Abstract
Streptomyces sp. GET02.ST and Achromobacter sp. GET02.AC were isolated together from the gut of the wharf roach, Ligia exotica, inhabiting the intertidal zone of the west coast of Korea. The co-cultivation of these two strains significantly induced the production of two new metabolites, ligiamycins A (1) and B (2), which were barely detected in the single culture of Streptomyces sp. GET02.ST. The planar structures of ligiamycins A (1) and B (2) were elucidated as new decalins coupled with amino-maleimides by the analysis of various spectroscopic data, including nuclear magnetic resonance (NMR), ultraviolet (UV), and mass (MS) data. The assignment of two nitrogen atoms in amino-maleimide in 1 was accomplished based on 1H-15N heteroatom single quantum coherence spectroscopy (HSQC) NMR experiments. The relative configurations of the ligiamycins were determined using rotating frame Overhauser effect spectroscopy (ROESY) NMR data, and their absolute configurations were deduced by comparing their experimental and calculated optical rotations. Ligiamycin A (1) displayed antibacterial effects against Staphylococcus aureus and Salmonella enterica, while ligiamycin B (2) exhibited mild cell cytotoxicity against human colorectal cancer cells.
Keywords: co-culture, Streptomyces, Achromobacter, structure elucidation, Ligia exotica, optical rotation, cytotoxicity, antibacterial, decalin, maleimide
1. Introduction
Marine microorganisms physically and chemically interact with neighboring microbes. These interactions are regarded as a driving force to produce bioactive secondary metabolites with pharmaceutical potential [1]. Even though mimicking such complex microbial interactions is challenging, co-culturing two microbial strains in a single culture vessel has been continuously utilized to induce the production of previously unreported bioactive natural products, which has contributed to increasing the chemical diversity of marine microbial compounds [2].
The co-cultivation of the marine fungus Pestalotia sp. with the bacterial strain Thalassospira sp. CNJ328 produced a new chlorinated benzophenone antibiotic, pestalone, as the first natural product by co-culture [3]. The strain Thalassospira sp. CNJ328 also triggered the production of new cytotoxic diterpenoids, libertellenones A–D from the marine fungus Libertella sp. [4]. The production of cyclic lipopeptides, emericellamides A and B, was enhanced by 100 times in the co-culture of the fungus Emericella sp. from a marine green alga and the marine bacterium Salinispora arenicola [5]. However, these early co-culture experiments among marine microbes did not consider ecological relevance much and randomly mixed two different strains. Relatively recently, it has been realized that co-culturing symbiotic or ecologically relevant microbes more efficiently induces the production of new metabolites that are not produced in single cultures. Microscale co-culture experiments with symbiotic Micromonosporaceae strains from marine sponges and ascidians have revealed that 18.5% of 65 co-cultured strains produce unique metabolites, demonstrating the efficiency of this approach [6]. Subsequent chemical studies have discovered a new multi-glycosylated antibiotic, keyicin, from a co-culture of symbiotic Micromonospora and Rhodococcus associated with marine invertebrates [7]. A mixed culture of marine bacterial strains isolated together from the same marine sediment sample yielded a cyclic depsipeptide, dentigerumycin E, with a polyketide and nonribosomal peptide hybrid origin [8]. The induction of antibacterial metabolites has been observed by co-culturing Micromonospora sp. and Actinokinespora sp. associated with sea sponges [9]. The co-cultivation of mangrove endophytically symbiotic fungus Trichoderma sp. with marine pathogenic bacterium Acinetobacter johnsonii has led to the discovery of new sesquiterpenoids, microsphaeropsisins B and C [10].
Among marine invertebrates, which have been considered to harbor microbes potentially producing bioactive metabolites [11], wharf roaches belonging to order Isopoda in the subphylum Crustacea, have not been chemically explored in much detail. In particular, Ligia spp. are globally distributed wharf roaches living on rocks on the seashore [12]. As Ligia spp. feed on phytoplanktons and degraded plants in intertidal zones, of which the environments vary extremely by tides and host diverse beneficial and pathogenic microorganisms, they could also be associated with microorganisms potentially biosynthesizing biologically and structurally unique compounds. Previous chemical studies reported that new aspochalasins and cyclopeptide metabolites were produced by Ligia-associated Aspergillus sp. Z4 as a rare chemical study example for Ligia spp., indicating the chemical potential of wharf roach-associated microbes [13,14,15].
In this work, we co-cultivated and chemically analyzed microorganisms isolated from the gut of Ligia exotica, collected in an intertidal mudflat on the west coast of the Korean peninsula, and found that the co-culture of Streptomyces sp. GET02.ST and Achromobacter sp. GET02.AC produced distinct chemical profiles compared to the individual cultures of these strains. Further large culture and chromatographic separation of the distinct metabolites enabled us to elucidate their structures and to evaluate their biological activity. Herein, we report the isolation, structure determination, and bioactivities of these compounds, ligiamycins A (1) and B (2), from the co-culture of the gut strains of Ligia exotica.
