Abstract
We report the identification and characterization of a new Drosophila clock-regulated gene, takeout (to). to is a member of a novel gene family and is implicated in circadian control of feeding behavior. Its gene expression is down-regulated in all of the clock mutants tested. In wild-type flies, to mRNA exhibits daily cycling expression but with a novel phase, delayed relative to those of the better-characterized clock mRNAs, period and timeless. The E-box-containing sequence in the to promoter shows impressive similarities with those of period and timeless. However, our results suggest that the E box is not involved in the amplitude and phase of the transcriptional cycling of to. The circadian delayed transcriptional phase is therefore most likely the result of indirect regulation through unknown transcription factors.
Circadian (∼24-h) behavioral and physiological rhythms are manifest in virtually all organisms. Our understanding of the underlying molecular rhythms comes largely from genetic investigations of five different classes of organisms: plants (28), photosynthetic bacteria (17), Neurospora (8), Drosophila (32), and mice (44, 47). Recent progress has reinforced the negative feedback regulation of transcription, originally proposed for Drosophila (14, 14a, 15a, 50), as a central theme of circadian rhythms in these organisms (9). In particular, Drosophila clocks display conservation with mammalian clocks. At the sequence level, many Drosophila clock components have one or more mammalian homologs, which are suggested to play similar roles in mammalian rhythms. This further validates Drosophila as an animal model system for the study of circadian rhythms.
The first Drosophila clock component identified was the period (per) gene (3, 20, 31). Biochemical and genetic data suggested a transcriptional autoregulatory feedback loop involving PER (14, 14a, 15a, 50). The second essential pacemaker component, timeless (tim), was subsequently identified, and both per and tim reciprocally autoregulate at the transcriptional level (29, 39). TIM dimerizes with PER (10, 24, 51), and the interaction is suggested to be important for the posttranscriptional regulation and nuclear entry of both proteins (35, 48). Although their precise biochemical functions are not certain, PER and TIM probably function directly in the negative regulation of transcription (7, 22). In contrast, the biochemical functions of the recently identified clock genes Clock (Clk) and cycle (cyc) are apparent from their primary sequences (1, 7, 34). Both CLK and CYC belong to the basic helix-loop-helix (bHLH)–PAS (Per-Arnt-Sim) transcription factor family, members of which are involved in a wide range of other life processes. For example, the mammalian ARNT-AHR heterodimer is involved in xenobiotic resistance (37), and the Drosophila SIM-TANGO heterodimer is involved in embryonic development of the central nervous system midline cells (41).
In the Drosophila mutants Clkjrk and cyc01 (1, 34), the rate of transcription of the two major clock components, per and tim, is very low. Both mammalian and Drosophila CLK and CYC were found to bind the Drosophila per 21-bp E-box-containing sequence in yeast one-hybrid assays (7, 11). Binding was also shown by DNA mobility shift assays (23). CLK-CYC was also found to activate transcription from promoters containing four copies of the 18-bp E-box-containing sequence from both the per and tim promoters in Drosophila cell culture, and the activation was dramatically reduced by an E-box central 2-bp mutation (7).
Recent studies have suggested that the CLK-CYC heterodimer may directly regulate circadian output genes as well as central clock genes. The neuropeptide arginine vasopressin is synthesized and released in a circadian manner from suprachiasmatic nucleus neurons. It is involved in peripheral salt and water balance (38) and also has some distinct effects within the central nervous system (16). The vasopressin peptide rhythm was found to be transcriptionally regulated (6). More recently, the vasopressin mRNA rhythm was shown to be abolished in Clock/Clock mice (18). An E-box-containing sequence in its promoter was found to be necessary for the CLK-BMAL1-mediated transcriptional activation in cell culture, suggesting that CLK-BMAL1 may directly regulate vasopressin transcription (18). Another output gene, dbp, which encodes a basic-leucine zipper transcription factor (25), is down-regulated in Clock/Clock mice, and CLK is found in a protein complex that binds to an E box within the dbp first intron (31a). There is therefore strong evidence that the CLK–BMAL1–E-box complex is relevant to output gene as well as clock gene transcription (11, 18). This indicates that CLK-BMAL1 acts directly on this E box.
In this paper, we report the identification, characterization, and transcriptional regulation of a novel clock-regulated output gene, takeout (to). to was identified through a subtractive hybridization that enriched genes differentially expressed in cyc01 and wild-type flies. to mRNA levels are undetectable in cyc01 and Clkjrk mutant flies, and to transcription cycles with a delayed phase compared to that of per and tim. Functional analysis (36) shows that TO is involved in an output pathway which conveys temporal and food status information to feeding-relevant metabolisms and activities. Our data here suggest that a prominent E box in the to promoter is not involved in its temporal transcriptional regulation. Therefore, there is most likely an indirect regulation by CLK and CYC which gives rise to the delayed to transcriptional phase relative to per and tim.
MATERIALS AND METHODS
Fly strains.
The strain of wild-type flies used was Canton-S. cyc01;ry506,per01,yw;tim01,per01;tim01;ry506, and yw;Clkjrk flies were used for analysis. Genomic DNA of the flies was tested with PCR for the presence of to.
Constructs and transgenic flies.
The lacZ constructs were generated by inserting SalI/SalI or XhoI/XhoI promoter fragments (Fig. 1A) into the XhoI-cut pBglII-lacZ reporter vector (11). The to21-lacZ XhoI/XhoI insert sequence is CCGCTCGAGGCAGCTCACGTGATGGAACTCGAGCGG, and the to21e-lacZ sequence is CCGCTCGAGGCAGCTCAgcTGATGGAACTCGAGCGG (lowercase letters indicate mutation changes). Likewise, the per21-lacZ XhoI/XhoI insert sequence is CCGCTCGAGCCGCCGCTCACGTGGCGAACTCTCGAG, and the per21e-lacZ sequence is CCGCTCGAGCCGCCGCTCAgcTGGCGAACTCTCGAG. to80x3-lacZ was generated by synthesizing the 80 bp around the E box with SalI on one side and XhoI on the other. Since SalI and XhoI have compatible ends, multimers of the 80 bp were generated by cutting, washing, ligating, and then cutting with both restriction enzymes. Trimers were selected after running on an agarose gel and then ligated to the XhoI-cut pBglII-lacZ reporter vector. The per 69x3-lacZ construct was made likewise.
FIG. 1.
lacZ reporter yeast constructs (A) and luciferase reporter constructs and flies (B). Phs, heat shock promoter.
The luciferase constructs were generated in the CaSpeR4 vector. They contain a basal heat shock promoter (CaSpeR-hs43-lacZ; GenBank accession number X81643) ligated to SalI/KpnI of the luciferase (42). The XhoI site at the 5′ end of the basal heat shock promoter allowed insertion of the different SalI/XhoI and SalI/SalI promoter pieces (Fig. 1B). Note that SalI and XhoI have compatible ends, and ligation of the two overhangs destroys the restriction site. The reporter constructs were then used to generate germ line transformants by injecting yw;Ki pP [ry+ Δ2-3]/+.
PCR-based cDNA subtraction and screening.
