Abstract
Recent evidence suggests that certain LEF/TCF family members act as repressors in the absence of Wnt signaling. We show here that repression by LEF1 requires histone deacetylase (HDAC) activity. Further, LEF1 associates in vivo with HDAC1, and transcription of a model LEF1-dependent target gene is modulated by the ratio of HDAC1 to β-catenin, implying that repression by LEF1 is mediated by promoter-targeted HDAC. Consistent with this hypothesis, under repression conditions the promoter region of a LEF1 target gene is hypoacetylated. By contrast, when the reporter is activated, its promoter becomes hyperacetylated. Coexpression of β-catenin with LEF1 and HDAC1 results in the formation of a β-catenin/HDAC1 complex. Surprisingly, the enzymatic activity of HDAC1 associated with β-catenin is attenuated. Together, these findings imply that activation of LEF1-dependent genes by β-catenin involves a two-step mechanism. First, HDAC1 is dissociated from LEF1 and its enzymatic activity is attenuated. This first step yields a promoter that is inactive but poised for activation. Second, once HDAC1-dependent repression has been overridden, β-catenin binds LEF1 and the β-catenin–LEF1 complex is competent to activate the expression of downstream target genes.
The LEF1 transcription factor and its homologs (TCF1, TCF3, TCF4, dTCF, and pop1) transduce Wnt signals during development and the genesis of colon cancer (8, 14, 18, 28, 29, 38, 47). Wnt-stimulated transcriptional activation by the LEF/TCF family is mediated by a bipartite transcriptional activator composed of a LEF/TCF family member and β-catenin. The rate-limiting step in the formation of this dimeric transcription factor appears to be the nuclear accumulation of β-catenin. In the absence of Wnt signal, β-catenin is localized to the cytoplasm, where it is phosphorylated by glycogen synthase kinase 3β (GSK3β) and rapidly degraded. Phosphorylation of β-catenin by GSK3β is thought to occur within a multiprotein complex containing the adenomatous polyposis coli tumor suppressor protein and axin. Wnt signaling regulates β-catenin turnover by inactivating cytoplasmic GSK3β, resulting in the stabilization of β-catenin. Stabilized β-catenin accumulates and translocates to the nucleus, where it interacts with an N-terminal region of members of the LEF/TCF family.
LEF/TCF proteins were originally identified as transcriptional activators. However, a growing body of evidence indicates that LEF/TCF proteins also function as transcriptional repressors in the absence of Wnt signals (4). For example, in the early Xenopus laevis embryo, XTCF3 represses transcription of the Wnt-responsive homeobox gene siamois when Wnt signals are not present and activates siamois expression in cells receiving Wnt signals (7). Genetic studies of the Drosophila melanogaster LEF/TCF homologue dTCF (pangolin) and the Caenorhabditis elegans LEF/TCF homolog pop1 suggest that these transcription factors also repress the transcription of downstream target genes in the absence of Wnt signals. Thus, this feature of LEF/TCF function is highly conserved (44, 45, 51).
Recent studies have shown that several transcriptional repressors function by recruiting corepressor complexes to DNA (1, 21, 42, 53). For example, the mammalian Mad family of repressors interacts with the mSin3A corepressor (20, 32), while the Saccharomyces cerevisiae repressor UME6 interacts with the yeast ortholog of mSin3A (26). Another class of repressors in D. melanogaster utilizes the corepressor Groucho (41). The mechanisms by which these corepressors facilitate transcriptional repression are distinct, but all the corepressors appear to act on the underlying chromatin template. Both mSin3A and Sin3p are part of large multiprotein complexes (20, 27, 57) that contain histone deacetylases (HDAC), and HDAC enzymatic activity is required for Sin3-dependent repression. Groucho interacts with the hypoacetylated amino-terminal tails of histone H3 (40). The mechanism by which the interaction of Groucho corepressors with H3 drives transcriptional repression is unclear, but a chromatin-mediated mechanism is strongly implicated.
In D. melanogaster and X. laevis, certain LEF/TCF family members interact genetically and biochemically with Groucho corepressors (11, 34, 46). However, not all members of the LEF/TCF family interact with Groucho/TLE proteins. Among these are human LEF1, murine TCF3, and murine TCF4 (46). Furthermore, XTCF3 requires XCtBP to function as a transcriptional repressor (6). Reasoning that additional mechanisms for LEF/TCF-mediated repression might exist, we tested LEF1 for interaction with other corepressor molecules. Here, we present evidence that LEF1 represses transcription by recruiting HDAC activity to DNA. In addition, our experiments suggest that β-catenin interacts with HDAC1 in a LEF1-dependent fashion and that the enzymatic activity of β-catenin-bound HDAC1 is reduced compared to that of unbound HDAC1. We propose that transcriptional activation by LEF1–β-catenin involves the attenuation of HDAC activity and dissociation of HDAC1 from LEF1 by β-catenin.
MATERIALS AND METHODS
Cell culture, luciferase assays, and antibodies.
293 cells were maintained in Dulbecco minimal essential medium–10% calf serum (HyClone) supplemented with penicillin and streptomycin. For luciferase assays, 2 × 105 cells were plated in 60-mm-diameter dishes and transfected by calcium phosphate in triplicate. Typically, 200 ng of siamois or FLASH promoters, 400 ng of pcDNA3 β-catenin, 500 ng of pcDNA3HDAC1, 1 μg of pME18LEF1 or mutant derivatives of LEF1, and 20 ng of β-galactosidase control plasmid were used as indicated in the figure legends. In the experiments where the amount of either β-catenin or HDAC1 expression vectors was varied, the total amount of vector DNA was kept constant by including the appropriate empty expression vector. At 24 h after transfection, the cells were harvested and luciferase and β-galactosidase assays were carried out as specified by the manufacturer (Promega, Tropix). Data shown are from representative experiments that were done in triplicate, and the error bars indicate the standard error of the mean. Treatments with the deacetylase inhibitor trichostatin A (TSA) were performed using the drug at 100 ng/ml for 8 h. FLAG monoclonal antibodies, FLAG polyclonal antibodies, MYC monoclonal antibodies, and β-catenin antibodies were purchased from Sigma, Zymed, Santa Cruz, and Transduction Labs, respectively. mSin3A and HDAC1 antibodies were as described previously (20, 50). Immunoprecipitations were performed as described previously (2).
Cloning.
pME18LEF and pME18ΔN67LEF1 were constructed by amplifying LEF1 cDNA by PCR and cloning the products in frame with the FLAG tag of pME18 (31). pcDNA3-HDAC1 was constructed by cloning the BamHI fragment from pBJ5HDAC1 (50) into pcDNA3 (Invitrogen). The HDAC1-LEF1 chimera was constructed by mutating the termination codon, which follows a FLAG epitope, in HDAC1 to leucine and fusing this molecule in frame to full-length LEF1. The deletion of the C-terminal activation domain of β-catenin was constructed by introducing a stop codon immediately after the last armadillo repeat. The template β-catenin molecule, Glu-β-catenin, has one copy of the Glu-Glu epitope. Point mutations were generated using the Quickchange mutagenesis kit (Stratgene). MYC-LEF was constructed by subcloning an EcoRI-XbaI fragment from pME18LEF into pCS2MYC6tag.
HDAC Assays.
Assays were performed using washed immunoprecipitates as described (50) with acetate-labeled histones extracted from in vivo-labeled HeLa cells (9). Quantitation of the immunologically detectable HDAC1 was performed using a Lumimager from Boehringer Mannheim. This instrument measures the light generated by chemiluminescence and is linear over 4 orders of magnitude.
ChIP.
