Abstract
Delta1, Jagged1, and Jagged2, commonly designated Delta/Serrate/LAG-2 (DSL) proteins, are known to be ligands for Notch1. However, it has been less understood whether they are ligands for Notch receptors other than Notch1. Meanwhile, ligand-induced cleavage and nuclear translocation of the Notch protein are considered to be fundamental for Notch signaling, yet direct observation of the behavior of the Notch molecule after ligand binding, including cleavage and nuclear translocation, has been lacking. In this report, we investigated these issues for Notch2. All of the three DSL proteins bound to endogenous Notch2 on the surface of BaF3 cells, although characteristics of Jagged2 for binding to Notch2 apparently differed from that of Delta1 and Jagged1. After binding, the three DSL proteins induced cleavage of the membrane-spanning subunit of Notch2 (Notch2TM), which occurred within 15 min. In a simultaneous time course, the cleaved fragment of Notch2TM was translocated into the nucleus. Interestingly, the cleaved Notch2 fragment was hyperphosphorylated also in a time-dependent manner. Finally, binding of DSL proteins to Notch2 also activated the transcription of reporter genes driven by the RBP-Jκ-responsive promoter. Together, these data indicate that all of these DSL proteins function as ligands for Notch2. Moreover, the findings of rapid cleavage, nuclear translocation, and phosphorylation of Notch2 after ligand binding facilitate the understanding of the Notch signaling.
The Notch family of proteins consists of transmembrane receptors that play a critical role in the determination of cell fate (1, 14, 60). Multiple Notch homologs have been described in higher vertebrates, including Notch1 through Notch4 in rodents and humans (9, 27, 30, 46, 56, 61, 62), although only one Notch gene has been identified in Drosophila melanogaster (63). The basic structure of the Drosophila and mammalian Notch proteins comprises 29 to 36 epidermal growth factor (EGF)-like repeats and three copies of a Lin-12/Notch/Glp motif in the extracellular region and cdc10/Ankyrin repeats and a PEST-containing domain in the intracellular region. Much information concerning the function of Notch has been provided by experiments using truncated proteins consisting only of the intracellular domain of Notch, which have constitutive transducing activity (33, 52). Studies with the truncated form of Notch1 provide evidence that Notch1 regulates differentiation in various types of cells (26, 39, 42) and the T-cell lineage decision (45, 47, 59). Finding that the truncated forms of Notch1 and Notch2 inhibit myeloid differentiation in response to different cytokines has revealed a functional diversity between the two Notch molecules (3). Recent analyses of phenotype in Notch1 and Notch2 knockout mice have also supported that these molecules have an individual role (5, 16, 53).
The functional form of the mammalian Notch receptor is a heterodimeric molecule composed of two cleavage products associated at the cell surface (4). Truncated forms of Notch that consist only of the intracellular domain localize predominantly in the nucleus (13, 26, 33, 52). It has therefore been proposed that ligand binding to Notch induces cleavage to cause the release of the intracellular domain of Notch, which subsequently translocates into the nucleus, where it activates the transcription of genes such as HES-1 in cooperation with RBP-Jκ (18, 19, 43). Ligand-dependent cleavage of an intracellular domain of Drosophila Notch or mammalian Notch1 has been previously demonstrated in vivo and in tissue culture (24, 47), but nuclear translocation has not been demonstrated directly. While ligand-dependent nuclear translocation of the Notch intracellular domain has been shown using an in vivo reporter assay (51), it was uncertain whether it was a result of the proteolytic cleavage. For better understanding of the Notch signaling, it is desirable to trace behavior of the Notch molecule after ligand stimulation in an experimental system capable of observing both cleavage and nuclear translocation.
Two proteins, Delta and Serrate, characterized by a common structure, a Delta/Serrate/Lag-2 (DSL) domain, have been shown to be natural ligands for the Notch receptor in Drosophila (12, 28). Serrate can compensate for loss-of-function mutations of Delta, at least in part (15). Similarly, vertebrate counterparts of the two Drosophila genes, Delta and Serrate, have been identified: Delta-like-1 (Delta1) in mice and chickens (2, 17) and Delta-like-3 (Dll3) in mice (8); Delta2 in Xenopus laevis (20); and the potential orthologs of Serrate, Jagged1, and Jagged2 in rats, mice, and humans (32, 36, 37, 49, 50, 57). It is known that Delta1, Jagged1, and Jagged2 are expressed at different sites and time points during embryogenesis (10, 31, 49, 58), that their phenotypes in knockout mice are different from each other (6, 21, 53, 64), and that each of the three DSL proteins can transduce, at least, a Notch1 signal (19, 32, 36, 37). However, it has been poorly understood whether any of the mammalian DSL proteins act as a ligand for Notch receptors other than Notch1, except a report showing that Jagged1 inhibits myocytic differentiation of C2C12 cells through exogenous Notch2 (41). Owing to the existence of a functional diversity between the Notch receptors as described above, further investigation of the receptor-ligand relationships between the DSL proteins and the Notch receptors has been required for full understanding of the biology of the Notch system in mammals.
We report here that the three DSL proteins, Delta1, Jagged1, and Jagged2, act as functional ligands for Notch2. Our results show that binding of these ligands to Notch2 rapidly triggers cleavage of Notch2 and that the cleaved fragment translocates into the nucleus. Unlike Delta1 or Jagged1, Jagged2 exhibits unique characteristics in binding to Notch2, suggesting the existence of a mechanism regulating its binding to Notch2 at the cell surface. Furthermore, we reveal that Notch2 is hyperphosphorylated in response to binding of DSL proteins.
MATERIALS AND METHODS
Plasmid constructions.
cDNA for mouse Delta1 was a kind gift from A. Gossler. cDNA for mouse Jagged1 was isolated as described (50). cDNA for mouse Jagged2 was originally isolated from a mouse embryo cDNA library. The sequence of the entire coding region was verified, and fragments were assembled. cDNA for mouse Notch2 was as described elsewhere (50). Soluble Jagged1 fusion with an Fc portion of human immunoglobulin G (IgG) (Fc) or a Flag(His)6 sequence and soluble Notch2 tagging with a Flag(His)6 sequence were as described previously (50). To generate soluble Delta1 and Jagged2, their cDNAs were truncated at the codon CAT corresponding to histidine (amino acid 535) and at the codon GGT corresponding to glycine (amino acid 1083), respectively. cDNA for Fc or a Flag(His)6 sequence was fused to the 3′ end of the truncated cDNAs, respectively. To generate full-length DSL proteins and Notch2 protein with a Flag(His)6 tag, a Flag(His)6 sequence was fused in frame to the 3′-end coding region of mouse Delta1, Jagged1, Jagged2, and Notch2 cDNAs. These cDNAs were constructed in an expression vector pTraserCMV (Clontech).
Antibodies.
For Western blot analyses using the Flag(His)6-tagged proteins, an anti-Flag monoclonal antibody (M2; Eastman Kodak) was used at a dilution of 1:15,000. An alkaline phosphatase-conjugated anti-mouse secondary antibody (Promega) was used at a dilution of 1:5,000. For the cell-binding assay using Fc-fused proteins, a phycoerythrin (PE)-conjugated anti-human Fc antibody (Chimicon) was used at a dilution of 1:200. A rabbit anti-Notch2 polyclonal antibody was described elsewhere (50) and used at a dilution of 1:1,000 for immunoprecipitation. For Western blot analyses of Notch2, an anti-Notch2 monoclonal antibody (bhN6) (65) recognizing the intracellular domain of Notch2 was used at a dilution of 1:20. An anti-insulin receptor β (IRβ) antibody (Santa Cruz), an anti-Jun D antibody (Santa Cruz), and an anti-insulin receptor substrate-2 antibody (Upstate Biotechnology) were used at a dilution of 1:500.
Cell culture.
BaF3 was maintained in RPMI medium supplemented with 10% fetal bovine serum (FBS) and 0.5 ng of recombinant mouse interleukin-3 (a gift from Kirin Brewery, Takasaki, Japan) per ml. CHO ras clone-I [CHO(r)] (22) was maintained in alpha-minimal essential medium containing 10% FBS.
Preparation of soluble fusion proteins.
Soluble Jagged1 proteins [sJ1-Fc and sJ1-Flag(His)6] were prepared as described previously (50). The same protocol was used to prepare soluble Delta1 proteins [sD1-Fc and sD1-Flag(His)6] and the Jagged2 protein (sJ2-Fc).
Cell-binding assay.
