Abstract
Saccharomyces cerevisiae carries ∼150 ribosomal DNA (rDNA) copies in tandem repeats. Each repeat consists of the 35S rRNA gene, the NTS1 spacer, the 5S rRNA gene, and the NTS2 spacer. The FOB1 gene was previously shown to be required for replication fork block (RFB) activity at the RFB site in NTS1, for recombination hot spot (HOT1) activity, and for rDNA repeat expansion and contraction. We have constructed a strain in which the majority of rDNA repeats are deleted, leaving two copies of rDNA covering the 5S-NTS2-35S region and a single intact NTS1, and whose growth is supported by a helper plasmid carrying, in addition to the 5S rRNA gene, the 35S rRNA coding region fused to the GAL7 promoter. This strain carries a fob1 mutation, and an extensive expansion of chromosomal rDNA repeats was demonstrated by introducing the missing FOB1 gene by transformation. Mutational analysis using this system showed that not only the RFB site but also the adjacent ∼400-bp region in NTS1 (together called the EXP region) are required for the FOB1-dependent repeat expansion. This ∼400-bp DNA element is not required for the RFB activity or the HOT1 activity and therefore defines a function unique to rDNA repeat expansion (and presumably contraction) separate from HOT1 and RFB activities.
In most eukaryotic organisms, the ribosomal RNA genes (rDNA) are present in long tandem repeats at one or a few chromosomal loci, the nucleolar organizers, and function in the synthesis of rRNA. In the yeast Saccharomyces cerevisiae, approximately 150 rDNA tandem repeats are located on chromosome XII. A single unit of rDNA consists of two transcribed genes (5S and 35S rRNA genes) and two nontranscribed regions (NTS1 and NTS2) (Fig. 1). The 35S rRNA gene is transcribed by RNA polymerase I (Pol I), yielding the 35S rRNA, which is then processed into mature 18S, 5.8S, and 25S rRNAs, while the 5S rRNA gene is transcribed by Pol III. Two DNA elements related to DNA replication, the origin of replication (ARS) and the replication fork barrier (RFB), are located in NTS2 and NTS1, respectively. During each round of DNA replication, a bidirectional replication is initiated at, on the average, one in five ARS sites (2, 21). The RFB located near the end of the 35S rRNA gene allows the progression of the replication fork in the direction of 35S rRNA transcription but not in the opposite direction (2, 3, 19). The RFB site overlaps the E element of HOT1 (17, 35). (Actually, two closely spaced sites, RFB1 and RFB2, are present in this region [37], but we call these sites collectively the RFB site in this paper.) HOT1 was originally discovered as a DNA element that stimulates genetic exchanges at nearby regions when inserted at a non-rDNA site (17). Two elements were subsequently identified as essential for HOT1 activity: the I element, which corresponds to the Pol I promoter region, and the E element, which overlaps the enhancer for Pol I transcription originally identified by Elion and Warner (6). Thus, HOT1 activity appears to be causally related to stimulation of transcription by Pol I.
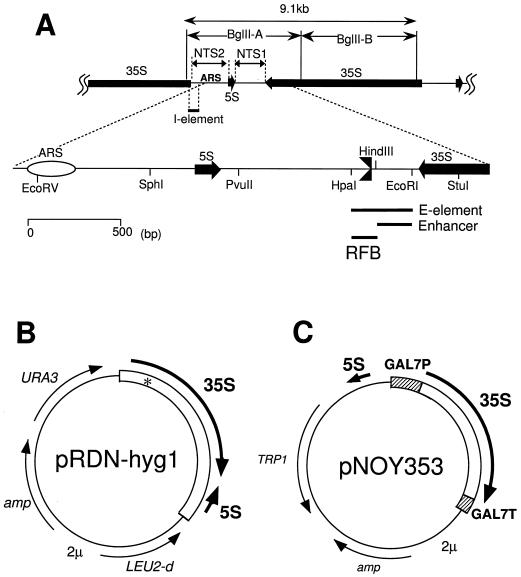
FIG. 1.
(A) Structure of rDNA repeats in S. cerevisiae. The locations of the 35S and 5S rRNA genes (with the direction of transcription indicated by arrows), the two nontranscribed spacer regions (NTS1 and NTS2), ARS (replication origin), and the HOT1 I-element are shown in the upper part. BglII A and B DNA fragments are also shown. NTS1 and its surrounding regions are expanded. Three solid bars represent the HOT1 E-element, Pol I Enhancer, and RFB (the replication fork blocking site, also indicated by  ). (B) Structure of pRDN-hyg1 (4). This plasmid carries a single copy of rDNA repeats obtained by cutting the repeats with SmaI (hence the copy starting from −206 and ending at −207; the numbering is with respect to the Pol I transcription start site as +1). There is a mutation in the 18S rRNA gene (indicated as an asterisk) which makes the ribosome hygromycin B resistant. (C) Structure of pNOY353. This plasmid carries the 7.5-kb BamHI-XhoI fragment, which contains GAL7-35S rDNA (the 35S rRNA coding region fused to the GAL7 promoter as described by Nogi et al. [23]) inserted between the BamHI and SalI sites of the pTV3 vector (27). This plasmid also contains the 1,085-bp PvuII-EcoRV fragment carrying the 5S rRNA gene (see panel A) inserted in the SmaI site upstream of the GAL7 promoter. The 35S rRNA coding region contains up to the HindIII site, +6935. Thus, the Pol I enhancer is present but the region from HindIII to PvuII in NTS1 and the Pol I promoter region (from −1 to the EcoRV site at +8757 or the 381-bp region) are absent.
). (B) Structure of pRDN-hyg1 (4). This plasmid carries a single copy of rDNA repeats obtained by cutting the repeats with SmaI (hence the copy starting from −206 and ending at −207; the numbering is with respect to the Pol I transcription start site as +1). There is a mutation in the 18S rRNA gene (indicated as an asterisk) which makes the ribosome hygromycin B resistant. (C) Structure of pNOY353. This plasmid carries the 7.5-kb BamHI-XhoI fragment, which contains GAL7-35S rDNA (the 35S rRNA coding region fused to the GAL7 promoter as described by Nogi et al. [23]) inserted between the BamHI and SalI sites of the pTV3 vector (27). This plasmid also contains the 1,085-bp PvuII-EcoRV fragment carrying the 5S rRNA gene (see panel A) inserted in the SmaI site upstream of the GAL7 promoter. The 35S rRNA coding region contains up to the HindIII site, +6935. Thus, the Pol I enhancer is present but the region from HindIII to PvuII in NTS1 and the Pol I promoter region (from −1 to the EcoRV site at +8757 or the 381-bp region) are absent.
The total number of rDNA repeats per genome varies greatly depending on the organism. For a given organism, the repeat number appears to be maintained at an appropriate level, e.g., approximately 150 per haploid genome for S. cerevisiae. However, variations of the repeat numbers were observed quite often, and most organisms appear to have the ability to alter repeat numbers in response to changes in intra- as well as extracellular conditions. For example, we have previously shown that the absence of an essential subunit of Pol I triggers a gradual decrease in the number of rDNA repeats to about half the normal level and reintroduction of the missing Pol I gene induces a gradual increase of the number of repeats back to the original level (18). By analogy to this observation, one can imagine that a harmful deletion of a significant fraction of rDNA repeats by homologous recombination could be repaired by the ability of cells to expand repeat numbers, as was in fact observed for Drosophila bobbed mutations (26, 34). In addition, it was recently discovered that yeast mutants defective in the Pol I transcription factor UAF give rise to variants that are able to grow by transcribing chromosomal rDNA repeats by Pol II and that the switch to growth using the Pol II system is accompanied by a large expansion of rDNA repeats up to approximately 400 (25, 36). In this case, the repeat expansion clearly represents an adaptation process to growth without the intact Pol I system. Thus, although an extensive recombination activity in rDNA repeats may be harmful to cells, as discussed in connection with cell aging and SIR2-dependent gene silencing in yeast cells (5, 11, 29), some limited and regulated recombinational activities within rDNA repeats appear to be important for cellular adaptation and repeat number maintenance, in addition to their well-discussed role in the maintenance of sequence homogeneity among many rRNA genes. However, although extensive studies were carried out on the mechanism of recombination within rDNA repeats in Drosophila and yeast and some specific models were proposed (7, 18, 33, 39; for studies on Drosophila, see the review in reference 12), actual molecular mechanisms unique to rDNA have remained largely unknown.