293 cells (1 × 107 to 5 × 107) were transfected by the calcium phosphate method with the following plasmids: 3 μg of pTOPFLASH, 10 μg of pcDNAβ-catenin, 30 μg of pME18LEF1 Flag, or 91 μg of pcDNA3HDAC1 or pcDNA3. After 24 h, the cells were cross-linked by adding formaldehyde to the culture to a final concentration of 1% and incubated at room temperature for 10 min. Cross-linked cells were washed twice in 1× phosphate-buffered saline and harvested in 1× phosphate-buffered saline plus protease inhibitors by centrifugation at 400 × g for 4 min. The cells were lysed in 200 μl of sodium dodecyl sulfate (SDS) lysis buffer (1% SDS, 10 mM EDTA, 50 mM Tris-HCl [pH 8.1]) and sonicated on ice by three 10-s pulses on power setting 1 with a microtip sonicator (Misonix Inc.). Cell debris was removed by centrifugation at 23,000 × g for 10 min at 4°C. The supernatant was diluted 10-fold in immunoprecipitation (IP) buffer (0.01% SDS, 1.1% Triton X-100, 1.2 mM EDTA, 16.7 mM Tris-HCl [pH 8.1], 16.7 mM NaCl). Samples were precleared by addition of 30 μl of preblocked protein A-Sepharose (Sigma) by incubation for 1 h with rotation at 4°C. The supernatant was subsequently divided into two parts. To half of the supernatant, 10 μl of anti-acetylated histone H4 chromatin (ChIP) grade antibody (Upstate Biotechnology) was added. Immunoprecipitate with or without antibody were incubated overnight with rotation at 4°C. Immune complexes were harvested by incubation with preblocked protein A-Sepharose for 2 h and subsequent centrifugation. The beads were washed twice in IP buffer, once in wash 3 buffer (0.25 M LiCl, 1% NP-40, 1% sodium deoxycholate, 1 mM EDTA, 10 mM Tris-HCl [pH 8.1]), and twice in Tris-EDTA. Immune complexes were eluted by two 15-min incubations at room temperature in 250 μl of 1% SDS in 0.1 M NaHCO3 with constant rotation. The beads were collected by centrifugation, and NaCl was added to the supernatant to a final concentration of 200 mM. Cross-links were reversed by incubation at 65°C for 4 h. Samples were treated with proteinase K for 1 h at 45°C, phenol-chloroform extracted, and back-extracted. DNA was ethanol precipitated and assayed by PCR using primers specific to the pTOP-FLASH-pFOPFLASH promoter region. The following primer pair was used to assay ChIP samples: 5′ AGTCGCGGTTGGAGTAGTAG 3′ and 5′ CATGTCTGGATCCTCTAGAGTCG 3′. PCR was carried out at an MgCl2 concentration of 3 mM and an annealing temperature of 55°C. The linear range of the PCR was determined for each sample. ChIP products could be detected between 25 and 30 cycles. PCR products were detected by ethidium bromide staining using a Bio-Rad Geldoc system. The PCR assays were repeated two to six times for each sample.
RESULTS
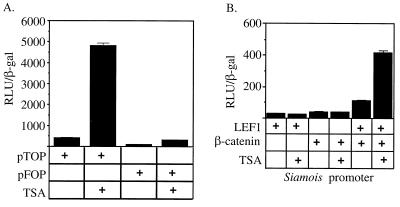
To determine whether LEF1-dependent transcriptional repression requires HDAC activity, we transfected 293 cells with two reporter genes shown previously to be LEF1 responsive and tested whether the HDAC inhibitor TSA could stimulate their activity. We used the synthetic pTOPFLASH promoter, which has four multimerized LEF1 binding sites cloned upstream of a segment of the Fos promoter, or, as a negative control, the pFOPFLASH promoter, which has mutant LEF1 binding sites (29), We also tested the promoter of the siamois gene that is a direct target for regulation by the Xenopus LEF/TCF factor XTCF3 (7). 293 cells were used for this study because they express both wild-type adenomatous polyposis coli protein and β-catenin, and thus β-catenin levels are properly regulated (43). 293 cells also contain endogenous LEF1 (43) and HDACs (data not shown). We reasoned that if HDACs were involved in LEF1-dependent repression, the activity of the pTOPFLASH and siamois reporters should be sensitive to TSA treatment. As expected, both pTOPFLASH and siamois responded similarly to transfected LEF1 and β-catenin (data not shown). However, their response to TSA was markedly different. TSA treatment stimulated pTOP-FLASH approximately 11.5-fold above background, while the mutant promoter lacking LEF/TCF binding sites was stimulated only about 3.5-fold, suggesting that repression by endogenous LEF/TCF requires HDAC activity (Fig. 1A). Furthermore, the stimulation of the pTOPFLASH reporter by TSA did not require transfection of either LEF1 or β-catenin (data not shown). By contrast, TSA was unable to activate the siamois promoter unless LEF1 and β-catenin were both coexpressed (Fig. 1B). Transcriptional stimulation by TSA required the LEF/TCF binding sites in the siamois promoter since no activation was observed from a promoter lacking these binding sites (data not shown). Thus, LEF1 appears to act as a repressor on both the synthetic pTOPFLASH reporter and the native siamois reporter, and this transcriptional repression depends on LEF1 DNA binding and HDAC activity.
FIG. 1.
LEF-dependent reporters are derepressed by deacetylase inhibitors. Luciferase activity was measured from cells either untreated or following an 8-h treatment with the specific HDAC inhibitor TSA. (A) pTOPFLASH reporter constructs containing binding sites for LEF/TCF family members or the pFOP-FLASH reporter lacking LEF/TCF binding sites. (B) Expression vectors encoding LEF1 and β-catenin were cotransfected with the siamois promoter in the combinations indicated by the plus signs.
Previous experiments suggested that deacetylase inhibitors can overcome a repressive chromatin barrier and render the template poised for activation but that actual induction of transcription requires DNA-bound activators (20, 32). The pTOPFLASH promoter alone was stimulated by TSA, but TSA stimulation of the siamois promoter required LEF and β-catenin. Therefore, it is likely that endogenous activators were bound to the pTOPFLASH promoter, probably to the segment of the Fos promoter, and that no other endogenous activators can interact with the siamois promoter in 293 cells.
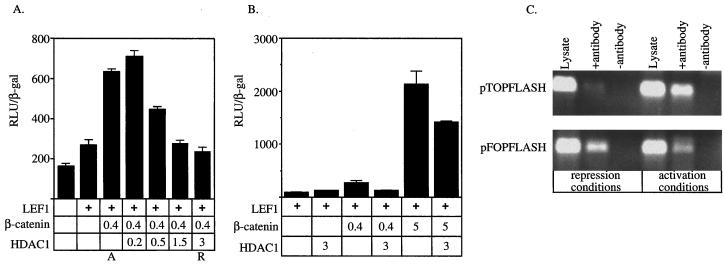
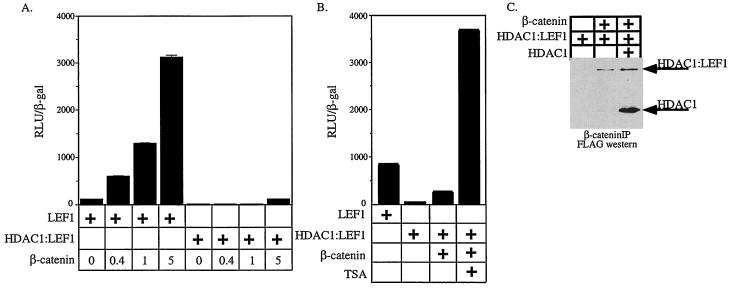
These results suggest that transcription from LEF/TCF-dependent reporters is controlled by the relative abundance of repressor and activator molecules. To test this hypothesis, we next asked whether increasing the deacetylase activity by transfecting HDAC1 could repress LEF1–β-catenin transactivation. Cotransfection of LEF1 with β-catenin resulted in approximately fourfold activation of the pTOPFLASH reporter (activation conditions in Fig. 2A). Increasing the amount of HDAC1 expression plasmid resulted in dose-dependent repression of LEF1–β-catenin transactivation, nearly to the level of the unstimulated reporter (repression conditions in Fig. 2A). Cotransfection of HDAC1 does not dramatically affect the activity of the CMV-β expression plasmid used for normalization (data not shown), suggesting that increasing the HDAC concentration in cells does not nonspecifically inhibit gene expression. Thus, HDAC1 can repress LEF1–β-catenin transactivation, suggesting that HDAC1 can act as a corepressor for LEF1.
