Abstract
The TSC13/YDL015c gene was identified in a screen for suppressors of the calcium sensitivity of csg2Δ mutants that are defective in sphingolipid synthesis. The fatty acid moiety of sphingolipids in Saccharomyces cerevisiae is a very long chain fatty acid (VLCFA) that is synthesized by a microsomal enzyme system that lengthens the palmitate produced by cytosolic fatty acid synthase by two carbon units in each cycle of elongation. The TSC13 gene encodes a protein required for elongation, possibly the enoyl reductase that catalyzes the last step in each cycle of elongation. The tsc13 mutant accumulates high levels of long-chain bases as well as ceramides that harbor fatty acids with chain lengths shorter than 26 carbons. These phenotypes are exacerbated by the deletion of either the ELO2 or ELO3 gene, both of which have previously been shown to be required for VLCFA synthesis. Compromising the synthesis of malonyl coenzyme A (malonyl-CoA) by inactivating acetyl-CoA carboxylase in a tsc13 mutant is lethal, further supporting a role of Tsc13p in VLCFA synthesis. Tsc13p coimmunoprecipitates with Elo2p and Elo3p, suggesting that the elongating proteins are organized in a complex. Tsc13p localizes to the endoplasmic reticulum and is highly enriched in a novel structure marking nuclear-vacuolar junctions.
The sphingolipids are essential components of eukaryotic cells that have been implicated in a large number of cellular processes, including signaling, secretion, Ca2+ homeostasis, and heat stress response (46). Sphingolipids consist of ceramide linked through either a glucosyl or phosphodiester bond to a polar head group. The ceramide moiety is comprised of a fatty acid joined in amide linkage to a long-chain base (LCB). In Saccharomyces cerevisiae, the fatty acid is a hydroxylated C26 very long chain fatty acid (VLCFA), the LCB is usually phytosphingosine, and the polar head group is an inositolphosphoryl moiety that can be further decorated by mannosylation and a second inositolphosphorylation (Fig. 1; for a review, see reference 13). The importance of the VLCFAs is highlighted by the observation that mutants unable to synthesize LCBs are inviable, but growth can be restored by second-site mutations in the SLC1 gene that result in the synthesis of a novel class of VLCFA-containing inositolglycerophospholipids (36), which structurally mimic sphingolipids.
FIG. 1.
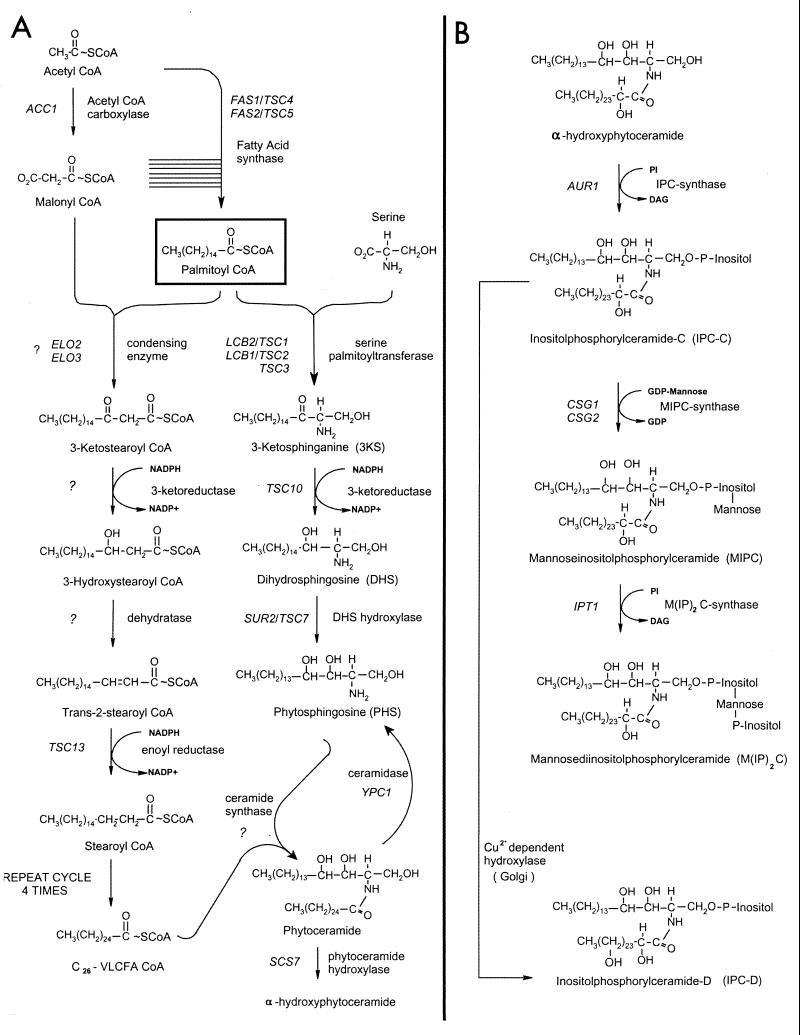
Pathways of fatty acid elongation, LCB, and sphingolipid synthesis in S. cerevisiae. Palmitoyl-CoA is synthesized from acetyl-CoA and malonyl-CoA by soluble FAS. Palmitoyl-CoA is elongated to a C26 VLCFA by a membrane-associated fatty acid elongating system (A, left branch). Each cycle of elongation requires four successive reactions and lengthens the growing fatty acid by two carbon units; condensation of malonyl-CoA with the acyl-CoA substrate, reduction of the 3-ketoacyl-CoA, dehydration of the 3-hydroxyacyl-CoA, and reduction of the trans-2,3-acyl-CoA. Although the intermediates and the product of the elongation cycle are shown as CoA derivatives, this has not yet been experimentally confirmed. The organization of the elongating enzymes with respect to each other is unknown. The LCBs are synthesized by the pathway shown in the right branch of panel A; the pathway for the conversion of ceramide to mature sphingolipids is shown in panel B.
The majority of cellular long-chain fatty acids have 16 or 18 carbons and are synthesized by the soluble fatty acid synthase (FAS) complex that is comprised of two multifunctional subunits encoded by the FAS2 (α-subunit) and FAS1 (β-subunit) genes (28, 52, 53, 65). While the bulk of the cellular fatty acids are synthesized by FAS, the VLCFAs are synthesized by membrane-associated fatty acid elongating systems. Although fatty acid elongation has been extensively assayed in microsomal fractions from mammalian cells, the component enzymes have resisted purification, and until recently none of the genes encoding the enzymes had been identified (reviewed in reference 7). The elongating systems catalyze four reactions, lengthening the fatty acid by two carbons in each cycle of elongation (Fig. 1). Malonyl coenzyme A (malonyl-CoA), synthesized by acetyl-CoA carboxylase (ACC), provides the 2-carbon unit both for de novo long-chain fatty acid synthesis by FAS and for elongation of the long-chain fatty acids to the VLCFAs (Fig. 1). In contrast to acc1 null mutants (21), strains defective in the FAS1 or FAS2 gene can be rescued by supplementation of the growth medium with long-chain fatty acids (44). This indicates that malonyl-CoA is required for an essential process other than de novo long-chain fatty acid synthesis, most likely the elongation of the long-chain fatty acids to the VLCFAs. Conditional mutations of acc1 result in severe membrane phenotypes at the nuclear envelope-nuclear pore complex under restrictive conditions (48) and also affect vacuolar membrane morphology and inheritance, possibly through altered fatty acylation of the vacuolar membrane protein Vac8p (47).
Despite the ubiquitous presence of VLCFAs and the evidence that they are essential for viability, little is known about the precise roles of the VLCFAs or about the molecular nature of the fatty acid elongating enzymes. Candidates for yeast cells defective in the elongating system were identified in two independent screens. These screens took advantage of the observation that fas2 mutants will grow in medium supplemented with myristate (C14) as long as this fatty acid can be converted to the long-chain (C16 and C18) and very long chain (typically C26) fatty acids by the elongating systems. An additional mutation in the fas2 background rendering cells unable to grow on myristate but able to grow on palmitate (C16) identified the ELO1 gene (14, 59). Subsequently, Elo1p was demonstrated to be required for efficient elongation of myristate to palmitate. The ELO1 gene has two structural and functional homologs in S. cerevisiae, FEN1/ELO2 and SUR4/ELO3. Elo2p is involved in elongation of fatty acids up to C22 and C24, while Elo3p has a broader substrate specificity and is required for conversion of C24 to C26 (37). Neither Elo2p nor Elo3p confers essential functions by itself; however, elo2Δ elo3Δ double mutants are inviable (37).
The yeast ELO3/SUR4 and/or ELO2/FEN1 genes have also been identified in a number of other genetic screens (17, 19, 56), including one for suppressors of the Rvs (reduced viability upon starvation) phenotype of cells lacking the RVS161 gene (12). Rvs161p is homologous to amphiphysin, a vesicle-associated protein that participates in endocytosis in mammalian cells (10, 57, 58), and a mutant allele of rvs161 was recovered in a screen for endocytic mutants in S. cerevisiae (35). Interestingly, inactivation of the ELO2 or ELO3 gene also bypasses the requirement for Snc V-SNAREs, further suggesting a role for VLCFAs in membrane trafficking (11).
VLCFAs are predominantly present in sphingolipids and the phosphatidylinositol (PtdIns) moiety of glucosylphosphatidylinositol (GPI)-anchored proteins. Sphingolipids in yeast are assembled in the endoplasmic reticulum (ER) and then modified and matured in the Golgi; they are major lipid constituents of the plasma membrane (22). The csg1 and csg2 mutants are defective in sphingolipid synthesis at the step of inositolphosphorylceramide (IPC) mannosylation (Fig. 1); they therefore accumulate high levels of IPC, which is correlated with sensitivity to 10 mM Ca2+ in the growth medium (3, 68). Many mutations that suppress the Ca2+ sensitivity of the csg2 mutant cells reside in genes required for IPC synthesis (15). Interestingly, mutations that suppress the multiple phenotypes of the rvs161Δ mutant (named sur mutants) (12, 43, 56) identify genes that overlap extensively with the collection of sphingolipid synthesis mutants that were identified as suppressors of the csg2 Ca2+ sensitivity. The SUR1 gene is allelic to CSG1 (3). The SUR2 gene encodes the hydroxylase that converts dihydrosphingosine to phytosphingosine; therefore, sur2 mutants synthesize ceramides and sphingolipids with dihydrosphingosine rather than phytosphingosine as the LCB (20) (Fig. 1). The sur4/elo3 mutant is deficient in sphingolipid synthesis because it makes reduced levels of VLCFA (11, 14, 37, 59). We identified both the SUR4/ELO3 and FEN1/ELO2 genes in the csg2 suppressor screen because null mutations in either gene reduce IPC accumulation and thereby reverse Ca2+ sensitivity of the csg2Δ mutant. This observation suggested that other genes required for fatty acid elongation might be identified in our suppressor collection.
In this report, we identify and characterize the TSC13 gene and provide evidence that it encodes the enoyl reductase component of the elongating system required for the synthesis of VLCFAs. Tsc13p interacts physically and genetically with other components of the elongation machinery and is localized in the ER but is highly enriched at the interface between the nucleus and the vacuole, marking a novel subdomain of the ER, the nuclear-vacuolar junction.
MATERIALS AND METHODS
Media, strains, and genetic manipulations.
The yeast strains used in this study are listed in Table 1. Yeast media were prepared and cells were grown according to standard procedures (54).
TABLE 1.
