Abstract
Structural maintenance of chromosomes (SMC) family proteins play critical roles in structural changes of chromosomes. Previously, we identified two human SMC family proteins, hCAP-C and hCAP-E, which form a heterodimeric complex (hCAP-C–hCAP-E) in the cell. Based on the sequence conservation and mitotic chromosome localization, hCAP-C–hCAP-E was determined to be the human ortholog of the Xenopus SMC complex, XCAP-C–XCAP-E. XCAP-C–XCAP-E is a component of the multiprotein complex termed condensin, required for mitotic chromosome condensation in vitro. However, presence of such a complex has not been demonstrated in mammalian cells. Coimmunoprecipitation of the endogenous hCAP-C–hCAP-E complex from HeLa extracts identified a 155-kDa protein interacting with hCAP-C–hCAP-E, termed condensation-related SMC-associated protein 1 (CNAP1). CNAP1 associates with mitotic chromosomes and is homologous to Xenopus condensin component XCAP-D2, indicating the presence of a condensin complex in human cells. Chromosome association of human condensin is mitosis specific, and the majority of condensin dissociates from chromosomes and is sequestered in the cytoplasm throughout interphase. However, a subpopulation of the complex was found to remain on chromosomes as foci in the interphase nucleus. During late G2/early prophase, the larger nuclear condensin foci colocalize with phosphorylated histone H3 clusters on partially condensed regions of chromosomes. These results suggest that mitosis-specific function of human condensin may be regulated by cell cycle-specific subcellular localization of the complex, and the nuclear condensin that associates with interphase chromosomes is involved in the reinitiation of mitotic chromosome condensation in conjunction with phosphorylation of histone H3.
Structural maintenance of chromosomes (SMC) family proteins play critical roles in various nuclear events that require structural changes of chromosomes, including mitotic chromosome organization, DNA recombination and repair, and global transcriptional repression (for reviews, see references 9, 12, and 18). The SMC proteins are conserved in eukaryotes as well as in prokaryotes, underscoring their essential roles in the cell. The impairment of SMC function in both prokaryotes and eukaryotes leads to mitotic chromosome segregation defects, suggesting a critical function for SMC family proteins in mitotic chromosome dynamics.
The protein structure of SMC family members is reminiscent of a myosin-like motor protein; it contains conserved head and tail regions with a nucleotide-binding site in the N terminus and a coiled-coil central domain. At least four SMC family proteins are conserved in eukaryotes. For example, the SMC family gene products termed Smc1, Smc2, Smc3, and Smc4 in Saccharomyces cerevisiae are equivalent to Xenopus SMC1 (XSMC1), Xenopus chromosome-associated protein E (XCAP-E), XSMC3, and XCAP-C, and human SMC1 (hSMC1), hCAP-E, hSMC3, and hCAP-C, respectively (9, 12, 18, 22). XCAP-C and XCAP-E form a heterodimeric complex (XCAP-C–XCAP-E), which is part of the condensin multiprotein complex shown to be required for mitotic chromosome condensation in an in vitro embryonic Xenopus extract system (11). The hCAP-E and hCAP-C proteins also form a stable complex (hCAP-C–hCAP-E), which is the human ortholog of XCAP-C–XCAP-E as determined by its amino acid sequence similarity with XCAP-C–XCAP-E and specific localization to mitotic chromosomes (22). However, the presence of a higher-order complex equivalent to Xenopus condensin has not been demonstrated in human cells.
The mechanism of SMC-mediated chromosome condensation in the cell is not well understood. The studies using purified Xenopus condensin complex revealed that the complex utilizes its ATPase activity and introduces writhe in naked supercoiled plasmid DNA (15, 16). Although this may explain the basic mechanism of condensation, condensation of chromatin fibers in the cell at the correct stage in the cell cycle most likely requires additional highly regulated molecular events. For example, it has been demonstrated that the mitosis-specific phosphorylation of condensin components by Cdc2 kinase is required for the function of Xenopus condensin in chromosome condensation (14). The presence of histones on DNA is also an important factor that most likely influences condensin function. Phosphorylation of a specific serine residue in the histone H3 tail is initiated from pericentromeric regions of chromosomes at the end of G2 phase and spreads over the entire chromosome, closely correlating with mitotic chromosome condensation (8). It was shown recently that this phosphorylation is required for proper condensation and segregation of chromosomes (24). The role of this phosphorylation at the molecular level is not understood. A possible recruitment of condensation factors, such as the condensin complex, by this modified H3 tail has been suggested. However, no direct evidence of such an interaction has been demonstrated.
In human cells, the hCAP-C–hCAP-E heterodimeric complex is expressed throughout the cell cycle, suggesting the complex is regulated posttranslationally in order to perform its mitosis-specific role (22). To address the mechanism and regulation of hCAP-C–hCAP-E function, cellular factors that interact with hCAP-C–hCAP-E were purified by coimmunoprecipitation with the endogenous hCAP-C–hCAP-E from HeLa cells. Here we report the identification of the condensation-related SMC-associated protein 1 (CNAP1), which forms a complex with hCAP-C–hCAP-E. CNAP1 was found to be the human homolog of Xenopus condensin component XCAP-D2 (10, 14), suggesting the presence of a human condensin complex equivalent to the Xenopus condensin. To understand the cell cycle-specific regulation of human condensin, comparative immunolocalization analyses of CNAP1 and hCAP-C–hCAP-E and biochemical analysis of the complex at different cell cycle stages were performed using HeLa cells. The results revealed that human condensin is present throughout the cell cycle, but its subcellular localization is cell cycle regulated. The majority of the interphase condensin complex is sequestered in the cytoplasm, while a subpopulation of the complex was found to remain on chromosomes as distinct foci in the interphase nucleus. Biochemical studies revealed that the condensin complex interacts with histone H3 in a DNA-independent manner, suggesting that the chromosome association of condensin is at least partly mediated by the interaction with core histones. Importantly, condensin forms larger foci that colocalize with clusters of phosphorylated histone H3 (phosphor-H3) on locally condensed chromatin during the G2/M transition. This is the first evidence that demonstrates a direct link between the condensin complex and mitotic phosphorylation of histone H3 and suggests that the interphase nuclear condensin plays a critical role in reinitiation of mitotic chromosome condensation together with phosphor-H3. In this paper, the human condensin complex has been identified and systematically characterized during the cell cycle, providing the basis for understanding SMC-mediated mitotic chromosome condensation in the cell.
MATERIALS AND METHODS
Cell lines.
HeLa cells were grown in Dulbecco's modified Eagle medium (DMEM; Sigma Chemical Co.) supplemented with 10% fetal bovine serum, l-glutamate, and penicillin-streptomycin.
Antibody.
Rabbit polyclonal antibodies raised against recombinant polypeptides corresponding to the middle subdomains of hCAP-C and hCAP-E expressed in Escherichia coli were described previously (22). Antibodies were also raised against bacterially expressed amino-terminal (amino acids 1 to 241) and carboxyl-terminal (amino acids 997 to 1401) subdomains of CNAP1. Antibodies were subsequently affinity purified using antigen affinity columns. Similarly, a guinea pig antibody against the middle domain of hCAP-E was prepared. The specificity of each antibody was verified by Western blot analysis of HeLa cell extracts. Rabbit polyclonal antibody specific for the phosphor-H3 tail (Upstate Biotechnology, Lake Placid, N.Y.) was used for colocalization and interaction studies. Human CREST autoimmune serum that contains a mixture of antibodies against the centromeric proteins CENP-A, -B, and -C was used to visualize centromeric regions (kindly provided by W. R. Brinkley at Baylor College of Medicine, Houston, Tex.) (2, 5, 19). Goat anti-rabbit immunoglobulin G (IgG) antibody conjugated with Cy3 (Jackson Laboratories, West Grove, Pa.), horse anti-guinea pig IgG antibody conjugated with fluorescein (Vector Laboratories, Burlingame, Calif.), and goat anti-human IgG antibody conjugated with Cy3 (Jackson Laboratories) or fluorescein (Vector Laboratories) were used as secondary antibodies.
