Abstract
The titin gene (TTN) is associated with several diseases, including inherited arrhythmias. Most of these diagnoses are attributed to rare TTN variants encoding truncated forms, but missense variants represent a diagnostic challenge for clinical genetics. The proper interpretation of genetic data is critical for translation into the clinical setting. Notably, many TTN variants were classified before 2015, when the American College of Medical Genetics and Genomics (ACMG) published recommendations to accurately classify genetic variants. Our aim was to perform an exhaustive reanalysis of rare missense TTN variants that were classified before 2015, and that have ambiguous roles in inherited arrhythmogenic syndromes. Rare missense TTN variants classified before 2015 were updated following the ACMG recommendations and according to all the currently available data. Our cohort included 193 individuals definitively diagnosed with an inherited arrhythmogenic syndrome before 2015. Our analysis resulted in the reclassification of 36.8% of the missense variants from unknown to benign/likely benign. Of all the remaining variants, currently classified as of unknown significance, 38.3% showed a potential, but not confirmed, deleterious role. Most of these rare missense TTN variants with a suspected deleterious role were identified in patients diagnosed with hypertrophic cardiomyopathy. More than 35% of the rare missense TTN variants previously classified as ambiguous were reclassified as not deleterious, mainly because of improved population frequencies. Despite being inconclusive, almost 40% of the variants showed a potentially deleterious role in inherited arrhythmogenic syndromes. Our results highlight the importance of the periodical reclassification of rare missense TTN variants to improve genetic diagnoses and help increase the accuracy of personalized medicine.
Keywords: sudden cardiac death, inherited arrhythmogenic syndromes, TTN, genetics, interpretation
1. Introduction
Inherited arrhythmogenic syndromes (IASs) are a group of genetic diseases encompassing channelopathies, principally, long QT syndrome (LQTS), short QT syndrome (SQTS), Brugada syndrome (BrS), and catecholaminergic polymorphic ventricular tachycardia (CPVT); and cardiomyopathies, mainly hypertrophic cardiomyopathy (HCM), dilated cardiomyopathy (DCM), arrhythmogenic cardiomyopathy (ACM), and left ventricular non-compacted cardiomyopathy (LVNC) [1]. IASs are characterized by malignant arrhythmias leading to ventricular fibrillation and sudden cardiac death (SCD), which is sometimes the first manifestation of disease [2]. Clinical guidelines recommend genetic testing of patients diagnosed with an IAS. If a definite pathogenic variant is identified as a cause of disease, genetic analysis should also be performed in their relatives who, despite being asymptomatic, may harbor the deleterious genetic alteration and the risk of SCD [3,4].
In 2015, the American College of Medical Genetics and Genomics (ACMG) published updated recommendations for the accurate assessment of rare variants [5], helping to define the pathogenicity of a genetic variant and proper clinical translation [6]. However, a lack of data impedes the suitable classification of many rare variants that remain of uncertain/ambiguous significance (VUS) [7]. These ambiguities leave families with inconclusive genetic diagnoses, which are not helpful in clinical decision-making [4,8]. In these cases, the ambiguous variant is disregarded in the diagnosis, and only medical and family histories are referenced for risk assessment and clinical management [5]. This situation occurs with many rare missense variants located in the TTN gene.
The TTN gene encodes the largest human protein, titin [9]. Rare variants in this 363-exon gene are associated with several musculoskeletal and cardiac diseases, including IASs [10]. Many definite deleterious TTN variants are truncated; few studies have reported missense variants. The roles of these rare missense TTN variants remain unclear and challenging to determine in IASs, mainly because of a lack of functional data. The use of the ACMG recommendations and new available data on global population frequencies may change prior classifications. The limited studies conducted in the past few years support a periodic reclassification of rare variants [11,12,13,14,15], especially if classified with an ambiguous role and not following the ACMG guidelines [16], because of the importance of understanding these variants before interpretation in a clinical setting. Here, we used currently available data to reanalyze and reclassify rare missense TTN variants that were previously classified as VUS in IASs.
2. Materials and Methods
2.1. Study Cohort
Our retrospective study included 193 patients with a definitive clinical diagnosis of an inherited arrhythmia, as determined following the available guidelines [17] (clinical assessment of all the patients was performed between 2011 and 2015, both years included). After comprehensive genetic analysis using a next-generation sequencing (NGS) approach, all the patients received an inconclusive genetic diagnosis explaining the origin of the disease. All the patients carried a rare missense variant in the TTN gene only, and the variant was classified as having an unknown/ambiguous role, according to the guidelines available at the time [18]. Suspected cases with an inconclusive clinical diagnosis were not included in the present study to avoid bias. All cases carrying a rare variant in any of the genes analyzed, classified as definitely or potentially causative of disease, were not included following the same approach. Genetic analysis was approved by the Ethics Committee of Hospital Josep Trueta (Girona, Catalonia, Spain), following the World Medical Association Declaration of Helsinki. Clinical and genetic data on all the patients were anonymized and kept confidential. Written informed consent was obtained from all the patients before genetic analysis.
2.2. Genetic Analysis
A resequencing custom-made panel including the most prevalent genes associated with inherited arrhythmias up until 2015 was used (ACTC1, ACTN2, ANK2, CACNA1C, CACNB2, CASQ2, CAV3, CRYAB, CSRP3, DES, DMD, DSC2, DSG2, DSP, EMD, FBN1, GLA, GPD1L, HCN4, JPH2, JUP, KCNE1, KCNE2, KCNH2, KCNJ2, KCNQ1, LAMP2, LDB3, LMNA, MYBPC3, MYH6, MYH7, MYL2, MYL3, MYOZ2, PDLIM3, PKP2, PLN, PRKAG2, RYR2, SCN4B, SCN5A, SGCA, SGCB, SGCD, TAZ, TCAP, TGFB3, TGFBR2, TNNC1, TNNI3, TNNT2, TPM1, TTN, and VCL). All the gene isoforms are described in Ensembl 75 (www.ensembl.org/, accessed on 5 January 2022), linked to a RefSeq code (www.ncbi.nlm.nih.gov/refseq/, accessed on 5 January 2022) or CCDS (www.ncbi.nlm.nih.gov/CCDS/, accessed on 5 January 2022). Bioinformatic analysis included adaptor and low-quality base trimming of the FASTQ files. Trimmed reads were mapped with GEM III. The output was sorted, and uniquely and properly mapped read pairs were selected. Variant calling from the cleaned BAM files was performed with SAMtools v.1.2 and an ad hoc-developed script. The final annotation steps provided information included in public databases. Identified uncommon (minor allele frequency (MAF) < 1%) genetic variants were confirmed by Sanger sequencing. The exons and exon–intron boundaries of each gene were amplified in both directions.
All sequences obtained up until 2015 were reanalyzed with updated software (SeqScape v2.7, Applied Biosystems). No new rare variants were identified in any of the analyzed genes, including the TTN gene. The original classification compared rare missense variants in TTN with DNA sequences from 200 healthy Spanish individuals (individuals not related to any index case and of the same ethnicity) as control cases, contrasted with HapMap (www.hapmap.ncbi.nlm.nih.gov/, accessed on 5 January 2022), the 1000 Genomes project (www.1000genomes.org/, accessed on 5 January 2022), and the Exome Variant Server (EVS) (www.evs.gs.washington.edu/EVS/, accessed on 5 January 2022), available in 2015. Sequence variants were described following the Human Genome Variation Society (HGVS) rules (www.hgvs.org/, accessed on 5 January 2022). Currently, all rare missense variants in TTN were contrasted in the Genome Aggregation Database (gnomAD) (www.gnomad.broadinstitute.org/, accessed on 5 January 2022). All rare missense TTN variants were also consulted in ClinGen (www.clinicalgenome.org/, accessed on 5 January 2022), VarSome (www.varsome.com/, accessed on 5 January 2022), the SCD-associated Variants Annotation Database (SVAD) (www.svad.mbc.nctu.edu.tw/, accessed on 5 January 2022), CardioClassifier (www.cardioclassifier.org/, accessed on 5 January 2022), InterVar (www.wintervar.wglab.org/, accessed on 5 January 2022), CardioVAI (www.cardiovai.engenome.com/, accessed on 5 January 2022), and CardioBoost (www.cardiodb.org/cardioboost/, accessed on 5 January 2022).
2.3. Data Sources
An exhaustive review of all the available data, up until December 2021, on each rare missense variant in the TTN gene was performed independently by three authors; afterwards, the reviews were compared and verified. Data were collected from the Human Gene Mutation Database (HGMD) (www.hgmd.org, accessed on 5 January 2022), ClinVar (www.ncbi.nlm.nih.gov/clinvar/intro/, accessed on 5 January 2022), the National Center for Biotechnology Information single-nucleotide polymorphism (SNP) database (www.ncbi.nlm.nih.gov/SNP, accessed on 5 January 2022), Index Copernicus (www.en.indexcopernicus.com), Google Scholar (www.scholar.google.es, accessed on 5 January 2022), Springer Link (www.link.springer.com, accessed on 5 January 2022), Science Direct (www.sciencedirect.com, accessed on 5 January 2022), the Excerpta Medica Database (www.elsevier.com/solutions/embase-biomedical-research, accessed on 5 January 2022), and the IEEE Xplore Digital Library (www.ieeexplore.ieee.org/Xplore/home.jsp, accessed on 5 January 2022).
