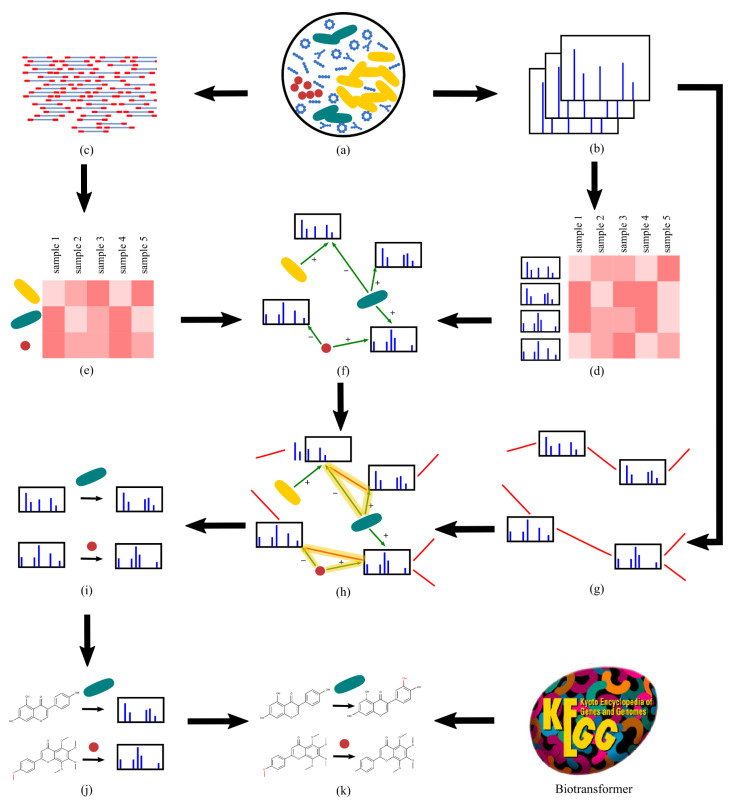
Figure 1.
TransDiscovery framework for discovering novel biotransformations of human dietary ingredients by (a) the gut microbiome. Starting with (b) the mass spectral data of small gut molecules and (c) metagenomics data of gut microbes, the pipeline includes the following steps: extracting (d) molecular and (e) microbial features from raw data, (f) constructing an association network [26,27,28] of molecular and microbial features (edges shown in green), (g) constructing a molecular network [30] (edges shown in red), (h) integrating associations and the molecular network, (i) extracting candidate biotransformations as golden triangles, (j) identifying substrates of biotransformations with an in silico database search with Dereplicator+ [31], and (k) characterizing molecular products of known biotransformations using in silico predictions of BioTransformer [29]. Note that in steps (f,h–k), the nodes can represent either strains or enzymes. In steps (f,h), the plus and minus labels indicate that the substrate is negatively correlated with the microbial feature and the product is positively correlated with the microbial feature.