Dai uses FRET spectroscopy to show that the epilepsy-related potassium channels KCNQ2/3 are recruited to lipid-raft membrane domains to form a signaling complex with the Alzheimer’s disease–related protein BACE1, a process that is dependent on the palmitoylation of BACE1.
Abstract
β-Secretase 1 (β-site amyloid precursor protein [APP]-cleaving enzyme 1, BACE1) plays a crucial role in the amyloidogenesis of Alzheimer’s disease (AD). BACE1 was also discovered to act like an auxiliary subunit to modulate neuronal KCNQ2/3 channels independently of its proteolytic function. BACE1 is palmitoylated at its carboxyl-terminal region, which brings BACE1 to ordered, cholesterol-rich membrane microdomains (lipid rafts). However, the physiological consequences of this specific localization of BACE1 remain elusive. Using spectral Förster resonance energy transfer (FRET), BACE1 and KCNQ2/3 channels were confirmed to form a signaling complex, a phenomenon that was relatively independent of the palmitoylation of BACE1. Nevertheless, palmitoylation of BACE1 was required for recruitment of KCNQ2/3 channels to lipid-raft domains. Two fluorescent probes, designated L10 and S15, were used to label lipid-raft and non-raft domains of the plasma membrane, respectively. Coexpressing BACE1 substantially elevated FRET between L10 and KCNQ2/3, whereas the BACE1-4C/A quadruple mutation failed to produce this effect. In contrast, BACE1 had no significant effect on FRET between S15 probes and KCNQ2/3 channels. A reduction of BACE1-dependent FRET between raft-targeting L10 probes and KCNQ2/3 channels by applying the cholesterol-extracting reagent methyl-β-cyclodextrin (MβCD), raft-disrupting general anesthetics, or pharmacological inhibitors of palmitoylation, all supported the hypothesis of the palmitoylation-dependent and raft-specific localization of KCNQ2/3 channels. Furthermore, mutating the four carboxyl-terminal cysteines (4C/A) of BACE1 abolished the BACE1-dependent increase of FRET between KCNQ2/3 and the lipid raft–specific protein caveolin 1. Taking these data collectively, we propose that the AD-related protein BACE1 underlies the localization of a neuronal potassium channel.
Introduction
This paper concerns a regulatory interaction of an Alzheimer’s-related enzyme and a potassium (K+) channel. Alzheimer’s disease (AD), the most prevalent form of dementia, is characterized by elevated amounts of amyloid plaque in the brain (Perl, 2010; Weller and Budson, 2018). β-Secretase 1 (BACE1) is one of the major proteases that cleave the amyloid precursor protein (APP), leading to the accumulation of amyloid β (Aβ) fragments (Vassar et al., 2014). A single mutation in APP that compromises the ability of BACE1 to cleave APP can reduce Aβ generation and slow the progression of AD (Di Fede et al., 2009; Jonsson et al., 2012). Therefore, inhibition of BACE1 has been proposed as a promising therapeutic strategy to treat AD (Vassar and Kandalepas, 2011). Indeed, mice with BACE1 gene knockout, BACE1−/−, do not exhibit Aβ plaques or develop AD; however, they do show debilitating changes in behavior tests, including spatial memory deficits and sensorimotor impairments (Cole and Vassar, 2007; Kobayashi et al., 2008). Furthermore, BACE1−/− mice exhibit neuronal deficits including axonal organization defects in the hippocampus (Ou-Yang et al., 2018), myelination deficits (Hu et al., 2006), and altered synaptic function and neurotransmitter metabolism (Lombardo et al., 2019). In addition, BACE1−/− mice exhibit epileptic seizures, suggesting a possible direct role of BACE1 in regulating neuronal voltage-gated Na+ and K+ channels (Kim et al., 2007; Lehnert et al., 2016).
Candidate ion channels for regulation by BACE1 include the M-type K+ channels, which consist of KCNQ1–5 (Kv7.1–7.5) subunits (Brown and Adams, 1980; Hessler et al., 2015). Two channel genes, KCNQ2 and KCNQ3, are widely expressed in the central nervous system (Wang et al., 1998); they form a heteromeric channel important for transducing sympathetic stimuli, dampening excessive neuronal excitability, reducing excitatory postsynaptic potentials, and controlling neuronal firing (Abbott, 2020; Hernandez et al., 2008). KCNQ2/3 channels are abundantly present in the hippocampus and are critical for hippocampal excitability (Carver et al., 2020; Klinger et al., 2011; Shah et al., 2002; Yue and Yaari, 2004). In 2015, BACE1 was reported to interact with KCNQ2/3 channels directly to potentiate M-currents in the hippocampus (Hessler et al., 2015). Using patch-clamp electrophysiology, immunoprecipitation, and proximity-ligation assay, BACE1 was found to function as an auxiliary subunit of KCNQ2/3 channels, apparently independent of its proteolytic activity (Hessler et al., 2015). Since KCNQ channel genes are well known for their involvement in several types of epilepsy (Biervert et al., 1998; Singh et al., 1998), this research seemingly provides a novel linkage between KCNQ2/3 channels and the epileptic phenotypes seen in BACE1−/− mice. Not surprisingly, it was also found that AD patients are more susceptible to developing epilepsy (Vossel et al., 2017).
The enzymatic activity of BACE1 as an APP protease occurs in membrane lipid raft domains (Cordy et al., 2003; Ehehalt et al., 2003). Lipid rafts are dynamic membrane microdomains containing sphingolipids, cholesterol, and certain proteins (Cornell et al., 2020; Matsubara et al., 2021; Nahmad-Rohen et al., 2020; Pavel et al., 2020; Robinson et al., 2019; Simons and Ikonen, 1997). Lipid rafts are also recognized as detergent-resistant membrane fractions, structurally more ordered than non-raft areas; they are signaling hubs orchestrating many cellular events (Lingwood and Simons, 2010; Pike, 2004). Lipid-raft localization of BACE1 biases the processing of APP in the amyloidogenic direction, generating Aβ (Ehehalt et al., 2003; Wang et al., 2021). When APPs are outside of the raft domains, they are preferentially cleaved by the nonamyloidogenic α-secretase (Cordy et al., 2006; Kojro et al., 2001; Wang et al., 2021). Similar to other raft-localizing proteins such as tyrosine kinases, G protein α-subunits, and purinergic P2X7 receptors, BACE1 uses palmitoylation, in this case, of four cysteines at its carboxyl-terminal end for specific membrane anchoring (Linder and Deschenes, 2007; Vetrivel et al., 2009). However, it is not well understood where the nonenzymatic activities of BACE1 localize, especially for regulating KCNQ channels. These channels do not have known palmitoylation sites, but are reported to interact with palmitoylated scaffold proteins, including A-kinase anchoring proteins (AKAP79 in human) and ankyrin-G (Chung et al., 2006; He et al., 2012; Keith et al., 2012; Zhang and Shapiro, 2016). Furthermore, previous research suggested a spatial overlap in the subcellular localization of BACE1 and KCNQ channels; KCNQ channels are thought to be localized in axons and axon initial segments via ankyrin-G, whereas BACE1 is abundant in axons and implicated functionally in the myelination of axons (Buggia-Prevot et al., 2014; Chung et al., 2006; Hu et al., 2006).
To understand the regulation of KCNQ2/3 channels by BACE1, we looked for localization of the BACE1-KCNQ complex in the lipid rafts of cultured cells and provided further evidence supporting the interaction of BACE and KCNQ2/3. We used FRET to demonstrate the proximity of membrane proteins and measured FRET, in a spectral manner, based on the sensitized emission of FRET acceptor after exciting the FRET donor. Using engineered fluorescent probes derived from short lipidated sequences of protein kinases Lck and Src to label lipid raft and non-raft domains, respectively (Chichili and Rodgers, 2007; Myeong et al., 2021; Rodgers, 2002), we show that KCNQ channels are localized to lipid rafts. This specific localization of KCNQ channels is dependent on the palmitoylation of BACE1. We provide a strategy to determine the localization of proteins in specific lipid microdomains, which could be broadly applied to answer similar questions of other membrane protein complexes.
Materials and methods
Cell culture, molecular biology, and reagents
The tsA-201 cells were cultured, maintained and transfected as previously reported (Dai et al., 2016). The mBACE1 construct was obtained from Addgene. The fluorescent L10 and S15 constructs were made in the lab of Dr. Byung-Chang Suh at Daegu Gyeongbuk Institute of Science and Technology (Myeong et al., 2021). The caveolin 1 construct was from Suzanne F. Scarlata (Worcester Polytechnic Institute, Worcester, MA). Point mutations were made using Quickchange II XL Site-Directed Mutagenesis kit (Agilent Technologies). The human KCNQ2 and human KCNQ3 constructs were fused with sequence of enhanced YFP at the carboxyl-terminal end. The sequences of the DNA constructs were confirmed by fluorescence-based DNA sequencing (Genewiz). The pANAP plasmid (Addgene) contained the orthogonal tRNA/aminoacyl-tRNA synthetase specific to L-Anap. L-Anap (AsisChem) was dissolved in ethanol as a 10-mM stock, stored at −20°C, and diluted 500-fold into the culture medium. 2-Bromopalmitate (2-BP) was delivered with fatty acid–free BSA (A6003; Sigma-Aldrich) at a stock concentration of 1 mM albumin with 5 mM 2-BP, which was diluted 50-fold into the culture medium, generating a final concentration of 0.1 mM 2-BP.
