Abstract
Cyclin-dependent kinase 7 (CDK7) is the catalytic subunit of the metazoan CDK-activating kinase (CAK), which activates CDKs, such as CDC2 and CDK2, through phosphorylation of a conserved threonine residue in the T loop. Full activation of CDK7 requires association with a positive regulatory subunit, cyclin H, and phosphorylation of a conserved threonine residue at position 170 in its own T loop. We show that threonine-170 of CDK7 is phosphorylated in vitro by its targets, CDC2 and CDK2, which also phosphorylate serine-164 in the CDK7 T loop, a site that perfectly matches their consensus phosphorylation site. In contrast, neither CDK4 nor CDK7 itself can phosphorylate the CDK7 T loop in vitro. The ability of CDC2 or CDK2 and CDK7 to phosphorylate each other but not themselves implies that each kinase can discriminate among closely related sequences and can recognize a substrate site that diverges from its usual preferred site. To understand the basis for this paradoxical substrate specificity, we constructed a chimeric CDK with the T loop of CDK7 grafted onto the body of CDK2. Surprisingly, the hybrid enzyme, CDK2-7, was efficiently activated in cyclin A-dependent fashion by CDK7 but not at all by CDK2. CDK2-7, moreover, phosphorylated wild-type CDK7 but not CDK2. Our results suggest that the primary amino acid sequence of the T loop plays only a minor role, if any, in determining the specificity of cyclin-dependent CAKs for their CDK substrates and that protein-protein interactions involving sequences outside the T loop can influence substrate specificity both positively and negatively.
Full activation of cyclin-dependent kinases (CDKs) requires the binding of a positive regulatory subunit or cyclin and the phosphorylation of a threonine residue on a conserved loop—the activation segment or T loop—of the catalytic subunit by a CDK-activating kinase (CAK) (reviewed in references 20 and 32). The major CAK in metazoan cells is itself a CDK containing CDK7 as its catalytic subunit (10, 29, 34, 45). That CDK7 activates CDKs in vivo was confirmed in Drosophila melanogaster; flies with a temperature-sensitive CDK7 had a defect in CDC2 phosphorylation, resulting in a block to mitosis at the nonpermissive temperature (25).
The T-loop region of CDK7 has a threonine residue, threonine-170 (T170), in the same location as and within a sequence context similar to that for the activating threonines of other CDKs. T170 is required for activation of dimeric complexes of CDK7 with its physiologic partner cyclin H in vitro and in vivo (13, 27) and for the basal kinase activity associated with monomeric CDK7 (28), probably reflecting the phosphorylation of T170 by a putative CAK-activating kinase (CAKAK) (12). What regulatory function, if any, T-loop phosphorylation of CDK7 serves in vivo is unknown. The major form of CDK7 in the cell is a ternary complex of CDK7, cyclin H, and the RING finger protein MAT1 (7, 12, 47). MAT1 stabilizes the cyclin H-CDK7 complex and can bypass the need for T170 phosphorylation in vitro; trimeric CDK7-cyclin H-MAT1 complexes formed with CDK7 bearing the T170-to-alanine (T170A) mutation appear to be fully active towards CDK2 (12).
Several lines of evidence suggest that phosphorylation-dependent activation of CDK7 and its relatives in lower eukaryotes occurs in vivo. CDK7 and cyclin H form a stable complex when overexpressed together in mammalian or insect cells or in budding yeast; stable association requires phosphorylation of T170 (12, 27; unpublished observations). In both amphibian (24) and mammalian (P. Jin and D. O. Morgan, unpublished observations) cells, T170 is a major site of CDK7 phosphorylation in vivo. In fission yeast, the ortholog of CDK7, Mcs6, can be activated in vitro by another kinase, Csk1, through T-loop phosphorylation (17, 26); this presumably explains the reduced Mcs6-associated kinase activity observed in strains deleted for csk1 (17, 30). The physiologic importance of this activation is unclear, however, because Csk1 also directly activates Cdc2, the major CDK in fission yeast (26). In budding yeast, the CDK7 family member Kin28 is not a CAK but is a target for T-loop phosphorylation by another enzyme, Cak1 (8, 21). Recent studies have shown that T-loop phosphorylation dramatically stimulates the kinase activity of Kin28 in vivo (8, 21) but is not essential for viability (21).
Active CDK2-cyclin A complexes promote CDK7-cyclin H assembly in vitro in T170-dependent fashion, suggesting that CDK7 and its targets might participate in positive feedback loops of activating phosphorylation (12). Direct phosphorylation of T170 by a CDK has never been demonstrated, however, and a mechanism whereby CDC2 or CDK2 phosphorylates CDK7 on another site to promote autophosphorylation on T170 by CDK7 itself was considered possible. The T loops of CDC2, CDK2, and CDK7 are all quite similar, so it was not obvious from primary sequence comparisons why CDK7 was able to activate both CDC2 and CDK2 (13) (as well as the much more divergent CDK4 [29] and -6 [1]) but was unable to autoactivate. Conversely, the sequence surrounding T170 of CDK7 bears no resemblance to the consensus phosphorylation site for CDC2 and CDK2, Ser/Thr-Pro-X-Lys/Arg (where X is any residue) (2, 18, 31, 46). Phosphorylation of T170 would represent a significant departure from the normal substrate specificity of the prototypic CDKs.
In this report, we confirm that T170 is a direct target for phosphorylation by CDC2 and CDK2 but not by CDK4 or CDK7 in vitro. In addition, both CDC2 and CDK2 phosphorylate S164 within a CDK consensus phosphorylation site also present in the T loop of CDK7; the two phosphorylations, however, occur independently of one another. We constructed a chimeric CDK with the T loop of CDK7 grafted onto the body of CDK2 to show that sequences outside the T loop are primarily responsible both for promoting T-loop phosphorylation by CAK, independent of the primary amino acid sequence surrounding the phosphorylation site, and for preventing autophosphorylation by CDK2, even when its T loop contains a perfect consensus CDK2 phosphorylation site.
MATERIALS AND METHODS
Baculoviruses.
The construction of recombinant baculoviruses encoding untagged CDK7 (wild type and a catalytically inactive mutant, K41A), cyclin H, and MAT1-His has been described previously (12, 13). Viruses encoding CDK7 fused to a carboxyl-terminal hemagglutinin (HA) epitope have also been described. In this study, we used wild-type CDK7-HA (13); CDK7-HA(T170A), a mutant in which the activating threonine-170 residue is mutated to alanine (13); CDK7-HA(S164A), in which the consensus phosphorylation site for CDK family kinases, serine-164, was mutated to alanine by site-directed mutagenesis (23) with the oligonucleotide 5′-GCTCTATTGGGcgcCCCAAAAGATTTGGC-3′ (mutagenic nucleotides in lowercase); and CDK7-HA(S164A/T170A), in which both phosphorylation sites were changed to alanines with the oligonucleotide 5′-CAACCTGATGTGcATAAGCTCTATTGGGcgcCCCAAAAGATTTGGC-3′.
In order to create the baculovirus encoding the CDK2-7 mutant, we synthesized a double-stranded oligonucleotide corresponding to the NheI-BstEII fragment of the CDK2 cDNA with the desired mutations to the CDK7 sequence (see Fig. 4A), 5′ CTAGCAGACTTTGGATAGCCAaAtCTTTTGGgagCCCcaaTagagGCTTAtACaCATcAGGTG 3′ (sense strand) and 5′GTCACCACCTgATGtGTaTAAGctctAttgGGGctcCCAAAAGaTtTGGCTAGTCCAAAGTCT 3′ (antisense strand).
FIG. 4.
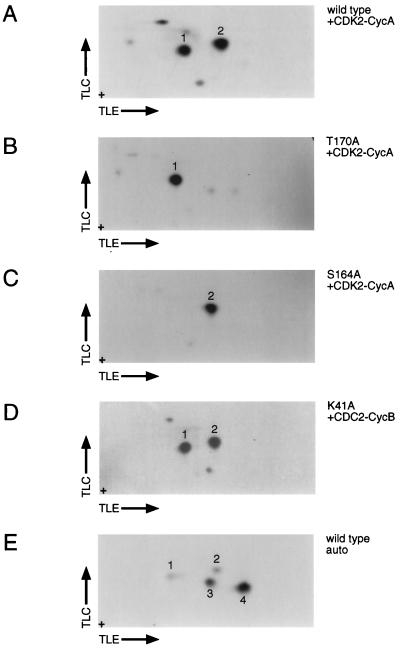
CDC2 and CDK2 phosphorylate CDK7 on S164 and T170. Tryptic phosphopeptide mapping was performed as described in Materials and Methods. (A) Phosphorylation of CDK7 by activated CDK2-cyclin A complexes (as described for Fig. 2A, lane 2) results in strong phosphorylation of two tryptic peptides (1 and 2). These spots comigrate with the two major phosphopeptides produced by phosphorylation of CDK7 in vivo (data not shown). (B) The map generated by phosphorylation of CDK7(T170A) in vitro lacks spot 2. (C) The map generated by phosphorylation of CDK7(S164A) in vitro lacks spot 1. (D) Phosphorylation of the CDK7 kinase-dead mutant, K41A, in vitro. Both spots 1 and 2, corresponding to phosphorylation of S164 and T170, respectively, are generated with pure CDC2-cyclin B in vitro. (E) Phosphopeptide map produced by autophosphorylation of wild-type CDK7 (in the presence of stoichiometric amounts of cyclin H). Autophosphorylation reactions were performed with fourfold more radiolabeled ATP and required two- to fourfold-longer autoradiographic exposures than did reactions with CDC2 and CDK2. For all maps, thin-layer electrophoresis (TLE) was from left (+ electrode) to right (− electrode) in the first dimension, followed by ascending thin-layer chromatography (TLC) in the second dimension. Samples were spotted at the origin (+).
