Abstract
In adult patients, the treatment outcome of acute lymphoblastic leukemia (ALL) remains suboptimal. Here, we used an ex vivo drug testing platform and comprehensive molecular profiling to discover new drug candidates for B-ALL. We analyzed sensitivity of 18 primary B-ALL adult patient samples to 64 drugs in a physiological concentration range. Whole-transcriptome sequencing and publicly available expression data were used to examine gene expression biomarkers for observed drug responses. Apoptotic modulators targeting BCL2 and MDM2 were highly effective. Philadelphia chromosome–negative (Ph–) samples were sensitive to both BCL2/BCL-W/BCL-XL-targeting agent navitoclax and BCL2-selective venetoclax, whereas Ph-positive (Ph+) samples were more sensitive to navitoclax. Expression of BCL2 was downregulated and BCL-W and BCL-XL upregulated in Ph+ ALL compared with Ph– samples, providing elucidation for the observed difference in drug responses. A majority of the samples were sensitive to MDM2 inhibitor idasanutlin. The regulatory protein MDM2 suppresses the function of tumor suppressor p53, leading to impaired apoptosis. In B-ALL, the expression of MDM2 was increased compared with other hematological malignancies. In B-ALL cell lines, a combination of BCL2 and MDM2 inhibitor was synergistic. In summary, antiapoptotic proteins including BCL2 and MDM2 comprise promising targets for future drug studies in B-ALL.
INTRODUCTION
The treatment of acute lymphoblastic leukemia (ALL) has evolved rapidly during the recent decade.1 The expanding use of modern sequencing techniques has elaborated the genetic background of ALL and helped to define new subtypes, such as Philadelphia chromosome–like subtype of B-cell ALL (Ph-like ALL).2 Novel immunotherapeutic approaches, such as bispecific T-cell engaging antibodies, antibody-drug conjugates, and chimeric antigen receptor T-cell therapy, have achieved encouraging results in relapsed or refractory ALL.3–5 In addition, other novel targeted therapies, such as the third generation tyrosine kinase inhibitor (TKI) ponatinib and allosteric inhibitor asciminib (ABL001) are being introduced to Ph-like ALL and Philadelphia chromosome–positive (Ph+) ALL.6,7 Despite this progress, a significant fraction of adult patients still succumbs to leukemia or treatment-related events. Especially the treatment of elderly ALL patients remains challenging, as intensive chemotherapy regimens or allogeneic hematopoietic stem cell transplantation (alloHSCT) are frequently not suitable for nonfit patients.8 There is an urgent need for less toxic, yet effective treatment alternatives.
Our aim was to understand the potential value of new or repurposed drugs in the treatment of adult B-cell ALL using a well-established ex vivo drug sensitivity and resistance testing (DSRT) platform that included 64 selected drugs in 5 different concentrations. We also tested the most interesting drug combinations in selected B-ALL cell lines. In addition, whole-transcriptome sequencing (RNAseq) and publicly available expression data was used to dissect the molecular background underlying drug responses.
MATERIALS AND METHODS
Patients
We obtained diagnostic-phase, pretreatment bone marrow (BM) samples from the Finnish Hematology Registry and Clinical Biobank (FHRB, fhrb.fi) with appropriate ethics approval. All available diagnostic-phase samples that represented adult B-ALL were requested from the FHRB. We used viably frozen BM mononuclear cells (MNC) for DSRT analysis and BM MNC pellets for RNAseq. More details are found in Suppl. Methods. Patient characteristics are listed in Suppl. Table S1, as well as results from Archer FusionPlex fusion gene screening. The study was conducted in accordance with the Declaration of Helsinki and the Helsinki University Hospital Ethical Committee.
Drug sensitivity and resistance testing
Initially we received 31 samples from the FHRB, of which 13 samples (Ph+ ALL, n = 3; Philadelphia chromosome negative [Ph–] ALL, n = 10) were discarded due to low cell viability. Altogether, 18 adult B-ALL samples were subjected to DSRT analysis (Ph+ ALL, n = 10; Ph– ALL, n = 8). A custom drug plate with 64 different drugs in 5 different concentrations covering a 10 000-fold concentration range was designed for DSRT. A detailed design of the custom drug plate can be found in Suppl. Table S2. An elaborate DSRT protocol has been published earlier,9 and the experiment was performed accordingly and as indicated in the Suppl. Methods. Cell viability readouts were used to calculate drug sensitivity score (DSS).10 DSS measures the area under the dose response curve and takes into account both drug efficacy and potency. Drugs with DSS scores >10 were considered effective and DSS > 20 as highly effective.
Target addiction scoring
Since drug responses are complex events and can result from interplay of multiple factors, DSRT data were also analyzed using a targeted addiction score (TAS) approach that combines drug sensitivity profiles with drug-target interactions and covers both known on-target and off-target effects of the tested drugs.11 Details of TAS analysis are described in Suppl. Methods.
Whole-transcriptome sequencing and data analysis
As 2 of the DSRT patients did not have BM MNC pellets for RNA extraction, 16 samples (Ph+ ALL, n = 9; Ph– ALL, n = 7) were analyzed using RNAseq. Construction of RNAseq libraries and processing of RNAseq data were performed as previously described,12 and as indicated in Suppl. Methods.
Archer FusionPlex Pan-Heme Kit
We analyzed 7 of the patient samples with Archer FusionPlex Pan-Heme Kit (ArcherDX, Boulder, CO) to characterize fusion genes in them. Six of these 7 samples were analyzed with RNAseq, as well. In addition to point mutations, the FusionPlex Pan-Heme Kit can identify both known and novel fusions in fusion panel target genes as well as deletions in IKZF1. The details of this assay are found in Suppl. Methods.
