Abstract
Hyperthermophiles, typically defined as organisms with growth optima ≥80°C, are dominated by the Archaea. Proteins that support life at the extremes of temperatures often retain substantial biotechnological and commercial value, but the recombinant expression of individual hyperthermophilic proteins is commonly complicated in non-native mesophilic hosts due to differences in codon bias, intracellular solutes and the requirement for accessory factors that aid in folding or deposition of metal centers within archaeal proteins. The development of versatile protein expression and facilitated protein purification systems in the model, genetically tractable, hyperthermophilic marine archaeon Thermococcus kodakarensis provides an attractive platform for protein expression within the hyperthermophiles. The assortment of T. kodakarensis genetic backgrounds and compatible selection markers allow iterative genetic manipulations that facilitate protein overexpression and expedite protein purifications. Expression vectors that stably replicate both in T. kodakarensis and Escherichia coli have been validated and permit high-level ectopic gene expression from a variety of controlled and constitutive promoters. Biologically relevant protein associations can be maintained during protein purifications to identify native protein partnerships and define protein interaction networks. T. kodakarensis thus provides a versatile platform for the expression and purification of thermostable proteins.
1. Introduction
Well-regulated and robust bacterial- and fungal-based recombinant gene expression systems are commonly employed to overproduce and, ultimately, purify proteins in large quantities for biochemical and structural characterization (Kim, Yoo, & Kang, 2015; Rosano & Ceccarelli, 2014). In choosing a recombinant expression system, parameters such as cost, yield and ease of purification are considered. Additional parameters, including the assembly of metal-cluster binding sites, proper post-translational modifications, introduction of disulfide-linkages, protein secretion, post-translation N- and C-terminal processing, intein excision, the availability of specific chaperones, unique genetic codes, insertion into membranes of specific compositions and the availability of essential cofactors limit the choice of production host or demand overexpression in the native host (Duong-Ly & Gabelli, 2014; Peleg & Unger, 2012). Protein expression systems have been widely established and commercialized in several mesophilic species, including Escherichia coli, yeasts and insect cells (with baculoviruses). While these systems often permit production of cross-species, and occasionally cross-Domain recombinant protein expression, there remain instances wherein the canonical protein expression platforms fail to retain the necessary parameters for optimal protein expression. An increasing class of such proteins originate in the Archaea, wherein proteins often meant to survive the extremes of pressures, temperature, salinity and pH have unique expression and folding requirements that are not easily replicated outside of the archaeal Domain.
The dearth of genetic systems for most archaeal lineages limits controlled protein overexpression in most archaeal clades, but a few systems have been established that permit the controlled overexpression of proteins in archaeal cell lysates or whole cells (Santangelo, Cubonová, & Reeve, 2008a; Takemasa, Yokooji, Yamatsu, Atomi, & Imanaka, 2011). Archaeal-derived in vitro translation systems, often-linked to in vitro transcription systems, provide a route to produce archaeal proteins in minute quantities but are impractical for moderate- or large-scale protein production. Many efforts have thus focused on the few genetically tractable clades of Archaea to develop techniques to aid in protein overexpression and to facilitate rapid and efficient purification of the desired protein products (Farkas, Picking, & Santangelo, 2013). Controlled and inducible expression systems have long been established in halophilic (salt-loving) archaeal species (see Haque, Paradisi, & Allers, 2020; Martínez-Espinosa, 2020) but the near-saturating intracellular salinities often impair protein folding of non-halophilic proteins and impede downstream protein processing in almost all cases. Several methanogenic archaeal lineages have been manipulated to provide controlled and inducible protein expression platforms (Guss, Rother, Zhang, Kulkarni, & Metcalf, 2008), but none have been developed at scale, as the growth rates and complicated growth conditions of many methanogens limit the scale of culture growth. The remaining archaeal cell-based expression systems are found in species that grow at high (>60 °C, thermophilic) or very high (>80 °C, hyperthermophilic) temperatures. The thermophilic protein-expression systems established for the Sulfolobales (see Schocke, Bräsen, & Siebers, 2019) and the hyperthermophilic protein expression systems established for the Thermococcales (Catchpole, Gorlas, Oberto, & Forterre, 2018; Farkas, Chung, DeBarry, Adams, & Westpheling, 2011; Santangelo, Cubonová, & Reeve, 2008a; Speed, Burkhart, Picking, & Santangelo, 2018; Walker & Santangelo, 2015) provide unique platforms for expression of thermostable proteins with a broad range of applications.
Hyperthermophilic archaea have significant potential as host systems to invigorate new biotechnological innovations and conversion chemistries (Adams & Kelly, 1998; Counts et al., 2017; Crosby et al., 2019; Straub et al., 2018; Zeldes et al., 2015). Protein expression in hyperthermophilic archaeal systems motivates both basic and industrial efforts for greener chemistries, particularly toward the production of enzymes that survive the high temperatures common to many manufacturing processes. Microbial contamination is reduced or effectively eliminated by carrying out enzymatic conversions at high temperatures, and the increased half-life of thermotolerant or thermostable enzymes often reduces total enzyme requirements and costs (Kumar, Dangi, Shukla, Baishya, & Khare, 2019; Suresh et al., 2021). Thermostable enzymes, particularly amylases and xylanases, are employed in bulk in biomass degradation and processing (Kumar et al., 2019) as well as in pulp and fiber productions (Bhardwaj, Kumar, & Verma, 2019) and starch liquefaction processes (de Souza & de Oliveira Magalhães, 2010). Thermostable proteases are in high-demand for bulk addition to laundry detergents, for leather processing, as rennins for cheese-making, as meattenderizers’ and for baking and brewing platforms (Razzaq et al., 2019). Thermostable lipases are employed in high-temperature, low-water-content solvent systems for hydrolysis activities, transesterification and ester synthesis (Kumar, Dhar, Kanwar, & Arora, 2016). Thermostable enzymes are more diversely used in smaller scale molecular studies, including PCR, next-generation sequencing techniques and manipulations of nucleic acids. Finally, many thermostable proteins are stably folded and display reduced long-range intramolecular dynamics at ambient and/or mesophilic conditions, properties that often aid or inform structural or biophysical studies.
Thermococcus kodakarensis has emerged as a common model species for the study of biological processes at high temperatures and as an expression platform for thermostable enzymes (Atomi & Reeve, 2019). T. kodakarensis is an ecologically pervasive, fast-growing, anaerobic, heterotroph first isolated from a solfatara on Kodakara island, Japan (Atomi, Fukui, Kanai, Morikawa, & Imanaka, 2004; Morikawa, Izawa, Rashid, Hoaki, & Imanaka, 1994) that can be grown to high cell densities with economical media requirements. The development of highly reproducible, accurate genomic and ectopic manipulation strategies for T. kodakarensis permits rational and iterative strain construction (Santangelo, Cubonová, & Reeve, 2010; Sato, Fukui, Atomi, & Imanaka, 2003, 2005), controlled protein overexpression (Santangelo, Cubonová, & Reeve, 2008a; Takemasa et al., 2011), and the introduction of protein tags or epitopes (Burkhart, Febvre, & Santangelo, 2019; Li et al., 2011; Li, Santangelo, Cubonová, Reeve, & Kelman, 2010) that aid in rapid and cost-effective protein purifications. T. kodakarensis thus provides a readily available and versatile platform for the expression of thermostable proteins for functional studies (Farkas et al., 2013) and a relatively untapped source of thermostable enzymes. Here, we outline current methodologies for expression and purification of native and recombinant proteins which have been developed for T. kodakarensis. These procedures take advantage of the natural competency of T. kodakarensis cells, reliable culturing techniques and commercialized approaches for affinity chromatography. We first introduce the general procedures that permit genetic manipulation of T. kodakarensis, then detail genomic-, then plasmid-based protein expression and purification systems.
