Abstract
Fighting against the emergent coronavirus disease (COVID-19) remains a big challenge at the front of the world communities. Recent research has outlined the potential of various medicinal herbs to counteract the infection. This study aimed to evaluate the interaction of artemisinin, a sesquiterpene lactone extracted from the Artemisia genus, and its derivatives with the SARS-CoV-2 main protease. To assess their potential use against COVID-19, the interactions of the main active principle of Artemisia with the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) main protease (Mpro) was investigated through in silico probing. Our results showed that artemesinin and its derivatives manifested good oral absorption and bioavailability scores (0.55). They potently bound to the Mpro site of action—specifically, to its Cys145 residue. The selected compounds established two to three conventional hydrogen bonds with binding affinities ranging between −5.2 and −8.1 kcal/mol. Furthermore, artemisinin interactions with angiotensin converting enzyme 2 (ACE2) were dependent on the ACE2 allelic variants. The best score was recorded with rs961360700. A molecular dynamic simulation showed sufficient stability of the artemisinin–Mpro complex on the trajectory of 100 ns simulation frame. These binding interactions, together with drug-likeness and pharmacokinetic findings, confirmed that artemisinin might inhibit Mpro activity and explain the ethnopharmacological use of the herb and its possible antiviral activity against SARS-CoV-2 infection inducing COVID-19. Nevertheless, it interacted differently with the various ACE2 allelic variants reported to bind with the SARS-CoV-2 spike protein.
Keywords: artemisinin, SARS-CoV-2, COVID-19, ACE2 allelic variants, computational approach, main protease (Mpro), molecular dynamics
1. Introduction
The coronavirus infectious disease (COVID-19), due to the emergent severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection, has caused over 2.236 million deaths in the world. Although great efforts have been spent to fight COVID-19, finding specific and efficacious treatments is still a central challenge for medical and scientific communities [1,2], and different strategies have been envisaged to combat the disease [3]. As urgently required, newly developed anti-SARS-CoV-2 vaccines are being delivered with unprecedented clinical phase cascade overtum. While showing first-to-second-phase effectiveness in controlling the disease, their secondary outcomes are still debated, specifically within the emergent RNA vaccine type [4,5]. Within the rapid mutability and tremendous appearance of new variants of the SARS-CoV-2 virus, vaccines might lose their specificities and performance [6,7]. In our opinion, classical treatments, like the antimalarial hydroxychloroquine that has been efficaciously used by several infectious centers and has been approved in treating the disease by many authors [8,9,10], are sought as ideal for relieving the infection and saving lives. Alternative medicine also showed great potential in fighting COVID-19 [11,12].
COVID-19 results from a SARS-coronavirus infection that has a single-stranded genome encoding for two specific functional enzymes, a papain-like protease (PL-pro) and a 3C-like protease (3CL-pro) that is also called Main protease (Mpro). Both enzymes are translated by the host-cell ribosomes. These two enzymes govern a proteolytic process producing 16 nonstructural proteins in need for the virus replication. PL-pro cuts the viral RNA-expressed polyprotein into three sites and the later accomplishes the cleaving process at 11 different sites [13]. The Main virus protease (Mpro) is a 306 amino acid chain with a 3-chymotrypsin-like protease activity that comprises three domains. Its substrate binding site is settled in a pocket region formed by two domains and presents a catalytic dyad (Cys145/His41) [14,15]. Its activation is processed by transferring a proton from Cys145 to His41, an acylation–diacylation intermediate state and interactions with peptide substrates [16]. Due to its importance in viral replication, Mpro is sought as an ideal target for developing new drugs inhibiting coronaviruses—in particular, the endemic emergent COVID-19 [17,18,19]. Recent data show proof of the antiviral effects of various herbal medicines that constitute a potential chemical scaffold to inhibit this enzyme [20,21,22,23,24]. Other investigations focused on targeting the papain-like protease of SARS-CoV-2 [25,26,27]. Among these medicinal plants, Artemisia shrubs (Asteraceae family), which were assigned as safe in several clinical trials [28,29,30,31], were considered with great interest for fighting COVID-19 [32]. These herbs have been used by several populations to cure influenza, cough, and respiratory distress [33]. In an experimental model of lung microbial infection, Yang and colleagues (2015) [34] showed that the essential oil of Artemisia efficiently inhibits microbial replication and biofilm formation that perturbs the pulmonary architecture; it was suggested that such effects might be a consequence of both their antiviral and antimicrobial activities. Molecular screening revealed the presence of several chemicals in the herb extracts that exhibit virucidal activity, such as artemisinin, rutin, and kaempferol. In particular, artemisinin, a sesquiterpene lactone, and its derivatives were shown to efficiently treat malaria. They also exert substantial antiviral activity [35] and were also shown to bind the SARS-CoV-2 spike protein at Lys353, thereby inhibiting the spike protein binding to angiotensin-converting enzyme 2 (ACE2) receptors of the host cells [36]. Nevertheless, their efficiency remains dependent on ACE2 receptor polymorphism. Several studies reported that SARS-CoV-2 mediates ACE2 receptor and serine protease TMPRSS2 for cellular entry and S protein priming, respectively [1,37,38]. Further investigations have proven that artemisinin derivatives interact with various cellular receptors, such as the VEGFR-1 [39,40], glucocorticoid [41] and probably the ACE2 receptors [42,43]. Taken together, these experimental findings suggest the potential of this medicinal plant genus to alleviate or neutralize COVID-19 through multiple pathway mechanisms.
In the current work, the inhibitory potential of five artemesinin derivatives on Mpro was investigated using in silico probing methods and comparisons to chloroquine as a reference drug. Molecular dynamics (MD) was carried for 100 ns as a validation approach for the complex artemisinin–SARS-CoV-2 Mpro. The molecular interactions of artemisinin with the ACE2 allelic variants, which have been proven to interact with coronaviruses, were also assessed. Furthermore, the ADMET properties, the drug-likeness, and the pharmacokinetic properties of these compounds were assessed to clarify the myth or the reality about Artemisia herbs use against COVID-19.
2. Results
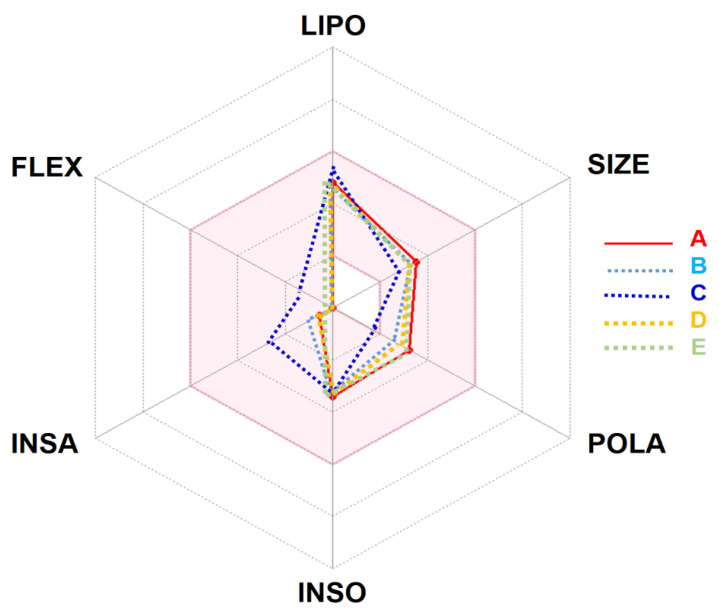
This in silico ADMET study revealed that the selected compounds possessed the same physicochemical properties, lipophilicity, drug-likeness, and pharmacokinetics. The studied compounds manifested a good oral absorption and bioavailability score (0.55) together with an acceptable TPSA value and consensus Log Po/w. The likeness behavior and properties allow the Artemisia studied compounds to be orally absorbed and, thus, fall in the colored area of the bioavailability radar/polygon (Figure 1).