FIG. 2.
Reciprocal regulation of LEF-dependent reporters by HDAC1 and β-catenin. (A and B) Expression from the pTOPFLASH promoter was tested in the presence of different combinations of expression vectors encoding LEF1, β-catenin, and HDAC1 as indicated by the plus signs. (A) Cells were transfected with the amount (in micrograms) of HDAC expression plasmid indicated. A and R denote the combinations and amounts of expression vectors referred to as activation and repression conditions, respectively. (B) Cells were transfected with 0.4 or 5 μg of β-catenin expression plasmid in the presence or absence of 3 μg of HDAC1 expression plasmid as indicated. (C) Chromatin was immunoprecipitated using antiserum specific for acetylated amino-terminal tails of histone H4 from cells transfected under repression conditions and activation conditions. The pTOPFLASH and pFOPFLASH DNAs were detected by PCR using primers (see Materials and Methods) that spanned the wild-type or mutant LEF binding sites. “Lysate” represents 1:50 of the amount of lysate used in the immunoprecipitations and controls for DNA recovery. The data shown are representative of 4 independent experiments.
We next tested whether nuclear accumulation of β-catenin could overcome transcriptional repression imposed by HDAC1. We transfected increasing amounts of β-catenin expression construct into 293 cells along with constant amounts of LEF1 and HDAC1 expression vectors (Fig. 2B). To ensure that the template was initially repressed, we transfected 3 μg of HDAC1 expression construct, which was sufficient to completely counteract activation by β-catenin in the previous experiment (Fig. 2A). HDAC1 did not repress transcription when cotransfected with LEF1 alone, suggesting that the reporter was maximally repressed by endogenous HDAC(s) (Fig. 2B). Transfection of increasing amounts of β-catenin expression plasmid resulted in dose-dependent transcriptional activation. Cotransfection of the HDAC1 expression plasmid completely blocked activation by small amounts of β-catenin and partially blocked activation by larger amounts of β-catenin (Fig. 2B). Together, these experiments suggest that LEF/TCF-dependent transcription depends on the relative abundance and/or activities of HDAC1 and β-catenin.
Transcriptional repression and activation are correlated with hypoacetylation and hyperacetylation, respectively, of the amino-terminal tails of histones that comprise the promoter-proximal nucleosomes (21, 42). Because LEF1 required HDAC activity to repress transcription and HDAC1 could block activation by β-catenin–LEF1 complexes, we determined whether LEF1 directly targets chromatin. To do this, we examined the acetylation status of the pTOPFLASH promoter using antiserum specific for acetylated histone H4 and chromatin immunoprecipitation (ChIP). Repression and activation conditions (Fig. 2) were established by transfecting HEK293 cells with pTOPFLASH or, as a control, pFOPFLASH and with different combinations of expression vectors. DNA recovered by ChIP was detected by PCR using primers spanning the four LEF binding sites in the promoter. Equivalent amounts of pTOP-FLASH or pFOPFLASH DNA were present in each of the lysates prepared from cells transfected under activation or repression conditions (Fig. 2C). Under each condition, the amount of precipitated pFOPFLASH was equivalent and is a measure of the background acetylation level of the promoter in the absence of any LEF1 binding. Under repression conditions, less pTOPFLASH was precipitated than pFOPFLASH. Under activation conditions, more pTOPFLASH were precipitated than pFOPFLASH. Therefore, these results demonstrate that this LEF1-dependent target is hypoacetylated when repressed and hyperacetylated when activated. The observed effects on template acetylation status require LEF1 binding sites in the promoter. Because LEF1 has no known enzymatic activity, these results suggest that template hypoacetylation and hyperacetylation result from LEF1-bound HDACs and histone acetyltransferases (HATs), respectively, modifying promoter-proximal nucleosomes.
HDAC1 associates with LEF1.
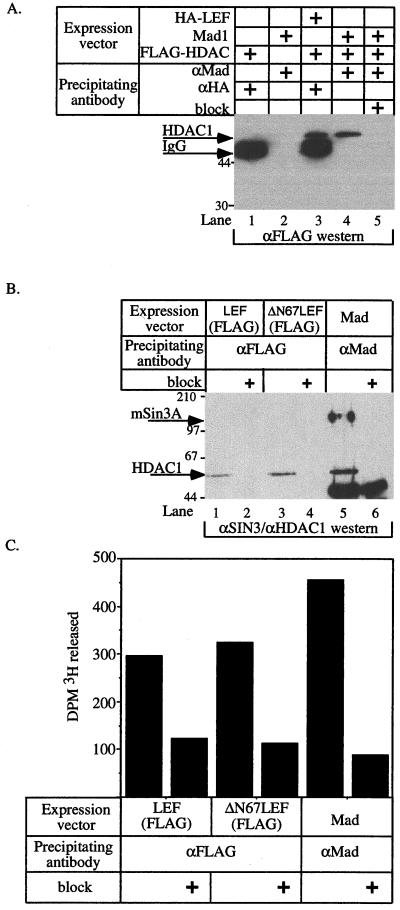
To further test the hypothesis that LEF1 can act as a transcriptional repressor via an associated HDAC, we asked whether LEF1 interacts with HDAC1 or an HDAC-containing corepressor complex. We transfected 293 cells with hemagglutinin (HA)-tagged LEF1 and FLAG-tagged HDAC1 and immunoprecipitated LEF1-associated proteins with anti-HA monoclonal antibody. As a positive control, the amount of HDAC1 that coimmunoprecipitated with the HDAC-dependent transcriptional repressor Mad1 was also determined. In this experiment, LEF1 and Mad1 were abundantly and uniformly expressed under the different transfection combinations (data not shown). HDAC1 was detected in the HA immunoprecipitate only when HDAC1 and LEF1 were coexpressed, suggesting that the interaction is specific (Fig. 3A, compare lanes 1 and 3). Furthermore, the amount of HDAC1 protein that coimmunoprecipitated with LEF1 was nearly identical to that immunoprecipitated with Mad1 (compare lanes 3 and 4). In subsequent experiments (Fig. 4), we wished to detect LEF1 and HDAC1 simultaneously using the same monoclonal antibody, so we constructed an expression vector encoding FLAG-tagged LEF1 (FLAG-LEF). To test for an interaction between FLAG-LEF1 and endogenous HDAC1, we transfected 293 cells with either FLAG-LEF1, a FLAG-tagged amino-terminal deletion of LEF1 (ΔN67LEF1) that is unable to interact with β-catenin, or Mad1 as a positive control. Mad1 is known to interact with the mSin3A-HDAC corepressor complex (20, 32), and, as expected, transfected Mad1 specifically recovered both HDAC1 and mSin3A (Fig. 3B, lane 5). Endogenous HDAC1 was specifically retained by both wild-type and mutant LEF1 molecules, but no mSin3A was recovered (Fig. 3B, lanes 1 and 3). These data demonstrate that LEF1 can interact with endogenous HDAC1 independently of the mSin3A corepressor. The ability of ΔN67LEF1 to interact with HDAC1 indicates that HDAC1 and β-catenin do not compete for binding to LEF1. Finally, both LEF1 molecules and Mad1 coimmunoprecipitate HDAC activity (Fig. 3C), which demonstrates that the LEF1-associated HDAC1 is enzymatically active and presumably capable of repressing transcription.
FIG. 3.
LEF1 and HDAC1 associate in vivo. (A and B) 293 cells were transfected with expression vectors encoding the indicated proteins in the combinations denoted by plus signs. After 24 h, cell extracts were prepared and proteins were precipitated with the indicated antisera. “block” indicates incubation of the antiserum with cognate peptide. Proteins associated with LEF1 were detected by Western blotting using antiserum specific for the FLAG epitope (A) or antisera specific for mSin3A and HDAC1 (B). Arrows mark HDAC1 and mSin3A. (C) The deacetylase activity associated with LEF1, ΔN67LEF1, or Mad1. ΔN67LEF lacks its amino-terminal β-catenin-binding domain. Western blotting of cell extracts prepared from the various transfected cell populations indicated abundant and equivalent expression from the transfected cDNAs (data not shown).