2. Results and Discussion
2.1. Co-Culture of Streptomyces sp. and Achromobacter sp. from Ligia exotica
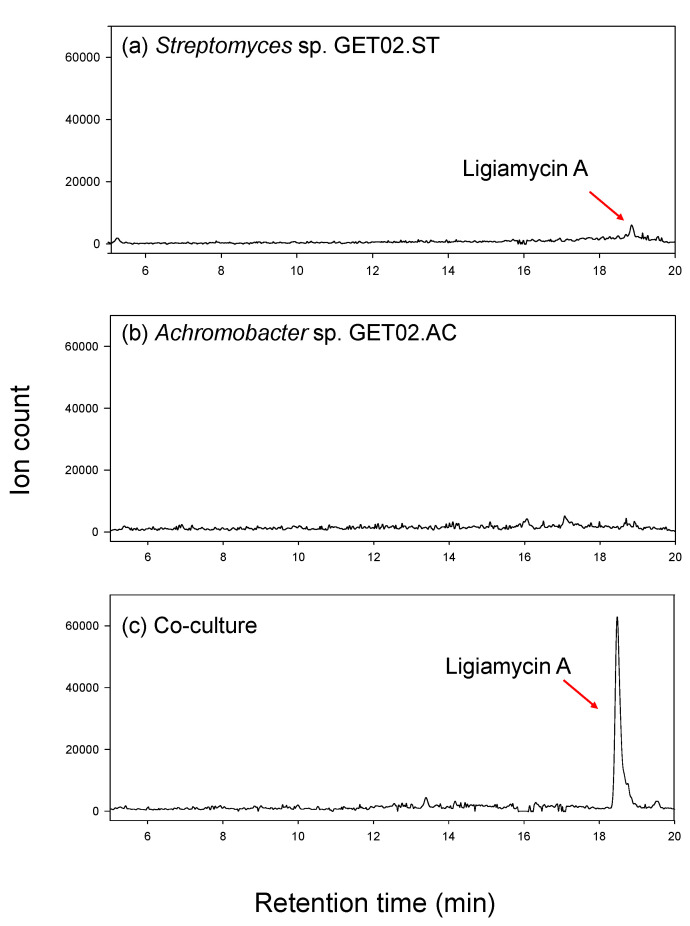
When cultivated in a pure single culture of the strain, Streptomyces sp. GET02.ST, isolated from the gut of Ligia exotica, produced a distinguished metabolite, ligiamycin A (1) with [M + H]+ at m/z 317 and ultraviolet (UV) absorption (λmax at 250 and 351 nm). However, the production level was minuscule, and this compound was barely identified by the liquid chromatography (LC)/mass spectrometry (MS) analysis. For further characterization of the compound, we applied the co-culture strategy and enhanced the yield of 1. Interestingly, when we co-cultured Streptomyces sp. GET02.ST with Achromobacter sp. GET02.AC, which was also isolated from the gut of L. exotica, the productivity of 1 was increased by approximately 24 times that of the pure culture of Streptomyces sp. GET02.ST (Figure 1). Moreover, an additional derivative of 1, ligiamycin B (2), which was hardly detected in the pure cultured medium of GET02.ST, was identified in the LC/MS profile of the co-cultured (with Achromobacter sp. GET02.AC) medium (Figure S18). The significant enhancement of the yields enabled subsequent chromatographic purification and structure elucidation of 1 and 2.
Figure 1.
LC/MS traces (ion counts) of ion extraction for the ion [M + H]+ at m/z 317 (ligiamycin A) (a) in a pure culture of Streptomyces sp. GET02.ST, (b) in a pure culture of Achromobacter sp. GET02.AC, and (c) in a co-culture. The LC/MS analyses were acquired with 10–100% aqueous CH3CN over 20 min.
2.2. Structural Elucidation
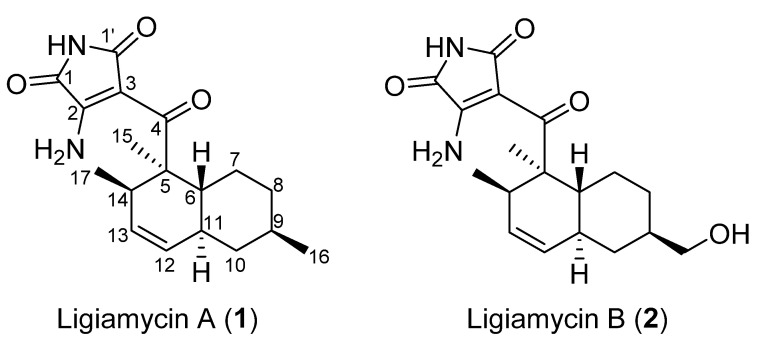
Ligiamycin A (1) (Figure 2) was isolated as a white powder, and the molecular formula of compound 1 was deduced to be C18H24N2O3, with eight degrees of unsaturation, by the positive high-resolution electrospray ionization mass spectrometry (HR-ESI-MS) analysis ([M + H]+ at m/z 317.1861, calculated as 317.1860) (Figure S15). The 13C and multiplicity-edited heteronuclear single quantum coherence spectroscopy (multiplicity-edited HSQC) nuclear magnetic resonance (NMR) spectra of 1 indicated the existence of three carbonyl carbons (δC 200.0, 169.8, and 165.1), two fully substituted olefinic carbons (δC 157.8 and 98.7), two olefinic methine carbons (δC 130.2 and 129.0), eight aliphatic sp3 carbons (δC 50.1, 42.1, 39.3, 38.0, 35.7, 34.6, 32.9, and 27.2), and three methyl groups (δC 22.5, 18.5, and 14.8). A comprehensive analysis of the 1H NMR spectrum in combination with the multiplicity-edited HSQC NMR spectrum showed that 1 possesses two olefinic protons (δH 5.52 and 5.31), four methine protons (δH 3.06, 1.75, 1.55, and 1.48), six methylene protons (δH 1.88, 1.77, 1.70, 1.00, 0.88, and 0.78), three methyl groups (δH 1.27, 0.88, and 0.68), and three heteroatom-bound protons (δH 10.6, 9.16, and 8.90) (Table 1). The two double bonds and three carbonyl groups accounted for five unsaturation degrees, thereby suggesting that ligiamycin A (1) contains three rings.
Figure 2.
Structures of ligiamycins A (1) and B (2).
Table 1.
1H, 13C, and 15N NMR data for ligiamycins A (1) and B (2) in DMSO-d6.