An important gene required for rDNA repeat expansion and contraction discovered in the yeast system is FOB1 (18). FOB1 was originally identified as the gene required for both replication fork-blocking activity (RFB activity) at the RFB site within the rDNA repeats and HOT1 activity in a recombination test system outside the rDNA repeats (20). Using the Pol I-dependent rDNA repeat expansion-contraction assay system mentioned above, it was subsequently demonstrated that FOB1 is required for efficient rDNA expansion and contraction (18). In addition, mutation in the FOB1 gene was found to reduce the frequency of the formation of extrachromosomal rDNA circles from the rDNA repeats (5) as well as the frequency of actual recombination as assayed by the use of a marker gene integrated within rDNA repeats (K. Johzuka and T. Horiuchi, unpublished experiments). Because FOB1-dependent replication fork blocking takes place at the RFB site (3, 19) and because pausing of replication is known, at least in bacterial systems, to stimulate both DNA double-strand breakage (22) and recombination (13, 14, 28), we have previously proposed that FOB1-dependent rDNA repeat expansion and contraction takes place as a result of FOB1-dependent replication fork blocking at the RFB site, presumably involving double-strand breakage and repair of the break via gene conversion, as illustrated in Fig. 2 (see the legend for further explanation). If this proposal is correct, that is, if the FOB1-dependent replication fork block is in fact the cause of the stimulation of rDNA repeat expansion and contraction by FOB1, the RFB site located near the end of the 35S rRNA gene should be essential for this expansion and contraction process. To examine this question and to find whether any other DNA cis elements surrounding the RFB site are required for repeat expansion and contraction, we have developed a system in which these questions can be studied by mutational analysis. Obviously, the presence of redundant rDNA copies makes the mutational analysis very difficult. We have constructed a yeast strain in which the majority of rDNA repeats are deleted, leaving two copies of rDNA covering the 5S-NTS2-35S regions and a single intact NTS1 region in between and whose growth is supported by a multicopy helper plasmid which does not carry the intact NTS1. Using this strain, initial mutational analyses were carried out. We have found that the RFB site is in fact essential for FOB1-dependent rDNA repeat expansion. We have also found that in addition to the RFB region, the adjacent ∼400-bp region in NTS1 is required for the efficient repeat expansion but the Pol I transcription enhancer region is apparently not required. The ∼530-bp region, which combines the RFB region with the newly identified ∼400-bp region, is now called EXP (for expansion of rDNA repeats). The requirement of the new DNA cis element(s) independent of the RFB site can now define a new function(s) which is involved in the rDNA repeat expansion independent of the RFB activity.
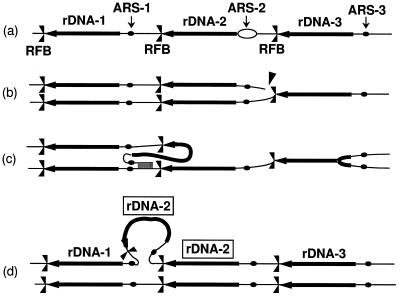
FIG. 2.
The fork block-dependent recombination model for rDNA repeat expansion and contraction. The positions of ARS and RFB are shown as solid dots and  , respectively. Individual lines represent chromatids with double-stranded DNA. In this model, DNA replication starts from one of the ARS sites (ARS-2) bidirectionally (a). In the yeast rDNA repeats, about one in five ARS sites is used as an active origin (2, 21). A rightward replication fork is arrested at the RFB site, and this arrest is supposed to stimulate a double-strand break of DNA at a nearby site (indicated by an arrowhead in row b). A strand invasion at a homologous duplex (a downstream sister chromatid near ARS-1 in this example) takes place (c), and a new replication fork is formed. The new replication fork meets with the leftward replication fork from the upstream site, resulting in formation of two sister chromatids, one of which gains an extra copy of rDNA, indicated as boxed rDNA-2 (d). If the strand invasion is at a site in a upstream repeat (e.g., near ARS-3), a loss, rather than a gain, of an rDNA repeat is expected. This model was proposed previously to explain the observed strong dependence of rDNA repeat expansion and contraction on FOB1 (18).
, respectively. Individual lines represent chromatids with double-stranded DNA. In this model, DNA replication starts from one of the ARS sites (ARS-2) bidirectionally (a). In the yeast rDNA repeats, about one in five ARS sites is used as an active origin (2, 21). A rightward replication fork is arrested at the RFB site, and this arrest is supposed to stimulate a double-strand break of DNA at a nearby site (indicated by an arrowhead in row b). A strand invasion at a homologous duplex (a downstream sister chromatid near ARS-1 in this example) takes place (c), and a new replication fork is formed. The new replication fork meets with the leftward replication fork from the upstream site, resulting in formation of two sister chromatids, one of which gains an extra copy of rDNA, indicated as boxed rDNA-2 (d). If the strand invasion is at a site in a upstream repeat (e.g., near ARS-3), a loss, rather than a gain, of an rDNA repeat is expected. This model was proposed previously to explain the observed strong dependence of rDNA repeat expansion and contraction on FOB1 (18).
MATERIALS AND METHODS
Media, strains, and plasmids.
SD is a synthetic glucose medium (16). SGal is the same as SD except that 2% glucose is replaced by 2% galactose. Both SD and SGal were supplemented appropriately with amino acids and bases to satisfy nutritional requirements and also to retain unstable plasmids (16).
Yeast strains and plasmids used in this study are listed in Table 1. Disruption of FOB1 was described previously (18). Plasmid pTAK101 was constructed by inserting the FOB1 gene amplified by PCR (20) into the BamHI site of YEplac181 (8). TAK200 was constructed from NOY408-1b as previously described (4, 24) and is described in Results.
TABLE 1.
Yeast strains and plasmids used in this study
| Designation | Genotype and comments | Reference |
|---|---|---|
| Strains | ||
| NOY408-1b | MATa ade2-1 ura3-1 his3-11 trp1-1 leu2-3,112 can1-100 pNOY102 | 23 |
| TAK200 | Same as NOY408-1b except for deletion of rDNA repeats, leaving two copies (Fig. 3), and for the presence of pRDN-hyg1 instead of pNOY102 | |
| TAK201 | Same as TAK200 except for the presence of fob1Δ::HIS3, and pNOY353 instead of pRDN-hyg1 | |
| Plasmids | ||
| pRDN-hyg1 | Multicopy plasmid carrying rdn-hyg1, rdn-ani1, leu2-d, URA3, 2 μm, and Ampr | 4 |
| pNOY353 | Multicopy plasmid carrying GAL7-35S rDNA, 5S rDNA, TRP1, 2 μm, and Ampr | 24 |
| YEplac181 | Multicopy plasmid vector carrying LEU2 and 2 μm | 8 |
| pTAK101 | YEplac181 carrying FOB1 |
NTS1 mutants A to G were constructed from TAK201 by gene replacement (16). The region (∼1.1 kb) covering the NTS1 and the 5S RNA gene was subdivided into seven segments, A to G (see Fig. 3 and below), and each segment was replaced individually with the 1,162-bp HindIII fragment containing URA3 as follows. Two DNA sequences of approximately 500 bp that flank a segment to be replaced were amplified by PCR using DNA prepared from TAK201. Each of the primers used for PCR had recognition sites at the 5′ ends, one for BamHI (distal primer) and the other for PinAI (proximal primer, i.e., the primer containing the site to be used for connection to the URA3 fragment). The two PCR products were digested with these two enzymes and cloned together into the pUC18 vector at the BamHI site. A DNA fragment consisting of the 1,162-bp HindIII fragment containing URA3 and additional PinAI sites at both ends was constructed by PCR, cleaved with PinAI, and inserted at the PinAI site between the two 500-bp flanking sequences in pUC18 in the orientation that would make URA3 and the 5S rRNA gene face the same direction. The resultant recombinant plasmid was digested with BamHI. The fragment containing URA3 and the two flanking sequences was separated from the vector portion and then introduced into TAK201 by transformation for replacement of the pertinent segment with URA3. PCR was used to confirm the positions and the size of the insert expected from the correct replacement. The positions of the segments replaced by URA3 are as follows (using the conventional rDNA repeat numbering system, starting with +1 at the site of the start of Pol I transcription and increasing in the direction of 35S rRNA transcription): G, 6750 to 6934; F, 6935 to 7063; E, 7064 to 7193; D, 7209 to 7326; C, 7327 to 7462; B, 7463 to 7712; and A, 7713 to 7895. (A gap of 15 bp is present between the E and D segments but is irrelevant to the experimental design and the conclusion.)