The oligonucleotides were annealed and then ligated with CDK2 cDNA (containing a carboxyl-terminal HA tag) digested with NheI and BstEII. The specificity of mutagenesis was confirmed by direct sequencing, and the CDK2-7–HA coding sequence was used to construct a baculovirus by standard methods (13, 33).
Protein purification.
Purification of cyclin H-His from bacteria (13) and of CDK7, cyclin H, and MAT1-His from lysates of insect cells infected with recombinant baculoviruses (11) has been previously described. We used an abbreviated version of the CDK7 purification scheme to obtain partially purified wild-type and kinase-deficient CDK7. The ATP-agarose chromatography used to purify wild-type CDK7 to homogeneity (11) was omitted, because the catalytically inactive K41A mutant failed to bind this resin efficiently (R. P. Fisher and D. O. Morgan, unpublished observations). Instead, both the wild-type and mutant proteins were purified by chromatography on DEAE-Sepharose Fast Flow (Pharmacia), HiTrap SP (Pharmacia), and Superose 12 (Pharmacia). Both proteins appear to be monomeric, with an apparent size in Superose 12 gel filtration chromatography of ∼40 kDa (data not shown). After the gel filtration step, proteins were concentrated by adsorption to a 1-ml HiTrap SP column in buffer C (11) plus 50 mM NaCl (followed by step elution with buffer C plus 400 mM NaCl), frozen in liquid N2, and stored at −80°C. Based on Coomassie blue staining of sodium dodecyl sulfate (SDS)-polyacrylamide gels, we estimate that both wild-type and mutant CDK7 are ∼80% pure (data not shown).
Monomeric CDK2 and cyclin A were purified from baculovirus-infected insect cell lysates as previously described (6, 37). Pure CDK2-cyclin A complexes activated by CAK-mediated phosphorylation (41) were generously provided by A. Russo and N. Pavletich of Memorial Sloan-Kettering Cancer Center. Pure complexes of CDC2 with glutathione transferase (GST)-cyclin B and of CDK4 with cyclin D2 were the kind gift of G. Bollag (Onyx Pharmaceuticals). CDK2-7 was purified by the CDK2 protocol previously described (37).
Synchronization of HeLa cells.
HeLa cells were grown in minimal medium supplemented with 5% fetal calf serum on plastic dishes and synchronized in different cell cycle intervals by standard methods (14, 38). Briefly, cells arrested at the G1/S boundary were obtained by a double thymidine block; logarithmically growing cells were treated with 2 mM thymidine for 12 to 14 h, followed by a recovery period of 9 h in drug-free medium, followed by a second treatment with 2 mM thymidine for 12 to 14 h prior to harvest. A population of cells synchronized in G2 was generated by the double thymidine block, followed by release into drug-free medium for 7 to 9 h before harvest. To obtain cells arrested in mitosis, cells treated once for 12 to 14 h in 2 mM thymidine were released into medium containing 50 ng of nocodazole/ml and incubated for 18 to 24 h. Mitotic cells that were detached or loosely adherent to the dish were then harvested. To obtain cells in G1, cells arrested in mitosis with nocodazole were harvested by centrifugation, washed extensively, and released into drug-free medium. After 4 to 6 h, cells remaining in mitosis were removed by shakeoff and adherent cells were harvested. To assess the efficiency of arrest and synchronization, parallel cultures were analyzed by flow cytometry.
Preparation of HeLa cell lysates.
Cells were harvested by scraping (G1, S, and G2 populations) or by shakeoff (mitotic cells), washed with phosphate-buffered saline, and lysed by resuspension in ∼0.2 ml of lysis buffer per 150-mm dish: 25 mM HEPES (pH 7.4), 150 mM NaCl, 0.1% Triton X-100, 1 mM EDTA, 1 mM dithiothreitol (DTT), 50 mM NaF, 80 mM β-glycerophosphate, 0.1 mM Na3VO4, 0.5 mM phenylmethylsulfonyl fluoride, 2 μg of aprotinin/ml, and 1 μg of leupeptin/ml. Resuspended cells were vortexed briefly, incubated 10 min on ice, vortexed again, and centrifuged for 20 min at 14,000 rpm in a microcentrifuge (Eppendorf) at 4°C. The supernatant was transferred to a fresh tube, frozen in liquid nitrogen, and stored at −80°C. To resolve CDK7 phosphoisoforms, we used SDS-polyacrylamide gels containing the cross-linker piperazine diacrylamide (PDA) and raised the pH of the separating gel buffer to 9.2 (22, 25).
Phosphorylation of CDKs in vitro.
Autophosphorylation of CDK7 was examined by incubating 1 μg of pure CDK7, alone or in combination with equimolar amounts of pure cyclin H and/or MAT1-His, in 30 μl of kinase mix containing 10 mM HEPES (pH 7.4), 150 mM NaCl, 10 mM MgCl2, 50 μM unlabeled ATP, and 2.5 to 10 (depending on the experiment) μCi of [γ-32P]ATP. In some experiments, phosphorylation of CDK7 by other CDKs was assayed by a simple modification of this protocol; pure, active complexes of CDC2, CDK2, and CDK4 were added to reactions containing pure or partially pure wild-type or kinase-deficient CDK7. We also used HA-tagged CDK7-cyclin H complexes immobilized on protein A-Sepharose (Sigma) with monoclonal antibody (MAb) 12CA5 (BAbCO) to activate mixtures of CDC2 or CDK2 and cyclin A or B that had been purified separately, exactly as described previously (6, 37). Aliquots (200 ng) of CDK-cyclin complexes thus activated were added to kinase mixes containing 1 μg of pure CDK7 (with or without 1 μg of pure cyclin H) and labeled ATP, as described above.
Tryptic phosphopeptide mapping.
A further modification of this protocol was made in order to generate tryptic phosphopeptide maps of the two phosphorylation site mutants of CDK7 after phosphorylation by CDK2-cyclin A complexes in vitro. Pure CDK2 and cyclin A were mixed and activated by immobilized trimeric (CDK7-HA–cyclin H–MAT1-His) CAK complexes. An aliquot (200 ng) of the activated complex was then transferred to a second tube containing CDK7-HA(T170A) or CDK7-HA(S164A) immunoprecipitated from a baculovirus-infected insect cell lysate with MAb 12CA5 and protein A-Sepharose and the radioactive kinase mix described above. Reactions were terminated by the addition of fourfold-concentrated SDS-polyacrylamide gel electrophoresis (PAGE) sample buffer, and labeled proteins were denatured by boiling and separated by SDS-PAGE.
CDK2 and CDK2-7 were labeled by incubating 1 μg of either pure protein with 300 ng of CDK7-cyclin H in the presence of 10 mM MgCl2, 50 μM unlabeled ATP, and 10 μCi of [γ-32P]ATP. In order to label wild-type CDK7 (see Fig. 6), 1 μg of CDK2 and 1.5 μg of cyclin A were activated with 300 ng of CDK7-cyclin H in the presence of 10 mM MgCl2 and 1 mM unlabeled ATP. Sf9 lysate containing CDK7-HA (100 μg of total protein) was immunoprecipitated with MAb 16B12 (BAbCO), washed, and incubated with 250 ng of activated CDK2-cyclin A in the presence of 10 mM MgCl2 and 20 μCi of [γ-32P]ATP.
FIG. 6.
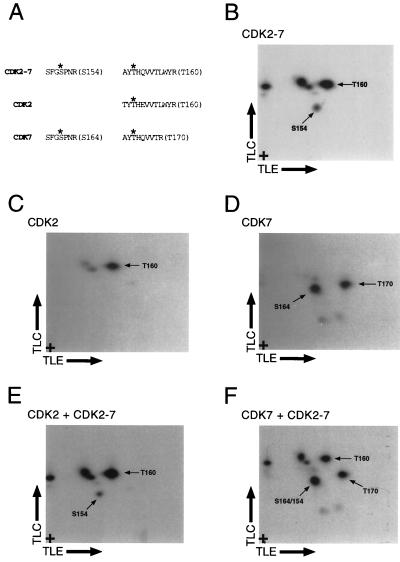
CDK2-7 is phosphorylated by CDK7-cyclin H on both S154 and T160. TLC, thin-layer chromatography. TLE, thin-layer electrophoresis. (A) A diagram of expected T-loop phosphopeptides derived from the labeling and tryptic digestion of CDK2, -7, and -2-7. The residues expected to be targets for phosphorylation are indicated with asterisks, and the peptides are named according to the identity and number of this residue (in parentheses at right). The S154 peptide of CDK2-7 and the S164 peptide of CDK7 are identical and therefore comigrate. Although they are not identical, the T160 peptides of CDK2-7 and CDK2 should also comigrate; the two amino acid differences in the peptides are not predicted to have any effect on their mobility in either dimension (3). (B) The tryptic phosphopeptide map of CDK2-7 labeled by CDK7-cyclin H has two spots that correspond to labeling on both S154 and T160 in the T loop (indicated by labels). Three additional spots appear, which presumably represent phosphorylation at non-T-loop residues (see text). (C) Phosphorylation of CDK2 by CDK7-cyclin H generates one major spot that corresponds to phosphorylation on T160. (D) Phosphorylation of CDK7 by activated CDK2-cyclin A yields two major spots, which correspond to phosphorylation at S164 and T170 (Fig. 3). (E) Mixing of labeled CDK2-7 and CDK2 reveals comigration of the T160 peptides of each of these CDKs, indicating that CDK2-7 is in fact phosphorylated on T160. The spot corresponding to phosphorylation on the S154 peptide, which has no counterpart in the map derived from wild-type CDK2, is visible, as are three spots presumed to be due to be phosphorylation outside the T loop. (F) Mixing of CDK7 and CDK2-7 samples generates a major spot of increased intensity that corresponds to comigration of the S154 and S164 phosphopeptides. Two spots corresponding to the T160 and T170 phosphopeptides, which are not expected to comigrate, are visible, as are the spots previously ascribed to non-T-loop phosphorylation.