| Ligiamycin A (1) a | Ligiamycin B (2) b | |||
|---|---|---|---|---|
| Position | δC/δN, Type | δH, Mult (J in Hz) | δC, Type | δH, Mult (J in Hz) |
| 1 | 165.1, C | 165.2, C | ||
| 1’ | 169.8, C | 169.9, C | ||
| 2 | 157.8, C | 157.8, C | ||
| 3 | 98.7, C | 98.7, C | ||
| 4 | 200.0, C | 200.0, C | ||
| 5 | 50.1, C | 50.1, C | ||
| 6 | 39.3, CH | 1.55, t (10.0) | 39.6, CH | 1.56, t (10.0) |
| 7a | 27.2, CH2 | 1.88, m | 26.8, CH2 | 1.92, m |
| 7b | 0.88, m c | 0.86, m | ||
| 8a | 35.7, CH2 | 1.70, m | 30.3, CH2 | 1.75, m |
| 8b | 1.00, ddd (16.0, 12.5, 3.5) | 0.99, m | ||
| 9 | 32.9, CH | 1.48, m | 40.9, CH | 1.47, m |
| 10a | 42.1, CH2 | 1.77, m | 36.8, CH2 | 1.84, m |
| 10b | 0.78, m | 0.76, m | ||
| 11 | 38.0, CH | 1.75, m | 37.7, CH2 | 1.73, m |
| 12 | 129.0, CH | 5.31, dd (10.0, 2.0) | 129.1, CH | 5.33, dd (10.0, 2.0) |
| 13 | 130.2, CH | 5.52, m | 130.2, CH | 5.52, m |
| 14 | 34.6, CH | 3.06, m | 34.6, CH | 3.06, m |
| 15 | 14.8, CH3 | 1.27, s | 14.8, CH3 | 1.28, s |
| 16 | 22.5, CH3 | 0.88, m c | 66.5, CH2 | 3.22, d (6.0) |
| 17 | 18.5, CH3 | 0.68, d (7.0) | 18.5, CH3 | 0.68, d (6.5) |
| 1,1’-NH | 146.8 d, NH | 10.6, br s | 10.6, br s | |
| 2-NH2a | 103.9 d, NH2 | 9.16, s | 9.16, s | |
| 2-NH2b | 8.90, br s | 8.89, br s | ||
| 16-OH | 4.36, br s | |||
a 1H and 13C NMR data were recorded at 800 and 200 MHz, respectively. b 1H and 13C NMR data were recorded at 850 and 225.5 MHz, respectively. c Overlapping signals. d The chemical shifts of 15N in the groups of N were determined based on the 1H-15N HSQC NMR spectrum.
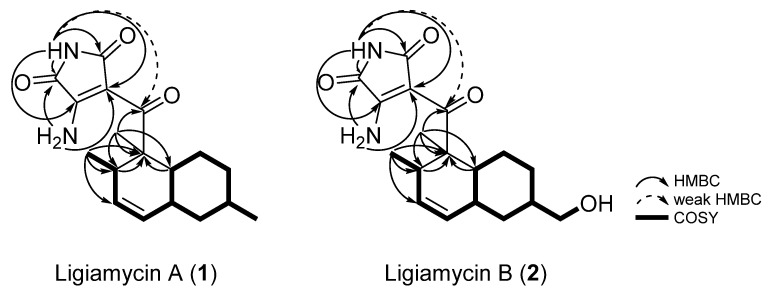
Comprehensive analyses of 1H, 13C, and correlation spectroscopy (COSY) NMR spectra was performed to identify partial structures of 1. The doublet methyl group protons (δH 0.68) at C-17 (δC 18.5) correlated with H-14 (δH 3.06), assigning the C-17 methyl group bound to C-14 (δC 34.6). The H-14/H-13 (δH 5.52) COSY correlation connected C-14 and C-13 (δC 130.2). The 10 Hz coupling constant between H-13 and H-12 (δH 5.31) indicated C-13–C-12 (δC 129.0) connectivity inside a six-membered ring. C-11 (δC 38.0) was then assigned adjacent to C-12 by H-12/H-11 (δH 1.75) COSY coupling. H-11 correlated with H2-10 (δH 1.77 and 0.78) and H-6 (δH 1.55), extending the chain to C-10 (δC 42.1) and C-6 (δC 39.3). Furthermore, H-9 (δH 1.48) displayed COSY correlations with H2-8 (δH 1.70 and 1.00) and H3-16 (δH 0.88), attaching the methylene C-8 (δC 35.7) and the methyl C-16 (δC 22.5) groups to C-9 (δC 32.9). A H2-8/H2-7 (δH 1.88 and 0.88) COSY correlation located C-7 (δC 27.2) next to C-8. H-6, which showed a COSY correlation with H-11, as mentioned, also correlated with H2-7, constructing a methylcyclohexane moiety. This substructure accounted for one of the predicted three rings. The other six-membered ring, which was deduced by 3JH12H13 (10 Hz), was elucidated as a dimethylcyclohexene (C-6, C-7, C-11, C-12 (δC 129.0), C-13 (δC 130.2), and C-14 with the two methyl groups C-15 and C-17) based on the HMBC correlations from the singlet methyl protons H3-15 (δH 1.27) to C-6, C-5 (δC 50.1), and C-14 and from H-6 to C-5 and H-14 to C-5. This connectivity was also confirmed by the H3-17/C-5 heteronuclear coupling. Thus, the two identified six-membered rings were coupled as a decalin structure (Figure 3).
Figure 3.
Key HMBC and COSY correlations of ligiamycins A (1) and B (2).
As the elucidated decalin moiety was composed of C13H21, C5H3N2O3 remained to construct the full structure of ligiamycin A (1). An analysis of the 1H-15N HSQC NMR spectrum identified that all three exchangeable protons were bound at nitrogen. Among them, two protons, 2-NH2a (δH 9.16) and 2-NH2b (δH 8.90), were connected to the same nitrogen at δN 103.8, forming a primary amine group (Figure S8). On the contrary, the remaining proton (1,1’-NH) at δH 10.60 was revealed as an amide proton based on the chemical shift (δN 146.8) of its 1-bond nitrogen and HMBC correlations from 1,1’-NH to two carbonyl carbons (C-1 and C-1’; δC 165.1 and 169.8).
Based on HMBC correlations from 1,1’-NH (δH 10.60) to C-2 (δC 157.8) and C-3 (δC 98.7) and from 2-NH2 to C-1 and C-3, the last remaining ring structure was elucidated as 3-amino-maleimide (3-amino-1H-pyrrole-2,5-dione). The established two substructures, decalin and 3-amino-1H-pyrrole-2,5-dione, were assembled through C-4 (δC 200.0) based on a HMBC correlation from H3-15 to C-4 and a weak HMBC correlation from 1,1’-NH to C-4, determining the full planar structure of ligiamycin A (1) (Figure 3).
