Abstract
We previously characterized the SLS1 gene in the yeast Yarrowia lipolytica and showed that it interacts physically with YlKar2p to promote translocation across the endoplasmic-reticulum membrane (A. Boisramé, M. Kabani, J. M. Beckerich, E. Hartmann, and C. Gaillardin, J. Biol. Chem. 273:30903–30908, 1998). A Y. lipolytica Kar2p mutant was isolated that restored interaction with an Sls1p mutant, suggesting that the interaction with Sls1p could be nucleotide and/or conformation dependent. This result was used as a working hypothesis for more accurate investigations in Saccharomyces cerevisiae. We show by two-hybrid an in vitro assays that the S. cerevisiae homologue of Sls1p interacts with ScKar2p. Using dominant lethal mutants of ScKar2p, we were able to show that ScSls1p preferentially interacts with the ADP-bound conformation of the molecular chaperone. Synthetic lethality was observed between ΔScsls1 and translocation-deficient kar2 or sec63-1 mutants, providing in vivo evidence for a role of ScSls1p in protein translocation. Synthetic lethality was also observed with ER-associated degradation and folding-deficient kar2 mutants, strongly suggesting that Sls1p functions are not restricted to the translocation process. We show that Sls1p stimulates in a dose-dependent manner the binding of ScKar2p on the lumenal J domain of Sec63p fused to glutathione S-transferase. Moreover, Sls1p is shown to promote the Sec63p-mediated activation of Kar2p's ATPase activity. Our data strongly suggest that Sls1p could be the first GrpE-like protein described in the endoplasmic reticulum.
Protein translocation across the endoplasmic reticulum (ER) membrane is the first step of the secretory pathway in eukaryotic cells and may occur either cotranslationally or posttranslationally (for reviews, see references 13, 33, and 58). In the cotranslational pathway, the signal peptide of a nascent secretory polypeptide is recognized when emerging from the ribosome by the signal recognition particle (SRP), thereby causing a translational arrest or pausing. Targeting to the ER membrane is ensured by an interaction of SRP with its receptor (21, 22), and the ribosome-nascent chain complex is then transferred to the translocon, a multicomponent complex that forms an aqueous pore through the ER membrane (66). Three to four heterotrimers composed of the Sec61α, Sec61β, and Sec61γ proteins (Sec61p, Sbh1p, and Sss1p, respectively in Saccharomyces cerevisiae) oligomerize to form the pore (26). In mammal cells, the TRAM protein is also present as a core component of the translocon and is required for the translocation of many but not all secretory polypeptides (23). Translation proceeds, and the newly synthesized polypeptide is translocated through the channel formed by the tight junction between the ribosome and the translocation pore (2).
In S. cerevisiae, an SRP-independent translocation pathway was described and is essential in this yeast (27, 29, 57). Secretory polypeptides are entirely synthesized in the cytosol and maintained in a translocation-competent conformation by members of the 70-kDa class of heat-shock-cognate (Hsc70s) molecular chaperones (14). Targeting to the ER membrane is ensured by signal peptide recognition by Sec62p (16), a subunit of the heterotetrameric Sec62-Sec63p complex (also containing Sec71p and Sec72p proteins) that associates with the Sec61 complex to form a seven-component Sec complex (55). Translocation of the polypeptide through the channel requires Kar2p, a lumenal member of the 70-kDa class of heat shock proteins (Hsp70s) (termed BiP or GRP78 in mammalian cells), which was recently shown to act as a molecular ratchet, binding to the translocating peptide and preventing it from moving backward to the cytosol (43). Kar2p was also shown to be required in the cotranslational pathway, probably via the same mechanism (8).
Kar2p is involved in protein translocation, in folding, in ER-associated degradation (ERAD), and in the maintenance of the permeability barrier between the ER and the cytosol by sealing the pore through a direct or indirect interaction with its lumenal face (20, 25, 43). This functional diversity of Kar2p relies on the intrinsic properties of this class of molecular chaperones, as well as on its interaction with regulatory proteins. As for all members of the Hsp70/Hsc70 family, Kar2p is composed of three domains, a 44-kDa regulatory N-terminal ATPase domain, a 18-kDa peptide binding pocket, and a 10-kDa C-terminal domain (11, 47). Binding and release of substrate peptides is regulated by ATP; both are fast in the ATP-bound form and slow in the ADP-bound form (54, 62). The weak ATPase activity of the Hsp70 is stimulated by members of the DnaJ family, which share a common 70-amino-acid J domain. ATP hydrolysis is followed by a conformational change that stabilizes the interaction of the substrate peptide; the C-terminal domain is predicted to form a lid over the peptide-binding pocket (69). Nucleotide exchange, which induces peptide release, is stimulated by GrpE in Escherichia coli (39) and Mge1p in mitochondria (46), and it does not seem to be a limiting step in the other eukaryotic cellular compartments, since only the BAG1 protein was identified as an ADP-ATP exchanger for the cytosolic mammalian Hsc70 (30). In the yeast ER, the transmembrane protein Sec63p (17, 60) bears a lumenal J domain, and both genetic (63) and biochemical (9, 12, 40, 41, 44) data show that this protein binds to Kar2p, recruiting it to the translocon and activating its ATPase activity. The thermosensitive sec63-1 mutant that contains a point mutation in the J domain was shown to be defective in binding to Kar2p; it accumulates untranslocated preproteins in the cytosol in a way similar to kar2 mutants. ER microsomes prepared from several kar2 mutants and from a sec63-1 strain are defective for both post- and cotranslational translocation in vitro (8, 59). Kar2p and Sec63p are required for peptide release from the initial recognition complex at the cytosolic face of the ER and for completion of preprotein passage through the translocation channel (40).
Our previous studies in the yeast Yarrowia lipolytica revealed that the cotranslational pathway is essential in this organism. Inactivation of the genes encoding the 7S RNA component of SRP is lethal (28), whereas deletion of YlSRP54 and YlSEC65 results in very slow growth (38, 61), suggesting that SRP, as in Schizosaccharomyces pombe (7) and in contrast to S. cerevisiae, plays an essential function in Y. lipolytica. The purification of ribosome-associated membrane proteins, which are indicative of cotranslational translocons, showed that more than 75% of the Sec61 complex was associated with the ribosomes (i.e., the cotranslational translocon), whereas only 30% was in the ribosomal pellet fraction in S. cerevisiae, further demonstrating the respective importance of cotranslational or posttranslational translocation in each organism (6). Genetic screens in Y. lipolytica led to the cloning of several new genes (4, 5, 42). Among these, the SLS1 gene was identified as a mutation that led to synthetic lethality in combination with a thermosensitive 7S RNA mutation (5). The SLS1 gene product was shown to be an ER resident protein, and its disruption affected growth at high temperature and translocation of the secreted reporter protein AEP (alkaline extracellular protease). Immunoprecipitation and two-hybrid experiments showed that Sls1p is located in proximity with the translocon and interacts with the ATPase domain of Kar2p (6, 34). This interaction was shown to be required for efficient translocation of AEP. Indeed, the sls1.5 mutant unable to bind to Kar2p was defective in AEP synthesis and translocation, suggesting that the function(s) of Sls1p rely on its ability to bind to Kar2p (6). The finding of a new Hsp70 cofactor further demonstrates that the modulation and specificity of the chaperone's activity is ensured by several classes of proteins.
We demonstrate here by two-hybrid and genetic approaches that the S. cerevisiae Sls1p homologue interacts with Kar2p to promote protein translocation across the ER membrane. The S. cerevisiae SLS1 gene (ScSLS1) is not essential but genetic interactions with kar2 and sec63 mutants show an in vivo role of ScSls1p not only in protein translocation but also in ERAD and folding. We show that the interaction between ScSls1p and ScKar2p depends on the conformational state of the chaperone protein. In vitro binding assays show that Sls1p stimulates recruitment of Kar2p by Sec63p and promotes the Sec63p-mediated ATPase activation, highlighting the physiological importance of nonessential partners in such complex mechanisms as protein translocation and folding.
MATERIALS AND METHODS
Strains and media.
The E. coli strains used were DH5α [endA1 hsdR17 supE44 thi-1 recA1 gyrA relA1 Δ(lacZYA-argF)U169 deoR (φ80 dlacΔ[lacZ° M15])], BL21 [F− ompT hsdS (rB−, mB−) gal), and XL1red [andA1 gyrA96 thi-1 hsdR17 supE44 relA1 lac mutD5 mutS mutT Tn10 (Tetr)] (Stratagene). E. coli strains were grown in Luria-Bertani LB or 2×YT medium supplemented with ampicillin for plasmid selection (1). Yeast strains used in this study are described in Table 1. Yeasts were grown on YPD medium or on YNB minimal medium as described earlier (34).
TABLE 1.
Yeast strains used in this study
| Strain | Genotype | Reference |
|---|---|---|
| PJ69-4A | MATa trp1-901 leu2-3,112 ura3-52 his3-200 gal4Δ gal80Δ GAL2-ADE2 LYS2::GAL1-HIS3 met2::GAL7-lacZ | 32 |
| RSY801 | MATa ade2-101 leu2-3,113 ura3-52 | 62 |
| RSY586 | MATa ade2-101 leu2-3,113 ura3-52 kar2-159 | |
| RSY578 | MATa ade2-101 leu2-3,113 ura3-52 trp1-1 kar2-113 | |
| RSY579 | MATa ade2-101 leu2-3,113 ura3-52 trp1-1 kar2-203 | |
| MS111 | MATa ade2-101 leu2-3,112 ura3-52 kar2-1 | 10 |
| MS193 | MATa ade2-101 leu2-3,112 ura3-52 kar2-133 | |
| RSY151 | MATα leu2-3,112 pep4-3 ura3-52 sec63-1 | 59 |
DNA manipulation techniques.
Standard techniques were used (1). Restriction enzymes were used according to the manufacturer's instructions (Gibco-BRL and Biolabs). Ready-To-Go PCR beads (Pharmacia Biotech) and Crocodile III thermocycler (Appligene Oncor) were used for PCR analyses. Sequencing was done as described earlier (34).
Cloning of ScSLS1, ScKAR2, and construction of Scsls1.5.
The gene encoding ScSLS1 (YOL031c) was amplified by PCR of genomic DNA. The oligonucleotides used were ScSls1α (5′-CGCGGGATCCCATCTGGAGGCGAAATC-3′) and ScSls1β (5′-CGCGGATCCTATGAGCCATGGGGTTGC-3′), which allowed cloning in the pBluescript SK(−) vector (Stratagene) via BamHI sites (underlined). The cloned PCR product was sequenced, and it contained residues 30 to 413 (after the signal sequence cleavage site and before the ER retention signal RDEL). The Scsls1.5 mutant was obtained by in vitro site-directed (37) deletion of amino acids 365 to 369 (FLNWL) using the Scsls1.5 oligonucleotide (5′-CGATCAACAAAGGG/GCGCAACAATGTAAAGC-3′). The sequence encoding ScKAR2 was obtained by PCR of genomic DNA with oligonucleotides ScKar2α (5′-CGCGGATCCTAGTTAGAGGTGCCGATG-3′) and ScKar2β (5′-CAGGGATCCCATCGTCATCTTCATCTTC-3′). Amplified product was cloned in pBluescript SK(−) (Stratagene) via BamHI sites (underlined), sequenced, and found to contain amino acids 39 to 674.