Binding of each soluble DSL protein to the surface of a pro-B cell line BaF3 was performed as previously described (50). Briefly, 3 × 105 BaF3 cells were incubated with 33 nM concentrations of various soluble DSL proteins in cell-binding buffer (phosphate-buffered saline [PBS] containing 2% FBS, 100 μg of CaCl2 per ml, and 0.05% NaN3) at 37°C after blocking with 5 μl of rabbit serum. After 15 min of incubation, the cells were washed three times with the cell-binding buffer and further incubated with a PE-conjugated anti-hIgG antibody. The cells were then analyzed using FACSCaliber (Becton Dickinson Immunocytometry Systems).
Coprecipitation using the soluble DSL proteins.
A total of 107 BaF3 cells was subjected to cell-binding assay in a buffer containing 20 mM HEPES (pH 7.5), 150 mM NaCl, 100 μg of CaCl2 per ml, and a 7 nM concentration of the soluble DSL-Fc. Disuccinyl glutarate (DSG; Pierce), a cross-linking reagent, was then added to the DSL-Fc-bound BaF3 at a final concentration of 20 μM, followed by further incubation for 30 min at room temperature. After the cross-linking reaction, the cells were solubilized in a TNE buffer containing 20 mM Tris-Cl (pH 7.4), 150 mM NaCl, 1.0% NP-40, 5 μg of aprotinin per ml, and 1 mM EDTA for 30 min at 4°C. The lysates were precipitated with protein G-bound beads and then washed four times with TNP buffer and boiled in the sodium dodecyl sulfate sample buffer under a reducing condition. Notch2TM precipitated with each DSL protein was detected by a Western blot probed with an anti-Notch2 antibody.
Cell-cell association assay.
CHO(r) cells with or without various exogenous DSL proteins were inoculated at 106 in a 12-well plate. After overnight culture, 106 BaF3 cells were spread over the CHO(r) cells. After 15 min of coculture at 37°C, BaF3 cells which did not anchor to the CHO(r) cell layer were collected by very gently swirling the plate and washing the wells once gently with RPMI medium. The population obtained through this procedure was determined to be nonadhered BaF3. Next, PBS containing 2 mM EGTA was added to the wells, and BaF3 cells adhering to the CHO(r) cell layer were allowed to dissociate by tapping the plate. These BaF3 cells together with additional cells collected by washing with RPMI medium were determined as adhered BaF3. The cell number in each fraction was then determined.
Coprecipitation using membrane-bound DSL proteins.
DSL-CHO(r) cells with various exogenous DSL proteins were inoculated at 6 × 106 in a 10-cm plate. After overnight culture, 2 × 107 of BaF3 cells were added and allowed to bind to DSL-CHO(r) in a buffer containing 20 mM HEPES (pH 7.5), 150 mM NaCl, and 100 μg of CaCl2 per ml at room temperature for 15 min. DSG was then added to the mixture at a final concentration of 20 μM, followed by further incubation for 30 min. Following the cross-linking reaction, the cells were solubilized with a TNE buffer, and the lysates then immunoprecipitated with an anti-Flag monoclonal or anti-Notch2 polyclonal antibody.
Transient-transcription assay.
Two kinds of cells were transiently transfected with various reporter plasmids and used as target cells: BaF3 and CHO(r) stably expressing or not expressing exogenous full-length Notch2 (fN2-CHO). CHO(r) cells stably expressing or not expressing various exogenous full-length DSL proteins were therefore used as stimulators. BaF3 cells at 2 × 105 were transfected with a TP1-luciferase reporter plasmid, pGa981-6 (40), by an electroporation method under conditions of 250 V and 960 μF. After electropolation, the transfected BaF3 cells were spread over a monolayer of stimulator cells inoculated at 2 × 105 in a 24-well plate for 24 h prior to coculture and then incubated for 24 h. The BaF3 cells were recovered and used for luciferase assay. When CHO(r) cells were used as a target, 4 × 104 CHO(r) cells with or without exogenous Notch2 were inoculated into a 24-well plate and transfected with either of the three reporter plasmids, pGa981-6, pHES1-luc, and pHES5-luc (40, 54, 55), by a liposome-based method (SuperFect; Qiagen). Following transfection, stimulator CHO(r) cells at 5 × 104 were added and cocultured for 40 h. The mixture of two kinds of CHO(r) cells was then used for the luciferase assay.
Preparation of membrane-cytosol-rich and nucleus-rich fractions from BaF3.
After coculturing with the stimulator CHO(r) cells, BaF3 was collected and suspended in 10 mM HEPES (pH 7.4), 10 mM NaCl, 1.5 mM MgCl2, 1 mM EDTA, 1 mM phenylmethylsulfonyl fluoride, 5 μg of aprotinin per ml, and 10 μg of pepstatin per ml (Sol A). The chilled cell suspension was then homogenized using a homogenizer (Dounce) and pelleted out by centrifugation. The supernatant was mixed with an equivalent volume of ice-chilled Sol B (identical to Sol A but with 500 mM NaCl and 1% NP-40) and allowed to stand on ice for 30 min. After centrifugation at 15,000 rpm for 10 min, the clear supernatant was preserved and used as a membrane-cytosol-rich fraction. The pellet of the homogenized cell suspension was suspended in Sol C (identical to Sol A but with 500 mM NaCl), gently rotated for 30 min at 4°C and centrifuged at 15,000 rpm for 10 min. The clear supernatant was collected as a nucleus-rich fraction. To adjust the contents in the fraction to the membrane-cytosol-rich fraction, an equivalent volume of Sol D (identical to Sol A but with 1% NP-40) was added. These two fractions were used for further immunoprecipitation with a polyclonal anti-Notch2 antibody.
Treatment with alkaline phosphatase.
Protein G-bound beads carrying Notch2 fragments precipitated from the membrane-rich and nucleus-rich fractions were washed with alkaline phosphatase buffer, suspended in the same buffer, and then divided into two aliquots. Bacterial alkaline phosphatase (Takara) was added to one of the two aliquots. Both aliquots were incubated at 37°C for 30 min and then analyzed by Western blotting with bhN6 antibody as probe.
RESULTS
Binding of full-length DSL proteins to Notch2 on the cell surface.
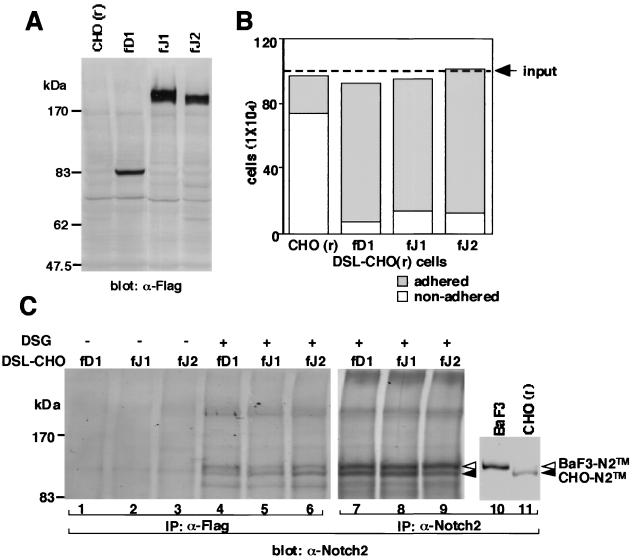
We generated three types of CHO(r) cell lines expressing the full-length DSL proteins, which were tagged in frame with a Flag sequence at the C terminus (Fig. 1A). These lines were designated as fD1-CHO, fJ1-CHO, and fJ2-CHO. We first tested the physical binding of the three DSL-CHO(r) lines to BaF3 cells by a cell-cell association assay in which the number of adhering and nonadhering BaF3 cells was counted after overlay onto the DSL-CHO(r) cell layer. Results showed that most of the overlaid BaF3 cells anchored to the CHO(r) cells expressing the full-length DSL protein within 10 min (Fig. 1B). In contrast, most of the BaF3 cells overlaid on the parental CHO(r) cells did not adhere to the monolayer (Fig. 1B). Among the three DSL-CHO(r) cells, fD1-CHO had the strongest BaF3-anchoring capacity, followed by fJ2-CHO and fJ1-CHO. The binding of all DSL-CHO(r) to BaF3 was Ca2+ dependent (data not shown).
FIG. 1.