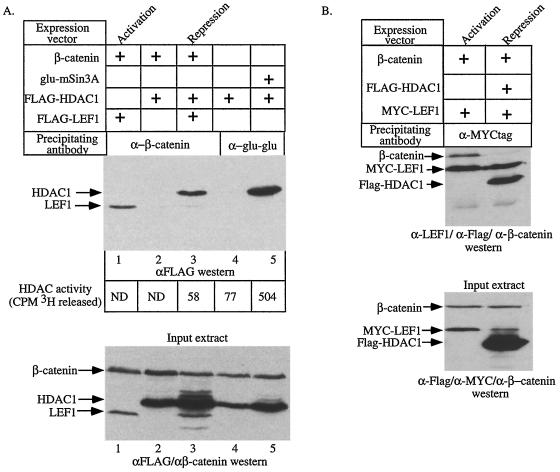
FIG. 4.
β-Catenin can interact with HDAC1 and attenuate HDAC activity. (A) 293 cells were transfected with expression vectors encoding the indicated cDNAs. The amount of FLAG-tagged LEF1 or FLAG-tagged HDAC1 associated with β-catenin or mSin3A in each extract was detected by Western blotting for the FLAG epitope following immunoprecipitation with antisera specific for β-catenin or the Glu-Glu epitope on mSin3A (top panel). The values at the bottom of the top panel are the HDAC activities determined for each immunoprecipitation. ND, not determined. Western blotting of whole-cell extracts shows that the transfected HDAC1, LEF1, and β-catenin proteins are expressed to similar levels (bottom panel). The top and bottom panels are aligned so that the IPs shown in a given lane were performed from the cell extracts shown in the lane directly below. (B) 293 cells were transfected with expression vectors encoding the indicated cDNAs. The proteins associated with MYC-LEF1 were detected with a mixture of antibodies specific for the FLAG epitope, LEF1, and β-catenin (top panel). Western blotting of whole-cell extracts shows that the transfected HDAC1, LEF1, and β-catenin proteins are expressed to similar levels (bottom panel). The panels are aligned as in panel A. In both panels, “Activation” and “Repression” mark the lanes showing experiments performed under activation and repression conditions, respectively.
β-catenin and HDAC1 interact to modulate LEF1 transcriptional activity.
Data presented in Fig. 2 suggest that the transcriptional activity of LEF1 is regulated by the relative amounts of β-catenin and HDAC1 present in the nucleus. To investigate how LEF1 switches from being a repressor to an activator, we tested whether LEF1 could form a tripartite complex with β-catenin and HDAC1 or whether binding of either the coactivator or corepressor to LEF1 was mutually exclusive. We transfected 293 cells using the repression and activation conditions shown in Fig. 2A. 293 cells were also transfected with β-catenin plus HDAC1 and, as an additional control, Glu-Glu epitope-tagged mSin3A plus FLAG-HDAC1. Proteins associated with β-catenin and mSin3A were collected by IP with antisera specific for β-catenin and the Glu-Glu tag, respectively, and subjected to Western blotting for the FLAG epitope on HDAC1 and LEF1. Under activation conditions, β-catenin associates with LEF1 as previously shown (3) (Fig. 4A, top, lane 1). Under repression conditions, very little LEF1 associated with β-catenin, but we detected an unexpected interaction between β-catenin and HDAC1 (Fig. 4A top, compare lanes 1 and 3). The levels of LEF1 were similar in all transfected cell extracts (Fig. 4A, bottom, compare lanes 1 and 3), suggesting that under repression conditions, HDAC1–β-catenin complexes form at the expense of LEF1–β-catenin complexes. Interaction between HDAC1 and β-catenin was detected only when LEF1 was also coexpressed, suggesting that LEF1 is required for this interaction. Finally, as expected, HDAC1 specifically associated with epitope-tagged mSin3A.
Analysis of HDAC assays performed in parallel showed that no HDAC activity coimmunoprecipitated with β-catenin from cells expressing β-catenin, HDAC1, and LEF1 (Fig. 4A top, CPM 3H released), even though these β-catenin immunoprecipitates contained easily detectable levels of HDAC1 protein (Fig. 4A, top). To ensure that the activity of the HDAC1 associated with β-catenin was not below the sensitivity of detection for the deacetylase assay, we determined the relative amount of HDAC protein in the β-catenin and mSin3A immunoprecipitates. Approximately 2.5-fold more HDAC1 immunoprecipitated with mSin3A (Fig. 4A top, lane 5) than immunoprecipitated with β-catenin (lane 3), but the mSin3A-associated HDAC1 was approximately 10 times more active than that associated with β-catenin (Fig. 4A, lane 5). Furthermore, deacetylase activity measurements of control immunoprecipitations (lane 4) showed that β-catenin-associated HDACs had only background deacetylase activity. As a control for HDAC activity in the lysates, the supernatants from the β-catenin immunoprecipitates were reimmunoprecipitated with antibodies against mSin3A. These immunoprecipitates contained equivalent amounts of HDAC activity (data not shown), demonstrating that the HDAC associated with β-catenin was not inactivated by a contaminant in the β-catenin antibody preparation or for another technical reason. These results demonstrate that the β-catenin-associated HDAC1 activity is partially or totally inactivated relative to that associated with mSin3A.
Since β-catenin and HDAC1 can interact under repression conditions, it is possible that HDAC1 is no longer bound to LEF1. Instead, repression might result because β-catenin–LEF1 complexes do not form. To test this, we determined which proteins associated with LEF1 under repression conditions. Cells were transfected using activation and repression conditions with a MYC-tagged LEF1 molecule (MYC-LEF). The composition of MYCtag immunocomplexes was determined by Western blot analysis using a combination of polyclonal antisera against FLAG (to detect FLAG-HDAC1), β-catenin, and LEF1. Consistent with data shown in Fig. 4A, MYCtag immunoprecipitation of cells transfected under repression conditions showed little or no β-catenin associated with LEF1 (Fig. 4B, top). Strikingly, under repression conditions HDAC1 still associated with LEF1. Thus, the interaction of β-catenin with HDAC1 under repression conditions was not sufficient to remove all of the HDAC1 from LEF1, and repression is likely to be mediated by LEF1-HDAC1 complexes rather than a lack of β-catenin–LEF1 complexes.
These data suggest that transcriptional activation by LEF1 normally occurs when β-catenin accumulates in the nucleus to a threshold level that is sufficient to completely dissociate HDAC1 from LEF1. β-Catenin above this threshold amount would then be free to form a transcriptionally active complex with LEF1. As such, LEF1-dependent transactivation should not occur if β-catenin is unable to remove HDAC1 from LEF1. To test this model, we transfected 293 cells with an HDAC-LEF1 chimera or wild-type LEF1, pTOPFLASH, and increasing amounts of β-catenin. As expected, increasing amounts of β-catenin resulted in a dose-dependent activation of the reporter in the presence of LEF1 (Fig. 5A). In contrast, the HDAC1-LEF1 chimera almost completely blocked coactivation by β-catenin, mediating only weak transactivation at the highest levels of β-catenin tested. Wild-type LEF1 and HDAC1-LEF1 were expressed at similar levels, and both could bind β-catenin. β-Catenin activated the pTOPFLASH promoter in the presence of HDAC1-LEF1 if cells were treated with TSA (Fig. 5B), indicating that the chimeric molecule retains all known activities of wild-type LEF1. These data are consistent with a model whereby β-catenin must interact with and remove LEF1-bound HDAC1 in order to function as a coactivator for LEF1.
FIG. 5.
HDAC1-LEF1 chimeras cannot be activated by β-catenin. (A and B) 293 cells were transfected with expression constructs encoding the proteins as indicated and the pTOPFLASH reporter. The amount of β-catenin transfected, in micrograms, is indicated. In panel B, cells transfected with HDAC1-LEF1 and β-catenin were analyzed following an 8-h treatment with TSA. (C) Cells were transfected with the indicated expression constructs. Immunoprecipitation were performed from nuclear extracts with anti-β-catenin and HDAC1, and HDAC1-LEF1 was detected by Western blotting.