Classification/Interpretation
All rare missense variants in TTN were classified following both the clinical [17] and genetic [18] recommendations available up until 2015. Currently, all the rare missense variants in TTN are classified following the ACMG standards and guidelines [5], including updates; the PM2 item in the ACMG classification was considered fulfilled if the MAF in the relevant population databases was ≤0.01% [19]. The vast majority of reported pathogenic variants in IASs are very rare (MAF < 0.005%) [20], consistent with all the missense TTN variants currently classified as likely pathogenic/pathogenic (LP/P), showing an extremely rare frequency (MAF < 0.001%) (www.gnomad.broadinstitute.org/ and www.clinicalgenome.org/, accessed on 5 January 2022). The variants classified by 2021 as VUS, supported by the available current MAF and a definite association with TTN disease, VUS-likely benign (VUS-LB) (MAF > 0.001%, and no definite TTN IAS association) and VUS-LP (no MAF or MAF < 0.001% and certain TTN IAS association) subgroups, were studied to clarify their potentially ambiguous role in clinical practice. Genetic data were independently evaluated and classified by five authors, specialists in the genetics of inherited arrhythmias, to avoid bias. All investigators agreed on a final classification of all the rare missense TTN variants included in this study.
3. Results
3.1. Cohort
Our retrospective study included 193 patients with a definitive clinical diagnosis of any IAS: 96 cases of HCM (49.74%), 30 cases of DCM (15.54%), 22 cases of LQTS (11.39%), 19 cases of BrS (9.84%), 17 cases of ARVC (8.8%), 5 cases of CPVT (2.59%), 2 cases of SQTS (1.03%), and 2 cases of LVNC (1.03%) (Table 1 and Table 2). Patients were diagnosed between 2011 and 2015, according to the clinical guidelines available at the time. The analysis of the clinical data has not changed, and IAS was definitively diagnosed.
Table 1.
Genetic data of rare missense TTN variants. ACMG American College of Medical Genetics and Genomics, ARVC arrhythmogenic right ventricular tachycardia, B benign, BrS Brugada syndrome, ClinVar Clinical Variant, CPVT catecholaminergic polymorphic ventricular tachycardia, dbSNP Single Nucleotide Polymorphism database, DCM dilated cardiomyopathy, EVS Exome Variant Server, GnomAD Genome Aggregation Database, HCM hypertrophic cardiomyopathy, LB likely benign, LQTS long QT syndrome, LP likely pathogenic, LVNC left ventricular non-compacted, NA not available, P pathogenic, SQTS short QT syndrome, VUS variant of uncertain significance.
| Disease | Exon | Nucleotide | Protein | dbSNP/ ClinVar -2015- |
EVS -2015- (EA/AA/All) (%) | dbSNP/ ClinVar -2021- |
GnomAD -2021- Alleles (%) | 2021 Score |
|---|---|---|---|---|---|---|---|---|
| BrS | 17 | c.3100G>A | p.Val1034Met | rs142951505/VUS | 0.1163/0.0/0.0769 | rs142951505/B | 203/250400 (0.081%) | B |
| BrS | 37 | c.8938G>A | p.Ala2980Thr | rs72647885/VUS | 0.0/0.3177/0.1076 | rs72647885 /LB |
93/250936 (0.037%) | B |
| BrS | 77 | c.19712G>A | p.Arg6571Gln | rs372804810 | 0.0122/0.0/0.0084 | rs372804810/VUS | 10/248156 (0.004%) | VUS-LB |
| BrS | 88 | c.22940A>G | p.Asn7647Ser | rs146057575/VUS | 0.0121/0.0/0.0083 | rs146057575/LB | 91/248780 (0.03%) | B |
| BrS | 138 | c.30010G>A | p.Val10004Ile | Novel | NA | rs996345107/VUS | NA | VUS |
| BrS | 222 | c.44643C>A | p.Asn14881Lys | Novel | NA | rs1165942763 | 4/246250 (0.001%) | VUS |
| BrS | 231 | c.47350G>A | p.Val15784Ile | Novel | NA | rs769334194 | 7/248094 (0.002%) | VUS |
| BrS | 244 | c.50303A>T | p.Asn16768Ile | rs371851242 | 0.0121/0.0/0.0083 | rs371851242 | 8/248688 (0.003%) | VUS |
| BrS | 252 | c.53992G>A | p.Val17998Ile | Novel | NA | rs764777213/VUS | 19/247880 (0.007%) | LB |
| BrS | 274 | c.68941G>A | p.Gly22981Ser | rs181166140 | T = 0.0014/2 | rs181166140/B | 91/247998 (0.03%) | B |
| BrS | 274 | c.71630G>A | p.Arg23877His | Novel | NA | rs764254441/LB | 10/248100 (0.004%) | VUS-LB |
| BrS | 274 | c.71887G>A | p.Glu23963Lys | Novel | NA | rs772211147/VUS | 2/248206 (0.0008%) | VUS |
| BrS | 274 | c.74000T>C | p.Leu24667Pro | Novel | NA | Novel | NA | VUS |
| BrS | 274 | c.75148A>T | p.Thr25050Ser | Novel | NA | Novel | NA | VUS |
| BrS | 274 | c.75359G>A | p.Arg25120His | rs185002960 | 0.048/0.076/0.057 | rs185002960/B | 853/248882 (0.3%) | B |
| BrS | 274 | c.78119T>C | p.Leu26040Pro | Novel | NA | Novel | NA | VUS |
| BrS | 277 | c.80297A>G | p.Asp26766Gly | Novel | NA | Novel | NA | VUS |
| BrS | 279 | c.80711T>C | p.Ile26904Thr | Novel | NA | Novel | NA | VUS |
| BrS | 306 | c.98638C>T | p.Arg32880Trp | Novel | NA | rs530453291/LB | 11/247068 (0.004%) | VUS-LB |
| CPVT | 50 | c.12278A>G | p.Asn4093Ser | Novel | NA | rs752924679/ VUS |
15/246972 (0.006%) | LB |
| CPVT | 81 | c.20890G>A | p.Glu6964Lys | rs190192954 | 0.0/0.051/0.016 | rs190192954/ VUS |
11/248320 (0.004%) | LB |
| CPVT | 121 | c.28322C>T | p.Ala9441Val | Novel | NA | rs794729407/ VUS |
NA | VUS |
| CPVT | 172 | c.33199G>A | p.Ala11067Thr | rs191699632 | 0.0/0.110/0.03 | rs191699632/ VUS |
10/156128 (0.006%) | LB |
| CPVT | 287 | c.84973A>G | p.Lys28325Glu | rs370541682 | 0.0/0.025/0.008 | rs370541682/ VUS |
7/248784 (0.002%) | VUS-LB |
| LQT | 23 | c.4090G>A | p.Ala1364Thr | Novel | NA | rs1171035485 | 5/250708 (0.0019%) | VUS |
| LQT | 104 | c.26753C>T | p.Thr8918Met | rs200593368/VUS | 0.12/0.075/0.105 | rs200593368/ LB |
105/249288 (0.042%) | B |
| LQT | 176 | c.34216G>A | p.Val11406Ile | rs373881831/VUS | 0.0121/0.026/0.016 | rs373881831/ VUS |
10/248362 (0.004%) | VUS-LB |
| LQT | 179 | c.34820T>G | p.Phe11607Cys | Novel | NA | rs575813993 | 12/248322 (0.004%) | VUS-LB |
| LQT | 179 | c.34952G>A | p.Ser11651Asn | Novel | NA | rs748928207 | 1/248508 (0.0004%) | VUS |
| LQT | 180 | c.35188G>A | p.Glu11730Lys | Novel | NA | rs778634417/ VUS |
4/248512 (0.001%) | VUS |
| LQT | 230 | c.46697A>G | p.Asn15566Ser | Novel | NA | rs971483896 | NA | VUS |
| LQT | 242 | c.49774G>A | p.Val16592Ile | rs200778464/VUS | 0.0122/0.026/0.016 | rs200778464/ VUS |
23/247908 (0.009%) | B |
| LQT | 249 | c.51698G>A | p.Gly17233Asp | rs202206216 | NA | rs202206216/ LB |
46/248146 (0.01%) | B |
| LQT | 252 | c.53434C>A | p.Leu17812Met | rs201167216 | 0.0/0.262/0.083 | rs201167216/ LB |
67/247466 (0,02%) | B |
| LQT | 252 | c.54868A>G | p.Thr18290Ala | rs200689750/VUS | 0.109/0.078/0.099 | rs200689750/ LB |
101/248280 (0.04%) | B |
| LQT | 253 | c.55552G>A | p.Gly18518Arg | Novel | NA | rs755518802 | 1/247294 (0.0004%) | VUS |
