Abstract
Rhizosphere fungi have the beneficial functions of promoting plant growth and protecting plants from pests and pathogens. In our preliminary study, rhizosphere fungus JP-NJ4 was obtained from the soil rhizosphere of Pinus massoniana and selected for further analyses to confirm its functions of phosphate solubilization and plant growth promotion. In order to comprehensively investigate the function of this strain, it is necessary to ascertain its taxonomic position. With the help of genealogical concordance phylogenetic species recognition (GCPSR) using five genes/regions (ITS, BenA, CaM, RPB1, and RPB2) as well as macro-morphological and micro-morphological characters, we accurately determined the classification status of strain JP-NJ4. The concatenated phylogenies of five (or four) gene regions and single gene phylogenetic trees (ITS, BenA, CaM, RPB1, and RPB2 genes) all show that strain JP-NJ4 clustered together with Talaromyces brevis and Talaromyces liani, but differ markedly in the genetic distance (in BenA gene) from type strain and multiple collections of T. brevis and T. liani. The morphology of JP-NJ4 largely matches the characteristics of genes Talaromyces, and the rich and specific morphological information provided by its colonies was different from that of T. brevis and T. liani. In addition, strain JP-NJ4 could produce reduced conidiophores consisting of solitary phialides. From molecular and phenotypic data, strain JP-NJ4 was identified as a putative novel Talaromyces fungal species, designated T. nanjingensis.
Keywords: rhizosphere beneficial fungi, Pinus massoniana, genealogical concordance phylogenetic species recognition, one new taxon (Talaromyces nanjingensis sp. nov)
1. Introduction
Rhizosphere fungi play roles in promoting plant growth and protecting plants from pests and pathogens. Phosphate-solubilizing fungi (PSF) are an important group of such fungi. Phosphate-solubilizing microbes in soil include PSF [1] and phosphate-solubilizing bacteria (PSB) [2]. Fungi and bacteria have their own advantages in adaptation in different environments. The variety and quantity of PSB were more than that of PSF [3], and the research studies on them are still in progress. Common PSF include Aspergillus, Penicillium, Trichoderma, and some mycorrhizal fungi. Phosphate-solubilizing fungi can be applied to a variety of crop ecosystems. For example, Aspergillus niger and Penicillium chrysogenum promote the growth and nutrient uptake of groundnut (Arachis hypogaea) [4]. Inoculation with the PSF Aspergillus niger significantly increases growth, root nodulation, and yield of soybean plants [5]. Phosphate-solubilizing fungi can also be applied to forest ecosystems. The fungal suspension and extracellular metabolites of Penicillium guanacastense have shown to increase the shoot length and root crown diameter of Pinus massoniana seedlings [6].
Penicillium is one of the most common genera of fungi worldwide. It is widely distributed in nature and primarily functions in breaking down organic matter to provide nutrients for its growth [7,8]. Since Link (1809) introduced the species concept of Penicillium [9] and Dierckx [10] introduced the subgenus classification system of Penicillium, studies on Penicillium have become increasingly popular. At the beginning of the 20th century, an early system of classification and identification based on colony characteristics and conidiophore branching patterns was proposed. The genus Talaromyces was first introduced by Benjamin (1955) as the sexual state of the genus Penicillium [11]. Stolk and Samson (1972) divided Talaromyces into four sections based on differences in their asexual states [12]. Later, more advanced and novel classification schemes based on conidiophore structure, branching pattern, and phialide shape, as well as strain growth characteristics, emerged. Pitt (1979) classified Penicillium into four subgenera: Aspergilloides, Biverticillium, Furcatum, and Penicillium, which contain 10 sections and 21 series. Since then, the modern concept of Penicillium sensu lato has emerged [13].
With the popularization of DNA-based phylogenetic studies of fungi, it has been gradually recognized that the subgenus Biverticillium within the genus Penicillium sensu lato is phylogenetically separate from other subgenera of Penicillium and is closely related to Talaromyces, the previously mentioned sexual morph of Penicillium. The subgenera Aspergilloides, Furcatum, and Penicillium originated from Penicillium sensu lato, together with the genus Eupenicillium, and other species now fall within Penicillium sensu stricto, whereas subgenus Biverticillium is synonymized under the current genus Talaromyces [14,15]. Today, section Talaromyces is not limited to sexual species, but it still contains most of the sexually reproducing species in the genus Talaromyces. Yilmaz et al. (2014) proposed a new sectional classification for the genus Talaromyces, placing the 88 accepted species into seven sections, namely, Bacillispori, Helici, Islandici, Purpurei, Subinflati, Talaromyces, and Trachyspermi [15]. Talaromyces flavus (Klöcker) Stolk and Samson (= T. vermiculatus (P.A. Dang.) C.R. Benj.) has always been the typus of genus in the Talaromyces and Talaromyces section Talaromyces through many revisions of the genus Talaromyces [12,13,15]. The current latest concept of species in Talaromyces section Talaromyces is consistent with what Stolk and Samson (1972) described. Stolk and Samson (1972) introduced the Talaromyces section to include species that produce yellow ascomata, which can occasionally be white, creamish, pinkish, or reddish and yellow ascospores. Conidiophores are usually biverticillate-symmetrical, with some species having reduced conidiophores with solitary phialides. Phialides are usually acerose, with a small proportion of species having wider bases [12]. Section Talaromyces species are commonly isolated from soil, indoor environments, humans with talaromycosis and food products. Common species include T. flavus, T. funiculosus, T. macrosporus, T. marneffei, T. pinophilus, and T. purpurogenus.
Micromorphological features such as asexual sporulation structures (e.g., conidiophore) and sexual sporulation structures (e.g., cleistothecium) were of great significance for taxonomy. The branching pattern of conidiophores, namely the type of penicillus, is an important reference index for the traditional classification methods of Penicillium and Talaromyces fungi. The branching pattern generally includes Monoverticillate, Biverticillate, Terverticillate, Quaterverticillate, and Conidiophores with solitary phialides and Divaricate [13,16,17,18]. Although the classification of Penicillium (and Talaromyces) based on these branching patterns is not completely consistent with the classification status of Penicillium (and Talaromyces) in modern taxonomy, an accurate description of these morphological and structural characteristics is still considered important. The important micromorphology characteristics of Penicillium and Talaromyces fungi include the following: all components of conidiophore (stipes, ramus, ramulus, metula, and phialide) and the sizes, wall texture/ornamentation, color of conidium, ascocarp, ascus, ascospore, and sclerotium. The penicillus includes four parts: ramus, ramulus, metula, and phialide. Sclerotium is produced only under certain conditions; if there are any, observe and record it.
A breakthrough period in the rapid development of classification systems came with the advent of DNA sequencing technology in the 1990s. The identification of Penicillium-group and filamentous fungi began to shift from observation of morphological characteristics to molecular phylogeny. Morphological features are the physical structures with which an organism operates and adapts to its environment, and some features may differ or may be affected by specific factors in the surrounding environment. The effects of medium preparation, inoculation techniques, and culture conditions can be minimized by using strictly standardized protocols [19,20,21]. Morphological identification still plays an irreplaceable role in the fine identification of strains, and a polyphasic approach using both techniques was finally adopted.
In Penicillium, Talaromyces, and many other genera of ascomycetes, internal transcribed spacer (ITS) sequences have been used to classify strains into species complexes or sections, as well as for species identification [15,18]. Due to the limitations of species barcoding based on the ITS region, secondary barcodes or identification markers are often required to identify isolated strains to the species level. Secondary barcodes should be easily amplified, able to distinguish closely related species, and come with a complete reference dataset (including representative gene sequences of all species). The following barcodes can generally be used for the identification of Talaromyces species. The Internal Transcribed Spacer (ITS) rDNA sequence is accepted as the official barcode for fungi [22]. β-tubulin (BenA) is used for the accurately identification of Penicillium species and can also be applied to Talaromyces species [15,18]. Trees have been constructed using other DNA barcode markers (Calmodulin (CaM), DNA-dependent RNA polymerase II (beta) largest subunit (RPB1), and DNA-dependent RNA polymerase II (beta) second largest subunit (RPB2)). Among these, CaM, RPB1, and RPB2 exhibit the same potential as BenA and can be used as secondary barcodes for species identification. In recent years, usage of the CaM gene has gradually increased, and its reference dataset has become relatively complete. RPB1 and RPB2 have the added advantage of lacking introns in the amplicon, allowing for robust and easy alignment when used for phylogenetic analysis, but they may be difficult to amplify. At present, the reference dataset for the RPB2 gene of Talaromyces species is fairly robust, whereas that for the RPB1 gene is still being improved. During phylogenetic tree construction, in addition to the reference sequences of ex-types, other multiple collections from the same species should be considered to cover possible sequence variations. Comparing ITS, BenA, CaM, RPB1, and RPB2 sequences from a suspected new species with sequences of the same markers in related species can help to determine whether a species is new via genealogical concordance phylogenetic species recognition (GCPSR) [23]. This approach, which involved multigene phylogeny, morphological descriptions using macro-morphological and micro-morphological characters and analysis of extrolites, has been used to develop the polyphasic species concept of filamentous fungi such as Penicillium and Talaromyces.
In our preliminary study, rhizosphere fungus JP-NJ4 was obtained from Masson pine rhizosphere soil and screened for phosphate solubilization and plant growth promotion [24]. Fungus JP-NJ4 has the potential to be used as an ecofriendly soil amendment for forestry and farming. With the aid of internal transcribed spacer (ITS) sequences, this strain was preliminarily identified as Penicillium pinophilum (which is now classified in the genus Talaromyces and has been renamed Talaromyces pinophilus). However, the variability of ITS sequences is insufficient to distinguish among closely related species [22]. To comprehensively investigate the function of fungus JP-NJ4, the classification status of this strain was investigated further. The identification process for strain JP-NJ4 involved many standard strains (type strains) that are currently stored at the Central Bureau of Fungal Cultures (Centraalbureau Voor Schimmelcultures (CBS)), which is part of the Royal Netherlands Academy of Arts and Sciences and was founded in 1904 by the Association Internationale des Botanistes [25]. Currently, CBS is one of the largest mycological research centers in the world, with more than 60,000 species in cultivation, including the type strains of many filamentous fungus and yeast species. Here, after reviewing the literature and observing the characteristics of fungus JP-NJ4, this strain was identified and described by referring to the standard research method (GCPSR) recommended in previous international research on filamentous fungal species such as Penicillium and Talaromyces, etc.
2. Materials and Methods
2.1. Source of the Strain
The strain JP-NJ4 was a phosphate-solubilizing fungus isolated from rhizosphere soil of Pinus massoniana (yellow brown soil) in the back mountain of Nanjing Forestry University. The strain is now stored in the China Center for Type Culture Collection (CCTCC) (http://www.cctcc.org, accessed on 18 January 2022). Holotype with the preservation number M 2012167 was stored in a metabolically inactive state by cryopreservation [26,27].
2.2. DNA Extraction, PCR Amplification, and Sequencing of Strain JP-NJ4
Strain JP-NJ4 was cultured on malt extract agar (MEA) culture medium at 25 °C for 7–14 days. Genomic DNA was extracted and purified according to the method of Cubero et al. [28], and the extract was stored at −20 °C. The DNA barcode markers required for the identification of JP-NJ4 strain included the ITS region and BenA, CaM, RPB1, and RPB2 genes [29,30,31,32,33,34,35,36,37,38]. The primers needed for the amplification of these genes are shown in Table S1. All primers and polymerase chain reaction (PCR) amplification sequences needed for the experiment were synthesized and sequenced by the Shanghai Sangon Company (http://www.sangon.com, accessed on 18 January 2022).
In this study, a 50.0 μL DNA amplification thermal cycling reaction mixture system was selected, and the formula of 20.0 μL reaction system was also provided. The volumes of the components in the system are as follows: premix Taq™ solution 25.0 μL, DNA template (10 ng/μL) 2.5 μL, forward primer 2.5 μL, reverse primer 2.5 μL and dd H2O 17.5 μL for 50.0 μL system; premix Taq™ solution 10.0 μL, DNA template (10 ng/μL) 1.5 μL, forward primer 1.0 μL, reverse primer 1.0 μL, and dd H2O 6.5 μL for 20.0 μL system. Premix Taq™ (Ex Taq ™ Version 2.0 plus dye) is a 2x concentration mixed reagent of DNA polymerase, buffer mixture, and dNTP mixture required for PCR reactions purchased from Takara company (https://takara.company.lookchem.cn/, accessed on 18 January 2022). The concentration of the ingredients in the Premix Taq™ solution is as follows: Ex Taq Buffer (2×conc.) with Mg2+ at a concentration of 4mM (mmol/L); highly efficient amplification DNA polymerase (TaKaRa Ex Taq) at a concentration of 1.25 U/25 μL; the dNTP (deoxy-ribonucleoside triphosphate) Mixture (2×conc.), with a concentration of 0.4 mM (mmol/L) for each base; additional pigment markers (Tartrazine/Xylene Cyanol FF), specific gravity additaments, and stabilizers were included. The reagent is stored at −20 °C. The total amount of DNA template can be 10–100 ng, and it can be added according to the experimental requirements. The concentration of primer prepared in accordance with the operational guidelines is 100 μmol/L, diluted 10 times to 10 μmol/L for use.
The DNA amplification thermal cycling programs for each gene is as follows: Standard PCR was selected for general ITS, BenA, and CaM, with initial denaturing 94 °C for 5 min, cycles 35 of denaturation 94 °C for 45 s, annealing 55 °C (52 °C) for 45 s, elongation 72 °C for 60 s, final elongation 72 °C for 7 min, and rest period 10 °C, ∞. Touch-down PCR was selected for RPB1, with 5 cycles of 30 s denaturation at 94 °C, followed by primer annealing for 30 s at 51 °C, and elongation for 1 min at 72 °C; followed by 5 cycles with annealing for 30 s at 49 °C and 30 cycles for 30 s at 47 °C, finalized with an elongation for final 10 min at 72 °C, rest period 10 °C, ∞ (the denaturation and elongation conditions of the second and third cycles are the same as those of the first cycle). Touch-up PCR (= step-up PCR) was selected for RPB2, with initial denaturing 94 °C for 5 min, followed by 5 cycles of 45 s denaturation at 94 °C, primer annealing for 45 s at 50 °C (48 °C), and elongation for 1 min at 72 °C; followed by 5 cycles with annealing for 45 s at 52 °C (50 °C) and 30 cycles for 45 s at 55 °C (52 °C), finalized with an elongation for final 7 min at 72 °C, rest period 10 °C, ∞ (the denaturation and elongation conditions of the second and third cycles are the same as those of the first cycle). The values in parentheses refer to alternative reaction conditions.
2.3. Phylogenetic Tree Construction of Strain JP-NJ4
Sequences of five genes from strain JP-NJ4 have been sequenced and deposited in GenBank (Table 1). By conducting a Basic Local Alignment Search Tool (BLAST) search in National Center for Biotechnology Information (NCBI) database (https://www.ncbi.nlm.nih.gov, accessed on 18 January 2022), the results showed the best matched DNA sequences for each gene/region. In order to make phylogenetic trees, the type strains of Talaromyces species were added. For monogenic and polygenic phylogeny, ITS, BenA, CaM, RPB1, and RPB2 sequence data were compared and aligned using ClustalW software included in the MEGA package version 6.0.6 [39]. All datasets (DNA sequences) were concatenated in MEGA and the BioEdit Sequence Alignment Editor software (Version 7.0.9.0) [40]. The aligned data sets were analysed using both Maximum Likelihood (ML) and Bayesian inference (BI) methods, and ML phylogenetic trees were constructed for each gene/region and concatenated polygenic sequences. According to the results of Akaike Information Criterion (AIC) calculated in MEGA package, the best model for ML phylogenetic tree construction is selected. The ML analysis is performed, and the trees were constructed by calculating the initial tree (constructed by the BioNJ method), selecting the Nearest-Neighbour-Interchange (NNI) option for the following heuristic search. Bootstrap analysis was performed on 1000 repetitions to calculate the support at the node. Bayesian Inference phylogenies were inferred using PhyloSuite v1.2.1 [41]. ModelFinder was used to select the best-fit model (2 parallel runs, 2,000,000 generations) using Bayesian Information Criterion (BIC) for BI [42]. The sample frequency was set at 100, with 25% of trees removed as burn-in. Bayesian inference posterior probabilities (BIpp) values and bootstrap values are labelled on nodes.
Table 1.
Collection numbers of strains, isolation details and GenBank accession numbers of the five genes/region used for phylogenetic analysis of the strain JP-NJ4.