Given that β-catenin can inactivate the enzymatic activity of HDAC1, it is surprising that it cannot overcome repression by the HDAC1-LEF1 chimera. However, β-catenin can interact with HDAC1 only when LEF1 is coexpressed (Fig. 4A). In addition, β-catenin can interact with free HDAC1 in the presence of the HDAC1-LEF1 chimera (Fig. 5C). Together these data are consistent with a model where HDAC1 binds LEF1 or the LEF1 portion of the HDAC1-LEF1 fusion and is recognized and removed by β-catenin. We propose that β-catenin cannot recognize the HDAC1 portion of the HDAC1-LEF1 fusion and for this reason cannot overcome transcriptional repression by the chimera (see Discussion).
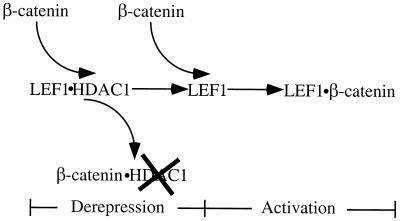
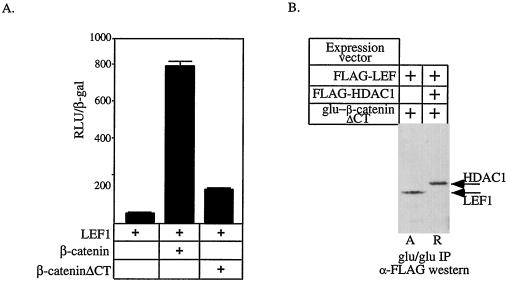
Our data suggest that LEF1-dependent reporter genes are activated in two separate steps (Fig. 6). We hypothesize that the dissociation and attenuation of LEF1-bound HDAC by β-catenin results in a template that is derepressed and poised for activation. Subsequent binding of β-catenin to LEF1 on this derepressed template accounts for true activation. In support of this model, derepression of an HDAC-dependent barrier to transcription with TSA on the siamois promoter is not sufficient to yield gene activation but requires the activator β-catenin (Fig. 1B). To determine whether activation per se was needed for derepression, we tested whether a mutant β-catenin lacking its C-terminal (β-cateninΔCT) transcriptional activation domain (22, 54) could still interact with LEF1 and dissociate HDAC1 from LEF1. As predicted, β-cateninΔCT lacks potent transactivation function (Fig. 7A). β-CateninΔCT retains its LEF1 interaction domain and, as expected, interacted with LEF1 under activation conditions (Fig. 7B). Furthermore, like wild-type β-catenin, β-cateninΔCT formed complexes with HDAC1 but not with LEF1 under repression conditions. Therefore, β-cateninΔCT can interact with HDAC1 and remove it from LEF1, suggesting that it is able to derepress the template but that because it lacks its activation domain, only modest levels of activation are observed. As such, transcriptional activation per se is not required for derepression.
FIG. 6.
Model for β-catenin-dependent gene activation. See the text for details.
FIG. 7.
β-Catenin activates transcription in two steps. (A) The activity of a pTOPFLASH was determined by transfecting 293 cells with expression vectors encoding LEF1, β-catenin, and β-cateninΔCT in the indicated combinations. (B) 293 cells were transfected with the indicated expression constructs. Proteins complexed to β-cateninΔCT were identified by immunoprecipitation using a monoclonal antibody that recognizes the Glu-Glu epitope on the plasmid-expressed β-catenin protein. Associated LEF1 or HDAC1 was detected by probing Western blots for their FLAG epitopes. In each cell extract, FLAG-LEF1 was expressed to similar levels (data not shown). A and R mark the lanes showing experiments performed under activation and repression conditions, respectively.
DISCUSSION
We have demonstrated that human LEF1 requires HDAC activity to repress transcription and that it interacts in vivo with HDAC1. These results support the model that LEF1 represses transcription by targeting HDAC-containing corepressor complexes to DNA. Histone deacetylases are thought to mediate transcriptional repression by altering the acetylation state of promoter-proximal histones. Consistent with this model, we found that under repression conditions, the pTOPFLASH promoter is hypoacetylated, demonstrating that LEF1-associated HDACs are active on the underlying nucleosomal template. It is likely that the hypoacetylated template is less accessible to other transcriptional activators or the basal transcription machinery, resulting in reduced rates of transcriptional initiation (16, 30). However, since nuclear proteins other than histones are also acetylated (5, 17, 25, 35, 39, 48), the involvement of nonhistone substrates in LEF1-dependent transcriptional repression cannot be ruled out.
We show that LEF1, unlike the Mad family of repressors, associates with HDAC1 independently of mSin3A. LEF1 may interact directly with HDAC1, or the interaction may be indirect, perhaps via the recently described HDAC- and ATPase-containing NuRD complex (52, 55, 56, 58) or via a novel corepressor. Our attempts to coimmunoprecipitate endogenous LEF1-HDAC1 complexes have not been successful, leaving open the formal possibility that the detected interaction resulted from overexpression of LEF1. However, we think that this is unlikely, because overexpressed LEF1 can immunoprecipitate both endogenous HDAC1 and HDAC activity at a level comparable to that observed with overexpressed Mad1 (Fig. 3B and C). Furthermore, both pTOPFLASH and siamois promoters were derepressed by TSA (Fig. 1) and pTOP-FLASH was hypoacetylated under repression conditions (Fig. 2C) strongly implicating promoter-targeted HDAC as a feature of LEF1-dependent repression. LEF1 can interact with HDAC1, but other LEF/TCF family members interact with different corepressors. For example, human TCF1, D. melanogaster dTCF, and X. laevis TCF-3 (XTCF-3) require Groucho-related transcriptional corepressors for function (11, 46). Interestingly, XTCF-3 also uses the X. laevis homolog of CtBP (XCtBP) as a corepressor (6). In agreement with a previous report (46), we have found no evidence that LEF1 interacts with Groucho-related proteins (data not shown). In contrast, Levanon et al. detected an interaction between LEF1 and Groucho/TLE (34). These contrasting observations may reflect differences in experimental procedures or variations in LEF1 interactions in different cell types. Other TCF factors, namely, mTCF3 and mTCF4, do not interact with Groucho/TLE proteins (46); these might represent additional LEF/TCF factors that use HDAC1 as a corepressor. Given the number and variety of corepressors utilized by the LEF/TCF family and the recent findings that other repressors interact with multiple corepressors (6, 37), it is possible that LEF1 interacts with other corepressors in addition to HDAC1.
Our data are consistent with a two-step model for activation of LEF-dependent target genes by the dimeric β-catenin–LEF1 activator (Fig. 6). The derepression step in the activation process occurs when nuclear β-catenin rises to a level sufficient to remove HDAC1 from LEF1 and attenuate its activity. Derepression yields a template that is bound only by LEF1. This derepressed template is capable of being activated, but in the absence of activators, only basal levels of transcription result. As β-catenin levels continue to rise, it forms the dimeric LEF1–β-catenin activator. We refer to this step in the activation process as “true activation.” Such coupling of derepression of a chromatin-based barrier to transcription (hypoacetylated nucleosomes produced by LEF1-bound HDAC1) with true activation is analogous to a model originally proposed for the in vitro activation of transcription on nucleosomal templates (33).
The repression conditions used in our experiments are ones in which both HDAC1 and β-catenin are present in the nucleus, but the ratio of HDAC1 to β-catenin is such that activation cannot occur (Fig. 2A). These conditions may be similar to those encountered in a cell that is accumulating nuclear β-catenin in response to Wnt signals. Under repression conditions, β-catenin is detected primarily in a β-catenin–HDAC1 complex, with very little β-catenin being associated with LEF1 (Fig. 5A). β-Catenin and HDAC1 interact only in the presence of LEF1, suggesting that β-catenin interacts with the repressive LEF1-HDAC1 complex and triggers the dissociation of HDAC1 from LEF1. HDAC1 associated with LEF1 under repression conditions (Fig. 4B), suggesting that β-catenin does not exceed the threshold required to dissociate all HDAC1-LEF1 complexes and that the template is still repressed by an HDAC1-dependent mechanism.