Yeast strains used in this study
| Strain | Genotype |
|---|---|
| TDY2037 | matα lys2 ura3-52 trp1Δ leu2Δ |
| TDY2038 | matα csg2::LEU2 lys2 ura3-52 trp1Δ leu2Δ |
| TDY2050 | mata tsc13-1 csg2::LEU2 ade2-101 ura 3-52 trp1Δ leu2Δ |
| TDY2055 | mata elo2::URA3 csg2::LEU2 his4-619 ura3-52 trp1Δ leu2Δ lys2 |
| TDY2056 | mata elo3::URA3 csg2::LEU2 his4-619 ura3-52 trp1Δ leu2Δ lys2 |
| TDY2051 | matα tsc13-1 ade2-101 ura3-52 trp1Δ leu2Δ lys2 |
| TDY2053 | mata elo2::URA3 his4-619 ura3-52 trp1Δ leu2Δ |
| TDY2054 | mata elo3::URA3 his4-619 ura3-52 trp1Δ leu2Δ |
| TDY2057 | matα elo2::TRP1 tsc13-1 ade2-101 ura3-52 trp1Δ leu2Δ lys2 |
| TDY2058 | matα elo3::TRP1 tsc13-1 ade2-101 ura3-52 trp1Δ leu2Δ |
| FY1679a | mata/α ura3-52/ura3-52 leu2Δ1/+trp1Δ63/+his3Δ200/+ |
| DTY10A | mata leu2-3,112 can1-100 ura3-1 ade2-1 his3-11,15 (TRP1+) |
| DTY004 | mata elo2::HIS3 leu2-3, 112 can1-100 ura3-1 ade2-1 |
| SEY002b | matα acc1ts (mtr7) lys? leu2 ade2 his3 |
| SEY003c | matα aac1csleu2-3,112 ura3-1 ade2-1 his3-11,15 can1-100 |
| SEY004d | mata acc1-2150 lys? leu2 ade2 ura3 |
| BJ9367e | matα pep3::TRP1 ura3-52 leu2-Δ1 his3-Δ200 trp1 |
| RW9065f | mata ade2 his3-Δ200 lys2-801 ura3-52 met? {pGAL1-HMG1} |
| CSY10A | mata elo3::HIS3 leu2-3,112 can1-10 ura3-1 ade2-1 |
| CSY10Aelo2/HA-ELO | mata elo2::HIS3 can1-100 ura3-1 ade2-1 {pADH1-HA-ELO2-LEU2} |
| CSY10Aelo3/HA-ELO3 | mata elo3::HIS3 can1-100 ura3-1 ade2-1 {pADH1-HA-ELO3-LEU2} |
| CSY10Aelo2/GFP-ELO2 | mata elo2::HIS3 can1-100 ura3-1 ade2-1 {pGAL1-GFP-ELO2-LEU2} |
Provided by J. Hegemann.
Provided by A. Tartakoff.
Provided by H. Klein.
Provided by E. Schweizer.
Provided by E. Jones.
Provided by R. Wright.
Cloning and disruption of the TSC13 gene.
The wild-type TSC13 gene was cloned from a YCp50-based genomic library (45) based on its ability to complement the ts (temperature-sensitive) phenotype of TDY2050 (tsc13-1 csg2::LEU2) (2). Two plasmids that conferred temperature resistance were recovered, and sequence analysis showed that they shared a segment of chromosome IV containing YDL015c and YDL016c. A 1,500-bp NotI and SalI-ended PCR fragment containing only the YDL015c open reading frame with 370 bp of upstream and 205 bp of downstream flanking sequence was generated using primers 12200 and 12201 (Table 2). The fragment was cloned into pRS316 (55), and the resulting plasmid (pTSC13-316) was found to complement the ts phenotype of TDY2050. A disrupting allele of YDL015c/TSC13 was constructed by placing an EcoRI-ended PCR fragment carrying the TRP1-selectable marker between the MunI (located 42 bp upstream of the start codon) and EcoRI (located in codon 72) sites of YDL015c.
TABLE 2.
Sequences of oligonucleotides used for cloning, tagging, and disrupting TSC13, ELO2, and ELO3
| Oligonucleotide | Sequencea |
|---|---|
| 12201 | 5′-GGCCGCGGCCGCGGGCAACTTAGATTTATCCG-3′ |
| 12200 | 5′-GGCCGTCGACGTACTTACATATTTGCATAGT-3′ |
| 12449 | 5′-TTTGAATTTAATTTGAAA-ATGCCTAGGATCACCATAAAAAGC-3′ |
| 12450 | 5′-GCTTTTTATGGTGATCCTAGGCATTTTCAAATTAAATTCAAA-3′ |
| ELO2F-SAL1 | 5′-CGCAGGTCGACCATGAATTCACTCGTTACTC-3′ |
| ELO2R | 5′-TTCACACAGGAAACAG-3′ |
| ELO3F-SAL1 | 5′-CGCAGGTCGACCATGAACACTACCACATCTAC-3′ |
| ELO3R | 5′-GTCCTGCGATTTTATTCG-3′ |
| 9041 | 5′-GGCCGGATCCTCCAGACTGTGAATAAAC-3′ |
| 9042 | 5′-GGCCGGATCCGTATCACTACTCCTCACTGT-3′ |
| 9043 | 5′-GGCCGGATCCGTACGTATTCACATGTCCTG-3′ |
| 9044 | 5′-GGCCGGATCCTAGACATGACTGTCGAAAGG-3′ |
Restriction sites referred to in the text are underlined.
Epitope tagging of Tsc13p, Elo2p, and Elo3p.
A three-copy Myc epitope (Myc3) was introduced at the amino terminus of Tsc13p. To accomplish the tagging, an AvrII site was first introduced after the start codon of TSC13 on plasmid pTSC13-316 by QuikChange mutagenesis (Stratagene, La Jolla, Calif.), using the complementary mutagenic primers 12449 and 12450 (Table 2). A SpeI-ended fragment carrying the Myc3 cassette was generated by PCR using a Bluescript-based Myc3-containing plasmid (gift of Dan TerBush, Department of Biochemistry, Uniformed Services University of the Health Sciences) and ligated into the AvrII site. The NotI-to-XhoI fragment carrying the Myc-tagged TSC13 allele was ligated into pRS426 to generate the MYC-TSC13-426 plasmid. This Myc-tagged TSC13 allele complemented the temperature sensitivity of the tsc13 mutant, demonstrating that it is functional.
The hemagglutinin epitope (HA)-tagged Elo3p and HA-tagged Elo2p constructs were made by inserting SalI-ended restriction fragments extending from the start codon of Elo2p (or Elo3p) to a site 128 bp (for ELO2) or 1,115 bps (for ELO3) downstream of the stop codon into the SalI site of plasmid pADH-HA. The SalI fragments were generated using the PCR primers listed in Table 2 with vector pCRELO2 (37) as template for ELO2 and strain DTY10A (Table 1) genomic DNA for ELO3. The resulting plasmids, pADH-HA-ELO2 and pADH-HA-ELO3, contained the full-length ELO genes with an HA tag fused at the amino terminus, under control of the ADH1 promoter. The HA-tagged Elo2p and Elo3p restored normal VLCFA synthesis to the elo2 and elo3 mutants, respectively.
Disrupting the ELO2 and ELO3 genes.
A BamHI-ended PCR fragment extending from 280 bp upstream of the start codon of ELO3 to 180 bp past the stop codon was generated using primers 9041 and 9042 (Table 2) and ligated into pUC19. The resulting plasmid was digested with MscI to release a 140-bp fragment (encoding codons 45 to 91 of ELO3), and an XhoI linker was ligated in at the deletion junction. A SalI-ended TRP1 fragment was ligated into the XhoI site to generate the disrupting allele, which was liberated from plasmid pUC19 by digestion with KpnI and SalI. For the ELO2 gene, a BamHI-ended PCR fragment extending from 160 bp upstream of the start codon to 170 bp past the stop codon was generated using primers 9043 and 9044 (Table 2) and ligated into pUC19. An EcoRI-ended TRP1 fragment was ligated between the MunI sites of ELO2, thereby replacing codons 92 to 183 with the selectable marker. The disrupting allele was liberated from the plasmid by digestion with KpnI and SalI.
Ceramide and LCB analysis.
Ceramides were extracted and analyzed by thin-layer chromatography (TLC) as previously described (20). LCBs were extracted, separated by TLC, and visualized using ninhydrin as described elsewhere (2).
Fatty acid analysis.
Cells (5 × 108) in mid-logarithmic-stage growth were harvested and resuspended in 100 μl of distilled H2O along with 25 μg of linoleic acid (18:2) as an internal standard for extraction of the fatty acids. Fatty acid methyl esters were prepared by HCl methanolysis as previously described (59), resuspended in 50 μl of hexane, and stored at −20°C. Gas chromatography (GC) was performed using a Varian 3400CX chromatograph and a Supelcowax TM10 column, and data were collected and analyzed using Class-VP Chromatography Data System version 4.1 software (Shimadzu Scientific Instruments) as described elsewhere (6).
Elongase assays.
Microsomes were prepared from the wild-type or tsc13-1 mutant cells as previously described (18). By using the purified microsomes for the elongation assays, the background of acyl-CoA-independent incorporation of malonyl-CoA into fatty acids (as a result of the soluble FAS activity) was reduced to less than 10% of the acyl-CoA-dependent incorporation of malonyl-CoA into fatty acids. Total elongase activity was measured in a volume of 200 μl containing 50 mM Tris (pH 7.5), 1 mM MgCl2, 150 μM Triton X-100, 1 mM NADPH, 1 mM NADH, 10 mM β-mercaptoethanol, 40 μM acyl-CoA acceptor (either palmitoyl-CoA, stearoyl-CoA, or eicosanoyl-CoA), and 60 μM [2-14C]malonyl-CoA (0.05 μCi/ml) at 37°C. The reaction was initiated by the addition of 0.3 to 1.0 mg of microsomal protein. Protein concentrations were determined using the Bio-Rad protein assay reagent (Bio-Rad Laboratories, Hercules, Calif.). For assays of only the condensing activity, the NADPH and NADH were omitted. At various times, the reaction was terminated by adding 200 μl of 5 M KOH–10% methanol MeOH and heating at 80°C for 1 h. Following addition of 200 μl of 10 N H2SO4, fatty acids were recovered by two 1.5-ml extractions into hexane. The extracted fatty acids were resolved by silica gel TLC using hexane-diethyl ether-acetic acid (30:70:1) as the developing solvent. The radiolabeled fatty acids were detected and quantified using a PhosphorImager SI (Molecular Dynamics, Inc., Sunnyvale, Calif.).
Immunoprecipitation.
Microsomes were prepared from strains containing Tsc13p-Myc and either Elo2p-HA or Elo3p-HA as previously described (18). The microsomes were solubilized at 1 mg/ml with 2 mM sucrose monolaurate (Roche Diagnostics, Indianapolis, Ind.) for 10 min, and the high-speed (105 × g, 30 min) supernatant was collected. The supernatant (150 μl) was incubated with 3 μl of the precipitating antibody for 2 h and then with 20 μl of protein A-Sepharose (125 mg/ml; Sigma) for 2 h. The precipitates were washed three times with 600 μl of 50 mM HEPES (pH 7.5) and resuspended in 150 μl of sodium dodecyl sulfate (SDS) loading buffer; a 10-μl sample was subjected to SDS-polyacrylamide gel electrophoresis (PAGE) on an 8% gel. Following transfer of the separated proteins to nitrocellulose, the blots were blocked in 0.1 M Tris (pH 7.5)–0.15 M NaCl–0.1% Tween 20–5% dry milk. Tsc13p-Myc was detected with horseradish peroxidase (HRP)-conjugated monoclonal anti-Myc antibodies (from InVitrogen) at 1/5,000. Elo2p-HA and Elo3p-HA were detected using HRP-conjugated monoclonal anti-HA antibodies (from Boehringer Mannheim) at 1/1,000. The bound antibodies were detected by the ECL Western blotting detection system (Amersham Pharmacia Biotech).
Construction of Tsc13p-GFP by chromosomal fusion.
Plasmid pMK199-GA5-yEGFP-kanMX6 containing a five-copy glycine-alanine (GA5) linker fused to yeast enhanced green fluorescent protein (yEGFP) and the kanMX6 resistance marker, was used as the template for PCRs to amplify the integration cassette (64). Using the two primers pYDL015cGA5→ and pYDL015cMX6← (Table 3) a 2,550-bp fragment was generated by PCR. The upstream primer contained 30 nucleotides homologous to the GA5 linker 5′ to the yEGFP sequence of the template plasmid and 46 nucleotides corresponding to the 3′ end of the YDL015c coding sequence (excluding the stop codon). The downstream primer contained 26 nucleotides homologous to the kanMX6 sequence and 47 nucleotides homologous to the chromosomal sequence downstream of the YDL015c reading frame. PCR amplification was performed in a 25-μl standard reaction mix containing 1× Ex-Taq buffer (as specified by the supplier [Boehringer, Mannheim, Germany]), 300 ng of plasmid DNA, 100 pmol of each primer per μl, and 2.5 mM deoxynucleoside triphosphates. After the initial denaturation step at 94°C for 5 min, the PCR was started by the addition of 2 U of Takara Ex-Taq polymerase (Boehringer). The fragment was amplified during eight cycles of 30 s at 94°C, 30 s at 54°C, and 150 s at 72°C, 25 cycles of 30 s at 94°C and 180 s at 72°C, and a final elongation step of 12 min at 72°C. The resulting PCR fragment was purified by using a QIAquick PCR purification kit (Qiagen). About 0.5 to 1 μg of the PCR product was used for yeast transformation into diploid wild-type strain FY1679, and transformants were selected on plates containing Geneticin (G418; 200 μg/ml; Calbiochem). After restreaking on G418 plates, positive transformants were verified by colony PCR using the primers pYDL015c→, kanMX←, and pGFP4← (Table 3). Growth tests verified that the C-terminal chromosomal fusion of TSC13 with green fluorescent protein (GFP) rendered cells fully viable and phenotypically indistinguishable from wild-type cells.