| Species Name | Collection Number | Substrate and Origin | GenBank Accession Number | ||||
|---|---|---|---|---|---|---|---|
| ITS | BenA | CaM | RPB1 | RPB2 | |||
| strain JP-NJ4 | M 2012167 | Rhizosphere soil from Pinus massoniana; Nanjing, Jiangsu, China | MW130720 | MW147759 | MW147760 | MW147761 | MW147762 |
| Talaromyces brevis | CBS 141833 (T) = DTO 349-E7 |
Soil; Beijing, China | MN864269 | MN863338 | MN863315 | MN863328 | |
| DTO 307-C1 | Soil; Zonguldak, Turkey | MN864270 | MN863339 | MN863316 | MN863329 | ||
| CBS 118436 = DTO 004-D8 |
Soil; Maroc | MN864271 | MN863340 | MN863317 | MN863330 | ||
| Talaromyces liani | CBS 225.66 (T) | Soil; China | JN899395 | JX091380 | KJ885257 | JN680280 | KX961277 |
| CBS 118434 | Soil in orchid garden; Sanur, Bali, Indonesia | KM066208 | KM066139 | MK451683= KP453744 | - | - | |
| CBS 118885 | Soil of pepper field; DaeJeon, Korea | KM066210 | KM066138 | - | - | - | |
| NRRL 1009 | Derived from Biourge 368 | MH793030 | MH792902 | MH792966 | - | MH793093 | |
| NRRL 1014 | = 1009 | MH793031 | MH792903 | MH792967 | - | MH793094 | |
| NRRL 1015 | = 1009 | MH793032 | MH792904 | MH792968 | - | MH793095 | |
| NRRL 1019 | USA, Arizona, isol ignotae, KD Butler, 1936. | MH793033 | MH792905 | MH792969 | - | MH793096 | |
| NRRL 3380 | China, isol ex soil, = CBS 225.66 | MH793037 | MH792909 | MH792973 | - | MH793100 | |
| NRRL 28778 | Brazil, isol ex soil, RW Jackson, 1956. | MH793047 | MH792919 | MH792983 | - | MH793110 | |
| NRRL 28834 | India, isol ignotae | MH793048 | MH792920 | MH792984 | - | MH793111 | |
| CMV011D7 | Passiflora edulis; South Africa | - | MK451201 | - | - | - | |
| KUC21412 | Mudflat; South Korea | MN518409 | MN531288 | - | - | - | |
| DTO 058F2 | Heat tretaed corn kernels; the Netherlands | KM066209 | KM066140 | - | - | - | |
| Talaromyces aculeatus | CBS 289.48 (T) = NRRL2129 |
Textile; USA | KF741995 | KF741929 | KF741975 = JX140684 = MH792972 | - | KM023271 |
| CBS 282.92 | Soil in secondary forest; Brazil | KF741981 | KF741914 | KF741946 | - | - | |
| CBS 290.65 | Nut; South Africa | KF741982 | KF741915 | KF741948 | - | - | |
| CBS 563.92 | Stem of Dicymbe Altsonii; French Guiana | KF741986 | KF741920 | KF741963 | - | - | |
| CBS 136673 = IBT14255 |
Weathering wood stakes; Palmerston North, New Zealand | KF741990 | KF741927 | KF741970 | - | - | |
| Talaromyces adpressus | NRRL 6014 | Peanuts; Unknown | MH793039 | MH792911 | MH792975 | - | MH793102 |
| NRRL 62466 | Peanuts; Unknown | MH793088 | MH792961 | MH793025 | - | MH793152 | |
| CBS 140620 | Indoor air; China | KU866657 | - | - | - | KU867001 | |
| DTO 317-G4 | Indoor air; China | - | KU866844 | KU866741 | - | - | |
| CMV011C5 | Soil; South Africa | MK450741 | MK451191 | MK451673 | - | - | |
| Talaromyces aerugineus | CBS 350.66 (T) | Debris; United Kingdom | AY753346 = NR 147420 | KJ865736 | KJ885285 | JN121657 | JN121502 |
| Talaromyces albobiverticillius | CBS 133440 (T) = Penicillium albobiverticillium isolate 900890701 |
Decaying leaves of a broad-leaved tree; Taiwan | HQ605705 = KF114734 | KF114778 | KJ885258 | KF114753 | KM023310 |
| CBS 133441 | Decaying leaves of a broad-leaved tree; Taiwan | KF114733 | KF114777 | - | KF114755 | - | |
| Talaromyces allahabadensis | CBS 453.93 (T) | Cultivated soil; Allahabad, India | KF984873 | KF984614 = JX494298 |
KF984768 | JN680309 | KF985006 |
| CBS 178.81 | Crepis zacintha; Alicante, Spain; Type of Penicillium zacinthae | KF984863 | KF984612 | KF984767 | - | KF985004 | |
| CBS 441.89 | Seed groud; Denmark | KF984872 | KF984613 | KF984759 | - | KF985005 | |
| CBS 137397 = DTO245E3 |
House dust; Mexico | KF984864 | KF984605 | KF984761 | - | KF984998 | |
| CBS 137399 = DTO267H6 |
House dust; Thailand | KF984866 | KF984607 | KF984762 | - | KF984997 | |
| Talaromyces amestolkiae | CBS 132696 (T) = DTO179F5 |
House dust; South Africa | JX315660 = NR 120179 | JX315623 | KF741937 = JX315650 | JX315679 | JX315698 |
| DTO179E4 | House dust; South Africa | KJ775706 | KJ775199 | JX140685 | - | - | |
| DTO179F1 | House dust; South Africa | KJ775707 | KJ775200 | JX140686 | - | - | |
| DTO179F6 | House dust; South Africa | KJ775708 | KJ775201 | - | - | - | |
| Talaromyces angelicus | KACC 46611 (T) = CNU 100013 = DTO303E2 |
Dried roots of Angelica gigas; Pyeongchang, Korea | KF183638 | KF183640 | KJ885259 | - | KX961275 |
| FMR 15489 | Unknown | LT899791 | LT898316 | LT899773 | - | LT899809 | |
| FMR 15490 | Unknown | LT899792 | LT898317 | LT899774 | - | LT899810 | |
| Talaromyces apiculatus | CBS 312.59 (T) | Soil; Japan | JN899375 = NR 121530 | KF741916 = JX091378 |
KF741950 | JN680293 | KM023287 |
| CBS 548.73 | Soil; Suriname | KF741985 | KF741919 | KF741962 | - | - | |
| CBS 101366 | Soil; Hong Kong, China | KF741977 | KF741910 | KF741932 | - | - | |
| Talaromyces argentinensis | NRRL 28750 (T) | Soil; Unknown | MH793045 = NR 165525 | MH792917 | MH792981 | - | MH793108 |
| NRRL 28758 | Soil; Unknown | MH793046 | MH792918 | MH792982 | - | MH793109 | |
| Talaromyces assiutensis | CBS 147.78 (T) | Soil; Egypt | JN899323 | KJ865720 | KJ885260 | JN680275 | KM023305 |
| CBS 645.80 | Gossypium; India; Type of Talaromyces gossypii | JN899334= NR 147423 | KF114802 | - | JN680317 | - | |
| CBS 116554 | Pasteurised canned strawberries; the Netherlands | KM066167 | KM066124 | MK451674 | - | - | |
| CBS 118440 | Soil; Fes, Marocco | KM066168 | KM066125 | MK451675 | - | - | |
| Talaromyces atricola | CBS 255.31 (T) | Unknown | KF984859 | KF984566 | KF984719 | - | KF984948 |
| Talaromyces atroroseus | CBS 133442 (T) | House dust; South Africa | KF114747 = NR 137815 | KF114789 | KJ775418 | KF114763 | KM023288 |
| DTO267I1 | House dust; Thailand | KJ775716 | KJ775209 | - | - | - | |
| DTO270D5 | House dust; Mexico | KJ775734 | KJ775227 | - | - | - | |
| DTO270D6 | House dust; Mexico | KJ775735 | KJ775228 | - | - | - | |
| Talaromyces aurantiacus | CBS 314.59 (T) | Soil; Georgia | JN899380 = NR103681.2 | KF741917 | KF741951 | JN680294 | KX961285 |
| Talaromyces australis | IBT14256 (T) | Unknown | KF741991 = NR 147431 | KF741922 | KF741971 | - | - |
| IBT14254 | Unknown | KF741989 | KF741923 | KF741969 | - | - | |
| MDL18159 | Bronchoscopy; USA | MK601840 | MK626507 | - | MK626517 | - | |
| Talaromyces austrocalifornicus | CBS 644.95 (T) | Soil; California, USA | JN899357 = NR 137079 | KJ865732 | KJ885261 | JN680316 | - |
| Talaromyces bacillisporus | CBS 296.48 (T) | Leaf; New York, USA | JN899329 | AY753368 | KJ885262 | JN121634 | JF417425 |
| CBS 102389 | Sludge of anaerobic pasteurised organic household waste; Sweden |
KM066179 | KM066135 | - | - | - | |
| CBS 110774 | Rye bread; the Netherlands | KM066180 | KM066136 | - | - | - | |
| CBS 116927 | Soil; the Netherlands | KM066181 | KM066137 | - | - | - | |
| Talaromyces bohemicus | CBS 545.86 (T) | Peloids for balneological purposes; Czech Republic | JN899400 = NR 137081 |
KJ865719 | KJ885286 | JN121699 | JN121532 |
| Talaromyces boninensis | CBS 650.95 (T) | Peloids for balneological purposes; Czech Republic | JN899356 = NR 145157 |
KJ865721 | KJ885263 | JN680319 | KM023276 |
| Talaromyces brunneus | CBS 227.60 (T) | Milled rice imported into Japan; Thailand | JN899365 = NR 111688 |
KJ865722 = JX494296 |
KJ885264 | JN680281 | KM023272 |
| Talaromyces calidicanius | CBS 112002 (T) | Soil; Nantou County, Taiwan | JN899319 = HQ149324 = NR 103665.2 |
HQ156944 | KF741934 = JX140688 |
JN899305 | KM023311 |
| ACCC:39162 | Luffa; Beijing; China | KY225703 | KY225714 | - | KY225712 | - | |
| ACCC:39164 | Cucumber; Beijing; China | KY225702 | KY225715 | - | KY225711 | - | |
| Talaromyces californicus | NRRL 58168 (T) | Air sample; Unknown | MH793056 = NR 165527 | MH792928 | MH792992 | - | MH793119 |
| NRRL 58177 | Air sample; Unknown | MH793057 | MH792929 | MH792993 | - | MH793120 | |
| NRRL 58207 | Air sample; Unknown | MH793058 | MH792930 | MH792994 | - | MH793121 | |
| NRRL 58221 | Air sample; Unknown | MH793059 | MH792931 | MH792995 | - | MH793122 | |
| NRRL 58661 | Air sample; Unknown | MH793060 | MH792932 | MH792996 | - | MH793123 | |
| Talaromyces cecidicola | CBS 101419 (T) = Penicillium cecidicola strain DAOM 233329 = Penicillium cecidicola isolate KAS504 |
Cynipid insect galls on Quercus pacifica twigs; Oregon, USA | AY787844 = MH862736 | FJ753295 | KJ885287 | - | KM023309 |
|
Talaromyces
cellulolyticus |
Y-94 = FERM: BP-5826 |
Unknown; A synonym of Talaromyces pinophilus | AB474749 | AB773823 | - | AB856422 | - |
| Talaromyces chloroloma | DAOM 241016 (T) = Penicillium sp. CMV-2008a isolate Pen389 = Penicillium sp. CMV-2008a isolate CV389 |
Fynbos soil; Western Cape, South Africa | FJ160273 | GU385736 | KJ885265 | - | KM023304 |
| DTO 180-F4 = Penicillium sp. CMV-2008a isolate CV390 = Penicillium sp. CMV-2008a isolate Pen390 |
Fynbos soil; South Africa | FJ160272 | GU385737 | - | - | - | |
| DTO 182-A5 = CV785 = CV0785 |
Air sample; Malmesbury, South Africa | JX091485 | JX091597 | JX140689 | - | MK450871 | |
| Talaromyces cinnabarinus | CBS 267.72 (T) | Soil, Japan | JN899376 | AY753377 | KJ885256 | JN121625 | JN121477 |
| CBS 357.72 | Soil, Japan | KM066178 = MH860496 = AY753347 | KM066134 = AY753376 |
- | - | - | |
|
Talaromyces
cnidii |
KACC 46617 (T) = DTO 303-E1 = CNU 100149 |
Dried roots of Cnidium officinale; Jecheon, Korea | KF183639 | KF183641 | KJ885266 | - | KM023299 |
| DTO 269-H8 | House dust; Thailand | KJ775724 | KJ775217 | KJ775426 | - | - | |
| DTO 270-A4 | House dust; Thailand | KJ775729 | KJ775222 | KJ775430 | - | - | |
| DTO 270-A8 | House dust; Thailand | KJ775730 | KJ775223 | KJ775431 | - | - | |
| DTO 270-B7 | House dust; Thailand | KJ775731 | KJ775224 | KJ775432 | - | - | |
| Talaromyces coalescens | CBS 103.83 (T) | Soil under Pinus sp.; Spain | JN899366 = NR 120008 |
JX091390 | KJ885267 | - | KM023277 |
| Talaromyces columbinus | NRRL 58811 (T) | Air; Loisiana, USA | KJ865739 = NR 147433 | KF196843 | KJ885288 | - | KM023270 |
| CBS 137393 = DTO 189-A5 |
Chicken feed (Unga); Nairobi, Kenya | KF984794 | KF984659 | KF984671 | - | KF984897 | |
| NRRL 58644 | Air; Maryland, USA | KF196899 | KF196842 | KF196880 | - | KF196987 | |
| NRRL 62680 | Corn grits; Illinois, USA | KF196901 | KF196844 | KF196882 | KF196949 | KF196988 | |
| Talaromyces convolutus | CBS 100537 (T) | Soil; Kathmandu, Nepal | JN899330 = NR 137157 | KF114773 | - | JN121553 | JN121414 |
| Talaromyces dendriticus | CBS 660.80 (T) | Eucalyptus pauciflora leaf litter; New South Wales, Australia | JN899339 | JX091391 | KF741965 | JN121714 | KM023286= JN121547 |
| DAOM 226674 = Penicillium dendriticum isolate KAS849 |
Doryanthes excelsa spathes; Mangrove Mountain, New South Wales, Australia | AY787842 | FJ753293 | - | - | - | |
| DAOM 233861 = Penicillium dendriticum isolate KAS1190 |
Unindentified insect gall on Eucalyptus leaf; Kalnura, New South Wales, Australia | AY787843 | FJ753294 | - | - | - | |
| DTO 183-G3 = CV2026 |
Mite; Struisbaai, South Africa | JX091486 | JX091619 | JX140692 | - | MK450872 | |
|
Talaromyces
derxii |
CBS 412.89 (T) | Cultivated soil; Japan | JN899327 = NR 145152 |
JX494306 | KF741959 | JN680306 | KM023282 |
| Talaromyces diversus | CBS 320.48 (T) | Leather; USA | KJ865740 | KJ865723 | KJ885268 | JN680297 | KM023285 |
| DTO 133-A7 | House dust; Thailand | KJ775701 | KJ775194 | - | - | - | |
| DTO 133-E4 | House dust; Thailand | KJ775702 | KJ775195 | - | - | - | |
| DTO 133-I6 | Lotus tea; produced in Vietnam, imported to the Netherlands | KJ775700 | KJ775193 | - | - | - | |
| DTO 244-E6 | House dust; New Zealand | KJ775712 | KJ775205 | - | - | - | |
| Talaromyces domesticus | NRRL 58121 | Floor swab; Unknown | MH793055 | MH792927 | MH792991 | - | MH793118 |
| NRRL 62132 | Exposed cloth; Unknown | MH793066 | MH792938 | MH793002 | - | MH793129 | |
| Talaromyces duclauxii | CBS 322.48 (T) | Canvas; France | JN899342 = NR 121526 |
JX091384 | KF741955 | JN121643 | JN121491 |
| Talaromyces emodensis | CBS 100536 (T) | Soil; Kathmandu, Nepal | JN899337 = NR 137077 | KJ865724 | KJ885269 | JN121552 | JF417445 |
| Talaromyces erythromellis | CBS 644.80 (T) | Soil from creek bank; New South Wales | JN899383 | HQ156945 | KJ885270 | JN680315 | KM023290 |
| Talaromyces euchlorocarpius | PF 1203 (T) = DTO 176I3 = CBM-FA-0942 |
Soil; Yokohama, Japan | AB176617 | KJ865733 | KJ885271 | - | KM023303 |
| Talaromyces flavovirens | CBS 102801 (T) | Dead leaves of Quercus ilex; Parque del Retiro, Madrid, Spain | JN899392 | JX091376 | KF741933 | - | KX961283 |
| DAOM236381 | Leaves of Quercus suber; port de la Selva, Girona, Spain | JX013912 | JX091373 | - | - | - | |
| DAOM236382 | Leaves of Quercus suber; Selva de Mar, Girona, Spain | JX013913 | JX091374 | - | - | - | |
| DAOM236383 | Leaves of Quercus suber; Barraca d’en Rabert, Paau, Girona, Spain | JX013914 | JX091377 | - | - | - | |
| DAOM236384 | Leaves of Quercus suber; Xovar, Alt Palacia, Valencia | JX013915 | JX091375 | - | - | - | |
|
Talaromyces
flavus |
CBS 310.38 (T) | Unknown; New Zealand | JN899360 | JX494302 | KF741949 = FJ530982 | JN121639 | JF417426 |
| CBS 437.62 | Compost; Bonn, Germany | KM066202 | KM066156 | - | - | - | |
|
Talaromyces
francoae |
CBS 113134 (T) | Leaf litter; Colombia | NR 154940 | - | - | - | - |
| DTO 056D9 | Leaf litter; Colombia | KX011510 | KX011489 | KX011501 | - | - | |
| Talaromyces funiculosus | CBS 272.86 (T) | Lagenaria vulgaris; India | JN899377 = NR 103678.2 |
JX091383 | KF741945 | JN680288 | KM023293 |
| CBS 171.91 | Unknown | KM066193 | KM066162 | MK451679 | - | MK450873 | |
| CBS 883.70 | Unknown; Java | KM066196 | KM066163 | MK451680 | - | MK450874 | |
| CBS 884.70 | Unknown; Java | KM066195 | KM066164 | MK451681 | - | MK450875 | |
| CBS 885.71 | Air; Java, Jakarta | KM066194 | KM066165 | - | - | MK450876 | |
|
Talaromyces
fuscoviridis |
CBS 193.69 (T) | Unknown | KF741979 = NR 153227 | KF741912 | KF741942 | - | - |
| NRRL 66370 | Unknown | MH793092 | MH792965 | MH793029 | - | MH793156 | |
| Talaromyces galapagensis | CBS 751.74 (T) | Shaded soil under Maytenus obovate; Galapagos Islands, Isla, Santa Cruz, Ecuador | JN899358 = NR 147426 |
JX091388 = KF114770 |
KF741966 | JN680321 | KX961280 |
| NRRL 13068 | Maytenus obovata | MH793042 | MH792914 | MH792978 | - | MH793105 | |
| Talaromyces hachijoensis | IFM 53624 (T) = PF 1174 = CBM-FA-0948 |
Soil; Hachijojima, Japan | AB176620 | - | - | - | - |
|
Talaromyces
helicus |
CBS 335.48 (T) | Soil; Sweden | JN899359 = NR 147427 | KJ865725 | KJ885289 | JN680300 | KM023273 |
| CBS 134.67 | Green house soil under Lycopersicon esculentum; Wageningen, the Netherlands | KM066176 | KM066133 | - | - | - | |
| CBS 550.72 | Saline soil; Vallee de la Seille, France | KM066177 = MH860565 | KM066132 | - | - | - | |
| CBS 649.95 = Talaromyces barcinensis |
Unknown | JN899349 = MH862547 = NR 137078 | KJ865737 | - | JN680318 | - | |
| CBS 652.66 | Unknown | JN899335 | KJ865738 | - | JN680320 | - | |
| Talaromyces indigoticus | CBS 100534 (T) | Soil; Japan | JN899331 = NR 137076 | JX494308 | KF741931 | JN680323 | KX961278 |
| Talaromyces intermedius | CBS 152.65 (T) | Allauvial pasture and swamp soil; Nottingham, England | JN899332 = NR 145154 | JX091387 | KJ885290 | JN680276 | KX961282 |
|
Talaromyces
islandicus |
CBS 338.48 (T) | Unknown; Cape Town, South Africa | KF984885 | KF984655 = JX494293 |
KF984780 | JN121648 | KF985018 = JN121495 |
| CBS 165.81 | spice mixture used in sausage making industry; Spain; Type of Penicillium aurantioflammiferum | KF984883 | KF984653 | KF984778 | - | KF985016 | |
| CBS 394.50 | Kapok fibre; unkown | KF984886 | KF984656 | KF984781 | - | KF985019 | |
| CBS 117284 | Wheat flour; the Netherlands | KF984882 | KF984652 | KF984777 | - | KF985015 | |
|
Talaromyces
kabodanensis |
DI16-149 | Unknown | - | - | LT795598 | - | LT795599 |
|
Talaromyces
kendrickii |
IBT13593 (T) | Unknown | KF741987 = NR 147430 | KF741921 | KF741967 | - | - |
| IBT14128 | Unknown | KF741988 | KF741925 | KF741968 | - | - | |
| CBS 100105 | Unknown | KF741976 | KF741909 | KF741930 | - | - | |
| CBS 133088 | Unknown | KF741978 | KF741911 | KF741939 | - | - | |
| Talaromyces loliensis | CBS 643.80 (T) | Rye grass (Lolium); New Zealand | KF984888 | KF984658 | KF984783 | JN680314 | KF985021 |
| CBS 172.91 | Soil; New Zealand | KF984887 | KF984657 | KF984782 | - | KF985020 | |
| Talaromyces louisianensis | NRRL 35823 (T) | Air sample; Unknown | MH793052= NR 165526 | MH792924 | MH792988 | - | MH793115 |
| NRRL 35826 | Air sample; Unknown | MH793053 | MH792925 | MH792989 | - | MH793116 | |
| NRRL 35928 | Air sample; Unknown | MH793054 | MH792926 | MH792990 | - | MH793117 | |
| Talaromyces macrosporus | CBS 317.63 (T) | Apple juice; Stellenbosch, South Africa | JN899333 = NR 145155 | JX091382 | KF741952 | JN680296 | KM023292 |
| CBS 117.72 | Cotton fabric; USA | KM066188 | KM066148 | - | - | - | |
| CBS 131.87 | Faecal pellet of grasshopper; Malaysia | KM066191 | KM066147 | - | - | - | |
| CBS 353.72 | Tentage; New Guinea | KM066189 | KM066149 | - | - | - | |
| DTO 077-C5 | Pine apple concentrate; the Netherlands | KM066192 | KM066150 | - | - | - | |
| DTO 105-C4 | Unknown | KM066190 | KM066146 | - | - | - | |
| BCC 14364 | Unknown | AY753345 | AY753373 | - | - | - | |
| AS3.6680 | Unknown | - | - | AY678608 | - | - | |
| Talaromyces malicola | NRRL 3724 (T) | Soil under apple tree; Unknown | MH909513= NR 165531 | MH909406 | MH909459 | - | MH909567 |
