Abstract
Nidogen 1 is a highly conserved protein in mammals, Drosophila melanogaster, Caenorhabditis elegans, and ascidians and is found in all basement membranes. It has been proposed that nidogen 1 connects the laminin and collagen IV networks, so stabilizing the basement membrane, and integrates other proteins, including perlecan, into the basement membrane. To define the role of nidogen 1 in basement membranes in vivo, we produced a null mutation of the NID-1 gene in embryonic stem cells and used these to derive mouse lines. Homozygous animals produce neither nidogen 1 mRNA nor protein. Surprisingly, they show no overt abnormalities and are fertile, their basement membrane structures appearing normal. Nidogen 2 staining is increased in certain basement membranes, where it is normally only found in scant amounts. This occurs by either redistribution from other extracellular matrices or unmasking of nidogen 2 epitopes, as its production does not appear to be upregulated. The results show that nidogen 1 is not required for basement membrane formation or maintenance.
The nidogens form a family of related proteins, which in addition to the original mammalian nidogen, nidogen 1 (16) or entactin 1 (6), also includes a second member, nidogen 2 (14) or entactin 2 (13), and several related species from ascidians (23), Caenorhabditis elegans (12) and Drosophila melanogaster (19).
Nidogen 1, the best-described member of this family is, together with perlecan, laminin, and collagen IV, a ubiquitous component of basement membranes (32). First identified from a basement membrane-secreting cell line (4) and the murine EHS tumor (31), nidogen 1 comprises three globular domains, G1 to G3, with G1 and G2 connected by a flexible link and G2 and G3 connected by a rod-like domain (10). When isolated under nondenaturing conditions, nidogen 1 is bound noncovalently to laminin (24) by the G3 domain that has been demonstrated to interact with high affinity with the LE4 module in the short arm of the laminin γ1 chain (17, 25). As nidogen 1 has also been shown to bind to collagen IV by its G2 domain (3, 10), it has been proposed to be crucial in linking the laminin and collagen IV networks. In vitro nidogen 1 binds to perlecan and fibulins (32) and has therefore been considered to play a key role in the stabilization of the basement membrane. Nidogen 1 is highly susceptible to protease degradation (8, 18, 28), and its destruction may be the initial step in the breakdown of the basement membrane needed in tissue remodeling (1).
Disruption of the laminin-nidogen 1 interaction in organ cultures by use of antibodies against the laminin LE4 domain impaired branching morphogenesis in the kidney or salivary gland and induced a distortion of the basement membrane (9, 11). These effects could be counteracted by epidermal growth factor, which increased the production of nidogen 1 in the mesenchyme (11).
Nidogen 2, as described by Kohfeldt et al. (14), conserves all the domains of nidogen 1 and interacts with collagens I and IV as well as perlecan. However, its binding to the LE4 motif of the laminin γ1 chain is markedly weaker than that seen for nidogen 1, and it does not interact with fibulins. Nidogen 2 is present in most basement membranes but has in some a different distribution than nidogen 1, particularly in those surrounding striated muscles.
Based on the in vitro binding data, the lack of nidogen 1 is expected to affect the structure of all basement membranes. We therefore decided to generate nidogen 1-deficient mouse lines in order to define its role in basement membrane formation in vivo.
MATERIALS AND METHODS
Production of the targeting construct.
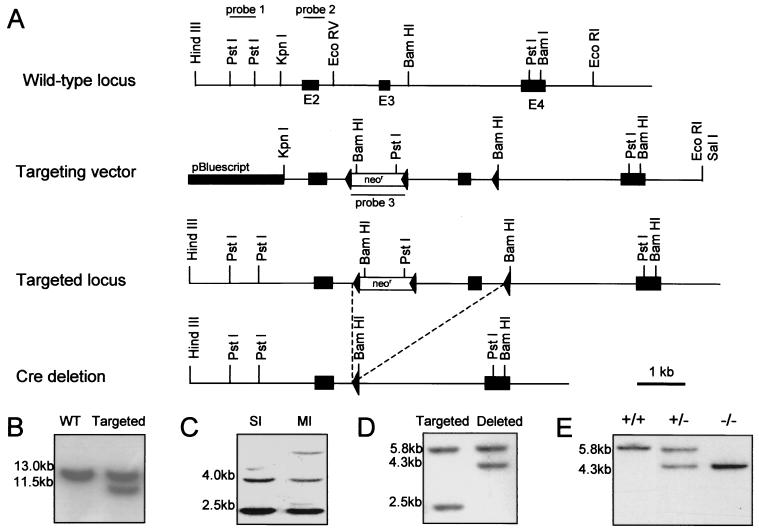
A lambda FIX II genomic library (Stratagene) of the 129SVJ mouse line was screened using a DNA fragment corresponding to exons 2 to 4 of the mouse NID-1 cDNA (7). Six individual clones were isolated and mapped. The targeting construct in pBluescript II KS (Stratagene) contains a 7-kb KpnI/EcoRI genomic fragment comprising 0.8 kb upstream of exon 2 to 1 kb downstream of exon 4. The phosphoglycerate kinase-driven neomycin resistance (neoR) cassette flanked by loxP sites (kindly provided by W. Müller, Institute for Genetics, University of Cologne) was inserted into the unique EcoRV site located 950 bp upstream of exon 3 and the third loxP site into the BamHI site located 300 bp downstream of exon 3 (see Fig. 1A).