Figure 1.
Bioavailability radar of Artemisia campestris selected compounds (A: artemisinin, B: acetate of dihydroartemisinin, C: artemisinic aldehyde, D: deoxyartemisinin, and E: dihydroartemisinin) based on physicochemical indices ideal for oral bioavailability. LIPO, Lipophilicity: −0.7 < XLOGP3 < þ 5; SIZE, Molecular size: 150 g/mol < mol. wt. < 500 g/mol; POLA, Polarity: 20 Å2 < TPSA <130 Å2; INSO, Insolubility: 0 < Log S (ESOL) < 6; INSA, Insaturation: 0.25 < Fraction Csp3 < 1; FLEX, Flexibility: 0 < Number of rotatable bonds < 9. The colored zone is the suitable physicochemical space for oral bioavailability. Note: the selected compounds stand in the pink area, which indicates the ideal and suitable compounds’ bioavailability.
The table also displayed also high gastrointestinal (GI) absorption and blood–brain barrier (BBB) permeant. These selected Artemisia compounds were predicted to inhibit the CYP1A2 enzyme outside of the screened cytochrome P450 isoforms: CYP 1A2, CYP2C19, CYP2C9, CYP2D6, and CYP3A4. However, these compounds are not suitable for transdermal delivery (Log Kp = −5.96) (Table 1). Furthermore, Table 1 shows that the assessed compounds are not P-gp substrates, which means that there will be no problems regarding their excretion.
Table 1.
Absorption, distribution, metabolism, excretion, and toxicity (ADMET) properties of Artemisinin and its selected derivative compounds.
| Entry | Compounds | ||||
|---|---|---|---|---|---|
| A | B | C | D | E | |
| Properties/Lipophilicity/Drug-likeness | |||||
| Molecular weight (g × mol−1) | 282.33 | 266.33 | 218.33 | 266.33 | 284.35 |
| Num. heavy atoms | 20 | 19 | 16 | 19 | 20 |
| Num. arom. heavy atoms | 0 | 0 | 0 | 0 | 0 |
| Fraction Csp3 | 0.93 | 0.87 | 0.67 | 0.93 | 1.00 |
| Num. rotatable bonds | 0 | 0 | 2 | 0 | 0 |
| Num. H-bond acceptors | 5 | 4 | 1 | 4 | 5 |
| Num. H-bond donors | 0 | 0 | 0 | 0 | 1 |
| Molar Refractivity | 70.38 | 69.71 | 69.24 | 69.29 | 71.34 |
| TPSA (Ų) | 53.99 | 36.92 | 17.07 | 44.76 | 57.15 |
| Consensus Log Po/w | 2.49 | 2.72 | 3.44 | 2.58 | 2.25 |
| Lipinski’s Rule. | Yes | Yes | Yes | Yes | Yes |
| Bioavailability Score | 0.55 | 0.55 | 0.55 | 0.55 | 0.55 |
| PAINS | 0 alert | 0 alert | 0 alert | 0 alert | 0 alert |
| Pharmacokinetics | |||||
| GI absorption | High | High | High | High | High |
| BBB permeant | Yes | Yes | Yes | Yes | Yes |
| P-gp substrate | No | No | No | No | No |
| CYP1A2 inhibitor | Yes | Yes | No | Yes | Yes |
| CYP2C19 inhibitor | No | No | Yes | No | No |
| CYP2C9 inhibitor | No | No | Yes | No | No |
| CYP2D6 inhibitor | No | No | No | No | No |
| CYP3A4 inhibitor | No | No | No | No | No |
| Log Kp (cm/s) | −5.96 | −6.05 | −5.01 | −5.90 | −5.91 |
A: artemisinin, B: acetate of dihydroartemisinin, C: artemisinic aldehyde, D: deoxyartemisinin, and E: dihydroartemisinin.
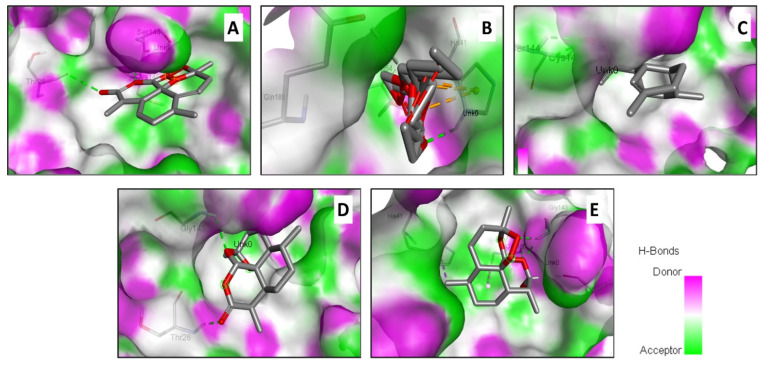
In an attempt to rationalize the antiviral effect, a molecular docking assay was carried out between artemisinin derivatives encountered in Artemisia extract and the Mpro of SARS-CoV-2 inducing COVID-19 (Figure 2).
Figure 2.
Illustration of the H-bond acceptor and donor interactions and the 3D-binding conformation of the Artemisia campestris selected compounds ((A) artemisinin, (B) acetate of dihydroartemisinin (C) artemisinic aldehyde, (D) deoxyartemisinin, and (E) dihydroartemisinin) docked to the pocket region of the SARS-CoV-2 main protease (6LU7).
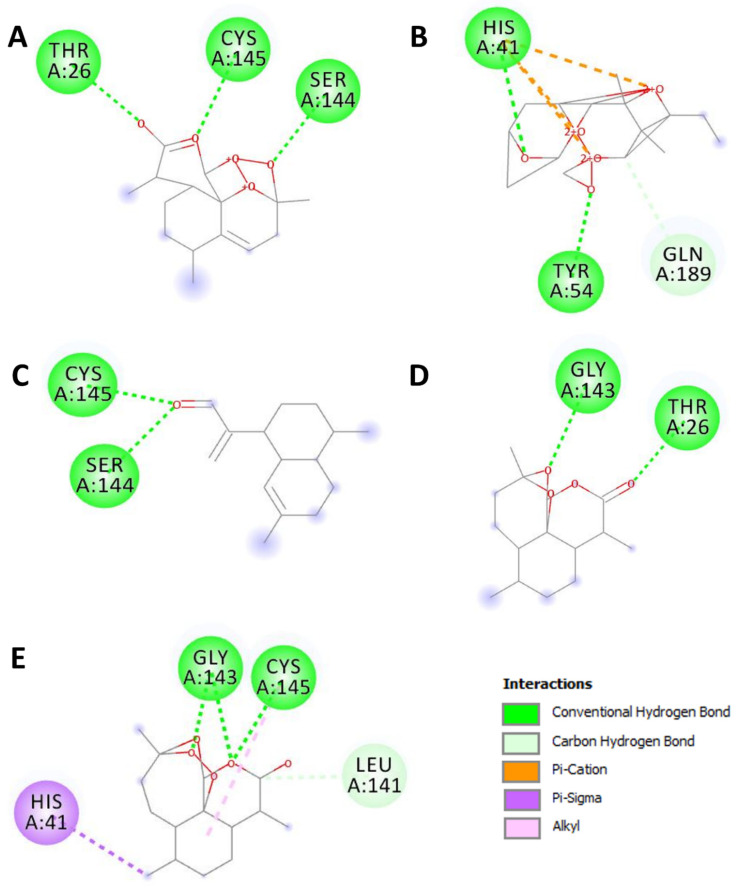
Table 2 exhibits the binding results. All the assessed compounds showed negative binding energy ranging from −5.2 to −8.1 kcal/mol. The highest binding score was found with the acetate of dihydroartemisinin (−8.1), followed by artemisinin (−7.2). The latter exhibited the highest number of conventional hydrogen bonds, similar to dihydroartemisinin and the reference compound. Regardless of artemisinic aldehyde, all the herb selected compound showed better affinities than the referenced drug: chloroquine (Table 2). These hydrogen bounds are represented in Figure 3.
