Abstract
Aerobic respiration is a key energy-producing pathway in many prokaryotes and virtually all eukaryotes. The final step of aerobic respiration is most commonly catalyzed by heme-copper oxidases embedded in the cytoplasmic or mitochondrial membrane. The majority of these terminal oxidases contain a prenylated heme (typically heme a or occasionally heme o) in the active site. In addition, many heme-copper oxidases, including mitochondrial cytochrome c oxidases, possess a second heme a cofactor. Despite the critical role of heme a in the electron transport chain, the details of the mechanism by which heme b, the prototypical cellular heme, is converted to heme o and then to heme a remain poorly understood. Recent structural investigations, however, have helped clarify some elements of heme a biosynthesis. In this review, we discuss the insight gained from these advances. In particular, we present a new structural model of heme o synthase (HOS) based on distance restraints from inferred coevolutionary relationships and refined by molecular dynamics simulations that is in good agreement with the experimentally determined structures of HOS homologs. We also analyze the two structures of heme a synthase (HAS) that have recently been solved by other groups. For both HOS and HAS, we discuss the proposed catalytic mechanisms and highlight how new insights into the heme binding site locations shed light on previously obtained biochemical data. Finally, we explore the implications of the new structural data in the broader context of heme trafficking in the heme a biosynthetic pathway and heme-copper oxidase assembly.
Keywords: cytochrome c oxidase, cytochrome c oxidase assembly, heme a synthase, heme o synthase, heme-copper oxidases, heme oxidation, heme trafficking, intramembrane aromatic prenyltransferase
Introduction
For practically all aerobic forms of life, respiration is a key pathway for producing useable cellular energy in the form of ATP. Aerobic respiration is the metabolic process in which electrons are transferred from intermediate redox cofactors (such as NADH and FADH2) to a membrane-localized electron transport chain and finally to O2, the terminal electron acceptor. The flow of electrons through the electron transport chain is coupled to movement of protons across the membrane from the N-side (negative side) to the P-side (positive side), generating the chemiosmotic gradient that drives ATP production. To allow for continued flow of electrons through the electron transport chain, the terminal oxidase accepts electrons from the chain while catalyzing the four-electron reduction of O2 to H2O (Ferguson-Miller and Babcock 1996). Two unrelated superfamilies of terminal oxidases are known, the bd quinol oxidases and the heme-copper oxidases. As their respective names suggest, the bd oxidases use hemes b and d as cofactors, while the heme-copper oxidases have a bimetallic heme-copper active site, as well as additional heme and copper cofactors. While bd oxidases are only found in certain bacterial and archaeal phyla, heme-copper oxidases are found in all three domains of life (Garcia-Horsman et al. 1994; Schafer et al. 1999; Borisov et al. 2011; Borisov and Siletsky 2019; Refojo et al. 2019).
Heme-copper oxidases have two heme cofactors with different axial ligand coordination and spin states (Figure 1A). One of these hemes is a low-spin, bis-histidine axially ligated heme that transfers electrons to the active site. In contrast, the active site heme is high-spin and only has one axial histidine ligand (Garcia-Horsman et al. 1994; Ferguson-Miller and Babcock 1996; Wikstrom et al. 2018). Depending on the specific heme-copper oxidase, different types of hemes with different porphyrin ring substituents occupy the low-spin and high-spin heme binding sites. The low-spin heme binding site may be occupied by heme a or b, while the high-spin site may be occupied by heme a, b, or o (Garcia-Horsman et al. 1994). In addition to the low-spin and high-spin heme binding sites that are common to all heme-copper oxidases, some oxidases have additional heme binding sites that precede the low-spin and high-spin hemes in the oxidase’s electron transfer pathway. The heme type found in these sites is invariably heme c (heme b that is covalently crosslinked to the protein via thioether linkages to cysteine residues) (Pereira et al. 2001). By convention, when naming oxidases, the hemes are listed in the order of electron flow through the oxidase: heme c (where present) is listed first, followed by the low-spin heme, and ending with the high-spin (active site) heme, which is designated with a subscript “3.” Although the heme-copper oxidase superfamily can be divided into three phylogenetically distinct families (A, B, and C), heme types do not strictly correlate with phylogeny: All family-C oxidases are cbb3 cytochrome c oxidases, while family A includes mitochondrial-like aa3 oxidases and bo3 oxidases, and family B includes ba3 oxidases as well as a different subset of aa3 oxidases (Pereira et al. 2001; Pereira et al. 2008). While C-type oxidases and B-type oxidases have only been identified in bacteria or bacteria and archaea, respectively, A-type oxidases are present in all three domains of life and account for the majority of the heme-copper oxidase sequences that have been identified thus far (Garcia-Horsman et al. 1994; Schafer et al. 1999; Hemp 2008; Borisov and Siletsky 2019; Refojo et al. 2019). Furthermore, although many prokaryotes have branched electron transport chains with multiple types of terminal oxidases, the only type of terminal oxidase present in eukaryotic mitochondria is an aa3 cytochrome c oxidase from family A (Pereira et al. 2001; Pereira et al. 2008; Refojo et al. 2019). Given the widespread role of heme a as a cofactor for terminal heme-copper oxidases, heme a biosynthesis is a critical component of primary metabolism for many aerobic organisms, and is essential for energy production in higher eukaryotes.
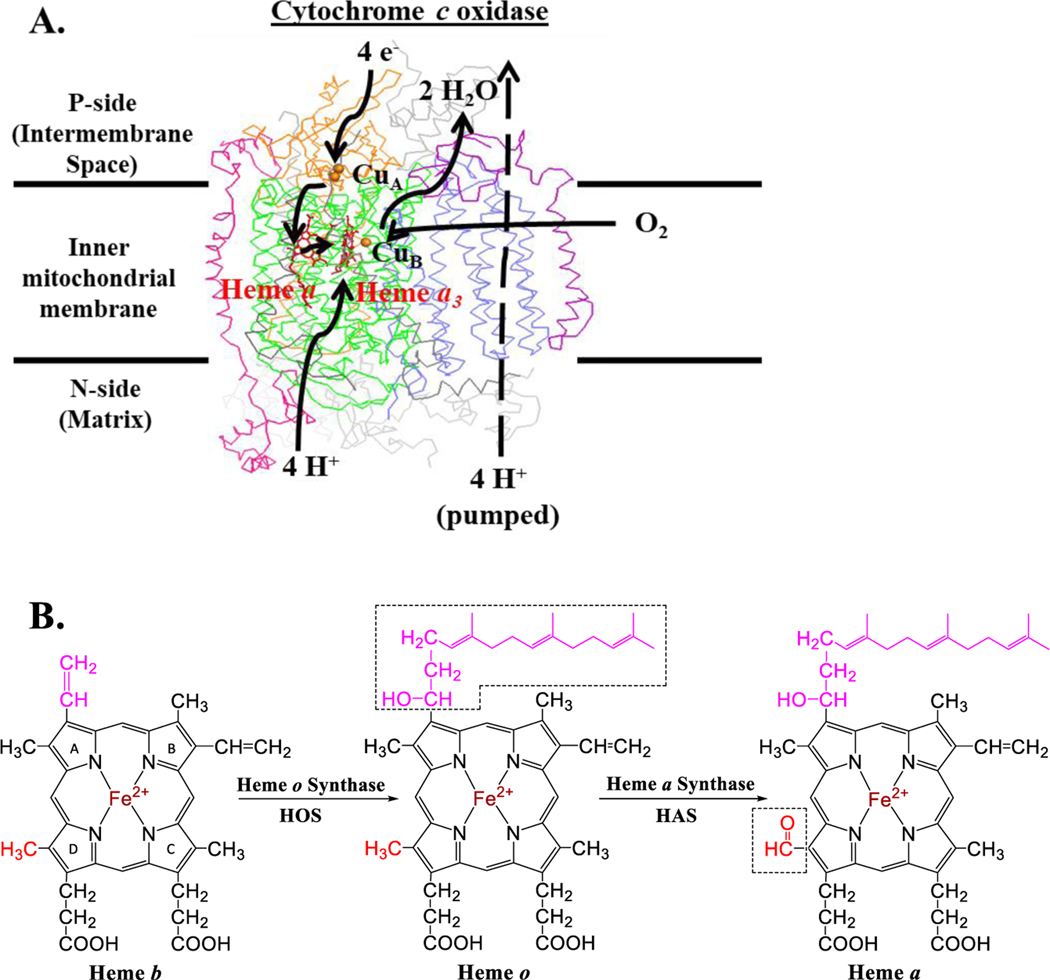
Figure 1. Heme a, a cofactor of cytochrome c oxidase, is synthesized from heme b via two enzymatically catalyzed steps.
A. Heme-copper oxidases catalyze the 4-electron reduction of O2 to H2O with concomitant proton pumping. The 11-subunit S. cerevisiae cytochrome c oxidase (a mitochondrial-type aa3 oxidase) is shown. Subunit I (which binds low-spin heme a, high-spin heme a3, and CuB) is shown in green. Subunit II (which includes the binuclear CuA site) is shown in orange, and subunit III is shown in slate. Nuclear-encoded subunits Cox5a and Cox13 are shown in hot pink and purple, respectively. The remaining 5 nuclear-encoded subunits are shown in gray. Electrons are transferred from cytochrome c (not shown) to the CuA site, then to heme a, and finally to the binuclear active site (heme a3 and CuB) where O2 is reduced to H2O. Although varied proton pumping stoichiometries have been observed for isolated heme-copper oxidases from different families, family A oxidases (such as the one shown) have the greatest proton pumping capacities, with pumped H+/e− ratios approaching 1 (Pereira et al. 2008). This figure was prepared using a previously published homology model of S. cerevisiae cytochrome c oxidase based on the bovine crystal structure (Maréchal et al. 2012). B. Summary of heme o and heme a biosynthesis. Heme o is synthesized from heme b by the conversion of the vinyl group from pyrrole ring A (C2 position) to a hydroxyethylfarnesyl moiety (both shown in pink). Heme a is synthesized from heme o by the oxidation of the pyrrole ring D methyl group (C8 position) to an aldehyde (shown in red).
Hemes a and o are chemically modified derivatives of the prototypical heme, heme b. Hemes o and a can be synthesized from heme b via sequential enzyme-catalyzed reactions (Figure 1B). First, heme b’s vinyl group from pyrrole ring A (the C2 position in Fischer nomenclature) is converted to a hydroxyethylfarnesyl moiety, producing heme o (Puustinen and Wikstrom 1991). This reaction is catalyzed by heme o synthase (HOS), a prenyltransferase (Saiki et al. 1992; Saiki, Mogi, Ogura, et al. 1993; Svensson et al. 1993; Glerum and Tzagoloff 1994; Mogi et al. 1994). Then, in organisms requiring heme a, heme a synthase (HAS) catalyzes the oxidation of the pyrrole ring D methyl (C8 position) to a formyl group (Svensson et al. 1993; Mogi et al. 1994; Svensson and Hederstedt 1994; Svensson et al. 1996; Barros et al. 2001; Brown et al. 2002; Brown KR et al. 2004). The only known function for heme a is to serve as a cofactor for heme-copper terminal oxidases, and, in some acidophilic archaea, as a cofactor for the cytochrome b-like subunit of quinol reductases (analogous to mitochondrial complex III) (Garcia-Horsman et al. 1994; Lübben and Morand 1994; Lübben et al. 1994; Bandeiras et al. 2009; Castelle et al. 2015). Heme o serves either as a precursor for heme a synthesis or as a terminal oxidase cofactor (or both, in some prokaryotic species) (Sone and Fujiwara 1991; Matsushita, Ebisuya, Adachi 1992; Matsushita, Ebisuya, Ameyama, et al. 1992; Garcia-Horsman et al. 1994; Sone et al. 1994; Auer et al. 1995; Peschek et al. 1995; Schröter et al. 1998; Contreras-Zentella et al. 2003).
The biosynthesis of hemes o and a appears to be a universally conserved process that uses heme b as the initial porphyrin substrate. All known genes encoding HOS belong to the same sub-family of the aromatic intramembrane prenyltransferase (UbiA) superfamily (Mogi et al. 1994; Li 2016). Similarly, all identified HAS-encoding genes belong to one family (Cox15/CtaA family) (Hederstedt 2012; He et al. 2016). Although alternate forms of heme o (hemes oT, oP1, oP2) and heme a (heme aS) have been identified in archaea and one bacterial species, the deviations from the “normal” prenylated heme structures are minimal and are located on pyrrole ring A at the C2 position (Lübben and Morand 1994; Lübben et al. 1994; Castelle et al. 2015). In most cases, genes for canonical HOS and HAS can be identified within the genomes of the species in question (Table 1) (Lewin and Hederstedt 2006; Hederstedt 2012). This association suggests that the modified hemes are synthesized by a canonical HOS that uses an alternate prenyl donor or a slightly different reaction mechanism instead of by an unrelated enzyme (see the section entitled “Heme o synthase”). Oxidation of the methyl group on pyrrole ring D of the heme o variant would then be catalyzed by a canonical HAS.
Table 1.
Genes encoding HOS (CtaB) and HAS (CtaA) from selected prokaryotic species that synthesize alternate prenylated hemes. The genes for B. subtilisHOS and HAS (which have been biochemically characterized and which synthesize standard heme oand heme a, respectively) are included for reference. All genes encoding HOS listed include highly conserved residues that are expected to be required for activity. All genes encoding HAS listed include the characteristic highly conserved histidines, with the exception of the HAS sequences discussed below in (f).
| Organism | Prenylated heme(s) |
HOS | HAS | References | ||
|---|---|---|---|---|---|---|
| Genea | TM#b | Genea | TM#b | |||
| Bacteria | ||||||
| Bacillus subtilis | a |
ctaO
(ctaB1) c |
9 | (Svensson et al. 1993; Svensson and Hederstedt 1994; Throne-Holst and Hederstedt 2000; Mogi 2009a) | ||
|
ctaB
(ctaB2) c |
9 | ctaA | 8 | |||
| Thermus thermophilus | a s | ctaAB | 17d | ctaAB | 17d | (Lübben and Morand 1994; Lyons et al. 2012) |
| Archaea | ||||||
| Sulfolobus acidocaldarius | a s | ctaB | 9 |
ctaA (SACI_RS02090) |
8 | (Lübben and Morand 1994; Lübben et al. 1994) |
| Acidianus ambivalens (Desulfurolobus ambivalens) Halobacterium salinarum | a s | ctaB | 9 |
ctaA (D1866_00290) |
8 | (Lübben and Morand 1994; Bandeiras et al. 2009) |
| a s | ctaAB | 13e | ctaAB | 13e | (Lübben and Morand 1994) | |
| ccp | 8f | |||||
| Natronomonas phaoronis | a s | ctaAB1 | 13e | ctaAB1 | 13e | (Lübben and Morand 1994; Scharf et al. 1997) |
| ctaAB2 | 13e | ctaAB2 | 13e | |||
| ctaA | 8f | |||||
| Thermoplasma acidophilum Pyrobaculum aerophilum | o T | ctaB | 9 | NA | (Lübben and Morand 1994) | |
| oP1, oP2, as | ctaB1 | 9 | Unknown | (Lübben and Morand 1994) | ||
| ctaB2 | 9 | |||||
Fused genes encoding HAS and HOS from T. thermophilus, H. salinarum, and N. phaoronis were renamed ctaAB. The gene names from Genbank have also been included for genes that are currently not annotated as ctaA in Genbank.
The number (or predicted number) of transmembrane (TM) helices (TM#) is listed. In some cases, the predicted number of TM helices from Uniprot has been corrected by the authors after manual inspection of sequence alignments.
CtaO is a paralog of CtaB that can complement a ctaB deletion mutant (Throne-Holst and Hederstedt 2000). The alternate gene names ctaB1 and ctaB2 are from Uniprot.
The gene encoding HAS (TM1–8) is fused to the gene encoding HOS (TM9–17).
The gene encoding a shortened HAS variant (TM1–4) is fused to the gene encoding HOS (TM5–13).
The genes encoding these HAS sequences are missing one (H. salinarum ccp) or both (N. phaoronis ctaAB1 and ctaAB2) highly conserved C-terminal histidine residues. However, these species also contain a 4-TM version of HAS fused to HOS.
While the structures and biochemical reactions of all eight enzymes that catalyze the steps of the most common heme b biosynthetic pathway have been studied extensively (Heinemann et al. 2008; Swenson SA et al. 2020), prenylated heme biosynthesis is not as well understood. This is partly due to the fact that HOS and HAS are both integral membrane proteins that can be difficult to overexpress and purify. However, recent progress has been made in the structural characterization of both enzymes. The first crystal structures from the prenyltransferase superfamily that HOS belongs to have been solved (Cheng and Li 2014; Huang et al. 2014), and we have used these new structural data and a coevolution-based machine learning technique to model the structure of HOS. The structure of HAS from two different species has also been determined independently by X-ray crystallography and cryo-electron microscopy (cryo-EM), and the substrate-bound structure of HAS was modelled on the basis of the crystal structure (Niwa et al. 2018; Zeng et al. 2020). The new structural information about HOS and HAS provides fresh insights into the structure-function paradigm and mechanism of these enzymes, and also raises new questions about intracellular transfer of hemes b, o, and a.
Heme o synthase
Heme o properties and function
Heme o is synthesized from heme b by the conversion of the vinyl group of pyrrole ring A (C2 vinyl) to a hydroxyethylfarnesyl group (Figures 1B, 2) (Puustinen and Wikstrom 1991; Puustinen 1992; Saiki, Mogi, Ogura, et al. 1993; Mogi et al. 1994). While the reduction potentials of hemes b and o are similar, the addition of the lipid tail produces a porphyrin that can bind a protein more tightly. A direct comparison of these two hemes binding to the same synthetic heme protein maquette shows the hydroxyethylfarnesyl moiety increases the binding affinity of heme o by a factor of 700 (ferrous state) and 40,000 (ferric state) relative to heme b (Zhuang, Reddi, et al. 2006). The hydroxyl group of the hydroxyethylfarnesyl moiety for the high-spin (active site) heme is located in one of the proton channels in family-A heme-copper oxidases and has been proposed to participate in proton delivery to the active site (Iwata et al. 1995; Abramson et al. 2000; Wang et al. 2005). In addition, this hydroxyl group forms a hydrogen bond with the tyrosine residue that is crosslinked to one of the histidine ligands of copper in the oxidase’s binuclear active site, possibly allowing for modulation of the tyrosine pKa in aa3 oxidases (Buschmann et al. 2010; Sharma et al. 2011; Wikstrom et al. 2018). Interestingly, in cbb3 oxidases, a slightly different arrangement of the active site may compensate for the absence of this hydroxyl group (Buschmann et al. 2010; Wikstrom et al. 2018). Thus, the hydroxyethylfarnesyl tail of heme o (and heme a) seems to play a major role in enhancing the heme’s affinity for its oxidase and may also play a role in catalysis.