Bulk phosphorylation of CDKs was visualized by autoradiography of the dried gels and quantified by liquid scintillation counting of excised gel bands. To detect phosphorylation on specific residues, labeled proteins were transferred electrophoretically to polyvinylidene difluoride membranes (Immobilon-P; Whatman), autoradiographed and stained with Ponceau S to locate and excise labeled CDK, and processed for tryptic phosphopeptide mapping essentially as described earlier (3, 14). Phosphoamino acid analysis was performed on both major phosphopeptides of CDK7, which were recovered by scraping from the chromatography plate, according to published methods (3).
CAK assays.
Activation of CDK complexes was assayed essentially as previously described (13). To measure activation of CDK2-7, 500 ng of CDK2-7 was mixed with 750 ng of cyclin A and 100 ng of CDK7-cyclin H in the presence of Mg-ATP, immunoprecipitated with MAb 16B12, and tested for kinase activity with 5 μg of GST–carboxyl-terminal domain (CTD) substrate.
The ability of CDK2-7 to phosphorylate CDK2 was assayed in a similar manner. Instead of GST-CTD substrate, the activated CDK2-7 was incubated with either 1 μg of CDK2 and 1.5 μg of cyclin A or 5 μg of histone H1 (to confirm activation). To measure direct phosphorylation of CDK2-7 by CDK7-cyclin H, we incubated 1 μg of CDK2-7 (or wild-type CDK2 as a control) and 1.5 μg of cyclin A for 30 min in a 30-μl reaction mixture containing 10 mM HEPES (pH 7.4), 150 mM NaCl, 1 mM DTT, 10 mM MgCl2, 50 μM ATP, and 2 μCi of [γ-32P]ATP, with or without 100 ng of pure CDK7-cyclin H. Radiolabeled products were detected as above. To activate CDK2-cyclin A complexes prior to testing their ability to phosphorylate CDK2-7, we immunoprecipitated Csk1-HA (26) from 10 μg of appropriately infected Sf9 cell lysate with MAb 16B12. The beads were washed and incubated with 1 μg of CDK2, 1.5 μg of cyclin A, 1 mM ATP, and 10 mM MgCl2 in 30 μl of total volume. An aliquot (3 μl) of supernatant containing activated CDK2-cyclin A was then removed and tested for kinase activity towards CDK2-7 or histone H1 (5 μg per reaction) as described above. To assess CDK2-7 activity towards CDK7, 1 μg of CDK2-7 or CDK2 was incubated with or without 1.5 μg of cyclin A in the absence or presence of 300 ng of CDK7-cyclin H in a mixture containing 10 mM MgCl2 and 1 mM ATP. CDK7-HA was immunoprecipitated from 50 μg of Sf9 lysate with MAb 16B12 and incubated with 100 ng of activated CDK2 or CDK2-7 in the presence of labeled ATP, as described above.
CAKAK assay.
Phosphorylation-dependent activation of dimeric CDK7-cyclin H complexes in vitro was assayed as previously described (12), with minor modifications. Insect cell lysates (50 μg of total protein) containing ∼1 μg of either CDK7-HA(S164A) or CDK7-HA(S164A/T170A) were incubated with 1 μg of pure cyclin H in an activation mixture containing 10 mM HEPES (pH 7.4), 10 mM MgCl2, 1 mM ATP, 1 mM DTT, and either 100 to 500 ng of pure activated CDK2-cyclin A (41) or 50 μg of HeLa cell lysate protein. After a 15-min incubation at 25°C, CDK7 complexes were recovered on protein A-Sepharose by immunoprecipitation with MAb 16B12. Immunoprecipitates were washed three times with 10 mM HEPES (pH 7.4), 150 mM NaCl, 0.1% Triton X-100, and 10 mM EDTA; washed twice with 10 mM HEPES (pH 7.4), 150 mM NaCl, and 1 mM DTT; and then tested for CAK activity as previously described (12, 13) with unphosphorylated CDK2-cyclin A complexes (19; kind gift of Russo and Pavletich, Memorial Sloan-Kettering Cancer Center) as the substrate. In indicated control reactions, 1 μg of MAT1-His was included in the activation mix to stabilize (and activate) CAK independent of phosphorylation. Phosphorylated CDK2 was detected by autoradiography of the dried gels and quantified with a PhosphorImager (Molecular Dynamics).
Immunodepletion of HeLa cell extracts.
Mitotic HeLa cell extracts were subjected to two rounds of immunodepletion with antibodies to CDC2 (polyclonal antibody C-19; Santa Cruz), to CDK2 (polyclonal antibody M2; Santa Cruz), or to GST (mock depletion). For each round of immunodepletion, anti-CDC2 (60 μg), anti-CDK2 (10 μg), or anti-GST (30 μg) was preadsorbed to 25 μl of protein A-Sepharose beads in the presence of 20 mg of bovine serum albumin/ml. HeLa extract (250 μg of total protein) was added to antibody-containing beads, incubated at 4°C for 1 h, and cleared by centrifugation. Immunoprecipitation was repeated with the supernatant from the first round, and the beads from both rounds were combined for a subsequent kinase assay. Immunoblotting performed with 10% of the twice-depleted extract confirmed complete removal of CDC2 and CDK2 with the appropriate antibodies and no detectable losses due to cross-reactivity or nonspecific adsorption. The remainder of the depleted extract (∼90%) was then used for CDK7 activation assays.
Insect cell lysate (100 μg/reaction) containing CDK7-HA(S164A) was incubated for 2 h at 4°C with protein A-Sepharose beads (50 μl/reaction) plus MAb 16B12. The beads were washed as described above and were aspirated dry. We added either depleted or mock-depleted extract (∼200 μg each) and incubated the beads for 30 min at room temperature in the presence of 10 mM MgCl2 and 1 mM ATP. Pure cyclin H (1 μg) was added, and the mixtures were incubated for an additional 30 min. The beads were then washed and assayed as described above (see “CAKAK assay”), except that the substrate was 5 μg of GST-CTD per reaction.
To measure CAKAK activity of immunoprecipitated HeLa CDKs, we incubated the beads containing immune complexes with 100 μg of CDK7-HA(S164A) lysate for 30 min at room temperature in the presence of 10 mM MgCl2 and 1 mM ATP. Cyclin H (1 μg) was added, and the beads were incubated for another 30 min. The beads were recovered by centrifugation, washed, and used to perform a histone H1 kinase assay as previously described (13). The supernatant was added to 50 μl of protein A-Sepharose beads plus MAb 16B12 and incubated at 4°C for 2 h. The beads containing activated CDK7-cyclin H complexes were recovered by centrifugation, washed, and assayed for CTD kinase activity as described above. Phosphorylation was detected by autoradiography of dried gels and was quantified either by liquid scintillation counting or by scanning with a PhosphorImager.
RESULTS
Phosphorylation of CDK7 in vitro.
The CDK7 homolog in Xenopus laevis is phosphorylated at two major sites in vivo (24) and in egg extracts in vitro (35). The mammalian protein is phosphorylated at the same two sites: S164 (corresponding to serine-170 of Xenopus CDK7) and T170 (corresponding to threonine-176 of Xenopus CDK7) in COS-7 cells (unpublished observations). Both S164 and T170 lie within the T loop. A comparison of human CDK T loops (Fig. 1) reveals that T170 of CDK7 is in the same position as and in a sequence context similar to that of threonine-160 (T160), the activating residue (and CAK target site) of CDK2. S164, which has no obvious corresponding residue in CDC2 or CDK2 (Fig. 1), is in a context that matches the consensus sequence for sites phosphorylated by CDC2 (CDK1) and CDK2 (2, 18, 31, 46).
FIG. 1.
Alignment of CDK T-loop sequences. The activation segments or T-loop sequences of five mammalian CDKs are compared by aligning the DFG and APE motifs (underlined), which are conserved among protein kinases (15). Sites of phosphorylation are shown in boldface and include the activating threonine residue present in all five T loops (T170 in CDK7) and the consensus CDC2/CDK2 phosphorylation site, serine-164, which is unique to CDK7.
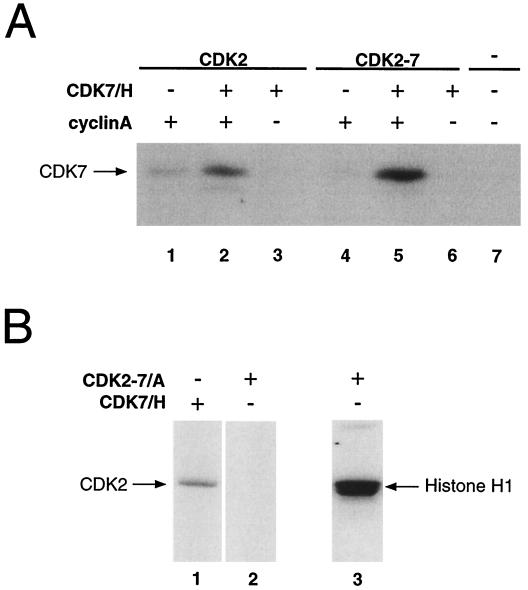
We observed labeling of CDK7 in assays containing pure recombinant CAK and either CDK2-cyclin A or CDC2-cyclin B complexes as the substrates (unpublished observations). We therefore tested directly whether CDKs could phosphorylate CDK7 in vitro. As shown in Fig. 2A, pure CDK2-cyclin A (lanes 1 to 3) and CDC2-cyclin B (lanes 4 to 6) efficiently phosphorylated CDK7 in vitro. Phosphorylation of CDK7 by CDC2 and CDK2 did not require the CDK7 binding partner, cyclin H (which is itself phosphorylated weakly by both kinases). In fact, phosphorylation of CDK7 was somewhat more intense in the absence of cyclin H (Fig. 2A, compare lanes 1 and 3 and lanes 4 and 6). Labeling of CDK7 in these reactions did not require the catalytic activity of CDK7 itself; we observed similar levels of incorporation whether the substrate was wild-type CDK7 or the catalytically inactive Lys-41-to-Ala (K41A) mutant (13) (Fig. 2B, compare lanes 2 to 4 with lanes 9 to 11).