FIG. 1.
Targeted disruption of the NID-1 gene and generation of chimeras. (A) A restriction map and exon-intron structure (top), the targeting vector (middle), and the mutant allele after homologous recombination and Cre recombinase-mediated deletion (bottom) are shown. Exon numbers are indicated. (B) BamHI-digested DNA from ES clones analyzed with probe 1. The wild-type (WT) and targeted alleles generate 13- and 11.5-kb fragments, respectively. (C) PstI-digested DNA from ES cells analyzed with probe 3 revealed two bands of 2.5 and 4 kb for single integration (SI) of the targeting construct and additional bands in the case of multiple integrations (MI). (D) Southern blot of genomic DNA from ES clones after transient transfection with the Cre recombinase expression plasmid analyzed with probe 2. Hybridization of PstI-digested DNA yields for the targeted allele bands of 5.8 and 2.5 kb and, after Cre recombinase-mediated deletion of the neoR cassette and exon 3, a shift of the smaller band to 4.3 kb. (E) BamHI-digested tail DNA of the offspring from NID-1+/− mice matings hybridized with probe 2 show that homozygous animals were born.
Disruption of the NID-1 gene in ES cells and generation of chimeric mice.
E14 embryonic stem (ES) cells were grown under standard ES cell conditions. ES cells were transfected by electroporation with the SalI-linearized targeting construct, and colonies were selected for resistance to G418. Surviving clones were screened for homologous recombination by Southern blotting. The BamHI-digested DNA was analyzed with the external probe 1 (Fig. 1B). In cases of correct integration, the wild-type 13-kb fragment was reduced to 11.5 kb. Cointegration of the third loxP site was analyzed by PCR, and single insertion was demonstrated by hybridization of PstI-digested DNA with the neoR probe (probe 3), resulting in two fragments of 2.5 and 4 kb (Fig. 1C). For Cre recombinase-mediated deletion of the neoR cassette and the third exon, the correctly targeted ES clones were transiently transfected with the plasmid pCrePac encoding Cre recombinase and the gene for puromycin N-acetyltransferase (30). After puromycin selection for 48 h, the cells were trypsinized and replated. Clones were picked and expanded, and DNA was extracted for Southern blotting. PstI-digested DNA was probed with internal probe 2; in nontransfected cells, this yielded two fragments of 5.8 and 2.5 kb. After Cre recombinase-mediated deletion of both the neoR cassette and exon 3, the smaller band was shifted to 4.3 kb (Fig. 1D).
Two independent ES cell lines were used to generate germ line chimeras as previously described (29). Briefly, the ES cells were injected into C57BL/6-derived blastocystes. Male chimeric animals were bred to C57BL/6 females, and germline transmission was shown by coat color and Southern blotting (Fig. 1E). Heterozygous animals were mated together to obtain nidogen 1-null offsprings.
Analysis of nidogen expression.
Tissue samples from wild-type and homozygous mice were isolated for RNA and protein analysis. To obtain protein, tissue was homogenized in extraction buffer (50 mM Tris-HCl [pH 7.5]), 1% Triton X-100, 10 mM EDTA, 100 mM NaCl, 10 ng of leupeptin per ml, 1 ng of pepstatin per ml, 1 mM benzamidin, 0.5 mM phenylmethylsulfonyl fluoride [PMSF]). After being centrifuged, the supernatants (30 μg/lane) were fractionated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) on 5 to 12% gradient gels under reducing conditions for detection of nidogen 1 and laminin 1 and under nonreducing conditions for detection of nidogen 2. After the proteins were transferred to nitrocellulose, the membranes were blocked overnight in Tris-buffered saline with 5% skimmed-milk powder and incubated with polyclonal rabbit sera either directed against nidogen 1 (sera diluted 1:1,000) (10), nidogen 2 (diluted 1:500) (14), or the laminin 1-nidogen 1 complex (diluted 1:1,000) (15). For detection, a horseradish peroxidase-conjugated swine anti-rabbit antibody (diluted 1:2,000) was used, followed by development with the ECL system (Amersham Pharmacia Biotech). The protein concentration of the supernatants was determined with a commercial assay (Bio-Rad). As a control for the amount of protein blotted, the membranes were stained with Ponceau S solution (Sigma).
