene causative of CAKUT and thus the diagnosis of CAKUT could be established using ES in 2% to 12%.24–28 The comparability of these studies to available pediatric studies is however limited because of very diverse inclusion criteria and an often unrecognizable congenital onset of the disease. A major caveat of molecular genetic diagnostics is the problem that mild extrarenal (syndromic) manifestations can easily be overlooked during the initial clinical evaluation.8 Reverse phenotyping is an approach in which specific clinical features are interrogated in a subsequent clinical examination once a likely molecular genetic diagnosis has been established.29 This method has been shown to detect causative genetic variants in genes that cause syndromic Mendelian diseases, including renal diseases such as pediatric-onset steroid-resistant nephrotic syndrome,30,31 renal cystic ciliopathies,32 and also CAKUT.15
Another major weakness of ES is the long list of potentially deleterious variants after initial filtering. This is a particular problem in CAKUT, in which 25 of the 39 primary CAKUT genes (Supplemental Tables 1 and 2) and 61 of the 135 syndromic genes with facultative CAKUT (Supplemental Table 3) are inherited in an autosomal dominant manner, necessitating to consider heterozygous variants, which are much more abundant in ES results than biallelic recessive variants. Because rare syndromic (extrarenal) phenotypes are often clinically very specific and scarce,15 they offer a strong independent route of confirmation because of (1) their rarity, (2) their specificity, and (3) the fact that they are initially missed, giving an opportunity for testing an unbiased (reverse) hypothesis, when contacting the clinician on the basis of the genetic finding resulting from ES evaluation.
We hypothesized that reverse phenotyping might increase the diagnostic yield of ES in families with CAKUT. Therefore, we examined an international cohort of 731 CAKUT-affected families using ES and systematically attempted reverse phenotyping in equivocal cases, ie, in patients in whom the a priori phenotype was not sufficiently explained by the variant detected by ES. We show here that reverse phenotyping facilitated identification of 19 of 83 (22.9%) genetic variants within the total number of 83 of 731 (11.4%) families with CAKUT in whom we detected the likely causative variant by ES.
Materials and Methods
Enrollment of families with CAKUT
This study was approved by the institutional review board of the University of Michigan and of Boston Children’s Hospital as well as the institutional review boards of institutions where families were recruited. In total, 731 unrelated families with at least 1 individual affected by CAKUT were enrolled between September 2005 and October 2019 after obtaining informed consent (822 affected individuals; a total of 1362 individuals, including unaffected relatives). The clinical diagnosis of CAKUT was made by the index patient’s pediatric nephrologist or urologist on the basis of clinical examination and medical imaging studies. CAKUT was defined as any abnormality of number, size, shape, or anatomical position of the kidneys or upper genitourinary tract (from ureteropelvic to ureterovesical junction) or lower genitourinary tract (from ureterovesical junction to external urethral orifice), including at least 1 of the following manifestations: renal agenesis, renal hypodysplasia, multicystic dysplastic kidney, hydronephrosis, ureteropelvic junction obstruction, hydroureter, vesicoureteral reflux, ectopic or horseshoe kidney, duplex collecting system, ureterovesical junction obstruction, neurogenic bladder, epispadias/hypospadias, posterior urethral valves, and cryptorchidism. Although isolated CAKUT in an individual was defined as CAKUT representing the only phenotypic organ manifestation reported, syndromic CAKUT was defined as the presence of at least 1 extrarenal (syndromic) feature in addition to CAKUT. A family was defined as affected by syndromic CAKUT if at least 1 CAKUT-affected individual presented with at least 1 extrarenal feature. Unaffected relatives were enrolled, where available, to enable determination of segregation of genetic variants within the family. Where feasible, the enrolling physician was requested to screen reportedly asymptomatic relatives by renal ultrasound imaging to rule out a nonovert presentation of CAKUT such as unrecognized unilateral renal agenesis.
ES and variant calling
Genomic DNA was subjected to ES at the Yale Center for Mendelian Genomics, followed by variant calling, homozygosity mapping, and family analysis. ES data was then examined for potentially deleterious variants in any of the 23 known isolated CAKUT genes (Supplemental Table 1) and the 16 known syndromic CAKUT genes (Supplemental Table 2), followed by an additional analysis for variants in the 135 genes, in which variants are known to cause syndromes with facultative CAKUT, if mutated (Supplemental Table 3), and in 46 genes known to cause, if mutated, renal cystic ciliopathies as frequent phenocopies of CAKUT (Supplemental Table 4). Assessment of deleteriousness of the remaining variants was performed using our previously published criteria6 and as further outlined in Supplemental Methods.
Reverse phenotyping
In patients in whom ES resulted in a likely CAKUT-causing variant of a gene that is known to cause syndromic CAKUT or a syndrome with facultative CAKUT, the enrolling physician was contacted and requested to conduct reverse phenotyping, ie, to query for additional and specific signs/symptoms by history taking, physical examination, or advanced diagnostic measures in the affected individual and their relatives, if applicable. The specific phenotypic details that the physician was prompted to search for were taken from OMIM and the primary literature reports (see Supplemental Tables 1–4 for references) on the gene in which a likely deleterious variant was detected. In case an in-person examination could not be performed, we resorted to telehealth consultations or reports from previous notes. After returning the result of reverse phenotyping, the genotype–phenotype correlation was reassessed and the final diagnostic verdict was made.
Results
Clinical characteristics of 731 families with CAKUT
We enrolled an international cohort of 822 individuals with CAKUT from 731 unrelated families and subjected their DNA samples to ES, including 540 reportedly unaffected relatives (1362 individuals in total). Clinical characteristics are listed in Table 1. We observed a predominance of male individuals with CAKUT (475/822; 58%). Extrarenal manifestations (syndromic CAKUT) were present in 153 of 822 (19%) enrollees with CAKUT (Table 1). Using homozygosity mapping, we detected significant homozygosity by descent (≥60 Mb)33 in 106 of 822 (13%) affected individuals (ie, 101/731 [14%] families). For 77 of 822 (9%) affected individuals, CKD was reported at enrollment or in the clinical course. The phenotypic spectrum of CAKUT was broad. In total, 113 of 822 (14%) affected individuals had unilateral renal agenesis, a severe form of CAKUT.8
Table 1.