| LQT | 261 | c.57988A>C | p.Lys19330Gln | Novel | NA | rs775738687 | 3/245480 (0.001%) | VUS |
| LQT | 264 | c.58988G>A | p.Arg19663His | rs200971254/VUS | 0.0/0.28/0.0901 | rs200971254/ LB |
85/247816 (0.03%) | B |
| LQT | 265 | c.59276A>G | p.Tyr19759Cys | Novel | NA | rs757701692 | 1/248264 (0.0004%) | VUS |
| LQT | 272 | c.61160G>C | p.Gly20387Ala | rs201381085 | 0.0366/0.0/0.0252 | rs201381085/ LB |
63/243090 (0.02%) | B |
| LQT | 274 | c.62185A>G | p.Arg20729Gly | Novel | NA | rs779474257 | 1/246798 (0.0004%) | VUS |
| LQT | 274 | c.67908A>T | p.Arg22636Ser | Novel | NA | Novel | NA | VUS |
| LQT | 274 | c.78614C>T | p.Thr26205Ile | Novel | NA | Novel | NA | VUS |
| LQT | 306 | c.94583C>A | p.Thr31528Asn | rs375002174 | 0.0121/0.0/0.0082 | rs375002174/ VUS |
13/247980 (0.005%) | LB |
| LQT | 306 | c.95173A>G | p.Lys31725Glu | rs72629783/VUS | 0.0242/0.0/0.0166 | rs72629783/ LB |
54/249004 (0.02%) | B |
| LQT | 310 | c.99813T>G | p.Ser33271Arg | Novel | NA | rs776981475/ VUS |
6/248908 (0.002%) | VUS-LB |
| SQT | 2 | c.208G>A | p.Val70Met | Novel | NA | rs772248060/ VUS |
12/251358 (0.0047%) | LB |
| SQT | 311 | c.100160C>T | p.Thr33387Ile | Novel | NA | rs370267738/ VUS |
NA | VUS |
| ARVC | 2 | c.289G>A | p.Val97Met | rs185921345 | 0.034/0.022/0.030 | rs185921345/ B |
599/251394 (0.23%) | B |
| ARVC | 27 | c.5138G>A | p.Arg1713Lys | Novel | NA | rs890589931 | NA | VUS-LP |
| ARVC | 33 | c.8095T>G | p.Ser2699Ala | rs373857878 | 0.011/0.0/0.007 | rs373857878/ VUS |
2/251202 (0.00079%) | VUS-LP |
| ARVC | 46 | c.10754A>C | p.Gln3585Pro | rs375177753 | 0.024/0.0/0.016 | rs375177753/ VUS |
28/248890 (0.011%) | LB |
| ARVC | 202 | c.40051T>C | p.Tyr13351His | Novel | NA | Novel | NA | VUS-LP |
| ARVC | 210 | c.41606T>A | p.Val13869Asp | Novel | NA | rs767768313/ VUS |
5/247480 (0.002%) | VUS |
| ARVC | 211 | c.41709G>T | p.Trp13903Cys | rs202094100/VUS | 0.0839/0.0/0.057 | rs202094100/ B |
86/247226 (0.03%) | B |
| ARVC | 217 | c.43010G>A | p.Arg14337His | rs191539637 | 0.060/0.0/0.041 | rs191539637/ B |
97/247328 (0.03%) | B |
| ARVC | 224 | c.45148C>T | p.Arg15050Cys | rs201213901/VUS | 0.191/0.0/0.129 | rs201213901/ B |
245/248616 (0.09%) | B |
| ARVC | 257 | c.56694G>T | p.Leu18898Phe | Novel | NA | rs1194410087 | 1/214938 (0.0004%) | VUS-LP |
| ARVC | 257 | c.56950A>G | p.Ile18984Val | rs201247592 | 0.0/0.133/0.041 | rs201247592/ LB |
27/243056 (0.01%) | B |
| ARVC | 274 | c.65321T>C | p.Ile21774Thr | Novel | NA | Novel | NA | VUS-LP |
| ARVC | 274 | c.66898A>G | p.Ile22300Val | rs72646898 | 0.024/0.0/0.016 | rs72646898/ LB |
58/247298 (0.02%) | B |
| ARVC | 274 | c.75560A>G | p.Lys25187Arg | Novel | NA | Novel | NA | VUS-LP |
| ARVC | 278 | c.80386G>A | p.Gly26796Ser | rs183013408/VUS | 0.048/0.0/0.032 | rs183013408/ LB |
99/248324 (0.03%) | B |
| ARVC | 282 | c.81625G>A | p.Glu27209Lys | Novel | NA | Novel | NA | VUS-LP |
| ARVC | 295 | c.88720G>A | p.Val29574Ile | Novel | NA | rs779663332/ VUS |
20/246836 (0.008%) | LB |
| DCM | 15 | c.2629C>A | p.Pro877Thr | Novel | NA | rs751640052/ VUS |
NA | VUS-LP |
| DCM | 28 | c.6587G>A | p.Cys2196Tyr | Novel | NA | rs878854326/ VUS |
1/250806 (0.00039%) | VUS-LP |
| DCM | 38 | c.9247G>A | p.Glu3083Lys | Novel | NA | Novel | NA | VUS-LP |
| DCM | 49 | c.11902G>A | p.Gly3968Arg | rs377754692 | 0.01/0,0/0,01 | rs377754692/ VUS |
7/248948 (0.0028%) | VUS |
| DCM | 55 | c.13484C>T | p.Thr4495Ile | Novel | NA | rs751087281/ VUS |
NA | VUS-LP |
| DCM | 81 | c.20920A>G | p.Ser6974Gly | rs72648980/VUS | 0.0/0.7105/0.2241 | rs72648980/ LB |
155/248460 (0.06%) | B |
| DCM | 113 | c.27659G>A | p.Arg9220Gln | Novel | NA | rs727504757/ VUS |
6/174408 (0.003%) | VUS |
| DCM | 155 | c.31802A>C | p.Lys10601Thr | rs371380876 | 0.0123/0.0/0.0085 | rs371380876/ VUS |
12/248564 (0.004%) | VUS-LB |
| DCM | 206 | c.40614G>A | p.Met13538Ile | Novel | NA | rs762621076 | 2/245318 (0.0008%) | VUS-LP |
| DCM | 225 | c.45392G>A | p.Arg15131His | rs72646808/VUS | 0.3044/0.0533/0.2256 | rs72646808/ B |
457/247686 (0.1%) | B |
| DCM | 228 | c.46444C>G | p.Arg15482Gly | Novel | NA | rs55734111/ VUS |
NA | VUS-LP |
| DCM | 237 | c.48559C>T | p.Arg16187Cys | rs143659933 | 0.0121/0.0/0.0082 | rs143659933 | 11/247830 (0.004%) | VUS-LB |
| DCM | 242 | c.49738A>G | p.Met16580Val | rs188185141 | 0.0603/0.0/0.0411 | rs188185141/ LB |
123/248042 (0.04%) | B |
| DCM | 249 | c.51830G>A | p.Arg17277His | rs201457934 | 0.0122/0.0/0.0083 | rs201457934/ LB |
26/248278 (0.01%) | B |
| DCM | 261 | c.57893G>A | p.Ser19298Asn | Novel | NA | Novel | NA | VUS-LP |
| DCM | 261 | c.57968C>T | p.Pro19323Leu | Novel | NA | rs397517662/ VUS |
12/246570 (0.004%) | VUS-LB |
| DCM | 264 | c.58943G>A | p.Arg19648His | Novel | NA | Novel rs753717922 | 7/247984 (0.002%) | VUS |
| DCM | 274 | c.62042T>G | p.Val20681Gly | Novel | NA | Novel | NA | VUS-LP |
| DCM | 274 | c.65012T>A | p.Met21671Lys | Novel | NA | rs750298083/ VUS |
8/175928 (0.004%) | VUS |
| DCM | 274 | c.73195G>A | p.Val24399Ile | Novel | NA | rs1257567608 | NA | VUS-LP |
| DCM | 274 | c.73501C>G | p.Pro24501Ala | Novel | NA | rs770542451 | 1/246988 (0.0004%) | VUS-LP |
| DCM | 274 | c.73967A>G | p.Asn24656Ser | rs368443217/VUS | 0.0122/0.0/0.0084 | rs368443217/ VUS |
20/243932 (0.008%) | B |
| DCM | 274 | c.74942C>T | p.Ala24981Val | Novel | NA | rs749950083 | NA | VUS-LP |
| DCM | 274 | c.78758G>C | p.Gly26253Ala | Novel | NA | Novel | NA | VUS-LP |
| DCM | 277 | c.80081T>C | p.Val26694Ala | Novel | NA | rs1053653300/VUS | 2/248614 (0.0008%) | VUS-LP |
| DCM | 283 | c.81985C>T | p.Leu27329Phe | rs180798672 | A=0.0005/1 | rs180798672/ VUS |
5/248826 (0.002%) | VUS |
| DCM | 291 | c.87711C>A | p.Phe29237Leu | Novel | NA | rs587780983/ VUS |
10/248118 (0.004%) | VUS-LB |
| DCM | 306 | c.95905C>T | p.Arg31969Trp | Novel | NA | rs753957126 | 14/249090 (0.005%) | VUS-LB |
| DCM | 306 | c.96068G>A | p.Arg32023Gln | Novel | NA | rs778021095/ VUS |
3/248866 (0.001%) | VUS-LP |
| DCM | 306 | c.98408A>G | p.Asn32803Ser | Novel | NA | rs1200663369 | 1/248986 (0.0004%) | VUS-LP |
| HCM | 5 | c.758C>T | p.Thr253Ile | Novel | NA | rs945095401 | NA | VUS-LP |
| HCM | 15 | c.2611G>T | p.Val871Leu | rs72647861 | NA | rs72647861/ VUS |
18/251176 (0.0071%) | LB |
| HCM | 19 | c.3241G>A | p.Ala1081Thr | rs55914517/VUS | 0.1047/0.0454/0.0846 | rs55914517/ LB |
168/251122 (0.069%) | B |
| HCM | 21 | c.3716A>G | p.Tyr1239Cys | Novel | NA | rs794729569 | 2/250772 (0.0007%) | VUS-LP |