TABLE 3.
Sequences of oligonucleotides used for constructing Tsc13p-GFP and Opi3p-GFP
| Oligonucleotide | Sequencea |
|---|---|
| pYDL015cGA5→ | 5′-caaaaagtatcataccagaagagcattcttgattccatttgtatttGGAGCAGGTGCTGGTGCTGGTGCTGGAGCA-3′ |
| pYDL015cMX6← | 5′-gctaatatctctttaccttgcatttgggcatgttgcaaacaggaggatcaATCGATGAATTCGAGCTCGTTTAAAC-3′ |
| pYDL015c→ | 5′-gtaacgctaagatccgtgtc-3′ |
| pOPI3GA5→ | 5′-ccttttactgccatgatctacgctaaccgtgataaggccaaaaagaatatgGGAGCAGGTGCTGGTGCTGGTGCTGGAGCA-3′ |
| pOPI3MX6← | 5′-cggtaatagcataggcttctaacattatagaatatatagaaatagagcactcaATCGATGAATTCGAGCTCGTTTAAAC-3′ |
| pOPI3→ | 5′-ggatgagagagtcaccggc-3′ |
| kanMX-reverse← | 5′-ctagcggatctgccggtagag-3′ |
| pGFP4← | 5′-ggtcaatttaccgtaagt-3′ |
Lowercase, coding gene sequence; italic, 3′ untranslated region gene sequence; uppercase lightface, GA5 linker sequence (30 bp); uppercase boldface, kanMX6 sequence (26 bp); underlined, stop codon.
Construction of Opi3p-GFP by chromosomal fusion.
The Opi3p-GFP (a bona fide ER protein; phospholipid-N-methyltransferase 24) chromosomal fusion was constructed and verified as described above for Tsc13p-GFP, using the primer pairs listed in Table 3. PCR products were transformed into the diploid wild-type strain FY1679, and transformants were selected on YPD plates containing Geneticin (200 μg/ml). After restreaking on G418 plates, positive transformants were verified by colony PCR using the primers pOP13→, kanMX←, and pGFP4← (Table 3).
Construction of Elo2p-GFP.
The GFP-fused Elo2p construct was made by inserting the SalI-flanked restriction fragment containing ELO2 (described above) into the SalI site of vector pGAL1-GFP. The resulting plasmid encoded a chimeric protein consisting of a GFP domain fused in frame to the amino terminus of the full-length Elo2 protein. The GFP-Elo2p fusion repaired the altered VLCFA phenotype associated with the elo2 mutant.
Fluorescence microscopy.
Microscopy was performed on a Leica TCS 4d confocal microscope equipped with an Ar-Kr laser, an acousto-optical tunable filter for laser wavelength selection and attenuation, a 500-nm beam splitter/dichroic mirror, and a 525/50-nm band-pass filter for GFP detection. For double-labeling experiments with the vacuolar dye FM4-64 [N](3-triethylammoniumpropyl)-4-(p-diethylaminophenylhexatrienyl)] pyridinium dibromide Molecular Probes, Eugene, Oreg.), an additional dichroic 525-nm beam splitter was used in the emission path, and 525/50-nm band-pass and 590-nm long-pass filters were used for simultaneous GFP and FM4-64 detection, respectively. The objective was a 100×/1.4-numerical-aperture lens, and transmission was recorded using differential interference contrast (DIC; Nomarski) optics. Images were routinely acquired using 4-8 × line averaging mode (ScanWare; Leica Microsystems). Cells were stained with FM4-64 (Molecular Probes) (63) and immobilized for microscopic inspection as described elsewhere (26). Images were edited using Adobe Photoshop 5.0 and NIH Image 1.62.
RESULTS
Isolation of the tsc13 mutants and cloning of the TSC13 gene.
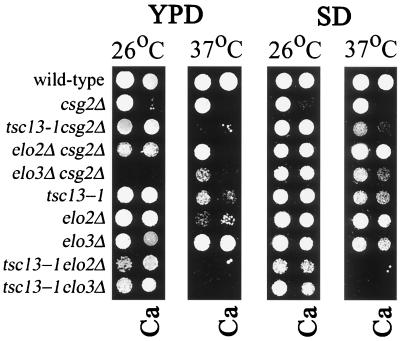
The csg2Δ mutant fails to mannosylate IPC and consequently accumulates high levels of IPC (3, 68). These lipid alterations result in a Ca2+-sensitive phenotype; therefore, suppressor mutants that are able to grow on Ca2+ identify genes required for IPC synthesis. Twenty-one TSC complementation groups were identified in a screen for mutants that had acquired a single mutation that conferred both suppression of Ca2+ sensitivity and temperature sensitivity (2). Genetic linkage analysis demonstrated that in most cases both phenotypes were caused by the same mutation. Three independent mutations in the TSC13 gene were identified in this screen. The TDY2050 mutant, containing the tsc13-1 allele, was chosen for further analysis and was found to have phenotypes (Fig. 2) similar to those observed in mutants lacking either the ELO2 or ELO3 gene. As shown in Fig. 2, disruption of either the ELO2 or the ELO3 gene also suppressed the Ca2+-sensitive phenotype of the csg2Δ mutant grown to an optical density at 600 nm (OD600) of 0.1 on SD minimal medium. On rich (YPD) medium, the elo3 csg2 double mutant grew poorly at 37°C and failed to grow at 26°C. The ts phenotype caused by the tsc13-1 mutation was more severe in strains lacking the CSG2 gene (Fig. 2). Furthermore, disruption of either the ELO2 or ELO3 gene exacerbated the ts phenotype caused by the tsc13-1 mutation (Fig. 2). The genetic interactions between tsc13 and elo2 and with elo3 suggested that Tsc13p might be required for VLCFA synthesis and function in the same pathway as Elo2p and Elo3p.
FIG. 2.
A mutation in the TSC13 gene suppresses the Ca2+ sensitivity of the csg2Δ mutant and confers temperature-sensitive lethality. The elo2Δ and elo3Δ mutations also suppress the Ca2+ sensitivity of the csg2Δ mutant, and the tsc13-1 mutation displays synthetic growth phenotypes with the elo2Δ and elo3Δ mutations. The indicated strains (Table 1) were grown in SD medium to an OD600 of 0.1 and then diluted 1/100 into the wells of a microtiter plate. The cells were transferred to plates containing the indicated medium with or without 50 mM CaCl2 (Ca), and the plates were incubated at 26°C for 3 days or 37°C for 2 days.
The wild-type TSC13 gene was cloned based on its ability to complement the ts phenotype of the TDY2050 (tsc13-1) mutant. Subcloning experiments demonstrated that the YDL015c open reading frame was responsible for complementation (see Materials and Methods). A URA3-marked wild-type YDL015c allele was found to be tightly linked to the tsc13-1 mutant allele, demonstrating that the complementing YDL015c gene corresponds to the wild-type TSC13 gene. A diploid heterozygous for the tsc13::TRP1 disruption (see Materials and Methods) was sporulated, and tetrads were dissected. Each tetrad gave rise to two viable tryptophan-requiring spore colonies, confirming the previously reported (60) finding that the TSC13/YDL015c gene is essential for viability.
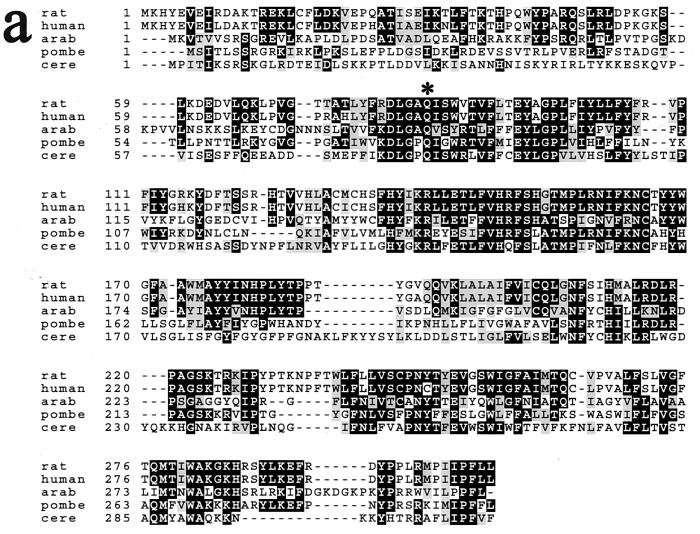
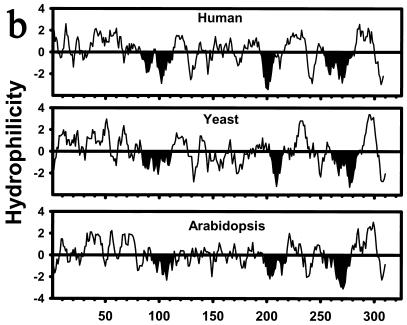
Tsc13p is evolutionarily conserved and has similarity to a steroid reductase.
The TSC13 gene is predicted to encode a protein of 310 amino acids with significant homology (35% identity, 50% similarity) over its entire length to evolutionarily conserved proteins of previously unknown function called the SC2 proteins (Fig. 3a). The SC2 gene, originally identified in a screen for cDNAs that encode rat synaptic glycoproteins, was found to be expressed at high levels in the brain and at lower levels in other tissues (23). The pattern of hydrophobicity and the positions of potential membrane-spanning domains in the Tsc13p/SC2 proteins have been conserved throughout evolution (Fig. 3b), and Tsc13p behaves as an integral membrane protein (discussed below). As previously noted (23) for the SC2 protein, Tsc13p has homology in the carboxy-terminal 150 amino acids (29% identical, 45% similar) to steroid-5-α-reductase, which catalyzes the reduction of testosterone to dihydrotestosterone. Steroid-5-α-reductase, like the enoyl reductase of the fatty acid elongating system, catalyzes the reduction of a double bond that is α,β to a carbonyl group. The discovery that the tsc13 mutants are deficient in VLCFA synthesis raised the possibility that Tsc13p catalyzes the last step in each cycle of elongation, reduction of the trans-2,3-enoyl-CoA intermediate (Fig. 1).
FIG. 3.
The Tsc13p and SC2 proteins are members of an evolutionarily conserved family of proteins. (a) Alignment of rat, human, Arabidopsis thaliana (arab), Schizosaccharomyces pombe (pombe), and S. cerevisiae (cere) homologs. The tsc13-1 mutant allele has a substitution of lysine for the highly conserved glutamine at residue 81 of the S. cerevisiae protein (marked with an asterisk). (b) All of the Tsc13p/SC2 protein homologs have similar hydrophilicity profiles. Segments of the proteins with the potential to span the membrane are indicated in black.
The tsc13 mutant accumulates LCBs as well as ceramides that have fatty acids with chain lengths of less than 26 carbons.
In contrast to wild-type cells, the elo2Δ and elo3Δ mutant cells accumulated high levels of LCBs (Fig. 4 and reference 37). The tsc13 mutant also accumulated free LCBs, and this phenotype was even more severe in tsc13-1 elo2Δ and tsc13-1 elo3Δ double mutants than in the single mutants (Fig. 4). The accumulated LCBs, primarily phytosphingosine but also dihydrosphingosine and small amounts of 3-ketosphinganine, are normal intermediates in the LCB biosynthetic pathway. Thus, the mutant phenotype may reflect reduced partitioning of the LCBs into ceramides and sphingolipids because of a defect in VLCFA synthesis in the mutants.
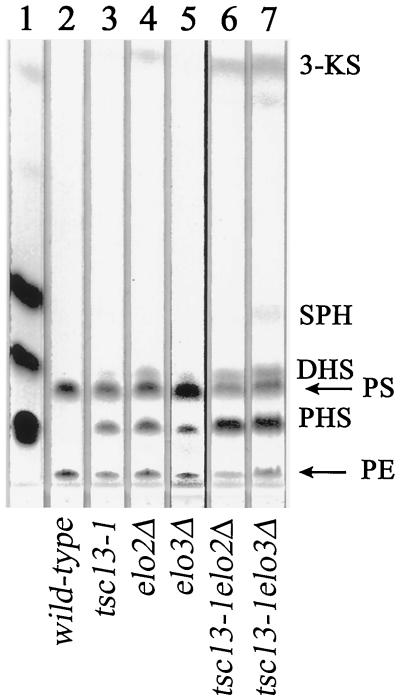
FIG. 4.