2. Genetic systems for Thermococcus kodakarensis
The Thermococcales family is distributed globally and represents a well-studied branch within the Euryarchaeota that includes a growing list of species divided among three genera: the Pyrococcus, Paleococcus and Thermococcus (Schut et al., 2014). T. kodakarensis was initially classified as Pyrococcus kodakaraensis strain KOD1 but was reclassified as T. kodakarensis strain KOD1 after genome re-sequencing and phylogenetic analysis of the 16S ribosomal RNA (note that the original species designation was also changed from kodakaraensis to kodakarensis) (Fukui et al., 2005). T. kodakarensis is naturally competent and, through homologous recombination, readily integrates exogenous DNA into its genome (Čuboňováa et al., 2012; Sato et al., 2003). The natural competency of T. kodakarensis contributes to its utility as a platform for protein expression. Transformation procedures are not arduous and clonal populations can be selected and maintained on a variety of media using a range of selectable markers (Farkas et al., 2013). Specialized strains (Table 1) permit selections based on the restoration of prototrophy (Fukuda, Morimoto, Imanaka, & Fujiwara, 2008; Santangelo et al., 2010; Sato et al., 2003, 2005) and resistance to toxic nucleotide analogs (Santangelo et al., 2010; Sato et al., 2003) (Table 2). Complementary counter-selective pressures permit unlimited reuse of selective markers and iterative strain constructions, inclusive of both genomic modifications and plasmid retention (Hileman & Santangelo, 2012; Sato et al., 2003). While T. kodakarensis is not known to harbor any naturally occurring plasmids, cryptic mini plasmids from related members of the Thermococcales have been modified to stably replicate in T. kodakarensis (Catchpole et al., 2018; Santangelo, Cubonová, & Reeve, 2008a). The development of genetic techniques has led to the emergence of T. kodakarensis as a versatile host for protein expression.
Table 1.
T. kodakarensis strains and compatible selection strategies.
| Strain | Genotype | Medium requirements | Available selection strategies | Reference(s) |
|---|---|---|---|---|
| KOD1 | Wild type | Rich or minimal medium | HMG-CoA reductase and citrulline | Morikawa et al. (1994) and Fukui et al. (2005) |
| KU216 | ΔpyrF | Rich or minimal medium with uracil | HMG-CoA reductase and citrulline. Transformation with DNA encoding pyrF confers uracil prototrophy | Sato et al. (2005) |
| KUW1 | ΔpyrF; ΔtrpE | Minimal medium with uracil and tryptophan | HMG-CoA reductase and citrulline. Transformation with DNA encoding pyrF or trpE confers uracil or tryptophan prototrophy, respectively | Sato et al. (2005) |
| KUWH1 | ΔpyrF; ΔtrpE; ΔhisD | Minimal medium with uracil, tryptophan and histidine | HMG-CoA reductase and citrulline. Transformation with DNA encoding pyrF, trpE, or hisD confers uracil, tryptophan, or histidine prototrophy, respectively | Sato et al. (2005) |
| KW128 | ΔpyrF; ΔtrpE::pyrF | Rich medium or minimal medium with tryptophan | HMG-CoA reductase and citrulline. Compatible with MAE using tryptophan (selection) and 5-FOA (counter-selection) | Sato et al. (2005) |
| TS517 | ΔpyrF; ΔtrpE::pyrF; ΔHPRT | Rich medium or minimal medium with tryptophan | HMG-CoA reductase and citrulline. Compatible with MAE using tryptophan (selection) and 6MP (counter-selection) | Santangelo et al. (2010) |
| TS559 | ΔpyrF; ΔtrpE::pyrF; ΔpdaD; ΔHPRT | Rich medium with agmatine or minimal medium with tryptophan and agmatine | HMG-CoA reductase and citrulline. Compatible with MAE using agmatine (selection) and 6MP (counter-selection) | Santangelo et al. (2010) |
| TS900 | ΔpyrF; ΔtrpE; ΔpdaD; ΔHPRT | Rich medium with agmatine or minimal medium with tryptophan and agmatine | HMG-CoA reductase and citrulline. Compatible with MAE using (i) agmatine (selection) and 6MP (counter-selection); (ii) uracil (selection) and 5-FOA (counter-selection) | Unpublished |
Table 2.
Selectable markers for T. kodakarensis.
| Selection | Genetic background |
Selection requirements |
Compatible strains |
Reference(s) |
|---|---|---|---|---|
| Tryptophan | ΔtrpE (ΔTK0254) | Minimal medium lacking tryptophan | KUW1, KUWH1, KW128 | Sato et al. (2003) and Sato et al. (2005) |
| Simvastatin, mevinolin | None | Requires overexpression of HMG-CoA reductase (PF1848) | Any strain | Matsumi, Manabe, Fukui, Atomi, and Imanaka (2007) and Santangelo et al. (2008b) |
| Agmatine | ΔpdaD (ΔTK0149) | Minimal or rich medium lacking agmatine supplementation | TS559, TS900 | Fukuda et al. (2008) and Santangelo et al. (2008b) |
| Histidine | ΔhisD (ΔTK0244) | Minimal medium lacking histidine | KUWH1 | Sato et al. (2005) |
| 6-Methyl purine (6MP) | ΔHPRT (ΔTK0664) | Minimal medium containing 6MP | TS517, TS559, TS900 | Santangelo et al. (2010) |
| Uracil, 5-fluorooratic acid (5-FOA) | ΔpyrF (ΔTK2276) | Minimal medium lacking uracil and supplemented with 5-FOA, respectively | KU216, KW128, TS517, TS559, TS900 | Sato et al. (2003) |
| Citrulline, aspartate | + argG (PF0207), + argH (PF0208) | Minimal medium lacking citrulline and aspartate | Any strain | Atomi et al. (2004) |
2.1. Genetic backgrounds and selectable markers
The choice of selection strategy and host genotype dictate initial plans for protein expression and purification within T. kodakarensis. When using T. kodakarensis strains with WT or near WT genotypes, only two validated selection markers exist that do not require specialized genotypic backgrounds. The first is based on hydroxymethylglutaryl coenzyme A (HMG-CoA) reductase activity that is essential for archaeal isoprenoid-based lipid biosynthesis (Cabrera, Bolds, Shields, Havel, & Watson, 1986). HMG-CoA is inhibited with simvastatin or mevinolin (a simvastatin analog) (Matsumi et al., 2007) and overexpression of the P. furiosus HMG-CoA reductase via a constitutive promoter provides T. kodakarensis strains resistance to x~8 μM simvastatin (Matsumi et al., 2007) and >30μM mevinolin Santangelo et al. (2008b). The second universal selective pressure is based on amino acid auxotrophies. Unlike many other Thermococcus species, T. kodakarensis lacks an arginine biosynthetic pathway and requires arginine supplementation for growth (Atomi et al., 2004; Fukuda et al., 2008). The P. furiosus genes PF0207 and PF0208 encode argininosuccinate synthase and argininosuccinate lyase, respectively, which together catalyze the synthesis of arginine from citrulline or aspartate (Farkas et al., 2013; Santangelo & Reeve, 2010). T. kodakarensis strains that incorporate and express donor DNA carrying both PF0207 and PF0208 can grow with citrulline or aspartate in the absence of arginine in minimal medium (Santangelo & Reeve, 2010). These selection markers are compatible with all known strains; however, counter selection strategies independent of genotypic background have not yet been developed.
It is far more common to direct protein expression in one of several strains (Table 1) that have been constructed to allow auxotrophic-prototrophic selection of genomic integration of donor DNA by homologous recombination or retention of autonomously replicating plasmids (Catchpole et al., 2018; Gehring, Sanders, & Santangelo, 2017; Santangelo, Cubonová, & Reeve, 2008a). Auxotrophic strains that incorporate donor DNA restoring prototrophy provide positive selection strategies. Counter selection strategies lead to the excision or loss of donor DNA, as the activity of counter selectable markers result in death of all cells still containing the counterselectable marker. KU216 lacks the orotidine-5′-phosphate decarboxylase (pyrF) gene and displays uracil auxotrophy in minimal medium (Sato et al., 2005). KU216 also confers resistance to 5-fluoroorotic acid (5-FOA), allowing counter selection for cells that have excised pyrF encoded donor DNA. Building from KU216, stains KUW1 and KUWH1 additionally lack anthranilate synthase (TK0254), encoded by the trpE locus, conferring tryptophan auxotrophy (Sato et al., 2005). KUWH1 further lacks hisD, providing histidine auxotrophy. Strain KW128 lacks pyrF and the trpE locus is replaced with pyrF, leading to tryptophan auxotrophy (Sato et al., 2005). The absence of the amino acid corresponding to the induced auxotrophy provides a positive selection strategy based on the restoration of prototrophies provided in the donor DNA.
Multiple strains derived from KU216 were constructed to allow for sequential selection and counter selection of genomically integrated DNA. Such strains are useful for genetic manipulations via markerless allelic exchange (MAE) and are the backbone of genetic techniques in T. kodakarensis. Strain TS517 is genotypically similar to KW128 but with an additional deletion of hypoxanthine/guanine phosphoribosyltransferase (TK0664), a purine scavenging protein (Santangelo et al., 2010). Strain TS559 additionally lacks the pyruvoyl-dependent arginine decarboxylase (TK0149) which leads to agmatine auxotrophy. Integration of donor DNA encoding TK0149 results in agmatine prototrophy. Similar to TS517, ejection of donor DNA carrying TK0664 is selected for by isolating mutants resistant to 6-methylpurine (6MP) (Gehring, Sanders, & Santangelo, 2017). Lastly, strain TS900 was derived from TS559 and is cleanly deleted for pyrF (unpublished). Selection and counter selection of transformants in TS900 are identical to that of TS559. The strains described here and their compatible selection strategies allow targeted mutagenesis of chromosomal DNA.