Table 2.
Binding affinity, conventional hydrogen bonds and interacting residues of the main Artemisia campestris docked compounds into the SARS-CoV-2 Mpro (6LU7).
| Compound Name | Binding Affinity (kcal/mol) | Intermolecular Interactions | ||
|---|---|---|---|---|
| Conventional Hydrogen Bonds | Interacting Residues in the Pocket Region of SARS-CoV-2 Mpro |
Closest Residue (Distance, Å) | ||
| Artemisinin | −7.2 | 3 | Thr26, Ser144, Cys145 | Ser144 (2.436) |
| Acetate of dihydroartemisinin | −8.1 | 2 | His41, Tyr54, Gln189 | Tyr54 (2.743) |
| Artemisinic Aldehyde | −5.2 | 2 | Ser144, Cys145 | Cys145 (2.374) |
| Deoxyartemisinin | −6.5 | 2 | Thr26, Gly143 | Thr26 (2.206) |
| Dihydroartemisinin | −6.6 | 3 | His41, Leu141, Gly143, Cys145 | Gly143 (1.980) |
| Chloroquine (Reference) | −5.3 | 3 | Leu27, His41, Leu141, Ser144, Cys145, Met165, Glu166 | (2.815) |
Bold amino acids: interacting with the correspondent compound via conventional H-bonds. Underlined amino acids: same interacting residues as for the reference compound.
Figure 3.
2D diagrams of the closest interactions between the Artemisia campestris selected compounds ((A) artemisinin, (B) acetate of dihydroartemisinin, (C) artemisinic aldehyde, (D) deoxyartemisinin, and (E) dihydroartemisinin) and the main protease (Mpro) of SARS-CoV-2 (6LU7) inducing COVID-19.
A molecular docking analysis was also used to predict the binding affinities between artemisinin and the seventeen ACE2 variants reported to bind with the coronavirus spike protein. The binding affinities ranged between −4.9 and −8.2 kcal/mol for rs766996587 and rs961360700, respectively (Table 3).
Table 3.
Binding affinity, conventional hydrogen bonds, and interacting residues of the different ACE2 domains reported to bind with the coronavirus spike protein and artemisinin.
| ACE2 Allelic Variant |
Intermolecular Interactions | |
|---|---|---|
| Interacting Residues in the Pocket Region of ACE2 Variant (Distance, Å) | Binding Affinity (kcal/mol) | |
| rs4646116 | Conventional H-Bond: Thr41 (2.219) | −6.2 |
| Pi Cation: Trp37 (4.044) | ||
| Alkyl: Arg34 (4.897) | ||
| Pi-Alkyl: Trp37 (4.394) | ||
| rs73635825 | Conventional H-Bond: Ser13 (2.274), Ser13 (2.874) | −5.4 |
| Pi-Cation: Trp16 (4.328), Trp16 (4.514), Trp16 (4.105) | ||
| rs146676783 | Conventional H-Bond: Asn53 (2.258), Ala41 (3.359), Asn53 (3.104) | −5.2 |
| Carbon H-Bond: Ala41 (2.892) | ||
| rs76289235 | Conventional H-Bond: Tyr26 (2.163) | −5.7 |
| Pi-Cation: Tyr26 (4.375) | ||
| rs14393283 | Conventional H-Bond: Trp33 (2.921), Gln30 (3.372) | −5.8 |
| Attractive Charge: Glu17 (3.790), Glu17 (3.195) | ||
| Carbon H-Bond: Trp33 (3.338) | ||
| rs766996587 | Conventional H-Bond: Asn53 (2.254), Asn53 (2.917), Asn53 (3.017) | −4.9 |
| rs1348114695 | Conventional H-Bond: Arg21 (2.428) | −5.7 |
| Alkyl: Ile18 (4.943) | ||
| Pi-Alkyl: Phe12 (4.405) | ||
| rs961360700 | Conventional H-Bond: Thr3 (2.162), Gly33 (3.377) | −8.2 |
| rs755691167 | Conventional H-Bond: Gln54 (2.346) | −6.7 |
| Alkyl: Val58 (3.886) | ||
| rs1316056737 | Conventional H-Bond: Lys37 (2.363), Lys37 (2.588), Lys37 (2.632), Phe35 (3.267) | −5.1 |
| Alkyl: Lys23 (4.193) | ||
| rs781255386 | Conventional H-Bond: Arg25 (2.494), Lys30 (2.829) | −5.1 |
| rs1299103394 | Conventional H-Bond: Ser10 (2.222), Asp13 (3.259) | −5.4 |
| Carbon H-Bond: Ser10 (3.414) | ||
| Alkyl: Cys16 (4.680), Val24 (4.002) | ||
| rs759134032 | Conventional H-Bond: Ser57 (2.127) | −5.7 |
| Carbon H-Bond: Leu29 (3.680) | ||
| rs1238146879 | Conventional H-Bond: Tyr5 (3.056) | −5.4 |
| Pi-Cation: Tyr5 (3.898) | ||
| rs778500138 | Conventional H-Bond: Leu37 (2.078), Ser38 (2.958), Asn36 (3.219) | −5.6 |
| Alkyl: Leu35 (4.335) | ||
| rs1396769231 | Conventional H-Bond: Ser48 (2.586), Arg49 (2.670), Leu51 (2.417) | −5.5 |
| rs1016777825 | Conventional H-Bond: His34 (2.831) | −5.4 |
| Alkyl: Pro21 (4.570), Val27 (4.102), Ile29 (5.295) | ||
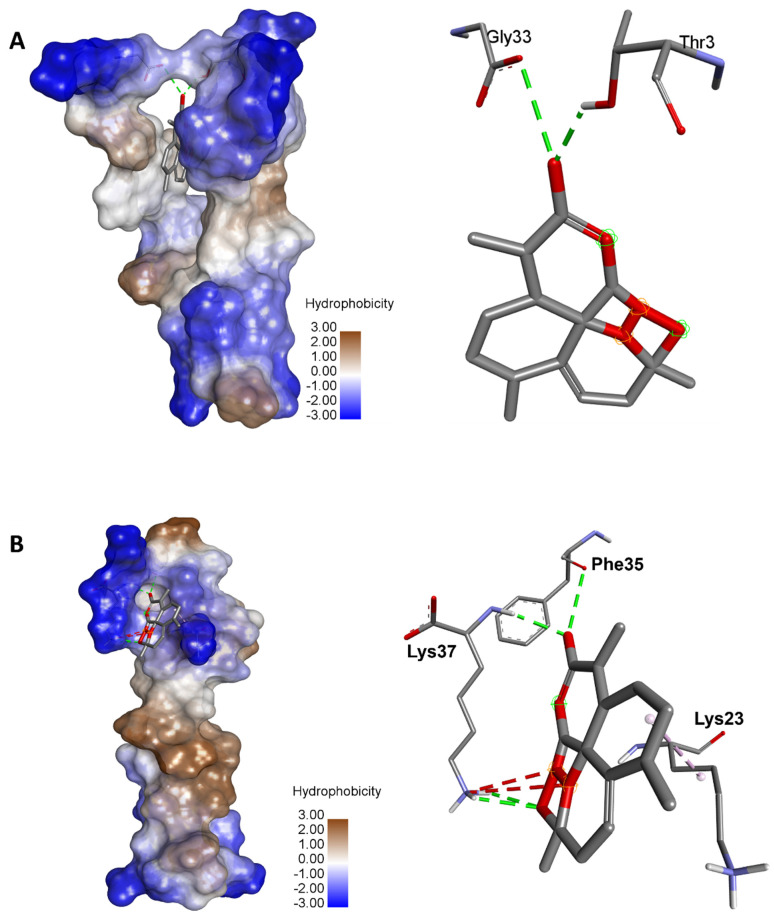
The binding energies were supported with a network of conventional H-bond, carbon H-bond, attractive charge, alkyl, Pi-alkyl, and Pi-cation. The complex of the variant rs1316056737 artemisinin showed the highest number of conventional H-bonds (n = 4). Artemisinin was H-bonded to key amino acids: once to Phe35 and three times to Lys37 (Figure 4). The acetate of dihydroartemisinin, which exhibited the highest binding score with SARS-CoV-2 Mpro (−8.1 kcal/mol), also showed acceptable binding scores and interactions with the different ACE2 variants reported to bind with coronaviruses. The results are shown in Figure S1 (Supplementary File). The acetate of dihydroartemisinin was associated with negative binding energy (between −6.5 and −4.3 kcal/mol) for the seventeen targeted ACE2 variants. The best binding energy was predicted for the variant rs961360700 followed by rs755691167, with −6.5 and −5.6 kcal/mol, respectively.