FIG. 2.
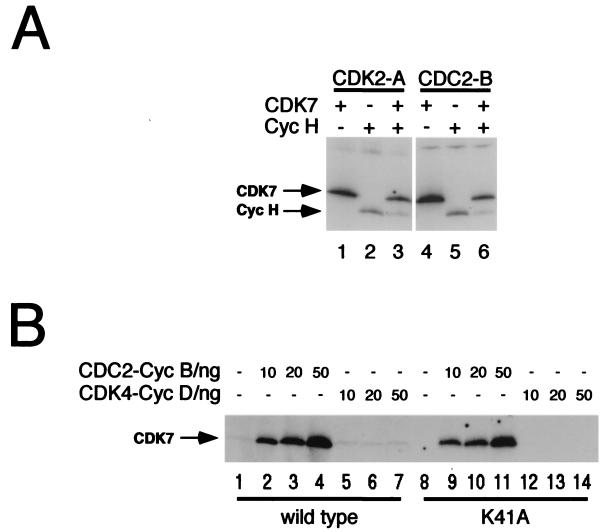
Phosphorylation and activation of CDK7 in vitro. (A) CDK2-cyclin A (lanes 1 to 3) and CDC2-cyclin B (lanes 4 to 6) complexes were formed by mixing the pure subunits in vitro and were activated by incubation with HA-tagged CDK7-cyclin H immobilized on protein A-Sepharose. A 100-ng aliquot of each was used to phosphorylate 1 μg of pure CDK7 either in the absence (lanes 1 and 4) or in the presence (lanes 3 and 6) of 1 μg of pure bacterial cyclin H-His (incubated without CDK7 in lanes 2 and 5). Arrows at left indicate mobilities of CDK7 and cyclin H-His. Bands were excised from the gel, and incorporation was quantified by liquid scintillation counting; in the absence of cyclin H, activated CDC2 transfers ∼1.5 mol of phosphate per mol of CDK7 protein (data not shown). (B) Pure CDK7 (1 μg), either wild type or the catalytically inactive K41A mutant, was phosphorylated efficiently by pure CDC2-cyclin B (lanes 2 to 4 and 9 to 11) but not by pure CDK4-cyclin D (lanes 5 to 7 and 12 to 14). Both CDC2 and CDK4 efficiently phosphorylated a GST-retinoblastoma protein substrate (data not shown).
In contrast to CDC2-cyclin B and CDK2-cyclin A, which phosphorylated CDK7 efficiently (and roughly equally), active CDK4-cyclin D complexes were unable to phosphorylate CDK7 above the background level in vitro (Fig. 2B, lanes 5 to 7 and 12 to 14). To determine the phosphorylation state of CDK7 at different points in the cell cycle when different CDK-cyclin complexes are active, we ran SDS-polyacrylamide gels containing the cross-linking agent PDA to resolve CDK7 isoforms (22, 25). As shown in Fig. 3A, phosphorylation of both T170 and S164 in vitro by CDK2-cyclin A markedly increased the electrophoretic mobility of CDK7 (Fig. 3A, lane 2). Phosphorylation of T170 alone caused an intermediate shift (Fig. 3A, lane 4), whereas phosphorylation of S164 alone did not produce a consistent, detectable change in mobility (Fig. 3A, lane 6). We next looked at the steady-state distribution of CDK7 among different isoforms in synchronized HeLa cell extracts (Fig. 3B). The form that was phosphorylated on both S164 and T170 predominated throughout the cell cycle. The unphosphorylated form was more abundant in the extract from asynchronous cells; in other experiments, the distribution of isoforms in asynchronous cells appeared more similar to that in synchronized populations, with the doubly phosphorylated form predominating (unpublished observations). There appear to be no major fluctuations, however, in steady-state levels or distribution of CDK7 T-loop phosphorylation in HeLa cells arrested or synchronized in different phases of the cell cycle with drug treatments.
FIG. 3.
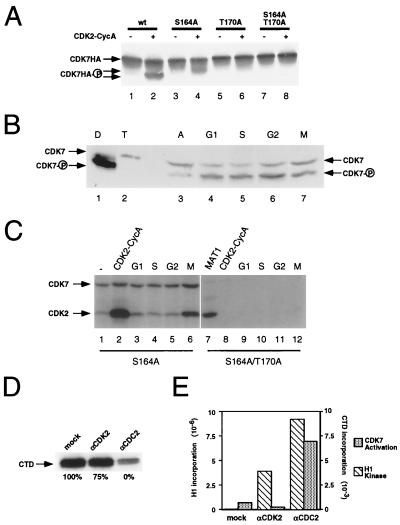
Cell cycle analyses of CDK7 phosphorylation states and activating kinases. (A) T-loop phosphorylation of CDK7 increases its electrophoretic mobility. Insect cell lysates containing ∼1 μg of HA-tagged wild-type CDK7 (wt; lanes 1 and 2), CDK7(S164A) (lanes 3 and 4), CDK7(T170A) (lanes 5 and 6), or CDK7(S164A/T170A) (lanes 7 and 8) were incubated without (odd-numbered lanes) or with (even-numbered lanes) 100 ng of active CDK2-cyclin A in the presence of 1 mM ATP. Proteins were denatured and subjected to SDS-PAGE in gels containing the cross-linker PDA and were transferred to nitrocellulose membranes. CDK7 was detected by immunoblotting with MAb 16B12 specific for the HA epitope. (B) CDK7 is phosphorylated on both T170 and S164 throughout the cell cycle. Extracts prepared from asynchronous (A) HeLa cells (lane 3) and from cells synchronized in G1, S, G2, or M phase were subjected to SDS-PAGE in PDA-containing gels and analyzed by immunoblotting with anti-CDK7 antibody. The mobilities of the unphosphorylated (CDK7) and singly and doubly phosphorylated (CDK7-P) CDK7 isoforms are indicated at left; CDK7-cyclin H dimeric complexes (D) purified after double infection of insect Sf9 cells contain the doubly phosphorylated form predominantly whereas CDK7-cyclin H-MAT1 trimeric complexes (T) produced by triple infection contain only the unphosphorylated form. (C) CAKAK peaks at mitosis in HeLa cells. Lysate containing ∼1 μg of CDK7-HA(S164A) (lanes 1 to 6) or CDK7-HA(S164A/T170A) (lanes 7 to 12) was mixed with 1 μg of pure cyclin H and incubated without further protein addition (lane 1) or with 200 ng of activated CDK2-cyclin A (lanes 2 and 8), 1 μg of pure MAT1-His (lane 7), or 50 μg of lysate from HeLa cells in G1 (lanes 3 and 9), S (lanes 4 and 10), G2 (lanes 5 and 11), or M phase (lanes 6 and 12). CDK7-cyclin H complexes were then immunoprecipitated and tested for the ability to phosphorylate CDK2 (indicated by lower arrow at left). CDK7 (higher arrow) was also phosphorylated in the reaction. (D) Immunodepletion of CDK2 from the mitotic extract slightly reduces CDK7-activating kinase, whereas immunodepletion of CDC2 reduces CDK7 activation to the background level observed in the absence of extract. Mitotic extract was subjected to two rounds of immunodepletion with antibodies to CDK2 (αCDK2), CDC2 (αCDC2), or GST (mock). Depleted extracts were incubated with CDK7-HA(S164A) immunoprecipitated from Sf9 lysate and 1 μg of pure cyclin H in the presence of MgCl2 and ATP. The CDK7-cyclin H complexes were then assayed for activity towards the CTD of RNA polymerase II. Activity was quantified by liquid scintillation counting, and residual activity was expressed as a percentage of activity in mock-depleted extract (mock) (defined as 100%), as indicated below each lane. (E) CDC2 activates CDK7 more efficiently than does CDK2. Mitotic extracts were immunodepleted of CDC2 or CDK2 as described for panel D. The immunoprecipitated kinases were incubated with lysates containing CDK7-HA(S164A) and pure cyclin H in the presence of MgCl2 and ATP. The CDK7 complexes were then immunoprecipitated and assayed for activity towards the CTD. The beads containing the immunoprecipitated CDC2 or CDK2 were then washed and assayed for activity towards histone H1. Activity was quantified by PhosphorImager scanning.
To investigate further whether the phosphorylation-dependent activation of CDK7 could be regulated in cell cycle-dependent fashion, we tested extracts of HeLa cells arrested or synchronized at different positions in the cell cycle for the ability to activate CDK7-cyclin H complexes in vitro (Fig. 3C). To ensure that activation was dependent on T170 phosphorylation, we used the S164A mutant form of CDK7 and performed duplicate assays with the S164A/T170A mutant. Only the single mutant, which retains an intact T170, can be activated by the phosphorylated CDK2-cyclin A complex (Fig. 3C, lane 2). The double phosphorylation site mutant is completely refractory to CDK2 (Fig. 3C, lane 8) but is activated when MAT1 is included in the preincubation to stabilize a ternary complex (Fig. 3C, lane 7).
CAKAK activity was virtually undetectable in extracts prepared from HeLa cells in S or G2 (Fig. 3C, lanes 4 and 5) but rose dramatically in cells arrested in mitosis with nocodazole (Fig. 3C, lane 6). CAKAK activity in the mitotic extract was approximately 10-fold higher than in a G1 extract (Fig. 3C, lane 3) (data not shown). We suspect that the decrease upon exit from mitosis is even sharper, because our G1 cell population, derived from nocodazole-arrested cells that were allowed to reattach to culture dishes in drug-free medium, consistently contained 10 to 20% cells with a G2/M DNA content by flow cytometry (data not shown). None of the extracts could activate the S164A/T170A mutant CDK7 (Fig. 3C, lanes 9 to 12). Because pure MAT1 can activate the double mutant (Fig. 3C, lane 7), activation by the extracts therefore cannot be due to the presence of free MAT1.
