Abstract
Cse4p is an evolutionarily conserved histone H3-like protein that is thought to replace H3 in a specialized nucleosome at the yeast (Saccharomyces cerevisiae) centromere. All known yeast, worm, fly, and human centromere H3-like proteins have highly conserved C-terminal histone fold domains (HFD) but very different N termini. We have carried out a comprehensive and systematic mutagenesis of the Cse4p N terminus to analyze its function. Surprisingly, only a 33-amino-acid domain within the 130-amino-acid-long N terminus is required for Cse4p N-terminal function. The spacing of the essential N-terminal domain (END) relative to the HFD can be changed significantly without an apparent effect on Cse4p function. The END appears to be important for interactions between Cse4p and known kinetochore components, including the Ctf19p/Mcm21p/Okp1p complex. Genetic and biochemical evidence shows that Cse4p proteins interact with each other in vivo and that nonfunctional cse4 END and HFD mutant proteins can form functional mixed complexes. These results support different roles for the Cse4p N terminus and the HFD in centromere function and are consistent with the proposed Cse4p nucleosome model. The structure-function characteristics of the Cse4p N terminus are relevant to understanding how other H3-like proteins, such as the human homolog CENP-A, function in kinetochore assembly and chromosome segregation.
The centromere in budding yeast consists of conserved centromere DNA elements CDEI, CDEII, and CDEIII, which span about 125 bp of the centromere DNA (CEN) in each of the 16 Saccharomyces cerevisiae chromosomes (12, 13). Mutations altering any of these DNA elements cause defective chromosome segregation. The effects of CDE mutations range from relatively mild increases in mitotic chromosome loss (2- to 20-fold in CDEI mutants) to complete loss of centromere function (single point mutation in the central CCG triplet of CDEIII) (11, 14, 16, 21, 31). The centromere DNA is the site of assembly of the kinetochore, a protein complex that connects the chromosome to the mitotic spindle. To date, at least 10 different S. cerevisiae kinetochore proteins have been identified, some of which bind directly to CEN DNA. A homodimer of Cbf1p binds to CDEI (2, 4, 8, 9, 32) and induces a bend in the DNA that enhances centromere function (24, 36). The CBF3 complex contains four essential protein subunits (p110, p64, p58, and p23), which assemble specifically on CDEIII DNA (26, 27). The CBF3-CDEIII complex is required but by itself is not sufficient to confer centromere function and accurate chromosome segregation (11, 14). The A+T-rich CDEII DNA, which has an intrinsic bend even in the absence of bound protein, is necessary in addition to CDEIII to form a functional kinetochore microtubule binding site (25). Candidate CDEII DNA binding proteins include Mif2p, an essential centromere protein containing an “AT-hook” motif (34), and Cse4p (44). In addition, Cse4p, Mif2p, and Cbf1p make connections to CBF3-CDEIII through interactions with the Ctf19p-Mcm21p-Okp1p complex at the centromere (37). Ctf19p may also be involved in regulating kinetochore-microtubule interactions (19). It is proposed that kinetochore assembly initiates with formation of the CBF3-CDEIII complex, which then recruits additional kinetochore proteins, including Cse4p, to the centromere (33, 37).
Yeast centromere DNA is organized into a distinctive chromatin structure (6). Current models propose that two copies of the histone H3-like protein Cse4p replace both copies of H3 in a centromere-specific nucleosome (23, 35). The idea that Cse4p is incorporated into specialized centromeric nucleosomes is consistent with the histone-like biochemical properties of the Cse4p protein (44) and with the results of Cse4p mutational analysis (23). In addition, a mutation in either Cse4p or H4 (hhf1-20) disrupts chromatin structure at the centromere (35, 42). Overexpression of Cse4p, but not that of H3, suppresses the conditional growth, chromosome missegregation, and defective centromeric chromatin phenotypes of hhf1-20 mutant cells, providing strong evidence that Cse4p interacts with H4 at the centromere (35). Genetic interactions between cse4 and other kinetochore genes have been detected; Baker et al. (3) isolated a mutant cse4 allele that is synthetic lethal with cep1Δ (cbf1Δ), and Ortiz et al. (37) showed a two-hybrid interaction between Cse4p and Ctf19p. Recently, Cse4p was implicated in the association of cohesin with the centromere (47), a further indication that Cse4p is an integral part of a higher-order chromatin structure at the budding yeast centromere.
Centromere H3-like proteins are evolutionarily conserved. In addition to Cse4p, fly (Cid), worm (HCP-3), and mammalian (CENP-A) homologs have been identified (7, 17, 46). Homology between these proteins is limited to the H3-like histone fold domain (HFD) and ranges from 34 to 57% identical amino acids (17). Although originating from different organisms, Cse4p-, CENP-A–, and HCP-3–green fluorescent protein (GFP) fusions all preferentially localize to pericentric heterochromatin when expressed in Drosophila cells while Cse4p and HCP-3 preferentially localize to heterochromatic sites (including centromeres) in human cells (17). All of the centromere H3-like proteins have distinctly different N termini with little homology to known proteins (17, 44). Studies of CENP-A show that the HFD, but not the N terminus, is required for centromere targeting (46). A similar conclusion has been drawn for the Cse4p N terminus (37). However, the Cse4p N terminus has at least one essential function, since deletion of the first 50 amino acids is lethal to the cell (23). By analogy to the known folded structure of H3 in the nucleosome, the HFD of Cse4p would assemble into the octamer with the N terminus of the protein extending away from the core between the wrapped DNA helices (28). In this conformation, the Cse4p N terminus would be available to interact with proteins involved in critical centromere functions, including kinetochore protein recruitment and assembly, kinetochore-microtubule connections, and sister chromatid cohesion. The Cse4p N terminus may also be a target for posttranslational modifications, as observed for the N termini of the standard core histones (50). Such modifications may be required for the integrity of kinetochore structure or for other centromere functions, such as checkpoint signaling.
Here we report a comprehensive mutagenesis study of the N-terminal 130 amino acids of Cse4p. Our results show that the Cse4p N terminus contains an essential region of 33 amino acids which performs a function distinct from that of the HFD. Surprisingly, the position of the essential N-terminal domain relative to the HFD is highly flexible although the N terminus must be physically linked to the HFD for the protein to function. We present genetic and biochemical evidence that Cse4p proteins interact directly in vivo. In addition, we describe results supporting interactions between the N terminus of Cse4p and the newly identified centromere protein complex Mcm21p-Ctf19p-Okp1p. Our results reinforce the proposed Cse4p-nucleosome model and provide new information with implications for understanding the function of centromere H3-like proteins and how they are targeted to the centromere DNA.
MATERIALS AND METHODS
Yeast strains and genetic methods.
The yeast strains used in this study are shown in Table 1. YC190 and YC121 contain the integrated cse4-39 and cse4-542 alleles, respectively. Integration was accomplished by recloning the mutant alleles into integrating vector pRS304 or pRS306 (41) and then deleting the HFD segments by cleaving with BstBI and SalI and religating. The resulting truncated cse4 constructs were linearized by NdeI and then transformed into a wild-type yeast strain. Plasmid integration resulted in a genetic duplication at the CSE4 locus consisting of a full-length mutant cse4 allele and a 3′-truncated (nonfunctional) copy of the endogenous wild-type allele. The expected genome changes were confirmed by diagnostic PCR.
TABLE 1.
Strains used in this study
| Strain | Genotype | Source or reference |
|---|---|---|
| KC100 | MATα ade2-101 his3-11,15 leu2-3 lys2-801 trp1Δ901 ura3-52 cse4::HIS3 [pCL1] (CSE4 in YCp50) | 23 |
| YC190 | MATα ade2-101 ade3-24 his3-11,15 leu2-3 lys2-801 trp1Δ901 ura3-52 cse4-39::TRP1 | This study |
| YC191 | MATa ade2-101 ade3-24, his3-11,15 leu2-3 lys2-801 trp1Δ901 ura3-52 cse4-39::TRP1 | This study |
| YC121 | MATa ade2-101 his3-11,15 leu2-3 trp1Δ901 ura3-52 cse4-542::URA3 | This study |
| YC320 | MATa ade2-101 ade3-24 leu2-3 ura3-52 cep1Δ | This study |
| YC222 | MATα ade2-101 his3-11,15 leu2-3 trp1Δ901 ura3-52 mcm21Δ::HIS3 | This study |
| YC220 | MATα ade2-101 his3-11,15 leu2-3 trp1Δ901 ura3-52 mcm21Δ::HIS3 cse4-39::TRP1 [p31A] | This study |
| YMB120 | MATa ade2-101 his3-Δ200 leu2-Δ1 lys2-801 trp1Δ901 ura3-52 spt4Δ2::HIS3 CFVII-URA3-SUP11 | 5 |
| YPH1314 | MATα ade2-101 his3-Δ200 leu2-Δ1 lys2-801 trp1Δ61 ura3-52 ctf19Δ1::HIS3 | 19 |
| 301-2BΔ22 | MATa his4 leu2 trp1 ura3 mcm22-Δ1::URA3 | 38 |
| YJL158 | MATa ade2-101 his3-Δ200 leu2-Δ1 lys2-801 trp1Δ63 ura3-52 okp1-5::TRP1 cyh2 (CF CEN6 URA3 SUP11 CYH2) | 37 |
| PKY031 | MATa ade2-101 his3 leu2 lys2 trp1 ura3 cac2::hisG-URA3-hisG | 22 |
| PKY034 | MATa ade2-101 his3 leu2 lys2 trp1 ura3 cac3::hisG-URA3-hisG | 22 |
Yeast genetic manipulations were carried out using standard procedures (40). The media and growth conditions used were as previously described (39). Transformations were performed by the lithium acetate procedure (40). The restrictive temperatures for temperature-sensitive (Ts) and cold-sensitive (Cs) mutations were 38 and 15°C, respectively.