Total RNA was isolated from tissues by homogenization in RNAzol reagent (Wak-Chemie) according to the supplier's protocol. For Northern hybridization, 20 μg of total RNA was separated in a 1% agarose gel under denaturing conditions and then blotted onto a nylon membrane. As a control for the amount and integrity of the RNA blotted, the membrane was stained with methylene blue (0.4% in 0.2 M sodium acetate [pH 5.2]). Filters were then hybridized according to published protocols with 32P-labeled full-length nidogen 1 cDNA (16).
Immunofluorescence staining of tissues.
Organs were isolated and sectioned as described before (29). Immunostaining was performed using polyclonal rabbit antisera directed against nidogen 1 (10), nidogen 2 (14), laminin 1 (15), and perlecan (27). Immunoreactivity was detected with a Cy3-conjugated goat anti-rabbit immunoglobulin G polyclonal serum (Jackson Immunodiagnostics).
Electron microscopy and immunogold labeling of tissues.
The kidneys and the hind leg muscles of 6-week-old wild-type and nidogen 1−/− mice were dissected. For morphological investigation, 2-mm2 kidney tissue fragments were isolated; for immunogold histochemistry, 1-mm2 specimens of the soleus muscle were isolated. Fixation, preparation, and staining for morphology have been described before (21). For immunogold histochemistry, anti-nidogen 2 polyclonal serum (diluted 1:200) was used. Antibody labeling was carried out as described previously (21). To exclude nonspecific binding, control sections were incubated with the pure gold solution or with the gold-coupled secondary antibodies alone. All controls were negative.
RESULTS
Generation of mice with a disrupted NID-1 gene.
A targeting vector was constructed in which the exon 3 of the NID-1 gene (7) was flanked with loxP sites (Fig. 1A). Of 250 G418 resistant ES cell clones analyzed, 23 had undergone recombination at the NID-1 locus. This is shown by the appearance of an 11.5-kb band with intensity equal to that of the wild-type band of 13 kb (Fig. 1B). A correct and single integration was proven by the use of an internal probe (Fig. 1C). Of these 23 clones, only 2 showed cointegration of the third downstream-located loxP site. These ES cell clones were used for Cre recombinase-mediated deletion of the neoR cassette and exon 3. Of 200 clones isolated after transient transfection with the Cre recombinase-containing plasmid, 5 had undergone this deletion event (Fig. 1D). Hybridization of PstI-digested DNA with probe 2 yielded the expected shift of the 2.5-kb band obtained with the targeted allele to a 4.3 kb band after deletion of the neoR cassette and exon 3. These clones were used to generate chimeric males that transmitted the mutant allele to their progeny. Mice heterozygous for the mutation in the NID-1 gene were identified by Southern blot analysis of PstI-digested tail DNA (Fig. 1E). Heterozygous mice appeared normal and were indistinguishable from their wild-type littermates. To obtain homozygous animals for the NID-1 mutation, heterozygous mice were intercrossed (Fig. 1E).
Nidogen 1-deficient mice are viable.
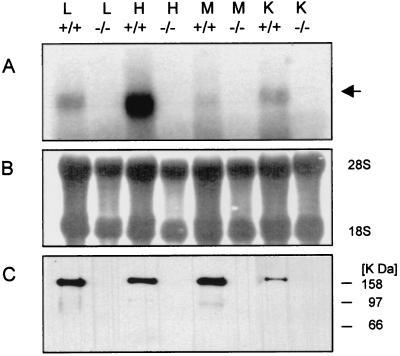
Homozygous animals were born and appeared phenotypically normal when compared to their heterozygous and wild-type littermates. Among the 273 viable offspring from heterozygous matings, expected Mendelian ratios were seen: 137 (50%) were heterozygotes, 64 (24%) were wild type, and 72 (26%) were homozygotes, showing that the mutation in the NID-1 gene does not cause an embryonic lethality. Further, the homozygous mutant mice do not show any overt anatomical abnormality and are fertile. Based upon the genomic organization of the mouse NID-1 gene (7), deletion of exon 3 should result in a frameshift mutation, introducing multiple-stop codons and so disrupting the NID-1 transcripts. In wild-type animals, a protein band of 150 kDa corresponding to nidogen 1 could be detected after immunoblotting with a nidogen 1-specific antibody, while no signal, either of 150 kDa or any truncated form, was seen in any of the organs of homozygous mutant animals. Northern hybridization failed to reveal a band corresponding to the full-length nidogen message in the mutant animals or any shorter transcript, suggesting that the loss of the third exon presumably led to destabilization of the NID-1 mRNA. In wild-type controls, the 6-kb nidogen message was evident (Fig. 2). These results show that the mutation resulted in a null allele.
FIG. 2.