Wild-type and cyc01 mutant flies of the same age were entrained at 25°C in a 12-h light–12-h dark (LD) cycle for 2 days before being collected at zeitgeber time 15. (ZT15; zeitgeber time is the time in hours in a 12-h light–2-h dark cycle, where ZT0 is lights on and ZT12 is lights off). Frozen heads were isolated, and total RNA was extracted using TRIzol reagent (GibcoBRL). Poly(A)+ RNA was prepared using the PolyATract mRNA isolation system (Promega). cDNAs were prepared from 2 μg of the poly(A)+ mRNA from each sample and were hybridized according to the Clontech protocol. Conditions of the PCR selection were optimized by monitoring the subtraction efficiency by measuring the level of controls: tim, which is rare and differentially expressed in the two subtraction samples; and rhodopsin, which is abundant and equally expressed. The level of the controls in the subtracted and unsubtracted PCR products was measured by slot blot hybridization. Subtracted PCR products were then cloned into NotI-digested pBluescript II KS+ and used to transform Escherichia coli DH5α (UltraMAX DH5α-FT competent cells; GibcoBRL). Transformation efficiency of 2 × 107 CFU/μg was obtained. Clones were randomly picked, and their plasmid inserts were amplified by PCR. The PCR products were transferred to HyBond-N+ membranes using a dot blot minifold (Schleicher & Schuell, Inc.). The membranes were hybridized to 32P- or fluorescein-labeled subtracted and unsubtracted PCR products. Clones that showed substantial differential expression in the two populations of probes were further examined on Northern blots.
RNA extraction and analyses.
For Northern blots, total and poly(A)+ RNAs were prepared as described above. One-microgram samples of poly(A)+ mRNA were loaded on formaldehyde gels and then transferred onto nylon membranes. 32P-labeled probes were prepared by random priming of gel-purified fragments using Prime-It II (Stratagene). Prehybridization (∼1 h) and hybridization (∼16 h) were performed at 65°C in 10 ml of Church buffer (0.5 M NaHPO4 [pH 7.2], 7% sodium dodecyl sulfate, 1% bovine serum albumin, 1 mM EDTA). The membranes were washed in washing buffer (0.2× SSC [1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate] 0.1% SDS) twice briefly at room temperature and then twice at 60°C for 20 min before being exposed to film either at room temperature or at −80°C with an intensifying screen, depending on the strength of the signals. RNase protection assays were performed as described by Marrus et al. (27). The RNA probe protects nucleotide (nt) 529 to 839 of the cDNA region.
mRNA in situ hybridization.
Flies were entrained to LD cycles. Frozen sections (10 μm) of adult heads and bodies were cut, and in situ hybridization was performed as described by Hasan (14b). Digoxigenin-labeled riboprobes were prepared from the full-length to and per cDNAs and were hydrolyzed prior to use. All hybridization and washes were performed at 65°C.
Genomic and cDNA library screening.
A Drosophila genomic library in EMBL3 was generously provided by Ron Blackman. Two Drosophila adult head cDNA libraries were screened. Seven clones were sequenced, and a full-length sequence of 1,064 nt was obtained. While this report was under review, the Drosophila genome sequence was assembled and annotated. The to genomic sequence is confirmed by genomic scaffold accession number AE003751, the CG11853 gene. The λZAPII cDNA library generated with half oligo(dT) and half random primers was generously provided by Thomas Schwarz; the directionally cloned λEXLX(−) cDNA library generated with oligo(dT) primers alone was generously provided by Bruce Hamilton (30). Molecular techniques were performed using standard protocols (2).
Nuclear run-on assay.
Nuclear run-on assays were performed as described by So and Rosbash (40). PCR products covering the full-length genomic region (∼1.4 kb) were used as probe.
Yeast one-hybrid assay.
The yeast one-hybrid assay was performed as described elsewhere (7), using the constructs described above and in Fig. 1A.
Luminescence monitoring and analysis.
Luminescence monitoring was performed as described elsewhere (5). The readings were taken every 30 min, and data were analyzed according to the I-and-A software documentation (30a). The food was prepared with 1% Bacto Agar, 5% sucrose, and 15 mM luciferin.
RESULTS
to is a novel clock-regulated gene.
To identify novel genes involved in circadian rhythms, we performed a PCR-based cDNA subtraction and screening (Clontech; see Materials and Methods) whereby poly(A)+ RNA from heads of cycle null mutant (cyc01) flies was subtracted from wild-type RNA. We speculated that genes regulated by this transcription factor are inessential and that some of them are related to circadian behavior. Therefore, the aim was to find genes under circadian regulation by identifying genes differentially expressed in wild-type and cyc01 mutant flies. After screening 108 subtracted clones, we identified three different novel genes that are down-regulated in cyc01 flies. Here we present the cloning, characterization, and transcriptional regulation of one of them, to.
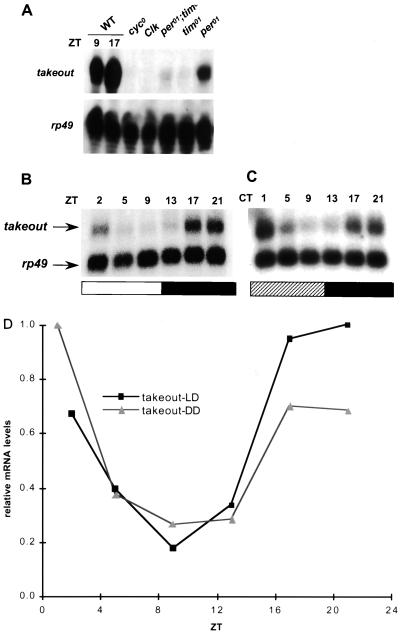
to mRNA expression is down-regulated in cyc01 flies and in all other circadian mutants tested (Fig. 2A). Its level is undetectable in cyc01 and Clkjrk mutants, as measured by RNase protection and Northern blotting. In contrast, there is detectable to mRNA in all other genotypes tested, though it is substantially lower than that in wild-type flies. As there is little or no functional CLK-CYC heterodimer in the cyc01 and Clkjrk backgrounds, the simplest way to explain this observation is that to is directly regulated by CLK and CYC. The higher to transcription in per01, tim01, and per01 tim01 double-mutant flies is presumably due to residual functional CLK-CYC heterodimer in these backgrounds (22; L. Sarov-Blat, unpublished data). per01 flies reproducibly showed a higher level of to expression than tim01, indicating that PER and TIM may differentially regulate to expression. However, the mechanism underlying this difference is still unknown. When mRNA levels at different time points were measured, to did not show a significant cycling pattern in the clock mutants tested (data not shown).
FIG. 2.
to mRNA expression. (A) RNase protection assay showing that to mRNA is down-regulated in clock mutants. WT denotes wild type; rp49 is an internal control. The gel is overexposed to show the low-level signal in per01, tim01, and double-mutant flies. (B and C) Northern blots showing the to mRNA cycling profiles in LD (B) and constant dark (C) conditions. To obtain comparable signals, the rp49 probe was about half the length of and 1/40 lower in specific activity than the to probe. Open and filled bars represent the time when light was on (ZT0 to ZT12) and off (ZT12 to ZT24), respectively; the hatched bar represents the subjective day (circadian time [CT] 0 to 12). (D) Quantification of to mRNA levels in panels B and C. The to mRNA signals were normalized to the rp49 signals. DD, constant dark.
to mRNA levels cycle with a novel phase in the head.