Public databases
Microarray expression data were obtained from the European Bioinformatics Institute’s ArrayExpress database (http://www.ebi.ac.uk/arrayexpress).13 We used E-MTAB-5035 accession that includes 96 Ph– and 41 Ph+ adult B-ALL patients measured with Affymetrix Human Genome U133 Plus 2.0 Microarray. In addition, we analyzed publicly available transcriptome sequencing data from St Jude PeCan Data Portal (https://pecan.stjude.cloud) from a study published by Gu et al.14 The analyzed cohort comprised of 670 adult and adolescent patients, including 83 Ph+ ALL cases. Median age in the cohort was 40 years (range 16–79). We included only subgroups with more than 20 patients for the analyses. For comparing cell type prediction results, we analyzed data from HEMAP pre-B-ALL cohort (hemap.uta.fi).15 The analyzed HEMAP cohort included 1300 patients with pre-B-ALL, the vast majority being pediatric cases. In addition, we analyzed MDM2 expression in all HEMAP samples.
Cell lines and drug combination testing
To test the most interesting drug combinations, we analyzed selected B-ALL cell lines, that represented BCR-ABL1-positive ALL (NALM-21), BCR-ABL1-negative B-ALL (Kasumi-2), and BCR-ABL1-like ALL (MHH-CALL-4). The tested combinations were dasatinib+venetoclax, dasatinib+navitoclax, dasatinib+idasanutlin, venetoclax+idasanutlin, and navitoclax+idasanutlin. The treatment of the cell lines is described in detail in Suppl. Methods, and the concentrations of the tested combinations are found in Suppl. Table S3. Each compound was tested in 5 different concentrations covering a 10 000-fold concentration range.
Bioinformatics and statistical analyses
Details of bioinformatics and statistical analyses are found in Suppl. Methods and in corresponding figure legends.
RESULTS
Genetic characteristics of the B-ALL patient cohort used in the drug testing
Our RNAseq cohort included 9 Ph+ ALL and 7 Ph– ALL patients. Analysis of the RNAseq data verified BCR-ABL1 fusion genes in all known Ph+ patients. In principal component analysis, Ph+ samples clustered together in the first variance component along with samples from Ph– patients Pt_4 and Pt_1 (Suppl. Figure S1A). In Pt_4, RNAseq revealed a MEF2D-CSF1R fusion gene, a fusion known in ALL to activate tyrosine kinase pathway and to cause a Ph-like transcriptional signature.16 Pt_1, in turn, was identified to carry a FLT3 kinase point mutation Y842H based on transcript variant analysis. In B-ALL, FLT3 mutations are classified to Ph-like subtype explaining the clustering together with Ph+ samples. All key mutations are depicted in Figure 1, and additional variants in commonly mutated genes in leukemia that were called from RNAseq are shown in Suppl. Figure S1B. RNAseq quality control values are assembled to Suppl. Table S4. FusionPlex Pan-Heme Kit results are listed in Suppl. Table S1.
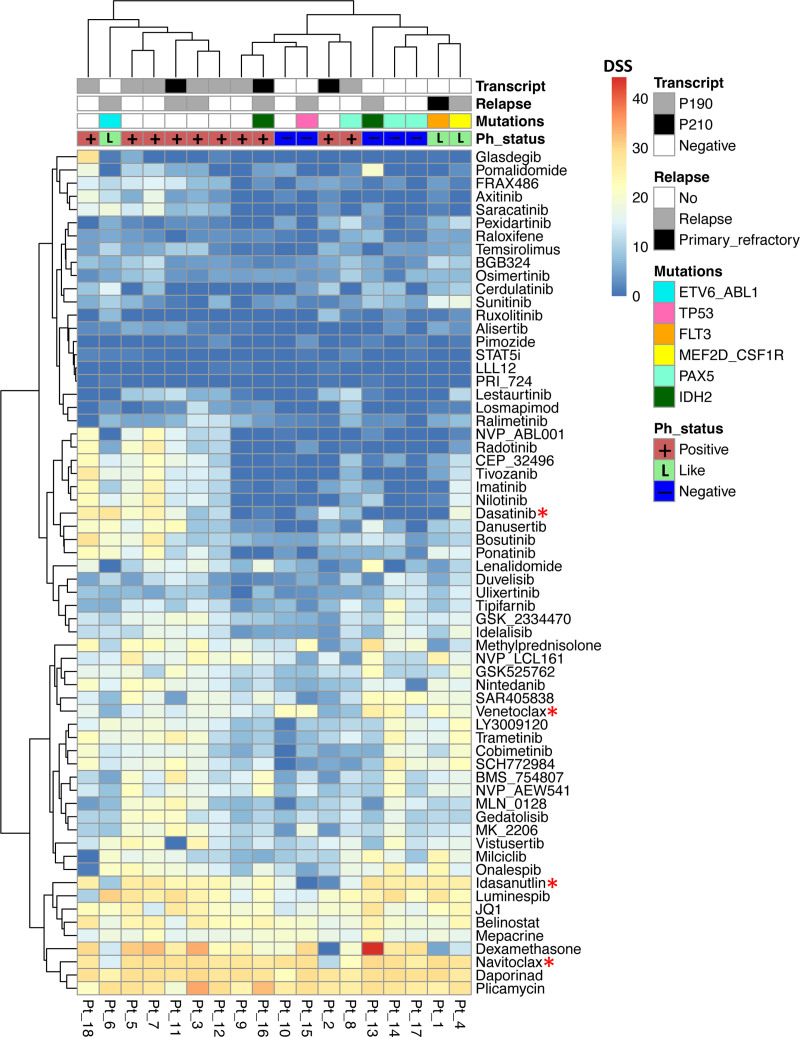
Figure 1.
Heatmap showing the custom drug plate results. Philadelphia chromosome-positive patients (n = 10) are annotated in red, Philadelphia chromosome-negative patients in blue (n = 5), and Philadelphia-like patients in light green (n = 3). In addition, BCR-ABL1 transcript type, relapse status, and key mutations defined from RNAseq data are shown in additional tracks. Drug potency and efficacy are color-coded according to DSS. If DSS >10, drug has efficacy, and if DSS >20, drug has excellent efficacy. Clustering by Euclidean distance measurement and complete clustering method. Drugs that were used in combination assays are marked with a red asterisk. DSS = drug sensitivity score.