FIG. 3.
(A) Analysis of the size of chromosome XII by CHEF electrophoresis. Eight independent hygromycin-resistant mutants (lanes 1 to 8), as well as the control strain, NOY408-1b, (lane WT), were examined. The left panel shows chromosome patterns revealed by staining with EtBr. The right panel shows an autoradiogram obtained after hybridization with an rDNA probe (probe 3 in Fig. 5C). Size markers (lane M) are made up of Hansenula wingei chromosomes (Bio-Rad). (B) Analysis of the sizes of rDNA repeats by field inversion gel electrophoresis. DNA samples prepared from mutants 7 and 8 (those shown in lanes 7 and 8 in panel A, respectively) were digested with BamHI, subjected to the electrophoresis, and analyzed by hybridization using an rDNA probe (probe 3 in Fig. 5C). A band seen in lane 8 which corresponds to the size expected from two copies of rDNA is indicated by an arrowhead. Lane M is the 5-kb ladder provided by Bio-Rad. Because the amounts of the marker DNA molecules were much larger than the amount of fragment containing two copies of rDNA, nonspecific hybridization of the probe to the markers took place, providing positions of the markers conveniently on the same autoradiogram. (C) Structures of two rDNA repeats remaining in strain TAK201 and the NTS1-5S region subjected to the mutational analysis (expanded below). Seven segments, A to G, replaced by URA3 individually in mutants A to G are indicated. The precise positions of each segment are given in Materials and Methods.
Determination of the copy number of rDNA repeats.
In the rDNA repeat expansion experiments, the number of rDNA repeats was determined after ∼45 generations of growth. The number of generations was estimated based on the observation that a single colony with a diameter of 1 mm contained ∼2 × 105 cells and the consequent assumption that cells in colonies of this size corresponded to progeny 18 generations from the individual ancestor cells. The FOB1 gene was introduced into the control strain (TAK201) and NTS1 substitution mutants A to G by transformation using pTAK101. Colonies 1 mm in diameter were picked from Leu+ selection plates and restreaked on the same plates, and the same-sized colonies were taken to inoculate the supplemented SGal medium. Cells were then grown for nine generations before being harvested, thus making a total of ∼45 generations after the introduction of the FOB1 gene. Control transformation was done using the vector plasmid YEplac181, and Leu+ transformants (“vector transformants”) were subjected to the same processes. DNA was then isolated, digested with BglII, subjected to agarose gel electrophoresis (1% agarose), and analyzed by Southern hybridization using probes for rDNA (probe 2 for mutants A to C and probe 1 for D to G [see Fig. 5C]) and for MCM2 (a 1.4-kb fragment prepared by PCR) as described previously (30). Ratios of rDNA to MCM2 were quantified, and the rDNA copy numbers were calculated by comparing these ratios (rDNA/MCM2) with the corresponding ratio obtained for TAK201, which contained two copies of rDNA. The amounts of radioactive probes hybridized were determined by phosphorimager analysis (BAS2000; Fujifilm).
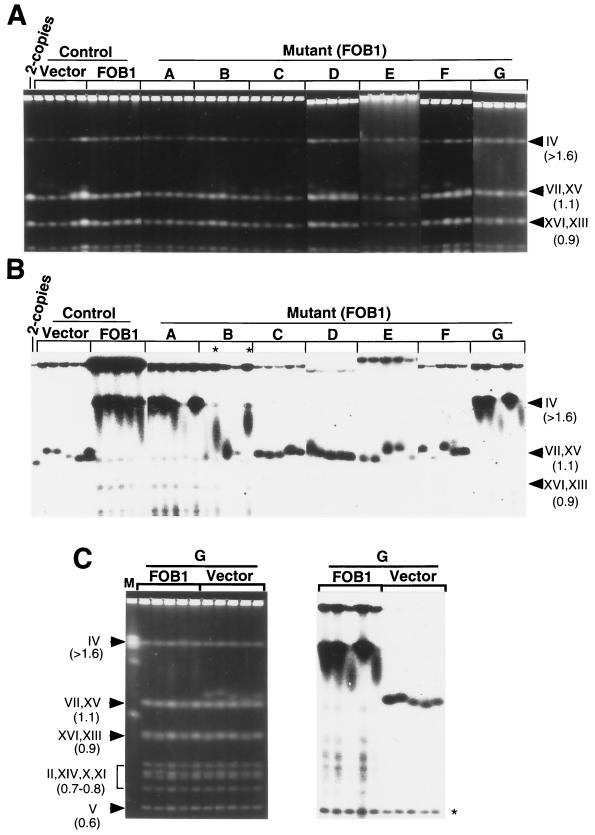
FIG. 5.
Expansion of rDNA repeats observed in FOB1 transformants of NTS1 substitution mutants. (A) The DNA samples analyzed in the experiments in Fig. 4A and B were digested with BglII and analyzed by Southern hybridization using rDNA-specific probes, probe 2 for the left panel and probe 1 for the right panel (the probes are indicated in panel C). The gels were also analyzed using a probe specific for a single-copy gene, MCM2, as a reference. (B) The numbers of rDNA repeats was calculated for each transformant, and the values for five independent transformants derived from each mutant and control strain were averaged. The results are shown as bars, and standard deviations are indicated as lines. (C) Summary of the mutational analysis indicating the region (EXP) essential for FOB1-dependent rDNA repeat expansion. The locations of segments A to G as well as probes 1, 2, and 3 used for hybridization are shown together with pertinent restriction sites in this region.
Other methods.
Samples for contour-clamped homogeneous electric field (CHEF) electrophoresis were prepared as described previously (31). Electrophoresis was carried out in a 0.8% agarose gel with 0.5× Tris-borate-EDTA (TBE) buffer, using CHEF-DRII (Bio-Rad, Richmond, Calif.) with a pulse time of 300 to 900 s and 100 cV for 68 h at 14°C. For the experiment in Fig. 4, a 1% agarose gel was used and the conditions for electrophoresis were altered to a pulse time of 60 to 120 s, 200 cV, and 40 h at 14°C. The gel was then stained with 1 μg of ethidium bromide (EtBr) per ml for 30 min at room temperature, photographed, and then subjected to Southern hybridization analysis (30). RFB activity was analyzed using two-dimensional (2D) gel electrophoresis as described previously (1). For field inversion gel electrophoresis, samples were prepared as previously described (31), digested with BamHI, and subjected to gel electrophoresis in a 1% agarose gel with 0.5× TBE buffer, using FIGE Mapper (Bio-Rad). The conditions used included a switch time ramp of 0.4 to 2.0 s (linear shape), 180 cV (forward), 120 cV (reverse), and 20 h at 14°C. The gel was then stained with 1 μg of EtBr per ml for 30 min at room temperature, photographed, and subjected to Southern hybridization analysis (30).
FIG. 4.