| Talaromyces marneffei | CBS 388.87 (T) | Bamboo rat (Rhizomys sinensis); Vietnam | JN899344 = NR 103671.2 | JX091389 | KF741958 | JN899298 | KM023283 |
| CBS 108.89 | Human (male); China | KM066187 | KM066157 | - | - | - | |
| CBS 122.89 | Male AIDS patient after travel to Indonesia | KM066183 | KM066161 | - | - | - | |
| CBS 135.94 | Haemoculture; Nonthaburi, Thailand | KM066184 | KM066158 | - | - | - | |
| CBS 549.77 | Man spleen; unknown | KM066185 | KM066159 | - | - | - | |
| CBS 119456 | Male blood; Thailand | KM066186 | KM066160 | - | - | - | |
| Talaromyces mimosinus | CBS 659.80 (T) | Soil from creek bank, New South Wales | JN899338 | KJ865726 | KJ885272 | JN899302 | - |
| NRRL 13069 = NRRL 13609 (BenA) |
Unknown | KX946911 | KX946880 | KX946897 | - | KX946926 | |
| Talaromyces minioluteus | CBS 642.68 (T) | Unknown | JN899346 = NR 121527 |
KF114799 | KJ885273 | JN121709 | JF417443 |
| CBS 137.84 = Penicillium samsonii strain CBS137.84 |
Fruit, damaged by insect; Valladolid, Spain | KM066171 | KF114798 | - | JN680273 | - | |
| CBS 270.35 | Zea mays; Castle Rock, Virginia, USA; Type of Penicillium purpurogenum var. rubrisclerotium | KM066172 | KM066129 | - | JN680287 | - | |
| Talaromyces muroii | CBS 756.96 (T) | Soil; Taiwan | JN899351 = NR 103672.2 | KJ865727 | KJ885274 | JN680322 | KX961276 |
| CBS 261.55 | Clematis; Boskoop, the Netherlands | KM066200 | KM066153 | - | - | - | |
| CBS 283.58 | Jute potato bag, treated with copper oxide ammonia; unknown | KM066197 | KM066151 | - | - | - | |
| CBS 284.58 | Unknown; the Netherlands | KM066199 | KM066152 | - | - | - | |
| CBS 351.61 | Chicken crop; the Netherlands | KM066198 | KM066155 | - | - | - | |
| CBS 889.96 | Dung of sheep; Papua New Guinea | KM066201 | KM066154 | - | - | - | |
| Talaromyces oumae-annae | CBS 138208 (T) = DTO 269-E8 |
House dust; South Africa | KJ775720 = NR 147432 | KJ775213 | KJ775425 | - | KX961281 |
| CBS 138207 = DTO 180-B4 |
House dust; South Africa | KJ775710 | KJ775203 | KJ775421 | - | - | |
| Talaromyces palmae | CBS 442.88 (T) | Chrysalidocarpus lutescens seed; Wageningen, the Netherlands | JN899396 | HQ156947 | KJ885291 | JN680308 | KM023300 |
| Talaromyces panamensis | CBS 128.89 (T) | Soil; Barro Colorado Island, Panama | JN899362 | HQ156948 = JX091386 |
KF741936 = JX140695 |
JN899291 | KM023284 |
| Talaromyces paucisporus | PF 1150 (T) = IFM 53616 = CBM-FA-0944 |
Soil; Aso-machi, Japan | AB176603 | - | - | - | - |
|
Talaromyces
piceus = Talaromyces piceae? |
CBS 361.48 (T) | Unknown | KF984792 | KF984668 | KF984680 | - | KF984899 |
| CBS 116872 | Production plant; the Netherlands | KF984788 | KF984660 | KF984678 | - | KF984903 | |
| CBS 132063 | Straw used in horse stable; the Netherlands | KF984789 | KF984665 | KF984674 | - | KF984904 | |
| CBS 137363 = DTO58D1 |
Pectin; unknown | KF984787 | KF984664 | KF984677 | - | KF984902 | |
| CBS 137377 = DTO178F3 |
House dust; South Africa | KF984784 | KF984661 | KF984676 | - | KF984900 | |
| Talaromyces pinophilus | CBS 631.66 (T) | PVC; France | JN899382 = NR 111691 | JX091381 | KF741964 | JN680313 | KM023291 |
| CBS 173.91 | Unknown; USA | KM066206 | KM066141 | - | - | - | |
| CBS 235.94 | Unknown; USA | KM066204 | KM066145 | - | - | - | |
| CBS 269.73 | Unknown; Germany | KM066207 | KM066144 | KM520392= MK451686 | - | - | |
| CBS 440.89 | Zea mays; India | KM066203 | KM066143 | - | - | - | |
| CBS 762.68 | Rhizosphere; India; Type of Penicillium korosum | JN899347 | JX494301 | - | - | - | |
| CBS 101709 | Soil; Japan | KM066205 | KM066142 | KM520391 = MK451685 |
- | - | |
| DTO183-I6 = CV2460 |
Protea repens infructescense; Struisbaai, South Africa | JX091488 | JX091621 | JX140697 | - | MK450878 | |
| NRRL 1060 | Seed; Unknown | MH909460 | MH909351 | MH909407 | - | MH909514 | |
| NRRL 3503 | Radio set; Unknown | MH909462 | MH909353 | MH909409 | - | MH909516 | |
| NRRL 5200 | Unknown; Type of Penicillium korosum | MH909464 | MH909355 | MH909411 | - | MH909518 | |
| NRRL 13016 | Dung ball; Unknown | MH909466 | MH909357 | MH909413 | - | MH909520 | |
| NRRL 62103 | Canvas cloth; Unknown | MH909482 | MH909373 | MH909429 | - | MH909535 | |
| NRRL 62172 | Wheat; Unknown | MH909492 | MH909383 | MH909439 | - | MH909545 | |
| ATCC 11797 | Unknown | KU729085 | KU896999 | - | - | - | |
| CABI IMI114933 | Unknown; France | KC962105 | KC992266 | - | - | - | |
|
Talaromyces
pittii |
CBS 139.84 (T) | Clay soil under poplar trees; Spain | JN899325 = NR 103667.2 | KJ865728 | KJ885275 | JN680274 | KM023297 |
| Talaromyces pratensis | NRRL 62170 (T) | Unknown | MH793075 = NR 165529 |
MH792948 | MH793012 | - | MH793139 |
| NRRL 13548 | Corn; Unknown | MH793044 | MH792916 | MH792980 | - | MH793107 | |
| NRRL 62126 | River water; Unknown | MH793065 | MH792937 | MH793001 | - | MH793128 | |
| Talaromyces primulinus | CBS 321.48 (T) | Unknown; USA | JN899317 = NR 145151 | JX494305 | KF741954 | JN680298 | KM023294 |
| Talaromyces proteolyticus | CBS 303.67 (T) | Granite soil; Ukraine | JN899387 = NR 103685.2 | KJ865729 | KJ885276 | JN680292 | KM023301 |
| Talaromyces pseudostromaticus | CBS 470.70 (T) | Feather of Hylocichla fuscescens; Minnesota, USA | JN899371 | HQ156950 | KJ885277 | JN899300 | KM023298 |
| Talaromyces ptychoconidium | DAOM 241017 (T) = DTO 180-E7 = CV2808 = Penicillium sp. CMV-2008c isolate CV319 = Penicillium sp. CMV-2008c isolate Pen322 |
Fynbos soil; Malmesbury, South Africa | FJ160266 | GU385733 | JX140701 | - | KM023278 |
| DTO 180-E9 = Penicillium sp. CMV-2008c isolate Pen319 = Penicillium sp. CMV-2008c isolate CV322 |
Fynbos soil; Malmesbury, South Africa | FJ160267 | GU385734 | - | - | MK450879 | |
| DTO 180-F1 = Penicillium sp. CMV-2008c isolate CV323 |
Fynbos soil; Malmesbury, South Africa | GQ414762 | GU385735 | - | - | - | |
| Talaromyces purpureus | CBS 475.71 (T) | Soil; France | JN899328 = NR 145153 | GU385739 | KJ885292 | JN121687 | JN121522 |
| Talaromyces purpurogenus | CBS 286.36 (T) | Parasitic on a culture of Aspergillus oryzae; Japan | JN899372 = NR 121529 |
JX315639 | KF741947 = JX315655 |
JN680271 | JX315709 |
| CBS 184.27 | Soil; Lousiana, USA | JX315665 = MH854924 |
JX315637 | JX315658 | JX315684 = JN680270 |
- | |
| CBS 122434 | Unknown | JX315663 | JX315640 | JX315659 | JX315682 | - | |
| CBS 132707 = DTO189A1 |
Moulded field corn; Wisconsin, USA | JX315661 | JX315638 | JX315642 | JX315680 | - | |
| Talaromyces rademirici | CBS 140.84 (T) | Air under willow tree; Valladolid, Spain | JN899386 = NR 103684.2 |
KJ865734 | - | - | KM023302 |
| Talaromyces radicus | CBS 100489 (T) | Root seadling; New South Wales | KF984878 | KF984599 | KF984773 | - | KF985013 |
| CBS 100488 | Wheat root; New South Wales | KF984877 | KF984598 | KF984772 | - | KF985012 | |
| CBS 100490 | Wheat root; New South Wales | KF984879 | KF984600 | KF984774 | - | KF985014 | |
| CBS 137382 = DTO181D5 |
Fynbos soil; South Africa | KF984875 | KF984602 | KF984775 | - | KF985009 | |
| DTO181D4 | Fynbos soil; South Africa | KF984880 | KF984601 | KF984770 | - | KF985008 | |
| DTO181D7 | Fynbos soil; South Africa | KF984881 | KF984603 | KF984771 | - | KF985010 | |
| Talaromyces ramulosus | DAOM 241660 (T) = CV2837 = CV113 |
Soil; Malmesbury, South Africa | EU795706 | FJ753290 | JX140711 | - | KM023281 |
| DTO 181-E3 = CV314 = CV0314 | Mite; Stellenbosch, South Africa | JX091494 | JX091626 | JX140706 | - | - | |
| DTO 181-F6 = CV394 = CV0394 |
Protea repens infructescense; Stellenbosch, South Africa | JX091495 | JX091629 | JX140707 | - | - | |
| DTO 182-A3 = CV735 = CV0735 |
Protea repens infructescense; Stellenbosch, South Africa | JX091496 | JX091630 | JX140708 | - | - | |
| DTO 182-A6 = CV787 = CV0787 |
Air, Malmesbury; South Africa | JX091497 | JX091631 | JX140709 | - | - | |
| DTO 183-A7 = CV1426 |
Protea repens infructescense; Malmesbury, South Africa | JX091493 | JX091632 | JX140710 | - | - | |
| Talaromyces rotundus | CBS 369.48 (T) | Cardboard; Norway | JN899353 | KJ865730 | KJ885278 | - | KM023275 |
|
Talaromyces
ruber |
CBS 132704 (T) = DTO193H6 |
Air craft fuel tank; United Kingdom | JX315662 = NR 111780 |
JX315629 | KF741938 | JX315681 | JX315700 |
| CBS 196.88 | Unknown | JX315666 = JN899312 |
JX315627 | JX315657 | JN680278 = JX315685 | - | |
| CBS 237.93 | Unknown | JX315667 | JX315628 | JX315656 | JX315686 = JN899306 | - | |
| CBS 370.48 | Currency paper; Washington, USA | JX315673 | JX315630 | JX315649 | JX315692 | - | |
| CBS 868.96 | Unknown | JX315677 | JX315631 | JX315643 | JX315696 = JN899309 | - | |
| Talaromyces rubicundus | CBS 342.59 (T) | Soil; Georgia | JN899384 | JX494309 | KF741956 | JN680301 | KM023296 |
| Talaromyces rugulosus | CBS 371.48 (T) | Roating potato tubers (Solanum tuberosum), USA | KF984834 | KF984575 = JX494297 |
KF984702 | JN680302 | KF984925 |
| CBS 344.51 | Unknown; Japan; Type of Penicillium echinosporum | KF984858 | KF984574 | KF984701 | - | KF984924 | |
| CBS 137366 = DTO61E8 |
Air sample, beer producing factory; Kaulille, Belgium; Type of Penicillium chrysitis | KF984850 | KF984572 | KF984700= JX140720 |
- | KF984922 | |
| NRRL 1053 | Unknown | KF984848 | KF984577 | KF984710 | - | KF984945 | |
| NRRL 1073 | decaying twigs; France; Type of Penicillium tardum and Penicillium elongatum | KF984832 | KF984579 | KF984711 | - | KF984927 | |
| Talaromyces ryukyuensis | NHL 2917 (T) = DTO 176-I6 = strain: NHL2917 |
Soil; Naha, Japan | AB176628 = NR147414 | - | - | - | - |
| Talaromyces sayulitensis | CBS 138204 (T) = DTO 245-H1 |
House dust; Mexico | KJ775713 | KJ775206 | KJ775422 | - | - |
| CBS 138205 = DTO 245-H2 |
House dust; Mexico | KJ775714 | KJ775207 | KJ775423 | - | - | |
| CBS 138206 = DTO 245-H3 |
House dust; Mexico | KJ775715 | KJ775208 | KJ775424 | - | - | |
| NRRL 1064 | Corn; Unknown | MH793034 | MH792906 | MH792970 | - | MH793097 | |
| NRRL 6420 | Corn; Unknown | MH793041 | MH792913 | MH792977 | - | MH793104 | |
| FMR 15842 | Unknown | - | LT898325 | - | - | - | |
| BEOFB2600m | Unknown; Serbia | MH630050 | MH780060 | - | - | - | |
| BEOFB2601m | Unknown; Serbia | MH630051 | MH780061 | - | - | - | |
| Talaromyces scorteus | CBS 340.34 (T) = NRRL 1129 |
Military equipment; Japan | KF984892 = NR153234 = KF196908 | KF984565 = KF196851 | KF984684 = KX946895 | KF196953 | KF984916 = KF196961 |
| CBS 233.60 | Milled Californian rice; Japan; Type of Talaromyces phialosporus | KF984895 | KF984562 = HQ156949 | KF984683 | JN680282 | KF984917 | |
| CBS 499.75 | Unknown; Nigeria | KF984894 | KF984563 | KF984685 | - | KF984918 | |
| CBS 500.75 | Unknown; Sierra Leone | KF984896 | KF984564 | KF984687 | - | KF984919 | |
| DTO 270-A6 | House dust; Thailand | KF984893 | KF984561 | KF984686 | - | KF984915 | |
| Talaromyces siamensis | CBS 475.88 (T) | Forest soil; Thailand | JN899385 = NR 103683.2 | JX091379 | KF741960 | - | KM023279 |
| DTO 269-I3 | House dust; Thailand | KJ775726 | KJ775219 | KJ775428 | - | - | |
| Talaromyces solicola | CBS 133445 (T) = DAOM 241015 = Penicillium sp. CMV-2008d isolate Pen193 = Penicillium sp. CMV-2008d isolate CV191 |
Soil; Malmesbury, South Africa | FJ160264 | GU385731 | KJ885279 | - | KM023295 |
| CBS 133446 | Soil; Malmesbury, South Africa | KF114730 | KF114775 | - | - | - | |
| Talaromyces stipitatus | CBS 375.48 (T) | Decaying wood; Louisiana, USA | JN899348 = NR 147424 |
KM111288 | KF741957 | JN680303 | KM023280 |
| NBRC 100533 | Unknown | - | AB773824 | - | AB856423 | - | |
|
Talaromyces
stollii |
CBS 408.93 (T) | AIDS patient; the Netherlands | JX315674 = NR 111781 |
JX315633 | JX315646 | JX315693 | JX315712 |
| CBS 169.91 | Unknown substrate; South Africa | JX315664 | JX315634 | JX315647 | JX315683 | - | |
| CBS 265.93 | Bronchoalveolar lavage of patient after lung transplantation (subclinical); France |
JX315670 | JX315635 | JX315648 | JX315689 | - | |
| CBS 581.94 | Unknown | JX315675 | JX315632 | JX315645 | JX315694 | - | |
| CBS 624.93 | Ananas camosus cultivar; Martinique | JX315676 | JX315636 | JX315644 = JX965209 | JX315695 = JX965281 | JX965315 | |
| NRRL 1768 | USA, Georgia, isol ex peanut, RJ Cole, 1974. | - | - | - | - | MH793098 | |
| NRRL 62122 | Unknown | - | - | - | - | MH793127 | |
| NRRL 62160 | Unknown | - | - | - | - | MH793136 | |
| NRRL 62163 | Unknown | - | - | - | - | MH793137 | |
| NRRL 62165 | Soil; Unknown | - | - | - | - | MH793138 | |
| NRRL 62171 | Unknown | - | - | - | - | MH793140 | |
| NRRL 62227 | Corn; Unknown | - | - | - | - | MH793144 | |
| Talaromyces subinflatus | CBS 652.95 (T) | Copse soil; Japan | JN899397 = NR 137080 | KJ865737 = JX494288 | KJ885280 | JN899301 | KM023308 |
| Talaromyces tardifaciens | CBS 250.94 (T) | Paddy soil; Bhaktapur, Nepal | JN899361 | KC202954 = KF984560 | KF984682 | JN680283 | KF984908 |
| Talaromyces thailandensis | CBS 133147 (T) | Soil; Thailand | JX898041 = NR 147428 | JX494294 | KF741940 | JX898043 | KM023307 |
| Talaromyces trachyspermus | CBS 373.48 (T) | Unknown; USA | JN899354 = NR 147425 |
KF114803 | KJ885281 | JN121664 | JF417432 |
| CBS 116556 | Pasteurised canned strawberries; Germany | KM066170 | KM066126 | MK451694 | - | - | |
| CBS 118437 | Soil; Marocco | KM066169 | KM066127 | MK451695 | - | - | |
| CBS 118438 | Soil; Marocco | KM066166 | KM066128 | MK451696 | - | - | |
| Talaromyces tratensis | CBS 113146 (T) = CBS 133146 (RPB1)? |
Soil; Trat, Thailand | KF984891 | KF984559 | KF984690 | JX898042 | KF984911 |
| CBS 137400 = DTO 270-F5 |
House dust; Mexico | KF984889 | KF984557 | KF984688 | - | KF984909 | |
| CBS 137401 = NRRL1013 |
Carbonated beverage; Washington D.C., USA | KF984890 | KF984558 | KF984689 | - | KF984910 | |
| Talaromyces tumuli | NRRL 62151 (T) | Soil; Unknown | MH793071= NR 165528 | MH792944 | MH793008 | - | MH793135 |
| NRRL 6013 | Unknown | MH793038 | MH792910 | MH792974 | - | MH793101 | |
| NRRL 62469 | Peanut; Unknown | MH793089 | MH792962 | MH793026 | - | MH793153 | |
| NRRL 62471 | Peanut; Unknown | MH793090 | MH792963 | MH793027 | - | MH793154 | |
| F-3 | Unknown | MT434004 | - | - | - | - | |
| Talaromyces ucrainicus | CBS 162.67 (T) | Unknown | JN899394 = NR 153205 |
KF114771 | KJ885282 | JN680277 | KM023289 |
| CBS 127.64 | soil treated with cyanamide; Germany; Type of Talaromyces ohiensis | KM066173 | KF114772 | - | JN680272 | - | |
| CBS 583.72A | Soil; Japan | KM066174 | KM066130 | - | - | - | |
| CBS 583.72C | Soil; Japan | KM066175 | KM066131 | - | - | - | |
| Talaromyces udagawae | CBS 579.72 (T) | Soil; Misugimura, Japan | JN899350 = NR 145156 | KF114796 | KX961260 | JN680310 | - |
| Talaromyces unicus | CBS 100535 (T) | Soil; Taiwan | JN899336 = NR 157429 | KJ865735 | KJ885283 | JN680324 | - |
| Talaromyces varians | CBS 386.48 (T) | Cotton yarn; England | JN899368 = NR 111689 |
KJ865731 | KJ885284 | JN680305 | KM023274 |
| Talaromyces veerkampii | CBS 500.78 (T) | Unknown | KF741984 = NR 153228 |
KF741918 | KF741961 | - | KX961279 |
| NRRL 6095 | Unknown | MH793040 | MH792912 | MH792976 | - | MH793103 | |
| NRRL 62286 | Wheat flour; Unknown | MH793085 | MH792958 | MH793022 | - | MH793149 | |
| IBT18366 | Unknown | KF741993 | KF741924 | KF741973 | - | - | |
| CMV005D6 | Soil; South Africa | MK450751 | MK451043 | - | - | - | |
| Talaromyces verruculosus | NRRL 1050 (T) = CBS 388.48 |
Soil; Texas, USA | KF741994 | KF741928 | KF741974 | - | KM023306 |
| CBS 254.56 | Unknown; Yangambi, Zaire | KF741980 | KF741913 | KF741944 | - | - | |
| DTO 129-H4 | House dust; Thailand | KJ775698 | KJ775191 | KJ775419 | - | - | |
| DTO 129-H5 | House dust; Thailand | KJ775699 | KJ775192 | KJ775420 | - | - | |
| AX2101 I | Metallic surface; Para, Brazil | KJ413368 | KJ413340 | - | - | KJ476428 | |
| Talaromyces viridis | CBS 114.72 (T) = Sagenoma viride |
Soil; Australia | AF285782 = MH860406 = NR160136 | JX494310 | KF741935 | JN121571 | JN121430 |
| Talaromyces viridulus | CBS 252.87 (T) | Soil from bank of creek floading into Little river; New South Wales | JN899314 = NR103663.2 |
JX091385 | KF741943 | JN680284 = JN121620 | JF417422 |
| Talaromyces wortmannii | CBS 391.48 (T) | Soil; Denmark | KF984829 | KF984648 | KF984756 | JN121669 | KF984977 = JF417433 |
| CBS 319.63 | Unknown | KF984828 | KF984651 | KF984755 | - | KF984961 | |
| CBS 385.48 = NRRL 1048 |
coconut matting; Johannesburg, South Africa; Type of Talaromyces variabilis | KF196915 | KF196853 = JX494295 | KF196878 | JN680304 | KF196975 = KX657552 | |
| CBS 895.73 | Unkown; Japan | KF984811 | KF984626 | KF984737 | - | KF984982 | |
| CBS 137376 = DTO 176-I7 |
soil; Japan; Type of Talaromyces sublevisporus | KF984800 | KF984632 | KF984724 | - | KF984979 | |
| NRRL 2125 = DTO 278-E7 |
Weathering canvas; Panama | KF984797 | KF984635 | KF984731 | - | KF984991 | |
| Talaromyces xishaensis | HMAS 248732 (T) | China | NR147445 | - | - | - | - |
| - | China | KU644580 | KU644581 | KU644582 | - | - | |
| Talaromyces yelensis | CBS 138210 (T) = DTO 268-E5 |
House dust; Micronesia | KJ775717 | KJ775210 | KP119162 | - | KP119164 |
| CBS 138209 = DTO 268-E7 |
House dust; Micronesia | KJ775719 = NR 145183 | KJ775212 | KP119161 | - | KP119163 | |
The result of NCBI standard nucleotide blast is considered preferentially; moreover, the aim of adding type strains genus Talaromyces is to make the phylogenetic tree more plentiful. Genus and species in the columns are represented by bold Italic. T indicates ex type. Sect. Talaromyces; sect. Helici; sect. Purpurei; sect. Trachyspermi; sect. Bacillispori; sect. Subinflati; sect. Islandici.