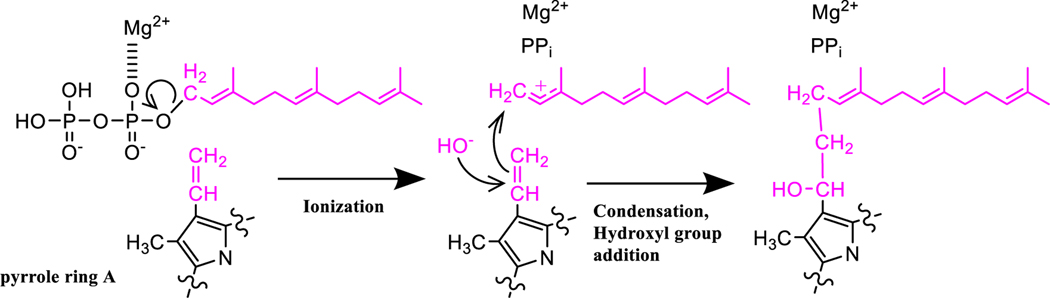
Figure 2. Proposed reaction mechanism for the conversion of heme b to heme o.
HOS catalyzes the transfer of a farnesyl moiety (in pink) from farnesyl diphosphate to the vinyl group of heme b (in pink) at pyrrole ring A. For simplicity, only pyrrole ring A of the heme is shown. Ionization of farnesyl diphosphate yields a farnesyl cation intermediate stabilized by delocalization across C′1-C′3. Interactions with Mg2+ ions (only one shown here) make the pyrophosphate a better leaving group and allow ionization to proceed. The vinyl group then attacks the carbocation intermediate to form a new C-C bond (condensation), and a hydroxyl group is added to C1 to produce heme o. A basic residue in the active site could promote the formation of a hydroxyl group prior to the attack of C1, or water could attack C1 and subsequently be deprotonated. The timing of the main three steps (ionization, condensation, and hydroxyl group addition) has not been established for HOS. For example, ionization and condensation may be concerted. Additionally, condensation may precede addition of the hydroxyl group, or these two steps may occur simultaneously. For heme variants oT and oP1, the hydroxyethylfarnesyl group is replaced by an ethylprenyl group (C15 or C20, respectively), indicating that the catalytic activity of HOS in certain archaea differs slightly from the canonical HOS mechanism shown here (Lübben and Morand 1994).
HOS mechanism
HOS, which is located either in the prokaryotic cytoplasmic membrane or the eukaryotic inner mitochondrial membrane, belongs to the UbiA superfamily of intramembrane aromatic prenyltransferases (Nobrega et al. 1990; Saiki, Mogi, Ogura, et al. 1993). These integral membrane proteins transfer polyprenyl groups from polyprenyl diphosphates of varying chain length (XPP) to a variety of aromatic (and some non-aromatic) substrates, including heme b, chlorophyll, and the ubiquinone precursor p-hydroxybenzoate (PHB) (Li 2016). In the general catalytic mechanism for this superfamily, the XPP substrate is cleaved to generate a polyprenyl carbocation, followed by condensation with the aromatic substrate and elimination of a proton to generate the prenylated product. For example, PHB octaprenyltransferase (UbiA) catalyzes the transfer of an octaprenyl group from octaprenyldiphosphate to the meta position of PHB. This reaction requires the presence of divalent metal cofactors, such as Mg2+, presumably to facilitate phosphoester bond cleavage by making the pyrophosphate a better leaving group (Melzer and Heide 1994; Wessjohann and Sontag 1996; Bräuer et al. 2008; Li 2016). The proposed ionization-condensation-elimination mechanism for the UbiA superfamily is based on biochemical characterization of the structurally related soluble isoprenyl pyrophosphate synthases (IPPS), which also rely on divalent metal cations for pyrophosphate ionization (Poulter and Rilling 1978; Hosfield et al. 2004; Kavanagh et al. 2006). While this mechanism is often drawn as three distinct steps, the available experimental data for IPPS would also be consistent with the first two steps (ionization and condensation) occurring in a concerted fashion, without the formation of a distinct carbocation intermediate (Poulter and Rilling 1978). There is, however, evidence for a carbocation intermediate in an unrelated family of soluble aromatic prenyltransferases, where the cation intermediate appears to be stabilized by interactions with active site tyrosine residues and with the aromatic substrate via cation-π interactions (Luk and Tanner 2009; Yang Y et al. 2012; Rudolf and Poulter 2013; Rudolf et al. 2013; Bayse and Merz 2014). Regardless of whether phosphoester bond cleavage and C-C bond formation occur sequentially or concurrently, the final step in the general catalytic mechanism of intramembrane aromatic prenyltransferases is elimination of a proton and rearomatization of the prenyl acceptor. One key difference between the prototypical UbiA reaction and the reaction catalyzed by HOS is that instead of proton abstraction, HOS catalyzes the addition of a hydroxyl group at the C1 position of the attacking vinyl. At present, the mechanism for this hydroxyl group addition is unknown, although simultaneous hydroxyl group addition and condensation has been proposed (Saiki, Mogi, Ogura, et al. 1993; Mogi et al. 1994).
Interestingly, archaea synthesize modified versions of heme o with the usual hydroxyethylfarnesyl moiety replaced by an ethylprenyl group (hemes oT and oP1) or a hydroxyethylgeranylgeranyl group (heme oP2) (Lübben and Morand 1994). The lack of a hydroxyl group in hemes oT and oP1 suggests that for these hemes, heme o synthesis proceeds via the more common ionization-condensation-elimination mechanism, with elimination of a proton from C2 reforming the double bond between C1 and C2. However, the C1-C2 double bond could also be formed via dehydration of the hydroxyprenyl group (Mogi et al. 1994). Synthesis of hemes with longer (C20) prenyl groups (oP1, oP2, and aS) is thought to be due to the higher availability of geranylgeranyl pyrophosphate in some archaea (Lübben and Morand 1994).
Only one in vitro activity assay has been reported for HOS. This assay was performed using cytoplasmic membrane vesicles prepared from E. coli overexpressing the native E. coli HOS (CyoE, referred to as EcHOS) (Saiki, Mogi, Ogura, et al. 1993). Heme o synthesis was observed upon addition of hemin (heme b), farnesyl diphosphate, Mg2+ or Ca2+, and a reducing agent to the membrane vesicles. Reduction of the heme iron is presumably required to increase the electron density of the heme’s vinyl groups and thus enable the vinyl C2 to perform a nucleophilic attack (Saiki, Mogi, Ogura, et al. 1993). This part of the mechanism is similar to the proposed mechanism for the biosynthesis of cytochrome c, which also involves nucleophilic attack by the vinyl groups of the substrate heme (Kranz et al. 2009). Although studying HOS via in vitro activity assays is complicated by the difficulties associated with overexpressing a membrane protein, in vivo activity assays have been used to study the importance of key residues in different domains of HOS (Saiki, Mogi, Hori, et al. 1993). Combined with insight gained from the available structures of UbiA prenyltransferases and a new structural model of HOS (Cheng and Li 2014; Huang et al. 2014; Li 2016; Heo and Feig 2020), a picture of how HOS binds cofactors and substrates is beginning to emerge.
Overview of aromatic intramembrane prenyltransferase (UbiA) superfamily structures
To date, two intermembrane prenyltransferases from hyperthermophilic archaea have been crystallized: Aeropyrum pernix UbiA (ApUbiA) and Archaeoglobus fulgidus UbiA (AfUbiA) (Cheng and Li 2014; Huang et al. 2014). The native substrates have not been identified for either enzyme, but ApUbiA appears to be most closely related to the PHB prenyltransferase (UbiA) sub-family, while AfUbiA is not closely related to any well-characterized sub-families (Li 2016). The overall architectures of ApUbiA and AfUbiA are very similar. Both have nine transmembrane (TM) helices. TM1–4 and TM5–8 form four-helical bundles that are related to each other by pseudo-twofold symmetry, suggesting an ancient gene duplication and fusion (Huang et al. 2014). This pair of four-helical bundles surrounds a central cavity on the cytoplasmic (negative) side of the membrane. A cytoplasmic domain composed of the helix-loop linker between TM2–3 (HL23) and a second helix-loop linker between TM6–7 (HL67) is positioned over the central cavity (Figure 3A). Each of these linkers contains an aspartate-rich motif. Conserved residues in this extramembrane domain (including the aspartate residues) are involved in coordinating two Mg2+ ions, either directly or indirectly. While the stoichiometric ratio of Mg2+ ions to UbiA monomer has not been determined, XPP-bound structures show two Mg2+ ions positioned on either side of the pyrophosphate group, and binding assays have confirmed that the Mg2+-coordinating residues are required for XPP binding. Other conserved, polar residues that face the central cavity also participate in pyrophosphate coordination. While the overall structure does not change much between the apoprotein and substrate-bound forms, this cytoplasmic region becomes more ordered when both Mg2+ and the XPP substrate are bound, restricting solvent access to the central cavity; the ability to protect the active site from water would be critical for a reaction mechanism involving a polyprenyl carbocation intermediate (Cheng and Li 2014; Huang et al. 2014). Additionally, a strictly conserved tyrosine residue positioned near the XPP binding site could potentially stabilize a carbocation intermediate via cation-π interactions in a manner similar to that of soluble aromatic prenyltransferases (Yang Y et al. 2012; Bayse and Merz 2014; Huang et al. 2014).
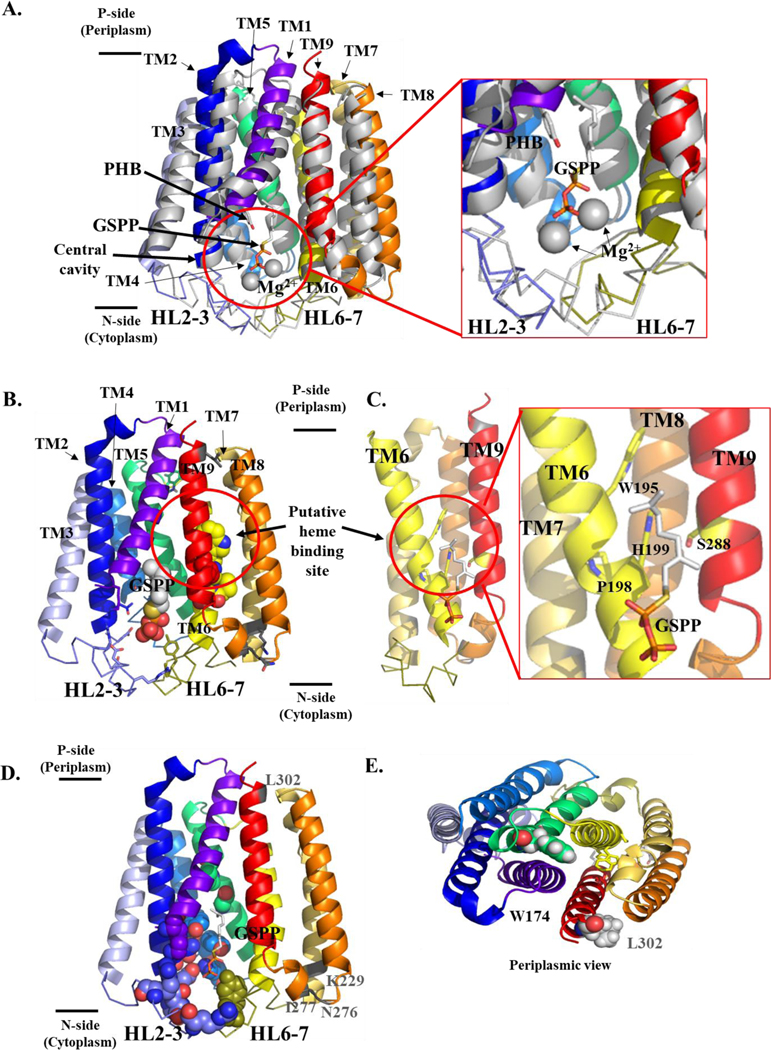
Figure 3. Structural model of B. subtilis HOS showing the cytoplasmic central cavity and predicted heme binding site.
A. Superimposition of substrate-bound ApUbiA, shown in gray, (PDB 4OD5) (Cheng and Li 2014) and the BsHOS model (colored according to secondary structure). The prenyl acceptor, PHB, (gray and red) and the prenyl donor analog, GSPP (gray, red, and orange) are shown as sticks. Mg2+ ions are shown as gray spheres. BsHOS model coloring: TM1 purple-blue, TM2 blue, HL2–3 slate (ribbon), TM3 light blue, TM4 marine, TM5 lime green, TM6 yellow, HL6–7 deep olive (ribbon), TM7 yellow-orange, TM8 orange, TM9 red. The N-terminus (aa 1–28) of BsHOS is not shown. B. BsHOS model viewed from the plane of the membrane. Placement of GSPP (shown as spheres) was based on the superimposition of the BsHOS model with the ApUbiA structure as shown in panel A. The critical, central cavity-facing residues are shown as sticks (colors corresponding to secondary structure). The extramembrane residues (charged in EcHOS, varying levels of conservation) are shown as gray sticks. The putative heme binding site between TM6 (yellow) and TM9 (red) is circled. Putative heme binding site residues are shown as spheres (yellow and red). C. The model is rotated 90° to show the putative heme binding site adjacent to the central cavity. For clarity, TM1–5 have been removed. Critical residues in the putative heme binding site are shown as yellow sticks, and GSPP is shown as sticks. D. View of the BsHOS model from the TM plane, highlighting critical residues that face the central cavity as well as charged, extramembrane residues that do not face the central cavity but are critical in EcHOS. Key central cavity residues are shown as spheres. The backbone positions of four “charged” extramembrane key residues are shown in gray and labeled in gray. The substrate analog GSPP is shown as sticks. E. Periplasmic view of W174, a critical residue that does not face the central cavity. W174 is shown in green spheres. L302 (analogous to the critical charged, extramembrane residue D282 in EcHOS) is shown as gray spheres. GSPP – geranyl thiolopyrophosphate; PHB – p-hydroxybenzoate; HL – helix-loop; TM – transmembrane helix.
Deeper in the membrane, the central cavity becomes more hydrophobic, reflecting its role as the prenyl tail binding site. Of course, because many members of the UbiA superfamily use very long chain polyprenyl diphosphates (up to C60) as substrates, the entire prenyl tail cannot fit within the central cavity (Li 2016). Since ApUbiA and AfUbiA have only been crystallized with short chain XPP substrates, the exact location of the longer-chain polyprenyl binding site is not known. However, in ApUbiA, the hydrophobic region of the central cavity has an opening to the lipid bilayer between TM1 and TM9 that could accommodate longer chain polyprenoids (Cheng and Li 2014). In AfUbiA, different placement of TM9 blocks direct access to the central cavity but also creates a longer hydrophobic tunnel between TM8 and 9 that connects the central hydrophobic pocket to the bilayer and could serve as a binding site for long polyprenyl tails (Huang et al. 2014). In addition to these alternate sites for prenyl tail binding, there are differences in the site where the prenyl acceptor binds. In ApUbiA, a small basic pocket within the first four-helix bundle and adjacent to the hydrophobic portion of the central cavity forms a binding site for PHB, with conserved residue R43 interacting with the substrate’s carboxyl group (Cheng and Li 2014). In AfUbiA, this pocket lacks any basic residues, and the prenyl tail from the XPP analog binds here in the crystal structure (Huang et al. 2014). Thus, some of the key differences between the A. pernix and A. fulgidus structures arise either from different placement of TM9 or the chemical environment of the prenyl acceptor binding site. These differences likely reflect differences in substrate specificity (Li 2016).
HOS structural model and HOS mutagenesis
We constructed a structural model for Bacillus subtilis HOS (BsHOS) and refined this model via molecular dynamics simulations (Heo and Feig 2020). The initial model was generated by trRosetta, a new high-accuracy machine learning method that predicts interresidue distances and orientations for each pair of amino acid residues in a protein sequence based on evolutionary relationships inferred from multiple sequence alignments (Yang et al. 2020). The rationale for this approach is that residues that co-vary are likely to interact in three-dimensional space and are therefore positioned close to each other in the model (Yang et al. 2020). The initial model generated by trRosetta was refined by physics-based molecular dynamics simulations (Heo and Feig 2020) to generate the final structural model of BsHOS shown in Figure 3. Although earlier investigators had reported hydropathy plots for HOS and predicted anywhere from 7 to 9 TM helices, our new model indicates that HOS has nine transmembrane helices like the UbiA homologs that have been crystallized (Saiki, Mogi, Hori, et al. 1993; Antonicka, Leary, et al. 2003; Mogi 2009a). The HOS model suggests that the overall fold of HOS is similar to the known structures of the UbiA superfamily, with a central cavity on the cytoplasmic side of the membrane that is capped by the cytoplasmic linkers bearing the aspartate-rich motifs. This cytoplasmic N-terminus/periplasmic C-terminus orientation is in agreement with the experimentally predicted topology for EcHOS (Chepuri and Gennis 1990). The majority of the conserved residues in HOS face the central cavity, including several residues that are likely to interact with farnesyl diphosphate. For illustrative purposes, the position of the uncleavable XPP analog geranyl thiolopyrophosphate (GSPP) in the ApUbiA crystal structure was used to guide the positioning of GSPP in the HOS model (Figure 3B-D). (ApUbiA was chosen as the guide because this enzyme is more closely related to the HOS sub-family than AfUbiA) Li 2016).
The most extensive mutagenesis study of a HOS gene was an alanine scanning mutagenesis experiment in which 40 residues from EcHOS were substituted. Of these 40 residues, 23 were determined to be necessary for in vivo activity (Table 2) (Saiki, Mogi, Hori, et al. 1993). These critical residues can be divided into three different categories based on their predicted position in our new HOS model: residues that face the central cavity, residues that are located in or near inter-helical loops but do not face the central cavity, and TM residues that do not face the central cavity. The majority of these critical residues (14 out of 23) fall into the first category, i.e., residues that face the central cavity (7 in the cytoplasmic linkers HL2–3 and HL6–7, and 7 in various TM helices) (Figure 3D, Table 2). All 14 of these residues are conserved in HOS sequences, and the majority of them are conserved across sequences of the superfamily as well. Most residues in this category are charged or polar and are likely involved in coordinating Mg2+ or pyrophosphate. The number of critical, conserved residues in this region highlights the importance and the invariance of Mg2+ and pyrophosphate binding. Moreover, since our model was developed without taking into account the mutagenesis data, the positioning of these conserved residues at the central cavity active site indicates that the HOS model is in good agreement with the available experimental data.
Table 2.