Clinical characteristics of the 822 affected individuals from 731 families with CAKUT included in the study
| Characteristics | n (N = 822) | % |
|---|---|---|
|
| ||
| Sex | ||
| Female | 347 | 42 |
| Male | 475 | 58 |
| Extrarenal manifestations (syndromic CAKUT) | ||
| Yes | 153 | 19 |
| No | 669 | 81 |
| Homozygosity by descent | ||
| ≥60 Mb | 106 | 13 |
| <60 Mb | 716 | 87 |
| Reportedly CKD at enrollment or in the clinical course | ||
| Yes | 77 | 9 |
| No or not mentioned | 745 | 91 |
| CAKUT phenotype | ||
| Isolated distal CAKUT (eg, PUV) | 41 | 5 |
| Distala CAKUT with additional proximal phenotype | 87 | 11 |
| Proximalb unilateral CAKUT | 349 | 42 |
| Proximal bilateral concordant CAKUT | 213 | 26 |
| Proximal bilateral discordant CAKUT | 105 | 13 |
| CAKUT phenotype without detailed specification | 27 | 3 |
| Unilateral renal agenesis | 113 | 14c |
CAKUT, congenital anomalies of the kidney and urinary tract; CKD, chronic kidney disease; PUV, posterior urethral valve.
Distal CAKUT: from ureterovesical junction to external urethral orifice.
Proximal CAKUT: from kidney to ureterovesical junction.
Unilateral renal agenesis is listed here as a separate distinctive feature (unilateral renal agenesis vs no unilateral renal agenesis) because it is recognized as a severe form of CAKUT in respect to renal development.8 Because these 113 families may have exhibited additional CAKUT phenotypes, they are additionally listed in the CAKUT phenotype section of this table under Proximal unilateral CAKUT, Proximal bilateral concordant CAKUT, or Proximal bilateral discordant CAKUT.
Detection of likely deleterious variants in 85 of 588 (14.5%) families with isolated CAKUT
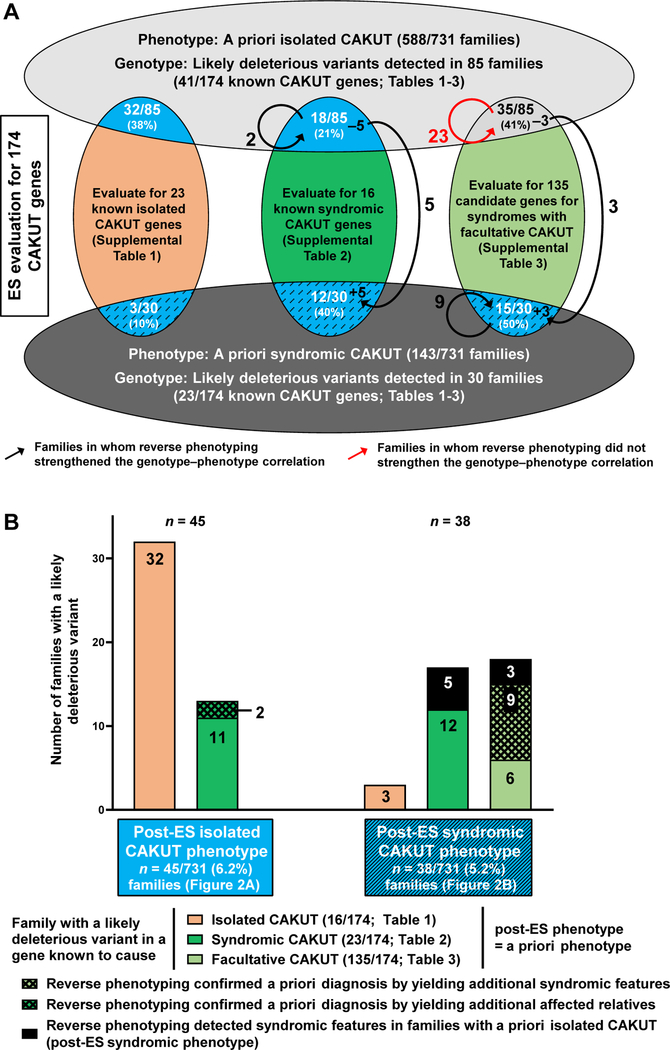
We first analyzed ES data from the 588 of 731 families with an a priori clinical phenotype of isolated CAKUT (Figure 1A, upper horizontal oval) for likely deleterious variants within the 23 genes known to cause isolated CAKUT (Supplemental Table 1), the 16 genes known to cause syndromic CAKUT (Supplemental Table 2), and the 135 candidate genes known to cause syndromes with facultative CAKUT (Supplemental Table 3). We identified a likely deleterious variant in 85 of 588 (14.5%) families with isolated CAKUT (Figure 1A, intersecting segments of upper horizontal oval and the 3 vertical ovals). These 85 variants were detected in 41 of the 174 genes in question. For the 588 families with isolated CAKUT (Figure 1A, upper horizontal oval), we hypothesized to identify likely deleterious variants predominantly in the 23 genes known to cause isolated CAKUT, if mutated (Supplemental Table 1), which was the case for 32 of 85 (38%) families (Figure 1A, intersecting segment of upper horizontal oval and red vertical oval). We also identified likely deleterious variants in the 16 genes known to cause syndromic CAKUT, if mutated (Supplemental Table 2), in 18 of 85 (21%) families with isolated CAKUT (Figure 1A, intersecting segment of upper horizontal oval and dark green vertical oval). In addition, we detected likely deleterious variants in the 135 candidate genes for syndromes with facultative CAKUT (Supplemental Table 3) in 35 of 85 (41%) families. Because variants in those genes have been previously reported to cause multiorgan syndromes with CAKUT involvement in <50% of cases (Supplemental Table 3), we hypothesized that reverse phenotyping may facilitate detection of initially overlooked extrarenal features and strengthen the genotype–phenotype correlation toward a sufficient diagnostic level.
Figure 1. Procedural flowsheet for detection of likely deleterious variants and effect of reverse phenotyping in 731 families with CAKUT.