Two-hybrid assays.
Plasmids pAS2ΔΔ-YlSLS1 and pACT2-YlKAR2 were the same as described previously (6). The sequences encoding ScSLS1 and Scsls1.5 cloned in pBluescript SK(−) (see above) were recovered by BamHI digestion, gel electrophoresis, and purification with the Qiagen gel extraction kit. Recovered inserts were directly cloned in the BamHI site of pGAD-C2 (32) to obtain an in-frame AD-Gal4-ScSLS1 or AD-Gal4-Scsls1.5 fusion. The same method was used to clone the sequence encoding ScKAR2 in pGBD-C2 (32). The plasmids pMR2619, pMR2620, and pMR2618 encoding the G246D, G247D, and G274D kar2 mutants (45), respectively, were digested by KpnI and AflII to obtain a 400-bp fragment containing the mutations. The wild-type KpnI-AflII fragment was removed from the plasmid pGBD-C2-ScKAR2 and replaced by the mutation-containing fragments. Various combinations of these plasmids were introduced in the two-hybrid PJ69-4A strain (32), and expression of the reporter genes was determined as described by Boisrame et al. (6) and Kabani et al. (34). β-Galactosidase activity was determined as described elsewhere (36); given values are an average of two measurements on each of three independent clones.
Disruption of ScSLS1.
The ScSLS1 gene, with promoter and terminator regions, was amplified by PCR using oligonucleotides ScSls1P (5′-GCGCGTTACATAAATCGATAG-3′) and ScSls1T (5′-CGTTCAGCATGCATATAACT-3′) and cloned into the ClaI and SphI sites (underlined) of pBluescript SK(−) vector. The region encoding amino acids 40 to 260 of ScSLS1 was deleted by BglII restriction and replaced by the URA3 marker obtained by digesting pFL61 plasmid (48) with BglII. The resulting plasmid was digested by ClaI and SphI to generate a disruption fragment that was used to transform the wild-type and mutant strains (see Results). ΔScsls1 strains were selected on minimal medium without uracil and checked for the replacement of wild-type gene by the URA3-disrupted copy by PCR and Southern blotting.
In vivo assessment of translocation by immunodetection.
Strains were grown at 20°C (kar2-113 and sec63-1) or 25°C (kar2-1, kar2-133, and kar2-203) in yeast-peptone-dextrose (YPD) (with 0.003% adenine) to mid-log exponential phase. An aliquot with an optical density at 600 nm (OD600) of 3 was taken, and cultures were shifted for 2 h at a semipermissive temperature (26 or 34°C, respectively) before another aliquot of 3 OD600 was obtained. Samples were centrifuged, and cell pellets were resuspended in 500 μl of TE (see below) before the addition of 50 μl of NaOH (1.85 M) on ice. After a 10-min incubation, 50 μl of 50% trichloroacetic acid was added, and samples were kept on ice for 1 h and then centrifuged at 4°C at 15,000 rpm (Sigma 2-MK rotor). Protein pellets were resuspended in 70 μl of TE: 1 M Tris (2:1) plus 70 μl of twofold-concentrated sample buffer (100 mM Tris-Cl, pH 6.8; 4 mM EDTA; 4% sodium dodecyl sulfate [SDS]; 20% glycerol; 0.002% bromophenol blue). The mixture was heated at 95°C for 10 min. Then, 15 μl of each sample was separated by SDS-polyacrylamide gel electrophoresis (PAGE) and transferred to a nitrocellulose membrane. After incubation in TBS-Tween 20 plus 5% skim milk (Difco), membranes were incubated with primary antibodies (antibodies to pre-pro-α-factor [ppαF], carboxypeptidase Y [CPY], Gas1p, and ScKar2p; all gifts from the Rosine Haguenauer-Tsapis laboratory), washed, and incubated with anti-rabbit immunoglobulin G (Fc) alkaline phosphatase-conjugated antibodies (Promega). Detection was done using nitroblue tetrazolium and BCIP (5-bromo-4-chloro-3-indolylphosphate) reagents from Promega according to the manufacturer's instructions.
Purification of wild-type or mutant ScSls1p.
Sequences encoding ScSls1p and ScSls1.5p were cloned in the BamHI site of pGEX-5X-1 plasmid (Pharmacia Biotech) and transformed in BL21 E. coli strain. Then, 50 ml of culture was grown in 2×YT containing 50 μg of ampicillin per ml overnight at 28°C and diluted in 1 liter of the same medium. After 2 h at 28°C, IPTG (isopropyl-β-d-thiogalactopyranoside) was added to a final concentration of 0.2 mM, and cells were grown for an additional 3 h. Cells were harvested and washed once in water and once in phosphate-buffered saline (PBS; pH 7.4)–2 mM EDTA, and the cell pellet was then frozen at −20°C. The cell pellet was thawed and resuspended in ∼20 ml of sonication buffer (PBS, pH 7.4; 2 mM EDTA; 1 mM β-mercaptoethanol; 1 mM phenylmethylsulfonyl fluoride [PMSF]; 1 μg of leupeptin per ml; 1 μg of pepstatin A per ml; 1 mM benzamidine). The cells were sonicated (Branson sonifier 250) for 30 s three to four times at a high setting, with 2 min on ice between sonications. Sonicated cells were centrifuged at 12,000 rpm in a Sorvall SA600 rotor for 10 min, and the supernatant was centrifuged at 30,000 rpm in a Beckman 70 Ti rotor to obtain a clearer lysate. This lysate was loaded on a 2-ml glutathione-Sepharose 4B column (Pharmacia Biotech) equilibrated in sonication buffer plus 1% Triton X-100. The column was washed sequentially with 30 ml of (i) sonication buffer; (ii) sonication buffer plus 500 mM KCl; (iii) 50 mM Tris-Cl (pH 7.5)–10 mM magnesium acetate–200 mM potassium acetate–2 mM ATP, and (iv) PBS glutathione S-transferase (GST)–ScSls1p (or GST-ScSls1.5p) was eluted with 10 ml of elution buffer (50 mM Tris-Cl, pH 8.0; 10 mM reduced glutathione; 5% glycerol). Then, 1-ml fractions were collected. A total of 5 μl of each fraction was spotted onto a nitrocellulose membrane and stained with amido black (Sigma). Peak fractions were pooled, dialyzed against 10 mM Tris-Cl (pH 8.0)–5% glycerol, and frozen at −80°C. SDS-PAGE followed by Coomassie brilliant blue staining showed that GST fusion proteins were purified to near homogeneity. GST alone was purified with the same protocol from BL21 strain transformed with pGEX-5X-1.
To obtain untagged versions of ScSls1p and ScSls1.5p, Factor Xa (Pharmacia Biotech) was used to cleave off the GST tag bound to the glutathione-Sepharose column according to the manufacturer's instructions. After elution of the proteins, Factor Xa was removed with benzamidine-Sepharose 6B (Pharmacia Biotech). ScSls1p and ScSls1.5p were then dialyzed and frozen as described above.
Purification of wild-type or mutant hexahistidine-tagged ScKar2p.
Wild-type ScKar2p, G246D, G247D, and G274D mutants were purified from BL21 E. coli strain respectively transformed with pMR2623, pMR2619, pMR2620, and pMR2618 (45). Plasmids encoding His-tagged T59G and T249G mutants were from T. A. Rapoport's laboratory (50). Purification of the His6-tagged proteins was carried out as described earlier (45) except that the protease inhibitors used were 1 mM PMSF, 1 μg of leupeptin per ml, 1 μg of pepstatin A per ml, and 1 mM benzamidine (in a 500× stock solution).
Purification of GST-tagged Sec63J.
GST-63J (12) was purified from BL21 on a glutathione-Sepharose 4B column (Pharmacia Biotech). Cells were grown overnight at 28°C in 2×YT containing 50 μg of ampicillin per ml then diluted in 1 liter of the same medium. After 2 h, 0.2 mM IPTG was added, and the cells were allowed to grow for an additional 3 h. Cells were harvested, washed once in PBS (pH 7.4)–2 mM EDTA plus proteases inhibitors (see above), and kept at −20°C. The cell pellet was thawed on ice and resuspended in ∼20 ml of sonication buffer (PBS, pH 7.4; 2 mM EDTA; 2 mM EDTA; protease inhibitors). The cells were then treated as described above for GST-ScSls1p purification, and the cell lysate was applied to a 2-ml glutathione-Sepharose 4B column equilibrated in sonication buffer plus 1% Triton X-100. The column was washed as described for the GST-ScSls1p preparation except the second wash was done with sonication buffer plus 1 M KCl–0.1% Triton X-100. GST-63J was eluted with 10 ml of elution buffer (50 mM Tris-Cl, pH 8.0; 10 mM reduced glutathione; 10% glycerol), and 1-ml fractions were collected. Then, 5 μl of each fraction was spotted onto a nitrocellulose membrane and stained with amido black (Sigma). Peak fractions were pooled, dialyzed against dialysis buffer (20 mM HEPES, pH 6.8; 75 mM potassium acetate; 250 mM sorbitol; 5 mM magnesium acetate; 10% glycerol), and frozen in small aliquots at −80°C. SDS-PAGE, followed by Coomassie brilliant blue staining, showed that GST fusion proteins were purified to near homogeneity.
GST binding assays.
The GST pulldown assays were performed essentially as described by Corsi and Schekman (12). A total of 10 μg of GST-ScSls1p (or GST-ScSls1.5p) or 3 μg of GST-63J or equivalent amounts of GST was incubated with 20 μl of glutathione-Sepharose 4B (50% slurry) in GST-binding buffer (20 mM HEPES, pH 6.8; 100 mM KCl; 5 mM MgCl2; 0.1% NP-40; 2% glycerol; 1 mM dithiothreitol [DTT]; 1 mM EDTA; 1 mM PMSF) and rotated for 1 h at 4°C in a total volume of 100 μl. Reactions were centrifuged for 2 min at 15,000 rpm (Sigma 2-MK rotor), supernatant was removed, and the pellet washed three times with 100 μl of GST-binding buffer. When indicated, 2 μg of ScKar2p (or dominant lethal mutants), 2 μg of ScSls1p (or ScSls1.5p), 1 mM nucleotide (ATP or ADP), and GST-binding buffer were added to the pellet to a final volume of 100 μl. The reactions were rotated for 2 h at 4°C and centrifuged for 2 min at 15,000 rpm (Sigma 2-MK rotor). The supernatant was collected, and the pellet was washed three times with 100 μl of GST-binding buffer. SDS-PAGE sample buffer was added to the supernatant and pellet fractions, and they were then analyzed by SDS-PAGE using 8% polyacrylamide gels, followed by Coomassie brilliant blue staining. Quantification of proteins on stained gels was performed by scanning densitometry with NIH Image 1.61 software.
