Supplemental Digital Content is available in the text.
Keywords: artificial intelligence, big data, electronic health records, inflammatory bowel disease, natural language processing
Background
The impact of relapses on disease burden in Crohn’s disease (CD) warrants searching for predictive factors to anticipate relapses. This requires analysis of large datasets, including elusive free-text annotations from electronic health records. This study aims to describe clinical characteristics and treatment with biologics of CD patients and generate a data-driven predictive model for relapse using natural language processing (NLP) and machine learning (ML).
Methods
We performed a multicenter, retrospective study using a previously validated corpus of CD patient data from eight hospitals of the Spanish National Healthcare Network from 1 January 2014 to 31 December 2018 using NLP. Predictive models were created with ML algorithms, namely, logistic regression, decision trees, and random forests.
Results
CD phenotype, analyzed in 5938 CD patients, was predominantly inflammatory, and tobacco smoking appeared as a risk factor, confirming previous clinical studies. We also documented treatments, treatment switches, and time to discontinuation in biologics-treated CD patients. We found correlations between CD and patient family history of gastrointestinal neoplasms. Our predictive model ranked 25 000 variables for their potential as risk factors for CD relapse. Of highest relative importance were past relapses and patients’ age, as well as leukocyte, hemoglobin, and fibrinogen levels.
Conclusion
Through NLP, we identified variables such as smoking as a risk factor and described treatment patterns with biologics in CD patients. CD relapse prediction highlighted the importance of patients’ age and some biochemistry values, though it proved highly challenging and merits the assessment of risk factors for relapse in a clinical setting.
Introduction
Crohn’s disease (CD) is a chronic inflammatory disease of the gastrointestinal tract characterized by alternating periods of remission and relapse [1–3]. Patients with CD experience a variety of symptoms that may include localized abdominal pain, chronic diarrhea, weight loss, fatigue, anxiety, and depression [2–6]. Due to complications associated with progressive bowel damage, individuals with CD can require abdominal resections associated with ileostomy or colostomy, and further surgery for perianal complications, including strictures, fistulas, and abscesses [3,7,8]. Thus, the burden for patients with CD is physical, emotional, and economic [8–12], posing a major challenge for healthcare systems worldwide [9,13].
The etiology of CD is only partially known and complex in nature. Existing evidence indicates that the pathological inflammation of the intestinal tissue in CD is mediated by an aberrant mucosal immune response to enteric bacterial flora [1–3,14]. This response is most likely caused by the interaction between genetic susceptibility and environmental factors (e.g., smoking, drugs such as NSAIDs or antibiotics, and urban environment) [2,3,15,16].
Despite recent efforts toward a better understanding of the pathophysiology and diagnosis of CD, the therapeutic options available for these patients are still far from optimal [17]. The traditional approach to treatment in CD was based on a ‘step-wise’ paradigm [1]; patients with CD are often treated with pharmacological agents such as corticosteroids, immunomodulators, and biologics aimed at treating inflammation and related complications while achieving or maintaining remission [2]. In addition, a key aspect of patient management involves the early identification of upcoming relapses to avoid cumulative tissue damage. Because the clinical situation observed at a given timepoint does not anticipate future disease activity [18], predictive models for CD relapses and other complications must take into account large amounts of heterogeneous data, including but not limited to gut microbiota dynamics [19], blood-based and molecular biomarkers [20,21], and standard laboratory results [22,23].
From a clinical standpoint, complex and relatively low-prevalence diseases, such as CD, are best understood using large, population-based registries with available follow-up information [24,25]; a prominent data source with these characteristics is patients’ electronic health records (EHRs). EHRs are growingly available and contain heterogeneous information resulting from medical examinations, diagnosis, prescriptions, and procedures, as well as laboratory testing [26]. Crucially, most of the information in EHRs is unstructured [27,28], including imaging results or the valuable free-text clinical notes written down by physicians and other health professionals in their routine practice. Recent advances in the realms of natural language processing (NLP) and machine learning (ML) are now enabling access to the free-text, unstructured information in EHRs and have yielded valuable contributions in specific clinical populations [29,30], epidemiology [31], and healthcare resource use [32]. However, the application of these cutting-edge technologies is only beginning to be explored in CD [33–35]. Though several studies use ML models to identify risk factors related to CD or NLP to better define cases, our study is the first to combine NLP and ML for a predictive model of relapse, to the best of our knowledge.
In light of the above discussion, here, we used NLP and ML techniques to access and analyze the free-text clinical information contained in the EHRs of a large series of patients with CD in selected hospitals within the Spanish National Healthcare System. Our main objectives were to (1) describe clinical characteristics and current medical management with biologics of patients with CD and (2) generate a data-driven predictive model for the occurrence of relapses.
Methods
Data source
This study was conducted within the scope of the PREMONITION-CD project, sponsored by Takeda and was formally approved by the Madrid Institutional Review Board in May 2018. It was registered in ClinicalTrials.gov with the identifier number NCT03668249. This was a multicenter, retrospective study using data from the EHRs of eight participating tertiary hospitals within the Spanish National Healthcare Network (Fig. 1). Data from the EHRs were collected using NLP for the period between 1 January 2014 and 31 December 2018 (except for one participating site with electronic data available from 2013 to 2017) and were obtained from all available departments (including inpatient hospital, outpatient hospital, and emergency room) for virtually all types of services provided in each participating hospital. The study database was fully anonymized and contained no personal information from patients.
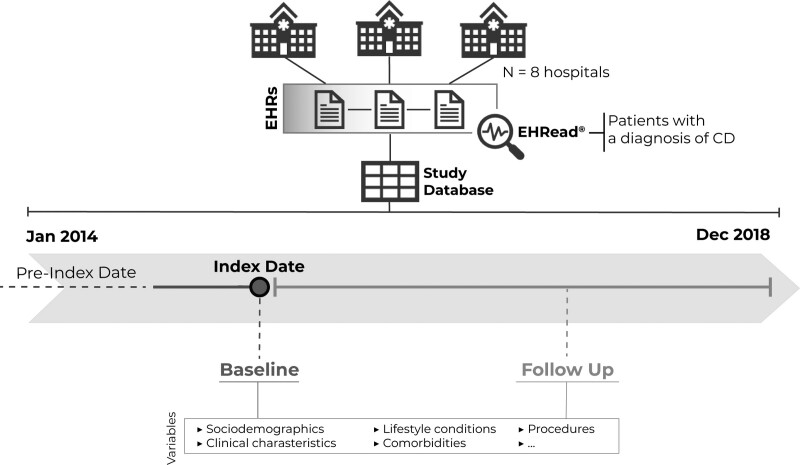
Fig. 1.