Binding of full-length DSL proteins to Notch2 on the BaF3 cell surface. (A) Generation of three kinds of DSL-CHO(r) cell lines expressing full-length Delta1, Jagged1, and Jagged2. Expression of each full-length DSL protein was verified by Western blot analysis. fD1, fD1-CHO; fJ1, fJ1-CHO; fJ2, fJ2-CHO. (B) Binding of three DSL-CHO(r) cells to BaF3 was examined in a cell-cell association assay. adhered, BaF3 which adhered to CHO cells; nonadhered, BaF3 which did not adhere to CHO cells. (C) Binding of three membrane-bound DSL proteins to Notch2 on the BaF3 cell surface was verified by the methods described for coprecipitation using membrane-bound DSL proteins in Materials and Methods in the absence (lanes 1 to 3) or presence (lanes 4 to 9) of the cross-linking reagent DSG. To identify the Notch2 protein fragments, these lysates were precipitated with an anti-Notch2 rabbit polyclonal antibody (lanes 7 to 9). To show the size difference between BaF3-Notch2TM and CHO-Notch2TM, lysates of these cells were separately precipitated with an anti-Notch2 rabbit polyclonal antibody (lanes 10 and 11). BaF3-Notch2TM, BaF3-derived Notch2TM; CHO-Notch2TM, CHO(r)-derived Notch2TM.
In our previous work, we showed that Notch2 is the major protein detected among Notch family proteins expressed in BaF3 (50). It was therefore expected that Notch2 would be responsible for the adhesion of BaF3 to the DSL-CHO(r) cells through DSL proteins present on their cell surface. To verify this, immunoprecipitation with an anti-Flag antibody against the lysates of BaF3 and each DSL-CHO(r), which were allowed to cross-link after binding, was performed (Fig. 1C). Results showed that the BaF3-derived, intracellular domain-containing membrane-spanning subunit of Notch2 (Notch2TM) was precipitated by all of the DSL proteins in the presence of a cross-linking reagent (Fig. 1C, cf. lanes 1 and 4, lanes 2 and 5, and lanes 3 and 6). The amount of coprecipitated BaF3-derived Notch2 fragment differed among DSL proteins, with Delta1 being greatest, followed by Jagged2 and Jagged1 in this order. The order corresponds to the result of the cell-cell association assay.
Binding of soluble DSL proteins to Notch2 on the cell surface.
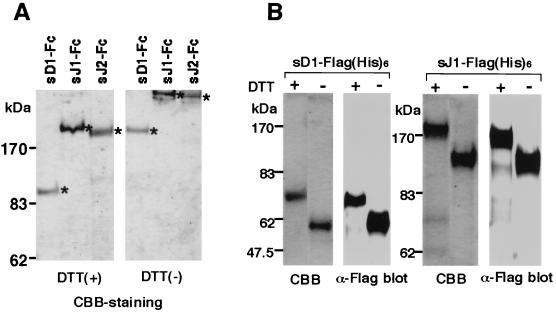
To better understand the binding of the DSL proteins to Notch2, several experiments were further performed using their soluble forms. We recently established a system for assessing the binding of Jagged1 to live cells using the soluble Jagged1 protein (sJ1) (cell-binding assay) in which it was shown that sJ1 protein binds to Notch2 on the BaF3 cell surface (50). The full-length extracellular domains of Delta1 (sD1) and Jagged2 (sJ2), in addition to Jagged1, were tagged with an Fc portion of hIgG (Fc) or a Flag(His)6 sequence at the C terminus to generate sD1-Fc, sJ1-Fc, sJ2-Fc, sD1-Flag(His)6 and sJ1-Flag(His)6. These proteins were produced in a stable expression system using Chinese hamster ovary Ras [CHO(r)] cells. Proteins were purified using protein G- or Ni-bound beads. The purity of the three Fc-fused and two Flag-tagged proteins was confirmed by Coomassie brilliant blue staining to be >95 and >90%, respectively (Fig. 2). The difference seen in their migration position under reducing and non-reducing conditions indicated that sD1-Fc, sJ1-Fc, and sJ2-Fc were dimerized at the Fc portion (Fig. 2A).
FIG. 2.
Preparation of soluble DSL proteins comprising a full-length extracellular region tagged with Fc or Flag(His)6. Three kinds of Fc-fused DSL proteins and two kinds of Flag(His)6-tagged DSL proteins produced in CHO(r) cells were purified with protein G or Ni beads, respectively. Integrity and purity were verified by Coomassie brilliant blue (CBB) staining in reducing and nonreducing conditions for sD1-Fc, sJ1-Fc, and sJ2-Fc (A) and by CBB staining and Western blot for sD1-Flag(His)6 and sJ1-Flag(His)6 (B).
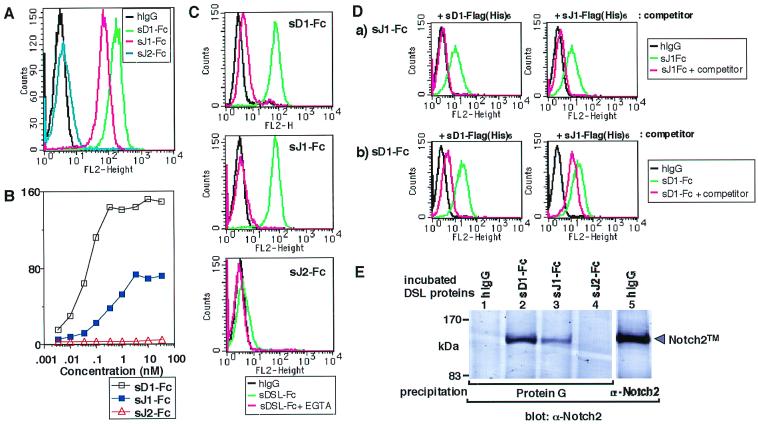
We tested the binding of the Fc-fused soluble DSL proteins to BaF3 by the cell-binding assay. Results showed that each DSL-Fc protein bound to BaF3 in a concentration-dependent manner (Fig. 3A and B), with sD1-Fc having the highest affinity, followed by sJ1-Fc and sJ2-Fc in that order (Fig. 3B). The binding activity of sJ2-Fc was obviously weak and irrelevant to the efficient binding capacity in the membrane-bound form of Jagged2 (cf. Fig. 1 and Fig. 3B). The addition of EGTA abolished the binding of both sD1-Fc and sJ2-Fc, as shown previously in the case of sJ1-Fc (50), indicating that these interactions are dependent on Ca2+ (Fig. 3C). The binding of sJ1-Fc to BaF3 was cancelled by the addition of a 500-fold molar excess of sD1-Flag(His)6 used as a competitor (Fig. 3Da). A 500-fold molar excess of sJ1-Flag(His)6 inversely reduced binding of sD1-Fc to BaF3, though in an incomplete fashion (Fig. 3Db). These results suggest that Delta1 shares the same binding site with Jagged1 in the Notch2 receptor on the cell surface.
FIG. 3.
Soluble Delta1, Jagged1, and Jagged2 protein binding to Notch2 present on the cell surface. (A) Three DSL-Fc proteins were allowed to bind to BaF3 at the same molar concentration (33 nM). As a control, the same concentration of hIgG was used. (B) Binding of increasing concentrations of three DSL proteins to BaF3. The extent of fluorescence brightness that gives the highest frequency (vertical axis) was plotted against the concentration of sD1-Fc, sJ1-Fc, and sJ2-Fc (horizontal axis). (C) Requirement of Ca2+ in binding of soluble DSL proteins to BaF3. BaF3 was incubated with each DSL protein in the absence (green) or presence (red) of 2 mM EGTA. As a control, hIgG was incubated with the cells in the absence of EGTA (black). (D) Self-displacement and reciprocal displacement of soluble Delta1 and soluble Jagged1. (a) Displacement of sJ1-Fc binding to BaF3 by a 500-fold molar excess of sD1-Flag(His)6 or sJ1-Flag(His)6. (b) Displacement of sD1-Fc binding to BaF3 by a 500-fold molar excess of sD1-Flag(His)6 or sJ1-Flag(His)6. (E) Coprecipitation analysis. DSL-Fc (lane 1) and hIgG (lane 5) in cell-binding buffer were allowed to bind to BaF3. Protein G beads were then added directly to the BaF3 lysate to precipitate DSL-Fc-containing complex (lanes 1 to 4). To identify Notch2 protein fragments, the BaF3 lysate was precipitated with an anti-Notch2 rabbit polyclonal antibody (lanes 5). These precipitates were analyzed by a Western blot probing with a Notch2-specific monoclonal antibody. Size marker protein positions are shown on the left. Bands of approximate sizes of 120 kDa represent the membrane-spanning subunit (Notch2TM). fN2, full-length Notch2; Protein G, precipitation with Protein G alone.