Expression analysis in tissues from homozygous animals (−/−) and their wild-type littermates (+/+). Total RNA and protein were isolated from lung (L), heart (H), skeletal muscle (M), and kidney (K). Total RNA (20 μg/lane) was hybridized with the 32P-labeled full-length nidogen 1 cDNA (A). The nidogen 1-specific message is indicated. Methylene blue staining of 28S and 18S rRNA is shown as a control for equal loading (B). Immunoblot analysis of protein extracts (30 μg/lane) after SDS-PAGE with a polyclonal nidogen 1-specific antiserum. The gel was calibrated by globular marker proteins (in kilodaltons) as indicated (C).
Basement membranes form in the absence of nidogen 1.
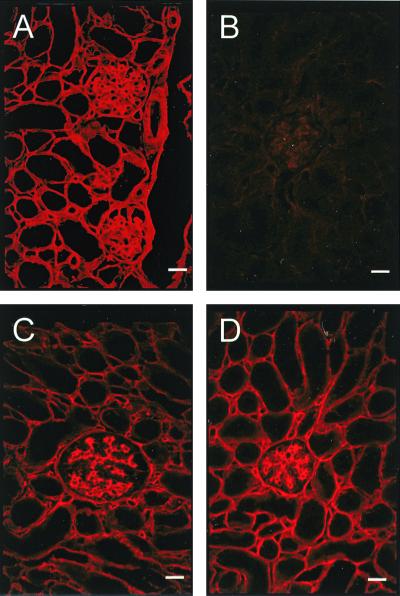
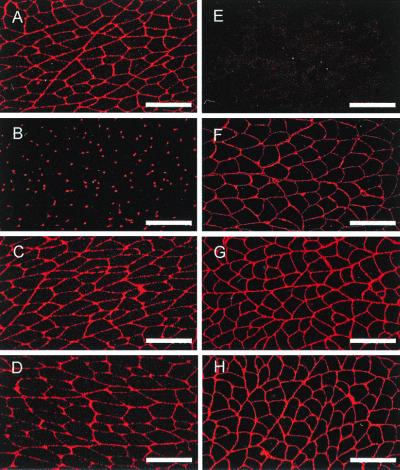
To analyze the molecular composition of the basement membrane, sections of various organs, including kidney (Fig. 3), skeletal muscle (Fig. 4), and heart (Fig. 5), were immunostained for the main basement membrane proteins. In all tissues of homozygous mutant mice, no signal was observed for nidogen 1, and the staining for laminin 1 or perlecan was unchanged compared with that for wild-type littermates (Fig. 4). In most organs (for example, in the kidney [Fig. 3]), nidogen 2 staining is found in all basement membranes, showing no alterations upon the loss of nidogen 1. In contrast, nidogen 2-specific antibodies in striated muscles detected a marked change. In normal skeletal muscle, staining for nidogen 2 is weak in the basement membranes surrounding the sarcomeres, while the endothelial basement membranes stain strongly (Fig. 4B). In the nidogen 1−/− animals, however, nidogen 2 appeared abundant in both sites with a staining pattern reminiscent of that of nidogen 1 in the control animals (Fig. 4F). Similar results were obtained for heart tissue (Fig. 5), with a more intense nidogen 2 staining of the basement membranes surrounding cardiocytes than in sections from wild-type littermates. Immunogold histochemistry using nidogen 2 antibodies gave similar results with a marked increase in the labeling of both the basement membrane and the extracellular matrix surrounding myocytes in the nidogen 1-null animals (Fig. 6).
FIG. 3.
Immunofluorescence staining for components of the basement membrane in frozen kidney sections from wild-type (left panels) and nidogen 1-deficient (right panels) animals. The primary polyclonal antibodies used were against nidogen 1 (A and B) and nidogen 2 (C and D). Bar, 50 μm.
FIG. 4.
Immunofluorescence staining for basement membrane components in frozen soleus muscle sections from wild-type (left panels) and nidogen 1-deficient (right panels) animals. The primary polyclonal antibodies used were against nidogen 1 (A and E), nidogen 2 (B and F), laminin 1 (C and G), and perlecan (D and H). Bar, 50 μm.
FIG. 5.
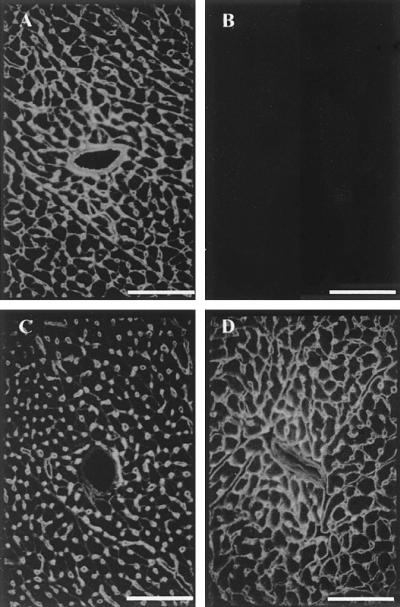
Immunofluorescence staining of frozen heart sections from wild-type (upper panels) and homozygous (lower panels) animals. The primary polyclonal antibodies used were against nidogen 1 (A and B) and nidogen 2 (C and D). Bar, 50 μm.