Ex vivo drug testing assay revealed Pan-ALL and subtype-specific drug responses
In the ex vivo drug sensitivity testing on primary leukemic cells glucocorticoids (especially dexamethasone), BCL2 family inhibitors venetoclax and navitoclax, belinostat (histone deacetylase inhibitor), daporinad (nicotinamide phosphoribosyltransferase [NAMPT] inhibitor), idasanutlin (MDM2 inhibitor), JQ1 (bromodomain and extra-terminal motif inhibitor), luminespib (HSP90 inhibitor), and plicamycin (antineoplastic antibiotic) were broadly effective (Figure 1 and Suppl. Figure S2). For the other drugs, including TKIs, we found great variability with only individual patients being sensitive, reflecting the diverse molecular background of ALL.
We applied TAS analysis to our drug testing data to obtain more information of the druggable protein targets behind the observed drug responses. TAS analysis was well in accordance with the observed drug responses as histone deacetylase, NAMPT, BCL-XL (BCL2L1), MDM2, and HSP90 class gene targets were highly addicted. Target genes identified by TAS were highly expressed throughout the samples (Suppl. Figure S3).
Sensitivity to tyrosine kinase inhibitors clustered patients in the drug data
In the heatmap analysis, patients were grouped into 2 main clusters. This clustering was mainly driven by ABL1-targeting TKIs which clustered together among all tested drugs (Figure 1). The clustering was not clearly affected by patient age, survival, or somatic mutations by visual inspection. Four Ph+ patients, Pt_2, Pt_8, Pt_9, and Pt_16, were unexpectedly resistant to ABL1-targeting TKIs. Pt_8 had borderline cell viability after 72 hours incubation and Pt_2 had relatively low blast count (60%) in the diagnostic sample, which may explain the moderately low TKI sensitivities in these 2 samples. Pt_9 and Pt_16 had high blast counts, good cell viability, and did not share any common features besides Ph+. Of these 4 patients, only Pt_16 has relapsed, whereas the other 3 have remained in remission.
In addition, a Ph– like ALL sample with an ETV6-ABL1 fusion (Pt_6) was sensitive to several ABL1-targeting TKIs (dasatinib DSS = 27.6, ponatinib DSS = 20.1, nilotinib DSS = 15.3, bosutinib DSS = 19.9, axitinib DSS = 12.1), similarly as Ph-like Pt_4 with MEF2D-CSF1R fusion (dasatinib DSS = 18.1, imatinib DSS = 15.2, ponatinib DSS = 15.2, nilotinib DSS = 15.4, bosutinib DSS = 10.9). The third Ph– like ALL patient sample with FLT3 kinase point mutation (Pt_1) was sensitive to broad-spectrum kinase inhibitor ponatinib (DSS = 16) and to FLT3 inhibitor sunitinib (DSS = 15.7).
MDM2 inhibitor idasanutlin and BCL2 inhibitors navitoclax and venetoclax were highly effective in B-ALL samples
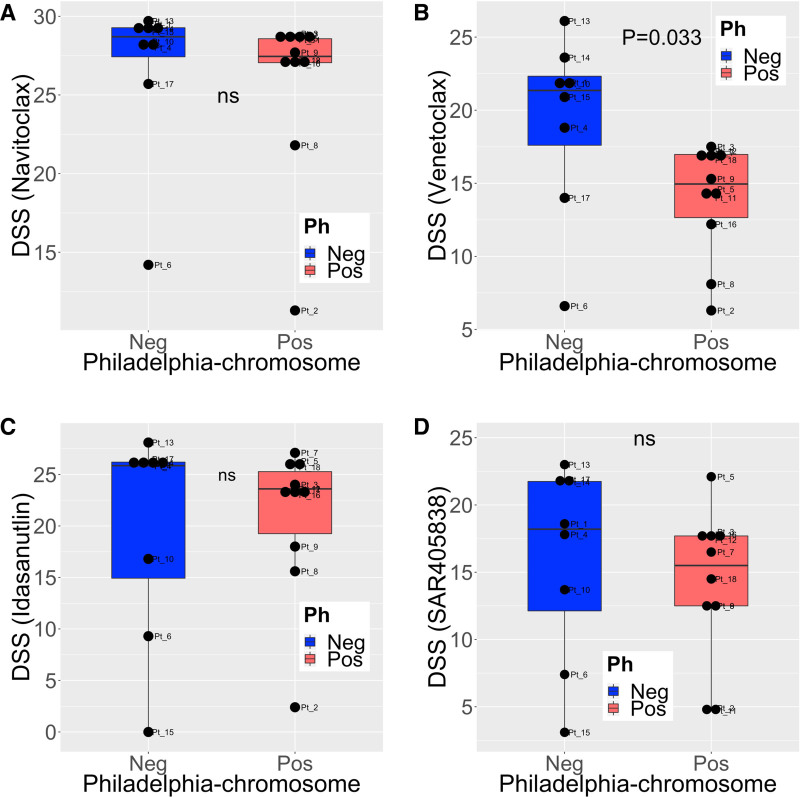
Almost all patient samples (16/18, 89%) were highly sensitive (DSS > 20) to navitoclax, a BCL2, BCL-XL, and BCL-W inhibitor (Figure 2A). BCL2-inhibitor venetoclax was also highly effective (DSS > 20) in 28% (5/18) of the samples, and moderately effective (DSS > 10) in 83% (15/18) samples. Unexpectedly, Ph+ samples were markedly less sensitive to venetoclax than Ph– samples (Mann-Whitney U test, P = 0.033) (Figure 2B).
Figure 2.
The efficacy of BCL2 and MDM2 inhibitors in Philadelphia chromosome-positive and negative samples. DSS for (A) navitoclax, (B) venetoclax, (C) idasanutlin, and (D) MDM2-inhibitor SAR405838 in Philadelphia chromosome-positive (n = 10) and negative (n = 8) ALL patient samples. Group differences tested with Mann-Whitney U test. DSS = drug sensitivity scores; Ns, not significant.
Another apoptosis-promoting drug, MDM2 inhibitor idasanutlin showed high efficacy (DSS > 20) in 67% of the patient samples (12/18), and 83% (15/18) responded to the drug (DSS > 10). Patient sample with a TP53 mutation (Pt_15) was resistant to the drug, as expected based on the mode of action of the drug (Figure 1 and Suppl. Figure S1B). No difference in responses was observed between Ph+ and Ph– ALL patients (Figure 2C). Another MDM2 inhibitor SAR405838 was less effective than idasanutlin (DSS>20 in 22% [4/18 samples] and DSS>10 in 78% (14/18 samples); Figure 2D).
