Abstract
Hepatitis B virus (HBV) infection remains a worldwide health problem and no eradicative therapy is currently available. Host T cell immune responses have crucial influences on the outcome of HBV infection, however the development of therapeutic vaccines, T cell therapies and the clinical evaluation of HBV-specific T cell responses are hampered markedly by the lack of validated T cell epitopes. This review presented a map of T cell epitopes functionally validated from HBV antigens during the past 33 years; the human leukocyte antigen (HLA) supertypes to present these epitopes, and the methods to screen and identify T cell epitopes. To the best of our knowledge, a total of 205 CD8+ T cell epitopes and 79 CD4+ T cell epitopes have been defined from HBV antigens by cellular functional experiments thus far, but most are restricted to several common HLA supertypes, such as HLA-A0201, A2402, B0702, DR04, and DR12 molecules. Therefore, the currently defined T cell epitope repertoire cannot cover the major populations with HLA diversity in an indicated geographic region. More researches are needed to dissect a more comprehensive map of T cell epitopes, which covers overall HBV proteome and global patients.
Keywords: hepatitis B virus, T cell epitope, HLA restriction
1. Introduction
Hepatitis B virus (HBV) infection still disseminates across the world and causes the most common and fatal liver diseases including acute liver failure, chronic hepatitis, liver cirrhosis (LC), and hepatocellular carcinoma (HCC) [1,2]. Nucleoside analogs and/or interferon are widely utilized antiviral drugs, which can effectively suppress virus replication, decrease serum HBV DNA to undetectable levels, mitigate liver fibrosis, and reduce HCC risk [3,4,5], however cannot eliminate the virus in patients. Recurrence after therapy discontinuation is emerging to be a common etiology of morbidity and mortality in patients with chronic HBV infection [6].
Numerous researches have demonstrated the important influence of HBV-specific T cell responses on virus clearance [7], disease progression [8,9,10], antiviral efficacy [11,12], and recurrence [13,14,15], particularly the CD8+ T cells, which act as vital effector cells to kill virus-infected hepatocytes and secret cytokines. Patients with acute-resolving HBV infection show robust HBV-specific CD8+ T cell responses, while the patients with chronic HBV infection present a phenomenon termed CD8+ T cell functional exhaustion with multifactorial heterogeneity [9], and differs depending on the targeted antigen for HLA-A02 restricted epitopes located in the core antigen versus polymerase [16]. Furthermore, the heterogeneity of HBV-specific T cells also responds differently to therapeutic stimuli [17]. Therefore, T cells specific for HBV not only are the potential markers for monitoring the effects of antiviral therapy and predicting the recurrence [18], but also are the promising modulators in specific immunotherapy. Identifying the T cell epitopes as many as possible from HBV antigens will greatly contribute to the design and development of epitope-based and T cell-based therapies and the detection of host HBV-specific T cell immunity. Although a systematic review of T cell epitopes in HBV antigens was reported in 2008 [19], an updated map of the T cell epitopes is urgently needed.
Here, this review comprehensively collected the CD8+ T cell epitopes and CD4+ T cell epitopes defined from HBV proteome during the past 33 years. Information resources are the English language journals collected in Pubmed, Scopus, Embase, SinoMed, and Google Scholar databases. The latest online search was conducted on October 8, 2021. “T cell epitopes” and “HBV or hepatitis B virus” were used as specific searching terms. An initial search identified 451 studies from multiple databases and manual searches. All articles were imported to Endnote software X8 (Thompson and Reuters, Philadelphia, PA, USA) and 121 duplicates were removed. In total, 330 studies from 1988 to 2021 were collected. Then, 233 articles were filtered out after abstract and full-text screening, according to the exclusion criteria below: (1) not related to the screening or identification of T cell epitopes; (2) just using in silico prediction or molecular structure bioinformatic analysis rather than satisfactory cell functional experiments, tetramer staining, binding assay, stabilization assay, or immunization; (3) with incomplete information regarding epitopes sequences. Finally, 97 articles were analyzed and referenced in this review.
2. Polymorphism of HLA Alleles and Association with HBV Infection
Human leukocyte antigens (HLA) are coded by human major histocompatibility complex and have multiple important functions. In particular, they present antigenic peptides (T cell epitopes) in the form of peptide/HLA complex to T cell receptors onto specific T cells by which to initiate the adaptive immune response. HLA class I molecules (classically HLA-A, -B, and -C) are constitutively expressed onto almost all nucleated cells with distinct levels and present antigenic peptides to specific CD8+ T cells, while HLA class II molecules (classically HLA-DR, -DQ and -DP) are mainly expressed onto professional antigen-presenting cells (APCs, including monocytes, macrophages, dendritic cells and B cells) and present peptides to specific CD4+ T cells. In virus infection, HLA class I molecules expressed by virus-infected cells present the viral endogenous epitope peptides to specific CD8+ T cells, thus initiating the naive CD8+ T cells to activate, proliferate and differentiate to cytotoxic T lymphocytes (CTLs). The resulting CTLs mediate the cytolysis of virus-infected cells by Fas/FasL, TNF/TNFL, and perforin/granzyme pathways [20]. HLA class II molecules expressed by APCs present exogenous viral peptides to CD4+ helper T cells, thus eliciting the naive CD4+ T cells to differentiate into effective Th1 or Th2 cells. The former help virus-specific CD8+ T cells activation and the latter help virus-specific B cells differentiate to plasma cells and produce antibodies [21]. However, HLA molecules are highly polymorphic in the general population. As of October 2021, a total of 24,284 alleles have been described at HLA class I and class II loci in the global populations, including 6921 HLA-A, 8181 HLA-B, 6779 HLA-C, 3801 HLA-DRB1, 2033 HLA-DQB1, and 1862 HLA-DPB1 alleles, according to the International Immunogenetics Information Project/HLA database (IMGT; www.ebi.ac.uk/imgt/hla/stats.html, accessed on 11 November 2021). HLA allotypes are distinctive from individual to individual, and each HLA allotype presents distinctive antigenic peptides, thus leading to different immune patterns in different individuals against the same pathogen such as HBV [22,23]. Among the different ethnic populations in different geographic regions, the distribution of prevalent HLA alleles is markedly different. For an instance, 13 kinds of predominant HLA-A allotypes (each allotype has a gene frequency of >1% in Chinese herd) gather a total HLA-A allele frequency of around 95.5% in the Chinese population while 94%, 83%, 80%, 70% and 63% in Northeast Asia, Southeast Asia, Europe, South America, and North America populations, respectively (http://www.allelefrequencies.net, accessed on 11 November 2021).
Consequently, some alleles of HLA molecules have increasingly been linked to the occurrence of the indicated diseases, which are usually associated with abnormal immune function and genetic tendency [24,25]. Although the association of HLA alleles with HBV infection is not well clarified, a few studies have indicated that HLA-DRB1*13 and HLA-DRB1*07 are related to susceptibility to chronic HBV infection, and DRB1*15 is negatively related to persistence to chronic HBV infection in the populations of Africans [26], Europeans [27], Koreans [28] and Northwestern Chinese [29]. In addition, HLA-A*33 is closely associated with susceptibility to persisting HBV infection, and HLA-DRB1*13 is closely related to protection against persisting HBV infection in an Iranian population [30]. A*0301 and DRB1*1302 are relevant to viral clearance and B*08 is associated with viral persistence in Caucasians [31]. However, although the correlation between HBV infection and HLA alleles has been studied for several decades, in accordance with what we described in the above review, it often has conflicting results. These variations partly result from host HLA polymorphism in different races and regions [32,33]. Further studies should be explored in different regions to reduce the heterogeneity of results.
3. HBV Proteome and the Approaches Identifying T Cell Epitopes
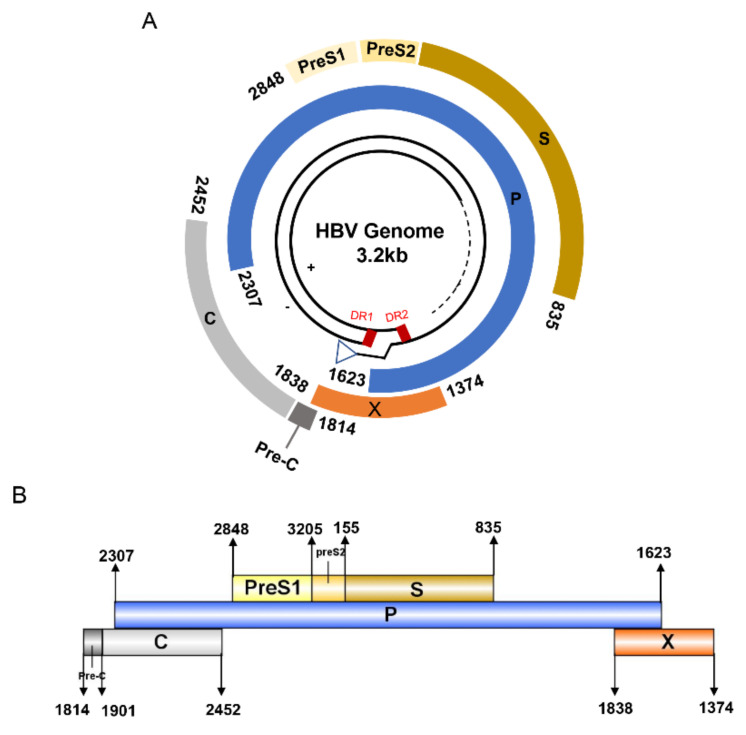
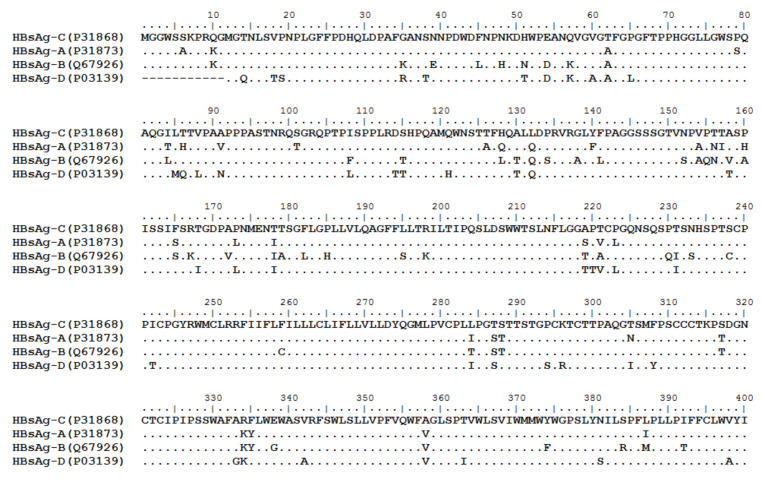
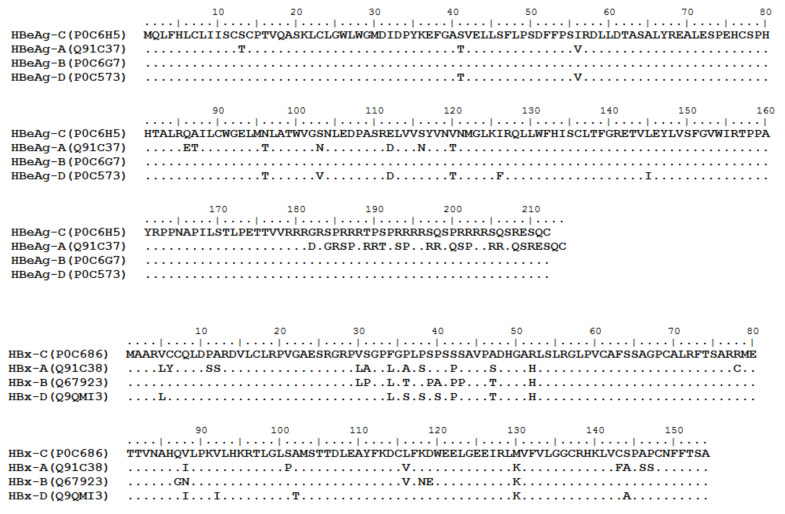
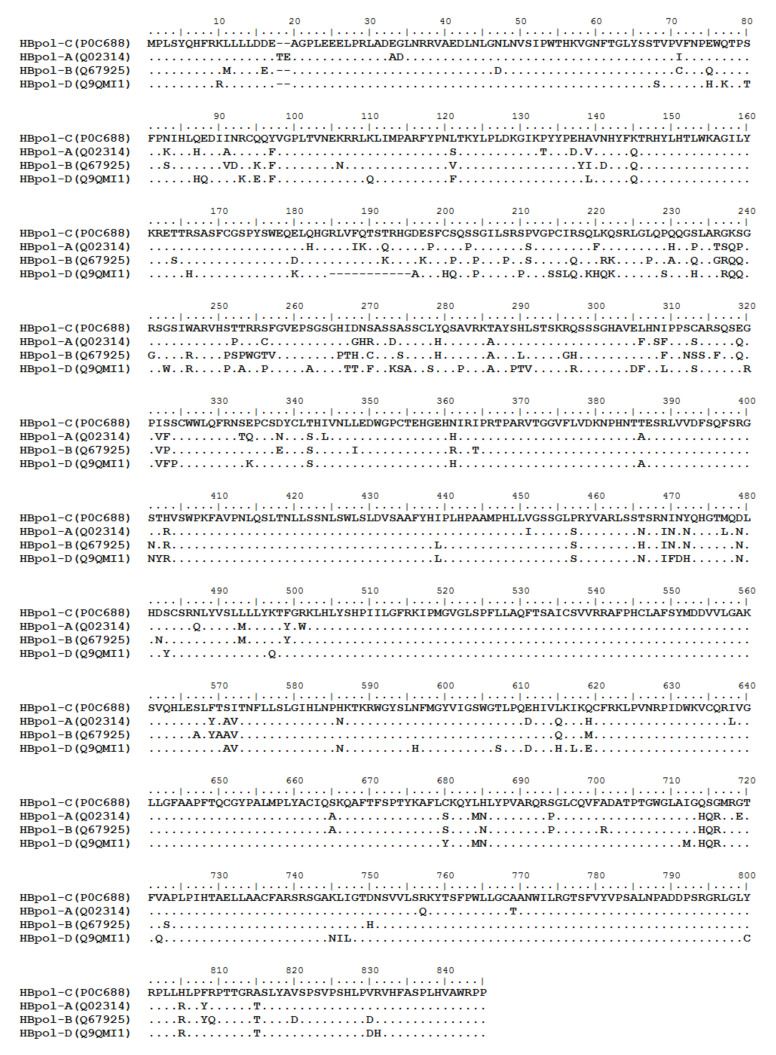
HBV is one of the smallest viruses with a genome length of 3.2 Kb [34]. Its genome contains four open reading frames (ORFs) coding four partially overlapping proteins as displayed in Figure 1: (1) preS/S ORF encodes large (L), middle (M), and small (S) surface antigens (HBsAg). HBsAg is being widely investigated in clinical fields and quantified as a diagnostic marker of HBV infection as it can reflect the level of covalently closed circular DNA (cccDNA) and intrahepatic HBV DNA in chronic infection [35,36]. (2) Pre-core/core ORF encodes hepatitis B e antigen (HBeAg), core antigen (HBcAg) or in combination core-related antigen (HBcrAg). HBeAg has long been advocated as a serum marker for guiding the clinical practice of chronic hepatitis B virus [37,38]. HBcrAg has been demonstrated more recently as a potential surrogate marker of cccDNA [39]. (3) X ORF encodes HBx antigen (HBxAg), which plays an important role in virus genome transcription and is correlated with liver cancer. The expression of HBxAg in HBV-associated HCC patients is significantly higher than other viral proteins [40]. (4) P ORF encodes the viral DNA polymerase (HBpol), which is responsible for the replication of the viral genome and is an effective target for the therapeutic intervention of chronic HBV infection [41]. Human HBV strains occur in nine genotypes A-I, and its major HBV surface antigen (HBsAg) has several immune protective conformational B cell epitopes a, d or y, w1–4 or r [42]. The entire amino acid sequences of each protein from different genotypes were obtained from the UniProt database and aligned in Figure 2.