The tsc13-1, elo2Δ, and elo3Δ mutants accumulate high levels of free LCBs. LCBs were extracted from 10 OD600 units of the indicated cells, separated by TLC, and visualized by ninhydrin staining. The LCB standards sphingosine (SPH), dihydrosphingosine (DHS), phytosphingosine (PHS), and 3-ketosphingosine (3-KS) were spotted in lane 1 (positions indicated). The ninhydrin-reactive species that migrate just below DHS and near the origin (indicated by the arrows) are phosphatidylserine (PS) and phosphatidylethanolamine (PE).
A significant fraction of the ceramides from elo2Δ, elo3Δ, and tsc13-1 mutant cells displayed reduced TLC mobilities indicative of increased hydrophilicity in comparison to the C-ceramide (containing phytosphingosine and α-OH-C26) present in wild-type cells (Fig. 5a). The accumulation of the relatively hydrophilic ceramides was more severe in the tsc13-1 elo2Δ and tsc13-1 elo3Δ double mutants than in the single mutants. The mobility of these ceramide species suggested that they were likely to be hydroxylated on the α-carbon of the fatty acid and to contain fatty acids of chain lengths shorter than C26. To test this hypothesis, we disrupted the SCS7 gene encoding the enzyme required for the α-hydroxylation of the VLCFA (20, 34) and compared the ceramides present in the single and double mutants. The increased mobility of the ceramides in scs7Δ elo2Δ and scs7Δ elo3Δ double mutants confirmed that Scs7p (Figure 5b) hydroxylates the ceramides in the mutants.
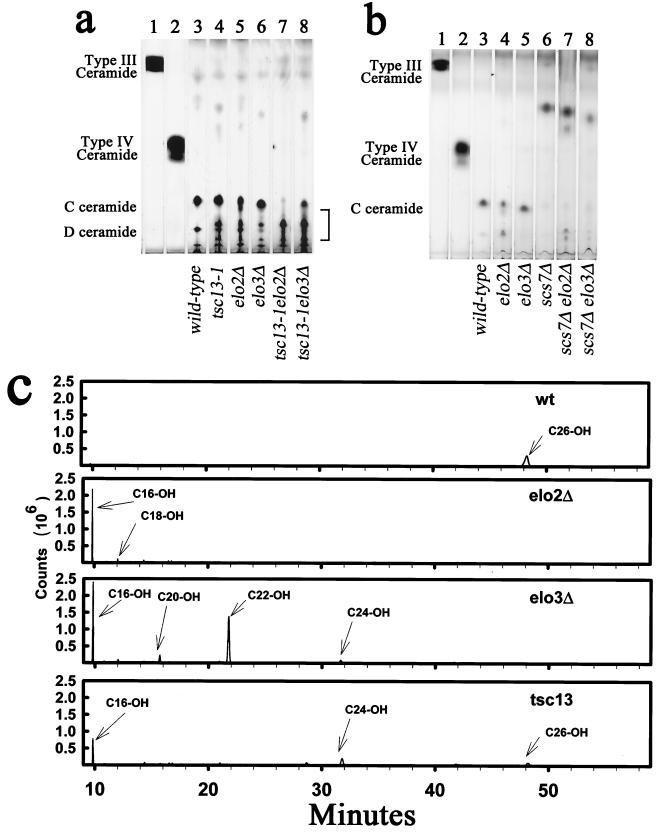
FIG. 5.
The tsc13-1, elo2Δ, and elo3Δ mutants accumulate ceramides with chain lengths shorter than C26. (a) Ceramides were extracted from 10 OD600 units of the indicated cells, separated by TLC, and visualized by charring. Bovine ceramide type III (consisting of sphingosine and C16 fatty acid) and type IV (consisting of sphingosine and α-OH-C16 fatty acid) standards are present in lanes 1 and 2. The predominant ceramide in wild-type cells (C-ceramide) consists of phytosphingosine and α-OH-C26 fatty acid. The D-ceramide is presumed to contain dihydroxy-VLCFA and phytosphingosine. (b) Ceramides from the indicated mutants were analyzed as for panel a. Deletion of SCS7 prevented hydroxylation of the fatty acid on the α-carbon, causing the ceramides to become more hydrophobic. (c) Ceramides purified from wild-type (wt) and tsc13-1, elo2Δ, and elo3Δ mutant cells were hydrolyzed to generate fatty acid methyl esters, and the extracted methyl esters were subjected to GC-mass spectrometry. The hydrophilic ceramides in the mutants (purified from the region of the TLC below C-ceramide as indicated by the bracket in panel a) contained α-hydroxylated fatty acids with chain lengths of less than C26. A mock extraction of the silica gel was conducted, and the background spectrum was subtracted.
The unhydroxylated ceramides in the scs7Δ elo2Δ and scs7Δ elo3Δ double mutants also displayed altered ceramide mobilities consistent with increased hydrophilicity. To investigate whether this was indeed due to shorter chain lengths of the fatty acids, the ceramides were purified from the TLC plates and hydrolyzed, and the methyl esters of the liberated fatty acids were analyzed by GC-mass spectrometry. As shown in Figure 5c, the mutant strains accumulated ceramides containing fatty acids with less than 26 carbons. The net ceramide levels do not appear to be appreciably lower than in wild-type cells (Fig. 5a). However, while the C24-containing ceramides are inositolphosphorylated, the resulting IPCs are not mannosylated (37). Furthermore, the hydrophilic ceramides containing fatty acids with chain lengths shorter than C24 apparently are not inositolphosphorylated since hydrophilic IPCs do not accumulate in the elongase mutants (data not shown). Taken together, these data suggest that the accumulation of LCB in the elo2, elo3, and tsc13 mutants may reflect, at least in part, reduced incorporation of the LCBs into the mature sphingolipids, presumably due to impaired availability of VLCFAs.
The tsc13 mutant has a defect in VLCFA synthesis.
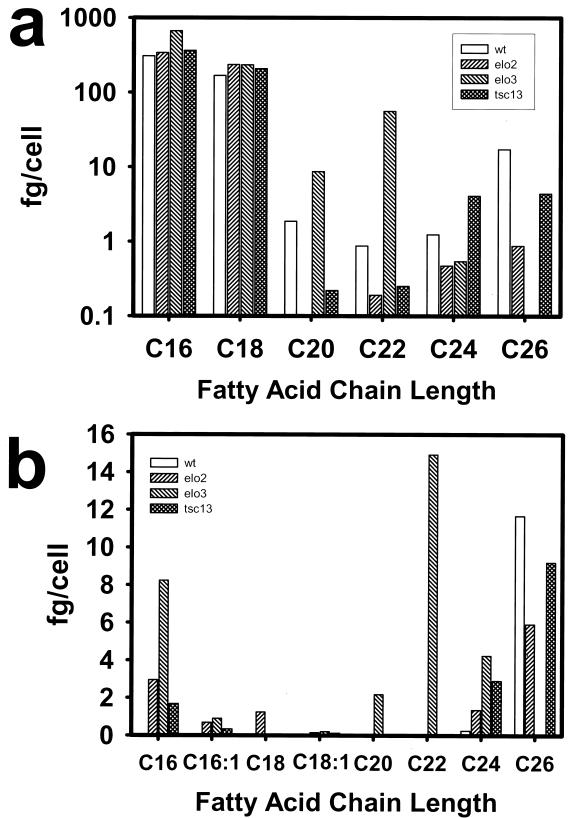
The fatty acid composition of the mutant cells confirmed a deficiency in VLCFA synthesis. The tsc13 mutant cells had greatly (∼25%) reduced levels of 26:0 (Fig. 6a), which is the predominant species found in wild-type cells, and an approximate fourfold increase in the 24:0 intermediate species. By comparison, the elo3Δ mutant lacked 26:0 and accumulated high levels of C22 and C20 intermediates, while the elo2Δ mutant was deficient in the synthesis of all VLCFA species. The tsc13, elo2Δ, and elo3Δ mutants also exhibited striking differences from wild type in their hydroxy fatty acid profiles (Fig. 6b). In particular, the three mutants accumulated high levels of OH-16:0, and the elo2Δ mutant also accumulated significant levels of OH-18:0. OH-C16 and OH-C18 fatty acids were not detected in wild-type cells. The three mutants also displayed high levels of OH-24:0, a minor species in the wild type, and reduced levels of OH-26:0 (particularly elo3), which is the most abundant hydroxylated fatty acid in the wild type. As indicated previously, the elo3 mutant accumulated high levels of 20:0 and 22:0, which was not found in either the tsc13 or elo2 mutant.
FIG. 6.
The elongase mutants are deficient in synthesis of VLCFAs and accumulate hydroxylated fatty acids with chain lengths shorter than C26. Fatty acid methyl esters were derived from wild-type (wt) and tsc13-1, elo2Δ, and elo3Δ mutant cells and analyzed by GC. The mass of each fatty acid species per cell was determined by comparison to an internal standard that was added to the cells prior to the methanolysis reaction. (a) Mass of total nonhydroxylated fatty acids in cells (log scale); (b) mass of total hydroxylated fatty acids.
Previously, a conditional mtr7 mutant, defective in ACC (ACC1) activity was found to contain significantly reduced levels of C26 fatty acid (21, 48, 49). It was suggested that malonyl-CoA-dependent chain elongation was limiting in this mutant under restrictive conditions. Consistent with a role for Tsc13p in fatty acid elongation, mtr7 tsc13-1 double mutants were nonviable even under conditions permissive for both single mutants. A similar synthetic lethality was observed between elo2 mtr7 and elo3 mtr7 double mutants; synthetic lethality of elo2, elo3, and tsc13-1 with acc1 mutants was not allele specific (data not shown). ACC activity can be specifically inhibited by the polyketide fungicide soraphen A (48, 50, 62). The tsc13, elo2, and elo3 mutants were found to be hypersensitive to soraphen A (data not shown). Taken together, these data suggest that reduced production of malonyl-CoA in acc1 mutants or in the presence of soraphen A in addition to compromised fatty acid elongation activity is detrimental to the cell.
The tsc13 mutant has reduced fatty acid elongation activity.
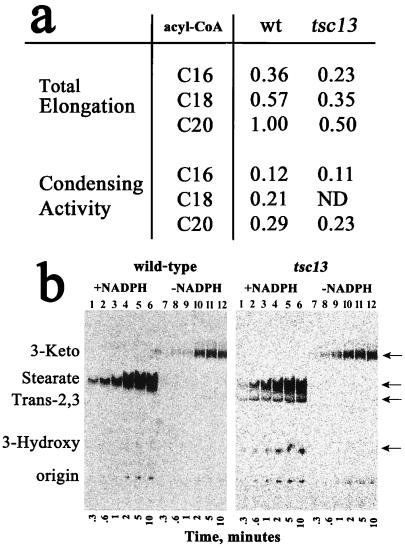
The TSC13 gene, unlike the ELO2 and ELO3 genes, is essential for viability, and there are no TSC13 homologous genes in the S. cerevisiae genome. Elo2p and Elo3p have partially redundant functions and are believed to participate in the same step of the elongation cycle, with preferences for acyl-CoA substrates of different chain lengths (37, 59). However, Tsc13p is likely to be required for catalyzing a step in each cycle of fatty acid elongation for acyl-CoA substrates of all chain lengths. To address which step of elongation might be affected by the tsc13 mutation, microsomes were prepared from the tsc13 mutant cells and assayed for elongase activity in vitro. For these experiments, the incorporation of [2-14C]malonyl-CoA into elongated acyl-CoA products was measured. The first step of the elongation cycle is the condensation of malonyl-CoA with an acyl-CoA (e.g., palmitoyl-CoA) to form a 3-ketoacyl-CoA intermediate (Fig. 1). Omitting pyridine nucleotide from the assay mix prevents the reduction of the 3-ketoacyl-CoA intermediate, thereby allowing the first step of elongation (condensation) to be measured. The condensation activity measured in wild-type or tsc13-1 microsomes was very similar when palmitoyl-CoA (or eicosanoyl-CoA) was used as a substrate (Fig. 7a). However, the total elongation activity measured using microsomes from the tsc13 mutant cells was about 50% of that in wild-type microsomes for each of the acyl-CoA substrates tested (Fig. 7a). These results indicate that Tsc13p catalyzes a step in elongation subsequent to the condensation step.