CSE4 mutants and epitope tagging.
All mutant cse4 alleles were constructed by sequential PCR. The template for both reactions was plasmid pCSE4 DNA consisting of wild-type CSE4 in the polylinker of pRS314 (44). First, a PCR was carried out using a primer (Integrated DNA Technology) containing the desired mutation and a primer complementary to polylinker sequences of the vector (T7 or T3 promoter). This PCR product was then used as a “megaprimer,” along with the T7 or T3 primer from the opposite end of the polylinker, for the second PCR. The product of the second PCR was digested with BamHI and SalI and reinserted into pRS314. All mutations were confirmed by sequencing. For those cse4 alleles which showed growth phenotypes, the entire mutant cse4 gene was sequenced.
Two forms of hemagglutinin (HA)-tagged CSE4 were used. CSE4HA, which has three HA repeats inserted at CSE4 codon 83 (engineered NotI site), was described by Stoler et al. (44). CSE4HAn, constructed for this study, has the triple HA epitope inserted immediately after the initiating methionine in the N terminus. In CSE4HAn, an EcoRI site was incorporated into the junction between the HA and CSE4 sequences. The HAn versions of mutant cse4 alleles were constructed by replacing the wild-type CSE4 sequence of CSE4HAn with the mutant cse4 sequence, obtained by PCR using an upstream primer carrying an EcoRI site and a downstream polylinker primer. GFP-tagged CSE4 (CSE4GFP) was obtained by replacing the triple-HA segment of CSE4HA with DNA encoding GFP-Bex1 (1). The GFP-Bex1 insert was generated by PCR using primers designed with in-frame EagI sites, destroying the NotI site, and introducing an SpeI site at the 5′ side. CSE4SpA was constructed by replacing the GFP segment of CSE4GFP with DNA encoding the ZZ domain of protein A, obtained by PCR amplification of plasmid pYES2 (Invitrogen), retaining the unique SpeI and NotI sites. The cse4-39SpA allele was made by replacing the BamHI-SpeI fragment of CSE4SpA with the BamHI-XbaI fragment of cse4-39. The cse4Δ55GFP and cse4-107GFP alleles were made by replacing the BamHI-SpeI or NotI-SalI fragment of CSE4GFP with the BamHI-XbaI fragment of cse4Δ55 or the NotI-SalI fragment of cse4-107HA, respectively. The constructs were sequenced to confirm that the epitope tag was inserted properly in frame and that no undesired mutations were present.
Analyses of cse4 gene expression and function.
The ability of mutant cse4 alleles to complement a cse4 null allele was tested using a plasmid shuffle assay described previously (23). Plasmids carrying the cse4 mutant alleles were transformed into a cse4::HIS3 strain (KC100) harboring wild-type CSE4 on a single-copy URA3 plasmid. Transformants were plated on 5-fluoroorotic acid (5-FOA) medium to select for loss of the URA3-CSE4 plasmid. 5-FOA-resistant cells, lacking the wild-type CSE4 plasmid, must rely on the mutant cse4 allele for viability. Three KC100 transformants for each cse4 allele were picked and suspended in 1.5 ml of water. The optical density at 660 nm (OD660) of the cell suspension was adjusted to 0.5. Six-microliter volumes of this suspension and of a 10-fold dilution were plated on medium lacking tryptophan and on 5-FOA medium, and the plates were photographed after 2 to 3 days at 30°C. Those cse4 mutant alleles that could complement cse4Δ::HIS3 were tested further for conditional growth phenotypes at 15 and 38°C.
Tests for interallelic complementation were carried out using KC100 carrying the same or different cse4 alleles on separate TRP1 and URA3 vectors (pRS314 and pRS316, respectively). Cells were suspended in 1.5 ml of water, the cell suspension was adjusted to an OD660 of 1.0 and serial threefold dilutions were made. Six microliters of each dilution was plated on double-selective medium lacking tryptophan and uracil, incubated at 30 and 38°C, and photographed after 3 and 5 days of incubation, respectively.
Cellular levels of Cse4p were analyzed by immunoblotting as described by Keith et al. (23), except that cells were boiled for 10 min and sodium dodecyl sulfate (SDS)–10% polyacrylamide gel electrophoresis (PAGE) was used to resolve the proteins. Quantitative mitotic chromosome loss rate assays were carried out exactly as described by Keith et al. (23). For the qualitative chromosome loss assays, cells were plated directly onto indicator medium and incubated at 30°C for 5 days. The sectoring of colonies of mutant cells was visually compared to that of wild-type cells (18).
Cse4p coprecipitation and CHIP.
Yeast cells carrying the cse4-39SpA and cse4-107HA alleles on plasmids pRS314 and pRS316, respectively, were grown in double-selective medium at 30 or 38°C to a concentration of 107/ml. About 109 cells (100 ml) were collected and washed once with 5 ml of cold water and once with 5 ml of cold buffer A (20 mM Tris-HCl [pH 8.0], 5 mM MgCl2, 1 mM dithiothreitol, 0.2% Triton X-100, 1 mM phenylmethylsulfonyl fluoride, 150 mM potassium acetate). The cell pellet was resuspended in 250 μl of buffer A, and 150 μl of glass beads was added. Breakage was achieved by vortexing on a shaker for 30 min at 4°C, and the whole-cell extract was obtained after centrifugation for 7 min at 25,000 × g. After removal of 5% of the extract for analysis, 25 μl of pre-equilibrated immunoglobulin G (IgG)-Sepharose beads (Pharmacia) was added to the remainder and the mixture was incubated for 2 h at 4°C. The beads were collected by centrifugation, washed four times for 5 min (each time) with 1 ml of buffer A, and resuspended in 100 μl of SDS sample buffer. Ten to 15% of the beads and 0.5% of the total extract were used for immunoblot analysis as described above.
Chromatin immunoprecipitation (CHIP) analysis was carried out as previously described (45). Briefly, about 109 cells were treated in situ with 1% formaldehyde for 1 h. Cross-linked chromatin was then prepared (450 μl) and sonicated to fragment chromosomal DNA. Immunoprecipitation was conducted with 400 μl of the chromatin solution using 20 μl of polyclonal rabbit anti-GFP (α-GFP) (Pharmacia) or monoclonal mouse α-HA (Pharmacia) antibody. The remaining 50 μl, not subject to immunoprecipitation, was used to prepare the total chromatin samples. PCR was performed with 1/50 of the total chromatin or 1/10 of the immunoprecipitated chromatin using the primers described by Ortiz et al. (37). About one-third of each PCR product was analyzed on an agarose gel stained with ethidium bromide. Cells used in the CHIP assay were grown in dropout medium to 2 × 107/ml (30°C). For the 38°C samples, an overnight culture grown at 30°C was diluted into prewarmed medium to a density of 6 × 105 cells/ml and grown at 38°C to a density of 107 cells/ml.
Microscopy.
Cells carrying GFP-tagged proteins were grown in YEPD medium to a density of 0.5 × 107 to 1 × 107/ml. The cells were fixed by the addition of 1/10 volume of 37% formaldehyde and continued incubation for 45 min under growth conditions. The fixed cells were washed once in standard phosphate-buffered saline (PBS), incubated on ice for 15 min in ethanol containing 4′,6′-diamidino-2-phenylindole (DAPI) at 1 μg/ml, washed twice with PBS, resuspended in PBS, and kept on ice. Microscopy was performed with a Nikon microscope equipped with epifluorescence optics and a charge-coupled device camera (Santa Barbara Instrument Group). Collected images were adjusted for contrast and brightness and colorized using Adobe Photoshop.
Isolation of dosage suppressors.
KC100 cells with plasmid-borne cse4-23 providing the only source of Cse4p were transformed with a high-copy-number URA3 yeast genomic library (10). Transformants were selected directly at 38°C, the nonpermissive temperature for cse4-23. A small portion of transformed cells was incubated at 30°C to estimate the total number of transformants (approximately 22,000). After 7 days, 85 colonies were obtained, 78 of which grew when restreaked and incubated at 38°C. Total DNA was prepared from the 78 candidates, and 13 of them were found to contain wild-type CSE4 by PCR. Library plasmids were recovered from 61 of the remaining 65 candidates and tested for the ability to suppress the Ts phenotype of cse-23 upon retransformation. Twenty-six clones remained positive and were analyzed by restriction enzyme digestion and DNA sequencing, which revealed 15 different genomic DNA inserts. When tested for the ability to rescue cse4-39, another N-terminal mutation, four groups were positive, one having an insert of 8.3 kb corresponding to nucleotides 1098917 to 1107259 of chromosome IV. Subcloning mapped the suppressor function to a 2.3-kb HindIII-XbaI fragment containing only the MCM21 open reading frame (ORF) (YDR318W). MCM21 function required at least 500 bp of DNA upstream of YDR318W, confirming a recent report that the gene contains an intron and that Mcm21p translation starts at an ATG triplet 434 bp upstream of the YDR318W ORF annotated in the S. cerevisiae genome database (37).
Two-hybrid analysis.
The two-hybrid vectors and host strain of James et al. (20) were used. The CSE4 coding region and its mutant forms were inserted into pAD-C2 to produce Cse4p-Gal4 activation domain (AD) fusion proteins (Cse4p-AD), while the CTF19 coding region was inserted into pBD-C2 to yield a Ctf19p-Gal4 DNA binding domain (BD) fusion protein (Ctf19p-BD). In both cases, the inserts were obtained by PCR, incorporating BamHI and SalI sites into the primers for cloning into the polylinker region of the vectors. Cse4p-AD and Ctf19p-BD plasmids were cotransformed into strain PJ69-4A, which carries a GAL1-HIS3 reporter gene. Three transformants were picked and suspended in 1.5 ml of water. The OD660 of the cell solution was determined and adjusted to 1.0, and then a series of fivefold dilutions was made. Six microliters of each dilution was plated on complete minimal medium and on minimal medium lacking histidine and supplemented with 0.5 mM 3-aminotriazole (to suppress growth due to basal HIS3 transcription). The plates were incubated at 30°C and photographed after 5 days.