A. Of 731 families, 588 with a priori diagnosis of isolated CAKUT (upper horizontal oval) and 143 families with a priori diagnosis of syndromic CAKUT (lower horizontal oval) were subjected to ES. ES data were interrogated for variants in the 23 genes known to cause isolated CAKUT (red oval; Supplemental Table 1), 16 genes known to cause syndromic CAKUT (dark green oval; Supplemental Table 2), and 135 candidate genes for syndromes with facultative CAKUT (light green oval; Supplemental Table 3), if mutated. Intersections of the ovals indicate the number of families in which a likely deleterious variant was detected. Black arrows depict the effect of reverse phenotyping on confirming the genotype–phenotype correlation, leading to, (1) reallocation of 8 (5 + 3) families from an a priori isolated CAKUT phenotype to a post-ES syndromic CAKUT phenotype (semicircular black arrows); (2) detection of additional and more specific features for the syndrome post-ES in families with an a priori syndromic CAKUT phenotype (lower circular arrow; 9 families), which was confirmatory of syndromic CAKUT; and (3) confirmation of isolated CAKUT in families with the a priori phenotype of isolated CAKUT by revealing additional evidence (upper circular black arrow; 2 families). Detailed information on the yield of reverse phenotyping in these families is available in Supplemental Table 5. The red arrow denotes 23 of 35 families with isolated CAKUT, which had a likely deleterious variant in 1 of the 135 candidate genes for a syndrome with facultative CAKUT (Supplemental Table 3), in which no extrarenal features could be detected by reverse phenotyping, rendering the variants unlikely to be the cause of CAKUT (details in Supplemental Table 7). B. Summary of molecular diagnosis of isolated vs syndromic CAKUT resulting from ES and reverse phenotyping from Figure 1A. The 45 of 731 (6.2%) families with a post-ES diagnosis of isolated CAKUT and detection of a likely monogenic cause of their disease (compared with Figure 2A) consist of 32 families with detection of a likely causative variant in a gene known to cause isolated CAKUT (left panel; orange bar) and 13 families with detection of a likely causative variant in a gene known to cause syndromic CAKUT (dark green bar), of which 2 have been confirmed by reverse phenotyping (cross-hatched segment of dark green bar). The 38 of 731 (5.2%) families with post-ES syndromic CAKUT and detection of a likely monogenic cause of their disease (compared with Figure 2B) consist of 3 families with detection of a likely causative variant in a gene known to cause isolated CAKUT (left panel; orange bar); 17 families with detection of a likely causative variant in a gene known to cause syndromic CAKUT (central bar), of which 5 have been reallocated from a priori isolated CAKUT by reverse phenotyping (black segment); and 18 families with detection of a likely causative variant in a candidate gene for a syndrome with facultative CAKUT (right bar), of which 3 have been reallocated from a priori isolated CAKUT by reverse phenotyping (black segment); 9 families revealed additional syndromic features (cross-hatched segment of light green bar). CAKUT, congenital anomalies of the kidneys and urinary tract; ES, exome sequencing.
Reverse phenotyping facilitates assessment of variant pathogenicity
Previous evidence indicated that in families who present with isolated CAKUT to the enrolling physician, (mild) extrarenal features may be overlooked and missed because of incomplete penetrance and broad phenotypic heterogeneity of syndromic CAKUT (see Introduction).
We therefore used reverse phenotyping as an independent and specific criterion for assessment of variant deleteriousness (see Material and Methods). In 10 of 85 CAKUT families (11.8%) with a priori isolated CAKUT, reverse phenotyping revealed additional rare clinical features after ES, which were not detected during the initial clinical examination at enrollment. We interpreted the reverse detection of the specific finding on the basis of the detected variants as confirmatory for the candidate of the detected variant for each of those genes (Figure 1A, black arrows; Supplemental Table 5). This led to reallocation of 8 (5 + 3) families with an a priori isolated CAKUT phenotype to a post-ES syndromic CAKUT phenotype (Figure 1A, black semicircular arrows). In 2 families, reverse phenotyping revealed additional affected relatives, who were later confirmed to share the variant with the index patient, thereby strengthening the genotype–phenotype correlation based on familial segregation (Figure 1A, black circular arrow).
As an example of reverse phenotyping that confirmed a specific molecular genetic diagnosis, the index individual from family B3947 was enrolled with the a priori phenotype of bilateral hydronephrosis, left-sided vesicoureteral reflux grade 5, and left hydroureter. By ES, we detected a likely deleterious heterozygous variant in PTPN11 (Supplemental Table 6), a gene in which variants cause Noonan syndrome (OMIM 163950), a multiorgan syndrome with facultative CAKUT (Supplemental Table 3). The variant we detected has been previously reported to be causative of Noonan syndrome.34 However, owing to the a priori phenotype, which was limited to isolated CAKUT, a specific genotype–phenotype correlation that would allow to make a final diagnosis was lacking (Figure 1A, upper right intersecting segment). We approached the enrolling physician to perform reverse phenotyping in his patient to detect the presence of features of Noonan syndrome. The following extrarenal features were detected: microcephaly, ptosis, myopia, widely spaced teeth, and pectus carinatum (Supplemental Table 5). Because these features, when occurring together, are fairly specific for and frequent in Noonan syndrome,34 we changed this individual’s post-ES phenotype from isolated CAKUT to syndromic CAKUT most likely because of the variant in PTPN11 (Figure 1A, right black semicircular arrow, which reallocates the family from isolated CAKUT to syndromic CAKUT).
Conversely, among the 35 families with a priori isolated CAKUT and a variant detected in a candidate gene for a syndrome with facultative CAKUT (Figure 1A, upper right intersecting segment), reverse phenotyping confirmed the absence of extrarenal features in 23 of 35 (65.7%) families after ES (Figure 1A, red semicircular arrow), rendering these variants even more unlikely to represent the cause of CAKUT in these families. In the remaining 9 families within this group, reverse phenotyping could not be accomplished because of loss of follow-up. We therefore rejected the assumed molecular genetic diagnosis suggested by ES in these 32 (23 + 9) families with isolated CAKUT listed in Supplemental Table 7 and in turn considered these families’ ES results as inconclusive (included in Figure 2D).