Quantification of FRET and spectroscopic analysis
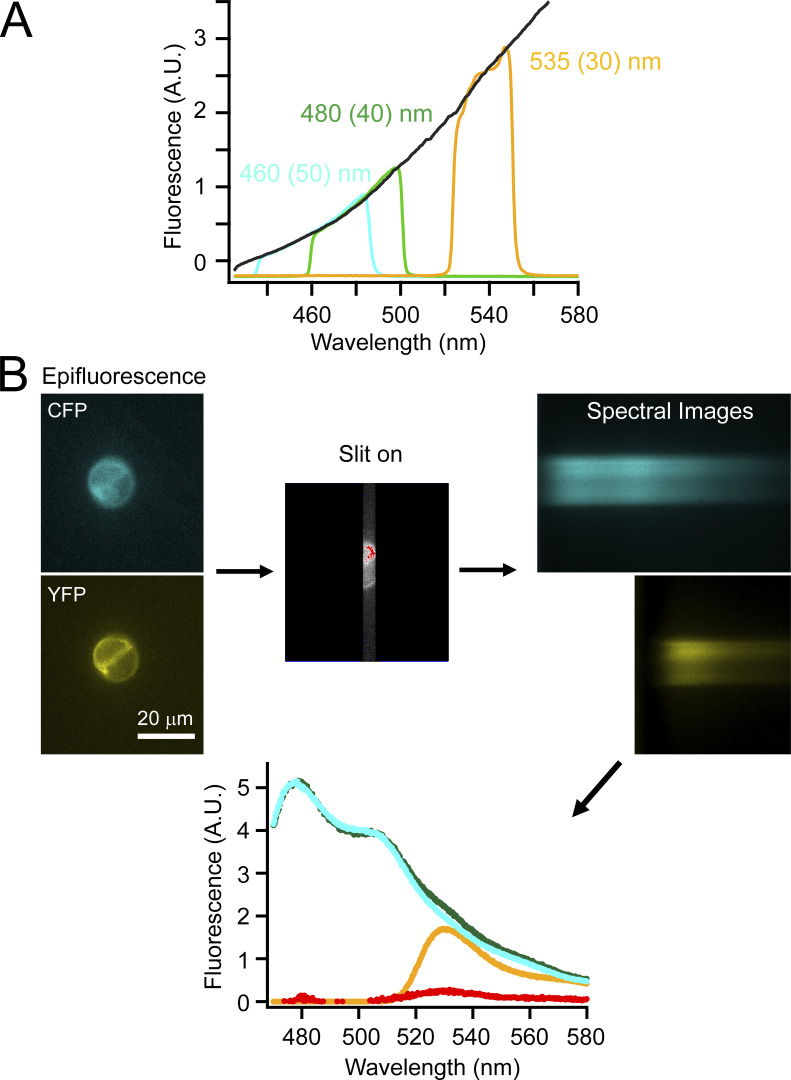
A Nikon Eclipse TE2000-E inverted microscope with a 60×, 1.2-NA water-immersion objective was used for fluorescence measurements. Epifluorescence recording was performed with wide-field excitation using a Lambda LS Xenon Arc lamp (Sutter Instruments). For spectral measurements, images were collected by a spectrograph (model 2150i, 300 g/mm grating, blaze = 500 nm; Acton Research Corp.) mounted between the output port of the microscope and an Evolve 512 EMCCD camera (Photometrics) as previously reported (Dai et al., 2019). A filter cube containing a 376-nm (centered)/30-nm (width) excitation filter was used for L-Anap. CFP was excited with a filter cube containing a 425-/40-nm excitation filter, and YFP was excited with a filter cube containing a 490-/10-nm excitation filter. Emission filters were removed from the filter cubes while the spectrograph functioned similarly to an emission filter to only measure the fluorescence with the light wavelength from 424 to 583 nm (Fig. S1 A). The dichroic mirror cutoffs were ∼450 nm for the CFP filter cube, 514 nm for the YFP filter cube, and 400 nm for the Anap filter cube. CFP or YFP excitation light (not shown in figures) were also detected in the spectral images because the dichroic could not block all the excitation light, but they did not interfere with the acquired fluorescence emission spectra of interest. Spectral images were collected with exposure times of 200–500 ms depending on the fluorescence intensity of the individual cell using the Evolve 512 EMCCD camera and MetaMorph software (Molecular Devices). The exposure time was kept the same for recording donor-emission and acceptor-emission spectra in the same FRET experiment. The spectral data were analyzed with ImageJ/Fiji (National Institutes of Health) and IgorPro (Wavemetrics). The background fluorescence spectrum was subtracted using areas outside of the cell of interest. All microscopic experiments were done at room temperature.
Figure S1.
Procedure of performing spectral measurements of cultured cells. (A) Calibration of spectral measurements using different emission filters with tungsten light illumination. (B) Flow-chart illustration of an example of measuring spectral FRET using CFP-tagged and YFP-tagged constructs transfected into tsA cells.
FRET donor-only (Anap or CFP) constructs were used to obtain the averaged donor emission spectrum excited by donor excitation wavelength. We then normalized this template emission of donor alone, called Fdonor alonedonor, to the peak intensity of the FCFP/YFPCFP or FAnap/YFPAnap spectrum for each experiment. For the labeling used throughout the article, the part in superscript always means the property of the excitation light, specific either to the donor or to the acceptor, whereas the part in subscript means the fluorophores expressed for the experiment, either donor/acceptor both present, or donor alone or acceptor alone. For instance, FCFP/YFPCFP indicates the emission spectrum generated after exciting the CFP when both CFP- and YFP-containing constructs are expressed. The normalized Fdonor alonedonor was then subtracted from either the FCFP/YFPCFP or the FAnap/YFPAnap spectrum in each case, generating the spectrum FFRET, according to the following equations:
| (1) |
This subtraction was for correcting the bleed-through of the donor emission into the spectral range for the acceptor emission. Ratio A was then calculated as
| (2) |
for the Anap/YFP pair or for the CFP/YFP pair. The denominator of ratio A is the spectrum generated by using the excitation light at the YFP excitation wavelength. In addition, two emission spectra were collected from cells that were transfected only with YFP-containing constructs (in the absence of FRET donors): FYFPCFP or FYFPAnap, which was the emission spectrum using the donor (CFP or Anap) excitation, and FYFPYFP, which was the emission spectrum using the acceptor (YFP) excitation. Ratio A0 was calculated using
| (3) |
for the Anap/YFP FRET pair or for the CFP/YFP FRET pair. Ratio A0 measures the efficiency of the crosstalk of FRET, which contains the fraction of the direct excitation of FRET acceptors by the donor-excitation light. The other component of such crosstalk, which is the direct excitation of FRET donors by the acceptor-excitation light, is negligible for the FRET pairs used in this article. Ratio A0 is a constant for each FRET pair; it is 0.06 for the Anap/YFP pair or 0.136 for the CFP/YFP pair under our experimental conditions. The FRET efficiency to measure was linearly proportional to the difference between ratio A and ratio A0; ratio A − ratio A0 > 0 indicated a detectable FRET signal (Erickson et al., 2001; Takanishi et al., 2006). The apparent FRET efficiency (Eapp) was calculated as follows:
| (4) |
where εA and εD are the molar extinction coefficients of acceptors and donors at the donor excitation. The values of εA/εD were estimated as 0.10 for the CFP/YFP pair and 0.24 for the Anap/YFP pair. The properties of extinction coefficient (molar absorptivity) were from previously measured values and from a website (http://www.fpbase.org): 17,500 cm−1 · M−1 for L-Anap; 83,400 cm−1 · M−1 for eYFP; and 32,500 cm−1 · M−1 for eCFP. According to the known spectrum of the YFP absorption, the ratio of YFP absorption at the donor excitation relative to the peak YFP absorption was 0.05 for the Anap/YFP pair and 0.04 for the CFP/YFP pair.
In our experiments, the apparent FRET efficiency Eapp correlates positively with the ratio of the FRET donor-to-acceptor peak fluorescence intensity: F = Fdonor/Facceptor. At very high F, the Eapp is expected to saturate and reach a steady-state value, which should correspond to the maximal FRET efficiency Emax (Takanishi et al., 2006). In theory, for the sensitized-emission type of FRET,
| (5) |
where DA and A indicate concentrations of the donor–acceptor pairs and the free acceptors, respectively (Takanishi et al., 2006). The quantitative amount of DA and A cannot be measured in our case. Nevertheless, in the extreme situation where all the acceptors are paired with the donors, the value of A would be 0, and Eapp equals Emax. This Eq. 5 is in contrast to the comparable equation when performing FRET experiments by monitoring the donor dequenching after acceptor photobleaching (Takanishi et al., 2006); in that specific case, Eapp would correlate negatively with F. In practice, no matter which method is used to perform FRET, the apparent FRET efficiency is always smaller than the real FRET efficiency Emax.
Assuming random pairings between the donors and acceptors based on a simplified model of protein interactions (Bykova et al., 2006), and inspired by Eq. 5, the apparent FRET efficiency Eapp can also be expressed using
| (6) |
where C is the concentration ratio of the total donor versus the total acceptor, which is dependent on the expression level of proteins: C = Cdonor/Cacceptor. An S factor (S = Sdonor/Sacceptor) links the molar ratio C to the fluorescence ratio F of the measured FRET donor-to-acceptor peak intensity: F = C × S. Rearranging Eq. 6 using F = C × S generates the following equation for fitting the relationship between the apparent FRET efficiency and F:
| (7) |
Here, the coefficient S is dependent on the optical properties of fluorophores, the recording system, the percentage of fluorophores that pair in close distance for FRET, and the efficiency of FRET. In general, higher Emax often leads to dimmer donor fluorescence and brighter acceptor fluorescence, and a resulting smaller S. We compared FRET at different conditions using the Emax values obtained after curve fitting. The fits were evaluated using the χ2 goodness-of-fit test, and all the χ2 values were <0.1. Eq. 7 is relatively model independent and serves as a simple way to predict FRET efficiency. In addition, this equation is mathematically identical to a nonlinear saturable isotherm that was previously used for fitting the FRET data with unfixed donor/acceptor ratio (Zacharias et al., 2002): Eapp = Emax × [F/(K + F)], where K is analogous to the dissociation constant for ligand binding (Kenworthy and Edidin, 1998).