RESULTS
Alanine scanning mutagenesis identifies an important region in the Cse4p N terminus.
The N terminus of Cse4p contains several clusters of charged amino acids. To investigate the functional significance of these charged amino acids, groups of charged residues were changed to alanines and the resulting alleles were tested for Cse4p function in vivo. Haploid cells with a disrupted CSE4 gene (cse4Δ null) and carrying wild-type CSE4 on a URA3 plasmid to maintain viability were transformed with low-copy-number (CEN) plasmids carrying different mutated cse4 genes. The transformants were plated on agar medium containing 5-FOA to select for loss of the CSE4-URA3 plasmid and to obtain cells that were completely dependent on the function of the mutant cse4-encoded protein. In these 5-FOAr cells, the effects of the cse4 mutations on cell growth and chromosome segregation were assessed without interference from the wild-type protein. Cells that cannot lose the URA3-CSE4 plasmid, and therefore cannot grow on 5-FOA medium, carry plasmids with lethal cse4 mutations. Nineteen different alanine scanning mutations (cse4-21 through cse4-39, Fig. 1A) were tested for the ability to rescue cse4 null cells in the plasmid shuffle assay. Most of the alanine substitution mutants exhibited wild-type phenotypes (data not shown). However, cse4-23 (R44A, R46A, and K49A) caused a leaky Ts growth phenotype and the cse4-22 mutant (D36A and R37A) showed a small increase in chromosome missegregation (data not shown). The cse4-22 and cse4-23 mutations were combined to generate a new cse4-39 alanine substitution mutant (D36A, R37A, R44A, R46A, and K49A) which grew at 30°C but exhibited both Ts and Cs growth phenotypes (see Fig. 7A). These results suggested that a region of the N terminus encompassing cse4-39 is important for the function of Cse4p.
FIG. 1.
Mutagenesis of the Cse4p N terminus. (A) Alanine scanning mutations. The N-terminal 135 amino acids of wild-type Cse4p are shown. Numbers identifying each cse4 mutation (cse4-21 to -39) are above the sequence at the positions where the charged residues (R, D, K, and E) indicated by brackets were changed to alanine for each allele. Mutant alleles that exhibit growth and/or chromosome loss phenotypes are denoted by asterisks. (B) N-terminal deletions. The N terminus of Cse4p is depicted as a thick line terminating at an open box representing the HFD, which starts at amino acid 135. Numbers above the lines indicate the residues remaining in the construct, except for alleles 541 to 548, where the residues deleted are indicated. The cse4-encoded mutant proteins initiate with methionine and continue with the amino acids indicated. The viability and growth phenotypes of the mutants are given on the right: +, growth at all of the temperatures tested (30, 15, and 38°C); ts, no growth at 38°C; cs, no growth at 15°C; −, inviable (unable to complement the cse4 null mutation).
FIG. 7.
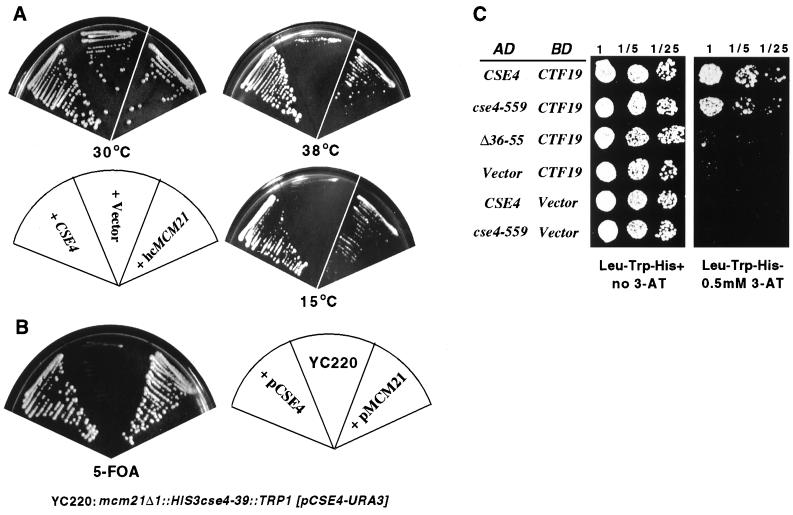
Cse4p N terminus interaction with components of the Mcm21p-Ctf19p-Okp1p complex. (A) Suppression of phenotypes due to cse4-39 by high-copy MCM21. The cse4-39 mutant yeast strain was transformed with a high-copy-number plasmid carrying MCM21 and tested for growth at 30, 38, and 15°C. (B) Synthetic lethality between mcm21Δ and cse4-39. Cells from strain YC220 carrying cse4-39 and a disrupted mcm21 gene (mcm21Δ::HIS3) depend on a CSE4-URA3 plasmid for viability and therefore failed to grow on 5-FOA medium (middle sector). The 5-FOA sensitivity was relieved by introduction of either an MCM21 plasmid (right sector) or a CSE4 plasmid (left sector). (C) Two-hybrid analysis of Cse4p-Ctf19p interactions. Two-hybrid reporter cells (PJ69-4A) were cotransformed with plasmids expressing all or part of Cse4p fused to the GAL4 AD and a plasmid carrying full-length Ctf19p fused to the GAL4 BD. Different AD fusion constructs were derived from the cse4 alleles shown at the left and tested with BD-Ctf19p. The designation Vector indicates that no CSE4 or CTF19 sequences are present in those clones. Transformants were plated in a series of fivefold dilutions spotted on the media indicated. Interaction between AD and BD fusion proteins allowed growth on 3-aminotriazole medium.
Deletion mutagenesis further delineates a 33-amino-acid region essential and sufficient for wild-type Cse4p function.
As a complement to the alanine scanning mutagenesis, we also performed a systematic deletion analysis of the Cse4p N terminus. The first set of nested deletions retained the Cse4p initiating methionine but removed successively larger segments of the N terminus (cse4Δ15 to cse4Δ129, Fig. 1B). Results of plasmid shuffle assays showed that, in agreement with previous results (23), removing residues 2 to 27 from the N terminus (cse4Δ27) was without apparent effect, but removing residues 2 to 55 (cse4Δ55) was lethal (Fig. 1B and 2A).
FIG. 2.
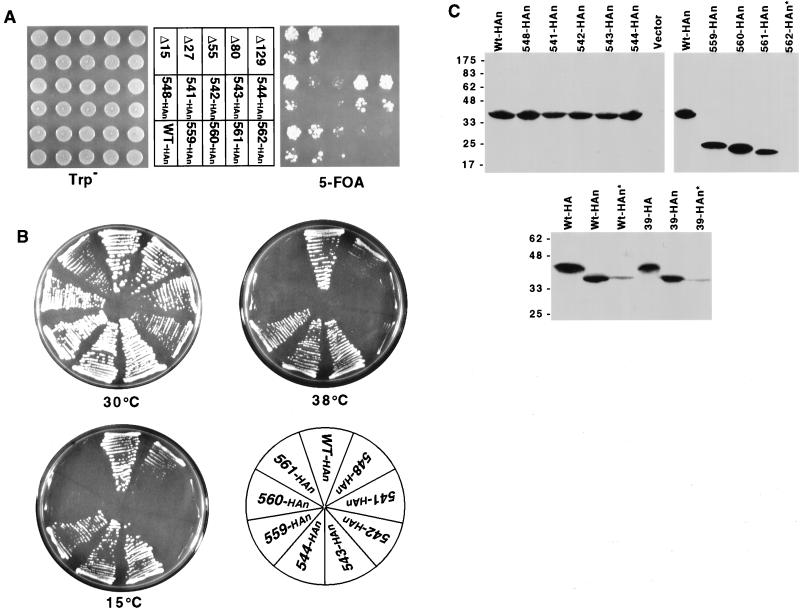
Phenotypes of Cse4p N-terminal mutants. (A) Plasmid shuffle test (see Materials and Methods) showing the ability of various cse4 alleles (middle) to complement the cse4 null lethality. HAn indicates HA epitope-tagged alleles. The bottom of each pair of spots is a 10-fold dilution of the cell suspension used for the top spots. Only yeast strains carrying nonlethal cse4 mutations can grow on 5-FOA medium (right side). (B) Viable cse4 alleles were tested for conditional growth phenotypes. Plates were photographed after 3 days at 30°C, 5 days at 38°C, and 14 days at 15°C, respectively. (C) Immunoblot analysis of cse4 mutant proteins. Immunoblots of representative viable and lethal cse4 mutants are shown in the top two blots. The positions of protein molecular size markers (kilodaltons) are indicated at the left. Proteins were extracted from the cse4 mutant strains indicated above the lanes. The Vector lane contains proteins from cells carrying a vector without a cse4 insert. Asterisks denote cases in which the cells carried an untagged wild-type (Wt) CSE4 allele in addition to the tagged cse4 mutant allele. The HA- and HAn-tagged alleles differ in the location of the HA tag, either at amino acid 83 (Cse4HA) or at the extreme N terminus (Cse4HAn).