Key residues in HOS. The residues listed are critical for activity in E. coliHOS (EcHOS), as demonstrated by loss of in vivoHOS function when substituted with alanine (Saiki, Mogi, Hori, et al. 1993). The residues can be grouped into four topological categories on the basis of our new structural model of B. subtilisHOS (BsHOS). The predicted functions of residues facing the central cavity were assigned based on the alignment of BsHOS with A. pernixUbiA (ApUbiA) (Cheng and Li 2014). TM – transmembrane helix; HL23 – Helix-loop between TM2 and TM3.
| Category | Location | BsHOS residuea |
Echos residuea |
Predicted function |
References |
|---|---|---|---|---|---|
| Central-cavity facing | |||||
| TM1 | K30 b | K 11 b | |||
| TM2 | N 81 | N 57 | PPi coordination | (Cheng and Li 2014) | |
| HL23 | D 85 | D 61 | Mg2+ coordination | (Cheng and Li 2014) | |
| HL23 | D 87 | D 63 | |||
| HL23 | D 89 | D 65 | Mg2+ coordination | (Cheng and Li 2014) | |
| HL23 | R 94 | R 70 | PPi coordination | (Cheng and Li 2014) | |
| HL23 | R 98 | R 74 | PPi coordination | ||
| TM4 | Y 143 b | Y 120 b | |||
| TM4 | Y 147 | Y 124 | PPi coordination; carbocation stabilization | (Cheng and Li 2014; Huang et al. 2014) | |
| TM4 | K 152 | K 129 | PPi coordination | (Cheng and Li 2014) | |
| TM5 TM5 |
G 166 P 169 | G 143 P 146 | |||
| HL67 | D 210 | D 187 | Mg2+ coordination | (Cheng and Li 2014) | |
| HL67 | Y 211 b | Y 188 b | |||
| Periplasmic TM5-TM6 interface | |||||
| TM5 | W 174b | Y 151b | |||
| Heme binding site | |||||
| TM6 | W 195 b | W 172 b | |||
| TM6 | P 198 b | P 175 b | |||
| TM6 | H 199 b | H 176 b | Heme b (substrate) ligand | (Mogi 2009a) | |
| TM9 | S 288 b | S 268 b | |||
| Charged, extramembrane | |||||
| TM7 | K229c | K 206c | |||
| HL89 | N 276 | D 256 | |||
| HL89 | I 277 | D 257 | |||
| TM9 | L 302 | D 282 |
Residues in bold are highly conserved in HOS.
Residues at this position are only conserved in the HOS sub-family.
There is a preference for a charged residue at this position in the UbiA superfamily.
In addition to the 14 conserved residues that face the central cavity, there are 4 critical residues in EcHOS predicted to be in or near inter-helical loops in the C-terminal four-helical bundle (Figure 3D-E, Table 2). All four of these residues are charged in EcHOS. K206 (K229 in BsHOS) is predicted to be near the N-terminal end of TM7, at a position where there is a preference for positively charged residues in the superfamily. D256 and D257 (N276 and I277 in BsHOS) are predicted to be in the cytoplasmic loop between TM8 and TM9. Neither position is strongly conserved, but nonpolar residues seem to be excluded from these positions. Finally, D282 (L302 in BsHOS) is predicted to be at the periplasmic end of TM9, a site where there is a preference for negatively charged residues, with aspartate predominating. The specific functions of these four charged residues remain unclear, as they appear to be too far from the central cavity to be involved in pyrophosphate or Mg2+ binding. However, their extramembrane locations serves to confirm that the overall topology of the model is likely accurate.
The remaining residues that lead to abolished activity when substituted with alanine in EcHOS are conserved residues in TM helices that do not face the central cavity. These residues are all located in the C-terminal four helical bundle (B. subtilis numbering listed first, followed by E. coli numbering in parentheses): W174 (Y151), W195 (W172), P198 (P175), H199 (H176), and S288 (S268). W174 is not very close to the other residues. It is located near the periplasmic end of TM5 (facing TM6) (Figure 3E, Table 2). Overall, there is a preference for an aromatic residue at this position in the superfamily, but its role is unclear.
The other four residues are clustered together and are only conserved in the HOS sub-family. W195, P198, and H199 are all located on TM6 (Figure 3B-C, Table 2). P198 faces the center of the second four helical bundle, while W195 and H199 face TM9. H199 is directly across from S288 on TM9; both residues are adjacent to the central cavity. The position of H199 in the model (Figure 3C) is particularly intriguing since this residue is only strictly conserved in the HOS sub-family and is thought to be the ligand for heme b (the prenyl acceptor). Consistent with this hypothesis, isolated BsHOS (overexpressed in E. coli) has a Soret peak that would be typical of histidine ligation (Mogi 2009a). Furthermore, the H199A BsHOS variant is inactive in vivo even though the expression level of the substituted protein is similar to that of the wild-type enzyme (Mogi 2009a). Assuming H199 is the heme ligand, this would place the heme binding site partly within the second four helical bundle, likely between TM6 and TM9. In the current structural model, the Nδ of H199 is approximately 8 Å from the closest carbon atoms in the GSPP prenyl tail, indicating that a heme ligated by H199 would be reasonably close to the active site. It should be noted that the model does not possess enough space between TM6 and TM9 for heme to bind. This crowdedness is due to the fact that the method used for constructing the model places residues that coevolve in close proximity to each other but does not take into account the possibility that a substrate (such as heme) could be located between coevolving residues (Heo and Feig 2020).
The most obvious difference between the structural model of HOS and the known crystal structures of related proteins is the location of the prenyl acceptor binding site. As mentioned above, ApUbiA is most closely related to PHB prenyltransferases, and a substrate-bound form has been crystallized with electron density that matches PHB in a basic pocket within the N-terminal four-helical bundle (Cheng and Li 2014; Li 2016). The arginine (R43 in ApUbiA) that makes the pocket basic appears to interact with the carboxylate of PHB. This arginine is only conserved within the PHB prenyltransferase sub-family. When substituted, it completely abolishes PHB binding, supporting the notion that the basic binding pocket is the true substrate binding site for this sub-family. On the other hand, in BsHOS, the prenyl acceptor site (heme binding site) seems to be located within the C-terminal four-helical bundle, on the opposite side of the central cavity. Most of the highly conserved residues unique to the HOS sub-family are located on TM6, including H199 (Figure 3C). Because there are few sequence motifs that are indicative of a heme binding site, it can be hard to predict exactly which residues will form a heme binding pocket. However, heme binding pockets tend to be enriched in aromatic residues that can form stacking interactions with the heme (Li et al. 2011). In our model, W195, which is strictly conserved in HOS sequences, is positioned at the interface between TM6 and TM9 just above H199. Together, this observation, the presence of a strictly conserved histidine on TM6 (H199 in BsHOS) that could serve as a heme ligand, and a highly conserved serine (S288) facing H199 strongly suggest that heme b binds between TM6 and TM9, adjacent to the central cavity. As discussed above, the crystal structures of ApUbiA and AfUbiA indicate that TM9 placement defines prenyl tail binding locations. In HOS, TM9 seems to serve an additional role, forming part of the prenyl acceptor binding pocket.
The effects of several mutations in genes encoding eukaryotic HOS have also been studied. HOS from the pathogenic yeast Aspergillus fumigatus has been substituted at three positions in HL2–3: E230A, D234A, and R243Q (Li et al. 2020). The latter two HOS variants exhibit decreased in vivo activity, as would be expected since the analogous EcHOS variants (D65A and R74A) are also inactive (Saiki, Mogi, Hori, et al. 1993). The first conserved aspartate of the aspartate-rich motif is replaced by glutamate in A. fumigatus; interestingly, substituting E230 with alanine does not affect in vivo HOS activity (Li et al. 2020). In contrast, the aspartate at this position is required for activity not only for EcHOS (D61A is inactive), but also for other E. coli octaprenyltransferases (UbiA and MenA) from the UbiA superfamily (Saiki, Mogi, Hori, et al. 1993; Cheng and Li 2014; Huang et al. 2014). Additionally, in AfUbiA, the corresponding aspartate ligates a Mg2+ ion, and in both of the archaeal homologs that have been crystallized, the analogous aspartate-to-alanine variants are essentially unable to bind XPP (Cheng and Li 2014; Huang et al. 2014). The tolerance of the fungal HOS for the substitution of a conserved glutamate to alanine in HL2–3 could perhaps indicate a species-specific difference in HL2–3 arrangement or Mg2+ coordination.
In contrast to prokaryotes, which often have alternate respiratory pathways, respiration in humans depends solely on cytochrome c oxidase and therefore also on heme o biosynthesis. Therefore, mutations that completely abolish HOS activity are most likely embryonic lethal in humans and thus have not been observed. However, four single amino acid substitutions have been identified in human HOS that retain a residual level of activity, yet are still severe enough to cause diseases due to the loss of cytochrome c oxidase function (Valnot et al. 2000; Antonicka, Leary, et al. 2003). Two of these substitutions map to residues within the cytoplasmic loops that sit over the central cavity and are relatively (P225) and highly (D336) conserved, respectively. (The corresponding residues in BsHOS are P99 and D210). The other two substitutions, T196 and N204, are located in TM2. T196 is predicted to be located on the IMS (intramembrane space) side of TM2 facing the center of the N-terminal four-helical bundle. The disease-causing mutation in humans yields a T196K variant; the introduction of a positively charged residue at this position may be destabilizing, as has been demonstrated for the analogous substitution in the S. cerevisiae HOS (T188K) (Khalimonchuk et al. 2012). N204K, on the other hand, is predicted to be on the matrix side of TM2, facing the central cavity. Interestingly, the analogous substitution in S. cerevisiae, N196K, is active and can actually rescue the stability or activity of other point mutations affecting HOS (Bestwick et al. 2010; Khalimonchuk et al. 2012). Additional data are needed to understand the discrepancy between substituting this position in human HOS versus yeast HOS. However, the dramatic effects produced by mutations in both species highlight the importance of residues that face the central cavity.
Heme a synthase
Heme a properties and function
Heme a synthase (HAS) catalyzes the oxidation of the C8 methyl group of heme o (pyrrole ring D) to a formyl group (Figures 1B, 4). This is the second and final step of the heme a biosynthetic pathway (Svensson et al. 1993; Mogi et al. 1994; Barros et al. 2001). The conversion of pyrrole ring D’s methyl substituent to an electron-withdrawing formyl group increases the redox potential of heme a relative to hemes b and o. For example, the reduction potential of heme a bound to a heme protein maquette is 179 mV more positive than that of heme o bound to the same maquette (Zhuang, Reddi, et al. 2006). This increase in redox potential is due to a decrease in the binding affinity of ferric (Fe3+) heme a relative to ferrous (Fe2+) heme a. The destabilization of ferric heme a’s binding affinity is, however, offset by the presence of the hydroxyethylfarnesyl substituent, which, as discussed above for heme o, substantially increases the binding affinity of hemes o and a for a heme protein maquette relative to heme b. Functionally, in an aa3 terminal oxidase, the high midpoint reduction potential of heme a (+380 mV) allows it to accept electrons from relatively poor electron donors such as cytochrome c (midpoint redox potential = +260 mV) (Tsudzuki and Wilson 1971; Myer et al. 1979; Ferguson-Miller and Babcock 1996; Zhuang, Amoroso, et al. 2006). Thus, the two modified porphyrin ring substituents of heme a appear to act in concert to provide a heme cofactor that has a relatively high reduction potential, yet can still bind tightly to a heme-copper oxidase.
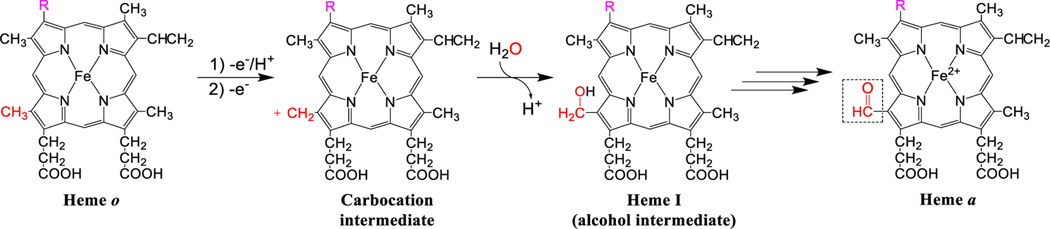
Figure 4. Overview of the proposed reaction mechanism for the conversion of heme o to heme a.
The C-8 methyl group (shown in red) loses a proton and two electrons, generating a carbocation. An oxygen atom from water (shown in red) traps this carbocation, yielding heme I, an alcohol intermediate. A second oxidation step ultimately converts the alcohol into an aldehyde, generating heme a.
HAS mechanism
HAS oxidizes the methyl group of pyrrole ring D to an aldehyde (Figures 4 and 5) (Svensson et al. 1993; Mogi et al. 1994; Svensson and Hederstedt 1994; Svensson et al. 1996; Barros et al. 2001; Brown et al. 2002; Brown KR et al. 2004). Like HOS, HAS is an integral membrane protein located in the cytoplasmic membrane of prokaryotes or in the inner mitochondrial membrane of eukaryotes (Glerum and Tzagoloff 1994; Svensson and Hederstedt 1994). A single-turnover in vitro activity assay with membranes isolated from E. coli overexpressing HAS was reported, which indicated that HAS could convert heme o to heme a in the presence of reductant (Sakamoto et al. 1999); however, no successful in vitro activity assay has been described for purified HAS. Most HAS activity assays published to date have been performed in vivo using cells in which HAS was overproduced. For example, HAS was heterologously overexpressed in E. coli, which lacks a native gene for HAS, and the substrate, heme o, was provided either by E. coli’s native HOS, or by overexpressing HOS (Svensson and Hederstedt 1994; Brown et al. 2002; Brown KR et al. 2004; Mogi 2009a). Expression of B. subtilis HAS (BsHAS) in E. coli demonstrated that HAS can catalyze the successive oxidation of the C8 methyl group of heme o to an alcohol, aldehyde, and carboxylate (Brown et al. 2002). The aldehyde product is heme a, and the alcohol and carboxylate products are presumed to be an intermediate (heme I) (Figure 4) and an overoxidized product (heme II), respectively. Synthesis of heme a (and hemes I and II) was also shown to depend on the presence of oxygen (Brown et al. 2002). Because HAS is a heme-binding, oxygen-dependent protein, it was speculated that the oxygen atom from heme a’s aldehyde group was initially derived from O2 using a cytochrome P450-like monooxygenase mechanism (Svensson et al. 1996; Sakamoto et al. 1999; Barros et al. 2001; Brown et al. 2002). A similar oxygen-activating mechanism has been reported for chlorophyll a oxygenase, a nonheme iron monooxygenase which also catalyzes the oxidation of a methyl group from a pyrrole ring (Schneegurt and Beale 1992; Porra et al. 1993; Oster et al. 2000). Isotope labeling studies, however, indicate that O2 is not the source of the oxygen in the formyl group of heme a or in the hydroxyl group of heme I. Instead, the oxygen atom added at the C8 position is most likely derived from water. Therefore, although cytochrome P450 and HAS are both oxygen-activating enzymes, it seems most likely that HAS uses an electron transfer mechanism in which O2 activation yields a high-valent iron-oxo species that serves as an oxidant but not as an oxygen donor (Figure 5) (Brown KR et al. 2004). While cytochromes P450 also activate O2 to a high-valent iron-oxo species, in P450-catalyzed reactions, this species oxidizes the substrate via hydrogen atom abstraction, typically transferring the oxygen atom from the high-valent iron-oxo species directly to the substrate via a hydroxyl radical recombination mechanism (Sono et al. 1996). In contrast, HAS uses a catalytic mechanism that is more similar to that of peroxidases, in which the high-valent iron-oxo species oxidizes the substrate via electron transfer instead of hydrogen atom abstraction.
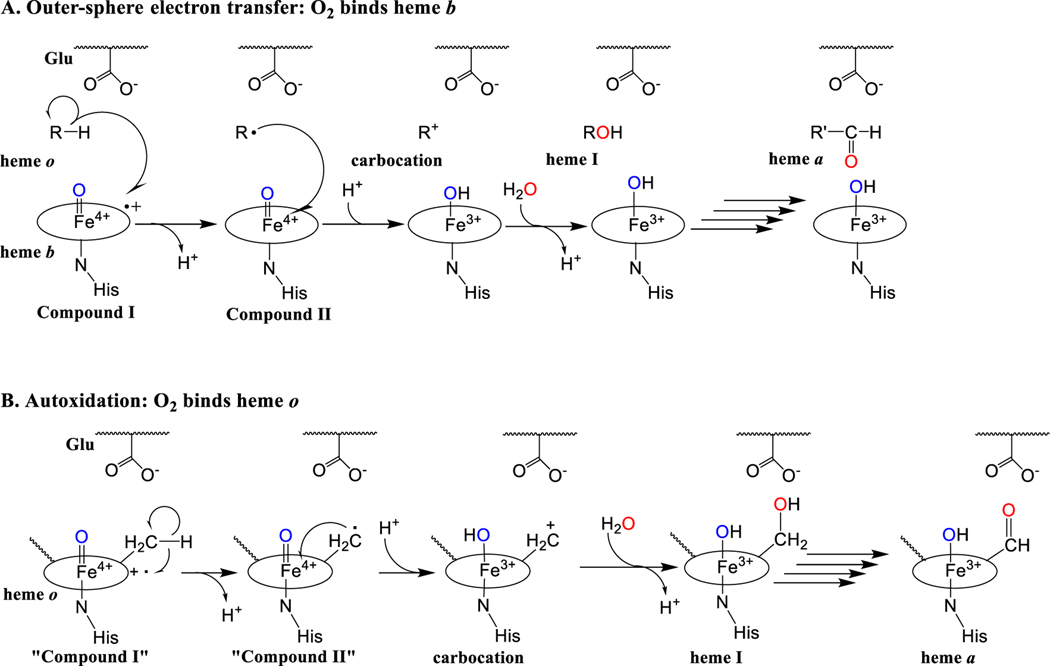
Figure 5. Possible mechanisms of oxygen activation by HAS.
In HAS, O2 activation may occur at heme b (A) or heme o (B). Either proposed mechanism would likely require the displacement of a histidine ligand to allow O2 to bind. In both mechanisms, substrate oxidation is shown as two successive one-electron transfer steps instead of hydrogen atom abstraction because the substrate (the C8 methyl group of heme o) is not positioned appropriately relative to the ferryl group in compound I to allow hydrogen atom abstraction. A. Outer-sphere electron transfer. O2 activation leads to the formation of a high-valent iron-oxo species (compound I, far left) that removes an electron from the C8 methyl group of heme o (shown as R-H). This leads to the formation of a radical intermediate and compound II. Compound II then removes another electron from the substrate radical, forming a carbocation intermediate. Water traps the carbocation intermediate to form heme I. This process can then be repeated with heme I as the substrate to form a geminal diol (not shown), which readily dehydrates to form the aldehyde in heme a, the final product. A conserved glutamate positioned near the heme o binding site is proposed to stabilize the carbocation intermediate. B. Autoxidation mechanism. Heme o activates O2 to form a compound I-like species. “Compound I” of heme o then oxidizes its own C8 methyl group using the general mechanism described in mechanism A.