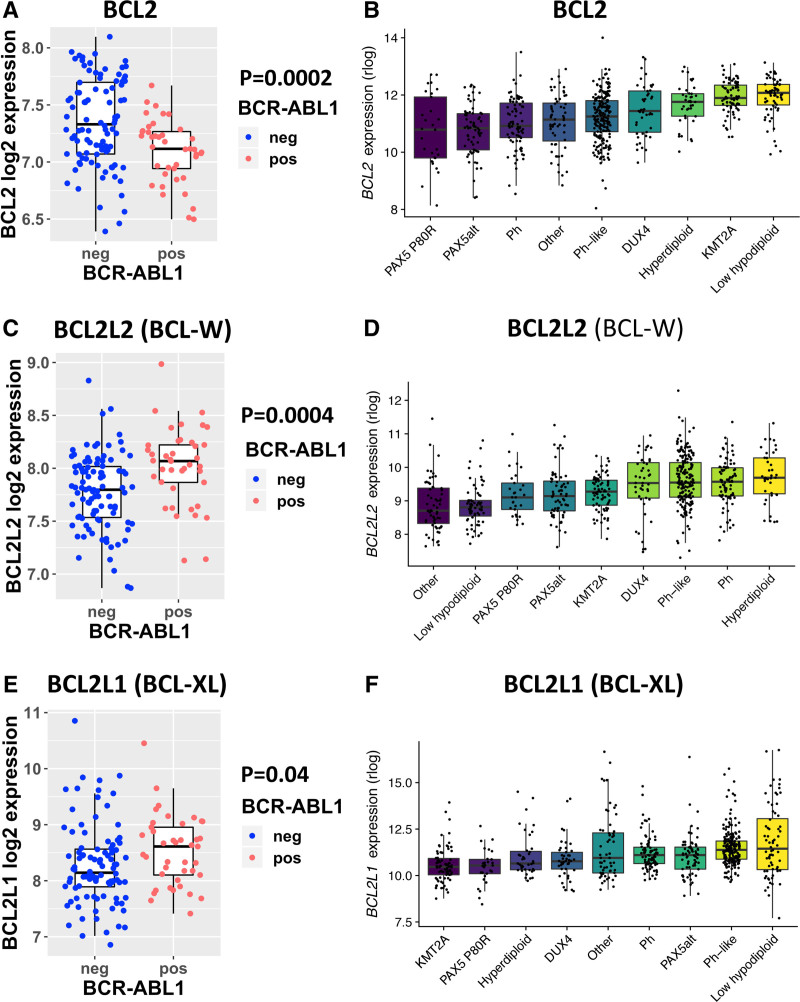
BCL-W was upregulated and BCL2 downregulated in Ph+ ALL, driving the responses to venetoclax and navitoclax
To explore the biological determinants behind the differences in the BCL2-inhibitor responses between Ph– and Ph+ ALL patients, we analyzed 2 large publicly available gene expression datasets.13,14 BCL2 expression was downregulated in Ph+ patients compared with Ph– patients in both E-MTAB-5035 (P = 0.0002) and B-ALL 1988 cohorts (Figure 3A, B). BCL-W expression was higher (P = 0.0004) in Ph+ patients in the E-MTAB-5035 cohort and trended towards higher expression in the B-ALL 1988 cohort (Figure 3C, D). There was also a trend for BCL-XL upregulation in Ph+ patients in both cohorts (Figure 3E, F). MDM2 expression was significantly higher in Ph+ patients in both cohorts (Suppl. Figure S4). When exploring MDM2 expression in an additional database (HEMAP),15 MDM2 was clearly overexpressed in B-ALL compared with other hematological malignancies, supporting the strong idasanutlin responses in our drug panel (Suppl. Figure S5).
Figure 3.
The expression of BCL2, BCL-W, and BCL-XL in Philadelphia chromosome-positive and negative samples. The expression of BCL2 in (A) E-MTAB-5035 and (B) B-ALL 1988 datasets, the expression of BCL-W in (C) E-MTAB-5035 and (D) B-ALL 1988 datasets, and the expression of BCL-XL in (E) E-MTAB-5035 and (F) B-ALL 1988 datasets. In E-MTAB-5035 cohort comparison between Philadelphia chromosome-positive and negative patients with Mann-Whitney U test. In the B-ALL 1988, cohort subgroups were visualized and annotated as in the original data. Only subgroups with >20 patients were included.