Table 1 summarised the information of type strains and other related strains, including collection numbers, source and location of strains, and GenBank accession numbers of five genes/regions (ITS barcode and four auxiliary molecular markers: BenA, CaM, RPB1, and RPB2) used for phylogenetic analysis of strain JP-NJ4. According to the information of Samson et al. (2011) and Yilmaz et al. (2014) [14,15], the type strains and other related strains were selected. The current sectional classification information of Talaromyces species was also marked in Table 1.
2.4. Observation on the Morphological Characteristics of Strain JP-NJ4
Important features used to describe the large group of Penicillium and its related fungi are as follows: Macromorphology, including colony texture, mycelium growth and color, shape, color, abundance, and texture of conidia, the presence and color of soluble pigments and exudates, the reverse color of the colony and the acid production of the strain on creatine sucrose agar (CREA) [43], etc. Micromorphology, including asexual sporulation structures (e.g., conidiophore) and sexual sporulation structures (e.g., cleistothecium), etc. To comprehensively investigate the growth of strain JP-NJ4 on different media, we formulated the following supplemented-medium types (see Table S2) from common media, these media can be used to observe other taxonomic characteristics of strains.
Czapek yeast autolysate (CYA) [13] and malt extract agar (MEA) [8] are two standard media recommended for species identification of Penicillium and related filamentous fungi. Czapek’s agar (CZ) [16] CZ is the medium used by Raper and Thom (1949) and Ramírez (1982) in taxonomic studies [44]; this also includes Blakeslee’s Malt extract agar (MEAbl) of Blakeslee (1915) [45]; yeast extract sucrose agar (YES) [43]; and YES as the recommended medium for the analysis of species’ extracellular secretions (extrolites). Oatmeal agar (OA) [8] and Hay infusion agar (HAY) medium [46] were also included. Sexual reproduction of fungal strains most often occurs on OA and HAY media, which can provide valuable information for taxonomy. Use oatmeal/flakes for OA and dry straw for HAY. Creatine sucrose agar (CREA) is the production of acid that can be observed by color reactions (ranging from purple to yellow) in CREA, which are often useful for distinguishing closely related species. Dichloran 18% Glycerol agar (DG18) [47] and Czapek Yeast Autolysate agar with 5% NaCl (CYAS) [18] were used. DG18 and CYAS were used to detect the growth rate of the strain under low water activity.
Preparation for macromorphology observation: The strain JP-NJ4 was inoculated in Potato dextrose agar (PDA) medium and cultured at 15 °C for 25 days to collect conidia. Conidia were washed with distilled deionized water (dd H2O) and diluted with a semi-solid agar solution containing 0.2% agar and 0.05% Tween 80 to prepare the conidia suspension, which was stored at 4 °C for standby use [13]. Conidia suspension was extracted with a micropipette (Eppendorf) and inoculated in three points (1 μL per point) [18]. All media were incubated at a constant temperature of 25 °C for 7 days; each formula of medium is shown in Table S2. In addition, the Czapek Yeast Autolysate agar (CYA) was cultured at 30 °C and 37 °C, and Malt Extract agar (MEA) was cultured at 30 °C, and the data were recorded for the species identification of strain JP-NJ4. After 7 and 14 days of strain culture, the criss-cross method was used to measure the colony diameter.
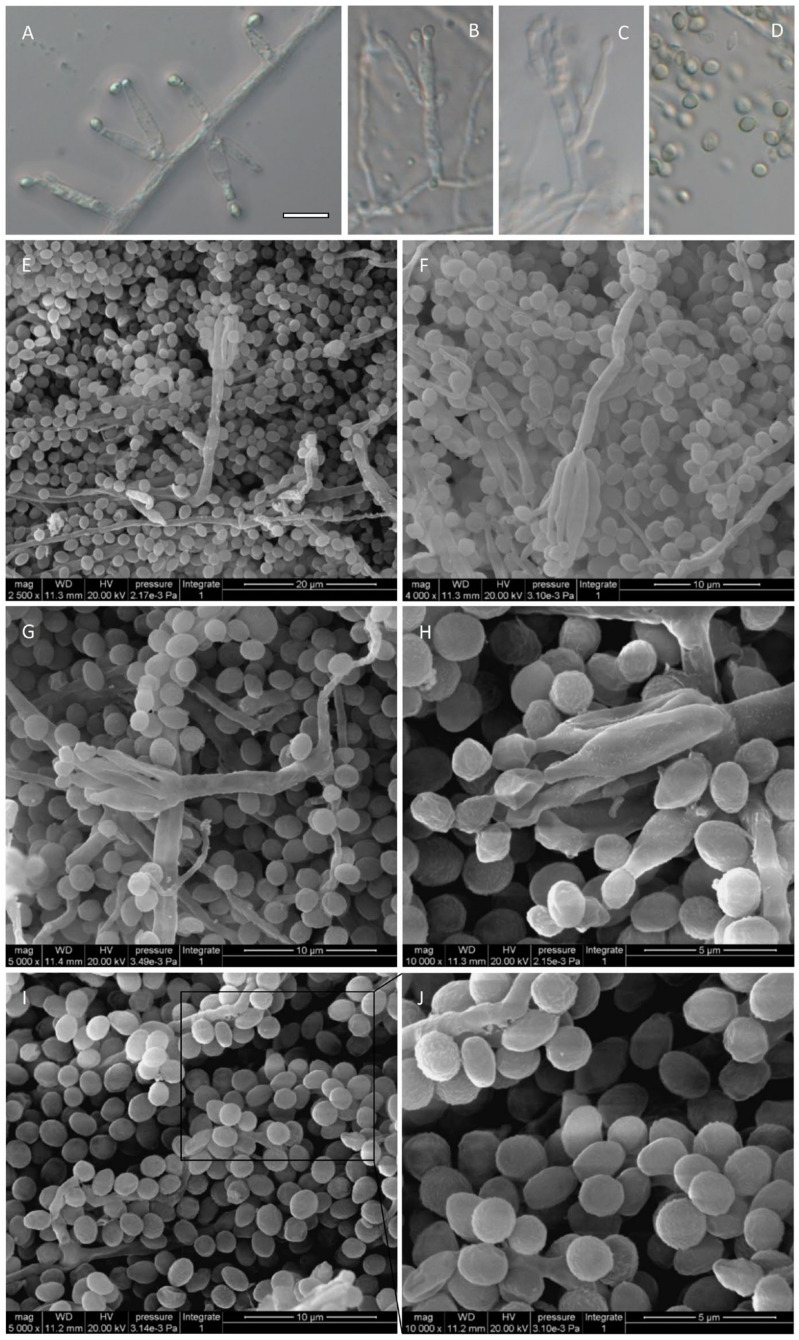
Preparation for the micromorphology observation: Colonies of strain JP-NJ4 cultured on MEA for one to two weeks in a dark environment at 25 °C were used for micromorphology observation, and OA and HAY medium were used when ascomata were not observed on MEA. OA and HAY media are often used for the observation of ascocarp, ascus and ascospore [31,48,49,50] and may be cultured for up to 3 weeks if required for ascocarp production. Then, the colonies used for micromorphology observation were rinsed with 2mL 0.1 mol/L phosphate-buffered saline (PBS) three times. Lactic acid (60%) was used as the fixative. Since most species produce large amounts of hydrophobic conidia, 70% ethanol is usually used to flush out excess conidia and prevent air from getting trapped in lactic acid between the slide and cover. The characteristics of strain JP-NJ4 were observed with a compound microscope (Axio Imager M2.0; Zeiss, Germany) equipped with a digital camera (AxioCam HRc; Zeiss, Germany). The colonies were dehydrated in a graded ethanol solution and dried with liquid carbon dioxide at a critical point (EmiTech K850). After gold spraying (Hitachi E-1010), the Micromorphology of strain JP-NJ4 (conidiophore, conidium, ascocarp, ascus, and ascospore) was observed by scanning electron microscope (SEM) (FEI Quanta 200, FEI, USA).
3. Results
3.1. Taxonomy of Strain JP-NJ4
From the molecular and phenotypic data, it can be inferred that the strain JP-NJ4 belongs to Talaromyces. We identified it as a putative new species (new taxon) here [27,51].
Taxonomy
Talaromyces nanjingensis X.R. Sun, X.Q. Wu and W. Wei, sp. nov. (this study).
MycoBank (No: MB837590).
Etymology: Latin, ‘nanjingensis’ refers to Nan jing, the name of the city where the species originated.
Typus (Type strain): China, Jiangsu, Nanjing, on the rhizosphere soil from Pinus massoniana, 11 April 2011, W. Wei, deposited in China Center for Type Culture Collection (CCTCC) (Collection number. CCTCC M 2012167) (http://www.cctcc.org, accessed on 18 January 2022). Holotype: CCTCC M 2012167. Culture ex-holotype: CCTCC M 2012167.
Distribution: Area of Nanjing, China.
Habitat: Rhizosphere soil from Pinus massoniana.
ITS barcode: MW130720. (Alternative markers for identification: BenA = MW147759; CaM = MW147760; RPB1 = MW147761; RPB2 = MW147762).
In: Talaromyces section Talaromyces
Colony diam, 7 d (mm): CZ 29–33; CYA 25 °C 25–29; CYA 30 °C 30–37; CYA 37 °C 21–31; MEA 25 °C 31–33; MEA 30 °C 35–41; MEAbl (34–43); OA 38–44; DG18 15–18; CYAS No growth; YES 30–40; CREA 18–24; HAY No growth.
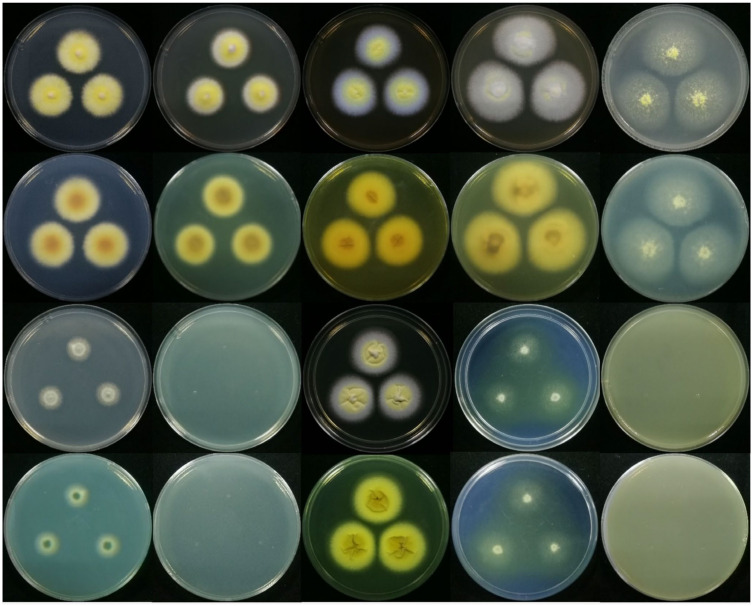
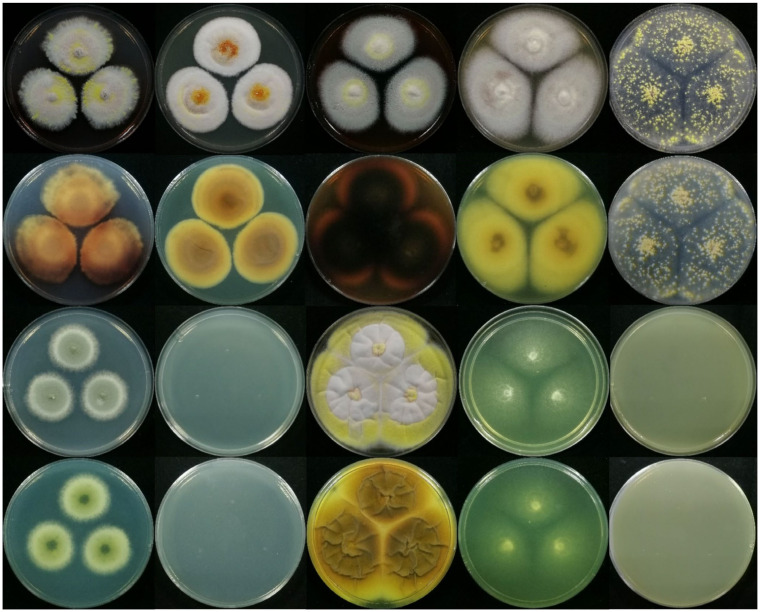
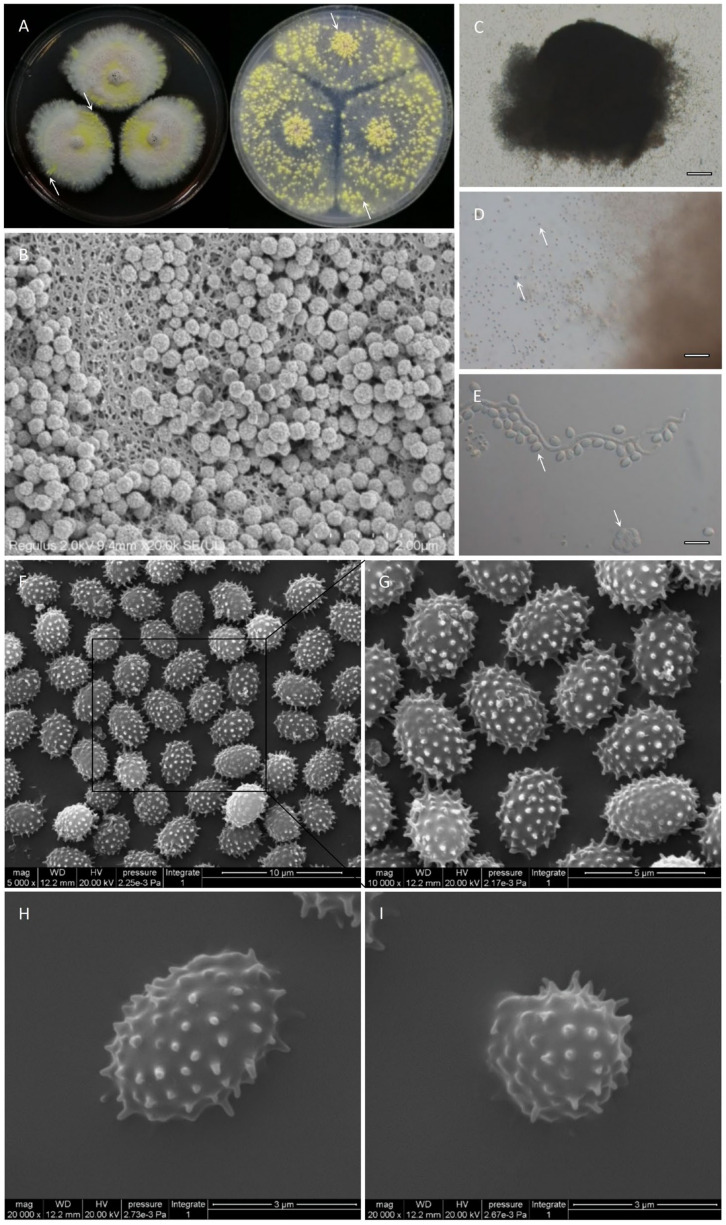
Colony characters: The top and reverse colony morphology of the strain Talaromyces nanjingensis in different media was described. CYA 25 °C, 7 d: top colonies raised at the centre, yellow and margins white; margins low, plane, entire; texture velvety to floccose; sporulation absent to sparse; a small amount of yellow and orange soluble pigments present at 25, 30 and 37 °C; exudates absent; reverse centre pastel yellow (2D4) to pale yellow (1A4); 25 °C, 14 d: top colonies centre pale yellow (1A4) and margins white; the amount of orange exudates present on colonies centre; 30 °C, 14 d: top colonies white, pastel yellow and pinkish-red; a small amount of orange exudates present on colonies centre; formation of yellow ascomata; 37 °C, 14 d: top colonies greyish green (28C5); orange exudates abundant on the gully formed by colony bulge. MEA 25 °C, 7 d: top colonies low, plane; margins low, plane, entire (2–3 mm); white and yellow; texture velvety to floccose; sporulation sparse to moderately dense, conidia greyish-green (26B4-26C4); weak yellow and orange soluble pigments present; exudates absent; reverse light orange to light yellow (5A5-4A5); 25 °C, 14 d: top colonies centre pale yellow (1A4) and margins greyish-yellow (1B4); reverse dark brown (9F8) centre fading into reddish-brown (9E8) to greyish orange (5C5) at margins; a large amount of red soluble pigments present; 30 °C, 14 d: formation of yellow ascomata. YES 25 °C, 7 d: top colonies raised at the centre, sulcate; margins low, plane, entire (3–4 mm); light yellow (4A5) and margins white; texture velvety to floccose; sporulation absent, conidia dull green to greyish green (25D4-25D5); soluble pigments absent; exudates absent; reverse centre pastel yellow and margins white; 25 °C, 14 d: top colonies centre white and margins light yellow (4A5); reverse deep yellow and deep orange (4A8-5A8) to light yellow (2A5); a small amount of yellow soluble pigments present. DG18 25 °C, 7 d: top colonies slightly raised at the centre, plane; margins low, plane, entire (2 mm); pastel green (28A4) and margins white; texture floccose; sporulation moderately dense, conidia greyish green to dull green (25D5-25E4); soluble pigments absent; exudates absent; reverse centre dark green (28F5) and margins white; 25 °C, 14 d: top colonies centre greyish green (28C5) and margins white, reverse pale light green (1B3) to white. OA 25 °C, 7 d: top colonies raised at the centre, plane, formation of yellow ascomata (abundant at 25 °C, 14 d); margins low, plane, entire (2–3 mm); white and yellow; sporulation absent; soluble pigments absent; exudates absent; reverse pastel yellow (2D4). CREA 25 °C, 7 d: acid production present strong; 25 °C, 14 d: acid production present very strong; mycelia all weak at 7 d and 14 d.
Micromorphology: Conidiophores monoverticillate and biverticillate; it also produces reduced conidiophores consisting of solitary phialides. Stipes smooth-walled, 20–100 × 2.5–3 μm; branches 8–20 μm; metulae two to five, divergent, 7–16 × 2.5–3 μm; phialides acerose, two to five per metulae, 6–8 × 2–3 μm; Conidia smooth, globose to subglobose, 2–3 × 3 μm, sometimes ovoid, 3 × 3–3.5 μm. Ascomata mature after one week of incubation on OA, two weeks of incubation on CZ at 25 °C and on CYA and MEA at 30 °C. Ascomata yellow, globose to subglobose, 300–950 × 300–1000 μm, Asci, which are irregular in shape and size depending on the number of ascospores inside them, 10–12 × 8–10 μm; Ascospores, the shape and size are uniform and stable, broadly ellipsoidal, spiny, 3.5–5 × 2–3 μm.
Distinguishing characters: Talaromyces nanjingensis produces relatively fast-growing colonies (Colony diam (mm)) on MEA (31–33), CYA (25–29) and YES (30–40) at 25 °C (faster at 30 °C, MEA 35–41, CYA 30–37), as well as the fastest-growing colonies on MEAbl (34–43) and OA (38–44) at 25 °C. It produces yellow ascomata on CZ and OA media with spiny ellipsoidal ascospores, similar to those of T. austrocalifornicus, T. flavovirens, T. flavus, T. macrosporus, T. muroii, T. thailandensis, and T. tratensis. On colony size at 25 °C on CYA and MEA after 7 d (CYA 25–29; MEA 31–33), T. nanjingensis is more similar to T. aculeatus, T. angelicus, T. dendriticus, T. indigoticus, T. panamensis, T. varians, and T. siamensis. According to the phylogenetic tree, T. nanjingensis and T. liani are clustered together. T. nanjingensis produces yellow ascomata, whereas T. brevis and T. liani produce yellow to orange and yellow to orange-red ascomata on OA medium, respectively. Talaromyces nanjingensis, T. brevis and T. liani both have ellipsoidal ascospores. Talaromyces nanjingensis grows more faster and produces more acid on CREA than T. brevis and T. liani.
