Abstract
Progress of molecular meningioma characterization (*courtesy of Ralf Ketter, Homburg, Germany).
![]()
Keywords: meningioma
While meningiomas represent the most frequent primary intracranial tumor and, therefore, play an important role in daily clinical practice for neurosurgeons, neurooncologists, and neuropathologists, this tumor type has not been covered within the well‐recognized format of a mini‐symposium in Brain Pathology. Now, this gap is going to be filled, and the timing cannot be better. Both the understanding of the molecular alterations acting in meningiomagenesis, as well as the classification of the tumors by combining conventional histological features with a site of tumor growth, somatic mutations, and epigenetic characteristics has expanded our understanding of the biological landscape of this tumor previously regarded as “boring” to investigate and simple to treat. This evolution is reflected by the just recently updated WHO classification of brain tumors which includes molecular features as an important layer to classify and grade meningiomas [1] (Table 1).
TABLE 1.
Current WHO classification of meningioma and frequently associated molecular alterations
| Meningioma subtype | CNS WHO Grade | Frequent molecular alteration | Typical Methylation group (14) |
|---|---|---|---|
| (Meningioma, NOS) | |||
| Meningothelial | 1 | AKT1/TRAF7, SMO | Ben‐2 |
| Fibrous | 1 | NF2, 22q del | Ben‐1 |
| Transitional | 1 | NF2, 22q del | Ben‐1 |
| Psammomatous | 1 | NF2, 22q del | Ben‐1 |
| Angiomatous | 1 | Trisomy 5 | Ben‐3 |
| Microcystic | 1 | Trisomy 5 | Ben‐3 |
| Secretory | 1 | KLF4/TRAF7 | Ben‐2 |
| Lymphoplasmacyte‐rich | 1 | (None) | |
| Metaplastic | 1 | Trisomy 5 | Ben‐3 |
| Chordoid | 2 | Various | |
| Clear cell | 2 | SMARCE1 | Separate group |
| Rhabdoid | 2 | BAP1 | mal (if “true” rhabdoid) |
| Papillary | 2 | PBRM1 | mal (if “true” papillary) |
| Atypical | 2 | Enriched for int‐A/int‐B | |
| Brain invasive | 2 | ||
| Anaplastic | 3 | CDKN2A/B, TERT | mal |
Initiated with the description of the loss of chromosomal material on 22q [2] and the identification of NF2 alterations in about 50% of sporadic meningiomas in the 1990s [3], the field got boosted by the identification of several recurrent somatic mutations taking advantage of next‐generation sequencing (NGS) techniques, resulting in the denomination of a non‐NF2 driven meningioma group [4]. Besides mutations, genome‐wide methylation profiling studies proposed epigenetic subtyping with clinical relevance [5]. For the first time, classification systems integrating various layers of information, for example, morphological, molecular, and clinical data were established, providing a reproducible system for risk assessment to the patients and clinicians. The most recent include refinements and extensions incorporating large data sets derived from genetics, epigenetics, and proteomics [6, 7], while the practical use of these classification systems is subject to further evaluation (Figure 1).
FIGURE 1.
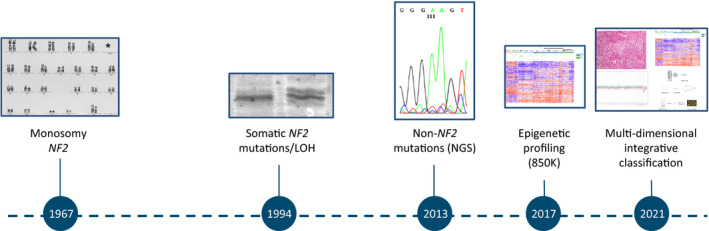
Progress of molecular meningioma characterization (*courtesy of Ralf Ketter, Homburg, Germany)
The current mini‐symposium aims to harness the molecular findings in meningioma for more practical aspects. The series is opened with a review article written by Norbert Galldiks and colleagues, summarizing the state‐of‐the‐art in the rapidly evolving field of radiogenomics. Preoperative assessment based on sophisticated image analyses using artificial intelligence approaches which address molecular characteristics might help to adjust operative strategies and reduce perioperative morbidity. Additionally, radiogenomic assessment of recurrent meningiomas may guide the decision for the optimal treatment strategy, that is, the decision between watch and wait strategy, irradiation including gammaknife or radiosurgery, and re‐operation.