2.2. Targeted mutagenesis of chromosomal DNA
T. kodakaraensis is naturally competent for DNA uptake and incorporates donor DNA into its genome by homologous recombination (Sato et al., 2003, 2005), allowing targeted chromosomal mutations for increased protein expression (Takemasa et al., 2011), placement of extracellular secretion signals (Takemasa et al., 2011), gene deletions (Gehring et al., 2017; Ishino et al., 2011; Santangelo, Cuboňová, & Reeve, 2011), epitope- or affinity-purification (Burkhart et al., 2019; Li et al., 2010), or allelic substitutions (Sanders et al., 2020). Genome manipulations are currently possible using MAE where transformation with plasmid DNAs that carry a positive selection marker (i.e.: TK0254 (trpE), TK2276 (pyrF) or TK0149 (pdaD)) are integrated into the genome resulting in tryptophan, uracil and agmatine prototrophic transformants, respectively (Fig. 1) (Gehring, Sanders, & Santangelo, 2017; Santangelo et al., 2010; Sato et al., 2005). After selection and confirmation of the intermediate genotype, growth of confirmed intermediate-genome containing cells in the presence of tryptophan, uracil, or agmatine permits a small percentage of cells to spontaneously remove the integrated plasmid sequences via homologous recombination (Sato et al., 2003). These unique cells containing the desired final genotypes are now able to survive in the presence of 5-FOA or 6MP due to the loss of the counterselectable marker (Santangelo et al. (2008b); Sato et al., 2003). MAE, sometimes referred to as pop-in/pop-out, permits a tailored approach to expression and purification of proteins in T. kodakarensis.
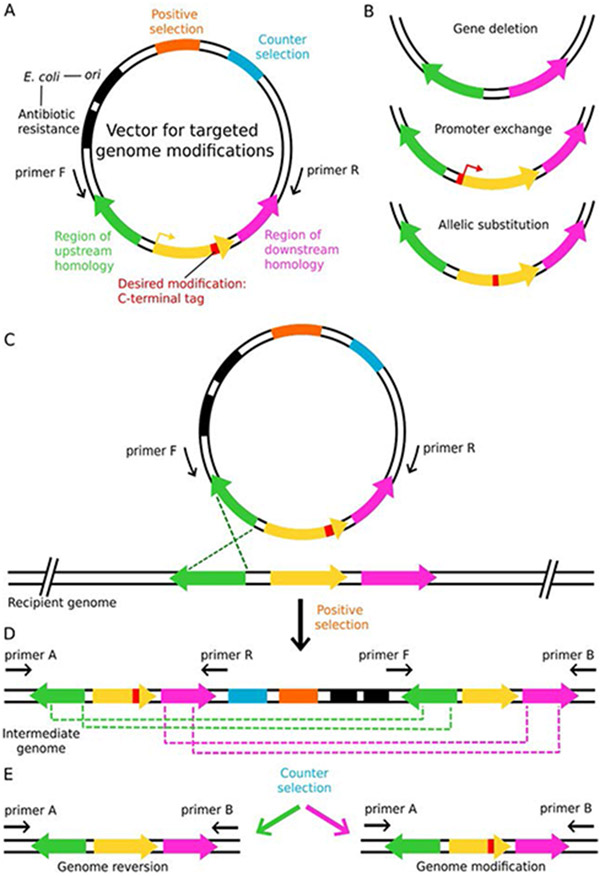
Fig. 1.
Sequential homologous recombination events permit modifications of chromosomal DNA. A vector designed for genomic integration (A) includes replication and selection elements for E. coli (black) but only selection (orange) and counter selection (blue) elements for T. kodakarensis. Regions of upstream (green) and downstream (pink) homology to the target locus sequences (yellow, with red modification) permit the vector to integrate adjacent to the target locus. Possible genomic modifications including (B) gene deletion(s), promoter exchange and allelic substitutions can be generated using identical pop-in/pop-out techniques. Genomic integration is mediated by either the region of upstream or downstream homology (C), and the first recombination event (pop-in) is selected for using a positive selection marker (D). In this example, the region with upstream homology (green) is the recombination point. The intermediate genome is verified using primers pairs that have homology to the genome and the plasmid separately (primers A/F and primers R/B). The second recombination event, wherein the plasmid is excised from the genome (pop-out), is selected for using a counter selectable marker (E); there is an equal probability of recombination at the green or pink loci. In this example, recombination between the green loci yields the parent genome while recombination between the pink loci results in the desired modification. Depending on the modification, the final genome can be verified using PCR and amplicon or whole genome sequencing.
Targeted chromosomal alterations are achieved by transforming a vector containing the desired modification flanked by several hundred bases (i.e.: >500bp) of up- and downstream homology (Fig. 1A) (Gehring, Sanders, & Santangelo, 2017). Routine modifications including N- or C-terminal epitope- or affinity-tag additions, promoter exchange, gene deletions and allelic substitutions (Fig. 1A and B) are encoded on a vector that has both a selectable and counter selectable marker compatible with MAE in T. kodakarensis (Fig. 1A and Table 2). Components for autonomous replication and selection in E. coli are included; however, no T. kodakarensis origin of replication (ori) is necessary, as the plasmid is expected to integrate into the genome. After vector transformation, either the up- or downstream regions homologous to the target loci will integrate into the genome (Fig. 1C). Prototrophic selection of plasmid integration (pop-in) via homologous recombination is mediated by the positive selectable marker (Table 2). The intermediate genome is confirmed via PCR using two primer sets (primers A/R and primers F/B, Fig. 1D). Primers A and B are external to the site of plasmid integration and are not homologous to any region on the integrative vector, while primers F and R have homology to the plasmid. Spontaneous recombination of the plasmid out of the genome (pop-out) is selected for using a counter selectable marker (Fig. 1E and Table 2) and has a ~50% chance of reverting to the parent genome (Fig. 1E, green) or including the modification (Fig. 1E, pink). Confirmation of the final genotype can be achieved through PCR analysis and/or Sanger sequencing of the target loci (Hileman & Santangelo, 2012). Whole genome sequencing should be considered as off target recombination events are possible. While modification of promoter or expression sequences (i.e.: Shine-Delgarno sequences) may alter protein expression, many changes to chromosomal coding sequences are specifically designed to retain native protein expression. Ectopic shuttle vectors which stably replicate in T. kodakarensis and E. coli are often more practical for high level protein expression.
2.3. Autonomously replicating plasmids
Plasmids that autonomously replicate and express selectable markers in both T. kodakaraensis and E. coli can be easily manipulated and confirmed in E. coli, then transferred to T. kodakaraensis where manipulation of plasmids is more complicated. Shuttle vectors, when modified to encode expression cassettes under the control of a variety of constitutive promoters (Table 3 and Fig. 2, purple), provide a mechanism for high levels of ectopic expression. The cell surface glycoprotein (csg, TK0895) is a highly expressed gene in T. kodakarensis, and therefore the Pcsg promoter, among others, can direct high level expression when inserted into the multiple cloning site (Fig. 2, green) (Kanai et al., 2015). Where an exogenous promoter is preferred, the Phmtb promoter from Methanothermobacter thermautotrophicus is compatible with T. kodakarensis expression machinery Santangelo et al. (2008b). In the related species, Pyrococcus furiosus, some level of inducible expression can be achieved from the PcipA promoter (Basen et al., 2012). With the exception of a riboswitch-mediated expression system, no single-gene controlled expression system currently exists in T. kodakarensis (Speed et al., 2018). The endogenous Pfbpase promoter can be activated by the addition of elemental sulfur (S) but like the PcipA promoter, such large scale changes to growth conditions alter the expression of many genes. Select changes to the ribosome binding site (RBS, 5′-AGGTGA) can act as another point to control protein expression levels (Santangelo, Cubonová, & Reeve, 2008a).
Table 3.