To define precisely the essential domain between residues 27 and 55, we made the following series of small deletions spanning the region: cse4-548 (Δ28-35), cse4-541 (Δ36-40), cse4-542 (Δ41-45), cse4-543 (Δ46-50), and cse4-544 (Δ51-55) (Fig. 1B). Two of these cse4 N-terminal mutants, those with the cse4-541 and cse4-542 deletions, were viable in the plasmid shuffle assay but formed very small colonies on 5-FOA medium that were distinctly different from the normal-size colonies produced by the deletion cse4-548 and cse4-543 mutants (Fig. 2A). Upon restreaking, the cse4-541 and cse4-542 mutant cells grew slower than control strains at 30°C and were Ts and Cs (Fig. 2B). One deletion mutant, the cse4-548 mutant, grew well at 30°C but slowly at 38 and 15°C. These results were consistent with those of the alanine scanning mutagenesis and conclusively identified the region located between residues 28 and 46 in the Cse4p N terminus as being important for wild-type function. Alanine scanning mutagenesis of individual residues in this region failed to identify any single amino acid whose replacement with alanine resulted in lethality. However, the double replacement of asparagine 35 with alanine and arginine 44 with glycine resulted in a severe slow-growth phenotype at 30°C (data not shown). These two residues are adjacent to and within the critical region defined by the cse4-39 mutation.
A third set of nested deletion mutations was constructed to test for functionally important sequences in the Cse4p N terminus proximal to the HFD (cse4-556, -557, -558, and -555; Fig. 1B). By analogy to the structure of H3 in the nucleosome (28), Cse4p residues 130 to 135 form the junction of the N terminus and HFD and are positioned to exit the octamer and pass between the two DNA helices wrapped around the histone core. To avoid altering the junction region, the HFD-proximal cse4 deletion mutants were designed to have common endpoints at residue 130 (Fig. 1B). Throughout the rest of this report, we will consider residues 130 to 135 to be part of the HFD. All of these cse4 N-terminal deletion mutants grew like the wild type, revealing that the 70 amino acids proximal to the Cse4p HFD are not required for Cse4p function (data not shown).
Having found that residues 2 to 27 and 51 to 129 in the Cse4p N terminus are dispensable, we next identified the minimal sequences sufficient to provide the essential N-terminal function. Cse4-559p contained amino acids 28 to 60 fused directly to the HFD (Fig. 1B) and was functional at all of the temperatures tested (Fig. 2B). Additional deletion of amino acids 51 to 60 (cse4-560) or 28 to 35 (cse4-561) impaired but did not abolish function. The cse4-560 and cse4-561 mutants, while viable, exhibited conditional growth phenotypes and consistently formed small colonies on 5-FOA medium at the permissive temperature (Fig. 2A and B). The region of overlap between cse4-560 and cse4-561 did not confer the essential N terminus function (cse4-562, Fig. 2A). We concluded that amino acids 28 to 60 are sufficient to provide the essential function of the Cse4p N terminus, and we have named this region the essential N-terminal domain (END).
Expression of cse4 N terminus mutant proteins.
Expression of the mutant cse4 proteins was assayed by immunoblotting. With one possible exception, cse4 mutant phenotypes could not be attributed simply to altered protein expression levels. As shown in Fig. 2C, alleles cse4-541, -542, -560, and -561, all of which cause detectable growth and/or segregation phenotypes, produce protein at levels comparable to that of the wild-type allele. The exceptional case is the lethal allele cse4-562, which did not express a detectable level of protein. The presence of wild-type Cse4p, necessary where the mutant Cse4p was nonfunctional, resulted in significantly reduced steady-state levels of the mutant proteins (asterisks, Fig. 2C). Interestingly, the location of the epitope tag affected the migration of the protein in the SDS-gel. When the triple-HA tag was inserted at residue 83, the resulting Cse4pHA protein migrated with an apparent molecular weight of 43,000, in agreement with previous results (23, 44). When the same tag was fused to the extreme N terminus, epitope-tagged Cse4p migrated with an apparent molecular weight of 37,000.
Mutations in the N terminus of Cse4p cause increased chromosome missegregation.
Cse4p plays a critical role in centromere function during mitosis, and mutations in the HFD cause increased chromosome loss rates (23, 35, 44). To determine if chromosome segregation is impaired in strains carrying Cse4p N-terminal mutations, we screened the collection of Cse4p N-terminal alanine scanning and deletion mutants for increased chromosome missegregation levels using a qualitative colony color sectoring assay (18). The frequencies of sectored colonies made by diploid cse4 null cells expressing different cse4 N-terminal mutant proteins were visually compared to those of cells expressing wild-type CSE4. We assigned tentative chromosome loss phenotypes to 3 of the 19 alanine scanning mutants (cse4-39, cse4-22, and cse4-23), two partial END deletions (cse4-541 and cse4-542), and two END boundary mutants (cse4-560 and cse4-561). Chromosome loss rates were quantified for three of these mutants (cse4-39, cse4-541, and cse4-542), all of which exhibited increases in chromosome loss rates of 12- to 14-fold (Table 2). In contrast, an only 1.7-fold increase in chromosome loss was found for the Cse4-559p protein, which contains the minimal END fused to the HFD.
TABLE 2.
Increased chromosome loss in Cse4p N-terminal mutants
| CSE4 allele | No. of chromosome loss events/cell division (103)a |
|---|---|
| CSE4 | 0.17 (1) |
| cse4-39 | 2.42 (14) |
| cse4-541 | 2.28 (13) |
| cse4-542 | 2.00 (12) |
| cse4-559 | 0.29 (1.7) |
The values in parentheses are fold increases relative to wild-type CSE4.
Is the Cse4p N terminus posttranslationally modified?
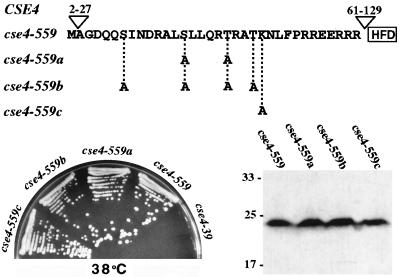
The N-terminal tails of standard core histones are posttranslationally modified, and some modifications are critical for histone function in transcriptional regulation, cell division, and chromatin structure (48, 50). The Cse4HA protein migrates on SDS-PAGE with an apparent molecular mass (43 kDa) much larger than that predicted for the HA-tagged protein (31 kDa) (23, 44), suggesting that Cse4p is modified in vivo. The Cse4p N-terminal amino acid sequence contains several potential posttranslational modification sites. As most of the Cse4p N terminus is dispensable, it is unlikely that modification of any of these nonessential residues is functionally critical; however, the END contains two serines, two threonines, and one lysine, potential sites of phosphorylation (serine or threonine) or acetylation (lysine). To test this possibility, we changed these residues to alanines in three different cse4 alleles (cse4-559a to -559c, Fig. 3). In no case did the mutations affect the growth of the cells or the expression levels or migration of the proteins on SDS-gels (Fig. 3). We concluded that neither phosphorylation nor acetylation of residues within the END is essential for Cse4p function, although other possible modifications are not excluded. The modification mutants were also screened for chromosome segregation defects using the visual colony sectoring assay. The cse4-559a and cse4-559b mutants exhibited a detectable increase in chromosome loss (approximately two- to threefold) compared to the parent (cse4-559), while the sectoring phenotype of cse4-559c was identical to that of cse4-559. The relatively mild chromosome segregation defects observed could be due either to the lack of modification or to alteration of the END structure caused by the mutations.
FIG. 3.
Mutational analysis of possible posttranslational modification sites in the Cse4p END. (Top) The initiating methionine and the 33-amino-acid END sequence in the N terminus of Cse4-559p. Potential phosphorylation (S or T) or acetylation (K) sites were changed to alanine codons, generating the three new alleles shown. (Bottom left) Plate showing growth phenotypes at 38°C (5 days) of strains carrying the indicated HAn-tagged cse4 mutant alleles as their sole source of Cse4p. All strains grew well at 30°C (data not shown). (Bottom right) Immunoblot showing the expression of cse4-encoded mutant proteins. The values on the left are molecular sizes in kilodaltons.
Evidence for a functional multimeric complex containing at least two molecules of Cse4p.
Current genetic evidence is consistent with the idea that Cse4p binds to H4 and forms H4-Cse4p dimers, which assemble into H4-Cse4p–Cse4p-H4 tetramers and ultimately into (H4-Cse4p)2(H2A-H2B)2 octamers (35). In the case of a cell expressing two mutant cse4 alleles, this model predicts that two types of Cse4p octamers would form: homotypic complexes containing two molecules of the same mutant cse4 protein and heterotypic complexes containing one each of the two different mutant cse4 proteins. If the different cse4 mutations affect different biochemical activities of the protein, wild-type function might be partially or fully recovered through the formation of the mixed (i.e., heterotypic) complex (interallelic complementation).