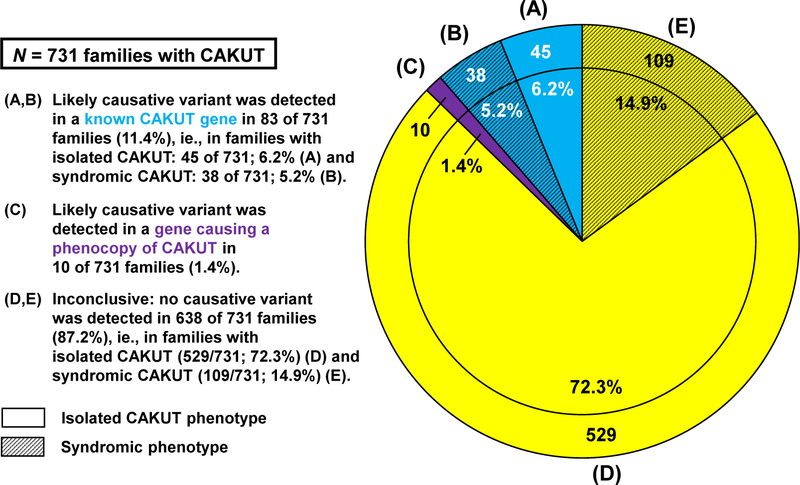
Figure 2. Results from exome sequencing evaluation of 731 families with CAKUT for likely causative variants in known CAKUT-causing genes and for phenocopies of CAKUT.
Exome sequencing (ES) data of 822 affected individuals with CAKUT from 731 unrelated families were analyzed for likely causative variants in the genes known to cause, if mutated, isolated CAKUT (23 genes, Supplemental Table 1), syndromic CAKUT (16 genes, Supplemental Table 2), and a syndrome with facultative CAKUT (135 genes, Supplemental Table 3) and for genes that represent a phenocopy of CAKUT (46 genes, Supplemental Table 4). The pie chart summarizes the findings for all 731 families, which is divided into the following subgroups. A. The clear blue segment denotes 45 of 731 (6.2%) families with isolated CAKUT, in which a likely causative variant in 1 of the genes known to cause CAKUT, if mutated, was detected. B. The hatched blue segment denotes 38 of 731 (5.2%) families with syndromic CAKUT, in which a likely causative variant in 1 of the genes known to cause CAKUT, if mutated, was detected. C. The purple segment denotes 10 of 731 (1.4%) families with CAKUT, in which a likely causative variant in a gene causing a phenocopy of CAKUT was detected. D. The clear yellow segment denotes 529 of 731 (72.3%) families with isolated CAKUT and inconclusive ES evaluation. E. The hatched yellow segment denotes 109 of 731 (14.9%) families with syndromic CAKUT and inconclusive ES evaluation. The outer ring segments denote the absolute number of families out of all 731 families with CAKUT; the inner circle segments show percentage of families from total (731 families = 100%). CAKUT, congenital anomalies of the kidneys and urinary tract.
Detection of variants likely causative of isolated CAKUT in 6.2% of families
As shown in Figure 1B (left panel), in 588 families with isolated CAKUT, we detected likely deleterious variants in a total of 45 families (6.2%), ie, variants in isolated CAKUT genes in 32 families and variants in syndromic CAKUT genes in 13 families with isolated CAKUT. Of 18 families with a priori isolated CAKUT, 5 had to be reallocated to a post-ES syndromic phenotype after reverse phenotyping (Figure 1A, semicircular arrows). These 45 of 731 (6.2%) families with isolated CAKUT, in whom we made a molecular genetic diagnosis, are depicted in Figure 2A.
Detection of likely deleterious variants in 30 of 143 (21.0%) families with syndromic CAKUT
Next, we analyzed ES data from 143 of 731 families with an a priori clinical phenotype of syndromic CAKUT (Figure 1A, lower horizontal oval) for likely deleterious variants within the 23 genes known to cause isolated CAKUT (Supplemental Table 1), the 16 genes known to cause syndromic CAKUT (Supplemental Table 2), and the 135 candidate genes for syndromes with facultative CAKUT (Supplemental Table 3). We identified a likely deleterious variant in 30 of 143 (21.0%) families (Figure 1A, intersecting segments of lower horizontal oval and the 3 vertical ovals). These 30 variants were detected in 23 of the 174 genes in question. On the basis of the a priori phenotype (syndromic CAKUT), we hypothesized that we would identify likely deleterious variants predominantly in the genes known to cause syndromic CAKUT (Supplemental Table 2) and the candidate genes for syndromes with facultative CAKUT (Supplemental Table 3), if mutated. From the total of 30 variants detected, 3 (10%) were detected in isolated CAKUT genes (Figure 1A, intersecting segment of lower horizontal oval and red vertical oval), 12 (40%) were detected in syndromic CAKUT genes (Figure 1A, intersecting segment of lower horizontal oval and dark green vertical oval), and 15 (50%) occurred in the candidate genes for syndromes with facultative CAKUT (Figure 1A, intersecting segment of lower horizontal oval and light green vertical oval).
On the basis of our previous observations in isolated CAKUT, we hypothesized that reverse phenotyping could reveal concealed (mild) syndromic features likewise in families with syndromic CAKUT. We performed reverse phenotyping and identified additional rare features in 9 of 15 families (60%), thereby confirming a syndromic genotype–phenotype correlation (Figure 1A, lower circular arrow; Supplemental Table 5).
Detection of variants likely causative of syndromic CAKUT in 5.2% of families
As shown in Figure 1A, in 143 families with syndromic CAKUT, we detected likely deleterious variants in a total of 30 families. In addition, those 8 (5 + 3) families with a priori isolated CAKUT, in whom we identified a syndromic phenotype after reverse phenotyping, had to be reallocated to the subset of syndromic families (Figure 1A, black semicircular arrows), thereby leading to the identification of a molecular genetic diagnosis in 38 of 731 (5.2%) families with CAKUT (Figure 2B and Figure 1B, right panel).