The distances r between the donors and acceptors were estimated using the Förster equation, Emax = 1/[1 + (r/R0)6], and assuming the Förster radius R0, at which the FRET efficiency is 50%, to be 50 Å for the CFP/YFP FRET pair. The R0 value for the L-Anap/YFP pair was calculated as 47 Å as follows:
| (8) |
where C is the scaling factor, J is the overlap integral of the Anap emission spectrum and the YFP absorption spectrum, Q is the quantum yield of L-Anap (∼0.3 for L-Anap near the cytoplasmic side of the membrane; Dai et al., 2019), η is the index of refraction, and κ2 is the orientation factor. η was assumed to be 1.33, and κ2 was assumed to be 2/3.
Online supplemental material
Fig. S1 shows the procedure of performing spectral measurements of cultured cells. Fig. S2 shows a schematic illustration of L-Anap incorporation to BACE1.
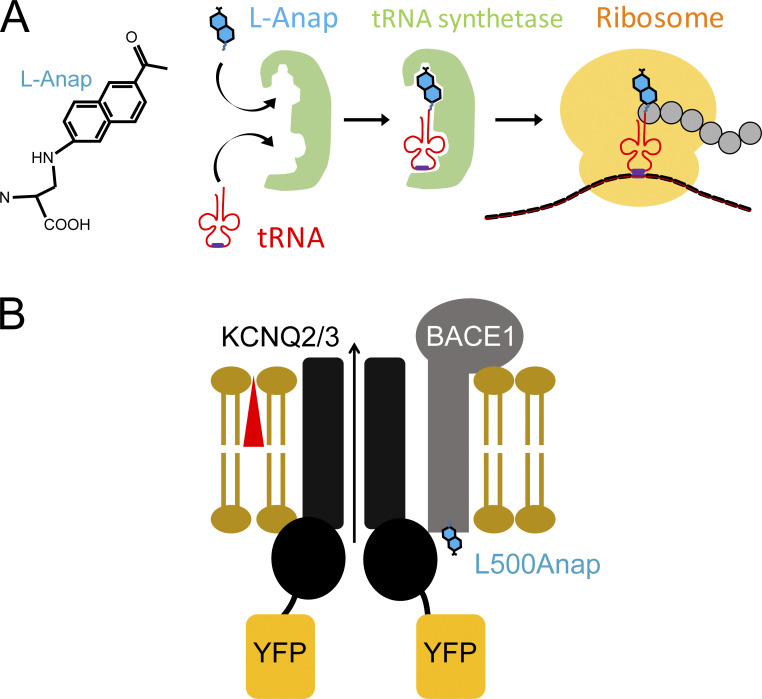
Figure S2.
Schematic illustration of L-Anap incorporation to BACE1. (A) Structure of L-Anap and the amber stop-codon suppression strategy for incorporating L-Anap. (B) Cartoon showing FRET between BACE1-L500Anap and KCNQ2/3-YFP.
Results
This article reports the specific localization of neuronal KCNQ2/3 channels in membrane microdomains (lipid rafts), controlled by formation of a complex with BACE1.
FRET between KCNQ2/3 and BACE1 is consistent with their close assembly
BACE1 interacts with KCNQ2/3 channels directly, demonstrated by previous research using protein coimmunoprecipitation and other biochemical methods (Hessler et al., 2015). Because some KCNQ channels can assemble with single-transmembrane auxiliary subunits, e.g., KCNE for cardiac KCNQ1 channels (Abbott, 2020; Sun and MacKinnon, 2020), BACE1, which is also a single-transmembrane protein, has been considered by analogy as a novel auxiliary subunit for KCNQ2/3 channels (Hessler et al., 2015). Atomic details of the interaction between BACE1 and KCNQ2/3 are largely unknown. Here, we further tested this interaction using FRET between a fluorescent noncanonical amino acid, L-Anap (Chatterjee et al., 2013; Kalstrup and Blunck, 2013; Fig. S2), incorporated into the carboxyl-terminus of a mouse BACE1 and a YFP fused to the carboxyl-terminus of both human KCNQ2 and KCNQ3 subunits, all expressed in a cell line. KCNQ2 and KCNQ3 subunits form heteromeric channel complexes and were expressed together throughout this article in the tsA201 cell line derived from human embryonic kidney cells.
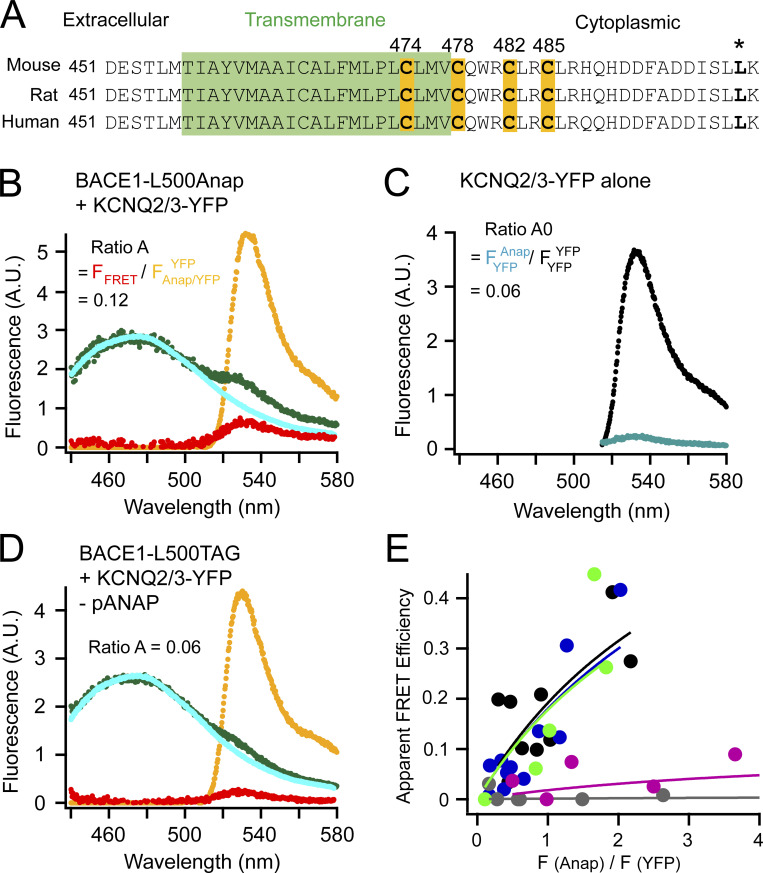
BACE1 consists of an extracellular (amino-terminal) proteolytic domain, one helical transmembrane domain, and a short intracellular (carboxyl-terminal) region (Fig. 1 A). Four cysteines, one near the cytoplasmic end of the transmembrane domain (C474) and three within the intracellular region (C478, C482, and C485) are S-palmitoylated, conferring the lipid raft localization of BACE1. Mutating these cysteines is reported to reduce the raft localization of BACE1 and amyloid burden but does not abolish the processing of APP by BACE1 (Andrew et al., 2017; Vetrivel et al., 2009). The DXXLL motif in the carboxyl-terminus of BACE1 (Fig. 1 A) is involved in the endocytosis and retrograde transport of BACE1 to the trans-Golgi network (Pastorino et al., 2002; Toh et al., 2018). Mutating this motif increases the plasma membrane localization of BACE1 (Pastorino et al., 2002). In addition, the transmembrane and carboxyl-terminal regions of BACE1 are highly conserved (almost identical) among different species.
Figure 1.
FRET between L-Anap incorporated in BACE1 and YFP-tagged KCNQ2/3 channels. (A) Sequence alignment of mouse, rat, and human BACE1 proteins highlighting the four cysteines for palmitoylation and the site of leucine 500 (*). The transmembrane region is in green. (B) Representative emission spectra used to calculate FRET between BACE1-L500Anap and KCNQ2/3-YFP. The cyan (donor only) and green spectra (both donor and acceptor) are generated by the Anap excitation wavelength; the yellow spectrum (with both donor and acceptor) by YFP excitation; the red spectrum is the subtraction of the cyan spectrum from the green spectrum. The ratio of the peak intensity of the red spectrum versus that of the yellow spectrum (ratio A = FFRET/FAnap/YFPYFP) after subtracting the crosstalk (ratio A0 = FYFPAnap/FYFPYFP shown in C) is proportional to the amount of FRET detected. (C) Representative emission spectra of KCNQ2/3-YFP-only cells using direct YFP excitation (large spectrum, FYFPYFP) and using excitation for Anap (small spectrum, FYFPAnap) for calculating ratio A0. (D) Representative emission spectra showing the negative control condition when the BACE1-L500TAG and KCNQ2/3-YFP were transfected but in the absence of the pANAP plasmid. (E) Relationship between the apparent FRET efficiency and the ratio of the peak intensity (at ∼475 nm) of the green spectrum divided by that of the yellow spectrum (at 530 nm) as in B or D in the following conditions shown in different colors: no pANAP (gray); the following conditions all with pANAP: wild-type BACE1 (black), mutant BACE1-4C/A (blue), with added 2-BP (green), and with KCNH instead of KCNQ2/3 (magenta). Each point represents data from one cell.
Considering the small size of the intracellular carboxyl-terminal part of BACE1, fusing a large GFP-derived fluorescent protein might perturb the natural properties of BACE1, so a fluorescent noncanonical amino acid, L-Anap, was incorporated at the carboxyl-terminal end of BACE1. An amber stop-codon (TAG) suppression strategy (Chatterjee et al., 2013) was used to incorporate L-Anap at the leucine 500 (L500) position within the DXXLL motif of BACE1 (Fig. S1, asterisk). To express BACE1-L500Anap in tsA-201 cells, cells were transiently cotransfected with the BACE1-L500TAG cDNA and a plasmid pANAP encoding the orthogonal amber suppressor tRNA/aminoacyl-tRNA synthetase (aaRS) pair for L-Anap and incubated overnight with culture medium supplemented with 20 μM of the membrane-permeable methyl-ester form of L-Anap (Gordon et al., 2018; Zagotta et al., 2016). When the translation machinery arrives at the TAG stop-codon site, translation continues by including L-Anap and avoiding mRNA chain termination. This strategy has been previously shown to achieve satisfactory incorporation of L-Anap into exogenously expressed recombinant membrane proteins (Dai et al., 2019; Dai et al., 2021; Dai and Zagotta, 2017). In our experiments, cells grown without applying pANAP plasmid (tRNA/aaRS), but with BACE1-L500TAG cDNA transfected and L-Anap in media were used as negative controls for the FRET experiments.