ATPase assays.
Kar2p (1 μM) was incubated with 200 μM ATP and 0.1 μCi of [α-32P]ATP (3,000 Ci/mmol; ICN) in ATPase buffer (50 mM HEPES, pH 7.4; 50 mM NaCl; 10 mM DTT; 2 mM MgCl2) for 10 min at room temperature in a total volume of 20 μl. When indicated, GST-63Jp (2 μM), ScSls1p (4 μM), ScSls1.5p (4 μM), RCMLA (4 μM; Sigma), and α-lactalbumine (α-Lact; 4 μM; Sigma) were present. Reactions were stopped on ice, and 1 μl was spotted in triplicate onto polyethyleneimine cellulose thin-layer chromatography (TLC) plates (Sigma). Plates were developed in 1 M formic acid and 1 M lithium chloride (1:1) (64), and conversion of [α-32P]ATP to [α-32P]ADP was determined with a PhosphorImager (Molecular Dynamics). For single-turnover ATPase assays, ScKar2p (5 μM) was incubated with 100 μCi of [α-32P]ATP for 10 min at room temperature. The [α-32P]ScKar2 complex was purified from free nucleotide on G-50 microspin columns (Amersham Pharmacia Biotech) and kept on ice. Assays contained [α-32P]ScKar2 (1 μM), cold ATP (100 μM) and, when indicated, GST-63Jp (1 μM) and either ScSls1p or ScSls1.5p (4 μM) in ATPase buffer in a total volume of 100 μl. Reactions were incubated at room temperature, and 20-μl aliquots were obtained at various times and purified from free nucleotide on G-50 microspin columns. Then, 3 μl from each reaction was spotted in triplicate on polyethyleneimine TLC plates (Sigma) and developed as described above. Plates were analyzed and quantified by using a PhosphorImager with the ImageQuant software (Molecular Dynamics).
RESULTS
Identification of a mutation in YlKar2p that restores interaction with YlSls1.5p in a two-hybrid assay.
We previously showed that YlSls1p interacts with the ATPase domain of YlKar2p to promote protein translocation across the ER membrane in Y. lipolytica (6, 34). A strain bearing the sls1.5 mutation was affected in the cotranslational translocation of the reporter protein AEP, and YlSls1.5p lost its ability to interact with YlKar2p, as shown by two-hybrid and coimmunoprecipitation assays. To further investigate the relationships between YlSls1p and YlKar2p, mutations in YlKar2p were screened for their ability to restore interaction with YlSls1.5p in a two-hybrid assay.
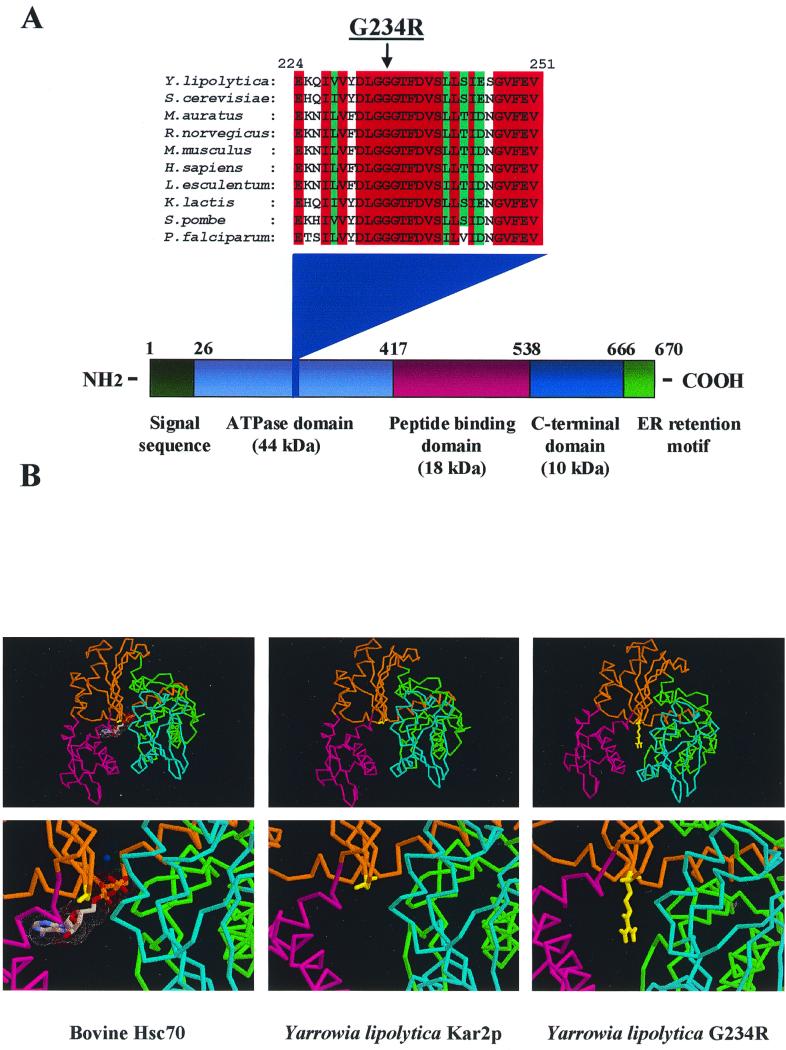
A pool of randomly mutagenized pACT2-YlKAR2 plasmid (6) was obtained by propagation in the XL1red E. coli strain (Stratagene) and introduced into the two-hybrid PJ69-4A yeast strain (32) bearing the plasmid pAS2ΔΔ-Ylsls1.5. A [His+ Ade+] transformant was selected, and the corresponding pACT2-Ylkar2* vector was isolated. The Ylkar2* insert was cloned de novo in pACT2 to confirm the interaction, and chimeras between the wild-type and mutant YlKAR2 strains were generated to identify the position of the mutation (data not shown). The latter was located in the ATPase domain of YlKar2p, and sequencing identified a change of glycine 234 to arginine (Fig. 1A). As shown in Fig. 1A, this mutation affects a highly conserved residue in the ATPase domain of Kar2p. Comparison between the three-dimensional structure of the ATPase domain of bovine Hsc70 (18) and the putative YlKar2p ATPase domain three-dimensional structure modeled by SWISS-MODEL (24, 56) shows that glycine 234 (glycine 202 in bovine Hsc70) is located in the central nucleotide binding cleft, close to the bound nucleotide (Fig. 1B). Three-dimensional modeling of the G234R mutation suggests a position of the side chain of G234 pointing into the nucleotide binding cleft (Fig. 1B). One could speculate that this large side chain may affect ATP binding and/or hydrolysis. In agreement with this hypothesis, several mutations in various Hsp70 or Hsc70 were already isolated by others either at the corresponding glycine or at other residues located into the ATP binding cleft (45, 68). These mutants were shown to be defective in ATP binding and ATP-dependent conformational change (68) and to be lethal dominant in S. cerevisiae (45). Therefore, the interaction of YlSls1p and YlKar2p could be ATP and/or conformation dependent. These results were used as a working hypothesis for further genetic and biochemical experiments in S. cerevisiae, where a variety of well-characterized mutants are available.
FIG. 1.
Mutation of glycine 234 to arginine can affect ATP binding and/or hydrolysis. (A) Multiple alignment showing the conserved position of glycine 234 among Kar2p related proteins; residue numbers correspond to the YlKar2p, and domains were deduced from sequence comparison between YlKar2p and bovine Hsc70. (B) Three-dimensional structure of the ATPase domain of bovine Hsc70 (18) and wild-type and G234R YlKar2p (as determined by using SWISS-MODEL [24, 56]). Subdomain colors are as described previously (18). Bound ATP (with associated magnesium and sodium ions) in bovine Hsc70 are represented; glycine 202 (bovine Hsc70), glycine 234 (YlKar2p), and arginine 234 (G234R) are depicted in yellow.
Disruption of ScSLS1 lead to synthetic lethality in combination with translocation-deficient kar2 and sec63 mutants.
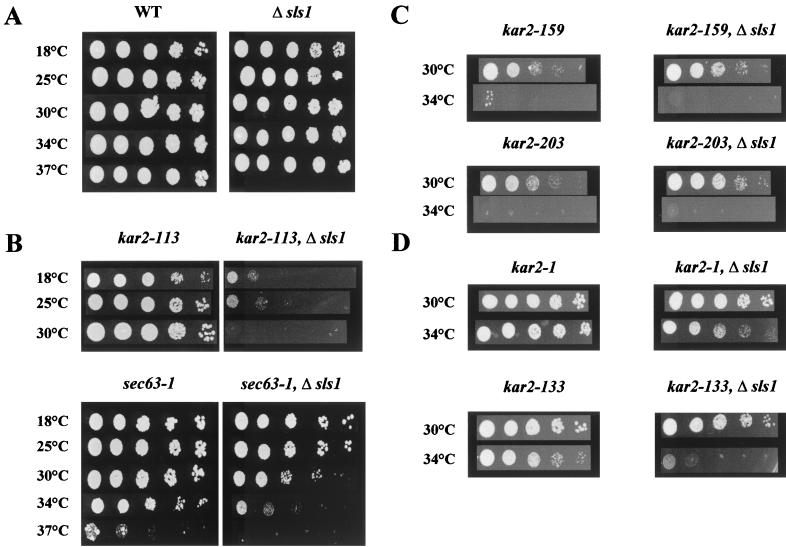
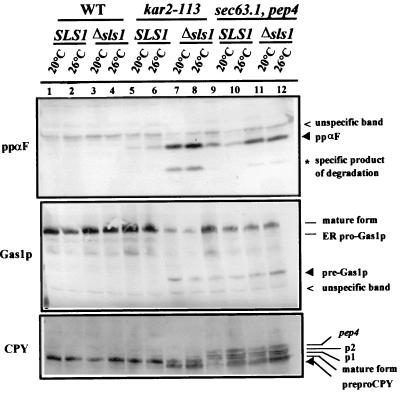
We first wanted to check whether ScSls1p (YOL031cp) is functional in vivo in S. cerevisiae, playing a similar role in protein translocation as in Y. lipolytica. Disruption of ScSLS1 in various wild-type genetic backgrounds had no detectable effect on the growth rates nor on any of the physiological processes assessed, i.e., translocation and secretion (data not shown). We then examined the effect of combining the disruption of ScSLS1 with several well-characterized thermosensitive mutations in the ScKAR2 and SEC63 genes. Indeed, genetic interactions, either synthetic lethality or extragenic suppression, have been successfully used to show the involvement of two genes in the same biological process (5, 35, 42). The kar2-113, kar2-159, and kar2-203 mutants were shown to be affected to different extents in protein translocation (8). We also checked the genetic interactions between ΔScsls1 and the sec63-1 mutant (51), which bears a point mutation in the luminal J domain of Sec63p and that is affected in the translocation process (59). ScSLS1 was disrupted in each of these mutants, and the growth of the resulting double mutants was assessed at various temperatures. As shown in Fig. 2B, the growth of the kar2-113 ΔScsls1 and sec63-1 ΔScsls1 double mutant strains was greatly reduced at 18 and 30°C, respectively, compared to the wild type and the single mutants (Fig. 2A). A subtle synthetic lethal phenotype was detected and demonstrates a physiological role of Sls1p in S. cerevisiae. To further characterize the double mutants, we assessed the translocation of several reporter proteins with well-known processing pathways: CPY, Gas1p, and ppαF (52). As shown in Fig. 3, when the wild-type or Δsls1 strains were cultivated at a permissive temperature or shifted for 2 h at a semipermissive temperature, no accumulation of the cytoplasmic form of each reporter protein was detectable (lanes 1 to 4). When the same experiment was performed with kar2-113 or sec63-1 single mutants, little or no cytoplasmic forms of CPY, ppαF, or Gas1p were detectable at a permissive temperature, but when shifted at 26°C these precursor forms slightly accumulated in the cytosol (lanes 5, 6, 9, and 10; black arrowheads). In the double mutants lacking ScSLS1, greater amounts of precursors accumulated in the cytosol even when the cells were cultured at 20°C (compare the black arrowheads in lanes 7 and 8 to lanes 5 and 6 and in lanes 11 and 12 to lanes 9 and 10, respectively). The observed accumulation of the P1 (ER) and P2 (Golgi) forms of CPY in the sec63-1 and sec63-1 Δsls1 strains probably results from a lack of a maturating enzyme, whose translocation is also blocked in these mutants.