Study design and timeline. For each patient in the database, the Index Date (i.e., Baseline) was defined as the timepoint when diagnostic criteria for CD is first identified. All available EHRs before January 2014 were handled to extract information regarding the clinical history of patients (dotted line). The follow-up period ranged from the index date to the end of the study period or the last data point available. Data from patients’ EHRs were extracted and organized with the EHRead technology. See the Methods section for further details. CD, Crohn’s disease; EHR, electronic health record.
Study design
As shown in Fig. 1, data are presented in relation to two main time frames, namely Index Date and Follow Up. Index Date (also referred to as Baseline) was defined as the timepoint when diagnostic criteria for CD were first identified; this timepoint could occur before study onset. Analyses were performed for the time window comprising the 5-year post-baseline - this time window coincides with the longest follow-up period (Fig. 1). Additional information about the patient’s medical history was also collected before baseline (i.e., pre-index date; Fig. 1).
Participants
The study sample included pediatric and adult patients in the source population with a documented diagnosis of CD. Patients were identified based on clinical diagnosis as included in the unstructured, free-text information in the EHRs.
EHRead
To extract and analyze the unstructured, free-text information in patients’ EHRs, we used Savana’s EHRead technology [36–40]. Based on NLP, ML, and deep learning methodologies, this technology enables the extraction of information from all types of EHRs and the subsequent normalization of the extracted clinical entities to a unique terminology. Savana’s custom body of terminology was originally built from SNOMED CT and includes more than 400 000 medical concepts, acronyms, and laboratory parameters amassed over the course of 5 years of free-text mining. The terminology entities detected in the patients’ records are later classified based on sections contained in the EHRs (e.g., demographics, medical history, medications, diagnoses, etc.), hospital service where the data were originally captured, and other clinical specifications.
The extent to which EHRead correctly identified records that mention CD and predefined associated variables (i.e., CD-related relapse and ‘vedolizumab’) was calculated according to previously published procedures [41,42]. Briefly, given the lack of clinically coded data in Spanish, our evaluation required the development of an annotated corpus, otherwise known as the ‘gold standard’. The gold standard consists of a series of documents marked up by expert physicians with any metadata tag related to CD and CD-related variables. This corpus is compared against EHRead’s output, and performance is calculated in terms of the standard metrics of accuracy (P), recall (R), and their harmonic mean F-score. For the evaluated clinical terms, all F-scores were above 0.80, thus indicating that CD and associated variables were accurately identified in patients’ EHRs.
Data analyses
All categorical variables (e.g., medication use, surgical procedures) are shown in frequency tables, whereas continuous variables (e.g., age) are described using summary tables that include the mean, SD, median, minimum, maximum, and quartiles for each variable. Given the descriptive nature of the present study, no tests were performed to assess possible statistically significant differences in the distribution of categorical or numerical variables.
Predictive model
All patients included in the descriptive study were also included in the predictive one, with a random 70% assigned to training and 30% to validation. The results reported correspond to the performance of the fitted models on this validation set. The models aimed at identifying the clinical factors that prelude future relapses in patients with CD were built as follows.
Variables
In all cases, the dependent variables considered were the occurrence/absence of relapses (i.e., binary variable). Because relapses are not always registered as such in EHRs, we also inferred relapses from hospitalizations that coincided with drug administration associated with CD relapse. Tens of thousands of independent variables were used to train the predictive model. These variables include all available information captured in EHRs (e.g., family records, medical history, surgeries, treatments, hospitalizations, etc.), with special focus on previously used variables in CD studies (e.g., substance use, phenotype, age at diagnosis, and additional information about CD-related complications).
Temporality
The models aimed at predicting the occurrence of relapses in the near future; we considered a 3-month time window for all models.
Machine learning
Predictive models were generated using three different algorithms, namely logistic regression, decision trees, and random forests.
-
(1)
Logistic regression. In this model, the logarithm addressing the odds for the occurrence of a relapse/complication is a linear combination of the independent variables. In this case, the weights of the regression reflect the relative importance of each independent variable in predicting the desired outcome. We applied this model to our data to assess the importance of the variables when considered independently.
-
(2)
Decision trees. This algorithm was used to classify patients according to whether they will experience a relapse/complication or not, based on their individual characteristics and medical history. Classification trees can show interactions between variables and enable the identification of any important subgroups that could determine disease prognosis.
-
(3)
Random forests. Unlike classification trees, which apply the learning algorithm to the full dataset, random forests focus on a random sample of the data. Instead of attempting to fit the data into one single large ‘tree’, they fit a very large number of trees on random samples of the data (hence the name random forest). Random forests efficiently deal with issues such as collinearity of variables, can uncover relationships or patterns buried in large datasets, and also provide intuitive measures regarding the relative importance of the variables in predicting disease outcomes.
Model performance
To evaluate the predictive power of the models, we provide standard measures of performance, including the following:
-
(1)
The confusion matrix shows, for each row, the number of positives (i.e., presence of relapse) and negatives (i.e., absence of relapse) in our database. In this matrix, columns represent the positives and negatives that were predicted by the model. The elements in the main diagonal show the elements that were correctly predicted, while the off-diagonal terms show the false negatives (element 2,1) and false positives (1,2).
-
(2)
Precision: Proportion of true positives over positives obtained by the model, calculated as the fraction of patients that our model correctly identified to have an upcoming relapse over the total number of patients that the model predicted to have a relapse or complication.
-
(3)
Recall: Fraction of patients that the model correctly identified to have a relapse over the total number of patients that actually experienced a relapse.
-
(4)
Accuracy: Proportion of correct predictions calculated as the number of true positives and true negatives divided by the total amount of predictions.
-
(5)
F-score: Used to account for a possible imbalance between precision and recall; it is the harmonic mean of these two measures.
Ethical considerations
The present study was classified as a ‘post-authorization study’ (EPA-OD) by the Spanish Agency of Medicines and Health Products and was approved by the regional institutional review board. This study was conducted in compliance with legal and regulatory requirements and followed generally accepted research practices described in the ICH Guideline for Good Clinical Practice, the Helsinki declaration in its latest edition, Good Pharmacoepidemiology Practices, and applicable local regulations. Because we collected data retrospectively and handled all clinical information in an aggregate, anonymized, and irreversibly dissociated manner, regulations regarding informed patient consent do not apply to the present study.
Results
EHRs from 2 242 730 patients were processed from eight participating hospitals. We identified a total of 5938 patients with CD; 43.4% (n = 2575) of patients had already been diagnosed at the onset of the study (i.e., before the index date), whereas 56.6% of patients (n = 3363) were newly diagnosed within the study period. Patients’ demographics are shown in Table 1. Notably, nearly 40% (n = 1364) of patients were smokers, and 29% (n = 1010) were former smokers (Table 1). However, information about tobacco use was only collected in 58% of the analyzed EHRs at baseline, and only 15% of the documents contained information about alcohol use.
Table 1.