In the well-established mechanism for plant heme peroxidases (such as horseradish peroxidase), peroxide binds end-on at the distal side of the active site heme (heme b). Cleavage of the O-O bond is catalyzed by the electron-donating “push” of the proximal heme histidine ligand and the “pull” of a distal histidine, which acts as a general acid/base catalyst to facilitate proton removal from the proximal oxygen and protonation of the distal oxygen (Sono et al. 1996; Hiner et al. 2002; Poulos 2010). Heterolytic O-O bond cleavage produces a high-valent iron-oxo species known as compound I, an Fe(IV)=O porphyrin cation radical. Subsequent stepwise electron donation from an external electron donor (often an aromatic substrate) reduces the porphyrin ring and then the heme iron, with protonation of the second oxygen atom leading to its release as water (Hiner et al. 2002; Poulos 2010). In the case of HAS, two alternative modified versions of this mechanism can be envisioned (Brown KR et al. 2004), in which compound I formation is initiated by O2 binding to the heme iron instead of peroxide binding (Figure 5). Reduction of the oxyferrous complex to the peroxo state requires the donation of two electrons from an external source. In yeast, these electrons seem to be provided by mitochondrial ferredoxin (Yah1) and ferredoxin dehydrogenase (Arh1), while in B. subtilis, reducing equivalents may be provided by the quinol pool (Svensson and Hederstedt 1994; Barros et al. 2001; Barros and Tzagoloff 2002). Once the peroxo state is reached, the remaining two electrons needed to cleave the O-O bond and generate a compound I-like species could be provided by the heme iron and porphyrin ring as in the peroxidase mechanism. Compound I is a very potent oxidant that could remove an electron from the C8 methyl group of heme o, converting the methyl group into a carbon-centered radical and reducing compound I to compound II. Compound II, which is still a potent oxidant, could then remove a second electron from the radical to form a carbocation. Finally, water could trap the carbocation to produce heme I, the hydroxylated intermediate. This entire process could then be repeated, oxidizing a second C-H bond to convert heme I to the corresponding geminal diol, which could spontaneously dehydrate to form an aldehyde (i.e., generate heme a). This proposed mechanism is consistent with the available isotope-labeling data on HAS, but the details have yet to be confirmed experimentally (Brown et al. 2002; Brown KR et al. 2004).
One of the unanswered questions about heme a synthase’s reaction mechanism is whether O2 binds to the heme o substrate or to heme b, which usually co-purifies with HAS and is thought to serve as a cofactor (Figure 5) (Svensson and Hederstedt 1994; Svensson et al. 1996; Hederstedt et al. 2005; Mogi 2009b; Hederstedt 2012; Zeng et al. 2020). Since both hemes appear to be low-spin hexacoordinate in the resting state of the enzyme (Svensson et al. 1996), it is not clear which heme is the site of O2 binding and subsequent compound I formation. Arguments can be made for both scenarios. As discussed below, the two hemes are likely close enough to each other to allow for electron transfer between them. Thus, it is feasible that O2 could bind heme b and form a “true” compound I that could remove electrons from the target methyl on heme o via outer sphere electron transfer (Brown KR et al. 2004). This is the mechanism typically employed by peroxidases (Figure 5A) (Poulos 2010). Alternatively, O2 could bind heme o and form a prenylated compound I-like Fe(IV)=O porphyrin cation radical that catalyzes the oxidation of its own methyl substituent. In this case, heme b would serve strictly in an electron transfer capacity (Figure 5B) (Brown KR et al. 2004). This autoxidation mechanism is similar to the proposed mechanism for the modification of the heme cofactor in mammalian peroxidases (Colas et al. 2002; Ortiz de Montellano 2008). In this family of peroxidases, the active site heme is proposed to generate compound I and autocatalytically modify its methyl substituent. This reaction step generates a carbocation at the methyl position that is normally trapped by a nearby glutamate side chain to form an ester crosslink in the isolated form of the enzyme. Substitution of the crosslinking glutamate with aspartate, however, prevents this crosslink from forming, presumably because the shorter residue is too far away from the heme. Instead, the carbocation intermediate is trapped by water, yielding a hydroxylated heme similar to the heme I intermediate generated by HAS (Colas et al. 2002; Ortiz de Montellano 2008). Thus, precedence in the literature exists for both compound I-catalyzed autoxidation as well as for outer sphere electron transfer. These two mechanisms cannot be distinguished based on the currently available data.
Overview of HAS topology and mutagenesis
HAS is a multipass integral membrane protein located in the prokaryotic cell membrane or the eukaryotic inner mitochondrial membrane. Heme a synthases from all three domains of life belong to the same family, termed the Cox15/CtaA family (Mogi et al. 1994; Barros et al. 2001; Hederstedt 2012; He et al. 2016). There are a few phylogenetically diverse groups from archaea (Pyrobaculum) (Table 1) and lower-order eukaryotes (rhodophyte and glaucophyte algae along with jakobids) where a sequence encoding HAS has not been identified in the genome despite the presence (or predicted presence) of an aa3 terminal oxidase (Lübben and Morand 1994; He et al. 2016). However, given the low sequence similarity between distantly related HAS proteins, this does not completely rule out the possibility that a gene encoding a member of the Cox15/CtaA family is present in these organisms. Indeed, sequences encoding HAS in Sulfolobaceae genomes have only recently been discovered (Degli Esposti et al. 2020).
Most identified HAS sequences possess eight transmembrane helices. Sequence homology between the N-terminal half of HAS (TM 1–4) and the C-terminal half (TM 5–8) suggests that the standard 8-TM topology resulted from duplication and fusion of an ancient 4-TM gene (Svensson and Hederstedt 1994). Recent phylogenetic analysis suggests that this ancestral gene may have coded for a protein that contained a 4-TM domain with two conserved histidines (Domain of Unknown Function 420) (Degli Esposti et al. 2020). The TM helices of HAS are connected with short loops, with the exception of the N-terminal periplasmic loop between TM1–2 (L1–2) and the corresponding C-terminal loop between TM5–6 (L5–6) (Figure 6A) (Hederstedt et al. 2005; Mogi 2009b; Hederstedt 2012; Swenson S et al. 2016). HAS sequences can be broadly divided into two types based on the presence (type 1) or absence (type 2) of a pair of conserved cysteine residues in L1–2. Type-1 HAS is found in archaea and several bacterial clades, while type-2 HAS is found in other bacterial clades and in all eukaryotes (with only one exception) (Hederstedt 2012; He et al. 2016). Type-1 HAS proteins can be further subdivided into three sub-types (A-C) (Lewin and Hederstedt 2016). Type-1A HAS representatives are found in certain archaea, and possess only one four-helical bundle that includes the pair of conserved cysteine residues in L1–2. These truncated HAS proteins appear to function as dimers (Lewin and Hederstedt 2006; Hederstedt 2012). Together, types 1B, 1C, and type 2 comprise the majority of known HAS sequences and have 8 TM helices. These types have a cysteine pair in both L1–2 and L5–6 (class B), only in L1–2 (class C), or in neither elongated loop (type 2)1 (He et al. 2016; Lewin and Hederstedt 2016). The most well-characterized HAS, BsHAS, is type 1B. Mutagenesis of the gene encoding BsHAS indicates that the less highly conserved C-terminal cysteine pair (C191, C197) is not required for activity, while the N-terminal cysteine pair is important for activity in type-1 HAS (C35, C42) (Mogi 2009b; Hederstedt 2012; Lewin and Hederstedt 2016). (B. subtilis numbering is used throughout this section.)
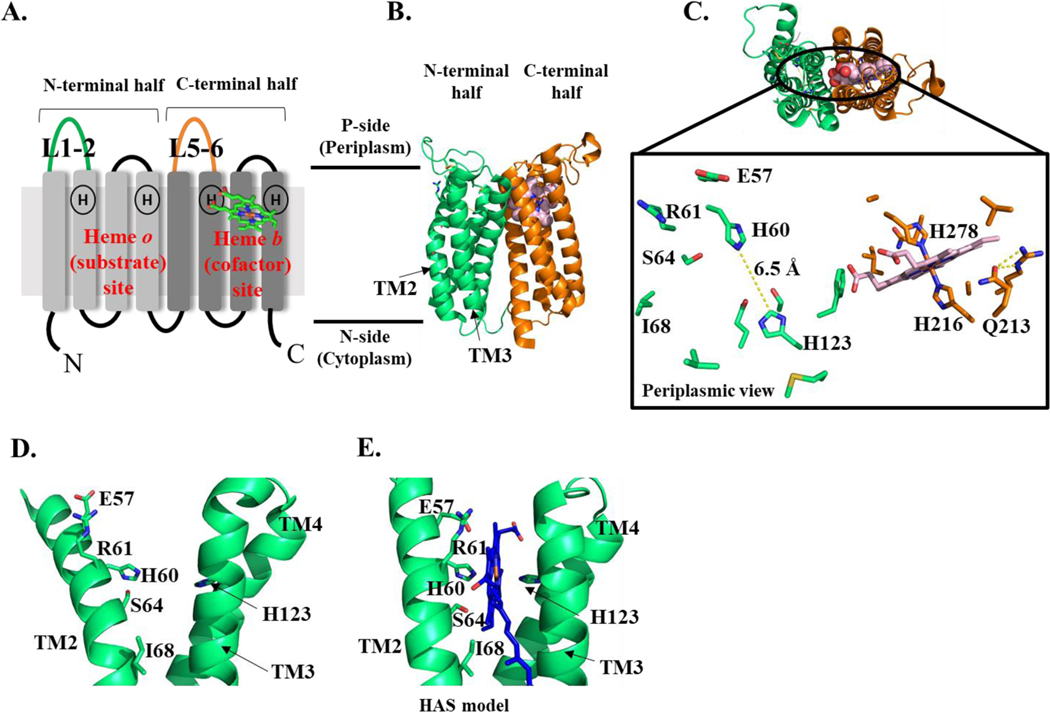
Figure 6. HAS topology (A), crystal structure (B-D), and substrate-bound model (E).
A. Diagram showing the 8 TM helices of HAS and the position of the two heme binding sites, each with two highly conserved His residues. The elongated loops connecting TM1 and 2 (L1–2) and TM5 and 6 (L5–6) are colored green and orange, respectively. B. Crystal structure of B. subtilis HAS showing heme b in the C-terminal heme binding site and the empty N-terminal heme binding site. The N-terminal four-helical bundle is shown in green; the C-terminal four-helical bundle is shown in orange and heme b is shown in light pink (spheres). C. This structure is rotated 90° to show the top-down view from the periplasm. Top: TM helices are shown as a cartoon; heme b is shown as spheres (light pink). Bottom: conserved residues in the N-terminal and C-terminal heme binding sites. Heme b is shown as light pink sticks. Residues discussed in the text are labeled, with the exception of G65. D-E. The N-terminal heme binding site of BsHAS with (D) and without (E) heme o. D. The crystal structure shown in B is rotated 90° to show the empty heme binding site. E. A model based on the crystal structure of BsHAS is shown with TM2 straightened and heme o placed into the N-terminal heme binding site. Figures 6B-D and 6E were prepared using a previously published crystal structure (PDB 6IED) and a previously published model (model PDB file: pnas.1813346115.sd01), respectively (Niwa et al. 2018).
In addition to the cysteinyl pair that is conserved in type-1 HAS, there are only a few residues that are highly conserved across all HAS proteins (Hederstedt 2012; He et al. 2016). Most notably, there are four highly conserved histidine residues in HAS, two in the N-terminal half (H60 and H123, located on TM2 and TM4) and two at corresponding positions in the C-terminal half (H216 and H278, located on TM6 and TM8) (Figure 6A) (Hederstedt et al. 2005; Mogi 2009b; Hederstedt 2012). All four of these histidines are present in almost all known genes encoding HAS (Table 1) (Mogi 2009a; Hederstedt 2012; He et al. 2016). Early work with BsHAS isolated from B. subtilis showed the presence of two bis-histidine axially-ligated hemes, heme a (presumably unreleased product) and heme b (the putative cofactor) (Svensson and Hederstedt 1994; Svensson et al. 1996)2. Therefore, it was postulated that all four conserved histidine residues could act as heme ligands (Svensson et al. 1996; Sakamoto et al. 1999; Hederstedt et al. 2005; Mogi 2009b).
Each highly conserved histidine residue in BsHAS has been individually substituted. These histidine variants were overexpressed and purified in either E. coli (Mogi 2009b) or B. subtilis (Hederstedt et al. 2005). As has been discussed in a previous review (Hederstedt 2012), these mutagenesis studies did not fully clarify which histidine ligated which type of heme because all of the stable mutants still copurified both with prenylated heme(s) and heme b (Hederstedt et al. 2005; Mogi 2009b).
However, the mutagenesis data did show that all four histidines are necessary for full HAS activity. Similarly, substitution of each of the analogous histidine residues in eukaryotic HAS resulted in a complete loss of in vivo activity (Bareth et al. 2013; Swenson S et al. 2016; Merli et al. 2017). Careful analysis of the BsHAS variant properties suggests that the histidines have unique roles. For example, substitution of an N-terminal histidine did not produce the same result as altering its C-terminal counterpart. Substitution of either N-terminal histidine (H60 or H123) to a non-heme-ligating residue (A or L) completely abolishes activity, although this does not prevent heme o from binding to HAS. In contrast, substitution of the C-terminal histidines (H216 or H278) to non-ligating residues sometimes resulted in partially active HAS. However, changing the C-terminal histidines (and some other conserved C-terminal residues) also destabilized BsHAS in most cases (Hederstedt et al. 2005; Mogi 2009b; Hederstedt 2012).
In addition to this discrepancy between the N-terminal and C-terminal substitutions, the data obtained from expressing BsHAS in B. subtilis indicated that the two histidines belonging to the same half of HAS also seem to have distinct roles. For example, substituting the N-terminal histidine H123 with methionine, an alternate heme ligand, completely abolishes activity, but substituting the other N-terminal histidine for methionine (H60M) yields an enzyme that is partially active but co-purifies with a lower level of heme a than observed for the wild-type HAS (Hederstedt et al. 2005; Hederstedt 2012). Overall, these data indicate that while all four histidine residues are important for proper HAS function, they seem to serve unique roles, supporting the hypothesis that HAS has two heme binding sites with different functions.
Besides the histidine variants mentioned above, the only point mutations associated with eukaryotic HAS that have been experimentally characterized are mimics of disease-causing mutations in humans. As discussed above for HOS, substitutions that completely abolish HAS activity are presumably embryonic lethal. However, three missense mutations have been identified in human HAS that significantly decrease HAS activity and result in very severe diseases associated with low levels of cytochrome c oxidase activity (Antonicka, Mattman, et al. 2003; Oquendo et al. 2004; Bugiani et al. 2005; Alfadhel et al. 2011; Miryounesi et al. 2016; Swenson S et al. 2016). One of these mutations results in the substitution of L139 with valine (human numbering) (Miryounesi et al. 2016). L139 is predicted to map to L1–2, the elongated loop between TM1 and TM2 where several highly conserved residues are located. The second disease-causing alteration, yielding the S344P variant, is located at the N-side (matrix) end of TM6 and destabilizes HAS, although the explanation for this instability is unclear (Swenson S et al. 2016). The position of the third known disease-causing point alteration is in the P-side (IMS) loop connecting TM3 and TM4. This loop is very short in BsHAS (~4 amino acids), but longer in eukaryotic HAS (~10–14 amino acids), where the residue in question, R217, appears to be fairly well conserved (Swenson S et al. 2016). Expression of the variant human protein in human fibroblasts and of the analogous substitution in S. cerevisiae HAS indicates that this change inactivates HAS without affecting stability (Swenson S et al. 2016). In the absence of additional structural information about type-2 HAS, the role of this amino acid residue is unclear.
The remaining highly conserved residues in HAS are clustered on the periplasmic (IMS) side of the TM helices or in the elongated periplasmic loops, L1–2 and L5–6. As discussed above, the N- and C-terminal halves of HAS exhibit low sequence homology, and the pattern of highly conserved residues from the two halves of the protein are therefore fairly well correlated (Svensson and Hederstedt 1994; Hederstedt et al. 2005; Hederstedt 2012). For example, the conserved histidine from TM2 is preceded by a glutamate and followed by an arginine, forming the motif EXXHR, while the analogous C-terminal histidine from TM6 is part of a very similar motif, Q/HXXHR. However, both the number of conserved residues and the conservation level of most of these residues is higher in the N-terminal half of HAS than in the C-terminal half. This discrepancy is most evident for L1–2 versus L5–6, but also holds true for the transmembrane domain. However, prior to the determination of the HAS structure, it was not clear how these relatively minor differences in the sequence motifs of the N- and C-terminal halves of HAS resulted in a protein with two functionally distinct heme binding sites.
HAS structure
Recently, the structures of two different type-1 bacterial HASs have been solved. The structure of BsHAS was determined by X-ray crystallography at 2.2 Å resolution (Figure 6B-C) (Niwa et al. 2018), and the structure of HAS from the hyperthermophilic bacterium Aquifex aeolicus (AaHAS) was solved by cryo-EM at 4.2 Å resolution (Zeng et al. 2020). Both structures show the same overall fold, and a homology model for AaHAS based on the BsHAS crystal structure fit well within the cryo-EM density for the TM helices. AaHAS forms a trimer, which will be discussed briefly. Here we focus primarily on the crystal structure of BsHAS and on the accompanying substrate-bound model.
The structure of BsHAS reveals that the N- and C-terminal halves of HAS each form a four-helical bundle (Figure 6B) (Niwa et al. 2018). These bundles are pseudo-symmetrically related, as was predicted based on sequence homology between the two halves of HAS (Hederstedt et al. 2005; Hederstedt 2012; Swenson S et al. 2016). Heme b is bound within the C-terminal bundle, near the periplasmic side of the membrane. Consistent with previous optical and electron paramagnetic resonance analysis, heme b is ligated by two of the conserved histidine residues with nearly perpendicular imidazole planes (H216 and H278) and with an Fe-His distance of ~2 Å for each (Svensson et al. 1996; Zoppellaro et al. 2009) (Figure 6C). The cryo-EM structure of AaHAS also shows density in the center of the C-terminal four-helical bundle that is likely a heme (Zeng et al. 2020). The corresponding N-terminal heme binding site, where the other two conserved histidine residues (H60 and H123) are located, is empty in the crystal structure (Niwa et al. 2018) and also appears to be empty in the cryo-EM structure (Zeng et al. 2020) (Figure 6C). This empty heme binding site is likely where the substrate, heme o, binds. In the crystal structure, the two elongated periplasmic loops (L1–2 and L5–6) are positioned above the N- and C-terminal heme binding sites, respectively. All of the highly conserved residues in HAS are clustered around the two heme binding sites (Figure 6C).
Although the overall fold of the N-terminal heme binding site matches that of the C-terminal heme binding site, the conformation of TM2 deviates from its C-terminal counterpart, TM6. TM2 is bent at a semi-conserved glycine (G65), which places the N-terminal histidine ligands (H60 and H123) too far apart (6.5Å) to provide bis-histidine axial heme ligation (Figure 6C) (Niwa et al. 2018). Since bis-histidine axial ligation is expected for prenylated substrate/product hemes (hemes o/a), heme o binding was modeled by straightening TM2 using the conformation of TM6 as a guide (Niwa et al. 2018). This modification allows heme o to be ligated by both H60 and H123 in the model (Figure 6E).
The determination of a crystal structure showing heme b tightly bound to the C-terminal four-helical bundle provides strong support for the hypothesis that HAS possesses both a cofactor heme binding site and a substrate heme binding site (Niwa et al. 2018). Identification of the C-terminal four-helical bundle site as the binding site for heme b and the N-terminal bundle as the substrate binding site also seems to explain the discrepancy in stability between the N- and C-terminal BsHAS mutants (Hederstedt et al. 2005; Mogi 2009b; Hederstedt 2012). Because the N-terminal four-helical bundle has to bind substrate and release product, it seems likely that this domain exhibits some conformational flexibility, especially at TM2, which is predicted to move toward the center of the four-helical bundle after substrate binding (Niwa et al. 2018). This built-in flexibility may allow the N-terminal half of HAS to tolerate mutations that perturb its structure in the heme binding site. In contrast, the cofactor heme binding site is more likely to be rigid, as there is no need for the cofactor to be released. Thus, it seems logical that disruptions to the heme b binding site may compromise protein stability. Interestingly, the structure of AaHAS supports this idea. AsHAS forms heat-stable trimers with the C-terminal four-helical bundle of each monomer at the core of the trimer. This observation suggests that the C-terminal half of HAS is fairly rigid (Zeng et al. 2020).