Coimmunoprecipitation.
HeLa cell extracts were prepared as previously described (25). Antibody was prebound to protein A-Sepharose beads (Amersham-Pharmacia Biotech). Cell extracts were then added to antibody-protein A beads and incubated for 3 h at 4°C. The antibody-protein complex was washed in HEMG buffer (25 mM HEPES [pH 7.6], 12.5 mM MgCl2, 0.1 mM EDTA, 10% glycerol) containing 0.1 and 1.0 M KCl in the presence of 0.1% Nonidet P-40. Bound proteins were eluted from the beads by washing with 2 M guanidine-HCl. Proteins were precipitated with trichloroacetic acid (TCA), and the recovered pellet was washed with acetone before resuspension in sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) sample buffer. Precipitated proteins were separated by SDS-PAGE and analyzed by silver staining or Western blotting. For a large-scale purification of the complex, antibody was cross-linked to protein A beads and subjected to multiple rounds of immunopurification. The washing conditions were the same as described above. The bound materials were eluted with glycine, neutralized, and immediately dialyzed against HEMG buffer with 0.1 M KCl. Dialyzed samples from each immunopurification were pooled and TcA precipitated prior to preparative SDS-PAGE.
Peptide sequencing.
After the immunopurified hCAP-C–hCAP-E-containing complex was resolved by SDS-PAGE, the proteins were transferred to nitrocellulose membrane, visualized by Ponceau S staining, and excised. Each polypeptide species on the membrane was subjected to trypsin digestion, and digested polypeptides were separated by reverse-phase high-pressure liquid chromatography and sequenced as described previously (22).
Western blot analysis.
HeLa cell extracts or immunoprecipitated protein complexes were subjected to SDS-PAGE and then transferred to nitrocellulose membranes as described previously (22). The membranes were blocked with 5% milk or 3% bovine serum albumin (BSA)–0.05% Tween 20 in phosphate-buffered saline (PBS). The primary antibody was incubated in 3% BSA–0.05% Tween 20 in PBS for 1 h to overnight, followed by three washes in PBS–0.05% Tween 20. The secondary antibody conjugated with alkaline phosphatase (Promega, Madison, Wis.) was incubated in 3% BSA–0.05% Tween 20 in PBS for 1 h at room temperature. The filter was then washed three times in PBS–0.05% Tween 20 before development.
Synchronization of HeLa cells.
HeLa cells were synchronized by double thymidine block in combination with nocodazole as described previously (1, 17, 26), with slight modification. Briefly, cells were incubated in 2 mM thymidine for 17 h, rinsed, and incubated in DMEM for 9 h. The 2 mM thymidine was added again for 15 h, and cells were rinsed and grown in DMEM for an appropriate length of time for each cell cycle stage. The efficiency of synchronization at S phase by double thymidine block was assessed by fluorescence-activated cell sorting analysis (data not shown). The percentage of cells subsequently entering M phase synchronously at 13 h after the release from thymidine was more than 80%, indicating proper synchronization (data not shown). In combination with a transient nocodazole treatment, HeLa cells were synchronized at G1, S, G2, and M phases.
Immunofluorescent staining.
Cells were grown on glass coverslips in 24-well plates until 50 to 70% confluent. Mitotic chromosome spreads were prepared essentially as described elsewhere (22). The coverslips were washed in PBS twice and then fixed with 4% paraformaldehyde at 4°C or acetone at −20°C. Cells and spreads were subjected to immunofluorescent staining as described previously (22). For DNA detection, 2,6-diamidinophenylindole (DAPI) was used. The coverslips were then rinsed in distilled H2O and mounted onto slides with antifade (0.055 mM p-phenylenediamine dihydrochloride in PBS with glycerol added to 90%) (13) or Prolong (Molecular Probes, Eugene, Oreg.). Image analysis was performed using a Zeiss Axioplan 2 microscope with a Photometrics Sensys charge-coupled device camera. J/C Cittert-type iterative digital deconvolution was performed using the Zeiss KS 300 program. Confocal image analysis was performed using a Bio-Rad MRC 1024UV laser scanning confocal microscope equipped with a krypton-argon mixed-gas laser with excitation at 488 nm and 568-nm wavelength beam and an argon ion water-cooled UV laser with excitation wavelength of 363 nm. Transmitted images were collected using the transmitted light device and collected in three separate colors. The confocal device is attached to a Nikon DiaPhot inverted microscope. Magnification was achieved using a 60× PlanApo NA 1.4 objective. Images were collected by averaging 12 images using the Kalman filter.
In situ cell extraction.
Cell extractions were performed essentially according to Fey et al. (6). HeLa cells were grown on coverslips coated with polylysine. The cells were washed with PBS and cytoskeleton (CSK) buffer [10 mM piperazine-N,N′-bis(2-ethanesulfonic acid) (PIPES; pH 7.0), 100 mM NaCl, 300 mM sucrose, 3 mM MgCl2]. Cells were extracted using CSK buffer with 0.5% Triton X-100 for 5 min at 4°C to remove soluble cytoplasmic proteins. Cells were next treated with an extraction buffer (42.5 mM Tris-HCl [pH 8.3], 8.5 mM NaCl, 2.6 mM MgCl2, 1% Tween 20, 0.5% deoxycholic acid) for 5 min at 4°C to remove cytoskeletal proteins. The cells were then treated with CSK buffer (containing 2 mM CaCl2 and 2 mM MgCl2), 0.5% Triton X-100, and DNase I (100 μg/ml; Worthington, Freehold, N.J.) for 30 min at 37°C. The cells were subsequently washed with 0.25 M ammonium sulfate in CSK buffer. In some cases, cells were treated with 2 M NaCl before or after DNase I treatment. The proteins resistant to these extractions are defined to be associated with the nuclear matrix (4, 6). At each step, subsets of coverslips were fixed with 4% paraformaldehyde for 30 min at 4°C and stained with antibody as described above. Immunofluorescence images of the cells from each extraction step were collected with the same exposure time.
RESULTS
Identification of CNAP1, a 155-kDa protein associated with hCAP-C–hCAP-E.
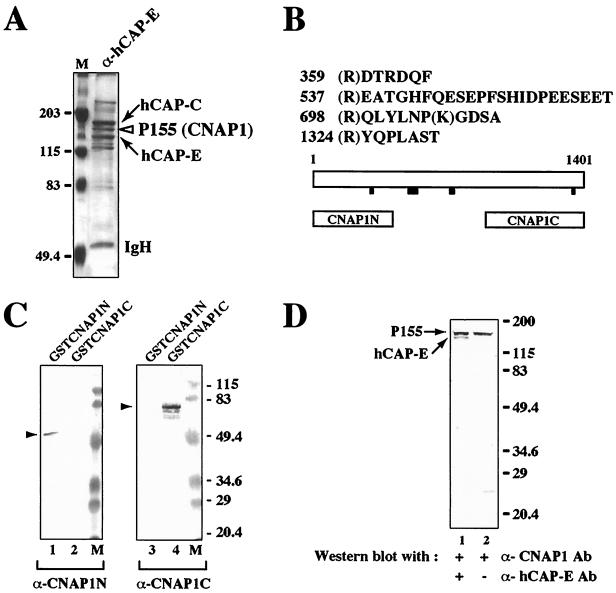
The endogenous protein complex containing hCAP-C–hCAP-E from HeLa nuclear extracts was isolated using an anti-hCAP-E antibody affinity column. Under stringent immunoprecipitation conditions including a 1 M salt wash with detergent (see Materials and Methods), several polypeptide species were copurified with the hCAP-C–hCAP-E complex (Fig. 1A). The trypsin-digested peptides derived from a protein species of 155 kDa were subjected to microsequencing analysis (Fig. 1B). A BLAST search identified matching sequences in a human cDNA encoding a protein of unknown function designated KIAA0159 (GenBank accession number D63880) (20). The cDNA encodes an open reading frame of 1,401 amino acids with a calculated molecular mass of 157,122 Da and a pI of 6.11. This protein specifically interacts with the condensation-related SMC complex hCAP-C–hCAP-E and thus was designated CNAP1. This cDNA product is homologous to XCAP-D2, a subunit of the Xenopus condensin complex (14), as well as to pEg7, the same protein identified independently based on its function in chromosome condensation in Xenopus egg extracts (3).