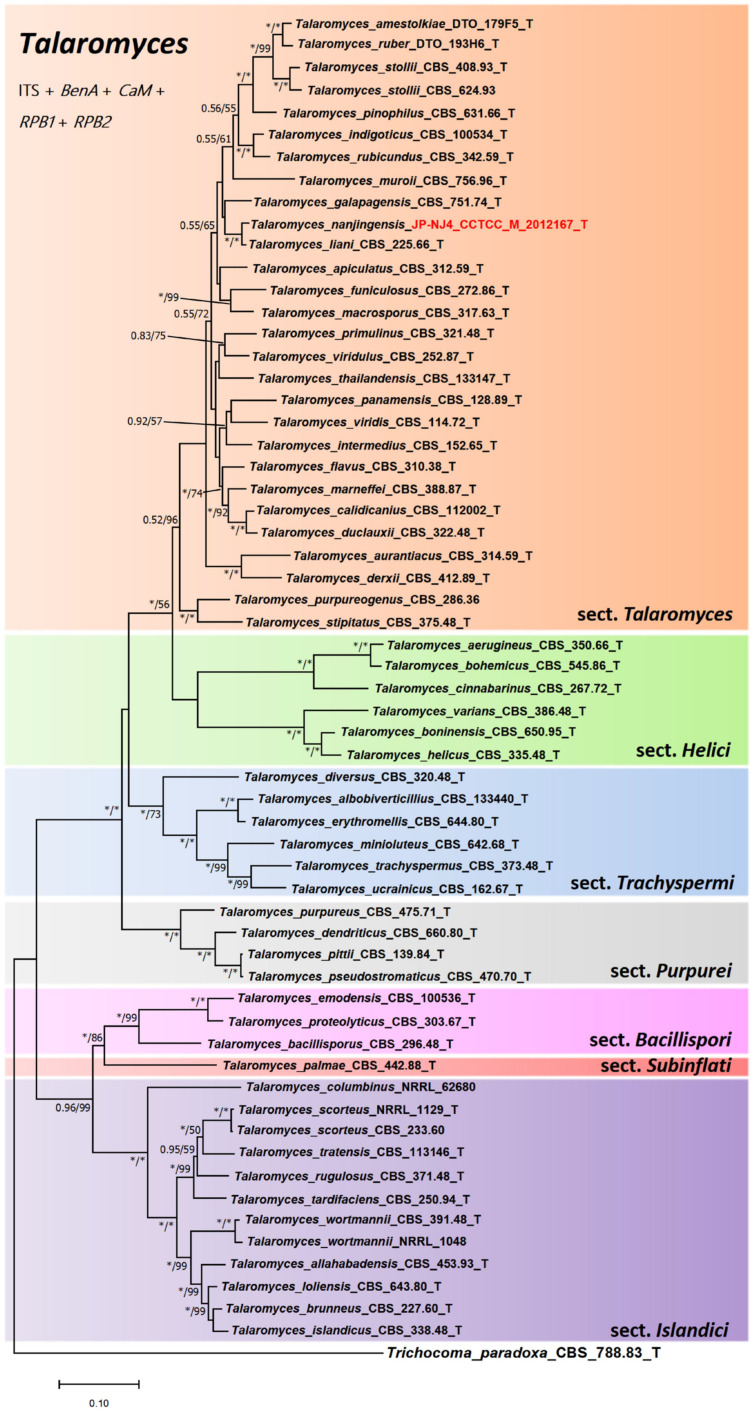
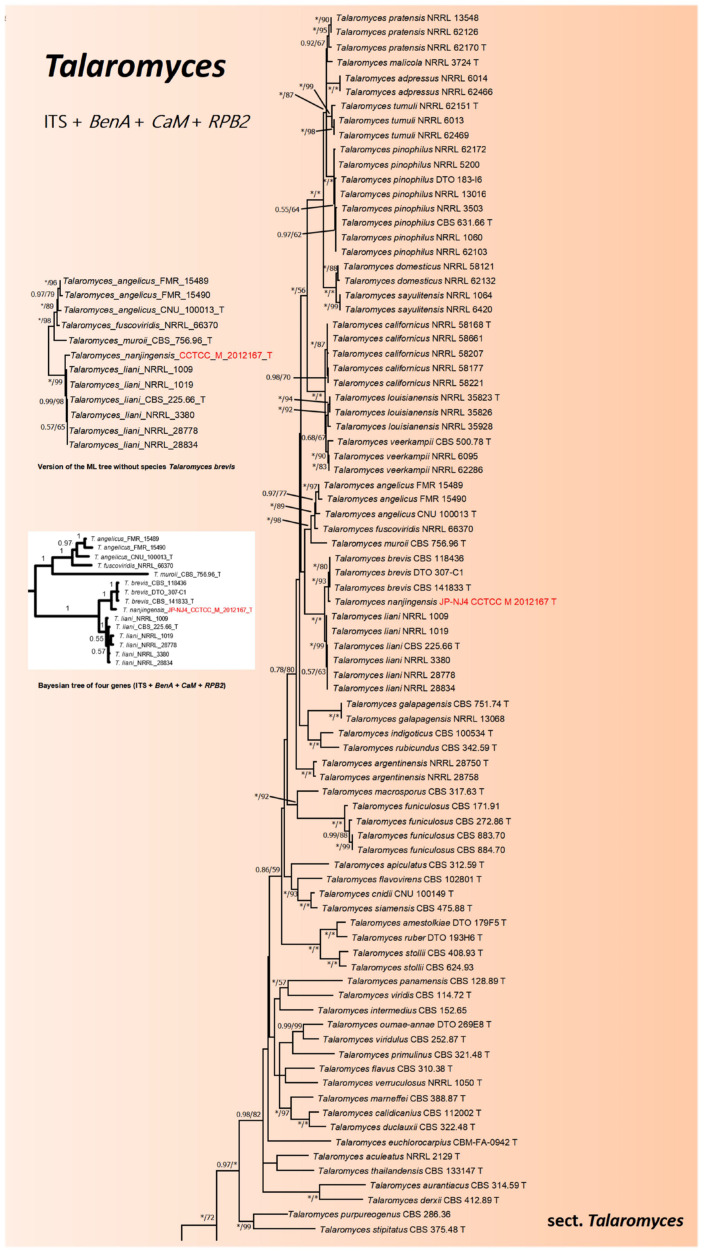
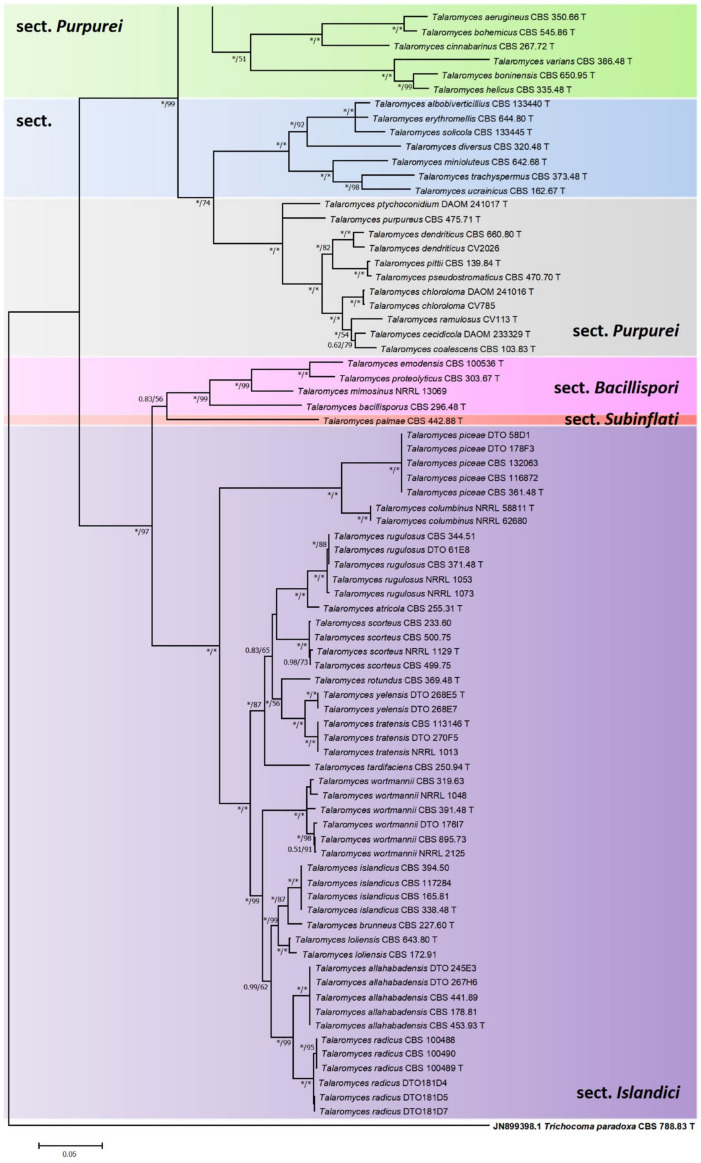
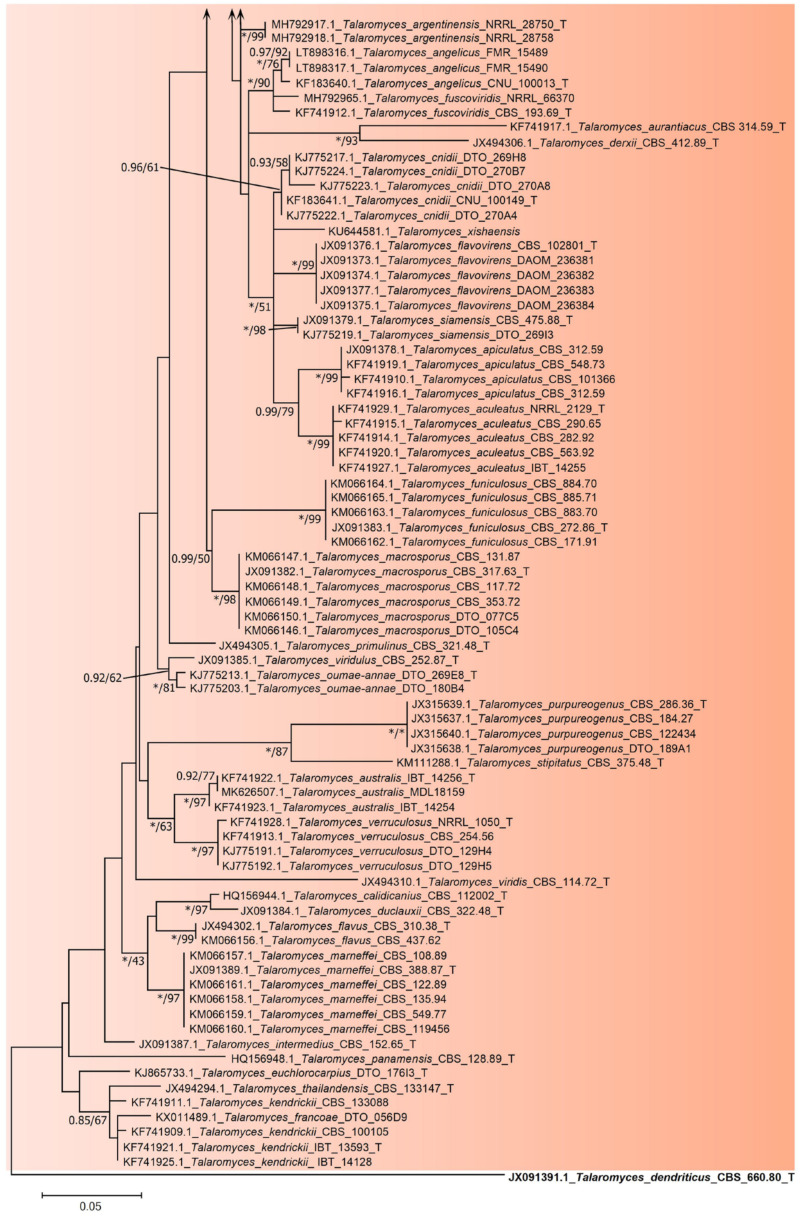
3.2. Phylogeny-Based Species Identification
With the help of concatenated phylogenetic trees based on five gene regions, including the internal transcribed spacer region, BenA, CaM, RPB1, and RPB2, we investigated the taxonomic position of strain JP-NJ4. Figure 1, Figure 2 and Figure 3 and Figures S1–S4 show the phylogenetic relationships among strain JP-NJ4 and representative species of Talaromyces. Concatenated phylogenetic trees of five (ITS, BenA, CaM, RPB1, and RPB2) and four (ITS, BenA, CaM, and RPB2) gene regions and individual phylogenetic trees of each gene region were constructed using the maximum-likelihood method. Talaromyces dendriticus (CBS_660.80_T) was chosen as an out-group for Talaromyces section Talaromyces. Trichocoma paradoxa (CBS_788.83_T) was chosen as the out-group for the Talaromyces genus. Bootstrap values obtained from 1000 replications are shown at the nodes of the tree, and bootstrap support lower than 50 is not shown. In the multi-gene phylogenetic analysis (five gene region), strain JP-NJ4 clustered with T. liani (Figure 1) in Talaromyces section Talaromyces (orange area), with bootstrap values of 100% (BIpp = 1). The concatenated phylogeny of five gene region shows that strain JP-NJ4 and T. liani differ in their genetic distance from other species of Talaromyces. Phylogenetically, the results of four genes indicate that strain JP-NJ4 T. nanjingensis is close to T. brevis, with bootstrap values of 93% (BIpp = 1) (Figure 2).
Figure 1.
Combined phylogeny of the ITS, BenA, CaM, RPB1, and RPB2 gene regions of species from Talaromyces. Maximum likelihood tree of strain JP-NJ4 was constructed. Trichocoma paradoxa (CBS_788.83_T) was chosen as out-group. Support in nodes is indicated above branches and is represented by posterior probabilities (BI analysis) and bootstrap values (ML analysis). Full support (1.00/100%) is indicated with an asterisk (*). Bootstrap values lower than 50 is hidden. Best-fit model of Bayesian Inference phylogeny according to BIC: SYM+I+G4; best-fit model of Maximum likelihood phylogeny according to AIC: Kimura 2-parameter (K2) +G+I; alignment, 444 (ITS) + 294 (BenA) + 489 (CaM)+ 491 (RPB1) + 677 (RPB2) = 2395 bp. Scale bar: 0.10 substitutions per nucleotide position. T indicates ex type. The strain with red font is the strain JP-NJ4 to be identified.
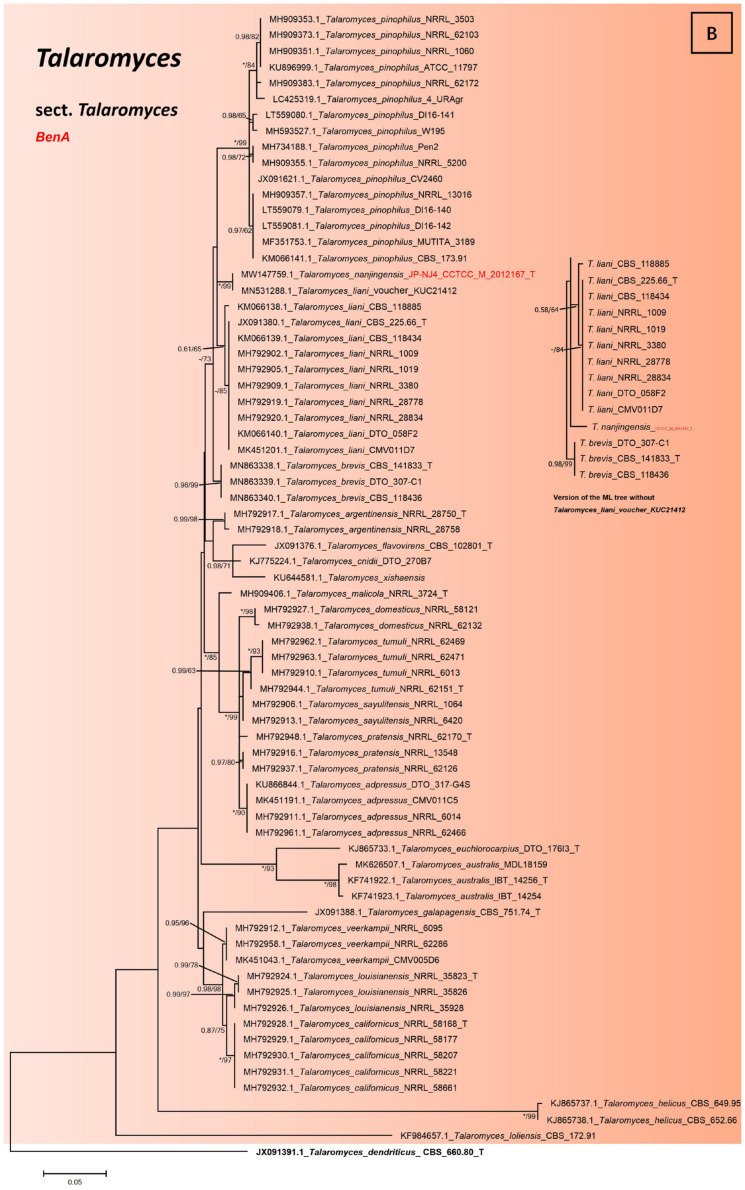
Figure 2.
Combined phylogeny of the ITS, BenA, CaM, and RPB2 gene regions of species from Talaromyces. Maximum likelihood tree of strain JP-NJ4 was constructed. Trichocoma paradoxa (CBS_788.83_T) was chosen as out-group. Support in nodes is indicated above branches and is represented by posterior probabilities (BI analysis) and bootstrap values (ML analysis). Full support (1.00/100%) is indicated with an asterisk (*). Bootstrap values lower than 50 is hidden. Best-fit model of Bayesian Inference phylogeny according to BIC: SYM+I+G4; best-fit model of Maximum likelihood phylogeny according to AIC: Kimura 2-parameter (K2) +G+I; alignment, 439 (ITS) + 284 (BenA) + 482 (CaM) + 677 (RPB2) = 1882 bp. Scale bar: 0.05 substitutions per nucleotide position. T indicates ex type. The strain with red font is the strain JP-NJ4 to be identified.
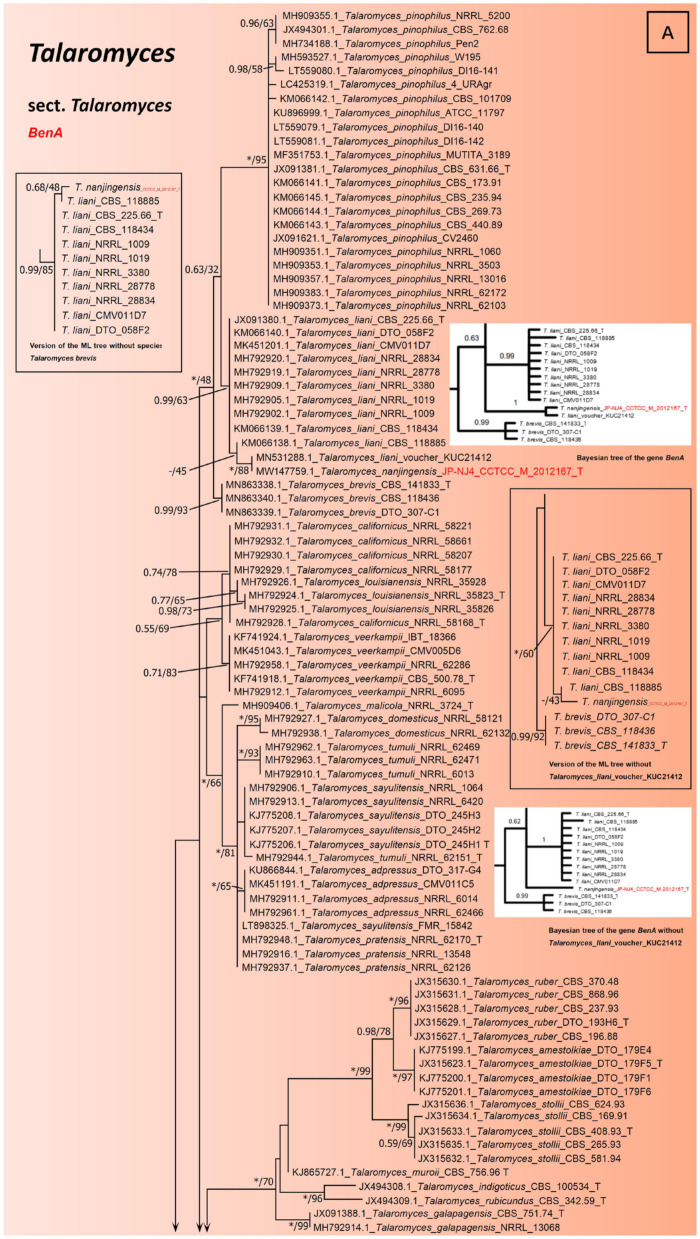
Figure 3.