FIG. 7.
The tsc13 mutant cells have normal condensation activity but are deficient in total fatty acid chain elongation. (a) Fatty acid elongation activities in wild-type (wt) and tsc13 mutant microsomes were compared using C16-, C18-, and C20-CoAs as substrates by measuring the incorporation of radiolabeled malonyl-CoA into hexane-extractable fatty acids. The assays were conducted in the absence of NADPH or NADH to measure condensation activity and in the presence of NADPH or NADH to measure total elongation. Activities are normalized to the overall activity measured with wild-type microsomes, using C20-CoA as the substrate. ND, not determined. The assays were conducted in triplicate, and the results were averaged. The variation was less than 7%. (b) The elongation assays were conducted as for panel a, using C16-CoA as the substrate and either wild-type (left) or tsc13 mutant (right) microsomes. The reactions were stopped at the indicated times, and the fatty acids were extracted and separated by TLC. The reactions were conducted with (lanes 1 to 6) or without (lanes 7 to 12) NADPH or NADH. The positions of the 3-ketostearate (3-Keto), stearate, trans-2,3-stearate (Trans-2,3), and 3-hydroxystearate (3-Hydroxy) intermediates were determined by running the standards on the TLC plate and charring after exposure to PhosphorImager screens.
To investigate the elongation defect in the tsc13-1 mutant further, time courses of the condensation reaction and the total elongation reaction were conducted, and the products were analyzed by TLC using conditions that resolved the four intermediates of fatty acid elongation. Elongation intermediates did not accumulate during the elongation reactions catalyzed by the wild-type microsomes (Fig. 7b), which is consistent with previous studies of the rat microsomal elongating systems demonstrating that the condensation reaction is rate limiting (5). However, when microsomes prepared from the tsc13 mutant were used in the assay, accumulation of the trans-2,3-stearoyl and 3-hydroxystearoyl intermediates was observed. This is consistent with the defect in Tsc13p causing reduced activity of the trans-2,3-enoyl-CoA reductase leading to accumulation of the trans-2,3-stearoyl-CoA. Because the dehydratase step of elongation is reversible (25), accumulation of trans-2,3-stearoyl-CoA would be expected to cause the observed accumulation of 3-hydroxystearoyl-CoA as well. Based on its homology to steroid-5-α-reductase, Tsc13p may directly catalyze the reduction of the enoyl intermediate in each cycle of fatty acid elongation.
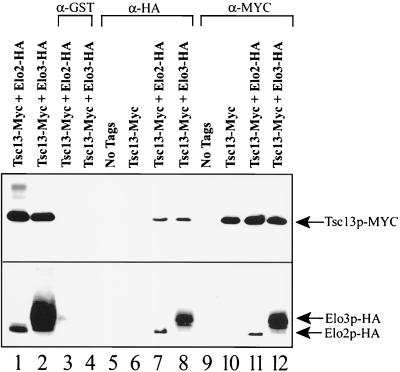
Tsc13p coimmunoprecipitates with Elo2p and Elo3p.
The enzymes responsible for fatty acid elongation have not yet been purified, and virtually nothing is known about their molecular organization. For example, it is not known whether they associate into a complex that processively elongates fatty acids, or whether the acyl-CoA intermediates diffuse from one membrane-associated elongating enzyme to the next. To address whether Tsc13p associates with Elo2p and Elo3p, we attempted protein coimmunoprecipitation. The plasmid carrying the Myc-tagged TSC13 allele (MYC-TSC13-426) was transformed into TDY2058 (Table 1) independently or in combination with a plasmid carrying either HA-tagged Elo3p or HA-tagged Elo2p. Tsc13p, Elo2p, and Elo3p (as their Myc- or HA-tagged versions, respectively) were all found to display detergent solubilization properties indicating that they are tightly associated integral membrane proteins (data not shown). Antibodies to the Myc epitope coimmunoprecipitated Elo2p-HA or Elo3p-HA along with Tsc13p-Myc, and vice versa (Fig. 8), from solubilized microsomal preparations. These data suggest that Tsc13p forms complexes with Elo2p and with Elo3p. It is not yet known whether Elo2p and Elo3p coexist in the same Tsc13p-containing complexes. Although it appears that Elo3p is more abundant than Elo2p (Fig. 8, lower panel, compare lanes 1 and 2), the tagged proteins are being expressed by the ADH1 promoter on plasmids. Furthermore, the strain (TDY2058) harboring the tagged proteins lacks the wild-type ELO3 gene but has the wild-type ELO2 gene, and the level of either tagged protein is reduced when the corresponding endogenous wild-type protein is present (data not shown).
FIG. 8.
Tsc13p coimmunoprecipitates with Elo2p and with Elo3p. Microsomes were prepared from strain TDY2058 cells containing no tagged proteins (lanes 5 and 9) or the same cells containing only Myc-tagged Tsc13p (lanes 6 and 10), Myc-tagged Tsc13p and HA-tagged Elo2p (lanes 1, 3, 7, and 11) or Myc-tagged Tsc13p and HA-tagged Elo3p (lanes 2, 4, 8, and 12); 10 μg of total microsomal protein was loaded in lanes 1 and 2. The microsomes were solubilized, and the 100,000 × g supernatant was used for immunoprecipitation experiments using anti-GST (lanes 3 and 4), anti-HA (lanes 5 to 8), or anti-Myc (lanes 9 to 12) antibodies. The immunoprecipitated proteins were separated by SDS-PAGE (12% gel) and analyzed by immunoblotting with HRP-conjugated anti-Myc (top panel) or anti-HA (bottom panel) antibodies.
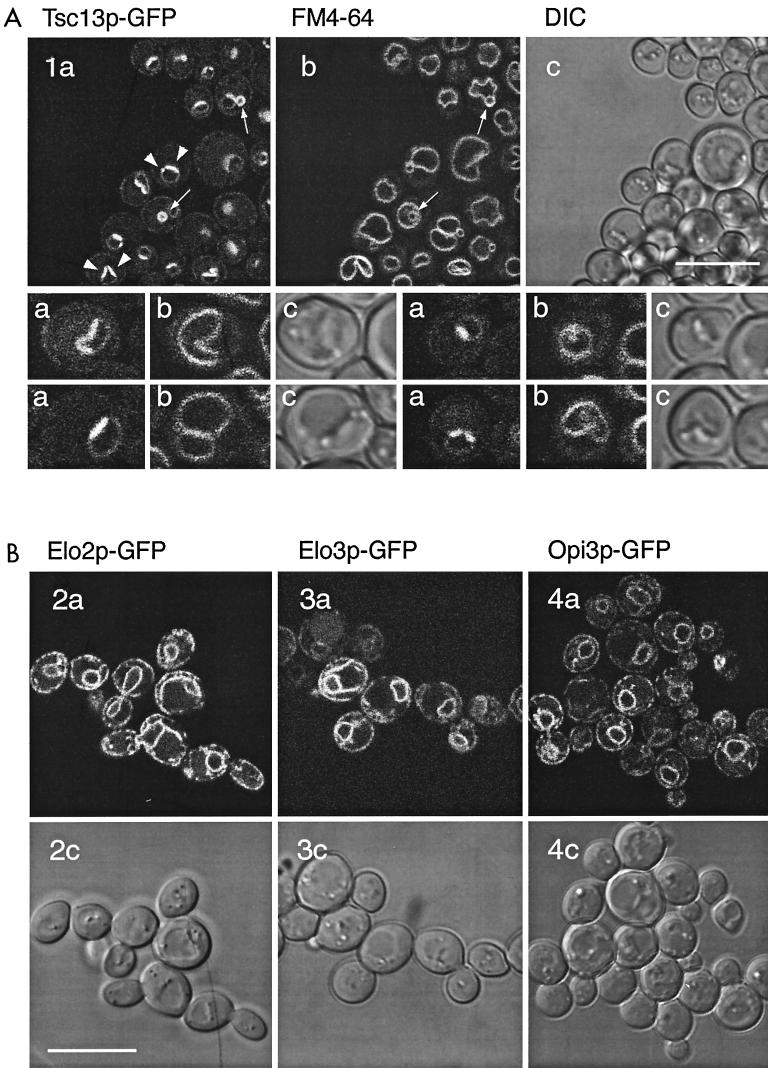
Tsc13p localizes in the ER and is enriched at sites of vacuole-nuclear envelope interaction.
Based on cell fractionation of the epitope-tagged proteins, Elo2p, Elo3p (reference 14 and this study), and Tsc13p are present in the microsomal membrane fraction, suggesting localization to the ER. To analyze Tsc13p subcellular localization in greater detail, Tsc13p was C-terminally tagged with GFP by chromosomal fusion. The localization of Tsc13p-GFP, demonstrated to be a functional protein, revealed a perinuclear and peripheral staining consistent with association with the ER (Fig. 9A). A similar pattern of staining was observed in cells expressing a chromosomal Elo3p-GFP fusion (Fig. 9B, 2a and c). Elo2p-GFP, expressed from an episomal plasmid under control of the GAL1 promoter, showed a staining pattern indistinguishable from that of Elo3p-GFP (Fig. 9B, 3a and c), and Opi3p-GFP, which was used as a control for the ER (24) (Fig. 9B, 4a and c). Interestingly, however, in addition to the typical nuclear rim-ER staining exhibited by Elo2p, Elo3p, or Opi3p, Tsc13p-GFP was highly enriched in structures reminiscent of the recently described nuclear-vacuolar junctions (39) that are contact sites between the nucleus and the vacuole (Fig. 9A). The intensity of the Tsc13p-GFP signal and enrichment of Tsc13p to the sites of nuclear-vacuolar interaction increased during growth on complete medium. Furthermore, vesicles that contain Tsc13p-GFP in the membrane appeared closely associated to, or even enclosed by, FM4-64-labeled vacuolar membrane structures (Fig. 9A). These structures appeared more frequently as cells entered stationary phase.
FIG. 9.
(A) C-terminally tagged Tsc13p-GFP shows a typical ER localization pattern and, in addition, is highly enriched in a region of the nuclear envelope adjacent to the vacuole. Selected cells are shown at higher magnification in the lower panel. (B) Elo2p-GFP, Elo3p-GFP, and Opi3p-GFP localize to the ER; i.e., around the nucleus and at the cell periphery. a, GFP fluorescence; b, FM4-64-labeled vacuolar membranes; c, DIC transmission images. Each scale bar is 10 μm.
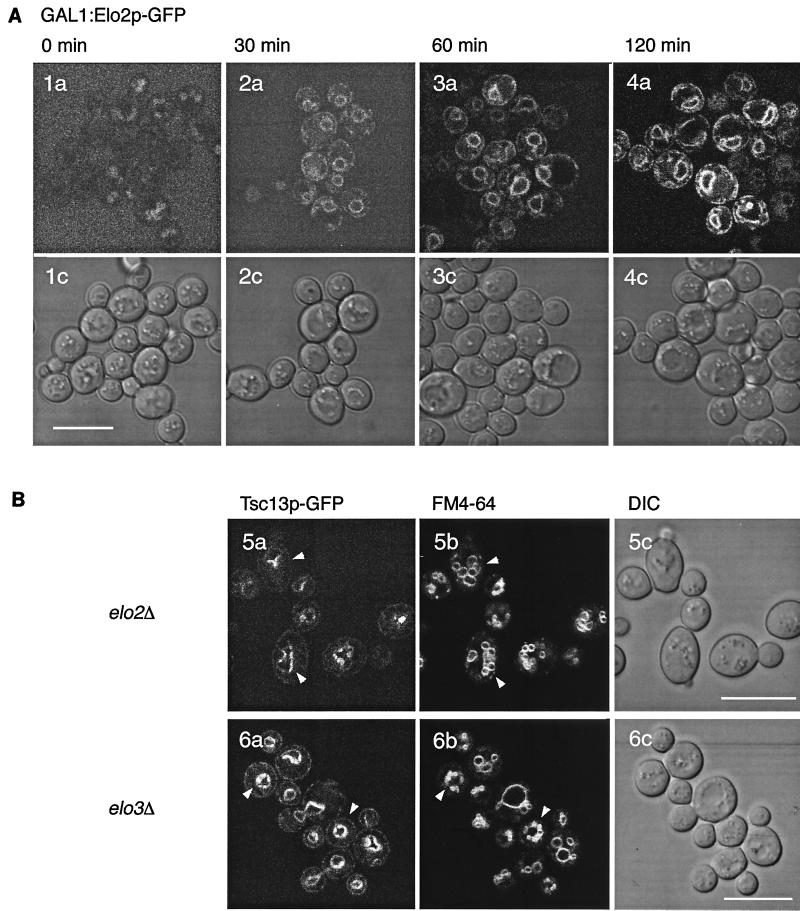
Although Elo2p, Elo3p, and Tsc13p appear to be involved in the same pathway and may physically interact, as demonstrated above, their localization patterns are different. The codon adaptation indices for Elo2p (0.239) and Elo3p (0.372) are similar to that of Tsc13p (0.240 9]), which may indicate similar expression levels (8). To analyze whether apparently homogeneous ER staining of Elo2p or Elo3p is due to increased abundance that obscures a specific labeling pattern, we have modulated expression levels of one of these proteins by virtue of GAL1-controlled expression. Figure 10A shows a time course of derepression of a plasmid-borne Elo2p-GFP after a shift from glucose to galactose medium. Even at very low expression levels, Elo2p-GFP distribution in the ER was indistinguishable from the wild-type pattern; under no conditions did we observe a pattern reminiscent of Tsc13p-GFP, suggesting that Tsc13p localization to nuclear-vacuolar junctions does not depend on Elo2p.