FIG. 2.
Disruption of ScSLS1 lead to synthetic lethality in a kar2 or sec63-1 context. ScSLS1 was disrupted in various kar2 or sec63 contexts. Cells (5 μl) from mid-log-phase cultures were spotted onto YPD and allowed to grow at the indicated temperatures.
FIG. 3.
Disruption of ScSLS1 enhances the translocation defect of the kar2-113 and sec63-1 mutants. The indicated strains were checked for their ability to translocate several reporter proteins (ppαF, Gas1p, and CPY). Strains were grown at 20°C to mid-log phase and then shifted to 26°C for 2 h. Equivalent amounts of total extracts were prepared from samples taken at each stage and then resolved by SDS-PAGE. Proteins were transferred to nitrocellulose membranes and blotted with antibodies against the reporter proteins. The different detectable forms resulting from processing through the secretory pathway are indicated. Accumulation of the cytoplasmic precursors of each protein (black arrowhead) is indicative of a translocation defect.
Interestingly, when ScSLS1 is disrupted in the kar2-159 and kar2-203 mutants, no synthetic lethal effect was observed (Fig. 2C), nor did accumulation of the reporter protein cytoplasmic precursors occur (data not shown). This allele specificity could be explained by the fact that neither Kar2-159p nor Kar2-203p can bind to ATP-agarose, whereas Kar2-113p displays normal ATP binding and hydrolysis rates (8). Indeed, if Sls1p's function is tightly linked to the nucleotide binding and/or hydrolysis rates of Kar2p (see below), mutants defective in one or both of these two properties would be predicted to be insensible to the presence or absence of Sls1p.
These results show in vivo that ScSls1p is involved in protein translocation and that its function is directly linked to Kar2p and Sec63p, two well-characterized ER components that were shown to be required for co- and posttranslational translocation (8).
Disruption of ScSLS1 leads to synthetic lethality in ERAD-specific mutants.
ScSLS1 was also disrupted in the kar2-1 and kar2-133 mutants that are not affected in the translocation process but that display defects in protein folding and ERAD (10). As shown in Fig. 2D, the kar2-1 Δsls1 and kar2-133 Δsls1 mutants grew much less than the corresponding single mutants at a semipermissive temperature (34°C). This result suggested an involvement of ScSls1p in another essential process in the ER: protein quality control and degradation. A recent report from the P. Walter and J. S. Weissman laboratories demonstrated that disruption of the PER100 gene (identical to ScSLS1) lead to a subtle ERAD defect. In the per100 mutant, CPY*, a constitutively misfolded soluble secretory protein rapidly degraded in a wild-type strain, was stabilized in a similar way to that in ERAD-deficient alleles of KAR2 (67). To determine whether, in the absence of ScSls1p, a defect in protein translocation could be observed in the kar2-1 and kar2-133 mutants, Western blots were performed on the single and double mutants at 25°C (permissive temperature) and after a 2-h shift at 34°C (semipermissive temperature). No cytoplasmic precursor could be detected (data not shown), confirming that ScKar2p action in protein translocation and ERAD differs (10).
ScSls1p and ScKar2p interact in a two-hybrid assay.
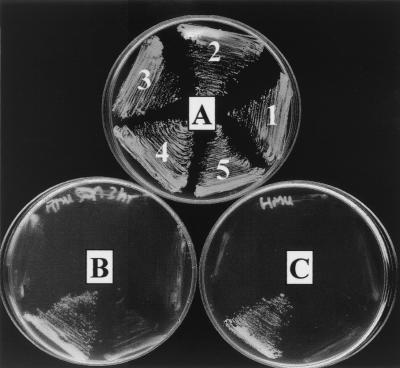
Then we checked the ability of ScSls1p to interact with ScKar2p in a two-hybrid assay. The sequences encoding ScSls1p and ScKar2p were cloned in pGAD-C2 and pGBD-C2, respectively (32). The plasmids were then introduced into the PJ69-4A strain and tested for activation of reporter genes. As shown in Fig. 4, controls could not grow on YNB lacking leucine, tryptophan, and adenine plus aminotriazole (5 mM) or on YNB lacking leucine, tryptophan, and adenine alone (sectors 1 and 2), whereas a strain bearing both plasmids (sector 4) could grow on these media, indicating that, as in Y. lipolytica, ScSls1p and ScKar2p interact physically. We then constructed the Scsls1.5 mutant by in vitro mutagenesis (see Materials and Methods), cloned the mutated sequence in pGAD-C2, and checked for reporter gene activation in the presence of pGBD-C2 or pGBD-C2-ScKAR2. Neither combination allowed growth on YNB lacking leucine, tryptophan, and adenine plus aminotriazole (5 mM) or on YNB lacking leucine, tryptophan, and adenine alone (sectors 3 and 5 in Fig. 4), indicating that ScSls1.5p, like the related Y. lipolytica mutant, is unable to bind to ScKar2p in a two-hybrid assay.
FIG. 4.
ScSls1p and ScKar2p interact in a two-hybrid assay. PJ69-4A strain was cotransformed with the following plasmid combinations: 1, pGAD-C2 and pGBD-C2-ScKAR2; 2, pGAD-C2-ScSLS1 and pGBD-C2; 3, pGAD-C2-Scsls1.5 and pGBD-C2; 4, pGAD-C2-ScSLS1 and pGBD-C2-ScKAR2; and 5, pGAD-C2-Scsls1.5 and pGBD-C2-ScKAR2. Samples were then plated on minimal medium lacking leucine and tryptophan (A); minimal medium lacking leucine, tryptophan, and adenine (B); or minimal medium lacking leucine, tryptophan, and histidine but with 5 mM 3-aminotriazole (C).
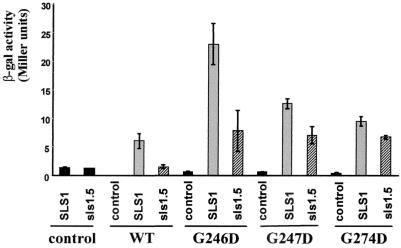
Dominant lethal mutations in ScKar2p restore interaction with ScSls1.5p and increase binding to ScSls1p.
We then wanted to test our hypothesis that mutants that fail to undergo ATP-dependent conformational change would be able to restore the interaction with ScSls1.5p in the two-hybrid assay, as is the case for the YlG234R mutant. The sequences encoding G246D, G247D, and G274D mutations (45) were cloned in pGBD-C2 and introduced in PJ69-4A with either pGAD-C2, pGAD-C2-ScSLS1, or pGAD-C2-Scsls1.5. All fusion proteins (wild type or mutants) were expressed in equal amounts, as checked by Western blot analysis (data not shown). We then measured the β-galactosidase activity for each combinations, and the results are depicted in Fig. 5. As expected, the β-galactosidase activities in a strain coexpressing AD-ScSls1.5p and DBD-ScKar2p were very low and were similar to those for controls, whereas a strain coexpressing AD-ScSls1p and DBD-ScKar2p displays higher β-galactosidase activity. When (AD) ScSls1.5p is coexpressed with any (DBD) ScKar2p mutants, the β-galactosidase activity increased significantly (Fig. 5), indicating that the ScKar2p mutations indeed can restore interaction with ScSls1.5p. Interestingly, β-galactosidase activity increased further (Fig. 5) when ScSls1p was coexpressed with the ScKar2p lethal dominant mutants, indicating a better affinity of the mutants for ScSls1p than of the wild type for ScKar2p. The dominant lethal mutants display very low levels of ATP binding and are blocked in an ADP-bound-like conformation (45), corresponding to low on and off rates of peptide binding (50). Sls1p may then interact with Kar2p-ADP, either bound to its DnaJ partner (i.e., Sec63p) (12), to a substrate peptide (62), or to both.
FIG. 5.
Dominant lethal mutations in ScKar2p restore interaction with ScSls1.5p in a two-hybrid assay. PJ69-4A strain was cotransformed with the indicated combinations of plasmids, and the β-galactosidase activity was assessed as described earlier (34). Reported values are an average of two measurements on three independent clones.
In vitro binding assays.