By ES and reverse phenotyping, a likely monogenic cause of CAKUT can be detected in 11.4% of families
In summary, by ES and reverse phenotyping, we identified the likely monogenic cause of CAKUT in 83 of 731 (11.4%) families (Figure 2A and 2B). In 19 of these 83 (22.9%) families, assignment of disease causation was directly facilitated by reverse phenotyping (Figure 1B, black and hatched subsegments; Supplemental Table 5). Detailed clinical data and information on variants are listed in Supplemental Table 6. Of these 83 variants, 12 (14%) were predicted to result in a truncated protein (ie, a premature stop of translation, frameshift, or obligatory splice site disruption), and 16 (19%) variants had previously been reported as disease-causing in the literature, including 8 variants that are categorized as pathogenic in ClinVar (Supplemental Table 6). Novel, ie, previously unreported, nontruncating heterozygous variants in dominant genes without proven de novo status are difficult to assess in terms of definite pathogenicity and would therefore be classified as variants of unknown significance in a clinical diagnostic setting. We therefore additionally labeled 56 families to whom this pertains (Supplemental Table 6). Among the 37 genes in which these deleterious variants were identified, GREB1L was the most frequent with likely deleterious variants in 10 families. Furthermore, in descending order, variants in the following genes were accountable for CAKUT in at least 5 families: EYA1, ROBO2, PAX2, SRGAP1, and SALL1 (Supplemental Table 6). We detected recurrent variants in EYA1 (families B1481 and B3542) and PAX2 (families B1677, B3358, and B3775).
Detection of likely phenocopies of CAKUT in 1.4% of families
Pathogenic variants in renal cystic ciliopathy genes have been previously shown to act as phenocopies of CAKUT.6 We thus interrogated families, in whom no likely deleterious variants in the a priori CAKUT genes (Supplemental Tables 1–3) were detected, for variants in 46 genes known to cause renal cystic ciliopathies (Supplemental Table 4). We detected likely causative variants in 10 of 731 (1.4%) families with CAKUT (Figure 2C). Strikingly, 7 of 10 (70%) variants were predicted to cause a truncation of the protein, and 7 of 10 (70%) variants had previously been reported as disease-causing in the literature (details on families and variants are outlined in Supplemental Table 8). In 5 of 10 (50%) families, reverse phenotyping strengthened the genotype–phenotype correlation by clarification of the CAKUT phenotype and detection of extrarenal features (Supplemental Table 5).
The zygosity of detected variants correlates with measured homozygosity by descent
To test for consistency of identified likely causative variants within pedigrees, we correlated the zygosity of variants with the relative amount of homozygosity by descent (Table 2). Concordant with the high number of queried genes that are inherited in an autosomal dominant fashion, we identified predominantly heterozygous variants (77/93; 83%) across the cohort. As expected, these heterozygous variants were enriched in families with low amounts of homozygosity by descent. In contrast, all 5 of 93 (5%) likely deleterious homozygous variants occurred in families with significant amounts of homozygosity by descent (≥ 60 Mb). Conversely, in 4 of the 5 of 93 (5%) families, in whom we identified compound heterozygous variants, homozygosity by descent was below our threshold of 60 Mb, as expected.33 Across the cohort, we detected 5 of 93 (5%) variants that were found to be de novo. We were able to determine segregation of the identified variant within at least 1 parent of the index individual in 53 of 93 (57%) families.
Table 2.
Distribution of likely CAKUT-causing variants detected by exome sequencing by their zygosity
| Verdict | No. of Families | Autosomal Recessive Homozygous | Autosomal Recessive Compound Heterozygous | Autosomal Dominant Heterozygous | Autosomal Dominant De Novo | X-Linked | Sum |
|---|---|---|---|---|---|---|---|
|
| |||||||
| Solved | 83/731a | 2(2/2)/93 | 4(1/4)/93 | 71(11/71)/93 | 5(2/5)/93 | 1(1/1)/93 | 83/93c |
| Phenocopy | 10/731b | 3(3/3)/93 | 1(0/1)/93 | 6(1/6)/93 | 0/93 | 0/93 | 10/93d |
| SUM | 93/731b | 5/93 | 5/93 | 77/93 | 5/93 | 1/93 | 93e |
The 93 likely deleterious variants that were detected by exome sequencing are broken down by their zygosity. Superscript numbers indicate the number of families (of total) with relevant homozygosity by descent (≥60 Mb). Detailed information about each variant is available in Supplemental Tables 6 and 8.
Total number of genes with variants detected was 37 of 174 (Supplemental Tables 1–3).
Total number of genes with variants detected was 8 of 46 (Supplemental Table 4).
Number of families with verdict “Solved” of total number of families.
Number of families with verdict “Phenocopy” of total number of families.
Total number of families in whom a likely deleterious variant was detected.
Discussion
We here performed ES and reverse phenotyping in a cohort of 731 families with CAKUT. We detected a likely monogenic cause of CAKUT in 11.4% of families (Figure 2). We found that reverse phenotyping facilitated assignment of likely causative variants by detection of rare specific syndromic extrarenal phenotypes a posteriori on the basis of the likely causative variant detected by ES in 22.9% of families (Figure 1). In addition, we showed that renal cystic ciliopathies cause phenocopies of CAKUT in 1.4% of families.
Enrollment of an international cohort of 731 families with CAKUT
To our knowledge, this study represents, so far, the largest and geographically most diverse CAKUT cohort studied using ES. This cohort is >3-fold larger than our previous study on monogenic causes of CAKUT6 (Table 1). Another recently published study by Rao et al19 showed a diagnostic rate of 17% in a pediatric cohort of 159 Chinese individuals with CAKUT. However, detailed clinical information on these individuals is not available, which complicates comparability with our study. In a similar study, Lei et al21 applied ES on 163 Chinese families with a prenatal diagnosis of CAKUT and identified causative variants in 12.3% of cases, whereof 8% of variants occurred in CAKUT genes, and 4.3% of variants occurred in genes causative of renal cystic ciliopathies. Adult cohorts previously studied using ES to a significant extent involved indistinct phenotypes, such as CKD of unknown cause, which to their nature harbor a higher likelihood of being caused by nonmonogenic conditions.25,26,35 In a study from our group, in which ES was applied to adults with CKD from 114 families, CAKUT was found to be the underlying disease in 12% and thus was the most frequent cause.24
Reverse phenotyping helps discern likely causative variants from inconclusive findings
Analysis of ES data yielded a high number of likely deleterious variants in genes previously reported to cause syndromes with facultative CAKUT in families with a priori isolated CAKUT (Figure 1A). These 135 distinct monogenic genes make up 78% of the total 174 genes we considered in our analysis, and 61 of the 135 monogenic syndromes are inherited in an autosomal dominant manner (Supplemental Table 3). Hence, we had to consider an abundance of heterozygous variants, which are called in ES evaluation much more frequently than biallelic variants, rendering the identification of false positive findings likely. Particularly, for many of these genes, the pathophysiological basis is not fully understood and is based on few cases with extensive multiorgan phenotypes (Supplemental Table 3).