| HCM | 24 | c.4396T>C | p.Phe1466Leu | rs151310601 | 0.1279/0.0681/0.1076 | rs151310601/ LB |
173/250944 (0.068%) | B |
| HCM | 27 | c.5740G>A | p.Ala1914Thr | rs118161093 | T=0.0009/2 | rs118161093/ LB |
35/251134 (0.013%) | B |
| HCM | 37 | c.8920A>G | p.Met2974Val | Novel | NA | rs993689796 | NA | VUS-LP |
| HCM | 43 | c.10162C>T | p.Arg3388Trp | Novel | NA | rs758680640/ LB |
5/250482 (0.0019%) | VUS |
| HCM | 49 | c.12014A>G | p.Gln4005Arg | Novel | NA | Novel | NA | VUS-LP |
| HCM | 51 | c.12581A>G | p.Gln4194Arg | rs190636272/VUS | 0.0964/0.0254/0.0736 | rs190636272/ LB |
147/247774 (0.059%) | B |
| HCM | 52 | c.12814G>T | p.Ala4272Ser | rs72648940/VUS | 0.1339/0.0/0.092 | rs72648940/ LB |
132/248410 (0.053%) | B |
| HCM | 53 | c.13075C>G | p.Leu4359Val | Novel | NA | Novel | NA | VUS-LP |
| HCM | 62 | c.15418C>A | p.Pro5140Thr | rs72648953/VUS | 0.2188/0.0267/0.1586 | rs72648953/ B |
738/242524 (0.3%) | B |
| HCM | 66 | c.16609G>A | p.Glu5537Lys | rs72648958/VUS | 0.1461/0.0/0.1003 | rs72648958/ LB |
248/245308 (0.1%) | B |
| HCM | 77 | c.19723G>C | p.Glu6575Gln | rs201420077 | 0.1097/0.0/0.0755 | rs201420077/ LB |
45/248270 (0.018%) | B |
| HCM | 78 | c.19943T>G | p.Ile6648Arg | rs201181445 | 0.0122/0.0/0.0084 | rs201181445 | 4/244710 (0.001%) | VUS-LP |
| HCM | 78 | c.20030A>G | p.Lys6677Arg | Novel | NA | Novel | NA | VUS-LP |
| HCM | 80 | c.20375C>T | p.Ser6792Leu | rs200598509/VUS | 0.0242/0.0/0.0165 | rs200598509/ VUS |
18/248776 (0.0072%) | LB |
| HCM | 81 | c.20887G>A | p.Asp6963Asn | Novel | NA | rs756813056/ VUS |
8/248344 (0.003%) | VUS-LB |
| HCM | 82 | c.21220G>A | p.Val7074Ile | rs200103997 | 0.0847/0.0256/0.0658 | rs200103997/ LB |
146/248762 (0.058%) | B |
| HCM | 88 | c.22762A>G | p.Ile7588Val | rs72648989 | 0.0981/0.0/0.0678 | rs72648989/ LB |
190/245890 (0.07%) | B |
| HCM | 89 | c.23203A>C | p.Asn7735His | rs376982715/VUS | 0.012/0.0/0.0082 | rs376982715/ LB |
34/248706 (0.013%) | B |
| HCM | 96 | c.25117G>A | p.Glu8373Lys | rs144025230 | T=0.0009/2 | rs144025230/ VUS |
10/249144 (0.004%) | LB |
| HCM | 98 | c.25582G>A | p.Val8528Met | Novel | NA | rs563073635/ LB |
14/249214 (0.0056%) | LB |
| HCM | 114 | c.27707A>C | p.His9236Pro | Novel | NA | Novel | NA | VUS-LP |
| HCM | 119 | c.28179A>C | p.Glu9393Asp | Novel | NA | Novel | NA | VUS-LP |
| HCM | 127 | c.28888G>A | p.Glu9630Lys | Novel | NA | Novel | NA | VUS-LP |
| HCM | 138 | c.29974C>T | p.Leu9992Phe | Novel | NA | rs967680270 | NA | VUS-LP |
| HCM | 138 | c.30040G>A | p.Glu10014Lys | Novel | NA | rs778130826 | 1/240058 (0.0004%) | VUS-LP |
| HCM | 152 | c.31502C>T | p.Pro10501Leu | Novel | NA | Novel | NA | VUS-LP |
| HCM | 153 | c.46630G>C | p.Ala15544Pro | Novel | NA | Novel | NA | VUS-LP |
| HCM | 184 | c.35996G>C | p.Arg11999Thr | Novel | NA | rs555652524 | 3/240444 (0.001%) | VUS-LP |
| HCM | 197 | c.38683G>A | p.Gly12895Arg | rs200042932/VUS | 0.0365/0.0271/0.0336 | rs200042932/ LB |
72/246698 (0.02%) | B |
| HCM | 204 | c.40364C>T | p.Ser13455Phe | Novel | NA | rs949595987 | NA | VUS-LP |
| HCM | 210 | c.41471C>A | p.Ala13824Glu | Novel | NA | rs750310775/ VUS |
1/247347 (0.0004%) | VUS-LP |
| HCM | 227 | c.45932T>C | p.Ile15311Thr | Novel | NA | rs760585965 | 4/246104 (0.001%) | VUS-LP |
| HCM | 230 | c.46801C>A | p.Pro15601Thr | Novel | NA | Novel | NA | VUS-LP |
| HCM | 230 | c.47036T>C | p.Met15679Thr | rs200585270 | NA | rs200585270/ LB |
80/248418 (0.03%) | B |
| HCM | 234 | c.47651C>G | p.Ser15884Cys | Novel | NA | rs567515550 | 3/244826 (0.001%) | VUS-LP |
| HCM | 240 | c.49369G>A | p.Val16457Ile | rs181957743 | T=0.0009/2 | rs181957743/ LB |
45/244382 (0.01%) | B |
| HCM | 245 | c.50516C>G | p.Thr16839Ser | Novel | NA | Novel | NA | VUS-LP |
| HCM | 245 | c.50693G>C | p.Gly16898Ala | rs201922910 | 0.0361/0.0/0.0245 | rs201922910/ LB |
38/248508 (0.01%) | B |
| HCM | 249 | c.51661G>A | p.Asp17221Asn | Novel | NA | rs1352997069/VUS | 9/247586 (0.003%) | VUS-LB |
| HCM | 250 | c.51953T>G | p.Val17318Gly | Novel | NA | rs755949982/ VUS |
13/243972 (0.005%) | LB |
| HCM | 251 | c.52301A>G | p.Asp17434Gly | rs199512049/VUS | 0.1099/0.0/0.0756 | rs199512049/ LB |
149/248058 (0.06%) | B |
| HCM | 252 | c.52562T>C | p.Val17521Ala | Novel | NA | rs886039085/ VUS |
4/227726 (0.001%) | VUS-LP |
| HCM | 252 | c.53272G>A | p.Ala17758Thr | rs370995867 | 0.0121/0.0/0.0083 | rs370995867/ LB |
30/246926 (0.01%) | B |
| HCM | 252 | c.55343G>C | p.Ser18448Thr | Novel | NA | rs1169239147/VUS | NA | VUS-LP |
| HCM | 254 | c.55885A>G | p.Ile18629Val | rs72646855/VUS | 0.0726/0.0258/0.0577 | rs72646855/ LB |
144/247742 (0.05%) | B |
| HCM | 262 | c.58382G>A | p.Arg19461His | rs72646868/VUS | 0.0122/0.0/0.0083 | rs72646868/ VUS |
13/247276 (0.005%) | LB |
| HCM | 263 | c.58522T>A | p.Trp19508Arg | Novel | NA | Novel | NA | VUS-LP |
| HCM | 264 | c.58946T>G | p.Phe19649Cys | Novel | NA | rs764330098/ VUS |
2/248030 (0.0008%) | VUS-LP |
| HCM | 267 | c.59659G>A | p.Val19887Ile | Novel | NA | rs759848349 | 4/246976 (0.001%) | VUS-LP |
| HCM | 267 | c.59894C>T | p.Thr19965Ile | Novel | NA | Novel | NA | VUS-LP |
| HCM | 268 | c.60393G>C | p.Gln20131His | Novel | NA | rs727504520/ VUS |
13/248510 (0.005%) | LB |
| HCM | 269 | c.60596C>T | p.Pro20199Leu | Novel | NA | rs775953148/ VUS |
7/246890 (0.002%) | VUS-LB |
| HCM | 274 | c.62924A>T | p.Asp20975Val | Novel | NA | Novel | NA | VUS-LP |
| HCM | 274 | c.63604G>A | p.Asp21202Asn | Novel | NA | rs747260758 | 14/246618 (0.005%) | LB |
| HCM | 274 | c.63644G>A | p.Arg21215His | rs145504744 | T=0.0009/2 | rs145504744/ VUS |
11/246464 (0.004%) | VUS |
| HCM | 274 | c.64831G>A | p.Val21611Ile | Novel | NA | rs1400549364 | 1/168286 (0.0005%) | VUS-LP |
| HCM | 274 | c.68123C>A | p.Thr22708Asn | Novel | NA | Novel | NA | VUS-LP |
| HCM | 274 | c.69946G>A | p.Glu23316Lys | Novel | NA | rs747699837 | 4/248188 (0.001%) | VUS-LP |
| HCM | 274 | c.70579C>G | p.Pro23527Ala | Novel | NA | rs912485960 | 1/248792 (0.0004%) | VUS-LP |
| HCM | 274 | c.70972G>C | p.Glu23658Gln | Novel | NA | rs754878387/ LB |
18/248186 (0.007%) | LB |
| HCM | 274 | c.71161C>T | p.Pro23721Ser | Novel | NA | rs1030641242 | NA | VUS-LP |
| HCM | 274 | c.72422T>C | p.Ile24141Thr | Novel | NA | rs757763969 | 1/248214 (0.0004%) | VUS-LP |
| HCM | 274 | c.72559T>C | p.Phe24187Leu | rs200181804/VUS | 0.0243/0.0/0.0166 | rs200181804/ LB |
36/248728 (0.01%) | B |
| HCM | 274 | c.72722G>A | p.Gly24241Asp | Novel | NA | rs532522359 | 7/248186 (0.002%) | VUS-LB |
| HCM | 274 | c.72997A>G | p.Ile24333Val | rs201562505 | 0.1/0/0.09 | rs201562505/ B |