The predicted movement of TM2 upon heme o binding in BsHAS may represent a generally applicable method for substrate binding in HAS. In both the crystal structure and the substrate-bound model, there is a fairly large lateral opening between TM2 and TM3 that connects the N-terminal heme binding site to the bilayer. In the model, the hydroxyethylfarnesyl tail of heme o protrudes through this opening (Figures 6E, 7). (In contrast, the C-terminal four-helical bundle has a much smaller lateral opening.) On the basis of their model, Niwa et al. (2018) predicted that hydrogen bonding and hydrophobic interactions between residues on TM2 and TM3 are important for drawing heme o into the heme binding site. Three residues from TM2 seem to be the most important side chains for this mode of substrate binding in BsHAS: S64 and I68 (Figure 6C), which presumably interact with the hydroxyl and the polyprenyl portions of the hydroxyethylfarnesyl moiety of heme o, and G65, the location of the kink in TM2 (Niwa et al. 2018). Interestingly, although G65 is only conserved in approximately 30% of HAS sequences, glycine occurs frequently in HAS sequences at the positions corresponding to S64 and I68 in BsHAS. These glycines may either substitute for G65 in providing the kink in TM2 or interact with the lipid portion of heme o’s hydroxyethylfarnesyl moiety. This sequence specificity suggests that all HAS proteins may share BsHAS’s strategy of moving TM2 to allow substrate entry.
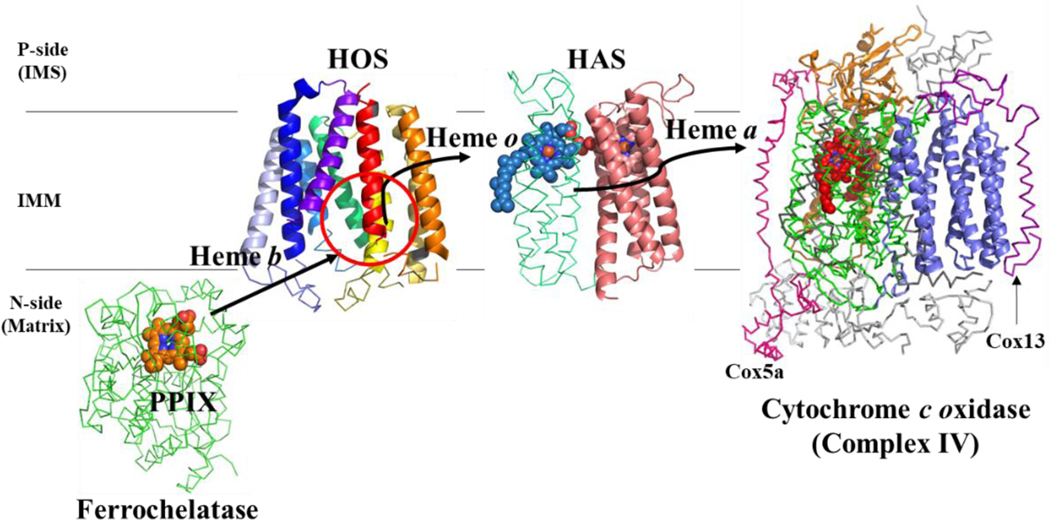
Figure 7. Proposed heme trafficking route in the eukaryotic heme a biosynthetic pathway.
Heme must be transferred from ferrochelatase to HOS, from HOS to HAS, and finally from HAS to cytochrome c oxidase. Note that these transfer steps could involve an unknown heme chaperone (see text). Proteins shown: Membrane-associated (eukaryotic) ferrochelatase (green ribbon) is shown in complex with protoporphyrin IX (PPIX) (orange spheres). The porphyrin plane is roughly parallel to the membrane lipids, and pyrrole rings A and D are closest to the membrane. Note that while mammalian ferrochelatase can dimerize, only one monomer is shown here (Burden et al. 1999; Wu et al. 2001; Medlock et al. 2007). (Ferrochelatase structure (human): PDB E343K) (Medlock et al. 2007). HOS model (this study) is shown with the proposed heme binding site circled in red (BsHOS). HAS model (Niwa et al. 2018) showing heme o (blue spheres) bound in the N-terminal heme binding site on the P-side (periplasmic or IMS side) of the membrane. This model is based on the crystal structure of BsHAS, which has an empty N-terminal heme binding site (model PDB file: pnas.1813346115.sd01). Cytochrome c oxidase is depicted as in Figure 1A, except that hemes a and a3 are shown as red spheres. Subunit I (Cox1) is shown in green ribbon; the other core subunits are shown as orange and slate cartoons. The nuclear-encoded subunits are shown as gray ribbons, except for Cox5a (hot pink) and Cox13 (purple). IMS – intermembrane space; IMM – inner mitochondrial membrane; P-side – positive side; N-side – negative side.
The substrate-bound model of BsHAS also has some interesting catalytic implications. First, the model indicates that the substrate and cofactor hemes are positioned close to each other, with their propionates facing each other (Figure 7). This orientation suggests that direct electron transfer between the two hemes is possible (Niwa et al. 2018), which is necessary for either version of the proposed catalytic mechanism (Figure 5) (Brown KR et al. 2004). Straightening TM2 also positions E57 (Figure 6C), a highly conserved glutamate, near the C8 methyl group of heme o. Because this methyl group is oxidized during catalysis, the proximity strongly suggests that E57 plays a role in the oxidation of the C8 methyl (Niwa et al. 2018). In light of the putative peroxidase-like mechanism for HAS, it seems plausible that the negatively charged glutamate could stabilize a carbocation intermediate that forms during the oxidation of the C8 methyl group, in a manner similar to the proposed mechanism for the glutamate-to-aspartate mammalian peroxidase variant discussed above (Colas et al. 2002; Brown KR et al. 2004; Ortiz de Montellano 2008). It should be noted, however, that both the outer sphere electron transfer mechanism and the autoxidation mechanism include the formation of a carbocation intermediate, so the placement of E57 cannot be used to unambiguously distinguish between the two mechanisms. In the C-terminal heme-binding site, a glutamine (Q213) replaces E57 (Niwa et al. 2018). This nonpolar residue likely cannot stabilize a carbocation intermediate as effectively as glutamate, providing another line of evidence that the N-terminal heme binding site is the substrate binding site.
Ever since it was first observed that HAS co-purifies with heme b and heme a, investigators have speculated that HAS possesses distinct substrate and cofactor heme binding sites (Svensson and Hederstedt 1994; Svensson et al. 1996), although experimental evidence in support of this hypothesis was tenuous prior to structural determination. TM1–4 and TM5–8 were also correctly predicted to form homologous four-helical bundles. The four-helical bundle structure of each half of HAS, and the characterization of both heme b and heme a as low-spin, hexacoordinate hemes is reminiscent of certain electron transfer heme proteins, such as cytochrome b from complex III, supporting the idea that one of these heme groups is involved in electron transfer from an external electron donor (Svensson et al. 1996; Barros et al. 2001; Brown BM et al. 2004; Zoppellaro et al. 2009; Niwa et al. 2018). The other heme is expected to bind and activate O2 during catalysis, requiring displacement of one of the histidine ligands. However, it is not clear from the substrate-bound model which histidine is displaced, due to the nearly equivalent positioning of the histidine residues in both heme binding sites. This question will have to be addressed by future mechanistic studies. The structures of BsHAS and AaHAS also suggest a possible mechanism for substrate entry and product exit from the N-terminal heme binding site based on the apparent conformational flexibility of this domain (Niwa et al. 2018; Zeng et al. 2020). This observation raises intriguing questions about how prenylated hemes transfer to and from HAS, which will be discussed in the final section of this review.
Heme trafficking in the heme a biosynthetic pathway
Overview of prenylated heme trafficking
A successful heme a biosynthetic pathway depends not only on the synthesis of heme o and heme a, but also on proper heme trafficking throughout the course of this biosynthetic pathway. Specifically, heme must be transferred from the upstream proteins involved in heme b synthesis and/or trafficking to HOS, from HOS to HAS, and finally to the heme-copper oxidase recipient (Figure 7). In general, intracellular heme trafficking is mediated by proteins, such as heme chaperones or transporters, or by lipid vesicle transport. This delivery process allows the intracellular concentration of “free” heme to be kept very low, which protects the cell from oxidative damage that might otherwise be caused by non-protein-bound heme (Donegan et al. 2019; Swenson SA et al. 2020). Since heme a has a higher redox potential than heme b (and thus is a better oxidant), controlling heme a transport seems to be particularly important in terms of protecting the cell from oxidative damage (Myer et al. 1979; Zhuang, Amoroso, et al. 2006; Khalimonchuk et al. 2007). However, intracellular heme trafficking in general, and trafficking of prenylated hemes in particular, is still poorly understood (Hannappel et al. 2012; Donegan et al. 2019; Swenson SA et al. 2020). The recent structural advances that have been made for HOS and HAS have provided some missing pieces of the puzzle, but many questions remain. Here, we discuss the currently available information on heme transfer through the heme a biosynthetic pathway, with an emphasis on the new structural data.
Transfer of heme b to HOS
The heme a biosynthetic pathway is one of several competing routes for newly synthesized heme b. Heme b, the prototypical cellular heme, is used as a cofactor without further modification in a wide array of proteins with different sub-cellular locations, including cytochrome b of the cytochrome bc1 complex (complex III in eukaryotes), the cytochromes P450, and hemoglobin and myoglobin. In addition, heme b is also the precursor for biosynthetic pathways that modify the heme, such as for cytochrome c maturation and heme a synthesis. Thus, heme b must ultimately be distributed to many proteins and locations throughout the cell. In the case of heme a biosynthesis, heme b must be trafficked from the enzyme that catalyzes the final step of heme b biosynthesis to HOS.
The final step in heme b biosynthesis is most commonly catalyzed by ferrochelatase, although more recently, two alternate heme b biosynthetic pathways with different intermediates and terminal enzymes (coproheme decarboxylase (ChdC) or AdoMet-dependent heme synthase (AhbD)) have been identified in gram-positive bacteria or in certain archaea and sulfate-reducing bacteria (Heinemann et al. 2008; Bryant et al. 2020). Ferrochelatase inserts iron into the porphyrin ring of its substrate, protoporphyrin IX (PPIX). Prokaryotic ferrochelatases are soluble proteins located in the cytoplasm, whereas eukaryotic ferrochelatases are localized to the mitochondrial matrix and contain a hydrophobic domain that allows them to interact with the matrix side of the inner mitochondrial membrane (Figure 7) (Heinemann et al. 2008). The heme binding site in HOS almost certainly faces the matrix in eukaryotes; therefore, direct transfer of heme b from ferrochelatase to HOS may be possible. Because product release is the rate-limiting step in the ferrochelatase reaction, it has been suggested that ferrochelatase’s interactions with the apoprotein form of hemoproteins could cause a conformational change in ferrochelatase, thereby leading to heme transfer (Donegan et al. 2019). Ferrochelatase could interact with HOS in a similar manner to deliver heme b. Although there is currently no experimental evidence to substantiate this particular interaction, many potential interaction partners for ferrochelatase have been identified by affinity purification of murine ferrochelatase and mass spectrometry analysis of its interactome (Piel et al. 2019). Because ferrochelatase also interacts with putative heme chaperones (PGRMC1 and PGRMC2), it is also possible that one of these chaperones (or a currently unidentified chaperone) transfers heme b to HOS (Piel et al. 2019).
Transfer of heme b to HAS
Because HAS utilizes heme b as a cofactor, delivery of heme b to HAS is also required. Both of the heme binding sites in HAS are on the P-side (IMS side or periplasmic side) of the membrane, which would suggest that heme b must traverse the membrane to be inserted into HAS. In terms of membrane solubility, hemes are amphipathic molecules with a fairly nonpolar porphyrin ring and two negatively charged propionates. In theory, heme could cross the membrane unaided if the propionates are protonated or their negative charge is shielded by cations. Due to the oxidizing capacity of heme, however, it is generally assumed that heme transfer across membranes is mediated by proteins to protect the cells from oxidative damage (Reddi and Hamza 2016). In light of these considerations, it seems likely that heme b is delivered to HAS either from the N-side (matrix or cytosolic side) of the membrane via interactions with ferrochelatase (or ChdC or AhbD) or a heme chaperone, or heme b could be transported across the membrane and delivered by a chaperone from the P-side. Both of these heme delivery routes have also been proposed for the hemylation of cytochrome b of the bc1 complex, another inner mitochondrial membrane hemoprotein (Kim et al. 2012). Understanding the hemylation of HAS and other membrane proteins will require further investigation.
Transfer of heme o to from HOS to HAS
After HOS converts heme b to heme o, heme o must be transferred to HAS (Figure 7). (Delivery of heme o to prokaryotic terminal oxidases will be discussed briefly below.) The fact that HOS precedes HAS in the heme a biosynthetic pathway seems to be an important strategy in heme trafficking. Conversion of the vinyl group on pyrrole ring A to a hydroxyethylfarnesyl moiety makes heme o much more hydrophobic than heme b, effectively sequestering heme o in the membrane. Adding the lipid tail first and then oxidizing the methyl group on pyrrole ring D to synthesize heme a prevents misincorporation of a high-potential heme (i.e., heme with a formyl substituent) into water-soluble proteins (Zhuang, Reddi, et al. 2006).
Although heme o is lipid-soluble, the location of the heme bindings sites of HOS and HAS on opposite sides of the membrane seems to require the movement of the negatively charged propionates from heme o through the lipid bilayer, an energetically unfavorable process (Figure 7) (Kim et al. 2012). There are at least three different scenarios one can envision to overcome this problem and transfer heme o from HOS to HAS. First, HOS could release heme o into the membrane, and heme o could diffuse into the active site of HAS. As discussed above, this process would require shielding the negatively charged propionates from the hydrophobic environment of the lipid bilayer via protonation or interaction with metal cations (Reddi and Hamza 2016). Alternatively, HOS and HAS could interact to transfer substrate. This interaction would also shield heme o’s propionates from the lipid environment, and it has the added benefit of protecting the lipids in the bilayer from oxidative damage mediated by heme o. Finally, heme o delivery to HAS could be mediated by a chaperone or transporter. However, a heme o chaperone/transporter has never been identified despite decades of research on prokaryotic and eukaryotic heme-copper oxidases and the genes required for their assembly (Brown BM et al. 2004). Therefore, it seems more likely that heme o transfer from HOS to HAS is accomplished without the aid of additional proteins.
Prokaryotic HOS and HAS have been shown to interact when heterologously overexpressed in E. coli. Specifically, these experiments demonstrated that both type-1 (B. subtilis) HAS and type-2 (R. sphaeroides) HAS coimmunoprecipitate with BsHOS and R. sphaeroides HOS, respectively (Brown BM et al. 2004). This result suggests that in prokaryotes, HOS and HAS may interact directly to transfer heme o. An interaction between eukaryotic HOS and eukaryotic HAS (also type-2), however, has never been demonstrated experimentally, despite several investigations into the protein-protein interactions of both HOS and HAS in S. cerevisiae (Barros and Tzagoloff 2002; Khalimonchuk et al. 2012; Bareth et al. 2013; Swenson S et al. 2016; Taylor et al. 2017; Herwaldt et al. 2018). Therefore, if HOS and HAS interact in eukaryotes, this interaction is likely transient.
Although difficult to detect experimentally, the physiologically relevant interaction of HOS and HAS is an intriguing possibility. As discussed above, heme o entry into HAS likely requires conformational changes in HAS that widen a lateral opening into the N-terminal heme binding site of HAS. While it seems clear that the heme binding site in HOS is located between two TM helices adjacent to the central cavity, it is less clear how heme o is released from HOS. It has been predicted that in closely related HOS homologs (PHB prenyltransferases), the polyprenyl tail protrudes into the lipid bilayer through an opening between TM1 and TM9 (analogous to the purple-blue and red helices shown in Figures 3 and 7) (Cheng and Li 2014). Thus, heme o could exit “tail-first” by traveling through the central cavity. This substrate release process would require a conformational change to allow the porphyrin ring access to the lipid bilayer. Alternatively, heme o might be able to exit “propionate-first” through the C-terminal four-helical bundle if the appropriate conformational change were made to release heme o from the heme binding site. In either case, it could be imagined that interactions with HAS facilitate this conformational change. Elucidating the details of a potentially transient HOS-HAS interaction (or lack thereof) will probably require a combination of in vivo imaging techniques and structural and computational approaches.
Transfer of prenylated hemes (hemes o and a) to subunit I of terminal heme-copper oxidases
Heme a’s ultimate fate is insertion into either aa3 or ba3 heme-copper terminal oxidases. Specifically, one or two heme a molecules must be inserted into the largest oxidase subunit, termed subunit I in mitochondrial-like aa3 oxidases (from class A) (Figure 7). Subunit I is one of the core oxidase subunits and also binds a copper cofactor (CuB) in addition to the two heme cofactors. Insertion of these metal cofactors into subunit I is part of the larger process of heme-copper oxidase biogenesis, a complex process that involves the sequential assembly of the multiple protein subunits of the oxidase.
Heme-copper oxidase assembly has thus far been studied most extensively for family-A heme-copper oxidases from model organisms, such as the bo3 quinol oxidase from E. coli (Stenberg et al. 2007; Price and Driessen 2010; Palombo and Daley 2012), the cytochrome c oxidase from Paracoccus denitrificans (Hannappel et al. 2012; Schimo et al. 2017), and the mitochondrial aa3 oxidase from S. cerevisiae (Kim et al. 2012; Timón-Gómez et al. 2018). Assembly of prokaryotic oxidases involves a minimal set of assembly factors (Price and Driessen 2010; Hannappel et al. 2012), while the assembly of eukaryotic cytochrome c oxidase, also known as complex IV, is much more complicated (Kim et al. 2012; Timón-Gómez et al. 2018). This added layer of complexity is due to the fact that eukaryotes have to coordinate the assembly of three core mitochondrial-encoded subunits and multiple nuclear-encoded subunits that are unique to eukaryotes (Capaldi 1990; Tsukihara et al. 1996; Pereira et al. 2001; Timón-Gómez et al. 2018). Correct assembly of eukaryotic cytochrome c oxidase involves up to 30 additional assembly factors that typically interact with one or more structural subunits to form subassembly complexes at specific stages of oxidase assembly (Kim et al. 2012; Timón-Gómez et al. 2018). The steps of oxidase assembly, the sequence of these steps, and the number of assembly factors involved varies among eukaryotes and especially between eukaryotes and prokaryotes. Despite these differences, insertion of the heme and copper cofactors into subunit I usually occurs fairly early in the assembly of both prokaryotic and eukaryotic heme-copper terminal oxidases (Hannappel et al. 2012; Kim et al. 2012; Timón-Gómez et al. 2018).
