Abstract
Telomere repeat sequences cap the ends of eucaryotic chromosomes and help stabilize them. At interstitial sites, however, they may destabilize chromosomes, as suggested by cytogenetic studies in mammalian cells that correlate interstitial telomere sequence with sites of spontaneous and radiation-induced chromosome rearrangements. In no instance is the length, purity, or orientation of the telomere repeats at these potentially destabilizing interstitial sites known. To determine the effects of a defined interstitial telomere sequence on chromosome instability, as well as other aspects of DNA metabolism, we deposited 800 bp of the functional vertebrate telomere repeat, TTAGGG, in two orientations in the second intron of the adenosine phosphoribosyltransferase (APRT) gene in Chinese hamster ovary cells. In one orientation, the deposited telomere sequence did not interfere with expression of the APRT gene, whereas in the other it reduced mRNA levels slightly. The telomere sequence did not induce chromosome truncation and the seeding of a new telomere at a frequency above the limits of detection. Similarly, the telomere sequence did not alter the rate or distribution of homologous recombination events. The interstitial telomere repeat sequence in both orientations, however, dramatically increased gene rearrangements some 30-fold. Analysis of individual rearrangements confirmed the involvement of the telomere sequence. These studies define the telomere repeat sequence as a destabilizing element in the interior of chromosomes in mammalian cells.
Several kilobases of short repeated sequences—TTAGGG in vertebrates—make up the DNA component of telomeres, which cap the ends of eucaryotic chromosomes (9). These sequences serve as binding sites for a collection of proteins that compensate for progressive losses due to replication (9), protect the ends from nuclease degradation and end-to-end fusion (11, 71), and give rise to a unique chromatin structure (70). Telomeric proteins play additional roles in chromosome attachment to the nuclear matrix (44) and in the separation of telomeres at mitosis and meiosis (13, 40). Thus, the telomere sequence mediates a complicated interplay of proteins and processes.
Telomeres influence replication, gene expression, and recombination in their vicinity. Activation of replication origins is delayed or abolished near telomeres in mitotically dividing Saccharomyces cerevisiae (21, 26, 57, 69), and replication timing is shifted from middle to late for the breakpoint region adjacent to a repaired telomere in human cells (53). Genes near telomeres in S. cerevisiae (28), Schizosaccharomyces pombe (52), Drosophila melanogaster (43), and Trypanosoma brucei (33, 61) are transcriptionally repressed. Near telomeres in mammalian cells, selectable genes with strong promoters are not affected, whereas genes driven by weak promoters may be slightly repressed (8, 14). During meiosis in S. cerevisiae, ectopic recombination is significantly greater between inserts near telomeres than it is between more centrally located inserts (27), although recombination between directly repeated LEU2 gene segments was unaffected by proximity to the telomere (55). In humans, meiotic recombination is elevated near telomeres (5, 39). By contrast, molecular and cytological studies of meiosis in grasshoppers show reduced recombination near telomeres (48).
Telomere repeats are not confined to the ends of chromosomes but are also found at discrete intrachromosomal sites in many eucaryotic species (1, 6, 19, 56). It is thought that these interstitial telomere repeats arose as the result of chromosome rearrangements in the course of genome evolution (34, 67), a view supported by occasional observation of aberrant chromosomes that have telomere repeats at the site of rearrangement (58). Like repeats at telomeres, interstitial repeats also appear to influence aspects of DNA metabolism in their vicinity. Cytogenetic studies in mitotically dividing cells have linked interstitial telomere repeats with sites of spontaneous and radiation-induced chromosome rearrangements (10, 17, 54, 66), chromosome fragility (12, 50), and unstable rearrangements known as jumping translocations (16, 36, 72). In meiotic cells in the Armenian hamster, an interstitial telomere repeat was a site of frequent chiasma formation, consistent with a hotspot for homologous recombination (4). DNA molecules injected into the macronucleus of Paramecium primaurelia preferentially integrate by illegitimate recombination in or near interstitial telomere repeats (37).
Because interstitial telomere sequences are uncharacterized for length, purity, and repeat orientation and because interstitial repeats are not all hotspots for rearrangement (10), several studies introduced defined telomeric sequences into the genome. In S. cerevisiae, insertion of 49 bp of telomeric sequence at the HIS4 locus stimulated meiotic homologous recombination and the formation of nearby meiosis-specific double-strand DNA breaks (22, 74). In mitotic yeast cells, homologous recombination between 300-bp duplications of telomeric sequence occurred at roughly the same frequency as that between the same length of unique sequence, except in the vicinity of the telomere, where telomere repeat recombination was reduced 10-fold (68). Overexpression of the telomere-binding protein Rap1p eliminated repression of recombination near telomeres and stimulated recombination at interior telomeric repeats, indicating that some telomere-repeat-binding proteins recognize interstitial sequences (68). Finally, at several locations in the S. cerevisiae genome, telomere repeats repress transcription of nearby genes (68).
In mammalian cells, telomere repeat sequences have been introduced to fragment chromosomes and to generate minichromosomes (7, 23–25, 29, 32, 35, 41, 49). Random integration of plasmids carrying telomere repeats adjacent to a selectable marker generated selected colonies with a newly seeded telomere next to the marker at a frequency of 20% in Chinese hamster ovary (CHO) cells (23) and 70% in HeLa cells (29). Surprisingly, the majority of such clones carried duplications or other rearrangements at the site of chromosome truncation (14, 24, 32). The role of telomere sequence in chromosome truncation and terminal rearrangement—beyond its capacity to seed new telomeres—is unclear. Cytogenetic analysis of three human cell lines with randomly integrated telomere-repeat-containing plasmids showed that two were highly unstable, but the instability was due not to telomere sequence (20) but rather to random integration, which commonly generates ongoing rearrangements (47, 59).
To assess the effects of interstitial telomere sequence on several aspects of DNA metabolism, we used site-specific recombination to insert 800 bp of functional vertebrate telomere sequence in two orientations into the second intron of the adenosine phosphoribosyltransferase (APRT) gene in CHO cells. Site-specific recombination avoids the inherent instability of many random integrants (20, 47, 59), and targeting to the APRT locus allows us to make comparisons with previous results (63–65). These cell lines allowed us to test the effects of telomere sequence on gene expression, homologous recombination, gene rearrangements, and chromosome truncation.
MATERIALS AND METHODS
Construction of vectors.
Targeting vectors were constructed from previously described vectors (63, 65), which contained the herpesvirus TK gene, the bacterial GPT gene, and an APRT− gene truncated at the 3′ end of the last exon. To generate targeting vector pAK30 containing telomere sequence in the CCCTAA orientation, telomere sequence from the Sty11 plasmid, kindly provided by Titia de Lange (29), was subcloned into a polylinker adjacent to a FLP recombinaton target (FRT) site, and the pair were then cloned into the polylinker in pGS89. To construct targeting vector pAK50 containing telomere in the TTAGGG orientation, telomere sequence was cloned into the polylinker adjacent to the FRT site in pGS101. Orientations of the telomere sequence are indicated by the sequence of the repeat in the mRNA-like strand of the DNA. Targeting vectors pAK30 and pAK50 were checked for the presence and correct orientation of telomere sequence by restriction digestion of surrounding polylinker sequence and by sequence analysis. In both cases, the telomere sequence consisted predominantly of TTAGGG repeats, with interspersed TTGGGG repeats common at the nonseeding (TA-rich) end but rare at the seeding (G-rich) end. (Telomerase adds new telomere repeats to the 3′ end of the G-rich strand, which we refer to as the seeding end because of its ability to serve as a substrate for addition of telomere repeats.) The telomere sequences in pAK30 and pAK50 were identical to that in Sty11 except that both were missing one TTGGGG repeat at the nonseeding end. Targeting vectors containing the I-SceI recognition site were constructed by inserting a synthetic I-SceI site into a restriction site in the polylinker adjacent to the seeding end of the telomere sequence.