Figure 4.
Illustration of the 3D complexes of ACE2 and artemisinin (left) and the artemisinin interactions with the amino acid residues (right). (A) Variant 8 of ACE2 (rs961360700) with the highest binding energy (–8.2 kcal/mol). (B) Variant 10 of ACE2 (rs1316056737) with the highest number of hydrogen bonds (n = 4).
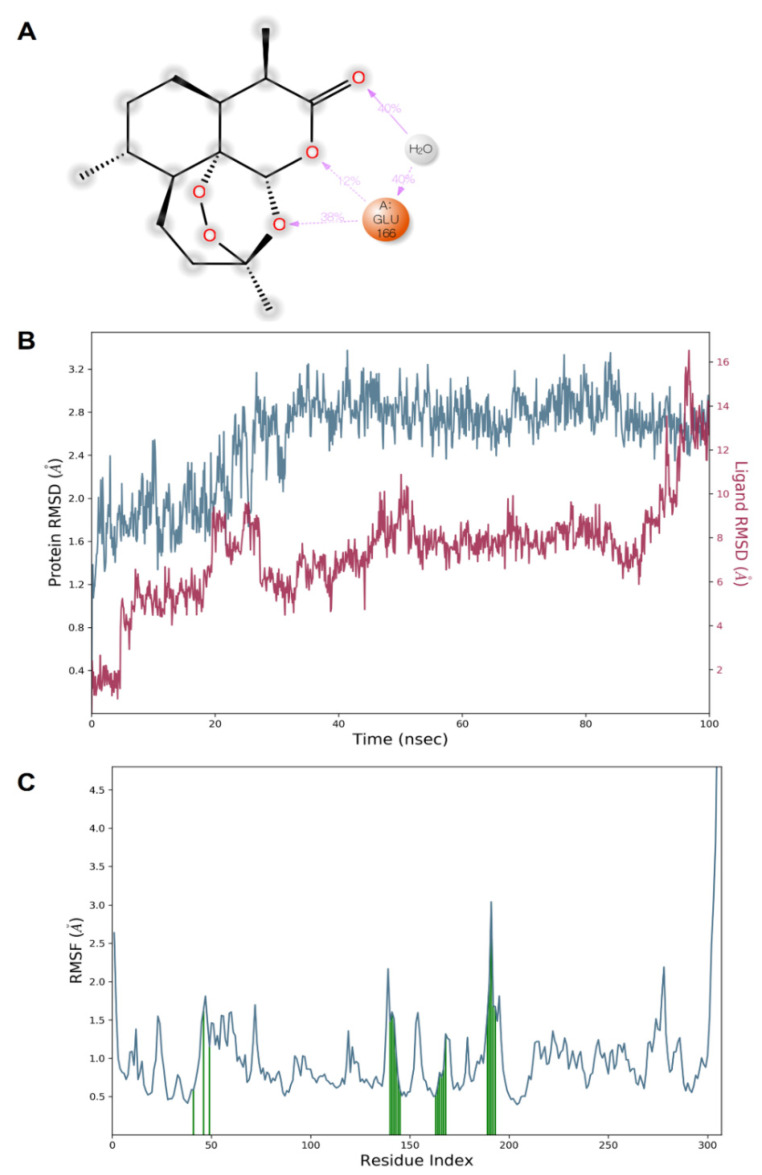
The dynamic simulation results for both artemisinin bound to the target (6LU7) showed that the complex was stable. The complex structure did not fall apart and remained bound throughout the simulations. The protein–ligand complex stability was further evaluated by measuring the root mean square deviation (RMSD) and root mean square fluctuations (RMSFs) to determine the fluctuation/thermal motion in the protein residues during the simulation. The RMSD plot for 6LU7 is shown in Figure 5 and gives an overview of the protein conformational perturbation during the binding. The RMSD analysis showed a little fluctuate, probably to lower the binding affinity at the 6LU7-binding site. The RMSD plot revealed that, after 20 ns, the system attained equilibrium and then oscillated further with RMSD of 0.4 Å and 2.8 Å, respectively. The fluctuations in the 6LU7 protein residues were analyzed by backbone atom motions and local changes in the secondary structure elements. The RMSF plot (Figure 5) shows that, despite both inhibitors showing the same binding amino acid residues, larger backbone residue fluctuations were seen for a complex system of artemisinin.
Figure 5.
Illustration of the molecular interactions of artemisinin within the pocket region of SARS-CoV-2 Mpro (A). Plots of RMSD (B) and RMSF (C) values.
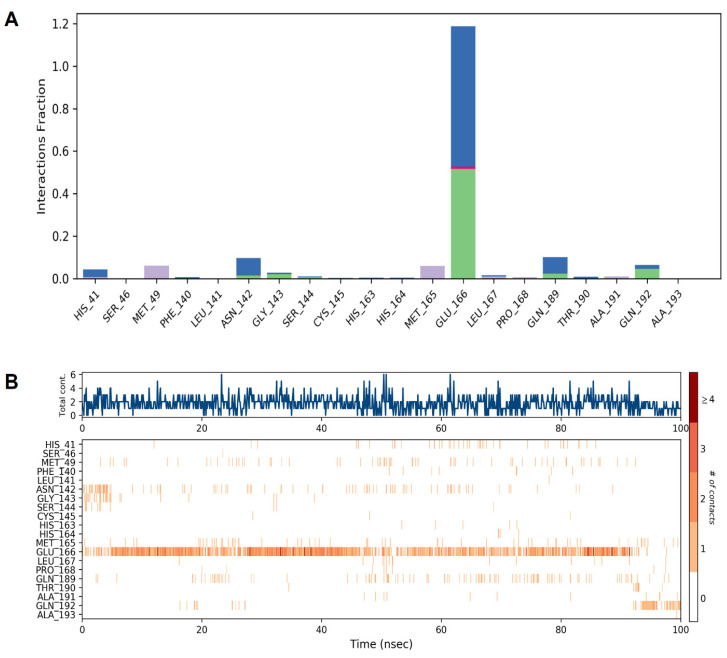
The docking and MD simulation results revealed that multiple bonds (H-bond or Halogen bond) have been formed with the receptor, and its stability was further analyzed in a dynamic simulation trajectory. The dynamic simulation trajectory revealed that both the inhibitors retained their Halogen bond with a backbone atom of the Glu166 residue. The high frequency of interactions with the Glu166 residue is shown in Figure 6.