403/248414 (0.1%) | B |
| HCM | 274 | c.73675A>C | p.Ile24559Leu | Novel | NA | rs1325322358 | 1/245806 (0.0004%) | VUS-LP |
| HCM | 274 | c.74571G>C | p.Gln24857His | Novel | NA | Novel | NA | VUS-LP |
| HCM | 274 | c.75283T>G | p.Ser25095Ala | Novel | NA | Novel | NA | VUS-LP |
| HCM | 274 | c.75465G>T | p.Glu25155Asp | Novel | NA | Novel | NA | VUS-LP |
| HCM | 274 | c.76732G>A | p.Val25578Ile | rs371366196 | 0.0243/0.0/0.0166 | rs371366196/ VUS |
10/245028 (0.004%) | LB |
| HCM | 274 | c.77758G>A | p.Val25920Ile | rs377264123 | 0.012/0.0/0.0082 | rs377264123/ LB |
17/248104 (0.006%) | LB |
| HCM | 282 | c.81646G>T | p.Val27216Phe | Novel | NA | Novel | NA | VUS-LP |
| HCM | 286 | c.84247C>T | p.Pro28083Ser | Novel | NA | rs927540266 | NA | VUS-LP |
| HCM | 287 | c.85076T>A | p.Ile28359Lys | Novel | NA | rs531432790/ LB |
157/248510 (0.06%) | B |
| HCM | 287 | c.85081G>A | p.Ala28361Thr | Novel | NA | rs1192451781/VUS | 1/248372 (0.0004%) | VUS-LP |
| HCM | 287 | c.85301G>T | p.Ser28434Ile | rs180975448 | 0.0846/0.0/0.0575 | rs180975448/ LB |
159/248444 (0.06%) | B |
| HCM | 287 | c.85877A>G | p.Tyr28626Cys | Novel | NA | rs543223589/ VUS |
12/248882 (0.004%) | LB |
| HCM | 288 | c.86578C>T | p.Arg28860Cys | Novel | NA | rs190282707/ VUS |
6/248622 (0.002%) | VUS-LB |
| HCM | 291 | c.87632T>C | p.Ile29211Thr | Novel | NA | rs1460359915/VUS | 1/248674 (0.0004%) | VUS-LP |
| HCM | 292 | c.87995T>A | p.Phe29332Tyr | Novel | NA | Novel | NA | VUS-LP |
| HCM | 295 | c.89159C>G | p.Pro29720Arg | Novel | NA | Novel | NA | VUS-LP |
| HCM | 296 | c.89236A>G | p.Lys29746Glu | Novel | NA | Novel | NA | VUS-LP |
| HCM | 296 | c.89395C>T | p.Arg29799Cys | rs202064385/VUS | 0.0243/0.0/0.0166 | rs202064385/ LB |
110/248626 (0.04%) | B |
| HCM | 299 | c.90118A>G | p.Arg30040Gly | Novel | NA | Novel | NA | VUS-LP |
| HCM | 303 | c.91623T>A | p.Asp30541Glu | Novel | NA | Novel | NA | VUS-LP |
| HCM | 304 | c.92444T>C | p.Val30815Ala | Novel | NA | rs1237202129 | NA | VUS-LP |
| HCM | 306 | c.93637A>G | p.Ile31213Val | Novel | NA | rs1029128193 | NA | VUS-LP |
| HCM | 306 | c.93829T>C | p.Tyr31277His | Novel | NA | rs1227515280/VUS | 1/248712 (0.0004%) | VUS-LP |
| HCM | 306 | c.94693A>C | p.Ile31565Leu | Novel | NA | rs1328046092 | 1/249066 (0.0004%) | VUS-LP |
| HCM | 306 | c.95708G>A | p.Arg31903Gln | rs149391616 | T = 0.0028/5 | rs149391616/ LB |
80/248208 (0.03%) | B |
| HCM | 306 | c.96710G>A | p.Arg32237Gln | rs115150240/VUS | 0.0121/0.0/0.0083 | rs115150240/ LB |
33/247298 (0.013%) | B |
| HCM | 308 | c.99357G>C | p.Lys33119Asn | Novel | NA | rs113190638 | 1/247918 (0.0004%) | VUS-LP |
| LVNC | 62 | c.15430G>A | p.Val5144Ile | Novel | NA | rs550617268/ LB |
12/246042 (0.0048%) | VUS-LB |
| LVNC | 274 | c.64442T>C | p.Leu21481Pro | rs56399205 | 0.0975/0.0266/0.0752 | rs56399205/ B |
140/248224 (0.05%) | B |
Table 2.
Ratio changes and disease. Rare missense TTN variants (percentage). ARVC arrhythmogenic right ventricular tachycardia, B benign, BrS Brugada syndrome, CPVT catecholaminergic polymorphic ventricular tachycardia, DCM dilated cardiomyopathy, HCM hypertrophic cardiomyopathy, LB likely benign, LQTS long QT syndrome, LP likely pathogenic, LVNC left ventricular non-compacted, NA not available, P pathogenic, SQTS short QT syndrome, VUS variant of uncertain significance.
| 2015 | 2021 | |||||
|---|---|---|---|---|---|---|
| VUS | B | LB | VUS-LB | VUS | VUS-LP | |
| BrS | 19 (9.84%) | 5 (2.59%) | 1 (0.51%) | 3 (1.55%) | 10 (5.18%) | 0 |
| CPVT | 5 (2.59%) | 0 | 3 (1.55%) | 1 (0.51%) | 1 (0.51%) | 0 |
| LQTS | 22 (11.39%) | 8 (4.14%) | 1 (0.51%) | 3 (1.55%) | 10 (5.18%) | 0 |
| SQTS | 2 (1.03%) | 0 | 1 (0.51%) | 0 | 1 (0.51%) | 0 |
| ARVC | 17 (8.8%) | 7 (3.62%) | 2 (1.03%) | 0 | 1 (0.51%) | 7 (3.62%) |
| DCM | 30 (15.54%) | 5 (2.59%) | 0 | 5 (2.59%) | 5 (2.59%) | 15 (7.77%) |
| HCM | 96 (49.74%) | 25 (12.95%) | 12 (6.21%) | 5 (2.59%) | 2 (1.03%) | 52 (26.94%) |
| LVNC | 2 (1.03%) | 1 (0.51%) | 0 | 1 (0.51%) | 0 | 0 |
| 193 | 51 (26.42%) | 20 (10.36%) | 18 (9.84%) | 30 (15.54%) | 74 (38.34%) | |
| 122 (63.21%) | ||||||
| 193 | ||||||
3.2. Reanalysis and Reclassification
In 2015, all the patients included in our study carried only one rare missense heterozygous variant in the TTN gene. No patients carried any other rare alteration (including copy number variants) in any of the genes analyzed at that time. All the variants were then classified with an ambiguous role in IASs, because of a lack of available data, leaving all the cases without a conclusive genetic diagnosis.
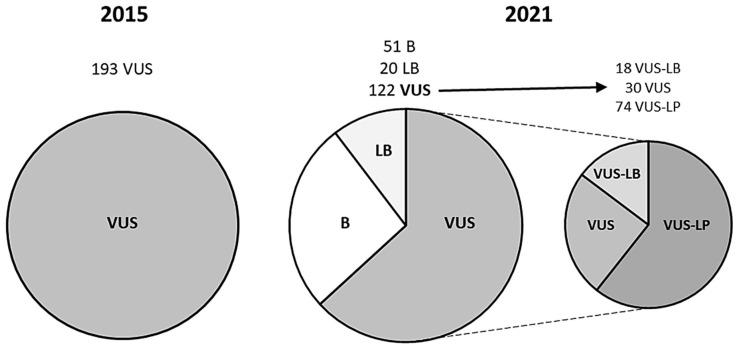
We performed a reanalysis that included all the available data and followed the ACMG guidelines. The reclassification showed that 71 (36.78%) rare missense TTN variants changed from their previous VUS classification. Twenty variants (10.36%) downgraded to LB, and 51 (26.42%) to benign (B). None of the rare missense variants in TTN upgraded to LP or P. Importantly, all the modifications of the first classification to LB and B were because of increased MAF from the previous classification to the present. Hence, 122 (63.22%) rare missense TTN variants remain classified as VUS strictly following the current ACMG guidelines (Table 1 and Table 2; Figure 1).
Figure 1.
Reclassification of rare missense TTN variants. B benign, LB likely benign, LP likely pathogenic, P pathogenic, VUS variant of uncertain significance.
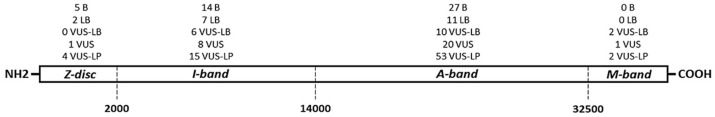
For the detection of MAF, 57 (29.53%) variants did not show any population frequency in the present analysis, in contrast to 125 (64.76%) in 2015. In addition, 36 (0.56%) variants showed a MAF of <0.001%, but none did in 2015. All rare missense TTN variants without a MAF, or with a very low MAF, identified during the present analysis, remained classified as VUS because of a lack of functional data or any other additional studies, which impedes a more accurate classification following the ACMG guidelines. We developed three subgroups within the VUS category. Eighteen (9.32%) variants were reclassified as VUS-LB and 74 (38.34%) as VUS-LP. The remaining 30 (15.54%) rare missense TTN variants remain as VUS (Table 1 and Table 2; Figure 1). In assessing the distribution of rare missense TTN variants throughout the gene, we identified that the variants were mainly located in the A-band zone of the TTN gene, especially the variants within the VUS-LP subgroup (53 variants, 27.46%) (Figure 2).