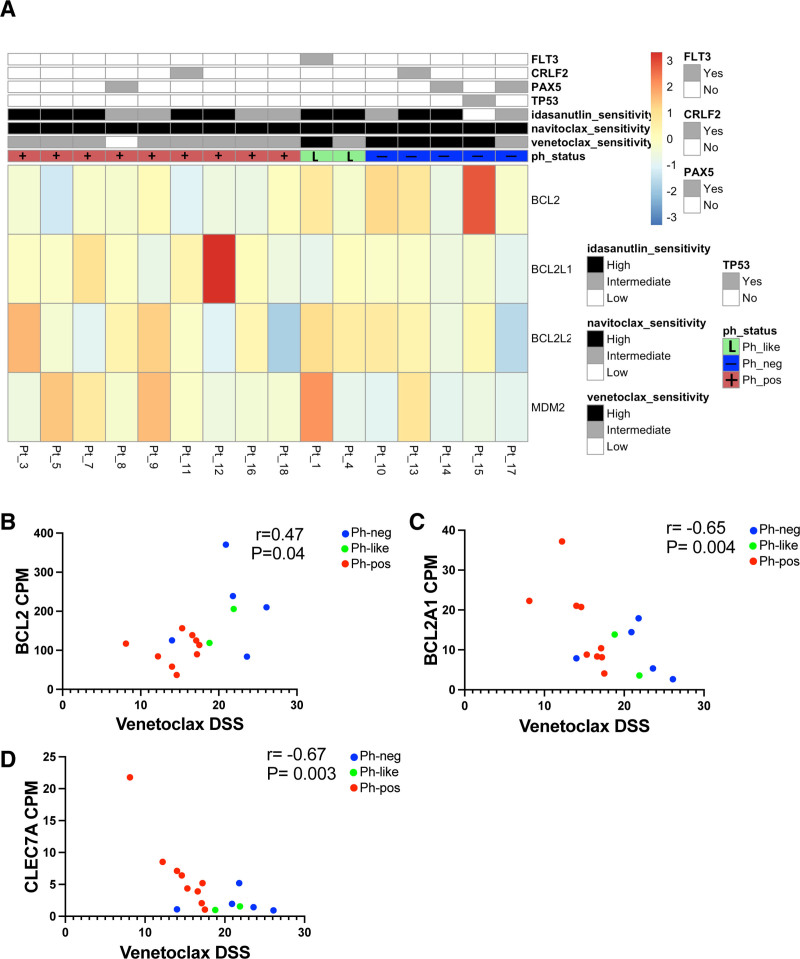
Figure 4A summarizes the gene expression of the known venetoclax, navitoclax, and idasanutlin drug targets in our patient samples and the corresponding drug sensitivity. BCL2 expression correlated with venetoclax sensitivity (Figure 4B), further confirming the observations seen in the public expression data. However, the expression of BCL-XL (BCL2L1) and BCL2L2 (BCL-W) did not clearly correlate with Ph-status, possibly caused by the small size of our cohort, which would not detect subtle differences. The expression of BCL2A1 and CLEC7A correlated inversely with the sensitivity to venetoclax (Figure 4C, D), as previously has been reported in acute myeloid leukemia (AML).17 As idasanutlin was effective in nearly all of our samples, the differences in MDM2 expression were more difficult to detect.
Figure 4.
The gene expression of known BCL2 inhibitor and MDM2 inhibitor drug targets, and the correlation of BCL2, BCL2A1, and CLEC7A expression and venetoclax sensitivity. (A) Heatmap showing gene expression levels of known venetoclax, navitoclax, and idasanutlin drug targets combined with drug sensitivities. Categorical drug sensitivities were coded according to DSS. If DSS is >20, sensitivity is high, if DSS is between 10 and 20 sensitivity is intermediate, and if DSS is <10 sensitivity is low. Philadelphia chromosome-positive patients (n = 9) are annotated in red, Philadelphia chromosome-negative patients in blue (n = 5), and Philadelphia-like patients in light green (n = 2). In addition, key mutations are shown in an additional track. Non-parametric Spearman correlation between the expression of (B) BCL2, (C) BCL2A1, and (D) CLEC7A (CPM counts) and venetoclax sensitivity (DSS). DSS = drug sensitivity score.
The results from differential gene expression and gene set enrichment analyses can be found from the Suppl. Material. Ph-like patients were included in the analysis, but the comparison was made between Ph+ and true Ph– patients. Altogether 242 protein-coding genes were differentially expressed (q < 0.05), (Suppl. Table S5; Suppl. Figure S6). All enriched pathways from Enrichr databases related to cancer, cell signaling, and kinase enrichment are listed in the Suppl. Table S6.
Venetoclax and idasanutlin showed synergy in B-ALL cell lines
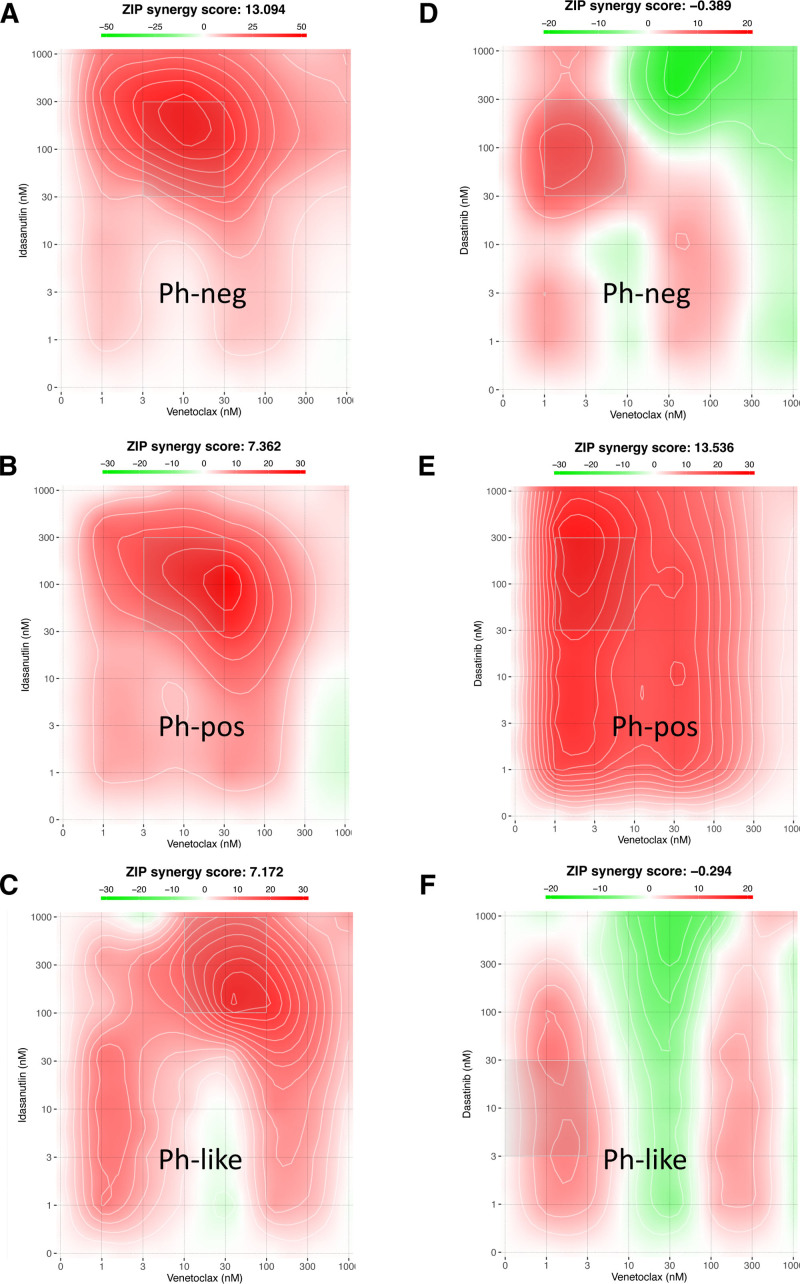
To study the interactions between the most relevant compounds in more detail, we tested several 2-drug combinations between BCL2 inhibitors venetoclax and navitoclax, MDM2 inhibitor idasanutlin, and BCR-ABL1 TKI dasatinib in 3 human cell lines representing Ph+, Ph–, and Ph-like ALL. The combination of venetoclax and idasanutlin was synergistic in all cell lines. Of the other combinations, a combination of navitoclax and idasanutlin was synergistic in Ph– ALL cell line Kasumi-2, and the combinations of dasatinib with venetoclax, navitoclax, and idasanutlin were synergistic in Ph+ ALL cell line NALM-21. Synergy scores from all combinations as well as single compound DSS values are listed in Suppl. Table S7, and the visualization of venetoclax and idasanutlin and venetoclax and dasatinib synergy score maps are shown in Figure 5. The combination score matrixes of these combinations are displayed in Suppl. Figure S7.