A targeting vector containing HPRT DNA was constructed by ligating an 800-bp PCR fragment (from bases 14928 to 15730 of the human HPRT intron 2) into the SalI and NotI sites in the polylinker at the EcoRI site in pGS101, via SalI and NotI sites in the PCR primers. The targeting vector containing HPRT DNA with a central I-SceI site was constructed by recombinant PCR. The outside primers were the same as above, and the inside primers created an I-SceI site.
Vectors for random integration were constructed by modification of plasmid pGS36, which carried the wild-type APRT gene with an adjacent GPT gene and 4.5 kb of upstream sequences. A HindIII-XhoI fragment from pAK30 or pAK50, which includes the upstream sequences, the GPT gene, and a segment of the APRT gene containing exons 1 through the middle of exon 3 and the telomere sequences, was used to replace the corresponding segment of pGS36 to generate plasmids pAK301 and pAK501. These vectors were linearized at the unique HindIII site prior to transfection.
Construction of cell lines.
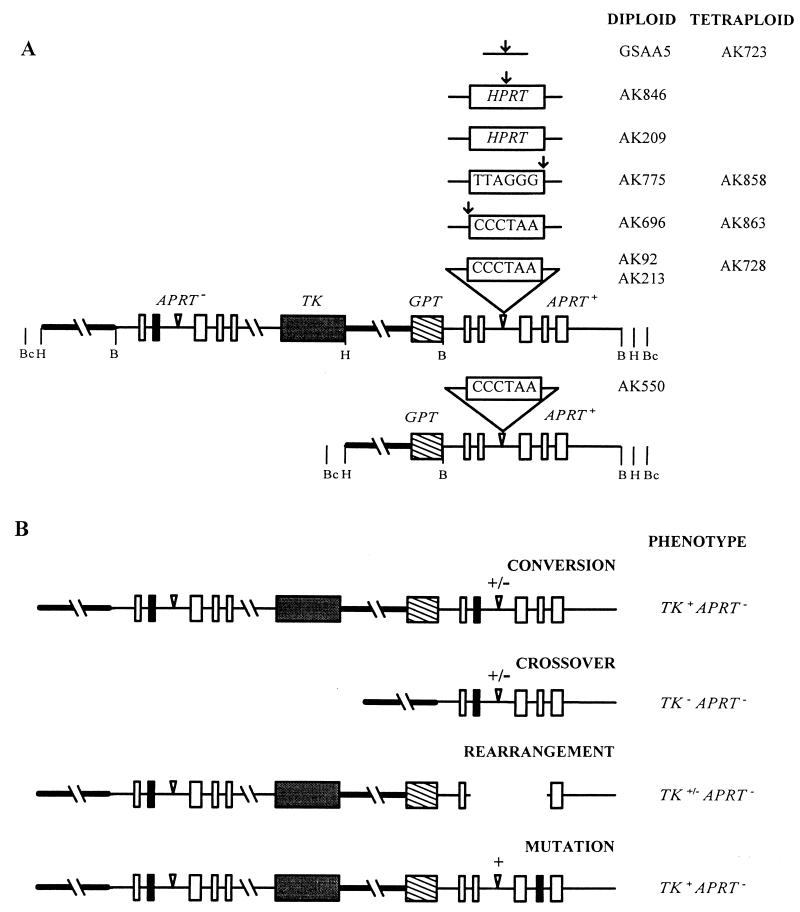
FLP recombinase-mediated site-specific recombination between the engineered FRT sites in the vectors and in the endogenous APRT gene on the chromosome was carried out as described previously (46). The APRT gene in the RMP41 cell line (46) carries a nonreverting point mutation that eliminates the EcoRV site in exon 2 (63). Site-specific recombination generated the tandemly duplicated gene structures shown in Fig. 1. The upstream APRT gene carries two mutations: the point mutation in exon 2 and the truncation of the 3′ end. The downstream, functional APRT gene carries the telomere sequence or HPRT DNA in intron 2. Cell line AK550 was derived from cell line AK213 by selection for TK− APRT+ colonies arising by homologous recombination (63).
FIG. 1.
Molecular structures of the substrates at the APRT locus in diploid and tetraploid cell lines and of the products isolated in various selections. (A) Inserted sequences are shown above their common site in the second intron of the downstream, functional APRT gene (the five exons of APRT are shown as boxes). Vertical arrows indicate locations of the I-SceI recognition sites. Inverted triangles indicate the positions of the FRT recognition sequences. The upstream copy of APRT is nonfunctional by virtue of a truncated fifth exon and a mutation in exon 2 (filled box). The upstream and downstream copies share 6.8 kb of homology: 4.5 kb upstream of the APRT gene (thick line) and 2.3 kb of homology within the gene itself. Cleavage sites for the restriction enzymes BamHI (B), HindIII (H), and BclI (Bc) that were used in Southern analyses are indicated. An additional BamHI site in AK775 and AK858 is located in the polylinker adjacent to the TA end of the telomere sequence (not shown). The hybridization probe corresponds to the downstream BamHI fragment that encompasses the APRT gene, but included no inserted sequences. (B) Products were distinguished based on Southern patterns after BamHI and HindIII cleavage and PCR analysis (63–65). Conversions have a structure like the parental tandem duplication, except that some lose the insert as part of the conversion process (status of the insert is indicated by +/−). Conversions were shown to contain the EcoRV mutation in exon 2 (filled box) by PCR amplification across the exon (Fig. 4A) followed by incubation with EcoRV. Crossovers have a single copy of the APRT gene whose size depends on whether the insert was retained or lost. Rearrangements yield a Southern pattern that does not correspond to conversions or crossovers (Fig. 3); they were subjected to further Southern and PCR analyses (Fig. 4A). Mutations were identical to conversions by Southern analysis but were shown not to contain the EcoRV mutation by PCR analysis. They are assumed to carry point mutations or small deletions elsewhere in the APRT gene; however, they have not been further characterized.
Tetraploid cell lines were constructed by cell fusion (2) between the tandem duplication cell lines carrying telomere sequence and a ouabain-resistant derivative of T2S24 (51), in which exons 1 and 2 of the APRT gene are deleted. Fused cells were selected by growth in medium containing 1 mM ouabain and ALASA (25 μM alanosine, 50 μM azaserine, 100 μM adenine) (2) and shown to be tetraploid by fluorescence-activated cell sorting analysis.
Structures of all cell lines were verified by Southern analysis following digestion with restriction enzymes diagnostic for the predicted structure. The parental cell line and selected diploid and tetraploid cell lines were shown to contain active telomerase by telomeric repeat amplification protocol assays (38).
Cell culture, fluctuation analysis, and transfection.
Cell lines were maintained in Dulbecco's modified Eagle medium supplemented with amino acids and 10% fetal calf serum. Selections were carried out as previously described (62). APRT+ cells were selected by growth in ALASA medium. APRT− cells were selected by growth in medium made with 10% dialyzed fetal calf serum and supplemented with 400 μM 8-aza-adenine. TK− APRT− cells were selected by growth in APRT− selection medium supplemented with 0.3 μM fluoroiodoarabinosyluridine.
Fluctuation analysis (42, 45) was carried out using 12 parallel cultures grown from initial populations of 50 to 100 cells for each rate determination, as described previously (63). The numbers of APRT− or TK− APRT− colonies in parallel cultures were used to calculate rates by the method of the median (42). A single colony was picked from each parallel culture to ensure that all analyzed colonies arose independently.