Ligiamycin B (2) (Figure 1) was isolated as a white powder, and its molecular formula was deduced as C18H24N2O4 having eight degrees of unsaturation based on the positive high-resolution fast atom bombardment mass spectrometry (HR-FAB-MS) analysis ([M + H]+ at m/z 333.1816, calculated as 333.1814) (Figure S16). Interpretation of the HSQC NMR spectrum allowed for all one-bond 1H-13C assignments. By comparing the 1D and 2D NMR spectra of 1 and 2, ligiamycin B (2) has one less doublet methyl group than ligiamycin A (1), but instead possesses one heteroatom-bound methylene group (δH 3.22/δC 66.5) and one exchangeable proton at δH 4.36. An analysis of the COSY NMR spectrum of 2 revealed that this proton (δH 4.36) was bound to the oxygen atom attached to C-16 based on the COSY signal of H2-16 (δH 3.22)/16-OH (Figure 3). The substitution of this hydroxy group affected the 13C chemical shifts. Compared to the 13C chemical shifts of 1, C-9 (δC 40.9) resonated in a lower field by the β-effect, whereas C-8 (δC 30.3) and C-10 (δC 36.8) appeared in a higher field by the γ-effect of this substitution. The full structure of 2 was elucidated as a C-16 hydroxy analogue of 1 by COSY and HMBC correlations (Figure 3).
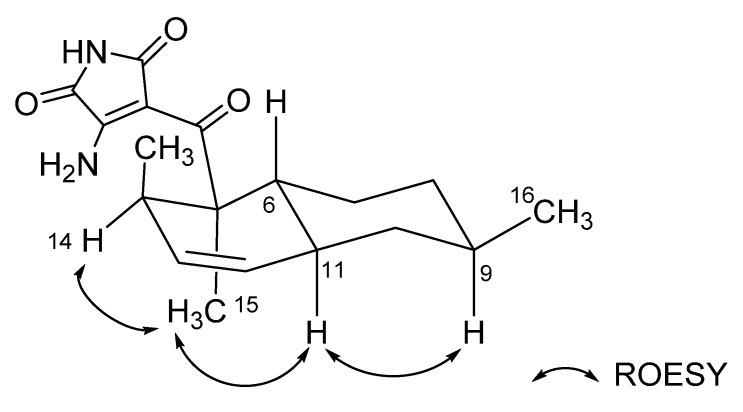
The relative configurations of the decalin structure in ligiamycin A (1) were determined by an analysis of the rotating frame Overhauser effect spectroscopy (ROESY) NMR spectrum (Figures S6 and S7). H-9/H-11, H-11/H3-15, and H3-15/H-14 ROESY correlations indicated that H-9, H-11, H-14, and H3-15 are oriented to the same phase, which elucidated 5R*, 6R*, 9R*, 11S*, and 14R* configurations along with trans-decalin conformation for 1 (Figure 4 and Figure S17a). The relative configurations of ligiamycin B (2) were also established as 5R*, 6R*, 9R*, 11S*, and 14R* by its ROESY correlations, analogous to those of 1 (Figure S17b).
Figure 4.
Key ROESY correlations of ligiamycin A (1).
For the purpose of deducing the absolute configurations of the ligiamycins, we attempted electronic circular dichroism (ECD) calculations [16]. Energy-minimized structures of the two possible enantiomers (5R/6R/9R/11S/14R and 5S/6S/9S/11R/14S) of 1 and 2 were constructed, and their ECD calculations were performed. However, the experimental ECD spectra of 1 and 2 did not display consistency with any calculated ECD spectra of 1 and 2 (Figures S19 and S20). Then, we conducted optical rotation calculations [17,18,19,20]. The calculated value of optical rotation of the 5R, 6R, 9R, 11S, and 15R enantiomer had a negative optical rotation value of − 65, whereas a positive optical rotation value ( + 66) was calculated for the opposite enantiomer. Based on the measured optical rotation value of 1 ( − 78), the absolute configurations of 1 were deduced as 5R, 6R, 9R, 11S, and 14R (Table 2). In the same manner, the absolute configurations of ligiamycin B (2) were inferred as 5R, 6R, 9R, 11S, and 14R (Table 2).
Table 2.
Experimental and calculated optical rotation of ligiamycins A (1) and B (2).
| Sample | Optical Rotation | ||
|---|---|---|---|
| Experimental | Calculated of 5R, 6R, 9R, 11S, 14R |
Calculated of 5S, 6S, 9S, 11R, 14S |
|
| Ligiamycin A (1) | −78 | −65 | +66 |
| Ligiamycin B (2) | −67 | −64 | +64 |
2.3. Biological Evaluation
We evaluated the antibacterial (Staphylococcus aureus ATCC25923, Enterococcus faecalis ATCC19433, Enterococcus faecium ATCC19434, Klebsiella pneumoniae ATCC10031, Salmonella enterica ATCC14028, and Escherichia coli ATCC25922) and antifungal (Aspergillus fumigatus HIC6094, Trichophyton rubrum NBRC9185, Trichophyton mentagrophytes IFM40996, and Candida albicans ATCC10231) activities of ligiamycins. Ligiamycin A (1) showed moderate inhibitory effects against S. aureus and S. enterica, whereas ligiamycin B (2) exhibited weak or no inhibitory effects when using ampicillin and tetracycline as positive control compounds, implying that the presence of a hydroxy group in 2 might play a negative role in antibacterial effects. Both ligiamycins A (1) and B (2) did not show any significant inhibitory effects (MIC > 128 μg/mL) against the tested fungal strains (C. albicans, A. fumigatus, T. rubrum, and T. mentagrophytes) when using amphotericin B as a positive control compound (Table 3).
Table 3.
Inhibitory effects of ligiamycins against bacterial strains.
| Sample | MIC (μg/mL) | |||||
|---|---|---|---|---|---|---|
| Gram-Positive | Gram-Negative | |||||
| S. aureus | E. faecalis | E. faecium | K. pneumonia | S. enterica | E. coli | |
| Ligiamycin A (1) | 16 | >128 | >128 | >128 | 16 | >128 |
| Ligiamycin B (2) | 64 | >128 | >128 | >128 | >128 | >128 |
| Ampicillin | 0.13 | 0.25 | 0.5 | 128 | 0.28 | 32 |
| Tetracycline | 0.13 | 0.25 | 0.13 | 0.5 | 0.25 | 0.5 |
In our cell proliferation assay against the human carcinoma cell lines SNU638 (human gastric cancer cells), SK-HEP-1 (human liver cancer cells), A549 (human lung cancer cells), HCT116 (human colorectal cancer cells), and MDA-MB-231 (human breast cancer cells), ligiamycin A (1) did not show cytotoxicity, whereas ligiamycin B (2), which bears a hydroxy group (16-OH), displayed moderate cell cytotoxicity against HCT116 cancer cells (20.1 μM) (Table 4).
Table 4.