Analysis of the size of chromosome XII in FOB1 transformants of NTS1 substitution mutants by CHEF electrophoresis. (A and B) Five independent FOB1 transformants derived from each of mutants A to G and from the control strain (TAK201) were analyzed along with five vector transformants of the control strain after ∼45 generations. The reference TAK201, which had two rDNA repeats without expansion, was also analyzed (lane 2-copies). (A) Chromosomal patterns revealed by staining with EtBr; (B) autoradiograms obtained after hybridization with an rRNA probe (probe 3 in Fig. 5C). (C) Analysis of five FOB1 transformants and five vector transformants derived from mutant G by hybridization using a URA3 probe. The left panel shows chromosomal patterns revealed by staining with EtBr. The right panel shows an autoradiogram obtained after hybridization with the URA3 probe. The position of chromosome V carrying the native URA3 gene is indicated by an asterisk. On the right sides of the gels in panels A and B and on the left side in panel C, the positions of chromosomes and their sizes (in megabases) are indicated.
RESULTS
Construction of a strain with two rDNA repeats.
Most of the yeast rDNA repeats can be deleted using a method described by Chernoff et al. (4). Plasmid pRDN-hyg1 carries a single rDNA repeat with a recessive hygromycin resistance mutation in the 18S rRNA gene (Fig. 1B). This plasmid was first introduced into a control strain, NOY408-1b, using URA3 for selection. The resultant strain was then subjected to a hygromycin resistant selection. Because the wild-type allele is dominant to the mutant hyg1 allele, hygromycin-resistant mutants selected in this way are expected to have lost most of the chromosomal rDNA repeats by recombinational events. Because the rDNA repeats (∼150 copies of the 9.1-kb repeat or ∼1.4 Mb) represent a large fraction of the total length of chromosome XII (1.05 Mb of non-rDNA regions plus 1.4 Mb rDNA repeats, or ∼2.5 Mb), degrees of reduction in rDNA repeat numbers can be assessed by measuring the length of chromosome XII in these hygromycin-resistant mutants by CHEF electrophoresis. Eight mutants were analyzed in this way, and the result is shown in Fig. 3A. Compared to the control strain (lane WT), a large reduction in the length of chromosome XII was evident for all the mutants analyzed and the remaining rDNA repeat numbers were estimated to be less than 20 in these mutants. We selected mutants 7 and 8 (Fig. 3A, lanes 7 and 8) and determined the copy numbers of their chromosomal rDNA repeats more precisely. Field inversion gel electrophoresis was carried out after digestion of their chromosomal DNA with BamHI. As shown in Fig. 3B (lane 8), mutant 8 showed a band of approximately 57 kb. Knowing the DNA sequences of non-rDNA flanking the rDNA repeats, including the BamHI sites closest to the rDNA, we can calculate that this value matches that for the presence of two rDNA copies, as shown in Fig. 3C. Mutant 7 failed to show any significant signal (Fig. 3B, lane 7) and may have lost the rDNA repeats completely. No further analysis was done on this mutant.
We selected mutant 8 (TAK200) for subsequent studies of rDNA repeat expansion. It should be noted that the end of the intact rDNA repeats at the right border (telomere proximal) is near the end of the 5S rRNA gene and that their left end is within the RFB region according to the GenBank sequence information (9). We determined the sequences around these two boundaries on DNA isolated from strain TAK200 and confirmed that they are identical to those in the data bank. Thus, TAK200 contains two intact 35S rRNA genes, two intact NTS2 regions, and a single intact NTS1 region (Fig. 3C). We note that the portion of the RFB region remaining at the left border is not sufficient to cause replication fork blocking, as judged from the results of previous experiments (19), leaving the single intact RFB in the middle for the RFB function in this strain. To prevent FOB1-dependent repeat expansion, thus stabilizing the two-copy state, the FOB1 gene of TAK200 was disrupted by replacement with HIS3. In addition, plasmid pRDN-hyg1 was replaced by pNOY353. This plasmid contains the 35S rDNA fused to the GAL7 promoter and, in addition, the 1.1-kb PvuII-EcoRV fragment carrying the 5S rRNA gene and lacks most of NTS1 (i.e., the segment between HindIII and PvuII which includes the RFB site [see the legend to Fig. 1]). This helper plasmid was used to minimize repairs of mutations to be introduced in the chromosomal NTS1 by gene conversion, which might take place when a single intact rDNA repeat is present on a helper plasmid like pRDN-hyg1. The resulting fob1-disrupted strain carrying pNOY353 (TAK201) was used for mutational analysis of rDNA repeat expansion. As expected from the limited number (two copies) of the chromosomal rDNA repeats and the GAL7-dependent rRNA synthesis through the helper plasmid, growth of TAK201 was galactose dependent.
Mutational analysis of the NTS1 region to identify cis elements required for rDNA repeat expansion.
A systematic mutational analysis of the NTS1 region was done by dividing this region (and the 5S rRNA coding region) into seven segments (A to G) (Fig. 3C) (see Materials and Methods) and replacing each of them with the URA3 gene, creating seven NTS1 substitution mutants (mutants A to G). Segment F corresponds to the 129-bp HindIII-HpaI region which contains the RFB site. Segment G roughly corresponds to the 190-bp EcoRI-HindIII region which was originally defined as the Pol I transcription enhancer element (6). The URA3 gene was placed in the same direction as the 5S rRNA gene. Expected mutational alterations in these mutant strains were confirmed by digestion of their DNA with BamHI followed by Southern analysis, which showed no increase of rDNA repeat numbers, and by PCR analysis, which showed correct replacements of each segment with URA3 (data not shown).
The seven NTS1 mutant strains, A to G, as well as the original control strain, TAK201, were transformed with a plasmid, pTAK101, which carries the wild-type FOB1 gene, to induce a FOB1-dependent expansion of rDNA repeats (18). Transformants were selected using LEU2 on the plasmid on supplemented SGal plates which did not contain tryptophan or leucine. Five independent transformants were picked for each strain and purified by streaking on the SGal plates. Single colonies were then inoculated in liquid SGal medium with the same supplements, and the cells were grown for 18 h. Including colony formation twice on the plates and the following growth in the liquid medium, it was estimated that approximately 45 generations had occurred since the FOB1 gene was introduced into these strains (see Materials and Methods). The size of chromosome XII was then analyzed by CHEF electrophoresis. The gels were stained with EtBr (Fig. 4A) and subjected to hybridization with an rDNA-specific probe and autoradiography (Fig. 4B). In the original strain (TAK201), which had two copies of rDNA, the band of chromosome XII overlapped those of chromosomes VII and XV in the EtBr-stained gel (Fig. 4A and B, lane 2-copies). This result was expected because the calculated size of chromosome XII in the original strain is 1.05 Mb, which is similar to the size (1.09 Mb) of chromosomes VII and XV. In contrast, chromosome XII in five transformants derived from the control strain (TAK201), which grew for 45 generations after introduction of the FOB1 gene, was much larger than that of the two-copy control, and this was the case for all five independent transformants, as can be seen from the autoradiogram in Fig. 4B (lanes Control, FOB1). Each sample appears to represent a heterogeneous mixture of cells carrying chromosomes XII with different sizes, displaying smears rather than defined bands. For this reason, no defined bands corresponding to chromosome XII were observed for these samples on the EtBr-stained gel (Fig. 4A). The observed extensive expansion of rDNA repeats in these transformants requires the presence of the FOB1 gene. No such expansion was observed for the five control cultures derived from five independent Leu+ transformants, which were formed on introduction of the vector DNA without FOB1 (Fig. 4A and B, lanes control, Vector). However, some small increases in the length of chromosome XII were clearly seen relative to the original two-copy rDNA strain, and the extents of the increases varied depending on the transformants obtained independently. The bands of chromosome XII were relatively homogeneous in sizes and could be seen even in the EtBr-stained gel. The observed FOB1-independent increase in rDNA repeat numbers is discussed below.