Figure 1.
The circular (A) and linear (B) diagram of HBV genome.
Figure 2.
Homologous analysis of HBsAg, HBeAg, HBx and HBpol proteins from HBV C, A, B, and D genotypes. The entire amino acid sequences of each protein from different HBV genotypes were obtained from the UniProt database, aligned and used for in silico prediction of HBV antigen T cell epitopes presented by HLA-A allotypes.
The process of T cell epitope identification begins with the selection of candidate epitope peptides. The first strategy is using overlapping peptides (OLPs) spanning the entire proteome or selected antigens of interest (peptide scanning). Chen et al. expanded HBV-specific T cells in vitro by co-culturing the overlapping peptide pools spanning the entire sequence of HBV genotypes B and C and the peripheral blood mononuclear cells (PBMCs) from patients with chronic HBV infection, followed by the detection of T cell response in each co-culture using IFN-γ enzyme-linked immunospot (IFN-γ ELISpot) assay, IFN-γ intracellular staining and flow cytometry [43]. However, peptide scanning is a high-cost and laborious method due to a large amount of OLPs spanning overall HBV proteins. For CD8+ T cell epitopes, HBsAg, HBeAg, HBx and HBpol contain 131, 68, 49, and 279 OLPs, respectively, when overlapping 6 amino acids in each 9-mer peptide. An alternative strategy is to focus on the in silico predicted T cell epitopes binding to the indicated HLA supertypes as calculated by multiple epitope prediction tools and algorithms. Brinck-Jensen et al. predicted 20 HBV-specific epitopes using combined in silico methods and evaluated for the immunogenicity of these epitopes through exposure to patients’ PBMCs by IFN-γ ELISpot [44]. More recently, a similar in silico approach was also employed to assess all previously verified HBx- and HBpol-derived epitopes and to predict novel HLA-binding peptides for 6 HLA supertypes. Then, a part of reported epitopes were chosen for experimental validation. A total of 13 HLA binders derived from HBx and 33 binders from HBpol were described across HLA subtypes by this strategy [45]. Predicted epitopes are based on the indicated HLA restrictions and limit the number of research objects with diverse HLA subtypes to a reasonable range, yet the inaccuracy of theoretical prediction may omit some real-world epitopes.
The methodologies to validate the immunogenicity of candidate epitope peptides have been improved remarkably over the last two decades. Different assays are utilized for the detection of peptide-induced T cell response or peptide-specific T cells with individual advantages and disadvantages in terms of practicability, cost, sensitivity, function evaluation. The following approaches are currently widely used, such as cytotoxicity assay, proliferation assay, intracellular cytokine staining (ICS), ELISpot/FluoroSpot, and peptide-MHC multimers staining (tetramers, pentamers, or dextramers). The cytotoxicity assay was initially performed to validate CD8+ T cell epitopes by co-culturing patients’ PBMCs with target cells labeled with Chromium-51, after the PBMCs were stimulated by the indicated candidate epitope peptides [46]. Additionally, lymphocyte proliferation assay is mostly applied to CD4+ T cell epitopes validation. The PBMCs from HBV-infected or HBV-vaccinated individuals were co-cultured with HBV-derived peptides for several days and 3H-thymidine pulses were administered eventually followed by quantifying the incorporated radioactivity [47]. One more common approach currently utilized is ICS or ELISpot/FluoroSpot. Patient’s PBMCs are in vitro or ex vivo stimulated with the candidate epitope peptides and simultaneously cytokine release is blocked followed by ICS and flow cytometry to define whether CD4+ T cells or CD8+ T cells activation [48]. The ELISpot or FluoroSpot technology enables the detection of single activated cells among one million PBMCs. The accuracy, sensitivity, reproducibility and durability have led to its widespread applications in researches and the broad prospects in the clinical detection of antigen-specific T cells [49,50]. An issue encountered with ELISpot, FluoroSpot, ICS, and related assays is that they may ignore T cells that produce different cytokines or trace cytokines during the window of time of the assay (e.g., Follicular helper CD4+ T cells generally produce very low amounts of cytokines). Peptide-MHC tetramer staining has been the gold standard to quantify antigen-specific T cells with high sensitivity and precision, thus is often used to identify T cell epitopes in many researches. However, the preparation of peptide-MHC tetramers or multimers is high-cost, complicated, and time-consumption [51,52]. A pioneering study focused on all possible peptides of the entire HBV genome and 484 unique HLA-A1101-restricted epitopes predicted by NetMHC algorithms were validated using mass cytometry and multiplex peptide-tetramers staining [53]. Many researchers also have established a transgenic mouse model to map HLA-restricted epitopes. Ru et al. developed and immunized HLA-A2/DP4 mice with epitopes derived from HBsAg to identify four new HLA-DP4-restricted epitopes [54]. Besides cellular functional experiments, peptide-HLA molecule binding and stabilization assays were commonly used to identify epitopes. Pan et al. defined 16 HBV epitopes by analyzing the different binding affinities of candidate epitope peptides with HLA-A3303 using RMA-S cells binding and stability assay. More recently, Ferretti et al. used a high-throughput genome-wide screening technology to identify the target cells expressing candidate epitopes productively recognized by T cells (T-Scan) and determined 29 epitopes in SARS-CoV-2 for the six most prevalent HLA types [55]. Chikata et al. employed immunocapture and liquid chromatography mass spectrometry (LC-MS) subsequent to pre-treatment of the target protein to disrupt its three-dimensional structure to characterize HIV-1 epitope peptides on a large scale presented by HLA-C1202 [56]. A variety of epitope assay strategies have been utilized with their own features and potential.
4. Defined T Cell Epitopes in HBV Proteins during the Past 33 Years
Table 1 collected the CD8+ T cell epitopes and CD4+ T cell epitopes defined from HBV proteome during the past 33 years and displayed their HLA restrictions and the methods used to validate their immunogenicity. Notably, we performed manual management in this review, only the epitopes of 8–14 or 12–25 amino acids in length presented by HLA class I molecules or class II molecules are displayed since they reflect the standard size of the peptide-binding groove of HLA molecules. According to the previous report, if the epitope peptides are too short or long, the experiment tends to represent false positives instead of the result caused by the binding of peptide and HLA molecule [57].
Table 1.
List of CD4+ T cell epitopes and CD8+ T cell epitopes validated from HBV proteins.
| Sequence | Protein | Position | Reference | HLA Restriction | Method to Screen Candidate Epitopes | Method to Validate the Candidate Epitopes |
|---|---|---|---|---|---|---|
| MQLFHLCLI | Core | 1–8 | [61] | A*0201 | Predicted | Binding assay; ELISpot; Cytotoxicity assay; CTL assay |
| KEFGASVEL(L) | Core | 7–15/16 | [62] | A*0206, B*4001 | Predicted | ELISpot; ICS; Binding assay |
| EFGASVELL | Core | 8–16 | [63] | A*0201, A*0207 | overlapping | ICS; ELISpot |
| FLPSDFFPS | Core | 18–26 | [64] | A*0201 | Predicted | ICS; Tetramer staining |
| FLPSDFFPSV | Core | 18–27 | [45,65,66,67,68,69,70,71,72,73,74,75,76,77,78,79] | A*02, A*0201, A*0202, A*0203, A*0206, A*6802, A*0301, A*0207 | overlapping | Immunization of mice; Cytotoxicity assay; CTL assay; Binding assay; Tetramer staining; ELISA |
| LPSDFFPSV | Core | 19–27 | [74,80,81,82,83] | B*3501, B*51, B*5301, B*5401, B*07, B*51, A*0201 | overlapping | Binding assay; CTL assay; Cytotoxicity assay; Tetramer staining |
| FFPSIRDLL | Core | 23–31 | [84] | A*24 | Predicted | Tetramer staining |
| DLLDTASALY | Core | 39–48 | [81] | A*0101, A*2902, A*3002 | Predicted | Binding assay; Immunization of mice; ELISpot |
| DFFPSIRDL | Core | 51–59 | [85] | A*2402 | Predicted | ELISpot |
| LCWGELMNL | Core | 60–68 | [86] | A*0201 | Predicted | Stabilization assay; ELISpot assay |
| ELMNLATWV | Core | 64–72 | [87] | A*02 | Predicted | Binding assay; ELISpot; Cytotoxicity assay |
| SYVNMNMGL | Core | 87–95 | [88] | A*2402 | Predicted | Binding assay; CTL assay |
| SYVNTNMGL | Core | 87–95 | [89] | A*02 | Predicted | Tetramer staining |
| YVNVNMGLK | Core | 88–96 | [63] | A*1101 | overlapping | ICS; ELISpot |
| MGLKFRQL | Core | 93–100 | [90] | A*0201 | Predicted | Immunization of mice; FACS |
| LLWFHISCL | Core | 101–108 | [43] | A*0201 | Predicted | Proliferation assay; ICS; Cytotoxicity assay |
| LWFHISCLTF | Core | 101–110 | [85] | A*2402, A*2301 | Predicted | ELISpot |
| HISCLTFGR | Core | 104–112 | [91,92] | A*33 | Predicted | Cytotoxicity assay; ICS; Tetramer staining |
| CLTFGRETV | Core | 107–115 | [93] | A*02 | Predicted | Tetramer staining |
| EYLVSFGVW | Core | 117–125 | [81,84,88] | A*2402, A*2407, A*2301 | Predicted | Stabilization assay; CTL assay; Cytotoxicity assay; Tetramer staining; Binding assay; Immunization of mice; ELISpot |
| YLVSFGVWI | Core | 118–126 | [43] | A*0201 | Predicted | Proliferation assay; ICS; Cytotoxicity assay |
| LVSFGVWIR | Core | 119–127 | [91] | A*33 | Predicted | Stabilization assay; ELISpot; Cytotoxicity assay; Immunization of mice |
| GLKILQLL | Core | 123–130 | [82] | B*08 | overlapping | ICS; Tetramer staining |
| AYRPPNAPI | Core | 131–139 | [94] | A*0201 | Predicted | ELISpot; Cytotoxicity assay |
| LTFGRETVLEN | Core | 137–147 | [95] | A*0101, A*02, A*2902, A*3002 | Predicted | ELISpot |
| ILSTLPETTV | Core | 139–148 | [75] | A*02 | Predicted | CTL assay |
| STLPETTVVR | Core | 141–150 | [74,96] | A*11, A*6801, A*02 | overlapping | Binding assay; CTL assay; Cytotoxicity assay; ELISpot |
| STLPETTVVRR | Core | 141–151 | [17,76,81,92,97] | A*31, A*68, A*02, A*0201, A*6801, A*03, A*11, A*3101, A*0201 | overlapping | Cytotoxicity assay; Immunization of mice; CTL assay; Binding assay; ELISpot |
| TLPETTVVRR | Core | 142–151 | [63] | A*1101 | overlapping | ICS; ELISpot |
| GVWIRTPPA | Core | 152–160 | [98] | A*0201 | Predicted | ELISpot |
| STLPETAVVRR | Core | 170–180 | [9] | A*1101 | Predicted | Proliferation assay; Tetramer staining |
| RTQSPRRR | Core | 196–203 | [9] | A*1101 | Predicted | Proliferation assay; Tetramer staining |
| RTQSPRRRR | Core | 196–204 | [9] | A*1101 | Predicted | Proliferation assay; Tetramer staining |
| RSQSPRRRRSK | Core | 196–206 | [9] | A*1101 | Predicted | Proliferation assay; Tetramer staining |
| RLCCQLDPA | HBx | 4–12 | [99] | A*0201 | Predicted | Binding assay; ELISpot; Cytotoxicity assay |
| AYFKDCVFKDW | HBx | 6–16 | [45] | A*2402 | Predicted | ELISA |
| QLDPARDVL | HBx | 8–16 | [45,65,73,99,100,101] | A*0201 | Predicted | ELISpot |
| VLCLRPVGA | HBx | 15–23 | [45,99,102] | A*0201 | Predicted | ELISpot |
| RGRPVSGPF | HBx | 26–34 | [85] | A*2402 | Predicted | ELISpot |
| PVSGPFGPL | HBx | 29–37 | [100] | A*0201 | Predicted | Immunization of mice; CTL assay; Cytotoxicity assay |
| AVPADHGAHL | HBx | 44–53 | [100] | A*0201 | Predicted | Immunization of mice; CTL assay; Cytotoxicity assay |
| HLSLRGLPV | HBx | 52–60 | [65,99,100,101,102,103] | A*0201, A*02 | Predicted | Cytotoxicity assay; Immunization of mice; CTL assay; Binding assay; ELISpot |
| LPVCAFSSA | HBx | 58–66 | [45] | B*0702 | Predicted | ELISA |
| AFSSAGPCALRF | HBx | 62–73 | [45] | A*2402 | Predicted | ELISA |
| ALRFTSARR | HBx | 70–78 | [45] | A*0301 | Predicted | ELISA |
| ALRFTSARRM | HBx | 70–79 | [100] | A*0201 | Predicted | Immunization of mice; CTL assay; Cytotoxicity assay |
| NAHQILPKV | HBx | 84–92 | [99] | A*0201 | Predicted | Binding assay; ELISpot; Cytotoxicity assay |
| (K)VLHKRTLGL | HBx | 91/92–100 | [65,100,102] | A*0201 | Predicted | Cytotoxicity assay; Binding assay; ELISpot; Tetramer staining |
| VLHKRTLGL | HBx | 92–100 | [99,101,104] | A*0201, A*02 | Predicted | Binding assay; ELISpot; Cytotoxicity assay; Proliferation assay; ELISpot; ICS |
| TLGLAAMST | HBx | 97–105 | [100] | A*0201 | Predicted | Binding assay; ELISpot; Cytotoxicity assay |
| GLSAMSTTDL | HBx | 99–108 | [99,100,104] | A*0201, A*02 | Predicted | Binding assay; ELISpot; Cytotoxicity assay |