The Anap emission spectrum overlaps well with the excitation spectrum of YFP, making them a practical FRET pair (Chatterjee et al., 2013; Dai et al., 2019) with an estimated Förster radius R0 of 47 Å. A high FRET would indicate close proximity of the two proteins. FRET between L-Anap and YFP was determined by calculations from the emission spectra of both fluorophores measured using a spectrometer attached to the microscope (see Materials and methods). This type of spectrum-based FRET directly measures the degree of the sensitized emission of the FRET acceptor after exciting the FRET donor. The sensitized-emission method also includes corrections for bleed-through and crosstalk due to the spectral properties of FRET donors and acceptors. Cells that expressed BACE1-L500TAG alone were used to generate a template L-Anap emission spectrum (spectrum in cyan, Fig. 1 B) to correct for the bleed-through of the Anap emission in the wavelength range for YFP emission. In addition, cells that expressed only KCNQ2/3-YFP channels (FRET acceptor alone) were used to determine a crosstalk efficiency for FRET: the ratio of peak intensities of a YFP emission spectrum by the Anap excitation versus the YFP emission directly excited by the YFP excitation. This ratio, called ratio A0, was a constant, which equaled 6% for the Anap/YFP pair under our experimental conditions (Fig. 1 C). The 6% crosstalk efficiency had to be subtracted from the FRET spectrum. To determine the FRET spectrum, the fluorescence spectrum was measured when both labeled BACE1 and KCNQ2/3-YFP channels were present (Fig. 1 B, spectrum in green). The cyan Anap template spectrum in Fig. 1 B was then scaled and subtracted from the green spectrum, yielding the resulting FFRET spectrum (spectrum in red). This subtraction was to correct for the above bleed-through. Furthermore, the peak intensity of the FFRET spectrum was then divided by the peak intensity of the YFP emission spectrum directly excited by the YFP excitation (spectrum in yellow), generating the ratio called ratio A. This noticeable difference between the values of ratio A (Fig. 1 B) and ratio A0 (Fig. 1 C) was proportional to the apparent FRET efficiency detected, suggesting that a population of Anap-labeled BACE1 molecules was in close proximity to KCNQ2/3-YFP channels. Finally, as a negative-control experiment, fluorescence spectra were measured when both the BACE1-L500TAG and KCNQ2/3-YFP channels were cotransfected, but in the absence of the pANAP plasmid (Fig. 1 D). In this case, ratio A had a value similar to ratio A0, suggesting that the nonspecific FRET level was negligible between the free L-Anap in the cell and the KCNQ2/3-YFP.
The calculated apparent FRET efficiency (Eapp) varied with the relative amount of L-Anap-incorporated BACE1 compared with the KCNQ2/3-YFP expressed in each cell (see Materials and methods). Eapp correlated positively with the ratio of Anap-versus-YFP peak fluorescence intensity, and in theory would be saturable to a true FRET efficiency (Emax) at high values of this fluorescence ratio (Takanishi et al., 2006; Fig. 1 E). Such a positive correlation is typical for ensemble FRET with variable fluorophore stoichiometries (Takanishi et al., 2006; Zacharias et al., 2002). With a low expression of the FRET donor BACE1-L500Anap compared with YFP, this relationship looked more linear. Note that the fluorescence ratio F(Anap)/F(YFP) in Fig. 1 E is not equal to the molar ratio of Anap versus YFP, but rather is also dependent on our optical apparatus and on the FRET efficiency, since FRET decreases Anap fluorescence intensity while increasing that of YFP. Essentially, there were different populations of fluorescent molecules affecting the apparent FRET: donors alone (D), acceptors alone (A), and donors paired with acceptors (DA). Because detailed information about stoichiometry, contact site, and affinity of the BACE1 and KCNQ2/3 interaction was lacking, fitting the relationship between the F(Anap)/F(YFP) ratio and the apparent FRET efficiency using published models with model-dependent assumptions would be unsuitable or oversimplified (Bykova et al., 2006; Cheng et al., 2007). Therefore, a nonlinear saturable isotherm was used for an empirical fitting of the data (see Materials and methods; Zacharias et al., 2002). Using this fit, evaluated by χ2 tests, the Emax was estimated at a value of 0.88 between BACE1-L500Anap and KCNQ2/3-YFP, indicating an upper limit of the apparent FRET efficiency (Fig. 1 E). This Emax predicts a distance of 34 Å based on the Förster equation, consistent with a close assembly. Later in the article, we also used this spectral FRET analysis for the CFP and YFP pairs.
The FRET efficiency between BACE1-L500Anap and KCNQ2/3-YFP did not change apparently when the four palmitoylatable cysteines in BACE1 were mutated to alanines (4C/A) or when cells were incubated with the palmitoylation inhibitor 2-BP (Davda et al., 2013; 0.1 mM 2-BP; see conditions indicated by different colors in Fig. 1 E and its legend). Negative-control cells lacking the transfected pANAP plasmid showed negligible FRET (Emax = 0.006), even though visible Anap fluorescence was present due to nonspecific entry of L-Anap to cells. Furthermore, another voltage-gated potassium channel, KCNH (a zebrafish ELK; Dai and Zagotta, 2017), which was considered to be structurally similar to KCNQ but to have no reported interaction with BACE1, showed a smaller FRET (Emax = 0.1). These results, together with previously published work (Hessler et al., 2015), suggest that BACE1 and KCNQ2/3 channels form a protein complex that is relatively independent of the palmitoylation of BACE1.
BACE1-dependent proximity between KCNQ2/3 and the raft protein caveolin 1
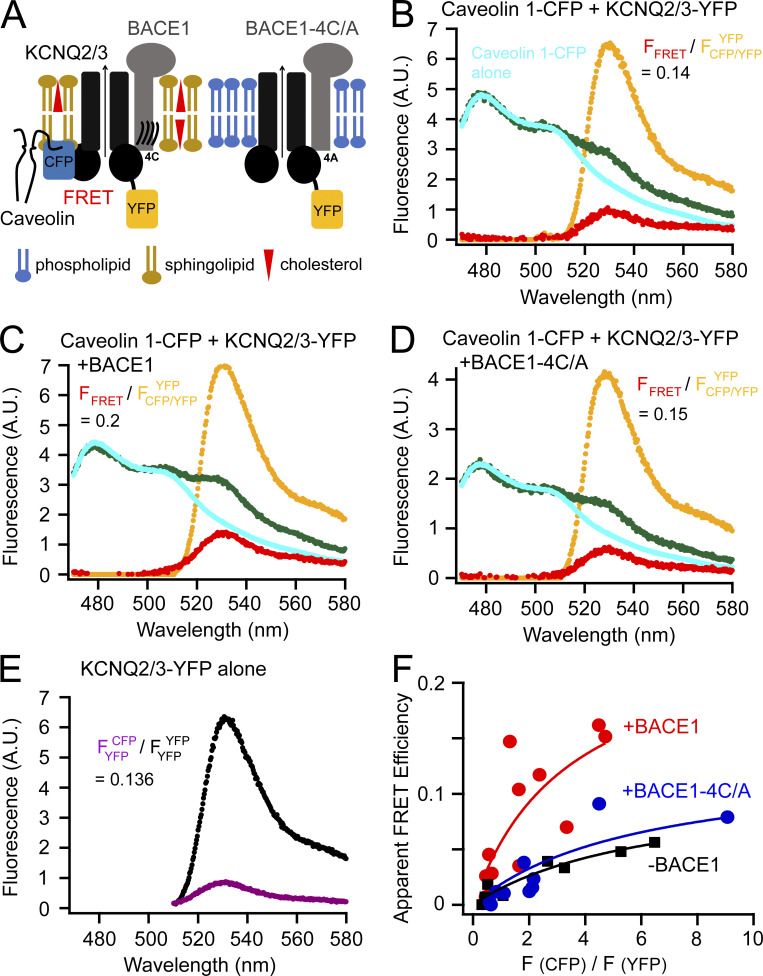
If KCNQ2/3 forms a complex with BACE1, is it localized to lipid rafts? We used the protein caveolin 1, a principal component of caveolae localized to lipid rafts (Li et al., 1996; Williams and Lisanti, 2004; Zacharias et al., 2002), to answer this question. Caveolin 1 often dimerizes at its amino-terminal region and uses the palmitoylation of three cysteines at its carboxyl-terminus to target to lipid rafts (Williams and Lisanti, 2004; Zacharias et al., 2002). A carboxyl-terminal CFP-tagged caveolin 1 was coexpressed with KCNQ2/3-YFP channels in tsA cells, and their FRET was measured using spectral FRET. If BACE1 could recruit KCNQ2/3 channels closer to the lipid-raft microdomains, a higher FRET between KCNQ2/3-YFP and caveolin 1–CFP would be expected (Fig. 2 A). Again, using the same strategy as above, ratio A0 for the CFP/YFP pair was determined to be 0.136 under our experimental conditions (Fig. 2 E), and the apparent FRET efficiency increased as the ratio of CFP/YFP fluorescence increased (Fig. 2 F). The relationship between the apparent FRET efficiency and the CFP/YFP ratio was also fitted empirically using the saturable nonlinear isotherm (Fig. 2 F) to estimate the upper limit of FRET efficiency. Without BACE1, the FRET between caveolin 1–CFP and KCNQ2/3-YFP was noticeable (Emax = 0.13; Fig. 2, B and F). Lacking additional evidence of direct complex formation, it is possible that caveolin 1 and KCNQ2/3 might not interact directly like BACE1 with KCNQ2/3. Instead, they might FRET nonspecifically because of a sufficiently high local density (Takanishi et al., 2006; Zacharias et al., 2002). However, with the coexpression of BACE1, there was a large increase in the FRET intensity (Emax = 0.25; Fig. 2, C and F). In contrast, with coexpression of the BACE1-4C/A mutant construct, there was no increase in FRET, and the relationship between the apparent FRET efficiency and the donor/acceptor intensity ratio superimposed on that seen without BACE1 (Emax = 0.11; Fig. 2, D and F). Comparison of these conditions was best done at a fixed ratio of F(CFP)/F(YFP), so the representative FRET spectra chosen in Fig. 2, B–D, had a similar ratio of CFP/YFP peak fluorescence intensity. Together, these results suggest that, with the help of BACE1, overexpressed KCNQ2/3 channels were drawn to lipid-raft domains where they could FRET more efficiently with raft-specific caveolin 1. Endogenous BACE1 is reported to be present in human embryonic kidney cells (based on the Human Protein Atlas: http://proteinatlas.org), but in our overexpression system, there may have been too little endogenous BACE1 or other scaffold proteins to occlude the actions of additional overexpressed BACE1.