In wild-type flies, to mRNA levels exhibit a daily fluctuation in both cycling LD and constant dark conditions (Fig. 2B to D). The cycling in free-running conditions indicates that this property is a function of the endogenous clock rather than light driven. The amplitude (peak-to-trough ratio) of cycling is about 5, significantly smaller than those of per (∼10) and tim (>10) mRNA expression. Interestingly, from several Northern blot analyses and RNase protection assays, the mRNA levels peak at about ZT17 to ZT20, a 2- to 5-h phase delay with respect to the per and tim cycling profiles.
Since to expression is down-regulated in the clock mutants, it was of interest to learn if the regulation is directly via CLK and CYC. To determine the localization of to expression, mRNA in situ hybridization to fly head sections was performed. For comparison, clock-expressing cells in the brain were also visualized by assaying tim expression as well as Clk and cyc expression. The in situ results showed very similar expression patterns at each gene's high time points (Fig. 3A). All four genes are expressed throughout the brain cortex, especially in the region between the optic lobe and the central complex, where the lateral neurons are located. They are also expressed in photoreceptor cells, although the to photoreceptor signal is generally lower than that in the brain cortex. However, to was not observed in other tim-expressing cells such as the glial cells in the optic lobe, the central body in the central complex, and the proboscis (data not shown). In general, the tim expression pattern is very similar to that of Clk and cyc. In bodies, the to expression pattern is a subset of the tim expression pattern (36). The data here suggest that to is expressed in a significant subset of clock-expressing cells in the head (see Discussion). However, due to the low resolution of mRNA in situ hybridization, we cannot rule out the possibility that to is expressed in cells adjacent to clock cells.
FIG. 3.
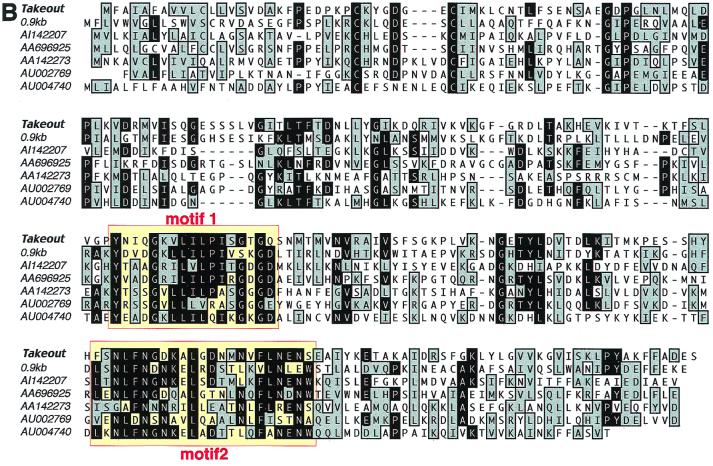
(A) to mRNA is colocalized with tim and exhibits cycling expression in adult head sections. Shown are in situ hybridizations of antisense digoxigenin RNA probes for to, tim, Clk, and cyc, frontal sections of adult head at ZT13 and ZT23. Arrows point to where the lateral neurons are located. The mRNA expression of to and tim cycles with a different phase. Expression of both to and tim was detected in the photoreceptors and brain cortex, especially in the optic lobe regions. tim has broader expression, e.g., in the glial cells and in the central body of the central complex. Clk and cyc have the same expression pattern as tim. In situ hybridizations using sense RNA probes did not show obvious signals. (B) Alignment of the to family members. Boxes indicate the signature motifs that define the to family. Out of the 10 family members identified so far, 7 that have full-length or almost full-length conceptual translation sequences are shown. Five of the members, including to and 0.9kb (GenBank accession number AL024485), AA696925, and AA142273, are from Drosophila melanogaster. Two, including AI142207, are from Manduca sexta. Three, including AU002769 and AU004740, are from Bombyx mori. The original EST sequences were obtained from the BDGP/HHMI Drosophila EST project. The EST sequences shown here are refinements from the different clone sequences, and accession numbers of the representative ones are used. Black background with white letters indicates identity; grey background with black letters indicates similarity.
to is a member of a novel family.
The full-length sequence of the 1,064-nt to gene was obtained through screening of two Drosophila adult head cDNA libraries (see Materials and Methods). The sequence was later confirmed with the expressed sequence tag (EST) clones derived from an adult Drosophila head cDNA library (BDGP/HHMI Drosophila EST project; accession numbers AI403166 for the 5′ sequence and AI107005 for the 3′ sequence). A BLAST search against the nonredundant database using the open reading frame indicates that to is a novel gene. It has sequence similarity with a Drosophila gene called 0.9kb (26) (see Discussion) and a group of EST clones from insects (Fig. 3B). They form a novel protein family with approximately 250 amino acid residues. Sequence similarity extends throughout the entire protein, with two stretches of highly conserved regions defined as motifs 1 and 2. These regions were used during database searching as criteria for defining family members. Family members have been found only in insects, and sequence analysis suggests a ligand-binding function (see Discussion).
to mRNA cycling is transcriptionally regulated.
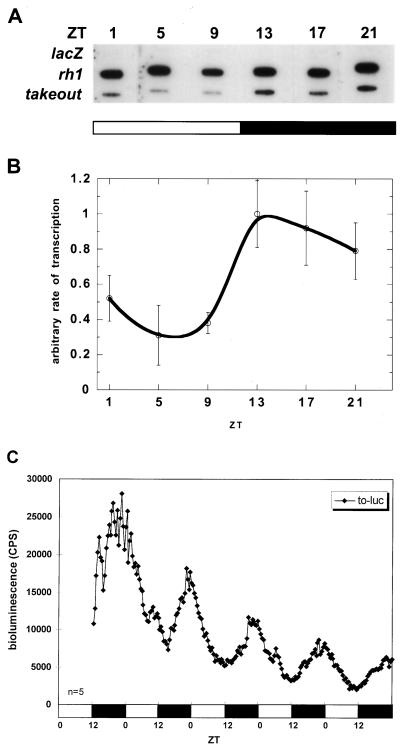
To determine the level of regulation of to mRNA cycling, the in vivo transcription rate was measured by a nuclear run-on assay. The results show that the to transcription rate exhibits a daily fluctuation (Fig. 4A and B). The peak of transcription is about 3 to 4 h in advance of the mRNA peak (Fig. 2B to D), as expected for a gene regulated at the level of transcription (40). Consistent with the mRNA comparisons, the transcription profile of to is delayed by about 3 to 4 h with respect to the well-characterized per and tim profiles (40). This is most apparent by comparing the rising phase of transcription: per and tim transcription starts to rise at about ZT5 (40), whereas to transcription starts at about ZT9.
FIG. 4.