Figure 2.
Representative diagram of titin protein and missense variants analyzed. Titin extends from the Z-disk of the sarcomere (N-terminus) to the M-band (C-terminus). The central part of the protein contains the I-band region (I) and the A-band region (A). B benign, LB likely benign, LP likely pathogenic, P pathogenic, VUS variant of uncertain significance.
We also analyzed by the type of IAS. In 2015, we identified 19 and 22 VUS linked with BrS and LQTS patients, respectively; the current reanalysis indicated that 10 variants in each disease (52.63% and 45.45%, respectively) remain as VUS following the ACMG guidelines. All other variants decreased in their potential to be deleterious. For CPVT, SQTS, and LVNC, the present analysis classified most variants as having decreased potential to be deleterious compared with the previous classification in 2015. In contrast, the present study classified 7 (41.17%) of the 17 VUS in ARVC patients as having an increased potential to be deleterious (VUS-LP) compared with the previous classification in 2015. In DCM patients, of the 30 variants categorized as VUS in 2015, 15 (50%) increased to VUS-LP with the current classification. Similarly, in HCM patients, 96 variants were classified as VUS in 2015, and 52 (54.16%) of these variants increased to VUS-LP following the current ACMG guidelines (Table 1 and Table 2).
4. Discussion
An accurate genetic diagnosis is crucial before clinical translation in IASs [3,17] Early identification of a pathogenic genetic alteration is critical in therapeutic management, reducing the risk of malignant arrhythmias and SCD [4]. The current ACMG recommendations help in obtaining an accurate genetic classification [5], but implicit stringency and a lack of sufficient scientific data lead to many rare variants remaining categorized with an ambiguous role, especially in TTN, a large and relatively understudied gene. A VUS does not provide conclusive data of a cause of an IAS but also cannot be disregarded [16]. Continuous genetic and clinical improvements may change a previous classification, highlighting the importance of periodically revising and clarifying the roles of VUS [21]. To our knowledge, our study is the first reanalysis focused on rare missense TTN variants that were previously classified as VUS. In agreement with recent studies published by our group [14,16], IAS variants that were not classified following the recommendations at the time should be immediately reclassified, especially if they were previously interpreted as VUS.
In comparison to 2015, more than 30% of rare missense TTN variants changed from their previous VUS classification; all decreasing their ambiguous roles to benign roles, at least in IASs. It is important to note that despite no variants being reclassified as having increased pathogenic potential in IASs, the fact that previous VUS are now classified with a benign role is critical, as these variants can be disregarded as the main cause of disease in each patient. Therefore, reducing the ambiguity of a VUS is an important advantage of a comprehensive reanalysis. From a medico-legal point of view, regular reclassification of the variants can be considered as a standard of care; being a tool for the prevention of malpractice claims: an outdated classification may lead to the risk of missed diagnosis (if the pathogenic significance of a variant is not assessed, potentially life-saving preventive interventions are not indicated) or misdiagnosis (if a benign variant is misinterpreted, useless, expensive and potentially invasive preventive interventions can be indicated) [22]. However, no interpretation should be made on its possible role in phenotype modification, and no impact on clinical management is justified according to the current clinical guidelines [3,4]. Consequently, for each patient with an IAS, rare missense TTN variants should be comprehensively analyzed, considering all the available data, so that they can be properly interpreted in a personalized approach [23]. It should be mandatory to update MAF in public databases, which are periodically improved and may definitively change a previously ambiguous classification, as occurs in all of our reclassified rare missense TTN variants. This approach, focused on population frequency, could help in a quick reclassification on the part of rare variants in TTN, but also in other genes associated with rare diseases. However, further studies should be performed to confirm the appropriate method for each disease/gene.
In our study, most of the rare missense TTN variants remain classified as VUS (63.22%). To shed light on the ambiguity implicit in VUS, subgroups were categorized according to extremely rare MAF for definite pathogenic variants in IASs [20] and the definite association of TTN with IASs diagnosed in each patient. Disease-specific phenotypes significantly increase the accuracy of classification and reinforce the need for clinical data in genetic diagnoses, aiding VUS interpretation [12]. Hence, of the 122 (63.21%) rare missense TTN variants that remain as VUS, 9.32% potentially decrease their ambiguous role (VUS-LB), 15.54% remain as VUS, and 38.74% increase their potential pathogenic role (VUS-LP). Despite all the variants remaining as VUS following the ACMG guidelines, currently available data suggest, but do not confirm, a tendency towards a benign (VUS-LB) or deleterious (VUS-LP) role in IASs. It is crucial to state that our classification should also be performed in other centers, and in large cohorts, to corroborate these VUS sub-classifications. Importantly, no clinical translation and implementation/modification of therapeutic measures should be performed according to our suggested categorization of VUS-LB or VUS-LP. Functional studies, together with a complete family segregation, may corroborate the definite role of a rare missense TTN variant. Our classification, despite not being conclusive, may help to select families in order to perform functional studies, especially if focused on human induced pluripotent stem cell technology. Unfortunately, for the VUS analyzed in our study, neither a complete family segregation nor a functional analysis is available. These studies will be crucial to confer a real risk of rare missense TTN variants in IASs [24], distinguishing real pathogenic variants from the majority of VUS that will not play a causative role in IASs. Therefore, a lack of definitive data classifies all these variants as VUS, following the current ACMG guidelines.
4.1. Channelopathies
First, it is important to highlight the fact that few studies have suggested a potential role of rare TTN variants as phenotypic modifiers in BrS, LQTS, as well as sudden arrhythmogenic death syndrome (SADS) and sudden unexplained arrhythmogenic syndrome (SUDS) [25,26,27], but no definite causative association to any inherited channelopathy is widely accepted to date. Our results are in accordance with those suggesting the non-deleterious roles of rare missense TTN variants in inherited channelopathies. In BrS, LQTS, and SQTS, nearly 50% of rare missense TTN variants currently remain as VUS following the ACMG guidelines. All other variants decrease in their deleterious roles. For CPVT, most variants demonstrate a decrease in their potential deleterious roles or remain as VUS. Therefore, clinical translation of rare missense TTN variants in inherited channelopathies should be taken into account, as no conclusive deleterious role has been identified to date, but the variants that remain classified as VUS following the ACMG recommendations should not be disregarded until clarifying their roles.
4.2. Cardiomyopathies
Rare genetic alterations in the TTN gene are associated with inherited cardiomyopathies, mainly DCM [10,28]. However, most reported deleterious variants are “radical” (nonsense and indels) [9,29,30], and only a few studies have examined the pathogenicity of missense TTN variants in inherited cardiomyopathies [31,32,33,34,35]. The clinical role of rare missense TTN variants remains unclear because practically all reported missense TTN variants have been classified as VUS following the ACMG recommendations, because of a lack of additional clinical, genetic, and functional studies concerning no radical TTN variants. Hence, to our knowledge, no more than 30 rare missense TTN variants have been definitively classified as LP or P to date. In our cohorts of DCM patients, 50% of variants previously classified as VUS increased to VUS-LP, suggesting a potential pathogenicity despite a lack of data, which impedes the ability to obtain a definite role in clinical practice.
Patients diagnosed with HCM due to rare truncated TTN variants have been reported [36,37,38], but, as also occurs in other IASs, rare missense TTN variants have not been deeply analyzed and thus, currently remain as VUS following the ACMG recommendations. In our study, 54.16% of variants classified previously as VUS in HCM now increase to VUS-LP, suggesting potential pathogenicity. However, a lack of sufficient accurate data impedes the identification of a conclusive role that may help in clinical translation.
For ARVC, despite some articles suggesting that TTN variants potentially cause disease [39,40], a definite association with ARVC or as a phenotype modifier is a current matter of debate [41,42]. In our study, most rare missense TTN variants were reclassified as having no deleterious role in ARVC. However, 41.17% of variants that were previously classified as VUS showed a tendency to have a potentially damaging role. This potential pathogenic predisposition is because of the extremely rare frequency in global population databases, despite which no conclusive role can be assumed following the current ACMG recommendations. We believe that potentially deleterious roles should not be disregarded a priori for any of these rare missense TTN variants, and future studies may help to clarify if they have a role in ARVC.
In the past few years, some studies have focused on truncated TTN variants as potential causes of LVNC [43,44,45]. However, to our knowledge, no studies investigating the role of rare missense TTN variants in LVNC have been published to date. Our study showed that rare missense TTN variants previously classified as VUS in LVNC decrease to a benign role. Therefore, a definite association of rare missense TTN variants with LVNC should be confirmed, and its role in clinical translation should be interpreted with caution in each patient.
Limitations
The reclassification of rare missense TTN variants has some limitations that should be mentioned. Firstly, the patients included in our cohort may carry additional rare variants in other genes not included in our panel. Secondly, a lack of available functional and clinical data for many rare missense variants in TTN impedes a more accurate interpretation, causing many variants to remain categorized with ambiguous roles in IASs. Thirdly, there is a lack of family segregation, which is critical to clarify the role of a rare variant in IASs. Despite this fact, and taking into account that variant classification is subject to inherent intra- and interlaboratory differences [46], the use of available MAF helps to discern a high percentage of the genetic background that is not damaging in IASs, as our results show. However, to strongly support our results, additional studies should be performed in other IAS cohorts carrying missense variants in the TTN gene. Concerning the prevalence/incidence in population, it is also important to remark that cardiovascular diseases are the main cause of deaths worldwide [47]. Focusing on the younger population, IASs are the main current cause of death. Finally, the economic cost of a comprehensive reanalysis has not been yet assessed, as well as who should assume this cost. This is a controversial point that should be deeply analyzed because each country has a unique health system.