Figure 5.
Combination score visualizations of the venetoclax—idasanutlin and venetoclax—dasatinib drug combinations. Venetoclax-idasanutlin combination score map in (A) Ph– (Kasumi-2), (B) Ph+ (NALM-21), and (C) Ph-like (MHH-CALL-4) cell lines and venetoclax—dasatinib combination score map in (D) Ph– (Kasumi-2), (E) Ph+ (NALM-21), and (F) Ph-like (MHH-CALL-4) cell lines. ZIP combination score >5 denotes synergistic effect. Ph– = Philadelphia chromosome negative; Ph+ = Philadelphia chromosome positive.
DISCUSSION
Here, we analyzed primary B-ALL patient samples using a well-established ex vivo drug testing platform and whole-transcriptome sequencing. This information was combined with publicly available expression data to define gene expression biomarkers for ex vivo drug responses. The BCL2 inhibitors venetoclax and navitoclax showed promising efficacy in our drug panel. BCL2 is an antiapoptotic protein, which is located on the outer mitochondrial membrane. Venetoclax and navitoclax inhibit the action of BCL2, thereby allowing proapoptotic proteins to activate.18 Malignant cells can overexpress BCL2 and become BCL2-dependent for their survival, which offers an opportunity to target cancer cells without harming the normal tissues.19
In preclinical and early clinical studies, BCL2 family inhibitors have shown efficacy in various hematological malignancies and solid tumors.18,20 Currently, venetoclax is the only FDA-approved BCL2 inhibitor, which is used for the treatment of chronic lymphocytic leukemia and AML. Venetoclax has been effective in patient-derived ALL xenografts, and importantly, ex vivo response has been reported to correlate with in vivo responses.21–23 In preclinical trials, especially combination strategies, such as with inotuzumab ozogamicin and dexamethasone, seem promising.24 Several clinical trials investigating venetoclax in different drug combinations in relapsed or refractory ALL are ongoing (clinicaltrials.gov). Our results showed that venetoclax was significantly more effective in Ph– patients, whereas navitoclax showed more uniform potency. With differential gene expression analysis of BCL family genes, we were able to show that BCL2 was downregulated and BCL-W upregulated in Ph+ ALL in comparison to Ph– ALL. In addition to BCL2, navitoclax targets BCL-W and BCL-XL, whereas venetoclax is BCL2-selective,25,26 offering an explanation why navitoclax was more effective in Ph+ samples than venetoclax.
Platelets express prosurvival protein BCL-XL, and dose-limiting thrombocytopenia has previously limited the use of navitoclax in solid tumors.27 We hypothesize, that especially in combinations with other agents, navitoclax may be used in the treatment of acute leukemias, where the management of transient thrombocytopenia constitutes a routine part of all treatment protocols. Navitoclax showed better efficacy in Ph+ ALL samples compared with venetoclax. This would suggest a rationale for combining navitoclax with TKIs. In a phase I study, low-dose navitoclax in combination with venetoclax and chemotherapy in relapsed or refractory ALL was well tolerated with preliminarily promising efficacy in a heavily pretreated population (NCT03181126).28
Wild-type p53, encoded by TP53, is a tumor suppressor, that is mutated in more than half of all solid malignancies. In hematological malignancies, however, TP53 mutations are relatively rare,29 and in ALL, TP53 mutations are found in approximately 15% of new cases.30 Even without mutations, the function of wild-type p53 in cancer is often suppressed by an increased amount of regulatory protein MDM2, leading to impaired cancer cell apoptosis.31,32 It is noteworthy that MDM2 inhibitor idasanutlin, was effective in nearly all of our samples. Idasanutlin blocks the interaction of MDM2 and p53, leading to restoration of wild-type p53 function and enhanced cancer cell apoptosis.33 Monotherapy with BCL2 inhibitors can easily lead to overexpression of antiapoptotic MCL1 and subsequent drug resistance.34 Activation of TP53 promotes degradation of MCL1, offering a rationale for combining MDM2 and BCL2 inhibitors. Combination treatment with MDM2 and BCL2 inhibitors has already shown synthetic lethality in resistant AML mouse models.35 Idasanutlin is currently being tested in combination with either chemotherapy or venetoclax in a phase 1/2 clinical trial for relapsed or refractory (R/R) AML and R/R ALL (NCT04029688, clinicaltrials.gov).
We tested a combination of BCL2 inhibitor venetoclax and MDM2 inhibitor idasanutlin in 3 human cell lines representing Ph+, Ph–, and Ph-like ALL. In all 3 cell lines, this combination was synergistic.