Next, to confirm whether the soluble form of DSL proteins bound to BaF3 through Notch2, a coprecipitation experiment was performed using protein G against the DSL-Fc proteins in the binding complex. Results showed that the Notch2TM fragments were most strongly coprecipitated with sD1-Fc, followed by sJ1-Fc (Fig. 3E). The Notch2TM fragments were undetectable in the precipitate with sJ2-Fc (Fig. 3E). These data closely reflect the results of the cell-binding assay as shown in Fig. 3A and B.
All DSL proteins act as ligands for Notch2: induction of cleavage, hyperphosphorylation, and nuclear accumulation of the intracellular domain of Notch2.
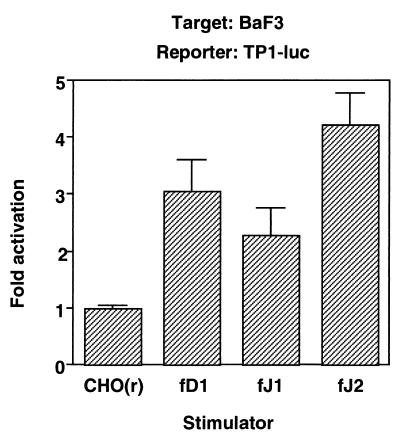
Next, we sought to determine whether the full-length DSL proteins were able to transduce a signal through Notch2. We performed a transient reporter assay, in which BaF3 was transiently transfected with a reporter plasmid, PGa981-6, and then cocultured with the parental or full-length DSL protein-expressing CHO(r) lines. pGa981-6 is a reporter plasmid containing multiple repeats of an RBP-Jκ-binding sequence from the TP1 promoter of Epstein-Barr virus (40). The results showed that luciferase activities were similarly induced by stimulation with any DSL protein (Fig. 4).
FIG. 4.
Transduction of a Notch signal in BaF3 by full-length DSL proteins. BaF3 cells at 2 × 105 were transiently transfected with the PGa981-6 plasmid and were spread over a monolayer of CHO(r) cells stably expressing or not expressing various exogenous full-length DSL proteins. fD1, fD1-CHO; fJ1, fJ1-CHO; fJ2, fJ2-CHO.
Concerning the mechanism of activation of a Notch receptor by a ligand, it is generally accepted that the ligand binding to Notch induces cleavage, causing the release of the intracellular domain of Notch which then translocates into the nucleus (24, 48, 51). From this point of view, we further evaluated their abilities to transduce a Notch2 signal. To see ligand-dependent nuclear translocation of Notch2, BaF3 was lysed and separated into membrane-cytosol-rich and nucleus-rich protein fractions (MC fraction and N fraction, respectively) after coculturing with the parental-CHO(r) and DSL-CHO(r) lines, and Notch2 in each fraction was detected by Western blotting. We confirmed that the MC and N fractions were correctly fractionated using antibodies against proteins representing each fraction (Fig. 5A).
FIG. 5.
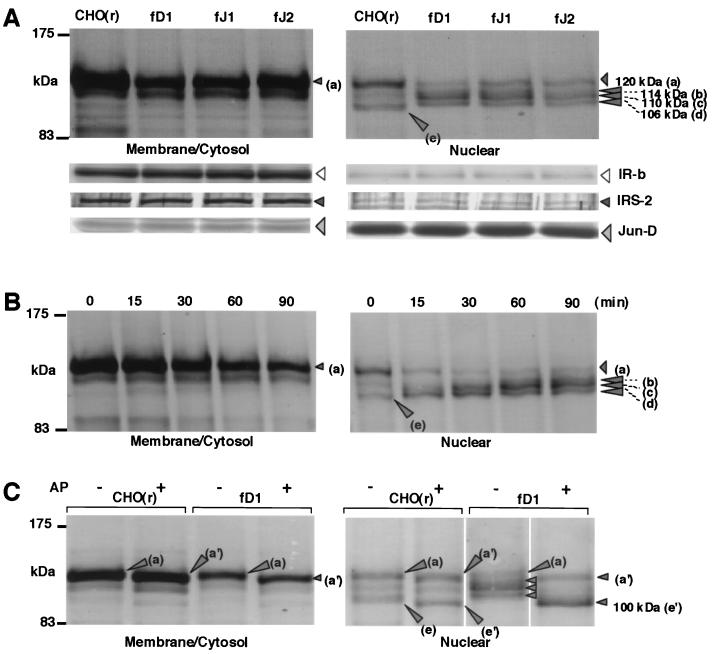
Induction of cleavage, hyperphosphorylation, and nuclear accumulation of the intracellular domain of Notch2 by DSL proteins. (A) Cleavage of the membrane-spanning subunit of Notch2 and nuclear accumulation of intracellular domain of Notch2 by stimulation with various full-length DSL proteins. BaF3 cells were collected 1.5 h after coculture with CHO(r) lines stably expressing or not expressing various exogenous full-length DSL proteins. MC and N fractions were prepared as described in Materials and Methods, and in each fraction Notch2 fragments containing an intracellular domain were analyzed by Western blot using an anti-Notch2 antibody after immunoprecipitation. As controls for correct fractionation of the MC and N fractions, antibodies against IRβ for membrane proteins, insulin receptor substrate-2 (IRS-2) for cytosolic proteins, and Jun-D for nuclear proteins were used for each fraction in Western blot analysis. (B) Time-course analysis of Notch2 after stimulation with fD1-CHO. (C) Effect on Notch2 fragments of alkaline phosphatase (AP) treatment. From each MC and N fraction prepared from BaF3 after coculture with wild-type CHO(r) and fD1-CHO, Notch2 fragments containing the intracellular domain were immunoprecipitated. The samples were then treated by alkaline phosphatase and subjected to Western blotting.
Notch2TM was shown as a 120-kDa band (Fig. 5, a) in the MC fraction. Coculture of BaF3 with the parental CHO(r) did not change Notch2 behavior in either the MC or the N fraction (data not shown). The amount of Notch2TM, however, was significantly reduced in the MC fraction when BaF3 was cocultured with any of the three DSL-CHO(r) cells (Fig. 5A, left). In contrast, at least three new Notch2 fragments of 114 kDa (b), 110 kDa (c), and 106 kDa (d) became visible in the N fraction upon stimulation with any of the DSL proteins (Fig. 5A, right). A small amount of Notch2TM (band a) was also seen in the N fraction and was considered to be a contamination into the N fraction. (Fig. 5A, cf. left and right). A 104-kDa band (e) shown in the N fraction disappeared upon stimulation with any of the DSL proteins (Fig. 5A, right).
Time-course analysis after stimulation with fD1-CHO cells showed a decrease in the amount of Notch2TM (a) and an increase in that of nucleus-accumulating Notch2 fragments (b to d) in a time-dependent manner (Fig. 5B). It was further shown that the proportion of the amount of the three Notch2 fragments (b to d) within the N fraction changed in a time-dependent manner, which was demonstrated by the intensity shifted from the smaller to larger size (Fig. 5B). These biochemical dynamics were observed within 15 min after the start of reaction and reached a plateau at 1 h (Fig. 5B).
To investigate whether the size difference among bands b to d was due to the different phosphorylation status of a single species, we added alkaline phosphatase to both fractions after precipitation. On coculture with the wild-type CHO(r) cells, both Notch2TM (a) and the nuclear Notch2 fragment (e) shifted to slightly smaller species of 117 kDa (a′) and 100 kDa (e′), respectively, in the presence of alkaline phosphatase [Fig. 5C, lanes CHO(r)]. This indicates that both proteins (bands a and e) were slightly phosphorylated without exogenous ligand stimulation. Using fD1-CHO cells in this coculture system, all of the three nucleus-accumulating Notch2 fragments (b to d) shifted to a single 100-kDa protein, apparently represented by the same band as band e′ (Fig. 5C, right, lanes fD1-CHO). This suggests that the three Notch2 fragments were hyperphosphorylated forms of the 104-kDa phosphoprotein.
Activation of the transcription by DSL proteins through exogenous Notch2.