Promoters compatible with gene expression in T. kodakarensis.
| Promoter | Gene ID | Origin | Description | Reference(s) |
|---|---|---|---|---|
| Phmtb | TM0254 | Methanothermobacter thermautotrophicus | Constitutive; histone B | Santangelo, Cubonová, James, and Reeve (2007) |
| Pcap | TK2279 | Thermococcus kodakarensis | Constitutive; CDP-alcohol phosphatidyltransferase | Rodrigues et al. (2007) |
| Pgdh | TK1431 | Thermococcus kodakarensis | Constitutive; glutamate dehydrogenase | Santangelo et al. (2008b) |
| Pcsg | TK0895 | Thermococcus kodakarensis | Constitutive; cell surface glycoprotein | Takemasa et al. (2011) |
| Pfbpase | TK2164 | Thermococcus kodakarensis | Inducible; fructose-1,6-bisphosphatase. Involvement in central metabolism hinders tight regulation | Sato et al. (2004) |
| PcipA | PF0190 | Pyrococcus furiosus | Inducible; membrane-bound glycoprotein, cold-induced protein A. Induction via heat/cold shock or other stress has significant biological effects | Basen, Sun, and Adams (2012) |
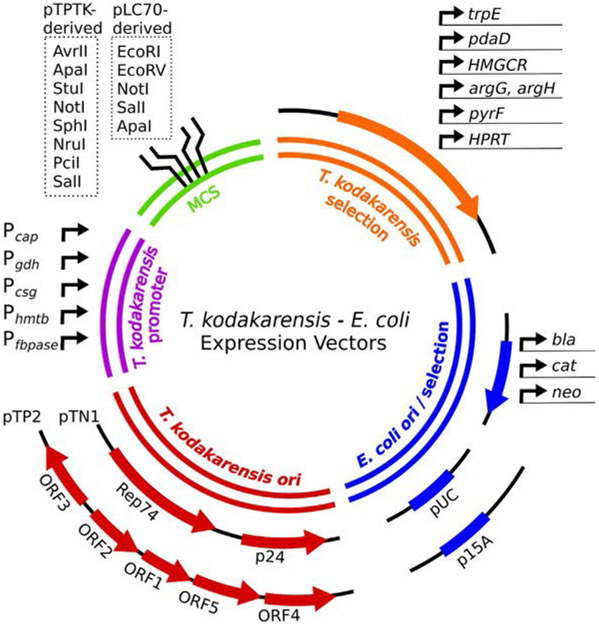
Fig. 2.
Autonomously replicating expression vectors permit stable replication and gene expression in T. kodakarensis and E. coli. Constitutive or inducible promoters (purple) drive high levels of ectopic gene expression of sequences cloned into the multiple cloning site (MCS, green). Selections in T. kodakarensis (orange) rely on restorations of prototrophies (agmatine, tryptophan, arginine, uracil), statin resistance (mevinolin, simvastatin) or resistance to toxic nucleoside analogs (6MP, 5-FOA). Expression vectors retain either a pUC or p15A origin (ori) and an antibiotic selection cassette(s) for replication in E. coli (blue): bla (ampicillin resistance); cat (chloramphenicol resistance); neo (kanamycin resistance). The entire pTN1 or pTP2 plasmid sequences (red) permit autonomous replication in T. kodakarensis.
No naturally occurring plasmids exist in T. kodakarensis (Krupovic, Gonnet, Hania, Forterre, & Erauso, 2013). All T. kodakarensis–E. coli shuttle vectors (Table 4) rely on one of two plasmids isolated from close relatives in the Thermococcales for replication (Fig. 2, red). The cryptic mini plasmid pTN1 isolated from T. nautilus encodes a rolling-circle replication initiator protein (Rep74) and a p24 protein, although p24 is not essential (Soler et al., 2007). The fusion of pTN1 to E. coli vectors provide the minimal replicative sequences for both species and selectable markers for E. coli, but not T. kodakarensis (Santangelo, Cubonová, & Reeve, 2008a). Analogous to pTN1, pTP2 from Thermococcus prieurii contains five putative open reading frames, one of which encodes a rolling circle replication gene that supports plasmid replication in T. kodakarensis (Catchpole et al., 2018). Both pTN1- and pTP2-based plasmids can be maintained within the same cell, providing two distinct and simultaneous means of selection. T. kodakarensis is polyploid, retaining ~7–19 genomes (Spaans, van der Oost, & Kengen, 2015). pTN1-based plasmids have been measured to exist at a copy number of ~3 copies per chromosome (Santangelo, Cubonová, & Reeve, 2008a) while pTP2-based plasmids have been observed at ~50 copies per cell (Gorlas, Krupovic, Forterre, & Geslin, 2013) (Table 4). Stable replication of plasmids in E. coli is dependent on a pUC or p15A origin (Fig. 2, blue), with a copy number of ~500–700 and ~20–30 per cell, respectively (Table 4) (Catchpole et al., 2018; Del Solar, Giraldo, Ruiz-Echevarría, Espinosa, & Díaz-Orejas, 1998; Lin-Chao, Chen, & Wong, 1992; Santangelo, Cubonová, & Reeve, 2008a).
Table 4.
T. kodakarensis–E. coli shuttle vectors.
| Plasmid | Size (bp) |
T. kodakarensis copy number |
E. coli copy number |
T. kodakarensis selection(s) |
E. coli selection(s) | Description | Genetic background |
Reference |
|---|---|---|---|---|---|---|---|---|
| pLC64 | 9034 | ~3/chromosome | ~500–700/cell | Tryptophan | Ampicillin, kanamycin | pTN1 + pCR2.1-TOPO::Pcap-TrpE | ΔtrpE (ΔTK0254) | Santangelo et al. (2008b) |
| pLC70 | 10495 | ~3/chromosome | ~500–700/cell | Statins, tryptophan | Ampicillin, kanamycin | pTN1 + pCR2.1-TOPO::Pcap-TrpE; Pgdh-HMG-CoA | ΔtrpE (ΔTK0254) | Santangelo et al. (2008b) |
| pLC71 | 9190 | ~3/chromosome | ~500–700/cell | Statins, tryptophan | Ampicillin, kanamycin | pTN1::Δp24 + pCR2.1-TOPO::Pcap-TrpE; Pgdh-HMG-CoA | ΔtrpE (ΔTK0254) | Santangelo et al. (2008b) |
| pTS543 | 8196 | ~3/chromosome | ~500–700/cell | Agmatine | Ampicillin, kanamycin | pTN1 + pCR2.1-TOPO::PhmtB-PdaD | ΔpdaD (ΔTK0149) | Santangelo et al. (2010) |
| pTPTK1 | 5489 | ~50/cell | ~20–30/cell | Statins | Chloramphenicol | pTP2 + pBAD33::Pgdh-HMG-CoA | Any strain | Catchpole et al. (2018) |
| pTPTK2 | 5444 | ~50/cell | ~20–30/cell | Tryptophan | Chloramphenicol | pTP2 + pBAD33::Pcap-TrpE | ΔtrpE (ΔTK0254) | Catchpole et al. (2018) |
| pTPTK3 | 4710 | ~50/cell | ~20–30/cell | Agmatine | Chloramphenicol | pTP2 + pBAD33::Ppdad-PdaD | ΔpdaD (ΔTK0149) | Catchpole et al. (2018) |
Naturally occurring replicative plasmids (i.e.: pTP2 and pTN1; Fig. 2, red) have been modified to contain cassettes for expression of selection markers in both T. kodakaraensis and E. coli (Table 4). Ampicillin, chloramphenicol and kanamycin resistance genes (bla, cat and neo, respectively; Fig. 2, blue) have previously been used for selection in E. coli, while a variety of prototrophic and statin-resistance strategies have been validated in T. kodakarensis (Fig. 2, orange) (Catchpole et al., 2018; Santangelo, Cubonová, & Reeve, 2008a). The pLC plasmid series (e.g.: pLC64, pLC70, pLC71 and pTS543) can provide statin resistance or restore tryptophan or agmatine prototrophy to transformants (Santangelo et al., 2007, 2010; Santangelo, Cubonová, & Reeve, 2008a). The pLC64 plasmid was constructed by ligating together pTN1 and the commercially available pCR2.1-TOPO. pLC64 contains the trpE gene under control of the constitutive promoter, Pcap (Rodrigues et al., 2007). Plasmids pLC70 and then pLC71 were derived by adding HMG-CoA from P. furiosus under the T. kodakarensis Pgdh promoter, providing an additional mechanism for statin resistance. Several restriction enzyme sites are available for digestion at the MCS (Fig. 2, green).
The pTPTK plasmid series (e.g.: pTPTK1, pTPTK2, pTPTK3) are a result of the ligation of pTP2 and pBAD33 and have been modified to enable statin-resistance or restore tryptophan or agmatine prototrophy under a native or constitutive promoter (Fig. 2, purple) (Catchpole et al., 2018). Restriction enzyme recognition sites are present in the multiple cloning site; however, StuI and SalI digestion should be avoided in pTPTK2 as their recognition sequences are present in the trpE cassette. With the exception of agmatine prototrophy, all other prototrophic selection strategies require cells to be grown in minimal medium. Use of all prototrophic selective markers requires a mutant genetic background that induces auxotrophy for the selection nutrient (Santangelo et al., 2008b; Santangelo, Cubonová, & Reeve, 2010; Sato et al., 2003, 2005).