Demographics, clinical characteristics, and medication use
| N = 5938 | |
|---|---|
| Demographics | |
| Sex, n (%) | |
| Female | 3034 (51.1) |
| Male | 2904 (48.9) |
| Age (years)a | |
| N | 5934 |
| Mean (SD) | 48.3 (18.3) |
| Median | 46.3 |
| (Q1-Q3) | (35.0-61.0) |
| Adults (≥18 years old), n (%) | 5782 (97.37) |
| Children (<18 years old), n (%) | 152 (2.56) |
| Missing | 4 |
| Substance useb | |
| Tobacco use, n (%) | 3465 |
| Ex | 1010 (29.15) |
| No | 1091 (31.5) |
| Yes | 1364 (39.36) |
| Missing | 2473 |
| Alcohol use, n (%) | 893 |
| Ex | 81 (9.1) |
| No | 324 (36.3) |
| Yes | 492 (55.1) |
| Missing | 5045 |
| Disease characteristicsb,c | |
| Location, n (%) | |
| L1 ileal | 913 (54.0) |
| L2 colonic | 199 (11.8) |
| L3 ileocolonic | 507 (30.0) |
| L4 isolated upper diseased | 73 (4.3) |
| Missing | 4246 |
| Behavior, n (%) | |
| B1 nonstricturing, nonpenetrating: inflammatory | 589 (48.8) |
| B2 stricturing | 356 (29.5) |
| B3 penetrating | 262 (21.7) |
| Missing | 4731 |
Age at registered Crohn’s disease diagnosis.
Subjects with missing values are not included in percentage calculations.
Based on the Montreal Classification.
L4 is a modifier that can be added to L1–L3 when concomitant upper gastrointestinal disease is present.
Using the Montreal Classification [43] as a reference, we analyzed the location and behavior of CD at baseline (Table 1). Among patients with available information (approximately 30% of total patients), the most common location was ileal (L1), in 54% (n/n = 913/1692) of patients. Regarding disease behavior, nearly half of patients (48.8%; n/n = 589/1207) suffered from inflammatory (nonstricturing, nonpenetrating) CD (B1).
Regarding family history, our analyses revealed that this variable was poorly registered in the EHRs, as it was only available for 34% of the patients (n = 2042). The most common conditions (up to 37.1%; n = 757) were related to neoplasms (including cysts and polyps), of which the most frequent were malignant tumor of the large intestine, neoplasm of the large intestine, carcinoma of the stomach, and malignant tumor of the breast. Gastrointestinal and hepatobiliary disorders were also highly prevalent (27.3%; n = 557), where inflammatory disorder of the digestive tract and disorder of the lower gastrointestinal tract were predominant (Table 2).
Table 2.
Family history
| N (%)a | |
|---|---|
| Crohn’s disease | 401 (6.8) |
| Other medical conditions and diseases | |
| Blood and lymphatic system disorders | 87 (1.5) |
| Cardiovascular disorders | 421 (7.1) |
| Disorder of cardiovascular system | 151 (2.5) |
| Myocardial infarction | 76 (1.3) |
| Structural disorder of heart | 67 (1.1) |
| Cerebrovascular disease | 45 (0.8) |
| Others | 189 (3.2) |
| Congenital, familial, and genetic disorders | 31 (0.5) |
| Ear and labyrinth disorders | 12 (0.2) |
| Endocrine disorders | 339 (5.7) |
| Diabetes mellitus | 148 (2.5) |
| Disorder of endocrine system | 108 (1.8) |
| Disorder of thyroid gland | 63 (1.1) |
| Others | 79 (1.3) |
| Eye disorders | 69 (1.2) |
| Gastrointestinal and hepatobiliary disorders | 557 (9.4) |
| Inflammatory disorder of digestive tract | 136 (2.3) |
| Disorder of lower gastrointestinal tract | 97 (1.6) |
| Colitis | 75 (1.3) |
| Viral hepatitis | 54 (0.9) |
| Disorder of rectum | 34 (0.6) |
| Others | 293 (4.9) |
| General disorders | 22 (0.4) |
| Immune system disorders | 88 (1.5) |
| Infections and infestations | 82 (1.4) |
| Injury, poisoning, and procedural complications | 13 (0.2) |
| Metabolism and nutrition disorders | 130 (2.2) |
| Disorder of lipoprotein and/or lipid metabolism | 35 (0.6) |
| Others | 103 (1.7) |
| Musculoskeletal and connective tissue disorders | 183 (3.1) |
| Neoplasms, benign (incl. cysts and polyps) | 109 (1.8) |
| Neoplasms, malignant (incl. cysts and polyps) | 457 (7.7) |
| Malignant tumor of breast | 190 (3.2) |
| Malignant neoplasm of intraabdominal organ | 47 (0.8) |
| Primary malignant neoplasm of colon | 33 (0.6) |
| Neoplastic disease | 32 (0.5) |
| Others | 250 (4.2) |
| Neoplasms, unspecified (incl. cysts and polyps) | 757 (12.7) |
| Malignant tumor of large intestine | 251 (4.2) |
| Neoplasm of large intestine | 137 (2.3) |
| Carcinoma of stomach | 107 (1.8) |
| Neoplasm of lung | 96 (1.6) |
| Neoplasm of breast | 68 (1.1) |
| Neoplasm of prostate | 42 (0.7) |
| Neoplasm of ovary | 36 (0.6) |
| Others | 286 (4.8) |
| Nervous system disorders | 196 (3.3) |
| Cerebral degeneration presenting primarily with dementia | 41 (0.7) |
| Others | 158 (2.7) |
| Pregnancy, puerperium, and perinatal conditions | 37 (0.6) |
| Psychiatric disorders | 72 (1.2) |
| Renal and urinary disorders | 56 (0.9) |
| Reproductive system and breast disorders | 50 (0.8) |
| Respiratory, thoracic, and mediastinal disorders | 171 (2.9) |
| Bronchial hyperreactivity/hyperresponsiveness | 33 (0.6) |
| Others | 146 (2.5) |
| Skin and subcutaneous tissue disorders | 162 (2.7) |
| Acquired disorder of keratinization | 52 (0.9) |
| Others | 116 (2) |
| Social circumstances | 5 (0.1) |
| Surgical and medical procedures | 13 (0.2) |
Percentage based on the total number of patients. All medical terms were obtained from the standardized SNOMED CT glossary.
Management and treatment of Crohn’s disease
Table 3 summarizes the number of patients assigned to different procedures and surgical interventions aimed at treating CD during the follow-up period. Colonoscopy and esophagogastroduodenoscopy were the most frequent types of endoscopy procedures in 28.7% (n = 1705) and 11.3% (n = 673) of patients, respectively. A total of 43.1% of patients (n = 2558) underwent imaging procedures, including diagnostic radiography of abdomen (31.7%; n = 1883), computed tomography of abdomen (18.0%; n = 1068), and ultrasonography of abdomen (16.4%; n = 975). Surgical interventions were documented in 38.1% of patients (n = 2260). The most common surgeries during follow up were intestinal structure excision, colectomy, and gastrointestinal and digestive anastomosis.