| AMSTTDLEA | HBx | 102–110 | [99] | A*0201 | Predicted | Binding assay; ELISpot; Cytotoxicity assay |
| STTDLEAYFK | HBx | 104–113 | [45] | A*1101 | Predicted | ELISA |
| DLEAYFKDCL | HBx | 107–116 | [100] | A*0201 | Predicted | Immunization of mice; CTL assay; Cytotoxicity assay |
| CLFKDWEEL | HBx | 115–123 | [99,100,102] | A*0201 | Predicted | Immunization of mice; CTL assay; Cytotoxicity assay; Binding assay; ELISpot |
| ELGEEIRLKV | HBx | 122–131 | [100] | A*0201 | Predicted | Immunization of mice; CTL assay; Cytotoxicity assay |
| EIRLKVFVL | HBx | 126–134 | [100] | A*0201 | Predicted | Immunization of mice; CTL assay; Cytotoxicity assay |
| VLGGCRHKL | HBx | 133–141 | [99,101] | A*0201, A*02 | Predicted | Binding assay; ELISpot; Cytotoxicity assay; ELISpot |
| VLGGCRHKL(V) | HBx | 133–141/142 | [98] | A*0201 | Predicted | Immunization of mice; CTL assay; Cytotoxicity assay |
| LLDDEAGPL | Pol | 13–21 | [105,106] | A*0201 | Predicted | Binding assay; Immunization of mice; CTL assay; Cytotoxicity assay |
| PLEEELPRL | Pol | 20–28 | [105,106] | A*0201 | Predicted | Binding assay; Immunization of mice; CTL assay; Cytotoxicity assay |
| DLNLGNLN | Pol | 40–48 | [106] | A*0201 | Predicted | Binding assay; Immunization of mice; CTL assay; Cytotoxicity assay |
| NLGNLNVSI | Pol | 42–50 | [106] | A*0201 | Predicted | Binding assay; Immunization of mice; CTL assay; Cytotoxicity assay |
| NVSIPWTHK | Pol | 47–55 | [9,74,81] | A*03, A*11, A*6801, A*0301, A*1101 | Predicted | Stabilization assay; ELISpot; Cytotoxicity assay; Immunization of mice; Proliferation assay; Tetramer staining; Binding assay |
| KVGNFTGLY | Pol | 55–63 | [45,74] | A*0301, A*03, A*11 | Predicted | Binding assay; CTL assay; Cytotoxicity assay; ELISA |
| GLYSSTVPV | Pol | 61–69 | [73,105,106] | A*0201 | Predicted | Binding assay; Immunization of mice; CTL assay; Cytotoxicity assay; Tetramer staining |
| LYSSTVPVF | Pol | 62–70 | [79] | A*24 | Predicted | ELISpot |
| STVPCFNPK | Pol | 65–73 | [9] | A*1101 | Predicted | Proliferation assay; Tetramer staining |
| TVPCFNPK | Pol | 66–73 | [9] | A*1101 | Predicted | Proliferation assay; Tetramer staining |
| PSFPHIHLK | Pol | 77–85 | [9] | A*1101 | Predicted | Proliferation assay; Tetramer staining |
| QYVGPLTVN | Pol | 94–102 | [85] | A*2402 | Predicted | ELISpot |
| YLHTLWKAGI | Pol | 147–156 | [65] | A*02 | Predicted | ELISpot assay; Tetramer staining |
| (H)TLWKAGILYK | Pol | 149/150–159 | [81] | A*03 | Predicted | Binding assay; Immunization of mice; ELISpot |
| HTLWKAGILYK | Pol | 149–159 | [74,76,98] | A*03, A*11, A*3101, A*3301, A*6801, A*02, A*11 | Predicted | Immunization of mice; Cytotoxicity assay; Binding assay; CTL assay |
| TLWKAGILY(K) | Pol | 150–158/159 | [74] | A*03, A*11 | Predicted | Binding assay; CTL assay; Cytotoxicity assay |
| RSASFCGSPY | Pol | 164–173 | [45] | A*1101 | Predicted | ELISA |
| ASFCGSPYSW | Pol | 166–175 | [45,62,63] | A*2402, B*5801 | overlapping | ELISA; ELISpot; ICS |
| SFCGSPYSW | Pol | 167–175 | [45] | A*2402 | Predicted | ELISA |
| ASFCGSPY | Pol | 166–173 | [81,95,107] | A*0101, A*2902, A*3002 | overlapping | Binding assay; Immunization of mice; ELISpot; Tetramer staining |
| SPYSWEQEL | Pol | 171–179 | [17] | A*0201, B*3501 | Predicted | Tetramer staining |
| QSSGILSR | Pol | 200–207 | [9] | A*1101 | Predicted | Proliferation assay; Tetramer staining |
| GILPRSSVGPR | Pol | 205–215 | [9] | A*1101 | Predicted | Proliferation assay; Tetramer staining |
| CLHQSAVRK | Pol | 274–282 | [45] | A*0301, A*1101 | Predicted | ELISA |
| KTAYSHLSTSK | Pol | 283–293 | [9] | A*1101 | Predicted | Proliferation assay; Tetramer staining |
| SSARSQSER | Pol | 310–318 | [9] | A*1101 | Predicted | Proliferation assay; Tetramer staining |
| CLSLIVNLL | Pol | 338–346 | [65] | A*02 | Predicted | ELISpot assay; Tetramer staining |
| TPARVTGGV | Pol | 354–362 | [45] | B*0702 | Predicted | ELISA |
| TPARVTGGVF | Pol | 354–363 | [45] | B*0702 | Predicted | ELISA |
| RVTGGVFLV | Pol | 357–365 | [45] | A*0201 | Predicted | ELISA |
| VTGGVFLVDK | Pol | 358–367 | [45] | A*1101, A*03 | Predicted | ELISA |
| RIPRTPSRV | Pol | 361–369 | [65] | A*02 | Predicted | ELISpot assay; Tetramer staining |
| TPARVTGGVF | Pol | 365–374 | [74,76,108] | B*0702, B*3501, A*03, B*07, A*02, B*51 | Predicted | Immunization of mice; Cytotoxicity assay; Binding assay; CTL assay |
| RVTGGVFLVDK | Pol | 368–378 | [74] | A*11 | Predicted | Binding assay; CTL assay; Cytotoxicity assay |
| VTGGVFLVDK | Pol | 369–378 | [74] | A*03, A*11 | Predicted | Binding assay; CTL assay; Cytotoxicity assay |
| FLVDKNPHNT | Pol | 374–383 | [62] | A*0203 | Predicted | ELISpot; ICS; Binding assay |
| LVVDFLHQFSR | Pol | 377–386 | [9] | A*1101, A*3301, A*6801 | Predicted | Proliferation assay; Tetramer staining; Binding assay; Immunization of mice; ELISpot; CTL assay; Cytotoxicity assay |
| SRLVVDFSQF | Pol | 386–395 | [63] | B*1301 | overlapping | ICS; ELISpot |
| VVDFSQFSR | Pol | 389–397 | [74,91] | A*11, A*6801, A*33 | Predicted | Stabilization assay; ELISpot; Cytotoxicity assay; Binding assay; Immunization of mice; CTL assay |
| SWPKFAVPNL | Pol | 392–401 | [45] | A*2402 | Predicted | ELISA |
| WPKFAVPNL | Pol | 393–401 | [45] | B*0702 | Predicted | ELISA |
| FAVPNLQSL | Pol | 396–404 | [45] | A*0201 | Predicted | ELISA |
| NLQSLTNLL | Pol | 411–419 | [105,106] | A*0201 | Predicted | Cytotoxicity assay; Immunization of mice; Binding assay; CTL assay |
| LLSSNLSWL | Pol | 418–426 | [65,105,106] | A*0201 | Predicted | Cytotoxicity assay; Immunization of mice; Binding assay; CTL assay; ELISpot; Tetramer staining |
| NLSWLSLDV | Pol | 422–430 | [101,105,106] | A*0201, A*02 | Predicted | Cytotoxicity assay; Immunization of mice; Binding assay; CTL assay; ELISpot |
| LSLDVSAAFY | Pol | 426–435 | [81] | A*0101, A*2902, A*3002 | Predicted | Binding assay; Immunization of mice; ELISpot |
| HPAAMPHLL | Pol | 440–448 | [74] | B*0702 | Predicted | Binding assay; CTL assay; Cytotoxicity assay |
| HLLVGSSGL | Pol | 446–454 | [105,106] | A*0201 | Predicted | Cytotoxicity assay; Immunization of mice; Binding assay; CTL assay |
| GLPRYVARL | Pol | 453–461 | [65,71,73,74,81,92,93,100,101,106,109,110,111] | A*0201, A*0202, A*0203, A*02, A*0207 | Predicted | Cytotoxicity assay; Immunization of mice; Binding assay; CTL assay; ELISpot; Tetramer staining |
| RIINNQHR | Pol | 466–473 | [9] | A*1101 | Predicted | Proliferation assay; Tetramer staining |
| RNLYVSLLL | Pol | 484–492 | [85] | A*2402 | Predicted | ELISpot |
| NLYVSLLLL | Pol | 485–493 | [65,106] | A*0201, A*02 | Predicted | Cytotoxicity assay; Immunization of mice; Binding assay; CTL assay; ELISpot; Tetramer staining |
| KLHLYSHPI | Pol | 500–508 | [45,62,93,101,106] | A*0201, A*02, A*0203, B*0801 | Predicted | Cytotoxicity assay; Immunization of mice; Binding assay; CTL assay; ELISpot; Tetramer staining; ELISA |
| HLYSHPIIL | Pol | 502–510 | [65,105,112,113,114] | A*0201, A*02, A*0203 | overlapping | Cytotoxicity assay; Immunization of mice; Binding assay; ELISpot; Tetramer staining |
| IPMGVGLSP | Pol | 504–512 | [45] | B*0702 | Predicted | ELISA |
| ILGFRKIPM | Pol | 509–517 | [45] | B*0801 | Predicted | ELISA |
| FLLAQFTSAI | Pol | 524–533 | [65,101] | A*0201, A*02 | Predicted | ELISpot; Tetramer staining |
| LLAQFTSAI | Pol | 525–533 | [65,101,106] | A*0201, A*02 | Predicted | Cytotoxicity assay; Immunization of mice; Binding assay; ELISpot; Tetramer staining |
| SAICSVVRR | Pol | 531–539 | [74] | A*11, A*3301, A*6801 | Predicted | Binding assay; CTL assay; Cytotoxicity assay |
| SVVRRAFPH | Pol | 535–542 | [9] | A*1101 | Predicted | Proliferation assay; Tetramer staining |
| FFPHCLAFSYM | Pol | 539–550 | [81] | B*07 | Predicted | Binding assay; Immunization of mice; ELISpot |
| FPHCLAFSYM | Pol | 540–550 | [74] | B*0702, B*3501, B*51, B*5301, B*5401 | Predicted | Binding assay; CTL assay; Cytotoxicity assay |
| YMDDVVLG | Pol | 549–556 | [81] | A*0201, A*0202, A*0203, A*0206, A*6802 | Predicted | Binding assay; Immunization of mice; ELISpot |
| YMDDVVLGA | Pol | 549–557 | [45,71,72,99,101,114,115,116] | A*0201, A*02, A*0101 | overlapping | Cytotoxicity assay; Immunization of mice; Binding assay; ELISpot; CTL assay; ELISA |
| YMDDVVLGAK | Pol | 549–558 | [74] | A*03 | Predicted | Binding assay; CTL assay; Cytotoxicity assay |
| FLLSLGIHL | Pol | 573–581 | [71,73,74,81,93,106,108,110,116,117,118,119,120] | A*02, A*0201, A*0206, A*0202 | Predicted | Cytotoxicity assay; Immunization of mice; Binding assay; ELISpot; CTL assay; Tetramer staining |
| SLNFMGYVI | Pol | 592–600 | [106] | A*0201 | Predicted | Binding assay; Immunization of mice; CTL assay; Cytotoxicity assay |
| PVNRPIDWK | Pol | 612–620 | [9] | A*1101 | Predicted | Proliferation assay; Tetramer staining |
| PVNRPIDWK | Pol | 623–631 | [74] | A*03, A*11 | Predicted | Binding assay; CTL assay; Cytotoxicity assay |
| CGYPALMPLY | Pol | 638–647 | [45] | A*2402 | Predicted | ELISA |
| GYPALMPLY | Pol | 639–647 | [45] | A*2402 | Predicted | ELISA |
| YPALMPLYA | Pol | 651–659 | [74] | B*0702, B*3501, B*51, B*5401 | Predicted | Binding assay; CTL assay; Cytotoxicity assay |
| YPALMPLSA | Pol | 651–659 | [62] | B*5401 | Predicted | ELISpot; ICS; Binding assay |
| ALMPLYACI | Pol | 653–661 | [71,74,93,106] | A*0201, A*0202, A*0203, A*0204, A*0206, A*02 | Predicted | Cytotoxicity assay; Immunization of mice; Binding assay; ELISpot; CTL assay; Tetramer staining |
| QAFTFSPTYK | Pol | 665–674 | [74,113] | A*03, A*11, A*6801 | Predicted | Cytotoxicity assay; Binding assay; CTL assay |
| VFADATPTGW | Pol | 686–695 | [45] | A*2402 | Predicted | ELISA |
| GLCQVFADA | Pol | 692–700 | [45] | A*0201 | Predicted | ELISA |
| LPIHTAELL | Pol | 712–720 | [45] | B*0702 | Predicted | ELISA |
| PLPIHTAEL | Pol | 722–730 | [106] | A*0201 | Predicted | Binding assay; Immunization of mice; CTL assay; Cytotoxicity assay |
| IIGTDNSVV | Pol | 744–752 | [65] | A*0201 | Predicted | ELISpot assay; Tetramer staining |
| RKYTSFPWLL | Pol | 744–753 | [45] | A*2402 | Predicted | ELISA |
| KYTSFPWLLG | Pol | 745–754 | [45] | A*2402 | Predicted | ELISA |
| GTDNSVVLSR | Pol | 746–755 | [74] | A*11 | Predicted | Binding assay; CTL assay; Cytotoxicity assay |
| KYTSFPWLL | Pol | 756–764 | [63,81,84,88,93] | A*24, A*2301, A*2402 | overlapping | Cytotoxicity assay; Immunization of mice; Binding assay; ELISpot; CTL assay; Tetramer staining; ICS; ELISA |
| LLGCAANWI | Pol | 763–771 | [65,106] | A*0201 | Predicted | Cytotoxicity assay; Immunization of mice; Binding assay; ELISpot; CTL assay; Tetramer staining |
| WILRGTSFV | Pol | 770–778 | [65,105] | A*0201, A*02 | Predicted | Immunization of mice; Binding assay; ELISpot; Tetramer staining |
| ILRGTSFVYV | Pol | 771–780 | [65,71] | A*0201, A*02 | Predicted | Cytotoxicity assay; ELISpot; Tetramer staining |
| DPSRGRLGL | Pol | 789–797 | [74] | B*0702 | Predicted | Binding assay; CTL assay; Cytotoxicity assay |
| RLGLSRPLL | Pol | 794–802 | [106] | A*0201 | Predicted | Binding assay; Immunization of mice; CTL assay; Cytotoxicity assay |
| GLSRPLLRL | Pol | 796–804 | [65] | A*02 | Predicted | ELISpot assay; Tetramer staining |
| LVYRPTTGR | Pol | 804–812 | [9] | A*1101 | Predicted | Proliferation assay; Tetramer staining |
| SLYADSPSV | Pol | 814–822 | [65,71,73,90,93,106,114,116] | A*0201, A*02 | Predicted | Cytotoxicity assay; Immunization of mice; Binding assay; ELISpot; CTL assay; Tetramer staining; FACS |
| FLLTRILTI | S | 20–28 | [66,67,68,77,100,121] | A*0201 | Predicted | ICS; Tetramer staining; Cytotoxicity assay; Degranulation assay |
| PLGFFPDH | S | 21–28 | [122] | A*11 | Predicted | ELISpot |
| NLLGWSPQA | S | 73–81 | [63] | A*0201, A*0207 | overlapping | ICS; ELISpot |
| LTTVPAASLLA | S | 85–95 | [95] | A*02 | Predicted | ELISpot |
| TTSTGPCK | S | 115–122 | [9] | A*1101 | Predicted | Proliferation assay; Tetramer staining |
| LLDPRVRGL | S | 131–139 | [75] | A*02 | Predicted | CTL assay |
| AILSKTGDPV | S | 160–169 | [116] | A*02 | Predicted | Tetramer staining |
| FLGPLLVLQA | S | 182–190 | [62,63,107] | C*0801 | overlapping | Cytotoxicity assay; Binding assay; ELISpot; Tetramer staining; |
| VLQAGFFL | S | 188–195 | [62] | C*0801 | Predicted | ELISpot; ICS; Binding assay |
| VLQAGFFLL | S | 188–196 | [65,73,101,116,123] | A*0201, A*02 | Predicted | Cytotoxicity assay; Immunization of mice; Binding assay; ELISpot; CTL assay; Tetramer staining |
| SWWTSLNFL | S | 192–200 | [85] | A*2402 | Predicted | ELISpot |