The effects of substitution mutations (A to G) on FOB-dependent rDNA repeat expansion were examined in the same way, that is, by introducing the FOB1 gene by transformation and analyzing the size of chromosome XII after ∼45 generations of growth. Five independent FOB1 transformants were analyzed for each mutant, together with five independent vector transformants. The results for the FOB1 transformants are shown in Fig. 4A and B. The results for the vector transformants are not shown except for those derived from mutant G (Fig. 4C; see below), but all the vector transformants showed only a limited increase in the size of chromosome XII, as with the vector transformants derived from TAK201 mentioned above and those derived from mutant G. For FOB1 transformants, an efficient expansion of rDNA, that is, a large increase in the size of chromosome XII, was clearly observed for all the transformants derived from mutant A and G (Fig. 4B, lanes A and G). Some of them (one transformant of A and two transformants of G), however, showed lower degrees of expansion compared to others with the same mutation (A or G) or those without mutation (mentioned above). For mutant B, the extent of expansion was reduced significantly. However, two of the five FOB1 transformants (marked with asterisks in Fig. 4B) showed a clear expansion and two others showed a smear, suggesting that at least some fractions of heterogeneous cell populations had started repeat expansion (Fig. 4B, lanes B).
For the other mutants, C, D, E, and F, no significant FOB1-dependent repeat expansion was observed. Only a limited increase in size was observed (Fig. 4A and B, lanes C, D, E, and F), and the patterns of chromosome XII bands shown by five FOB1 transformants for each of these mutants were similar to those seen for vector transformants of the control strain, TAK201, or vector transformants of mutant G; that is, the bands were relatively homogeneous and could be recognized above the 1.1-Mb bands of chromosome VII and XV in the EtBr-stained gel (Fig. 4A). The absence of FOB1-dependent expansion was expected for mutant F because the RFB region has been completely replaced by the URA3 gene in this mutant, and the model shown in Fig. 2 predicted this result. The results obtained for mutants C, D, and E demonstrate that there are DNA elements in these regions which are required for efficient FOB1-dependent expansion of rDNA repeats.
The URA3 gene fragment which has replaced segments A to G individually in the NTS1 mutants A to G was found to undergo repeat expansion processes together with adjacent rDNA. In the experiment in Fig. 4, we rehybridized the same filter (A to G) with a URA3-specific probe after stripping the rDNA probe and obtained patterns of chromosome XII sizes similar to those shown in Fig. 4B (data not shown except for mutant G as an example in Fig. 4C). Hybridization with the URA3 probe revealed bands of chromosome V, which carries a single copy of the native URA3 gene (asterisk in Fig. 4C). Comparison of the intensities of chromosome XII bands with those revealed by the single-copy URA3 show strong coamplification of URA3 in FOB1 transformants of mutant G and limited coamplification in vector transformants of mutant G.
In the CHEF electrophoresis experiments described above, it is difficult to obtain accurate estimates of the degree of rDNA repeat expansion. First, the conditions of electrophoresis were chosen to improve the resolution of chromosomal bands with different sizes at a region near 1.1 Mb, which made resolution of bands of 1.5 Mb or higher difficult (compare lane M in Fig. 4C with lane M in Fig. 3A). Second, significant fractions of chromosome XII failed to enter the gel, presumably reflecting the difficulty of obtaining complete release of this large chromosome from cellular components and/or debris resistant to enzyme digestion during sample preparation. Therefore, we determined the extent of increase of rDNA repeat numbers by Southern hybridization after digestion of DNA with BglII. Specific probes used to detect rDNA were probe 2 for mutant strains A to C and probe 1 for mutant strains D to G (indicated in Fig. 5C). A single-copy gene, MCM2, was also analyzed as a reference by using a suitable hybridization probe. The results obtained are shown in Fig. 5A for a single FOB1 transformant taken from each group of mutants as well as single FOB1 and single vector transformants of the control strain that were subjected to the rDNA repeat expansion process. First, it should be noted that the BglII fragment detected for the transformants derived from the control strain had a size of 4.6 kb (Fig. 5A, arrowhead marked rDNA), corresponding to the BglII-A fragment shown in Fig. 1A. The bands detected for the mutants were larger [Fig. 5A, arrowhead marked rDNA (URA3)], and no heterogeneity was observed for each mutant band. The larger sizes reflect the differences between the sizes of each region deleted (120 to 250 bp) and the size of the URA3 fragment inserted (1.1 kb). The results demonstrate that each repeating unit in rDNA after extensive expansion (mutants A, B, and G [sample B shown in Fig. 5A was the one with a large expansion]) or after limited expansion (mutants C, D, E, and F) contained URA3; that is, URA3 was coamplified with the remaining rDNA in the expansion process.
The number of rDNA repeats in the samples was determined by first measuring the intensities of bands, calculating the ratios of the rDNA to MCM2 signals for each sample, and then comparing these ratios to the ratio obtained for the reference two-copy rDNA strain. This Southern analysis was repeated with the remaining four FOB1 transformants of the mutants (A to G) and the control strain, as well as four vector transformants of the control strain. Averages of the values for five independent transformants obtained in this way were then calculated, and the results are shown in Fig. 5B. It is evident that replacing regions C, D, E, and F with URA3 abolished the efficient FOB1-dependent repeat expansion. Repeat numbers were less than 10 in all these cases and were not larger than the small increases observed for the vector transformants of the control strain. In contrast, replacement of the A, B, and G segments with URA3 still allowed FOB1-dependent expansion, although the extent of expansion appeared to be less than that observed for the control strain. In summary, the experiments described in this section demonstrate that the DNA region covering segments C to F is required for FOB1-dependent rDNA repeat expansion. We call this region EXP (for “expansion of rDNA repeats”) (Fig. 5C).
Effects of NTS1 substitution mutations on replication fork-blocking activity.
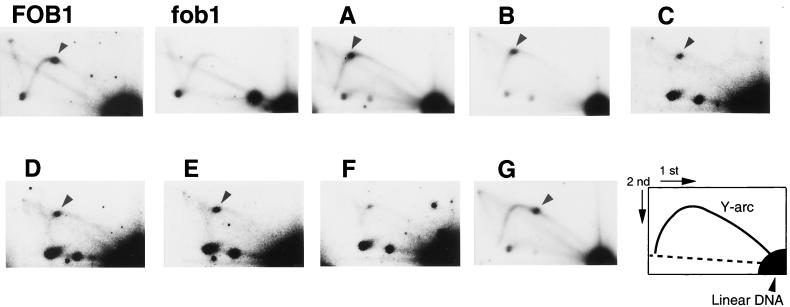
Although deletion analysis was previously carried out to define the region (the RFB region) required for RFB activity, the analysis was done by using artificial plasmid systems (3, 19) rather than in the context of the native rDNA locus on chromosome XII. To examine the relationship between the DNA elements required for rDNA repeat expansion and those required for RFB activity, we used the 2D electrophoresis method (1) and analyzed cultures of the five FOB1 transformants of the mutants and the FOB1 and vector transformants of the control strain (those used in the experiment in Fig. 5A) for accumulation of intermediates of replication arrested at the RFB site. DNA was isolated from cells growing exponentially in galactose medium, digested with BglII, and subjected to 2D gel electrophoresis followed by hybridization using a rDNA probe (probe 3 in Fig. 5C). The results are shown in Fig. 6. The control FOB1 transformant culture showed a spot (indicated by an arrowhead) corresponding to the replication fork intermediate arrested at the RFB site (panel FOB1). The vector transformant culture did not show such a spot (panel fob1), as expected from the previous work (20). For mutants A through E, a spot was observed at a position which is shifted slightly to the left from the position of the spot seen for the control FOB1 cells (see the position of spots indicated by arrowheads in panels A through E relative to the position in panel FOB1). This small shift to the left is consistent with the increase in the size of the BglII A fragment caused by the URA3 substitution (as mentioned above in connection with the results in Fig. 5A) combined with the expectation that the increase is in the replicated “branch” region of the Y-shaped intermediate formed at the site.
FIG. 6.