Whereas copper chaperones are necessary for copper insertion in both prokaryotic and eukaryotic heme-copper oxidases, only two potential heme a chaperones have been identified, and they are not universally conserved among organisms with aa3 or ba3 oxidases. One of these chaperones, CbaX, is only found in the bacterial Thermaceae group. CbaX has been implicated in heme a insertion into the ba3 oxidase (family B) from T. thermophilus, but this protein has not yet been fully characterized (Werner et al. 2010). The putative heme a chaperone Surf1 (Shy1 in yeast) has been studied more extensively. Genes encoding Surf1/Shy1 have been identified in eukaryotes, proteobacteria, and, more recently, in actinobacteria. The importance of Surf1/Shy1 in cytochrome c oxidase assembly has been confirmed in several model organisms with family A oxidases, including P. denitrificans, S. cerevisiae, and Corynebacterium glutamicum, as well as in humans (Niebisch and Bott 2001; Pereira et al. 2008; Timón-Gómez et al. 2018; Davoudi et al. 2019). The specific function of Surf1/Shy1, however, is still debated, and it may play slightly different roles in prokaryotes and eukaryotes. In almost all Surf1-containing species studied thus far, residual levels of cytochrome c oxidase activity remain even when Surf1/Shy1 is truncated or deleted, indicating that a small amount of subunit I is still hemylated correctly in the absence of this putative chaperone (Smith et al. 2005; Hannappel et al. 2011, 2012; Timón-Gómez et al. 2018). Heterologous coexpression of P. denitrificans Surf1, HOS, and HAS in E. coli indicated that Surf1 could bind heme a, and incubation of P. denitrificans HAS with Surf1 demonstrated that HAS can transfer heme a to Surf1 in vitro (Bundschuh et al. 2009; Hannappel et al. 2012). However, mutation of the histidine predicted to ligate heme in Shy1 from S. cerevisiae indicated that heme a binding was not required for the assembly factor’s function (Hannappel et al. 2012; Bareth et al. 2013). Instead, Shy1 seems to be important for stabilizing a subunit I-containing subassembly complex that forms at the time of heme a insertion in yeast (Khalimonchuk et al. 2007; Pierrel et al. 2008; Khalimonchuk et al. 2012; Bareth et al. 2013). Surf1c also seems to associate with subunit I-containing subassembly complexes in P. denitrificans (Schimo et al. 2017). Therefore, although Surf1/Shy1 is required for efficient maturation of subunit I in eukaryotes and some prokaryotes, it may not be a true heme a chaperone that shuttles heme a between HAS and subunit I.
The apparent lack of a heme a chaperone in many prokaryotic species suggests that in these species, heme a transfer to subunit I may occur via a direct interaction between HAS and subunit I. As discussed at the beginning of this section, the alternatives to this hypothesis are: 1) Heme a is released from HAS and diffuses through the membrane to subunit I unaided; 2) Another, as-yet unidentified heme a chaperone accepts heme a from HAS and transfers it to subunit I. Because the heme binding sites in HAS and subunit I are both located closer to the P-side (periplasmic/IMS side) of the membrane, a transporter is probably not required to translocate the propionate groups of heme a through the membrane (Kim et al. 2012). Therefore, heme a could, in theory be released directly into the lipid bilayer. Heme a can also spontaneously bind synthetic four-helical bundles that mimic the structure of heme-copper oxidases, suggesting that from a thermodynamic standpoint, a chaperone is not necessarily required to facilitate the binding of heme a to subunit I (Zhuang, Amoroso, et al. 2006). The most common argument against unassisted heme a diffusion in the lipid bilayer is that heme a transfer must be controlled to protect the lipid bilayer from heme a, a prooxidant (Zhuang, Reddi, et al. 2006; Khalimonchuk et al. 2007; Donegan et al. 2019).
An attractive alternative hypothesis is that HAS delivers heme a directly to subunit I. This hypothesis is supported by the observation that overexpression of HAS often results in copurification of HAS and its unreleased product, heme a. This phenomenon has been reported not only for heterologous overexpression of HAS in E. coli (Sakamoto et al. 1999; Lewin and Hederstedt 2006; Hannappel et al. 2011; Hederstedt 2012), but also for lower-level, more physiologically relevant expression conditions in B. subtilis (Svensson et al. 1996). These results have led to the suggestion that in addition to synthesizing heme a, HAS functions as a heme a reservoir/chaperone, remaining bound to heme a until interactions with subunit I (or a subunit-I containing subassembly complex) trigger its release (Hederstedt 2012). At present, the most obvious conformational change in HAS that would lead to heme a release would be movement of TM2 away from the center of the four-helical bundle where heme a binds, which would effectively remove one of the histidine ligands from the heme binding site (Niwa et al. 2018). Of course, the existence of novel, uncharacterized prokaryotic heme a chaperones that trigger heme a release from HAS cannot be ruled out.
Heme-copper oxidases with heme o in the active site require proper heme o insertion into subunit I. However, there is generally less information available about the assembly of these oxidases. Studies of quinol bo3 assembly in E. coli indicate that both heme b and heme o are inserted into subunit I post-translationally (Stenberg et al. 2007; Price and Driessen 2010). Efficient hemylation only seems to occur after subunit I has formed a complex with two other subunits (III and IV), possibly because interactions with these subunits induce a conformational change in subunit I that allows for the formation of the heme binding pockets (Palombo and Daley 2012). No heme o chaperone has been identified, and the precise process governing the insertion of heme b into the low-spin heme binding pocket and heme o into the high-spin heme binding pocket is unknown. However, it seems likely that the structural difference between heme b and heme o (i.e., the presence of a hydroxyethylfarnesyl tail on heme o) helps guide the insertion of each heme into the correct site.
Incorporation (or misincorporation) of alternate heme types into the same oxidase can occur in some prokaryotes, indicating that the assembly process for these oxidases is more flexible than that of their highly-regulated eukaryotic counterparts. For example, heme o can be misincorporated into the low-spin site of the bo3 quinol oxidase in E. coli strains overexpressing their native HOS, resulting in a functional oo3 oxidase (Puustinen 1992; Garcia-Horsman et al. 1994). For a handful of prokaryotic species, functional switching between heme o and heme a as cofactors for the same oxidase has also been reported (Sone and Fujiwara 1991; Matsushita, Ebisuya, Adachi 1992; Matsushita, Ebisuya, Ameyama, et al. 1992; Garcia-Horsman et al. 1994; Sone et al. 1994; Auer et al. 1995; Peschek et al. 1995; Schröter et al. 1998; Contreras-Zentella et al. 2003). Interestingly, the switch from heme a to heme o occurs under microaerobic conditions, although it is not clear if this switch is regulated transcriptionally or if low O2 availability prevents heme a synthesis.
Taken together, these results suggest that either HOS or HAS may interact with subunit I or subunit I-containing assembly intermediates in these organisms to deliver heme, although the existence of an unidentified heme o chaperone or heme a chaperone in these organisms cannot be ruled out. A bifunctional chaperone is also possible, as exemplified by a mutant version of P. denitrificans Surf1 that can bind both heme o and heme a, although this variant does not seem to support insertion of either heme into its cognate oxidase (Hannappel et al. 2011). In summary, the process of heme a transfer from HAS and insertion into heme-copper terminal oxidases seems to vary across species. While heme a insertion into subunit I may not require a heme a chaperone in all organisms, this process does seem to be tightly regulated, especially in eukaryotes. Future work will be needed to clarify the role of potential heme a chaperones (or lack thereof) and the details of heme a transfer and insertion into subunit I.
Eukaryotic HOS oligomers and HAS oligomers and their potential role in cytochrome c oxidase assembly
HOS and HAS form distinct oligomeric, high-molecular weight complexes in S. cerevisiae. There is also some evidence that HAS is part of high-molecular weight complexes in mammals (Swenson S et al. 2016), P. denitrifican (Schimo et al. 2017), and the hyperthermophilic bacterium A. aeolicus, where purified HAS was recently shown to form a trimer (Zeng et al. 2020). These complexes migrate well above the expected monomeric weights for HOS and HAS in non-denaturing blue native PAGE (BN-PAGE) gels, and have also been detected via density gradient centrifugation (Khalimonchuk et al. 2010; Khalimonchuk et al. 2012; Bareth et al. 2013; Swenson S et al. 2016; Herwaldt et al. 2018; Swenson SA et al. 2020). In S. cerevisiae, both HOS complexes and HAS complexes have been shown to be primarily homo-oligomeric (Khalimonchuk et al. 2012; Swenson S et al. 2016; Herwaldt et al. 2018). Furthermore, the formation and stability of HOS homo-oligomers does not depend on the presence of HAS and vice versa (Khalimonchuk et al. 2012; Bareth et al. 2013; Swenson S et al. 2016). Formation of HOS and HAS oligomers also doesn’t depend on the catalytic activity of their respective monomers (Khalimonchuk et al. 2012; Swenson S et al. 2016). For example, mutation of the histidine ligands in HAS does not prevent HAS oligomerization (Bareth et al. 2013; Swenson S et al. 2016). However, in situations where HOS or HAS oligomerization is lost, catalytic activity is also lost, even when expression levels of HOS and HAS remain fairly constant (Khalimonchuk et al. 2012; Swenson S et al. 2016; Taylor et al. 2017). Thus, the physiological role of these oligomers has remained elusive.
Studies of both the HOS oligomers and HAS oligomers in S. cerevisiae suggest that these complexes may play a role in cytochrome c oxidase assembly beyond cofactor synthesis. The molecular weights of HOS homo-oligomers and HAS homo-oligomers are similar to that of various cytochrome c oxidase subassembly complexes, and formation of these homo-oligomers seems to be influenced by the presence of assembly factors associated with subunit I (Cox1) maturation (Pierrel et al. 2008; Bestwick et al. 2010; Khalimonchuk et al. 2012; Swenson S et al. 2016; Taylor et al. 2017). In particular, both HOS and HAS have been shown to either interact with or affect the formation of complexes containing Shy1, although neither HOS nor HAS is a stoichiometric part of these subassembly complexes (Khalimonchuk et al. 2012; Bareth et al. 2013). Overall, these data suggests that HOS and HAS oligomers either interact with or indirectly influence the stability of cytochrome c oxidase subassembly complexes that form prior to Cox1 hemylation (Kim et al. 2012). For HAS, these interactions could involve the transfer of heme a to Cox1.
Surprisingly, HAS has also been found to interact with the respiratory supercomplexes in S. cerevisiae (Herwaldt et al. 2018). Respiratory supercomplexes have been identified in eukaryotes, bacteria, and archaea (Berry and Trumpower 1985; Sone et al. 1987; Lübben et al. 1994; Iwasaki, Matsuura, et al. 1995; Iwasaki, Wakagi, et al. 1995; Cruciat et al. 2000; Schägger and Pfeiffer 2000; Kao et al. 2016; Milenkovic et al. 2017; Davies et al. 2018; Wiseman et al. 2018). These supercomplexes include, at minimum, two different respiratory oxidoreductase complexes. Although the role of the supercomplexes is still debated, their presence in all three domains of life, albeit with slightly different architectures, suggests they are physiologically relevant (Milenkovic et al. 2017; Davies et al. 2018).
High-resolution cryo-EM structures of supercomplexes from several species, including S. cerevisiae, have now been solved (Mileykovskaya et al. 2012; Gu et al. 2016; Letts et al. 2016; Milenkovic et al. 2017; Davies et al. 2018; Wiseman et al. 2018; Hartley et al. 2019; Rathore et al. 2019; Berndtsson et al. 2020; Hartley et al. 2020). Supercomplexes from S. cerevisiae are composed of the cytochrome bc1 dimer (complex III) flanked by either one or two cytochrome c oxidase complexes (complex IV) (Mileykovskaya et al. 2012; Hartley et al. 2019; Rathore et al. 2019; Berndtsson et al. 2020; Hartley et al. 2020). The complex IV subunit Cox5a (or Cox5b, the hypoxic isoform) and multiple complex III subunits form the interface between complex III and complex IV (Hartley et al. 2019; Rathore et al. 2019; Berndtsson et al. 2020; Hartley et al. 2020). Specifically, cryo-EM structures show that Cox5a interacts directly with Cor1 in the matrix and with cytochrome c1 and Qcr6 in the IMS, and also seems to indirectly interact with other complex III subunits via bridging membrane lipids. Additionally, biochemical data indicate that each of these Cox5a-complex III interactions play a role in supercomplex formation, although the interaction between Cox5a and Cor1 in the matrix seems to be the most critical3 (Herwaldt et al. 2018; Berndtsson et al. 2020).
The observation that a small fraction of HAS co-migrates with the respiratory supercomplexes in BN-PAGE was unexpected. Because HAS is responsible for synthesizing heme a, which is inserted into subunit I (Cox1) fairly early in the process of cytochrome c oxidase assembly, the role of HAS in later assembly steps was not anticipated. Coimmunoprecipitation (CoIP) experiments, however, have verified that HAS interacts with the cytochrome c oxidase subunits Cox5a (an early-assembling subunit) and Cox13 (a late-assembling subunit) as well as with Cor1 from complex III (Figure 1) (Herwaldt et al. 2018). It should be noted that CoIP experiments can detect both direct protein-protein interactions and indirect interactions (i.e., interactions mediated by another protein), so this does not provide information about the specific location of the putative HAS-supercomplex interface. However, it was demonstrated that the interaction with the cytochrome bc1 complex persists in knockout strains where fully assembled complex IV is not present. While the physiological relevance of this interaction is unclear, it has been suggested that complex IV assembly and supercomplex formation can occur simultaneously via interactions of the complex IV subassembly complexes with complex III (Bianchi et al. 2004; Brandner et al. 2005; Mick et al. 2007; Lazarou et al. 2009). If this is the case, the small population of HAS that interacts with the supercomplexes may be dedicated specifically to the hemylation of supercomplex-associated cytochrome c oxidase.
Possible role of L1–2 from HAS in protein-protein interactions
The elongated extracellular loops of eukaryotic HAS may play a role in forming protein-protein interactions. As discussed in the “Overview of HAS topology and mutagenesis” section, HAS proteins from different organisms can be broadly divided into two categories, type 1 and type 2, with the defining feature of type-1 enzymes being the presence of a cysteinyl pair in the N-terminal periplasmic loop. This pair of cysteinyl residues is replaced by nonpolar residues in type-2 HAS (which includes all eukaryotic HAS), suggesting that the cysteines do not directly participate in the catalytic mechanism of HAS, but are perhaps important for structural reasons or for mediating species-specific protein-protein interactions (Hederstedt 2012; He et al. 2016). This loop is considerably longer in type-2 (eukaryotic) HAS than in type-1 HAS, which might facilitate additional protein-protein interactions not possible in type-1 HAS. One intriguing possibility is that this loop is responsible for mediating interactions with heme-copper oxidase assembly factors, although this hypothesis has yet to be explored experimentally.
Summary and concluding remarks
Heme a is essential for aerobic respiration in eukaryotes and many prokaryotes due to its role as a cofactor for the majority of known heme-copper terminal oxidases. The heme a biosynthetic pathway is a two-step process in which the prenylation and oxidation of specific sites on the porphyrin ring of heme b are catalyzed by HOS and HAS, respectively. The genes necessary for these modifications appear to be conserved in almost all species known to synthesize heme a, while some other prokaryotic species, such as E. coli, only possess the gene for HOS and utilize a heme o-containing terminal oxidase.
Our structural model of HOS and the recently published crystal structure of HAS (Niwa et al. 2018) have confirmed the probable location of heme binding sites in HOS and HAS. For both synthases, only a relatively small set of residues are strictly conserved. Some of these residues are either presumed to be metal ligands or to participate in the catalytic mechanism, suggesting that the general mechanisms of HOS and HAS are similar across species. Furthermore, the new structural data for HOS and HAS indicate that many of the residues previously identified as critical via site-directed mutagenesis either surround a heme binding site or, in the case of HOS, the farnesyl diphosphate binding site. Interestingly, the substrate heme binding site in HOS is most likely located on the negative side of the membrane, while both the substrate and cofactor heme binding sites of HAS are located on the positive side of the membrane. Thus, one of the unanswered questions about the heme a biosynthetic pathway is how heme o is transferred across the membrane from HOS to HAS (Kim et al. 2012).
More generally, the transfer of heme throughout the heme a biosynthetic pathway and the final insertion of heme a into the terminal oxidase remains unclear, as does the potential role of the protein-protein interactions of HOS and HAS in the heme trafficking process. For both enzymes, the availability of structural data will help guide the design of future experiments to probe each synthase’s respective catalytic mechanism, while the advent of new techniques for labeling and quantifying intracellular heme should prove invaluable in the study of heme trafficking (Swenson SA et al. 2020). The continued development of other techniques, such as cryo-EM and computational modeling, may also help define the protein-protein interaction interfaces of HOS and HAS.
Acknowledgments
This work was supported by the NIH NIGMS under grants R35 GM126948 and R01 GM101386.
The authors gratefully acknowledge Prof. Robert Hausinger for his critical reading of this manuscript.
Footnotes
The HOS model is available at https://github.com/feiglab/heme-o-synthase
Type-2 HAS can also be referred to as “class D” (Lewin and Hederstedt 2016).
HAS from different organisms also typically co-purifies with heme b and heme o or heme a when expressed in E. coli, although the heme type and heme/protein stoichiometry varies (Sakamoto et al. 1999; Lewin and Hederstedt 2006; Mogi 2009b; Hannappel et al. 2011; Zeng et al. 2020).
Appending a C-terminal epitope tag to Cox5a causes slight supercomplex destabilization, presumably because the tag disrupts interactions between Cox5a and cytochrome c1 or Qcr6 in the IMS. See Figure 5D in Herwaldt et al. 2018. The absence of cardiolipin (the bridging lipid that mediates Cox5a-complex III interactions) slightly destabilizes the supercomplexes, while mutagenesis of Cor1 indicates that Cor1-Cox5a interactions are necessary and sufficient for supercomplex formation (Berndtsson et al., 2020).
The authors declare that they have no conflicts of interest.