Our data are most consistent with HDAC1–β-catenin complexes forming at the expense of HDAC1-LEF1 during derepression. However, because under repression conditions we detected both HDAC1 and LEF1 associated with β-catenin (Fig. 4A), it is difficult to distinguish between an incomplete conversion of HDAC1-LEF to HDAC1–β-catenin complexes and the formation of HDAC1–LEF1–β-catenin ternary complexes. However, we have recently shown that β-catenin can remove HDAC1 from a LEF1 mutant that lacks its β-catenin interaction domain (data not shown), suggesting that the formation of ternary complexes is unlikely.
The derepression step of this two-step activation model requires that an HDAC-dependent repression mechanism be overcome. In our experiments, HDAC-mediated repression can be canceled by either the deacetylase inhibitor TSA or the removal and attenuation of LEF1-bound HDAC1 by β-catenin. The observation that HDAC1-LEF1 chimeras cannot be activated by β-catenin suggests that the physical association of β-catenin with HDAC1 and the subsequent disassociation of this complex from LEF1 are required for derepression and then activation. Further, our data suggest that derepression and activation are separate processes because a β-catenin mutant lacking an activation domain is able to associate with HDAC1.
Interestingly, β-catenin cannot activate the HDAC1-LEF1 chimeras, suggesting that it cannot interact with and inactivate the HDAC1 portion of the HDAC1-LEF1 chimera. We detected an interaction between β-catenin and HDAC1 only when LEF1 or HDAC1-LEF1 was coexpressed, suggesting that when HDAC1 is bound to LEF1, HDAC1 adopts a conformation that is recognized by β-catenin. Since the HDAC1 portion of the HDAC1-LEF1 fusion is in a different spatial orientation from HDAC1 simply bound to LEF1, it seems unlikely that the LEF1-fused HDAC1 would be capable of adopting the conformation necessary for interaction with β-catenin. Therefore, it is perhaps not surprising that β-catenin cannot inactivate HDAC1 in the context of a LEF1 fusion.
We observe that HDAC1 associated with β-catenin is enzymatically inactive (Fig. 4B), suggesting that attenuation of HDAC1 activity is an important step in the activation process. Therefore, we propose that to fully inactivate HDAC1 as a LEF corepressor, β-catenin must both physically sequester and attenuate the enzymatic activity of HDAC1. We have not directly tested whether attenuation of HDAC1 by β-catenin is necessary for β-catenin-dependent activation of LEF1 reporter genes. However, such a mechanism would ensure that HDAC1 was both physically removed from LEF1 and no longer capable of acting as a corepressor. We speculate that other transcriptional activators will function in part by blocking the activity of corepressor complexes or proteins.
It is most likely that the transcriptionally active species is a heterocomplex consisting of β-catenin and LEF1, as suggested previously (3, 7, 8, 10, 23, 24, 54). However, it is formally possible that the transcriptionally active species is a LEF1–HDAC (inactive)–β-catenin ternary complex. Our data also demonstrate that under activation conditions, LEF1-dependent target genes are hyperacetylated, implying that the LEF1–β-catenin dimeric activator is associated with HAT or a HAT-containing complex. We detected HAT activity in LEF1 immunoprecipitations from cells transfected under activation conditions (data not shown) but have not yet identified the HAT that is responsible for the LEF1-associated activity.
The finding that β-catenin-associated HDAC1 is inactive parallels the recent demonstration that the transcription factor Twist and the viral oncoprotein E1A can inhibit the acetyltransferase activity of both p300 and PCAF by binding to the HAT domains of these proteins (12, 19, 36). Together, these findings suggest that the regulation of both HAT and HDAC activities by nuclear proteins may be a common mechanism for modulating transcription. The molecular mechanism of HDAC1 inactivation by β-catenin is not yet known. The activity of immunoprecipitated HDAC1 is not inhibited by recombinant β-catenin armadillo repeats (data not shown), suggesting that either another region of the β-catenin molecule is required or another protein associated with β-catenin in vivo is the inhibitory factor.
The LEF/TCF family acts as transcriptional repressors in the absence of nuclear β-catenin. However, individual family members utilize different corepressors to perform this task. Interestingly, these different repression mechanisms all appear to target the underlying chromatin template. For instance, Groucho/TLE proteins have been shown to interact with the hypoacetylated amino-terminal tail of histone H3. The XTCF-3 corepressor CtBP appears to require HDAC activity for repressive function (15, 49), and we have shown that LEF1 repression also required HDACs. LEF1 lacks the carboxy-terminal sequences required for CtBP binding, making it unlikely that it interacts with HDACs via CtBP (6). The recent report showing that Drosophila RPD3 (a HDAC homolog) and Groucho interact genetically and physically (13) suggests that there may be functional overlap between these different corepressors. In support of this idea, we have detected a weak interaction between HDAC1 and the human Groucho homolog TLE in extracts from mammalian cells (data not shown). The fact that LEF/TCF proteins have adopted similar but distinct mechanisms of transcriptional repression emphasizes the necessity of tightly regulating Wnt signal target genes in the absence of Wnt signaling.
ACKNOWLEDGMENTS
We thank Kathryn Coulter, Jennifer Logan, and Andrew Thorburn for critically reading the manuscript; Marion Waterman for providing the human LEF1 cDNA; Stefano Stifani for providing Groucho/TLE antibodies; David Kimmelman for providing the siamois reporters; and Jacqueline Papkoff for providing the Glu-Glu-epitope-tagged β-catenin cDNA.
A.N.B. is supported by Cancer Center training grant 3P30CA42014. This work was supported by a pilot project grant from the Huntsman Cancer Institute. D.E.A. is a Scholar of the Leukemia and Lymphoma Society (formerly the Leukemia Society of America).