Maximum likelihood phylogeny of BenA gene regions for strain JP-NJ4 and other species classified in Talaromyces sect. Talaromyces. (A) Short sequence version with multiple species, alignment, BenA 316 bp. Best-fit model of Bayesian Inference phylogeny according to BIC: K80 (K2P) +I+G4; best-fit model of Maximum likelihood phylogeny according to AIC: Kimura 2-parameter (K2) +G; (B) Long sequence version with few species, alignment, BenA 391 bp. Best-fit model of Bayesian Inference phylogeny according to BIC: SYM+G4; best-fit model of Maximum likelihood phylogeny according to AIC: Kimura 2-parameter (K2) +G. Talaromyces dendriticus (CBS_660.80_T) was chosen as out-group. Support in nodes is indicated above branches and is represented by posterior probabilities (BI analysis) and bootstrap values (ML analysis). Full support (1.00/100%) is indicated with an asterisk (*). Missing data from the Bayesian tree are indicated with a dash (-). Scale bar: 0.05 substitutions per nucleotide position. T indicates ex type. The strain with red font is the strain JP-NJ4 to be identified.
Among the phylogenetic trees obtained from each DNA gene region, that of the ITS region was less clearly resolved; although most species formed monophyletic groups in the strict consensus trees, several had low bootstrap support values. The ITS sequence of strain JP-NJ4 clustered with those of nine other strains of T. liani and three strains of T. brevis (bootstrap = 32%, BIpp = 0.61) (Figure S1). The CaM sequence of strain JP-NJ4 clustered well with two strains of T. brevis (bootstrap = 65%, BIpp = 0.99) (one strain of T. brevis was deleted because its sequence was shorter) and seven other strains of T. liani (bootstrap = 99%, BIpp = 0.96) (Figure S2). The RPB1 sequence of strain JP-NJ4 clustered with that of the type strain of T. liani (CBS_225.66_T) (bootstrap = 85%, BIpp = 0.99) (Figure S3). The RPB2 sequence of strain JP-NJ4 clustered perfectly with three strains of T. brevis (bootstrap = 99%, BIpp = 1) (Figure S4). The phylogenetic tree of the CaM gene region shows that strain JP-NJ4 and T. liani differed little in their genetic distance from other species of Talaromyces. However, the phylogenetic trees of the BenA, RPB1, and RPB2 gene regions show that strain JP-NJ4 and T. liani differed markedly in their genetic distance from type strain of T. liani and other multiple collections of T. liani.
The result of single gene BenA indicate that T. nanjingensis and ‘T. liani’ (voucher KUC21412) are clustered together (Bootstrap 88%/ BIpp 1), and both of them have lower bootstrap values (Bootstrap 45%/ BIpp -) with T. liani (CBS_118885) and higher bootstrap values (Bootstrap 63%/BIpp 0.99) with nine other strains of T. liani at the node (Figure 3). This indicates that T. nanjingensis is still genetically different from its genetic relatives T. liani and T. brevis. The sequences of T. nanjingensis and ‘T. liani’ (voucher KUC21412) were significantly similar in BenA gene. However, T. nanjingensis and ‘T. liani’ (voucher KUC21412) differ from T. liani and T. brevis in BenA gene by more than ten bases, and half of their base arrangement pattern is similar to T. liani and the other half is similar to T. brevis (Figure S8a). This could mean they should be new species. It also explains the low bootstrap values. This is also due to the continued discovery of new species, filling gaps in the evolutionary trees, and the lack of some transitional species, of which the T. nanjingensis is one, which has characteristics common to both T. liani and T. brevis. T. nanjingensis is more similar to T. brevis in acid production. T. liani (CBS_118885) is the only acid-producing strain of T. liani that is genetically closest to T. nanjingensis. The phenotypic information of these species may also hint at evolutionary continuity. After a detailed search, we found that T. liani strain T2C1 is equivalent to ‘T. liani’ (voucher KUC21412) and ITS sequence was also obtained. ‘T. liani’ (voucher KUC21412) has at least two base differences with T. nanjingensis, T. liani, and T. brevis in the ITS region, and the front-end of the sequence is similar to that of T. liani and T. brevis (CBS_141833_T). In particular, it has a distinctive differential base A at the end of its sequence (Figure S8b). Therefore, ‘T. liani’ (voucher KUC21412) is also different from T. nanjingensis. ‘T. liani’ (voucher KUC21412) is described by Heo et al. as one of the microorganisms selected from intertidal mudflats and abandoned solar salterns that can produce bioactive compounds [52]. The strain voucher KUC21412 was described as ‘T. liani’ (with quotation marks) in this manuscript, as its current species identity may be in some doubt. ‘T. liani’ (voucher KUC21412) is a comparable species, although only ITS (MN518409.1) and BenA sequences (MN531288.1) have been submitted to NCBI, the quality of the sequences is reliable, which proves that the BenA sequence of T. nanjingensis is reliable. Although T. nanjingensis had little difference with T. brevis in ITS, CaM, RPB1, and RPB2 genes, it had great difference with T. brevis in BenA gene (Figure S8a). BenA is the secondary barcode with the highest reliability in filamentous fungi; thus, the classification results obtained by using this gene are relatively more referential and accurate [15,18]. According to the Bayesian tree (ITS + BenA + CaM + RPB2 and single gene BenA) and the ML tree with deletion of T. brevis and ‘T. liani’ (voucher KUC21412), it can be more obvious that T. nanjingensis has a long genetic distance from T. brevis and T. liani (the small diagrams in Figure 2 and Figure 3).
More obviously, the phylogenetic tree of BenA gene with long sequence version (Figure 3B) was better than that of the BenA gene with short sequence version (Figure 3A) to show the true classification status of strain JPNJ4. The results showed that strain JPNJ4 was quite different from T. brevis and T. liani in phylogeny (Figure 3B). The long sequence version of the phylogenetic tree (Figure 3B) only reduced or lost information on some other species, but this did not affect the accurate identification of JP-NJ4, because this version included T. brevis and T. liani. New species resources are undoubtedly important, as are innovations in identification methods and fine-delineation of species. Taxonomy of these species of Talaromyces are similar to the results obtained by Samson et al. (2011) and Yilmaz et al. (2014) [14,15]. These phylogenetic results suggest that strain JP-NJ4 is a potential novel species.
3.3. Species Identification Based on Macromorphology and Micromorphology
By combining these results with morphological observations, the taxonomic position of strain JP-NJ4 can be further elucidated. The macromorphology of strains, including the morphology and diameter of colonies on specific media, is an important trait for species identification. Based on the preliminary results on CYA and MEA, the macromorphology of the strain may be observed on several other culture media for more accurate identification.
We selected as many media as possible to observe the strains in more detail. Czapek (CZ) medium was used in early taxonomic studies of Penicillium and was selected for comparison with CYA medium. Blakeslee’s MEA, which has been widely used historically, was compared with the MEA culture medium used by the CBS-KNAW Fungal Biodiversity Centre in Utrecht (Table S2). Medium with the addition of hay (HAY) was compared with oatmeal agar for observing the sexual reproduction of fungal strain JP-NJ4. However, strain JP-NJ4 did not grow on this recommended medium.
To clearly observe the colony morphology of strain JP-NJ4 on various culture media, we obtained photographs with black and white background colors. In this paper, we provided colony morphology photographs of strain JP-NJ4 grown at 25 °C for 7 (Figure 4 and Figure S5) and 14 d (Figure 5 and Figure S6) on 10 different media, as well as image data for strain JP-NJ4 grown at 25 °C, 30 °C, and 37 °C in CYA medium (Figure S7).
Figure 4.
Macromorphological characters of strain JP-NJ4 (CCTCC M 2012167) (Inoculation at 25 °C for 7 days). Colonies from left to right: (the top two rows) CZ, CYA, MEA, MEAbl, OA, and the reverse side corresponding to these media; (the bottom two rows) DG18, CYAS, YES, CREA, HAY, and the reverse side corresponding to these media (the background color is black).
Figure 5.
Macromorphological characters of strain JP-NJ4 (Inoculation at 25 °C for 14 days). Colonies from left to right: (the top two rows) CZ, CYA, MEA, MEAbl, OA, and the reverse side corresponding to these media; (the bottom two rows) DG18, CYAS, YES, CREA, HAY, and the reverse side corresponding to these media (the background color is black).
Reverse colonies of Talaromyces species on CYA and MEA media commonly produce yellow or red soluble pigments. Numerous species of Talaromyces, including T. albobiverticillius, T. amestolkiae, T. atroroseus, T. cnidii, T. coalescens, T. marneffei, T. minioluteus, T. pittii, T. purpurogenus, T. ruber, and T. stollii produce soluble red pigments. In T. nanjingensis strain JP-NJ4, weak production of yellow and orange soluble pigments was observed on CYA and MEA in colonies grown at 25 °C for 7 d (Figure 4 and Figure S5). The reverse colony color on MEA was similar to those of T. albobiverticillius, T. minioluteus, and T. purpurogenus. Strong and stable red soluble pigment production occurred on MEA in colonies grown at 25 °C for 14 d (Figure 5 and Figure S6). The reverse colony color on MEA in colonies grown at 25 °C for 14 d was similar to those of T. amestolkiae, T. coalescens, and T. marneffei grown on MEA at 25 °C for 7 d. T. nanjingensis strain JP-NJ4 could produce acid on CREA at 25 °C for 7 d (present strong, the color reaction showed a marked shift from purple to yellow) and at 25 °C for 14 d (present very strong; the color reaction appears to be intense yellow).
The macromorphologies of T. nanjingensis strain JP-NJ4, T. liani, and T. brevis on various media were obviously different in terms of colony growth rate and mycelia color. In terms of colony growth rate, the difference between the three species on MEA and CREA medium was the greatest. In an ascending order of growth rate, we have MEA medium (25 °C, 7 d), T. nanjingensis strain JP-NJ4 (31–33), T. liani (35–45), and T. brevis (50–51); in addition, we also have CREA medium (25 °C, 7 d), T. liani (10–20), T. brevis (13–14), and T. nanjingensis strain JP-NJ4 (18–24). In terms of mycelia color, on OA medium (25 °C, 7 d), the three species were sequentially T. nanjingensis strain JP-NJ4 (white and yellow), T. brevis (primrose), and T. liani (white and yellow) according to the color of mycelia from light to dark; on CYA medium (25 °C, 7 d), the order was T. liani (white and pastel yellow), T. brevis (white and flesh) and T. nanjingensis strain JP-NJ4 (yellow and margins white). Other detailed data are shown in Table 2.
Table 2.
Morphological comparison of strain JP-NJ4 with Talaromyces liani and Talaromyces brevis.
| Morphological Characters | Species | ||||
|---|---|---|---|---|---|
| T. liani (Yilmaz et al., 2014) | Talaromyces Strain JP-NJ4 | T. brevis (Sun et al., 2020) | |||
| Macromorphological Characters | Ascomata | Present after 25 °C, 7 d on OA and MEA (at 30 °C abundant yellow ascomata) | Present after 25 °C, 7 d on OA, 25 °C, 14 d on CZ, and 30 °C, 14 d on CYA and MEA | Present after 25 °C, 7 d on OA | |
| Growth rate (mm) Diam (diameter), 7 d | CZ (25 °C) | Unknown | 29–33 | Unknown | |
| CYA (25 °C) | 20–30 | 25–29 | 30–31 | ||
| CYA (30 °C) | 25–37 | 30–37 | 28–30 | ||
| CYA (37 °C) | 20–25 | 21–31 | 25–26 | ||
| MEA (25 °C) | 35–45 | 31–33 | 50–51 | ||
| MEA (30 °C) | 50–55 | 35–41 | 57–60 | ||
| MEAbl (25 °C) | Unknown | 34–43 | Unknown | ||
| OA (25 °C) | 35–40 | 38–44 | 39–43 | ||
| DG18 (25 °C) | 10–17 | 15–18 | 13–15 | ||
| CYAS (25 °C) | No growth | No growth | No growth | ||
| YES (25 °C) | 35–40 | 30–40 | 42–43 | ||
| CREA (25 °C) | 10–20 | 18–24 | 13–14 | ||
| HAY (25 °C) | Unknown | No growth | Unknown | ||
| Colour of CYA reverse | Light orange and light yellow (5A5–4A5) | Centre pastel yellow (2D4) to pale yellow (1A4) | Ochreous (44) | ||
| Soluble pigment | Absent on CYA (in some isolates yellow) and MEA at 25 °C, 7 d | Weak yellow and orange soluble pigments present on CYA and MEA at 25 °C, 7 d; Strong red soluble pigments present on MEA at 25 °C, 14 d | Absent | ||
| MEA colony texture | Velvety and floccose | Velvety to floccose | Floccose | ||
| Acid production on CREA | Absent (in some isolates very weak) | Present strong | Present | ||
| Micromorphological Characters | Conidiophore | Present | Present | Present | |
| Conidiophore branching | Mono- to biverticillate | Monoverticillate to biverticillate, reduced conidiophores consisting of solitary phialides | Mono- to biverticillate | ||
| Conidium | Shape | Ellipsoidal | Globose to subglobose; (sometimes ovoid) | Subglobose to fusiform | |
| Size (μm) | 2.5–4(–4.5) × 2–3.5 | 2–3 × 3; (3 × 3–3.5) | 3–4(–5) × 2.5–3.5(–4.5) | ||
| Ornamentation | Smooth | Smooth | Smooth | ||
| Ascoma colour | Yellow to orange red | Yellow | Yellow to orange | ||
| Ascoma shape | Globose to subglobose | Globose to subglobose | Globose to subglobose | ||
| Ascoma size (μm) | 150–550 × 150–545 | 300–950 × 300–1000 | 400–550 × 400–550 | ||
| Asci size (μm) | 9–13 × 7.5–11 | 10–12 × 8–10 | Unknown | ||
| Ascospore | Shape | Broadly ellipsoidal | Broadly ellipsoidal | Ellipsoidal | |
| Size (μm) | 4–6 × 2.5–4 | 3.5–5 × 2–3 | 3.5–4.5 × 3–4 | ||
| Ridges | Absent | Absent | Absent | ||
| Ornamentation | Spiny | Spiny | Spiny | ||
Talaromyces species generally produce acerose phialides and ellipsoidal to fusiform conidia. T. nanjingensis strain JP-NJ4 produces reduced conidiophores consisting of solitary phialides (Figure 6A), most conidiophores are monoverticillate and biverticillate, and conidia are globose to subglobose and sometimes ovoid (Figure 6B–H). With the help of scanning electron microscopy, clearer pictures of conidia can be seen (Figure 6I,J). Talaromyces liani produces ellipsoidal conidia. Talaromyces brevis produces subglobose to fusiform conidia. In addition, some species of Talaromyces produce rough-walled, globose conidia, including T. aculeatus, T. apiculatus, and T. verruculosus (classified in sect. Talaromyces), as well as T. diversus and T. solicola (classified in sect. Trachyspermi).
Figure 6.
Micromorphological characters of JP-NJ4 (CCTCC M 2012167) (anamorphic stage) (inoculation for 1–2 wk on MEA). (A–D) Conidiophores and conidia, observed by optical microscope (Zeiss). (A) Reduced conidiophores consisting of solitary phialides. (B) Monoverticillate conidiophores. (C) Biverticillate conidiophores. (D) Conidia. (E–J) Conidiophores and conidia, observed by scanning electron microscope (SEM). (E–G) Conidiophores and conidia at different magnification (E. 2500×; F. 4000×; G. 5000×). (H) Phialides and conidia (10,000×). (I–J) Conidia. Scale bars: A = 10 μm, applies to A–D. E = 20 μm; F = 10 μm; G = 10 μm; H = 5 μm; I = 10 μm; J = 5 μm.
Many species of Talaromyces have the ability to produce ascomata (ascoma = ascocarp; plural, ascomata) (Figure 7A–D). Generally, ascomata are yellow, but some species produce green (T. derxii, T. euchlorocarpius, and T. viridis) or creamish white ascomata (T. assiutensis and T. trachyspermus). The size, shape, and ornamentation of ascospores can be used to distinguish among species of Talaromyces. In most species of Talaromyces, ascospores are broadly ellipsoidal and spiny, but T. bacillisporus and T. rotundus have spiny globose ascospores and T. tardifaciens produces smooth globose ascospores. The ascospores of strain JP-NJ4 and T. liani are broadly ellipsoidal and spiny, and the ascospores of T. brevis are ellipsoidal and spiny. The ascospore sizes of T. nanjingensis strain JP-NJ4, T. brevis, and T. liani differed, at 3.5–5 × 2–3 μm (Figure 7E–I), 3.5–4.5 × 3–4, and 4–6 × 2.5–4 μm, respectively. The ascospores of T. stipitatus have single equatorial ridges, whereas those of T. udagawae have numerous ornamented ridges, and T. helicus has smooth ascospores.
Figure 7.
Teleomorphic stage of JP-NJ4 (CCTCC M 2012167). (A) Colonies inoculated for 2 wk on CZ (left) and OA (right). (B–I) Micromorphological characters of JP-NJ4. (B) Primary ascomata collected from OA (inoculation for 1 wk), observed by scanning electron microscope (SEM). (C–D) A mature ascoma that is releasing asci and ascospores at different magnification ((C) 5×; (D) 20×), observed by optical microscope (Zeiss). (E) An ascus and ascospores (100×). (F–I) Ascospores, observed by SEM. Scale bars: B = 2 μm; C = 200 μm; D = 50 μm; E = 10 μm; F = 10 μm; G = 5 μm; H = 3 μm; I = 3 μm.
According to the phylogenetic results, T. nanjingensis strain JP-NJ4 belongs to the genus Talaromyces. The taxonomic status of this strain can be further determined through description of its morphological characters. Talaromyces liani [15] and Talaromyces brevis [53] are the two species most closely related to T. nanjingensis strain JP-NJ4 in terms of molecular phylogeny, and they were selected as the control group for morphological comparison (Table 2). Table 2 contains summaries of the general macro-morphological and micro-morphological characters observed, including the most important characters: growth rates on different media, production of ascomata and soluble pigments, and acid production on creatine sucrose agar.
4. Discussion
Talaromyces species have a cosmopolitan distribution and have been isolated from a wide range of substrates. Soil is their main habitat, but new species have been obtained from indoor air, dust, clinical samples, plants, leaf litter, honey, and pollen [18,54,55,56,57,58,59,60,61,62]. Talaromyces species have positive impacts in the medical field. The members of this genus can produce a variety of antibiotics and antibacterial substances, such as the rugulosin produced by T. rugulosus [11,63]. Other extrolites of the genus (e.g., erythroskyrine, etc.) have anti-tumor [64], anti-malignant cell proliferation (antiproliferative), and anti-oxidant properties [65]. Talaromyces fungi also have a strong ability to produce enzymes, including that of β-rutinosidase and phosphatase [66,67], endoglucanase and cellulase [68], cellulase [69,70,71], and others. These fungi have also been investigated for functions in plant disease resistance, such as T. flavus [72,73,74,75] and T. pinophilus [76]; moreover, this includes the plant growth promotion of T. pinophilus [77]. In the present study, fungal strain JP-NJ4, which was isolated from the rhizosphere soil of Pinus massoniana, exhibited abilities of phosphate solubilization and plant growth promotion [24], and it was identified as a novel species in genus Talaromyces, section Talaromyces, using the polyphasic approach in this manuscript.