FIG. 1.
Identification of a 155-kDa polypeptide as CNAP1. (A) Silver staining of the immunoprecipitated proteins from HeLa nuclear extracts using anti-hCAP-E antibody. The arrows indicate hCAP-C and hCAP-E. The open arrowhead indicates a 155-kDa protein (p155) corresponding to CNAP1. Positions of size markers (M) in panels A, C, and D are indicated in kilodaltons. IgH, immunoglobulin heavy chain (B) Peptide sequence analysis of p155. Numbers represent the amino acid residues in the CNAP1 protein. Black bars below the schematic diagram of CNAP1 protein indicate the positions of identified peptide sequences. The open boxes labeled CNAP1N and CNAP1C represent recombinant proteins corresponding to the N- and C-terminal regions of CNAP1 used to generate antibodies. (C) Western blot analysis of recombinant CNAP1 fusion proteins with antibodies specific for N- and C-terminal domains of CNAP1. Antibodies specific for CNAP1N and CNAP1C were used to test cross-reactivity of the two antibodies with GST-CNAP1N (lanes 1 and 3) and GST-CNAP1C (lanes 2 and 4). Lanes 2 and 3 demonstrate the lack of cross-reactivity between the two antibodies. Specific signals corresponding to the GST fusion proteins are indicated with arrowheads. (D) Detection of the endogenous CNAP1 by Western blot analysis of crude HeLa nuclear extracts using anti-CNAP1C antibody (Ab) (lanes 1 and 2). Lane 1 was further probed with anti-hCAP-E antibody to confirm the size of CNAP1 relative to that of hCAP-E.
Polyclonal antibodies were raised against bacterially expressed N- and C-terminal regions of CNAP1 and were affinity purified (Fig. 1B). The specificities of the antibodies were tested against the corresponding recombinant glutathione S-transferase (GST) fusion proteins and endogenous CNAP1 in crude HeLa nuclear extracts (Fig. 1C and D). No cross-reactivity was observed between the two antibodies, consistent with the lack of amino acid sequence homology between the two antigen fragments (Fig. 1C). Furthermore, both antibodies (anti-CNAP1C [Fig. 1D] and anti-CNAP1N [data not shown]) specifically recognized a single polypeptide of 155 kDa in crude HeLa nuclear extracts. These results demonstrate that CNAP1 cDNA indeed encodes a 155-kDa protein and that the two antibodies against CNAP1 are highly specific in human cells.
CNAP1 forms a complex with hCAP-C–hCAP-E and localizes to mitotically condensed chromosomes.
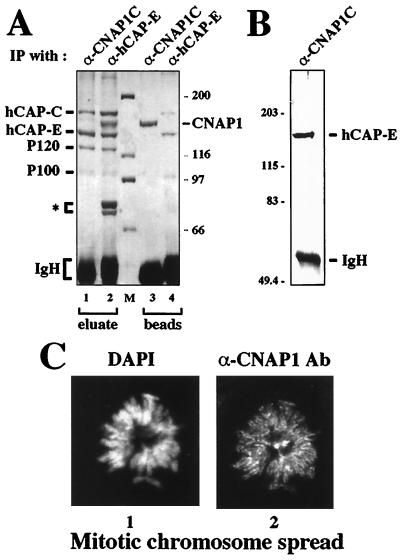
Having confirmed the specificity of the antibodies against CNAP1, we next tested the specificity of the interaction between CNAP1 and hCAP-C–hCAP-E by reciprocal immunoprecipitation. Antibody against CNAP1 specifically immunoprecipitated hCAP-C–hCAP-E from mitotic HeLa extracts in a stoichiometric manner (Fig. 2A). The interaction between hCAP-C and hCAP-E was highly stable and resistant to 2 M guanidine treatment (Fig. 2A, lane 4), whereas hCAP-C–hCAP-E dissociated from CNAP1 with this treatment (Fig. 2A, lanes 1 and 3), indicating that hCAP-C–hCAP-E is the core complex and CNAP1 associates with it. The ratio between hCAP-C–hCAP-E and CNAP1 was the same in reciprocal immunoprecipitation using either anti-CNAP1 or anti-hCAP-E antibody. This suggests that almost 100% of the molecules of hCAP-C–hCAP-E and CNAP1 are involved in complex formation in the cell (Fig. 2A, compare lanes 1 and 3 with lanes 2 and 4). The 2 M guanidine eluate from the anti-CNAP1 immunoprecipitation (Fig. 2A, lane 1) was subjected to Western blot analysis using anti-hCAP-E antibody, further confirming the identity of the coprecipitated protein (Fig. 2B). In addition to CNAP1, two other polypeptide species, P120 and P100, were immunoprecipitated by both antibodies against CNAP1 and hCAP-E (Fig. 2A, lanes 1 and 2). Their molecular weights suggest that P120 and P100 may correspond respectively to XCAP-G and XCAP-H found in the Xenopus condensin complex (10, 14). Taken together, these findings indicate that the protein complex containing hCAP-C–hCAP-E and CNAP1 is the human condensin complex.
FIG. 2.
Coimmunoprecipitation of the hCAP-C–hCAP-E complex with anti-CNAP1C antibody using mitotic HeLa extracts. (A) Silver staining of reciprocal coimmunoprecipitation. A pattern of immunoprecipitated (IP) proteins by anti-CNAP1C (lanes 1 and 3) is compared to that by anti-hCAP-E (lanes 2 and 4). Eluate, proteins that were eluted from beads with 2 M guanidine (see Materials and Methods); beads, proteins that remained on the beads after the elution; IgH, immunoglobulin heavy chain. Three polypeptides indicated by an asterisk are unique to anti-hCAP-E immunoprecipitation. Sizes of markers are indicated in kilodaltons. (B) Western blot analysis of the proteins immunoprecipitated with anti-CNAP1C antibody probed with anti-hCAP-E. (C) CNAP1 localization on mitotic chromosomes. Immunofluorescent staining of a HeLa mitotic chromosome spread using DAPI (panel 1) to visualize DNA and anti-CNAP1C antibody (Ab) to detect CNAP1 protein (panel 2). The bright spots in the center correspond to centrosomes.
Previously, we have shown that hCAP-C–hCAP-E localizes to mitotic chromosomes, consistent with the results observed with its Xenopus homolog (11, 22). Based on the specific interaction between hCAP-C–hCAP-E and CNAP1 in the mitotic extracts as described above, colocalization of the two were tested in HeLa cells. Immunofluorescent staining of metaphase spreads with anti-CNAP1 antibody revealed that CNAP1 specifically localizes to mitotic chromosomes (Fig. 2C). CNAP1 appears to distribute along the entire chromosome arm in a manner identical to hCAP-E (22). Centrosomes are also stained with this particular antibody (Fig. 2C, panel 2, center). However, unlike the consistent staining of mitotic chromosomes, centrosome staining was not reproducible by antibodies from different rabbits (data not shown). Therefore, the significance of centrosome staining is not clear. Taken together, the results show that hCAP-C–hCAP-E and CNAP1 not only form a complex in mitotic cells but also colocalize on mitotic chromosomes, confirming the notion that they are components of the human condensin complex.