Figure 2.
FRET between caveolin 1–CFP and KCNQ2/3-YFP channels. (A) Cartoon illustrating the probing of the localization of KCNQ2/3 channels using the lipid raft–specific protein caveolin 1. (B–D) Representative emission spectra for calculating FRET between caveolin 1–CFP and KCNQ2/3-YFP. The cyan and green spectra are generated by illumination of the CFP excitation wavelength; the yellow spectrum by YFP excitation; the red spectrum is the subtraction of the cyan spectrum from the green spectrum. The equations with the numbers shown indicate ratio A. (E) Representative emission spectrum of KCNQ2/3-YFP alone cells using direct YFP excitation versus the spectrum (smaller amplitude) from the same sample by the excitation for CFP. The number following the equation is ratio A0. (F) Relationship between the apparent FRET efficiency and the ratio of peak intensities of CFP versus YFP for FRET between KCNQ2/3 and caveolin 1 in the conditions with or without BACE1 or with BACE1-4C/A.
Using fluorescent probes that target lipid-raft and non-raft domains
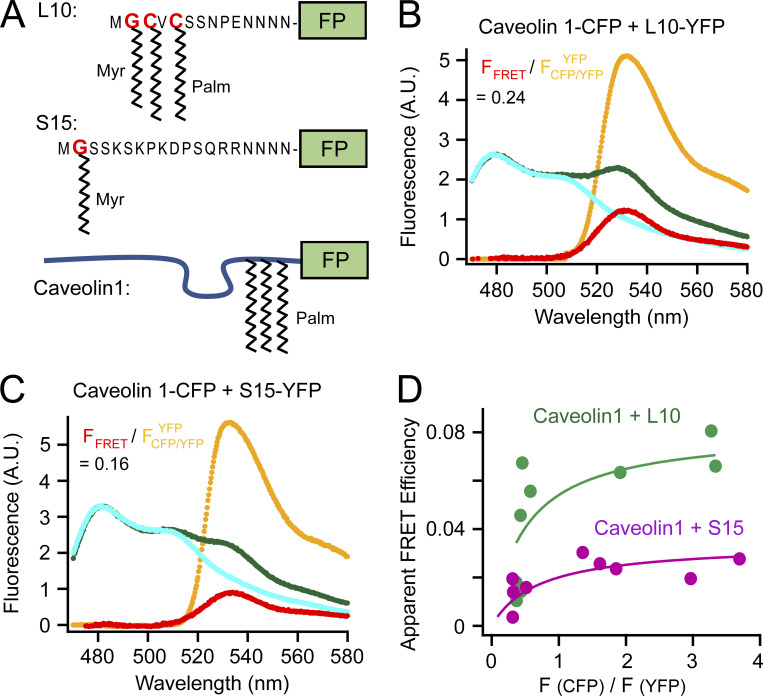
To further study the lipid raft localization of KCNQ2/3 channels, we turned to two fluorescent probes that can be targeted to liquid-ordered and -disordered domains at the plasma membrane (Fig. 3 A). The L10 probe derives from the first 10 amino acids of the amino terminus of Lck kinase, which contains two palmitoylation sites for localizing to the ordered lipid-raft domains (Chichili and Rodgers, 2007; Myeong et al., 2021; Rodgers, 2002). In contrast, the S15 probe derives from the first 15 amino acids from the amino terminus of Src kinase, which contains one myristoylation site, for localizing to the disordered non-raft domains (Chichili and Rodgers, 2007; Myeong et al., 2021; Rodgers, 2002).
Figure 3.
Fluorescent probes that target to lipid rafts and non-rafts. (A) Schematic illustration of L10, S15, and caveolin 1. Sites of myristoylation and palmitoylation are shown. The Poly-N sequence is the flexible linker connecting to a fluorescent protein (FP), CFP or YFP in this article. (B) Representative emission spectra for FRET between caveolin 1–CFP and L10-YFP. (C) Representative emission spectra for FRET between caveolin 1–CFP and S15-YFP. (D) Relationship between the apparent FRET efficiency and the ratio of peak intensity of CFP versus YFP for the FRET pairs shown in B and C.
Validation experiments were first performed comparing the FRET between the caveolin 1–CFP and L10-YFP pair to the FRET between the caveolin 1–YFP and S15-YFP FRET pair (Fig. 3, B–D). As expected, FRET between L10 and caveolin 1 was much higher than that between S15 and caveolin 1, consistent with the L10 probes localizing to raft microdomains where caveolin 1 is abundant, while S15 localize to non-raft areas (Myeong et al., 2021; Zacharias et al., 2002). Plotting the relationship between the apparent FRET efficiency versus the CFP/YFP ratio suggested a noticeable saturation curve, which might indicate a clustering of fluorescent molecules in a small area (Zacharias et al., 2002). Because lipid rafts are considered small (<250 nm in diameter), conventional fluorescence microscopy that is limited by the diffraction limit of light cannot visualize them directly. Our FRET method provides an alternative to measure the colocalization of proteins in small raft areas in a relatively high-throughput manner.
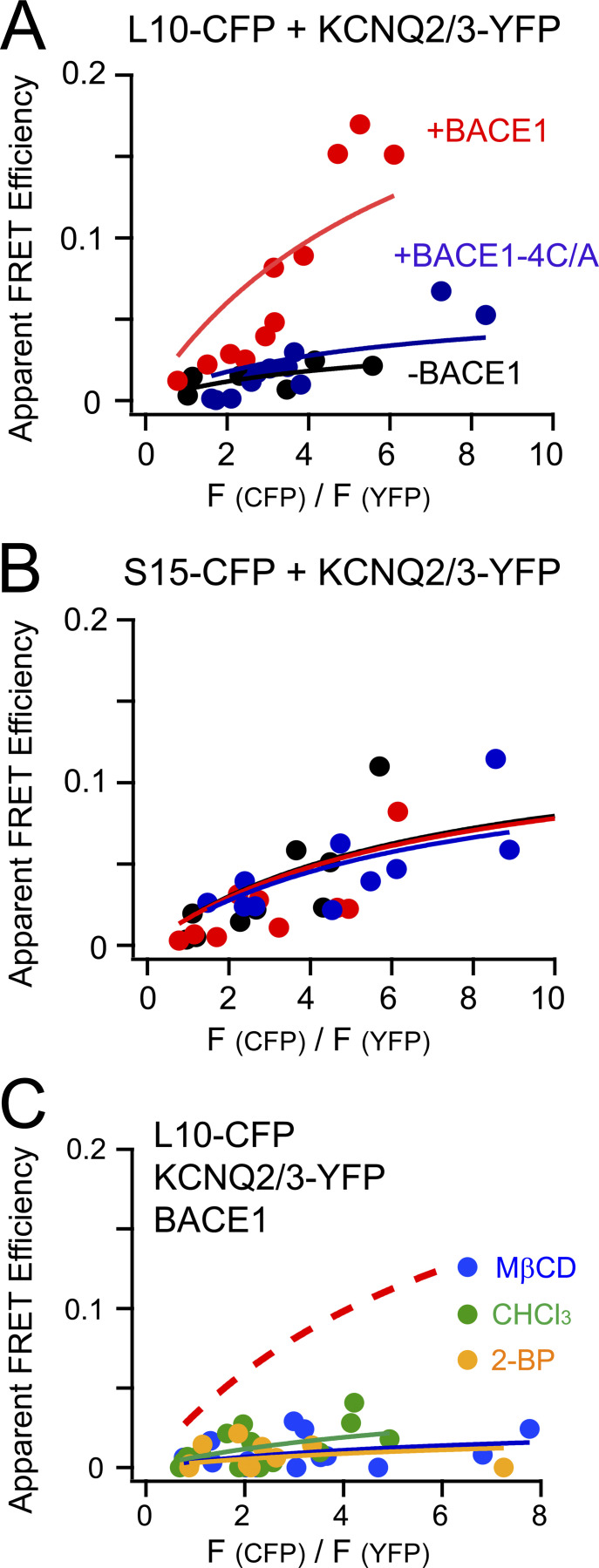
Next, L10 and S15 probes were used to test for possible colocalization with KCNQ2/3 channels and learn how it might be regulated by BACE1. L10-CFP probes were coexpressed with KCNQ2/3-YFP channels without BACE1. The FRET between L10-CFP and KCNQ2/3-YFP was small (Emax = 0.04; Fig. 4 A, black curve). Coexpression of BACE1 significantly elevated the FRET (Emax = 0.27, red curve), whereas coexpression of the mutant BACE1-4C/A did not (Emax = 0.06, blue curve). However, using the same experimental settings, neither BACE1 nor the mutant BACE1-4C/A construct had an effect on the FRET between S15-CFP and KCNQ2/3-YFP (Fig. 4 B). In addition, a higher FRET efficiency for KCNQ2/3-YFP with S15-CFP (Emax = 0.14) than with L10-CFP (Emax = 0.04)—without the coexpression of BACE1—suggests that KCNQ2/3-YFP channels by themselves were more distant from L10 than from S15. In other words, in the absence of BACE1, KCNQ2/3 might be distributed more in the non-raft areas, at least in our overexpression system, possibly due to their lack of known palmitoylation sites. Nevertheless, other fluorescence methods such as superresolution microscopy would help to further test this idea. In addition, our conclusion is predicated on the assumption that CFP fluorophores in L10 and S15 probes have comparable distances to the membrane and dipole orientations. To summarize, overexpressed KCNQ2/3 channels initially showed somewhat greater proximity with S15 than with L10 considering that endogenous BACE1 levels are low; however, overexpressing BACE1 apparently increased the localization of KCNQ2/3 channels to lipid rafts. With BACE1, KCNQ2/3 showed higher FRET with L10 than with S15, presumably due to a direct interaction between KCNQ and BACE1. Mutating the four cysteines in BACE1 to alanine eliminated this effect, corroborating a critical role of palmitoylation for the lipid raft localization of both BACE1 and its partner KCNQ2/3.