FIG. 10.
(A) Time course of GAL1P promoter-controlled Elo2p-GFP induction after shift from glucose to galactose-containing medium, determined at 0, 30, 60, and 120 min. The lower row shows corresponding DIC transmission images. The high background signal at 0 min is due to the high amplification settings on the microscope and the background fluorescence of the culture medium. (B) Tsc13p-GFP localizes to sites of nucleus-vacuole interaction in elo2 and elo3 mutants. elo2 and elo3 mutants show multilobed vacuoles; nevertheless, Tsc13p-GFP appears predominantly at sites between the nuclear envelope and vacuoles. a, GFP fluorescence; b, FM4-64 fluorescence; c, DIC transmission image.
Tsc13p-GFP localization is independent of Elo2p and Elo3p.
To further determine whether Tsc13p localization is dependent on the presence of either Elo2p or Elo3p, Tsc13p-GFP distribution was analyzed in elo2 or elo3 mutants, respectively (elo2 elo3 double mutants are nonviable). Labeling of cells with FM4-64 revealed that the vacuolar morphology was drastically altered in elo2 and elo3 mutants and displayed a multilobed structure, reminiscent of vac8 mutants (47, 66) (Fig. 10B). Despite the high fragmentation of vacuoles into smaller vesicles, the polarity of Tsc13p localization was still maintained and the protein was still present at nuclear-vacuolar junctions. No alteration of the Tsc13p distribution was observed in these mutants, compared to wild type, suggesting that neither Elo2p is Elo3p are required for polarized Tsc13p localization at the nuclear-vacuolar junction.
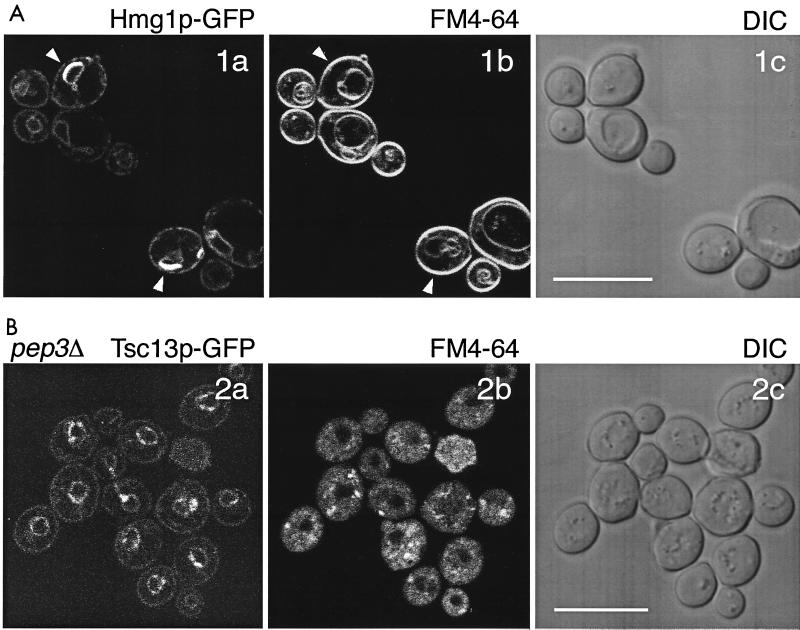
Expression of Tsc13p-GFP does not result in the formation of karmellae.
Some ER-resident proteins, e.g., Hmg1p (hydroxymethylglutaryl-CoA reductase), when overexpressed lead to the formation of karmellae, which are multiple stacks of ER membrane, highly enriched in these proteins (27, 40, 67). As a consequence, karmellae induced by Hmg1p-GFP appear as very bright structures on the surface of the nucleus, at regions located opposite the vacuole (27). As shown in Fig. 11A, the localization of karmellae, albeit similar in appearance on the nuclear surface, is clearly different from the localization pattern of Tsc13p-GFP in the nuclear-vacuolar contact sites. In addition, ER membranes labeled with DiOC6 (27) in strains expressing Tsc13p-GFP appeared indistinguishable from wild type, demonstrating that GFP tagging of Tsc13p does not result in the formation of karmellae. This was further confirmed by electron microscopy demonstrating lack of any membrane proliferations in Tsc13p-GFP-expressing strains (data not shown).
FIG. 11.
(A) Karmellae are distinctly different from the structures labeled by Tsc13p-GFP. Karmellae form upon overexpression of an Hmg1p-GFP construct (a, GFP fluorescence); orientation of karmellae localized on the nuclear envelope is always on the opposite site of the vacuole (b, FM4-64 staining). c, DIC transmission image. (B) Enrichment of Tsc13p-GFP to the sites of nucleus-vacuole interaction depends on the presence of vacuolar membranes. pep3 mutants contain vestigial vacuoles (b, FM4-64 staining); localization of Tsc13p-GFP appears dispersed on the surface of the nuclear membrane; polarization of the GFP signal is absent (a). c, DIC transmission image.
The polarized localization of Tsc13p-GFP depends on the presence of vacuoles.
Depending on growth phase, in the presence of nonfermentable carbon sources, or in elo2 and elo3 mutants, yeast vacuoles may split up into multiple smaller vesicles. Under these conditions, Tsc13p-GFP was consistently present in nuclear-vacuolar junctions. To test whether intact vacuoles are required to maintain this polarity, we have analyzed Tsc13p-GFP distribution in pep3 mutants which contain only vestigial nonacidic vacuoles (41, 42). As shown in Fig. 11B, the polarity of Tsc13p-GFP localization on the nuclear surface is lost in pep3 mutants but still appears somewhat clustered.
Taken together, the localization data suggest that Tsc13p is a component of the ER and that its enrichment at the nuclear-vacuolar interface is dependent on the presence of vacuolar membranes. The factors establishing this polarized localization pattern are still obscure but may be dispensable for Tsc13p to fulfill its essential function.
DISCUSSION
VLCFAs are essential cellular components that are predominantly present in sphingolipids, the lipid moiety of GPI anchors, and to some extent in a novel PtdIns species recently identified in nuclear membrane extracts (51). Based on nuclear membrane phenotypes associated with reduced synthesis of C26 in conditional acc1 mutants, a structural role for VLCFA in stabilizing the highly curved nuclear membrane at the nuclear pore complex was postulated (48, 51). The importance of VLCFAs for cell viability is further demonstrated by the fact that mutants unable to synthesize sphingolipids produce novel PtdIns species that contain C26 in the sn-2 position (30). Studies to determine the structures and organization of the enzymes responsible for VLCFA synthesis have only recently become possible. Multiple elongating systems within the same organism are apparently responsible for elongating acyl-CoA substrates with different chain lengths and degrees of unsaturation. The identification in S. cerevisiae of three structurally and functionally related elongase proteins, Elo1p, Elo2p, and Elo3p, that display different chain length preferences are consistent with this suggestion (37, 59). Since they control the abundance of fatty acids with different chain lengths, it seems likely that these proteins participate in the condensation step of elongation. However, the yeast Elo proteins have no homology to any of the condensing enzymes that have been characterized so far at the molecular level. For example, several genes encoding putative condensing enzymes of fatty acid elongation have been cloned from plants and have been demonstrated to stimulate VLCFA synthesis in heterologous expression systems (29, 32, 33). Interestingly, there are no homologs of these plant genes in S. cerevisiae, but there are homologs of the ELO genes in plants.
The characterization of mammalian homologs of the ELO genes has recently been undertaken (61). One mammalian homolog (Ssc1) was found to complement the sphingolipid deficiency of the elo3 mutant, and another (Cig30) reversed the phenotype of the elo2 mutant. Furthermore, Ssc1 mRNA levels were reduced in the brains of myelin-deficient mice that are known to have low fatty acid elongation activity.
In addition to the condensation reaction, a reduction step, a subsequent dehydration, and a second reduction are required for fatty acid chain elongation. The studies presented here demonstrate that the nonredundant TSC13 gene encodes a protein required for the enoyl reductase activity of fatty acid elongation. Based on the homology of Tsc13p to steroid-5-α-reductase, it is likely that Tsc13p is the enoyl reductase enzyme per se. Tsc13p is a member of a family of proteins that have been conserved from yeast to mammals. Substitution of the glutamine with lysine at residue 81 of Tsc13p is responsible for the ts phenotype conferred by the tsc13-1 mutant allele. This residue is conserved in all Tsc13p homologs in the database. The rat TSC13 homolog, SC2, was initially identified as a gene that is highly expressed in brain, an organ where VLCFA synthesis is known to occur at high rates.
For the synthesis of long-chain fatty acids up to C16, similar reactions are catalyzed by the multifunctional cytosolic FAS complex. Interestingly, none of the FAS domains has any homology to Elo2p, Elo3p, or Tsc13p. Furthermore, tsc13-1 mutants are not hypersensitive to diazaborin, a drug that inhibits bacterial enoyl-acyl carrier protein reductase (4), suggesting different reaction mechanisms for these enzymes.
The incorporation of VLCFA into sphingolipid is catalyzed by ceramide synthase, which has not been identified yet. In wild-type cells, VLCFAs are hydroxylated on C-2 by Scs7p after they are incorporated into ceramide (16, 20). Fatty acids with chain lengths shorter than C26 are not normally incorporated into ceramide and are thus not hydroxylated in wild-type cells. However, in mutants with VLCFA synthesis defects, fatty acids shorter than C26 are incorporated into ceramide and are thus subject to hydroxylation by Scs7p. It is interesting that the elo2Δ and tsc13-1 mutant cells contained similar levels of the hydroxylated C26 fatty acid as did wild-type cells, whereas the overall content of C26 was reduced. Thus, ceramide synthase appears to have a strong preference for C26 fatty acids but is also able to modify the fatty acids with chain lengths of less than C26 that accumulate in elo2 and tsc13-1 mutants. Since in wild-type cells none of the intermediates of the elongation reaction appear in hydroxylated form in ceramides, we suggest a metabolic channeling mechanism for VLCFA synthesis that would require a concerted action of the elongase complex, including Elo2, Elo3p, and Tsc13p, prior to incorporation of VLCFA into sphingolipid by ceramide synthase. Alternatively, synthesis of the C16- to C24-containing ceramides in the mutants could arise from the reversed activity of the ceramidase enzyme encoded by YPC1 (condensation of LCBs with free fatty acids 31) acids rather than from ceramide synthase) (Fig. 1). The reverse ceramidase reaction may be driven by the high levels of LCBs in the elongase mutants.
Ceramides in yeast are subject to inositolphosphorylation (an essential process) and subsequent mannosylation, which represents a nonessential modification to this class of lipids. It is interesting that the enzymes responsible for mannosylation of the IPC require a ceramide with a C26 fatty acid and that C24 ceramides accumulating in elongation mutants are not mannosylated. The chain length of the fatty acid is apparently also important for discrimination of IPC from PtdIns, since in slc1 mutants that incorporate C26 fatty acids into PtdIns this phospholipid becomes mannosylated. Thus, the availability of particular VLCFA species in a lipid appears to determine downstream lipid modification processes. Interestingly, recent studies using fatty acid elongation mutants defective in elo1 suggest that the availability of fatty acids with certain chain lengths could also be an important determinant of phospholipid composition (50). The molecular mechanisms involved, however, remain to be determined.