| FLLTRILTI | S | 194–202 | [54,74,76,81,90,93,94,101,108,111,114,116,119,120,123,124,125,126,127,128] | A*0201, A*0202, A*0203, A*0206, A*02 | overlapping | Cytotoxicity assay; Immunization of mice; Binding assay; ELISpot; CTL assay; Tetramer staining; ICS; FACS |
| IPQSLDSWWTSL | S | 202–213 | [129,130] | A*0201, A*02 | Predicted | Cytotoxicity assay; Immunization of mice; Binding assay; ELISpot |
| SILSPFLPLL | S | 207–216 | [131] | A*0201 | Predicted | Binding assay; ELISpot |
| NILSPFMPLL | S | 207–216 | [131] | A*0201 | Predicted | Binding assay; ELISpot |
| ILSPFMPLL | S | 208–216 | [131] | A*0201 | Predicted | Binding assay; ELISpot |
| TLSPFLPLL | S | 208–216 | [131] | A*0201 | Predicted | Binding assay; ELISpot |
| SWWTSLNFL | S | 208–216 | [84] | A*24 | Predicted | Tetramer staining |
| FLGGTPVCL | S | 215–223 | [95,116,123,125] | A*0201A*02, A*24 | Predicted | Cytotoxicity assay; Immunization of mice; Binding assay; ELISpot; CTL assay; Tetramer staining |
| SWLSLLVPF | S | 226–234 | [85] | A*2402 | Predicted | ELISpot |
| RWMCLRRFII | S | 236–245 | [85] | A*2402 | Predicted | ELISpot |
| CPGYRWMCL | S | 243–251 | [108] | B*07 | Predicted | Cytotoxicity assay |
| GYRWMCLRR | S | 245–253 | [91] | A*33 | Predicted | Stabilization assay; ELISpot; Cytotoxicity assay; Immunization of mice |
| RWMCLRRFII | S | 247–256 | [81] | A*2301, A*2402 | Predicted | Binding assay; Immunization of mice; ELISpot |
| ILLLCLIFL | S | 260–268 | [73,125] | A*0201 | Predicted | Cytotoxicity assay; Immunization of mice |
| LLLCLIFLL | S | 261–268 | [72] | A*02 | Predicted | Cytotoxicity assay |
| LLCLIFLLV | S | 262–269 | [65,115,123] | A*0201, A*02 | Predicted | Stabilization assay; ELISpot; Cytotoxicity assay; Tetramer staining; Immunization of mice |
| LCLIFLLVL | S | 263–271 | [85] | A*2402 | Predicted | ELISpot |
| (L)VLLDYQGML | S | 269/70–278 | [75] | A*0201 | Predicted | CTL assay |
| LLDYQGMLP | S | 271–279 | [123] | A*0201 | Predicted | Immunization of transgenic mice; Cytotoxicity assay; ELISpot; Binding assay |
| LLDYQGMLPV | S | 271–280 | [72,101,116,125] | A*02 | Predicted | ELISpot; Cytotoxicity assay; Binding assay; Tetramer staining |
| TSMFPSCCCTK | S | 305–315 | [9] | A*1101 | Predicted | Proliferation assay; Tetramer staining |
| IPIPSSWAF | S | 324–332 | [74,76,81,108] | B*0702, B*3501, B*51, B*5301, A*03, B*07, A*02, B*5101 | Predicted | ELISpot; Cytotoxicity assay; Immunization of mice; Binding assay; CTL assay |
| YLWEWASVR | S | 335–343 | [91] | A*33 | Predicted | Stabilization assay; ELISpot; Cytotoxicity assay; Immunization of mice |
| RFSWLSLLVPF | S | 343–353 | [81] | A*2301, A*2402 | Predicted | Binding assay; Immunization of mice; ELISpot |
| SWLSLLVPF | S | 345–353 | [84] | A*24 | Predicted | Tetramer staining |
| WLSLLVPFV | S | 346–354 | [71,72,73,74,75,76,99,105,108,117,118,120,123,132,133] | A*02, A*0201, A*0202, A*0203, A*0206, A*0207, A*04, A*6802 | Predicted | ELISpot; Cytotoxicity assay; Immunization of mice; Binding assay; Tetramer staining |
| LLVPFVQWFV | S | 349–358 | [93,101,111] | A*02 | Predicted | ICS; Degranulation assay; ELISpot; Tetramer staining |
| VGLSPTVWL | S | 358–366 | [85] | A*2402 | Predicted | ELISpot |
| GLSPTVWLS | S | 359–367 | [123] | A*0201 | Predicted | Immunization of transgenic mice; Cytotoxicity assay; ELISpot; Binding assay |
| GLSPTVWLSV | S | 359–368 | [72,73,90,93,105,111,114,116,124,125,128,130,134] | A*02, A*0201, A*0203, A*0207 | overlapping | Immunization of mice; FACS; CTL assay; ELISpot; Tetramer staining; Degranulation assay |
| VWLSVIWM | S | 364–371 | [90] | A*0201 | Predicted | Immunization of mice; FACS |
| (L)SVIWMMWYW | S | 366/367–375 | [62] | B*5801 | Predicted | ELISpot; ICS; Binding assay |
| SVIWMMWYW | S | 367–375 | [63,107] | B*5801 | overlapping | Tetramer staining; ICS; ELISpot |
| SIVSPFIPLL | S | 370–379 | [131] | A*0201 | Predicted | Binding assay; ELISpot |
| ILSPFLPLL | S | 371–379 | [131] | A*0201 | Predicted | Binding assay; ELISpot |
| MMWYWGPSLY | S | 371–380 | [74] | A*03 | Predicted | Binding assay; CTL assay; Cytotoxicity assay |
| NILSPFLPLL | S | 381–390 | [131] | A*0201 | Predicted | Binding assay; ELISpot |
| SILSPFLPLL | S | 381–390 | [77] | A*0201 | Predicted | ICS; Tetramer staining; |
| SIVSPFIPLL | S | 381–390 | [72,73,116,123] | A*02, A*0201 | Predicted | Immunization of mice; FACS; CTL assay; ELISpot; Tetramer staining |
| ILSPFLPLL | S | 382–390 | [75,90] | A*0201 | Predicted | Immunization of mice; FACS; CTL assay |
| IVSPFIPLL | S | 382–390 | [134] | A*0201 | Predicted | ELISA; Cytotoxicity assay |
| ILRSFIPLL | S | 382–390 | [95] | A*02, A*24 | Predicted | ELISpot |
| LLPIFFCLWV | S | 389–398 | [101] | A*02 | Predicted | ELISpot |
| DIDPYKEFGATVELL | Core | 2–16 | [135] | DRB1*0401 | overlapping | Proliferation assay; ICS |
| IDPYKEFGATVELLS | Core | 3–17 | [135] | DRB1*0401 | overlapping | Proliferation assay; ICS |
| DPYKEFGATVELLSF | Core | 4–18 | [135] | DRB1*0401 | overlapping | Proliferation assay; ICS |
| PYKEFGATVELLSFL | Core | 5–19 | [135] | DRB1*0401 | overlapping | Proliferation assay; ICS |
| YKEFGATVELLSFLP | Core | 6–20 | [135,136] | DRB1*0401, DRB1*1202 | overlapping | ICS; Proliferation assay |
| KEFGATVELLSFLPS | Core | 7–21 | [135] | DRB1*0401 | overlapping | Proliferation assay; ICS |
| EFGATVELLSFLPSD | Core | 8–22 | [135] | DRB1*0401 | overlapping | Proliferation assay; ICS |
| FGATVELLSFLPSDF | Core | 9–23 | [135] | DRB1*0401 | overlapping | Proliferation assay; ICS |
| GATVELLSFLPSDFF | Core | 10–24 | [135] | DRB1*0401 | overlapping | Proliferation assay; ICS |
| TVELLSFLPSDFFPS | Core | 12–26 | [135] | DRB1*0401 | overlapping | Proliferation assay; ICS |
| VELLSFLPSDFFPSV | Core | 13–27 | [135] | DRB1*0401 | overlapping | Proliferation assay; ICS |
| LLSFLPSDFFPSVRD | Core | 15–29 | [135] | DRB1*0401 | overlapping | Proliferation assay; ICS |
| LSFLPSDFFPSVRDL | Core | 16–30 | [135] | DRB1*0401 | overlapping | Proliferation assay; ICS |
| FLPSDFFPSVRD | Core | 18–29 | [137] | DPw4, DRB1*07 | Predicted | Cytotoxicity assay |
| RDLLDTASALYREALESPEH | Core | 28–47 | [138] | DRB1*07, DPw4 | overlapping | Proliferation assay |
| ALYREALESPEHCSP | Core | 36–50 | [136] | DRB1*1202 | overlapping | ICS |
| ALESPEHCSPHHTALRQAIL | Core | 41–60 | [139] | DRB1*13 | overlapping | Proliferation assay |
| EHCSPHHTALRQAIL | Core | 46–60 | [136] | DRB1*0803 | overlapping | ICS |
| PHHTALRQAILCWGELMTLA | Core | 50–69 | [81] | DRB1*07, DRB1*09, DRB1*11 | Predicted | Binding assay; Immunization of mice; ELISpot |
| HHTALRQAILCWGEL | Core | 51–65 | [136] | DRB1*1202 | overlapping | ICS |
| RQAILCWGELMNLAT | Core | 56–70 | [136] | DRB1*0803, DRB1*1202 | overlapping | ICS |
| LCWGELMTLATWVGVN | Core | 60–76 | [140] | DRB1*0101 | Predicted | Proliferation assay; ICS; Tetramer staining |
| MNLATWVGSNLEDPA | Core | 66–80 | [136] | DRB1*0803 | overlapping | ICS |
| LEDPASRELVVSYVN | Core | 76–90 | [136] | DRB1*1202 | overlapping | ICS |
| SRELVVSYVNVNMGL | Core | 81–95 | [136] | DRB1*0803 | overlapping | ICS |
| LEYLVSFGVWIRTPP | Core | 116–130 | [136] | DRB1*1202 | overlapping | ICS |
| EYLVSFGVWIRTPPA | Core | 117–131 | [138] | DRW52, DRB1*06 | overlapping | Proliferation assay |
| VSFGVWIRTPPAYRPPNAPI | Core | 120–139 | [81,138] | DRB1*01, DRB1*07, DRB1*11, DRB1*12, DRB1*13 | overlapping | Binding assay; Immunization of mice; ELISpot; Proliferation assay |
| NAPILSTLPETTVVR | Core | 136–150 | [136] | DRB1*0803 | overlapping | ICS |
| STLPETTVVRRRGRS | Core | 141–155 | [136] | DRB1*1202 | overlapping | ICS |
| STLPETTVVRRRGRSPRRRT | Core | 141–160 | [141] | DRB1*13 | Predicted | Proliferation assay; Cytotoxicity assay; ICS |
| PRRRTPSPRRRRSQS | Core | 156–170 | [136] | DRB1*0803 | overlapping | ICS |
| PPAYRPPNAPILSTL | Core | 158–172 | [135] | DRB1*0101 | overlapping | Proliferation assay; ICS |
| PAYRPPNAPIL | Core | 159–169 | [142] | DR52, DRw3 | overlapping | Proliferation assay; Cytotoxicity assay |
| PSPRRRRSQSPRRRR | Core | 161–175 | [136] | DRB1*0803 | overlapping | ICS |
| RRSQSPRRRRSQSRE | Core | 166–180 | [136] | DRB1*1202 | overlapping | ICS |
| YFKDCLFKDWEELGE | HBx | 111–125 | [143] | DRB1*1301 | overlapping | ELISpot; Binding assay; ICS |
| EIRLKVFVLGGCRHK | HBx | 126–140 | [143] | DRB1*0101, DRB1*0401, DRB1*1301, DRB5*0101 | overlapping | ELISpot; Binding assay; ICS |
| VFVLGGCRHKLVCAP | HBx | 131–145 | [143] | DRB1*1301 | overlapping | ELISpot; Binding assay; ICS |
| VGPLTVNEKRRLKLI | Pol | 96–111 | [113] | DRB1*0301 | Predicted | ELISpot; Cytotoxicity assay |
| RHYLHTLWKAGILYK | Pol | 145–160 | [113] | DRB1*0301, DRB1*07, DRB1*08, DRB1*09, DRB1*11, DRB1*12, DRB1*15 | Predicted | ELISpot; Cytotoxicity assay |
| ESRLVVDFSQFSRGN | Pol | 385–400 | [113] | DRB1*03, DRB1*04 | Predicted | ELISpot; Cytotoxicity assay |
| LQSLTNLLSSNLSWL | Pol | 412–427 | [113] | DRB1*01, DRB1*04, DRB1*07, DRB1*11, DRB1*12, DRB1*13, DRB1*15 | Predicted | ELISpot; Cytotoxicity assay |
| SSNLSWLSLDVSAAF | Pol | 420–435 | [113] | DRB1*01, DRB1*03, DRB1*04, DRB1*13 | Predicted | ELISpot; Cytotoxicity assay |
| LHLYSHPIILGFRKI | Pol | 501–516 | [113] | DRB1*01, DRB1*04, DRB1*11 | Predicted | ELISpot; Cytotoxicity assay |
| PFLLAQFTSAICSVV | Pol | 525–538 | [81] | DRB1*01, DRB1*04, DRB1*07, DRB1*08, DRB1*09, DRB1*11, DRB1*15, DRB5*01 | Predicted | Binding assay; Immunization of mice; ELISpot |
| KQCFRKLPVNRPIDW | Pol | 618–633 | [81,113] | DRB1*01, DRB1*04, DRB1*07, DRB1*13 | Predicted | Binding assay; Immunization of mice; ELISpot; Cytotoxicity assay |
| LCQVFADATPTGWGL | Pol | 649–664 | [81] | DRB1*03, DRB1*04, DRB1*07 | Predicted | Binding assay; Immunization of mice; ELISpot |
| KQAFTFSPTYKAFLC | Pol | 664–679 | [113] | DRB1*01, DRB1*04, DRB1*07, DRB1*08, DRB1*09, DRB1*11, DRB1*13, DRB1*15 | Predicted | ELISpot; Cytotoxicity assay |
| AANWILRGTSFVYVP | Pol | 676–691 | [81] | DRB1*07, DRB1*08, DRB1*09, DRB1*12, DRB1*13, DRB1*15 | Predicted | Binding assay; Immunization of mice; ELISpot |
| LCQVFADATPTGWGL | Pol | 694–709 | [113] | DRB1*03, DRB1*04 | Predicted | ELISpot; Cytotoxicity assay |
| AANWILRGTSFVYVP | Pol | 767–782 | [113] | DRB1*01, DRB1*07, DRB1*08, DRB1*09, DRB1*13, DRB1*15 | Predicted | ELISpot; Cytotoxicity assay |
| GTSFVYVPSALNPAD | Pol | 774–789 | [81] | DRB1*01, DRB1*04, DRB1*07, DRB1*08, DRB1*09, DRB1*11, DRB1*15, DRB5*01 | Predicted | Binding assay; Immunization of mice; ELISpot |
| AGFFLLTRILTIPQS | S | 17–31 | [144] | DRB1*07, DRB1*08, DRB1*11, DRB1*13 | Predicted | ELISpot; Proliferation assay |
| GFFPDHQLDPAF | S | 23–33 | [145] | DRB1*0405 | Predicted | Binding assay; FASC |
| TSLNFLGGSPVCLGQ | S | 37–51 | [144] | DRB1*01 | Predicted | ELISpot; Proliferation assay |
| GAFGPGFTPPHG | S | 61–72 | [145] | DRB1*0405 | Predicted | Binding assay; FASC |
| PICPGYRWMCLRRFI | S | 67–81 | [144] | DRB1*08, DRB1*11, DRB1*13 | Predicted | ELISpot; Proliferation assay |
| GWSPQAQGVLTT | S | 76–87 | [145] | DRB1*0405 | Predicted | ELISpot; Proliferation assay |
| MQWNSTTFHQTLQDPRVRGL | S | 109–134 | [47] | DRB1*01 | Predicted | Immunization of mice; Proliferation assay; ELISpot |
| TTFHQTLQDPRVRGL | S | 114–128 | [47] | DRB1*01 | Predicted | Immunization of mice; Proliferation assay; ELISpot |
| MQWNSTAFHQTLQDP | S | 109–123 | [146] | DRB1*02 | Predicted | Proliferation assay; Cytotoxicity assay |
| STLPETTVVRRRGRSPRRRT | S | 141–160 | [139] | DRB1*13 | overlapping | Proliferation assay |
| WASVRFSWLSLL | S | 165–176 | [147] | DRB1*11, DRB1*14 | Predicted | CTL assay; Proliferation assay |
| VPFVQWFVGLSPTVW | S | 177–191 | [144] | DRB1*11 | Predicted | ELISpot; Proliferation assay |
| QAGFFLLTRILTIPQS | S | 179–194 | [47] | DRB1*01 | Predicted | Immunization of mice; Proliferation assay; ELISpot |
| WLSVIWMMWYWGPSL | S | 191–205 | [136] | DRB1*1202 | overlapping | ICS |
| TSLNFLGGTTVCLGQ | S | 200–214 | [47] | DRB1*01 | Predicted | Immunization of mice; Proliferation assay; ELISpot |
| GPSLYSIVSPFIPLL | S | 202–216 | [144] | DRB1*07 | Predicted | ELISpot; Proliferation assay |
| LLPIFFCLWVYI | S | 215–226 | [147] | DRB1*07, DRB1*08, DRB1*14 | Predicted | CTL assay; Proliferation assay |
| PICPGYRWMCLRRFIIFL | S | 241–258 | [148] | DRB1*0201 | overlapping | Tetramer staining |
| FLLVLLDYQGMLP | S | 256–268 | [54] | DP4 | Predicted | Immunization of mice; Proliferation assay; ELISpot |
| WEWASARFSWLSL | S | 326–338 | [54] | DP4 | Predicted | Immunization of mice; Proliferation assay; ELISpot |