Inhibitory effects of ligiamycins on the proliferation of human cancer cell lines.
| IC50 (μM) | SNU638 | SK-HEP-1 | A549 | HCT116 | MDA-MB-23 |
|---|---|---|---|---|---|
| Ligiamycin A (1) | >50 | >50 | 42.0 | >50 | >50 |
| Ligiamycin B (2) | 46.2 | 34.6 | 25.9 | 20.1 | 23.7 |
| Etoposide | 2.17 | 0.24 | 0.35 | 0.66 | 2.64 |
3. Materials and Methods
3.1. General Experimental Procedure
Optical rotations of the ligiamycins were acquired using a JASCO P-2000 polarimeter (sodium light source, JASCO, Easton, PA, USA) with a 1 cm cell. CD and UV spectra were obtained by using Applied Photophysics Ltd., Chirascan Plus (Applied Photophysics, Leatherhead, Surrey, UK). Infrared (IR) spectra were obtained using JASCO, FT/IR-4200 (Thermo, Madison, CT, USA). All of the LC/MS data were collected by an Agilent Technologies 1200 series HPLC instrument (Agilent Technologies, Santa Clara, CA, USA) tandemly coupled with an Agilent Technologies 6130 quadrupole MS (Agilent Technologies, Santa Clara, CA, USA) equipped with an ESI source. 1D and 2D NMR spectra were acquired on Bruker Avance 800 and 850 MHz NMR spectrometers (Bruker, Billerica, MA, USA) located at the College of Pharmacy, Seoul National University and the National Center for Inter-University Research Facilities (NCIRF), Seoul National University, respectively. HR-ESI-MS for ligiamycin A (1) was collected by a high-resolution LC/MSMS spectrometer (Q-TOF 5600, AB SCIEX, Framingham, MA, USA) at the National Instrumentation Center for Environment Management (NICEM) in the College of Agriculture and Life Sciences, Seoul National University. HR-FAB-MS for ligiamycin B (2) was obtained using a gas chromatography high-resolution mass spectrometer (JMS-700, 6890 Series, JEOL, Akishima, Tokyo, Japan) at NCIRF, Seoul National University.
3.2. Collection and Bacterial Isolation of Wharf Roaches
The wharf roach specimens belonging to the marine isopod were collected at the mudflat experience center (36°8’19.02”, 126°34’58.0”) in Seocheon, Chungcheongnam-do, Korea, in April 2018. The specimens were morphologically identified as Ligia exotica [12]. The outer skins of the wharf roach specimens were sterilized by fresh ethanol (EtOH). The gut samples of the wharf roaches were isolated using a sterilized razor blade and suspended in 40 mL of sterilized distilled water. For the purpose of bacterial isolation from the gut of the wharf roaches, 400 µL of the suspension was dropped and spread on various isolation agar media (Table S1). GET02.ST and GET02.AC strains were isolated on modified K medium. The GET02.ST strain was identified as Streptomyces sp. (GenBank accession number: MZ675370), which was closest to Streptomyces sp. 4K301 (95% identity; GenBank accession number: MG770872.1). The GET02.AC strain was identified as Achromobacter sp. (GenBank accession number: MZ675369), which was most closely related to Achromobacter sp. CLC-M23 (99% identity; GenBank accession number: MH518245.1).
3.3. Large-Scale Co-Culture and Extraction
Both the GET02.ST and GET02.AC strains were cultivated in 50 mL of modified K medium (3 g of yeast extract, 2 g of glucose, 2 g of mannitol, 5 g of malt extract, 5 g of soluble starch, 5 g of soytone, 1 g of calcium carbonate, and 23 g of sea salt in 1 L of distilled water) in a 125 mL seed flask. After fermentation for 3 days on a rotary shaker at 170 rpm and 30 °C, 10 mL of the seed culture was inoculated in 250 mL of modified K medium in a 500 mL flask. After culturing for 4 days on a rotary shaker at 170 rpm and 30 °C, 7 mL of the GET02.ST strain and 3 mL of the GET02.AC strain were transferred together into 250 mL of modified K medium (48 × 250 mL; total volume of 12 L) for co-culture and then cultured for 8 days on a rotary shaker at 170 rpm and 30 °C. A total volume of 12 L of the whole co-culture was extracted with 27 L of ethyl acetate (EtOAc). The EtOAc and water layers were separated, and the residual water in the EtOAc layer was eliminated by adding anhydrous Na2SO4. The extract was concentrated in a rotary evaporator, obtaining 2 g of dry material.
3.4. Isolation and Purification of Ligiamycins
The crude extract was re-dissolved by methanol (MeOH) and then filtered by a 25HP045AN (ADVANTEC, Tokyo, Japan) syringe filter. The extract was directly injected into a reversed-phase high-performance liquid chromatography (HPLC) column (Phenomex Luna C18(2), 4.6 × 250 mm) with a step gradient solvent system (30–40% CH3CN-H2O gradient solvent system for 40 min; after 40 min, 60% CH3CN-H2O isocratic solvent system, flow rate 1 mL/min, and UV detection at 254 nm). Ligiamycins A (1) and B (2) were eluted at 59 and 29 min, respectively. 1 was further purified with the gradient solvent system (30–40% CH3CN-H2O gradient solvent system for 40 min, flow rate 1 mL/min, and UV detection at 254 nm) with the same reversed-phase HPLC column. Purified 1 (4 mg) was eluted at 37 min. 2 was also further chromatographed for purification with a gradient solvent system (55–77% CH3CN-H2O gradient solvent system for 40 min, flow rate 1 mL/min, and UV detection at 254 nm) with the same reversed-phase HPLC column. Purified 2 (2 mg) was eluted at 29 min.
3.4.1. Ligiamycin A (1)
White powder; − 78 (c = 0.1, MeOH); UV(MeOH) λmax (log ε) 250 (3.80), 351 (2.31) nm; IR (neat) νmax 3387, 2959, 1729, 1648, 1516, 1463, 1369, and 1055 cm−1; for 1H and 13C NMR spectral data, Table 1; HR-ESI-MS [M + H]+ m/z 317.18615 (calcd. for C18H25N2O3, 317.18597).