Table 3.
Procedures and surgical interventions during follow up
| N (%)a | |
|---|---|
| Endoscopy | 2161 (36.4) |
| Colonoscopy | 1705 (28.7) |
| Esophagogastroduodenoscopy | 673 (11.3) |
| Rectoscopy | 112 (1.9) |
| Rectosigmoidoscopy | 72 (1.2) |
| Missing | 3777 |
| Imaging | 2558 (43.1) |
| Diagnostic radiography of abdomen | 1883 (31.7) |
| CT of abdomen | 1068 (18.0) |
| Ultrasonography of abdomen | 975 (16.4) |
| CT of abdomen and pelvis | 744 (12.5) |
| MRI of abdomen | 682 (11.5) |
| Magnetic resonance enterography | 598 (10.1) |
| MRI of abdomen and pelvis | 24 (0.4) |
| Barium enema | 18 (0.3) |
| Missing | 3380 |
| Surgical interventions | 2260 (38.1) |
| Excision | 1550 (26.1) |
| Excision of intestinal structure | 933 (15.7) |
| Colectomy | 585 (9.9) |
| Gastrointestinal and digestive anastomosis | 561 (9.4) |
| Perianal region operations | 122 (2.1) |
| Colostomy | 138 (2.3) |
| Ileostomy operation | 183 (3.1) |
| Proctocolectomy | 53 (0.9) |
| Anal fistulectomy | 32 (0.5) |
| Small intestinal strictureplasty | 9 (0.2) |
| Missing | 3678 |
CT, computed tomography.
Percentage based on the total number of patients.
Regarding pharmacological management of CD, this study focused on the use of biologics. These analyses were performed on the subpopulation of biologics-treated patients in our database (n = 443). Specifically, we selected patients undergoing treatment with a biologic during the last year of the study period (i.e., 2018) and analyzed their therapeutical history with biologics since baseline. This methodology allowed us to obtain high-quality data in terms of quantity, homogeneity, and recency. In addition, we anticipated that the probability of receiving a prescription for any biologic would increase with time since diagnosis. Figure S2, Supplemental digital content 1, http://links.lww.com/EJGH/A730 and Table 4 display the sequencing patterns and flow of treatment with biologics across treatment lines. About half of the patients (46.7%; n/n = 207/443) in the first line (1L) were treated with adalimumab, followed by infliximab (43.3%; n/n = 192/443). Similarly, the percentage of patients treated with adalimumab in the second line (2L) was maintained (45.8%; n/n = 65/142), but the proportion of patients treated with infliximab decreased to 23.2% (n/n = 33/142). Most of the patients who switched treatment from adalimumab in the 1L changed to infliximab, and vice versa. Finally, time to treatment discontinuation was also evaluated in this subpopulation of patients treated with biologics (Figure S1, Supplemental digital content 1, http://links.lww.com/EJGH/A730).
Table 4.
Biologics treatment by treatment line during the last year of the follow-up period
| Line | Treatment | N = 443; N (%)a |
|---|---|---|
| 1L (N = 443) | Adalimumab | 207 (46.7) |
| Infliximab | 192 (43.3) | |
| Vedolizumab | 26 (5.9) | |
| Ustekinumab | 14 (3.2) | |
| Certolizumab pegol | 3 (0.7) | |
| Natalizumab | 1 (0.2) | |
| 2L (N = 142) | Adalimumab | 65 (45.8) |
| Infliximab | 33 (23.2) | |
| Ustekinumab | 26 (18.3) | |
| Vedolizumab | 14 (9.8) | |
| Certolizumab pegol | 4 (2.8) | |
| NAb | 301 | |
| 3L (N = 54) | Ustekinumab | 21 (38.8) |
| Vedolizumab | 20 (37) | |
| Infliximab | 5 (9.3) | |
| Adalimumab | 4 (7.4) | |
| Certolizumab pegol | 4 (7.4) | |
| NAb | 389 | |
| 4L (N = 22) | Ustekinumab | 11 (50) |
| Vedolizumab | 8 (36.4) | |
| Infliximab | 2 (9.1) | |
| Adalimumab | 1 (4.5) | |
| NAb | 421 | |
| 5L (N = 4) | Vedolizumab | 2 (50) |
| Ustekinumab | 1 (25) | |
| Certolizumab pegol | 1 (25) | |
| NAb | 439 |
Percentage based on the total number of biologics-treated patients during the selected window.
NA = not available, representing either patients who (1) continued treatment with the same biologic until the end of the study period, (2) discontinued the treatment and did not receive any other biologic, or (3) information regarding biologic treatment was not available in the electronic health records.
Predicting relapses in Crohn’s disease
As described above, one of the objectives of the study was to forecast whether a patient would suffer a relapse in a 3-month period. The percentage of patients with documented relapses after baseline until the end of the study period was 23.5% (n = 1393). The predictive models were adjusted to the data of 5938 patients, 19% of which suffered at least one relapse in the following 3 months from baseline. The number of variables that were included was over 25 000; the categories of variables included in the predictive models are shown in Table S1, Supplemental digital content 1, http://links.lww.com/EJGH/A730. The results of the predictive models are presented in Table 5. For all models tested, the false-positive and false-negative rates were approximately 50%, and accuracy was roughly 80%. As can be seen, the performance of the different ML models (logistic regression, decision trees, or random forests) was very similar. The relative importance for the variables in the random forest model can be found in Table 6, representing the 25 variables that were ranked as most important by the algorithm (a full list in Table S2, Supplemental digital content 1, http://links.lww.com/EJGH/A730). Those with the highest relative importance are past relapses and patients’ age, as well as leukocyte, hemoglobin, and fibrinogen levels. The clinical significance of the obtained metrics and selected variables is discussed below.
Table 5.
Predictive model for relapse risks at different timepoints
| Accuracy | Confusion matrixa | Precision | Recall | F-score | AUC | ||
|---|---|---|---|---|---|---|---|
| Decision tree, 3 months | 0.81 | 2191 | 281 | 0.50 | 0.50 | 0.50 | 0.84 |
| 288 | 291 | ||||||
| Logistic regression, 3 months | 0.82 | 2213 | 259 | 0.50 | 0.52 | 0.51 | 0.85 |
| 287 | 292 | ||||||
| Random forest, 3 months | 0.84 | 2295 | 177 | 0.46 | 0.60 | 0.52 | 0.88 |
| 309 | 270 | ||||||
| Random forest, 6 months | 0.84 | 2184 | 189 | 0.56 | 0.67 | 0.61 | 0.89 |
| 301 | 377 | ||||||
| Random forest, 1 year | 0.83 | 2089 | 186 | 0.59 | 0.71 | 0.65 | 0.90 |
| 318 | 458 | ||||||
| Random forest, 2 years | 0.83 | 1984 | 230 | 0.67 | 0.71 | 0.69 | 0.91 |
| 275 | 562 | ||||||
AUC, area under the curve.