The apparent restriction of CAKAK activity to mitosis is surprising, given the ability of CDK2—which is active throughout much of the S and G2 phases (38)—to activate CDK7 in vitro (12) (Fig. 3C, lane 2). To determine the relative contributions of CDC2 and CDK2 to CAKAK activity in the mitotic extract, we performed immunodepletion with antibodies to either CDK and measured residual activity of the depleted extracts in vitro. Immunoblotting was performed with aliquots of the depleted extracts to insure that the antibodies were specific and that all of the targeted protein was in fact removed (data not shown). In addition, a mock depletion was carried out with antibodies to GST.
Depletion of CDK2 from the extract produced at most a modest reduction (∼25%) in its ability to activate CDK7 (Fig. 3D). Clearing the extract of CDC2, on the other hand, caused the activity to drop to background levels. The small decrease in activity after CDK2 depletion, together with the complete removal of CAKAK activity by CDC2 depletion, suggests that CDC2 is the major CDK7-activating kinase in the extract and that the contribution of endogenous CDK2 is minor or even negligible. To get a better idea of their relative contributions, we directly assayed the ability to activate CDK7, relative to histone H1 kinase activity, of the two CDKs immunoprecipitated from the mitotic extract (Fig. 3E). Our results indicate that endogenous CDC2 is a more efficient activator of CDK7 than is CDK2. We therefore conclude that CDC2 is the major CAKAK in the mitotic extract, with only a minor contribution, if any, by CDK2. This presumably explains why S and G2 extracts—which contain high levels of active CDK2 but low levels of active CDC2 (14)—do not activate CDK7 efficiently in our assay (Fig. 3C).
Activating phosphorylation of CDK7 by CDC2 and CDK2.
The ability of pure CDK2-cyclin A to activate CDK7 in T170-dependent fashion (Fig. 3C, lane 2) strongly suggested a direct phosphorylation mechanism. Tryptic phosphopeptide mapping confirmed that both CDK2-cyclin A (Fig. 4A) and CDC2-cyclin B (Fig. 4D) phosphorylated CDK7 on both S164 and T170. The identity of the two major tryptic phosphopeptides generated by incubation with CDC2 or CDK2 was confirmed by analysis in vitro of CDK7 mutants in which either T170 or S164 was changed to alanine (Fig. 4B or C, respectively). The two phosphorylations were independent of one another; mutation of T170 to alanine had no effect on the phosphorylation of S164 (Fig. 4B), and phosphorylation of T170 was likewise unaffected by mutation of S164 (Fig. 4C). No significant differences were seen between the phosphorylation patterns produced by CDC2 and CDK2 (compare Fig. 4A and D). Addition of cyclin H had no consistent effect on the distribution of label among different phosphopeptides, although it did reduce the overall intensity of labeling (Fig. 2A).
The sequence surrounding T170 of CDK7 bears no resemblance to the phosphorylation site consensus recognized by CDC2 and CDK2 (Ser/Thr-Pro-X-Lys/Arg, where X is any residue [2, 18, 31, 46]). Moreover, the results shown for Fig. 4 indicated that CDC2 and CDK2 phosphorylate the T170 residue independently of the consensus S164 site. We investigated the possibility that CDK2 and CDC2 were not phosphorylating CDK7 directly but rather stimulating a cryptic autophosphorylation activity intrinsic to CDK7. We partially purified CDK7(K41A) and phosphorylated it in vitro with pure CDC2-cyclin B. The catalytically inactive CDK7 was also phosphorylated on both T170 and S164 by CDC2 (Fig. 4D); similar results were obtained with CDK2-cyclin A (data not shown). Thus, CDC2 and CDK2 must directly phosphorylate T170 in the T loop of CDK7. Activation of CDK7 by CDC2 and CDK2 is efficient: under optimal conditions (as described for Fig. 2A, lane 4), CDC2 can catalyze incorporation of ∼1.5 mol of phosphate per mol of CDK7 protein (data not shown). Indeed, phosphorylation of the nonconsensus site, T170, is consistently equivalent to phosphorylation of the consensus site, S164 (Fig. 4A and D), which serves as an internal control in our labeling reactions.
We consistently observed low levels of phosphorylation on CDK7 in the absence of other kinases (Fig. 2B). In contrast to phosphorylation that was dependent on the exogenous CDC2 or CDK2, the apparent autophosphorylation signal was abolished by the K41A mutation (Fig. 2B, compare lanes 1 and 8). Autophosphorylation by CDK7 in the presence of cyclin H alone is inefficient; the molar ratio of incorporated phosphate to CDK7 protein is much less than 1. Addition of MAT1 in stoichiometric amounts dramatically increased autophosphorylation (data not shown). However, virtually all of the increased labeling occurred on a site or sites distinct from T170 or S164, the identity of which remains unknown (corresponding to peptide 4 in Fig. 4E). We have also tested the ability of CDK7 complexes to autoactivate in trans in a CAKAK assay. Preassembled, active CDK7-cyclin H complexes were unable to activate CAK reconstituted from monomeric CDK7 and cyclin H subunits in vitro (unpublished observations). Thus, little or no autophosphorylation occurs in the T loop of CDK7 by either intra- or intermolecular mechanisms. We conclude that activating T-loop phosphorylation of CDK7 depends on an exogenous activating kinase and that this function can be provided in vitro by either CDC2 or CDK2.
The basis of substrate recognition by cyclin-dependent CAKs.
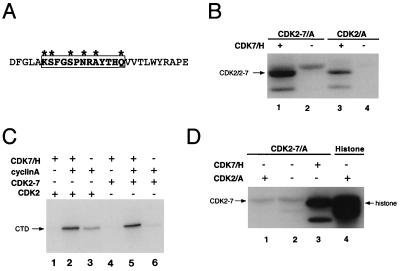
Our results suggested that CDK7 and its targets, CDC2 and CDK2, reciprocally activate each other by T-loop phosphorylation. In both cases, recognition of the relevant phosphorylation site within the T loop represents a departure from the orthodox, proline-directed mode of substrate recognition by CDKs (2, 18, 31, 46). To test whether the T-loop sequence itself directed the activation of CDK7 by CDK2, we replaced the wild-type CDK2 T-loop sequence with the analogous region of CDK7 (Fig. 5A). The mutant was expressed in both Sf9 insect cells and mammalian COS-7 cells at levels equal to those of wild-type CDK2, suggesting that no major structural alterations were introduced by the mutation (data not shown). We expected that, if phosphorylation depended primarily on the T-loop sequence, the chimeric enzyme CDK2-7 would not be activated by CDK7-cyclin H complexes and would instead be activated by CDK2-cyclin A. Indeed, we anticipated that the hybrid CDK might actually be capable of autoactivation, because its T loop contained a sequence phosphorylated by CDK2 when encountered in the context of CDK7.
FIG. 5.
Construction and phosphorylation in vitro of CDK2-7. (A) A schematic diagram of the CDK2-7 hybrid T loop. CDK2-7 was constructed by replacing the T loop of CDK2 with the analogous sequence from CDK7, a total of six amino acid changes. The region converted to the CDK7 sequence is indicated by the boxed residues, while the asterisks denote positions that actually differ between CDK2 and CDK7. (B) Pure CDK2-7 in a complex with cyclin A is efficiently phosphorylated by CDK7-cyclin H in vitro (lane 1) to a level significantly above that of background autophosphorylation (lane 2) and comparable to that of CAK-mediated phosphorylation of wild-type CDK2-cyclin A (lane 3). CDK2/2-7, CDK2 (lanes 3 and 4) and CDK2-7 (lanes 1 and 2). (C) Phosphorylation by CDK7-cyclin H activates CDK2-7. CTD kinase activity of CDK2-7 is dependent on association with cyclin A and activation by CDK7-cyclin H (compare lane 5 with lanes 4 and 6), as is that of wild-type CDK2 (compare lane 2 with lanes 1 and 3). (D) CDK2 cannot phosphorylate CDK2-7. CDK2-cyclin A activated by preincubation with CDK7-cyclin H is unable to phosphorylate CDK2-7-cyclin A (lane 1) above the background level presumably due to autophosphorylation (lane 2), even though it is active towards histone H1 (lane 4). As a control, CDK2-7 in complex with cyclin A was phosphorylated by CDK7-cyclin H (lane 3).
When pure CDK2-7 was mixed with pure cyclin A and incubated in vitro with pure CDK7-cyclin H complexes and [γ-32P]ATP, the CDK2-7 polypeptide was heavily phosphorylated (Fig. 5B, lane 1). In fact, we consistently observed more intense labeling of CDK2-7 than of wild-type CDK2 (Fig. 5B, compare lanes 1 and 3). There are two possible explanations, both of which are unexpected. Either CDK2-7 is an even better substrate for CDK7-cyclin H than is wild-type CDK2, or CDK7 phosphorylates more sites in CDK2-7 than in CDK2. In any case, CDK7-cyclin H efficiently phosphorylated CDK2-7 despite the change in its T-loop sequence.
CDK2-7 is activated by CDK7 in a cyclin-dependent manner.
We next asked whether phosphorylation of CDK2-7 by CDK7-cyclin H resulted in its enzymatic activation. The direct phosphorylation experiments (Fig. 5B) suggested that CDK2-7 was indeed phosphorylated by CDK7 within the T loop, because of the characteristically increased electrophoretic mobility of the labeled species. Indirect CAK assays (Fig. 5C), in which CDK2-7 was first incubated with CDK7-cyclin H and then tested for kinase activity towards the CTD of the RNA polymerase II large subunit, confirmed that CDK7 activates CDK2-7 in a cyclin A-dependent manner (Fig. 5C, compare lanes 5 and 6). This result strongly suggested that CDK7-cyclin H phosphorylated the T-loop threonine of CDK2-7, leading to its enzymatic activation, despite the mutations within the surrounding sequences.