Figure 4.
Using L10 and S15 to probe the localization of KCNQ2/3 channels. (A and B) Relationship between the apparent FRET efficiency and the ratio CFP/YFP for FRET between the L10-CFP (A) or S15-CFP (B) and the KCNQ2/3-YFP in the conditions with or without BACE1 or with BACE1-4C/A. (C) Relationship between the apparent FRET efficiency and the ratio CFP/YFP for FRET between L10-CFP and KCNQ2/3-YFP after treating cells with acutely applied 5 mM MβCD, 1 mM chloroform, and 0.1 mM 2-BP. The no-treatment control condition, which is the same fitted curve as in the red solid curve in A, is shown as the dashed curve for clarity.
Several additional pharmacological manipulations were applied to cells that showed high FRET between L10-CFP and KCNQ2/3-YFP in the presence of wild-type BACE1 (Fig. 4 C). We applied cholesterol-extracting methyl-β-cyclodextrin (MβCD) in the medium, which could efficiently decrease plasma membrane cholesterol levels and reduce the relative fraction of raft areas (Mahammad and Parmryd, 2015). MβCD application (5 mM for 5–10 min) decreased the FRET between L10-CFP and KCNQ2/3-YFP considerably, suggesting that this FRET is dependent on the domains as well as on cholesterol that helps to maintain the rafts (Fig. 4 C). Inhaled general anesthetics also disrupt lipid rafts at anesthetic concentrations (Pavel et al., 2020). We found that one general anesthetic, chloroform (1 mM), dramatically suppressed the FRET between L10-CFP and KCNQ2/3-YFP, similar to the effect of MβCD. In addition, 2-BP (0.1 mM), an inhibitor of palmitoylation, abolished the FRET, consistent with a role of palmitoylation in targeting membrane proteins to lipid microdomains.
Discussion
Using FRET as a spectroscopic distance ruler, we found that KCNQ2/3 channels are in proximity to lipid-raft markers and raft-specific proteins. We propose that neuronal KCNQ2/3 channels are localized to or near the ordered membrane microdomain lipid rafts. This localization is achieved by the interaction between KCNQ2/3 and BACE1 and is dependent on the S-palmitoylation of four cysteines in the BACE1 carboxyl terminus. Without BACE1, KCNQ2/3 channels at the plasma membrane might be distributed more in the disordered non-raft areas, at least in our heterologous expression system. Palmitoylation alters ion channel localization and affects KCNQ channels indirectly via their auxiliary protein BACE1 in this case. This type of ion channel regulation by S-palmitoylation is distinct from the direct lipidation and anchoring of ion-channel domains to lipid bilayers, thereby changing the gating rearrangements for the channel opening; for instance, palmitoylation of the P2X7 ATP receptor prevents its desensitization (McCarthy et al., 2019). However, this indirect regulation of KCNQ2/3 is reminiscent of the inhibition of the cardiac sodium/potassium pump by palmitoylation of the auxiliary phosphoprotein phospholemman (Howie et al., 2014).
Palmitoylation manifests an important way for localizing cell signaling ranging from cellular excitability to massive endocytosis (Hilgemann et al., 2018; Linder and Deschenes, 2007). For membrane proteins, S-palmitoylation is reversible and controlled by Asp-His-His-Cys acyltransferases (e.g., plasma membrane DHHC5), thought to be abundant in ordered lipid microdomains (Howie et al., 2014; Linder and Deschenes, 2007). The palmitoylation dependence of the KCNQ2/3 localization to rafts suggests that the channel function could also be indirectly affected by acyltransferases. Specifically, it is reasonable to predict that the KCNQ channel function could be modulated by cellular signals that are upstream of DHHC, like generation of reactive oxygen species (Gamper et al., 2006), opening of mitochondrial permeability transition pores, coenzyme A release, etc. (Hilgemann et al., 2018; Lin et al., 2013). Furthermore, in native neuronal environments, KCNQ2/3 channels are localized to axons by interacting with palmitoylated ankyrin G, which itself could possibly recruit KCNQ2/3 channels to lipid rafts (Chung et al., 2006; He et al., 2012). This ankyrin G interaction could also facilitate the interaction of KCNQ2/3 with BACE1. The possible synergistic effect of the palmitoylation of KCNQ2/3-associated proteins including ankyrin G, BACE1, and AKAP79 remains elusive.
This research provides a new function for the raft localization of BACE1, perhaps independent of the proteolytic or amyloidogenic features of BACE1. Previous research has shown that abolishing the raft localization of BACE1 reduces amyloid production and mitigates memory deficits in transgenic mouse models of AD (Andrew et al., 2017). Conversely, targeting BACE1 to lipid rafts by a glycosylphosphatidylinositol anchor elevates the β-site processing of APP (Cordy et al., 2003). In addition, a recent study shows that the targeted deletion of astrocyte cholesterol synthesis significantly reduces the amyloid burden in a mouse AD model, by decreasing the APP presence in lipid rafts where β- and γ-secretases are localized and functioning (Wang et al., 2021). Despite these findings, this role of the raft localization of BACE1 for APP processing is still debated; other research has suggested that the Aβ production was still present even in the absence of the palmitoylation-dependent targeting of BACE1 to lipid rafts (Vetrivel et al., 2009). Here, the raft localization of BACE1 for the KCNQ2/3 channel function could play a role in reducing neuronal electrical excitability (Carver et al., 2020), modulating afterhyperpolarization potentials (Gu et al., 2005; Tzingounis and Nicoll, 2008), and maintaining the proper propagation of nerve spikes (Yue and Yaari, 2004). It remains to be determined whether KCNQ2/3 channel activity affects the ability of BACE1 to cleave APP and generate Aβ peptides reciprocally.
Disruption of lipid rafts is proposed to underlie the mechanism for the medical effect of inhaled general anesthetics (Pavel et al., 2020). Lipid-soluble general anesthetics change the lipid organization of sphingomyelin- and cholesterol-rich rafts and displace phospholipase D from rafts to non-raft areas, which in turn generates phosphatidic acid that activates K2P (TREK) ion channels (Pavel et al., 2020). Here, applying chloroform was able to decrease the FRET between KCNQ2/3 with the raft marker L10, indicating that general anesthetics similarly displace KCNQ2/3 to non-raft areas and could possibly alter the activity of the channels. Future research will be needed to further understand the detailed structural mechanism and the electrophysiological effects underlying the modulation of KCNQ2/3 channels by BACE1, including whether and how signaling lipids such as PI(4,5)P2 and other AD-related molecular components could play regulatory roles.
Acknowledgments
Jeanne M. Nerbonne served as editor.
I thank Drs. Bertil Hille and William N. Zagotta (University of Washington School of Medicine) for support, sharing equipment, and advice on the manuscript; Dr. Donald W. Hilgemann (University of Texas Southwestern Medical Center) for helpful advice; Lea M. Miller for technical support; and Jongyun Myeong and other members of the Hille lab and the Zagotta lab for discussion and suggestions.
This work was supported by grants from the National Institutes of Health (R37NS08174 and R01EY010329).
The author declares no competing financial interests.
Author contributions: G. Dai conceptualized research, designed and performed experiments, analyzed data, and wrote the manuscript.