5. Conclusions
Limited studies focus on the clinical and functional data of missense variants in the TTN gene, and this lack of data impedes a proper variant classification, leading to many variants currently being classified as VUS. Updating the genetic data and the use of the ACMG criteria results in 36.78% of rare missense variants in TTN that were previously classified with an ambiguous role, changing their classification to having no deleterious roles. All the identified changes were mainly because of the new available data on global frequencies. Therefore, despite no conclusive damaging role having been established for any of the analyzed variants, the 38.34% of rare missense TTN variants remaining as VUS could play a potentially deleterious role in IASs, mainly in HCM. Further genotype-phenotype investigations should be performed in different diseases to decide the appropriate time period for a reanalysis of rare missense variants in the TTN gene.
Author Contributions
O.C., G.S.-B., A.O., J.B. and R.B. developed the concept; E.M.-B., O.C., A.F.-F., M.C., A.I., C.F.-C., S.C., J.C., E.A., A.P.-S., M.P., M.A., V.F., B.d.O., L.L., F.P., C.H., R.T. and S.G. acquired, pre-processed, and analyzed the data; Investigation, M.V.-P., J.C. and C.H.; O.C., E.M.-B. and G.S.-B. prepared the manuscript. O.C., A.O., J.B. and R.B. supervised the study. All authors contributed to the manuscript revision. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by Obra Social “La Caixa Foundation” (LCF/PR/GN16/50290001 and LCF/PR/GN19/50320002). CIBERCV is an initiative of the ISCIII, Spanish Ministry of Economy and Competitiveness. Funders had no role in the study design, data collection, data analysis, interpretation, or writing of the report.
Informed Consent Statement
Not applicable.
Conflicts of Interest
The authors declare that they have no competing interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Scrocco C., Bezzina C.R., Ackerman M.J., Behr E.R. Genetics and genomics of arrhythmic risk: Current and future strategies to prevent sudden cardiac death. Nat. Rev. Cardiol. 2021;18:774–784. doi: 10.1038/s41569-021-00555-y. [DOI] [PubMed] [Google Scholar]
- 2.Martinez H.R., Beasley G.S., Miller N., Goldberg J.F., Jefferies J.L. Clinical Insights into Heritable Cardiomyopathies. Front. Genet. 2021;12:663450. doi: 10.3389/fgene.2021.663450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Priori S.G., Blomstrom-Lundqvist C. 2015 European Society of Cardiology Guidelines for the management of patients with ventricular arrhythmias and the prevention of sudden cardiac death summarized by co-chairs. Eur. Heart J. 2015;36:2757–2759. doi: 10.1093/eurheartj/ehv316. [DOI] [PubMed] [Google Scholar]
- 4.Musunuru K., Hershberger R.E., Day S.M., Klinedinst N.J., Landstrom A.P., Parikh V.N., Prakash S., Semsarian C., Sturm A.C., American Heart Association Council on Genomic and Precision Medicine et al. Genetic Testing for Inherited Cardiovascular Diseases: A Scientific Statement from the American Heart Association. Circ. Genom. Precis. Med. 2020;13:e000067. doi: 10.1161/HCG.0000000000000067. [DOI] [PubMed] [Google Scholar]
- 5.Richards S., Aziz N., Bale S., Bick D., Das S., Gastier-Foster J., Grody W.W., Hegde M., Lyon E., Spector E., et al. Standards and guidelines for the interpretation of sequence variants: A joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet. Med. 2015;17:405–423. doi: 10.1038/gim.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Walsh R., Lahrouchi N., Tadros R., Kyndt F., Glinge C., Postema P.G., Amin A.S., Nannenberg E.A., Ware J.S., Whiffin N., et al. Enhancing rare variant interpretation in inherited arrhythmias through quantitative analysis of consortium disease cohorts and population controls. Genet. Med. 2021;23:47–58. doi: 10.1038/s41436-020-00946-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ghouse J., Skov M.W., Bigseth R.S., Ahlberg G., Kanters J.K., Olesen M.S. Distinguishing pathogenic mutations from background genetic noise in cardiology: The use of large genome databases for genetic interpretation. Clin. Genet. 2018;93:459–466. doi: 10.1111/cge.13066. [DOI] [PubMed] [Google Scholar]
- 8.Ackerman M.J. Genetic purgatory and the cardiac channelopathies: Exposing the variants of uncertain/unknown significance issue. Heart Rhythm. 2015;12:2325–2331. doi: 10.1016/j.hrthm.2015.07.002. [DOI] [PubMed] [Google Scholar]
- 9.Eldemire R., Tharp C.A., Taylor M.R.G., Sbaizero O., Mestroni L. The Sarcomeric Spring Protein Titin: Biophysical Properties, Molecular Mechanisms, and Genetic Mutations Associated with Heart Failure and Cardiomyopathy. Curr. Cardiol. Rep. 2021;23:121. doi: 10.1007/s11886-021-01550-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tharp C.A., Haywood M.E., Sbaizero O., Taylor M.R.G., Mestroni L. The Giant Protein Titin’s Role in Cardiomyopathy: Genetic, Transcriptional, and Post-translational Modifications of TTN and Their Contribution to Cardiac Disease. Front. Physiol. 2019;10:1436. doi: 10.3389/fphys.2019.01436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Denham N.C., Pearman C.M., Ding W.Y., Waktare J., Gupta D., Snowdon R., Hall M., Cooper R., Modi S., Todd D., et al. Systematic Re-evaluation of SCN5A Variants Associated with Brugada Syndrome. J. Cardiovasc. Electrophysiol. 2018;30:118–127. doi: 10.1111/jce.13740. [DOI] [PubMed] [Google Scholar]
- 12.Bennett J.S., Bernhardt M., McBride K.L., Reshmi S.C., Zmuda E., Kertesz N.J., Garg V., Fitzgerald-Butt S., Kamp A.N. Reclassification of Variants of Uncertain Significance in Children with Inherited Arrhythmia Syndromes is Predicted by Clinical Factors. Pediatr. Cardiol. 2019;40:1679–1687. doi: 10.1007/s00246-019-02203-2. [DOI] [PubMed] [Google Scholar]
- 13.Vallverdu-Prats M., Alcalde M., Sarquella-Brugada G., Cesar S., Arbelo E., Fernandez-Falgueras A., Coll M., Perez-Serra A., Puigmule M., Iglesias A., et al. Rare Variants Associated with Arrhythmogenic Cardiomyopathy: Reclassification Five Years Later. J. Pers. Med. 2021;11:162. doi: 10.3390/jpm11030162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sarquella-Brugada G., Fernandez-Falgueras A., Cesar S., Arbelo E., Coll M., Perez-Serra A., Puigmule M., Iglesias A., Alcalde M., Vallverdu-Prats M., et al. Clinical impact of rare variants associated with inherited channelopathies: A 5-year update. Hum. Genet. 2021 doi: 10.1007/s00439-021-02370-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen C.J., Lu T.P., Lin L.Y., Liu Y.B., Ho L.T., Huang H.C., Lai L.P., Hwang J.J., Yeh S.S., Wu C.K., et al. Impact of Ancestral Differences and Reassessment of the Classification of Previously Reported Pathogenic Variants in Patients with Brugada Syndrome in the Genomic Era: A SADS-TW BrS Registry. Front. Genet. 2018;9:680. doi: 10.3389/fgene.2018.00680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Campuzano O., Sarquella-Brugada G., Fernandez-Falgueras A., Coll M., Iglesias A., Ferrer-Costa C., Cesar S., Arbelo E., Garcia-Alvarez A., Jorda P., et al. Reanalysis and reclassification of rare genetic variants associated with inherited arrhythmogenic syndromes. EBioMedicine. 2020;54:102732. doi: 10.1016/j.ebiom.2020.102732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ackerman M.J., Priori S.G., Willems S., Berul C., Brugada R., Calkins H., Camm A.J., Ellinor P.T., Gollob M., Hamilton R., et al. HRS/EHRA expert consensus statement on the state of genetic testing for the channelopathies and cardiomyopathies: This document was developed as a partnership between the Heart Rhythm Society (HRS) and the European Heart Rhythm Association (EHRA) Europace. 2011;13:1077–1109. doi: 10.1093/europace/eur245. [DOI] [PubMed] [Google Scholar]
- 18.Richards C.S., Bale S., Bellissimo D.B., Das S., Grody W.W., Hegde M.R., Lyon E., Ward B.E. Molecular Subcommittee of the ALQAC: ACMG recommendations for standards for interpretation and reporting of sequence variations: Revisions 2007. Genet. Med. 2008;10:294–300. doi: 10.1097/GIM.0b013e31816b5cae. [DOI] [PubMed] [Google Scholar]