With the drug sensitivity testing and molecular profiling, we identified targetable Ph-like lesions in 3 of the study patients. These alterations were not detected in the clinical cytogenetics or targeted fusion screens that were used at diagnosis. Patient Pt_6 had ETV6-ABL1 fusion and was sensitive to selected TKIs in the drug testing. The patient did not achieve remission with first induction, remained minimal residual disease positive, and relapsed and died soon after alloHSCT. Patient Pt_1 had FLT3 point mutation Y842H in tyrosine kinase domain and died of primary refractory disease 6 months after the diagnosis. Patient Pt_4 had MEF2D-CSF1R fusion and was correspondingly sensitive to TKIs in our drug screen. The patient relapsed in less than a year and died of a relapse 2 years after the diagnosis. None of these patients had received TKIs or FLT3 inhibitors as part of their treatment protocols, because Ph-like status was not recognized.36,37
Although ex vivo drug testing of primary cancer cells has proven useful as a preclinical tool in evaluating drug candidates for clinical testing,38 and even in aiding treatment selection in relapsed/refractory patients,39 it is not well-suited for predicting responses to cytotoxic agents, nor drugs requiring a long exposure time for effect (eg, hypomethylating or differentiation-inducing drugs). Using flow cytometric and other single cell–based readouts may partially overcome these.40
In conclusion, the combination of ex vivo drug testing and molecular profiling is a powerful tool for identifying novel effective and actionable therapies for ALL patients. Targeting the apoptosis pathway by inhibiting antiapoptotic proteins (BCL2, BCL-XL, BCL-W) and p53 with BCL2 and MDM2 inhibitors in combination with established ALL drugs is a highly promising strategy for improving survival and reducing treatment-related toxicity.
ACKNOWLEDGMENTS
We thank laboratory technicians Laura Turunen and Maria Nurmi in the Institute for Molecular Medicine Finland for technical support with DSRT, and Minna Suvela, a laboratory technician in the Institute for Molecular Medicine Finland, for technical support with RNAseq. We thank Krister Wennerberg and Heikki Kuusanmäki in Institute for Molecular Medicine Finland for valuable comments regarding DSRT assay design and data analysis. We are grateful to the members of the Hematology Research Unit Helsinki and Institute for Molecular Medicine Finland for discussions and technical help. The samples of this project were provided by the Finnish Hematology Registry and Clinical Biobank (FHRB) with appropriate ethics approval (Dnro 202/06.01.00/2013). We thank all the patients for their generous participation. The FHRB Biobank is supported by the Finnish Association of Hematology, the Finnish Red Cross Blood Service, Institute for Molecular Medicine Finland, and the participating hospitals in Finland.
AUTHOR CONTRIBUTIONS
HH, SA-A, SM, and KP did conception and design. HH, SA-A, MK, BY, SP, OD, CH, VS, and SK did collection and assembly of data. All authors participated in data analysis and interpretation; article writing; and final approval of article. MK and SA-A have contributed equally to this work. SM and KP shared senior authorship of this article.
DISCLOSURES
HH and SAA have received research funding from Incyte. SM and KP have received research funding and honoraria from BMS, Novartis, and Pfizer (not related to this study). All the other authors have no conflicts of interest to disclose.
SOURCE OF FUNDING
This study was supported by the Doctoral Programme in Clinical Research in the University of Helsinki and personal grants (HH) from Incyte Nordic Grant, Emil Aaltonen Foundation, Ida Montin Foundation, Blood Disease Research Foundation, Finnish Hematology Association, Finnish Medical Foundation, Biomedicum Foundation, (SA-A) Incyte Nordic Grant, (SM) Finnish Cancer Institute, Finnish Cancer Organizations, Sigrid Juselius Foundation, Signe and Ane Gyllenberg Foundation, Relander Foundation, and state funding for university-level health research in Finland. VS has received funding from the European research Council under the Europeans Unions Horizon 2020 Research and Innovation program grant Agreement 694354.
Supplementary Material
Footnotes
Supplemental digital content is available for this article.
REFERENCES
- 1.Bassan R, Bourquin JP, DeAngelo DJ, Chiaretti S. New approaches to the management of adult acute lymphoblastic Leukemia. J Clin Oncol. 2018;36:3504–3519. [DOI] [PubMed] [Google Scholar]
- 2.Harvey RC, Tasian SK. Clinical diagnostics and treatment strategies for Philadelphia chromosome-like acute lymphoblastic leukemia. Blood Adv. 2020;4:218–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kantarjian H, Jabbour E. Incorporating immunotherapy into the treatment strategies of B-cell adult acute lymphoblastic leukemia: the role of blinatumomab and inotuzumab ozogamicin. Am Soc Clin Oncol Educ Book. 2018;38:574–578. [DOI] [PubMed] [Google Scholar]
- 4.Park JH, Rivière I, Gonen M, et al. Long-term follow-up of CD19 CAR therapy in acute lymphoblastic leukemia. N Engl J Med. 2018;378:449–459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Frey NV, Shaw PA, Hexner EO, et al. Optimizing chimeric antigen receptor T-cell therapy for adults with acute lymphoblastic leukemia. J Clin Oncol. 2020;38:415–422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Frisch A, Ofran Y. How I diagnose and manage Philadelphia chromosome-like acute lymphoblastic leukemia. Haematologica. 2019;104:2135–2143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jabbour E, DerSarkissian M, Duh MS, et al. Efficacy of ponatinib versus earlier generation tyrosine kinase inhibitors for front-line treatment of newly diagnosed philadelphia-positive acute lymphoblastic leukemia. Clin Lymphoma Myeloma Leuk. 2018;18:257–265. [DOI] [PubMed] [Google Scholar]
- 8.Aldoss I, Forman SJ, Pullarkat V. Acute lymphoblastic leukemia in the older adult. J Oncol Pract. 2019;15:67–75. [DOI] [PubMed] [Google Scholar]
- 9.Pemovska T, Kontro M, Yadav B, et al. Individualized systems medicine strategy to tailor treatments for patients with chemorefractory acute myeloid leukemia. Cancer Discov. 2013;3:1416–1429. [DOI] [PubMed] [Google Scholar]