| WLSLLVPFVQWFVGL | S | 335–349 | [149] | DRB1*0101 | Predicted | Immunization of mice; Pentamer staining; ELISpot; ICS; Cytotoxicity assay |
| SLLVPFVQWFVGLSPTVWLSV | S | 337–357 | [47] | DRB1*01 | Predicted | Immunization of mice; Proliferation assay; ELISpot |
| SVRFSWLSLLVPFVQWF | S | 343–357 | [148] | DRB1*0201 | overlapping | Tetramer staining |
| VGLSPTVWLSVI | S | 347–358 | [54] | DP4 | Predicted | Immunization of mice; Proliferation assay; ELISpot |
| GLSPTVWLSVIW | S | 348–359 | [149] | DRB1*0101 | Predicted | Immunization of mice; Pentamer staining; ELISpot; ICS; Cytotoxicity assay |
| TVWLSVIWMMWYW | S | 352–364 | [54] | DP4 | Predicted | Immunization of mice; Proliferation assay; ELISpot |
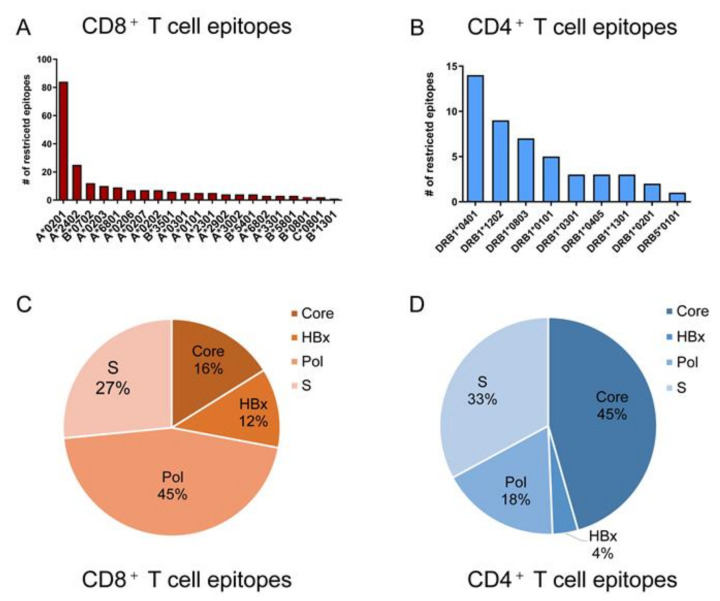
Overall, 82 and 19 studies reported the epitopes presented by HLA class I molecules and class II molecules, respectively, and totally contained 284 unique epitopes including 205 CD8+ T cell epitopes and 79 CD4+ T cell epitopes (Table 1). Of these, 121 (59.0%) CD8+ T cell epitopes are restricted by HLA-A0201, A2402 or B0702 (Figure 3A), which are common supertypes in Caucasians and less predominant in Asia and Africa [58,59]. The remainder are restricted mainly by 12 HLA-A, 5 HLA-B and 1 HLA-C supertypes. For the CD4+ T cell epitopes, the majority of currently described restrictions apply to 8 DRB1 supertypes (Figure 3B). The cumulative frequency of the HLA-A supertypes described in Figure 3A was highest in Europe (66.6%), followed by Asia (53.1%), Africa (50.7%), and North America (52.3%) while the HLA-B supertypes showed an accumulative frequency of 32.7% in Europe, 20.1% in Asia, 19.2% in Africa and 18.8% in North America. The DRB1 supertypes in Figure 3B displayed little difference in the cumulative gene frequency in Europe (30.5%), Asia (32.2%), Africa (31.1%) and North America (34.1%). (Data from http://www.allelefrequencies.net/, assessed on 11 November 2021). Obviously, the 284 validated T cell epitopes of HBV cannot cover the major populations in an indicated geographic region. More T cell epitopes restricted by more HLA supertypes are urgently needed. Further efforts are required to identify more T cell epitopes restricted to the regional prevalent HLA supertypes, especially for the HLA alleles prevalent in Asian populations with a high HBV incidence [59,60].
Figure 3.
HLA restriction and protein distribution of validated CD4+ T cell epitopes and CD8+ T cell epitopes in HBV proteome. (A,B) displayed the number of CD8+ T cell epitopes and CD4+ T cell epitopes restricted by each HLA supertype, respectively. (C,D) showed the fraction of CD8+ T cell epitopes and CD4+ T cell epitopes in each HBV protein, respectively.
In addition, although the validated T cell epitopes have derived from all HBV proteins, the CD8+ T cell epitopes mainly distribute in HBpol and HBsAg (72%) (Figure 3C), while the majority of CD4+ T cell epitopes concentrate in HBeAg and HBsAg (78%) (Figure 3D). The biased distribution of epitopes in proteome may be caused partially by the different lengths of proteins (HBpol 843aa, HBsAg 400 aa, HBeAg 212 aa, HBx 154 aa) and the pitfalls of screening methods.
As displayed in the sixth column of Table 1, most studies used the in silico prediction strategy to screen the candidate CD8+ T cell epitopes (92% of studies) and CD4+ T cells epitopes (63% of studies). Relatively, overlapping peptides were more often used in selecting candidate CD4+ T cell epitopes (7 of 19 studies; 37%) than CD8+ T cell epitopes (7 of 82 studies; 8%), partially due to the lower accuracy and efficacy of predicting HLA class II molecule-binding epitopes as compared with class I molecule-binding epitopes.
5. Conclusions
Here, we have taken an effort to present a reliable and updated T cell epitope repertoire of HBV. We summarized the statistics of 205 unique CD8+ T cell epitopes and 79 unique CD4+ T cell epitopes that have been experimentally validated and reported during the past 33 years, corresponding restricting HLA-molecule, and the methods to screen candidate epitopes and validate candidate epitopes. We hope that this review will be used as a tool for the design and development of therapeutic vaccines and T cell detection kits for HBV-infected patients.
Abbreviations
HBV: Hepatitis B virus; HLA: human leukocyte antigen; LC: liver cirrhosis; HCC: hepatocellular carcinoma; CTL: cytotoxic T lymphocyte; ORF: open reading frame; cccDNA: covalently closed circular DNA; OLPs: overlapping peptides; PBMCs: peripheral blood mononuclear cells; IFN-γ ELISpot: IFN-γ enzyme-linked immunospot; ICS: intracellular cytokine staining.
Author Contributions
Conceptualization, C.S.; Data curation, Y.W. and Y.D.; Funding acquisition, C.S.; Writing—original draft, Y.W.; Writing—review and editing, C.S. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by Jiangsu Provincial Science and Technology Fund of China (BE2017714). The sponsors had no role in study design, data collection and analysis, preparation of the manuscript, or decision to submit the article for publication.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.World Health Organization . Global Hepatitis Report 2017. World Health Organization; Geneva, Switzerland: 2017. [Google Scholar]
- 2.Liu J., Liang W., Jing W., Liu M. Countdown to 2030: Eliminating hepatitis B disease, China. Bull. World Health Organ. 2019;97:230–238. doi: 10.2471/BLT.18.219469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rouviere C.P., Dousson C.B., Tavis J.E. HBV replication inhibitors. Antivir. Res. 2020;179:104815. doi: 10.1016/j.antiviral.2020.104815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Viganò M., Mangia G., Lampertico P. HBeAg-negative chronic hepatitis B: Why do I treat my patients with nucleos(t)ide analogues? Liver Int. 2014;34:120–126. doi: 10.1111/liv.12401. [DOI] [PubMed] [Google Scholar]
- 5.Buti M. HBeAg-positive chronic hepatitis B: Why do I treat my patients with Nucleos(t)ide Analogs? Liver Int. 2013;34:108–111. doi: 10.1111/liv.12392. [DOI] [PubMed] [Google Scholar]
- 6.Pérez-Cameo C., Pons M., Esteban R. New therapeutic perspectives in HBV: When to stop NAs. Liver Int. 2013;34:146–153. doi: 10.1111/liv.12398. [DOI] [PubMed] [Google Scholar]
- 7.Isogawa M., Tanaka Y. Immunobiology of hepatitis B virus infection. Hepatol. Res. 2014;45:179–189. doi: 10.1111/hepr.12439. [DOI] [PubMed] [Google Scholar]
- 8.Tseng T.-C., Huang L.-R. Immunopathogenesis of Hepatitis B Virus. J. Infect. Dis. 2017;216:S765–S770. doi: 10.1093/infdis/jix356. [DOI] [PubMed] [Google Scholar]
- 9.Cheng Y., Zhu Y.O., Becht E., Aw P., Chen J., Poidinger M., de Sessions P.F., Hibberd M.L., Bertoletti A., Lim S.G., et al. Multifactorial heterogeneity of virus-specific T cells and association with the progression of human chronic hepatitis B infection. Sci. Immunol. 2019;4:eaau6905. doi: 10.1126/sciimmunol.aau6905. [DOI] [PubMed] [Google Scholar]
- 10.Wang X., He Q., Shen H., Lu X.-J., Sun B. Genetic and phenotypic difference in CD8+ T cell exhaustion between chronic hepatitis B infection and hepatocellular carcinoma. J. Med. Genet. 2018;56:18–21. doi: 10.1136/jmedgenet-2018-105267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xia Y., Liang T.J. Development of Direct-acting Antiviral and Host-targeting Agents for Treatment of Hepatitis B Virus Infection. Gastroenterology. 2019;156:311–324. doi: 10.1053/j.gastro.2018.07.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tang T.J., Kwekkeboom J., Mancham S., Binda R.S., de Man R.A., Schalm S.W., Kusters J.G., Janssen H.L. Intrahepatic CD8+ T-lymphocyte response is important for therapy-induced viral clearance in chronic hepatitis B infection. J. Hepatol. 2005;43:45–52. doi: 10.1016/j.jhep.2005.01.038. [DOI] [PubMed] [Google Scholar]
- 13.Papatheodoridis G., Vlachogiannakos I., Cholongitas E., Wursthorn K., Thomadakis C., Touloumi G., Petersen J. Discontinuation of oral antivirals in chronic hepatitis B: A systematic review. Hepatology. 2016;63:1481–1492. doi: 10.1002/hep.28438. [DOI] [PubMed] [Google Scholar]
- 14.Lampertico P., Agarwal K., Berg T., Buti M., Janssen H.L.A., Papatheodoridis G.V., Zoulim F., Tacke F. EASL 2017 Clinical Practice Guidelines on the management of hepatitis B virus infection. J. Hepatol. 2017;67:370–398. doi: 10.1016/j.jhep.2017.03.021. [DOI] [PubMed] [Google Scholar]
- 15.Rivino L., Le Bert N., Gill U.S., Kunasegaran K., Cheng Y., Tan D.Z., Becht E., Hansi N.K., Foster G.R., Su T.-H., et al. Hepatitis B virus–specific T cells associate with viral control upon nucleos(t)ide-analogue therapy discontinuation. J. Clin. Investig. 2018;128:668–681. doi: 10.1172/JCI92812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schuch A., Alizei E.S., Heim K., Wieland D., Kiraithe M.M., Kemming J., Llewellyn-Lacey S., Sogukpinar Ö., Ni Y., Urban S., et al. Phenotypic and functional differences of HBV core-specific versus HBV polymerase-specific CD8+ T cells in chronically HBV-infected patients with low viral load. Gut. 2019;68:905–915. doi: 10.1136/gutjnl-2018-316641. [DOI] [PubMed] [Google Scholar]
- 17.Lang-Meli J., Neumann-Haefelin C., Thimme R. Immunotherapy and therapeutic vaccines for chronic HBV infection. Curr. Opin. Virol. 2021;51:149–157. doi: 10.1016/j.coviro.2021.10.002. [DOI] [PubMed] [Google Scholar]
- 18.Loomba R., Liang T.J. Hepatitis B Reactivation Associated With Immune Suppressive and Biological Modifier Therapies: Current Concepts, Management Strategies, and Future Directions. Gastroenterology. 2017;152:1297–1309. doi: 10.1053/j.gastro.2017.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Desmond C.P., Bartholomeusz A., Gaudieri S., Revill P., Lewin S.R. A systematic review of T-cell epitopes in hepatitis B virus: Identification, genotypic variation and relevance to antiviral therapeutics. Antivir. Ther. 2008;13:161–175. [PubMed] [Google Scholar]
- 20.Nitschke K., Luxenburger H., Neumann-Haefelin C., Kiraithe M.M., Thimme R. CD8+ T-Cell Responses in Hepatitis B and C: The (HLA-) A, B, and C of Hepatitis B and C. Dig. Dis. 2016;34:396–409. doi: 10.1159/000444555. [DOI] [PubMed] [Google Scholar]
- 21.Roche P.A., Furuta K. The ins and outs of MHC class II-mediated antigen processing and presentation. Nat. Rev. Immunol. 2015;15:203–216. doi: 10.1038/nri3818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Elahi S., Horton H. Association of HLA-alleles with the immune regulation of chronic viral infections. Int. J. Biochem. Cell Biol. 2012;44:1361–1365. doi: 10.1016/j.biocel.2012.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang L., Zou Z.-Q., Wang K. Clinical Relevance of HLA Gene Variants in HBV Infection. J. Immunol. Res. 2016;2016:9069375. doi: 10.1155/2016/9069375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Boeijen L.L., Hoogeveen R.C., Boonstra A., Lauer G.M. Hepatitis B virus infection and the immune response: The big questions. Best Pract. Res. Clin. Gastroenterol. 2017;31:265–272. doi: 10.1016/j.bpg.2017.05.003. [DOI] [PubMed] [Google Scholar]
- 25.Medzhitov R. Recognition of microorganisms and activation of the immune response. Nature. 2007;449:819–826. doi: 10.1038/nature06246. [DOI] [PubMed] [Google Scholar]
- 26.Thursz M., Kwiatkowski D., Allsopp C.E., Greenwood B.M., Thomas H.C., Hill A.V. Association between an MHC Class II Allele and Clearance of Hepatitis B Virus in the Gambia. N. Engl. J. Med. 1995;332:1065–1069. doi: 10.1056/NEJM199504203321604. [DOI] [PubMed] [Google Scholar]
- 27.Cotrina M., Buti M., Jardí R., Rodríguez-Frías F., Campins M., Esteban R., Guardia J. [Study of HLA-II antigens in chronic hepatitis C and B and in acute hepatitis B] Gastroenterología Hepatología. 1997;20:115–118. [PubMed] [Google Scholar]
- 28.Ahn S.H., Han K.-H., Park J.Y., Lee C.K., Kang S.-W., Chon C.Y., Kim Y.S., Park K., Kim D.K., Moon Y.M. Association between hepatitis B virus infection and HLA-DR type in Korea. Hepatology. 2000;31:1371–1373. doi: 10.1053/jhep.2000.7988. [DOI] [PubMed] [Google Scholar]
- 29.Yang G., Liu J., Han S., Xie H., Du R., Yan Y., Xu D., Fan D. Association between hepatitis B virus infection and HLA-DRB1 genotyping in Shaanxi Han patients in northwestern China. Tissue Antigens. 2007;69:170–175. doi: 10.1111/j.1399-0039.2006.00744.x. [DOI] [PubMed] [Google Scholar]
- 30.Ramezani A., Roshan M.R.H., Kalantar E., Eslamifar A., Banifazl M., Taeb J., Aghakhani A., Gachkar L., Velayati A.A. Association of human leukocyte antigen polymorphism with outcomes of hepatitis B virus infection. J. Gastroenterol. Hepatol. 2008;23:1716–1721. doi: 10.1111/j.1440-1746.2008.05482.x. [DOI] [PubMed] [Google Scholar]