REFERENCES
- 1.Ayer D E. Histone deacetylases: transcriptional repression with SINers and NuRDs. Trends Cell Biol. 1999;9:193–198. doi: 10.1016/s0962-8924(99)01536-6. [DOI] [PubMed] [Google Scholar]
- 2.Ayer D E, Eisenman R N. A switch from Myc:Max to Mad:Max heterocomplexes accompanies monocyte/macrophage differentiation. Genes Dev. 1993;7:2110–2119. doi: 10.1101/gad.7.11.2110. [DOI] [PubMed] [Google Scholar]
- 3.Behrens J, von Kries J P, Kuhl M, Bruhn L, Wedlich D, Grosschedl R, Birchmeier W. Functional interaction of beta-catenin with the transcription factor LEF-1. Nature. 1996;382:638–642. doi: 10.1038/382638a0. [DOI] [PubMed] [Google Scholar]
- 4.Bienz M. TCF: transcriptional activator or repressor? Curr Opin Cell Biol. 1998;10:366–372. doi: 10.1016/s0955-0674(98)80013-6. [DOI] [PubMed] [Google Scholar]
- 5.Boyes J, Byfield P, Nakatani Y, Ogryzko V. Regulation of activity of the transcription factor GATA-1 by acetylation. Nature. 1998;396:594–598. doi: 10.1038/25166. [DOI] [PubMed] [Google Scholar]
- 6.Brannon M, Brown J D, Bates R, Kimelman D, Moon R T. XCtBP is a XTcf-3 co-repressor with roles throughout Xenopus development. Development. 1999;126:3159–3170. doi: 10.1242/dev.126.14.3159. [DOI] [PubMed] [Google Scholar]
- 7.Brannon M, Gomperts M, Sumoy L, Moon R T, Kimelman D. A beta-catenin/XTcf-3 complex binds to the siamois promoter to regulate dorsal axis specification in Xenopus. Genes Dev. 1997;11:2359–2370. doi: 10.1101/gad.11.18.2359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cadigan K M, Nusse R. Wnt signaling: a common theme in animal development. Genes Dev. 1997;11:3286–3305. doi: 10.1101/gad.11.24.3286. [DOI] [PubMed] [Google Scholar]
- 9.Carmen A A, Rundlett S E, Grunstein M. HDA1 and HDA3 are components of a yeast histone deacetylase (HDA) complex. J Biol Chem. 1996;271:15837–15844. doi: 10.1074/jbc.271.26.15837. [DOI] [PubMed] [Google Scholar]
- 10.Cavallo R, Rubenstein D, Peifer M. Armadillo and dTCF: a marriage made in the nucleus. Curr Opin Genet Dev. 1997;7:459–466. doi: 10.1016/s0959-437x(97)80071-8. [DOI] [PubMed] [Google Scholar]
- 11.Cavallo R A, Cox R T, Moline M M, Roose J, Polevoy G A, Clevers H, Peifer M, Bejsovec A. Drosophila Tcf and Groucho interact to repress Wingless signalling activity. Nature. 1998;395:604–608. doi: 10.1038/26982. [DOI] [PubMed] [Google Scholar]
- 12.Chakravarti D, Ogryzko V, Kao H Y, Nash A, Chen H, Nakatani Y, Evans R M. A viral mechanism for inhibition of p300 and PCAF acetyltransferase activity. Cell. 1999;96:393–403. doi: 10.1016/s0092-8674(00)80552-8. [DOI] [PubMed] [Google Scholar]
- 13.Chen G, Fernandez J, Mische S, Courey A J. A functional interaction between the histone deacetylase Rpd3 and the corepressor groucho in Drosophila development. Genes Dev. 1999;13:2218–2230. doi: 10.1101/gad.13.17.2218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Clevers H, van de Wetering M. TCF/LEF factor earn their wings. Trends Genet. 1997;13:485–489. doi: 10.1016/s0168-9525(97)01305-x. [DOI] [PubMed] [Google Scholar]
- 15.Criqui-Filipe P, Ducret C, Maira S M, Wasylyk B. Net, a negative Ras-switchable TCF, contains a second inhibition domain, the CID, that mediates repression through interactions with CtBP and de-acetylation. EMBO J. 1999;18:3392–3403. doi: 10.1093/emboj/18.12.3392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Davie J R. Covalent modifications of histones: expression from chromatin templates. Curr Opin Genet Dev. 1998;8:173–178. doi: 10.1016/s0959-437x(98)80138-x. [DOI] [PubMed] [Google Scholar]
- 17.Gu W, Roeder R G. Activation of p53 sequence-specific DNA binding by acetylation of the p53 C-terminal domain. Cell. 1997;90:595–606. doi: 10.1016/s0092-8674(00)80521-8. [DOI] [PubMed] [Google Scholar]
- 18.Gumbiner B M. Carcinogenesis: a balance between beta-catenin and APC. Curr Biol. 1997;7:R443–R446. doi: 10.1016/s0960-9822(06)00214-4. [DOI] [PubMed] [Google Scholar]
- 19.Hamamori Y, Sartorelli V, Ogryzko V, Puri P L, Wu H Y, Wang J Y, Nakatani Y, Kedes L. Regulation of histone acetyltransferases p300 and PCAF by the bHLH protein twist and adenoviral oncoprotein E1A. Cell. 1999;96:405–413. doi: 10.1016/s0092-8674(00)80553-x. [DOI] [PubMed] [Google Scholar]
- 20.Hassig C A, Fleischer T C, Billin A N, Schreiber S L, Ayer D E. Histone deacetylase activity is required for full transcriptional repression by mSin3A. Cell. 1997;89:341–347. doi: 10.1016/s0092-8674(00)80214-7. [DOI] [PubMed] [Google Scholar]
- 21.Hassig C A, Schreiber S L. Nuclear histone acetylases and deacetylases and transcriptional regulation: HATs off to HDACs. Curr Opin Chem Biol. 1997;1:300–308. doi: 10.1016/s1367-5931(97)80066-x. [DOI] [PubMed] [Google Scholar]
- 22.Hecht A, Litterst C M, Huber O, Kemler R. Functional characterization of multiple transactivating elements in beta-catenin, some of which interact with the TATA-binding protein in vitro. J Biol Chem. 1999;274:18017–18025. doi: 10.1074/jbc.274.25.18017. [DOI] [PubMed] [Google Scholar]
- 23.Hsu S C, Galceran J, Grosschedl R. Modulation of transcriptional regulation by LEF-1 in response to Wnt-1 signaling and association with beta-catenin. Mol Cell Biol. 1998;18:4807–4818. doi: 10.1128/mcb.18.8.4807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huber O, Korn R, McLaughlin J, Ohsugi M, Herrmann B G, Kemler R. Nuclear localization of beta-catenin by interaction with transcription factor LEF-1. Mech Dev. 1996;59:3–10. doi: 10.1016/0925-4773(96)00597-7. [DOI] [PubMed] [Google Scholar]
- 25.Imhof A, Yang X J, Ogryzko V V, Nakatani Y, Wolffe A P, Ge H. Acetylation of general transcription factors by histone acetyltransferases. Curr Biol. 1997;7:689–692. doi: 10.1016/s0960-9822(06)00296-x. [DOI] [PubMed] [Google Scholar]
- 26.Kadosh D, Struhl K. Repression by Ume6 involves recruitment of a complex containing Sin3 corepressor and Rpd3 histone deacetylase to target promoters. Cell. 1997;89:365–371. doi: 10.1016/s0092-8674(00)80217-2. [DOI] [PubMed] [Google Scholar]
- 27.Kasten M M, Dorland S, Stillman D J. A large protein complex containing the yeast Sin3p and Rpd3p transcriptional regulators. Mol Cell Biol. 1997;17:4852–4858. doi: 10.1128/mcb.17.8.4852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Korinek V, Barker N, Moerer P, van Donselaar E, Huls G, Peters P J, Clevers H. Depletion of epithelial stem-cell compartments in the small intestine of mice lacking Tcf-4. Nat Genet. 1998;19:379–383. doi: 10.1038/1270. [DOI] [PubMed] [Google Scholar]
- 29.Korinek V, Barker N, Morin P J, van Wichen D, de Weger R, Kinzler K W, Vogelstein B, Clevers H. Constitutive transcriptional activation by a beta-catenin-Tcf complex in APC−/− colon carcinoma. Science. 1997;275:1784–1787. doi: 10.1126/science.275.5307.1784. [DOI] [PubMed] [Google Scholar]
- 30.Kuo M H, Allis C D. Roles of histone acetyltransferases and deacetylases in gene regulation. Bioessays. 1998;20:615–626. doi: 10.1002/(SICI)1521-1878(199808)20:8<615::AID-BIES4>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- 31.Laherty C D, Billin A N, Lavinsky R M, Yochum G S, Bush A C, Sun J M, Mullen T M, Davie J R, Rose D W, Glass C K, Rosenfeld M G, Ayer D E, Eisenman R N. SAP30, a component of the mSin3 corepressor complex involved in N-CoR-mediated repression by specific transcription factors. Mol Cell. 1998;2:33–42. doi: 10.1016/s1097-2765(00)80111-2. [DOI] [PubMed] [Google Scholar]