Cell cycle-specific subcellular localization of the human condensin complex.
It has been demonstrated that hCAP-C–hCAP-E is expressed throughout the cell cycle, suggesting that its mitosis-specific function is regulated posttranslationally (22). Understanding how human condensin function is suppressed in interphase will help us decipher the mechanism of the cell cycle-specific regulation of chromosome condensation. It is possible, for example, that hCAP-C–hCAP-E and CNAP1 may not be in the same complex during interphase. Therefore, complex formation and subcellular distribution patterns of hCAP-C–hCAP-E and CNAP1 were followed during the cell cycle using synchronized HeLa cells.
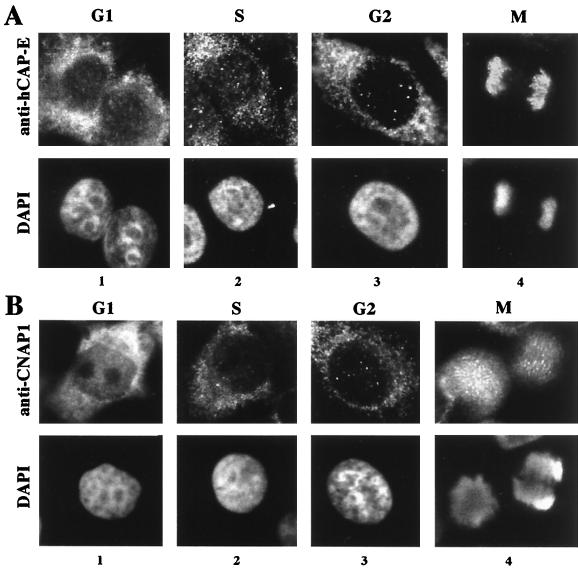
The subcellular distribution pattern of CNAP1 is similar to the pattern of hCAP-C–hCAP-E throughout the cell cycle (Fig. 3). The specificity of antibodies against hCAP-C and hCAP-E was demonstrated by Western blot analysis against crude HeLa extracts at various cell cycle stages in our previous study (22). As determined previously by reciprocal immunoprecipitation, almost 100% of hCAP-C and hCAP-E molecules are engaged in heterodimeric complex formation (22). Therefore, detection of either hCAP-C or hCAP-E can be interpreted as detection of the hCAP-C–hCAP-E complex. This was confirmed by costaining with antibodies against hCAP-C and hCAP-E (data not shown). The specificity of anti-CNAP1 antibody was further confirmed with immunodepletion of antibody with the CNAP1 antigen, which abolished the observed CNAP1 staining (data not shown). During M phase, hCAP-C–hCAP-E and CNAP1 both localize to condensed chromosomes, consistent with the result in Fig. 2C (Fig. 3, panels 4). A subpopulation of CNAP1 was also observed in the mitotic cytoplasm (Fig. 3B, panel 4). This was not detected with chromosome spreads, which lose cytoplasmic proteins during preparation (Fig. 2C). Furthermore, hCAP-E and hCAP-C were also detected in the cytoplasm in mitotic cells with longer exposure, indicating that a subpopulation of the complex is present in the mitotic cytoplasm. After cell division, both hCAP-C–hCAP-E and CNAP1 accumulate in the cytoplasm during G1 phase (Fig. 3, panels 1). Dissociation of hCAP-C–hCAP-E and CNAP1 from chromosomes was observed as early as late telophase (data not shown). The dissociated hCAP-C–hCAP-E and CNAP1 are diffusely present in the newly assembled nucleus during the telophase/G1 phase transition and appear to eventually relocalize to the cytoplasm during G1 phase (compare panels 1 in Fig. 3 with panel 1 in Fig. 6A, representing the early G1-phase cells, which have not fully flattened out). At this point, it is not clear whether the nuclear population of hCAP-C–hCAP-E and CNAP1 is actively exported to the cytoplasm at the M/G1 transition or the nuclear population becomes degraded and newly synthesized proteins accumulate in the cytoplasm. The majority of hCAP-C–hCAP-E and CNAP1 remains in the cytoplasm during S and G2 phases until prophase (Fig. 3, panels 2 and 3).
FIG. 3.
Analysis of the subcellular localization of the hCAP-C–hCAP-E complex and CNAP1 in HeLa cells at different cell cycle stages, as indicated at the top. The proteins are detected by immunofluorescent staining with antibody specific for hCAP-E, representing hCAP-C–hCAP-E (A) and CNAP1 (B). The top panels show immunofluorescent staining, and the bottom panels represent DAPI staining.
FIG. 6.
Immunofluorescent staining of the hCAP-C–hCAP-E complex in G1 (A)- and S (B)-phase-synchronized HeLa cells with in situ extraction and DNA digestion. The top panels show immunofluorescent staining with anti-hCAP-C antibody; the bottom panels indicate DNA visualized by DAPI staining. Each extraction step is indicated at the top.
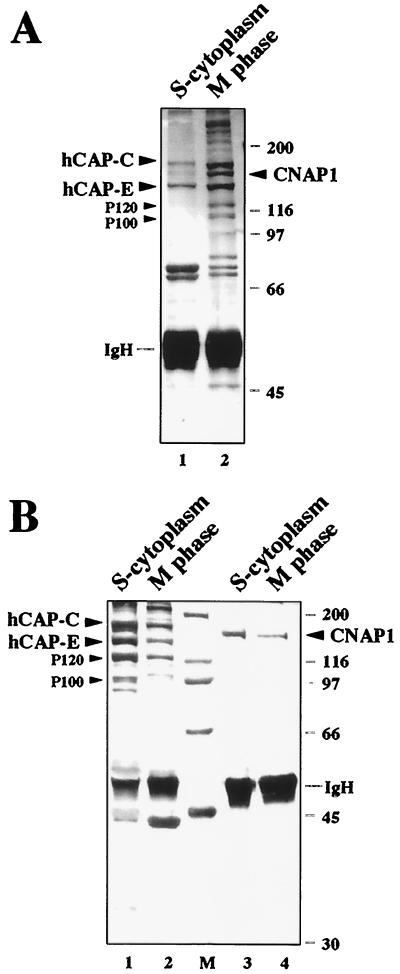
Next we tested whether hCAP-C–hCAP-E and CNAP1 are still in the same complex in the cytoplasm during interphase. The amount of hCAP-E and CNAP1 is consistent throughout the cell cycle, as confirmed by Western blot analysis (22) (data not shown). Anti-CNAP1 antibody immunoprecipitated hCAP-C–hCAP-E equally well from both S-phase cytoplasmic and mitotic extracts in a stoichiometric manner, suggesting that CNAP1 is in a complex with hCAP-C–hCAP-E throughout the cell cycle (Fig. 2A and 4B). Anti-hCAP-E antibody reciprocally coprecipitated CNAP1 both from mitotic extracts and, although less efficiently, from S-phase cytoplasmic extracts (Fig. 4A, compare lanes 1 and 2). The reason for the difference of the accessibility of the anti-hCAP-E antibody in the S-phase cytoplasm is not clear. Furthermore, crude cytoplasmic extracts were size fractionated using sucrose gradient ultracentrifugation to compare the protein peaks. The results showed that the peak of hCAP-C–hCAP-E coincides with the peak of CNAP1, indicating that the majority of both hCAP-C–hCAP-E and CNAP1 are in the same complex (data not shown). Taken together, these results indicate that hCAP-C–hCAP-E and CNAP1 are in a complex throughout the cell cycle and that subcellular distribution of the human condensin complex is regulated in a cell cycle-dependent manner.