This review is followed by a comprehensive study of Berghoff et al. which evaluates the practical use of methylation profiling compared to mutational analysis in a large series of clinically well‐characterized meningiomas. This data shows that both DNA methylation classification and panel‐based‐targeted mutational analysis improve the prognostic assessment and the identification of potential molecular alterations for targeted personalized therapy in meningiomas.
The paper of Berghoff and colleagues raises the question of how targeted molecular testing can be incorporated into neuropathological diagnostic. Therefore, the following article (Mawrin et al.) introduces a concise, time, and cost‐efficient NGS panel which is sufficient to detect relevant somatic mutations. The proposed panel covers all genetic alterations essential for exact classification (for instance, KLF4/TRAF7 for secretory meningiomas, SMARCE1 for clear cell meningiomas, or BAP1 in many rhabdoid meningiomas. Moreover, prognostically relevant alterations (TERT promoter mutations or loss of CDKN2A/B) can be detected as well. This amplicon‐based targeted meningioma panel might accelerate the introduction of genomic characterization in daily routine work.
Finally, a review with a special focus on brain‐invasive meningiomas covers the recent knowledge about the molecular mechanisms driving this process. While brain invasion has been established as a criterion for grade 2 meningioma, the relevance for future treatment decisions and recurrence rate estimation has been subject to discussion [8].
We hope that this mini‐symposium will stipulate further research into the biological underpinnings of meningiomas and their diagnostic or even therapeutic application.
CONFLICT OF INTEREST
There are no conflicts of interest to declare.
AUTHOR CONTRIBUTIONS
Felix Sahm: writing, artwork. Christian Mawrin: concept, writing, artwork.
Sahm F, Mawrin C. Introduction to the mini‐symposium “molecular neuropathology of meningioma”. Brain Pathol. 2022;32:e13055. 10.1111/bpa.13055
DATA AVAILABILITY STATEMENT
Does not apply.
REFERENCES
- 1. Louis DN, Perry A, Wesseling P, Brat DJ, Cree IA, Figarella‐Branger D, et al. The 2021 WHO classification of tumors of the central nervous system: a summary. Neuro‐oncology. 2021;23(8):1231–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Zang KD, Singer H. Chromosomal constitution of meningiomas. Nature. 1967;216(5110):84–5. [DOI] [PubMed] [Google Scholar]
- 3. Ruttledge MH, Sarrazin J, Rangaratnam S, Phelan CM, Twist E, Merel P, et al. Evidence for the complete inactivation of the NF2 gene in the majority of sporadic meningiomas. Nat Genet. 1994;6(2):180–4. [DOI] [PubMed] [Google Scholar]
- 4. Clark VE, Erson‐Omay EZ, Serin A, Yin J, Cotney J, Ozduman K, et al. Genomic analysis of non‐NF2 meningiomas reveals mutations in TRAF7, KLF4, AKT1, and SMO. Science. 2013;339(6123):1077–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Sahm F, Schrimpf D, Stichel D, Jones DT, Hielscher T, Schefzyk S, et al. DNA methylation‐based classification and grading system for meningioma: a multicentre, retrospective analysis. Lancet Oncol. 2017;18(5):682–94. [DOI] [PubMed] [Google Scholar]
- 6. Maas SLN, Stichel D, Hielscher T, Sievers P, Berghoff AS, Schrimpf D, et al. Integrated molecular‐morphologic meningioma classification: a multicenter retrospective analysis, retrospectively and prospectively validated. J Clin Oncol. 2021;39(34):3839–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Nassiri F, Liu J, Patil V, Mamatjan Y, Wang JZ, Hugh‐White R, et al. A clinically applicable integrative molecular classification of meningiomas. Nature. 2021;597(7874):119–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Perry A. The definition and role of brain invasion in meningioma grading: still controversial after all these years. Free Neuropathol. 2021;2(8):1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Does not apply.