- 31.Thio C.L., Thomas D.L., Karacki P., Gao X., Marti D., Kaslow R.A., Goedert J.J., Hilgartner M., Strathdee S.A., Duggal P., et al. Comprehensive Analysis of Class I and Class II HLA Antigens and Chronic Hepatitis B Virus Infection. J. Virol. 2003;77:12083–12087. doi: 10.1128/JVI.77.22.12083-12087.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fattovich G., Bortolotti F., Donato F. Natural history of chronic hepatitis B: Special emphasis on disease progression and prognostic factors. J. Hepatol. 2008;48:335–352. doi: 10.1016/j.jhep.2007.11.011. [DOI] [PubMed] [Google Scholar]
- 33.Rashidi S., Farhadi L., Ghasemi F., Sheikhesmaeili F., Mohammadi A. The potential role of HLA-G in the pathogenesis of HBV infection: Immunosuppressive or immunoprotective? Infect. Genet. Evol. 2020;85:104580. doi: 10.1016/j.meegid.2020.104580. [DOI] [PubMed] [Google Scholar]
- 34.Xu R., Hu P., Li Y., Tian A., Li J., Zhu C. Advances in HBV infection and replication systems in vitro. Virol. J. 2021;18:105. doi: 10.1186/s12985-021-01580-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lee J.M., Ahn S.H. Quantification of HBsAg: Basic virology for clinical practice. World J. Gastroenterol. 2011;17:283–289. doi: 10.3748/wjg.v17.i3.283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Buti M., Frías F.R., Esteban R. Cuantificación del antígeno de superficie del virus de la hepatitis B: Implicaciones clínicas. Med. Clin. 2012;138:483–488. doi: 10.1016/j.medcli.2011.04.024. [DOI] [PubMed] [Google Scholar]
- 37.Alexopoulou A. HBeAg negative variants and their role in the natural history of chronic hepatitis B virus infection. World J. Gastroenterol. 2014;20:7644–7652. doi: 10.3748/wjg.v20.i24.7644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ou J.-H. Molecular biology of hepatitis B virus e antigen. J. Gastroenterol. Hepatol. 1997;12:S178–S187. doi: 10.1111/j.1440-1746.1997.tb00499.x. [DOI] [PubMed] [Google Scholar]
- 39.Mak L.-Y., Wong D.K.-H., Cheung K.-S., Seto W.-K., Lai C.-L., Yuen M.-F. Review article: Hepatitis B core-related antigen (HBcrAg): An emerging marker for chronic hepatitis B virus infection. Aliment. Pharmacol. Ther. 2017;47:43–54. doi: 10.1111/apt.14376. [DOI] [PubMed] [Google Scholar]
- 40.Ali A., Abdel-Hafiz H., Suhail M., Al-Mars A., Zakaria M.K., Fatima K., Ahmad S., Azhar E., Chaudhary A., Qadri I. Hepatitis B virus, HBx mutants and their role in hepatocellular carcinoma. World J. Gastroenterol. 2014;20:10238–10248. doi: 10.3748/wjg.v20.i30.10238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Qi Y., Gao Z., Xu G., Peng B., Liu C., Yan H., Yao Q., Sun G., Liu Y., Tang D., et al. DNA Polymerase κ Is a Key Cellular Factor for the Formation of Covalently Closed Circular DNA of Hepatitis B Virus. PLoS Pathog. 2016;12:e1005893. doi: 10.1371/journal.ppat.1005893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gerlich W.H., Glebe D., Kramvis A., Magnius L.O. Peculiarities in the designations of hepatitis B virus genes, their products, and their antigenic specificities: A potential source of misunderstandings. Virus Genes. 2020;56:109–119. doi: 10.1007/s11262-020-01733-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chen Y., Yang D., Li S., Gao Y., Jiang R., Deng L., Frankel F.R., Sun B. Development of a Listeria monocytogenes-based vaccine against hepatocellular carcinoma. Oncogene. 2011;31:2140–2152. doi: 10.1038/onc.2011.395. [DOI] [PubMed] [Google Scholar]
- 44.Brinck-Jensen N.-S., Vorup-Jensen T., Leutscher P.D.C., Erikstrup C., Petersen E. Immunogenicity of twenty peptides representing epitopes of the hepatitis B core and surface antigens by IFN-γ response in chronic and resolved HBV. BMC Immunol. 2015;16:65. doi: 10.1186/s12865-015-0127-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.de Beijer M.T.A., Jansen D.T.S.L., Dou Y., van Esch W.J.E., Mok J.Y., Maas M.J.P., Brasser G., de Man R.A., Woltman A.M., Buschow S.I. Discovery and Selection of Hepatitis B Virus-Derived T Cell Epitopes for Global Immunotherapy Based on Viral Indispensability, Conservation, and HLA-Binding Strength. J. Virol. 2020;94:e01663-19. doi: 10.1128/JVI.01663-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shafer-Weaver K., Sayers T., Strobl S., Derby E., Ulderich T., Baseler M., Malyguine A. The Granzyme B ELISPOT assay: An alternative to the 51Cr-release assay for monitoring cell-mediated cytotoxicity. J. Transl. Med. 2003;1:14. doi: 10.1186/1479-5876-1-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pajot A., Michel M.-L., Bourgine M., Ungeheuer M.-N., Ojcius D., Deng Q., Lemonnier F.A., Lone Y.-C. Identification of novel HLA-DR1-restricted epitopes from the hepatitis B virus envelope protein in mice expressing HLA-DR1 and vaccinated human subjects. Microbes Infect. 2006;8:2783–2790. doi: 10.1016/j.micinf.2006.08.009. [DOI] [PubMed] [Google Scholar]
- 48.Freer G., Rindi L. Intracellular cytokine detection by fluorescence-activated flow cytometry: Basic principles and recent advances. Methods. 2013;61:30–38. doi: 10.1016/j.ymeth.2013.03.035. [DOI] [PubMed] [Google Scholar]
- 49.Ji N., Forsthuber T.G. Multiple Sclerosis. Volume 1304. Humana Press; New York, NY, USA: 2014. ELISPOT Techniques; pp. 63–71. [DOI] [PubMed] [Google Scholar]
- 50.Reynolds T.D., Moshkani S., Robek M.D. Hepatitis B Virus. Volume 1540. Humana Press; New York, NY, USA: 2017. An ELISPOT-Based Assay to Measure HBV-Specific CD8+ T Cell Responses in Immunocompetent Mice; pp. 237–247. Methods in Molecular Biology. [DOI] [PubMed] [Google Scholar]
- 51.Dolton G., Tungatt K., Lloyd A., Bianchi V., Theaker S.M., Trimby A., Holland C.J., Donia M., Godkin A., Cole D., et al. More tricks with tetramers: A practical guide to staining T cells with peptide-MHC multimers. Immunology. 2015;146:11–22. doi: 10.1111/imm.12499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Savage P., Millrain M., Dimakou S., Stebbing J., Dyson J. Expansion of CD8+ Cytotoxic T Cells in vitro and in vivo Using MHC Class I Tetramers. Tumor Biol. 2007;28:70–76. doi: 10.1159/000099152. [DOI] [PubMed] [Google Scholar]
- 53.Bertoletti A., Ferrari C. Adaptive immunity in HBV infection. J. Hepatol. 2016;64:S71–S83. doi: 10.1016/j.jhep.2016.01.026. [DOI] [PubMed] [Google Scholar]
- 54.Ru Z., Xiao W., Pajot A., Kou Z., Sun S., Maillere B., Zhao G., Ojcius D., Lone Y.-C., Zhou Y. Development of a Humanized HLA-A2.1/DP4 Transgenic Mouse Model and the Use of This Model to Map HLA-DP4-Restricted Epitopes of HBV Envelope Protein. PLoS ONE. 2012;7:e32247. doi: 10.1371/journal.pone.0032247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ferretti A.P., Kula T., Wang Y., Nguyen D.M., Weinheimer A., Dunlap G.S., Xu Q., Nabilsi N., Perullo C.R., Cristofaro A.W., et al. Unbiased Screens Show CD8+ T Cells of COVID-19 Patients Recognize Shared Epitopes in SARS-CoV-2 that Largely Reside outside the Spike Protein. Immunity. 2020;53:1095–1107.e3. doi: 10.1016/j.immuni.2020.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chikata T., Paes W., Akahoshi T., Partridge T., Murakoshi H., Gatanaga H., Ternette N., Oka S., Borrow P., Takiguchi M. Identification of Immunodominant HIV-1 Epitopes Presented by HLA-C*12:02, a Protective Allele, Using an Immunopeptidomics Approach. J. Virol. 2019;93:e00634-19. doi: 10.1128/JVI.00634-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Paul S., Croft N.P., Purcell A.W., Tscharke D.C., Sette A., Nielsen M., Peters B. Benchmarking predictions of MHC class I restricted T cell epitopes in a comprehensively studied model system. PLoS Comput. Biol. 2020;16:e1007757. doi: 10.1371/journal.pcbi.1007757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Palucka K., Banchereau J. Dendritic-Cell-Based Therapeutic Cancer Vaccines. Immunity. 2013;39:38–48. doi: 10.1016/j.immuni.2013.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lumley S., Noble H., Hadley M.J., Callow L., Malik A., Chua Y.Y., Duffey O.J., Grolmusova N., Kumar A., Ravenscroft S., et al. Hepitopes: A live interactive database of HLA class I epitopes in hepatitis B virus. Wellcome Open Res. 2016;1:9. doi: 10.12688/wellcomeopenres.9952.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Nayagam S., Thursz M., Sicuri E., Conteh L., Wiktor S., Low-Beer D., Hallett T. Requirements for global elimination of hepatitis B: A modelling study. Lancet Infect. Dis. 2016;16:1399–1408. doi: 10.1016/S1473-3099(16)30204-3. [DOI] [PubMed] [Google Scholar]
- 61.Zheng J., Ou Z., Lin X., Wang L., Liu Y., Jin S., Wu J. Analysis of epitope-based vaccine candidates against the E antigen of the hepatitis B virus based on the B genotype sequence: An in silico and in vitro approach. Cell. Immunol. 2018;329:56–65. doi: 10.1016/j.cellimm.2018.04.015. [DOI] [PubMed] [Google Scholar]
- 62.Rivino L., Tan A.T., Chia A., Kumaran E.A.P., Grotenbreg G.M., MacAry P., Bertoletti A. Defining CD8+ T Cell Determinants during Human Viral Infection in Populations of Asian Ethnicity. J. Immunol. 2013;191:4010–4019. doi: 10.4049/jimmunol.1301507. [DOI] [PubMed] [Google Scholar]
- 63.Tan A.T., Sodsai P., Chia A., Moreau E., Chng M.H.Y., Tham C.Y.L., Ho Z.Z., Banu N., Hirankarn N., Bertoletti A. Immunoprevalence and Immunodominance of HLA-Cw*0801-Restricted T Cell Response Targeting the Hepatitis B Virus Envelope Transmembrane Region. J. Virol. 2013;88:1332–1341. doi: 10.1128/JVI.02600-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hoogeveen R.C., Robidoux M.P., Schwarz T., Heydmann L., A Cheney J., Kvistad D., Aneja J., Melgaço J.G., A Fernandes C., Chung R.T., et al. Phenotype and function of HBV-specific T cells is determined by the targeted epitope in addition to the stage of infection. Gut. 2019;68:893–904. doi: 10.1136/gutjnl-2018-316644. [DOI] [PubMed] [Google Scholar]
- 65.Urbani S., Boni C., Amadei B., Fisicaro P., Cerioni S., Valli M.A., Missale G., Ferrari C. Acute phase HBV-specific T cell responses associated with HBV persistence after HBV/HCV coinfection. Hepatology. 2005;41:826–831. doi: 10.1002/hep.20614. [DOI] [PubMed] [Google Scholar]
- 66.Shen C., Xu T., Wu Y., Li X., Xia L., Wang W., Shahzad K.A., Zhang L., Wan X., Qiu J. Frequency and reactivity of antigen-specific T cells were concurrently measured through the combination of artificial antigen-presenting cell, MACS and ELISPOT. Sci. Rep. 2017;7:16400. doi: 10.1038/s41598-017-16549-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Tan A.T., Yang N., Krishnamoorthy T., Oei V., Chua A., Zhao X., Tan H.S., Chia A., Le Bert N., Low D., et al. Use of Expression Profiles of HBV-DNA Integrated Into Genomes of Hepatocellular Carcinoma Cells to Select T Cells for Immunotherapy. Gastroenterology. 2019;156:1862–1876.e9. doi: 10.1053/j.gastro.2019.01.251. [DOI] [PubMed] [Google Scholar]
- 68.Khakpoor A., Ni Y., Chen A., Ho Z.Z., Oei V., Yang N., Giri R., Chow J.X., Tan A.T., Kennedy P.T., et al. Spatiotemporal Differences in Presentation of CD8 T Cell Epitopes during Hepatitis B Virus Infection. J. Virol. 2019;93:e01457-18. doi: 10.1128/JVI.01457-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Bertoletti A., Chisari F.V., Penna A., Guilhot S., Galati L., Missale G., Fowler P., Schlicht H.J., Vitiello A., Chesnut R.C. Definition of a minimal optimal cytotoxic T-cell epitope within the hepatitis B virus nucleocapsid protein. J. Virol. 1993;67:2376–2380. doi: 10.1128/jvi.67.4.2376-2380.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Bertoletti A., Costanzo A., Chisari F., Levrero M., Artini M., Sette A., Penna A., Giuberti T., Fiaccadori F., Ferrari C. Cytotoxic T lymphocyte response to a wild type hepatitis B virus epitope in patients chronically infected by variant viruses carrying substitutions within the epitope. J. Exp. Med. 1994;180:933–943. doi: 10.1084/jem.180.3.933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Rehermann B., Fowler P., Sidney J., Person J., Redeker A., Brown M., Moss B., Sette A., Chisari F. The cytotoxic T lymphocyte response to multiple hepatitis B virus polymerase epitopes during and after acute viral hepatitis. J. Exp. Med. 1995;181:1047–1058. doi: 10.1084/jem.181.3.1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Rehermann B., Pasquinelli C., Mosier S.M., Chisari F.V. Hepatitis B virus (HBV) sequence variation of cytotoxic T lymphocyte epitopes is not common in patients with chronic HBV infection. J. Clin. Investig. 1995;96:1527–1534. doi: 10.1172/JCI118191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Sette A., Vitiello A., Reherman B., Fowler P., Nayersina R., Kast W.M., Melief C.J., Oseroff C., Yuan L., Ruppert J., et al. The relationship between class I binding affinity and immunogenicity of potential cytotoxic T cell epitopes. J. Immunol. 1994;153:5586–5592. [PubMed] [Google Scholar]