In experiments that used I-SceI to generate double-strand breaks, 15 μg of the expression vector for I-SceI, pCMVI-SceI (60), was introduced by LipofectAmine (Gibco/BRL) into subconfluent cultures on 100-mm-diameter plates as described previously (64).
Southern and Northern analyses, PCR analysis, and DNA sequencing.
Northern and Southern analyses were carried out using standard protocols (62). The probe for Southern analysis was the 3.9-kb BamHI fragment containing the entire APRT gene, labeled by random priming with [32P]dCTP. The probes for the Northern blot were a CHO APRT cDNA, kindly provided by Elliot Drobetsky, and GAPDH cDNA as an internal loading control. Quantification of RNA on Northern blots was performed by a PhosphorImager using Molecular Dynamics software. PCR analysis of the recombination products was carried out as previously described (63). The locations of PCR primers used for analysis of rearrangements are shown in Fig. 4A; their sequences are available on request. DNA sequencing was carried out using automated sequencing technology on targeting plasmids to confirm the orientation of the telomere sequence insert and on amplified PCR fragments to determine the sequences of the rearrangement junctions.
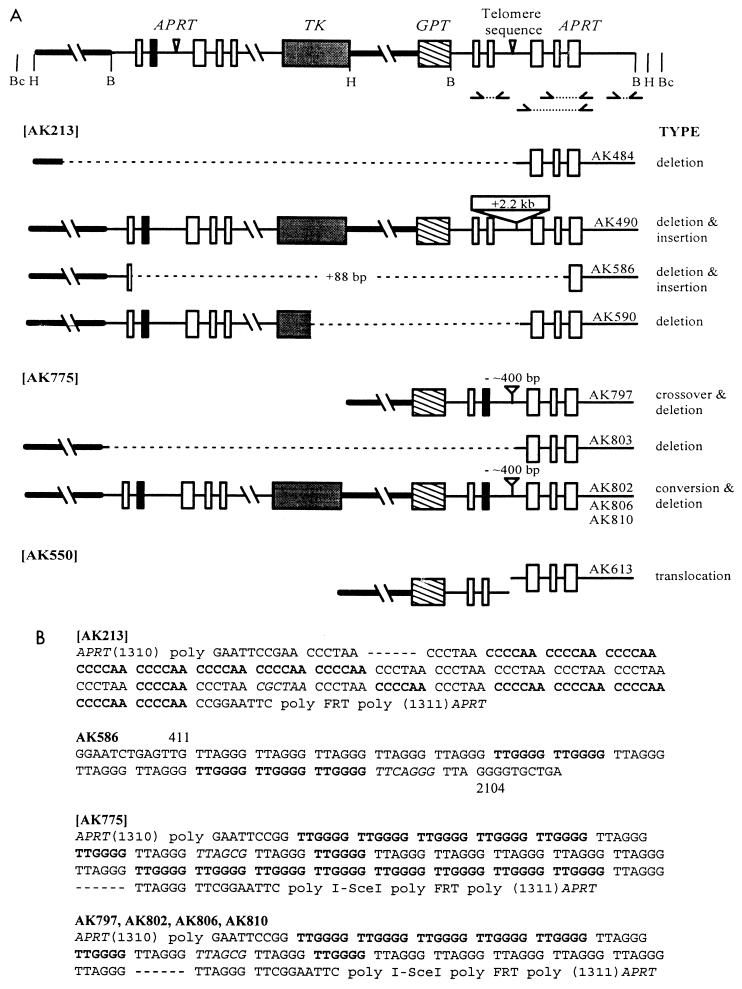
FIG. 4.
Molecular structures of rearrangements and sequences of some rearrangement junctions. (A) Locations of restriction enzyme and PCR primer sites are shown in the tandem duplication structure at the top. Parent cell lines from which the rearrangements arose are shown in brackets at the left. Sizes of deletions and insertions are estimates based on Southern and PCR analyses. Crossovers and conversions were confirmed by lack of cleavage of the PCR products across the second exon, indicating the presence of the EcoRV mutation. AK613 is designated a translocation because it gives two bands upon cleavage by BamHI, HindIII, or BclI; however, it could be an insertion of DNA that includes all three recognition sites. (B) Nucleotide sequences around the insertion point for the parental cell lines, AK213 and AK775, are shown along with the sequences of the rearrangement junctions. TTGGGG sequences (bold) are interspersed with TTAGGG sequences (lightface) and one TTAGCG sequence (italics) at the nonseeding (TA-rich) end of the telomere sequence. Dashes indicated the large number of predominantly TTAGGG sequences toward the seeding (G-rich) end of the telomere sequence. The site of insertion in APRT, nucleotide 1310, is indicated, as are the last nucleotides of APRT sequences that flank the insertion in AK586. The locations of polylinker (poly) sequences, FRT sites, and I-SceI sites are also indicated.
RESULTS
Effects of telomere sequence on the APRT+ phenotype.
To design the experiments described here, it was essential to know that telomere sequence in either orientation would not interfere with the ability of CHO cells to express the APRT+ phenotype. A functional APRT gene was necessary for our targeting strategy and for our loss-of-function assays for homologous recombination, gene rearrangement, and chromosome truncation. To address this question, we used plasmids that contained a GPT gene and a wild-type APRT gene, with or without a telomere repeat sequence in the second intron. We linearized plasmids pGS36 (no insert), pAK301 (CCCTAA), and pAK501 (TTAGGG) and transfected them into the APRT− cell line RMP41. Transfected cells were plated to recover APRT+ or GPT+ colonies arising by random integration of the plasmid DNA (Table 1). If the telomere sequence embedded in the middle of the APRT gene blocked its expression, we would have expected many fewer APRT+ colonies than GPT+ colonies. Because APRT+ and GPT+ colonies were recovered at roughly equal frequencies in transfections with each plasmid, we concluded that 800 bp of telomere sequence in the second intron did not affect the ability of the gene to express the APRT+ phenotype.
TABLE 1.
Relative transfection efficiencies of APRT genes with telomere sequence
| Plasmid (insert) | Transfection method | APRT+ frequency | GPT+ frequency | APRT+/GPT+ ratio |
|---|---|---|---|---|
| pGS36 (none) | Electroporation | 3.3 × 10−5 | 4.0 × 10−5 | 0.8 |
| pAK301 (CCCTAA) | Electroporation | 1.0 × 10−4 | 1.0 × 10−4 | 1.0 |
| pAK501 (TTAGGG) | Calcium phosphate | 4.0 × 10−3 | 5.4 × 10−3 | 0.7 |
Construction of cell lines and experimental rationale.
To test the effects of interstitial telomere sequence on gene expression, homologous recombination, gene rearrangements, and chromosome truncation, we constructed a variety of cell lines whose structures are shown in Fig. 1A. Targeting vectors carrying different DNA sequences adjacent to an FRT site in the second APRT intron were integrated via FLP-mediated site-specific recombination so that the inserted sequences were located in the downstream, APRT+ copy of the gene. The upstream APRT− copy of the gene carries a nonreverting point mutation and is truncated at its 3′ end. The structures of the tandem duplications in these cells lines is analogous to those we have used before (63, 65) and thus allow us to make direct comparisons with our previous results.
We chose to use 800 bp of telomere sequence because this length was sufficient to support telomere-associated chromosome fragmentation (TACF) in HeLa cells (29). Similar experiments in CHO cells demonstrated TACF using only 500 bp of telomere sequence (23). As a control for this length of insert, we constructed cell lines carrying an 800-bp fragment from intron 2 of the human HPRT gene (Fig. 1A). As a positive control to test the effects of double-strand breaks, we constructed parallel cell lines that carried the recognition site for endonuclease I-SceI at the seeding (G-rich) end of the telomere sequence or in the middle of the HPRT fragment (arrows in Fig. 1A).