FIG. 4.
Coimmunoprecipitation of human condensin complex from S-phase cytoplasmic and mitotic HeLa extracts. (A) Coimmunoprecipitation of the complex with anti-hCAP-E middle domain antibody from S-phase cytoplasmic (lane 1) and mitotic (lane 2) extracts. hCAP-C, hCAP-E, CNAP1, P120, and P100 are indicated. IgH, immunoglobulin heavy chain. (B) Coimmunoprecipitation of the complex with anti-CNAP1 antibody using S-phase cytoplasmic and mitotic extracts. Extracts used are indicated at the top. CNAP1 remained on beads after guanidine elution (lanes 3 and 4). In both panels, sizes are indicated in kilodaltons.
Involvement of the human condensin complex in the early stage of mitotic chromosome condensation together with phosphor-H3.
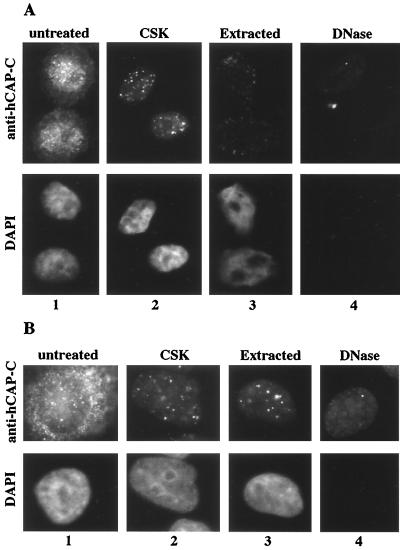
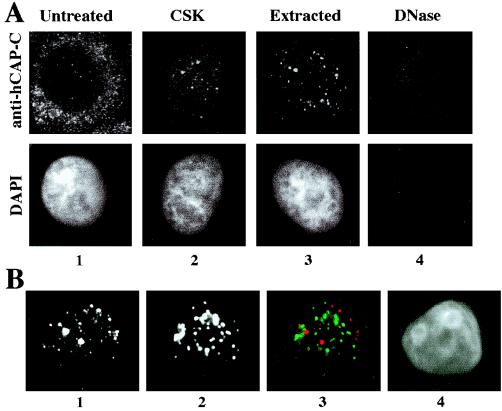
Interestingly, a subpopulation of hCAP-C–hCAP-E and CNAP1 was detected in the nucleus in G2 phase as distinct foci (Fig. 3, panels 3). The presence of nuclear foci of the condensin complex in interphase cells raises the possibility that a subpopulation of the condensin complex remains on chromosomes after mitosis and participates in the nucleation of chromosome condensation for the next cycle of mitosis. Therefore, we tested whether the foci observed in the interphase nuclei are localized on chromosomes or in the nucleoplasm. Stepwise in situ extraction of HeLa cells revealed that these condensin foci are associated with interphase chromosomes (Fig. 5A). This procedure includes the extraction of soluble proteins by detergent and the removal of chromosomal DNA by DNase I digestion. Cells were seeded on coverslips and synchronized at G2 phase prior to the extraction. The foci in the nucleus were detected by anti-hCAP-C antibody. Most of the cytoplasmic staining by anti-hCAP-C antibody was removed as soluble proteins were extracted, whereas the foci in the nucleus remained (Fig. 5A, panels 2 and 3). Cells were then treated with DNase I, which removed chromosomal DNA, as confirmed by the disappearance of DAPI staining (Fig. 5A, panel 4). The nuclear foci of hCAP-C were removed by this treatment, indicating that the nuclear foci of condensin are on chromosomes. Similar results were obtained using anti-hCAP-E and anti-CNAP1 antibodies, confirming the specificity of the signal (data not shown). These nuclear foci are not at the centromeric regions, as determined by costaining with CREST antibody, which detects the centromeric proteins CENP-A, -B, and -C (5) (Fig. 5B). Partial extraction revealed that the nuclear condensin foci are also present at G1 and S phases (Fig. 6), although the intensity of the G1 foci appears to decrease slightly in the second extraction, suggesting that the complex may bind to chromosomes somewhat differently during G1 phase (Fig. 6A, Extracted). Thus, while the majority of the complex relocalizes to the cytoplasm after mitosis, a subpopulation of human condensin remains on chromosomes at distinct sites until the onset of the next mitosis.
FIG. 5.
Nuclear foci of the human condensin complex. (A) Immunofluorescent staining of the hCAP-C–hCAP-E complex in G2-synchronized HeLa cells with in situ extraction and DNA digestion. The top panels show immunofluorescent staining of the hCAP-C–hCAP-E complex using anti-hCAP-C antibody; the bottom panel indicates DNA visualized by DAPI staining. The extraction steps are indicated at the top (see Materials and Methods). (B) Immunofluorescent costaining of the CSK-extracted cell with anti-hCAP-C and CREST antibodies. Panel 1, anti-hCAP-C; panel 2, CREST antibody; panel 3, merged image (red [anti-hCAP-C] and green [CREST]); panel 4, DAPI.
The above results support the hypothesis that these foci on chromosomes may be critical in the initiation of mitotic chromosome condensation, which occurs during G2 phase prior to breakdown of the nuclear membrane. To address this issue, we tested colocalization of hCAP-C–hCAP-E with phosphor-H3 during the G2/M transition. Phosphorylation of H3 occurs in a mitosis-specific manner and is required for condensation and segregation of chromosomes, although the precise role of the phosphorylated H3 tail is not clear (24). Phosphorylation is initiated from pericentromeric regions during late G2 phase and spreads throughout chromosomes as condensation proceeds, suggesting its direct role in condensation (8).
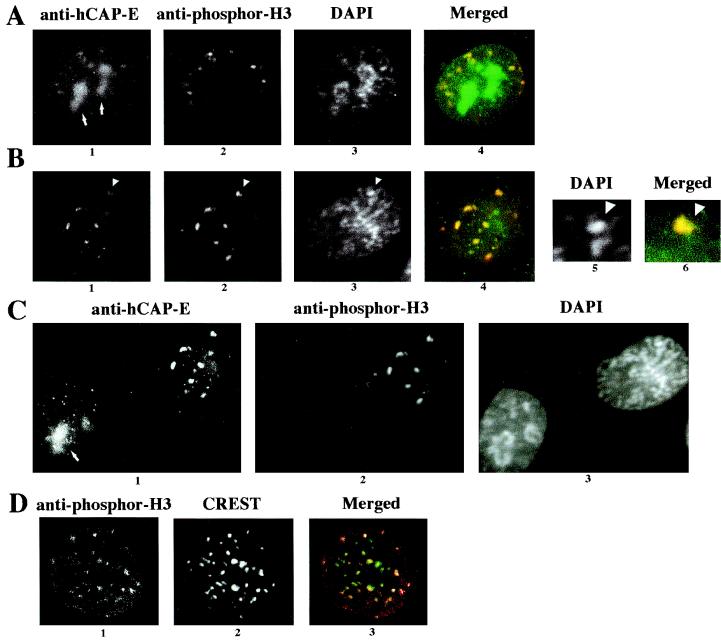
The foci of phosphor-H3 were observed only in late G2/early prophase HeLa cells (Fig. 7). Using our fixation protocol, clear visualization of the foci required the CSK treatment, which removes soluble cytoplasmic and cytoskeletal proteins (see Materials and Methods). However, similar foci were also observed in the cells fixed with 1% formaldehyde followed by Triton X-100 treatment as published previously (8) (data not shown). Furthermore, the observed phosphor-H3 foci after CSK treatment are confirmed to be at the centromeric regions by costaining with CREST antibody, which is consistent with published results (8) (Fig. 7D). These data suggest that the observed foci are physiologically relevant. The hCAP-C–hCAP-E and CNAP1 foci are originally not at the centromere or pericentromeric regions during interphase (Fig. 5B). However, we found that some of the foci are larger and colocalize with phosphor-H3 at late G2/early prophase, when local condensation is initiated but the mitotic chromosomes have not been completely formed (Fig. 7A and B). These foci appear to coincide with clusters of brighter DAPI staining that represent condensed DNA regions (Fig. 7B, panels 5 and 6). Interestingly, we also observed the condensin complex in the nucleolus localizing most prominently during G2 phase (Fig. 7A and C). Nucleolar localization was confirmed by costaining with antibody against B23, a nucleolus-specific protein (Santa Cruz Biotechnology, Santa Cruz, Calif.) (data not shown). In earlier interphase, the condensin foci do not colocalize with phosphor-H3, since no detectable staining was observed with anti-phosphor-H3 antibody, consistent with the notion that the H3 phosphorylation occurs in late G2 (Fig. 7C). These results indicate that human condensin, originally forming distinct foci on chromatin (not at the centromeres) during interphase, assembles into larger foci with phosphor-H3 at locally condensed chromosome regions around centromeres during the G2/M transition. During metaphase, once mitotic chromosome condensation is complete, both human condensin and phosphor-H3 coat the entire chromosome.