In a confusion matrix, the rows reflect the number of positives (i.e., presence of relapse) and negatives (i.e., absence of relapse), whereas columns indicate the positives and negatives that were predicted by the model. The elements in the main diagonal show the elements that were correctly predicted; off-diagonal terms show the false negatives (element 2,1) and false positives (1,2). See Methods section for further details regarding the calculation of each performance metric. See Table S2, Supplemental digital content 1, http://links.lww.com/EJGH/A730 for the relative importance of the most relevant variables included in the predictive model.
Table 6.
Relative importance of the 25 most relevant variables included in the predictive model for CD-related relapse
| Variable | Relative importance |
|---|---|
| Cumulative past flare | 0.358901 |
| Age | 0.029441 |
| Difference between event value and basal value leukocytes | 0.023361 |
| Difference between event value and basal value hemoglobin | 0.010256 |
| Increment respect to maximum normal value fibrinogen | 0.009158 |
| Disposition events - Past admissions | 0.008597 |
| Montreal Scale | 0.008082 |
| Proton pump inhibitors - A02BC | 0.007368 |
| Substance use findings (habits) - TOBACCO USE | 0.006733 |
| Acetic acid derivatives and related substances - M01AB | 0.006705 |
| Belladonna and derivatives in combination with analgesics - A03DB | 0.006624 |
| Evaluation procedures - CT of chest and abdomen | 0.005912 |
| methylprednisolone - H02AB04 | 0.005684 |
| ciprofloxacin - J01MA02 | 0.005551 |
| prednisone - H02AB07 | 0.005274 |
| Evaluation procedures - Source-specific culture | 0.004973 |
| Diagnostic procedures - Laboratory procedure | 0.004925 |
| Medical history - Infection due to Enterobacteriaceae | 0.004657 |
| Difference between event value and basal value CRP - 48 | 0.004641 |
| Sex | 0.004507 |
| mesalazine -A07EC02 | 0.004463 |
| Increment respect to maximum normal value leukocytes - 9 | 0.004463 |
| Evaluation procedures - Imaging of abdomen | 0.004428 |
| Increment respect to maximum normal value CRP - 48 | 0.004324 |
| Evaluation procedures - Blood gas measurement | 0.004311 |
CD, Crohn’s disease; CRP, C-reactive protein.
Discussion
Using NLP and ML to extract and analyze the clinical information captured in CD patients’ EHRs, our goals were to (1) describe clinical characteristics and current medical management with biologics of patients with CD and (2) generate a data-driven predictive model for the occurrence of relapses. Our results justify the use of this technology and data source to further explore the clinical features of CD, yet reflect some important caveats to overcome in future research.
Though numerous studies use big data analytics to assess coded or structured EHRs from CD patients, few instances of NLP-driven approaches from free-text EHRs or machine learning-driven predictive models are found for CD [44]. Recently, NLP was used in a large cohort of CD patients to assess biologic use and surgery rates [33]. Several previous studies have used NLP to improve the identification and case definition of patients with inflammatory bowel disease, separating them efficiently as ulcerative colitis or CD [35,45]. ML has also produced predictive models for risk factors regarding CD-related surgery [46] or azathioprine nonadherence in Chinese CD patients [47]. However, to the best of our knowledge, our study is the first to assess the predictability of relapse occurrence in CD.
Our study included nearly 6000 patients; a sample size substantially larger than most studies in CD to date and in a dataset not described previously, namely, Spanish patients [48,49]. Notably, the clinical characteristics of patients with CD reported here were in line with results using traditional methods [48,49], validating the use of our technology. Here, disease behavior (phenotype) was defined mostly as an inflammatory disease (i.e., nonstricturing and nonpenetrating disease) [48]. Regarding risk factors for CD, cigarette smoking is associated with CD onset, and postdiagnosis smoking leads to worse disease prognosis, as previously described [48,50,51]. In the present study, we found that only 30% of patients with CD were either nonsmokers or former smokers, though tobacco use information was missing for 40% of patients.
Although CD diagnosis has been associated with family medical history, only about 15% of patients with CD have family members with the disease [52,53]. In our sample, of the patients with information about family history, about 7% of patients had a documented family history of CD. Interestingly, we found a high incidence of gastrointestinal tumors as well as other gastrointestinal disorders in patients’ family history, suggesting that they may represent a risk factor for developing CD.
Treatment with biologics represents an important advancement in the management of patients with CD [54]. Here, we were able to document the treatment, treatment switches, and time to discontinuation in biologics-treated CD patients during a controlled, short period of time, though future research should aim at identifying the relationship between effective disease management across treatment lines.
As stated above, surgery is fairly common among patients with CD [48]. Previous studies reported that the probability of surgical resection after 1, 5, and 10 years from diagnosis is 9-15%, 25-30%, and 34-52%, respectively [48]. Nearly 70% of patients in our study were newly diagnosed within the 5-year study period. The remaining 30% had a disease evolution of more than 5 years (i.e., they were diagnosed before study onset). Thus, our results fall within the expected ranges, as around 38% of patients experienced a surgical procedure from baseline to the end of the study.
One of the main goals of this study was to identify the demographic and clinical variables that predict the occurrence of CD-related relapses in a time window of 3 months. Although critical to patient management [55], identifying risk factors that prelude CD-related complications or impact disease onset and evolution is still challenging [51]. Though we analyzed thousands of variables with our big data approach, the data captured in EHRs during routine clinical practice showed substantial missing information and inconsistencies that affected the clinical impact of the results. Overall and across models, the metrics obtained revealed that relapses can be predicted with an accuracy of over 80%, but with a false-positive/false-negative rate of around 50%.
Although the performance metrics of the predictive models were not optimal, some valuable insights were obtained regarding the relative importance of variables associated with CD relapses, namely age, past relapses and complications, and tobacco/substance abuse. In fact, a decision tree reflects the importance of cumulative relapses, obesity, imaging of abdomen or intake of belladonna and its derivatives in combination with analgesics in predicting a further relapse in a 3-month period (Figure S3, Supplemental digital content 1, http://links.lww.com/EJGH/A730). The highest-ranked variables in the model also included medical history, past surgeries, and laboratory results (i.e., fecal and blood tests), as disease-related treatments. Furthermore, the model identified problems related to malabsorption, such as some types of anemia (treated with iron bivalent preparations) and fistulization-related variables (e.g., urinary tract infection, genital infection, pain in male genitalia, and incontinence) as relevant predictors. Our predictive model also identified some unexpected factors, including problems in the respiratory tract (identified by the variable respiratory infection). The involvement of respiratory infections in CD prognosis is especially intriguing given that pulmonary manifestations with unknown origin have been documented in intestinal inflammatory diseases such as CD [56].