References
- Abramson J, Riistama S, Larsson G, Jasaitis A, Svensson-Ek M, Laakkonen L, Puustinen A, Iwata S, Wikström M. 2000. The structure of the ubiquinol oxidase from Escherichia coli and its ubiquinone binding site. Nature structural biology. 7(10):910–917. [DOI] [PubMed] [Google Scholar]
- Alfadhel M, Lillquist YP, Waters PJ, Sinclair G, Struys E, McFadden D, Hendson G, Hyams L, Shoffner J, Vallance HD. 2011. Infantile cardioencephalopathy due to a COX15 gene defect: report and review. American journal of medical genetics Part A. 155a(4):840–844. [DOI] [PubMed] [Google Scholar]
- Antonicka H, Leary SC, Guercin GH, Agar JN, Horvath R, Kennaway NG, Harding CO, Jaksch M, Shoubridge EA. 2003. Mutations in COX10 result in a defect in mitochondrial heme A biosynthesis and account for multiple, early-onset clinical phenotypes associated with isolated COX deficiency. Human molecular genetics. 12(20):2693–2702. [DOI] [PubMed] [Google Scholar]
- Antonicka H, Mattman A, Carlson CG, Glerum DM, Hoffbuhr KC, Leary SC, Kennaway NG, Shoubridge EA. 2003. Mutations in COX15 produce a defect in the mitochondrial heme biosynthetic pathway, causing early-onset fatal hypertrophic cardiomyopathy. American journal of human genetics. 72(1):101–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auer G, Mayer B, Wastyn M, Fromwald S, Eghbalzad K, Alge D, Peschek GA. 1995. Promiscuity of heme groups in the cyanobacterial cytochrome-C oxidase. Biochemistry and molecular biology international. 37(6):1173–1185. [PubMed] [Google Scholar]
- Bandeiras TM, Refojo PN, Todorovic S, Murgida DH, Hildebrandt P, Bauer C, Pereira MM, Kletzin A, Teixeira M. 2009. The cytochrome ba complex from the thermoacidophilic crenarchaeote Acidianus ambivalens is an analog of bc1 complexes. Biochimica et Biophysica Acta (BBA) - Bioenergetics. 1787(1):37–45. [DOI] [PubMed] [Google Scholar]
- Bareth B, Dennerlein S, Mick DU, Nikolov M, Urlaub H, Rehling P. 2013. The heme a synthase Cox15 associates with cytochrome c oxidase assembly intermediates during Cox1 maturation. Molecular and cellular biology. 33(20):4128–4137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barros MH, Carlson CG, Glerum DM, Tzagoloff A. 2001. Involvement of mitochondrial ferredoxin and Cox15p in hydroxylation of heme O. FEBS letters. 492(1–2):133–138. [DOI] [PubMed] [Google Scholar]
- Barros MH, Tzagoloff A. 2002. Regulation of the heme A biosynthetic pathway in Saccharomyces cerevisiae. FEBS letters. 516(1–3):119–123. [DOI] [PubMed] [Google Scholar]
- Bayse CA, Merz KM. 2014. Mechanistic insights into Mg2+-independent prenylation by CloQ from classical molecular mechanics and hybrid quantum mechanics/molecular mechanics molecular dynamics simulations. Biochemistry. 53(30):5034–5041. [DOI] [PubMed] [Google Scholar]
- Berndtsson J, Aufschnaiter A, Rathore S, Marin-Buera L, Dawitz H, Diessl J, Kohler V, Barrientos A, Büttner S, Fontanesi F et al. 2020. Respiratory supercomplexes enhance electron transport by decreasing cytochrome c diffusion distance. EMBO reports.e51015. [DOI] [PMC free article] [PubMed]
- Berry EA, Trumpower BL. 1985. Isolation of ubiquinol oxidase from Paracoccus denitrificans and resolution into cytochrome bc1 and cytochrome c-aa3 complexes. The Journal of biological chemistry. 260(4):2458–2467. [PubMed] [Google Scholar]
- Bestwick M, Khalimonchuk O, Pierrel F, Winge DR. 2010. The role of Coa2 in hemylation of yeast Cox1 revealed by its genetic interaction with Cox10. Molecular and cellular biology. 30(1):172–185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bianchi C, Genova ML, Parenti Castelli G, Lenaz G. 2004. The mitochondrial respiratory chain is partially organized in a supercomplex assembly: kinetic evidence using flux control analysis. The Journal of biological chemistry. 279(35):36562–36569. [DOI] [PubMed] [Google Scholar]
- Borisov VB, Gennis RB, Hemp J, Verkhovsky MI. 2011. The cytochrome bd respiratory oxygen reductases. Biochim Biophys Acta. 1807(11):1398–1413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borisov VB, Siletsky SA. 2019. Features of organization and mechanism of catalysis of two families of terminal oxidases: heme-copper and bd-type. Biochemistry (Mosc). 84(11):1390–1402. [DOI] [PubMed] [Google Scholar]
- Brandner K, Mick DU, Frazier AE, Taylor RD, Meisinger C, Rehling P. 2005. Taz1, an outer mitochondrial membrane protein, affects stability and assembly of inner membrane protein complexes: implications for Barth Syndrome. Molecular biology of the cell. 16(11):5202–5214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bräuer L, Brandt W, Schulze D, Zakharova S, Wessjohann L. 2008. A structural model of the membrane-bound aromatic prenyltransferase UbiA from E. coli. Chembiochem : a European journal of chemical biology. 9(6):982–992. [DOI] [PubMed] [Google Scholar]
- Brown BM, Wang Z, Brown KR, Cricco JA, Hegg EL. 2004. Heme O synthase and heme A synthase from Bacillus subtilis and Rhodobacter sphaeroides interact in Escherichia coli. Biochemistry. 43(42):13541–13548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown KR, Allan BM, Do P, Hegg EL. 2002. Identification of novel hemes generated by heme A synthase: evidence for two successive monooxygenase reactions. Biochemistry. 41(36):10906–10913. [DOI] [PubMed] [Google Scholar]
- Brown KR, Brown BM, Hoagland E, Mayne CL, Hegg EL. 2004. Heme A synthase does not incorporate molecular oxygen into the formyl group of heme A. Biochemistry. 43(27):8616–8624. [DOI] [PubMed] [Google Scholar]
- Bryant DA, Hunter CN, Warren MJ. 2020. Biosynthesis of the modified tetrapyrroles-the pigments of life. The Journal of biological chemistry. 295(20):6888–6925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bugiani M, Tiranti V, Farina L, Uziel G, Zeviani M. 2005. Novel mutations in COX15 in a long surviving Leigh syndrome patient with cytochrome c oxidase deficiency. Journal of medical genetics. 42(5):e28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bundschuh FA, Hannappel A, Anderka O, Ludwig B. 2009. Surf1, associated with Leigh syndrome in humans, is a heme-binding protein in bacterial oxidase biogenesis. The Journal of biological chemistry. 284(38):25735–25741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burden AE, Wu C, Dailey TA, Busch JL, Dhawan IK, Rose JP, Wang B, Dailey HA. 1999. Human ferrochelatase: crystallization, characterization of the [2Fe-2S] cluster and determination that the enzyme is a homodimer. Biochim Biophys Acta. 1435(1–2):191–197. [DOI] [PubMed] [Google Scholar]
- Buschmann S, Warkentin E, Xie H, Langer JD, Ermler U, Michel H. 2010. The structure of cbb3 cytochrome oxidase provides insights into proton pumping. Science (New York, NY). 329(5989):327–330. [DOI] [PubMed] [Google Scholar]
- Capaldi RA. 1990. Structure and assembly of cytochrome c oxidase. Archives of biochemistry and biophysics. 280(2):252–262. [DOI] [PubMed] [Google Scholar]
- Castelle CJ, Roger M, Bauzan M, Brugna M, Lignon S, Nimtz M, Golyshina OV, Giudici-Orticoni MT, Guiral M. 2015. The aerobic respiratory chain of the acidophilic archaeon Ferroplasma acidiphilum: a membrane-bound complex oxidizing ferrous iron. Biochim Biophys Acta. 1847(8):717–728. [DOI] [PubMed] [Google Scholar]
- Cheng W, Li W. 2014. Structural insights into ubiquinone biosynthesis in membranes. Science (New York, NY). 343(6173):878–881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chepuri V, Gennis RB. 1990. The use of gene fusions to determine the topology of all of the subunits of the cytochrome o terminal oxidase complex of Escherichia coli. The Journal of biological chemistry. 265(22):12978–12986. [PubMed] [Google Scholar]
- Colas C, Kuo JM, Ortiz de Montellano PR. 2002. Asp-225 and Glu-375 in autocatalytic attachment of the prosthetic heme group of lactoperoxidase. The Journal of biological chemistry. 277(9):7191–7200. [DOI] [PubMed] [Google Scholar]
- Contreras-Zentella M, Mendoza G, Membrillo-Hernández J, Escamilla JE. 2003. A novel double heme substitution produces a functional bo3 variant of the quinol oxidase aa3 of Bacillus cereus: purification and paratial characterization. The Journal of biological chemistry. 278(34):31473–31478. [DOI] [PubMed] [Google Scholar]
- Cruciat CM, Brunner S, Baumann F, Neupert W, Stuart RA. 2000. The cytochrome bc1 and cytochrome c oxidase complexes associate to form a single supracomplex in yeast mitochondria. The Journal of biological chemistry. 275(24):18093–18098. [DOI] [PubMed] [Google Scholar]
- Davies KM, Blum TB, Kühlbrandt W. 2018. Conserved in situ arrangement of complex I and III2 in mitochondrial respiratory chain supercomplexes of mammals, yeast, and plants. Proceedings of the National Academy of Sciences of the United States of America. 115(12):3024–3029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davoudi CF, Ramp P, Baumgart M, Bott M. 2019. Identification of Surf1 as an assembly factor of the cytochrome bc1-aa3 supercomplex of Actinobacteria. Biochimica et biophysica acta Bioenergetics. 1860(10):148033. [DOI] [PubMed] [Google Scholar]
- Degli Esposti M, Garcia-Meza V, Ceniceros Gómez AE, Moya-Beltrán A, Quatrini R, Hederstedt L. 2020. Heme A-containing oxidases evolved in the ancestors of iron oxidizing bacteria. Bio/Rxiv.2020.2003.2001.968255. [Google Scholar]
- Donegan RK, Moore CM, Hanna DA, Reddi AR. 2019. Handling heme: the mechanisms underlying the movement of heme within and between cells. Free radical biology & medicine. 133:88–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferguson-Miller S, Babcock GT. 1996. Heme/copper terminal oxidases. Chem Rev. 96(7):2889–2908. [DOI] [PubMed] [Google Scholar]
- Garcia-Horsman JA, Barquera B, Rumbley J, Ma J, Gennis RB. 1994. The superfamily of heme-copper respiratory oxidases. J Bacteriol. 176(18):5587–5600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glerum DM, Tzagoloff A. 1994. Isolation of a human cDNA for heme A:farnesyltransferase by functional complementation of a yeast cox10 mutant. Proceedings of the National Academy of Sciences. 91(18):8452–8456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu J, Wu M, Guo R, Yan K, Lei J, Gao N, Yang M. 2016. The architecture of the mammalian respirasome. Nature. 537(7622):639–643. [DOI] [PubMed] [Google Scholar]
- Hannappel A, Bundschuh FA, Ludwig B. 2011. Characterization of heme-binding properties of Paracoccus denitrificans Surf1 proteins. The FEBS journal. 278(10):1769–1778. [DOI] [PubMed] [Google Scholar]
- Hannappel A, Bundschuh FA, Ludwig B. 2012. Role of Surf1 in heme recruitment for bacterial COX biogenesis. Biochim Biophys Acta. 1817(6):928–937. [DOI] [PubMed] [Google Scholar]
- Hartley AM, Lukoyanova N, Zhang Y, Cabrera-Orefice A, Arnold S, Meunier B, Pinotsis N, Maréchal A. 2019. Structure of yeast cytochrome c oxidase in a supercomplex with cytochrome bc1. Nature structural & molecular biology. 26(1):78–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartley AM, Meunier B, Pinotsis N, Maréchal A. 2020. Rcf2 revealed in cryo-EM structures of hypoxic isoforms of mature mitochondrial III-IV supercomplexes. Proceedings of the National Academy of Sciences of the United States of America. 117(17):9329–9337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He D, Fu CJ, Baldauf SL. 2016. Multiple origins of eukaryotic cox15 suggest horizontal gene transfer from bacteria to jakobid mitochondrial DNA. Molecular biology and evolution. 33(1):122–133. [DOI] [PubMed] [Google Scholar]
- Hederstedt L. 2012. Heme A biosynthesis. Biochim Biophys Acta. 1817(6):920–927. [DOI] [PubMed] [Google Scholar]
- Hederstedt L, Lewin A, Throne-Holst M. 2005. Heme A synthase enzyme functions dissected by mutagenesis of Bacillus subtilis CtaA. J Bacteriol. 187(24):8361–8369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinemann IU, Jahn M, Jahn D. 2008. The biochemistry of heme biosynthesis. Archives of biochemistry and biophysics. 474(2):238–251. [DOI] [PubMed] [Google Scholar]
- Hemp JG, Robert 2008. Bioenergetics: energy conservation and conversion. In: Schäfer GnP, Harvey S, editor. Bioenergetics: Energy Conservation and Conversion. 2008 ed. Berlin: Springer. [Google Scholar]
- Heo L, Feig M. 2020. High-accuracy protein structures by combining machine-learning with physics-based refinement. Proteins. 88(5):637–642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herwaldt EJ, Rivett ED, White AJ, Hegg EL. 2018. Cox15 interacts with the cytochrome bc1 dimer within respiratory supercomplexes as well as in the absence of cytochrome c oxidase. The Journal of biological chemistry. 293(42):16426–16439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiner AN, Raven EL, Thorneley RN, García-Cánovas F, Rodríguez-López JN. 2002. Mechanisms of compound I formation in heme peroxidases. Journal of inorganic biochemistry. 91(1):27–34. [DOI] [PubMed] [Google Scholar]
- Hosfield DJ, Zhang Y, Dougan DR, Broun A, Tari LW, Swanson RV, Finn J. 2004. Structural basis for bisphosphonate-mediated inhibition of isoprenoid biosynthesis. The Journal of biological chemistry. 279(10):8526–8529. [DOI] [PubMed] [Google Scholar]
- Huang H, Levin EJ, Liu S, Bai Y, Lockless SW, Zhou M. 2014. Structure of a membrane-embedded prenyltransferase homologous to UBIAD1. PLoS biology. 12(7):e1001911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwasaki T, Matsuura K, Oshima T. 1995. Resolution of the aerobic respiratory system of the thermoacidophilic archaeon, Sulfolobus sp. strain 7. I: the archaeal terminal oxidase supercomplex is a functional fusion of respiratory complexes III and IV with no c-type cytochromes. The Journal of biological chemistry. 270(52):30881–30892. [DOI] [PubMed] [Google Scholar]
- Iwasaki T, Wakagi T, Isogai Y, Iizuka T, Oshima T. 1995. Resolution of the aerobic respiratory system of the thermoacidophilic archaeon, Sulfolobus sp. strain 7. II: Characterization of the archaeal terminal oxidase subcomplexes and implication for the intramolecular electron transfer. The Journal of biological chemistry. 270(52):30893–30901. [DOI] [PubMed] [Google Scholar]
- Iwata S, Ostermeier C, Ludwig B, Michel H. 1995. Structure at 2.8 A resolution of cytochrome c oxidase from Paracoccus denitrificans. Nature. 376(6542):660–669. [DOI] [PubMed] [Google Scholar]
- Kao WC, Kleinschroth T, Nitschke W, Baymann F, Neehaul Y, Hellwig P, Richers S, Vonck J, Bott M, Hunte C. 2016. The obligate respiratory supercomplex from Actinobacteria. Biochim Biophys Acta. 1857(10):1705–1714. [DOI] [PubMed] [Google Scholar]
- Kavanagh KL, Dunford JE, Bunkoczi G, Russell RG, Oppermann U. 2006. The crystal structure of human geranylgeranyl pyrophosphate synthase reveals a novel hexameric arrangement and inhibitory product binding. The Journal of biological chemistry. 281(31):22004–22012. [DOI] [PubMed] [Google Scholar]
- Khalimonchuk O, Bestwick M, Meunier B, Watts TC, Winge DR. 2010. Formation of the redox cofactor centers during Cox1 maturation in yeast cytochrome oxidase. Molecular and cellular biology. 30(4):1004–1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khalimonchuk O, Bird A, Winge DR. 2007. Evidence for a pro-oxidant intermediate in the assembly of cytochrome oxidase. The Journal of biological chemistry. 282(24):17442–17449. [DOI] [PubMed] [Google Scholar]
- Khalimonchuk O, Kim H, Watts T, Perez-Martinez X, Winge DR. 2012. Oligomerization of heme o synthase in cytochrome oxidase biogenesis is mediated by cytochrome oxidase assembly factor Coa2. The Journal of biological chemistry. 287(32):26715–26726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HJ, Khalimonchuk O, Smith PM, Winge DR. 2012. Structure, function, and assembly of heme centers in mitochondrial respiratory complexes. Biochim Biophys Acta. 1823(9):1604–1616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kranz RG, Richard-Fogal C, Taylor JS, Frawley ER. 2009. Cytochrome c biogenesis: mechanisms for covalent modifications and trafficking of heme and for heme-iron redox control. Microbiol Mol Biol Rev. 73(3):510–528, Table of Contents. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazarou M, Smith SM, Thorburn DR, Ryan MT, McKenzie M. 2009. Assembly of nuclear DNA-encoded subunits into mitochondrial complex IV, and their preferential integration into supercomplex forms in patient mitochondria. The FEBS journal. 276(22):6701–6713. [DOI] [PubMed] [Google Scholar]
- Letts JA, Fiedorczuk K, Sazanov LA. 2016. The architecture of respiratory supercomplexes. Nature. 537(7622):644–648. [DOI] [PubMed] [Google Scholar]
- Lewin A, Hederstedt L. 2006. Compact archaeal variant of heme A synthase. FEBS letters. 580(22):5351–5356. [DOI] [PubMed] [Google Scholar]
- Lewin A, Hederstedt L. 2016. Heme A synthase in bacteria depends on one pair of cysteinyls for activity. Biochim Biophys Acta. 1857(2):160–168. [DOI] [PubMed] [Google Scholar]
- Li T, Bonkovsky HL, Guo JT. 2011. Structural analysis of heme proteins: implications for design and prediction. BMC structural biology. 11:13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W. 2016. Bringing bioactive compounds into membranes: the UbiA superfamily of intramembrane aromatic prenyltransferases. Trends in biochemical sciences. 41(4):356–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Zhang Y, Zhang C, Wang H, Wei X, Chen P, Lu L. 2020. Mitochondrial dysfunctions trigger the calcium signaling-dependent fungal multidrug resistance. Proceedings of the National Academy of Sciences of the United States of America. 117(3):1711–1721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lübben M, Morand K. 1994. Novel prenylated hemes as cofactors of cytochrome oxidases: archaea have modified hemes A and O. The Journal of biological chemistry. 269(34):21473–21479. [PubMed] [Google Scholar]
- Lübben M, Warne A, Albracht SP, Saraste M. 1994. The purified SoxABCD quinol oxidase complex of Sulfolobus acidocaldarius contains a novel haem. Molecular microbiology. 13(2):327–335. [DOI] [PubMed] [Google Scholar]
- Luk LY, Tanner ME. 2009. Mechanism of dimethylallyltryptophan synthase: evidence for a dimethylallyl cation intermediate in an aromatic prenyltransferase reaction. Journal of the American Chemical Society. 131(39):13932–13933. [DOI] [PubMed] [Google Scholar]