- 10.Yadav B, Pemovska T, Szwajda A, et al. Quantitative scoring of differential drug sensitivity for individually optimized anticancer therapies. Sci Rep. 2014;4:5193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yadav B, Gopalacharyulu P, Pemovska T, et al. From drug response profiling to target addiction scoring in cancer cell models. Dis Model Mech. 2015;8:1255–1264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Adnan Awad S, Kankainen M, Ojala T, et al. Mutation accumulation in cancer genes relates to nonoptimal outcome in chronic myeloid leukemia. Blood Adv. 2020;4:546–559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Athar A, Füllgrabe A, George N, et al. ArrayExpress update—from bulk to single-cell expression data. Nucleic Acids Res. 2019;47:D711–D715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gu Z, Churchman ML, Roberts KG, et al. PAX5-driven subtypes of B-progenitor acute lymphoblastic leukemia. Nat Genet. 2019;51:296–307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pölönen P, Mehtonen J, Lin J, et al. HEMAP: an interactive online resource for characterizing molecular phenotypes across hematologic malignancies. Cancer Res. 2019;79:2466–2479. [DOI] [PubMed] [Google Scholar]
- 16.Gu Z, Churchman M, Roberts K, et al. Genomic analyses identify recurrent MEF2D fusions in acute lymphoblastic leukaemia. Nat Commun. 2016;7:13331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang H, Nakauchi Y, Köhnke T, et al. Integrated analysis of patient samples identifies biomarkers for venetoclax efficacy and combination strategies in acute myeloid leukemia. Nat Cancer. 2020;1:826–839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Adams JM, Cory S. The BCL-2 arbiters of apoptosis and their growing role as cancer targets. Cell Death Differ. 2018;25:27–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Certo M, Del Gaizo Moore V, Nishino M, et al. Mitochondria primed by death signals determine cellular addiction to antiapoptotic BCL-2 family members. Cancer Cell. 2006;9:351–365. [DOI] [PubMed] [Google Scholar]
- 20.Montero J, Letai A. Why do BCL-2 inhibitors work and where should we use them in the clinic? Cell Death Differ. 2018;25:56–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Diaz-Flores E, Comeaux EQ, Kim KL, et al. Bcl-2 is a therapeutic target for hypodiploid B-lineage acute lymphoblastic leukemia. Cancer Res. 2019;79:2339–2351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Seyfried F, Demir S, Hörl RL, et al. Prediction of venetoclax activity in precursor B-ALL by functional assessment of apoptosis signaling. Cell Death Dis. 2019;10:571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Frismantas V, Dobay MP, Rinaldi A, et al. Ex vivo drug response profiling detects recurrent sensitivity patterns in drug-resistant acute lymphoblastic leukemia. Blood. 2017;129:e26–e37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kirchhoff H, Karsli U, Schoenherr C, et al. Venetoclax and dexamethasone synergize with inotuzumab ozogamicin-induced DNA damage signaling in B-lineage ALL. Blood. 2021;137:2657–2661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Souers AJ, Leverson JD, Boghaert ER, et al. ABT-199, a potent and selective BCL-2 inhibitor, achieves antitumor activity while sparing platelets. Nat Med. 2013;19:202–208. [DOI] [PubMed] [Google Scholar]
- 26.van Delft MF, Wei AH, Mason KD, et al. The BH3 mimetic ABT-737 targets selective Bcl-2 proteins and efficiently induces apoptosis via Bak/Bax if Mcl-1 is neutralized. Cancer Cell. 2006;10:389–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mason KD, Carpinelli MR, Fletcher JI, et al. Programmed anuclear cell death delimits platelet life span. Cell. 2007;128:1173–1186. [DOI] [PubMed] [Google Scholar]
- 28.Pullarkat VA, Lacayo NJ, Jabbour E, et al. Venetoclax and navitoclax in combination with chemotherapy in patients with relapsed or refractory acute lymphoblastic leukemia and lymphoblastic lymphoma. Cancer Discov. 2021;11:1440–1453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Peller S, Rotter V. TP53 in hematological cancer: low incidence of mutations with significant clinical relevance. Hum Mutat. 2003;21:277–284. [DOI] [PubMed] [Google Scholar]
- 30.Stengel A, Schnittger S, Weissmann S, et al. TP53 mutations occur in 15.7% of ALL and are associated with MYC-rearrangement, low hypodiploidy, and a poor prognosis. Blood. 2014;124:251–258. [DOI] [PubMed] [Google Scholar]
- 31.Haupt Y, Maya R, Kazaz A, et al. Mdm2 promotes the rapid degradation of p53. Nature. 1997;387:296–299. [DOI] [PubMed] [Google Scholar]
- 32.Kubbutat MH, Jones SN, Vousden KH. Regulation of p53 stability by Mdm2. Nature. 1997;387:299–303. [DOI] [PubMed] [Google Scholar]
- 33.Shangary S, Wang S. Targeting the MDM2-p53 interaction for cancer therapy. Clin Cancer Res. 2008;14:5318–5324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lin KH, Winter PS, Xie A, et al. Targeting MCL-1/BCL-XL forestalls the acquisition of resistance to ABT-199 in acute myeloid leukemia. Sci Rep. 2016;6:27696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pan R, Ruvolo V, Mu H, et al. Synthetic lethality of combined Bcl-2 inhibition and p53 activation in AML: mechanisms and superior antileukemic efficacy. Cancer Cell. 2017;32:748–760.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Roberts KG, Gu Z, Payne-Turner D, et al. High frequency and poor outcome of philadelphia chromosome-like acute lymphoblastic leukemia in adults. J Clin Oncol. 2017;35:394–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tanasi I, Ba I, Sirvent N, et al. Efficacy of tyrosine kinase inhibitors in Ph-like acute lymphoblastic leukemia harboring ABL-class rearrangements. Blood. 2019;134:1351–1355. [DOI] [PubMed] [Google Scholar]
- 38.Pemovska T, Johnson E, Kontro M, et al. Axitinib effectively inhibits BCR-ABL1(T315I) with a distinct binding conformation. Nature. 2015;519:102–105. [DOI] [PubMed] [Google Scholar]
- 39.Malani D, Kumar A, Bruck O, et al. Implementing a functional precision medicine tumor board for acute myeloid leukemia. Cancer Discov. 2022;12:1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kuusanmäki H, Leppä AM, Pölönen P, et al. Phenotype-based drug screening reveals association between venetoclax response and differentiation stage in acute myeloid leukemia. Haematologica. 2020;105:708–720. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.