FIG. 6.
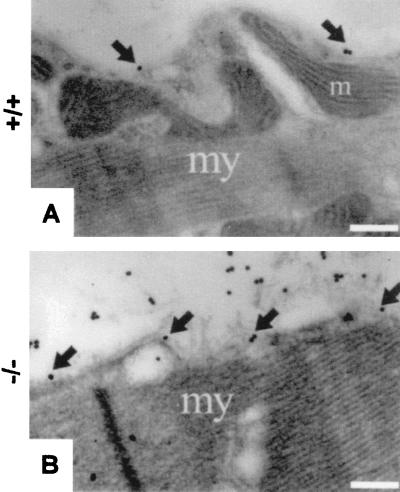
Immunogold histochemistry with antibodies against nidogen 2 on sections of soleus muscle from 6-week-old wild-type (A) and nidogen 1-deficient (B) animals. Note the sparse staining for nidogen 2 in the wild-type muscle basement membrane (arrows) and the strong staining within the basement membrane (arrows) and the extracellular matrix of the endomysium in the nidogen 1-deficient animals. my, myocyte; m, mitochondrion. Bars, 0.32 μm.
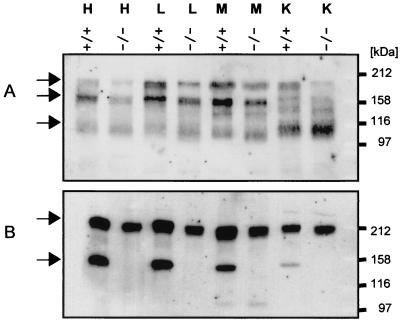
To investigate whether this change in signal intensity in NID-1−/− mice is due to an increased presence of nidogen 2, tissue extracts were analyzed by immunoblotting using a nidogen 2-specific antibody. As shown in Fig. 7A, three major bands of 200, 170, and approximately 110 kDa were detected, the two high-molecular-weight bands corresponding to the full-length protein and the protein after proteolytic cleavage in the link region (14), while the presence of further proteolytic products in tissue extracts has also been described (14). However, no significant differences in signal intensity were observed between wild-type tissues and those of nidogen−/− mice. As both nidogen isoforms bind to laminin, although with various affinities, we tested tissue extracts for the levels of laminin chains by immunoblotting using antibodies against the laminin 1-nidogen 1 complex. One band of approximately 200 kDa, corresponding to the laminin β1 and γ1 chains, was detected in all tissues; as well, a 150-kDa band, corresponding to nidogen 1, was found in the extracts from wild-type tissue (Fig. 7B). In agreement with the immunofluorescence data, there was no evident alteration in laminin present in the tissues.
FIG. 7.
Western blot analysis of tissue extracts from wild-type (+/+) and homozygous (−/−) animals. Tissue extracts from heart (H), lung (L), skeletal muscle (M), and kidney (K) were separated by SDS-PAGE under nonreducing conditions for detection of nidogen 2 (A) and under reduced conditions for detection of laminin chains (B). The blots were probed with antibodies directed against nidogen 2 or the laminin 1-nidogen 1 complex. Calibration shown on the right was carried out with globular proteins (in kilodaltons).
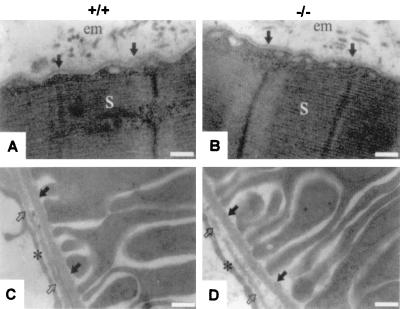
Nidogen 1 deficiency resulted in no ultrastructural changes in the basement membranes either in organs lacking a detectable nidogen 2 redistribution, such as kidney, or in striated muscles. Further, there was no evident change in the cellular architecture due to the absence of nidogen 1 (Fig. 8).
FIG. 8.
Electron micrographs of ultrathin sections of the hind limb soleus muscle (A and B) and the kidney cortex (C and D) from 6-week-old wild-type (+/+) and homozygous (−/−) animals. In panels A and B, the basement membrane of the myocyte is marked by arrows. em, endomysium; s, sarcomer. In panels C and D, the basement membrane of a proximal tubule is marked by solid arrows, and the endothelial basement membrane is indicated by open arrows. Endothelial cells are indicated by asterisks. Bar, 0.25 μm.