- 32.Laherty C D, Yang W M, Sun J M, Davie J R, Seto E, Eisenman R N. Histone deacetylases associated with the mSin3 corepressor mediate mad transcriptional repression. Cell. 1997;89:349–356. doi: 10.1016/s0092-8674(00)80215-9. [DOI] [PubMed] [Google Scholar]
- 33.Laybourn P J, Kadonaga J T. Role of nucleosomal cores and histone H1 in regulation of transcription by RNA polymerase II. Science. 1991;254:238–245. doi: 10.1126/science.254.5029.238. [DOI] [PubMed] [Google Scholar]
- 34.Levanon D, Goldstein R E, Bernstein Y, Tang H, Goldenberg D, Stifani S, Paroush Z, Groner Y. Transcriptional repression by AML1 and LEF-1 is mediated by the TLE/Groucho corepressors. Proc Natl Acad Sci USA. 1998;95:11590–11595. doi: 10.1073/pnas.95.20.11590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li Q, Herrler M, Landsberger N, Kaludov N, Ogryzko V V, Nakatani Y, Wolffe A P. Xenopus NF-Y pre-sets chromatin to potentiate p300 and acetylation-responsive transcription from the xenopus hsp70 promoter in vivo. EMBO J. 1998;17:6300–6315. doi: 10.1093/emboj/17.21.6300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li Q, Imhof A, Collingwood T N, Urnov F D, Wolffe A P. p300 stimulates transcription instigated by ligand-bound thyroid hormone receptor at a step subsequent to chromatin disruption. EMBO J. 1999;18:5634–5652. doi: 10.1093/emboj/18.20.5634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Meloni A R, Smith E J, Nevins J R. A mechanism for Rb/p130-mediated transcription repression involving recruitment of the CtBP corepressor. Proc Natl Acad Sci USA. 1999;96:9574–9579. doi: 10.1073/pnas.96.17.9574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Morin P J, Sparks A B, Korinek V, Barker N, Clevers H, Vogelstein B, Kinzler K W. Activation of beta-catenin-Tcf signaling in colon cancer by mutations in beta-catenin or APC. Science. 1997;275:1787–1790. doi: 10.1126/science.275.5307.1787. [DOI] [PubMed] [Google Scholar]
- 39.Munshi N, Merika M, Yie J, Senger K, Chen G, Thanos D. Acetylation of HMG I(Y) by CBP turns off IFN beta expression by disrupting the enhanceosome. Mol Cell. 1998;2:457–467. doi: 10.1016/s1097-2765(00)80145-8. [DOI] [PubMed] [Google Scholar]
- 40.Palaparti A, Baratz A, Stifani S. The Groucho/transducin-like enhancer of split transcriptional repressors interact with the genetically defined amino-terminal silencing domain of histone H3. J Biol Chem. 1997;272:26604–26610. doi: 10.1074/jbc.272.42.26604. [DOI] [PubMed] [Google Scholar]
- 41.Paroush Z, Finley R L, Jr, Kidd T, Wainwright S M, Ingham P W, Brent R, Ish-Horowicz D. Groucho is required for Drosophila neurogenesis, segmentation, and sex determination and interacts directly with hairy-related bHLH proteins. Cell. 1994;79:805–815. doi: 10.1016/0092-8674(94)90070-1. [DOI] [PubMed] [Google Scholar]
- 42.Pazin M J, Kadonaga J T. What's up and down with histone deacetylation and transcription? Cell. 1997;89:325–328. doi: 10.1016/s0092-8674(00)80211-1. [DOI] [PubMed] [Google Scholar]
- 43.Porfiri E, Rubinfeld B, Albert I, Hovanes K, Waterman M, Polakis P. Induction of a beta-catenin-LEF-1 complex by wnt-1 and transforming mutants of beta-catenin. Oncogene. 1997;15:2833–2839. doi: 10.1038/sj.onc.1201462. [DOI] [PubMed] [Google Scholar]
- 44.Riese J, Yu X, Munnerlyn A, Eresh S, Hsu S C, Grosschedl R, Bienz M. LEF-1, a nuclear factor coordinating signaling inputs from wingless and decapentaplegic. Cell. 1997;88:777–787. doi: 10.1016/s0092-8674(00)81924-8. [DOI] [PubMed] [Google Scholar]
- 45.Rocheleau C E, Downs W D, Lin R, Wittmann C, Bei Y, Cha Y H, Ali M, Priess J R, Mello C C. Wnt signaling and an APC-related gene specify endoderm in early C. elegans embryos. Cell. 1997;90:707–716. doi: 10.1016/s0092-8674(00)80531-0. [DOI] [PubMed] [Google Scholar]
- 46.Roose J, Molenaar M, Peterson J, Hurenkamp J, Brantjes H, Moerer P, van de Wetering M, Destree O, Clevers H. The Xenopus Wnt effector XTcf-3 interacts with Groucho-related transcriptional repressors. Nature. 1998;395:608–612. doi: 10.1038/26989. [DOI] [PubMed] [Google Scholar]
- 47.Rubinfeld B, Robbins P, El-Gamil M, Albert I, Porfiri E, Polakis P. Stabilization of beta-catenin by genetic defects in melanoma cell lines. Science. 1997;275:1790–1792. doi: 10.1126/science.275.5307.1790. [DOI] [PubMed] [Google Scholar]
- 48.Sakaguchi K, Herrera J E, Saito S, Miki T, Bustin M, Vassilev A, Anderson C W, Appella E. DNA damage activates p53 through a phosphorylation-acetylation cascade. Genes Dev. 1998;12:2831–2841. doi: 10.1101/gad.12.18.2831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sundqvist A, Sollerbrant K, Svensson C. The carboxy-terminal region of adenovirus E1A activates transcription through targeting of a C-terminal binding protein-histone deacetylase complex. FEBS Lett. 1998;429:183–188. doi: 10.1016/s0014-5793(98)00588-2. [DOI] [PubMed] [Google Scholar]
- 50.Taunton J, Hassig C A, Schreiber S L. A mammalian histone deacetylase related to the yeast transcriptional regulator Rpd3p. Science. 1996;272:408–411. doi: 10.1126/science.272.5260.408. [DOI] [PubMed] [Google Scholar]
- 51.Thorpe C J, Schlesinger A, Carter J C, Bowerman B. Wnt signaling polarizes an early C. elegans blastomere to distinguish endoderm from mesoderm. Cell. 1997;90:695–705. doi: 10.1016/s0092-8674(00)80530-9. [DOI] [PubMed] [Google Scholar]
- 52.Tong J K, Hassig C A, Schnitzler G R, Kingston R E, Schreiber S L. Chromatin deacetylation by an ATP-dependent nucleosome remodelling complex. Nature. 1998;395:917–921. doi: 10.1038/27699. [DOI] [PubMed] [Google Scholar]
- 53.Torchia J, Glass C, Rosenfeld M G. Co-activators and co-repressors in the integration of transcriptional responses. Curr Opin Cell Biol. 1998;10:373–383. doi: 10.1016/s0955-0674(98)80014-8. [DOI] [PubMed] [Google Scholar]
- 54.van de Wetering M, Cavallo R, Dooijes D, van Beest M, van Es J, Loureiro J, Ypma A, Hursh D, Jones T, Bejsovec A, Peifer M, Mortin M, Clevers H. Armadillo coactivates transcription driven by the product of the Drosophila segment polarity gene dTCF. Cell. 1997;88:789–799. doi: 10.1016/s0092-8674(00)81925-x. [DOI] [PubMed] [Google Scholar]
- 55.Wade P A, Jones P L, Vermaak D, Wolffe A P. A multiple subunit Mi-2 histone deacetylase from Xenopus laevis cofractionates with an associated Snf2 superfamily ATPase. Curr Biol. 1998;8:843–846. doi: 10.1016/s0960-9822(98)70328-8. [DOI] [PubMed] [Google Scholar]
- 56.Xue Y, Wong J, Moreno G T, Young M K, Cote J, Wang W. NURD, a novel complex with both ATP-dependent chromatin-remodeling and histone deacetylase activities. Mol Cell. 1998;2:851–861. doi: 10.1016/s1097-2765(00)80299-3. [DOI] [PubMed] [Google Scholar]
- 57.Zhang Y, Iratni R, Erdjument-Bromage H, Tempst P, Reinberg D. Histone deacetylases and SAP18, a novel polypeptide, are components of a human Sin3 complex. Cell. 1997;89:357–364. doi: 10.1016/s0092-8674(00)80216-0. [DOI] [PubMed] [Google Scholar]
- 58.Zhang Y, LeRoy G, Seelig H P, Lane W S, Reinberg D. The dermatomyositis-specific autoantigen Mi2 is a component of a complex containing histone deacetylase and nucleosome remodeling activities. Cell. 1998;95:279–289. doi: 10.1016/s0092-8674(00)81758-4. [DOI] [PubMed] [Google Scholar]