The unique localization of Tsc13p to a specific domain within the ER membrane at the nuclear-vacuolar interface raises many questions. It will be interesting to determine whether this domain of the ER membrane is the site where VLCFA synthesis occurs. It is possible that other enzymes required for sphingolipid synthesis, for example, the subunits of serine palmitoyltransferase or ceramide synthase, reside in this domain. Based on the experiments described here, this domain at the nuclear-vacuolar interface is not required for the function of Tsc13p, but its formation requires interaction with vacuolar membranes. Only recently, in studies on the localization of the vacuolar membrane protein Vac8p, a similar asymmetric protein distribution was observed at sites of vacuolar-vacuolar (38, 66) and vacuolar-nuclear envelope (47) interactions. Polarity of Vac8p to so-called nucleus-vacuole junctions requires the presence of Nvj1p in the nuclear envelope (39). Similarly, polarized localization of Nvj1p in the nuclear envelope depends on the presence of Vac8p. Whether Tsc13p requires Nvj1p or Vac8p for localization to sites of nucleus-vacuole interaction remains to be determined. However, localization of neither Nvj1p, Vac8p, nor Tsc13p to this domain is essential for viability, since both nvj1 or vac8 mutants and pep3 mutants deficient in vacuolar structures are viable. Furthermore, enrichment of Tsc13p to this domain is not dependent on C26 VLCFA since its localization is not altered in the elo mutants.
What could be the physiological role of the polarized localization of Tsc13p, or of the fatty acid elongation system, to sites of nuclear-vacuolar interaction? Due to the extended hydrophobic chain of VLCFAs, it might be necessary to provide a protein complex to accommodate these fatty acids in the membrane. Biochemical data as discussed above have already suggested a metabolic channeling mechanism for VLCFA synthesis. The role of vacuoles in this process is not clear. However, the appearance of vesicles highly enriched in Tsc13p that are budding off the nuclear membrane into the lumen of the vacuole when cells enter stationary phase indicates that a site of nuclear-vacuolar interaction may be a preferred site of nuclear envelope recycling. Thus, the striking nuclear membrane phenotype in mtr7 mutants defective in ACC, and thus in fatty acid chain elongation (48, 49), may be a consequence of perturbed nuclear membrane recycling to the vacuole.
VLCFAs are essential components of sphingolipids and, due to the extended hydrophobic chain, represent a major structural determinant for the membrane harboring these lipids. In mammalian cells, sphingolipids and cholesterol may assemble to form lipid rafts, which are characterized as detergent-insoluble membrane fractions. Very recently, lipid rafts have also been identified in yeast, and evidence suggests that certain secretory proteins, e.g., Pma1p and Gas1p, associate with rafts at the level of the ER (1). The basis for the formation of sphingolipid-cholesterol-enriched (detergent-insoluble) domains is not understood, but perhaps these membrane domains are synthesized in the ER membrane as a result of the enrichment of a specific class of lipid biosynthetic enzymes within a domain of the membrane. Thus, localized synthesis of VLCFAs and their incorporation into sphingolipids may lead to sequestering sterols to form detergent-insoluble membrane domains at specific sites—the nuclear-vacuolar interface of the ER. Furthermore, the possibility that a specific class of secretory vesicles buds off from this domain of the ER is raised.
ACKNOWLEDGMENTS
We thank E. Schweizer, A. Tartakoff, R. Wright, A. Jandrositz, E. Jones, J. Hegemann, H. Klein, and B. Winsor for providing yeast strains and plasmids, A. Kauschmann (BASF) for the gift of Soraphen A, G. Högenauer for the gift of diazaborin, and G. Gogg for excellent technical assistance. We also thank D. Goldfarb for helpful discussions about nuclear-vacuolar junctions and I. Kaizer and M. Veenhuis for electron microscopy analysis.
This work was supported by NIH grant GM51891 and NSF grant G171FL to T.D., NIH GM45768 to C.E.M. and the Austrian Science Fund, FWF (project F706), Oesterreichische Nationalbank (project P7273), BIO4-CT97-2294 (EUROFAN II essential genes) of the European Union, and the Austrian Ministry of Education, Science and Culture (EUROFAN II supplement project; AUSTROFAN) to S.D.K.
REFERENCES
- 1.Bagnat M, Keranen S, Shevchenko A, Simons K. Lipid rafts function in biosynthetic delivery of proteins to the cell surface in yeast. Proc Natl Acad Sci USA. 2000;97:3254–3259. doi: 10.1073/pnas.060034697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Beeler T, Bacikova D, Gable K, Hopkins L, Johnson C, Slife H, Dunn T. The Saccharomyces cerevisiae TSC10/YBR265w gene encoding 3-ketosphinganine reductase is identified in a screen for temperature-sensitive suppressors of the Ca2+-sensitive csg2Δ mutant. J Biol Chem. 1998;273:30688–30694. doi: 10.1074/jbc.273.46.30688. [DOI] [PubMed] [Google Scholar]
- 3.Beeler T J, Fu D, Rivera J, Monaghan E, Gable K, Dunn T M. SUR1 (CSG1/BCL21), a gene necessary for growth of Saccharomyces cerevisiae in the presence of high Ca2+ concentrations at 37°C, is required for mannosylation of inositolphosphorylceramide. Mol Gen Genet. 1997;255:570–579. doi: 10.1007/s004380050530. [DOI] [PubMed] [Google Scholar]
- 4.Bergler H, Wallner P, Ebeling A, Leitinger B, Fuchsbichler S, Aschauer H, Kollenz G, Hogenauer G, Turnowsky F. Protein EnvM is the NADH-dependent enoyl-ACP reductase (FabI) of Escherichia coli. J Biol Chem. 1994;269:5493–5496. [PubMed] [Google Scholar]
- 5.Bernert J T, Jr, Bourre J M, Baumann N A, Sprecher H. The activity of partial reactions in the chain elongation of palmitoyl-CoA and stearoyl-CoA by mouse brain microsomes. J Neurochem. 1979;32:85–90. doi: 10.1111/j.1471-4159.1979.tb04513.x. [DOI] [PubMed] [Google Scholar]
- 6.Choi J Y, Martin C E. The Saccharomyces cerevisiae FAT1 gene encodes an acyl-CoA synthetase that is required for maintenance of very long chain fatty acid levels. J Biol Chem. 1999;274:4671–4683. doi: 10.1074/jbc.274.8.4671. [DOI] [PubMed] [Google Scholar]
- 7.Cinti D L, Cook L, Nagi M N, Suneja S J. The fatty acid chain elongation system of mammalian endoplasmic reticulum. Prog Lipid Res. 1992;31:1–51. doi: 10.1016/0163-7827(92)90014-a. [DOI] [PubMed] [Google Scholar]
- 8.Coghlan A, Wolfe K H. Relationship of codon bias to mRNA concentration and protein length in Saccharomyces cerevisiae. Yeast. 2000;16:1131–1145. doi: 10.1002/1097-0061(20000915)16:12<1131::AID-YEA609>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- 9.Costanzo M C, Hogan J D, Cusick M E, Davis B P, Fancher A M, Hodges P E, Kondu P, Lengieza C, Lew-Smith J E, Lingner C, Roberg-Perez K J, Tillberg M, Brooks J E, Garrels J I. The yeast proteome database (YPD) and Caenorhabditis elegans proteome database (WormPD): comprehensive resources for the organization and comparison of model organism protein information. Nucleic Acids Res. 2000;28:73–76. doi: 10.1093/nar/28.1.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.David C, Solimena M, De Camilli P. Autoimmunity in stiff-man syndrome with breast cancer is targeted to the C-terminal region of human amphiphysin, a protein similar to the yeast proteins, Rvs167 and Rvs161. FEBS Lett. 1994;351:73–79. doi: 10.1016/0014-5793(94)00826-4. [DOI] [PubMed] [Google Scholar]
- 11.David D, Sundarababu S, Gerst J E. Involvement of long chain fatty acid elongation in the trafficking of secretory vesicles in yeast. J Cell Biol. 1998;143:1167–1182. doi: 10.1083/jcb.143.5.1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Desfarges L, Durrens P, Juguelin H, Cassagne C, Bonneu M, Aigle M. Yeast mutants affected in viability upon starvation have a modified phospholipid composition. Yeast. 1993;9:267–277. doi: 10.1002/yea.320090306. [DOI] [PubMed] [Google Scholar]
- 13.Dickson R C. Sphingolipid functions in Saccharomyces cerevisiae: comparison to mammals. Annu Rev Biochem. 1998;67:27–48. doi: 10.1146/annurev.biochem.67.1.27. [DOI] [PubMed] [Google Scholar]
- 14.Dittrich F, Zajonc D, Huhne K, Hoja U, Ekici A, Greiner E, Klein H, Hofmann J, Bessoule J J, Sperling P, Schweizer E. Fatty acid elongation in yeast—biochemical characteristics of the enzyme system and isolation of elongation-defective mutants. Eur J Biochem. 1998;252:477–485. doi: 10.1046/j.1432-1327.1998.2520477.x. [DOI] [PubMed] [Google Scholar]
- 15.Dunn T M, Gable K, Monaghan E, Bacikova D. Selection of yeast mutants in sphingolipid metabolism. Methods Enzymol. 2000;312:317–330. doi: 10.1016/s0076-6879(00)12918-0. [DOI] [PubMed] [Google Scholar]
- 16.Dunn T M, Haak D, Monaghan E, Beeler T J. Synthesis of monohydroxylated inositolphosphorylceramide (IPC-C) in Saccharomyces cerevisiae requires Scs7p, a protein with both a cytochrome b5-like domain and a hydroxylase/desaturase domain. Yeast. 1998;14:311–321. doi: 10.1002/(SICI)1097-0061(19980315)14:4<311::AID-YEA220>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- 17.el-Sherbeini M, Clemas J A. Cloning and characterization of GNS1: a Saccharomyces cerevisiae gene involved in synthesis of 1,3-β-glucan in vitro. J Bacteriol. 1995;177:3227–3234. doi: 10.1128/jb.177.11.3227-3234.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gable K, Slife H, Bacikova D, Monaghan E, Dunn T M. Tsc3p is an 80-amino acid protein associated with serine palmitoyltransferase and required for optimal enzyme activity. J Biol Chem. 2000;275:7597–7603. doi: 10.1074/jbc.275.11.7597. [DOI] [PubMed] [Google Scholar]
- 19.Garcia-Arranz M, Maldonado A M, Mazon M J, Portillo F. Transcriptional control of yeast plasma membrane H+-ATPase by glucose. Cloning and characterization of a new gene involved in this regulation. J Biol Chem. 1994;269:18076–18082. [PubMed] [Google Scholar]
- 20.Haak D, Gable K, Beeler T, Dunn T. Hydroxylation of Saccharomyces cerevisiae ceramides requires Sur2p and Scs7p. J Biol Chem. 1997;272:29704–29710. doi: 10.1074/jbc.272.47.29704. [DOI] [PubMed] [Google Scholar]
- 21.Hasslacher M, Ivessa A S, Paltauf F, Kohlwein S D. Acetyl-CoA carboxylase from yeast is an essential enzyme and is regulated by factors that control phospholipid metabolism. J Biol Chem. 1993;268:10946–10952. [PubMed] [Google Scholar]
- 22.Hechtberger P, Zinser E, Saf R, Hummel K, Paltauf F, Daum G. Characterization, quantification and subcellular localization of inositol-containing sphingolipids of the yeast, Saccharomyces cerevisiae. Eur J Biochem. 1994;225:641–649. doi: 10.1111/j.1432-1033.1994.00641.x. [DOI] [PubMed] [Google Scholar]
- 23.Johnston I G, Rush S J, Gurd J W, Brown I R. Molecular cloning of a novel mRNA using an antibody directed against synaptic glycoproteins. J Neurosci Res. 1992;32:159–166. doi: 10.1002/jnr.490320205. [DOI] [PubMed] [Google Scholar]
- 24.Kanipes M I, Henry S A. The phospholipid methyltransferases in yeast. Biochim Biophys Acta. 1997;1348:134–141. doi: 10.1016/s0005-2760(97)00121-5. [DOI] [PubMed] [Google Scholar]
- 25.Knoll A, Bessoule J J, Sargueil F, Cassagne C. Dehydration of 3-hydroxyacyl-CoA in brain very-long-chain fatty acid synthesis. Neurochem Int. 1999;34:255–267. doi: 10.1016/s0197-0186(99)00005-4. [DOI] [PubMed] [Google Scholar]