3.4.2. Ligiamycin B (2)
White powder; − 67 (c = 0.1, MeOH); UV(MeOH) λmax (log ε) 250 (3.38), 352 (1.92) nm; IR (neat) νmax 3386, 2923, 1724, 1651, 1515, 1463, 1356, and 1036 cm−1; for 1H and 13C NMR spectral data, Table 1; HR-FAB-MS [M + H]+ m/z 333.1816 (calcd. for C18H25N2O4, 333.1814).
3.5. ECD and Optical Rotation Calculation
The ECD calculation was performed as described previously [20]. The possible conformers of ligiamycins A (1) and B (2) were obtained based on the detailed analysis of ROESY spectroscopic data, and the energy-minimized conformational structures were calculated by Avogadro 1.2.0 [21]. All DFT calculations for ground state geometries of the ligiamycins were acquired by Turbomole 4.3.2. (TmoleX) [22] with the def-SVP set in all atoms at the density functional theory (DFT) (Table S2). ECD data of ligiamycins were obtained by overlapping each transition, where σ is the width of the band at 1/e height. ΔEi and Ri are the rotary strengths and excitation energies for transition i, respectively. For this study, σ was fixed at 0.10 Ev (Table S3). The calculated optical rotations of 1 and 2 corresponding to the optimized structures were obtained by TmoleX using DFT with functional B3-LYP and the basis set of def-SV(P) via TmoleX.
3.6. Cell Culture
All the cancer cell lines (SNU-638, SK-HEP-1, A549, HCT116, and MDA-MB-231 cancer cells) were obtained from the American Type Culture Collection (Manassas, VA, USA). SNU-638, A549, and HCT116 cells were cultured in Roswell Park Memorial Institute 1640 medium, and SK-HEP-1 and MDA-MB-231 cells were cultured in Dulbecco’s Modified Eagle’s medium. All media were supplemented with a penicillin–streptomycin mixture (10,000 units/mL sodium penicillin G and 10,000 μg/mL streptomycin) and 10% fetal bovine serum. They were incubated in a humidified incubator that contained 5% CO2 at 37 °C.
3.7. Cell Proliferation Assays
A sulforhodamine B (SRB) assay was performed for cell proliferation as described previously [23]. Cancer cells were seeded in 96-well plates and then incubated for 72 h with treatments of the ligiamycins. After 72 h, 10% trichloroacetic acid was used to fix the cells (30 min) and then washed with deionized water and dried overnight. Proteins were stained using a 0.4% SRB solution in 1% acetic acid; unbound dye was cleared using 1% acetic acid. After dissolving unbound dye with 10 mM Tris buffer (pH = 10.0), the absorbance of the cell solution was measured by a Versamax ELISA instrument (515 nm, Molecular Devices, LLC, San Jose, CA, USA). TableCurve 2D v5.01 software (Systant Software Inc., Richmond, CA, USA) was utilized to determine the IC50 values.
3.8. Antibacterial Activity Bioassays
The inhibitory activities of ligiamycins were tested against Gram-positive and Gram-negative bacteria in a similar way as described previously [24,25]. Gram-positive and Gram-negative bacteria were cultured in Mueller Hinton Broth (MHB) overnight. The cells were centrifuged and washed twice with sterilized distilled water. Ligiamycins A (1) and B (2) were dissolved in dimethyl sulfoxide (DMSO) separately and diluted with MHB for the purpose of preparing serial twofold dilutions in the range of 0.06–128 μg/mL. In each well of a 96-well plate, 190 μL of MHB containing the test compound (each 1 and 2) was mixed with 10 μL of broth containing the test bacterium (final concentration: 5 × 105 colony-forming units (cfu)/mL) adjusted to match the turbidity of a 0.5 MacFarland standard. The plates were incubated at 37 °C for 24 h. The MIC values were decided as the lowest concentration of the test compound that prevented cell growth. Ampicillin and tetracycline were used as control compounds.
4. Conclusions
The co-cultivation of marine wharf roach gut bacterial strains Streptomyces sp. GET02.ST and Achromobacter sp. GET02.AC induced the production of secondary metabolites from Streptomyces sp. GET02.ST by 24 times, leading to the discovery and structure determination of two new natural products, ligiamycins A (1) and B (2). The configurations of 1 and 2 were established by analyses of ROESY correlations and calculated optical rotations. Ligiamycins A (1) and B (2) displayed a unique structural feature by coupling a decalin with an amino maleimide moiety. Oxasetin, which was discovered from the fungus Vaginatispora aquatica, is structurally the most similar by bearing decalin and maleimide [26]. Dysidinoid A and the oxaleimides also have related structures to decalin and maleimide [27,28]. However, these compounds have hydroxy maleimide or different substitution patterns to decalin. The amino-maleimide substructure is extremely rare in nature. The only previously reported natural products with this moiety are hyperectine and isohyperectine from the plants Hypecoum erectum and Hypecoum leptocapum [29,30,31]. Therefore, ligiamycins A (1) and B (2) are the first amino-maleimide-bearing compounds from microorganisms. In addition, thus far, the combinations of decalin and maleimide have been reported only from fungi, highlighting that ligiamycins A (1) and B (2) are the first bacterial decalin–maleimide metabolites. Based on the structural similarity to oxaleimides, we proposed that ligiamycins were putatively biosynthesized through a polyketide synthase–nonribosomal peptide synthetase (PKS-NRPSs) hybrid pathway (Scheme S1) [28]. Ligiamycins A (1) and B (2) are biologically active with antibacterial activity and a weak cytotoxic effect against the tested cancer cell lines. It is still unclear how Achromobacter sp. GET02.AC elicits the production of the ligiamycins from Streptomyces sp. GET02.ST. Based on the previous reports regarding other co-cultures [32], cell–cell contact [4] or small molecule inducers [33] could be the eliciting mechanisms, but elucidating the role of Achromobacter sp. GET02.AC in this co-culture requires further comprehensive studies. Our finding about the ligiamycins demonstrates that the co-cultivation of ecologically relevant microbes enhances the production of otherwise neglected minor but structurally unique metabolites and provides an effective approach to discovering new bioactive compounds. Furthermore, utilizing the gut microflora of chemically under-investigated wharf roaches for exploring new chemical entities indicates that unexplored marine invertebrates such as wharf roaches could serve as sources to expand microbial chemical diversity.