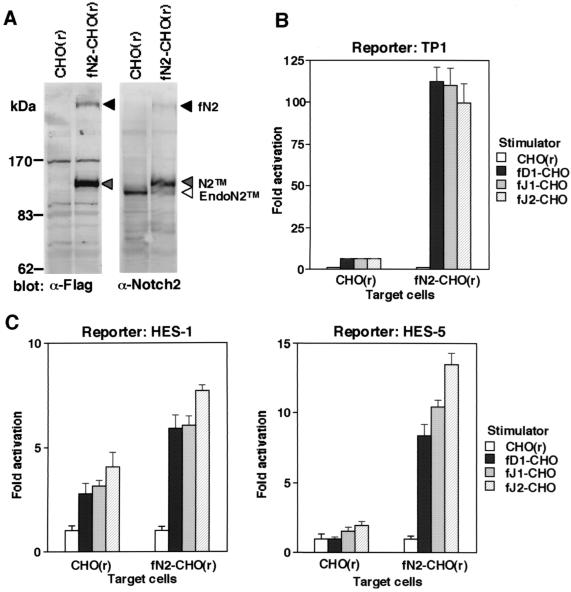
We further investigated the abilities of the three DSL proteins to transduce a signal through Notch2 in a transient reporter assay using full-length Notch2-overexpressing CHO(r) (fN2-CHO). Expression of exogenous Notch2 was detected at 120 kDa (N2TM) and at 300 kDa (fN2), which correspond to Notch2TM and the uncleaved full-length species, respectively (Fig. 6A). When the TP1-luciferase reporter plasmid was used, coculture of fN2-CHO with any of fD1-CHO, fJ1-CHO, and fJ2-CHO increased the luciferase activity by 100- to 120-fold compared to the activity in coculture with the parental CHO(r) cells (Fig. 6B, right). This result was reproducible with another fN2-CHO clone (data not shown). In contrast, when parental CHO(r) was used as a target instead of fN2-CHO, the luciferase activity increased only five- to sixfold with all DSL proteins (Fig. 6B, left). Similar results were obtained when we used reporter plasmids containing a promoter of the HES-1 and HES-5 genes, both of which contain a binding site for RBP-Jκ (Fig. 6C) (54, 55). Truncated proteins consisting only of the intracellular domain of Notch2 also activated the transcription of the reporter genes (data not shown).
FIG. 6.
Activation of the transcription by DSL proteins through exogenous Notch2. (A) Generation of CHO(r) expressing full-length Notch2 (fN2-CHO). Exogenous Notch2 in the fN2-CHO cells was verified by Western blot analyses using anti-Flag and anti-Notch2 monoclonal antibodies. fNotch2, unprocessed full-length Notch2; N2TM, membrane-spanning subunit of Notch2; EndoN2TM, endogenous Notch2TM fragment derived from CHO(r). (B and C) Reporter gene transactivation in CHO(r) with or without exogenous Notch2. CHO(r) cells with or without exogenous Notch2 were inoculated in a 24-well plate and transfected with three reporter plasmids: PGa981-6 (B), pHES1-luc, and pHES5-luc (C). The stimulators, CHO(r) lines stably expressing or not expressing various exogenous full-length DSL proteins, were then cocultured. In both panels B and C, the fold induction of luciferase activity in DSL-CHO(r) (mean of triplicate measurements with standard deviation) was calculated against that found in parental CHO(r) lines used as stimulators.
DISCUSSION
We show here that three mouse DSL proteins, Delta1, Jagged1, and Jagged2, function as ligands for Notch2. This conclusion was reached by studies on binding of the DSL proteins to Notch2 and observation of the ensuing signaling cascade, namely, cleavage of the Notch2 molecule, nuclear translocation of the cleaved Notch2 fragment and transactivation of a reporter gene driven by RBP-Jκ-responsive promoters. Unlike Delta1 or Jagged1, Jagged2 exhibited the unique characteristic that membrane-bound but not soluble Jagged2 efficiently bound to Notch2, suggesting the existence of a Jagged2-specific mechanism for binding to Notch2 at the cell surface. Another novel finding is hyperphosphorylation of the Notch2 fragment, which was rapidly induced by stimulation with the DSL proteins.
Binding features of DSL proteins to Notch2.
To obtain a better insight into binding characteristics of DSL proteins to Notch2, we used a membrane-bound form comprising the full-length and a soluble form of each DSL protein.
The membrane-bound forms of Delta1 and Jagged1 bound to Notch2 on the cell surface of BaF3, but the affinity was somewhat stronger for Delta1 (Fig. 1B and C). The higher binding affinity to Notch2 of Delta1 than Jagged1 was more obvious when soluble DSL proteins were compared (Fig. 3). However, we do not conclude that the affinity of Delta1 is invariably higher than that of Jagged1, since soluble Delta1 and Jagged1 equivalently coprecipitated Notch2 when CHO(r) cells were used instead of BaF3 cells (unpublished data). This discrepancy between the results in BaF3 and CHO(r) cells suggests the existence of a mechanism which modulates the binding of Delta1 and Jagged1 to Notch2 at the cell surface. In this respect, however, a further investigation would be required.
Jagged2 apparently differed from Delta1 and Jagged1 in that the binding activities of the membrane-bound and soluble forms were distinct: the binding activity of soluble Jagged2 was much lower (Fig. 3A and B) or hardly detectable (Fig. 3E), whereas the membrane-bound form of Jagged2 bound to Notch2 on the BaF3 cell surface as efficiently as that of Delta1 and Jagged1 (Fig. 1B and C). Soluble Jagged2 did not coprecipitate Notch2 expressed on the surface of CHO(r) or C2C12 cells (data not shown). The lack in the binding of soluble Jagged2 to Notch2 was not due to incorrect structure of soluble Jagged2, since it bound efficiently to the surface of CHO(r) and C2C12 cells that express Notch1 and Notch3 in addition to Notch2 (data not shown). Moreover, the cell-cell association assay using Jagged2 with a truncated intracellular domain showed that the intracellular domain does not play a significant role in full binding to Notch2 (data not shown). Taken together, these data suggest that expression on the cell surface is necessary for Jagged2 to be fully active in binding to Notch2, although the reason is unknown. One of the possibilities would be that Jagged2 may specifically need the support of a molecule(s) present on the cell surface for full binding to Notch2. In addition, we observed that soluble Jagged2 efficiently bound to soluble Notch1 and Notch3, with very little binding activity to soluble Notch2 (unpublished data). This observation suggests that the unique characteristics of Jagged2 binding described above may not be applicable to Notch1 or Notch3 but may be limited to Notch2.
DSL proteins as functional ligands for Notch2.
The conclusion that the three DSL proteins behave as ligands for Notch2 was drawn from our experimental results as follows. (i) All three DSL proteins activated reporter gene transcription through endogenous Notch receptors in BaF3 cells (Fig. 4), in which full-length Notch mRNAs other than that for Notch2 were not detected (50). (ii) In BaF3 cells, all three DSL proteins induced cleavage and nuclear translocation of the Notch2 molecule (Fig. 5). (iii) Exogenous Notch2 expressed on the CHO(r) cell surface transmitted a signal by the three DSL proteins to generate reporter gene transactivation (Fig. 6).
Interaction between the DSL proteins and Notch2 in vivo has been previously suggested by in situ hybridization analyses, in which a partially overlapping expression pattern was shown between the DSL proteins and Notch2 (35, 36). Integrating this information into our results, any one of the DSL proteins could be a natural ligand for Notch2 in vivo. In addition, given that they also function as ligands for Notch1 (19, 32, 36, 37), it is possible that they may be ligands for all Notch receptors. We have indeed found that they bind to soluble Notch3 protein (unpublished data). Is there then any selectivity for a Notch receptor among these ligands? In the experimental systems described here, we did not detect any major selectivity of these ligands for Notch2. However, we expect that the selectivity would be exhibited on the further involvement of Fringe proteins, a family of putative glycosyltransferases which may confer glycosyl chains to the Notch molecule, on the basis of findings that in the Drosophila Notch system Fringe inhibits Notch activation by Serrate but enhances that by Delta (11, 25, 44). Since three fringe genes are known to exist in mammals (5), regulation of the Notch-ligand system would be more complex. It is possible that a modulation mechanism exists for both the ligand side and the Notch receptor side, since the former has been discussed in the binding characteristics of Jagged2. These matters must be clarified before a full understanding of the Notch-ligand system can be obtained.
Ligand-dependent cleavage and nuclear translocation of Notch2.
Regarding the mechanism by which a Notch receptor is activated by a ligand, several studies have suggested that, upon ligand binding, the transmembrane subunit is cleaved to release the intracellular domain, which then translocates into the nucleus where it behaves as a transcriptional activator. Cleavage of an intracellular domain of Drosophila Notch or mammalian Notch1 in response to ligand has been previously demonstrated in vivo and in tissue culture (24, 48). Ligand-dependent nuclear translocation of the Notch intracellular domain has been also shown using a sensitive β-galactosidase reporter assay in Drosophila (51). However, evidence showing ligand-dependent nuclear translocation of Notch at the protein level has not been available. In addition, there has been no presentation in which the sequence of events, cleavage and the translocation, is clearly shown. In the present study, we succeeded in demonstrating these two events together in a coculture system using DSL-CHO(r) and BaF3 cells. Stimulation with all respective DSL proteins expressed in CHO(r) decreased the amount of Notch2TM in the MC fraction. In contrast, at least three Notch2-derived fragments were accumulated in the N fraction in considerable amounts (Fig. 5A).