- 26.Kohlwein, S. D. The beauty of the yeast: live cell microscopy at the limits of optical resolution. Microsc. Res. Technol. in press. [DOI] [PubMed]
- 27.Koning A J, Roberts C J, Wright R L. Different subcellular localization of Saccharomyces cerevisiae HMG-CoA reductase isozymes at elevated levels corresponds to distinct endoplasmic reticulum membrane proliferations. Mol Biol Cell. 1996;7:769–789. doi: 10.1091/mbc.7.5.769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kuziora M A, Chalmers J H, Jr, Douglas M G, Hitzeman R A, Mattick J S, Wakil S J. Molecular cloning of fatty acid synthetase genes from Saccharomyces cerevisiae. J Biol Chem. 1983;258:11648–11653. [PubMed] [Google Scholar]
- 29.Lassner M W, Lardizabal K, Metz J G. A jojoba β-ketoacyl-CoA synthase cDNA complements the canola fatty acid elongation mutation in transgenic plants. Plant Cell. 1996;8:281–292. doi: 10.1105/tpc.8.2.281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lester R L, Wells G B, Oxford G, Dickson R C. Mutant strains of Saccharomyces cerevisiae lacking sphingolipids synthesize novel inositol glycerophospholipids that mimic sphingolipid structures. J Biol Chem. 1993;268:845–856. [PubMed] [Google Scholar]
- 31.Mao C, Ruijuan X, Bielawska A, Obeid L. Cloning of an alkaline ceramidase from Saccharomyces cerevisiae. An enzyme with reverse (CoA-independent) ceramide synthase activity. J Biol Chem. 2000;275:6876–6884. doi: 10.1074/jbc.275.10.6876. [DOI] [PubMed] [Google Scholar]
- 32.Millar A A, Clemens S, Zachgo S, Giblin E M, Taylor D C, Kunst L. CUT1, an Arabidopsis gene required for cuticular wax biosynthesis and pollen fertility, encodes a very-long-chain fatty acid condensing enzyme. Plant Cell. 1999;11:825–838. doi: 10.1105/tpc.11.5.825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Millar A A, Kunst L. Very-long-chain fatty acid biosynthesis is controlled through the expression and specificity of the condensing enzyme. Plant J. 1997;12:121–131. doi: 10.1046/j.1365-313x.1997.12010121.x. [DOI] [PubMed] [Google Scholar]
- 34.Mitchell A G, Martin C E. Fah1p, a Saccharomyces cerevisiae cytochrome b5 fusion protein, and its Arabidopsis thaliana homolog that lacks the cytochrome b5 domain both function in the α-hydroxylation of sphingolipid-associated very long chain fatty acids. J Biol Chem. 1997;272:28281–28288. doi: 10.1074/jbc.272.45.28281. [DOI] [PubMed] [Google Scholar]
- 35.Munn A L, Stevenson B J, Geli M I, Riezman H. end5, end6, and end7: mutations that cause actin delocalization and block the internalization step of endocytosis in Saccharomyces cerevisiae. Mol Biol Cell. 1995;6:1721–1742. doi: 10.1091/mbc.6.12.1721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nagiec M M, Wells G B, Lester R L, Dickson R C. A suppressor gene that enables Saccharomyces cerevisiae to grow without making sphingolipids encodes a protein that resembles an Escherichia coli fatty acyltransferase. J Biol Chem. 1993;268:22156–22163. [PubMed] [Google Scholar]
- 37.Oh C S, Toke D A, Mandala S, Martin C E. ELO2 and ELO3, homologues of the Saccharomyces cerevisiae ELO1 gene, function in fatty acid elongation and are required for sphingolipid formation. J Biol Chem. 1997;272:17376–17384. doi: 10.1074/jbc.272.28.17376. [DOI] [PubMed] [Google Scholar]
- 38.Pan X, Goldfarb D S. YEB3/VAC8 encodes a myristylated armadillo protein of the Saccharomyces cerevisiae vacuolar membrane that functions in vacuole fusion and inheritance. J Cell Sci. 1998;111:2137–2147. doi: 10.1242/jcs.111.15.2137. [DOI] [PubMed] [Google Scholar]
- 39.Pan X, Roberts P, Chen Y, Kvam E, Shulga N, Huang K, Lemmon S, Goldfarb D S. Nucleus-vacuole junctions in Saccharomyces cerevisiae are formed through the direct interaction of Vac8p with Nvj1p. Mol Biol Cell. 2000;11:2445–2457. doi: 10.1091/mbc.11.7.2445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Parrish M L, Sengstag C, Rine J D, Wright R L. Identification of the sequences in HMG-CoA reductase required for karmellae assembly. Mol Biol Cell. 1995;6:1535–1547. doi: 10.1091/mbc.6.11.1535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Preston R A, Manolson M F, Becherer K, Weidenhammer E, Kirkpatrick D, Wright R, Jones E W. Isolation and characterization of PEP3, a gene required for vacuolar biogenesis in Saccharomyces cerevisiae. Mol Cell Biol. 1991;11:5801–5812. doi: 10.1128/mcb.11.12.5801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Preston R A, Reinagel P S, Jones E W. Genes required for vacuolar acidity in Saccharomyces cerevisiae. Genetics. 1992;131:551–558. doi: 10.1093/genetics/131.3.551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Revardel E, Bonneau M, Durrens P, Aigle M. Characterization of a new gene family developing pleiotropic phenotypes upon mutation in Saccharomyces cerevisiae. Biochim Biophys Acta. 1995;1263:261–265. doi: 10.1016/0167-4781(95)00124-y. [DOI] [PubMed] [Google Scholar]
- 44.Roggenkamp R, Numa S, Schweizer E. Fatty acid-requiring mutant of Saccharomyces cerevisiae defective in acetyl-CoA carboxylase. Proc Natl Acad Sci USA. 1980;77:1814–1817. doi: 10.1073/pnas.77.4.1814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rose M D, Novick P, Thomas J H, Botstein D, Fink G R. A Saccharomyces cerevisiae genomic plasmid bank based on a centromere-containing shuttle vector. Gene. 1987;60:237–243. doi: 10.1016/0378-1119(87)90232-0. [DOI] [PubMed] [Google Scholar]
- 46.Schneiter R. Brave little yeast, please guide us to Thebes: sphingolipid function in S. cerevisiae. Bioessays. 1999;21:1004–1010. doi: 10.1002/(SICI)1521-1878(199912)22:1<1004::AID-BIES4>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- 47.Schneiter R, Guerra C E, Lampl M, Tatzer V, Zellnig G, Klein H L, Kohlwein S D. A novel cold-sensitive allele of the rate-limiting enzyme of fatty acid synthesis, acetyl coenzyme A carboxylase, affects the morphology of the yeast vacuole through acylation of Vac8p. Mol Cell Biol. 2000;20:2984–2995. doi: 10.1128/mcb.20.9.2984-2995.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schneiter R, Hitomi M, Ivessa A S, Fasch E V, Kohlwein S D, Tartakoff A M. A yeast acetyl coenzyme A carboxylase mutant links very-long-chain fatty acid synthesis to the structure and function of the nuclear membrane-pore complex. Mol Cell Biol. 1996;16:7161–7172. doi: 10.1128/mcb.16.12.7161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schneiter R, Kohlwein S D. Organelle structure, function, and inheritance in yeast: a role for fatty acid synthesis? Cell. 1997;88:431–434. doi: 10.1016/s0092-8674(00)81882-6. [DOI] [PubMed] [Google Scholar]
- 50.Schneiter R, Tatzer V, Gogg G, Leitner E, Kohlwein S D. Elo1p-dependent carboxy-terminal elongation of C14:1Δ9 to C16:1Δ11 fatty acids in Saccharomyces cerevisiae. J Bacteriol. 2000;182:3655–3660. doi: 10.1128/jb.182.13.3655-3660.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schneiter R, Kohlwein S D. Identification of a very-long-chain fatty acid substituted phosphatidylinositol that possibly stabilizes highly curved membrane domains in yeast. Chem Phys Lipids. 1998;94:167. [Google Scholar]
- 52.Schweizer E, Werkmeister K, Jain M K. Fatty acid biosynthesis in yeast. Mol Cell Biochem. 1978;21:95–107. doi: 10.1007/BF00240280. [DOI] [PubMed] [Google Scholar]
- 53.Schweizer M, Lebert C, Holtke J, Roberts L M, Schweizer E. Molecular cloning of the yeast fatty acid synthetase genes, FAS1 and FAS2: illustrating the structure of the FAS1 cluster gene by transcript mapping and transformation studies. Mol Gen Genet. 1984;194:457–465. doi: 10.1007/BF00425558. [DOI] [PubMed] [Google Scholar]
- 54.Sherman F, Fink G R, Hicks J B. Methods in yeast genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1986. [Google Scholar]
- 55.Sikorski R S, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Silve S, Leplatois P, Josse A, Dupuy P H, Lanau C, Kaghad M, Dhers C, Picard C, Rahier A, Taton M, Le Fur G, Caput D, Ferrara P, Loison G. The immunosuppressant SR 31747 blocks cell proliferation by inhibiting a steroid isomerase in Saccharomyces cerevisiae. Mol Cell Biol. 1996;16:2719–2727. doi: 10.1128/mcb.16.6.2719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sivadon P, Bauer F, Aigle M, Crouzet M. Actin cytoskeleton and budding pattern are altered in the yeast rvs161 mutant: the Rvs161 protein shares common domains with the brain protein amphiphysin. Mol Gen Genet. 1995;246:485–495. doi: 10.1007/BF00290452. [DOI] [PubMed] [Google Scholar]
- 58.Sivadon P, Crouzet M, Aigle M. Functional assessment of the yeast Rvs161 and Rvs167 protein domains. FEBS Lett. 1997;417:21–27. doi: 10.1016/s0014-5793(97)01248-9. [DOI] [PubMed] [Google Scholar]
- 59.Toke D A, Martin C E. Isolation and characterization of a gene affecting fatty acid elongation in Saccharomyces cerevisiae. J Biol Chem. 1996;271:18413–18422. doi: 10.1074/jbc.271.31.18413. [DOI] [PubMed] [Google Scholar]
- 60.Tuller G, Prein B, Jandrositz A, Daum G, Kohlwein S D. Deletion of six open reading frames from the left arm of chromosome IV of Saccharomyces cerevisiae. Yeast. 1999;15:1275–1285. doi: 10.1002/(SICI)1097-0061(19990915)15:12<1275::AID-YEA456>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- 61.Tvrdik P, Westerberg R, Silve S, Asadi A, Jakobsson A, Cannon B, Loison G, Jacobsson A. Role of a new mammalian gene family in the biosynthesis of very long chain fatty acids and sphingolipids. J Cell Biol. 2000;149:707–718. doi: 10.1083/jcb.149.3.707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Vahlensieck H F, Pridzun L, Reichenbach H, Hinnen A. Identification of the yeast ACC1 gene product (acetyl-CoA carboxylase) as the target of the polyketide fungicide soraphen A. Curr Genet. 1994;25:95–100. doi: 10.1007/BF00309532. [DOI] [PubMed] [Google Scholar]
- 63.Vida T A, Emr S D. A new vital stain for visualizing vacuolar membrane dynamics and endocytosis in yeast. J Cell Biol. 1995;128:779–792. doi: 10.1083/jcb.128.5.779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wach A, Brachat A, Alberti-Segui C, Rebischung C, Philippsen P. Heterologous HIS3 marker and GFP reporter modules for PCR-targeting in Saccharomyces cerevisiae. Yeast. 1997;13:1065–1075. doi: 10.1002/(SICI)1097-0061(19970915)13:11<1065::AID-YEA159>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- 65.Wakil S J. Fatty acid synthase, a proficient multifunctional enzyme. Biochemistry. 1989;28:4523–4530. doi: 10.1021/bi00437a001. [DOI] [PubMed] [Google Scholar]
- 66.Wang Y X, Catlett N L, Weisman L S. Vac8p, a vacuolar protein with armadillo repeats, functions in both vacuole inheritance and protein targeting from the cytoplasm to vacuole. J Cell Biol. 1998;140:1063–1074. doi: 10.1083/jcb.140.5.1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wright R, Basson M, D'Ari L, Rine J. Increased amounts of HMG-CoA reductase induce “karmellae”: a proliferation of stacked membrane pairs surrounding the yeast nucleus. J Cell Biol. 1988;107:101–114. doi: 10.1083/jcb.107.1.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zhao C, Beeler T, Dunn T. Suppressors of the Ca2+-sensitive yeast mutant (csg2) identify genes involved in sphingolipid biosynthesis. Cloning and characterization of SCS1, a gene required for serine palmitoyltransferase activity. J Biol Chem. 1994;269:21480–21488. [PubMed] [Google Scholar]