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/md20020083/s1, Figures S1–S14: NMR spectra of ligiamycins; Figure S15: HR-ESI-MS data of ligiamycin A (1); Figure S16: HR-FAB-MS data of ligiamycin B (2); Figure S17: Energy-minimized conformations of (a) ligiamycin A (1) and (b) ligiamycin B (2); Figure S18: LC/MS traces (ion counts) of ion extraction for the ion [M-H]− at m/z 331 (ligiamycin B); Figures S19 and S20: Comparison of the experimental ECD data of ligiamycins with the calculated ECD data; Table S1: Composition of isolation agar media; Table S2: Cartesian coordinates of ligiamycins A (1) and B (2); Table S3: ECD calculations of ligiamycins A (1) and B (2); Scheme S1: Proposed biosynthetic pathway of the ligiamycins.
Author Contributions
Conceptualization, H.-J.L., J.S.A. and D.-C.O.; methodology, H.-J.L., J.S.A. and D.-C.O.; software, H.-J.L., J.S.A. and D.-C.O.; validation, H.-J.L., J.S.A. and D.-C.O.; formal analysis, H.-J.L., J.S.A., S.H., and D.-C.O.; investigation, H.-J.L., J.S.A., S.H. and D.-C.O.; resources, H.-J.L.; data curation, H.-J.L., J.S.A., E.S.B., E.C., S.H., S.-J.N., K.-B.O., S.K.L. and D.-C.O.; writing—original draft preparation, H.-J.L., J.S.A. and D.-C.O.; writing—review and editing, H.-J.L., J.S.A., E.S.B., E.C., S.-J.N., K.-B.O., S.K.L. and D.-C.O.; visualization, H.-J.L., J.S.A. and D.-C.O.; supervision, H.-J.L., J.S.A. and D.-C.O.; project administration, D.-C.O.; funding acquisition, S.-J.N., S.K.L. and D.-C.O. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by the National Research Foundation of Korea grants funded by the Korean Government (Ministry of Science and ICT) (2021R1A4A2001251 and 2020R1A2C2003518). This work was also supported by the Collaborative Genome Program of the Korea Institute of Marine Science and Technology Promotion (KIMST) funded by the Ministry of Oceans and Fisheries (MOF) (No. 20180430).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
All data are contained within this article and Supplementary Materials.
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Carroll A.R., Copp B.R., Davis R.A., Keyzers R.A., Prinsep M.R. Marine natural products. Nat. Prod. Rep. 2021;38:362–413. doi: 10.1039/D0NP00089B. [DOI] [PubMed] [Google Scholar]
- 2.Chen J., Zhang P., Ye X., Wei B., Emam M., Zhang H., Wang H. The structural diversity of marine microbial secondary metabolites based on co-culture strategy: 2009–2019. Mar. Drugs. 2020;18:449. doi: 10.3390/md18090449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cueto M., Jensen P.R., Kauffman C., Fenical W., Lobkovsky E., Clardy J. Pestalone, a new antibiotic produced by a marine fungus in response to bacterial challenge. J. Nat. Prod. 2001;64:1444–1446. doi: 10.1021/np0102713. [DOI] [PubMed] [Google Scholar]
- 4.Oh D.-C., Jensen P.R., Kauffman C.A., Fenical W. Libertellenones A–D: Induction of cytotoxic diterpenoid biosynthesis by marine microbial competition. Bioorg. Med. Chem. 2005;13:5267–5273. doi: 10.1016/j.bmc.2005.05.068. [DOI] [PubMed] [Google Scholar]
- 5.Oh D.-C., Kauffman C.A., Jensen P.R., Fenical W. Induced production of emericellamides A and B from the marine-derived fungus Emericella sp. in competing co-culture. J. Nat. Prod. 2007;70:515–520. doi: 10.1021/np060381f. [DOI] [PubMed] [Google Scholar]
- 6.Adnani N., Vazquez-Rivera E., Adibhatla S.N., Ellis G.A., Braun D.R., Bugni T.S. Investigation of interspecies interactions within marine Micromonosporaceae using an improved co-culture approach. Mar. Drugs. 2015;13:6082–6098. doi: 10.3390/md13106082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Adnani N., Chevrette M.G., Adibhatla S.N., Zhang F., Yu Q., Braun D.R., Nelson J., Simpkins S.W., McDonald B.R., Myers C.L., et al. Coculture of marine invertebrate-associated bacteria and interdisciplinary technologies enable biosynthesis and discovery of a new antibiotic, keyicin. ACS Chem. Biol. 2017;12:3093–3102. doi: 10.1021/acschembio.7b00688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shin D., Byun W.S., Moon K., Kwon Y., Bae M., Um S., Lee S.K., Oh D.-C. Coculture of marine Streptomyces sp. with Bacillus sp. produces a new piperazic acid-bearing cyclic peptide. Front. Chem. 2018;6:498. doi: 10.3389/fchem.2018.00498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hifnawy S.M., Hassan H.M., Mohammed R., Fouda M.M., Sayed A.M., Hamed A.A., AbouZid S.F., Rateb M.E., Alhadrami H.A., Abdelmohsen U.R. Induction of antibacterial metabolites by co-cultivation of two red-sea-sponge-associated actinomycetes Micromonospora sp. UR56 and Actinokinespora sp. EG49. Mar. Drugs. 2020;18:243. doi: 10.3390/md18050243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang L., Niaz S.I., Khan D., Wang Z., Zhu Y., Zhou H., Lin Y., Li J., Liu L. Induction of diverse bioactive secondary metabolites from the mangrove endophytic fungus Trichoderma sp. (Strain 307) by co-cultivation with Acinetobacter johnsonii (Strain B2) Mar. Drugs. 2017;15:35. doi: 10.3390/md15020035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rizzo C., Lo Giudice A. Marine invertebrates: Underexplored sources of bacteria producing biologically active molecules. Diversity. 2018;10:52. doi: 10.3390/d10030052. [DOI] [Google Scholar]
- 12.Kwon D.H. Terrestrial Isopoda (Crustacea) from Korea. Korean J. Syst. Zool. 1993;36:133–158. [Google Scholar]
- 13.Li X., Zhao Z., Ding W., Ye B., Wang P., Xu J. Aspochalazine A, a novel polycyclic aspochalasin from the fungus Aspergillus sp. Z4. Tetrahedron Lett. 2017;58:2405–2408. doi: 10.1016/j.tetlet.2017.04.071. [DOI] [Google Scholar]