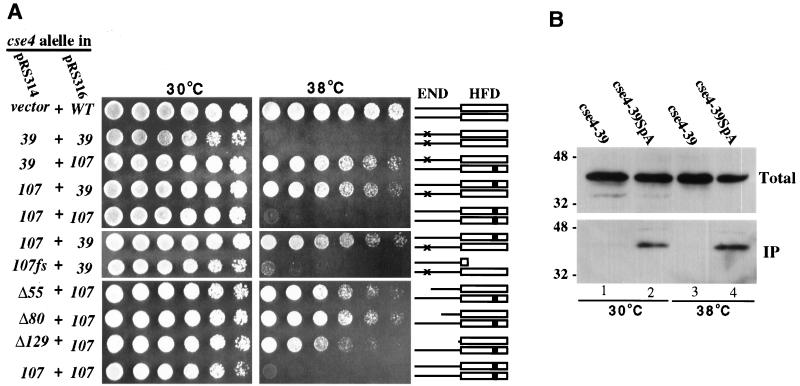
To test for interallelic complementation between cse4 N terminus and HFD mutations, we coexpressed the Ts alleles cse4-39 and cse4-107, which carry mutations in the END (Fig. 1A) and HFD (glutamine 219 changed to aspartate), respectively. Control cse4 null cells carrying either two cse4-39 plasmids or two cse4-107 plasmids failed to grow at 38°C. In contrast, the same yeast strain carrying both the cse4-39 plasmid and the cse4-107 plasmid could grow at 38°C, although not as well as cells expressing wild-type Cse4p (Fig. 4A). Apparently, the essential N-terminal function was provided by the wild-type N terminus of the cse4-107-encoded protein, while the wild-type HFD of the cse4-39-encoded protein conferred the essential HFD function, possibly through heterodimer formation with the defective HFD of Cse4-107p. Interallelic complementation was not observed when a frameshift mutation terminating translation between the N terminus and the HFD was introduced into the cse4-107 gene (Fig. 4A). Thus, the wild-type N terminus of the cse4-107 protein could not rescue the cse4-39 defect when separated from its HFD, consistent with the prediction that the HFD mediates the physical interactions between Cse4p proteins.
FIG. 4.
Genetic and biochemical evidence for Cse4p protein-protein interactions. (A) Interallelic complementation between cse4 END and HFD mutant alleles. The cse4 mutant alleles indicated at the left were expressed from low-copy-number pRS314 and pRS316 vectors and tested for interallelic complementation as described in Materials and Methods. Threefold serial cell dilutions were spotted left to right. The cse4-107fs frameshift mutation terminates translation of the cse4-107-encoded protein between the N terminus and the HFD. Cse4p dimers possibly formed are schematically shown at the right, with × denoting the END mutations in Cse4-39p and ■ indicating the HFD mutation in Cse4-107p. WT, wild type. (B) Coprecipitation of cse4-encoded mutant proteins. Extracts were prepared from cells coexpressing cse4-107HA and either protein A-tagged (lanes 2 and 4) or untagged (lanes 1 and 3) cse4-39. Protein A-tagged protein complexes were precipitated using IgG-Sepharose beads. Approximately 0.5% of the total extracts (Total) and 10 to 15% of the precipitated proteins (IP) were subjected to SDS-PAGE and immunoblot analysis using anti-HA antibodies. The values to the left are molecular sizes in kilodaltons.
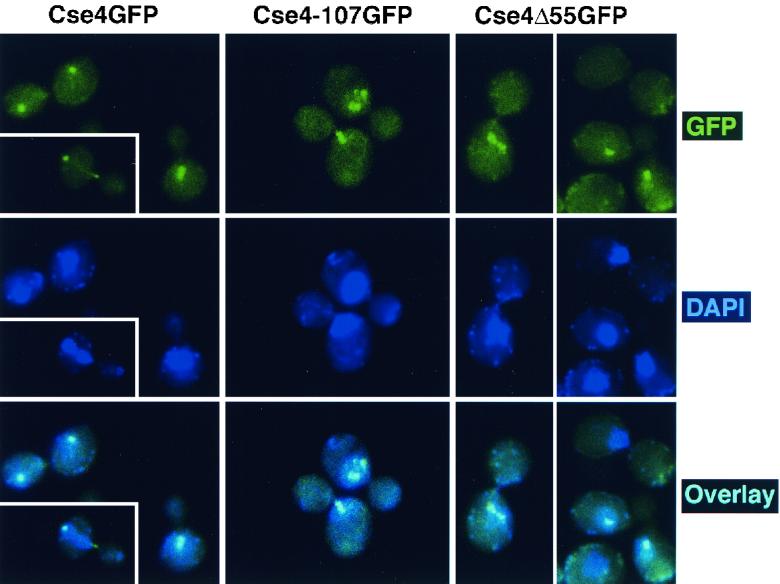
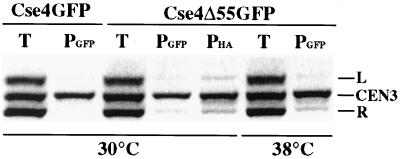
Expression of the Cse4p HFD alone was sufficient to rescue cse4-107 temperature sensitivity. Cells coexpressing one of the lethal cse4 N-terminal deletion mutants (cse4Δ55, cse4Δ80, or cse4Δ129) and the Ts cse4-107 allele were viable and grew at both 30 and 38°C (Fig. 4A). The cse4-107-encoded protein apparently provided the END function, while the mutant proteins with N-terminal deletions complemented the HFD function. This implies that only one functional Cse4p N terminus is required in each Cse4p nucleosome. To confirm that the N-terminally truncated cse4-encoded proteins were actually present in Cse4p complexes at functional centromeres, the cellular localization of Cse4Δ55p tagged with GFP (Cse4Δ55GFP) was determined in cells dependent on both Cse4-107p and Cse4Δ55GFP for viability. In agreement with previous indirect immunofluorescence analysis of Cse4HA (35), we found that wild-type Cse4GFP was localized as a single subnuclear focus in unbudded and small-budded cells and as a short bar or two distinct foci in preanaphase cells (Fig. 5, left panels). These localization patterns correspond to the clustered centromeres visualized in yeast nuclei when the centromeres are labeled by fluorescence in situ hybridization (15). In cells expressing Cse4-107GFP and grown at the permissive temperature, the mutant protein localizes like the wild-type protein (Fig. 5, middle panels) although the strain exhibits a mild growth defect and a high proportion of large-budded cells. When cse4-107 cells were rescued by the cse4Δ55GFP allele at 38°C, localization of Cse4Δ55GFP was very similar to that of Cse4-107GFP at 30°C (Fig. 5, right panels), supporting the idea that Cse4Δ55p is incorporated into complexes (presumably nucleosomes) at centromeres. Localization of Cse4Δ55GFP at the DNA level was further analyzed by CHIP. In cse4-107 mutant cells coexpressing cse4Δ55GFP, the GFP-tagged protein showed specific cross-linking to CEN DNA (Fig. 6) at both growth temperatures. Thus, by both biochemical and morphological criteria, Cse4Δ55GFP is properly localized to centromeres under these conditions.
FIG. 5.
Localization of GFP-tagged Cse4p proteins. Cells were fixed and prepared for microscopy as described in Materials and Methods. The top panels show GFP fluorescence, the middle panels show DAPI staining, and the bottom panels show the overlaid GFP and DAPI images. (Left) Yeast cells (KC100) carrying only wild-type Cse4GFP were grown at 30°C and shifted to 37°C for 5 h (three to four doublings) before fixation. (Middle) KC100 cells expressing only Cse4-107GFP grown at the permissive temperature (30°C). (Right) KC100 cells carrying cse4-107HA and cse4Δ55GFP on separate URA3 and TRP1 vectors, respectively, were streaked and grown at 38°C, the restrictive temperature for both mutations. A single colony was picked, inoculated into prewarmed medium, and grown for an additional 15 h (three to four doublings) at 38°C before fixation and microscopy.
FIG. 6.
CHIP of Cse4Δ55GFP. Cse4-107HA cells were transformed with plasmids carrying cse4GFP or cse4Δ55GFP. Transformants were grown at 30 or 38°C, and CHIP was performed as described in Materials and Methods. The ethidium bromide-stained gel shows the products of PCR using primers for CEN3 (CEN3, 243 bp), DNA 177 kbp to the right of CEN3 (R, 278 bp), and DNA 4 kbp to the left of CEN3 (L, 213 bp). The PCR template was total chromatin (T) or chromatin immunoprecipitated by α-GFP or α-HA antibody (PGFP and PHA, respectively).
Biochemical evidence supporting our interpretation that the interallelic complementation results from the formation of mixed complexes of Cse4p mutant proteins was obtained from coprecipitation assays using Cse4p proteins tagged with the divalent staphylococcal protein A analogue ZZ (SpA) (43). The Cse4SpA protein is fully functional in cse4 null cells and is efficiently precipitated from crude cell extracts by IgG-Sepharose beads. The cse4-39SpA allele was coexpressed with the HA-tagged cse4-107 allele in cse4 null cells. As previously observed, neither single mutant grew at 38°C but when expressed together, Cse4-39SpA and Cse4-107HA permitted growth at 38°C (data not shown). As shown in Fig. 4B, Cse4-107HA and Cse4-39SpA coprecipitated from extracts prepared from cells grown at both 30 and 38°C, demonstrating that the two mutant cse4 proteins formed a mixed complex in vivo. The fact that the heteromeric complexes were also observed at 30°C, a temperature at which both proteins are functional, suggests that complex formation is not an artifactual peculiarity of one or the other inactive mutant protein at the restrictive temperature.
Genetic interactions between cse4 N-terminal alleles and genes encoding subunits of the Mcm21p-Ctf19p-Okp1p complex.
To identify proteins that might potentially interact with the N terminus of Cse4p, we looked for dosage suppressors of the Ts phenotype of cse4-23 cells (see Materials and Methods). Among the candidate genes isolated in the primary screen was MCM21, a nonessential gene originally identified in a screen for minichromosome maintenance mutants (29). MCM21 in high copy number also suppressed the Ts and Cs phenotypes of cse4-39 (Fig. 7A). MCM21 suppression was selective for the END alleles; high-copy-number MCM21 did not suppress the Ts phenotype of the cse4-107 (glutamine 219 to aspartate) or cse4-1 (alanine 221 to threonine) HFD mutant (data not shown). A second genetic interaction, synthetic lethality, was also observed between mcm21 and cse4-39 (Fig. 7B; Table 3).
TABLE 3.