The fungal genera of Penicillium and Talaromyces have many similarities in morphology, such as asexual sporulation structures (e.g., conidiophore), the branching pattern of conidiophores, namely the type of penicillus, and sexual sporulation structures (e.g., cleistothecium). Mistakes are easily made when distinguishing between them. Therefore, we can use molecular methods to conduct preliminary identification of species in these genera. It should be noted, for the modern taxonomic identification of a species, morphological characteristics and molecular phylogenetic results are equally important. Professional recommendations regarding appropriate phylogenetic and morphological data in species delineation are necessary to avoid taxonomic discrepancies [78]. Phylogenetic trees of species are constructed using extensive data obtained through searches and literature review. Normally, the first step is to input a nucleotide sequence obtained through PCR and sequencing technology into the NCBI website for comparison using the nucleotide BLAST. Using the default settings, we obtained 100 sequences that are most similar to the target sequence. The purpose of this step is to roughly determine the genus of the unknown strain. In this paper, BLAST analysis was conducted using ITS, BenA, CaM, RPB1, and RPB2 sequences of strain JP-NJ4.
During the process of collecting and collating the sequences needed to build phylogenetic trees, we encountered the following problems. Among the sequences submitted to NCBI, for the same gene from the same strain of the same species, sequences were uploaded under multiple sequence numbers. By conducting BLAST analysis of the sequence and preliminary phylogenetic tree construction, we found that some of the sequences were consistent with the earliest submitted sequences, whereas others did not cluster with the type strains of their species. This difference may be due to misidentification by later sequence submitters or mislabeling of different strains as the same strain. Therefore, when selecting sequences to construct phylogenetic trees, if two sequences are obtained with differing base compositions, we used the sequences submitted earlier or those referenced in the authoritative literature. Only using validated sequences is also reliable. If the sequences were identical, they were all retained in the tables used to build the phylogenetic tree (Table 1).
The ITS region is the most commonly used molecular marker for fungal identification. In T. liani, NRRL 1014 and NRRL 1015 are equivalent to NRRL 1009, and the base sequences of the ITS region and the other four specific genes in the three strains are identical. Therefore, when constructing the ITS phylogenetic tree for strain JP-NJ4, NRRL 1009 was selected to represent all three strains [79]. In addition, nine other T. liani strains were added. By conducting sequence alignment analysis of the CaM gene, we found notable differences in the composition and arrangement of the bases in this gene among species in different sections of genus Talaromyces. This may result in the deletion of too many bases in order to ensure sequence alignment in the tree constructing of strain JP-NJ4 at the genus level, resulting in loss of information. Therefore, in order to ensure the length of a CaM sequence in tree construction and improve the accuracy of species identification, the CaM gene phylogenetic tree was constructed at the level of section Talaromyces. We further determined the taxonomic status of strain JP-NJ4 by evaluating the taxonomic relationships among these highly similar species within the genus Talaromyces. In addition, during the Alignment-Align process of ClustalW in MEGA software (Version 6.0 and 7.0), inaccuracies may be introduced into the alignment results when large differences exist among the sequences. Therefore, the best comparison results can be obtained through multiple repeated comparisons.
We also found that the gene sequences of RPB2 from some type strains of Talaromyces species could not be retrieved from the NCBI database. By performing comparison and analysis of the RPB2 sequences of other species in genus Talaromyces, we found that the gene sequence data of RNA polymerase (RNA polymerase gene, partial cds) downloaded for these type strains included the gene sequence of RPB2. Therefore, these RNA polymerase gene sequences can be used to complement the construction phylogenetic trees based on the RPB2 gene. Moreover, in previous studies of Penicillium and Talaromyces [14,15,18,31,80], the precedent of using the RNA polymerase gene sequence for constructing a RPB2 phylogenetic tree has been established (e.g., JX315698 Talaromyces amestolkiae DTO 179F5_T). Using this method, the taxonomic status of unknown species can be further refined. The sequences used for this analysis include the following: KX961275 Talaromyces angelicus Korean Agricultural Culture Collection (KACC) 46611, KX961285 T. aurantiacus CBS 314.59, KX961283 T. flavovirens CBS 102801, KX961280 T. galapagensis CBS 751.74, KX961278 T. indigoticus CBS 100534, KX961282 T. intermedius CBS 152.65, KX961276 T. muroii CBS 756.96, KX961281 T. oumae-annae CBS 138208, JX315712 T. stollii CBS 408.93, and KX961279 T. veerkampii CBS 500.78. Some specific genes, such as Translation elongation factor (Tef) and mitochondrial Cytochrome c oxidase 1 (Cox1), have not been universally used in Talaromyces, and relatively few sequences for these genes are available from the NCBI database. Currently, although phylogenetic trees of the genus Talaromyces constructed from these remain imprecise, the genes have been used for identifying Penicillium species [6].
When constructing phylogenetic trees, it is necessary to delete redundant and irrelevant sequences. In the BLAST comparison results, the sequences related to some species did not include the corresponding type strains. In such cases, the sequence information should be validated, as the sequences might have been misidentified (wrongly identified as another species). For example, in Table 1, species marked with a yellow background color did not cluster with the type strains of the corresponding species, and phylogenetic results indicate that these species may be new species—Talaromyces_stollii (blue font) (Figure S4). This discrepancy is due to the fact that not all sequences in the NCBI database have been verified. Therefore, type strains of these species should be added as references for molecular identification and construction of phylogenetic trees. Here, we selected sequences from the type strain of T. pinophilus and other related strains, and some sequences of T. pinophilus that were not relevant to our study were removed.
In addition, when building phylogenetic trees, if the sequences used to construct the tree are not sufficiently comprehensive, the strain to be identified will only cluster with the sequences of similar species, rather than the sequence of the closest species. This problem occurs because the sequences closest to that of the strain to be identified at the genetic level may not be included in the NCBI-BLAST results due to differences in the length of the uploaded sequences or differences in gene coverage, resulting in an inaccurate phylogenetic tree. Specifically, analysis of NCBI-BLAST results revealed that most sequences included only partial sequences of a gene (not all the bases of the gene). The uploaded gene sequences are inconsistent in length, and each sequence contains a different region of the full-length gene. These differences result in the common phenomenon of sequences that appear most similar in the alignment results not being those that are actually most similar to the destination sequence (i.e., the results are inaccurate).
In summary, in previous international research on filamentous fungal species such as Penicillium and Talaromyces, the standard research method (GCPSR) was recommended. This polyphasic approach, which involved multigene phylogeny, morphological descriptions using macro-morphological and micro-morphological characters. To build an accurate phylogenetic tree based on NCBI-BLAST sequences, it is essential to refer to sequences provided in the authoritative literature. For the gene sequences of type strains, the selected sequences should be validated or verified. Using ITS and four specific gene sequences in various Talaromyces species, we constructed two phylogenetic trees (tree 1: ITS, BenA, CaM, RPB1, and RPB2 (Figure 1); tree 2: ITS, BenA, CaM, and RPB2 (Figure 2)) based on combinations of multiple genes. In the genus Talaromyces, combinations of three or four genes are more common, whereas analyses of five genes have been rare. At present, ITS, BenA, CaM, RPB1, and RPB2 are the most authoritative and reliable genes for the identification of Talaromyces species. The preliminary phylogenetic tree construction results indicate that the species most closely related to strain JP-NJ4 is T. liani. The concatenated phylogenies of five (or four) gene regions and single gene phylogenetic tree (BenA, RPB1, and RPB2 genes) all also show that T. nanjingensis strain JP-NJ4 and T. liani clustered together but differ markedly in their genetic distance from type strain of T. liani and other multiple collections of T. liani. The morphology of JP-NJ4 (M 2012167) largely matches the characteristics of T. liani, but the rich and specific morphological information provided by its colonies was different from that of T. liani. In addition, strain JP-NJ4 could produce reduced conidiophores with solitary phialides. From molecular and phenotypic data, strain JP-NJ4 was identified as a putative novel Talaromyces fungal species, designated T. nanjingensis. T. nanjingensis also can produce yellow, orange, and red soluble pigments in their mycelium, including diffusing pigments, similar to other species of the genus [81,82]. Due to the rich and specific morphological information provided by colonies, additional colony morphology photographs of this strain growing at 25 °C for 14 days on 10 different media were captured. We believe that it is essential to apply this information as part of the general method of strain identification. Future research will focus on the ecological function of T. nanjingensis JP-NJ4 and its impacts on the environment in terms of ecological security will also be assessed.
The information of the culture preservation institutions involved is as follows (alphabetically):
ACCC: Agricultural Culture Collection of China.
ATCC: American Type Culture Collection, Manassas, VA, USA (WDCM 1) http://www.atcc.org/, accessed on 18 January 2022;
CABI: Centre for Agriculture and Bioscience International (International Mycological Institute, CABI Genetic Resource Collection).
CBS: culture collection of the CBS-KNAW Fungal Biodiversity Centre, Utrecht, Netherlands (WDCM 133) http://www.cbs.knaw.nl/databases/index.htm, accessed on 18 January 2022.
DTO: internal culture collection of CBS-KNAW Fungal Biodiversity Centre; IMI, CABI Genetic Resources Collection, Surrey, UK (WDCM 214) http://www.cabi.org/, accessed on 18 January 2022.
FERM: (Patent and Bio-Resource Center, National Institute of Advanced Industrial Science and Technology-AIST).
FMR: facultad de medicina, Universidad de Oviedo. 33071-Oviedo. Spain. Institute de Investigaciones Biomidicas C.S.I.C., Facultad de Medicina UAM, E-28029 Madrid, Spain.
HMAS: Fungarium of Institute of Microbiology.
IBT: culture collection of Center for Microbial Biotechnology (CMB) at Department of Systems Biology, Technical University of Denmark (WDCM 758) http://www.biocentrum.dtu.dk/, accessed on 18 January 2022.
MUCL: Mycotheque de l’Universite catholique de Louvain, Leuven, Belgium (WDCM 308).
NBRC: Biological Resource Center, NITE.
NRRL: ARS Culture Collection, U.S. Department of Agriculture, Peoria, Illinois, USA (WDCM 97) http://nrrl.ncaur.usda.gov/, accessed on 18 January 2022.
Acknowledgments
We appreciate editors and anonymous reviewers for providing constructive comments on the earlier versions of the manuscript.
Abbreviations
Notes: The abbreviations below are listed in the order in which they first appear in the manuscript: Genealogical concordance phylogenetic species recognition (GCPSR); Internal Transcribed Spacer rDNA area (ITS); β-tubulin (BenA); Calmodulin (CaM); DNA-dependent RNA polymerase II (beta) largest subunit (RPB1); DNA-dependent RNA polymerase II (beta) second largest subunit (RPB2); Talaromyces (T.); Penicillium (P.); Phosphate-solubilizing fungi (PSF); Phosphate-solubilizing bacteria (PSB); Centraalbureau Voor Schimmelcultures (CBS); The China Center for Type Culture Collection (CCTCC); Malt extract agar (MEA); Polymerase chain reaction (PCR); Basic Local Alignment Search Tool (BLAST); National Center for Biotechnology Information (NCBI); Maximum Likelihood (ML); Bayesian inference (BI); Akaike Information Criterion (AIC); Nearest-Neighbour-Interchange (NNI); Bayesian Information Criterion (BIC); Bayesian inference posterior probabilities (BIpp); Czapek stock solution (CSS); Trace elements stock solution (TESS); Czapek’s agar (CZ); Czapek Yeast Autolysate agar (CYA); Czapek Yeast Autolysate agar with 5% NaCl (CYAS); Blakeslee’s Malt extract agar (MEAbl); Dichloran 18% Glycerol agar (DG18); Yeast extract sucrose agar (YES); Oatmeal agar (OA); Creatine sucrose agar (CREA); Hay infusion agar (HAY); Potato dextrose agar (PDA); Phosphate-buffered saline (PBS); Scanning electron microscope (SEM); Translation elongation factor (Tef); mitochondrial Cytochrome c oxidase 1 (Cox1).
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/jof8020155/s1, Figure S1: Maximum likelihood phylogeny of ITS regions for strain JP-NJ4 and other species classified in Talaromyces sect. Talaromyces. Talaromyces dendriticus (CBS_660.80_T) was chosen as out-group. Support in nodes is indicated above branches and is represented by posterior probabilities (BI analysis) and bootstrap values (ML analysis). Full support (1.00/100%) is indicated with an asterisk (*). Bootstrap values lower than 50 is hidden. Best-fit model of Bayesian Inference phylogeny according to BIC: GTR+F+I+G4; Best-fit model of Maximum likelihood phylogeny according to AIC: Tamura 3-parameter (T92) +G+I; alignment, ITS 467 bp. Scale bar: 0.0020 substitutions per nucleotide position. T indicates ex type. The strain with red font is the strain JP-NJ4 to be identified. Figure S2: Maximum likelihood phylogeny of CaM gene regions for strain JP-NJ4 and other species classified in Talaromyces sect. Talaromyces. Talaromyces dendriticus (CBS_660.80_T) was chosen as out-group. Support in nodes is indicated above branches and is represented by posterior probabilities (BI analysis) and bootstrap values (ML analysis). Full support (1.00/100%) is indicated with an asterisk (*). Bootstrap values lower than 50 is hidden. Best-fit model of Bayesian Inference phylogeny according to BIC: SYM+I+G4; Best-fit model of Maximum likelihood phylogeny according to AIC: Kimura 2-parameter (K2) +G+I; alignment, CaM 475 bp. Scale bar: 0.05 substitutions per nucleotide position. T indicates ex type. The strain with red font is the strain JP-NJ4 to be identified. Figure S3: Maximum likelihood phylogeny of RPBI gene regions for strain JP-NJ4 and other species classified in Talaromyces sect. Talaromyces. Talaromyces dendriticus (CBS_660.80_T) was chosen as out-group. Support in nodes is indicated above branches and is represented by posterior probabilities (BI analysis) and bootstrap values (ML analysis). Full support (1.00/100%) is indicated with an asterisk (*). Bootstrap values lower than 50 is hidden. Support in nodes is indicated above branches and is represented by posterior probabilities (BI analysis) and bootstrap values (ML analysis). Full support (1.00/100%) is indicated with an asterisk (*). Best-fit model of Bayesian Inference phylogeny according to BIC: SYM+I+G; Best-fit model of Maximum likelihood phylogeny according to AIC: Kimura 2-parameter (K2) +G+I; alignment, RPBI 491 bp. Scale bar: 0.05 substitutions per nucleotide position. T indicates ex type. The strain with red font is the strain JP-NJ4 to be identified. Figure S4: Maximum likelihood phylogeny of RPB2 gene regions for strain JP-NJ4 and other species classified in Talaromyces sect. Talaromyces. Talaromyces dendriticus (CBS_660.80_T) was chosen as out-group. Support in nodes is indicated above branches and is represented by posterior probabilities (BI analysis) and bootstrap values (ML analysis). Full support (1.00/100%) is indicated with an asterisk (*). Bootstrap values lower than 50 is hidden. Best-fit model of Bayesian Inference phylogeny according to BIC: K80 (K2P) +I+G4; Best-fit model of Maximum likelihood phylogeny according to AIC: Kimura 2-parameter (K2) +G+I; alignment, RPB2 718 bp. Scale bar: 0.05 substitutions per nucleotide position. T indicates ex type. The strain with red font is the strain JP-NJ4 to be identified. Figure S5: Macromorphological characters of strain JP-NJ4 (CCTCC M 2012167) (Inoculation at 25 °C for 7 days). Colonies from left to right: (the top two rows) CZ, CYA, MEA, MEAbl, OA and the reverse side corresponding to these media; (the bottom two rows) DG18, CYAS, YES, CREA, HAY and the reverse side corresponding to these media (The background color is white). Figure S6: Macromorphological characters of strain JP-NJ4 (Inoculation at 25°C for 14 days). Colonies from left to right: (the top two rows) CZ, CYA, MEA, MEAbl, OA and the reverse side corresponding to these media; (the bottom two rows) DG18, CYAS, YES, CREA, HAY and the reverse side corresponding to these media. (The background color is white). Figure S7: Macromorphology of strain JP-NJ4 under different temperature and culture days, Colonies from left to right: (the top two rows) CYA 25 °C, 30 °C, 37 °C, inoculation for 7 days. CYA 25 °C, 30 °C, 37 °C, inoculation for 14 days. and the reverse side corresponding to these media (The background color is black); (the bottom two rows) (The background color is white). Figure S8: Base alignment and differences in specific genes of Talaromyces nanjingensis, T. brevis and T. liani. A. BenA gene, alignment, 348 bp; B. ITS region, alignment, 448 bp. Table S1: Primers for amplification and sequencing of ITS and specific genes in strain JP-NJ4. Table S2: Media required for the identification of strain JP-NJ4.
Author Contributions
Conceptualization: X.-R.S.; data curation: X.-R.S., M.-Y.X., W.-L.K. and F.W.; formal analysis: X.-R.S., M.-Y.X., W.-L.K. and F.W.; funding acquisition: X.-Q.W.; investigation: X.-R.S., W.-L.K., F.W., Y.Z. and X.-L.X.; methodology: X.-R.S.; project administration: X.-Q.W.; resources: X.-Q.W.; software: X.-R.S. and M.-Y.X.; supervision: X.-Q.W.; validation: X.-Q.W.; visualization: X.-R.S., M.-Y.X., Y.Z. and X.-L.X.; writing—original draft: X.-R.S.; writing—review and editing: D.-W.L. and X.-Q.W. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the National Key Research and Development Program of China (2017YFD0600104) and the Priority Academic Program Development of Jiangsu Higher Education Institutions (PAPD).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The materials are available as Supplementary materials (Tables S1 and S2; Figures S1–S8). Publicly available datasets were analyzed in this study. This data can be found here: https://www.ncbi.nlm.nih.gov/genbank, accessed on 18 January 2022; accession number ITS = MW130720, BenA = MW147759, CaM = MW147760, RPB1 = MW147761, RPB2 = MW147762.