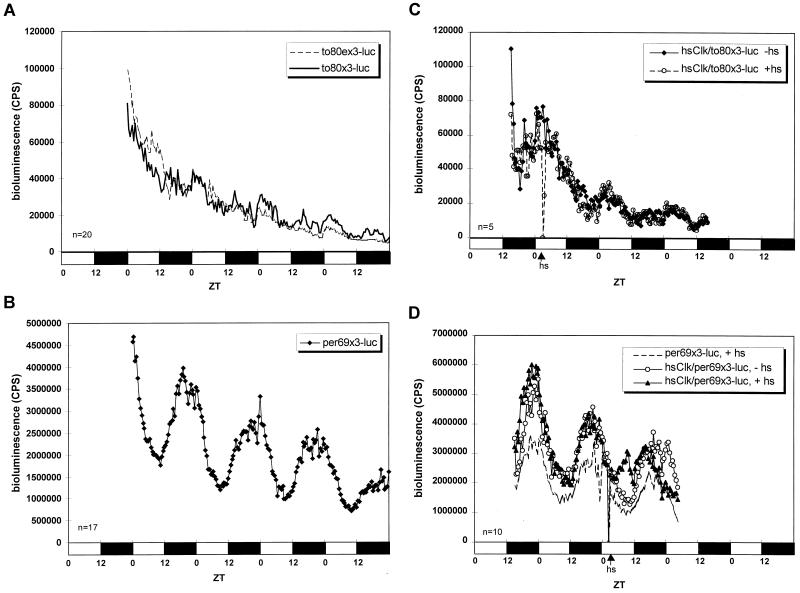
Rate of transcription in wild-type flies. (A) Representative blot from a nuclear run-on assay. Each column is an individual hybridization blot from flies entrained and collected at the time indicated above the blot. Each row shows hybridization signals from the genes indicated on the left. The lacZ gene is a negative control, and the rhodopsin gene (rh1) is an internal control that has constant transcription rates throughout the day. (B) Quantification of rate of to transcription. The run-on signals were normalized to rh1 signals. Averaged data are shown. Error bars indicate the standard deviation (n = 3 to 4). (C) Temporal luminescence from to-luc flies in LD. Average bioluminescence of representative lines is shown. Flies are heterozygous for the transgene. The number of flies used for the analysis is shown at the bottom left; open bars indicate lights on, and filled bars indicate lights off.
The phase delay was reproduced with a to 3.0-kb promoter driving a luciferase reporter gene in transgenic flies (to-luc [Fig. 4C]). A clear cycling of luminescence was exhibited by all six lines. The luminescence intensity is about two- to threefold higher than that of plo (luciferase driven by per promoter only [5, 42]), consistent with the estimated mRNA expression level difference between to and per observed in Northern blots and RNase protection assays (data not shown). The cycling peaks at about ZT23 and is about 5-h phase delayed compared to plo, which peaks at about ZT18 (5, 42). This strongly suggests that the regulatory information for the to phase of transcriptional cycling is contained within this 3.0-kb promoter fragment.
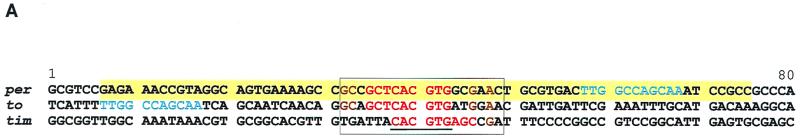
to promoter sequence contains an E box similar to those of the per and tim promoters.
The to promoter sequence revealed a remarkable sequence identity with the E-box region of the per and tim promoters (Fig. 5A). In particular, there is a 9-bp sequence identity around this E-box sequence. The other E-box sequences known in circadian genes usually share the 6-bp core sequence or the core sequence with an additional A (CACGTGA), which has shown to be strongly preferred by the mammalian BMAL1-MOP4 bHLH-PAS transcription factor heterodimer (15). In fact, the to and per promoters share 13 out of the 18 bp shown to be sufficient to drive transcriptional activation in S2 cells (7). This is also consistent with the fact that to mRNA is undetectable in cyc01 and Clkjrk mutants (Fig. 2A), suggesting that CLK-CYC regulates to transcription directly. This would be similar to per and tim transcriptional regulation, despite the phase difference.
FIG. 5.
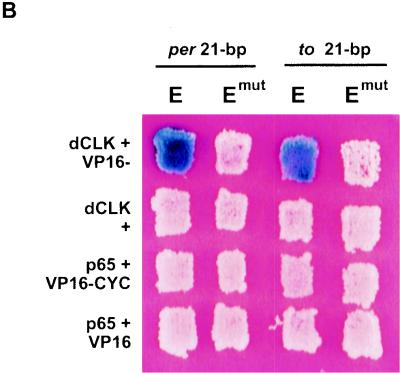
(A) The to promoter has sequence identity with per and tim promoters. In addition to the 9-bp sequence identity (in red) around the E box (underlined), there are adjacent identities (in brown) within the 18 bp (boxed) shown to be sufficient to drive transcriptional activation in S2 cells (7). The to 80-bp fragment shown here was used for the subsequent studies. The W box (in blue) is within the per 69-bp fragment (shaded in yellow) shown to be sufficient to drive transcriptional cycling in flies (12). This element, although conserved in per and to, was found not to be necessary for temporal regulation of these genes (data not shown). The per E box is at −528 from the transcriptional start; tim is at −678, and to is at −1143. (B) Yeast one-hybrid assays showing the binding of CLK and CYC to per and to upstream sequences. Shown are yeast patches expressing the indicated pairs of proteins (rows) and transformed with the indicated reporter constructs (columns). p65 (synaptotagmin) is a negative control (11). DNA binding results in the activation of the lacZ reporter gene, which in turn results in the blue coloration. Both CLK and CYC are required to bind the wild-type 21-bp E-box-containing sequence from per and to. They do not bind E-box-mutated (Emut) sequences (transversion mutations at the two middle base pairs of the core E-box, CAGCTG).
Binding of CLK and CYC to a 21-bp per E-box-containing fragment has been shown by a yeast one-hybrid assay (7). To determine if CLK and CYC also bind the to 21-bp E-box-containing sequence, similar yeast one-hybrid assays were performed (see Fig. 1A for constructs tested). Binding to the sequence is signaled by activation of the lacZ gene, resulting in blue color. As shown in Fig. 5B and similar to the per 21-bp control, both CLK and CYC are required to bind the wild-type to 21-bp sequence, but they do not bind to the identical sequence containing a mutated E box (CACGTG to CAGCTG, the same central 2-bp transversion mutation as in reference 7). The slight difference in the intensity of the blue coloration may be due to the different 2 μm plasmids used for the expression and reporter constructs.
Previous studies have shown that a 69-bp E-box-containing per upstream sequence fragment is sufficient to drive robust high-amplitude circadian cycling of reporter gene expression in flies (12). Since the to sequence and the yeast results suggest a similar to E box, we tested a comparable to 80-bp upstream sequence for its effect on in vivo transcription. The sequence was chosen so that the E box sits in the middle of the 80 bp (Fig. 5A). Because the nuclear run-on data indicate that the to transcription rate is lower than that of per and tim (reference 40 and Fig. 4A), a trimer of the 80-bp fragment (to80x3-luc) was assayed. At the same time, a mutated E-box reporter construct was also assayed (to80ex3-luc; the same 2-bp transversion mutations used in the yeast one-hybrid assays). As a control, a per 69-bp trimer (per69x3-luc) was examined in parallel.