- 74.Bertoni R., Sidney J., Fowler P., Chesnut R.W., Chisari F.V., Sette A. Human histocompatibility leukocyte antigen-binding supermotifs predict broadly cross-reactive cytotoxic T lymphocyte responses in patients with acute hepatitis. J. Clin. Investig. 1997;100:503–513. doi: 10.1172/JCI119559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lee H.G., Lim J.-S., Lee K.-Y., Choi Y.-K., Choe I.-S., Chung T.-W., Kim K. Peptide-specific CTL induction in HBV-seropositive PBMC by stimulation with peptides in vitro: Novel epitopes identified from chronic carriers. Virus Res. 1997;50:185–194. doi: 10.1016/S0168-1702(97)00068-3. [DOI] [PubMed] [Google Scholar]
- 76.Oseroff C., Sette A., Wentworth P., Celis E., Maewal A., Dahlberg C., Fikes J., Kubo R.T., Chesnut R.W., Grey H.M., et al. Pools of lipidated HTL-CTL constructs prime for multiple HBV and HCV CTL epitope responses. Vaccine. 1998;16:823–833. doi: 10.1016/S0264-410X(97)00264-8. [DOI] [PubMed] [Google Scholar]
- 77.Tham C.Y., Kah J., Tan A.T., Volz T., Chia A., Giersch K., Ladiges Y., Loglio A., Borghi M., Sureau C., et al. Hepatitis Delta Virus Acts as an Immunogenic Adjuvant in Hepatitis B Virus-Infected Hepatocytes. Cell Rep. Med. 2020;1:100060. doi: 10.1016/j.xcrm.2020.100060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Dou Y., Jansen D.T., Bosch A.V.D., de Man R.A., van Montfoort N., Araman C., van Kasteren S.I., Zom G.G., Krebber W.-J., Melief C.J., et al. Design of TLR2-ligand-synthetic long peptide conjugates for therapeutic vaccination of chronic HBV patients. Antivir. Res. 2020;178:104746. doi: 10.1016/j.antiviral.2020.104746. [DOI] [PubMed] [Google Scholar]
- 79.Bihl F.K., Loggi E., Chisholm J.V., Hewitt H.S., Henry L.M., Linde C., Suscovich T.J., Wong J.T., Frahm N., Andreone P., et al. Simultaneous assessment of cytotoxic T lymphocyte responses against multiple viral infections by combined usage of optimal epitope matrices, anti- CD3 mAb T-cell expansion and “RecycleSpot”. J. Transl. Med. 2005;3:20. doi: 10.1186/1479-5876-3-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Thimme R., Chang K.-M., Pemberton J., Sette A., Chisari F.V. Degenerate Immunogenicity of an HLA-A2-Restricted Hepatitis B Virus Nucleocapsid Cytotoxic T-Lymphocyte Epitope That Is Also Presented by HLA-B. J. Virol. 2001;75:3984–3987. doi: 10.1128/JVI.75.8.3984-3987.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Depla E., Van der Aa A., Livingston B.D., Crimi C., Allosery K., De Brabandere V., Krakover J., Murthy S., Huang M., Power S., et al. Rational Design of a Multiepitope Vaccine Encoding T-Lymphocyte Epitopes for Treatment of Chronic Hepatitis B Virus Infections. J. Virol. 2008;82:435–450. doi: 10.1128/JVI.01505-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Kefalakes H., Jochum C., Hilgard G., Kahraman A., Bohrer A.M., El Hindy N., Heinemann F.M., Verheyen J., Gerken G., Roggendorf M., et al. Decades after recovery from hepatitis B and HBsAg clearance the CD8+ T cell response against HBV core is nearly undetectable. J. Hepatol. 2015;63:13–19. doi: 10.1016/j.jhep.2015.01.030. [DOI] [PubMed] [Google Scholar]
- 83.Pacella I., Cammarata I., Martire C., Brancaccio G., Gaeta G.B., Barnaba V., Piconese S. CD8+ T cells specific to apoptosis-associated epitopes are expanded in patients with chronic HBV infection and fibrosis. Liver Int. 2021;41:470–481. doi: 10.1111/liv.14720. [DOI] [PubMed] [Google Scholar]
- 84.Kondo Y., Asabe S., Kobayashi K., Shiina M., Niitsuma H., Ueno Y., Kobayashi T., Shimosegawa T. Recovery of functional cytotoxic T lymphocytes during lamivudine therapy by acquiring multi-specificity. J. Med. Virol. 2004;74:425–433. doi: 10.1002/jmv.20194. [DOI] [PubMed] [Google Scholar]
- 85.Yamamiya D., Mizukoshi E., Kaji K., Terashima T., Kitahara M., Yamashita T., Arai K., Fushimi K., Honda M., Kaneko S. Immune responses of human T lymphocytes to novel hepatitis B virus-derived peptides. PLoS ONE. 2018;13:e0198264. doi: 10.1371/journal.pone.0198264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Zhang Y., Ren Y., Wu Y., Zhao B., Qiu L., Li X., Xu D., Liu J., Gao G.F., Meng S. The L60V Variation in Hepatitis B Virus Core Protein Elicits New Epitope-Specific Cytotoxic T Lymphocytes and Enhances Viral Replication. J. Virol. 2013;87:8075–8084. doi: 10.1128/JVI.00577-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Liu Q., Zheng Y., Yu Y., Tan Q., Huang X. Identification of HLA-A*0201-restricted CD8+ T-cell epitope C64–72 from hepatitis B virus core protein. Int. Immunopharmacol. 2012;13:141–147. doi: 10.1016/j.intimp.2012.03.018. [DOI] [PubMed] [Google Scholar]
- 88.Sobao Y., Sugi K., Tomiyama H., Saito S., Fujiyama S., Morimoto M., Hasuike S., Tsubouchi H., Tanaka K., Takiguchi M. Identification of hepatitis B virus-specific CTL epitopes presented by HLA-A*2402, the most common HLA class I allele in East Asia. J. Hepatol. 2001;34:922–929. doi: 10.1016/S0168-8278(01)00048-4. [DOI] [PubMed] [Google Scholar]
- 89.Zhou M., Xu Y., Lou Z., Cole D.K., Li X., Liu Y., Tien P., Rao Z., Gao G.F. Complex assembly, crystallization and preliminary X-ray crystallographic studies of MHC H-2Kdcomplexed with an HBV-core nonapeptide. Acta Crystallogr. Sect. D Biol. Crystallogr. 2004;60:1473–1475. doi: 10.1107/S0907444904013587. [DOI] [PubMed] [Google Scholar]
- 90.Riedl P., Wieland A., Lamberth K., Buus S., Lemonnier F., Reifenberg K., Reimann J., Schirmbeck R. Elimination of Immunodominant Epitopes from Multispecific DNA-Based Vaccines Allows Induction of CD8 T Cells That Have a Striking Antiviral Potential. J. Immunol. 2009;183:370–380. doi: 10.4049/jimmunol.0900505. [DOI] [PubMed] [Google Scholar]
- 91.Pan X., Ding H., Zhou X., Tien P. Identification of hepatitis B virus-specific CTL epitopes presented by HLA-A*33:03 in peripheral blood mononuclear cells from patients and transgenic mice. Biochem. Biophys. Res. Commun. 2014;449:135–140. doi: 10.1016/j.bbrc.2014.05.001. [DOI] [PubMed] [Google Scholar]
- 92.Khakoo S.I., Ling R., Scott I., Dodi A.I., Harrison T.J., Dusheiko G.M., A Madrigal J. Cytotoxic T lymphocyte responses and CTL epitope escape mutation in HBsAg, anti-HBe positive individuals. Gut. 2000;47:137–143. doi: 10.1136/gut.47.1.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Webster G.J.M., Reignat S., Brown D., Ogg G.S., Jones L., Seneviratne S.L., Williams R., Dusheiko G., Bertoletti A. Longitudinal Analysis of CD8+ T Cells Specific for Structural and Nonstructural Hepatitis B Virus Proteins in Patients with Chronic Hepatitis B: Implications for Immunotherapy. J. Virol. 2004;78:5707–5719. doi: 10.1128/JVI.78.11.5707-5719.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Chen A., Wang L., Zhang J., Zou L., Jia Z., Zhou W., Wan Y., Wu Y. H-2 Kd-Restricted Hepatitis B Virus-Derived Epitope Whose Specific CD8 + T Lymphocytes Can Produce Gamma Interferon without Cytotoxicity. J. Virol. 2005;79:5568–5576. doi: 10.1128/JVI.79.9.5568-5576.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Comber J.D., Karabudak A., Shetty V., Testa J.S., Huang X., Philip R. MHC Class I Presented T Cell Epitopes as Potential Antigens for Therapeutic Vaccine against HBV Chronic Infection. Hepat. Res. Treat. 2014;2014:860562. doi: 10.1155/2014/860562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Sun L., Zhang Y., Zhao B., Deng M., Liu J., Li X., Hou J., Gui M., Zhang S., Li X., et al. A new unconventional HLA-A2-restricted epitope from HBV core protein elicits antiviral cytotoxic T lymphocytes. Protein Cell. 2014;5:317–327. doi: 10.1007/s13238-014-0041-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Missale G., Redeker A., Person J., Fowler P., Guilhot S., Schlicht H.J., Ferrari C., Chisari F. HLA-A31- and HLA-Aw68-restricted cytotoxic T cell responses to a single hepatitis B virus nucleocapsid epitope during acute viral hepatitis. J. Exp. Med. 1993;177:751–762. doi: 10.1084/jem.177.3.751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Zhang Y., Wu Y., Deng M., Xu D., Li X., Xu Z., Hu J., Zhang H., Liu K., Zhao Y., et al. CD8 + T-Cell Response-Associated Evolution of Hepatitis B Virus Core Protein and Disease Progress. J. Virol. 2018;92:e02120-17. doi: 10.1128/JVI.02120-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Malmassari S., Lone Y.C., Zhang M., Transy C., Michel M.-L. In vivo hierarchy of immunodominant and subdominant HLA-A*0201-restricted T-cell epitopes of HBx antigen of hepatitis B virus. Microbes Infect. 2005;7:626–634. doi: 10.1016/j.micinf.2004.12.022. [DOI] [PubMed] [Google Scholar]
- 100.Ishioka G.Y., Fikes J., Hermanson G., Livingston B., Crimi C., Qin M., Del Guercio M.F., Oseroff C., Dahlberg C., Alexander J., et al. Utilization of MHC class I transgenic mice for development of minigene DNA vaccines encoding multiple HLA-restricted CTL epitopes. J. Immunol. 1999;162:3915–3925. [PubMed] [Google Scholar]
- 101.Gehring A., Ho Z.Z., Tan A.T., Aung M.O., Lee K.H., Tan K.C., Lim S.G., Bertoletti A. Profile of Tumor Antigen-Specific CD8 T Cells in Patients With Hepatitis B Virus-Related Hepatocellular Carcinoma. Gastroenterology. 2009;137:682–690. doi: 10.1053/j.gastro.2009.04.045. [DOI] [PubMed] [Google Scholar]
- 102.Guo Y.J., Zhu Y., Sun S.H. Identification and functional studies of HLA-A0201 restricted CTL epitopes in the X protein of hepatitis B virus. Acta Virol. 2011;55:107–115. doi: 10.4149/av_2011_02_107. [DOI] [PubMed] [Google Scholar]
- 103.Ding F.-X., Wang F., Lu Y.-M., Li K., Wang K.-H., He X.-W., Sun S.-H. Multiepitope peptide-loaded virus-like particles as a vaccine against hepatitis B virus-related hepatocellular carcinoma. Hepatology. 2009;49:1492–1502. doi: 10.1002/hep.22816. [DOI] [PubMed] [Google Scholar]
- 104.King T.H., Kemmler C.B., Guo Z., Mann D., Lu Y., Coeshott C., Gehring A.J., Bertoletti A., Ho Z.Z., Delaney W., et al. A Whole Recombinant Yeast-Based Therapeutic Vaccine Elicits HBV X, S and Core Specific T Cells in Mice and Activates Human T Cells Recognizing Epitopes Linked to Viral Clearance. PLoS ONE. 2014;9:e101904. doi: 10.1371/journal.pone.0101904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Van Der Burg S.H., Visseren M.J., Brandt R.M., Kast W.M., Melief C.J. Immunogenicity of peptides bound to MHC class I molecules depends on the MHC-peptide complex stability. J. Immunol. 1996;156:3308–3314. [PubMed] [Google Scholar]
- 106.Vitiello A., Sette A., Yuan L., Farness P., Southwood S., Sidney J., Chesnut R.W., Grey H.M., Livingston B. Comparison of cytotoxic T lymphocyte responses induced by peptide or DNA immunization: Implications on immunogenicity and immunodominance. Eur. J. Immunol. 1997;27:671–678. doi: 10.1002/eji.1830270315. [DOI] [PubMed] [Google Scholar]
- 107.Chang C.X.L., Tan A.T., Or M.Y., Toh K.Y., Lim P.Y., Chia A.S.E., Froesig T.M., Nadua K.D., Oh H.J., Leong H.N., et al. Conditional ligands for A sian HLA variants facilitate the definition of CD8+ T-cell responses in acute and chronic viral diseases. Eur. J. Immunol. 2013;43:1109–1120. doi: 10.1002/eji.201243088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Bertoni R., Sette A., Sidney J., Guidotti L.G., Shapiro M., Purcell R., Chisari F. Human class I supertypes and CTL repertoires extend to chimpanzees. J. Immunol. 1998;161:4447–4455. [PubMed] [Google Scholar]
- 109.Tangri S., Ishioka G.Y., Huang X., Sidney J., Southwood S., Fikes J., Sette A. Structural Features of Peptide Analogs of Human Histocompatibility Leukocyte Antigen Class I Epitopes That Are More Potent and Immunogenic than Wild-Type Peptide. J. Exp. Med. 2001;194:833–846. doi: 10.1084/jem.194.6.833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Sing G.K., Ladhams A., Arnold S., Parmar H., Chen X., Cooper J., Butterworth L., Stuart K., D’Arcy D., Cooksley W.G.E. A longitudinal analysis of cytotoxic T lymphocyte precursor frequencies to the hepatitis B virus in chronically infected patients. J. Viral Hepat. 2001;8:19–29. doi: 10.1046/j.1365-2893.2001.00260.x. [DOI] [PubMed] [Google Scholar]