3. Purifying proteins and identifying protein interactions
Recombinant protein expression in a genetically accessible, native hyperthermophile such as T. kodakarensis provides access to protein production and research applications in a novel setting. Hyperthermophilic protein expression can produce thermostable proteins in their native state, with biologically relevant associations and modifications (e.g.: in complex and/or containing post-translational modifications). Protein purifications via liquid chromatography (LC) can be applied (1) to purify proteins at industrial scale (typically from ectopic vectors) or (2) to describe native-level protein expression or identify protein–protein partnerships at research scale.
3.1. Protein production and purification
Ectopic expression vectors permit easy introduction and expression of heterologous genes and are commonly used for applications involving large-scale protein production. T. kodakarensis cultures grow rapidly at 85 °C, doubling every ~40min and will achieve a final cell density of ~108–9 cells/mL in rich media. Optical density (OD) at 600nm provides a convenient method to monitor cell growth with cultures reaching ~1.0–1.3 absorbance units at maximum cell densities. T. kodakarensis strains harboring the expression vector(s) may be cultured at a range of temperatures (~65–95 °C) and in media containing the necessary nutritional and selectable components using batch fermentation to yield large quantities of biomass. Growth conditions (e.g.: temperature) and media additives should be selected for the specific strain (Table 1) and the compatible genetic markers (Table 2). Industrial-scale protein purifications typically require greater culture volumes, and pilot trials at small scale are recommended for process development. Larger culture volumes (>10L) are ideally grown within a bioreactor. Protein production in minimal medium is possible, although culture yields are dramatically reduced resulting in minimal biomass. Cultures that have reached late exponential or early stationary growth phases are rapidly cooled and harvested, and the biomass is processed for protein purification. Cell pellets can be stored at −80 °C indefinitely prior to processing.
Cell lysis to separate cytoplasmic from membrane-bound or extracellular products is achieved by sonication in physiologically relevant buffers followed by centrifugation to pellet membranes, while retaining cytoplasmic proteins in the supernatant. Purification of the desired protein can be obtained by liquid chromatography, and conditions specific to the protein of interest should be considered (e.g.: a protein bound strongly to DNA may require elution at higher salinity or extreme pHs). Reverse phase, ion exchange, size exclusion or affinity chromatography can be employed for a variety of purification techniques. Purification efficacy can be assessed by SDS-PAGE and western blotting, if respective antibodies are available. The addition of epitope tags, such as the hemagglutinin epitope (HA) to the protein of interest can be utilized for immunodetection if protein-specific antibodies are not available. Tags which include a cleavage site can be used for structure specific applications (e.g.: crystallography).
3.2. Native protein-expression and retention of in vivo protein partnerships
Purification of tagged-protein complexes and retention of native protein interactions established in vivo permits identification of associated protein partnerships. Chromosomal MAE techniques can be used to introduce sequences to the genome that encode affinity-(His6) and epitope-(HA) tags to protein targets and result in retention of native promoters and native levels of target proteins within T. kodakarensis. Steady-state protein levels retain protein partnerships that can be identified through the rapid, gentle, co-purification of the tagged-target protein in association with its natural protein partners.
To maintain the greatest number of protein interactions, cultures should be chilled rapidly in an ice-water bath to preserve as many interactions in their native conformational state as possible. Subjecting cultures to dry ice-mediated cooling is an alternative when exceptionally delicate interactions (e.g.: protein–RNA associations) are of interest. Pelleted and resuspended cells can be sonicated briefly to disturb membranes while maintaining protein–protein interactions. The downstream chromatography must be completed as rapidly as possible to preserve the maximum number of protein partnerships. Purified fractions can be assessed for the presence of the tagged protein by SDS-PAGE and western blotting. Fractions containing the protein of interest are typically pooled, quantified for total protein and trypsin digested for peptide analysis on liquid chromatography tandem mass spectrometry (LC-MS/MS). The high-confidence peptide identification and low-input requirements offered by LC-MS/MS provides a simple and economical route to establish protein interactions in complex protein mixtures (Burkhart et al., 2019; Li et al., 2010).
In contrast to large-scale protein production strategies (Section 3.1), a single liter or less of T. kodakarensis culture grown in rich medium to mid-exponential phase is typically sufficient to identify protein-protein interaction networks. Meaningful interpretation of proteomics data requires reproducibility and biochemical validation. Repeated identification of target and co-purifying proteins, with a minimum of two unique peptides per protein, in multiple biological replicates (three are recommended) provides a high level of confidence in bona fide in vivo partnerships (Li et al., 2010). Reciprocal pulldowns and known protein associations (e.g.: established protein complexes) can be used to confirm interactions in vivo. Given that a minor number of T. kodakarensis proteins have a natural affinity for some purification matrices, care should be taken to ensure that co-purifying proteins are not present in identical purifications from near isogenic control strains (e.g.: the parental strain grown and processed under the same conditions).
4. Reagents, recipes and equipment
A list of reagents, recipes and equipment are provided for use in the protocols outlined in Section 5.
4.1. Reagents
S°; elemental sulfur; flowers of sulfur
Agmatine
Gelzan (Sigma-Aldrich)
6-methylpurine (Sigma-Aldrich)
Cysteine
Glutamic acid
Glycine
Arginine
Proline
Asparagine
Histidine
Isoleucine
Leucine
Lysine
Threonine
Tyrosine
Alanine
Methionine
Phenylalanine
Serine
Tryptophan
Aspartic acid
Glutamine
Valine
NiSO4
EDTA
Guanidine HCl
4.2. Recipes
- Trace minerals (1000 ×; 1L)
- 500 mg MnSO4·H2O
- 100mg CoCl2·6 H2O
- 100mg ZnSO4·7 H2O
- 10 mg CuSO4·5 H2O
- 10mg AlK(SO4)2·12 H2O
- 10mg H3BO3
- 10 mg Na2MoO4·2 H2O
- Artificial sea water containing yeast extract and tryptone (1.0 × ASW-YT; 1L)
- 5g tryptone
- 5g yeast extract
- 20g NaCl
- 3g MgCl2·6 H2O
- 6g MgSO4·7 H2O
- 800 mg (NH4)2SO4
- 160 mg NaHCO3
- 240 mg CaCl2·2 H2O
- 400 mg KCl
- 336 mg KH2PO4
- 40 mg NaBr
- 16 mg SrCl2·6 H2O
- 8 mg Fe(NH4)2(SO4)2·6 H2O
- 5 mL trace minerals
- 5g pyruvate (Optional)
- Polysulfides (500 ×; 15 mL)
- 10 g Na2S·9 H2O
-
3 g elemental sulfur*See Note 1.
- KOD1 Vitamins (200 ×; 1L)
- 200 mg niacin
- 80 mg biotin
- 200 mg pantothenate
- 200 mg lipoic acid
- 80 mg folic acid
- 200 mg p-aminobenzoic acid
- 200 mg thiamine
- 200 mg riboflavin
- 200 mg pyridoxine
- 200 mg cobalamin
- 20 amino acid mix (20 ×; 1L)
- 1 g cysteine
- 1 g glutamic acid
- 1 g glycine
- 500 mg arginine
- 500 mg proline
- 400 mg asparagine
- 400 mg histidine
- 400 mg isoleucine
- 400 mg leucine
- 400 mg lysine
- 400 mg threonine
- 400 mg tyrosine
- 300 mg alanine
- 300 mg methionine
- 300 mg phenylalanine
- 300 mg serine
- 300 mg tryptophan
- 200 mg aspartic acid
- 200 mg glutamine
-
200 mg valine*See Note 2.
- Solid-media plates (prepared for each transformation)
- 50mL 2 × media formulation (e.g.: ASW-YT), autoclave
- 50 mL water with 1 g Gelzan, autoclave
- Following separate autoclaving, immediately mix components a and b, 100 μL KOD1 vitamins, 200 μL polysulfides and any additional necessary components (i.e.: strain TS559 transformants rely on the absence of agmatine for positive selection)
- Mix anaerobically and pour ~25 mL into each of four glass petri dishes
- Lysis buffer (Buffer A)
- 10 mM Tris-HCl, pH 8
- 500 mM NaCl
- 10% (v/v) glycerol
- Elution buffer (Buffer B)
- 10 mM Tris-HCl, pH 8
- 100 mM NaCl
- 500 mM imidazole
- 10% (v/v) glycerol
4.3. Equipment
Microcentrifuge and microcentrifuge tubes
Floor-model centrifuge, compatible rotors and centrifuge tubes
Heating block
Anaerobic chambers (Coy Laboratory Products)
85 °C incubator
- Anaerobic microbiology culturing equipment
- Glass serum bottles
- Septa
- Aluminum crimp seals
-
Glass petri plates*See Note 3.