To determine if CDK2—a CAK for wild-type CDK7 in vitro—could activate the CDK2-7 chimera, we activated CDK2-cyclin A complexes by preincubation with immobilized HA-tagged Csk1 (26) and incubated them with pure CDK2-7–cyclin A (Fig. 5D). Although the CDK2-cyclin A complexes were active towards histone H1 (Fig. 5D, lane 4), they were unable to phosphorylate CDK2-7 measurably above the level of background autophosphorylation (Fig. 5D, compare lane 1 with lane 2). Thus, CDK2 recognized neither the noncanonical T170 phosphorylation site nor S164, which matches its consensus recognition sequence. This implies that structural features of CDK2-7 actually prevent CDK2 from phosphorylating sites that it would recognize in their normal context, i.e., CDK7. Interference appears to be specific to CDK2: both CDK7-cyclin H (Fig. 5C) and Csk1 (unpublished observations) activate CDK2-7 quite efficiently, suggesting that the T loop is accessible to certain kinases.
Fidelity of T-loop phosphorylation events in a CDK chimera.
Our results suggested that the overall protein context prevailed over the actual T-loop sequence in determining to which activating kinase the CDK was susceptible. Moreover, the inability of CDK2 to phosphorylate the CDK7 T loop out of its usual context implied that sequences outside the T loop could block as well as promote activating phosphorylation. We sought further evidence to support these rules by mapping the specific residues phosphorylated in the T loops of CDK2-7 and its two parental enzymes, CDK2 and CDK7, when they are activated. We labeled CDK2-7 (Fig. 6B) and CDK2 (Fig. 6C) in vitro with CDK7-cyclin H and labeled CDK7 with activated CDK2-cyclin A (Fig. 6D). We then analyzed the tryptic phosphopeptides generated in each reaction. Phosphorylation of CDK2-7 by CDK7 generated five discrete spots (Fig. 6B); three of the spots comigrated exactly with phosphopeptides from wild-type CDK2 phosphorylated in vitro by CDK7 (analyzed alone for Fig. 6C and mixed with CDK2-7 for Fig. 6E). The most prominent spot in the CDK2 tryptic phosphopeptide map corresponds to the T160-containing peptide derived from the T loop (14; data not shown); a precisely comigrating peptide is also the major labeled fragment of CDK2-7 (compare Fig. 6B, C, and E). Although the two predicted phosphopeptides differ in their amino acid compositions at two positions (Fig. 6A), neither change (Thr-to-Ala at position 1 and Glu-to-Gln at position 5) would be expected to affect mobility in either dimension under the conditions that we used (3).
To account for the other phosphopeptides in the map derived from CDK2-7, we compared them with the peptides derived from CDK7 labeled in vitro by CDK2 (analyzed alone for Fig. 6D and mixed with CDK2-7 for Fig. 6F). One peptide clearly comigrated with the previously identified (Fig. 4) S164-containing phosphopeptide of CDK7 (compare Fig. 6B, D, and F); the predicted tryptic peptides containing this residue are in fact identical in CDK7 and CDK2-7 (Fig. 6A). Of the three remaining spots generated from CDK2-7, two correspond to minor peptides derived from CDK2, as mentioned above. We believe that these represent labeling by CDK7 of nonphysiologic sites; we have previously observed such nonspecific labeling when monomeric, CDK2(T160A) was the substrate (Fisher and Morgan, unpublished observations). We used monomeric CDK2-7 as the substrate in this case to suppress the elevated level of autophosphorylation that occurs when CDK2-7 and cyclin A are mixed in the absence of CAK (Fig. 5B, lane 2). Thus, only one spot cannot be accounted for by comparison with the two parental enzymes. We conclude that CDK7-cyclin H phosphorylates both T160 and S154 within the T loop of the hybrid enzyme, CDK2-7. This argues that T-loop recognition by CDK7 is not principally dependent on the sequence of the T loop but rather on the overall protein context of the CDK in which it is embedded.
CDK2-7 retains the substrate specificity of CDK2.
Our failure to switch the substrate specificity of cyclin-dependent CAKs by swapping their target T loops implies that structural features outside the T loop are the principal determinants of CAK-CDK recognition. The revised model predicts that CDK2-7 would retain the ability to phosphorylate CDK7 but would be unable to phosphorylate CDK2 or to autoactivate. This is indeed the case, as shown by the ability of CDK2-7-cyclin A complexes activated by CDK7-cyclin H to phosphorylate CDK7 efficiently in vitro (Fig. 7A, lane 5). As in our other assays of CDK2-7-associated kinase activity (e.g., as shown in Fig. 5C), full catalytic activity of the hybrid CDK is dependent on both cyclin and an exogenous CAK, ruling out autoactivation by intramolecular phosphorylation. Although we have not specifically tested whether CDK2-7 can activate itself in trans, the hybrid enzyme is unable to phosphorylate wild-type CDK2 (Fig. 7B). Wild-type CDK2 and CDK2-7 differ only in their T-loop sequences (Fig. 5A), and CDK2-7 is able to phosphorylate the CDK7 T loop in its native context (Fig. 7A), so autoactivation by CDK2-7 through intermolecular phosphorylation seems unlikely. Taken together, in fact, our results suggest a general prohibition against autoactivation by CDKs, despite the ability of one CDK to activate another and the high degree of homology among CDK T-loop sequences.
FIG. 7.
CDK2-7 retains the substrate specificity of wild-type CDK2. (A) CDK2 (lanes 1 to 3) or CDK2-7 (lanes 4 to 6) was preincubated with combinations of cyclin A and CDK7-cyclin H indicated above each lane. Lane 7, control lacking any CDK-7-activating kinase. When activated by cyclin A and CAK, both CDK2 (lane 2) and CDK2-7 (lane 5) were capable of phosphorylating HA-tagged CDK7 immobilized on protein A-Sepharose beads. CDK2 demonstrated slight stimulation of its activity towards CDK7 by cyclin A alone (lane 1), while neither CDK2 nor CDK2-7 had any activity towards CDK7 when preincubated with CDK7-cyclin H alone (lanes 3 and 6). The CDK7-HA exhibited no visible autophosphorylation (lane 7), indicating that any signal is due to phosphorylation by CDK2 or CDK2-7. (B) CDK2-7–cyclin A activated by incubation with pure CDK7-cyclin H was unable to phosphorylate pure CDK2-cyclin A (lane 2), even though it was active towards histone H1 (lane 3). CDK2-cyclin A was directly phosphorylated by CDK7-cyclin H (lane 1).
DISCUSSION
A positive feedback loop of CDK activation?
The phosphorylation of CDK7 depends on a CAKAK. A physiologic CAKAK remains to be identified in metazoans; however, in HeLa cell culture, only cells arrested in prometaphase by nocodazole treatment contain detectable amounts of extractable CAKAK (Fig. 3C). This coincides with the peak of CDC2-cyclin B activity (38). Similar results have been reported for Xenopus CDK7; an extract from frog eggs arrested in mitosis with cytostatic factor or an interphase extract supplemented with cyclin A to activate endogenous CDKs was able to support activation and phosphorylation of CDK7 at enhanced rates relative to those for untreated interphase extracts (35). No exogenous sources of cyclin H or MAT1 were available for that study, and so the mechanism of CAK activation remained undetermined. Moreover, while activation in the Xenopus egg extract was dependent on an intact threonine-176 (analogous to T170 of mammalian CDK7), phosphorylation was not appreciably affected by mutation of this residue to alanine (35). We have demonstrated T170 dependence of CDK2-mediated activation of CDK7 (Fig. 3C) (12), as well as direct phosphorylation of T170 in vitro by CDC2 and CDK2 (Fig. 4), which are both active in mitotic extracts (38).
Does a positive feedback loop of reciprocal CDK-activating phosphorylation operate in vivo? CDK7 activates CDC2 in Drosophila (25), so the two kinases must interact directly. Although other kinases, perhaps analogous to fission yeast Csk1 (17, 26), might activate CDK7 in vivo, we have been unable to separate CAKAK activity from CDC2 and CDK2 upon fractionation of HeLa cell extracts by ion exchange or gel filtration chromatography (W. A. Barton and R. P. Fisher, unpublished observations). Our immunodepletion studies, moreover, seem to implicate CDC2 as the major CAKAK in extracts from HeLa cells arrested at mitosis (Fig. 3D and E). Reciprocal activation by CDK7 and CDC2 could contribute to the rapid activation of CDC2 that ensures the all-or-none onset of mitosis. We have not, however, detected dramatic fluctuations in the phosphorylation state of CDK7 as HeLa cells traverse the cell cycle in culture (Fig. 3B). Such a positive feedback loop, if it exists, might therefore involve only a fraction of CDK7 in the cell. Alternatively, increased rates of phosphorylation of the CDK7 T loop at specific points in the cell cycle might be balanced by the opposing action of T-loop phosphatases and thus fail to produce changes in the steady-state distribution of CDK7 phosphoisoforms.
CDKs that phosphorylate other CDKs.
The ability of two prototypic CDKs—mammalian CDC2 and CDK2—to activate CDK7 was unexpected. The sequence encompassing T170 of CDK7, Thr-His-Gln-Val, bears no apparent resemblance to the consensus phosphorylation site recognized by CDC2 and CDK2 (2, 18, 31, 46). The T loop of CDK7 does contain such a consensus sequence, Ser-Pro-Asn-Arg, which is a major site of phosphorylation in vivo (24; Jin and Morgan, unpublished observations) and is also phosphorylated by CDC2 or CDK2 in vitro (this work). Interestingly, CDK7 shows a similar, flexible substrate specificity by phosphorylating the conserved T-loop sequence Thr-His-Glu-Val, found in CDC2 and CDK2; the highly diverged variants Thr-Pro-Val-Val and Thr-Ser-Val-Val, found in the T loops of CDK4 and -6, respectively (1, 29); and the heptad repeat sequence of the RNA polymerase II CTD, Tyr-Ser-Pro-Thr-Ser-Pro-Ser (4, 9, 39, 43, 44). Even more remarkably, CDK7 appears to discriminate between the two serine residues within the CTD repeat, preferring Ser-5 to Ser-2, despite the high similarity of the surrounding residues to each other (36; unpublished observations). Perhaps most surprising is the inability of CDK7 to phosphorylate its own T loop efficiently, given that the sequence encompassing T170 differs only by a Glu-to-Gln change at the +3 position and by largely conservative changes at upstream residues from that of CDK2 (Fig. 1).