FIG. 7.
Colocalization analysis of the condensin complex with phosphor-H3. HeLa cells were treated with CSK buffer and costained with anti-hCAP-E, anti-phosphor-H3, and DAPI as indicated at the top. Colocalization of anti-hCAP-E (green) and anti-phosphor-H3 staining (red) is shown as a merged image (colocalization is shown in yellow). (A) Late-G2-phase cell. Nucleolus staining with hCAP-E is indicated by an arrows. (B) Early-prophase cell. The arrowheads in panels 1, 2, and 3 identify a site of locally condensed DNA, which is enlarged to show DAPI (panel 5) and the merged image of hCAP-E (green) and phosphor-H3 (red) (panel 6). (C) Comparison of early prophase (same as in panel B) and interphase cells. Staining is indicated at the top, and the nucleolus staining of anti-hCAP-E is indicated with an arrow. (D) Colocalization of phosphor-H3 and centromeric regions. The CSK-extracted cell was stained with anti-phosphor-H3 (panel 1) and CREST (panel 2); the merged image is shown in panel 3 (red [phosphor-H3] and green [CREST]). These images were captured with confocal microscopy.
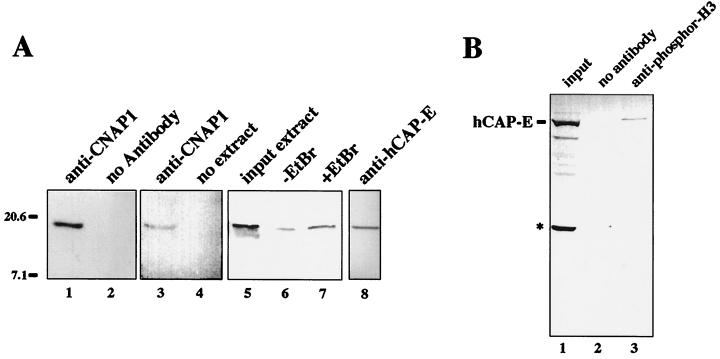
To substantiate the immunocolocalization results above, we next tested whether phosphor-H3 physically interacts with the condensin complex in mitotic cells. Coimmunoprecipitation was performed using mitotic HeLa extracts with anti-CNAP1 antibody. Although the majority of histones remain insoluble, small amounts of histones were extracted with 0.4 M salt extraction, and the immunoprecipitation was carried out at the same salt concentration. The immunoprecipitated materials were subjected to Western blot analysis using anti-phosphor-H3 antibody to detect the presence of phosphor-H3 (Fig. 8A). A specific signal corresponding to phosphor-H3 was detected in the 1 M salt eluate (Fig. 8A, lanes 1 and 3). As controls, protein A beads alone were incubated with the extracts, and anti-CNAP1 antibody on beads without extracts were also compared in the Western blot analysis (Fig. 8A, lanes 2 and 4, respectively). Furthermore, the presence or absence of ethidium bromide (EtBr) did not affect the coprecipitation, suggesting that the interaction is mediated by protein-protein interaction and not by DNA (Fig. 8A, lanes 6 and 7). The identity of the phosphor-H3 signal was further confirmed by antibody specific for general histone H3 (data not shown). Phosphor-H3 was also coimmunoprecipitated with antibody specific for hCAP-E, which we have shown copurifies the condensin complex (Fig. 2A, lane 2; Fig. 8A, lane 8). In contrast, anti-hCAP-C antibody, which immunoprecipitates the hCAP-C–hCAP-E heterodimeric complex but fails to coprecipitate other condensin components, most likely due to competition for the binding site with other condensin components, did not copurify phosphor-H3 (data not shown). This result suggests that the presence of some non-SMC component(s) of the condensin complex is required for the interaction with phosphor-H3. Finally, anti-phosphor-H3 antibody reciprocally immunoprecipitated condensin (Fig. 8B). These results demonstrate that human condensin and phosphor-H3 not only colocalize with each other but interact with each other in the cell. However, this interaction is apparently weaker than the interaction between the condensin components hCAP-C–hCAP-E and CNAP1, which is resistant to 1 M salt, suggesting that phosphor-H3 is not part of the condensin complex and the interaction may also be indirect. Similar coimmunoprecipitation was carried out using interphase extracts, which coprecipitated acetylated histone H3 but no detectable phosphor-H3 (data not shown). Therefore, condensin binding to histone H3 is not phosphorylation specific. This result is consistent with our observation that the earlier interphase cells lacking H3 phosphorylation still contain condensin speckles associated with chromatin (Fig. 6 and 7C). Taken together, these results suggest that the condensin association with chromosomes may be at least partly mediated by protein-protein interaction with core histones. Since the interaction of condensin with histone H3 is not necessarily limited to the phosphorylated form, the formation of the large condensation intermediate foci containing phosphor-H3 and condensin most likely requires an additional factor(s) and/or specific DNA sequence(s) or structure(s) as well as condensin component modification (e.g., phosphorylation). Nonetheless, these results strongly suggest that the nuclear population of human condensin in interphase cells plays an important role in the reinitiation of mitotic chromosome condensation together with phosphorylation of histone H3 by forming the condensation intermediate structure.
FIG. 8.
Coimmunoprecipitation of phosphor-H3 with the condensin complex. (A) Mitotic HeLa extracts were used for coimmunoprecipitation using anti-CNAP1 antibody (lanes 1, 3, 6, and 7). The immunoprecipitated materials eluted from antibody beads with 1 M salt were probed with anti-phosphor-H3 antibody in a Western blot analysis. Controls include protein A beads alone incubated with extract (lane 2) and protein A beads bound by antibody without extract (lane 4). In lane 7, EtBr was added to remove residual DNA contaminants from the extract prior to immunoprecipitation, which was compared to the sample without EtBr (lane 6) and the input extracts (lane 5). Lane 8 shows coimmunoprecipitation with anti-hCAP-E antibody probed with anti-phosphor-H3 antibody. (B) Reciprocal immunoprecipitation with anti-phosphor-H3 antibody probed with anti-hCAP-E antibody. The full-length hCAP-E is indicated. Input extract in lane 1 also shows the smaller degradation product of hCAP-E indicated by an asterisk, which was not coprecipitated with anti-phosphor-H3 (lane 3). Protein A beads alone did not coprecipitate hCAP-E from the extract (lane 2). In both panels, sizes are indicated in kilodaltons.