Sonicator
AKTA or similar liquid chromatography system
HiTrap chelating column (Cytiva)
- SDS-PAGE equipment
- Polyacrylamide Gels
- Running buffer
- Apparatus
- Compatible protein stains
- Western blotting equipment
- Membrane
- Transfer system
- Primary antibody (Invitrogen HA Tag Monoclonal Antibody, 26183)
- Secondary antibody
- Imaging system
5. Protocols
5.1. Targeted mutagenesis of chromosomal DNA
Markerless-modifications of T. kodakarensis chromosomal DNA typically rely on integration of a non-replicative plasmid harboring the desired modification into the genome, followed by excision of most plasmid sequences from the genome to yield a strain with a modified genomic sequence. Detailed instructions for the construction of integrative plasmids have been provided elsewhere (Gehring, Sanders, & Santangelo, 2017); herein we focus on the steps for transformation and confirmation of genomic modifications. All procedures are carried out anaerobically. For simplicity, we do not detail the specific selective and counter-selective procedures nor the exact media formulations of solid or liquid mediums, as the multitude of genetic techniques for T. kodakarensis provide many options depending on the host strain employed (Tables 1 and 2).
Transformation and selection for plasmid integration into the genome
Grow an MAE-compatible T. kodakarensis strain (Table 1) at 85 °C for 12 h in ASW-YT medium supplemented with S, KOD1 vitamins and any necessary additives. Harvest via centrifugation and resuspend cells in 3mL of 0.8 × ASW, transfer to a microcentrifuge tube, and place on ice.
Add ~ 2 μg of plasmid DNA (in <15 μL) to a 200 μL aliquot of concentrated cell suspension and incubate on ice for 30 min.
Heat shock the cell and plasmid mixture at 85 °C for 45 s using a heating block, then immediately incubate on ice for an additional 30 min.
Plate transformed cells on a selection medium and grow anaerobically at 85 °C for ~2–6 days.
Confirmation of appropriate plasmid integration and intermediate genome formation
-
5.
Colonies resulting from transformations are anticipated to have integrated the plasmid sequences into the genome. Cells from distinct colonies should be picked and inoculated in 5mL ASW-based medium containing KOD1 vitamins while ensuring maintenance of the positive selection(s) employed.
-
6.
5mL cultures are grown overnight anaerobically at 85 °C to yield sufficient biomass for 1 mL genomic DNA preparations following established procedures (Gehring, Sanders, & Santangelo, 2017).
-
7.
Perform diagnostic PCRs using genomic DNA.
*See Note 4.
Counter selection, confirmation of plasmid excision and final genome confirmation
-
8.
5mL ASW-based medium grown cultures of confirmed intermediate strains, supplemented to permit spontaneous excision of the selective and/or counterselective makers, should be grown overnight and the resulting cells spread on solid-medium containing the appropriate counter-selective pressures and allowed ~4 days of anaerobic growth at 85 °C.
-
9.
Resulting colonies are anticipated to have spontaneously excised the plasmid from their genomes and either retained the desired modification or reverted back to the parental genome (Fig. 1E). Cells from distinct colonies should be picked, inoculated in 5 mL ASW-based medium and grown anaerobically overnight at 85 °C to yield sufficient biomass for 1 mL genomic DNA preparations following established procedures (Gehring, Sanders, & Santangelo, 2017).
-
10.
Amplicons generated from primers pairs complementary to genomic sequences permit diagnostic confirmation of the desired final genotype. Depending on the desired modification, PCR followed by gel electrophoresis can be used for size discrimination of the target region. If the modification produces a size difference that is difficult to discern via PCR (i.e.: short sequences encoding affinity- or epitope-tags), differential restriction digestion patterns of the PCR amplicon originating from the region of interest may assist in identifying desired genotypes. When diagnostic PCRs indicate the presence of the desired modification, genotypes should be confirmed via Sanger sequencing of PCR amplicon(s) that span the target loci or whole genome sequencing (e.g.: Oxford Nanopore Sequencing).
5.2. Transformation of T. kodakarensis–E. coli shuttle vectors
*See Note 5.
Grow an MAE-compatible T. kodakarensis strain (Table 1) at 85 °C for 12 h in ASW-YT medium supplemented with S, KOD1 vitamins and any necessary additives. Harvest via centrifugation and resuspend cells in 3mL of 0.8 × ASW, transfer to a microcentrifuge tube and place on ice.
Add ~ 2 μg of plasmid DNA (in <15 μL) to a 200 μL aliquot of concentrated cell suspension and incubate on ice for 30 min.
Heat shock the cell and plasmid mixture at 85 °C for 45 s using a heating block, then immediately incubate on ice for an additional 30 min.
Spread transformed cells on a solid-medium compatible with the selection marker(s) (see Table 2). Incubate the plated cells anaerobically for ~2–6 days at 85 °C.
Pick cells from a discrete colony into 10 mL of the appropriate ASW-based medium required for the chosen selectable marker(s). Allow the culture to grow anaerobically at 85 °C overnight.
*See Note 6.
5.3. Ectopic protein expression
Inoculate 1L of ASW-based medium supplemented with S, KOD1 vitamins and any necessary additives with a 10 mL overnight culture.
*See Note 7.
-
2.
For expression reliant on constitutive promoters, cells should be harvested via centrifugation immediately prior to entry into the stationary phase to maximize cell density. For expression reliant on inducible promoters, culture conditions should be altered to permit protein expression when the optical density has reached OD600 ≈ 0.2–0.3, and cells should be harvested via centrifugation prior to reaching stationary growth phase.
*See Note 8.
5.4. Large-scale single protein purification
*See Note 9.
Cell lysis
Cell pellets are thawed (if frozen) and resuspended with ~3 mL/g of biomass in lysis buffer (Buffer A).
Sonicate the cell suspension on ice at half-maximal output for 10 s on/ 30 s off for a total of ~5–60 min. Suspension will be gray and homogenous when cells are sufficiently lysed. Avoid unnecessary sonication.
-
Generate a clarified cell lysate (CCL) by centrifugation (9000 × g for 20 min) to pellet membranes while retaining cytoplasmic proteins in the supernatant.
*See Note 10.
Purification via affinity chromatography
Prepare the AKTA chromatography unit (or similar) by ensuring all tubing is flushed and a fraction collector is prepared.
Charge HiTrap chelating column with 1M NiSO4.
Clean flow paths and HiTrap column with 5 column volumes of Buffer B followed by 5 column volumes of Buffer A.
-
Load CCL.
*See Note 11.
-
Wash the column with 5 column volumes of Buffer A.
*See Note 12.
-
Elute the target protein with an imidazole gradient and collect the eluant in small (~1 mL) fractions. Aliquots of each fraction can be resolved via SDS-PAGE and/or western blotting to identify fractions containing the target protein.
*See Note 13.
Flush the entire chromatography system with 5 column volumes Buffer B, followed by 10 column volumes of ultra-pure H2O. To regenerate the chelating column, first remove any remaining proteins with 5 column volumes of 6M guanidine HCl, then flush the column with H2O, and finally strip the bound Ni2+ from the column with 2 column volumes 0.5 M EDTA. Store the chromatography apparatus and HiTrap column in 20% ethanol.
5.5. Rapid and gentle purification to identify protein partnerships
*See Note 14.
Identification of in vivo protein partnerships
Chromatography fractions containing the tagged protein of interest are pooled, quantified, precipitated and trypsin-digested prior to the separation and identification of peptides via LC-MS/MS.
Peptides are identified using multidimensional protein identification technology (MuDPIT) and mapped to the T. kodakarensis proteome as described (Burkhart et al., 2019; Li et al., 2010).
6. Advantages and future perspectives
T. kodakarensis has emerged as a model species for the study of biological processes at high temperatures and as an expression platform for thermostable enzymes, owing mainly to the development of highly reproducible, accurate genomic and ectopic manipulation strategies (Atomi & Reeve, 2019). While numerous advantages of a native hyperthermophilic expression system are evident, the T. kodakarensis platform could be improved.