We believe that our studies of the CDK2/CDK7 hybrid (CDK2-7) may explain some of the seemingly quixotic substrate preferences of CDC2, CDK2, and CDK7. In fact, both CDK2 and CDK7 appear to be relatively insensitive to the amino acid sequence surrounding the phosphorylation sites of some substrates. Although CDK7 is not an established physiologic substrate of CDK2, CDC2 (and probably CDK2) is an important substrate for CDK7 in vivo (25), and our results suggest that a specific T-loop sequence is not required for CDK7-mediated activation of CDC2 and CDK2. We have not thoroughly investigated just how tolerant CDK7 (or CDK2) might be of divergence within the target T loop, although the ability to activate CDK4 (29) and CDK6 (1) suggests a high degree of flexibility. It will be interesting to test, for example, whether valines (or other hydrophobic residues) in the +3 and/or +4 positions are necessary for T-loop recognition by CDKs (Fig. 1). Clearly, however, no sequence within the T loop is sufficient to ensure phosphorylation.
CDK-mediated T-loop phosphorylation: a novel mode of substrate recognition?
The inability of CDK7 complexes to phosphorylate the CDK7 T loop, despite its close resemblance to other CDK7 targets, is a remarkable example of substrate specificity with possibly important biologic and evolutionary implications. For example, it necessitates a separate kinase to activate the enzyme. This may be a general rule for all CDKs; no CDK yet described is capable of autophosphorylation of its T loop, even though the requirement for T-loop phosphorylation has in at least two instances been circumvented by evolution, either natural (48) or artificial (5). That certain CDKs are capable of phosphorylating the T loops of other CDKs only deepens the mystery of why autoactivation appears to be strictly forbidden.
We attempted to get around this restriction by engineering a CDK2 mutant with the T loop of CDK7. We reasoned that if CDK2 could phosphorylate that T loop in its natural context (i.e., that of CDK7), it might also do so in its transplanted context. Unless the T-loop swap also affected substrate or cyclin-binding specificity, we could reasonably hope for a mutant, CDK2-7, capable of autoactivation. Instead, we generated a kinase that was still efficiently activated by CDK7-mediated T-loop phosphorylation. Thus, the inability of CDK7 to phosphorylate its own T loop must be due to interference by other structural features of the CDK7 complex. Likewise, CDK2 is prevented from phosphorylating its own T loop, even when it contains a perfect CDK2 consensus phosphorylation site, as is the case for CDK2-7.
Activation of CDKs by other CDKs represents an expansion of the substrate repertoire beyond the orthodox, proline-directed sites that they normally recognize. The relaxation of normal substrate recognition rules presumably requires protein-protein interactions involving surfaces remote from the active site of the enzyme or the target site of the substrate. Such interactions are clearly important for other enzyme-substrate interactions involving CDKs and their physiologic substrates (42). Our data suggest that contacts between CDKs and certain substrates might be strong enough and specific enough to override normal rules of substrate preference imposed (presumably) by the architecture of the enzyme active site. Perhaps in support of this notion, a quaternary complex containing CDK7-cyclin H and CDK2-cyclin A has been observed in vitro (40). Also suggestive of fundamentally different modes of substrate recognition is our recent finding that T170 phosphorylation stimulates the CTD kinase activity of the trimeric CDK7-cyclin H-MAT1 complex without affecting its CAK activity (S. Larochelle and R. P. Fisher, unpublished observations).
Can we identify the structural features that either direct or block productive interactions between the active site of one CDK-cyclin complex and the T loop of another? Are there other important, nonconsensus CDK substrates recognized in similar fashion? In the case of the CDK7 T loop, at least, phosphorylation at the nonconsensus site is as robust as phosphorylation directed by an adjacent site matching the consensus. It is interesting to note that the ability of CDK7 family members to activate CDKs is not universal and was either acquired during evolution by metazoan CDK7 and fission yeast Mcs6 or lost by budding yeast Kin28 (16, 20, 32). Mcs6, moreover, is both a general CTD kinase and a species-specific CAK able to activate fission yeast Cdc2 but not mammalian CDC2 or CDK2 (26). Thus, nonconserved determinants must direct Mcs6 to phosphorylate the T loop of Cdc2. We suggest that the ability to expand the substrate repertoire by an alternative mode of recognition could have facilitated the acquisition of CAK function—and the maintenance of specificity for the CTD—by CDK7 and its relatives.
ACKNOWLEDGMENTS
This work was supported by funding from the National Institute of General Medical Sciences (to R.P.F. and D.O.M.). R.P.F. was a Scholar of the Edward Mallinckrodt, Jr. Foundation.
We thank the National Cell Culture Center for growth of HeLa cells used in the early stages of this study. We also thank Karen Lee and Julia Saiz for kind gifts of Csk1 reagents and for much advice and assistance during the course of this work. We are grateful to Alicia Russo and Nikola Pavletich (Memorial Sloan-Kettering Cancer Center) for the kind gifts of CDK2-cyclin A complexes and to Gideon Bollag (Onyx Pharmaceuticals) for CDC2-cyclin B and CDK4-cyclin D. We thank Stéphane Larochelle for critical review of the manuscript and for helpful suggestions during the course of the work. We also thank Jeff Smith for assistance in preparing the manuscript.
REFERENCES
- 1.Aprelikova O, Xiong Y, Liu E T. Both p16 and p21 families of cyclin-dependent kinase (CDK) inhibitors block the phosphorylation of cyclin-dependent kinases by the CDK-activating kinase. J Biol Chem. 1995;270:18195–18197. doi: 10.1074/jbc.270.31.18195. [DOI] [PubMed] [Google Scholar]
- 2.Beaudette K N, Lew J, Wang J H. Substrate specificity characterization of a cdc2-like protein kinase purified from bovine brain. J Biol Chem. 1993;268:20825–20830. [PubMed] [Google Scholar]
- 3.Boyle W J, van der Geer P, Hunter T. Phosphopeptide mapping and phosphoamino acid analysis by two-dimensional separation on thin-layer cellulose plates. Methods Enzymol. 1991;201:110–149. doi: 10.1016/0076-6879(91)01013-r. [DOI] [PubMed] [Google Scholar]
- 4.Corden J L. Tails of RNA polymerase II. Trends Biochem Sci. 1990;15:383–387. doi: 10.1016/0968-0004(90)90236-5. [DOI] [PubMed] [Google Scholar]
- 5.Cross F R, Levine K. Molecular evolution allows bypass of the requirement for activation loop phosphorylation of the Cdc28 cyclin-dependent kinase. Mol Cell Biol. 1998;18:2923–2931. doi: 10.1128/mcb.18.5.2923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Desai D, Gu Y, Morgan D O. Activation of human cyclin-dependent kinases in vitro. Mol Biol Cell. 1992;3:571–582. doi: 10.1091/mbc.3.5.571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Devault A, Martinez A-M, Fesquet D, Labbé J-C, Morin N, Cavadore J-C, Dorée M. MAT1 (‘ménage à trois’), a new RING finger protein subunit stabilizing cyclin H-cdk7 complexes in starfish and Xenopus CAK. EMBO J. 1995;14:5027–5036. doi: 10.1002/j.1460-2075.1995.tb00185.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Espinoza F H E, Farrell A, Nourse J L, Chamberlin H M, Gileadi O, Morgan D O. Cak1 is required for Kin28 phosphorylation and activation in vivo. Mol Cell Biol. 1998;18:6365–6373. doi: 10.1128/mcb.18.11.6365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Feaver W J, Svejstrup J Q, Henry N L, Kornberg R D. Relationship of CDK-activating kinase and RNA polymerase II CTD kinase TFIIH/TFIIK. Cell. 1994;79:1103–1109. doi: 10.1016/0092-8674(94)90040-x. [DOI] [PubMed] [Google Scholar]
- 10.Fesquet D, Labbé J-C, Derancourt J, Capony J-P, Galas S, Girard F, Lorca T, Shuttleworth J, Dorée M, Cavadore J-C. The MO15 gene encodes the catalytic subunit of a protein kinase that activates cdc2 and other cyclin-dependent kinases (CDKs) through phosphorylation of Thr161 and its homologues. EMBO J. 1993;12:3111–3121. doi: 10.1002/j.1460-2075.1993.tb05980.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fisher R P. Reconstitution of mammalian CDK-activating kinase. Methods Enzymol. 1997;283:256–270. doi: 10.1016/s0076-6879(97)83021-2. [DOI] [PubMed] [Google Scholar]
- 12.Fisher R P, Jin P, Chamberlin H M, Morgan D O. Alternative mechanisms of CAK assembly require an assembly factor or an activating kinase. Cell. 1995;83:47–57. doi: 10.1016/0092-8674(95)90233-3. [DOI] [PubMed] [Google Scholar]