To distinguish likely disease alleles from inconclusive findings, we conducted reverse phenotyping, finding that it may confirm likely causality of alleles detected by ES in 22.9% of the 83 families with likely deleterious variants (Figure 1A and B). In contrast, by confirming the absence of extrarenal features in 23 families with a priori isolated CAKUT, reverse phenotyping showed to be critical in rejecting potential disease-causing variants resulting from ES, categorizing them as inconclusive findings because of an insufficient genotype–phenotype correlation (Supplemental Table 7). We found that the fraction of individuals in whom we detected a likely causative variant was high in syndromic compared with isolated CAKUT cases (Figure 2). This observation is consistent with previous results from our group6,15 and others.21,22 Reverse phenotyping shows to be particularly helpful in increasing the diagnostic yield of syndromic CAKUT. Efficient usage of reverse phenotyping in evaluating genetic renal diseases has also been shown for steroid-resistant nephrotic syndrome30 and renal cystic ciliopathies.32
Renal cystic ciliopathies cause phenocopies of CAKUT in 1.4% of cases
By interrogating ES data of families without a likely deleterious variant in a gene causative of CAKUT for variants in genes known to cause renal cystic ciliopathies (Supplemental Table 4), we detected phenocopies of CAKUT in 1.4% of families (Figure 2), Renal cystic ciliopathies have previously been shown to resemble CAKUT and vice versa.6 21 23 24 Because many of these diseases, such as Bardet-Biedl syndrome or Joubert syndrome, exhibit extrarenal features,13 we hypothesized that reverse phenotyping could strengthen the genotype-phenotype correlation, which was the case in 5 families (Supplemental Table 5).
Limitations of ES
ES is limited to detecting only variants occurring in coding exons and within intron–exon boundaries. Thus, a major limitation of our study is that potential noncoding variants, such as cryptic splice sites, may have systematically been missed. There are hints for noncoding variants to cause CAKUT36; however, there is only little evidence to date. Previous data implicate that approximately 90% of variants with large effects occur within protein-coding exons.37 In addition, copy number variants have been shown to cause CAKUT in up to 16.6% of cases.38–40
In conclusion, we showed that a likely monogenic cause of CAKUT can be detected using ES and reverse phenotyping in 11.4% of families. We suggest that reverse phenotyping be systematically applied to families with CAKUT to increase the diagnostic yield of molecular genetic diagnostics.
Supplementary Material
Acknowledgments
We thank all participating families and physicians for their contribution. F.H. is the William E. Harmon Professor of Pediatrics at Harvard Medical School. This research was supported by grants from the National Institutes of Health to F.H. (DK076683). Sequencing and data processing were performed by the Yale Centers for Mendelian Genomics funded by the National Human Genome Research Institute (U54 HG006504). S.Se. is supported by the Deutsche For-schungsgemeinschaft (DFG, German Research Foundation; 442070894). B.Z. is supported by the program of China Scholarships Council (grant 201908320472). N.M. is supported by funding from the National Institutes of Health (grant T32-DK007726). D.M.C. is funded by Health Research Board, Ireland (HPF-206-674), the International Pediatric Research Foundation Early Investigators’ Exchange Program, and the Amgen Irish Nephrology Society Specialist Registrar Bursary. She is also funded by the Eugen Drewlo Chair for Kidney Research and Innovation at the Schulich School of Medicine & Dentistry at Western University, London, Ontario, Canada. C.-H.W.W. is supported by funding from the National Institutes of Health (grant T32-GM007748) and the American College of Medical Genetics and Genomics Foundation (ACMG/Takeda Next-Generation Biochemical Genetics Award). F.B. was supported by the Deutsche For-schungsgemeinschaft (DFG, German Research Foundation; 404527522). V.K. was supported by the Deutsche For-schungsgemeinschaft (DFG, German Research Foundation; 403877094). A.C.O.-W. is supported by the National Institutes of Health F32 Ruth L. Kirschstein Postdoctoral Individual National Research Service Award (DK122766). A.J.M. is supported by National Institutes of Health Institutional K12 Child Health Research Center Career Development Award (5K12HD052896-13), ASN Foundation for Kidney Research (FP01025169), and Boston Children’s Hospital Manton Center for Orphan Disease Research Junior Faculty Award. F.H. and S.Sh. are supported by grants from the Begg Family Foundation. This research was also supported by the Isabella Forrest Julian Research Fund for Pediatric Post Kidney Transplant Research.
Footnotes
Conflict of Interest
F.H. is a cofounder and Scientific Advisory Committee member of and holds stocks in Goldfinch-Bio. All other authors declare no conflicts of interest.
Ethics Declaration
This study was approved by the institutional review board of the University of Michigan and of Boston Children’s Hospital as well as institutional review boards of institutions where families were recruited. Before inclusion, informed consent of each individual or their legal guardians was obtained.
Web Resources
ClinVar. National Center for Biotechnology Information, U.S. National Library of Medicine. Accessed June 1, 2021. http://www.ncbi.nlm.nih.gov/clinvar.
Clustal omega. Accessed June 1, 2021. http://www.ebi.ac.uk/Tools/msa/clustalo.
dbSNP. National Center for Biotechnology Information, U.S. National Library of Medicine. Accessed June 1, 2021. http://www.ncbi.nlm.nih.gov/snp.
Ensembl genome browser. EMBL-EBI. Accessed June 1, 2021. http://www.ensembl.org.
Exome variant server. NHLBI Exome Sequencing Project. Accessed June 1, 2021. http://evs.gs.washington.edu/EVS.
gnomAD browser. Genome Aggregation Database. Accessed June 1, 2021. http://gnomad.broadinstitute.org.