References
- Abbott, G.W. 2020. KCNQs: Ligand- and voltage-gated potassium channels. Front. Physiol. 11:583. 10.3389/fphys.2020.00583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrew, R.J., Fernandez C.G., Stanley M., Jiang H., Nguyen P., Rice R.C., Buggia-Prevot V., De Rossi P., Vetrivel K.S., Lamb R., et al. 2017. Lack of BACE1 S-palmitoylation reduces amyloid burden and mitigates memory deficits in transgenic mouse models of Alzheimer’s disease. Proc. Natl. Acad. Sci. USA. 114:E9665–E9674. 10.1073/pnas.1708568114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biervert, C., Schroeder B.C., Kubisch C., Berkovic S.F., Propping P., Jentsch T.J., and Steinlein O.K.. 1998. A potassium channel mutation in neonatal human epilepsy. Science. 279:403–406. 10.1126/science.279.5349.403 [DOI] [PubMed] [Google Scholar]
- Brown, D.A., and Adams P.R.. 1980. Muscarinic suppression of a novel voltage-sensitive K+ current in a vertebrate neurone. Nature. 283:673–676. 10.1038/283673a0 [DOI] [PubMed] [Google Scholar]
- Buggia-Prevot, V., Fernandez C.G., Riordan S., Vetrivel K.S., Roseman J., Waters J., Bindokas V.P., Vassar R., and Thinakaran G.. 2014. Axonal BACE1 dynamics and targeting in hippocampal neurons: A role for Rab11 GTPase. Mol. Neurodegener. 9:1. 10.1186/1750-1326-9-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bykova, E.A., Zhang X.D., Chen T.Y., and Zheng J.. 2006. Large movement in the C terminus of CLC-0 chloride channel during slow gating. Nat. Struct. Mol. Biol. 13:1115–1119. 10.1038/nsmb1176 [DOI] [PubMed] [Google Scholar]
- Carver, C.M., Hastings S.D., Cook M.E., and Shapiro M.S.. 2020. Functional responses of the hippocampus to hyperexcitability depend on directed, neuron-specific KCNQ2 K+ channel plasticity. Hippocampus. 30:435–455. 10.1002/hipo.23163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatterjee, A., Guo J., Lee H.S., and Schultz P.G.. 2013. A genetically encoded fluorescent probe in mammalian cells. J. Am. Chem. Soc. 135:12540–12543. 10.1021/ja4059553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng, W., Yang F., Takanishi C.L., and Zheng J.. 2007. Thermosensitive TRPV channel subunits coassemble into heteromeric channels with intermediate conductance and gating properties. J. Gen. Physiol. 129:191–207. 10.1085/jgp.200709731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chichili, G.R., and Rodgers W.. 2007. Clustering of membrane raft proteins by the actin cytoskeleton. J. Biol. Chem. 282:36682–36691. 10.1074/jbc.M702959200 [DOI] [PubMed] [Google Scholar]
- Chung, H.J., Jan Y.N., and Jan L.Y.. 2006. Polarized axonal surface expression of neuronal KCNQ channels is mediated by multiple signals in the KCNQ2 and KCNQ3 C-terminal domains. Proc. Natl. Acad. Sci. USA. 103:8870–8875. 10.1073/pnas.0603376103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole, S.L., and Vassar R.. 2007. The Alzheimer’s disease β-secretase enzyme, BACE1. Mol. Neurodegener. 2:22. 10.1186/1750-1326-2-22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordy, J.M., Hooper N.M., and Turner A.J.. 2006. The involvement of lipid rafts in Alzheimer’s disease. Mol. Membr. Biol. 23:111–122. 10.1080/09687860500496417 [DOI] [PubMed] [Google Scholar]
- Cordy, J.M., Hussain I., Dingwall C., Hooper N.M., and Turner A.J.. 2003. Exclusively targeting β-secretase to lipid rafts by GPI-anchor addition up-regulates β-site processing of the amyloid precursor protein. Proc. Natl. Acad. Sci. USA. 100:11735–11740. 10.1073/pnas.1635130100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornell, C.E., Mileant A., Thakkar N., Lee K.K., and Keller S.L.. 2020. Direct imaging of liquid domains in membranes by cryo-electron tomography. Proc. Natl. Acad. Sci. USA. 117:19713–19719. 10.1073/pnas.2002245117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai, G., Aman T.K., DiMaio F., and Zagotta W.N.. 2019. The HCN channel voltage sensor undergoes a large downward motion during hyperpolarization. Nat. Struct. Mol. Biol. 26:686–694. 10.1038/s41594-019-0259-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai, G., Aman T.K., DiMaio F., and Zagotta W.N.. 2021. Electromechanical coupling mechanism for activation and inactivation of an HCN channel. Nat. Commun. 12:2802. 10.1038/s41467-021-23062-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai, G., Yu H., Kruse M., Traynor-Kaplan A., and Hille B.. 2016. Osmoregulatory inositol transporter SMIT1 modulates electrical activity by adjusting PI(4,5)P2 levels. Proc. Natl. Acad. Sci. USA. 113:E3290–E3299. 10.1073/pnas.1606348113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai, G., and Zagotta W.N.. 2017. Molecular mechanism of voltage-dependent potentiation of KCNH potassium channels. Elife. 6:e26355. 10.7554/eLife.26355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davda, D., El Azzouny M.A., Tom C.T., Hernandez J.L., Majmudar J.D., Kennedy R.T., and Martin B.R.. 2013. Profiling targets of the irreversible palmitoylation inhibitor 2-bromopalmitate. ACS Chem. Biol. 8:1912–1917. 10.1021/cb400380s [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Fede, G., Catania M., Morbin M., Rossi G., Suardi S., Mazzoleni G., Merlin M., Giovagnoli A.R., Prioni S., Erbetta A., et al. 2009. A recessive mutation in the APP gene with dominant-negative effect on amyloidogenesis. Science. 323:1473–1477. 10.1126/science.1168979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehehalt, R., Keller P., Haass C., Thiele C., and Simons K.. 2003. Amyloidogenic processing of the Alzheimer β-amyloid precursor protein depends on lipid rafts. J. Cell Biol. 160:113–123. 10.1083/jcb.200207113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erickson, M.G., Alseikhan B.A., Peterson B.Z., and Yue D.T.. 2001. Preassociation of calmodulin with voltage-gated Ca2+ channels revealed by FRET in single living cells. Neuron. 31:973–985. 10.1016/s0896-6273(01)00438-x [DOI] [PubMed] [Google Scholar]
- Gamper, N., Zaika O., Li Y., Martin P., Hernandez C.C., Perez M.R., Wang A.Y., Jaffe D.B., and Shapiro M.S.. 2006. Oxidative modification of M-type K+ channels as a mechanism of cytoprotective neuronal silencing. EMBO J. 25:4996–5004. 10.1038/sj.emboj.7601374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon, S.E., Munari M., and Zagotta W.N.. 2018. Visualizing conformational dynamics of proteins in solution and at the cell membrane. Elife. 7:e37248. 10.7554/elife.37248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu, N., Vervaeke K., Hu H., and Storm J.F.. 2005. Kv7/KCNQ/M and HCN/h, but not KCa2/SK channels, contribute to the somatic medium after-hyperpolarization and excitability control in CA1 hippocampal pyramidal cells. J. Physiol. 566:689–715. 10.1113/jphysiol.2005.086835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He, M., Jenkins P., and Bennett V.. 2012. Cysteine 70 of ankyrin-G is S-palmitoylated and is required for function of ankyrin-G in membrane domain assembly. J. Biol. Chem. 287:43995–44005. 10.1074/jbc.M112.417501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez, C.C., Zaika O., Tolstykh G.P., and Shapiro M.S.. 2008. Regulation of neural KCNQ channels: Signalling pathways, structural motifs and functional implications. J. Physiol. 586:1811–1821. 10.1113/jphysiol.2007.148304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hessler, S., Zheng F., Hartmann S., Rittger A., Lehnert S., Volkel M., Nissen M., Edelmann E., Saftig P., Schwake M., et al. 2015. β-Secretase BACE1 regulates hippocampal and reconstituted M-currents in a β-subunit-like fashion. J. Neurosci. 35:3298–3311. 10.1523/jneurosci.3127-14.2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilgemann, D.W., Dai G., Collins A., Lariccia V., Magi S., Deisl C., and Fine M.. 2018. Lipid signaling to membrane proteins: From second messengers to membrane domains and adapter-free endocytosis. J. Gen. Physiol. 150:211–224. 10.1085/jgp.201711875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howie, J., Reilly L., Fraser N.J., Vlachaki Walker J.M., Wypijewski K.J., Ashford M.L., Calaghan S.C., McClafferty H., Tian L., Shipston M.J., et al. 2014. Substrate recognition by the cell surface palmitoyl transferase DHHC5. Proc. Natl. Acad. Sci. USA. 111:17534–17539. 10.1073/pnas.1413627111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu, X., Hicks C.W., He W., Wong P., Macklin W.B., Trapp B.D., and Yan R.. 2006. BACE1 modulates myelination in the central and peripheral nervous system. Nat. Neurosci. 9:1520–1525. 10.1038/nn1797 [DOI] [PubMed] [Google Scholar]
- Jonsson, T., Atwal J.K., Steinberg S., Snaedal J., Jonsson P.V., Bjornsson S., Stefansson H., Sulem P., Gudbjartsson D., Maloney J., et al. 2012. A mutation in APP protects against Alzheimer’s disease and age-related cognitive decline. Nature. 488:96–99. 10.1038/nature11283 [DOI] [PubMed] [Google Scholar]
- Kalstrup, T., and Blunck R.. 2013. Dynamics of internal pore opening in Kv channels probed by a fluorescent unnatural amino acid. Proc. Natl. Acad. Sci. USA. 110:8272–8277. 10.1073/pnas.1220398110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keith, D.J., Sanderson J.L., Gibson E.S., Woolfrey K.M., Robertson H.R., Olszewski K., Kang R., El-Husseini A., and Dell’acqua M.L.. 2012. Palmitoylation of A-kinase anchoring protein 79/150 regulates dendritic endosomal targeting and synaptic plasticity mechanisms. J. Neurosci. 32:7119–7136. 10.1523/JNEUROSCI.0784-12.2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenworthy, A.K., and Edidin M.. 1998. Distribution of a glycosylphosphatidylinositol-anchored protein at the apical surface of MDCK cells examined at a resolution of <100 Å using imaging fluorescence resonance energy transfer. J. Cell Biol. 142:69–84. 10.1083/jcb.142.1.69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, D.Y., Carey B.W., Wang H., Ingano L.A., Binshtok A.M., Wertz M.H., Pettingell W.H., He P., Lee V.M., Woolf C.J., and Kovacs D.M.. 2007. BACE1 regulates voltage-gated sodium channels and neuronal activity. Nat. Cell Biol. 9:755–764. 10.1038/ncb1602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klinger, F., Gould G., Boehm S., and Shapiro M.S.. 2011. Distribution of M-channel subunits KCNQ2 and KCNQ3 in rat hippocampus. Neuroimage. 58:761–769. 10.1016/j.neuroimage.2011.07.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi, D., Zeller M., Cole T., Buttini M., McConlogue L., Sinha S., Freedman S., Morris R.G., and Chen K.S.. 2008. BACE1 gene deletion: Impact on behavioral function in a model of Alzheimer’s disease. Neurobiol. Aging. 29:861–873. 10.1016/j.neurobiolaging.2007.01.002 [DOI] [PubMed] [Google Scholar]