Effects of NTS1 mutations on RFB activity analyzed by 2D gel electrophoresis. DNA was prepared from FOB1 transformants of NTS1 substitution mutants (A to G) and FOB1 and vector transformants (panels FOB1 and fob1, respectively) derived from the control strain, TAK201. DNA was then digested with BglII and subjected to 2D agarose gel electrophoresis followed by Southern hybridization using a rDNA probe (probe 3 in Fig. 5C). Spots indicated by arrowheads show the accumulation of Y-shaped molecules at RFB sites. A schematic diagram of the positions of various Y-shaped replication intermediates is shown as a Y-arc in the bottom right panel.
For mutant F, a large reduction in the spot intensity was observed, as expected from deletion of the previously defined RFB site in the F segment. However, we noted the presence of a weak spot at approximately the same position as those seen for mutants A to E, that is, slightly left of that seen for the control culture (compare panel F with other panels). Therefore, it appears that a weak RFB activity remains in mutant F and that the (weak) replication fork arrest takes place soon after replication of the URA3 fragment inserted to replace the F segment, i.e., presumably in the G segment. Although further studies are required to establish this tentative conclusion, it is possible that we failed to detect this weak activity previously because the previous work was done with an artificial plasmid system, where the RFB activity was weaker than that observed in the chromosomal rDNA repeats (19).
When mutant G was analyzed, a clear spot was observed and its position was shifted to the right along the Y arc from the position observed for the control FOB1 cells (panel G). This shift is consistent with replication fork arrest occurring at the previously defined RFB site in the F segment, that is, an increase in the size of the unreplicated “stem” of the Y-shaped replication intermediate in mutant G relative to the intermediate in the control FOB1 cells.
The main conclusion obtained from the 2D gel analysis described above is that replacement of the C, D, or E segment with URA3 does not affect the RFB activity even though it abolishes the FOB1-dependent expansion of rDNA repeats, as described in the previous section. Some specific DNA element(s) exists in the region covering segments C, D, and E (and perhaps extending to F) which is involved in a function(s) separate from replication fork blocking, enabling the expansion of rDNA repeats.
DISCUSSION
Identification of a new DNA cis element(s) required for expansion of rDNA repeats.
We have constructed a yeast strain which carries only a single intact NTS1 region surrounded by two 5S-NTS2-35S regions on chromosome XII. The strain also carries a deletion in the FOB1 gene, and an efficient FOB1-dependent repeat expansion can be induced by introduction of the missing FOB1 gene. Using this system, we carried out a mutational analysis of the entire NTS1 region. We first confirmed the prediction based on the previously proposed model (Fig. 2) that the 129-bp HindIII-HpaI region (segment F) containing the RFB site is required for rDNA repeat expansion. Although this confirmation does not necessarily prove the model (see the discussion below), the results are at least consistent with the proposal that replication fork blocking is required for the efficient expansion and contraction of rDNA repeats (18).
Somewhat unexpectedly, we have discovered that the adjacent ∼400-bp region distal to the 35S rRNA gene (segments C, D, and E) is required for the FOB1-dependent repeat expansion even though it is not required for the RFB activity. Thus, this new cis element(s) defines a new function required for expansion of rDNA repeats. Since both expansion and contraction are largely FOB1 dependent (18), we think it likely that this new cis element, called EXP, is involved in both expansion and contraction, although the present experiments demonstrate only the requirement for expansion and not that for contraction. It should be noted that we define the EXP element (or region) as the DNA region required for repeat expansion, and this includes segment F, which contains the RFB region; regardless of whether replication fork blocking is really essential for repeat expansion, the RFB region is required for expansion and hence is included in the EXP region. If replication fork blocking is really essential for efficient FOB1-dependent repeat expansion, the EXP region would be functionally divided into two subregions or DNA elements, one required for replication fork blocking and the other required for another function, a function presumably involved in a step subsequent to replication fork blocking, and these two elements might or might not overlap in segment F.
Regarding the function of the EXP element that is independent of the RFB function, we have little information. As discussed previously (18, 25), there are two different kinds of factors which influence rDNA repeat expansion and contraction. One comprises protein factors which are involved in recombination processes, such as Fob1 protein and Sir2 protein (in addition to proteins used in recombination in general, such as RAD52 [T. Kobayashi, unpublished data]), and the other includes protein factors, such as Pol I and Pol I-specific transcription factors, which presumably do not participate in recombination processes but do participate in the maintenance of rDNA repeat numbers within a certain range, presumably by forming some specific nucleolar structures that include rDNA repeats. For example, in mutants defective in Pol I and growing by Pol II-dependent transcription of an artificial fusion gene, GAL7-35S rDNA, on a plasmid, rDNA repeat numbers are reduced to about half of the normal level (18). Another example is that of mutants defective in the transcription factor UAF and growing by transcribing chromosomal rDNA by Pol II, where average rDNA repeat numbers are increased to approximately 400 (25, 36). In both instances, average repeat numbers are substantially altered relative to the wild-type level but the cells retain the ability to expand and contract rDNA repeats and the populations show a significant heterogeneity of cells with different sizes of rDNA repeats. In mutants C, D, and E, which fail to expand rDNA repeats, a limited degree of expansion was observed but the repeat numbers appeared to be relatively homogeneous. Repeat numbers obtained were also different among five different transformants for a given mutation. Therefore, it appears that the EXP element defined here is involved in a FOB1-dependent recombination process(es) rather than influencing repeat numbers through some nucleolar structures. Since the replacements of each of segments C, D, and E (but not A, B, and G [see below]) with the URA3 sequence all abolish FOB1-dependent repeat expansion, we suspect that the EXP element may represents a site(s) for the binding of some specific protein factor(s) that is involved in a recombination process(es) unique to rDNA repeats, possibly the one(s) counteracting the function of other rDNA-specific chromatin proteins, such as Sir2 protein, that decrease the frequency of recombination within rDNA repeats.
Replacement of each of segments A, B, and G with URA3 allowed FOB1-dependent rDNA repeat expansion, but the extents of repeat number increase after 45 generations were significantly lower than for the control. Since the degrees of expansion were not uniform in five FOB1 transformants analyzed for each of these mutants, these mutations appear to reduce the rate of expansion rather than limiting the extent of expansion. It is possible that some DNA sequence elements contained in these segments play some specific (stimulatory) role in repeat expansion but are not essential. Alternatively, the presence of a Pol II gene, URA3, in each repeat that would attract nonnucleolar proteins including the Pol II transcription machinery may cause a nonspecific inhibition of the expansion process.
In a search of genomic DNA elements that would promote the amplification of a plasmid carrying the thymidine kinase gene in cultured mouse cells, Wegner et al. (38) isolated two DNA fragments (called muNTS1 and muNTS2) which were identified as two segments within the nontranscribed spacer region in rDNA repeats. The plasmids carrying these sequences were found to be integrated into some chromosome locations in the form of long tandem repeats. Because these two nontranscribed spacer fragments were highly AT rich, a likely possibility considered was that they might function as origins of replication. The fragments in the EXP region studied here contain some AT-rich sequences, but other fragments that were not required for expansion (A, B, and G) also contain equally AT-rich segments. In addition, the function of the EXP element in the yeast system is clearly not that of a replication origin. Whether there is any functional relationship between the mouse nontranscribed spacer elements and the yeast EXP element is presently unclear.
FOB1-independent limited increases of rDNA repeats.
The system we used to study cis elements for rDNA repeat expansion contains a single NTS1 surrounded by two copies of 5S-NTS2-35S repeats. The FOB1-dependent expansion model in Fig. 2 requires at least three tandem repeats for repeat expansion. Therefore, expansion in the present system must have initially used a different mechanism, such as an unequal crossing over between sister chromatids. A limited degree of FOB1-independent expansion was in fact observed in vector transformants of the control strain (and of NTS1 mutants). The RFB-independent limited expansion observed in FOB1 transformants of mutant F may also represent such a FOB1-independent repeat expansion. Although we have not studied mechanisms involved in such FOB1- and RFB-independent copy number increase, this process is presumably very inefficient because of the small numbers of repeats available as sites of recombination and the general reduction of recombination caused by protein components unique to rDNA chromatin, such as the Sir2 and Net1 proteins (10, 32). We expect that once copy numbers reach certain sizes, FOB1-dependent repeat number alterations will start to become dominant and the rate of expansion per genome may presumably become higher with increased copy numbers during the 45 generations used for the analysis, because an increase in repeat number will increase the total frequency of FOB1-dependent recombinational events. In addition, the direction of copy number changes in populations will be mostly toward expansion rather than contraction due to a selection for faster growth, at least until certain repeat numbers are reached (see below). Such considerations may explain the large differences among five independent FOB1 transformants that were derived from the same strain (e.g., mutant B) and had undergone the same transformation and subsequent subcultures (Fig. 4B, mutant B).