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Mehta P., Sharma R., Putatunda C., Walia A. Endophytic Fungi: Role in Phosphate Solubilization. In: Singh B., editor. Advances in Endophytic Fungal Research. Springer; Cham, Switzerland: 2019. pp. 183–209. [Google Scholar]
- 2.Zheng B.X., Ibrahim M., Zhang D.P., Bi Q.F., Li H.Z., Zhou G.W., Ding K., Penuelas J., Zhu Y.G., Yang X.R. Identification and characterization of inorganic-phosphate-solubilizing bacteria from agricultural fields with a rapid isolation method. AMB Express. 2018;8:47. doi: 10.1186/s13568-018-0575-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jin S.C., Chun-Mei D.U., Ping W.X., Guan H.Y., Bao-Xing X.U. Advance in Phosphorus-Dissolving Microbes. J. Microbiol. 2006;2:73–78. doi: 10.1016/S1872-2032(06)60050-4. (In Chinese with English abstract) [DOI] [Google Scholar]
- 4.Malviya J., Singh K., Joshi V. Effect of Phosphate Solubilizing Fungi on Growth and Nutrient Uptake of Ground nut (Arachis hypogaea) Plants. Adv. Biores. 2011;2:110–113. [Google Scholar]
- 5.Saxena J., Rawat J., Sanwal P. Enhancement of Growth and Yield of Glycine max Plants with Inoculation of Phosphate Solubilizing Fungus Aspergillus niger K7 and Biochar Amendment in Soil. Commun. Soil. Sci. Plan. 2016;47:2334–2347. doi: 10.1080/00103624.2016.1243708. [DOI] [Google Scholar]
- 6.Qiao H., Sun X.R., Wu X.Q., Li G.E., Li D.W. The phosphate-solubilising ability of Penicilium guanacastense and its effects on the growth of Pinus massoniana in phosphate limiting conditions. Biol. Open. 2019;8:11. doi: 10.1242/bio.046797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Taniwaki M.H., Hocking A.D., Pitt J.I., Fleet G.H. Growth and mycotoxin production by food spoilage fungi under high carbon dioxide and low oxygen atmospheres. Int. J. Food. Microbiol. 2009;132:100–108. doi: 10.1016/j.ijfoodmicro.2009.04.005. [DOI] [PubMed] [Google Scholar]
- 8.Samson R.A., Houbraken J., Thrane U., Frisvad J.C., Andersen B. Food and Indoor Fungi. CBS-KNAW Fungal Biodiversity Centre; Utrecht, The Netherlands: 2010. [Google Scholar]
- 9.Link H.F. Observationes in Ordines Plantarum Naturales. Mag. Ges. naturf. Freunde. 1809;3:1–42. [Google Scholar]
- 10.Dierckx R.P. Un essai de revision du genre Penicillium Link. Annales de la Société Scientifique Bruxelles. 1901;25:90–104. [Google Scholar]
- 11.Breen J., Dacre J.C., Raistrick H., Smith G. Studies in the biochemistry of micro-organisms. 95. Rugulosin, a crystalline colouring matter of Penicillium rugulosum Thom. Biochem. J. 1955;60:618–626. doi: 10.1042/bj0600618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stolk A.C., Samson R.A. The genus Talaromyces—Studies on Talaromyces and related genera II. Stud. Mycol. 1972;2:1–65. doi: 10.2307/3758303. [DOI] [Google Scholar]
- 13.Pitt J.I. The Genus Penicillium and Its Teleomorphic States Eupenicillium and Talaromyces. Academic Press Inc.; London, UK: 1979. [Google Scholar]
- 14.Samson R.A., Yilmaz N., Houbraken J., Spierenburg H., Seifert K.A., Peterson S.W., Varga J., Frisvad J.C. Phylogeny and nomenclature of the genus Talaromyces and taxa accommodated in Penicillium subgenus Biverticillium. Stud. Mycol. 2011;70:159–183. doi: 10.3114/sim.2011.70.04. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yilmaz N., Visagie C.M., Houbraken J., Frisvad J.C., Samson R.A. Polyphasic taxonomy of the genus Talaromyces. Stud. Mycol. 2014;78:175–341. doi: 10.1016/j.simyco.2014.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Raper K.B., Thom C. A Manual of the Penicillia. The Williams & Wilkins Company; Baltimore, MD, USA: 1949. [Google Scholar]
- 17.Thom C. The Penicillia. The Williams & Wilkins Company, Baltimore; MD, USA: 1930. [Google Scholar]
- 18.Visagie C.M., Houbraken J., Frisvad J.C., Hong S.B., Klaassen C.H., Perrone G., Seifert K.A., Varga J., Yaguchi T., Samson R.A. Identification and nomenclature of the genus Penicillium. Stud. Mycol. 2014;78:343–371. doi: 10.1016/j.simyco.2014.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Toru Okuda M.A.K., Seifert K.A., Ando K. Media and incubation effects on morphological characteristics of Penicillium and Aspergillus. In: Samson R.A., Pitt J.I., editors. Integration of Modern Taxonomic Methods for Penicillium and Aspergillus Classification. Harwood Academic Publishers; Amsterdam, The Netherlands: 2000. pp. 83–99. [Google Scholar]
- 20.Okuda T. Variation in colony characteristics of Penicillium strains resulting from minor variations in culture conditions. Mycologia. 1994;86:259–262. doi: 10.2307/3760646. [DOI] [Google Scholar]
- 21.Samson R.A., Pitt J.I. General Recommendations. In: Samson R.A., Pitt J.I., editors. Advances in Penicillium and Aspergillus Systematics. Plenum Press; London, UK: 1985. pp. 455–460. [Google Scholar]
- 22.Schoch C.L., Seifert K.A., Huhndorf S., Robert V., Spouge J.L., Levesque C.A., Chen W., Fungal Barcoding Consortium Nuclear ribosomal internal transcribed spacer (ITS) region as a universal DNA barcode marker for Fungi. Proc. Natl. Acad. Sci. USA. 2012;109:6241–6246. doi: 10.1073/pnas.1117018109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Taylor J.W., Jacobson D.J., Kroken S., Kasuga T., Geiser D.M., Hibbett D.S., Fisher M.C. Phylogenetic species recognition and species concepts in fungi. Fungal. Genet. Biol. 2000;31:21–32. doi: 10.1006/fgbi.2000.1228. [DOI] [PubMed] [Google Scholar]
- 24.Qiao H., Xiao-Qin W.U., Wang Z. Phosphate-solubilizing characteristic of a Penicillium pinophilum strain JP-NJ4. Microbiol. China. 2014;9:1741–1748. doi: 10.13344/j.microbiol.china.130828. (In Chinese with English abstract) [DOI] [Google Scholar]
- 25.Samson R.A., van der Aa H.A., de Hoog G.S. Centraalbureau voor Schimmelcultures: Hundred years microbial resource centre. Stud. Mycol. 2005;50:1–8. doi: 10.1023/B:MYCO.0000012225.79969.29. [DOI] [Google Scholar]
- 26.May T.W., Redhead S.A., Bensch K., Hawksworth D.L., Lendemer J., Lombard L., Turland N.J. Chapter F of the International Code of Nomenclature for algae, fungi, and plants as approved by the 11th International Mycological Congress, San Juan, Puerto Rico, July 2018. IMA Fungus. 2019;10:21. doi: 10.1186/s43008-019-0019-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Aime M.C., Miller A.N., Aoki T., Bensch K., Cai L., Crous P.W., Hawksworth D.L., Hyde K.D., Kirk P.M., Lucking R., et al. How to publish a new fungal species, or name, version 3.0. IMA Fungus. 2021;12:11. doi: 10.1186/s43008-021-00063-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cubero O.F., Crespo A., Fatehi J., Bridge P.D. DNA extraction and PCR amplification method suitable for fresh, herbarium-stored, lichenized, and other fungi. Plant. Syst. Evol. 1999;216:243–249. doi: 10.1007/BF01084401. [DOI] [Google Scholar]
- 29.De Hoog G.S., van den Ende A.H.G.G. Molecular diagnostics of clinical strains of filamentous Basidiomycetes. Mycoses. 1998;41:183–189. doi: 10.1111/j.1439-0507.1998.tb00321.x. [DOI] [PubMed] [Google Scholar]
- 30.Hong S.B., Cho H.S., Shin H.D., Frisvad J.C., Samson R.A. Novel Neosartorya species isolated from soil in Korea. Int. J. Syst. Evol. Microbiol. 2006;56:477–486. doi: 10.1099/ijs.0.63980-0. [DOI] [PubMed] [Google Scholar]
- 31.Houbraken J., Samson R.A. Phylogeny of Penicillium and the segregation of Trichocomaceae into three families. Stud. Mycol. 2011;70:1–51. doi: 10.3114/sim.2011.70.01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Houbraken J., Spierenburg H., Frisvad J.C. Rasamsonia, a new genus comprising thermotolerant and thermophilic Talaromyces and Geosmithia species. Anton. Leeuw. Int. J. G. 2012;101:403–421. doi: 10.1007/s10482-011-9647-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu Y.J., Whelen S., Hall B.D. Phylogenetic relationships among ascomycetes: Evidence from an RNA polymerse II subunit. Mol. Biol. Evol. 1999;16:1799–1808. doi: 10.1093/oxfordjournals.molbev.a026092. [DOI] [PubMed] [Google Scholar]
- 34.Lousie G.N., Donaldson G.C. Development of primer sets designed for use with the PCR to amplify conserved genes from Filamentous Ascomycetes. Appl. Environ. Microb. 1995;61:1320–1330. doi: 10.1002/bit.260460112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Masclaux F., Gueho E., de Hoog G.S., Christen R. Phylogenetic relationships of human-pathogenic Cladosporium (Xylohypha) species inferred from partial LS rRNA sequences. J. Med. Vet. Mycol. 1995;33:327–338. doi: 10.1080/02681219580000651. [DOI] [PubMed] [Google Scholar]
- 36.Peterson S.W., Vega F.E., Posada F., Nagai C. Penicillium coffeae, a new endophytic species isolated from a coffee plant and its phylogenetic relationship to P. fellutanum, P. thiersii and P. brocae based on parsimony analysis of multilocus DNA sequences. Mycologia. 2005;97:659–666. doi: 10.1080/15572536.2006.11832796. [DOI] [PubMed] [Google Scholar]
- 37.Rivera K.G., Seifert K.A. A taxonomic and phylogenetic revision of the Penicillium sclerotiorum complex. Stud. Mycol. 2011;70:139–158. doi: 10.3114/sim.2011.70.03. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.White T., Bruns T., Lee S., Taylor F., White T., Lee S.H., Taylor L., Shawetaylor J. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis M.A.G., Sninsky D.H., John J., White J.J., Thomas J., editors. PCR Protocols: A Guide to Methods and Applications. Academic Press; Cambridge, MA, USA: 1990. [Google Scholar]
- 39.El-Esawi M.A., Witczak J., Abomohra A.E., Ali H.M., Elshikh M.S., Ahmad M. Analysis of the Genetic Diversity and Population Structure of Austrian and Belgian Wheat Germplasm within a Regional Context Based on DArT Markers. Genes. 2018;9:47. doi: 10.3390/genes9010047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hall T.A. BioEdit: A User-Friendly Biological Sequence Alignment Editor and Analysis Program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 1999;41:95–98. doi: 10.1021/bk-1999-0734.ch008. [DOI] [Google Scholar]
- 41.Zhang D., Gao F., Jakovlic I., Zou H., Zhang J., Li W.X., Wang G.T. PhyloSuite: An integrated and scalable desktop platform for streamlined molecular sequence data management and evolutionary phylogenetics studies. Mol. Ecol. Resour. 2020;20:348–355. doi: 10.1111/1755-0998.13096. [DOI] [PubMed] [Google Scholar]
- 42.Kalyaanamoorthy S., Minh B.Q., Wong T.K.F., von Haeseler A., Jermiin L.S. ModelFinder: Fast model selection for accurate phylogenetic estimates. Nat. Methods. 2017;14:587–589. doi: 10.1038/nmeth.4285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Frisvad J.C. Physiological criteria and mycotoxin production as AIDS in identification of common asymmetric penicillia. Appl. Environ. Microbiol. 1981;41:568–579. doi: 10.1128/aem.41.3.568-579.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ramírez C. Manual and Atlas of the Penicillia. Elsevier Biomedical Press; Amsterdam, The Netherlands: 1982. [Google Scholar]
- 45.Blakeslee A.F. Lindner’s roll tube method of separation cultures. Phytopathology. 1915;5:68–69. [Google Scholar]
- 46.David A. Hay Infusion. Tex. Sci. Teach. 1993;22:10. [Google Scholar]
- 47.Hocking A.D., Pitt J.I. Dichloran-glycerol medium for enumeration of xerophilic fungi from low-moisture foods. Appl. Environ. Microbiol. 1980;39:488–492. doi: 10.1128/aem.39.3.488-492.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Christensen M., Frisvad J.C., Tuthill D.E. Penicillium species diversity in soil and some taxonomic and ecological notes. In: Samson R.A., Pitt J.I., editors. Integration of Modern Taxonomic Methods for Penicillium and Aspergillus Classification. Harwood Academic Publishers; Amsterdam, The Netherlands: 2000. pp. 309–321. [Google Scholar]
- 49.Pitt J.I. An appraisal of identification methods for Penicillium species: Novel taxonomic criteria based on temperature and water relations. Mycologia. 1973;65:1135–1157. doi: 10.2307/3758294. [DOI] [PubMed] [Google Scholar]
- 50.Kong H.Z. Flora Fungorum Sinicorum. Volume 35. Science Press; Beijing, China: 2007. [Google Scholar]
- 51.Schoch C.L., Aime M.C., Beer W.D., Crous P.W., Miller A.N. Using standard keywords in publications to facilitate updates of new fungal taxonomic names. IMA Fungus. 2017;8:70–73. doi: 10.1007/BF03449466. [DOI] [Google Scholar]
- 52.Heo Y.M., Lee H., Kim K., Sun L.K., Kim J.J. Fungal Diversity in Intertidal Mudflats and Abandoned Solar Salterns as a Source for Biological Resources. Mar. Drugs. 2019;17:601. doi: 10.3390/md17110601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sun B.D., Chen A.J., Houbraken J., Frisvad J.C., Wu W.P., Wei H.L., Zhou Y.G., Jiang X.Z., Samson R.A. New section and species in Talaromyces. MycoKeys. 2020;68:75–113. doi: 10.3897/mycokeys.68.52092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Barbosa R.N., Bezerra J.D.P., Souza-Motta C.M., Frisvad J.C., Samson R.A., Oliveira N.T., Houbraken J. New Penicillium and Talaromyces species from honey, pollen and nests of stingless bees. Anton. Leeuw. Int. J. G. 2018;111:1883–1912. doi: 10.1007/s10482-018-1081-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chen A.J., Sun B.D., Houbraken J., Frisvad J.C., Yilmaz N., Zhou Y.G., Samson R.A. New Talaromyces species from indoor environments in China. Stud. Mycol. 2016;84:119–144. doi: 10.1016/j.simyco.2016.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Crous P.W., Wingfield M.J., Burgess T.I., Hardy G., Gene J., Guarro J., Baseia I.G., Garcia D., Gusmao L.F.P., Souza-Motta C.M., et al. Fungal Planet description sheets: 716–784. Persoonia. 2018;40:240–393. doi: 10.3767/persoonia.2018.40.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Guevara-Suarez M., Sutton D.A., Gene J., Garcia D., Wiederhold N., Guarro J., Cano-Lira J.F. Four new species of Talaromyces from clinical sources. Mycoses. 2017;60:651–662. doi: 10.1111/myc.12640. [DOI] [PubMed] [Google Scholar]
- 58.Peterson S.W., Jurjevic Z. New species of Talaromyces isolated from maize, indoor air, and other substrates. Mycologia. 2017;109:537–556. doi: 10.1080/00275514.2017.1369339. [DOI] [PubMed] [Google Scholar]
- 59.Rodriguez-Andrade E., Stchigel A.M., Terrab A., Guarro J., Cano-Lira J.F. Diversity of xerotolerant and xerophilic fungi in honey. IMA Fungus. 2019;10:20. doi: 10.1186/s43008-019-0021-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sang H., An T.J., Kim C.S., Shin G.S., Sung G.H., Yu S.H. Two novel Talaromyces species isolated from medicinal crops in Korea. J. Microbiol. 2013;51:704–708. doi: 10.1007/s12275-013-3361-9. [DOI] [PubMed] [Google Scholar]
- 61.Wang Q.M., Zhang Y.H., Wang B., Wang L. Talaromyces neofusisporus and T. qii, two new species of section Talaromyces isolated from plant leaves in Tibet, China. Sci. Rep. 2016;6:18622. doi: 10.1038/srep18622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yilmaz N., Lopez-Quintero C.A., Vasco-Palacios A., Frisvad J.C., Theelen B., Boekhout T., Samson R.A., Houbraken J. Four novel Talaromyces species isolated from leaf litter from Colombian Amazon rain forests. Mycol. Prog. 2016;15:1041–1056. doi: 10.1007/s11557-016-1227-3. [DOI] [Google Scholar]
- 63.Yamazaki H., Koyama N., Omura S., Tomoda H. New rugulosins, anti-MRSA antibiotics, produced by Penicillium radicum FKI-3765-2. Org. Lett. 2010;12:1572–1575. doi: 10.1021/ol100298h. [DOI] [PubMed] [Google Scholar]
- 64.Bladt T.T., Frisvad J.C., Knudsen P.B., Larsen T.O. Anticancer and antifungal compounds from Aspergillus, Penicillium and other filamentous fungi. Molecules. 2013;18:11338–11376. doi: 10.3390/molecules180911338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kumari M., Taritla S., Sharma A., Jayabaskaran C. Antiproliferative and Antioxidative Bioactive Compounds in Extracts of Marine-Derived Endophytic Fungus Talaromyces purpureogenus. Front. Microbiol. 2018;9:1777. doi: 10.3389/fmicb.2018.01777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Isbelia R., Louis B., Simard R.R., Phillipe T., Hani A. Characteristics of phosphate solubilization by an isolate of a tropical Penicillium rugulosum and two UV-induced mutants. Fems. Microbiol. Ecol. 1999;28:291–295. doi: 10.1111/j.1574-6941.1999.tb00584.x. [DOI] [Google Scholar]
- 67.Narikawa T., Shinoyama H., Fujii T. A β-Rutinosidase from Penicillium rugulosum IFO 7242 That Is a Peculiar Flavonoid Glycosidase. Biosci. Biotechnol. Bioch. 2000;64:1317–1319. doi: 10.1271/bbb.64.1317. [DOI] [PubMed] [Google Scholar]
- 68.Pol D., Laxman R.S., Rao M. Purification and biochemical characterization of endoglucanase from Penicillium pinophilum MS 20. Indian. J. Biochem. Biophys. 2012;49:189–194. doi: 10.1684/ecn.2012.0308. [DOI] [PubMed] [Google Scholar]
- 69.Fujii T., Hoshino T., Inoue H., Yano S. Taxonomic revision of the cellulose-degrading fungus Acremonium cellulolyticus nomen nudum to Talaromyces based on phylogenetic analysis. FEMS. Microbiol. Lett. 2014;351:32–41. doi: 10.1111/1574-6968.12352. [DOI] [PubMed] [Google Scholar]
- 70.Houbraken J., de Vries R.P., Samson R.A. Modern Taxonomy of Biotechnologically Important Aspergillus and Penicillium Species. Adv. Appl. Microbiol. 2014;86:199–249. doi: 10.1016/B978-0-12-800262-9.00004-4. [DOI] [PubMed] [Google Scholar]
- 71.Maeda R.N., Barcelos C.A., Santa Anna L.M., Pereira N., Jr. Cellulase production by Penicillium funiculosum and its application in the hydrolysis of sugar cane bagasse for second generation ethanol production by fed batch operation. J. Biotechnol. 2013;163:38–44. doi: 10.1016/j.jbiotec.2012.10.014. [DOI] [PubMed] [Google Scholar]
- 72.Fahima T., Henis Y. Proceedings of the Biological Control of Soil-Borne Plant Pathogens. CAB International; Wallingford, UK: 1997. Increasing of Trichoderma hamatum and Talaromyces flavus on root of healthy and useful hosts; pp. 296–322. [Google Scholar]
- 73.Naraghi L., Heydari A., Rezaee S., Razavi M. Biocontrol Agent Talaromyces flavus Stimulates the Growth of Cotton and Potato. J. Plant. Growth. Regul. 2012;31:471–477. doi: 10.1007/s00344-011-9256-2. [DOI] [Google Scholar]
- 74.Naraghi L., Heydari A., Rezaee S., Razavi M., Afshari-Azad H. Biological control of Verticillium wilt of greenhouse cucumber by Talaromyces flavus. Phytopathol. Mediterr. 2011;49:321–329. doi: 10.14601/Phytopathol_Mediterr-8450. [DOI] [Google Scholar]
- 75.Naraghi L., Heydari A., Rezaee S., Razavi M., Khaledi E.M. Biological control of tomato Verticillium disease by Talaromyces flavus. J. Plant Prot. Res. 2010;50:360–365. doi: 10.2478/v10045-010-0061-x. [DOI] [Google Scholar]
- 76.Abdel-Rahim I.R., Abo-Elyousr K.A.M. Talaromyces pinophilus strain AUN-1 as a novel mycoparasite of Botrytis cinerea, the pathogen of onion scape and umbel blights. Microbiol. Res. 2018;212–213:1–9. doi: 10.1016/j.micres.2018.04.004. [DOI] [PubMed] [Google Scholar]
- 77.Khalmuratova I., Kim H., Nam Y.J., Oh Y., Jeong M.J., Choi H.R., You Y.H., Choo Y.S., Lee I.J., Shin J.H., et al. Diversity and Plant Growth Promoting Capacity of Endophytic Fungi Associated with Halophytic Plants from the West Coast of Korea. Mycobiology. 2015;43:373–383. doi: 10.5941/MYCO.2015.43.4.373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Jeewon R., Hyde K.D. Establishing species boundaries and new taxa among fungi: Recommendations to resolve taxonomic ambiguities. Mycosphere. 2016;7:1669–1677. doi: 10.5943/mycosphere/7/11/4. [DOI] [Google Scholar]
- 79.Peterson S.W., Jurjevic Z. The Talaromyces pinophilus species complex. Fungal Biol. 2019;123:745–762. doi: 10.1016/j.funbio.2019.06.007. [DOI] [PubMed] [Google Scholar]
- 80.Houbraken J., Kocsube S., Visagie C.M., Yilmaz N., Wang X.C., Meijer M., Kraak B., Hubka V., Bensch K., Samson R.A., et al. Classification of Aspergillus, Penicillium, Talaromyces and related genera (Eurotiales): An overview of families, genera, subgenera, sections, series and species. Stud. Mycol. 2020;95:5–169. doi: 10.1016/j.simyco.2020.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Frisvad J.C., Yilmaz N., Thrane U., Rasmussen K.B., Houbraken J., Samson R.A. Talaromyces atroroseus, a new species efficiently producing industrially relevant red pigments. PLoS ONE. 2013;8:e84102. doi: 10.1371/journal.pone.0084102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Zaccarim B.R., de Oliveira F., Passarini M.R.Z., Duarte A.W.F., Sette L.D., Jozala A.F., Teixeira M.F.S., de Carvalho Santos-Ebinuma V. Sequencing and phylogenetic analyses of Talaromyces amestolkiae from amazon: A producer of natural colorants. Biotechnol. Prog. 2019;35:e2684. doi: 10.1002/btpr.2684. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The materials are available as Supplementary materials (Tables S1 and S2; Figures S1–S8). Publicly available datasets were analyzed in this study. This data can be found here: https://www.ncbi.nlm.nih.gov/genbank, accessed on 18 January 2022; accession number ITS = MW130720, BenA = MW147759, CaM = MW147760, RPB1 = MW147761, RPB2 = MW147762.