- 111.Tan A.T., Loggi E., Boni C., Chia A., Gehring A., Sastry K.S.R., Goh V., Fisicaro P., Andreone P., Brander C., et al. Host Ethnicity and Virus Genotype Shape the Hepatitis B Virus-Specific T-Cell Repertoire. J. Virol. 2008;82:10986–10997. doi: 10.1128/JVI.01124-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Zheng J., Xia Z., Xu Y., Ou Z., Lin X., Jin S., Liu Y., Wu J. Identification of novel hepatitis B virus therapeutic vaccine candidates derived from polymerase protein. Aging. 2021;13:14372–14384. doi: 10.18632/aging.203053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Mizukoshi E., Sidney J., Livingston B., Ghany M., Hoofnagle J.H., Sette A., Rehermann B. Cellular Immune Responses to the Hepatitis B Virus Polymerase. J. Immunol. 2004;173:5863–5871. doi: 10.4049/jimmunol.173.9.5863. [DOI] [PubMed] [Google Scholar]
- 114.Schirmbeck R., Riedl P., Fissolo N.M., Lemonnier F.A., Bertoletti A., Reimann J. Translation from Cryptic Reading Frames of DNA Vaccines Generates an Extended Repertoire of Immunogenic, MHC Class I-Restricted Epitopes. J. Immunol. 2005;174:4647–4656. doi: 10.4049/jimmunol.174.8.4647. [DOI] [PubMed] [Google Scholar]
- 115.Livingston B.D., Newman M., Crimi C., McKinney D., Chesnut R., Sette A. Optimization of epitope processing enhances immunogenicity of multiepitope DNA vaccines. Vaccine. 2001;19:4652–4660. doi: 10.1016/S0264-410X(01)00233-X. [DOI] [PubMed] [Google Scholar]
- 116.Lin C.-L., Tsai S.-L., Lee T.-H., Chien R.-N., Liao S.-K., Liaw Y.-F. High frequency of functional anti-YMDD and -mutant cytotoxic T lymphocytes after in vitro expansion correlates with successful response to lamivudine therapy for chronic hepatitis B. Gut. 2005;54:152–161. doi: 10.1136/gut.2003.032920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Maini M., Boni C., Ogg G.S., King A.S., Reignat S., Lee C.K., Larrubia J.R., Webster G.J., McMichael A.J., Ferrari C., et al. Direct ex vivo analysis of hepatitis B virus-specific CD8+ T cells associated with the control of infection. Gastroenterology. 1999;117:1386–1396. doi: 10.1016/S0016-5085(99)70289-1. [DOI] [PubMed] [Google Scholar]
- 118.Wu Y., Zhang J., Chen S., Chen A., Wang L., Li J., Zhao T., Zou L., Tang Y., Tingrong L., et al. Frequencies of epitope-specific cytotoxic T lymphocytes in active chronic viral hepatitis B infection by using MHC class I peptide tetramers. Immunol. Lett. 2004;92:253–258. doi: 10.1016/j.imlet.2004.01.002. [DOI] [PubMed] [Google Scholar]
- 119.Shi M., Qian S., Chen W.-W., Zhang H., Zhang B., Tang Z.-R., Zhang Z., Wang F.-S. Hepatitis B virus (HBV) antigen-pulsed monocyte-derived dendritic cells from HBV-associated hepatocellular carcinoma patients significantly enhance specific T cell responses in vitro. Clin. Exp. Immunol. 2006;147:277–286. doi: 10.1111/j.1365-2249.2006.03281.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Rehermann B., Lau D., Hoofnagle J.H., Chisari F.V. Cytotoxic T lymphocyte responsiveness after resolution of chronic hepatitis B virus infection. J. Clin. Investig. 1996;97:1655–1665. doi: 10.1172/JCI118592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Zhao X., Sankaran S., Yap J., Too C.T., Ho Z.Z., Dolton G., Legut M., Ren E.C., Sewell A.K., Bertoletti A., et al. Nonstimulatory peptide-MHC enhances human T-cell antigen-specific responses by amplifying proximal TCR signaling. Nat. Commun. 2018;9:2716. doi: 10.1038/s41467-018-05288-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Jin Y., Shih W.K., Berkower I. Human T cell response to the surface antigen of hepatitis B virus (HBsAg). Endosomal and nonendosomal processing pathways are accessible to both endogenous and exogenous antigen. J. Exp. Med. 1988;168:293–306. doi: 10.1084/jem.168.1.293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Loirat D., Lemonnier F.A., Michel M.-L. Multiepitopic HLA-A*0201-Restricted Immune Response Against Hepatitis B Surface Antigen After DNA-Based Immunization. J. Immunol. 2000;165:4748–4755. doi: 10.4049/jimmunol.165.8.4748. [DOI] [PubMed] [Google Scholar]
- 124.Loirat D., Mancini-Bourgine M., Abastado J., Michel M. HBsAg/HLA-A2 transgenic mice: A model for T cell tolerance to hepatitis B surface antigen in chronic hepatitis B virus infection. Int. Immunol. 2003;15:1125–1136. doi: 10.1093/intimm/dxg117. [DOI] [PubMed] [Google Scholar]
- 125.Vandepapelière P., Rehermann B., Koutsoukos M., Moris P., Garçon N., Wettendorff M., Leroux-Roels G. Potent enhancement of cellular and humoral immune responses against recombinant hepatitis B antigens using AS02A adjuvant in healthy adults. Vaccine. 2005;23:2591–2601. doi: 10.1016/j.vaccine.2004.11.034. [DOI] [PubMed] [Google Scholar]
- 126.Liu H.-G., Fan Z.-P., Chen W.-W., Yang H.-Y., Liu Q.-F., Zhang H., Tien P., Wang F.-S. A mutant HBs antigen (HBsAg)183–191 epitope elicits specific cytotoxic T lymphocytes in acute hepatitis B patients. Clin. Exp. Immunol. 2008;151:441–447. doi: 10.1111/j.1365-2249.2007.03570.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Zhang Z., Jin B., Zhang J.-Y., Xu B., Wang H., Shi M., Wherry E.J., Lau G.K., Wang F.-S. Dynamic decrease in PD-1 expression correlates with HBV-specific memory CD8 T-cell development in acute self-limited hepatitis B patients. J. Hepatol. 2009;50:1163–1173. doi: 10.1016/j.jhep.2009.01.026. [DOI] [PubMed] [Google Scholar]
- 128.Boni C., Laccabue D., Lampertico P., Giuberti T., Viganò M., Schivazappa S., Alfieri A., Pesci M., Gaeta G.B., Brancaccio G., et al. Restored Function of HBV-Specific T Cells After Long-term Effective Therapy With Nucleos(t)ide Analogues. Gastroenterology. 2012;143:963–973.e9. doi: 10.1053/j.gastro.2012.07.014. [DOI] [PubMed] [Google Scholar]
- 129.Schirmbeck R., Melber K., Mertens T., Reimann J. Selective stimulation of murine cytotoxic T cell and antibody responses by particulate or monomeric hepatitis B virus surface (S) antigen. Eur. J. Immunol. 1994;24:1088–1096. doi: 10.1002/eji.1830240512. [DOI] [PubMed] [Google Scholar]
- 130.Marsac D., Puaux A.-L., Rivière Y., Michel M.-L. In vivo induction of cellular and humoral immune responses by hybrid DNA vectors encoding simian/human immunodeficiency virus/hepatitis B surface antigen virus particles in BALB/c and HLA-A2-transgenic mice. Immunobiology. 2005;210:305–319. doi: 10.1016/j.imbio.2005.04.003. [DOI] [PubMed] [Google Scholar]
- 131.Liu Q., Tian Y., Li Y., Zhang W., Cai W., Liu Y., Ren Y., Liang Z., Zhou P., Zhang Y., et al. In vivo therapeutic effects of affinity-improved-TCR engineered T-cells on HBV-related hepatocellular carcinoma. J. Immunother. Cancer. 2020;8:e001748. doi: 10.1136/jitc-2020-001748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Bertoletti A., Sette A., Chisari F., Penna A., Levrero M., De Carli M., Fiaccadori F., Ferrari C. Natural variants of cytotoxic epitopes are T-cell receptor antagonists for antiviral cytotoxic T cells. Nature. 1994;369:407–410. doi: 10.1038/369407a0. [DOI] [PubMed] [Google Scholar]
- 133.Roy M.J., Wu M.S., Barr L.J., Fuller J.T., Tussey L.G., Speller S., Culp J., Burkholder J.K., Swain W.F., Dixon R.M., et al. Induction of antigen-specific CD8+ T cells, T helper cells, and protective levels of antibody in humans by particle-mediated administration of a hepatitis B virus DNA vaccine. Vaccine. 2000;19:764–778. doi: 10.1016/S0264-410X(00)00302-9. [DOI] [PubMed] [Google Scholar]
- 134.Riedl P., Bertoletti A., Lopes R., Lemonnier F., Reimann J., Schirmbeck R. Distinct, Cross-Reactive Epitope Specificities of CD8 T Cell Responses Are Induced by Natural Hepatitis B Surface Antigen Variants of Different Hepatitis B Virus Genotypes. J. Immunol. 2006;176:4003–4011. doi: 10.4049/jimmunol.176.7.4003. [DOI] [PubMed] [Google Scholar]
- 135.Feng I.-C., Koay L.-B., Sheu M.-J., Kuo H.-T., Sun C.-S., Lee C., Chuang W.-L., Liao S.-K., Wang S.-L., Tang L.-Y., et al. HBcAg-specific CD4+CD25+regulatory T cells modulate immune tolerance and acute exacerbation on the natural history of chronic hepatitis B virus infection. J. Biomed. Sci. 2007;14:43–57. doi: 10.1007/s11373-006-9129-z. [DOI] [PubMed] [Google Scholar]
- 136.Wang H., Luo H., Wan X., Fu X., Mao Q., Xiang X., Zhou Y., He W., Zhang J., Guo Y., et al. TNF-α/IFN-γ profile of HBV-specific CD4 T cells is associated with liver damage and viral clearance in chronic HBV infection. J. Hepatol. 2020;72:45–56. doi: 10.1016/j.jhep.2019.08.024. [DOI] [PubMed] [Google Scholar]
- 137.Desombere I., Gijbels Y., Verwulgen A., Leroux-Roels G. Characterization of the T cell recognition of hepatitis B surface antigen (HBsAg) by good and poor responders to hepatitis B vaccines. Clin. Exp. Immunol. 2000;122:390–399. doi: 10.1046/j.1365-2249.2000.01383.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Ferrari C., Bertoletti A., Penna A., Cavalli A., Valli A., Missale G., Pilli M., Fowler P., Giuberti T., Chisari F. Identification of immunodominant T cell epitopes of the hepatitis B virus nucleocapsid antigen. J. Clin. Investig. 1991;88:214–222. doi: 10.1172/JCI115280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Cao T., Desombere I., Vanlandschoot P., Sällberg M., Leroux-Roels G. Characterization of HLA DR13-restricted CD4+ T cell epitopes of hepatitis B core antigen associated with self-limited, acute hepatitis B. J. Gen. Virol. 2002;83:3023–3033. doi: 10.1099/0022-1317-83-12-3023. [DOI] [PubMed] [Google Scholar]
- 140.Raziorrouh B., Heeg M., Kurktschiev P., Schraut W., Zachoval R., Wendtner C., Wächtler M., Spannagl M., Denk G., Ulsenheimer A., et al. Inhibitory Phenotype of HBV-Specific CD4+ T-Cells Is Characterized by High PD-1 Expression but Absent Coregulation of Multiple Inhibitory Molecules. PLoS ONE. 2014;9:e105703. doi: 10.1371/journal.pone.0105703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Cao T., Meuleman P., Desombere I., Sällberg M., Leroux-Roels G. In Vivo Inhibition of Anti-Hepatitis B Virus Core Antigen (HBcAg) Immunoglobulin G Production by HBcAg-Specific CD4 + Th1-Type T-Cell Clones in a hu-PBL-NOD/SCID Mouse Model. J. Virol. 2001;75:11449–11456. doi: 10.1128/JVI.75.23.11449-11456.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Tsai S.-L., Chen P.-J., Yang P.-M., Liao T.-H., Sung J.-L., Lai M.-Y., Huang J.-H., Chang T.-H., Chen D.-S. Characterization of T cell clones specific to a determinant of hepatitis B virus core and e antigens in chronic type B hepatitis: Implication for a T cell mechanism of HBV immunopathogenesis. J. Biomed. Sci. 1994;1:105–118. doi: 10.1007/BF02257984. [DOI] [PubMed] [Google Scholar]
- 143.Malmassari S.L., Deng Q., Fontaine H., Houitte D., Rimlinger F., Thiers V., Maillere B., Pol S., Michel M.-L. Impact of hepatitis B virus basic core promoter mutations on T cell response to an immunodominant HBx-derived epitope. Hepatology. 2007;45:1199–1209. doi: 10.1002/hep.21594. [DOI] [PubMed] [Google Scholar]
- 144.Kruger A., Adams P., Hammer J., Böcher W.O., Schneider P.M., Rittner C., Hoehler T. Hepatitis B surface antigen presentation and HLA-DRB1*—Lessons from twins and peptide binding studies. Clin. Exp. Immunol. 2005;140:325–332. doi: 10.1111/j.1365-2249.2005.02765.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Kim J.-H., Park J.-H., Lee Y.-J., Cho E.-W., Bae Y.-S., Kim K.L. In Vitro Binding Analysis of Hepatitis B Virus preS-derived Putative Helper T-cell Epitopes to MHC Class II Molecules Using Stable HLA-DRB1*0405/-DRA*0101 Transfected Cells. IUBMB Life. 2000;50:379–384. doi: 10.1080/713803746. [DOI] [PubMed] [Google Scholar]
- 146.Barnaba V., Franco A., Alberti A., Balsano C., Benvenuto R., Balsano F. Recognition of hepatitis B virus envelope proteins by liver-infiltrating T lymphocytes in chronic HBV infection. J. Immunol. 1989;143:2650–2655. [PubMed] [Google Scholar]
- 147.Honorati M.C., Dolzani P., Mariani E., Piacentini A., Lisignoli G., Ferrari C., Facchini A. Epitope specificity of Th0/Th2 CD4+ T-lymphocyte clones induced by vaccination with rHBsAg vaccine. Gastroenterology. 1997;112:2017–2027. doi: 10.1053/gast.1997.v112.pm9178695. [DOI] [PubMed] [Google Scholar]
- 148.Chow I.-T., James E.A., Tan V., Moustakas A.K., Papadopoulos G.K., Kwok W.W. DRB1*12:01 presents a unique subset of epitopes by preferring aromatics in pocket. Mol. Immunol. 2012;50:26–34. doi: 10.1016/j.molimm.2011.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Bayard F., Malmassari S., Deng Q., Lone Y.-C., Michel M.-L. Hepatitis B virus (HBV)-derived DRB1*0101-restricted CD4 T-cell epitopes help in the development of HBV-specific CD8+ T cells in vivo. Vaccine. 2010;28:3818–3826. doi: 10.1016/j.vaccine.2010.03.026. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.