These results indicate the cleavage of Notch2TM and subsequent nuclear translocation of the cleaved component. Furthermore, the time-course analysis revealed that both events occurred within 15 min (Fig. 5B), suggesting that we observed their sequential progression. Ligand-dependent cleavage could indeed determine the nuclear translocation of Notch2 present on the cell surface, since Notch2 itself has nuclear localization signals in its intracellular domain. The nuclear translocation of the Notch2 intracellular domain should consecutively connect to the transcriptional activation of the downstream genes (18, 19, 43). The result of a transient reporter assay utilizing CHO(r) cells with or without exogenous Notch2 (Fig. 6C) raises the possibility that HES-1 and HES-5 are such immediate downstream genes of Notch2. Indeed, a previously reported time-course analysis of transcription of HES-1 after stimulation with Delta1 (29) was closely similar to that of nuclear translocation shown here. It is therefore anticipated that transcription is activated rapidly.
Treatment with alkaline phosphatase showed that the three translocated Notch2 fragments were derived from one protein species (Fig. 5C). This suggested that these fragments were generated by cleavage at a unique site, a notion in agreement with the result of experiments using Notch1 protein lacking most of the extracellular portion, which undergoes a unique proteolytic cleavage to release the intracellular domain (48). Presumably, a molecule belonging to the Presenillin family would also participate in the processing of Notch2, as is the case of Notch1 (7), since an inhibitor for Presenillin partially prevented Notch2 from proteolytic cleavage in response to ligand binding (unpublished data).
Ligand-dependent hyperphosphorylation of the Notch2 intracellular domain.
It has been described that the intracellular domain of Drosophila Notch is phosphorylated (24). However, the relationship between ligand stimulation and phosphorylation remains unknown. In this study, treatment with alkaline phosphatase revealed that the Notch2 fragment in the membrane and that in the nucleus were both phosphorylated at a basal level before stimulation (bands a and e in Fig. 5C). In the nucleus, the degree of phosphorylation was much increased, and the amount of Notch2 molecule was significantly increased when incubated with fD1-CHO(r), indicating that Notch2 fragment in the nucleus was hyperphosphorylated in a ligand-dependent fashion. In addition, the increase in the level of phosphorylation in the nucleus was time dependent (Fig. 5B), whereas Notch2 fragment in the MC fraction remained at a basal level of phosphorylation. It has been reported that, in a Drosophila embryo, highly phosphorylated forms of Notch fragment exist in the cytosolic fraction, while the translocated Notch fragment itself is not detected in the N fraction (24), a result inconsistent with our results. Given this observation in Drosophila and the fact that we observed a faint band in the MC fraction (Fig. 5B), it is more likely that phosphorylation of the cleaved Notch2 protein takes place outside the nucleus. Further investigation is required to determine where and how the phosphorylation takes place, particularly given that there are multiple phosphorylation sites in Notch2.
The reason for the discrepancy between our result and that in Drosophila regarding the localization of the phosphorylated Notch fragments is not clear. It could be explained by the proposal that translocation of the cleaved Notch fragments is facilitated by the increased ratio of Notch and Suppressor of Hairless [Su(H)] (the Drosophila counterpart of mammalian RBP-Jκ) (24), i.e., the Notch2/RBP-Jκ ratio in BaF3 might be higher than the Notch/Su(H) ratio in the whole body of the Drosophila embryo.
It is possible that the Notch2 fragment is phosphorylated by casein kinase II, since there are putative consensus sequences for this enzyme in the intracellular domain of Notch2. Since casein kinase II is known to regulate the transcriptional activity of transcription factors (34, 38), hyperphosphorylation of Notch2 may contribute to regulating the function of Notch2, such as transactivation of target genes.
With regard to a Notch2 fragment in the nucleus before ligand stimulation, the fragment may not be generated by constitutive activation through autocrine mechanism, since no DSL proteins are expressed in BaF3 cells (data not shown). Generation of this fragment could be explained by unknown mechanisms, which are yet to be elucidated.
In summary, we have concluded here that the three DSL proteins are ligands for Notch2, wherein binding, proteolytic cleavage, nuclear translocation, and hyperphosphorylation of Notch2 receptor are shown. We believe that these findings contribute to the further understanding of the Notch system.
ACKNOWLEDGMENTS
We thank S. Artavanis-Tsakonas for providing us with the bhN6 anti-Notch2 antibody, A. Gossler for mouse Delta1 cDNA, R. Kageyama for the pHES1-luc and pHES5-luc plasmids, T. Honjo for the pGa981-6 plasmid, and S. Shirahata for the CHO ras clone-I cells. We also thank G. Harris for his review of the manuscript.
This work was supported by grants-in-aid from the Ministry of Education, Science, Sport, and Culture of Japan, and the Ministry of Health and Welfare of Japan.
REFERENCES
- 1.Artavanis-Tsakonas S, Rand M D, Lake R J. Notch signaling: cell fate control and signal integration in development. Science. 1999;284:770–776. doi: 10.1126/science.284.5415.770. [DOI] [PubMed] [Google Scholar]
- 2.Bettenhausen B, Hrabe de Angelis M, Simon D, Guenet J L, Gossler A. Transient and restricted expression during mouse embryogenesis of Dll1, a murine gene closely related to Drosophila Delta. Development. 1995;121:2407–2418. doi: 10.1242/dev.121.8.2407. [DOI] [PubMed] [Google Scholar]
- 3.Bigas A, Martin D I, Milner L A. Notch1 and Notch2 inhibit myeloid differentiation in response to different cytokines. Mol Cell Biol. 1998;18:2324–2333. doi: 10.1128/mcb.18.4.2324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blaumueller C M, Qi H, Zagouras P, Artavanis-Tsakonas S. Intracellular cleavage of Notch leads to a heterodimeric receptor on the plasma membrane. Cell. 1997;90:281–291. doi: 10.1016/s0092-8674(00)80336-0. [DOI] [PubMed] [Google Scholar]
- 5.Cohen B, Bashirullah A, Dagnino L, Campbell C, Fisher W W, Leow C C, Whiting E, Ryan D, Zinyk D, Boulianne G, Hui C C, Gallie B, Phillips R A, Lipshitz H D, Egan S E. Fringe boundaries coincide with Notch-dependent patterning centres in mammals and alter Notch-dependent development in Drosophila. Nat Genet. 1997;16:283–288. doi: 10.1038/ng0797-283. [DOI] [PubMed] [Google Scholar]
- 6.Conlon R A, Reaume A G, Rossant J. Notch1 is required for the coordinate segmentation of somites. Development. 1995;121:1533–1545. doi: 10.1242/dev.121.5.1533. [DOI] [PubMed] [Google Scholar]
- 7.De Strooper B, Annaert W, Cupers P, Saftig P, Craessaerts K, Mumm J S, Schroeter E H, Schrijvers V, Wolfe M S, Ray W J, Goate A, Kopan R. A presenilin-1-dependent gamma-secretase-like protease mediates release of Notch intracellular domain. Nature. 1999;398:518–522. doi: 10.1038/19083. [DOI] [PubMed] [Google Scholar]