DISCUSSION
Regulation of chromosome condensation and decondensation during the cell cycle is essential for the normal life cycle of the cell. The condensin complex containing a heterodimeric SMC complex, XCAP-C–XCAP-E, has been demonstrated to be physically required for mitotic chromosome condensation in an in vitro Xenopus oocyte system. In this report, we describe the identification of a protein termed CNAP1 that is tightly associated with a human SMC protein complex, hCAP-C–hCAP-E, in the cell. The hCAP-C–hCAP-E heterodimer is the human ortholog of Xenopus XCAP-C–XCAP-E, and CNAP1 is homologous to Xenopus condensin component XCAP-D2. Thus, our results establish the presence of an equivalent condensin complex in mammalian cells. Regulation of condensin during the somatic cell cycle has not been investigated previously. Our immunolocalization and biochemical analyses comparing three of the condensin components, hCAP-C, hCAP-E, and CNAP1, demonstrate the cell cycle-specific behavior of the human condensin complex in somatic cells. The complex is present throughout the cell cycle but is sequestered in the cytoplasm during the interphase. Importantly, a subpopulation of the complex remains tightly associated with certain sites on chromosomes as foci throughout interphase. These foci become larger and colocalize with clusters of phosphor-H3 on condensed regions of chromosomes at the G2/M transition. These results suggest that the residual chromosome-associated condensin in the interphase nucleus participates in the reinitiation of mitotic chromosome condensation, perhaps by receiving a cell cycle-specific signal, and eventually recruits cytoplasmic condensin onto the neighboring chromosomes.
Human condensin complex containing CNAP1.
The Xenopus condensin complex was shown to contain five subunits, including the SMC family proteins XCAP-C and XCAP-E and the three non-SMC subunits XCAP-D2, XCAP-G, and XCAP-H (10). This complex is directly involved in mitotic chromosome condensation in Xenopus embryos. In our previous study, we discovered hCAP-C–hCAP-E, a human ortholog of the XCAP-C–XCAP-E complex, which associates with condensed chromosomes in mitotic human cells (22). In the present study, CNAP1 is identified as a protein that specifically forms a complex with hCAP-C–hCAP-E in human cells. CNAP1 is homologous to the Xenopus condensin component XCAP-D2 (14). Two other polypeptides (P120 and P100) that are copurified could correspond to XCAP-G and XCAP-H. Our results indicate the presence of a condensin complex in human somatic cells similar to the one in Xenopus embryos. Recently, the homologous condensin complex was identified in Schizosaccharomyces pombe and S. cerevisiae (7, 23). From these findings taken together, condensin appears to be conserved from yeast to humans despite the differences in the degree of chromosome condensation in these species.
In a Xenopus embryonic system, the function of the condensin complex appears to be regulated by mitosis-specific phosphorylation of several components of the complex (14). Xenopus condensin appears to be unphosphorylated during interphase. In somatic cells that undergo G1 and G2 phases, the regulation of mitosis-specific function of condensin may be different or more complex. Indeed, human condensin components are phosphorylated throughout the cell cycle at multiple sites, which most likely contributes to the fine-tuning of complex function (A. R. Ball, Jr., and K. Yokomori, unpublished data). Our studies identify the cell cycle-specific localization of the complex in the cytoplasm upon entering G1 phase. These results raise the possibility that the retention of the human condensin complex in the cytoplasm may be an important means to block the complex from acting on chromosomes prematurely during interphase. The study on the condensin complex in S. cerevisiae indicated that the condensin complex stays associated with chromosomes throughout the cell cycle (7). This is in contrast to the cell cycle-dependent relocalization of the complex in human cells (this report) and in S. pombe (23). Condensin function and regulation may differ in a budding yeast and higher eukaryotes despite the conservation of the complex components, presumably due to the differences in the complexity of the genome structure. The chicken hCAP-E homolog, ScII, was originally reported to loosely associate with the nucleus during interphase based on mass enucleation results (21). However, the immunolocalization analysis of ScII in the interphase cells was not provided in the study, and the reason for the apparent discrepancy between the results for chicken cells and for human cells is not clear. How the complex dissociates from chromosomes at the end of mitosis is not known. It will be important to investigate how relocalization of the complex is regulated. Despite the structural similarity within the conserved N- and C-terminal portions, the second SMC complex, hSMC1-hSMC3, which is involved in sister chromatid cohesion and metaphase progression, distributes differently in the cell (22) (H. C. Gregson et al., submitted for publication). This suggests that the interactions of other factors unique to each SMC complex are critical for specific subcellular localization.
Formation of large condensation intermediate foci containing condensin and phosphor-H3.
Whether there are specific sites on chromosomes for initiation of condensation and how condensation spreads over the entire chromosome are issues that remain unanswered. Phosphorylation of the histone H3 tail is a cell cycle-specific event initiated during the G2/M transition and is maintained until the end of mitosis, closely correlating with mitotic chromosome condensation. Histone H3 phosphorylation was recently shown to be required for proper condensation and segregation of chromosomes (24). Phosphorylation appears to spread from pericentromeric regions to the whole chromosome, which may reflect the spread of condensation (8). It was proposed that phosphor-H3 may be involved in the initiation of condensation by destabilizing local chromatin structure to allow easier access of condensation factors, or by directly recruiting factors required for condensation through its phosphorylated tail (24). However, the molecular event involving the phosphorylated tail is still not well understood.
Our study provides the first evidence that condensin and phosphor-H3 may be involved together in the chromosome condensation process. Our results demonstrate that the condensin complex and phosphor-H3 colocalize to the large foci coinciding with the locally condensed chromosomes during late G2/early prophase. Interestingly, the hCAP-C–hCAP-E foci are on chromosomes prior to the initiation of H3 phosphorylation that occurs at late G2 and do not initially localize to the centromeric regions. In addition, similar to the observation in S. cerevisiae (7), human condensin localizes to the nucleolus in a G2-phase-specific manner, in which phosphorylation of H3 is initially absent. Therefore, our results suggest that the human condensin has its own preferential binding sites on interphase chromosomes independent of phosphor-H3. Furthermore, our data provide evidence that condensin directly or indirectly interacts with histone H3 (regardless to its phosphorylation state) through protein-protein interaction, which may contribute to the association of condensin to chromosomes. It is not unreasonable to speculate that mitotic chromosome condensation is a multistep process requiring additional factors at each step. Therefore, the large condensation foci containing phosphor-H3 and condensin may be a higher-order architecture representing an intermediate step of condensation. Formation of such large foci most likely requires additional factors, since the interaction of condensin with histone H3 is not phosphorylation specific. Nonetheless, our results demonstrate that the human condensin complex remains associated with specific sites on chromosomes during interphase and participates in reinitiation of mitotic chromosome condensation together with phosphor-H3. Further investigation of the in vivo binding sites of the human condensin complex on interphase chromosomes as well as the requirement for the large foci formation with phosphor-H3 will be important to understand the role of human condensin in initiation of condensation in human cells.
ACKNOWLEDGMENTS
The first two authors contributed equally to this work.
We acknowledge W. R. Brinkley and C. D. Allis for kindly providing antibody for CREST antiserum and general histone H3, respectively. We thank M. Waterman, S. Sandmeyer, and A. R. Ball, Jr., for critical reading of the manuscript.
This work was supported in part by GM59150 from NIH to K.Y. K.Y. was a Leukemia Society of America Special Fellow.