- 13.Fisher R P, Morgan D O. A novel cyclin associates with MO15/CDK7 to form the CDK-activating kinase. Cell. 1994;78:713–724. doi: 10.1016/0092-8674(94)90535-5. [DOI] [PubMed] [Google Scholar]
- 14.Gu Y, Rosenblatt J, Morgan D O. Cell cycle regulation of CDK2 activity by phosphorylation of Thr160 and Tyr15. EMBO J. 1992;11:3995–4005. doi: 10.1002/j.1460-2075.1992.tb05493.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hanks S K, Quinn A M. Protein kinase catalytic domain sequence database: identification of conserved features of primary structure and classification of family members. Methods Enzymol. 1991;200:38–62. doi: 10.1016/0076-6879(91)00126-h. [DOI] [PubMed] [Google Scholar]
- 16.Harper J W, Elledge S J. The role of Cdk7 in CAK function, a retro-retrospective. Genes Dev. 1998;12:285–289. doi: 10.1101/gad.12.3.285. [DOI] [PubMed] [Google Scholar]
- 17.Hermand D, Pihlak A, Westerling T, Damagnez V, Vandenhaute J, Cottarel G, Mäkelä T P. Fission yeast Csk1 is a CAK-activating kinase (CAKAK) EMBO J. 1998;17:7230–7238. doi: 10.1093/emboj/17.24.7230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Holmes J K, Solomon M J. A predictive scale for evaluating cyclin-dependent kinase substrates. A comparison of p34cdc2 and p33cdk2. J Biol Chem. 1996;271:25240–25246. doi: 10.1074/jbc.271.41.25240. [DOI] [PubMed] [Google Scholar]
- 19.Jeffrey P D, Russo A A, Polyak K, Gibbs E, Hurwitz J, Massagué J, Pavletich N P. Mechanism of CDK activation revealed by the structure of a cyclinA-CDK2 complex. Nature. 1995;376:313–320. doi: 10.1038/376313a0. [DOI] [PubMed] [Google Scholar]
- 20.Kaldis P. The cdk-activating kinase (CAK): from yeast to mammals. Cell Mol Life Sci. 1999;55:284–296. doi: 10.1007/s000180050290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kimmelman J, Kaldis P, Hengartner C J, Laff G M, Koh S S, Young R A, Solomon M J. Activating phosphorylation of the kin28p subunit of yeast TFIIH by cak1p. Mol Cell Biol. 1999;19:4774–4787. doi: 10.1128/mcb.19.7.4774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kumagai A, Dunphy W G. Control of the Cdc2/cyclin B complex in Xenopus egg extracts arrested at a G2/M checkpoint with DNA synthesis inhibitors. Mol Biol Cell. 1995;6:199–213. doi: 10.1091/mbc.6.2.199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kunkel T A. Rapid and efficient site-directed mutagenesis without phenotypic selection. Proc Natl Acad Sci USA. 1985;82:488–492. doi: 10.1073/pnas.82.2.488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Labbé J-C, Martinez A-M, Fesquet D, Capony J-P, Darbon J-M, Derancourt J, Devault A, Morin N, Cavadore J-C, Dorée M. p40MO15 associates with a p36 subunit and requires both nuclear translocation and Thr176 phosphorylation to generate cdk-activating kinase activity in Xenopus oocytes. EMBO J. 1994;13:5155–5164. doi: 10.1002/j.1460-2075.1994.tb06845.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Larochelle S, Pandur J, Fisher R P, Salz H K, Suter B. Cdk7 is essential for mitosis and for in vivo Cdk-activating kinase activity. Genes Dev. 1998;12:370–381. doi: 10.1101/gad.12.3.370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee K M, Saiz J E, Barton W A, Fisher R P. Cdc2 activation in fission yeast depends on Mcs6 and Csk1, two partially redundant Cdk-activating kinases (CAKs) Curr Biol. 1999;9:441–444. doi: 10.1016/s0960-9822(99)80194-8. [DOI] [PubMed] [Google Scholar]
- 27.Mäkelä T P, Tassan J-P, Nigg E A, Frutiger S, Hughes G J, Weinberg R A. A cyclin associated with the CDK-activating kinase MO15. Nature. 1994;371:254–257. doi: 10.1038/371254a0. [DOI] [PubMed] [Google Scholar]
- 28.Martinez A-M, Afshar M, Martin F, Cavadore J-C, Labbé J-C, Dorée M. Dual phosphorylation of the T-loop in cdk7: its role in controlling cyclin H binding and CAK activity. EMBO J. 1997;16:343–354. doi: 10.1093/emboj/16.2.343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Matsuoka M, Kato J, Fisher R P, Morgan D O, Sherr C J. Activation of cyclin-dependent kinase-4 (CDK4) by mouse MO15-associated kinase. Mol Cell Biol. 1994;14:7265–7275. doi: 10.1128/mcb.14.11.7265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Molz L, Beach D. Characterization of the fission yeast mcs2 cyclin and its associated protein kinase activity. EMBO J. 1993;12:1723–1732. doi: 10.1002/j.1460-2075.1993.tb05817.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Moreno S, Nurse P. Substrates for p34cdc2: in vivo veritas? Cell. 1990;61:549–551. doi: 10.1016/0092-8674(90)90463-o. [DOI] [PubMed] [Google Scholar]
- 32.Morgan D O. Cyclin-dependent kinases: engines, clocks and microprocessors. Annu Rev Cell Dev Biol. 1997;13:261–291. doi: 10.1146/annurev.cellbio.13.1.261. [DOI] [PubMed] [Google Scholar]
- 33.O'Reilly D R, Miller L K, Luckow V A. Baculovirus expression vectors: a laboratory manual. W. H. New York, N.Y: Freeman and Co.; 1993. [Google Scholar]
- 34.Poon R Y C, Yamashita K, Adamczewski J P, Hunt T, Shuttleworth J. The cdc2-related protein p40MO15 is the catalytic subunit of a protein kinase that can activate p33cdk2 and p34cdc2. EMBO J. 1993;12:3123–3132. doi: 10.1002/j.1460-2075.1993.tb05981.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Poon R Y C, Yamashita K, Howell M, Ershler M A, Belyavsky A, Hunt T. Cell cycle regulation of the p34cdc2/p33cdk2-activating kinase p40MO15. J Cell Sci. 1994;107:2789–2799. doi: 10.1242/jcs.107.10.2789. [DOI] [PubMed] [Google Scholar]
- 36.Rickert P, Corden J L, Lees E. Cyclin C/CDK8 and cyclin H/CDK7/p36 are biochemically distinct CTD kinases. Oncogene. 1999;18:1093–1102. doi: 10.1038/sj.onc.1202399. [DOI] [PubMed] [Google Scholar]
- 37.Rosenblatt J, De Bondt H, Jancarik J, Morgan D O, Kim S-H. Purification and crystallization of human cyclin-dependent kinase 2. J Mol Biol. 1993;230:1317–1319. doi: 10.1006/jmbi.1993.1248. [DOI] [PubMed] [Google Scholar]
- 38.Rosenblatt J, Gu Y, Morgan D O. Human cyclin-dependent kinase 2 (CDK2) is activated during the S and G2 phases of the cell cycle and associates with cyclin A. Proc Natl Acad Sci USA. 1992;89:2824–2828. doi: 10.1073/pnas.89.7.2824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Roy R, Adamczewski J P, Seroz T, Vermuelen W, Tassan J-P, Schaeffer L, Nigg E A, Hoeijmakers J H J, Egly J-M. The MO15 cell cycle kinase is associated with the TFIIH transcription-DNA repair factor. Cell. 1994;79:1093–1101. doi: 10.1016/0092-8674(94)90039-6. [DOI] [PubMed] [Google Scholar]
- 40.Russo A A. Purification and reconstitution of cyclin-dependent kinase 2 in four states of activity. Methods Enzymol. 1997;283:3–12. doi: 10.1016/s0076-6879(97)83003-0. [DOI] [PubMed] [Google Scholar]
- 41.Russo A A, Jeffrey P D, Pavletich N P. Structural basis of cyclin-dependent kinase activation by phosphorylation. Nat Struct Biol. 1996;3:696–700. doi: 10.1038/nsb0896-696. [DOI] [PubMed] [Google Scholar]
- 42.Schulman B A, Lindstrom D L, Harlow E. Substrate recruitment to cyclin-dependent kinase 2 by a multipurpose docking site on cyclin A. Proc Natl Acad Sci USA. 1998;95:10453–10458. doi: 10.1073/pnas.95.18.10453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Serizawa H, Mäkelä T P, Conaway J W, Conaway R C, Weinberg R A, Young R A. Association of Cdk-activating kinase subunits with transcription factor TFIIH. Nature. 1995;374:280–282. doi: 10.1038/374280a0. [DOI] [PubMed] [Google Scholar]
- 44.Shiekhattar R, Mermelstein F, Fisher R P, Drapkin R, Dynlacht B, Wessling H C, Morgan D O, Reinberg D. Cdk-activating kinase (CAK) complex is a component of human transcription factor IIH. Nature. 1995;374:283–287. doi: 10.1038/374283a0. [DOI] [PubMed] [Google Scholar]
- 45.Solomon M J, Harper J W, Shuttleworth J. CAK, the p34cdc2 activating kinase, contains a protein identical or closely related to p40MO15. EMBO J. 1993;12:3133–3142. doi: 10.1002/j.1460-2075.1993.tb05982.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Songyang Z, Blechner S, Hoagland N, Hoekstra M F, Piwnica-Worms H, Cantley L C. Use of an oriented peptide library to determine the optimal substrates of protein kinases. Curr Biol. 1994;4:973–982. doi: 10.1016/s0960-9822(00)00221-9. [DOI] [PubMed] [Google Scholar]
- 47.Tassan J-P, Jaquenod M, Fry A M, Frutiger S, Hughes G, Nigg E A. In vitro assembly of a functional human cdk7/cyclin H complex requires MAT1, a novel 36 kD RING finger protein. EMBO J. 1995;14:5608–5617. doi: 10.1002/j.1460-2075.1995.tb00248.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tassan J-P, Jaquenoud M, Léopold P, Schultz S J, Nigg E A. Identification of human cyclin-dependent kinase 8, a putative protein kinase partner for cyclin C. Proc Natl Acad Sci USA. 1995;92:8871–8875. doi: 10.1073/pnas.92.19.8871. [DOI] [PMC free article] [PubMed] [Google Scholar]