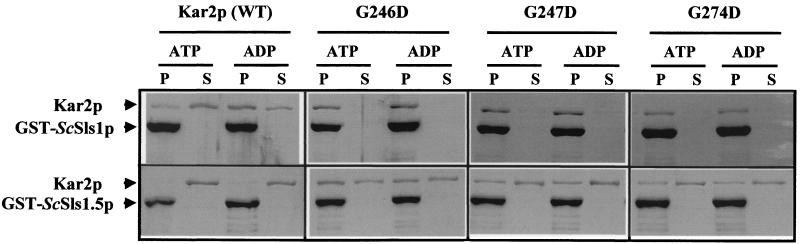
Since the results obtained with the two-hybrid approach could be in part due to the nonphysiological environment in the nucleus or to the non-native conformation of the hybrid proteins, we decided to confirm these interactions in vitro with purified tagged proteins. ScSls1p and ScSls1.5p were fused to GST and then purified to near homogeneity from the BL21 E. coli strain by affinity chromatography as described in Materials and Methods. His6-tagged versions of ScKar2p (wild type), G246D, G247D, and G274D were purified to near homogeneity from BL21 according to the method of McClellan et al. (45). We then examined ScKar2p proteins binding to GST-ScSls1p in pulldown assays (12). Purified wild-type and mutant forms of ScKar2p were incubated at 4°C for 2 h with GST-ScSls1p or GST-ScSls1.5p complexed with the glutathione matrix in the presence of 1 mM ATP or 1 mM ADP. As shown in Fig. 6, ScKar2p associates with GST-ScSls1p in the presence of 1 mM ADP but only barely in the presence of 1 mM ATP, while binding to ScSls1.5p is negligible (Table 2). We determined that ScKar2p does not bind to GST alone as reported previously (12). The possibility that ScKar2p is recognizing a population of misfolded GST fusion proteins is unlikely since no binding occurred with GST-ScSls1.5p, which should contain amounts of misfolded proteins similar to GST-ScSls1p. These results confirm the previously reported two-hybrid assays and show that the interaction between the two proteins is nucleotide dependent. The dominant lethal mutants associate with ScSls1p in the presence of either nucleotide (Fig. 6), with all the proteins being retained in the pellet fraction (Table 2). Interaction with ScSls1.5p was restored, although fewer proteins were bound than with GST-ScSls1p (Table 2). These results are in total agreement with the β-galactosidase activities in the two-hybrid assays; G246D gives the highest β-galactosidase activity and binds more efficiently with GST-ScSls1.5p than G247D or G274D, which display a lower β-galactosidase activity in the two-hybrid assay (Fig. 5).
FIG. 6.
GST binding assays. GST-ScSls1p, GST-ScSls1.5p, ScKar2p, G246D, G247D, and G274D were purified as described in Materials and Methods. GST-ScSls1p (or GST-ScSls1.5p) (10 μg) was prebound to glutathione-Sepharose, and ScKar2p (or dominant lethal mutants) (2 μg) and either ATP or ADP (1 mM) was added. After rotation at 4°C for 2 h, proteins associated with the pellet or remaining in the supernatant were resolved by SDS-PAGE (25% total) and visualized by Coomassie blue staining. Signals corresponding to GST-ScSls1p or ScKar2p are indicated.
TABLE 2.
Quantification of the ScKar2p (wild-type and mutant) fractions bound to GST-ScSls1p or GST-ScSls1.5p in pulldown assays expressed as a percentage of total inputa
| Fraction | % Total input
|
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Kar2p
|
G246D
|
G247D
|
G274D
|
T59G
|
T249G
|
|||||||
| ATP | ADP | ATP | ADP | ATP | ADP | ATP | ADP | ATP | ADP | ATP | ADP | |
| GST-Sls1p | 26 | 74 | 90 | 100 | 90 | 94 | 100 | 100 | 29 | 78 | 41 | 52 |
| GST-Sls1.5p | 0 | 8 | 65 | 55 | 37 | 34 | 35 | 26 | 30 | 29 | 0 | 0 |
GST binding assays were as in Fig. 6. Proteins bound to the glutathione pellet or remaining in the supernatant were quantified by scanning densitometry with NIH Image 1.61 software (values are an average of three independent experiments).
To more precisely understand the nucleotide and conformation dependence of the interaction between Sls1p and Kar2p, GST binding experiments were performed with the T59G and T249G Kar2p mutants (44). T59G is unable to undergo the conformational change following ATP binding, remaining in an ADP-bound conformation, whereas T249G binds ATP and undergoes the conformational change but fails to hydrolyze the bound nucleotide (68). T59G binds GST-ScSls1p in a way similar to that of the wild type, but significant binding was observed with GST-ScSls1.5p in a nucleotide-independent manner (Table 2), as is the case for the dominant lethal mutants. The T249G mutant binds GST-ScSls1p in nearly similar amounts in ATP or ADP but fails to bind to GST-ScSls1.5p (Table 2). This indicates that ATP hydrolysis per se is not required for the interaction of Sls1p and Kar2p, but the conformation of the chaperone seems to be critical and may regulate the binding and release of ScSls1p. It was not possible to perform the same in vitro experiments with Y. lipolytica proteins due to the high toxicity of YlKar2p in E. coli, thus preventing its easy purification (personal observations).
ScSls1p promotes binding of ScKar2p to Sec63p.
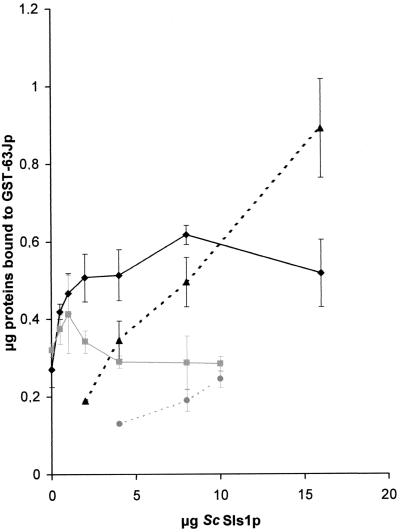
We next tested the effect of ScSls1p on the binding of Kar2p to Sec63p in GST pulldown assays (12). Untagged ScSls1p was purified after GST cleavage with Factor Xa (see Materials and Methods), along with a GST fusion protein containing the DnaJ domain of Sec63p, GST-63Jp (12). Increasing amounts of ScSls1p were added to binding reactions conducted in the presence of 1 mM ATP, since it was previously shown that Sec63p binding to Kar2p is strictly dependent on the presence of hydrolyzable ATP (12, 49). As shown in Fig. 7, ScSls1p stimulated ScKar2p binding to GST-63Jp up to two times, and maximum stimulation was achieved at a 6:1 (ScSls1p/ScKar2p) molecular ratio. When using ScSls1.5p in the same reactions, very low stimulation was achieved (Fig. 7), showing that the observed effect is specific and depends on a functional interaction between the two proteins. Interestingly, a significant proportion of ScSls1p was found in the pellet fraction in our binding assays (Fig. 7); quantitation by scanning densitometry predicted one to more than two molecules of ScSls1p bound per ScKar2p. Accounting for these data, native gel electrophoresis of purified ScSls1p suggested that this protein can be found as dimers or even as higher-order oligomers (unpublished results). ScSls1.5p displayed the same pattern as the wild type on the native gel, suggesting that the mutation does not cause major structural alterations of the protein. Since we could not observe any interaction between ScSls1p and the J domain of Sec63p by two-hybrid or in vitro binding assays (data not shown), ScSls1p should be retained through its interaction with ScKar2p. Indeed, very small amounts of ScSls1.5p were bound to the pellet fraction (Fig. 7), supporting the idea that the stimulation of GST-63Jp binding is tightly linked to a functional interaction with ScKar2p. This result can be a likely explanation for the stimulatory effect of Sls1p on protein translocation, more molecules of Kar2p being available in proximity with the translocon. However, since GST-63Jp is ultimately bound in the peptide binding pocket in the final Kar2-ADP-GST-63Jp complex (19, 49), we cannot exclude the possibility that ScSls1p could stimulate or stabilize the interaction of ScKar2p with substrate peptides and, more particularly, translocating peptides (see Discussion). To exclude the possibility that ScSls1p is recognized as a misfolded peptide in the final complex, similar binding reactions were conducted in the presence or absence of either ScSls1p, α-Lact (10 g), or its permanently misfolded form RCMLA (10 μg). Neither α-Lact nor RCMLA had any effect on the binding of ScKar2p and GST-63Jp, and neither one altered the stimulatory effect of ScSls1p (data not shown). Then, the increase of ScKar2p binding to GST-63Jp in the presence of ScSls1p seems to be specific, although the observed ternary complex might be only transient in vivo.
FIG. 7.
ScSls1p stimulates binding of ScKar2p to GST-63Jp. GST-63Jp (3 μg) was prebound to glutathione-Sepharose; increasing amounts of untagged ScSls1p (or ScSls1.5p) were added, along with ScKar2p (2 μg) and ATP (1 mM). Pulldown assays were done as in Fig. 6 except that all of the pellet fraction was loaded onto the gel. The amounts of pellet-associated ScKar2p in the presence of ScSls1p (⧫) or ScSls1.5p (■) and the amounts of pellet-associated ScSls1p (▴) or ScSls1.5p (●) were quantified as shown in Table 2 (average of three independent experiments).
ScSls1p stimulates the GST-63J activation of the ATPase activity of Kar2p.
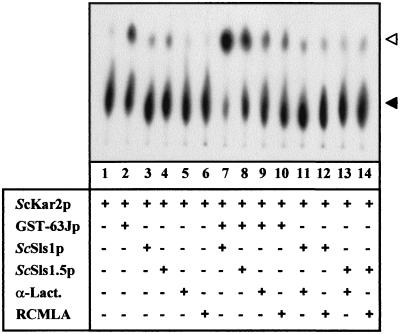
To test the effect of ScSls1p on the nucleotide binding and hydrolysis properties of ScKar2p, the ATPase activity was monitored under various conditions. ScKar2p (1 μM) was incubated in the presence of [α-32P]ATP for 10 min at room temperature, in the presence or absence of GST-63Jp (2 μM) and/or ScSls1p or ScSls1.5p (4 μM). The conversion of [α-32P]ATP to [α-32P]ADP was assessed by TLC as described previously (39), and the results are depicted in Fig. 8. ScKar2p alone has a very weak ATPase activity and therefore very small amounts of ATP were converted to ADP after the 10-min assays (Fig. 8, lane 1) and after 1 h (data not shown). When GST-63Jp is present, the ATPase activity of ScKar2p increases and a greater amount of ADP is detectable (Fig. 8, lane 2). After 1 h of incubation, nearly all the ATP is hydrolyzed (data not shown). While ScSls1p alone had no significant effect on the ATPase activity of ScKar2p (lane 3), the presence of both ScSls1p and GST-63J significantly increased the ATP hydrolysis (compare lane 7 to lane 2). ScSls1.5p had no effect alone (lane 3), but when GST-63Jp was present in the assay more ADP was detected (lane 8). However, the GST-63Jp mediated activation of the ATPase activity of ScKar2p was higher with ScSls1p than with ScSls1.5p (compare lanes 7 and 8, respectively). This suggests that a functional interaction between ScSls1p and ScKar2p is required for the stimulation of the ATPase activity by GST-63Jp. ScSls1.5p probably retained transient interaction ability with ScKar2p, allowing some ATPase activation.
FIG. 8.
ScSls1p promotes the ATPase activity of ScKar2p along with GST-63Jp. ScKar2p (1 μM) was incubated with cold ATP (200 μM) and [α-32P]ATP (0.1 μCi, 3,000 Ci/mmol). Where indicated, the following proteins were present in the assay: GST-63Jp (2 μM), ScSls1p (4 μM), ScSls1.5p (4 μM), α-Lact (4 μM), and RCMLA (4 μM). After 10 min of incubation at room temperature, 1 μl of each reaction mixture was spotted in triplicate onto polyethyleneimine TLC plates, and the conversion of [α-32P]ATP (black arrowhead) to [α-32P]ADP (white arrowhead) was analyzed by using a PhosphorImager (Molecular Dynamics).