HGMD Professional. Accessed June 1, 2021. http://www.hgmd.cf.ac.uk/ac.
Mouse genome informatics. The Jackson Laboratory. Accessed June 1, 2021. http://www.informatics.jax.org.
mutation t@sting. Mutation Taster. Accessed June 1, 2021. http://www.mutationtaster.org.
Online Mendelian Inheritance in Man (OMIM). Johns Hopkins University. Accessed June 1, 2021. http://www.omim.org.
PolyPhen2. Accessed June 1, 2021. http://genetics.bwh.harvard.edu/pph2
Sorting Intolerant From Tolerant (SIFT). Accessed June 1, 2021. https://sift.bii.a-star.edu.sg.
UCSC Genome Browser. University of California Santa Cruz Genomics Institute. Accessed June 1, 2021. http://genome.ucsc.edu/cgi-bin/hgGateway.
UniProt Consortium. Accessed June 1, 2021. http://www.uniprot.org.
VarSome. Accessed June 1, 2021. http://www.varsome.com.
Additional Information
The online version of this article (https://doi.org/10.1016/j.gim.2021.09.010) contains supplementary material, which is available to authorized users.
Data Availability
Depersonalized data that this study is based on are available from the corresponding author upon request.
References
- 1.Ichikawa I, Kuwayama F, Pope JC 4th, Stephens FD, Miyazaki Y. Paradigm shift from classic anatomic theories to contemporary cell biological views of CAKUT. Kidney Int. 2002;61(3):889–898. 10.1046/j.1523-1755.2002.00188.x. [DOI] [PubMed] [Google Scholar]
- 2.Costantini F. Genetic controls and cellular behaviors in branching morphogenesis of the renal collecting system. Wiley Interdiscip Rev Dev Biol. 2012;1(5):693–713. 10.1002/wdev.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vivante A, Hildebrandt F. Genetics of congenital anomalies of the kidneys and urinary tract. In: Barakat A, Rushton H, eds. Congenital Anomalies of the Kidney and Urinary Tract: Clinical Implications in Children. Springer Nature; 2016:303–322. [Google Scholar]
- 4.Becherucci F, Roperto RM, Materassi M, Romagnani P. Chronic kidney disease in children. Clin Kidney J. 2016;9(4):583–591. 10.1093/ckj/sfw047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bonthuis M, Cuperus L, Chesnaye NC, et al. Results in the ESPN/ERA-EDTA Registry suggest disparities in access to kidney transplantation but little variation in graft survival of children across Europe. Kidney Int. 2020;98(2):464–475. 10.1016/j.kint.2020.03.029. [DOI] [PubMed] [Google Scholar]
- 6.van der Ven AT, Connaughton DM, Ityel H, et al. Whole-exome sequencing identifies causative mutations in families with congenital anomalies of the kidney and urinary tract. J Am Soc Nephrol. 2018;29(9):2348–2361. 10.1681/ASN.2017121265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vivante A, Kohl S, Hwang DY, Dworschak GC, Hildebrandt F. Single-gene causes of congenital anomalies of the kidney and urinary tract (CAKUT) in humans. Pediatr Nephrol. 2014;29(4):695–704. 10.1007/s00467-013-2684-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.van der Ven AT, Vivante A, Hildebrandt F. Novel insights into the pathogenesis of monogenic congenital anomalies of the kidney and urinary tract. J Am Soc Nephrol. 2018;29(1):36–50. 10.1681/ASN.2017050561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Connaughton DM, Hildebrandt F. Personalized medicine in chronic kidney disease by detection of monogenic mutations. Nephrol Dial Transplant. 2020;35(3):390–397. 10.1093/ndt/gfz028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gribouval O, Gonzales M, Neuhaus T, et al. Mutations in genes in the renin-angiotensin system are associated with autosomal recessive renal tubular dysgenesis. Nat Genet. 2005;37(9):964–968. 10.1038/ng1623. [DOI] [PubMed] [Google Scholar]
- 11.Hwang DY, Kohl S, Fan X, et al. Mutations of the SLIT2-ROBO2 pathway genes SLIT2 and SRGAP1 confer risk for congenital anomalies of the kidney and urinary tract. Hum Genet. 2015;134(8):905–916. 10.1007/s00439-015-1570-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sanyanusin P, Schimmenti LA, McNoe LA, et al. Mutation of the PAX2 gene in a family with optic nerve colobomas, renal anomalies and vesicoureteral reflux. Nat Genet. 1995;9(4):358–364. [DOI] [PubMed] [Google Scholar]; Published correction appears in Nat Genet. 1996;13(1):129. 10.1038/ng0495-358 [DOI] [PubMed] [Google Scholar]
- 13.Hildebrandt F, Benzing T, Katsanis N. Ciliopathies. N Engl J Med. 2011;364(16):1533–1543. 10.1056/NEJMra1010172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hwang DY, Dworschak GC, Kohl S, et al. Mutations in 12 known dominant disease-causing genes clarify many congenital anomalies of the kidney and urinary tract. Kidney Int. 2014;85(6):1429–1433. 10.1038/ki.2013.508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vivante A, Hwang DY, Kohl S, et al. Exome sequencing discerns syndromes in patients from consanguineous families with congenital anomalies of the kidneys and urinary tract. J Am Soc Nephrol. 2017;28(1):69–75. 10.1681/ASN.2015080962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bekheirnia MR, Bekheirnia N, Bainbridge MN, et al. Whole-exome sequencing in the molecular diagnosis of individuals with congenital anomalies of the kidney and urinary tract and identification of a new causative gene. Genet Med. 2017;19(4):412–420. 10.1038/gim.2016.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Weber S, Moriniere V, Knüppel T, et al. Prevalence of mutations in renal developmental genes in children with renal hypodysplasia: results of the ESCAPE study. J Am Soc Nephrol. 2006;17(10):2864–2870. 10.1681/ASN.2006030277. [DOI] [PubMed] [Google Scholar]
- 18.Rasmussen M, Sunde L, Nielsen ML, et al. Targeted gene sequencing and whole-exome sequencing in autopsied fetuses with prenatally diagnosed kidney anomalies. Clin Genet. 2018;93(4):860–869. 10.1ll1/cge.13185. [DOI] [PubMed] [Google Scholar]