- 8.Dunwoodie S L, Henrique D, Harrison S M, Beddington R S. Mouse Dll3: a novel divergent Delta gene which may complement the function of other Delta homologues during early pattern formation in the mouse embryo. Development. 1997;124:3065–3076. doi: 10.1242/dev.124.16.3065. [DOI] [PubMed] [Google Scholar]
- 9.Ellisen L W, Bird J, West D C, Soreng A L, Reynolds T C, Smith S D, Sklar J. TAN-1, the human homolog of the Drosophila notch gene, is broken by chromosomal translocations in T lymphoblastic neoplasms. Cell. 1991;66:649–661. doi: 10.1016/0092-8674(91)90111-b. [DOI] [PubMed] [Google Scholar]
- 10.Felli M P, Maroder M, Mitsiadis T A, Campese A F, Bellavia D, Vacca A, Mann R S, Frati L, Lendahl U, Gulino A, Screpanti I. Expression pattern of notch1, 2 and 3 and Jagged1 and 2 in lymphoid and stromal thymus components: distinct ligand-receptor interactions in intrathymic T cell development. Int Immunol. 1999;11:1017–1025. doi: 10.1093/intimm/11.7.1017. [DOI] [PubMed] [Google Scholar]
- 11.Fleming R J, Gu Y, Hukriede N A. Serrate-mediated activation of Notch is specifically blocked by the product of the gene fringe in the dorsal compartment of the Drosophila wing imaginal disc. Development. 1997;124:2973–2981. doi: 10.1242/dev.124.15.2973. [DOI] [PubMed] [Google Scholar]
- 12.Fleming R J, Scottgale T N, Diederich R J, Artavanis-Tsakonas S. The gene Serrate encodes a putative EGF-like transmembrane protein essential for proper ectodermal development in Drosophila melanogaster. Genes Dev. 1990;4:2188–2201. doi: 10.1101/gad.4.12a.2188. [DOI] [PubMed] [Google Scholar]
- 13.Fortini M E, Rebay I, Caron L A, Artavanis-Tsakonas S. An activated Notch receptor blocks cell-fate commitment in the developing Drosophila eye. Nature. 1993;365:555–557. doi: 10.1038/365555a0. [DOI] [PubMed] [Google Scholar]
- 14.Greenwald I. LIN-12/Notch signaling: lessons from worms and flies. Genes Dev. 1998;12:1751–1762. doi: 10.1101/gad.12.12.1751. [DOI] [PubMed] [Google Scholar]
- 15.Gu Y, Hukriede N A, Fleming R J. Serrate expression can functionally replace Delta activity during neuroblast segregation in the Drosophila embryo. Development. 1995;121:855–865. doi: 10.1242/dev.121.3.855. [DOI] [PubMed] [Google Scholar]
- 16.Hamada Y, Kadokawa Y, Okabe M, Ikawa M, Coleman J R, Tsujimoto Y. Mutation in ankyrin repeats of the mouse Notch2 gene induces early embryonic lethality. Development. 1999;126:3415–3424. doi: 10.1242/dev.126.15.3415. [DOI] [PubMed] [Google Scholar]
- 17.Henrique D, Adam J, Myat A, Chitnis A, Lewis J, Ish-Horowicz D. Expression of a Delta homologue in prospective neurons in the chick. Nature. 1995;375:787–790. doi: 10.1038/375787a0. [DOI] [PubMed] [Google Scholar]
- 18.Jarriault S, Brou C, Logeat F, Schroeter E H, Kopan R, Israel A. Signalling downstream of activated mammalian Notch. Nature. 1995;377:355–358. doi: 10.1038/377355a0. [DOI] [PubMed] [Google Scholar]
- 19.Jarriault S, Le B O, Hirsinger E, Pourquie O, Logeat F, Strong C F, Brou C, Seidah N G, Isra I A. Delta-1 activation notch-1 signaling results in HES-1 transactivation. Mol Cell Biol. 1998;18:7423–7431. doi: 10.1128/mcb.18.12.7423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jen W C, Wettstein D, Turner D, Chitnis A, Kintner C. The Notch ligand, X-Delta-2, mediates segmentation of the paraxial mesoderm in Xenopus embryos. Development. 1997;124:1169–1178. doi: 10.1242/dev.124.6.1169. [DOI] [PubMed] [Google Scholar]
- 21.Jiang R, Lan Y, Chapman H D, Shawber C, Norton C R, Serreze D V, Weinmaster G, Gridley T. Defects in limb, craniofacial, and thymic development in Jagged2 mutant mice. Genes Dev. 1998;12:1046–1057. doi: 10.1101/gad.12.7.1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Katakura Y, Seto P, Ohashi H, Teruya K, Shirahata S. Cytotechnology. 1999;31:103–109. doi: 10.1023/A:1008048928053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kidd S, Baylies M K, Gasic G P, Young M W. Structure and distribution of the Notch protein in developing Drosophila. Genes Dev. 1989;3:1113–1129. doi: 10.1101/gad.3.8.1113. [DOI] [PubMed] [Google Scholar]
- 24.Kidd S, Lieber T, Young M W. Ligand-induced cleavage and regulation of nuclear entry of Notch in Drosophila melanogaster embryos. Genes Dev. 1998;12:3728–3740. doi: 10.1101/gad.12.23.3728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Klein T, Arias A M. Interactions among Delta, Serrate and Fringe modulate Notch activity during Drosophila wing development. Development. 1998;125:2951–2962. doi: 10.1242/dev.125.15.2951. [DOI] [PubMed] [Google Scholar]
- 26.Kopan R, Nye J S, Weintraub H. The intracellular domain of mouse Notch: a constitutively activated repressor of myogenesis directed at the basic helix-loop-helix region of MyoD. Development. 1994;120:2385–2396. doi: 10.1242/dev.120.9.2385. [DOI] [PubMed] [Google Scholar]
- 27.Kopan R, Weintraub H. Mouse notch: expression in hair follicles correlates with cell fate determination. J Cell Biol. 1993;121:631–641. doi: 10.1083/jcb.121.3.631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kopczynski C C, Alton A K, Fechtel K, Kooh P J, Muskavitch M A. Delta, a Drosophila neurogenic gene, is transcriptionally complex and encodes a protein related to blood coagulation factors and epidermal growth factor of vertebrates. Genes Dev. 1988;2:1723–1735. doi: 10.1101/gad.2.12b.1723. [DOI] [PubMed] [Google Scholar]
- 29.Kuroda K, Tani S, Tamura K, Minoguchi S, Kurooka H, Honjo T. Delta-induced Notch signaling mediated by RBP-J inhibits MyoD expression and myogenesis. J Biol Chem. 1999;274:7238–7244. doi: 10.1074/jbc.274.11.7238. [DOI] [PubMed] [Google Scholar]
- 30.Lardelli M, Williams R, Mitsiadis T, Lendahl U. Expression of the Notch 3 intracellular domain in mouse central nervous system progenitor cells is lethal and leads to disturbed neural tube development. Mech Dev. 1996;59:177–190. doi: 10.1016/0925-4773(96)00589-8. [DOI] [PubMed] [Google Scholar]
- 31.Lewis A K, Frantz G D, Carpenter D A, de Sauvage F J, Gao W Q. Distinct expression patterns of notch family receptors and ligands during development of the mammalian inner ear. Mech Dev. 1998;78:159–163. doi: 10.1016/s0925-4773(98)00165-8. [DOI] [PubMed] [Google Scholar]
- 32.Li L, Milner L A, Deng Y, Iwata M, Banta A, Graf L, Marcovina S, Friedman C, Trask B J, Hood L, Torok S B. The human homolog of rat Jagged1 expressed by marrow stroma inhibits differentiation of 32D cells through interaction with Notch1. Immunity. 1998;8:43–55. doi: 10.1016/s1074-7613(00)80457-4. [DOI] [PubMed] [Google Scholar]
- 33.Lieber T, Kidd S, Alcamo E, Corbin V, Young M W. Antineurogenic phenotypes induced by truncated Notch proteins indicate a role in signal transduction and may point to a novel function for Notch in nuclei. Genes Dev. 1993;7:1949–1965. doi: 10.1101/gad.7.10.1949. [DOI] [PubMed] [Google Scholar]
- 34.Lin A, Frost J, Deng T, Smeal T, al-Alawi N, Kikkawa U, Hunter T, Brenner D, Karin M. Casein kinase II is a negative regulator of c-Jun DNA binding and AP-1. Cell. 1992;70:777–789. doi: 10.1016/0092-8674(92)90311-y. [DOI] [PubMed] [Google Scholar]
- 35.Lindsell C E, Boulter J, diSibio G, Gossler A, Weinmaster G. Expression patterns of Jagged, Delta1, Notch1, Notch2, and Notch3 genes identify ligand-receptor pairs that may function in neural development. Mol Cell Neurosci. 1996;8:14–27. doi: 10.1006/mcne.1996.0040. [DOI] [PubMed] [Google Scholar]