In connection with the selective advantage of cells with increased rDNA repeat numbers, we note that rDNA repeat numbers (which are still less than 10) which are attained by the limited increase through the FOB1-independent mechanism are not sufficient for cell growth. We found that the vector transformants of the control strain were unable to form colonies on glucose plates after 45 generations of subculture while FOB1 transformants of control strains were able to form colonies on glucose (and to lose the helper plasmid, pNOY353). On the other hand, as emphasized previously (18), control cells with ∼40 rDNA repeats had growth rates identical to those with normal (i.e., ∼150) rDNA repeat numbers. Thus, expansion beyond ∼40 copies appears to be achieved not because of selective advantage but presumably because of the stability of a nucleolar structure(s) carrying rDNA repeat numbers close to ∼150.
In passing, we note that transformants of mutant G, which received FOB1, were able to expand rDNA repeats, although apparently not to the same extent as the control FOB1 transformants. The resultant strain lacks segment G, which was originally defined as the Pol I enhancer (6), in the expanded rDNA repeats except for the single copy at the leftmost end. However, this strain was able to form colonies on glucose plates and to lose the helper plasmid. Such a strain with rDNA repeats carrying mutation G and without the helper plasmid showed only a small decrease in growth rate in glucose medium compared to the control strain with the intact enhancer in all the rDNA repeats. The role of the enhancer element in Pol I transcription is a separate subject under current study.
Relationship between rDNA repeat expansion and recombination by HOT1.
The HOT1 element stimulates recombination between two nearby repeat sequences at a chromosome site outside the rDNA locus. HOT1 consists of two elements, the I element, which corresponds to the Pol I promoter, and the E element, which comprises segments F and G studied here. It has been assumed that HOT1 activity is responsible for recombinational events within rDNA repeats. The discovery that FOB1 is required for both HOT1 activity (20) and rDNA repeat expansion and contraction (18) has appeared to support this assumption. However, HOT1 activity requires active transcription by Pol I (15, 35) whereas recombinational events within rDNA repeats take place in the absence of their transcription (18). In addition, the present work has demonstrated clear differences in the cis elements required for stimulation of recombination between the two systems. First, segments C, D, and E are required for rDNA repeat expansion (see above) but not for HOT1 activity (35). Second, deletion (or substitution) of segment G abolishes HOT1 activity nearly completely (35) but reduces the extent and presumably the rate of rDNA repeat expansion only weakly (see above). (It should be noted that there is one copy of the intact G segment at the left border in mutant G used in the expansion experiments described in this paper. Thus, although we think it rather unlikely, we cannot eliminate the possibility that this single copy might play a role in recombination events responsible for repeat expansion.) The main features shared by the two systems are the requirement of segment F, which contains the RFB site, and the requirement of the intact FOB1 gene as mentioned above. Thus, the previous assumption may be incorrect and elucidation of the mechanisms of rDNA sequence homogenization as well as rDNA repeat expansion and contraction may have to depend on the use of systems designed within the native rDNA repeat locus. In addition to the present FOB1-induced repeat expansion system, we have previously described experimental systems in which the effects of various factors on the expansion and contraction of rDNA repeats can be studied (18, 25). These systems should be useful in studies not only of the mechanism but also of the physiological significance of rDNA repeat expansion and contraction.
After completion of the present work, a paper by Ward et al. (37) appeared, which has demonstrated that HOT1 activity can occur in the absence of replication fork blocking, even though both HOT1 and RFB activities requires FOB1. These workers also carried out mutational analysis within the F and G segments and found that some DNA elements are shared but others are required for one activity but not for the other. Thus, their conclusion that the FOB1 function is involved in two clearly different activities, HOT1 and RFB activities, is related to our conclusion that it is also required for two clearly separable activities, HOT1 and rDNA repeat expansion. Elucidation of the function(s) of the FOB1 gene product appears to be a key to solving the intriguing problem of relationships among these three activities. In addition, consideration of these new observations made by Ward et al. (37) and by the present study raises the question whether our previous proposal is really correct, that is, whether replication fork blocking is really the first step in rDNA expansion and contraction. Although available experimental results support this proposal, they have not proven it. Detailed mutational analysis of DNA sequence elements within the F segment may be helpful to settle this question. Regardless of the answer to this question, however, the discovery of the new DNA elements that are uniquely involved in rDNA repeat expansion (and presumably also in contraction) indicates the presence of an unexplored aspect(s) of recombinational mechanisms used in rDNA repeat structures that constitute the structurally and functionally essential component of the nucleolus.
ACKNOWLEDGMENTS
We thank S. Arfin for critical reading of the manuscript.
This work was supported in part by grants from the Ministry of Education, Science and Culture, Japan (to T.H. and T.K.), a grant from the Ministry of Health and Welfare, Japan (to T.K.), and a grant from the National Institutes of Health (to M.N.).
ADDENDUM IN PROOF
We replaced the G segment, still located at the left border of rDNA repeats in mutant G. In this mutant, the FOB1-dependent expansion of rDNA took place as well. Therefore, the G segment was not required for the expansion.
REFERENCES
- 1.Brewer B J, Fangman W L. The localization of replication origins on ARS plasmids in S. cerevisiae. Cell. 1987;51:463–471. doi: 10.1016/0092-8674(87)90642-8. [DOI] [PubMed] [Google Scholar]
- 2.Brewer B J, Fangman W L. A replication fork barrier at the 3′ end of yeast ribosomal RNA genes. Cell. 1988;55:637–643. doi: 10.1016/0092-8674(88)90222-x. [DOI] [PubMed] [Google Scholar]
- 3.Brewer B J, Lockshon D, Fangman W L. The arrest of replication forks in the rDNA of yeast occurs independently of transcription. Cell. 1992;71:267–276. doi: 10.1016/0092-8674(92)90355-g. [DOI] [PubMed] [Google Scholar]
- 4.Chernoff Y O, Vincent A, Liebman S W. Mutations in eukaryotic 18S ribosomal RNA affect translational fidelity and resistance to aminoglycoside antibiotics. EMBO J. 1994;13:906–913. doi: 10.1002/j.1460-2075.1994.tb06334.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Defossez P A, Prusty R, Kaeberlein M, Lin S J, Ferrigno P, Silver P A, Keil R L, Guarente L. Elimination of replication block protein Fob1 extends the life span of yeast mother cells. Mol Cell. 1999;3:447–455. doi: 10.1016/s1097-2765(00)80472-4. [DOI] [PubMed] [Google Scholar]
- 6.Elion E A, Warner J R. The major promoter element of rRNA transcription in yeast lies 2kb upstream. Cell. 1984;39:663–673. doi: 10.1016/0092-8674(84)90473-2. [DOI] [PubMed] [Google Scholar]