- Lyons JA, Aragão D, Slattery O, Pisliakov AV, Soulimane T, Caffrey M. 2012. Structural insights into electron transfer in caa3-type cytochrome oxidase. Nature. 487(7408):514–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maréchal A, Meunier B, Lee D, Orengo C, Rich PR. 2012. Yeast cytochrome c oxidase: a model system to study mitochondrial forms of the haem-copper oxidase superfamily. Biochim Biophys Acta. 1817(4):620–628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsushita K, Ebisuya H, Adachi O. 1992. Homology in the structure and the prosthetic groups between two different terminal ubiquinol oxidases, cytochrome a1 and cytochrome o, of Acetobacter aceti. The Journal of biological chemistry. 267(34):24748–24753. [PubMed] [Google Scholar]
- Matsushita K, Ebisuya H, Ameyama M, Adachi O. 1992. Change of the terminal oxidase from cytochrome a1 in shaking cultures to cytochrome o in static cultures of Acetobacter aceti. J Bacteriol. 174(1):122–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medlock AE, Dailey TA, Ross TA, Dailey HA, Lanzilotta WN. 2007. A pi-helix switch selective for porphyrin deprotonation and product release in human ferrochelatase. Journal of molecular biology. 373(4):1006–1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melzer M, Heide L. 1994. Characterization of polyprenyldiphosphate: 4-hydroxybenzoate polyprenyltransferase from Escherichia coli. Biochim Biophys Acta. 1212(1):93–102. [DOI] [PubMed] [Google Scholar]
- Merli ML, Cirulli BA, Menéndez-Bravo SM, Cricco JA. 2017. Heme A synthesis and CcO activity are essential for Trypanosoma cruzi infectivity and replication. The Biochemical journal. 474(14):2315–2332. [DOI] [PubMed] [Google Scholar]
- Mick DU, Wagner K, van der Laan M, Frazier AE, Perschil I, Pawlas M, Meyer HE, Warscheid B, Rehling P. 2007. Shy1 couples Cox1 translational regulation to cytochrome c oxidase assembly. The EMBO journal. 26(20):4347–4358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milenkovic D, Blaza JN, Larsson NG, Hirst J. 2017. The enigma of the respiratory chain supercomplex. Cell metabolism. 25(4):765–776. [DOI] [PubMed] [Google Scholar]
- Mileykovskaya E, Penczek PA, Fang J, Mallampalli VK, Sparagna GC, Dowhan W. 2012. Arrangement of the respiratory chain complexes in Saccharomyces cerevisiae supercomplex III2IV2 revealed by single particle cryo-electron microscopy. The Journal of biological chemistry. 287(27):23095–23103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miryounesi M, Fardaei M, Tabei SM, Ghafouri-Fard S. 2016. Leigh syndrome associated with a novel mutation in the COX15 gene. Journal of pediatric endocrinology & metabolism : JPEM. 29(6):741–744. [DOI] [PubMed] [Google Scholar]
- Mogi T. 2009a. Over-expression and characterization of Bacillus subtilis heme O synthase. Journal of biochemistry. 145(5):669–675. [DOI] [PubMed] [Google Scholar]
- Mogi T. 2009b. Probing structure of heme A synthase from Bacillus subtilis by site-directed mutagenesis. Journal of biochemistry. 145(5):625–633. [DOI] [PubMed] [Google Scholar]
- Mogi T, Saiki K, Anraku Y. 1994. Biosynthesis and functional role of haem O and haem A. Molecular microbiology. 14(3):391–398. [DOI] [PubMed] [Google Scholar]
- Myer YP, Saturno AF, Verma BC, Pande A. 1979. Horse heart cytochrome c: the oxidation-reduction potential and protein structures. The Journal of biological chemistry. 254(22):11202–11207. [PubMed] [Google Scholar]
- Niebisch A, Bott M. 2001. Molecular analysis of the cytochrome bc1-aa3 branch of the Corynebacterium glutamicum respiratory chain containing an unusual diheme cytochrome c1. Archives of microbiology. 175(4):282–294. [DOI] [PubMed] [Google Scholar]
- Niwa S, Takeda K, Kosugi M, Tsutsumi E, Mogi T, Miki K. 2018. Crystal structure of heme A synthase from Bacillus subtilis. Proceedings of the National Academy of Sciences of the United States of America. 115(47):11953–11957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nobrega MP, Nobrega FG, Tzagoloff A. 1990. COX10 codes for a protein homologous to the ORF1 product of Paracoccus denitrificans and is required for the synthesis of yeast cytochrome oxidase. The Journal of biological chemistry. 265(24):14220–14226. [PubMed] [Google Scholar]
- Oquendo CE, Antonicka H, Shoubridge EA, Reardon W, Brown GK. 2004. Functional and genetic studies demonstrate that mutation in the COX15 gene can cause Leigh syndrome. Journal of medical genetics. 41(7):540–544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortiz de Montellano PR. 2008. Mechanism and role of covalent heme binding in the CYP4 family of P450 enzymes and the mammalian peroxidases. Drug metabolism reviews. 40(3):405–426. [DOI] [PubMed] [Google Scholar]
- Oster U, Tanaka R, Tanaka A, Rüdiger W. 2000. Cloning and functional expression of the gene encoding the key enzyme for chlorophyll b biosynthesis (CAO) from Arabidopsis thaliana. The Plant journal : for cell and molecular biology. 21(3):305–310. [DOI] [PubMed] [Google Scholar]
- Palombo I, Daley DO. 2012. Heme incorporation into the cytochrome bo3 occurs at a late stage of assembly. FEBS letters. 586(23):4197–4202. [DOI] [PubMed] [Google Scholar]
- Pereira MM, Santana M, Teixeira M. 2001. A novel scenario for the evolution of haem-copper oxygen reductases. Biochim Biophys Acta. 1505(2–3):185–208. [DOI] [PubMed] [Google Scholar]
- Pereira MM, Sousa FL, Veríssimo AF, Teixeira M. 2008. Looking for the minimum common denominator in haem-copper oxygen reductases: towards a unified catalytic mechanism. Biochim Biophys Acta. 1777(7–8):929–934. [DOI] [PubMed] [Google Scholar]
- Peschek GA, Alge D, Fromwald S, Mayer B. 1995. Transient accumulation of heme O (cytochrome o) in the cytoplasmic membrane of semi-anaerobic Anacystis nidulans: evidence for oxygenase-catalyzed heme O/A transformation. The Journal of biological chemistry. 270(46):27937–27941. [DOI] [PubMed] [Google Scholar]
- Piel RB, 3rd, Dailey HA Jr., Medlock AE 2019. The mitochondrial heme metabolon: insights into the complex(ity) of heme synthesis and distribution. Molecular genetics and metabolism. 128(3):198–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierrel F, Khalimonchuk O, Cobine PA, Bestwick M, Winge DR. 2008. Coa2 is an assembly factor for yeast cytochrome c oxidase biogenesis that facilitates the maturation of Cox1. Molecular and cellular biology. 28(16):4927–4939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porra RJ, Schäfer W, Cmiel E, Katheder I, Scheer H. 1993. Derivation of the formyl-group oxygen of chlorophyll b from molecular oxygen in greening leaves of a higher plant (Zea mays). FEBS letters. 323(1–2):31–34. [DOI] [PubMed] [Google Scholar]
- Poulos TL. 2010. Thirty years of heme peroxidase structural biology. Archives of biochemistry and biophysics. 500(1):3–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poulter CD, Rilling HC. 1978. The prenyl transfer reaction: enzymatic and mechanistic studies of the 1’−4 coupling reaction in the terpene biosynthetic pathway. Accounts of Chemical Research. 11(8):307–313. [Google Scholar]
- Price CE, Driessen AJ. 2010. Biogenesis of membrane bound respiratory complexes in Escherichia coli. Biochim Biophys Acta. 1803(6):748–766. [DOI] [PubMed] [Google Scholar]
- Puustinen A. 1992. A novel type haem group of cytochrome o from Escherichia coli. Acta physiologica Scandinavica Supplementum. 607:265–268. [PubMed] [Google Scholar]
- Puustinen A, Wikstrom M. 1991. The heme groups of cytochrome o from Escherichia coli. Proceedings of the National Academy of Sciences. 88(14):6122–6126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rathore S, Berndtsson J, Marin-Buera L, Conrad J, Carroni M, Brzezinski P, Ott M. 2019. Cryo-EM structure of the yeast respiratory supercomplex. Nature structural & molecular biology. 26(1):50–57. [DOI] [PubMed] [Google Scholar]
- Reddi AR, Hamza I. 2016. Heme mobilization in animals: a metallolipid’s journey. Acc Chem Res. 49(6):1104–1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Refojo PN, Sena FV, Calisto F, Sousa FM, Pereira MM. 2019. Chapter six: the plethora of membrane respiratory chains in the phyla of life. In: Poole RK, editor. Advances in Microbial Physiology. Academic Press; p. 331–414. [DOI] [PubMed] [Google Scholar]
- Rudolf JD, Poulter CD. 2013. Tyrosine O-prenyltransferase SirD catalyzes S-, C-, and N-prenylations on tyrosine and tryptophan derivatives. ACS chemical biology. 8(12):2707–2714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudolf JD, Wang H, Poulter CD. 2013. Multisite prenylation of 4-substituted tryptophans by dimethylallyltryptophan synthase. Journal of the American Chemical Society. 135(5):1895–1902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saiki K, Mogi T, Anraku Y. 1992. Heme O biosynthesis in Escherichia coli: the cyoE gene in the cytochrome bo operon encodes a protoheme IX farnesyltransferase. Biochemical and biophysical research communications. 189(3):1491–1497. [DOI] [PubMed] [Google Scholar]
- Saiki K, Mogi T, Hori H, Tsubaki M, Anraku Y. 1993. Identification of the functional domains in heme O synthase: site-directed mutagenesis studies on the cyoE gene of the cytochrome bo operon in Escherichia coli. The Journal of biological chemistry. 268(36):26927–26934. [PubMed] [Google Scholar]
- Saiki K, Mogi T, Ogura K, Anraku Y. 1993. In vitro heme O synthesis by the cyoE gene product from Escherichia coli. The Journal of biological chemistry. 268(35):26041–26044. [PubMed] [Google Scholar]
- Sakamoto J, Hayakawa A, Uehara T, Noguchi S, Sone N. 1999. Cloning of Bacillus stearothermophilus ctaA and heme A synthesis with the CtaA protein produced in Escherichia coli. Bioscience, biotechnology, and biochemistry. 63(1):96–103. [DOI] [PubMed] [Google Scholar]
- Schafer G, Engelhard M, Muller V. 1999. Bioenergetics of the Archaea. Microbiol Mol Biol Rev. 63(3):570–620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schägger H, Pfeiffer K. 2000. Supercomplexes in the respiratory chains of yeast and mammalian mitochondria. The EMBO journal. 19(8):1777–1783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scharf B, Wittenberg R, Engelhard M. 1997. Electron transfer proteins from the haloalkaliphilic archaeon Natronobacterium pharaonis: possible components of the respiratory chain include cytochrome bc and a terminal oxidase cytochrome ba3. Biochemistry. 36(15):4471–4479. [DOI] [PubMed] [Google Scholar]
- Schimo S, Wittig I, Pos KM, Ludwig B. 2017. Cytochrome c oxidase biogenesis and metallochaperone interactions: steps in the assembly pathway of a bacterial complex. PloS one. 12(1):e0170037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneegurt MA, Beale SI. 1992. Origin of the chlorophyll b formyl oxygen in Chlorella vulgaris. Biochemistry. 31(47):11677–11683. [DOI] [PubMed] [Google Scholar]
- Schröter T, Winterstein C, Ludwig B, Richter O-MH. 1998. Expression of the Escherichia coli cyo operon in Paracoccus denitrificans results in a fully active quinol oxidase of unexpected heme composition. FEBS letters. 432(3):109–112. [DOI] [PubMed] [Google Scholar]
- Sharma V, Wikström M, Kaila VR. 2011. Stabilization of the peroxy intermediate in the oxygen splitting reaction of cytochrome cbb3. Biochim Biophys Acta. 1807(7):813–818. [DOI] [PubMed] [Google Scholar]
- Smith D, Gray J, Mitchell L, Antholine WE, Hosler JP. 2005. Assembly of cytochrome-c oxidase in the absence of assembly protein Surf1p leads to loss of the active site heme. The Journal of biological chemistry. 280(18):17652–17656. [DOI] [PubMed] [Google Scholar]
- Sone N, Fujiwara Y. 1991. Haem O can replace haem A in the active site of cytochrome c oxidase from thermophilic bacterium PS3. FEBS letters. 288(1–2):154–158. [DOI] [PubMed] [Google Scholar]
- Sone N, Ogura T, Noguchi S, Kitagawa T. 1994. Proton pumping activity and visible absorption and resonance Raman spectra of a cao-type cytochrome c oxidase isolated from the thermophilic bacterium Bacillus PS3. Biochemistry. 33(4):849–855. [DOI] [PubMed] [Google Scholar]
- Sone N, Sekimachi M, Kutoh E. 1987. Identification and properties of a quinol oxidase super-complex composed of a bc1 complex and cytochrome oxidase in the thermophilic bacterium PS3. The Journal of biological chemistry. 262(32):15386–15391. [PubMed] [Google Scholar]
- Sono M, Roach MP, Coulter ED, Dawson JH. 1996. Heme-containing oxygenases. Chem Rev. 96(7):2841–2888. [DOI] [PubMed] [Google Scholar]
- Stenberg F, von Heijne G, Daley DO. 2007. Assembly of the cytochrome bo3 complex. Journal of molecular biology. 371(3):765–773. [DOI] [PubMed] [Google Scholar]
- Svensson B, Andersson KK, Hederstedt L. 1996. Low-spin heme A in the heme A biosynthetic protein CtaA from Bacillus subtilis. European journal of biochemistry. 238(1):287–295. [DOI] [PubMed] [Google Scholar]
- Svensson B, Hederstedt L. 1994. Bacillus subtilis CtaA is a heme-containing membrane protein involved in heme A biosynthesis. J Bacteriol. 176(21):6663–6671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svensson B, Lübben M, Hederstedt L. 1993. Bacillus subtilis CtaA and CtaB function in haem A biosynthesis. Molecular microbiology. 10(1):193–201. [DOI] [PubMed] [Google Scholar]
- Swenson S, Cannon A, Harris NJ, Taylor NG, Fox JL, Khalimonchuk O. 2016. Analysis of oligomerization properties of heme a synthase provides insights into its function in eukaryotes. The Journal of biological chemistry. 291(19):10411–10425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swenson SA, Moore CM, Marcero JR, Medlock AE, Reddi AR, Khalimonchuk O. 2020. From synthesis to utilization: the ins and outs of mitochondrial heme. Cells. 9(3). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor NG, Swenson S, Harris NJ, Germany EM, Fox JL, Khalimonchuk O. 2017. The assembly factor Pet117 couples heme a synthase activity to cytochrome oxidase assembly. The Journal of biological chemistry. 292(5):1815–1825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Throne-Holst M, Hederstedt L. 2000. The Bacillus subtilis ctaB paralogue, yjdK, can complement the heme A synthesis deficiency of a CtaB-deficient mutant. FEMS microbiology letters. 183(2):247–251. [DOI] [PubMed] [Google Scholar]
- Timón-Gómez A, Nývltová E, Abriata LA, Vila AJ, Hosler J, Barrientos A. 2018. Mitochondrial cytochrome c oxidase biogenesis: recent developments. Seminars in cell & developmental biology. 76:163–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsudzuki T, Wilson DF. 1971. The oxidation-reduction potentials of the hemes and copper of cytochrome oxidase from beef heart. Archives of biochemistry and biophysics. 145(1):149–154. [DOI] [PubMed] [Google Scholar]
- Tsukihara T, Aoyama H, Yamashita E, Tomizaki T, Yamaguchi H, Shinzawa-Itoh K, Nakashima R, Yaono R, Yoshikawa S. 1996. The whole structure of the 13-subunit oxidized cytochrome c oxidase at 2.8 A. Science (New York, NY). 272(5265):1136–1144. [DOI] [PubMed] [Google Scholar]
- Valnot I, von Kleist-Retzow JC, Barrientos A, Gorbatyuk M, Taanman JW, Mehaye B, Rustin P, Tzagoloff A, Munnich A, Rötig A. 2000. A mutation in the human heme A:farnesyltransferase gene (COX10 ) causes cytochrome c oxidase deficiency. Human molecular genetics. 9(8):1245–1249. [DOI] [PubMed] [Google Scholar]
- Wang N, Zhao X, Lu Y. 2005. Role of heme types in heme-copper oxidases: effects of replacing a heme b with a heme o mimic in an engineered heme-copper center in myoglobin. Journal of the American Chemical Society. 127(47):16541–16547. [DOI] [PubMed] [Google Scholar]
- Werner C, Richter OM, Ludwig B. 2010. A novel heme a insertion factor gene cotranscribes with the Thermus thermophilus cytochrome ba3 oxidase locus. J Bacteriol. 192(18):4712–4719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wessjohann L, Sontag B. 1996. Prenylation of benzoic acid derivatives catalyzed by a transferase from Escherichia coli overproduction: method development and substrate specificity. Angewandte Chemie International Edition in English. 35(15):1697–1699. [Google Scholar]
- Wikstrom M, Krab K, Sharma V. 2018. Oxygen activation and energy conservation by cytochrome c oxidase. Chem Rev. 118(5):2469–2490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiseman B, Nitharwal RG, Fedotovskaya O, Schäfer J, Guo H, Kuang Q, Benlekbir S, Sjöstrand D, Ädelroth P, Rubinstein JL et al. 2018. Structure of a functional obligate complex III2IV2 respiratory supercomplex from Mycobacterium smegmatis. Nature structural & molecular biology. 25(12):1128–1136. [DOI] [PubMed] [Google Scholar]
- Wu CK, Dailey HA, Rose JP, Burden A, Sellers VM, Wang BC. 2001. The 2.0 A structure of human ferrochelatase, the terminal enzyme of heme biosynthesis. Nature structural biology. 8(2):156–160. [DOI] [PubMed] [Google Scholar]
- Yang J, Anishchenko I, Park H, Peng Z, Ovchinnikov S, Baker D. 2020. Improved protein structure prediction using predicted interresidue orientations. Proceedings of the National Academy of Sciences of the United States of America. 117(3):1496–1503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Miao Y, Wang B, Cui G, Merz KM Jr., 2012. Catalytic mechanism of aromatic prenylation by NphB. Biochemistry. 51(12):2606–2618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng H, Zhu G, Zhang S, Li X, Martin J, Morgner N, Sun F, Peng G, Xie H, Michel H. 2020. Isolated heme A synthase from Aquifex aeolicus is a trimer. mBio. 11(3). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhuang J, Amoroso JH, Kinloch R, Dawson JH, Baldwin MJ, Gibney BR. 2006. Evaluation of electron-withdrawing group effects on heme binding in designed proteins: implications for heme a in cytochrome c oxidase. Inorganic chemistry. 45(12):4685–4694. [DOI] [PubMed] [Google Scholar]
- Zhuang J, Reddi AR, Wang Z, Khodaverdian B, Hegg EL, Gibney BR. 2006. Evaluating the roles of the heme a side chains in cytochrome c oxidase using designed heme proteins. Biochemistry. 45(41):12530–12538. [DOI] [PubMed] [Google Scholar]
- Zoppellaro G, Bren KL, Ensign AA, Harbitz E, Kaur R, Hersleth H-P, Ryde U, Hederstedt L, Andersson KK. 2009. Review: studies of ferric heme proteins with highly anisotropic/highly axial low spin (S = 1/2) electron paramagnetic resonance signals with bis-histidine and histidine-methionine axial iron coordination. Biopolymers. 91(12):1064–1082. [DOI] [PMC free article] [PubMed] [Google Scholar]