In order to ensure that the observed effect was specific to ScSls1p, the same experiments were carried out with α-Lact and RCMLA. In our experimental conditions, neither α-Lact nor RCMLA significantly stimulated the ATPase activity of ScKar2p, either alone (lanes 5 and 6) or with GST-63Jp (lanes 9 and 10), ScSls1p (lanes 11 and 12), or ScSls1.5p (lanes 13 and 14). We also checked that any of the proteins used in these assays (except ScKar2p) had an intrinsic ATPase activity (data not shown). Therefore, ScSls1p and GST-63Jp conjointly activate the ATPase activity of ScKar2p, thereby enhancing the turnover of the molecular chaperone.
Influence of ScSls1p on the nucleotide binding properties of ScKar2p.
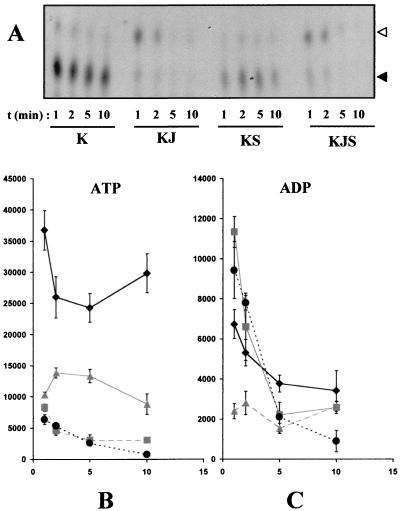
We have showed that the ATPase activity of ScKar2p is greatly induced by ScSls1p, but only when GST-63Jp was also present in the assay. A likely explanation would be that ScSls1p functions as a GrpE like protein and promotes nucleotide exchange. We then analyzed the effect of ScSls1p on the nucleotide binding and hydrolysis of ScKar2p in single-turnover ATPase assays. ScKar2p was preincubated for 10 min with [α-32P]ATP and rapidly purified from free nucleotide by gel filtration. The [α-32P]ScKar2p complex (1 μM) was then incubated at room temperature with cold ATP either alone or with GST-63Jp (1 μM), ScSls1p (4 μM), or both. At various times, aliquots were obtained and further purified from free nucleotide by rapid gel filtration. Bound nucleotide was determined by TLC, and the results are depicted in Fig. 9A. As expected, ScKar2p alone was mainly in the ATP-bound form, a result consistent with its weak ATPase activity (Fig. 9A, lanes K). When GST-63Jp was present in the assay, almost all of the bound ATP was converted to ADP within 1 min, a finding consistent with previous observations (Fig. 9A, lanes KJ). Interestingly, when ScSls1p was added, much less bound ATP was detected (Fig. 9A, lanes KS), suggesting either that it has been hydrolyzed or that it has been exchanged for cold ATP. Quantification by PhosphorImager analysis confirmed these observations and showed that, in the presence of ScSls1p, the amounts of bound ATP (Fig. 9B) and ADP (Fig. 9C) significantly decreased compared to the wild type. When both GST-63Jp and ScSls1p were added to the [α-32P]ScKar2p complex, no significant difference could be observed compared to GST-63Jp and [α-32P]ScKar2p alone (Fig. 9A, lanes KJS [Fig. 9B and C for quantification]). This is probably due to a very fast activation of the ATPase activity of ScKar2p by GST-63Jp in these conditions, with the ADP being rapidly released after hydrolysis. Taken together, these results show that the nucleotide binding properties of ScKar2p are influenced by ScSls1p.
FIG. 9.
Single-turnover ATPase assays. (A) A [α-32P]ScKar2 complex was formed after incubation of ScKar2p with 100 μCi of [α-32P]ATP for 10 min at room temperature and removal of free nucleotide by rapid gel filtration on microspin G-50 columns. [α-32P]ScKar2 (1 μM) was further incubated with or without GST-63Jp (1 μM) and/or ScSls1p (4 μM) in the presence of cold ATP (100 μM). Aliquots were obtained at 1, 2, 5, and 10 min and then separated from free nucleotide by gel filtration on microspin G-50 columns. Then, 3 μl from each reaction was spotted in triplicate onto polyethyleneimine TLC plates, and the conversion of [α-32P]ATP (black arrowhead) to [α-32P]ADP (white arrowhead) was analyzed using a PhosphorImager (Molecular Dynamics). K, J, and S represent ScKar2p, GST-63Jp, and ScSls1p, respectively. Quantification of ATP (B) and ADP (C) was performed with the ImageQuant software, and results were averaged from three independent experiments. Values were plotted as a function of time (K, diamonds; KS, triangles; KJ, squares; KJS, circles).
DISCUSSION
Our experiments describe the involvement of the SLS1 gene, previously characterized in the yeast Y. lipolytica, in the protein translocation process in S. cerevisiae. Disruption of the ScSLS1 gene in the model yeast had no detectable effect on viability, in contrast to the Y. lipolytica ΔYlsls1 mutant that is strongly impaired in growth at high temperature and in translocation of the reporter protein AEP (5). However, a synthetic lethal effect was observed when ScSLS1 was disrupted in the kar2-113 and sec63-1 mutants (Fig. 2), with enhanced defects in translocation of several reporter proteins (Fig. 3), indicating an involvement of ScSls1p in the translocation process. Disruption of ScSLS1 in a kar2-159 or a kar2-203 context did not result in a synthetic lethal phenotype (Fig. 2). Kar2-159p and Kar2-203p do not bind to ATP-agarose (8) but can still bind to ScSls1p (unpublished results), suggesting that this interaction is not functional; therefore, whether ScSls1p is present or not does not enhance the growth defect of the original mutant. A synthetic lethal phenotype was also observed with the kar2-1 and kar2-133 mutants (Fig. 2) that displayed defects in folding and ERAD, whereas protein translocation was not affected (10). This suggests that ScSls1p function is not restricted to the protein translocation process. Sls1p is more probably a partner of Kar2p in many if not all of the functions that the chaperone fulfills in the ER. Efforts to find other partners for YlSls1p by two-hybrid experiments were unsuccessful (34). In support to our findings, Travers et al. (67) have recently reported the identification of Per100p, which is identical to ScSls1p, in a wide range screen for unfolded protein response (UPR) target genes. They showed that Per100p is upregulated by the UPR, which is consistent with the observed increase of YlSls1p levels in cells that were treated with tunicamycin or heat shocked (unpublished results). Furthermore, these authors demonstrated a direct involvement of ScSls1p and Per100p in ERAD, since a constitutively misfolded form of CPY was stabilized in the per100 mutant in a way similar to that in ERAD-affected kar2 mutants.
A direct interaction between the ScKar2p and ScSls1p was shown by two-hybrid (Fig. 4) and in vitro binding assays (Fig. 6). The G234R mutant was isolated based on its ability to restore interaction with the YlSls1.5p mutant (6). Three-dimensional modeling predicted an inhibitory effect of this mutation on ATP binding (Fig. 1). Similar mutants were obtained in hamster BiP (68) and ScKar2p (45) and were shown to affect ATP binding and the conformational change following ATP hydrolysis. These mutants were shown to be blocked in an ADP-bound-like conformation that mediates stable interaction with substrate peptides. An attractive hypothesis was that Sls1p binding to Kar2p is ATP and/or conformation dependent and that the G234R is blocked in the preferential conformation for interaction with Sls1p. This hypothesis was supported by two-hybrid assays with three dominant lethal ScKar2p mutants (G246D, G247D, and G274D) (45). These mutants restored the interaction with ScSls1.5p and displayed higher β-galactosidase activity when assayed with ScSls1p than did wild-type ScKar2p (Fig. 5). The two-hybrid data were validated by in vitro binding assays (Fig. 6) and, taken together, confirmed that the conformation of Kar2p strongly affects binding to Sls1p, with preferential binding occurring when Kar2p is in the ADP-bound conformation (i.e., with the peptide-binding pocket occupied and closed by the C terminus lid [69]). Assays with T59G and T249G, two ScKar2p mutants affected in the conformational change following ATP binding, and in ATP hydrolysis, respectively (68) (Table 2), suggested that ATP hydrolysis is not necessary for the Sls1p binding but rather that the conformational change induced by ATP hydrolysis could regulate binding and release of Sls1p.
ScKar2p interacts with the lumenal J domain of Sec63p, and this interaction allows activation of Kar2p for peptide binding (8, 9, 12, 49). Pulldown experiments with purified GST-63Jp and ScKar2p confirmed the previously described (12, 45) ATP-dependent interaction between the chaperone and its DnaJ partner. The same reactions were conducted in the presence of increasing amounts of purified ScSls1p in the presence of ATP. A dose-dependent stimulation of ScKar2p binding to GST-63Jp was observed (Fig. 7), and a significant amount of ScSls1p remained bound to the glutathione pellet. Since ScSls1p does not interact with GST-63Jp (data not shown), the ScSls1p must remain bound to ScKar2p. This is not surprising since ScKar2p is in the ADP-bound conformation (i.e., the preferential conformation for binding with ScSls1p) in the final complex with GST-63Jp (12, 49). The observed stimulation of ScKar2p binding to GST-63Jp by ScSls1p could be the result of a stabilization of the [ScKar2p-ADP]-GST-63Jp complex by ScSls1p. It was shown that the interaction between Sec63p and ScKar2p depends on the presence of both the ATPase domain and the peptide-binding pocket (50) and that GST-63Jp, in the absence of another peptide, is ultimately bound to ScKar2p as a substrate peptide (19, 49). ScKar2p binds transiently with the J domain of Sec63p and is then rapidly transferred to a peptide substrate (either a translocating peptide or the J domain itself) (49). Based on our binding experiments, different hypotheses can be proposed. In one, ScSls1p could stabilize the transient interaction between ScKar2p and GST-63Jp to enhance the number of available activated ScKar2p's at the translocon. This interaction can prevent the transfer of GST-63Jp to the peptide-binding pocket in the absence of substrate peptide. A second hypothesis is that ScSls1p stabilizes the interaction of ScKar2p and GST-63Jp in a Hip-like manner (31). GST-63Jp would be recognized as a substrate peptide (bound to the peptide binding pocket of ScKar2p), and this enhanced association could promote the ratcheting function of ScKar2p.
Another attractive possibility is that ScSls1p acts as a nucleotide exchanger in a BAG1 manner (30). More ScKar2p-ATP complexes will then be available for binding with Sec63p, allowing a faster recycling of ScKar2p at the translocon. In support of this hypothesis, we showed that ScSls1p promotes the ATPase activity of ScKar2p when GST-63Jp is present (Fig. 8). This stimulatory effect was specific since neither α-Lact nor RCMLA had the same effect. Moreover, single-turnover assays showed that ScSls1p affects nucleotide binding to ScKar2p, since the α-32P-labeled nucleotide bound to ScKar2p seemed to be rapidly exchanged for a “cold” nucleotide (Fig. 9). This hypothesis does not exclude the possibility that ScSls1p will then stabilize the Sec63p-activated ScKar2p complex.