To test for chromosome truncation accompanied by the formation of new telomeres at the inserted telomere sequence, it was necessary to provide a second copy of the chromosome that carries the APRT gene. Since large portions of the APRT-containing chromosome are hemizygous (3), it was possible that chromosome truncation would eliminate an essential gene, rendering those cells nonviable. A second copy of the APRT-containing chromosome was provided by fusion with cell line T2S24, which carries a deletion in the APRT gene (51). A defective APRT gene on this second chromosome was critical for our analysis, which depends on selection for the APRT− phenotype.
Effects of telomere sequence on production of APRT mRNA.
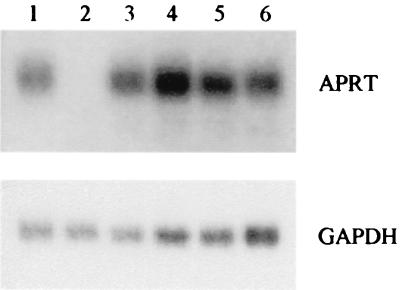
Although telomere sequence does not interfere with expression of the APRT+ phenotype (Table 1), it could still reduce mRNA levels substantially, since cells with only a few percent of wild-type Aprt enzyme activity are phenotypically APRT+ (18). To measure the effect of telomere sequence on production of APRT mRNA, we performed Northern analysis on RNA extracted from wild-type cells and from cells with targeted tandem duplication at the APRT locus (Fig. 2). The level of APRT mRNA relative to GAPDH mRNA is the same for control cell lines carrying a single copy of the APRT gene (lane 1), a tandem duplication with no insert (lane 3), and a tandem duplication with the HPRT insert (lane 4). Cell line AK213 with telomere sequence in the CCCTAA orientation expresses the same relative level of APRT mRNA as the control cell lines (lane 5). Cell line AK775, with telomere sequence in the TTAGGG orientation, expresses about half the amount of APRT mRNA, relative to GAPDH mRNA, as the other cell lines (lane 6).
FIG. 2.
Northern analysis of APRT expression in various cell lines. Lane 1, transcript levels in cell line RMP41 (20), which has a single copy of the APRT gene. Lane 2, transcript levels in cell line DELI26 (20), which has a tandem duplication structure but in which the downstream copy of APRT is lacking the promoter and 5′ half of the gene. Absence of a transcript in this strain indicates that no stable transcript is made from the truncated, upstream copy of APRT, which is identical to that in the other strains tested. Lane 3, transcript levels in cell line GSAA5, which carries I-SceI and FRT recognition sites. Lane 4, transcript levels in cell line AK209, which carries an HPRT insert. Lane 5, transcript levels in cell line AK213, which carries a CCCTAA insert. Lane 6, transcript levels in cell line AK775, which carries a TTAGGG insert. Hybridization to GAPDH serves as a loading control.
Since we did not know the transcriptional status of the upstream APRT gene fragment, we included in the analysis an additional cell line, DELI26, which carries a tandem duplication with the same structure except that the downstream gene is promoterless (46). The absence of APRT mRNA in this cell line (lane 2) indicates that the upstream fragment of the APRT gene does not yield a stable transcript. Thus, the detected transcripts in the other tandem-duplication cell lines must come exclusively from the downstream gene, which harbors the different sequences tested. From these experiments we conclude that the CCCTAA orientation of telomere sequence has no effect on APRT transcription and processing, whereas the TTAGGG orientation reduces mRNA production by about 50%.
Effects of telomere sequence on chromosome truncation.
The single functional APRT gene in CHO AT32 cells resides in a hemizygous region near the chromosome end and is transcribed toward the centromere (73). Truncation of the chromosome at the interstitial telomere sequence and the seeding of a new telomere, which could occur only in the CCCTAA orientation, would eliminate the 5′ end of the APRT gene along with more distal sequences. To render eliminated sequences nonessential, we fused tandemly duplicated cell lines to a cell line carrying an APRT deletion. In these tetraploid cell lines, APRT− colonies arose (presumably due to chromosome loss) at the same frequency in cell lines with no telomere sequence (AK723, [3.8 ± 1.8] × 10−4) and with a nonseeding, TTAGGG sequence (AK858, [4.9 ± 1.9] × 10−4). In two potentially seeding, CCCTAA cell lines (AK863 and AK728), APRT− colonies arose at similar frequencies (average, [6.6 ± 3.1] × 10−4). Thus, telomere sequence at the APRT locus does not cause chromosome truncation and the seeding of new telomeres at a frequency greater than 0.1%. Expression of I-SceI in these cell lines did not stimulate APRT− colony formation sufficiently above the level of chromosome loss to detect the seeding of new telomeres directly (data not shown).
Effects of telomere sequence on homologous recombination.
Although tandem duplications can give rise to APRT− cells in several ways (Fig. 1B), previous analysis of spontaneous events indicated that homologous recombination was dominant, accounting for about 95% of events, compared to 5% for mutations and <0.5% for rearrangements (63, 65). The same studies showed that TK− APRT− cells were generated entirely by homologous recombination (Fig. 1B). Thus, we measured the effects of telomere sequence on homologous recombination by measuring rates of production of APRT− and TK− APRT− phenotypes. Homologous recombination yields TK− APRT− cells by crossover (popout) recombination, which eliminates one copy of the APRT gene; it generates APRT− cells by crossover recombination and by gene conversion, in which the EcoRV mutation in the upstream copy is transferred to the downstream copy (Fig. 1B).
Tandem duplications carrying telomere sequence yielded TK− APRT− cells and APRT− cells at rates that were indistinguishable from those of cells carrying an HPRT insert or smaller inserts (Table 2), suggesting that homologous recombination was unaffected by telomere sequence in either orientation. Analysis of individual colonies by Southern blotting and PCR confirmed that the majority arose by homologous recombination (Fig. 3; Table 2). Among APRT− colonies from cell lines containing telomere sequence, the proportions of conversions and crossovers (18 versus 4) were similar to those observed previously (88 versus 17) (Table 2). In addition, crossovers that retained telomere sequence (13 of 24) or lost it (11 of 24) were generated in proportion to the lengths of homology flanking the telomere sequence, consistent with results for smaller inserts (63). Thus, analysis of neither the rates of recombination nor the nature of the products reveals any influence of telomere sequence.
TABLE 2.
Rates of TK− APRT− and APRT− colony formation and analysis of independent products
| Colony | Cell line(s) | Insert | Rate (10−7)b | No. of productsc
|
||||
|---|---|---|---|---|---|---|---|---|
| Crossover | Conversion | Rearrangement | Mutation | Total | ||||
| TK- APRT− | GSB, -C, -Ea | <200 | 1.2 ± 0.2 | 72 | 0 | 0 | 0 | 72 |
| AK209 | HPRT | 1.2 ± 0.4 | 12 | 0 | 0 | 0 | 12 | |
| AK92, AK213 | CCCTAA | 1.9 ± 0.5 | 11 | 0 | 2 | 0 | 13 | |
| AK775 | TTAGGG(I-SceI) | 1.3 | 9 | 0 | 1 | 0 | 10 | |
| APRT− | GSB, -C, -Ea | <200 | 15 ± 1 | 17 | 88 | 0 | 6 | 111 |
| AK209 | HPRT | 14 ± 4 | 0 | 11 | 0 | 0 | 11 | |
| AK92, AK213 | CCCTAA | 14± 3 | 2 | 12 | 3 | 7 | 24 | |
| AK775 | TTAGGG(I-SceI) | 16 | 2 | 6 | 4 | 0 | 12 | |
Rates and product analysis for the GSB, GSC, and GSE cell lines are from previous reports (63, 65). GSB cell lines contained a 114-bp insert carrying recognition sequences for the HO endonuclease and for the FLP recombinase; GSC cell lines carried no insert; GSE cell lines carried a 200-bp insert that contained a (GT)29 repeat.