- 14.Xu J., Zhao S., Yang X. A new cyclopeptide metabolite of marine gut fungus from Ligia oceanica. Nat. Prod. Res. 2014;28:994–997. doi: 10.1080/14786419.2014.902945. [DOI] [PubMed] [Google Scholar]
- 15.Li X., Ding W., Wang P., Xu J. Two novel aspochalasins from the gut fungus Aspergillus sp. Z4. Mar. Drugs. 2018;16:343. doi: 10.3390/md16100343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li X.-C., Ferreira D., Ding Y. Determination of absolute configuration of natural products: Theoretical calculation of electronic circular dichroism as a tool. Curr. Org. Chem. 2010;14:1678–1697. doi: 10.2174/138527210792927717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Polavarapu P.L. Optical rotation: Recent advances in determining the absolute configuration. Chirality. 2002;14:768–781. doi: 10.1002/chir.10145. [DOI] [PubMed] [Google Scholar]
- 18.Stephens P.J., Pan J.J., Devlin F.J., Cheeseman J.R. Determination of the absolute configurations of natural products using TDDFT optical rotation calculations: The iridoid oruwacin. J. Nat. Prod. 2008;71:285–288. doi: 10.1021/np070502r. [DOI] [PubMed] [Google Scholar]
- 19.Taniguchi T., Martin C.L., Monde K., Nakanishi K., Berova N., Overman L.E. Absolute configuration of actinophyllic acid as determined through chiroptical data. J. Nat. Prod. 2009;72:430–432. doi: 10.1021/np800665s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bae M., An J.S., Hong S.-H., Bae E.S., Chung B., Kwon Y., Hong S., Oh K.-B., Shin J., Lee S.K., et al. Donghaecyclinones A–C: New cytotoxic rearranged angucyclinones from a volcanic island-derived marine Streptomyces sp. Mar. Drugs. 2020;18:121. doi: 10.3390/md18020121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hanwell M.D., Curtis D.E., Lonie D.C., Vandermeersch T., Zurek E., Hutchison G.R. Avogadro: An advanced semantic chemical editor, visualization, and analysis platform. J. Cheminform. 2012;4:17. doi: 10.1186/1758-2946-4-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Balasubramani S.G., Chen G.P., Coriani S., Diedenhofen M., Frank M.S., Franzke Y.J., Furche F., Grotjahn R., Harding M.E., Hättig C., et al. TURBOMOLE: Modular program suite for ab initio quantum-chemical and condensed-matter simulations. J. Chem. Phys. 2020;152:184107. doi: 10.1063/5.0004635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Moon K., Cui J., Kim E., Riandi E.S., Park S.H., Byun W.S., Kal Y., Park J.Y., Hwang S., Shin D., et al. Structures and biosynthetic pathway of pulvomycins B–D: 22-membered macrolides from an estuarine Streptomyces sp. Org. Lett. 2020;22:5358–5362. doi: 10.1021/acs.orglett.0c01249. [DOI] [PubMed] [Google Scholar]
- 24.An J.S., Hong S.-H., Somers E., Lee J., Kim B.-Y., Woo D., Kim S.W., Hong H.-J., Jo S.-I., Shin J., et al. Lenzimycins A and B, metabolites with antibacterial properties from Brevibacillus sp. associated with the dung beetle Onthophagus lenzii. Front. Microbiol. 2020;11:599911. doi: 10.3389/fmicb.2020.599911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kim D.-G., Moon K., Kim S.-H., Park S.-H., Park S., Lee S.K., Oh K.-B., Shin J., Oh D.-C. Bahamaolides A and B, antifungal polyene polyol macrolides from the marine actinomycete Streptomyces sp. J. Nat. Prod. 2012;75:959–967. doi: 10.1021/np3001915. [DOI] [PubMed] [Google Scholar]
- 26.He H., Janso J.E., Yang H.Y., Singh M.P., Bernan V.S., Greenstein M., Carter G.T. Oxasetin, a new antibacterial polyketide produced by fungus Vaginatispora aquatica, HK1821. J. Antibiot. 2002;55:821–825. doi: 10.7164/antibiotics.55.821. [DOI] [PubMed] [Google Scholar]
- 27.Jiao W.-H., Li J., Liu Q., Xu T.-T., Shi G.-H., Yu H.-B., Yang F., Han B.-N., Li M., Lin H.-W. Dysidinoid A, an unusual meroterpenoid with anti-MRSA activity from the south China sea sponge Dysidea sp. Molecules. 2014;19:18025–18032. doi: 10.3390/molecules191118025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sato M., Dander J.E., Sato C., Hung Y.-S., Gao S.-S., Tang M.-C., Hang L., Winter J.M., Garg N.K., Watanabe K., et al. Collaborative biosynthesis of maleimide- and succinimide-containing natural products by fungal polyketide megasynthases. J. Am. Chem. Soc. 2017;139:5317–5320. doi: 10.1021/jacs.7b02432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Perel’son M.E., Aleksandrov G.G., Yakhontova L.D., Tolkachev O.N., Fesenko D.A., Komarova M.N., Esipov S.E. Alkaloids of Hypecoum erectum, the structure of hyperectine. Chem. Nat. Compd. 1984;20:592–598. doi: 10.1007/BF00580073. [DOI] [Google Scholar]
- 30.Yakhontova L.D., Yartseva I.V., Klyuev N.A., Tolkachev O.N. Structure of isohyperectine—An alkaloid from Hypecoum erectum. Chem. Nat. Compd. 1993;29:744–747. doi: 10.1007/BF00629644. [DOI] [Google Scholar]
- 31.Zhang G.-L., Rücker G., Breitmaier E., Mayer R. Alkaloids from Hypecoum leptocarpum. Phytochemistry. 1995;40:1813–1816. [Google Scholar]
- 32.Shank E.A., Kolter R. New developments in microbial interspecies signaling. Curr. Opin. Microbiol. 2009;12:205–214. doi: 10.1016/j.mib.2009.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rigali S., Titgemeyer F., Barends S., Mulder S., Thomae A.W., Hopwood D.A., van Wezel G.P. Feast or famine: The global regulator DasR links nutrient stress to antibiotic production by Streptomyces. EMBO Rep. 2008;9:670–675. doi: 10.1038/embor.2008.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data are contained within this article and Supplementary Materials.