Although genomic manipulation strategies in T. kodakarensis are simple and sufficiently straightforward to allow many researchers to generate large numbers of unique strains, genome modification systems that use CRISPR/Cas are not yet available for T. kodakarensis. Most archaeal species (including the Thermococcales) encode CRISPR elements (Fukui et al., 2005) and efforts to establish a thermostable CRISPR/Cas system for use in T. kodakarensis may accelerate strain construction. The development of additional cost-effective selection strategies, particularly non-prototrophic selections that could be universally deployed regardless of host genotype, would speed strain construction and add to the versatility of genetic techniques for T. kodakarensis. One of the most important tools for exploiting T. kodakarensis as an expression platform will be the development of a tightly controlled, inducible promoter system. Available riboswitch-mediated regulatory expression systems are useful and are commonly employed within the T. kodakarensis research community (Sanders et al., 2020; Speed et al., 2018) but lack the dynamic range desired for rapid and efficient control of protein expression.
Despite the challenges imposed by high temperature environments, the rapid development of tools for genetic manipulation for T. kodakarensis have provided the scientific community with a versatile protein expression platform conducive to the production and study of thermostable proteins. The combination of autonomously replicating ectopic expression systems, controlled protein overexpression and the ability to introduce genomic modifications makes T. kodakarensis a versatile platform for the expression of thermostable proteins and the evaluation of in vivo protein partnerships.
7. Notes
The powder is mixed with up to 15 mL H2O and heated until the solution is homogeneous and free of particulates.
Individual amino acids can be excluded to allow prototrophic selections.
Many plasticware items melt upon extended incubation at hyperthermophilic temperatures.
Amplicons generated from primer pairs complementary to genomic and integrated plasmid sequences permit diagnostic confirmation of the desired intermediate genotype(s). It is imperative that the primer sets used in the diagnostic PCRs include one primer that is complementary to genomic DNA sequences and a second primer complementary to sequences of the integrated plasmid (Fig. 1D). Such pairing ensures that resulting amplicons originate from a genomically integrated plasmid. Depending on how the plasmid integrated into the genome (either upstream or downstream to the modification site) two intermediate genomes are possible (Hileman & Santangelo, 2012). Whenever possible, identification of separate strains containing each potential intermediate genotype is desirable as each intermediate strain can yield the desired final genotype.
Methodologies for generating a T. kodakarensis–E. coli shuttle vector with selectable phenotypes and replicative origins are outlined elsewhere (Catchpole et al., 2018; Santangelo, Cubonová, & Reeve, 2008a).
Maintenance of a retained plasmid can be confirmed by growth on selective media, PCR, or by plasmid purification and transformation back into E. coli.
Cultures should be started with a 1:100 inoculum.
Secreted protein targets can be recovered from the culture media.
Both purification strategies (see Sections 5.4 and 5.5) are reliant on genetically modified strains encoding affinity and epitope tags on the protein of interest. Six-histidines (His6; affinity-tag) at the N- or C-terminus allow for protein purification using a chelating column charged with nickel. Bound proteins are eluted using an imidazole gradient. A hemagglutinin sequence (HA; epitope-tag) is encoded between the His6 and protein coding sequence to allow for easy identification of the target protein via western blotting. The following procedures should be done at 4 °C.
The CCL can be stored at 4 °C but should not be frozen if protein structure or function is of interest.
The His6 tagged protein will bind to the Ni2+-charged column.
The tagged protein should not be in the wash.
Subsequent purifications to increase protein purity may be required. Size exclusion often complements affinity chromatography and is recommended.
Epitope and affinity (HA and His6) tagged proteins can be rapidly purified through single-step chromatography to retain and identify in vivo protein partnerships. Cultures, clarified cell lysates and protein fractions are processed as described in Section 5.4 with care given to reducing sonication time, using gentle centrifugation, and proceeding as quickly as possible through all steps (Burkhart et al., 2019; Li et al., 2010).
Acknowledgments
We would like to thank members of the Santangelo lab for comments and improvements on this manuscript. KAS and SAW received financial support from a T32 training grant from the National Institutes of Health, GM132057.
Funding
This work was supported with funding (to TJS) from the National Science Foundation, grant EF-2022065, the US Department of Energy, grant DE-SC0014597, and the USA National Institutes of Health, GM100329.
References
- Adams MW, & Kelly RM (1998). Finding and using hyperthermophilic enzymes. Trends in Biotechnology, 16, 329–332. [DOI] [PubMed] [Google Scholar]
- Atomi H, Fukui T, Kanai T, Morikawa M, & Imanaka T (2004). Description of Thermococcus kodakaraensis sp. nov., a well studied hyperthermophilic archaeon previously reported as Pyrococcus sp. KOD1. Archaea, 1, 263–267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atomi H, & Reeve J (2019). Microbe profile: Thermococcus kodakarensis: the model hyperthermophilic archaeon. Microbiology, 165, 1166–1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basen M, Sun J, & Adams MWW (2012). Engineering a hyperthermophilic archaeon for temperature-dependent product formation. mBio, 3. e00053–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhardwaj N, Kumar B, & Verma P (2019). A Detailed overview of xylanases: An emerging biomolecule for current and future prospective. Bioresources and Bioprocessing, 6, 1–36. [Google Scholar]
- Burkhart BW, Febvre HP, & Santangelo TJ (2019). Distinct physiological roles of the three ferredoxins encoded in the hyperthermophilic archaeon Thermococcus kodakarensis. MBio, 10, e02807–e02818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabrera JA, Bolds J, Shields PE, Havel CM, & Watson JA (1986). Isoprenoid synthesis in Halobacterium halobium. Modulation of 3-hydroxy-3-methylglutaryl coenzyme a concentration in response to mevalonate availability. The Journal of Biological Chemistry, 261, 3578–3583. [PubMed] [Google Scholar]
- Catchpole R, Gorlas A, Oberto J, & Forterre P (2018). A series of new E. coli-Thermococcus shuttle vectors compatible with previously existing vectors. Extremophiles, 22, 591–598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Counts JA, et al. (2017). Physiological, metabolic and biotechnological features of extremely thermophilic microorganisms. Wiley Interdisciplinary Reviews. Systems Biology and Medicine, 9. 10.1002/wsbm.1377. PMID: 28206708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crosby JR, et al. (2019). Extreme thermophiles as emerging metabolic engineering platforms. Current Opinion in Biotechnology, 59, 55–64. [DOI] [PubMed] [Google Scholar]
- Čuboňováa L, et al. (2012). An archaeal histone is required for transformation of Thermococcus kodakarensis. Journal of Bacteriology, 194, 6864–6874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Souza PM, & de Oliveira Magalhães P (2010). Application of microbial α-amylase in industry—A review. Brazilian Journal of Microbiology, 41, 850–861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Del Solar G, Giraldo R, Ruiz-Echevarría MJ, Espinosa M, & Díaz-Orejas R (1998). Replication and control of circular bacterial plasmids. Microbiology and Molecular Biology Reviews, 62, 434–464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duong-Ly KC, & Gabelli SB (2014). Explanatory chapter: troubleshooting recombinant protein expression: General. Methods in Enzymology, 541, 209–229. [DOI] [PubMed] [Google Scholar]
- Farkas J, Chung D, DeBarry M, Adams MWW, & Westpheling J (2011). Defining components of the chromosomal origin of replication of the hyperthermophilic archaeon Pyrococcus furiosus needed for construction of a stable replicating shuttle vector. Applied and Environmental Microbiology, 77, 6343–6349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farkas JA, Picking JW, & Santangelo TJ (2013). Genetic techniques for the archaea. Annual Review of Genetics, 47, 539–561. [DOI] [PubMed] [Google Scholar]
- Fukuda W, Morimoto N, Imanaka T, & Fujiwara S (2008). Agmatine is essential for the cell growth of Thermococcus kodakaraensis. FEMS Microbiology Letters, 287, 113–120. [DOI] [PubMed] [Google Scholar]