Figure 6.
Illustration of the interaction fractions within the artemisinin and SARS-CoV-2 Mpro during a 100 ns simulation and the involved residues ((A) and (B), respectively).
3. Discussion
Since the COVID-19 fight is still an ongoing challenge, an in silico study with a focus on the molecular interactions of Artemisia sesquiterpene lactones with the Mpro of SARS-CoV-2 was performed. The five artemisinin derivatives showed negative binding energy. Such interactions suggest that the Artemisia antiviral effect [34] might involve Mpro inhibition. The highest binding energies was observed when using the acetate of dihydroartemisinin that presents a binding score of −8.1 kcal/mol. However, the best docking interaction to the Mpro pocket region was found with artemisinin and dihydroartemisinin that present an additional conventional hydrogen bond. Similar to chloroquine, both artemisinin and dihydroartemisinin interacted with the Cys145 residue. Cys145 was also found to establish a conventional H-bond with artemisinic aldehyde. Similar results were reported by Omar and colleagues (2020) [43]. These interactions confirmed that the antiviral effect of artemisinin and its derivates is possible. Nobel Prize Laureate, Professor Youyou Tu, discovered artemisinin, which was extracted from the Chinese Qinghao (Artemisia annua), and proved its efficacy and safety to eradicate malaria [44]. More recently, this ‘magic Qinghao’ revealed further therapeutic potential, including infections by SARS coronaviruses treatment. In fact, this plants genus (Artemisia) contains various chemicals that might act at different levels to cure COVID-19 in a synergetic fashion. Our in silico findings argue for such a hypothesis and give proof of artemisinin derivative inhibition of the main viral protease that is essential for SARS-CoV-2 replication.
Since Artemisia extracts are obviously safe for use, as reported in this study and previously reported for other chemicals [45,46], we foresee that they could be administrated by both oral and inhalation routes in order to directly target the SARS-CoV-2 that particularly colonizes in the respiratory tract. The fact that the herb contains compounds that directly interact with the viral spike protein and its host cell receptors and environment (notably, inflammatory cells and thrombocytes), the inhalation pathway of the plant extracts, in conjunction with their oral administration, is envisaged as a good approach to neutralize/inhibit the SARS-CoV-2 virus. The utilization of the total herb aqueous extract might have better pharmacological properties than separated molecules. In fact, it has been previously reported that the whole plant effect is usually much better than its separated active compounds [46,47]. Furthermore, our results confirmed the beneficial effects of phytochemical compounds [47,48,49,50].
The dynamic simulation results for artemisinin bound to the target (6LU7) exhibited a considerable stability of the complex. It was reported that, after 20 ns, the system attained equilibrium and then oscillated further, with RMSD of 0.4 Å and 2.8 Å, respectively. The fluctuations in the protein residues were analyzed by backbone atom motions and local changes in the secondary structure elements.
Our results revealed that multiple bonds (H-bond or Halogen bond) were formed with the receptor, and its stability was further analyzed in a dynamic simulation trajectory. The dynamic simulation trajectory revealed that both the inhibitors retained their Halogen bond with the backbone atom of the Glu166 residue. Our docking and dynamic simulation results corroborated with the earlier similar findings reported by de Olivera et al. [51].
The drug-likeness evaluation exhibited promising properties of the selected Artemisia compounds. This could explain the reported antioxidant, protective, and health-promoting effects of the plant [52,53,54]. In this study, the interactions of artemisinin with the different ACE2 variants that have been reported to bind with the coronavirus were also assessed. The in silico molecular docking results showed that artemisinin interacted differently with ACE2. While the binding energy was found to be negative for all the variants, its value was mediated by ACE2 polymorphism. It ranged between −4.9 and −8.1 kcal/mol. Together with the modes of the interactions, which are reported in Figure 4, these data confirmed the potential antiviral effect of artemisinin. Our results are comparable with those previously reported on the potential anti-COVID-19 effect of artemisinin, as well as its interaction with the SARS-CoV-2 active enzyme Mpro [55,56]. In fact, it has been reported that artemisinin showed similar interaction patterns to withaferin A, curcumin, and andrographolide with SARS-CoV-2 Mpro [56]. Likewise, artemisinin–thymoquinone hybrids were also proposed by de Oliveira et al. (2021) [51] as relevant interacting compounds with the active fraction of Mpro. The phytochemical properties of these compounds were associated in silico, and the docking results ascertained the antiviral effects of Artemisia herbs. Nevertheless, further in vivo assays might confirm these pharmacological potencies and the potential promising effects, particularly on SARS-CoV-2 infection inducing COVID-19.
4. Materials and Methods
4.1. In Silico ADMET and Pharmacokinetic Profiles
The physicochemical and pharmacokinetics properties of the selected Artemisia extracted compounds were estimated based on the ADMET (absorption, distribution, metabolism, excretion, and toxicity) properties, as previously described [47,57,58].
4.2. Genetic Variants of Human ACE2
The genetic variants of human ACE2 were retrieved from both gnomAD and Ensemble Genome Browser. Appropriate filters were applied to determine the coding region of the ACE2 variants. The corresponding seventeen protein sequences of the different variants, which have been previously reported to bind with SARS-CoV and SARS-CoV-2 [9,59,60], were selected for further approaches. The UniProt database and PDB-BLAST were used to identify the protein structures and their functional information. The RCSB protein data bank was then used to collect the seventeen protein variants, as previously reported [9].
4.3. Molecular Binding and Interactions
Five sesquiterpene lactones that originate from Artemisia herbs (artemisinin, acetate of dihydroartemisinin, artemisinic aldehyde, deoxyartemisinin, and dihydroartemisinin) were used to assess their possible antiviral effects. The chemical structures of these compounds and chloroquine (as a reference compound) were collected from PubChem. To this end, SARS-CoV-2 was selected, particularly the crystal structure of its main protease (Mpro), using the vina package. Later on, artemisinin was also used to assess its potential interactions with the different ACE2 variants reported to bind with the coronavirus, as previously described [9,48,49,61,62].
4.4. Molecular Dynamics (MD) Simulation
The MD simulation was run as previously described [63]. Briefly, the Desmond program (version 5.3) with an inbuilt OPLS3 force field was used to perform molecular dynamics (MD) simulations. The selected poses of the artemisinin and SARS-CoV-2 Mpro complex issued from molecular docking were selected for the dynamic simulation. The protein–ligand complex system was immersed in a predefined water model (TIP3P) as a solvent in an orthorhombic box of size 10 Å × 10 Å × 10 Å with periodic boundary conditions. Na+ and Cl− ions were added to neutralize the system until reaching a 0.15-M concentration. The system was relaxed, using a predefined protocol that consisted of Steepest Descent and the limited-memory Broyden-Fletcher-Goldfarb-Shanno (LBFGS) algorithms in a hybrid manner. The simulations were conducted with a constant temperature of 310.15 K using a Nosé-Hoover chain thermostat and Martyna-Tobias-Klein (MTTK) barostat methods 39 at 1 atm of pressure with the isotropic coupling type [64].