DISCUSSION
As nidogen 1 interacts in vitro with all other major basement membrane components, particularly collagen IV and laminin, a model for basement membrane deposition and organization has been suggested where the laminin gel and the collagen IV network are linked together by nidogen (32). As many cell culture studies with isolated laminin or laminin complexed with nidogen 1 have not shown any differences due to the presence of nidogen 1, it was believed that nidogen 1 plays a mainly structural role within the basement membrane. Further, embryonic basement membranes do not have a fully developed ultrastructural architecture until nidogen 1 becomes present (22). Given these observations, it has long been considered that nidogen 1 had a purely structural role, hence it was unexpected that mice lacking nidogen 1 produce normal basement membranes.
Absence of the laminin network induced by targeting the LAMC1 gene coding for the ubiquitous laminin γ1 chain prevents basement membrane formation (29), and the loss of less-ubiquitous laminin subunits (20, 26, 33) results in basement membranes which are often intrinsically weaker, leading to tissue abnormalities. As nidogen 1-null animals appear to develop normally and there is no evidence of structural deformity in the basement membrane, we assume that lack of nidogen 1 has no effect upon laminin assembly. The loss of perlecan results in a less-stable basement membrane in certain tissues (2, 5). Hence, it would appear that nidogen 1 is not essential for the retention of perlecan in the basement membrane. These results suggest either that nidogen 1 has no structural role in the basement membrane or that its absence is compensated for by other proteins. Interestingly, a C. elegans mutation was described recently where the loss of its single nidogen gene did not interfere with basement membrane formation but resulted in alterations in axonal patterning (12). This suggests that the nidogens may have nonstructural functions. This will have to be further elucidated in the nidogen 1−/− mice, in particular with respect to subtle neurological changes.
In nidogen 1-null mice, nidogen 2 staining is increased in certain basement membranes, particularly in cardiac and skeletal muscle, where it normally has only a limited expression. However, immunoblotting showed that the increase in the signal in the basement membrane was not due to more nidogen 2 present in the tissues. This suggests that in the absence of nidogen 1, there is either a redistribution of nidogen 2 from other extracellular sources into the basement membrane or an unmasking of nidogen 2 epitopes. If the latter, this could have occurred due to the unmasking of nidogen ligands which are preferentially bound by nidogen 1 or due to an upregulation of a binding protein in the basement membrane in response to the loss of nidogen 1.
While nidogen 1 and 2 are structurally related, in vitro binding studies show that they differ in their affinities for other basement membrane components. In particular, the interaction with the laminin γ1 chain is far weaker for nidogen 2 (14). Therefore, if nidogen 2 compensates for nidogen 1, this suggests that the in vitro binding data do not fully reflect the in vivo situation. Disrupting the laminin-nidogen 1 interaction by the use of antibodies against the LE4 domain of laminin leads to dramatic effects on organogenesis not seen upon the total removal of nidogen 1 (9, 11), again suggesting that the loss of nidogen 1 is compensated for by nidogen 2. However, the possibility that the bound antibody disturbed other proteins interacting with laminin close to the LE4 motif can not be excluded. The results presented here demonstrate that to identify the true roles of the nidogen family will require production of nidogen 2-null mouse lines, lines lacking both isoforms, and lines lacking the binding sites on relevant ligands (19).
ACKNOWLEDGMENTS
This work was supported by a grant from the German Research Foundation (FOR 265/2) and by the Bundesministerium für Bildung, Wissenschaft, Forschung, und Technologie (grant ZMMK-TV25).
We thank Christian Frie and Petra Weskamp (Institute for Biochemistry II) and Marion Reibetanz (Department of Dermatology) for excellent technical assistance. We are greatly indebted to R. Timpl and U. Mayer (Max Planck Institute for Biochemistry, Martinsried, Germany) for access to reagents and W. Müller (Institute for Genetics, University of Cologne) for helpful discussions.
M.M. and N.S. contributed equally to this work.