- 19.Kobayashi Y., Yang S., Nykamp K., Garcia J., Lincoln S.E., Topper S.E. Pathogenic variant burden in the ExAC database: An empirical approach to evaluating population data for clinical variant interpretation. Genome Med. 2017;9:13. doi: 10.1186/s13073-017-0403-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jordan D.M., Kiezun A., Baxter S.M., Agarwala V., Green R.C., Murray M.F., Pugh T., Lebo M.S., Rehm H.L., Funke B.H., et al. Development and validation of a computational method for assessment of missense variants in hypertrophic cardiomyopathy. Am. J. Hum. Genet. 2011;88:183–192. doi: 10.1016/j.ajhg.2011.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Salfati E.L., Spencer E.G., Topol S.E., Muse E.D., Rueda M., Lucas J.R., Wagner G.N., Campman S., Topol E.J., Torkamani A. Re-analysis of whole-exome sequencing data uncovers novel diagnostic variants and improves molecular diagnostic yields for sudden death and idiopathic diseases. Genome Med. 2019;11:83. doi: 10.1186/s13073-019-0702-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Grassi S., Campuzano O., Coll M., Brion M., Arena V., Iglesias A., Carracedo A., Brugada R., Oliva A. Genetic variants of uncertain significance: How to match scientific rigour and standard of proof in sudden cardiac death? Leg Med. 2020;45:101712. doi: 10.1016/j.legalmed.2020.101712. [DOI] [PubMed] [Google Scholar]
- 23.Eilbeck K., Quinlan A., Yandell M. Settling the score: Variant prioritization and Mendelian disease. Nat. Rev. Genet. 2017;18:599–612. doi: 10.1038/nrg.2017.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Blekhman R., Man O., Herrmann L., Boyko A.R., Indap A., Kosiol C., Bustamante C.D., Teshima K.M., Przeworski M. Natural selection on genes that underlie human disease susceptibility. Curr. Biol. 2008;18:883–889. doi: 10.1016/j.cub.2008.04.074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Campuzano O., Sarquella-Brugada G., Mademont-Soler I., Allegue C., Cesar S., Ferrer-Costa C., Coll M., Mates J., Iglesias A., Brugada J., et al. Identification of Genetic Alterations, as Causative Genetic Defects in Long QT Syndrome, Using Next Generation Sequencing Technology. PLoS ONE. 2014;9:e114894. doi: 10.1371/journal.pone.0114894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nunn L.M., Lopes L.R., Syrris P., Murphy C., Plagnol V., Firman E., Dalageorgou C., Zorio E., Domingo D., Murday V., et al. Diagnostic yield of molecular autopsy in patients with sudden arrhythmic death syndrome using targeted exome sequencing. Europace. 2016;18:888–896. doi: 10.1093/europace/euv285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Campuzano O., Sarquella-Brugada G., Fernandez-Falgueras A., Cesar S., Coll M., Mates J., Arbelo E., Perez-Serra A., Del Olmo B., Jorda P., et al. Genetic interpretation and clinical translation of minor genes related to Brugada syndrome. Hum. Mutat. 2019;40:749–764. doi: 10.1002/humu.23730. [DOI] [PubMed] [Google Scholar]
- 28.Azad A., Poloni G., Sontayananon N., Jiang H., Gehmlich K. The giant titin: How to evaluate its role in cardiomyopathies. J. Muscle Res. Cell Motil. 2019;40:159–167. doi: 10.1007/s10974-019-09518-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Herman D.S., Lam L., Taylor M.R., Wang L., Teekakirikul P., Christodoulou D., Conner L., DePalma S.R., McDonough B., Sparks E., et al. Truncations of titin causing dilated cardiomyopathy. N. Engl. J. Med. 2012;366:619–628. doi: 10.1056/NEJMoa1110186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jordan E., Peterson L., Ai T., Asatryan B., Bronicki L., Brown E., Celeghin R., Edwards M., Fan J., Ingles J., et al. An Evidence-Based Assessment of Genes in Dilated Cardiomyopathy. Circulation. 2021;144:7–19. doi: 10.1161/CIRCULATIONAHA.120.053033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Campuzano O., Sanchez-Molero O., Mademont-Soler I., Riuro H., Allegue C., Coll M., Perez-Serra A., Mates J., Pico F., Iglesias A., et al. Rare Titin (TTN) Variants in Diseases Associated with Sudden Cardiac Death. Int. J. Mol. Sci. 2015;16:25773–25787. doi: 10.3390/ijms161025773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Begay R.L., Graw S., Sinagra G., Merlo M., Slavov D., Gowan K., Jones K.L., Barbati G., Spezzacatene A., Brun F., et al. Role of Titin Missense Variants in Dilated Cardiomyopathy. J. Am. Heart Assoc. 2015;4:e002645. doi: 10.1161/JAHA.115.002645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hastings R., Villiers C.P.D., Hooper C., Ormondroyd L., Pagnamenta A., Lise S., Salatino S., Knight S.J., Taylor J.C., Thomson K.L., et al. Combination of Whole Genome Sequencing, Linkage, and Functional Studies Implicates a Missense Mutation in Titin as a Cause of Autosomal Dominant Cardiomyopathy with Features of Left Ventricular Noncompaction. Circ. Cardiovasc. Genet. 2016;9:426–435. doi: 10.1161/CIRCGENETICS.116.001431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Akinrinade O., Helio T., Deprez R.H.L., Jongbloed J.D.H., Boven L.G., Berg M.P.V.D., Pinto Y.M., Alastalo T.P., Myllykangas S., Spaendonck-Zwarts K.V., et al. Relevance of Titin Missense and Non-Frameshifting Insertions/Deletions Variants in Dilated Cardiomyopathy. Sci. Rep. 2019;9:4093. doi: 10.1038/s41598-019-39911-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jiang H., Hooper C., Kelly M., Steeples V., Simon J.N., Beglov J., Azad A.J., Leinhos L., Bennett P., Ehler E., et al. Functional analysis of a gene-edited mouse model to gain insights into the disease mechanisms of a titin missense variant. Basic Res. Cardiol. 2021;116:14. doi: 10.1007/s00395-021-00853-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lopes L.R., Zekavati A., Syrris P., Hubank M., Giambartolomei C., Dalageorgou C., Jenkins S., McKenna W., Plagnol V., Elliott P.M., et al. Genetic complexity in hypertrophic cardiomyopathy revealed by high-throughput sequencing. J. Med. Genet. 2013;50:228–239. doi: 10.1136/jmedgenet-2012-101270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang C., Zhang H., Wu G., Luo X., Zhang C., Zou Y., Wang H., Hui R., Wang J., Song L. Titin-Truncating Variants Increase the Risk of Cardiovascular Death in Patients with Hypertrophic Cardiomyopathy. Can. J. Cardiol. 2017;33:1292–1297. doi: 10.1016/j.cjca.2017.05.020. [DOI] [PubMed] [Google Scholar]
- 38.Chou C., Chin M.T. Pathogenic Mechanisms of Hypertrophic Cardiomyopathy beyond Sarcomere Dysfunction. Int. J. Mol. Sci. 2021;22:8933. doi: 10.3390/ijms22168933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Taylor M., Graw S., Sinagra G., Barnes C., Slavov D., Brun F., Pinamonti B., Salcedo E.E., Sauer W., Pyxaras S., et al. Genetic variation in titin in arrhythmogenic right ventricular cardiomyopathy—Overlap syndromes. Circulation. 2011;124:876–885. doi: 10.1161/CIRCULATIONAHA.110.005405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Brun F., Barnes C.V., Sinagra G., Slavov D., Barbati G., Zhu X., Graw S.L., Spezzacatene A., Pinamonti B., Merlo M., et al. Titin and desmosomal genes in the natural history of arrhythmogenic right ventricular cardiomyopathy. J. Med. Genet. 2014;51:669–676. doi: 10.1136/jmedgenet-2014-102591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chen K., Song J., Wang Z., Rao M., Chen L., Hu S. Absence of a primary role for TTN missense variants in arrhythmogenic cardiomyopathy: From a clinical and pathological perspective. Clin. Cardiol. 2018;41:615–622. doi: 10.1002/clc.22906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.James C.A., Syrris P., van Tintelen J.P., Calkins H. The role of genetics in cardiovascular disease: Arrhythmogenic cardiomyopathy. Eur. Heart J. 2020;41:1393–1400. doi: 10.1093/eurheartj/ehaa141. [DOI] [PubMed] [Google Scholar]
- 43.Waning J.I.V., Caliskan K., Hoedemaekers Y.M., Spaendonck-Zwarts K.Y.V., Baas A.F., Boekholdt S.M., Melle J.P.V., Teske A.J., Asselbergs F.W., Backx A., et al. Genetics, Clinical Features, and Long-Term Outcome of Noncompaction Cardiomyopathy. J. Am. Coll. Cardiol. 2018;71:711–722. doi: 10.1016/j.jacc.2017.12.019. [DOI] [PubMed] [Google Scholar]
- 44.Li S., Zhang C., Liu N., Bai H., Hou C., Song L., Pu J. Titin-truncating variants are associated with heart failure events in patients with left ventricular non-compaction cardiomyopathy. Clin. Cardiol. 2019;42:530–535. doi: 10.1002/clc.23172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Vershinina T., Fomicheva Y., Muravyev A., Jorholt J., Kozyreva A., Kiselev A., Gordeev M., Vasichkina E., Segrushichev A., Pervunina T., et al. Genetic Spectrum of Left Ventricular Non-Compaction in Paediatric Patients. Cardiology. 2020;145:746–756. doi: 10.1159/000510439. [DOI] [PubMed] [Google Scholar]
- 46.Amendola L.M., Jarvik G.P., Leo M.C., McLaughlin H.M., Akkari Y., Amaral M.D., Berg J.S., Biswas S., Bowling K.M., Conlin L.K., et al. Performance of ACMG-AMP Variant-Interpretation Guidelines among Nine Laboratories in the Clinical Sequencing Exploratory Research Consortium. Am. J. Hum. Genet. 2016;98:1067–1076. doi: 10.1016/j.ajhg.2016.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Oliva A., Flores J., Merigioli S., LeDuc L., Benito B., Partemi S., Arzamendi D., Campuzano O., Leung T.L., Iglesias A., et al. Autopsy investigation and Bayesian approach to coronary artery disease in victims of motor-vehicle accidents. Atherosclerosis. 2011;218:28–32. doi: 10.1016/j.atherosclerosis.2011.05.012. [DOI] [PubMed] [Google Scholar]