Flies transformed with to80x3-luc showed surprisingly weak cycling of luminescence (Fig. 6A). Although the cycling was observed in every line, it was not observed in all experiments (data not shown). Compared to the per69x3-luc control (Fig. 6B), not only was the cycling amplitude from the to80x3-luc flies much lower, but luciferase expression was much weaker (note the difference in scale between Fig. 6A and B). Therefore, the to E-box-containing 80-bp fragment does not drive robust transcriptional cycling in vivo. Moreover, the mutated version, to80ex3-luc, showed an identical weak cycling pattern, suggesting that this E box is not relevant to the transcriptional pattern. Consistent with this view, overexpression of CLK driven by a heat shock promoter had no detectable effect on to80x3-luc expression(Fig. 6C), whereas it clearly induced per69x3-luc expression (Fig. 6D). Note that all of these trimer promoter constructs, wild type and E-box mutated, resulted in higher luminescence than the monomer constructs, to-luc (Fig. 4C) and per-luc (plo in reference 5, 42). This is most likely due to cooperative activity of transcription factors that binds to these regulatory elements.
FIG. 6.
The 80-bp E-box-containing sequence in the to promoter is not sufficient for robust transcriptional cycling. (A) Comparison of the wild-type to80x3-luc flies with the mutated to80ex3-luc flies. Both showed a very weak cycling of luminescence. Representative lines and experiment are shown. Flies are heterozygous for the transgene. (B) The per69x3-luc flies showed an impressive rhythmicity of bioluminescence with strong intensity and robust cycling amplitude. (C) Overexpression of CLK in hsClk/to80x3-luc flies did not show an observable change in bioluminescence. hs, heat shock. (D) Overexpression of CLK in hsClk/per69x3-luc flies phase advanced the luciferase reporter gene expression. Heat shock of the per69x3-luc flies alone without the hsClk transgene is also shown as a control. Arrows indicate the time of a 30-min 37°C heat shock. Average bioluminescence of representative lines and experiments is shown. Numbers of flies used for the analyses are shown at the bottom left; open bars indicate lights on, and filled bars indicate lights off.
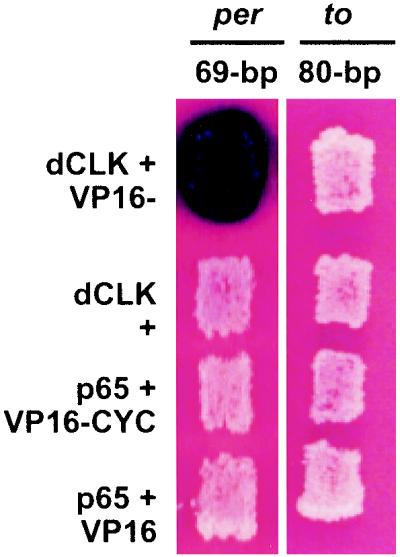
The failure to observe robust expression and transcriptional oscillation with the to 80-bp fragment suggested that the positive yeast two-hybrid result with the to 21-bp fragment was misleading. We therefore tested a larger to fragment in the yeast system. Consistent with the transgenic fly data (Fig. 6), binding of CLK and CYC to the to E box in yeast became undetectable when the E-box-containing sequence was extended from 21 bp (Fig. 5B) to 80 bp (Fig. 7). A similar negative result was observed with 1.5 and 3.0 kb of to upstream sequence (data not shown). On the other hand, binding of CLK and CYC to the per E box was unaffected by the increase in sequence length from 21 to 69 bp. This suggests that there is a major difference between the per and to E-box regions, which explains the different biological activities in flies.
FIG. 7.
Yeast one-hybrid assays showing that CLK and CYC bind a longer upstream sequence from the per promoter (69 bp) but not from the to promoter. Shown are yeast patches expressing the indicated pairs of proteins (rows) and transformed with the indicated reporter constructs (columns). DNA binding results in the activation of the lacZ reporter gene, which in turn results in the blue coloration.
The E box is not necessary for to transcription in S2 cells.
To provide yet another test of the to E box, we transformed S2 cells with a luciferase reporter gene driven by the 3.0-kb to promoter (to-luc). Luminescence was measured in the presence or absence of cotransfected CLK. There was a 3-fold induction of expression of to-luc by the Clk construct, much less than the 60- and 94-fold induction from the per and tim promoter fragments, respectively (Fig. 8A). Furthermore, an E-box deletion from the 3.0-kb to promoter did not diminish the level of transcriptional activation, unlike the E-box deletions from the per and tim promoters (data not shown). The results indicate that the to E box may not contribute to clock-regulated to transcription, suggesting that to transcription requires factors other than CLK and CYC.
FIG. 8.
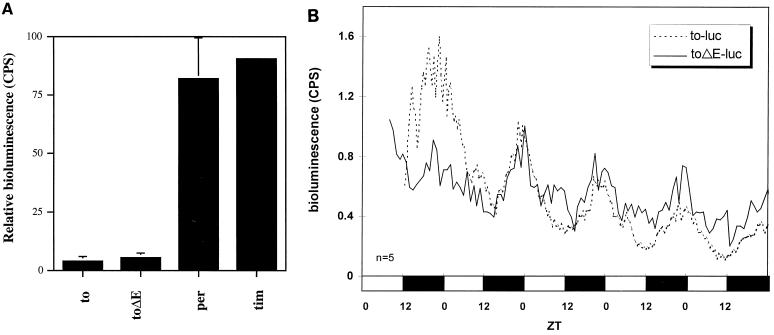
(A) Bioluminescence assays were performed on several promoter-luciferase fusion constructs in S2 cells. Cells were cotransformed with the 3-kb to promoter containing a wild-type or mutant E box and CLOCK-V5 to investigate CLK-CYC-dependent activation of the promoter. Full-length per and tim promoter-luciferase fusions are included as positive controls to demonstrate CLK-CYC-dependent activation on promoters of known circadian genes. The second peaks of the two sets of bioluminescent intensities are set to 1 for easier comparison, because the absolute intensities were slightly different. (B) Comparison of the temporal luminescence from the to-luc and toΔE-luc flies in LD. The 21-bp deletion that removes the E box (toΔE-luc) does not have an obvious effect on the cycling of bioluminescence. Average bioluminescence of representative lines is shown. The second peaks of the two sets of bioluminescence intensities are set to 1 for easier comparison, because the absolute intensities were slightly different. Flies are heterozygous for the transgene. The number of flies used for the analysis is shown at the bottom left; open bars indicate lights on, and filled bars indicate lights off.
The E box is not necessary for to transcription in vivo.
To test the in vivo role of the E box in to transcriptional cycling, transgenic flies carrying to-luc were compared with a 21-bp deletion version that removes the E box (toΔE-luc) [Fig. 1B]. Consistent with the S2 cell data (Fig. 8A), toΔE-luc flies also showed cycling of luminescence with amplitude and phase comparable to that of to-luc (Fig. 8B). This indicates that the E box is not required and that additional elements outside this 21-bp region are sufficient for cycling expression.
DISCUSSION
The amino acid sequence of the TO protein indicates that to is a member of a novel gene family, found only in insects. Although the search shown here failed to reveal any family members with a known biochemical function, a less restricted search identified more distant insect relatives (36). These include two hydrophobic ligand-binding proteins: hemolymph juvenile hormone-binding protein (45) and JP29 (49) from moths. The two ligand-binding proteins share homology throughout the complete sequence with the TO family (36). However, they lack the two motifs that define the family (Fig. 3B), implying that they form a superfamily with TO. The shared biochemical function of the superfamily is presumably to bind a hydrophobic ligand. However, the different family members may bind different ligands.