Determined by fluctuation analyses. For AK209, the rate is an average of two fluctuation analyses. One analysis for AK92 and two for AK213 were averaged. The rate for AK775 is from a single fluctuation analysis. The standard error of the mean is indicated.
The molecular structure of the APRT locus in individual colonies was determined by a combination of Southern and PCR analyses.
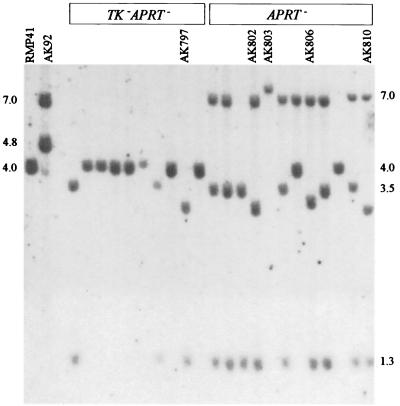
FIG. 3.
Southern analysis of TK− APRT− and APRT− colonies from AK775. DNAs from individual colonies were digested with BamHI and HindIII, and the fragments were resolved by electrophoresis and visualized by Southern blotting using a 32P-labeled BamHI fragment of the wild-type APRT gene as a probe. Crossovers that have lost the telomere sequence have a single band at 4.0 kb; crossovers that have retained the telomere sequence have bands at 3.5 and 1.3 kb. There is a BamHI site (not shown in Fig. 1A) at the TA end of the telomere sequence, so that the telomere sequence is in the 3.5-kb band. Conversions have the 7.0-kb band from the upstream copy of APRT. Conversions that have lost the telomere sequence have an additional band at 4.0 kb; conversions that retain the telomere sequence have two additional bands at 3.5 and 1.3 kb. Rearrangements have patterns that do not match these expectations; their identities are indicated at the top. Numbers at the sides indicate the lengths of fragments in kilobases. RMP41 carries a single copy of the APRT gene; AK92 carries a tandem duplication with telomere sequence in the CCCTAA orientation (it does not carry a BamHI site adjacent to the telomere sequence).
To determine whether stimulated recombination could be detected in the vicinity of telomere sequence, we expressed endonuclease I-SceI in cell lines that carried its recognition site alone, adjacent to the telomere sequence, or in the middle of the HPRT insert (Fig. 1A). I-SceI expression typically stimulates homologous recombination several hundred-fold in mammalian cells (60, 64). In all cases recombination was stimulated to similar levels (Table 3), much above the spontaneous rates (Table 2). Analysis of individual colonies confirmed that most arose by homologous recombination (data not shown). Since increased recombination could have been detected, we conclude that telomere sequence does not affect homologous recombination at APRT.
TABLE 3.
TK− APRT− and APRT− colony formation after treatment with endonuclease I-SceI
| Cell line | Insert | Frequency (10−4)a
|
|
|---|---|---|---|
| TK−APRT− | APRT− | ||
| GSAA5 | I-SceI | 1.3 ± 0.3 | 18 ± 7 |
| AK846 | HP(I-SceI)RT | 0.8 ± 0.2 | 9 ± 3 |
| AK696 | (I-SceI)CCCTAA | 5.7 ± 0.6 | 10 ± 2 |
| AK775 | TTAGGG(I-SceI) | 4.3 ± 0.2 | 6 ± 2 |
Values for GSAA5 are averages of six experiments; values for other cell lines are averages of three experiments. The standard error of the mean is indicated. Stimulation above spontaneous frequencies of TK− APRT− and APRT− cells in untreated populations averaged about 100-fold for each selection.
Effects of telomere sequence on gene rearrangements.
Our loss-of-function assay allows us to detect gene rearrangements in addition to homologous recombinants (Fig. 1B). Rearrangements had not previously been observed among spontaneous recombinants in wild-type CHO cells (63, 65). Thus, the most striking feature of the data in Table 2 is the presence of rearrangements, which were detected by their abnormal Southern blot patterns (Fig. 3). Among 59 colonies from tandem duplications carrying the telomere sequence, 10 were rearrangements. By contrast, none of 23 colonies from the HPRT insert were rearrangements. In experiments with inserts of less than 200 bp (63, 65), no rearrangements were detected among 183 analyzed colonies (Table 2). These numbers (10 of 59 for telomere sequence versus none of 206 for HPRT and small inserts) indicate that rearrangements were stimulated some 30-fold or more by interstitial telomere sequence. One additional rearrangement was found among 10 independent APRT− colonies isolated from cell line AK550, which had a single copy of the APRT gene (Fig. 1A).
PCR and Southern analyses of nine rearrangements showed that they included deletions, insertions, and a probable translocation and that all involved the telomere sequence (Fig. 4A). Five rearrangement junctions were successfully amplified by PCR and sequenced (Fig. 4B). In rearrangement AK586, APRT sequences around the original telomere sequence were deleted and replaced with a short telomere sequence, flipped with respect to the original orientation. The new telomere insert does not correspond to a known sequence in the original insert, and thus it may have been corrupted in the course of the rearrangement.
Rearrangements AK797, AK802, AK806, and AK810 each had lost some 400 bp of telomere sequence. Interspersed TTGGGG repeats at the nonseeding end of the telomere sequence allowed one end of the deletion to be mapped to a common region in all four rearrangements (Fig. 4B); however, their 3′ ends could not be positioned relative to the featureless repeats at the seeding end of the telomere sequence. Differences in lengths of Southern fragments and PCR products suggest that these telomere sequence deletions are not identical. Nevertheless, their similarity raised the possibility that they preexisted in the starting cell population. If their isolation in these experiments was coincidental, then they should be present in the starting population at roughly the same frequency as among the selected colonies, that is, at 15 to 20% (four isolates among 22 colonies). Sensitive Southern analysis, which could have detected about a 2% subpopulation, failed to reveal the band diagnostic for these telomere sequence deletions in the genomic DNA from the parental AK775 cell line (data not shown).
DISCUSSION
These studies clearly document that interstitial telomere repeat sequence of known purity, of specific length, and in either orientation confers instability at a defined site in a mammalian genome. By using the well-characterized APRT locus in CHO cells and a substrate design that allowed sensitive, simultaneous detection of homologous recombination and gene rearrangements, we have demonstrated that telomere sequence stimulates rearrangements some 30-fold above background, without noticeable effects on homologous recombination. Direct participation of telomere sequence in the detected rearrangements is supported by molecular analyses, which showed that every characterized rearrangement involved the telomere repeats. Previous studies using random integration of a 1.6-kb telomere sequence failed to detect repeat-induced instability by less sensitive cytogenetic methods (20). Thus, these studies define the telomere repeat sequence as a destabilizing element in the interior of a mammalian chromosome, providing direct support for previous correlations between interstitial telomere repeats and chromosome rearrangements (10, 12, 16, 17, 36, 50, 54, 66, 72).
The effects of telomeres on expression of nearby genes are dramatic in many lower eucaryotes (28, 33, 43, 52, 61) but weak or nonexistent in mammalian cells (8, 14). In yeast, interstitial telomere sequence also reduces expression of nearby genes (68). We have shown here that telomere sequence in an intron of the APRT gene has only a modest effect on expression of the gene (Fig. 2). Because only one orientation of the repeat (TTAGGG) reduced mRNA levels, it seems unlikely that the reduction is due to repeat-binding proteins, whose effects might be expected to be orientation independent. It may be that the repeated sequence in the template strand (3′-AATCCC) impedes RNA polymerase or that the repeated sequence in the nascent RNA (5′-UUAGGG) interferes with RNA processing. Further studies are required to resolve these possibilities.