Strengths and limitations
This study represents one of the first attempts to combine NLP and ML to explore the free-text, unstructured information from EHRs in a large set of CD patients. NLP-based EHR studies allow for collection of large amounts of data, longitudinal access to patients’ information, and exploration of unknown associations between clinical variables, which is not feasible with more traditional research methods. These advantages become especially relevant when studying complex diseases with low prevalence such as CD.
The results and conclusions of the present study should be interpreted in light of the following limitations. First, this study is based on the secondary use of data captured in patients’ records. Therefore, data quality depends on physicians’ criteria to jot down relevant patient information. EHRs of patients diagnosed before the study period might lack crucial demographic or clinical information previously recorded on paper. Missing and inaccurate information may have also impacted some key variables, as well as the metrics and clinical relevance of the predictive model for CD-related relapses. Second, unlike the a priori-designed data collection in classical trials, this was a data-driven, retrospective study, meaning we reuse existing clinical information. Also, we found a significant degree of variability across patients’ EHRs regarding the quality and quantity of available information, especially temporal heterogeneity. Third, drug availability during the study period should be considered regarding exposure to biologics, that is, dates of drug approval during the study period and the resulting limitations on prescription policies by site. Lastly, the technology used to extract and analyze the data is still under development. Undoubtedly, future studies will benefit from rapid technological advancements in the NLP system used, and from simultaneous analyses of unstructured and structured sources of information, improving the predicting power of the models.
Conclusion
When applied to patients’ EHRs, NLP and ML hold great potential to offer valuable insights into CD. In the present study, we were able to identify well-known and novel clinical characteristics in a large series of CD patients by exclusively analyzing the unstructured, free text captured in physicians’ notes during routine clinical practice. CD is a very complex disease and predicting relapses or other complications is highly challenging. Therefore, the risk factors identified in our predictive model, which are data-driven hypotheses, should be further explored and validated in controlled trials and in clinical settings. Clinical notes, as a data source, can provide key clinical information that is not found elsewhere - hence the importance of including full medical records in future studies to improve the quality of data analysis. Raising awareness among healthcare professionals on the importance of medical record completeness is crucial for improving patient care, conducting research, and managing healthcare resources [57–60].
Acknowledgements
We thank Laura Yebes, Carlos Del Río-Bermúdez, and Clara L. Oeste, Medical Writers at SAVANA, for writing and editing the manuscript, as well as preparing the figures. This study was sponsored by Takeda Farmacéutica España, S.A.
The Premonition-CD Study Group includes the following investigators: Carlos Castaño from HU Rey Juan Carlos, Madrid, Spain; Ángel Ponferrada Díaz from HU Infanta Leonor, Madrid, Spain; María Chaparro and María José Casanova from HU de La Princesa, Madrid, Spain; Felipe Ramos Zabala from HM Hospitales, Madrid, Spain; Almudena Calvache from HU Infanta Elena, Madrid, Spain; Fernando Bermejo from HU de Fuenlabrada, Madrid, Spain; Noemí Manceñido from HU Infanta Sofía, Madrid, Spain; Marta Calvo Moya from HU Puerta de Hierro, Majadahonda, Madrid, Spain.
Conflicts of interest
Dr. F. Gomollón has received educational grants from Janssen, MSD, Takeda, and Abbvie, and nonpersonal investigation grants from MSD, Janssen, Abbvie, Takeda, and Tilllots. Dr. J.P. Gisbert has served as a speaker, a consultant, and advisory member for, or has received research funding from, MSD, Abbvie, Hospira, Pfizer, Kern Pharma, Biogen, Takeda, Janssen, Roche, Sandoz, Celgene, Ferring, Faes Farma, Shire Pharmaceuticals, Dr. Falk Pharma, Tillotts Pharma, Chiesi, Casen Fleet, Gebro Pharma, Otsuka Pharmaceutical, and Vifor Pharma. Dr. I. Guerra has served as a speaker, a consultant, and advisory member for, or has received research funding from, Kern Pharma, Takeda and Janssen. Dr. R. Plaza has served as a speaker for Takeda and Janssen. Dr. M.I. Vera has served as a speaker, consultant, and advisory member for, or has received funding from, MSD, Abbvie, Pfizer, Ferring, Shire Pharmaceuticals, Takeda and Jannsen. Jesús Aparicio, Vicente Martínez, Ignacio Tagarro, Alonso Fernández-Nistal, and Carmen Montoto are employees at Takeda Farmacéutica España S.A. S. Lumbreras is an employee at Universidad Pontificia Comillas, Madrid. Dr. C. Maté is an employee at Medsavana S.L. The remaining authors have no conflicts of interest to declare.
Supplementary Material
Footnotes
Supplemental Digital Content is available for this article. Direct URL citations appear in the printed text and are provided in the HTML and PDF versions of this article on the journal's website, www.eurojgh.com.