The to ligand is unknown, but our recent data suggest that it may contribute to feeding-related functions. This is based on the to expression pattern in a few relevant body tissues. A to mutant strain also shows an unusual behavioral response and dies rapidly when subjected to starvation conditions. Finally, to mRNA levels increase in response to starvation, indicating that to expression is regulated by food availability as well as by the circadian pacemaker (36).
The only other family member with some relationship to circadian rhythms is the 0.9kb gene, initially identified as a gene adjacent to the per locus (3, 31). 0.9kb mRNA levels rise shortly before eclosion and decrease within a few hours after eclosion (26). As flies eclose under circadian clock control, the transcript is indirectly under circadian regulation but with only a single burst of expression at the beginning of the adult stage (26). This prior relationship with circadian rhythms therefore may be fortuitous. Preliminary assays on two other Drosophila EST clones in the to family also showed no obvious daily cycling expression by Northern blot analysis (data not shown). Therefore, the to family members may be regulated differently as well as bind different ligands and contribute to different physiological processes.
to cycling is due in large part to transcriptional regulation, as previously described for per and tim. However, the cycling apparently does not require a prominent cis-acting E box, unlike the transcription of per and tim. It is surprising that the to 80-bp and per 69-bp E-box-containing sequences, which have such striking nucleotide identities, are recognized so differently: the per 69-bp sequence is sufficient to drive robust transcriptional cycling, whereas the to counterpart is not (Fig. 5A). The to 80-bp fragment may be missing a separate sequence element required for CLK-CYC binding and activity, it may contain adjacent sequences that are inhibitory to strong binding and activity, or it may be missing an elusive E-box feature. The last possibility is consistent with indications that the core E-box sequence is necessary but not sufficient for potent binding of the mammalian CLK-containing heterodimer (15). The absence of a separate sequence element is consistent with experiments indicating that a single per 18-bp E-box sequence is not sufficient for cycling reporter gene expression in flies (P. Hardin, personal communication). We cannot exclude the possibility that the to 80-bp region contributes more directly to spatial or developmental regulation, as the per 69-bp region alone has shown to be sufficient to mediate proper developmental and spatial expression (13). But the weak CLK-CYC binding and activity suggests that the to E box is not a bona fide, relevant sequence important for to regulation. Moreover, toΔE-luc cycling is identical to to cycling.
There is the complication that a very modest cycling amplitude from the to 80-bp fragment is observed. This might reflect a cryptic, cycling (non-E-box) element within this 80 bp. The poor activity from the per 18-bp fragment may also reflect the lack of this element. Although there are some additional sequence similarities between the to 80-bp and per 69-bp fragments, we have been unable to define any contribution from non-E-box elements to the circadian transcriptional oscillations of to (data not shown). This points to a simple conclusion: the weak cycling amplitude from the to 80-bp element is irrelevant, making little or no contribution to circadian transcription. This hypothesis implies that the key cis-acting circadian elements lie elsewhere within the 3.0-kb promoter fragment, a conclusion consistent with our experiments (Fig. 8B). These elements may even include another, more relevant E box. However, a sequence comparison of the entire 3.0-kb to promoter (outside the 80 bp) with the per 69 bp also does not show any striking identities, including the lack of any additional E-box elements within the entire to genomic region (data not shown). These considerations suggest that to transcription is regulated by sequence elements different from those that govern per and tim transcription and only indirectly by CLK and CYC.
In the Drosophila system, the most dramatic phase differences are between the RNA profiles of Clk mRNA and cryptochrome mRNA on the one hand and those of per and tim on the other (2a, 43). Although it is not certain how this antiphase regulation takes place, it has been suggested that there is significant similarity with the canonical clock gene model but that PER and TIM might be positive regulators of Clk mRNA cycling (23). As they are negative regulators of their own transcription, this would explain the antiphase relationship. The much more modest phase delay of to transcription has not been previously reported for any clock gene or clock output gene. Our negative E-box results suggest a different explanation for the few-hour phase difference between per transcription and to transcription than for the antiphase genes, namely, that CLK-CYC regulation is entirely indirect and that there is another transcription factor or perhaps even more complicated regulatory features interposed between CLK and CYC on the one hand and the to promoter on the other.
Most clock-related RNA cycling previously reported exhibits a phase similar to that of the canonical per and tim patterns (33, 46). This includes output genes that appear directly hard-wired to the clock machinery, i.e., that are directly regulated by the CLK-CYC heterodimer or its mammalian equivalent, indistinguishably from the two canonical clock genes. In mammals, these include vasopressin and the transcription factor gene dbp, which then regulates a secondary set of output genes (21, 25, 31b). In flies, the transcription factor gene Vrille is regulated by CLK-CYC and probably regulates important downstream output genes (4). The Vrille-encoded protein or another putative transcription factor (e.g., see reference 33) could regulate to transcription. The time needed to accumulate active transcription factor would account for the lag between clock gene transcription and to transcription. The identification of the key cis-acting regions responsible for to transcription should help support this model and identify this putative clock-regulated transcription factor.
ACKNOWLEDGMENTS
We thank all members of Rosbash laboratory for helpful discussions and critical readings of the manuscript.
This work was supported by grants from the NSF Center for Biological Timing and the National Institutes of Health. R.A. was supported by an HHMI Postdoctoral Research Fellowship for Physicians and a Burroughs Wellcome Career Award.