5. Conclusions
The in silico investigation revealed a potent binding of artemisinin and its derivatives to the SARS-CoV-2 Mpro site of action and the stability of the complex along the assessed period of 100 ns through MD simulation. The in silico molecular docking, drug-likeness, and pharmacokinetics findings provided satisfactory evidence that artemesinin might inhibit the Mpro proteolytic process, which is a must for virus replication. It is thought that, using Artemisia extracts—in particular, through the inhalation pathway—is promising to fight COVID-19 and may justify its ethno–pharmaceutical use against the COVID-19 pandemic.
Supplementary Materials
The following supporting information can be downloaded at https://www.mdpi.com/article/10.3390/ph15020129/s1: Figure S1: Variation of the binding affinity of acetate of artemisinin with the 17 different variants of ACE2 proven to bind with coronaviruses (A). 3D illustration of acetate of artemisinin bound to the variant rs961360700 of ACE2, which exhibited the best free binding energy (−6.5 kcal/mol). Note the four conventional H-bonds (represented with green interrupted lines) and the involvement of three residues: Met11, Thr26 and Trp28.
Author Contributions
Conceptualization, R.B. and H.B.-N.; Data curation, S.E., J.M.A., and D.K.Y.; Formal analysis, M.S., S.E., A.J.S, M.A. and H.B.-N.; Investigation, M.S., W.S.H. and M.A.; Methodology, R.B., M.S. and H.B.-N.; Project administration, R.B.; Resources, A.J.; Software, R.B.; Supervision, H.N. and H.B.-N.; Validation, W.S.H., A.J.S., A.J. and A.M.E.S.; Visualization, W.S.H., A.J.S., J.M.A., and M.M.A.; Writing—original draft, H.B.-N. and R.B.; and Writing—review and editing, R.B. All authors have read and agreed to the published version of the manuscript.
Funding
This research was supported by grants from the Deanship of Scientific Research, University of Ha’il under project number RG-21 100.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Data sharing not applicable.
Conflicts of Interest
The authors declare no conflict of interests.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Badraoui R., Alrashedi M.M., El-May M.V., Bardakci F. Acute respiratory distress syndrome: A life threatening associated complication of SARS-CoV-2 infection inducing COVID-19. J. Biomol. Struct. Dyn. 2021;39:6842–6851. doi: 10.1080/07391102.2020.1803139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kumar S. Novel coronavirus (COVID-19) outbreak: Hope and search for effective therapeutic agent. Lett. Drug Des. Discov. 2020;17:940–942. doi: 10.2174/1570180817999200612095513. [DOI] [Google Scholar]
- 3.Sivaraman D., Pradeep P., Manoharan S.S., Bhat C.R., Leela K., Venugopal V. Current Strategies and Approaches in Combating SARS-CoV-2 Virus that Causes COVID-19. Lett. Drug Des. Discov. 2020;17:672–674. doi: 10.2174/157018081705200403092546. [DOI] [Google Scholar]
- 4.van Riel D., de Wit E. Next-generation vaccine platforms for COVID-19. Nat. Mater. 2020;19:810–812. doi: 10.1038/s41563-020-0746-0. [DOI] [PubMed] [Google Scholar]
- 5.Le T.T., Andreadakis Z., Kumar A., Román R.G., Tollefsen S., Saville M., Mayhew S. The COVID-19 vaccine development landscape. Nat. Rev. Drug Discov. 2020;19:305–306. doi: 10.1038/d41573-020-00151-8. [DOI] [PubMed] [Google Scholar]
- 6.Galloway S.E. Emergence of SARS-CoV-2b.1.1.7 lineage—United States, December 29, 2020–January 12, 2021. MMWR-Morb. Mortal. Wkly. Rep. 2021;70:95–99. doi: 10.15585/mmwr.mm7003e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lauring A.S., Hodcroft E.B. Genetic Variants of SARS-CoV-2—What Do They Mean? JAMA. 2021;325:529–531. doi: 10.1001/jama.2020.27124. [DOI] [PubMed] [Google Scholar]
- 8.Kumar R., Sharma A., Srivastava J.K., Siddiqui M.H., Uddin M.S., Aleya L. Hydroxychloroquine in COVID-19: Therapeutic promises, current status, and environmental implications. Environ. Sci. Pollut. Res. 2021;28:40431–40444. doi: 10.1007/s11356-020-12200-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Badraoui R., Adnan M., Bardakci F., Alreshidi M.M. Chloroquine and Hydroxychloroquine Interact Differently with ACE2 Domains Reported to Bind with the Coronavirus Spike Protein: Mediation by ACE2 Polymorphism. Molecules. 2021;26:673. doi: 10.3390/molecules26030673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Colson P., Rolain J.M., Lagier J.C., Brouqui P., Raoult D. Chloroquine and hydroxychloroquine as available weapons to fight COVID-19. Int. J. Antimicrob. Agents. 2020;55:105932. doi: 10.1016/j.ijantimicag.2020.105932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li J.G., Xu H. Chinese Medicine in Fighting against Covid-19: Role and Inspiration. Chin. J. Integr. Med. 2021;27:3–6. doi: 10.1007/s11655-020-2860-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang J., Xu C., Wong Y.K., He Y., Adegnika A.A., Kremsner P.G., Agnandji S.T., A Sall A., Liang Z., Qiu C., et al. Preparedness is essential for malaria-endemic regions during the COVID-19 pandemic. Lancet. 2020;395:1094–1096. doi: 10.1016/S0140-6736(20)30561-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Estrada E. Topological analysis of SARS CoV-2 main protease. Chaos. 2020;30:061102. doi: 10.1063/5.0013029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Patel D.V., Teli D.M., Kanhed A.M., Patel N.R., Shah B.S., Vora A.K., Chhabria M.T., Yadav M.R. Identification of Potential Mpro Inhibitors for the Treatment of COVID-19 by Targeted Covalent Inhibition: An In Silico Approach. Int. J. Quant. Struct. Prop. Relatsh. (IJQSPR) 2021;6:58–77. [Google Scholar]
- 15.Kanhed A.M., Patel D.V., Teli D.M., Patel N.R., Chhabria M.T., Yadav M.R. Identification of potential Mpro inhibitors for the treatment of COVID-19 by using systematic virtual screening approach. Mol. Divers. 2020;25:383–401. doi: 10.1007/s11030-020-10130-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Świderek K., Moliner V. Revealing the molecular mechanisms of proteolysis of SARS-CoV-2 M pro by QM/MM computational methods. Chem. Sci. 2020;11:10626–10630. doi: 10.1039/D0SC02823A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Biembengut Í.V., De Souza T.D.A.C. Coagulation modifiers targeting SARS-CoV-2 main protease Mpro for COVID-19 treatment: An in silico approach. Mem. Inst. Oswaldo Cruz. 2020;115:e200179. doi: 10.1590/0074-02760200179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dai W., Zhang B., Jiang X.M., Su H., Li J., Zhao Y., Xie X., Jin Z., Peng J., Liu F., et al. Structure-based design of antiviral drug candidates targeting the SARS-CoV-2 main protease. Science. 2020;368:1331–1335. doi: 10.1126/science.abb4489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang L., Lin D., Sun X., Curth U., Drosten C., Sauerhering L., Becker S., Rox K., Hilgenfeld R. Crystal structure of SARS-CoV-2 main protease provides a basis for design of improved α-ketoamide inhibitors. Science. 