- 19.Rao J, Liu X, Mao J, et al. Genetic spectrum of renal disease for 1001 Chinese children based on a multicenter registration system. Clin Genet. 2019;96(5):402–J10. 10.llll/cge.13606. [DOI] [PubMed] [Google Scholar]
- 20.Zhou X, Wang Y, Shao B, et al. Molecular diagnostic in fetuses with isolated congenital anomalies of the kidney and urinary tract by whole-exome sequencing. J Clin Lab Anal. 2020;34(ll):e23480. 10.1002/jcla.23480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lei TY, Fu F, Li R, et al. Whole-exome sequencing in the evaluation of fetal congenital anomalies of the kidney and urinary tract detected by ultrasonography. Prenat Diagn. 2020;40(10):1290–1299. 10.1002/pd.5737. [DOI] [PubMed] [Google Scholar]
- 22.Heidet L, Morinière V, Henry C, et al. Targeted exome sequencing identifies PBX1 as involved in monogenic congenital anomalies of the kidney and urinary tract. J Am Soc Nephrol. 2017;28(10):2901–2914. 10.1681/ASN.2017010043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mann N, Braun DA, Amann K, et al. Whole-exome sequencing enables a precision medicine approach for kidney transplant recipients. J Am Soc Nephrol. 2019;30(2):201–215. 10.1681/ASN.2018060575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Connaughton DM, Kennedy C, Shril S, et al. Monogenic causes of chronic kidney disease in adults. Kidney Int. 2019;95(4):914–928. 10.1016/j.kint.2018.10.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lata S, Marasa M, Li Y, et al. Whole-exome sequencing in adults with chronic kidney disease: a pilot study. Ann Intern Med. 2018; 168(2): 100–109. [DOI] [PMC free article] [PubMed] [Google Scholar]; Published correction appears in Ann Intern Med. 2018;168(4):308. 10.7326/M17-1319 [DOI] [Google Scholar]
- 26.Groopman EE, Marasa M, Cameron-Christie S, et al. Diagnostic utility of exome sequencing for kidney disease. N Engl J Med. 2019;380(2): 142–151. 10.1056/NEJMoal806891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mallett AJ, McCarthy HJ, Ho G, et al. Massively parallel sequencing and targeted exomes in familial kidney disease can diagnose underlying genetic disorders. Kidney Int. 2017;92(6):1493–1506. 10.1016/j.kint.2017.06.013. [DOI] [PubMed] [Google Scholar]
- 28.Mansilla MA, Sompallae RR, Nishimura CJ, et al. Targeted broadbased genetic testing by next-generation sequencing informs diagnosis and facilitates management in patients with kidney diseases. Nephrol Dial Transplant. 2021;36(2):295–305. 10.1093/ndt/gfzl73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schulze TG, McMahon FJ. Defining the phenotype in human genetic studies: forward genetics and reverse phenotyping. Hum Hered. 2004;58(3–4): 131–138. 10.1159/000083539. [DOI] [PubMed] [Google Scholar]
- 30.Landini S, Mazzinghi B, Becherucci F, et al. Reverse phenotyping after whole-exome sequencing in steroid-resistant nephrotic syndrome. Clin J Am Soc Nephrol. 2020;15(1):89–100. 10.2215/CJN.06060519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Becherucci F, Landini S, Cirillo L, Mazzinghi B, Romagnani P. Look alike, sound alike: phenocopies in steroid-resistant nephrotic syndrome. Int J Environ Res Public Health. 2020;17(22):8363. 10.3390/ijerph17228363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.de Goede C, Yue WW, Yan G, et al. Role of reverse phenotyping in interpretation of next generation sequencing data and a review of INPP5E related disorders. Eur J Paediatr Neurol. 2016;20(2):286–295. 10.1016/j.ejpn.2015.ll.012. [DOI] [PubMed] [Google Scholar]
- 33.Hildebrandt F, Heeringa SF, Rüschendorf F, et al. A systematic approach to mapping recessive disease genes in individuals from outbred populations. PLoS Genet. 2009;5(l):el000353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sarkozy A, Conti E, Seripa D, et al. Correlation between PTPN11 gene mutations and congenital heart defects in Noonan and LEOPARD syndromes. J Med Genet. 2003;40(9):704–708. 10.1136/jmg.40.9.704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Snoek R, van Jaarsveld RH, Nguyen TQ, et al. Genetics-first approach improves diagnostics of ESKD patients younger than 50 years. Nephrol Dial Transplant. Published online December 11, 2020. 10.1093/ndt/gfaa363. [DOI] [PubMed] [Google Scholar]
- 36.Dong S, Wang C, Li X, et al. Noncoding rare variants of TBX6 in congenital anomalies of the kidney and urinary tract. Mol Genet Genomics. 2019;294(2):493–500. 10.1007/s00438-018-1522-6. [DOI] [PubMed] [Google Scholar]
- 37.Lifton RP. Individual genomes on the horizon. N Engl J Med. 2010;362(13):1235–1236. 10.1056/NEJMe1001090. [DOI] [PubMed] [Google Scholar]
- 38.Sanna-Cherchi S, Kiryluk K, Burgess KE, et al. Copy-number disorders are a common cause of congenital kidney malformations. Am J Hum Genet. 2012;91(6):987–997. 10.1016/j.ajhg.2012.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Verbitsky M, Westland R, Perez A, et al. The copy number variation landscape of congenital anomalies of the kidney and urinary tract. Nat Genet. 2019;51(1):117–127. [DOI] [PMC free article] [PubMed] [Google Scholar]; Published correction appears in Nat Genet. 2019;51(4):764. 10.1038/s41588-018-0281-y [DOI] [Google Scholar]
- 40.Verbitsky M, Krithivasan P, Batourina E, et al. Copy number variant analysis and genome-wide association study identify loci with large effect for vesicoureteral reflux. J Am Soc Nephrol. 2021;32(4):805–820. 10.1681/ASN.2020050681. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Depersonalized data that this study is based on are available from the corresponding author upon request.