REFERENCES
- 1.Allada R, White N E, So W V, Hall J C, Rosbash M. A mutant Drosophila homolog of mammalian Clock disrupts circadian rhythms and transcription of period and timeless. Cell. 1998;93:791–804. doi: 10.1016/s0092-8674(00)81440-3. [DOI] [PubMed] [Google Scholar]
- 2.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. Brooklyn, N.Y: Greene Publishing Associates; 1994. [Google Scholar]
- 2a.Bae K, Lee C, Sidote D, Chuang K Y, Edery I. Circadian regulation of a Drosophila homolog of the mammalian Clock gene: PER and TIM function as positive regulators. Mol Cell Biol. 1998;18:6142–6151. doi: 10.1128/mcb.18.10.6142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bargiello T A, Jackson F R, Young M W. Restoration of circadian behavioural rhythms by gene transfer in Drosophila. Nature. 1984;312:752–754. doi: 10.1038/312752a0. [DOI] [PubMed] [Google Scholar]
- 4.Blau J, Young M W. Cycling vrille expression is required for a functional Drosophila clock. Cell. 1999;99:661–671. doi: 10.1016/s0092-8674(00)81554-8. [DOI] [PubMed] [Google Scholar]
- 5.Brandes C, Plautz J D, Stanewsky R, Jamison C F, Straume M, Wood K V, Kay S A, Hall J C. Novel features of Drosophila period transcription revealed by real-time luciferase reporting. Neuron. 1996;16:687–692. doi: 10.1016/s0896-6273(00)80088-4. [DOI] [PubMed] [Google Scholar]
- 6.Carter D A, Murphy D. Nuclear mechanisms mediate rhythmic changes in vasopressin mRNA expression in the rat suprachiasmatic nucleus. Mol Brain Res. 1992;12:315–321. doi: 10.1016/0169-328x(92)90133-v. [DOI] [PubMed] [Google Scholar]
- 7.Darlington T K, Wager-Smith K, Ceriani M F, Staknis D, Gekakis N, Steeves T D L, Weitz C J, Takahashi J S, Kay S A. Closing the circadian loop: CLOCK-induced transcription of its own inhibitors per and tim. Science. 1998;280:1599–1603. doi: 10.1126/science.280.5369.1599. [DOI] [PubMed] [Google Scholar]
- 8.Dunlap J C. Genetics and molecular analysis of circadian rhythms. Annu Rev Genet. 1996;30:579–601. doi: 10.1146/annurev.genet.30.1.579. [DOI] [PubMed] [Google Scholar]
- 9.Dunlap J C. Molecular bases for circadian clocks. Cell. 1999;96:271–290. doi: 10.1016/s0092-8674(00)80566-8. [DOI] [PubMed] [Google Scholar]
- 10.Gekakis N, Saez L, Delahaye-Brown A-M, Myers M P, Sehgal A, Young M W, Weitz C J. Isolation of timeless by PER protein interactions: defective interaction between timeless protein and long-period mutant PERL. Science. 1995;270:811–815. doi: 10.1126/science.270.5237.811. [DOI] [PubMed] [Google Scholar]
- 11.Gekakis N, Staknis D, Nguyen H B, Davis C F, Wilsbacher L D, King D P, Takahashi J S, Weitz C J. Role of the CLOCK protein in the mammalian circadian mechanism. Science. 1998;280:1564–1569. doi: 10.1126/science.280.5369.1564. [DOI] [PubMed] [Google Scholar]
- 12.Hao H, Allen D L, Hardin P E. A circadian enhancer mediates PER-dependent mRNA cycling in Drosophila melanogaster. Mol Cell Biol. 1997;17:3687–3693. doi: 10.1128/mcb.17.7.3687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hao H, Glossop N R, Lynos L, Morish B, Cheng Y, Helfrich-Forster C, Hardin P E. The 69 bp circadian regulatory sequence (CRS) mediates per-like developmental, spatial, and circadian expression and behavioral rescue in Drosophila. J Neurosci. 1999;19:987–994. doi: 10.1523/JNEUROSCI.19-03-00987.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hardin P E, Hall J C, Rosbash M. Feedback of the Drosophila period gene product on circadian cycling of its messenger RNA levels. Nature. 1990;343:536–540. doi: 10.1038/343536a0. [DOI] [PubMed] [Google Scholar]
- 14a.Hardin P E, Hall J C, Rosbash M. Circadian oscillations in period gene mRNA levels are transcriptionally regulated. Proc Natl Acad Sci USA. 1992;89:11711–11715. doi: 10.1073/pnas.89.24.11711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14b.Hasan G. Molecular cloning of an olfactory gene from Drosophila melanogaster. Proc Natl Acad Sci USA. 1990;87:9037–9041. doi: 10.1073/pnas.87.22.9037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hogenesch J B, Gu Y-Z, Jain S, Bradfield C A. The basic-helix-loop-helix-PAS orphan MOP3 forms transcriptionally active complexes with circadian and hypoxia factors. Proc Natl Acad Sci USA. 1998;95:5474–5479. doi: 10.1073/pnas.95.10.5474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15a.Huang Z J, Edery I, Rosbash M. PAS is a dimerization domain common to Drosophila period and several transcription factors. Nature. 1993;364:259–262. doi: 10.1038/364259a0. [DOI] [PubMed] [Google Scholar]
- 16.Ingram C D, Snowball R K, Mihai R. Circadian rhythms of neuronal activity in suprachiasmatic nucleus slices from the vasopressin-deficient Brattleboro rat. Neuroscience. 1996;75:635–641. doi: 10.1016/0306-4522(96)00274-6. [DOI] [PubMed] [Google Scholar]
- 17.Ishiura M, Kutsuna S, Aoki S, Iwasaki H, Andersson C R, Tanabe A, Golden S S, Johnson C H, Kondo T. Expression of a gene cluster kaiABC as a circadian feedback process in cyanobacteria. Science. 1998;281:1519–1523. doi: 10.1126/science.281.5382.1519. [DOI] [PubMed] [Google Scholar]
- 18.Jin X, Shearman L P, Weaver D R, Zylka M J, de Vries G J, Reppert S M. A molecular mechanism regulating rhythmic output from the suprachiasmatic circadian clock. Cell. 1999;96:57–68. doi: 10.1016/s0092-8674(00)80959-9. [DOI] [PubMed] [Google Scholar]
- 19.Katzenberg D, Young T, Finn L, Lin L, King D P, Takahashi J S, Mignot E. A Clock polymorphism associated with human diurnal preference. Sleep. 1998;21:569–576. doi: 10.1093/sleep/21.6.569. [DOI] [PubMed] [Google Scholar]
- 20.Konopka R J, Benzer S. Clock mutants of Drosophila melanogaster. Proc Natl Acad Sci USA. 1971;68:2112–2116. doi: 10.1073/pnas.68.9.2112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lavery D J, Lopez-Molina L, Margueron R, Fleury-Olela F, Conquet F, Schibler U, Bonfils C. Circadian expression of the steroid 15 alpha-hydroxylase (CYP2a4) and coumarin 7-hydroxylase (Cyp2a5) genes in mouse liver is regulated by the PAR leucine zipper transcription factor DBP. Mol Biol Cell. 1999;19:6488–6499. doi: 10.1128/mcb.19.10.6488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lee C, Bae K, Edery I. The Drosophila CLOCK protein undergoes daily rhythms in abundance, phosphorylation and interactions with the PER-TIM complex. Neuron. 1998;4:857–867. doi: 10.1016/s0896-6273(00)80601-7. [DOI] [PubMed] [Google Scholar]
- 23.Lee C, Bae K, Edery I. PER and TIM inhibit the DNA binding activity of a Drosophila CLOCK-CYC/dBMAL1 heterodimer without disrupting formation of the heterodimer: a basis for circadian transcription. Mol Cell Biol. 1999;19:5316–5325. doi: 10.1128/mcb.19.8.5316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee C, Parikh V, Itsukaichi T, Bae K, Edery I. Resetting the Drosophila clock by photic regulation of PER and a PER-TIM complex. Science. 1996;271:1740–1744. doi: 10.1126/science.271.5256.1740. [DOI] [PubMed] [Google Scholar]