In summary, our results demonstrate the involvement of a newly identified ER protein in the translocation process by its direct effect on the function of an Hsp70 family member. This protein was not identified in previous studies, probably because of its nonessential function in S. cerevisiae. This highlights the importance of studies carried on non-model organisms such as Y. lipolytica. Sls1p homologues were identified in human and mouse EST libraries and in the recently sequenced Drosophila melanogaster genome, showing that the role of Sls1p in higher eukaryotes and, more particularly, in humans could be more critical than it is in S. cerevisiae or even Y. lipolytica. Moreover, such proteins, which are nonessential in reconstituted in vitro systems, could be required for fine physiological regulation of complex mechanisms such as protein secretion, folding, or degradation, and defects in these proteins could be involved in several diseases. We still have to define in more detail the role of Sls1p in ERAD and/or folding with the genetic identification and analysis of specific mutants. Biochemical experiments are in progress to identify the precise role of Sls1p and to analyze more accurately its molecular relationships with Kar2p and Sec63p (or another DnaJ partner, such as Scj1p [3, 65] or Jem1p [53]).
Moreover, previous findings in Y. lipolytica (5) showed that overexpression of YlSls1p retained secretory proteins in the ER, delaying their transit through the secretory pathway. Similar effects were observed when hamster BiP was overexpressed in mammalian cells (15). Whether this effect is due to a chaperone-like function of Sls1p itself or relies on its association with Kar2p will have to be studied. This work supports previous findings that various classes of cofactors act to modulate the Hsp70s activity and to ensure their functional specificity.
ACKNOWLEDGMENTS
We thank Jeffrey L. Brodsky and Amie J. McClellan for strains, plasmids, and helpful discussion; Rosine Haguenauer-Tsapis for the gift of α-factor, CPY, and Gas1p antibodies and for helpful discussion; Tom A. Rapoport and Ian Collinson for T59G and T249G mutants; and David M. Ogrydziak and Anita Boisramé for critical reading of the manuscript.
This work was supported by a EEC BIOT4-CT96003 fellowship (M.K.).
REFERENCES
- 1.Ausebel F, Brent R, Kingston R, Moore D, Seidman J G, et al. Current protocols in molecular biology. New York, N.Y: Wiley Interscience; 1989. [Google Scholar]
- 2.Beckmann R, Bubeck D, Grassucci R, Penczek P, Verschoor A, Blobel G, Frank J. Alignment of conduits for the nascent polypeptide chain in the ribosome-Sec61 complex. Science. 1997;278:2123–2126. doi: 10.1126/science.278.5346.2123. [DOI] [PubMed] [Google Scholar]
- 3.Blumberg H, Silver P A. A homologue of the bacterial heat-shock gene DnaJ that alters protein sorting in yeast. Nature. 1991;349:627–630. doi: 10.1038/349627a0. [DOI] [PubMed] [Google Scholar]
- 4.Boisrame A, Beckerich J M, Gaillardin C. A mutation in the secretion pathway of the yeast Yarrowia lipolytica that displays synthetic lethality in combination with a mutation affecting the signal recognition particle. Mol Gen Genet. 1999;261:601–609. doi: 10.1007/s004380050002. [DOI] [PubMed] [Google Scholar]
- 5.Boisrame A, Beckerich J M, Gaillardin C. Sls1p, an endoplasmic reticulum component, is involved in the protein translocation process in the yeast Yarrowia lipolytica. J Biol Chem. 1996;271:11668–11675. doi: 10.1074/jbc.271.20.11668. [DOI] [PubMed] [Google Scholar]
- 6.Boisrame A, Kabani M, Beckerich J M, Hartmann E, Gaillardin C. Interaction of Kar2p and Sls1p is required for efficient co-translational translocation of secreted proteins in the yeast Yarrowia lipolytica. J Biol Chem. 1998;273:30903–30908. doi: 10.1074/jbc.273.47.30903. [DOI] [PubMed] [Google Scholar]
- 7.Brennwald P, Liao X, Holm K, Porter G, Wise J A. Identification of an essential Schizosaccharomyces pombe RNA homologous to the 7SL component of signal recognition particle. Mol Cell Biol. 1988;8:1580–1590. doi: 10.1128/mcb.8.4.1580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brodsky J L, Goeckeler J, Schekman R. BiP and Sec63p are required for both co- and posttranslational protein translocation into the yeast endoplasmic reticulum. Proc Natl Acad Sci USA. 1995;92:9643–9646. doi: 10.1073/pnas.92.21.9643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brodsky J L, Schekman R. A Sec63p-BiP complex from yeast is required for protein translocation in a reconstituted proteoliposome. J Cell Biol. 1993;123:1355–1363. doi: 10.1083/jcb.123.6.1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brodsky J L, Werner E D, Dubas M E, Goeckeler J L, Kruse K B, McCracken A A. The requirement for molecular chaperones during endoplasmic reticulum-associated protein degradation demonstrates that protein export and import are mechanistically distinct. J Biol Chem. 1999;274:3453–3460. doi: 10.1074/jbc.274.6.3453. [DOI] [PubMed] [Google Scholar]
- 11.Chappell T G, Konforti B B, Schmid S L, Rothman J E. The ATPase core of a clathrin uncoating protein. J Biol Chem. 1987;262:746–751. [PubMed] [Google Scholar]
- 12.Corsi A K, Schekman R. The lumenal domain of Sec63p stimulates the ATPase activity of BiP and mediates BiP recruitment to the translocon in Saccharomyces cerevisiae. J Cell Biol. 1997;137:1483–1493. doi: 10.1083/jcb.137.7.1483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Corsi A K, Schekman R. Mechanism of polypeptide translocation into the endoplasmic reticulum. J Biol Chem. 1996;271:30299–30302. doi: 10.1074/jbc.271.48.30299. [DOI] [PubMed] [Google Scholar]
- 14.Deshaies R J, Koch B D, Werner-Washburne M, Craig E A, Schekman R. A subfamily of stress proteins facilitates translocation of secretory and mitochondrial precursor polypeptides. Nature. 1988;332:800–805. doi: 10.1038/332800a0. [DOI] [PubMed] [Google Scholar]
- 15.Dorner A J, Wasley L C, Kaufman R J. Overexpression of GRP78 mitigates stress induction of glucose regulated proteins and blocks secretion of selective proteins in Chinese hamster ovary cells. EMBO J. 1992;11:1563–1571. doi: 10.1002/j.1460-2075.1992.tb05201.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dunnwald M, Varshavsky A, Johnsson N. Detection of transient in vivo interactions between substrate and transporter during protein translocation into the endoplasmic reticulum. Mol Biol Cell. 1999;10:329–344. doi: 10.1091/mbc.10.2.329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Feldheim D, Rothblatt J, Schekman R. Topology and functional domains of Sec63p, an endoplasmic reticulum membrane protein required for secretory protein translocation. Mol Cell Biol. 1992;12:3288–3296. doi: 10.1128/mcb.12.7.3288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Flaherty K M, DeLuca-Flaherty C, McKay D B. Three-dimensional structure of the ATPase fragment of a 70K heat-shock cognate protein. Nature. 1990;346:623–628. doi: 10.1038/346623a0. [DOI] [PubMed] [Google Scholar]
- 19.Gassler C S, Buchberger A, Laufen T, Mayer M P, Schroder H, Valencia A, Bukau B. Mutations in the DnaK chaperone affecting interaction with the DnaJ cochaperone. Proc Natl Acad Sci USA. 1998;95:15229–15234. doi: 10.1073/pnas.95.26.15229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gething M J. Role and regulation of the ER chaperone BiP. Semin Cell Dev Biol. 1999;10:465–472. doi: 10.1006/scdb.1999.0318. [DOI] [PubMed] [Google Scholar]
- 21.Gilmore R, Blobel G, Walter P. Protein translocation across the endoplasmic reticulum. I. Detection in the microsomal membrane of a receptor for the signal recognition particle. J Cell Biol. 1982;95:463–469. doi: 10.1083/jcb.95.2.463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gilmore R, Walter P, Blobel G. Protein translocation across the endoplasmic reticulum. II. Isolation and characterization of the signal recognition particle receptor. J Cell Biol. 1982;95:470–477. doi: 10.1083/jcb.95.2.470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gorlich D, Rapoport T A. Protein translocation into proteoliposomes reconstituted from purified components of the endoplasmic reticulum membrane. Cell. 1993;75:615–630. doi: 10.1016/0092-8674(93)90483-7. [DOI] [PubMed] [Google Scholar]
- 24.Guex N, Peitsch M C. SWISS-MODEL and the Swiss-Pdb Viewer: an environment for comparative protein modeling. Electrophoresis. 1997;18:2714–2723. doi: 10.1002/elps.1150181505. [DOI] [PubMed] [Google Scholar]
- 25.Hamman B D, Hendershot L M, Johnson A E. BiP maintains the permeability barrier of the ER membrane by sealing the lumenal end of the translocon pore before and early in translocation. Cell. 1998;92:747–758. doi: 10.1016/s0092-8674(00)81403-8. [DOI] [PubMed] [Google Scholar]
- 26.Hanein D, Matlack K E, Jungnickel B, Plath K, Kalies K U, Miller K R, Rapoport T A, Akey C W. Oligomeric rings of the Sec61p complex induced by ligands required for protein translocation. Cell. 1996;87:721–732. doi: 10.1016/s0092-8674(00)81391-4. [DOI] [PubMed] [Google Scholar]
- 27.Hann B C, Walter P. The signal recognition particle in S. cerevisiae. Cell. 1991;67:131–144. doi: 10.1016/0092-8674(91)90577-l. [DOI] [PubMed] [Google Scholar]
- 28.He F, Yaver D, Beckerich J M, Ogrydziak D, Gaillardin C. The yeast Yarrowia lipolytica has two, functional, signal recognition particle 7S RNA genes. Curr Genet. 1990;17:289–292. doi: 10.1007/BF00314874. [DOI] [PubMed] [Google Scholar]
- 29.High S. Protein translocation at the membrane of the endoplasmic reticulum. Prog Biophys Mol Biol. 1995;63:233–250. doi: 10.1016/0079-6107(95)00005-8. [DOI] [PubMed] [Google Scholar]