Chromosome truncation and the seeding of new telomeres were not detected above the background loss of chromosomes that is common in tetraploid cell lines (2), which places an upper limit of about 0.1% on the frequency of these events at the APRT locus. When these studies were initiated, the orientation of the APRT gene on the chromosome was unknown (73) and our particular arrangement of selectable markers did not allow us to distinguish between a lost chromosome and a truncated one. We have now reconfigured the markers to address this issue with more sensitivity. Nevertheless, the low frequency of chromosome truncation cannot account for the 20 to 70% truncation frequencies observed in TACF experiments (23, 29), suggesting that TACF is unlikely to occur by random integration followed by telomere sequence-induced breakage. It seems more likely that truncation observed in TACF experiments results from plasmid ligation to transient double-strand breaks or from random-integration-triggered rearrangements that are resolved when a break appears near the telomere sequence (14).
The effects of telomeres and telomere sequence on homologous recombination are varied, sometimes stimulating it (4, 5, 22, 27, 39, 74), sometimes inhibiting it (48, 68), and sometimes leaving it unaffected (55). At the APRT locus, telomere sequence does not detectably affect homologous recombination, as assessed by rates of recombination, proportions of crossovers and conversions, and distribution of exchanges. When double-strand breaks were deliberately introduced adjacent to telomere sequence, homologous recombination was stimulated to the same extent as in cell lines lacking telomere sequence. Thus, stimulated recombination could have been detected in the vicinity of the telomere sequence.
The inherent instability of interstitial telomere sequence likely contributes to the rearrangements observed in cancer cells subsequent to the chromosome fusions that occur when telomeres become critically short (15, 30, 31). The extraordinarily high instability (several percent) correlated with some naturally occurring interstitial telomere sequences (10, 12, 16, 17, 36, 50, 54, 66, 72) suggests that instability increases with telomere sequence length or with some undefined aspect of the arrangement or structure of the repeats. The approaches described here provide a means to quantify these undefined elements of telomere sequence-induced instability.
ACKNOWLEDGMENTS
We thank Gerald Adair and Olivia Perrera-Smith for advice about cell fusions, Dan Medina for help with telomerase assays, and Beth and Frank Chance for helpful discussions.
This investigation was supported by NIH grant GM38219 and by Department of Defense Breast Cancer Research Program grant DAMD17-97-1-7283 to J.H.W.
REFERENCES
- 1.Abuin M, Martinez P, Sanchez L. Localization of the repetitive telomeric sequence (TTAGGG)n in four salmonoid species. Genome. 1996;39:1035–1038. doi: 10.1139/g96-129. [DOI] [PubMed] [Google Scholar]
- 2.Adair G M, Thompson L H, Lindl P A. Six complementation classes of conditionally lethal protein synthesis mutants of CHO cells selected by 3H-amino acid. Somatic Cell Genet. 1978;4:27–44. doi: 10.1007/BF01546491. [DOI] [PubMed] [Google Scholar]
- 3.Adair G M, Stallings R L, Nairn R S, Siciliano M J. High-frequency structural gene deletion as the basis for functional hemizygosity of the adenine phosphoribosyltransferase locus in Chinese hamster ovary cells. Proc Natl Acad Sci USA. 1983;80:5961–5964. doi: 10.1073/pnas.80.19.5961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ashley T, Ward D C. A “hotspot” of recombination coincides with an interstitial telomeric sequence in the Armenian hamster. Cytogenet Cell Genet. 1993;62:169–171. doi: 10.1159/000133464. [DOI] [PubMed] [Google Scholar]
- 5.Ashley T. Mammalian meiotic recombination: a reexamination. Hum Genet. 1994;94:587–593. doi: 10.1007/BF00206950. [DOI] [PubMed] [Google Scholar]
- 6.Azzalin C M, Mucciolo E, Bertoni L, Giulotto E. Fluorescence in situ hybridization with a synthetic (TTAGGG)n polynucleotide detects several intrachromosomal telomere-like repeats on human chromosomes. Cytogenet Cell Genet. 1997;78:112–115. doi: 10.1159/000134640. [DOI] [PubMed] [Google Scholar]
- 7.Barnett M A, Buckle V J, Evans E P, Porter A C, Rout D, Smith A G, Brown W R. Telomere directed fragmentation of mammalian chromosomes. Nucleic Acids Res. 1993;21:27–36. doi: 10.1093/nar/21.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bayne R A L, Broccoli D, Taggart M H, Thomson E J, Farr C J, Cooke H J. Sandwiching of a gene within 12 kb of a functional telomere and alpha satellite does not result in silencing. Hum Genet. 1993;3:539–546. doi: 10.1093/hmg/3.4.539. [DOI] [PubMed] [Google Scholar]
- 9.Blackburn E H, Greider C W, editors. Telomeres. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1995. [Google Scholar]
- 10.Bouffler S D. Involvement of telomeric sequences in chromosomal aberrations. Mutat Res. 1998;404:199–204. doi: 10.1016/s0027-5107(98)00114-6. [DOI] [PubMed] [Google Scholar]
- 11.Bourgain F M, Katinka M D. Telomeres inhibit end to end fusion and enhance maintenance of linear DNA molecules injected into the Paramecium primaurelia macronucleus. Nucleic Acids Res. 1991;19:1541–1547. doi: 10.1093/nar/19.7.1541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Boutouil M, Fetni R, Qu J, Dallaire L, Richer C L, Lemieux N. Fragile site and interstitial telomere repeat sequences at the fusion point of a de novo (Y;13) translocation. Hum Genet. 1996;98:323–327. doi: 10.1007/s004390050216. [DOI] [PubMed] [Google Scholar]
- 13.Conrad M N, Dominguez A M, Dresser M E. Ndj1p, a meiotic telomere protein required for normal chromosome synapsis and segregation in yeast. Science. 1997;276:1252–1255. doi: 10.1126/science.276.5316.1252. [DOI] [PubMed] [Google Scholar]
- 14.Cooke H J. Non-programmed and engineered chromosome breakage. In: Blackborn E H, Greider C W, editors. Telomeres. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1995. pp. 219–242. [Google Scholar]
- 15.Counter C M, Hirte H W, Bacchetti S, Harley C B. Telomerase activity in human ovarian carcinoma. Proc Natl Acad Sci USA. 1994;91:2900–2904. doi: 10.1073/pnas.91.8.2900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cuthbert G, McCullough S, Finney R, Breese G, Brown N. Jumping translocation at 11q23 with MLL gene rearrangement and interstitial telomere sequences. Genes Chromosomes Cancer. 1999;24:295–298. doi: 10.1002/(sici)1098-2264(199904)24:4<295::aid-gcc1>3.0.co;2-8. [DOI] [PubMed] [Google Scholar]
- 17.Day J P, Limoli C L, Morgan W F. Recombination involving interstitial telomere repeat-like sequences promotes chromosomal instability in Chinese hamster cells. Carcinogenesis. 1998;19:259–265. doi: 10.1093/carcin/19.2.259. [DOI] [PubMed] [Google Scholar]
- 18.de Boer J G, Glickman B H. Mutational analysis of the structure and function of the adenine phosphoribosyltransferase enzyme of Chinese hamster. J Mol Biol. 1991;221:163–174. doi: 10.1016/0022-2836(91)80212-d. [DOI] [PubMed] [Google Scholar]