2020;368:409–412. doi: 10.1126/science.abb3405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.da Silva J.K.R., Figueiredo P.L.B., Byler K.G., Setzer W.N. Essential oils as antiviral agents, potential of essential oils to treat sars-cov-2 infection: An in-silico investigation. Int. J. Mol. Sci. 2020;21:3426. doi: 10.3390/ijms21103426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jo S., Kim S., Shin D.H., Kim M.S. Inhibition of SARS-CoV 3CL protease by flavonoids. J. Enzym. Inhib. Med. Chem. 2020;35:145–151. doi: 10.1080/14756366.2019.1690480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Narkhede R.R., Pise A.V., Cheke R.S., Shinde S.D. Recognition of natural products as potential inhibitors of COVID-19 main protease (Mpro): In-silico evidences. Nat. Prod. Bioprospect. 2020;10:297–306. doi: 10.1007/s13659-020-00253-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nguyen T.T.H., Woo H.J., Kang H.K., Kim Y.M., Kim D.W., Ahn S.A., Xia Y., Kim D. Flavonoid-mediated inhibition of SARS coronavirus 3C-like protease expressed in Pichia pastoris. Biotechnol. Lett. 2012;34:831–838. doi: 10.1007/s10529-011-0845-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ryu Y.B., Jeong H.J., Kim J.H., Kim Y.M., Park J.Y., Kim D., Nguyen T.T., Park S.J., Chang J.S., Park K.H., et al. Biflavonoids from Torreya nucifera displaying SARS-CoV 3CLpro inhibition. Bioorg. Med. Chem. 2010;18:7940–7947. doi: 10.1016/j.bmc.2010.09.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Arun P., Hussain R., Akhter Y., Rao M.V.B., Shyam P., Yellaboina S. Drug Re-purposing from SARS-CoV Led the Identification of Potential Candidate Drug Target and Alternate Drug Molecules against SARSCoV-2. Lett. Drug Des. Discov. 2020;17:1325–1327. doi: 10.2174/1570180817999200730190600. [DOI] [Google Scholar]
- 26.Kandeel M., Altaher A., Alnazawi M. Molecular dynamics and inhibition of MERS CoV papain-like protease by small molecule imidazole and aminopurine derivatives. Lett. Drug Des. Discov. 2019;16:584–591. doi: 10.2174/1570180815666180918161922. [DOI] [Google Scholar]
- 27.Kandeel M., Abdelrahman A.H., Oh-Hashi K., Ibrahim A., Venugopala K.N., Morsy M.A., Ibrahim M.A.A. Repurposing of FDA-approved antivirals, antibiotics, anthelmintics, antioxidants, and cell protectives against SARS-CoV-2 papain-like protease. J. Biomol. Struct. Dyn. 2020;39:5129–5136. doi: 10.1080/07391102.2020.1784291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cui Y., Zhao B., Huang Y., Chen Z., Liu P., Huang J., Lao L. Effects of moxa (Folium Artemisiae argyi) smoke exposure on heart rate and heart rate variability in healthy young adults: A randomized, controlled human study. Evid. Based Complement Altern. Med. 2013;2013:510318. doi: 10.1155/2013/510318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Munyangi J., Cornet-Vernet L., Idumbo M., Lu C., Lutgen P., Perronne C., Ngombe N., Bianga J., Mupenda B., Lalukala P., et al. Effect of Artemisia annua and Artemisia afra tea infusions on schistosomiasis in a large clinical trial. Phytomedicine. 2018;51:233–240. doi: 10.1016/j.phymed.2018.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 30.Yu C., Zhang N., Zhu W., Zhang Y., Yang J., Wang Y., Song X., Hu L., Wu Z., Liu Q., et al. Does Moxa Smoke Have Significant Effect on the Acupuncturist’s Respiratory System? A Population-Based Study. Evid. Based Complement Altern. Med. 2019;2019:4873235. doi: 10.1155/2019/4873235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wheeler J., Coppock B., Chen C. Does the burning of moxa (Artemisia vulgaris) in traditional Chinese medicine constitute a health hazard? Acupunct. Med. 2009;27:16–20. doi: 10.1136/aim.2009.000422. [DOI] [PubMed] [Google Scholar]
- 32.Kapepula P.M., Kabengele J.K., Kingombe M., Van Bambeke F., Tulkens P.M., Kishabongo A.S., Decloedt E., Zumla A., Tiberi S., Suleman F., et al. Artemisia spp. derivatives for COVID-19 treatment: Anecdotal use, political hype, treatment potential, challenges, and road map to randomized clinical trials. Am. J. Trop. Med. Hyg. 2020;103:960–964. doi: 10.4269/ajtmh.20-0820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Younis W., Asif H., Sharif A., Riaz H., Bukhari I.A., Assiri A.M. Traditional medicinal plants used for respiratory disorders in Pakistan: A review of the ethno-medicinal and pharmacological evidence. Chin. Med. 2018;13:48. doi: 10.1186/s13020-018-0204-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yang C., Hu D.H., Feng Y. Essential oil of Artemisia vestita exhibits potent in vitro and in vivo antibacterial activity: Investigation of the effect of oil on biofilm formation, leakage of potassium ions and survival curve measurement. Mol. Med. Rep. 2015;12:5762–5770. doi: 10.3892/mmr.2015.4210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Efferth T., Romero M.R., Wolf D.G., Stamminger T., Marin J.J., Marschall M. The antiviral activities of artemisinin and artesunate. Clin. Infect. Dis. 2008;47:804–811. doi: 10.1086/591195. [DOI] [PubMed] [Google Scholar]
- 36.Sehailia M., Chemat S. Antimalarial-agent artemisinin and derivatives portray more potent binding to Lys353 and Lys31-binding hotspots of SARS-CoV-2 spike protein than hydroxychloroquine: Potential repurposing of artenimol for COVID-19. J. Biomol. Struct. Dyn. 2020:1–11. doi: 10.1080/07391102.2020.1796809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hoffmann M., Kleine-Weber H., Schroeder S., Krüuger N., Herrler T., Erichsen S., Schiergens T.S., Herrler G., Wu N.-H., Nitsche A., et al. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181:271–280. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhou P., Yang X.-L., Wang X.-G., Hu B., Zhang L., Zhang W., Si H.-R., Zhu Y., Li B., Huang C.-L., et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Saeed M.E., Kadioglu O., Seo E.J., Greten H.J., Brenk R., Efferth T. Quantitative Structure–Activity Relationship and Molecular Docking of Artemisinin Derivatives to Vascular Endothelial Growth Factor Receptor 1. Anticancer Res. 2015;35:1929–1934. [PubMed] [Google Scholar]
- 40.Roy K., Ojha P.K. Advances in quantitative structure–activity relationship models of antimalarials. Expert Opin. Drug Discov. 2010;5:751–778. doi: 10.1517/17460441.2010.497812. [DOI] [PubMed] [Google Scholar]
- 41.Ginosyan S., Grabski H., Tiratsuyan S. In vitro and in silico Determination of the Interaction of Artemisinin with Human Serum Albumin. Mol. Biol. 2020;54:653–666. doi: 10.1134/S0026893320040056. [DOI] [PubMed] [Google Scholar]