REFERENCES
- 1.Alexander C M, Howard E W, Bissell M J, Werb Z. Rescue of mammary epithelial cell apoptosis and entactin degradation by a tissue inhibitor of metalloproteinases-1 transgene. J Cell Biol. 1996;135:1669–1677. doi: 10.1083/jcb.135.6.1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arikawa-Hirasawa E, Watanabe H, Takami H, Hassell J R, Yamada Y. Perlecan is essential for cartilage and cephalic development. Nat Genet. 1999;23:354–358. doi: 10.1038/15537. [DOI] [PubMed] [Google Scholar]
- 3.Aumailley M, Battaglia C, Mayer U, Nischt R, Timpl R, Fox J W. Nidogen mediates the formation of ternary complexes of basement membrane components. Kidney Int. 1993;43:7–12. doi: 10.1038/ki.1993.3. [DOI] [PubMed] [Google Scholar]
- 4.Carlin B, Jaffe R, Bender B, Chung A E. Entactin, a novel basal lamina-associated glycoprotein. J Biol Chem. 1981;256:5209–5214. [PubMed] [Google Scholar]
- 5.Costell M, Gustafsson E, Aszodi A, Mörgelin M, Bloch W, Hunziker E, Addicks K, Timpl R, Fässler R. Perlecan maintains the integrity of cartilage and some basement membranes. J Cell Biol. 1999;147:1109–1122. doi: 10.1083/jcb.147.5.1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Durkin M E, Chakravati S, Bartos B B, Liu S-H, Friedman R L, Chung A E. Amino acid sequence and domain structure of entactin. Homology with epidermal growth factor precursor and low density lipoprotein receptor. J Cell Biol. 1988;107:2749–2756. doi: 10.1083/jcb.107.6.2749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Durkin M E, Wewer U M, Chung A E. Exon organization of the mouse entactin gene corresponds to the structural domains of the polypeptide and has regional homology to the low-density lipoprotein receptor gene. Genomics. 1995;26:219–228. doi: 10.1016/0888-7543(95)80204-y. [DOI] [PubMed] [Google Scholar]
- 8.Dziadek M, Clements R, Mitrangas K, Reiter H, Fowler K. Analysis of degradation of the basement membrane protein nidogen, using a specific monoclonal antibody. Eur J Biochem. 1988;172:219–225. doi: 10.1111/j.1432-1033.1988.tb13876.x. [DOI] [PubMed] [Google Scholar]
- 9.Ekblom P, Ekblom M, Fecker L, Klein G, Zhang H-Y, Kadoya Y, Chu M-L, Mayer U, Timpl R. Role of mesenchymal nidogen for epithelial morphogenesis in vitro. Development. 1994;120:2003–2014. doi: 10.1242/dev.120.7.2003. [DOI] [PubMed] [Google Scholar]
- 10.Fox J W, Mayer U, Nischt R, Aumailley M, Reinhardt D, Wiedemann H, Mann K, Timpl R, Krieg T, Engel J, Chu M-L. Recombinant nidogen consists of three globular domains and mediates binding of laminin to collagen type IV. EMBO J. 1991;10:3137–3146. doi: 10.1002/j.1460-2075.1991.tb04875.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kadoya Y, Salmivirta K, Talts J F, Kadoya K, Mayer U, Timpl R, Ekblom P. Importance of nidogen binding to laminin γ1 for branching epithelial morphogenesis of the submandibular gland. Development. 1997;124:683–691. doi: 10.1242/dev.124.3.683. [DOI] [PubMed] [Google Scholar]
- 12.Kim S, Wadsworth W G. Positioning of longitudinal nerves in C. elegans by nidogen. Science. 2000;288:150–154. doi: 10.1126/science.288.5463.150. [DOI] [PubMed] [Google Scholar]
- 13.Kimura N, Toyoshima T, Kojima T, Shimane M. Entactin-2: a new member of basement membrane protein with high homology to entactin/nidogen. Exp Cell Res. 1998;241:36–45. doi: 10.1006/excr.1998.4016. [DOI] [PubMed] [Google Scholar]
- 14.Kohfeldt E, Sasaki T, Göhring W, Timpl R. Nidogen-2: a new basement membrane protein with diverse binding properties. J Mol Biol. 1998;282:99–109. doi: 10.1006/jmbi.1998.2004. [DOI] [PubMed] [Google Scholar]
- 15.Kücherer-Ehret A, Pottgießer J, Kreutzberg G W, Thoenen H, Edgar D. Developmental loss of laminin from the interstitial extracellular matrix correlates with decreased laminin gene expression. Development. 1990;110:1285–1293. doi: 10.1242/dev.110.4.1285. [DOI] [PubMed] [Google Scholar]
- 16.Mann K, Deutzmann R, Aumailley M, Timpl R, Raimondi L, Yamada Y, Pan T-C, Conway D, Chu M-L. Amino acid sequence of mouse nidogen, a multidomain basement membrane protein with binding activity for laminin, collagen IV and cells. EMBO J. 1989;8:65–72. doi: 10.1002/j.1460-2075.1989.tb03349.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mayer U, Nischt R, Pöschl E, Mann K, Fukuda K, Gerl M, Yamada Y, Timpl R. A single EGF-like motif of laminins is responsible for high affinity nidogen binding. EMBO J. 1993;12:1879–1885. doi: 10.1002/j.1460-2075.1993.tb05836.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mayer U, Zimmermann K, Mann K, Reinhardt D, Timpl R, Nischt R. Binding properties and protease stability of recombinant human nidogen. Eur J Biochem. 1995;227:681–686. doi: 10.1111/j.1432-1033.1995.tb20188.x. [DOI] [PubMed] [Google Scholar]