REFERENCES
- 1.Bootsma D, Budke L, Vos O. Studies on synchronous division of tissue culture cells initiated by excess thymidine. Exp Cell Res. 1964;33:301–309. doi: 10.1016/s0014-4827(64)81035-1. [DOI] [PubMed] [Google Scholar]
- 2.Brenner S, Pepper D, Berns M W, Tan E, Brinkley B R. Kinetochore structure, duplication, and distribution in mammalian cells: analysis by human autoantibodies from scleroderma patients. J Cell Biol. 1981;91:95–102. doi: 10.1083/jcb.91.1.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cubizolles F, Legagneux V, Le Guellec R, Chartrain I, Uzbekov R, Ford C, Le Guellec K. pEg7, a new Xenopus protein required for mitotic chromosome condensation in egg extracts. J Cell Biol. 1998;143:1437–1446. doi: 10.1083/jcb.143.6.1437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.de Belle I, Cai S, Kohwi-Shigematsu T. The genomic sequences bound to special AT-rich sequence-binding protein 1 (SATB1) in vivo in Jurkat T cells are tightly associated with the nuclear matrix at the bases of the chromatin loops. J Cell Biol. 1998;141:335–348. doi: 10.1083/jcb.141.2.335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Earnshaw W C, Rothfield N. Identification of a family of human centromere proteins using autoimmune sera from patients with scleroderma. Chromosoma. 1985;91:313–321. doi: 10.1007/BF00328227. [DOI] [PubMed] [Google Scholar]
- 6.Fey E G, Wan K M, Penman S. Epithelial cytoskeletal framework and nuclear matrix-intermediate filament scaffold: three-dimensional organization and protein composition. J Cell Biol. 1984;98:1973–1984. doi: 10.1083/jcb.98.6.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Freeman L, Aragon-Alcaide L, Strunnikov A. The condensin complex governs chromosome condensation and mitotic transmission of rDNA. J Cell Biol. 2000;149:811–824. doi: 10.1083/jcb.149.4.811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hendzel M J, Wei Y, Mancini M A, Van Hooser A, Ranalli T, Brinkley B R, Bazett-Jones D P, Allis C D. Mitosis-specific phosphorylation of histone H3 initiates primarily within pericentromeric heterochromatin during G2 and spreads in an ordered fashion coincident with mitotic chromosome condensation. Chromosoma. 1997;106:348–360. doi: 10.1007/s004120050256. [DOI] [PubMed] [Google Scholar]
- 9.Hirano T. SMC-mediated chromosome mechanics: a conserved scheme from bacteria to vertebrates? Genes Dev. 1999;13:11–19. doi: 10.1101/gad.13.1.11. [DOI] [PubMed] [Google Scholar]
- 10.Hirano T, Kobayashi R, Hirano M. Condensins, chromosome condensation protein complexes containing XCAP-C, XCAP-E, and a Xenopus homolog of the Drosophila Barren protein. Cell. 1997;89:511–521. doi: 10.1016/s0092-8674(00)80233-0. [DOI] [PubMed] [Google Scholar]
- 11.Hirano T, Mitchison T J. A heterodimeric coiled-coil protein required for mitotic chromosome condensation in vitro. Cell. 1994;79:449–458. doi: 10.1016/0092-8674(94)90254-2. [DOI] [PubMed] [Google Scholar]
- 12.Jessberger R, Frei C, Gasser S M. Chromosome dynamics: the SMC protein family. Curr Opin Genes Dev. 1998;8:254–259. doi: 10.1016/s0959-437x(98)80149-4. [DOI] [PubMed] [Google Scholar]
- 13.Johnson G D, Nogueira Araujo G M. A simple method of reducing the fading of immunofluorescence during microscopy. J Immunol Methods. 1981;43:349–350. doi: 10.1016/0022-1759(81)90183-6. [DOI] [PubMed] [Google Scholar]
- 14.Kimura K, Hirano M, Kobayashi R, Hirano T. Phosphorylation and activation of 13S condensin by Cdc2 in vitro. Science. 1998;282:487–490. doi: 10.1126/science.282.5388.487. [DOI] [PubMed] [Google Scholar]
- 15.Kimura K, Hirano T. ATP-dependent positive supercoiling of DNA by 13S condensin: a biochemical implication for chromosome condensation. Cell. 1997;90:625–634. doi: 10.1016/s0092-8674(00)80524-3. [DOI] [PubMed] [Google Scholar]
- 16.Kimura K, Rybenkov V V, Crisona N J, Hirano T, Cozzarelli N R. 13S condensin actively reconfigures DNA by introducing global positive writhe: implication for chromosome condensation. Cell. 1999;98:239–248. doi: 10.1016/s0092-8674(00)81018-1. [DOI] [PubMed] [Google Scholar]
- 17.Knehr M, Poppe M, Enulescu M, Eickelbaum W, Stoehr M, Schroeter D, Paweletz N. A critical appraisal of synchronization methods applied to achieve maximal enrichment of HeLa cells in specific cell cycle phases. Exp Cell Res. 1995;217:546–553. doi: 10.1006/excr.1995.1121. [DOI] [PubMed] [Google Scholar]
- 18.Koshland D, Strunnikov A. Mitotic chromosome condensation. Annu Rev Cell Dev Biol. 1996;12:305–333. doi: 10.1146/annurev.cellbio.12.1.305. [DOI] [PubMed] [Google Scholar]
- 19.Moroi Y, Peebles C, Fritzler M J, Steigerwald J, Tan E M. Autoantibody to centromere (kinetochore) in scleroderma sera. Proc Natl Acad Sci USA. 1980;77:1627–1631. doi: 10.1073/pnas.77.3.1627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nagase T, Seki N, Tanaka A, Ishikawa K, Nomura N. Prediction of the coding sequences of 40 new genes (KIAA0121-KIAA0160) deduced by analysis of cDNA clones from human cell line KG-1. DNA Res. 1995;2:167–174. doi: 10.1093/dnares/2.4.167. [DOI] [PubMed] [Google Scholar]
- 21.Saitoh N, Goldberg I G, Wood E R, Earnshaw W C. ScII: an abundant chromosome scaffold protein is a member of a family of putative ATPases with an unusual predicted tertiary structure. J Cell Biol. 1994;127:303–318. doi: 10.1083/jcb.127.2.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schmiesing J A, Ball A R, Gregson H C, Alderton J, Zhou S, Yokomori K. Identification of two distinct human SMC protein complexes involved in mitotic chromosome dynamics. Proc Natl Acad Sci USA. 1998;95:12906–12911. doi: 10.1073/pnas.95.22.12906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sutani T, Yuasa T, Tomonaga T, Dohmae N, Takio K, Yanagida M. Fission yeast condensin complex: essential roles of non-SMC subunits for condensation and Cdc2 phosphorylation of Cut3/SMC4. Genes Dev. 1999;13:2271–2283. doi: 10.1101/gad.13.17.2271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wei Y, Yu L, Bowen J, Gorovsky M A, Allis C D. Phosphorylation of histone H3 is required for proper chromosome condensation and segregation. Cell. 1999;97:99–109. doi: 10.1016/s0092-8674(00)80718-7. [DOI] [PubMed] [Google Scholar]
- 25.Yokomori K, Zeidler M P, Chen J-L, Verrijzer C P, Mlodzik M, Tjian R. Drosophila TFIIA directs cooperative DNA binding with TBP and mediates transcriptional activation. Genes Dev. 1994;8:2313–2323. doi: 10.1101/gad.8.19.2313. [DOI] [PubMed] [Google Scholar]
- 26.Zieve G W, Turnbull D, Mullins M, McIntosh J R. Production of large numbers of mitotic mammalian cells by use of the reversible microtubule inhibitor nocodazole. Exp Cell Res. 1980;126:397–405. doi: 10.1016/0014-4827(80)90279-7. [DOI] [PubMed] [Google Scholar]