- 42.Rolta R., Salaria D., Kumar V., Sourirajan A., Dev K. Phytocompounds of Rheum emodi, Thymus serpyllum and Artemisia annua inhibit COVID-19 binding to ACE2 receptor: In silico approach. Curr. Pharmacol. Rep. 2021;7:135–149. doi: 10.1007/s40495-021-00259-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Omar S., Bouziane I., Bouslama Z., Djemel A. In-silico identification of potent inhibitors of COVID-19 main protease (Mpro) and angiotensin converting enzyme 2 (ACE2) from natural products: Quercetin, hispidulin, and cirsimaritin exhibited better potential inhibition than hydroxy-chloroquine against COVID-19 main protease active site and ACE2. Biol. Med. Chem. 2020 doi: 10.26434/chemrxiv.12181404.v1. [DOI] [Google Scholar]
- 44.Saoudi M., Badraoui R., Bouhajja H., Ncir M., Rahmouni F., Grati M., Jamoussi K., El Feki A. Deltamethrin induced oxidative stress in kidney and brain of rats: Protective effect of Artemisia campestris essential oil. Biomed. Pharmacother. 2017;94:955–963. doi: 10.1016/j.biopha.2017.08.030. [DOI] [PubMed] [Google Scholar]
- 45.Tu Y. The discovery of artemisinin (qinghaosu) and gifts from Chinese medicine. Nat. Med. 2011;17:1217–1220. doi: 10.1038/nm.2471. [DOI] [PubMed] [Google Scholar]
- 46.Aggarwal B.B., Yuan W., Li S., Gupta S.C. Curcumin-free turmeric exhibits anti-inflammatory and anticancer activities: Identification of novel components of turmeric. Mol. Nutr. Food Res. 2013;57:1529–1542. doi: 10.1002/mnfr.201200838. [DOI] [PubMed] [Google Scholar]
- 47.Badraoui R., Rebai T., Elkahoui S., Alreshidi M., Veettil V.N., Noumi E., Al-Motair K.A., Aouadi K., Kadri A., De Feo V., et al. Allium subhirsutum L. as a Potential Source of Antioxidant and Anticancer Bioactive Molecules: HR-LCMS Phytochemical Profiling, In Vitro and In Vivo Pharmacological Study. Antioxidants. 2020;9:1003. doi: 10.3390/antiox9101003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Akacha A., Badraoui R., Rebai T., Zourgui L. Effect of Opuntia ficus indica extract on methotrexate-induced testicular injury: A biochemical, docking and histological study. J. Biomol. Struct. Dyn. 2020:1–11. doi: 10.1080/07391102.2020.1856187. [DOI] [PubMed] [Google Scholar]
- 49.Zammel N., Saeed M., Bouali N., Elkahoui S., Alam J.M., Rebai T., Kausar M.A., Adnan M., Siddiqui A.J., Badraoui R. Antioxidant and anti-Inflammatory effects of Zingiber officinale roscoe and Allium subhirsutum: In silico, biochemical and histological Study. Foods. 2021;10:1383. doi: 10.3390/foods10061383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mzid M., Badraoui R., Kedir S.B., Sahnoun Z., Rebai T. Protective effects of ethanolic extract of Urtica urens L. against toxicity of imidacloprid on bone remodeling in rats and antioxidant activities. Biomed. Pharmacother. 2017;91:1022–1041. doi: 10.1016/j.biopha.2017.05.023. [DOI] [PubMed] [Google Scholar]
- 51.De Olivera V.M., da Rocha M.N., Magalhães E.P., da Silva Mendes F.R., Marinho M.M., de Menezes R.R.P.P.B., Sampaio T.L., Dos Santos H.S., Martins A.M.C., Marinho E.S. Computational approach towards the design of artemisinin-thymoquinone hybrids against main protease of SARS-CoV-2. Future J. Pharm. Sci. 2021;7:185. doi: 10.1186/s43094-021-00334-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bakchiche B., Gherib A., Bronze M.R., Ghareeb M.A. Identification, quantification, and antioxidant activity of hydroalcoholic extract of Artemisia campestris from Algeria. Turk. J. Pharm. Sci. 2019;16:234. doi: 10.4274/tjps.galenos.2018.99267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Saoudi M., Badraoui R., Rahmouni F., Jamoussi K., El Feki A. Antioxidant and protective effects of Artemisia campestris essential oil against chlorpyrifos-induced kidney and liver injuries in rats. Front. Physiol. 2021;12:194. doi: 10.3389/fphys.2021.618582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pereira C.G., Barreira L., Bijttebier S., Pieters L., Marques C., Santos T.F., Rodrigues M.J., Varela J., Custodio L. Health promoting potential of herbal teas and tinctures from Artemisia campestris subsp. maritima: From traditional remedies to prospective products. Sci. Rep. 2018;8:4689. doi: 10.1038/s41598-018-23038-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ben Nasr H., Badraoui R. Approach of utilizing Artemisia herbs to treat covid-19. Braz. J. Pharm. Sci. 2022 in press. [Google Scholar]
- 56.Sudeep H.V., Gouthamchandra K., Shyamprasad K. Molecular docking analysis of Withaferin A from Withania somnifera with the Glucose regulated protein 78 (GRP78) receptor and the SARS-CoV-2 main protease. Bioinformation. 2020;16:411–417. doi: 10.6026/97320630016411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Daina A., Michielin O., Zoete V. SwissADME: A free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules. Sci. Rep. 2017;7:42717. doi: 10.1038/srep42717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hchicha K., Korb M., Badraoui R., Naïli H. A novel sulfate-bridged binuclear copper (II) complex: Structure, optical, ADMET and in vivo approach in a murine model of bone metastasis. New J. Chem. 2021;45:13775–13784. doi: 10.1039/D1NJ02388H. [DOI] [Google Scholar]
- 59.Li F., Li W., Farazan M., Harrison S.C. Structure of SARS coronavirus spike receptor-binding domain complex with receptor. Science. 2005;309:1864–1868. doi: 10.1126/science.1116480. [DOI] [PubMed] [Google Scholar]
- 60.Hussain M., Jabeen N., Raza F., Shabbir S., Baig A.A., Amanullah A., Aziz B. Structural variations in human ACE2 may influence its binding with SARS-CoV-2 spike protein. J. Med. Virol. 2020;92:1580–1586. doi: 10.1002/jmv.25832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Othman I.M.M., Gad-Elkareem M.A.M., Radwan H.A., Badraoui R., Aouadi K., Snoussi M., Kadri A. Synthesis, Structure-Activity Relationship and in silico Studies of Novel Pyrazolothiazole and Thiazolopyridine Derivatives as Prospective Antimicrobial and Anticancer Agents. ChemistrySelect. 2021;6:7860–7872. doi: 10.1002/slct.202101622. [DOI] [Google Scholar]
- 62.Badraoui R., Saeed M., Bouali N., Hamadou W.S., Elkahoui S., Alam M.J., Siddiqui A.J., Adnan M., Saoudi M., Rebai T. Expression profiling of selected immune genes and trabecular microarchitecture in breast cancer skeletal metastases model; effect of α–tocopherol acetate supplementation. Calcif. Tissue Int. 2022 doi: 10.1007/s00223-021-00931-3. in press. [DOI] [PubMed] [Google Scholar]
- 63.Saeed M., Shoaib A., Tasleem M., Alabdallah N.M., Alam M.J., El Asmar Z., Jamal Q.M.S., Bardakci F., Alqahtani S.S., Ansari I.A., et al. Assessment of antidiabetic activity of the shikonin by the allosteric inhibition of protein-tyrosine phosphate 1B (PTP1B) using state of art: An in silico and in vitro tactics. Molecules. 2021;26:3996. doi: 10.3390/molecules26133996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yadav D.K., Kumar S., Saloni , Misra S., Yadav L., Teli M., Sharma P., Chaudhary S., Kumar N., Choi E.H., et al. Molecular Insights into the Interaction of RONS and Thieno[3,2-c]pyran Analogs with SIRT6/COX-2: A Molecular Dynamics Study. Sci. Rep. 2018;8:4777. doi: 10.1038/s41598-018-22972-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data sharing not applicable.