- 19.Mayer U, Kohfeldt E, Timpl R. Structural and genetic analysis of laminin-nidogen interaction. Ann N Y Acad Sci. 1998;857:130–142. doi: 10.1111/j.1749-6632.1998.tb10113.x. [DOI] [PubMed] [Google Scholar]
- 20.Miner J H, Cunningham J, Sanes J R. Roles for laminin in embryogenesis: exencephaly, syndactyly, and placentopathy in mice lacking the laminin α5 chain. J Cell Biol. 1998;143:1713–1723. doi: 10.1083/jcb.143.6.1713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Miosge N, Heinemann S, Leissling A, Klenczar C, Herken R. Ultrastructural triple localization of laminin-1, nidogen-1, and collagen type IV helps elucidate basement membrane structure in vivo. Anat Rec. 1999;254:382–388. doi: 10.1002/(SICI)1097-0185(19990301)254:3<382::AID-AR9>3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
- 22.Miosge N, Quondamatteo F, Klenczar C, Herken R. Nidogen-1: expression and ultrastructural localization during the onset of mesoderm formation in the early mouse embryo. J Histochem Cytochem. 2000;48:229–237. doi: 10.1177/002215540004800208. [DOI] [PubMed] [Google Scholar]
- 23.Nakae H, Sugano M, Ishimori Y, Endo T, Obinata T. Ascidian entactin/nidogen. Implication of evolution by shuffling two kinds of cysteine-rich motifs. Eur J Biochem. 1993;213:11–19. doi: 10.1111/j.1432-1033.1993.tb17729.x. [DOI] [PubMed] [Google Scholar]
- 24.Paulsson M, Aumailley M, Deutzmann R, Timpl R, Beck K, Engel J. Laminin-nidogen complex. Extraction with chelating agents and structural characterization. Eur J Biochem. 1987;166:11–19. doi: 10.1111/j.1432-1033.1987.tb13476.x. [DOI] [PubMed] [Google Scholar]
- 25.Pöschl E, Fox J W, Block D, Mayer U, Timpl R. Two non-contiguous regions contribute to nidogen binding to a single EGF-like motif of the laminin γ1 chain. EMBO J. 1994;13:3741–3747. doi: 10.1002/j.1460-2075.1994.tb06683.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ryan M C, Lee K, Myashita Y, Carter W G. Targeted disruption of the LAMA3 gene in mice reveals abnormalities in survival and late stage differentiation of epithelial cells. J Cell Biol. 1999;145:1309–1323. doi: 10.1083/jcb.145.6.1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schulze B, Mann K, Battistutta R, Wiedemann H, Timpl R. Structural properties of recombinant domain III-3 of perlecan containing a globular domain inserted into an epidermal-growth-factor-like motif. Eur J Biochem. 1995;231:551–556. doi: 10.1111/j.1432-1033.1995.tb20731.x. [DOI] [PubMed] [Google Scholar]
- 28.Sires U L, Griffin G L, Broekelmann T J, Mecham R P, Murphy G, Chung A E, Welgus H G, Senior R M. Degradation of entactin by matrix metalloproteinases. J Biol Chem. 1993;268:2069–2074. [PubMed] [Google Scholar]
- 29.Smyth N, Vatansever H S, Murray P, Meyer M, Frie C, Paulsson M, Edgar D. Absence of basement membranes after targeting the LAMC1 gene results in embryonic lethality due to the failure of endoderm differentiation. J Cell Biol. 1999;144:151–160. doi: 10.1083/jcb.144.1.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Taniguchi M, Sanbo M, Watanabe S, Naruse I, Mishina M, Yagi T. Efficient production of Cre-mediated site-directed recombinants through the utilization of the puromycin resistance gene, pac: a transient gene-integration marker for ES cells. Nucleic Acids Res. 1998;26:679–680. doi: 10.1093/nar/26.2.679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Timpl R, Dziadek M, Fujiwara S, Nowack H, Wick G. Nidogen: a new, self-aggregating basement membrane protein. Eur J Biochem. 1983;15:455–465. doi: 10.1111/j.1432-1033.1983.tb07849.x. [DOI] [PubMed] [Google Scholar]
- 32.Timpl R, Brown J C. Supramolecular assembly of basement membranes. Bioessays. 1996;18:123–132. doi: 10.1002/bies.950180208. [DOI] [PubMed] [Google Scholar]
- 33.Xu H, Christmas P, Wu X R, Wewer U M, Engvall E. Defective muscle basement membrane and lack of M-laminin in the dystrophic dy/dy mouse. Proc Natl Acad Sci USA. 1994;91:5572–5576. doi: 10.1073/pnas.91.12.5572. [DOI] [PMC free article] [PubMed] [Google Scholar]