- 19.de la Sena C, Chowdhary B P, Gustavsson I. Localization of the telomeric (TTAGGG)n sequences in chromosomes of some domestic animals by fluorescence in situ hybridization. Hereditas. 1995;123:269–274. doi: 10.1111/j.1601-5223.1995.t01-1-00269.x. [DOI] [PubMed] [Google Scholar]
- 20.Desmaze C, Alberti C, Martins L, Pottier G, Sprung C N, Murnane J P, Sabatier L. The influence of interstitial telomeric sequences on chromosome instability in human cells. Cytogenet Cell Genet. 1999;86:288–295. doi: 10.1159/000015321. [DOI] [PubMed] [Google Scholar]
- 21.Dubey D D, Davis L R, Greenfeder S A, Ong L Y, Zhu J G, Broach J R, Newlon C S, Huberman J A. Evidence suggesting that the ARS elements associated with silencers of the yeast mating-type locus HML do not function as chromosomal DNA replication origins. Mol Cell Biol. 1991;11:5346–5355. doi: 10.1128/mcb.11.10.5346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fan Q Q, Xu F, White M A, Petes T D. Competition between adjacent meiotic recombination hotspots in the yeast Saccharomyces cerevisiae. Genetics. 1997;145:661–670. doi: 10.1093/genetics/145.3.661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Farr C, Fantes J, Goodfellow P, Cooke H. Functional reintroduction of human telomeres into mammalian cells. Proc Natl Acad Sci USA. 1991;88:7006–7010. doi: 10.1073/pnas.88.16.7006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Farr C J, Stevanovic M, Thomson E J, Goodfellow P N, Cooke H J. Telomere-associated chromosome fragmentation: applications in genome manipulation and analysis. Nat Genet. 1992;2:275–282. doi: 10.1038/ng1292-275. [DOI] [PubMed] [Google Scholar]
- 25.Farr C J, Bayne R A, Kipling D, Mills W, Critcher R, Cooke H J. Generation of a human X-derived minichromosome using telomere-associated chromosome fragmentation. EMBO J. 1995;14:5444–5454. doi: 10.1002/j.1460-2075.1995.tb00228.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ferguson B M, Fangman W L. A position effect on the time of replication origin activation in yeast. Cell. 1992;68:333–339. doi: 10.1016/0092-8674(92)90474-q. [DOI] [PubMed] [Google Scholar]
- 27.Goldman A S, Lichten M. The efficiency of meiotic recombination between dispersed sequences in Saccharomyces cerevisiae depends upon their chromosomal location. Genetics. 1996;144:43–55. doi: 10.1093/genetics/144.1.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gottschling D E, Aparicio O M, Billington B L, Zakian V A. Position effect at S. cerevisiae telomeres: reversible repression of Pol II transcription. Cell. 1990;63:751–762. doi: 10.1016/0092-8674(90)90141-z. [DOI] [PubMed] [Google Scholar]
- 29.Hanish J P, Yanowitz J L, de Lange T. Stringent requirements for the formation of human telomeres. Proc Natl Acad Sci USA. 1994;91:8861–8865. doi: 10.1073/pnas.91.19.8861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Harley C B, Kim N W, Prowse K R, Weinrich S L, Hirsch K S, West M D, Bacchetti S, Hirte H W, Greider C W, Wright W E, Shay J W. Telomerase, cell immortality and cancer. Cold Spring Harbor Symp Quant Biol. 1994;59:307–315. doi: 10.1101/sqb.1994.059.01.035. [DOI] [PubMed] [Google Scholar]
- 31.Hastie N D, Allshire R C. Human telomeres: fusion and interstitial sites. Trends Genet. 1989;5:326–331. doi: 10.1016/0168-9525(89)90137-6. [DOI] [PubMed] [Google Scholar]
- 32.Heller R, Brown K E, Burgtorf C, Brown W R A. Mini-chromosomes derived from the human Y chromosome by telomere directed chromosome breakage. Proc Natl Acad Sci USA. 1996;93:7125–7130. doi: 10.1073/pnas.93.14.7125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Horn D, Cross G A. A developmentally regulated position effect at a telomeric locus in Trypanosoma brucei. Cell. 1995;83:555–561. doi: 10.1016/0092-8674(95)90095-0. [DOI] [PubMed] [Google Scholar]
- 34.Ijdo J W, Baldini A, Ward D C, Reeders S T, Wells R A. Origin of human chromosome 2: an ancestral telomere-telomere fusion. Proc Natl Acad Sci USA. 1991;88:9051–9055. doi: 10.1073/pnas.88.20.9051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Itzhaki J E, Barnett M A, MacCarthy A B, Buckle V J, Brown W R, Porter A C. Targeted breakage of a human chromosome mediated by cloned human telomeric DNA. Nat Genet. 1992;2:283–287. doi: 10.1038/ng1292-283. [DOI] [PubMed] [Google Scholar]
- 36.Jewett T, Marnane D, Stewart W, Hayworth-Hodge R, Finklea L, Klinepeter K, Rao P N, Pettenati M J. Jumping translocation with partial duplications and triplications of chromosomes 7 and 15. Clin Genet. 1998;53:415–420. doi: 10.1111/j.1399-0004.1998.tb02757.x. [DOI] [PubMed] [Google Scholar]
- 37.Katinka M D, Bourgain F M. Interstitial telomeres are hotspots for illegitimate recombination with DNA molecules injected into the macronucleus of Paramecium primaurelia. EMBO J. 1992;11:725–732. doi: 10.1002/j.1460-2075.1992.tb05105.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kim N W, Piatyszek M A, Prowse K R, Harley C B, West M D, Ho P L C, Coviello G M, Wright W E, Weinrich S L, Shay J W. Specific association of human telomerase activity with immortal cells and cancer. Science. 1994;266:2011–2015. doi: 10.1126/science.7605428. [DOI] [PubMed] [Google Scholar]
- 39.Kipling D, Wilson H E, Thomson E J, Lee M, Perry J, Palmer S, Ashworth A, Cooke H J. Structural variation of the pseudoautosomal region between and within inbred mouse strains. Proc Natl Acad Sci USA. 1996;93:171–175. doi: 10.1073/pnas.93.1.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kirk K E, Harmon B P, Reichardt I K, Sedat J W, Blackburn E H. Block in anaphase chromosome separation caused by a telomerase template mutation. Science. 1997;275:1478–1481. doi: 10.1126/science.275.5305.1478. [DOI] [PubMed] [Google Scholar]
- 41.Kuroiwa Y, Shinohara T, Notsu T, Tomizuka K, Yoshida H, Takeda S, Oshimura M, Ishida I. Efficient modification of a human chromosome by telomere-directed truncation in high homologous recombination-proficient chicken DT40 cells. Nucleic Acids Res. 1998;26:3447–3448. doi: 10.1093/nar/26.14.3447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lea D E, Coulson C A. The distribution of the numbers of mutants in bacterial populations. J Genet. 1949;49:264–286. doi: 10.1007/BF02986080. [DOI] [PubMed] [Google Scholar]
- 43.Levis R, Hazelrigg T, Rubin G M. Effects of genomic position on the expression of transduced copies of the white gene of Drosophila. Science. 1995;229:558–561. doi: 10.1126/science.2992080. [DOI] [PubMed] [Google Scholar]
- 44.Luderus M E, van Steensel B, Chong L, Sibon O C, Cremers F F, de Lange T. Structure, subnuclear distribution, and nuclear matrix association of the mammalian telomeric complex. J Cell Biol. 1996;135:867–881. doi: 10.1083/jcb.135.4.867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Luria S E, Delbruck M. Mutations of bacteria from virus sensitivity to virus resistance. Genetics. 1943;28:491–511. doi: 10.1093/genetics/28.6.491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Merrihew R V, Sargent R G, Wilson J H. Efficient modification of the APRT gene by FLP/FRT site-specific targeting. Somat Cell Mol Genet. 1995;21:299–307. doi: 10.1007/BF02257465. [DOI] [PubMed] [Google Scholar]