- 30.Hohfeld J, Jentsch S. GrpE-like regulation of the hsc70 chaperone by the anti-apoptotic protein BAG-1. EMBO J. 1997;16:6209–6216. doi: 10.1093/emboj/16.20.6209. . (Erratum, 17:847, 1998.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hohfeld J, Minami Y, Hartl F U. Hip, a novel cochaperone involved in the eukaryotic Hsc70/Hsp40 reaction cycle. Cell. 1995;83:589–598. doi: 10.1016/0092-8674(95)90099-3. [DOI] [PubMed] [Google Scholar]
- 32.James P, Halladay J, Craig E A. Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics. 1996;144:1425–1436. doi: 10.1093/genetics/144.4.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Johnson A E, van Waes M A. The translocon: a dynamic gateway at the ER membrane. Annu Rev Cell Dev Biol. 1999;15:799–842. doi: 10.1146/annurev.cellbio.15.1.799. [DOI] [PubMed] [Google Scholar]
- 34.Kabani M, Boisrame A, Beckerich J M, Gaillardin C. A highly representative two-hybrid genomic library for the yeast Yarrowia lipolytica. Gene. 2000;241:309–315. doi: 10.1016/s0378-1119(99)00476-x. [DOI] [PubMed] [Google Scholar]
- 35.Kaiser C A, Schekman R. Distinct sets of SEC genes govern transport vesicle formation and fusion early in the secretory pathway. Cell. 1990;61:723–733. doi: 10.1016/0092-8674(90)90483-u. [DOI] [PubMed] [Google Scholar]
- 36.Kippert F. A rapid permeabilization procedure for accurate quantitative determination of beta-galactosidase activity in yeast cells. FEMS Microbiol Lett. 1995;128:201–206. doi: 10.1111/j.1574-6968.1995.tb07523.x. [DOI] [PubMed] [Google Scholar]
- 37.Kunkel T A, Roberts J D, Zakour R A. Rapid and efficient site-specific mutagenesis without phenotypic selection. Methods Enzymol. 1987;154:367–382. doi: 10.1016/0076-6879(87)54085-x. [DOI] [PubMed] [Google Scholar]
- 38.Lee I H, Ogrydziak D M. Yarrowia lipolytica SRP54 homolog and translocation of Kar2p. Yeast. 1997;13:499–513. doi: 10.1002/(SICI)1097-0061(199705)13:6<499::AID-YEA100>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- 39.Liberek K, Marszalek J, Ang D, Georgopoulos C, Zylicz M. Escherichia coli DnaJ and GrpE heat shock proteins jointly stimulate ATPase activity of DnaK. Proc Natl Acad Sci USA. 1991;88:2874–2878. doi: 10.1073/pnas.88.7.2874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lyman S K, Schekman R. Binding of secretory precursor polypeptides to a translocon subcomplex is regulated by BiP. Cell. 1997;88:85–96. doi: 10.1016/s0092-8674(00)81861-9. [DOI] [PubMed] [Google Scholar]
- 41.Lyman S K, Schekman R. Interaction between BiP and Sec63p is required for the completion of protein translocation into the ER of Saccharomyces cerevisiae. J Cell Biol. 1995;131:1163–1171. doi: 10.1083/jcb.131.5.1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mamoun C B, Beckerich J M, Gaillardin C. The TSR1 gene of Yarrowia lipolytica is involved in the signal recognition particle-dependent translocation pathway of secretory proteins. J Biol Chem. 1996;271:23895–23901. doi: 10.1074/jbc.271.39.23895. [DOI] [PubMed] [Google Scholar]
- 43.Matlack K E, Misselwitz B, Plath K, Rapoport T A. BiP acts as a molecular ratchet during posttranslational transport of prepro-alpha factor across the ER membrane. Cell. 1999;97:553–564. doi: 10.1016/s0092-8674(00)80767-9. [DOI] [PubMed] [Google Scholar]
- 44.Matlack K E, Plath K, Misselwitz B, Rapoport T A. Protein transport by purified yeast Sec complex and Kar2p without membranes. Science. 1997;277:938–941. doi: 10.1126/science.277.5328.938. . (Erratum, 277:1749.) [DOI] [PubMed] [Google Scholar]
- 45.McClellan A J, Endres J B, Vogel J P, Palazzi D, Rose M D, Brodsky J L. Specific molecular chaperone interactions and an ATP-dependent conformational change are required during posttranslational protein translocation into the yeast ER. Mol Biol Cell. 1998;9:3533–3545. doi: 10.1091/mbc.9.12.3533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Miao B, Davis J E, Craig E A. Mge1 functions as a nucleotide release factor for Ssc1, a mitochondrial Hsp70 of Saccharomyces cerevisiae. J Mol Biol. 1997;265:541–552. doi: 10.1006/jmbi.1996.0762. [DOI] [PubMed] [Google Scholar]
- 47.Milarski K L, Morimoto R I. Mutational analysis of the human HSP70 protein: distinct domains for nucleolar localization and adenosine triphosphate binding. J Cell Biol. 1989;109:1947–1962. doi: 10.1083/jcb.109.5.1947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Minet M, Dufour M E, Lacroute F. Complementation of Saccharomyces cerevisiae auxotrophic mutants by Arabidopsis thaliana cDNAs. Plant J. 1992;2:417–422. doi: 10.1111/j.1365-313x.1992.00417.x. [DOI] [PubMed] [Google Scholar]
- 49.Misselwitz B, Staeck O, Matlack K E, Rapoport T A. Interaction of BiP with the J-domain of the Sec63p component of the endoplasmic reticulum protein translocation complex. J Biol Chem. 1999;274:20110–20115. doi: 10.1074/jbc.274.29.20110. [DOI] [PubMed] [Google Scholar]
- 50.Misselwitz B, Staeck O, Rapoport T A. J proteins catalytically activate Hsp70 molecules to trap a wide range of peptide sequences. Mol Cell. 1998;2:593–603. doi: 10.1016/s1097-2765(00)80158-6. [DOI] [PubMed] [Google Scholar]
- 51.Nelson M K, Kurihara T, Silver P A. Extragenic suppressors of mutations in the cytoplasmic C terminus of SEC63 define five genes in Saccharomyces cerevisiae. Genetics. 1993;134:159–173. doi: 10.1093/genetics/134.1.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ng D T, Walter P. ER membrane protein complex required for nuclear fusion. J Cell Biol. 1996;132:499–509. doi: 10.1083/jcb.132.4.499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nishikawa S, Endo T. The yeast JEM1p is a DnaJ-like protein of the endoplasmic reticulum membrane required for nuclear fusion. J Biol Chem. 1997;272:12889–12892. doi: 10.1074/jbc.272.20.12889. [DOI] [PubMed] [Google Scholar]
- 54.Palleros D R, Reid K L, Shi L, Welch W J, Fink A L. ATP-induced protein-Hsp70 complex dissociation requires K+ but not ATP hydrolysis. Nature. 1993;365:664–666. doi: 10.1038/365664a0. [DOI] [PubMed] [Google Scholar]
- 55.Panzner S, Dreier L, Hartmann E, Kostka S, Rapoport T A. Posttranslational protein transport in yeast reconstituted with a purified complex of Sec proteins and Kar2p. Cell. 1995;81:561–570. doi: 10.1016/0092-8674(95)90077-2. [DOI] [PubMed] [Google Scholar]
- 56.Peitsch M C. ProMod and Swiss-Model: internet-based tools for automated comparative protein modelling. Biochem Soc Trans. 1996;24:274–279. doi: 10.1042/bst0240274. [DOI] [PubMed] [Google Scholar]
- 57.Rapoport T A, Matlack K E, Plath K, Misselwitz B, Staeck O. Posttranslational protein translocation across the membrane of the endoplasmic reticulum. Biol Chem. 1999;380:1143–1150. doi: 10.1515/BC.1999.145. [DOI] [PubMed] [Google Scholar]
- 58.Rapoport T A, Rolls M M, Jungnickel B. Approaching the mechanism of protein transport across the ER membrane. Curr Opin Cell Biol. 1996;8:499–504. doi: 10.1016/s0955-0674(96)80027-5. [DOI] [PubMed] [Google Scholar]
- 59.Rothblatt J A, Deshaies R J, Sanders S L, Daum G, Schekman R. Multiple genes are required for proper insertion of secretory proteins into the endoplasmic reticulum in yeast. J Cell Biol. 1989;109:2641–2652. doi: 10.1083/jcb.109.6.2641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sadler I, Chiang A, Kurihara T, Rothblatt J, Way J, Silver P. A yeast gene important for protein assembly into the endoplasmic reticulum and the nucleus has homology to DnaJ, an Escherichia coli heat shock protein. J Cell Biol. 1989;109:2665–2675. doi: 10.1083/jcb.109.6.2665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sanchez M, Beckerich J M, Gaillardin C, Dominguez A. Isolation and cloning of the Yarrowia lipolytica SEC65 gene, a component of the yeast signal recognition particle displaying homology with the human SRP19 gene. Gene. 1997;203:75–84. doi: 10.1016/s0378-1119(97)00496-4. [DOI] [PubMed] [Google Scholar]
- 62.Schmid D, Baici A, Gehring H, Christen P. Kinetics of molecular chaperone action. Science. 1994;263:971–973. doi: 10.1126/science.8310296. [DOI] [PubMed] [Google Scholar]
- 63.Scidmore M A, Okamura H H, Rose M D. Genetic interactions between KAR2 and SEC63, encoding eukaryotic homologues of DnaK and DnaJ in the endoplasmic reticulum. Mol Biol Cell. 1993;4:1145–1159. doi: 10.1091/mbc.4.11.1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Shlomai J, Kornberg A. A prepriming DNA replication enzyme of Escherichia coli. I. Purification of protein n′: a sequence-specific, DNA-dependent ATPase. J Biol Chem. 1980;255:6789–6793. [PubMed] [Google Scholar]
- 65.Silberstein S, Schlenstedt G, Silver P A, Gilmore R. A role for the DnaJ homologue Scj1p in protein folding in the yeast endoplasmic reticulum. J Cell Biol. 1998;143:921–933. doi: 10.1083/jcb.143.4.921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Simon S M, Blobel G. A protein-conducting channel in the endoplasmic reticulum. Cell. 1991;65:371–380. doi: 10.1016/0092-8674(91)90455-8. [DOI] [PubMed] [Google Scholar]
- 67.Travers K J, Patil C K, Wodicka L, Lockhart D J, Weissman J S, Walter P. Functional and genomic analyses reveal an essential coordination between the unfolded protein response and ER-associated degradation. Cell. 2000;101:249–258. doi: 10.1016/s0092-8674(00)80835-1. [DOI] [PubMed] [Google Scholar]
- 68.Wei J, Gaut J R, Hendershot L M. In vitro dissociation of BiP-peptide complexes requires a conformational change in BiP after ATP binding but does not require ATP hydrolysis. J Biol Chem. 1995;270:26677–26682. doi: 10.1074/jbc.270.44.26677. [DOI] [PubMed] [Google Scholar]
- 69.Zhu X, Zhao X, Burkholder W F, Gragerov A, Ogata C M, Gottesman M E, Hendrickson W A. Structural analysis of substrate binding by the molecular chaperone DnaK. Science. 1996;272:1606–1614. doi: 10.1126/science.272.5268.1606. [DOI] [PMC free article] [PubMed] [Google Scholar]