Synthetic lethal interactions of Cse4p N-terminal mutations
| Cross
|
Total no. of spores | No. of viable sporesa
|
Conclusionb | ||||
|---|---|---|---|---|---|---|---|
| mut1 | mut2 | Total | mut1 | mut2 | mut1 mut2 | ||
| cse4-542::URA3 | + | 140 | 127 | 63 | |||
| cse4-542::URA3 | cep1Δ | 76 | 48 | 14c | 14c | 0d | SL |
| cse4-39::TRP1 | spt4Δ::HIS3 | 76 | 60 | 24 | 28 | 11 | RSD |
| cse4-39::TRP1 | cac2::URA3 | 64 | 47 | 18 | 25 | 8 | RSD |
| cse4-39::TRP1 | cac3::URA3 | 60 | 48 | 20 | 19 | 11 | RSD |
| cse4-39::TRP1 | mcm21Δ::HIS3 | 80 | 51 | 16c | 13c | 0d | SL |
| cse4-542::URA3 | mcm22Δ::TRP | 72 | 53 | 16c | 18c | 0d | SL |
| cse4-39::TRP1 | ctf19Δ::HIS3 | 72 | 51 | 17c | 14c | 0d | SL |
| cse4-542::URA3 | okp1-5::TRP | 76 | 57 | 20c | 20c | 0d | SL |
If spore death were random, 50% of viable spores would be expected to carry a single mutation (mut1 or mut2). Likewise, 50% of mut1 spores would be expected to cosegregate mut2 and vice versa (none of the mutations is genetically linked to cse4).
SL, synthetic lethality; RSD, random spore death.
Recovery of mut1 or mut2 among viable spores was statistically significantly less than expected (chi-square test, P < 0.025).
Recovery of viable double mutants was statistically significantly less than expected (chi-square test, P < 0.005).
Since MCM21 encodes a subunit of the Mcm21p-Ctf19p-Okp1p centromere complex (37), we tested for genetic interactions between cse4 N-terminal mutations and other components of the complex, as well as other known centromere proteins. We observed synthetic lethal interactions of cse4-39 with ctf19Δ and mcm21Δ and of cse4-542 with okp1-5, cep1Δ, and mcm22Δ (Table 3). MCM22 is a nonessential gene whose function is not known that was isolated in the same genetic suppressor screen as MCM21 (29, 38). Synthetic lethality was not observed between cse4-39 and cac1 or cac2, genes encoding chromatin assembly factors (22), or spt4, which is involved in the structure of centromere chromatin (5). In addition to the observed synthetic lethality between cse4 END alleles and mcm21Δ, ctf19Δ, okp1-1, and cep1Δ, subsequent plasmid shuffle assays (similar to those of Fig. 7B) revealed that cse4-39 was synthetically lethal with ndc10csl, ctf13-30, and cep3csl, all genes encoding subunits of the CBF3 complex (data not shown). The ctf13-30 and cep3csl interactions were END specific, as these alleles were not synthetic lethal with cse4-107 and other HFD alleles (data not shown). Surprisingly, in contrast to high-copy-number suppression, synthetic lethal interactions between mcm21Δ and cse4 were not END specific; some HFD mutant alleles were also synthetic lethal with mcm21Δ (as tested by plasmid shuffle assay; data not shown).
Ortiz et al. (37) demonstrated an interaction between Cse4p and Ctf19p by yeast two-hybrid assay. We used a similar two-hybrid assay to test the dependence of the Cse4p-Ctf19p interaction on the Cse4p N terminus. Gal4 AD fusions of wild-type and mutant Cse4p proteins were coexpressed with Ctf19p fused to the Gal4 BD in yeast cells containing a HIS3 reporter. Cells expressing the full-length AD-Cse4p and BD-Ctf19p fusion proteins were His+ and grew well on indicator medium containing 3-aminotriazole, confirming Cse4p-Ctf19p interaction. Cells expressing the Ad–Cse4-559p fusion protein containing the minimal END attached to the HFD and BD-Ctf19p grew as well as cells expressing the wild-type AD-Cse4p and BD-Ctf19p proteins. However, when the END was deleted from the AD-Cse4p fusion (AD–Cse4Δ36-55p), cell growth was barely detectable (Fig. 7C). We observed no growth of cells coexpressing either the wild-type CSE4 or cse4-559 AD fusion with an empty BD vector. We concluded that Cse4p and Ctf19p interact in vivo and that the END of the N terminus is an important determinant of this protein-protein interaction.
DISCUSSION
Cse4p is novel among known histone H3-like proteins in having a unique 135-amino-acid-long N terminus extending from the conserved HFD homology region. Unlike the N terminus of yeast H3, which can be deleted without loss of cell viability (30), the Cse4p N terminus is essential (23). Here we have characterized the Cse4p N terminus by systematic mutagenesis, revealing important new information about its function. First, the Cse4p N terminus contains a spatially flexible 33-amino-acid domain, the END, that is essential for Cse4p function. Second, mutations affecting the END and HFD define distinct functions of the protein. Third, the END appears to be involved in the interaction between Cse4p and the Mcm21p-Ctf19p-Okp1p centromere complex. These results are consistent with the current hypothesis that Cse4p replaces H3 in a specialized nucleosome and mediates an essential interaction(s) with other components of centromeric chromatin (35).
An essential domain in the Cse4p N terminus.
The Cse4p N terminus contains a large proportion of charged amino acids, especially the region between residues 54 and 132, where 48% of the amino acids are charged. The N terminus also contains a putative bipartite nuclear localization signal between residues 115 and 132 (44), in addition to many possible posttranslational modification sites and a high concentration of serines within the first 22 amino acids. The combined mutagenesis results show that none of these features is critical; in fact, most of the Cse4p N terminus is dispensable. Only the region between amino acids 28 and 60, the domain we refer to as the END, is essential. Another region of possible functional importance includes residues 130 to 135, which, by analogy to the structure of the conventional nucleosome (28), would exit the core and pass between the two DNA helices wrapped around the histone octamer. Because of this, we avoided altering residues 130 to 135 in the mutagenesis study, although we now know that deletion of residues 130 to 135 does not cause growth or chromosome loss phenotypes in cells with wild-type centromeres (data not shown). This suggests that the positioning of the DNA gyres on the surface of the putative Cse4p-containing octamer is sufficiently flexible to accommodate totally different polypeptide chains passing between them. Interestingly, although the wild-type Cse4p N terminus is much longer than those of H3 and CENP-A, the Cse4p mutant protein Cse4-559p, which consists of the 33-amino-acid END fused directly to the HFD (including residues 130 to 135), confers wild-type function and is very similar in length to CENP-A and H3.
In mammals, H3 phosphorylation is required for the initiation of chromosome condensation (48) but phosphorylated H3 is excluded from the chromatin subjacent to the inner kinetochore plate (49). The centromere chromatin is also underacetylated in mammals (49). The nonphosphorylated, hypoacetylated chromatin zone corresponds to that region of the centromere associated with CENP-A. CENP-A lacks the phosphorylation epitope (serine 10) found in the N terminus of H3, as well as the acetylation sites associated with “transcriptional” acetylation (lysines 14, 18, and 23), implying that the lack of histone modification at mammalian centromeres is due to the displacement of H3 by CENP-A (49). Like the histone H3 tail, the N terminus of Cse4p contains several potential posttranslational modification sites. Defining the minimal END sequence allowed us to mutate all of the potential acetylation and phosphorylation sites present in the only region of the Cse4p N terminus that is essential. The lack of detectable growth phenotypes supports the conclusion that neither phosphorylation nor acetylation of the END is essential for Cse4p function. As suggested for CENP-A, it may be that one role of Cse4p is to establish a zone free of histone modifications at the yeast centromere. Two cse4 END modification mutants did exhibit mild chromosome segregation phenotypes; however, this cannot necessarily be attributed to the lack of modification, as the alanine substitutions could also alter the structure of the END. Conceivably, as is the case with histone H3, a Cse4p N terminus modification(s) could occur but be functionally redundant with a similar modification(s) on the N terminus of histone H4. We have not tested the effect of the END modification mutations in strains expressing H4 with an N-terminal mutation or deletion; however, the essential END function is unique to Cse4p, since END deletions are lethal in strains that are wild type for all of the standard core histones.
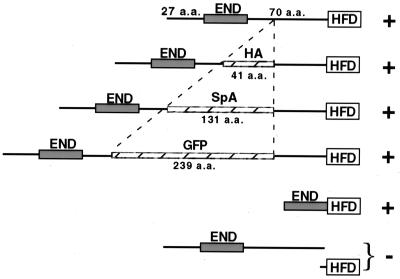
Whatever the essential role of the Cse4p END in centromere function, it is not appreciably compromised when the spacing between the END and the HFD is changed. Cse4p proteins in which the END is adjacent to (cse4-559) or separated by over 300 amino acids from (CSE4GFP) the HFD function essentially like wild-type Cse4p (Fig. 8). If the HFD of Cse4p is embedded in a core with the N terminus extending outward, then the END would be available for protein-protein or protein-DNA interactions with components in the surrounding centromeric chromatin. Given the observed flexibility in END-HFD spacing, the location of a potential interaction site could not be fixed in space relative to the DNA-bound HFD unless it was physically near the core, because of the close proximity of the END to the HFD in Cse4-559p. Alternatively, the END might function at a time when the HFD is not fixed in space, probably before Cse4p is incorporated into chromatin. It has also been noted that the CSE4 ORF contains an in-frame methionine located between the END and the HFD (residue 93). A protein initiated at this methionine would have almost the same length as H3. It is therefore possible that Cse4p is the evolutionary result of a gene fusion event through which H3, or a variant of H3, acquired the END function. That the END and the HFD behave to some degree independently, as suggested from the seeming lack of spacing constraints, is consistent with this idea.
FIG. 8.