- 25.Lopez-Molina L, Conquet F, Dubois-Dauphin M, Schibler U. The DBP gene is expressed according to a circadian rhythm in the suprachiasmatic nucleus and influences circadian behavior. EMBO J. 1997;16:6762–6771. doi: 10.1093/emboj/16.22.6762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lorenz L J, Hall J C, Rosbash M. Expression of a Drosophila mRNA is under circadian clock control during pupation. Development. 1989;107:869–880. doi: 10.1242/dev.107.4.869. [DOI] [PubMed] [Google Scholar]
- 27.Marrus S B, Zeng H, Rosbash M. Effect of constant light and circadian entrainment of pers flies: evidence for light-mediated delay of the negative feedback loop in Drosophila. EMBO J. 1996;15:6877–6886. [PMC free article] [PubMed] [Google Scholar]
- 28.Millar A J, Kay S A. The genetics of phototransduction and circadian rhythms in Arabidopsis. Bioessays. 1997;19:209–214. doi: 10.1002/bies.950190306. [DOI] [PubMed] [Google Scholar]
- 29.Myers M P, Wager-Smith K, Wesley C S, Young M W, Sehgal A. Positional cloning and sequence analysis of the Drosophila clock gene timeless. Science. 1995;270:805–808. doi: 10.1126/science.270.5237.805. [DOI] [PubMed] [Google Scholar]
- 30.Palazzolo M J, Hamilton B A, Ding D, Martin C H, Mead D A, Mierendorf R C, Raghavan K V, Meyerowitz E M, Lipshitz H D. Phage lambda cDNA cloning vectors for subtractive hybridization, fusion protein expression and Cre-loxP automatic plasmid subcloning. Gene. 1990;88:25–36. doi: 10.1016/0378-1119(90)90056-w. [DOI] [PubMed] [Google Scholar]
- 30a.Plautz J D, Kay S A. Synchronous real-time reporting of multiple cellular events. Methods Cell Biol. 1999;58:283–291. doi: 10.1016/s0091-679x(08)61961-5. [DOI] [PubMed] [Google Scholar]
- 31.Reddy P, Zehring W A, Wheeler D A, Pirrotta V, Hadfield C, Hall J C, Rosbash M. Molecular analysis of the period locus in Drosophila melanogaster and identification of a transcript involved in biological rhythms. Cell. 1984;38:701–710. doi: 10.1016/0092-8674(84)90265-4. [DOI] [PubMed] [Google Scholar]
- 31a.Ripperger J A, Shearman L P, Reppert S M, Schibler U. CLOCK, an essential pacemaker component, controls expression of the circadian transcription factor DBP. Genes Dev. 2000;6:679–689. [PMC free article] [PubMed] [Google Scholar]
- 31b.Reppert S M, Schwartz W J, Uhl G R. Arginine vasopressin: a novel peptide rhythm in cerebrospinal fluid. Trends Neurosci. 1987;10:76–80. [Google Scholar]
- 32.Rosbash M, Allada R, Dembinska M E, Guo W Q, Le M, Marrus S B, Qian Z, Rutila J E, Yaglom J, Zeng H. A Drosophila circadian clock. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1996. pp. 265–278. [PubMed] [Google Scholar]
- 33.Rouyer F, Rachidi M, Pikielny C, Rosbash M. A new clock gene regulated by the circadian clock in the Drosophila head. EMBO J. 1997;16:3944–3954. doi: 10.1093/emboj/16.13.3944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rutila J E, Suri V, Le M, So W V, Rosbash M, Hall J C. CYCLE is a second bHLH-PAS protein essential for circadian transcription of Drosophila period and timeless. Cell. 1998;93:805–814. doi: 10.1016/s0092-8674(00)81441-5. [DOI] [PubMed] [Google Scholar]
- 35.Saez L, Young M W. Regulation of nuclear entry of the Drosophila clock proteins PERIOD and TIMELESS. Neuron. 1996;17:911–920. doi: 10.1016/s0896-6273(00)80222-6. [DOI] [PubMed] [Google Scholar]
- 36.Sarov-Blat L, So W V, Liu L, Rosbash M. The Drosophila takeout gene is a novel link between circadian rhythms and feeding behavior. Cell. 2000;101:647–656. doi: 10.1016/s0092-8674(00)80876-4. [DOI] [PubMed] [Google Scholar]
- 37.Schmidt J V, Su G H T, Reddy J K, Simon M C, Bradfield C A. Characterization of a murine Ahr null allele: involvement of the Ah receptor in hepatic growth and development. Proc Natl Acad Sci USA. 1996;93:6731–6736. doi: 10.1073/pnas.93.13.6731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schrier R W. Vasopressin. New York, N.Y: Raven Press; 1985. [Google Scholar]
- 39.Sehgal A, Rothenfluh-Hilfiker A, Hunter-Ensor M, Chen Y, Myers M, Young M W. Circadian oscillations and autoregulation of timeless RNA. Science. 1995;270:808–810. doi: 10.1126/science.270.5237.808. [DOI] [PubMed] [Google Scholar]
- 40.So W V, Rosbash M. Post-transcriptional regulation contributes to Drosophila clock gene mRNA cycling. EMBO J. 1997;16:7146–7155. doi: 10.1093/emboj/16.23.7146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sonnenfeld M, Ward M, Nystrom G, Mosher J, Stahal S, Crews S. The Drosophila tango gene encodes a bHLH-PAS protein that is orthologous to mammalian Arnt and controls CNS midline and tracheal development. Development. 1997;124:4571–4582. doi: 10.1242/dev.124.22.4571. [DOI] [PubMed] [Google Scholar]
- 42.Stanewsky R, Jamison C F, Plautz J D, Kay S A, Hall J C. Multiple circadian-regulated elements contribute to cycling period gene expression in Drosophila. EMBO J. 1997;16:5006–5018. doi: 10.1093/emboj/16.16.5006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stanewsky R, Kaneko M, Emery P, Beretta M, Wager-Smith K, Kay S A, Rosbash M, Hall J C. The cryb mutation identifies cryptochrome as a circadian photoreceptor in Drosophila. Cell. 1998;95:681–692. doi: 10.1016/s0092-8674(00)81638-4. [DOI] [PubMed] [Google Scholar]
- 44.Tei H, Okamura H, Shigeyoshi Y, Fukuhara C, Ozawa R, Hirose M, Sakaki Y. Circadian oscillation of a mammalian homologue of the Drosophila period gene. Nature. 1997;389:512–516. doi: 10.1038/39086. [DOI] [PubMed] [Google Scholar]
- 45.Touhara K, Lerro K, Bonning B C, Hammock B D, Prestwich G D. Ligand binding by recombinant insect juvenile hormone binding protein. Biochem J. 1993;32:2068–2075. doi: 10.1021/bi00059a026. [DOI] [PubMed] [Google Scholar]
- 46.Van Gelder R N, Krasnow M A. A novel circadianly expressed Drosophila melanogaster gene dependent on the period gene for its rhythmic expression. EMBO J. 1996;15:1625–1631. [PMC free article] [PubMed] [Google Scholar]
- 47.Vitaterna M H, King D P, Chang A-M, Kornhauser J M, Lowrey P L, McDonald J D, Dove W F, Pinto L H, Turek F W, Takahashi J S. Mutagenesis and mapping of a mouse gene, Clock, essential for circadian behavior. Science. 1994;264:719–725. doi: 10.1126/science.8171325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Vosshall L B, Price J L, Sehgal A, Saez L, Young M W. Specific block in nuclear localization of period protein by a second clock mutation, timeless. Science. 1994;263:1606–1609. doi: 10.1126/science.8128247. [DOI] [PubMed] [Google Scholar]
- 49.Wojtasek H, Prestwich G D. Key disulfide bonds in an insect hormone binding protein: cDNA cloning of a juvenile hormone binding protein of Heliothis virescens and ligand binding by native and mutant forms. Biochemistry. 1995;34:5234–5241. doi: 10.1021/bi00015a037. [DOI] [PubMed] [Google Scholar]
- 50.Zeng H, Hardin P E, Rosbash M. Constitutive overexpression of the Drosophila period protein inhibits period mRNA cycling. EMBO J. 1994;13:3590–3598. doi: 10.1002/j.1460-2075.1994.tb06666.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zeng H, Qian Z, Myers M P, Rosbash M. A light-entrainment mechanism for the Drosophila circadian clock. Nature. 1996;380:129–135. doi: 10.1038/380129a0. [DOI] [PubMed] [Google Scholar]