- 7.Gangloff S, Zou H, Rothstein R. Gene conversion plays the major role in controlling the stability of large tandem repeats in yeast. EMBO J. 1996;15:1715–1725. [PMC free article] [PubMed] [Google Scholar]
- 8.Gietz R D, Sugino A. New yeast-Escherichia coli shuttle vectors constructed with in vitro mutagenized yeast genes lacking six-base pair restriction sites. Gene. 1988;74:527–534. doi: 10.1016/0378-1119(88)90185-0. [DOI] [PubMed] [Google Scholar]
- 9.Goffeau A, Barrell B G, Bussey H, Davis R W, Dujon B, Feldmann H, Gailbert F, Hoheisel J D, Jacq C, Johnston M, Louis E J, Mewes H W, Murakami Y, Philippsen P, Tettelin H, Oliver S G. Life with 6000 genes. Science. 1996;274:546–551. doi: 10.1126/science.274.5287.546. [DOI] [PubMed] [Google Scholar]
- 10.Gottlieb S, Esposito R E. A new role for a yeast transcriptional silencer gene, SIR2, in regulation of recombination in ribosomal DNA. Cell. 1989;56:771–776. doi: 10.1016/0092-8674(89)90681-8. [DOI] [PubMed] [Google Scholar]
- 11.Guarente L. Diverse and dynamic functions of the Sir silencing complex. Nature. 1999;23:281–285. doi: 10.1038/15458. [DOI] [PubMed] [Google Scholar]
- 12.Hawley R S, Marcus C H. Recombinational controls of rDNA redundancy in Drosophila. Annu Rev Genet. 1989;23:87–120. doi: 10.1146/annurev.ge.23.120189.000511. [DOI] [PubMed] [Google Scholar]
- 13.Horiuchi T, Fujimura Y. Recombinational rescue of the stalled DNA replication fork: a model based on analysis of an Escherichia coli strain with a chromosome region difficult to replicate. J Bacteriol. 1995;177:783–791. doi: 10.1128/jb.177.3.783-791.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Horiuchi T, Fujimura Y, Nishitani H, Kobayashi T, Hidaka M. The DNA replication fork blocked at the Ter site may be an entrance for the RecBCD enzyme into duplex DNA. J Bacteriol. 1994;176:4656–4663. doi: 10.1128/jb.176.15.4656-4663.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huang G S, Keil R L. Requirements for activity of the yeast mitotic recombination hotspot HOT1: RNA polymerase I and multiple cis-acting sequences. Genetics. 1995;141:845–855. doi: 10.1093/genetics/141.3.845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kaiser C, Michaelis S, Mitchell A. Methods in yeast genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1994. [Google Scholar]
- 17.Keil R L, Roeder G S. cis-acting recombination-stimulating activity in a fragment of the ribosomal DNA of S. cerevisiae. Cell. 1984;39:377–386. doi: 10.1016/0092-8674(84)90016-3. [DOI] [PubMed] [Google Scholar]
- 18.Kobayashi T, Heck D J, Nomura M, Horiuchi T. Expansion and contraction of ribosomal DNA repeats in Saccharomyces cerevisiae: requirement of replication fork blocking (Fob1) protein and the role of RNA polymerase I. Genes Dev. 1998;12:3821–3830. doi: 10.1101/gad.12.24.3821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kobayashi T, Hidaka M, Nishizawa M, Horiuchi T. Identification of a site required for DNA replication fork blocking activity in the rRNA gene cluster in Saccharomyces cerevisiae. Mol Gen Genet. 1992;233:355–362. doi: 10.1007/BF00265431. [DOI] [PubMed] [Google Scholar]
- 20.Kobayashi T, Horiuchi T. A yeast gene product, Fob1 protein, required for both replication fork blocking and recombinational hotspot activities. Genes Cells. 1996;1:465–474. doi: 10.1046/j.1365-2443.1996.d01-256.x. [DOI] [PubMed] [Google Scholar]
- 21.Linskens M H K, Huberman J A. Organization of replication of ribosomal DNA in Saccharomyces cerevisiae. Mol Cell Biol. 1988;8:4927–4935. doi: 10.1128/mcb.8.11.4927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Michel B, Ehrlich S D, Uzest M. DNA double-strand breaks caused by replication arrest. EMBO J. 1997;16:430–438. doi: 10.1093/emboj/16.2.430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nogi Y, Yano R, Nomura M. Synthesis of large rRNAs by RNA polymerase II in mutants of Saccharomyces cerevisiae defective in RNA polymerase I. Proc Natl Acad Sci USA. 1991;88:3962–3966. doi: 10.1073/pnas.88.9.3962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Oakes M L, Aris J P, Brockenbrough J S, Wai H, Vu L, Nomura M. Mutational analysis of the structure and localization of the nucleolus in the yeast Saccharomyces cerevisiae. J Cell Biol. 1998;143:23–34. doi: 10.1083/jcb.143.1.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Oakes M L, Siddiqi I, Vu L, Aris J P, Nomura M. Transcription factor UAF, expansion and contraction of ribosomal DNA (rDNA) repeats, and RNA polymerase switch in transcription of yeast rDNA. Mol Cell Biol. 1999;19:8559–8569. doi: 10.1128/mcb.19.12.8559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ritossa F M. Unstable redundancy of genes of ribosomal RNA. Proc Natl Acad Sci USA. 1968;60:509–516. doi: 10.1073/pnas.60.2.509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rose M D, Broach J R. Cloning genes by complementation in yeast. Methods Enzymol. 1991;194:195–230. doi: 10.1016/0076-6879(91)94017-7. [DOI] [PubMed] [Google Scholar]
- 28.Rothstein R, Michel B, Gangloff S. Replication fork pausing and recombination or “gimme a break.”. Genes Dev. 2000;14:1–10. [PubMed] [Google Scholar]
- 29.Rothstein R, Gangloff S. The shuffling of a mortal coil. Nat Genet. 1999;22:4–6. doi: 10.1038/8705. [DOI] [PubMed] [Google Scholar]
- 30.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 31.Smith C L, Klco S R, Cantor C R. Pulse-field gel electrophoresis and the technology of large DNA molecules. In: Dacis K E, editor. Genome analysis. Oxford, United Kingdom: IRL Press; 1988. pp. 41–112. [Google Scholar]
- 32.Straight A F, Shou W, Dowd G J, Turck C W, Deshaies R J, Johnson A D, Moazed D. Net1, a Sir2-associated nucleolar protein required for rDNA silencing and nucleolar integrity. Cell. 1999;97:245–256. doi: 10.1016/s0092-8674(00)80734-5. [DOI] [PubMed] [Google Scholar]
- 33.Szostak J W, Wu R. Unequal crossing over in the ribosomal DNA of Saccharomyces cerevisiae. Nature. 1980;284:426–430. doi: 10.1038/284426a0. [DOI] [PubMed] [Google Scholar]
- 34.Tartof K D. Unequal mitotic sister chromatid exchange as the mechanism of ribosomal RNA gene magnification. Proc Natl Acad Sci USA. 1974;71:1272–1276. doi: 10.1073/pnas.71.4.1272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Voelkel-Meiman K, Keil R L, Roeder G S. Recombination-stimulating sequences in yeast ribosomal DNA correspond to sequences regulating transcription by RNA polymerase I. Cell. 1987;48:1071–1079. doi: 10.1016/0092-8674(87)90714-8. [DOI] [PubMed] [Google Scholar]
- 36.Vu L, Siddiqui I, Lee B-S, Josaitis C A, Nomura M. RNA polymerase switch in transcription of yeast rDNA: role of transcription factor UAF (upstream activation factor) in silencing rDNA transcription by RNA polymerase II. Proc Natl Acad Sci USA. 1999;96:4390–4395. doi: 10.1073/pnas.96.8.4390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ward T R, Hoang M L, Prusty R, Lau C K, Keil R L, Fangman W L, Brewer B J. Ribosomal DNA replication fork barrier and HOT1 recombination hot spot: shared sequences but independent activities. Mol Cell Biol. 2000;20:4948–4957. doi: 10.1128/mcb.20.13.4948-4957.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wegner M, Zastrow G, Klavinius A, Schwender S, Muller F, Luksza H, Hoppe J, Wienberg J, Grummt F. Cis-acting sequences from mouse rDNA promote plasmid DNA amplification and persistence in mouse cells: implication of HMG-I in their function. Nucleic Acids Res. 1989;17:9909–9932. doi: 10.1093/nar/17.23.9909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zou H, Rothstein R. Holliday junctions accumulate in replication mutants via a RecA homolog-independent mechanism. Cell. 1997;90:87–96. doi: 10.1016/s0092-8674(00)80316-5. [DOI] [PubMed] [Google Scholar]