References
- 1.Baumgart DC, Sandborn WJ. Crohn’s disease. Lancet 2012; 380:1590–1605. [DOI] [PubMed] [Google Scholar]
- 2.Veauthier B, Hornecker JR. Crohn’s disease: diagnosis and management. Am Fam Physician 2018; 98:661–669. [PubMed] [Google Scholar]
- 3.Torres J, Mehandru S, Colombel JF, Peyrin-Biroulet L. Crohn’s disease. Lancet 2017; 389:1741–1755. [DOI] [PubMed] [Google Scholar]
- 4.Golan D, Gross B, Miller A, Klil-Drori S, Lavi I, Shiller M, et al. Cognitive function of patients with Crohn’s disease is associated with intestinal disease activity. Inflamm Bowel Dis 2016; 22:364–371. [DOI] [PubMed] [Google Scholar]
- 5.van Langenberg DR, Yelland GW, Robinson SR, Gibson PR. Cognitive impairment in Crohn’s disease is associated with systemic inflammation, symptom burden and sleep disturbance. United European Gastroenterol J 2017; 5:579–587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Barberio B, Zamani M, Black CJ, Savarino EV, Ford AC. Prevalence of symptoms of anxiety and depression in patients with inflammatory bowel disease: a systematic review and meta-analysis. Lancet Gastroenterol Hepatol 2021; 6:359–370 [DOI] [PubMed] [Google Scholar]
- 7.Loftus EV, Jr, Schoenfeld P, Sandborn WJ. The epidemiology and natural history of Crohn’s disease in population-based patient cohorts from North America: a systematic review. Aliment Pharmacol Ther 2002; 16:51–60. [DOI] [PubMed] [Google Scholar]
- 8.Panes J, Reinisch W, Rupniewska E, Khan S, Forns J, Khalid JM, et al. Burden and outcomes for complex perianal fistulas in Crohn’s disease: systematic review. World J Gastroenterol 2018; 24:4821–4834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Floyd DN, Langham S, Séverac HC, Levesque BG. The economic and quality-of-life burden of Crohn’s disease in Europe and the United States, 2000 to 2013: a systematic review. Dig Dis Sci 2015; 60:299–312. [DOI] [PubMed] [Google Scholar]
- 10.Kawalec P. Indirect costs of inflammatory bowel diseases: Crohn’s disease and ulcerative colitis. A systematic review. Arch Med Sci 2016; 12:295–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lichtenstein GR, Shahabi A, Seabury SA, Lakdawalla DN, Espinosa OD, Green S, et al. Lifetime economic burden of Crohn’s disease and ulcerative colitis by age at diagnosis. Clin Gastroenterol Hepatol 2020; 18:889–897.e10. [DOI] [PubMed] [Google Scholar]
- 12.Le Berre C, Ananthakrishnan AN, Danese S, Singh S, Peyrin-Biroulet L. Ulcerative colitis and Crohn’s disease have similar burden and goals for treatment. Clin Gastroenterol Hepatol 2020; 18:14–23. [DOI] [PubMed] [Google Scholar]
- 13.Bounthavong M, Li M, Watanabe JH. An evaluation of health care expenditures in Crohn’s disease using the United States Medical Expenditure Panel Survey from 2003 to 2013. Res Social Adm Pharm 2017; 13:530–538. [DOI] [PubMed] [Google Scholar]
- 14.Balfour Sartor R. Enteric microflora in IBD: pathogens or commensals? Inflamm Bowel Dis 1997; 3:230–235. [PubMed] [Google Scholar]
- 15.Soon IS, Molodecky NA, Rabi DM, Ghali WA, Barkema HW, Kaplan GG. The relationship between urban environment and the inflammatory bowel diseases: a systematic review and meta-analysis. BMC Gastroenterol 2012; 12:51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ananthakrishnan AN. Epidemiology and risk factors for IBD. Nat Rev Gastroenterol Hepatol 2015; 12:205–217. [DOI] [PubMed] [Google Scholar]
- 17.Wisniewski A, Danese S, Peyrin-Biroulet L. Evolving treatment algorithms in Crohn’s disease. Curr Drug Targets 2018; 19:782–790. [DOI] [PubMed] [Google Scholar]
- 18.Peyrin-Biroulet L, Reinisch W, Colombel JF, Mantzaris GJ, Kornbluth A, Diamond R, et al. Clinical disease activity, C-reactive protein normalisation and mucosal healing in Crohn’s disease in the SONIC trial. Gut 2014; 63:88–95. [DOI] [PubMed] [Google Scholar]
- 19.Braun T, Di Segni A, BenShoshan M, Neuman S, Levhar N, Bubis M, et al. Individualized dynamics in the gut microbiota precede Crohn’s disease flares. Am J Gastroenterol 2019; 114:1142–1151. [DOI] [PubMed] [Google Scholar]
- 20.Burakoff R, Pabby V, Onyewadume L, Odze R, Adackapara C, Wang W, et al. Blood-based biomarkers used to predict disease activity in Crohn’s disease and ulcerative colitis. Inflamm Bowel Dis 2015; 21:1132–1140. [DOI] [PubMed] [Google Scholar]
- 21.Parkes M, Noor NM, Dowling F, Leung H, Bond S, Whitehead L, et al. PRedicting Outcomes For Crohn’s dIsease using a moLecular biomarkEr (PROFILE): protocol for a multicentre, randomised, biomarker-stratified trial. BMJ Open 2018; 8:e026767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ghaly S, Murray K, Baird A, Martin K, Prosser R, Mill J, et al. High vitamin D-binding protein concentration, low albumin, and mode of remission predict relapse in Crohn’s disease. Inflamm Bowel Dis 2016; 22:2456–2464. [DOI] [PubMed] [Google Scholar]
- 23.Karoui S, Ouerdiane S, Serghini M, Jomni T, Kallel L, Fekih M, et al. Correlation between levels of C-reactive protein and clinical activity in Crohn’s disease. Dig Liver Dis 2007; 39:1006–1010. [DOI] [PubMed] [Google Scholar]
- 24.Dasari A, Shen C, Halperin D, Zhao B, Zhou S, Xu Y, et al. Trends in the incidence, prevalence, and survival outcomes in patients with neuroendocrine tumors in the United States. JAMA Oncol 2017; 3:1335–1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kiernan MC, Vucic S, Cheah BC, Turner MR, Eisen A, Hardiman O, et al. Amyotrophic lateral sclerosis. Lancet 2011; 377:942–955. [DOI] [PubMed] [Google Scholar]
- 26.Weber GM, Mandl KD, Kohane IS. Finding the missing link for big biomedical data. JAMA 2014; 311:2479–2480. [DOI] [PubMed] [Google Scholar]
- 27.Murdoch TB, Detsky AS. The inevitable application of big data to health care. JAMA 2013; 309:1351–1352. [DOI] [PubMed] [Google Scholar]
- 28.Kong HJ. Managing unstructured big data in healthcare system. Healthc Inform Res 2019; 25:1–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Goldstein BA, Navar AM, Pencina MJ, Ioannidis JP. Opportunities and challenges in developing risk prediction models with electronic health records data: a systematic review. J Am Med Inform Assoc 2017; 24:198–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zeiberg D, Prahlad T, Nallamothu BK, Iwashyna TJ, Wiens J, Sjoding MW. Machine learning for patient risk stratification for acute respiratory distress syndrome. PLoS One 2019; 14:e0214465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Moon KA, Pollak J, Hirsch AG, Aucott JN, Nordberg C, Heaney CD, et al. Epidemiology of Lyme disease in Pennsylvania 2006-2014 using electronic health records. Ticks Tick Borne Dis 2019; 10:241–50. [DOI] [PubMed] [Google Scholar]
- 32.Qiao Z, Sun N, Li X, Xia E, Zhao S, Qin Y. Using machine learning approaches for emergency room visit prediction based on electronic health record data. Stud Health Technol Inform 2018; 247:111–115. [PubMed] [Google Scholar]