- 36.Lindsell C E, Shawber C J, Boulter J, Weinmaster G. Jagged: a mammalian ligand that activates Notch1. Cell. 1995;80:909–917. doi: 10.1016/0092-8674(95)90294-5. [DOI] [PubMed] [Google Scholar]
- 37.Luo B, Aster J C, Hasserjian R P, Kuo F, Sklar J. Isolation and functional analysis of a cDNA for human Jagged2, a gene encoding a ligand for the Notch1 receptor. Mol Cell Biol. 1997;17:6057–6067. doi: 10.1128/mcb.17.10.6057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Manak J R, de Bisschop N, Kris R M, Prywes R. Casein kinase II enhances the DNA binding activity of serum response factor. Genes Dev. 1990;4:955–967. doi: 10.1101/gad.4.6.955. [DOI] [PubMed] [Google Scholar]
- 39.Milner L A, Bigas A, Kopan R, Brashem S C, Bernstein I D, Martin D I. Inhibition of granulocytic differentiation by mNotch1. Proc Natl Acad Sci USA. 1996;93:13014–13019. doi: 10.1073/pnas.93.23.13014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Minoguchi S, Taniguchi Y, Kato H, Okazaki T, Strobl L J, Zimber-Strobl U, Bornkamm G W, Honjo T. RBP-L, a transcription factor related to RBP-Jkappa. Mol Cell Biol. 1997;17:2679–2687. doi: 10.1128/mcb.17.5.2679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nofziger D, Miyamoto A, Lyons K M, Weinmaster G. Notch signaling imposes two distinct blocks in the differentiation of C2C12 myoblasts. Development. 1999;126:1689–1702. doi: 10.1242/dev.126.8.1689. [DOI] [PubMed] [Google Scholar]
- 42.Nye J S, Kopan R, Axel R. An activated Notch suppresses neurogenesis and myogenesis but not gliogenesis in mammalian cells. Development. 1994;120:2421–2430. doi: 10.1242/dev.120.9.2421. [DOI] [PubMed] [Google Scholar]
- 43.Ohtsuka T, Ishibashi M, Gradwohl G, Nakanishi S, Guillemot F, Kageyama R. Hes1 and Hes5 as notch effectors in mammalian neuronal differentiation. EMBO J. 1999;18:2196–2207. doi: 10.1093/emboj/18.8.2196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Panin V M, Papayannopoulos V, Wilson R, Irvine K D. Fringe modulates Notch-ligand interactions. Nature. 1997;387:908–912. doi: 10.1038/43191. [DOI] [PubMed] [Google Scholar]
- 45.Pui J C, Allman D, Xu L, DeRocco S, Karnell F G, Bakkour S, Lee J Y, Kadesch T, Hardy R R, Aster J C, Pear W S. Notch1 expression in early lymphopoiesis influences B versus T lineage determination. Immunity. 1999;11:299–308. doi: 10.1016/s1074-7613(00)80105-3. [DOI] [PubMed] [Google Scholar]
- 46.Reaume A G, Conlon R A, Zirngibl R, Yamaguchi T P, Rossant J. Expression analysis of a Notch homologue in the mouse embryo. Dev Biol. 1992;154:377–387. doi: 10.1016/0012-1606(92)90076-s. [DOI] [PubMed] [Google Scholar]
- 47.Robey E, Chang D, Itano A, Cado D, Alexander H, Lans D, Weinmaster G, Salmon P. An activated form of Notch influences the choice between CD4 and CD8 T cell lineages. Cell. 1996;87:483–492. doi: 10.1016/s0092-8674(00)81368-9. [DOI] [PubMed] [Google Scholar]
- 48.Schroeter E H, Kisslinger J A, Kopan R. Notch-1 signalling requires ligand-induced proteolytic release of intracellular domain. Nature. 1998;393:382–386. doi: 10.1038/30756. [DOI] [PubMed] [Google Scholar]
- 49.Shawber C, Boulter J, Lindsell C E, Weinmaster G. Jagged2: a serrate-like gene expressed during rat embryogenesis. Dev Biol. 1996;180:370–376. doi: 10.1006/dbio.1996.0310. [DOI] [PubMed] [Google Scholar]
- 50.Shimizu K, Chiba S, Kumano K, Hosoya N, Takahashi T, Kanda Y, Hamada Y, Yazaki Y, Hirai H. Mouse Jagged1 physically interacts with Notch2 and other notch receptors: assessment by quantitative methods. J Biol Chem. 1999;274:32961–32969. doi: 10.1074/jbc.274.46.32961. [DOI] [PubMed] [Google Scholar]
- 51.Struhl G, Adachi A. Nuclear access and action of notch in vivo. Cell. 1998;93:649–660. doi: 10.1016/s0092-8674(00)81193-9. [DOI] [PubMed] [Google Scholar]
- 52.Struhl G, Fitzgerald K, Greenwald I. Intrinsic activity of the Lin-12 and Notch Intracellular domains in vivo. Cell. 1993;74:331–345. doi: 10.1016/0092-8674(93)90424-o. [DOI] [PubMed] [Google Scholar]
- 53.Swiatek P J, Lindsell C E, del Amo F F, Weinmaster G, Gridley T. Notch1 is essential for postimplantation development in mice. Genes Dev. 1994;8:707–719. doi: 10.1101/gad.8.6.707. [DOI] [PubMed] [Google Scholar]
- 54.Takebayashi K, Akazawa C, Nakanishi S, Kageyama R. Structure and promoter analysis of the gene encoding the mouse helix-loop-helix factor HES-5. Identification of the neural precursor cell-specific promoter element. J Biol Chem. 1995;270:1342–1349. doi: 10.1074/jbc.270.3.1342. [DOI] [PubMed] [Google Scholar]
- 55.Takebayashi K, Sasai Y, Sakai Y, Watanabe T, Nakanishi S, Kageyama R. Structure, chromosomal locus, and promoter analysis of the gene encoding the mouse helix-loop-helix factor HES-1. Negative autoregulation through the multiple N box elements. J Biol Chem. 1994;269:5150–5156. [PubMed] [Google Scholar]
- 56.Uyttendaele H, Marazzi G, Wu G, Yan Q, Sassoon D, Kitajewski J. Notch4/int-3, a mammary proto-oncogene, is an endothelial cell-specific mammalian Notch gene. Development. 1996;122:2251–2259. doi: 10.1242/dev.122.7.2251. [DOI] [PubMed] [Google Scholar]
- 57.Valsecchi C, Ghezzi C, Ballabio A, Rugarli E I. JAGGED2: a putative Notch ligand expressed in the apical ectodermal ridge and in sites of epithelial-mesenchymal interactions. Mech Dev. 1997;69:203–207. doi: 10.1016/s0925-4773(97)00146-9. [DOI] [PubMed] [Google Scholar]
- 58.Vargesson N, Patel K, Lewis J, Tickle C. Expression patterns of Notch1, Serrate1, Serrate2 and Delta1 in tissues of the developing chick limb. Mech Dev. 1998;77:197–199. doi: 10.1016/s0925-4773(98)00138-5. [DOI] [PubMed] [Google Scholar]
- 59.Washburn T, Schweighoffer E, Gridley T, Chang D, Fowlkes B J, Cado D, Robey E. Notch activity influences the alphabeta versus gammadelta T cell lineage decision. Cell. 1997;88:833–843. doi: 10.1016/s0092-8674(00)81929-7. [DOI] [PubMed] [Google Scholar]
- 60.Weinmaster G. The ins and outs of notch signaling. Mol Cell Neurosci. 1997;9:91–102. doi: 10.1006/mcne.1997.0612. [DOI] [PubMed] [Google Scholar]
- 61.Weinmaster G, Roberts V J, Lemke G. A homolog of Drosophila Notch expressed during mammalian development. Development. 1991;113:199–205. doi: 10.1242/dev.113.1.199. [DOI] [PubMed] [Google Scholar]
- 62.Weinmaster G, Roberts V J, Lemke G. Notch2: a second mammalian Notch gene. Development. 1992;116:931–941. doi: 10.1242/dev.116.4.931. [DOI] [PubMed] [Google Scholar]
- 63.Wharton K A, Johansen K M, Xu T, Artavanis-Tsakonas S. Nucleotide sequence from the neurogenic locus notch implies a gene product that shares homology with proteins containing EGF-like repeats. Cell. 1985;43:567–581. doi: 10.1016/0092-8674(85)90229-6. [DOI] [PubMed] [Google Scholar]
- 64.Xue Y, Gao X, Lindsell C E, Norton C R, Chang B, Hicks C, Gendron-Maguire M, Rand E B, Weinmaster G, Gridley T. Embryonic lethality and vascular defects in mice lacking the Notch ligand Jagged1. Hum Mol Genet. 1999;8:723–730. doi: 10.1093/hmg/8.5.723. [DOI] [PubMed] [Google Scholar]
- 65.Zagouras P, Stifani S, Blaumueller C M, Carcangiu M L, Artavanis-Tsakonas S. Alterations in Notch signaling in neoplastic lesions of the human cervix. Proc Natl Acad Sci USA. 1995;92:6414–6418. doi: 10.1073/pnas.92.14.6414. [DOI] [PMC free article] [PubMed] [Google Scholar]