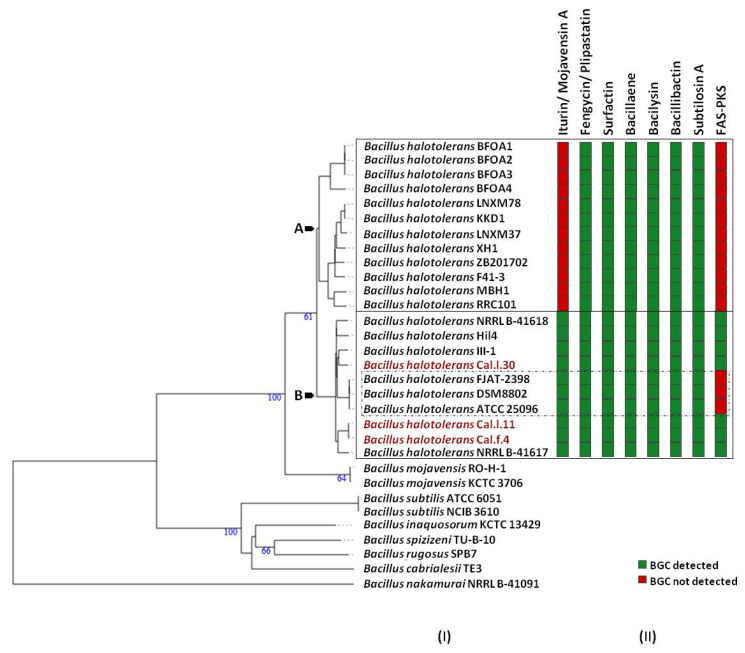
Figure 6.
The secondary metabolic gene clusters evolution. (I) Whole genome phylogeny of B. halotolerans strains highlighting the endophytic bacterial strains Cal.l.30, Cal.f.4 and Cal.l.11. The numbers above branches are pseudo-bootstrap support values from 100 replications. Inferred clusters are for B. halotolerans strains harboring (Cluster B) or lacking (Cluster A) the iturin/ mojavensin A BGC. (II) Number and distribution heatmap of the secondary metabolite biosynthetic gene clusters (iturin/ mojavensin A, fengycin/ plipastatin, surfactin, bacillaene, bacilysin, bacillibactin, subtilosin A and FAS-PKS) detected (green) or not (red) among B. halotolerans strains. B. halotolerans strains Cal.l.30, Cal.l.11 and Cal.f.4 are highlighted in red.