Flexible spacing between Cse4p END and HFD. The Cse4p END is spaced 70 amino acids (a.a.) away from the Cse4p HFD in the wild-type protein. This spacing can vary from zero (Cse4-559p) to 309 amino acids (Cse4GFP) without appreciably compromising Cse4p function (indicated by a plus sign); however, Cse4p function was not restored when the END and HFD peptides were expressed separately (indicated by a minus sign).
Cse4p END function is distinct from that of the HFD.
Current genetic data agree with the model in which Cse4p replaces H3 in a centromere-specific nucleosome (35). This model also predicts that the Cse4p HFD mediates Cse4p-Cse4p and Cse4p-H4 interactions in the histone octamer, which is supported by results of systematic HFD mutagenesis (23). Additional evidence that Cse4p proteins interact in vivo and probably form nucleosome-like structures is supplied by the interallelic complementation that we observed between END and HFD cse4 alleles. These results argue that functional heterotypic octamers can be assembled in which one of two Cse4p molecules is defective in the HFD and the other is defective in the END. Biochemical support for this interpretation is provided by the fact that coexpressed mutant cse4 proteins were copurified from cell extracts. The nucleosome model further predicts that the END is incorporated into centric chromatin by virtue of the attached HFD. Consistent with this model, introducing a frameshift mutation into Cse4p between the END and HFD abolishes interallelic complementation. The incorporation of Cse4p molecules into CEN chromatin in complemented cells is probably not random. If it were, half of the nucleosomes assembled would be homotypic (nonfunctional) and the probability of all 16 centromeres incorporating a functional, heterotypic octamer would be low, with severe effects on growth. Since this is not observed, functional heterotypic Cse4p nucleosomes are probably assembled preferentially. Alternatively, the centromere region may have multiple Cse4p nucleosomes, increasing the probability of at least one functional END at each centromere.
Besides providing genetic support for the nucleosome model, interallelic complementation between cse4 END and HFD alleles suggests that the Cse4p END and HFD have distinct functions: the HFD mediates nucleosome assembly, while the END is required for a different Cse4p function, such as nuclear localization, centromere targeting, or interactions with other kinetochore components. The END function could be required at a different time from that of the HFD, either before or after Cse4p dimer formation. Interestingly, cse4 alleles entirely lacking the N-terminal sequences can complement the cse4-107 HFD mutation, indicating that only one functional END per nucleosome is sufficient for Cse4p function at the centromere. The conclusion that the Cse4p END and HFD have different functions is also supported by genetic suppression and synthetic lethal analyses. The screen for high-copy-number suppressors of the cse4-23 END mutation yielded MCM21. In contrast, similar screens for high-copy-number suppressors of HFD mutants (e.g., cse4-1) consistently yielded SCM3 but not MCM21 (C. D. DeFalco et al., unpublished data). SCM3 does not suppress END mutants such as cse4-23 and cse4-39, while MCM21 does not suppress HFD mutants, including cse4-1 and cse4-107. Allele specificity is also observed with synthetic lethal gene interactions. The END allele cse4-39 is synthetic lethal with all of the centromere protein mutants tested, while cse4-107 and other HFD alleles are not synthetic lethal with ctf13 and cep3.
A network of kinetochore protein interactions involving the Cse4p END.
Recently, Ortiz et al. (37) reported that the Ctf19p-Mcm21p-Okp1p protein complex mediates protein-protein interactions at the yeast centromere. They showed by both two-hybrid analysis and coimmunoprecipitation that Ctf19p, Mcm21p, and Okp1p interact with each other, with one or more subunits of CBF3, with Mif2p, and with Cse4p. This network of protein-protein interactions potentially accounts for the localization of all known kinetochore components, including Cse4p, to the centromere. In addition, they demonstrated by CHIP that CDEIII is necessary and sufficient to localize the Ctf19p complex and Cse4p to the centromere.
Our results suggest that the Cse4p END is involved in the interaction(s) between Cse4p and the Ctf19p-Mcm21p-Okp1p complex. MCM21 is a dosage suppressor of END mutations, and a mutation in any of the three components of the Ctf19p-Mcm21p-Okp1p complex is synthetic lethal with a cse4 END mutation. Our two-hybrid results confirm the Ctf19p-Cse4p interaction (37) and further show that it is abolished when the END is deleted from the Cse4p-AD fusion protein, implying that the Cse4p END is required for this interaction. Protein-protein interactions between the END of Cse4p and other centromere proteins may be important for recruiting additional kinetochore proteins to the centromere or for localization of Cse4p itself. Although the Cse4p HFD alone (Cse4Δ55GFP) can be properly targeted to the centromere, as demonstrated by CHIP analysis, this should not be interpreted to mean that the END is not required for centromere localization. Indeed, results from the interallelic complementation and coprecipitation experiments suggest that Cse4Δ55GFP may be targeted by heterodimerization with another Cse4p molecule bearing an intact END (e.g., Cse4-107p or wild-type Cse4p).
An END in other H3-like centromere proteins?
The HFDs of centric histone variants are conserved from yeast to humans despite great divergence in the structures of the respective centromeres. While the HFD of CENP-A might be sufficient to target CENP-A to mammalian centromeres (46), a possible function for the CENP-A N terminus cannot be ruled out. N-terminally truncated CENP-A might lack an essential function and still be able to localize properly (e.g., Cse4Δ55GFP). A second possibility is that the CENP-A “END” became separated from the HFD during evolution and the END function is now supplied to human centromeres in trans by another, as yet unidentified, protein. The Cse4p END does not function in trans when expressed as a separate polypeptide (Fig. 8), possibly because the N terminus lacks targeting information or because the N-terminal peptide by itself is unstable. It would be interesting to determine if the Cse4p END function could be supplied in trans by fusing the Cse4p N terminus to another kinetochore protein, such as Cbf1p or the CBF3 components. Finally, it is also possible that the function of the Cse4p N terminus is unique to the “point” centromeres of budding yeast and that no comparable activity is necessary for the function of vertebrate kinetochores. Regardless of a possible cis or trans CENP-A END function, the similarities and differences between the centromere-specific CENP-A and Cse4p proteins continue to reveal features in common between the two very diverse kinetochore structures.
ACKNOWLEDGMENTS
We thank Jinyun Chen, Tom King, Ben Liu, and Wayne Decatur for technical advice and other members of the M.F.-H. laboratory for comments and suggestions. We are grateful to P. Hieter, J. Lechner, P. Sinha, M. A. Basrai, and P. D. Kaufman for yeast strains and plasmids and to Kellie Kosewski for constructing the GFP-tagged CSE4 allele.
This work was supported by grants to M.F.-H. from the National Institutes of Health (GM54766) and to R.E.B. from the National Science Foundation (MCB-9406050).
REFERENCES
- 1.Anderson M T, Tjioe I M, Lorincz M C, Parks D R, Herzenberg L A, Nolan G P. Simultaneous fluorescence-activated cell sorter analysis of two distinct transcriptional elements within a single cell using engineered green fluorescent proteins. Proc Natl Acad Sci USA. 1996;93:8508–8511. doi: 10.1073/pnas.93.16.8508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baker R E, Fitzgerald-Hayes M, O'Brien T C. Purification of the yeast centromere binding protein CP1 and a mutational analysis of its binding site. J Biol Chem. 1989;264:10843–10850. [PubMed] [Google Scholar]
- 3.Baker R E, Harris K, Zhang K. Mutations synthetically lethal with cep1 target S. cerevisiae kinetochore components. Genetics. 1998;149:73–85. doi: 10.1093/genetics/149.1.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baker R E, Masison D C. Isolation of the gene encoding the Saccharomyces cerevisiae centromere-binding protein CP1. Mol Cell Biol. 1990;10:2458–2467. doi: 10.1128/mcb.10.6.2458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Basrai M A, Kingsbury J, Koshland D, Spencer F, Hieter P. Faithful chromosome transmission requires Spt4p, a putative regulator of chromatin structure in Saccharomyces cerevisiae. Mol Cell Biol. 1996;16:2838–2847. doi: 10.1128/mcb.16.6.2838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bloom K S, Carbon J. Yeast centromere DNA is in a unique and highly ordered structure in chromosomes and small circular minichromosomes. Cell. 1982;29:305–317. doi: 10.1016/0092-8674(82)90147-7. [DOI] [PubMed] [Google Scholar]
- 7.Buchwitz B J, Ahmad K, Moore L L, Roth M B, Henikoff S. A histone H3-like protein in C. elegans. Nature. 1999;401:547–548. doi: 10.1038/44062. [DOI] [PubMed] [Google Scholar]
- 8.Cai M, Davis R W. Purification of a yeast centromere-binding protein that is able to distinguish single base-pair mutations in its recognition site. Mol Cell Biol. 1989;9:2544–2550. doi: 10.1128/mcb.9.6.2544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cai M, Davis R W. Yeast centromere binding protein CBF1, of the helix-loop-helix protein family, is required for chromosome stability and methionine prototrophy. Cell. 1990;61:437–446. doi: 10.1016/0092-8674(90)90525-j. [DOI] [PubMed] [Google Scholar]