- 47.Merrihew R V, Marburger K, Pennington S L, Roth D B, Wilson J H. High-frequency illegitimate integration of transfected DNA at preintegrated target sites in a mammalian genome. Mol Cell Biol. 1996;16:10–18. doi: 10.1128/mcb.16.1.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Miklos G L G, Nankivell R N. Telomeric satellite DNA functions in regulating recombination. Chromosoma. 1976;56:143–167. doi: 10.1007/BF00293113. [DOI] [PubMed] [Google Scholar]
- 49.Mills W, Critcher R, Lee C, Farr C J. Generation of an approximately 2.4 Mb human X centromere-based minichromosome by targeted telomere-associated chromosome fragmentation in DT40. Hum Mol Genet. 1999;8:751–761. doi: 10.1093/hmg/8.5.751. [DOI] [PubMed] [Google Scholar]
- 50.Musio A, Rainaldi G, Sbrana I. Spontaneous and aphidicolin-sensitive fragile site 3cen co-localizes with the (TTAGGG)n telomeric sequence in Chinese hamster cells. Cytogenet Cell Genet. 1996;75:159–163. doi: 10.1159/000134469. [DOI] [PubMed] [Google Scholar]
- 51.Nairn R S, Humphrey R M, Adair G M. Transformation of UV-hypersensitive Chinese hamster ovary cell mutants with UV-irradiated plasmids. Int J Radiat Biol. 1988;53:249–260. doi: 10.1080/09553008814550601. [DOI] [PubMed] [Google Scholar]
- 52.Nimmo E R, Cranston G, Allshire R C. Telomere-associated chromosome breakage in fission yeast results in variegated expression of adjacent genes. EMBO J. 1994;13:3801–3811. doi: 10.1002/j.1460-2075.1994.tb06691.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ofir R, Wong A C C, McDermid H E, Skorecki K L, Selig S. Position effect of human telomeric repeats on replication timing. Proc Natl Acad Sci USA. 1999;96:11434–11439. doi: 10.1073/pnas.96.20.11434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Park V M, Gustashaw T M, Wathen T M. The presence of interstitial telomeric sequences in constitutional chromosome abnormalities. Am J Hum Genet. 1992;50:914–923. [PMC free article] [PubMed] [Google Scholar]
- 55.Prado F, Agulera A. Role of reciprocal exchange, one-ended invasion crossover and single-strand annealing on inverted and direct repeat recombination in yeast: different requirements for the RAD1, RAD10, and RAD52 genes. Genetics. 1995;139:109–123. doi: 10.1093/genetics/139.1.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Regad F, Lebas M, Lescure B. Interstitial telomeric repeats within the Arabidopsis thaliana genome. J Mol Biol. 1994;239:163–169. doi: 10.1006/jmbi.1994.1360. [DOI] [PubMed] [Google Scholar]
- 57.Reynolds A E, McCarroll R M, Newlon C S, Fangman W L. Time of replication of ARS elements along yeast chromosome III. Mol Cell Biol. 1989;9:4488–4494. doi: 10.1128/mcb.9.10.4488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rossi E, Floridia G, Casali M, Danesino C, Chiumello G, Bernardi F, Magnani I, Papi L, Mura M, Zuffardi O. Types, stability, and phenotypic consequences of chromosome rearrangement leading to interstitial telomeric sequences. J Med Genet. 1993;30:926–931. doi: 10.1136/jmg.30.11.926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Roth D B, Wilson J H. Illegitimate recombination in mammalian cells. In: Kucherlapati R, Smith G R, editors. Genetic recombination. Washington, D.C.: American Society for Microbiology; 1988. pp. 621–654. [Google Scholar]
- 60.Rouet P, Smih F, Jasin M. Expression of a site-specific endonuclease stimulates homologous recombination in mammalian cells. Proc Natl Acad Sci USA. 1994;91:6064–6068. doi: 10.1073/pnas.91.13.6064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Rudenko G, Blundell P A, Dirks-Mulder A, Kieft R, Borst P. A ribosomal DNA promoter replacing the promoter of a telomeric VSG gene expression site can be efficiently switched on and off in T. brucei. Cell. 1995;83:547–553. doi: 10.1016/0092-8674(95)90094-2. [DOI] [PubMed] [Google Scholar]
- 62.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 63.Sargent R G, Merrihew R V, Nairn R, Adair G, Meuth M, Wilson J H. The influence of a (GT)29 microsatellite sequence on homologous recombination in the hamster APRT gene. Nucleic Acids Res. 1996;24:746–753. doi: 10.1093/nar/24.4.746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sargent R G, Brenneman M, Wilson J H. Repair of site-specific double-strand breaks on a mammalian chromosome by homologous and illegitimate recombination. Mol Cell Biol. 1997;17:267–277. doi: 10.1128/mcb.17.1.267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sargent R G, Rolig R L, Kilburn A E, Adair G M, Wilson J H, Nairn R S. Recombination-dependent deletion formation in mammalian cells deficient in the nucleotide excision repair gene ERCC1. Proc Natl Acad Sci USA. 1997;94:13122–13127. doi: 10.1073/pnas.94.24.13122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Slijepcevic P, Xiao Y, Natarajan A T, Bryant P E. Instability of CHO chromosomes carrying interstitial telomere sequences originating from Chinese hamster chromosome 10. Cytogenet Cell Genet. 1997;76:58–60. doi: 10.1159/000134516. [DOI] [PubMed] [Google Scholar]
- 67.Slijepcevic P. Telomeres and mechanisms of Robertsonian fusion. Chromosoma. 1998;107:136–140. doi: 10.1007/s004120050289. [DOI] [PubMed] [Google Scholar]
- 68.Stavenhagen J B, Zakian V A. Yeast telomeres exert a position effect on recombination between internal tracts of yeast telomeric DNA. Genes Dev. 1998;12:3044–3058. doi: 10.1101/gad.12.19.3044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Stevenson J B, Gottschling D E. Telomeric chromatin modulates replication timing near chromosome ends. Genes Dev. 1999;13:146–151. doi: 10.1101/gad.13.2.146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Tommerup H, Dousmansis A, de Lange T. Unusual chromatin in human telomeres. Mol Cell Biol. 1994;14:5777–5785. doi: 10.1128/mcb.14.9.5777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.van Steensel B, Smorgorzewska A, de Lange T. TRF2 protects human telomeres from end-to-end fusions. Cell. 1998;92:401–413. doi: 10.1016/s0092-8674(00)80932-0. [DOI] [PubMed] [Google Scholar]
- 72.Vermeesch J R, Petit P, Speleman F, Deuriendt K, Fryns J P, Marynen P. Interstitial telomeric sequences at the junction site of a jumping translocation. Hum Genet. 1997;99:735–737. doi: 10.1007/s004390050440. [DOI] [PubMed] [Google Scholar]
- 73.Wang P, Zhou R H, Zou Y, Jackson-Cook C K, Povirk L F. Highly conservative reciprocal translocations formed by apparent joining of exchanged DNA double-strand break ends. Proc Natl Acad Sci USA. 1997;94:12018–12023. doi: 10.1073/pnas.94.22.12018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Xu F, Petes T D. Fine-structure mapping of meiosis-specific double-strand DNA breaks at a recombination hotspot associated with an insertion of telomeric sequences upstream of the HIS4 locus in yeast. Genetics. 1996;143:1115–1125. doi: 10.1093/genetics/143.3.1115. [DOI] [PMC free article] [PubMed] [Google Scholar]