- Kojro, E., Gimpl G., Lammich S., Marz W., and Fahrenholz F.. 2001. Low cholesterol stimulates the nonamyloidogenic pathway by its effect on the α-secretase ADAM 10. Proc. Natl. Acad. Sci. U S A. 98:5815–5820. 10.1073/pnas.081612998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehnert, S., Hartmann S., Hessler S., Adelsberger H., Huth T., and Alzheimer C.. 2016. Ion channel regulation by β-secretase BACE1–enzymatic and non-enzymatic effects beyond Alzheimer’s disease. Channels (Austin). 10:365–378. 10.1080/19336950.2016.1196307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, S., Song K.S., and Lisanti M.P.. 1996. Expression and characterization of recombinant caveolin. Purification by polyhistidine tagging and cholesterol-dependent incorporation into defined lipid membranes. J. Biol. Chem. 271:568–573. [PubMed] [Google Scholar]
- Lin, M.J., Fine M., Lu J.Y., Hofmann S.L., Frazier G., and Hilgemann D.W.. 2013. Massive palmitoylation-dependent endocytosis during reoxygenation of anoxic cardiac muscle. Elife. 2:e01295. 10.7554/eLife.01295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linder, M.E., and Deschenes R.J.. 2007. Palmitoylation: Policing protein stability and traffic. Nat. Rev. Mol. Cell Biol. 8:74–84. 10.1038/nrm2084 [DOI] [PubMed] [Google Scholar]
- Lingwood, D., and Simons K.. 2010. Lipid rafts as a membrane-organizing principle. Science. 327:46–50. 10.1126/science.1174621 [DOI] [PubMed] [Google Scholar]
- Lombardo, S., Chiacchiaretta M., Tarr A., Kim W., Cao T., Sigal G., Rosahl T.W., Xia W., Haydon P.G., Kennedy M.E., and Tesco G.. 2019. BACE1 partial deletion induces synaptic plasticity deficit in adult mice. Sci. Rep. 9:19877. 10.1038/s41598-019-56329-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahammad, S., and Parmryd I.. 2015. Cholesterol depletion using methyl-β-cyclodextrin. Methods Mol. Biol. 1232:91–102. 10.1007/978-1-4939-1752-5_8 [DOI] [PubMed] [Google Scholar]
- Matsubara, T., IIjima K., Kojima T., Hirai M., Miyamoto E., and Sato T.. 2021. Heterogeneous ganglioside-enriched nanoclusters with different densities in membrane rafts detected by a peptidyl molecular probe. Langmuir. 37:646–654. 10.1021/acs.langmuir.0c02387 [DOI] [PubMed] [Google Scholar]
- McCarthy, A.E., Yoshioka C., and Mansoor S.E.. 2019. Full-length P2X7 structures reveal how palmitoylation prevents channel desensitization. Cell. 179:659–670.e613. 10.1016/j.cell.2019.09.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myeong, J., Park C.G., Suh B.C., and Hille B.. 2021. Compartmentalization of phosphatidylinositol 4,5-bisphosphate metabolism into plasma membrane liquid-ordered/raft domains. Proc. Natl. Acad. Sci. USA. 118:e2025343118. 10.1073/pnas.2025343118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nahmad-Rohen, A., Regan D., Masia F., McPhee C., Pope I., Langbein W., and Borri P.. 2020. Quantitative label-free imaging of lipid domains in single bilayers by hyperspectral coherent Raman scattering. Anal Chem. 92:14657–14666. 10.1021/acs.analchem.0c03179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ou-Yang, M.H., Kurz J.E., Nomura T., Popovic J., Rajapaksha T.W., Dong H., Contractor A., Chetkovich D.M., Tourtellotte W.G., and Vassar R.. 2018. Axonal organization defects in the hippocampus of adult conditional BACE1 knockout mice. Sci. Transl Med. 10. 10.1126/scitranslmed.aao5620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pastorino, L., Ikin A.F., Nairn A.C., Pursnani A., and Buxbaum J.D.. 2002. The carboxyl-terminus of BACE contains a sorting signal that regulates BACE trafficking but not the formation of total Aβ. Mol. Cell Neurosci. 19:175–185. 10.1006/mcne.2001.1065 [DOI] [PubMed] [Google Scholar]
- Pavel, M.A., Petersen E.N., Wang H., Lerner R.A., and Hansen S.B.. 2020. Studies on the mechanism of general anesthesia. Proc. Natl. Acad. Sci. USA. 117:13757–13766. 10.1073/pnas.2004259117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perl, D.P. 2010. Neuropathology of Alzheimer’s disease. Mt Sinai J. Med. 77:32–42. 10.1002/msj.20157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pike, L.J. 2004. Lipid rafts: Heterogeneity on the high seas. Biochem. J. 378:281–292. 10.1042/BJ20031672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson, C.V., Rohacs T., and Hansen S.B.. 2019. Tools for understanding nanoscale lipid regulation of ion channels. Trends Biochem. Sci. 44:795–806. 10.1016/j.tibs.2019.04.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodgers, W. 2002. Making membranes green: Construction and characterization of GFP-fusion proteins targeted to discrete plasma membrane domains. Biotechniques. 32:1044–1046. 10.2144/02325st05 [DOI] [PubMed] [Google Scholar]
- Shah, M., Mistry M., Marsh S.J., Brown D.A., and Delmas P.. 2002. Molecular correlates of the M-current in cultured rat hippocampal neurons. J. Physiol. 544:29–37. 10.1113/jphysiol.2002.028571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simons, K., and Ikonen E.. 1997. Functional rafts in cell membranes. Nature. 387:569–572. 10.1038/42408 [DOI] [PubMed] [Google Scholar]
- Singh, N.A., Charlier C., Stauffer D., DuPont B.R., Leach R.J., Melis R., Ronen G.M., Bjerre I., Quattlebaum T., Murphy J.V., et al. 1998. A novel potassium channel gene, KCNQ2, is mutated in an inherited epilepsy of newborns. Nat. Genet. 18:25–29. 10.1038/ng0198-25 [DOI] [PubMed] [Google Scholar]
- Sun, J., and MacKinnon R.. 2020. Structural basis of human KCNQ1 modulation and gating. Cell. 180:340–347.e349. 10.1016/j.cell.2019.12.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takanishi, C.L., Bykova E.A., Cheng W., and Zheng J.. 2006. GFP-based FRET analysis in live cells. Brain Res. 1091:132–139. 10.1016/j.brainres.2006.01.119 [DOI] [PubMed] [Google Scholar]
- Toh, W.H., Chia P.Z.C., Hossain M.I., and Gleeson P.A.. 2018. GGA1 regulates signal-dependent sorting of BACE1 to recycling endosomes, which moderates Aβ production. Mol. Biol. Cell. 29:191–208. 10.1091/mbc.e17-05-0270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzingounis, A.V., and Nicoll R.A.. 2008. Contribution of KCNQ2 and KCNQ3 to the medium and slow afterhyperpolarization currents. Proc. Natl. Acad. Sci. U S A. 105:19974–19979. 10.1073/pnas.0810535105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vassar, R., and Kandalepas P.C.. 2011. The β-secretase enzyme BACE1 as a therapeutic target for Alzheimer’s disease. Alzheimers Res. Ther. 3:20. 10.1186/alzrt82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vassar, R., Kuhn P.H., Haass C., Kennedy M.E., Rajendran L., Wong P.C., and Lichtenthaler S.F.. 2014. Function, therapeutic potential and cell biology of BACE proteases: Current status and future prospects. J. Neurochem. 130:4–28. 10.1111/jnc.12715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vetrivel, K.S., Meckler X., Chen Y., Nguyen P.D., Seidah N.G., Vassar R., Wong P.C., Fukata M., Kounnas M.Z., and Thinakaran G.. 2009. Alzheimer disease Aβ production in the absence of S-palmitoylation-dependent targeting of BACE1 to lipid rafts. J. Biol. Chem. 284:3793–3803. 10.1074/jbc.m808920200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vossel, K.A., Tartaglia M.C., Nygaard H.B., Zeman A.Z., and Miller B.L.. 2017. Epileptic activity in Alzheimer’s disease: Causes and clinical relevance. Lancet Neurol. 16:311–322. 10.1016/S1474-4422(17)30044-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, H., Kulas J.A., Wang C., Holtzman D.M., Ferris H.A., and Hansen S.B.. 2021. Regulation of beta-amyloid production in neurons by astrocyte-derived cholesterol. Proc. Natl. Acad. Sci. USA. 118. e2102191118. 10.1073/pnas.2102191118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, H.S., Pan Z., Shi W., Brown B.S., Wymore R.S., Cohen I.S., Dixon J.E., and McKinnon D.. 1998. KCNQ2 and KCNQ3 potassium channel subunits: Molecular correlates of the M-channel. Science. 282:1890–1893. 10.1126/science.282.5395.1890 [DOI] [PubMed] [Google Scholar]
- Weller, J., and Budson A.. 2018. Current understanding of Alzheimer’s disease diagnosis and treatment. F1000Res. 7:F1000. 10.12688/f1000research.14506.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams, T.M., and Lisanti M.P.. 2004. The caveolin proteins. Genome Biol. 5:214. 10.1186/gb-2004-5-3-214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yue, C., and Yaari Y.. 2004. KCNQ/M channels control spike afterdepolarization and burst generation in hippocampal neurons. J. Neurosci. 24:4614–4624. 10.1523/JNEUROSCI.0765-04.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zacharias, D.A., Violin J.D., Newton A.C., and Tsien R.Y.. 2002. Partitioning of lipid-modified monomeric GFPs into membrane microdomains of live cells. Science. 296:913–916. 10.1126/science.1068539 [DOI] [PubMed] [Google Scholar]
- Zagotta, W.N., Gordon M.T., Senning E.N., Munari M.A., and Gordon S.E.. 2016. Measuring distances between TRPV1 and the plasma membrane using a noncanonical amino acid and transition metal ion FRET. J. Gen. Physiol. 147:201–216. 10.1085/jgp.201511531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, J., and Shapiro M.S.. 2016. Mechanisms and dynamics of AKAP79/150-orchestrated multi-protein signalling complexes in brain and peripheral nerve. J. Physiol. 594:31–37. 10.1113/jphysiol.2014.287698 [DOI] [PMC free article] [PubMed] [Google Scholar]