- 33.Kurowski JA, Milinovich A, Ji X, Bauman J, Sugano D, Kattan MW, Achkar JP. Differences in biologic utilization and surgery rates in pediatric and adult Crohn’s disease: results from a large electronic medical record-derived cohort. Inflamm Bowel Dis 2020; 27:1035–1044. [DOI] [PubMed] [Google Scholar]
- 34.Gubatan J, Levitte S, Patel A, Balabanis T, Wei MT, Sinha SR. Artificial intelligence applications in inflammatory bowel disease: emerging technologies and future directions. World J Gastroenterol 2021; 27:1920–1935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ananthakrishnan AN, Cai T, Savova G, Cheng SC, Chen P, Perez RG, et al. Improving case definition of Crohn’s disease and ulcerative colitis in electronic medical records using natural language processing: a novel informatics approach. Inflamm Bowel Dis 2013; 19:1411–1420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hernandez Medrano ITG, J, Belda C, Urena A, Salcedo I, Espinosa-Anke L, Saggion H. Savana: re-using electronic health records with artificial intelligence. Int J Interact Multimed Artif Intel 2017; 4:8–12. [Google Scholar]
- 37.Graziani D, Soriano JB, Del Rio-Bermudez C, Morena D, Díaz T, Castillo M, et al. Characteristics and prognosis of COVID-19 in patients with COPD. J Clin Med 2020; 9:E3259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ancochea J, Izquierdo JL, Medrano IH, Porras A, Serrano M, Lumbreras S, et al. Evidence of gender differences in the diagnosis and management of COVID-19 patients: an analysis of electronic health records using natural language processing and machine learning. J Women Health 2020;In press. [DOI] [PubMed] [Google Scholar]
- 39.Izquierdo JL, Ancochea J, Soriano JB; Savana COVID-19 Research Group. Clinical characteristics and prognostic factors for intensive care unit admission of patients with COVID-19: retrospective study using machine learning and natural language processing. J Med Internet Res 2020; 22:e21801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Izquierdo JL, Almonacid C, González Y, Del Rio-Bermúdez C, Ancochea J, Cárdenas R, et al. The impact of COVID-19 on patients with asthma. Eur Respir J 2020; 57:2003142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Izquierdo JL, Morena D, González Y, Paredero JM, Pérez B, Graziani D, et al. Clinical management of COPD in a real-world setting. A big data analysis. Arch Bronconeumol (Engl Ed) 2021; 57:94–100. [DOI] [PubMed] [Google Scholar]
- 42.Canales L, Menke S, Marchesseau S, D’Agostino A, Del Rio-Bermudez C, Taberna M, Tello J. Assessing the performance of clinical natural language processing systems: development of an evaluation methodology. JMIR Med Inform 2021; 9:e20492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Silverberg MS, Satsangi J, Ahmad T, Arnott ID, Bernstein CN, Brant SR, et al. Toward an integrated clinical, molecular and serological classification of inflammatory bowel disease: report of a Working Party of the 2005 Montreal World Congress of Gastroenterology. Can J Gastroenterol 2005; 19 Suppl A:5A–36A. [DOI] [PubMed] [Google Scholar]
- 44.Seyed Tabib NS, Madgwick M, Sudhakar P, Verstockt B, Korcsmaros T, Vermeire S. Big data in IBD: big progress for clinical practice. Gut 2020; 69:1520–1532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tong Y, Lu K, Yang Y, Li J, Lin Y, Wu D, et al. Can natural language processing help differentiate inflammatory intestinal diseases in China? Models applying random forest and convolutional neural network approaches. BMC Med Inform Decis Mak 2020; 20:248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dong Y, Xu L, Fan Y, Xiang P, Gao X, Chen Y, et al. A novel surgical predictive model for Chinese Crohn’s disease patients. Medicine (Baltimore) 2019; 98:e17510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang L, Fan R, Zhang C, Hong L, Zhang T, Chen Y, et al. Applying machine learning models to predict medication nonadherence in Crohn’s disease maintenance therapy. Patient Prefer Adherence 2020; 14:917–926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Aniwan S, Park SH, Loftus EV, Jr. Epidemiology, natural history, and risk stratification of Crohn’s disease. Gastroenterol Clin North Am 2017; 46:463–480. [DOI] [PubMed] [Google Scholar]
- 49.Kayar Y, Baran B, Ormeci AC, Akyuz F, Demir K, Besisik F, Kaymakoglu S. Risk factors associated with progression to intestinal complications of Crohn disease. Chin Med J (Engl) 2019; 132:2423–2429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lichtenstein GR, Loftus EV, Isaacs KL, Regueiro MD, Gerson LB, Sands BE. ACG Clinical Guideline: management of Crohn’s disease in adults. Am J Gastroenterol 2018; 113:481–517. [DOI] [PubMed] [Google Scholar]
- 51.Maaser C, Langholz E, Gordon H, Burisch J, Ellul P, Ramirez VH, et al. European Crohn’s and colitis organisation topical review on environmental factors in IBD. J Crohns Colitis 2017; 11:905–920. [DOI] [PubMed] [Google Scholar]
- 52.Feuerstein JD, Cheifetz AS. Crohn disease: epidemiology, diagnosis, and management. Mayo Clin Proc 2017; 92:1088–1103. [DOI] [PubMed] [Google Scholar]
- 53.Gajendran M, Loganathan P, Catinella AP, Hashash JG. A comprehensive review and update on Crohn’s disease. Dis Mon 2018; 64:20–57. [DOI] [PubMed] [Google Scholar]
- 54.Cholapranee A, Hazlewood GS, Kaplan GG, Peyrin-Biroulet L, Ananthakrishnan AN. Systematic review with meta-analysis: comparative efficacy of biologics for induction and maintenance of mucosal healing in Crohn’s disease and ulcerative colitis controlled trials. Aliment Pharmacol Ther 2017; 45:1291–1302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Torres J, Bonovas S, Doherty G, Kucharzik T, Gisbert JP, Raine T, et al. ECCO Guidelines on therapeutics in Crohn’s disease: medical treatment. J Crohns Colitis 2020; 14:4–22. [DOI] [PubMed] [Google Scholar]
- 56.Lu DG, Ji XQ, Liu X, Li HJ, Zhang CQ. Pulmonary manifestations of Crohn’s disease. World J Gastroenterol 2014; 20:133–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hong CJ, Kaur MN, Farrokhyar F, Thoma A. Accuracy and completeness of electronic medical records obtained from referring physicians in a Hamilton, Ontario, plastic surgery practice: a prospective feasibility study. Plast Surg (Oakv) 2015; 23:48–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Del Rio-Bermudez C, Medrano IH, Yebes L, Poveda JL. Towards a symbiotic relationship between big data, artificial intelligence, and hospital pharmacy. J Pharm Policy Pract 2020; 13:75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lai FW, Kant JA, Dombagolla MH, Hendarto A, Ugoni A, Taylor DM. Variables associated with completeness of medical record documentation in the emergency department. Emerg Med Australas 2019; 31:632–638. [DOI] [PubMed] [Google Scholar]
- 60.Wu CHK, Luk SMH, Holder RL, Rodrigues Z, Ahmed F, Murdoch I. How do paper and electronic records compare for completeness? A three centre study. Eye (Lond) 2018; 32:1232–1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.