- 10.Carlson M, Botstein D. Two differentially regulated mRNAs with different 5′ ends encode secreted and intracellular forms of yeast invertase. Cell. 1982;28:145–154. doi: 10.1016/0092-8674(82)90384-1. [DOI] [PubMed] [Google Scholar]
- 11.Cumberledge S, Carbon J. Mutational analysis of meiotic and mitotic centromere function in Saccharomyces cerevisiae. Genetics. 1987;117:203–212. doi: 10.1093/genetics/117.2.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fitzgerald-Hayes M, Clarke L, Carbon J. Nucleotide sequence comparisons and functional analysis of yeast centromere DNAs. Cell. 1982;29:235–244. doi: 10.1016/0092-8674(82)90108-8. [DOI] [PubMed] [Google Scholar]
- 13.Fleig U, Beinhauer J D, Hegemann J H. Functional selection for the centromere DNA from yeast chromosome VII. Nucleic Acids Res. 1995;23:922–924. doi: 10.1093/nar/23.6.922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gaudet A, Fitzgerald-Hayes M. Alterations in the adenine-plus-thymine-rich region of CEN3 affect centromere function in Saccharomyces cerevisiae. Mol Cell Biol. 1987;7:68–75. doi: 10.1128/mcb.7.1.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Guacci V, Hogan E, Koshland D. Centromere position in budding yeast: evidence for anaphase A. Mol Biol Cell. 1997;8:957–972. doi: 10.1091/mbc.8.6.957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hegemann J H, Shero J H, Cottarel G, Philippsen P, Hieter P. Mutational analysis of centromere DNA from chromosome VI of Saccharomyces cerevisiae. Mol Cell Biol. 1988;8:2523–2535. doi: 10.1128/mcb.8.6.2523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Henikoff S, Ahmad K, Platero J S, van Steensel B. Heterochromatic deposition of centromeric histone H3-like proteins. Proc Natl Acad Sci USA. 2000;97:716–721. doi: 10.1073/pnas.97.2.716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hieter P, Mann C, Snyder M, Davis R. Mitotic stability of yeast chromosomes: a colony color assay that measures nondisjunction and chromosome loss. Cell. 1985;40:381–392. doi: 10.1016/0092-8674(85)90152-7. [DOI] [PubMed] [Google Scholar]
- 19.Hyland K M, Kingsbury J, Koshland D, Hieter P. Ctf19p: a novel kinetochore protein in Saccharomyces cerevisiae and a potential link between the kinetochore and mitotic spindle. J Cell Biol. 1999;145:15–28. doi: 10.1083/jcb.145.1.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.James P, Halladay J, Craig E A. Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics. 1996;144:1425–1436. doi: 10.1093/genetics/144.4.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jehn B, Niedenthal R, Hegemann J H. In vivo analysis of the Saccharomyces cerevisiae centromere CDEIII sequence: requirements for mitotic chromosome segregation. Mol Cell Biol. 1991;11:5212–5221. doi: 10.1128/mcb.11.10.5212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kaufman P D, Kobayashi R, Stillman B. Ultraviolet radiation sensitivity and reduction of telomeric silencing in Saccharomyces cerevisiae cells lacking chromatin assembly factor-I. Genes Dev. 1997;11:345–357. doi: 10.1101/gad.11.3.345. [DOI] [PubMed] [Google Scholar]
- 23.Keith K C, Baker R E, Chen Y, Harris K, Stoler S, Fitzgerald-Hayes M. Analysis of primary structural determinants that distinguish the centromere-specific function of histone variant Cse4p from H3. Mol Cell Biol. 1999;19:6130–6139. doi: 10.1128/mcb.19.9.6130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kent N A, Tsang J S H, Crowther D J, Mellor J. Chromatin structure modulation in Saccharomyces cerevisiae by centromere and promoter factor I. Mol Cell Biol. 1994;14:5229–5241. doi: 10.1128/mcb.14.8.5229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kingsbury J, Koshland D. Centromere-dependent binding of yeast minichromosomes to microtubules in vitro. Cell. 1991;66:483–495. doi: 10.1016/0092-8674(81)90012-x. [DOI] [PubMed] [Google Scholar]
- 26.Lechner J, Carbon J. A 240 kd multisubunit protein complex, CBF3, is a major component of the budding yeast centromere. Cell. 1991;64:717–725. doi: 10.1016/0092-8674(91)90501-o. [DOI] [PubMed] [Google Scholar]
- 27.Lechner J, Ortiz J. The Saccharomyces cerevisiae kinetochore. FEBS Lett. 1996;389:70–74. doi: 10.1016/0014-5793(96)00563-7. [DOI] [PubMed] [Google Scholar]
- 28.Luger K, Mader A W, Richmond R K, Sargent D F, Richmond T J. Crystal structure of the nucleosome core particle at 2.8A resolution. Nature. 1997;389:251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- 29.Maine G T, Sinha P, Tye B K. Mutants of S. cerevisiae defective in the maintenance of minichromosomes. Genetics. 1984;106:365–385. doi: 10.1093/genetics/106.3.365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mann R K, Grunstein M. Histone H3 N-terminal mutations allow hyperactivation of the yeast GAL1 gene in vivo. EMBO. 1992;11:3297–3306. doi: 10.1002/j.1460-2075.1992.tb05408.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McGrew J, Diehl B, Fitzgerald-Hayes M. Single base-pair mutations in centromere element III cause aberrant chromosome segregation in Saccharomyces cerevisiae. Mol Cell Biol. 1986;6:530–538. doi: 10.1128/mcb.6.2.530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mellor J, Jiang W, Funk M, Rathjen J, Barnes C A, Hinz T, Hegemann J H, Philippsen P. CPF1, a yeast protein which functions in centromeres and promoters. EMBO J. 1990;9:4017–4026. doi: 10.1002/j.1460-2075.1990.tb07623.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Meluh P, Koshland D. Budding yeast centromere composition and assembly as revealed by in vivo cross-linking. Genes Dev. 1997;11:3401–3412. doi: 10.1101/gad.11.24.3401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Meluh P B, Koshland D. Evidence that the MIF2 gene of Saccharomyces cerevisiae encodes a centromere protein with homology to the mammalian centromere protein, CENP-C. Mol Biol Cell. 1995;6:793–807. doi: 10.1091/mbc.6.7.793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Meluh P B, Yang P, Glowczewski L, Koshland D, Smith M M. Cse4p is a component of the core centromere of Saccharomyces cerevisiae. Cell. 1998;94:607–613. doi: 10.1016/s0092-8674(00)81602-5. [DOI] [PubMed] [Google Scholar]
- 36.Niedenthal R K, Sen-Gupta M, Wilmen A, Hegemann J H. Cpf1 protein induced bending of yeast centromere DNA element I. Nucleic Acids Res. 1993;21:4726–4733. doi: 10.1093/nar/21.20.4726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ortiz J, Stemmann O, Rank S, Lechner J. A putative protein complex consisting of Ctf19, Mcm21, and Okp1 represents a missing link in the budding yeast kinetochore. Genes Dev. 1999;13:1140–1155. doi: 10.1101/gad.13.9.1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Poddar A, Roy N, Sinha P. MCM21 and MCM22, two novel genes of the yeast Saccharomyces cerevisiae are required for chromosome transmission. Mol Microbiol. 1999;31:349–360. doi: 10.1046/j.1365-2958.1999.01179.x. [DOI] [PubMed] [Google Scholar]
- 39.Rose M D, Winston F, Hieter P. Methods in yeast genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1990. [Google Scholar]
- 40.Sherman F, Fink G, Hicks J B. Methods in yeast genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1983. [Google Scholar]
- 41.Sikorski R S, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Smith M M, Yang P, Santisteban M S, Boone P W, Goldstein A T, Megee P C. A novel histone H4 mutant defective for nuclear division and mitotic chromosome transmission. Mol Cell Biol. 1996;16:1017–1026. doi: 10.1128/mcb.16.3.1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stirling D A, Petrie A, Pulford D J, Paterson D T, Stark M J. Protein A-calmodulin fusions: a novel approach for investigating calmodulin function in yeast. Mol Microbiol. 1992;6:703–713. doi: 10.1111/j.1365-2958.1992.tb01519.x. [DOI] [PubMed] [Google Scholar]
- 44.Stoler S, Keith K C, Curnick K E, Fitzgerald-Hayes M. A mutation in CSE4, an essential gene encoding a novel chromatin-associated protein in yeast, causes chromosome nondisjunction and cell cycle arrest at mitosis. Genes Dev. 1995;9:573–586. doi: 10.1101/gad.9.5.573. [DOI] [PubMed] [Google Scholar]
- 45.Strahl-Bolsinger S, Hecht A, Luo K, Grunstein M. SIR2 and SIR4 interactions differ in core and extended telomeric heterochromatin in yeast. Genes Dev. 1997;11:83–93. doi: 10.1101/gad.11.1.83. [DOI] [PubMed] [Google Scholar]
- 46.Sullivan K F, Hechenberger M, Masri K. Human CENP-A contains a histone H3 related histone fold domain that is required for targeting to the centromere. J Cell Biol. 1994;127:581–592. doi: 10.1083/jcb.127.3.581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tanaka T, Cosma M P, Wirth K, Nasmyth K. Identification of cohesin association sites at centromeres and along chromosome arms. Cell. 1999;98:847–858. doi: 10.1016/s0092-8674(00)81518-4. [DOI] [PubMed] [Google Scholar]
- 48.Van Hooser A, Goodrich D W, Allis C D, Brinkley B R, Mancini M A. Histone H3 phosphorylation is required for the initiation, but not maintenance, of mammalian chromosome condensation. J Cell Sci. 1998;111:3497–3506. doi: 10.1242/jcs.111.23.3497. [DOI] [PubMed] [Google Scholar]
- 49.Van Hooser A A, Mancini M A, Allis C D, Sullivan K F, Brinkley B R. The mammalian centromere: structural domains and the attenuation of chromatin modeling. FASEB J. 1999;13(Suppl. 2):S216–S220. doi: 10.1096/fasebj.13.9002.s216. [DOI] [PubMed] [Google Scholar]
- 50.Wolffe A P, Hayes J J. Chromatin disruption and modification. Nucleic Acids Res. 1999;27:711–720. doi: 10.1093/nar/27.3.711. [DOI] [PMC free article] [PubMed] [Google Scholar]