- Fukui T, et al. (2005). Complete genome sequence of the hyperthermophilic archaeon Thermococcus kodakaraensis KOD1 and comparison with Pyrococcus genomes. Genome Research, 15, 352–363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gehring AM, et al. (2017). Genome replication in Thermococcus kodakarensis independent of Cdc6 and an origin of replication. Frontiers in Microbiology, 8, 2084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gehring AM, Sanders TJ, & Santangelo TJ (2017). Markerless gene editing in the hyperthermophilic archaeon Thermococcus kodakarensis. Bio-Protocol, 7(22), e2604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorlas A, Krupovic M, Forterre P, & Geslin C (2013). Living side by side with a virus: characterization of two novel plasmids from Thermococcus prieurii, a host for the spindle-shaped virus TPV1. Applied and Environmental Microbiology, 79, 3822–3828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guss AM, Rother M, Zhang JK, Kulkarni G, & Metcalf WW (2008). New methods for tightly regulated gene expression and highly efficient chromosomal integration of cloned genes for Methanosarcina species. Archaea, 2, 193–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haque RU, Paradisi F, & Allers T (2020). Haloferax volcanii for biotechnology applications: Challenges, current state and perspectives. Applied Microbiology and Biotechnology, 104, 1371–1382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hileman TH, & Santangelo TJ (2012). Genetics techniques for Thermococcus kodakarensis. Frontiers in Microbiology, 3, 195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishino S, et al. (2011). Biochemical and genetical analyses of the three mcm genes from the hyperthermophilic archaeon, Thermococcus kodakarensis. Genes to Cells, 16, 1176–1189. [DOI] [PubMed] [Google Scholar]
- Kanai T, et al. (2015). Overproduction of the membrane-bound [NiFe]-hydrogenase in Thermococcus kodakarensis and its effect on hydrogen production. Frontiers in Microbiology, 6, 847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H, Yoo SJ, & Kang HA (2015). Yeast synthetic biology for the production of recombinant therapeutic proteins. FEMS Yeast Research, 15, 1–16. [DOI] [PubMed] [Google Scholar]
- Krupovic M, Gonnet M, Hania WB, Forterre P, & Erauso G (2013). Insights into dynamics of mobile genetic elements in hyperthermophilic environments from five new Thermococcus plasmids. PLoS One, 8, e49044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S, Dangi AK, Shukla P, Baishya D, & Khare SK (2019).Thermozymes: Adaptive strategies and tools for their biotechnological applications. Bioresource Technology, 278, 372–382. [DOI] [PubMed] [Google Scholar]
- Li Z, Santangelo TJ, Cubonová L.’u., Reeve JN, & Kelman Z (2010). Affinity purification of an archaeal DNA replication protein network. MBio, 1. e00221–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, et al. (2011). A novel DNA nuclease is stimulated by association with the GINS complex. Nucleic Acids Research, 39, 6114–6123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin-Chao S, Chen WT, & Wong TT (1992). High copy number of the pUC plasmid results from a Rom/Rop-suppressible point mutation in RNA II. Molecular Microbiology, 6, 3385–3393. [DOI] [PubMed] [Google Scholar]
- Martínez-Espinosa RM (2020). Heterologous and homologous expression of proteins from haloarchaea: Denitrification as case of study. International Journal of Molecular Sciences, 21, 82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumi R, Manabe K, Fukui T, Atomi H, & Imanaka T (2007). Disruption of a sugar transporter gene cluster in a hyperthermophilic archaeon using a host-marker system based on antibiotic resistance. Journal of Bacteriology, 189, 2683–2691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morikawa M, Izawa Y, Rashid N, Hoaki T, & Imanaka T (1994). Purification and characterization of a thermostable thiol protease from a newly isolated hyperthermophilic Pyrococcus sp. Applied and Environmental Microbiology, 60, 4559–4566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peleg Y, & Unger T (2012). Resolving bottlenecks for recombinant protein expression in E. coli. Methods in Molecular Biology, 800, 173–186. [DOI] [PubMed] [Google Scholar]
- Razzaq A, Shamsi S, Ali A, Ali Q Sajjad M, Malik A, & Ashraf M (2019). Microbial proteases applications. Frontiers in Bioengineering and Biotechnology. 10.3389/fbioe.2019.00110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigues MV, et al. (2007). Bifunctional CTP:inositol-1-phosphate cytidylyltransferase/CDP-inositol:inositol-1-phosphate transferase, the key enzyme for di-myo-inositol-phosphate synthesis in several (hyper)thermophiles. Journal of Bacteriology, 189, 5405–5412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosano GL, & Ceccarelli EA (2014). Recombinant protein expression in Escherichia coli: Advances and challenges. Frontiers in Microbiology, 5, 172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders TJ, et al. (2020). FttA is a CPSF73 homologue that terminates transcription in Archaea. Nature Microbiology, 5, 545–553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santangelo TJ, et al. (2008b). Polarity in archaeal operon transcription in Thermococcus kodakaraensis. Journal of Bacteriology, 190, 2244–2248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santangelo TJ, Cubonová L.’u., James CL, & Reeve JN (2007). TFB1 or TFB2 is sufficient for Thermococcus kodakaraensis viability and for basal transcription in vitro. Journal of Molecular Biology, 367, 344–357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santangelo TJ, Cubonová L.’u., & Reeve JN (2008a). Shuttle vector expression in Thermococcus kodakaraensis: Contributions of cis elements to protein synthesis in a hyperthermophilic archaeon. Applied and Environmental Microbiology, 74, 3099–3104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santangelo TJ, Cubonová L.’u., & Reeve JN (2010). Thermococcus kodakarensis genetics: TK1827-encoded beta-glycosidase, new positive-selection protocol, and targeted and repetitive deletion technology. Applied and Environmental Microbiology, 76, 1044–1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santangelo TJ, Cuboňová L.’u., & Reeve JN (2011). Deletion of alternative pathways for reductant recycling in Thermococcus kodakarensis increases hydrogen production. Molecular Microbiology, 81, 897–911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santangelo TJ, & Reeve JN (2010). Genetic tools and manipulations of the hyperthermophilic heterotrophic archaeon Thermococcus kodakarensis. In Horikoshi K (Ed.), Extremophiles handbook (pp. 567–582). Tokyo: Springer. [Google Scholar]
- Sato T, Fukui T, Atomi H, & Imanaka T (2003). Targeted gene disruption by homologous recombination in the hyperthermophilic archaeon Thermococcus kodakaraensis KOD1. Journal of Bacteriology, 185, 210–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato T, Fukui T, Atomi H, & Imanaka T (2005). Improved and versatile transformation system allowing multiple genetic manipulations of the hyperthermophilic archaeon Thermococcus kodakaraensis. Applied and Environmental Microbiology, 71, 3889–3899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato T, Imanaka H, Rashid N, Fukui T, Atomi H, & Imanaka T (2004). Genetic evidence identifying the true gluconeogenic fructose-1,6-bisphosphatase in Thermococcus kodakaraensis and other hyperthermophiles. Journal of Bacteriology, 186(17), 5799–5807. 10.1128/JB.186.17.5799-5807.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schocke L, Bräsen C, & Siebers B (2019). Thermoacidophilic Sulfolobus species as source for extremozymes and as novel archaealplatform organisms. Current Opinion in Biotechnology, 59, 71–77. [DOI] [PubMed] [Google Scholar]
- Schut GJ, et al. (2014). The order thermococcales and the family thermococcaceae. In The prokaryotes: Other major lineages of bacteria and the archaea (pp. 363–383). [Google Scholar]
- Soler N, et al. (2007). The rolling-circle plasmid pTN1 from the hyperthermophilic archaeon Thermococcus nautilus. Molecular Microbiology, 66, 357–370. [DOI] [PubMed] [Google Scholar]
- Spaans SK, van der Oost J, & Kengen SWM (2015). The chromosome copy number of the hyperthermophilic archaeon Thermococcus kodakarensis KOD1. Extremophiles, 19, 741–750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Speed MC, Burkhart BW, Picking JW, & Santangelo TJ (2018). An archaeal fluoride-responsive riboswitch provides an inducible expression system for hyperthermophiles. Applied and Environmental Microbiology, 84, e02306–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Straub CT, et al. (2018). Biotechnology of extremely thermophilic archaea. FEMS Microbiology Reviews, 42, 543–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suresh A, et al. (2021). Recent advancements in the synthesis of novel thermostable biocatalysts and their applications in commercially important chemoenzymatic conversion processes. Bioresource Technology, 323, 124558. [DOI] [PubMed] [Google Scholar]
- Takemasa R, Yokooji Y, Yamatsu A, Atomi H, & Imanaka T (2011). Thermococcus kodakarensis as a host for gene expression and protein secretion. Applied and Environmental Microbiology, 77, 2392–2398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker JE, & Santangelo TJ (2015). Analyses of in vivo interactions between transcription factors and the archaeal RNA polymerase. Methods, 86, 73–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeldes BM, et al. (2015). Extremely thermophilic microorganisms as metabolic engineering platforms for production of fuels and industrial chemicals. Frontiers in Microbiology, 6, 1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar A, Dhar K, Kanwar SS and Arora PK (2016). Lipase catalysis in organic solvents: Advantages and applications, Biological Procedures Online, 18, 2, 10.1186/s12575-016-0033-2. [DOI] [PMC free article] [PubMed] [Google Scholar]