Abstract
Metal-on-metal (MoM) hip implants made of cobalt chromium (CoCr) alloy have shown early failure compared with other bearing materials. A consequence of the abnormal wear produced by these prostheses is elevated levels of cobalt in the blood of patients, which can lead to systemic conditions involving cardiac and neurological symptoms. In order to better understand the implications for patients with these implants, we carried out metal content and RNA-Seq analysis of excised tissue from rats treated intraperitonially for 28 days with low concentrations of cobalt. Cobalt blood levels in dosed rats were found to be similar to those seen in some patients with MoM implants (range: 4–38 μg/L Co in blood). Significant accumulation of cobalt was measured in a range of tissues including kidney, liver, and heart, but also in brain tissue. RNA-Seq analysis of neural tissue revealed that exposure to cobalt induces a transcriptional response in the prefrontal cortex (pref. cortex), cerebellum, and hippocampus. Many of the most up- and downregulated genes appear to correspond to choroid plexus transcripts. These results indicate that the choroid plexus could be the brain tissue most affected by cobalt. More specifically, the differentially expressed genes show a disruption of steroidogenesis and lipid metabolism. Several other transcripts also demonstrate that cobalt induces an immune response. In summary, cobalt exposure induces alterations in the brain transcriptome, more specifically, the choroid plexus, which is in direct contact with neurotoxicants at the blood–cerebrospinal fluid barrier.
Keywords: metal-on-metal (MoM) hip implants, cobalt, systemic cobaltism, neurotoxicity, RNA-Seq, RT-qPCR
1. Introduction
Hip arthroplasty procedures successfully lead to the reduction in pain and improved mobility in patients suffering from joint diseases such as osteoarthritis. However, a decade ago, consultants and regulatory bodies reported their concerns over the failures of certain models of metal-on-metal (MoM) hip implants, which resulted in the market withdrawal of the Articular Surface ReplacementTM (ASRTM) hip implant from DePuy Orthopaedics [1]. Recently, these concerns have been extended to all MoM implant models as it has been demonstrated that MoM implants induce adverse reaction to metal debris (ARMD), which includes metallosis, pseudotumours, aseptic lymphocytic vasculitis associated lesion (ALVAL), and even necrosis [2]. The metal debris is released from the bearing surface, and the taper junction in the case of the total hip replacements (THR), due to wear and corrosion of the metallic parts [3]. Metal ions eventually dissolve and are released into the bloodstream [4]. Co and Cr metal ions in blood are indicative of implant failure and tests for metal ion concentrations should be performed for all patients with MoM implants in the UK according to the MHRA updated guidelines [5]. The European Commission also recommends the monitoring of metal ion levels for MoM implant patients [6].
Neurological conditions thought to be caused by high levels of cobalt ions in blood have been demonstrated in several clinical reports in relation to patients with MoM implants [7,8,9,10,11]. These involve a range of symptoms such as cognitive decline, memory loss, and mood disturbances, in addition to other visual and auditory issues and peripheral neuropathy. There is little information about the actual effects of cobalt in the peripheral and the central nervous system, and the most appropriate response to diminish patients’ symptoms to date is to remove the implant as the source of cobalt ions through revision surgery [8,12,13]. However, the recovery process from cobaltism is not well documented and there is no unified action considered appropriate to treat these patients. Our lack of knowledge on cobalt actions in the body extends further since we also fail to consider the implications for asymptomatic patients with high levels of cobalt in the blood [14].
The effects of cobalt have been extensively researched in vitro with models such as astrocytes [15,16], neurons [17,18,19,20,21], and glial cells [22,23], which are valuable tools to understand the modes of action of cobalt toxicity in the brain. Nevertheless, the nature of in vitro work requires highly controlled experimental conditions, and this can diminish an organism’s complexity. Previous toxicogenomic analyses have demonstrated that in vitro systems fail to fully represent relevant processes occurring in rat liver tissues in vivo after exposure to toxic compounds [24]. Molecular and functional events occurring in vivo at the tissue and organ levels could be crucial mechanisms in the physiological response to cobalt. In this sense, transcriptomic applications have already been proven to be effective in toxicology, not only by understanding the mechanisms, but also by finding early end-points for the detection of toxicity and identification of biomarkers [25]. However, most in vivo cobalt studies have focused on the study of reactive oxygen species and the expression of hypoxia markers. Pregnant female rats dosed orally with 350 mg/L delivered pups with impaired levels of antioxidant proteins in the cerebrum and cerebellum [26]. Caltana et al. observed that the direct cortical injection of cobalt led to histological changes consistent with focal ischaemia involving neuronal and astrocyte morphological changes [27]. Another group applied cobalt dust directly into the dura mater [28] and discovered an elevated expression of proteins involved in thyroid transport and regulation of glycolysis. Nevertheless, it is difficult to establish the relevance of these studies for patients with MoM implants due to the high dosage of cobalt used in the animals, the different types of cobalt delivery methods, and the missing information on the resulting cobalt concentrations in blood or plasma. Our research mimicked the conditions that MoM implant patients endure to obtain a better representation of relevant cobalt toxic mechanisms for them.
The aim of this in vivo work was to identify how cobalt accumulated in tissues after long-term systemic circulation, and specifically answer whether cobalt concentration increased in the brain. For that, we carried out comprehensive time- and dose-response experiments to cobalt treatment. We also sought to understand the underlying molecular changes in the brain at the transcriptional level in laboratory rats after prolonged dose-response exposure. Research on MoM cobalt-induced systemic toxicity is scarce and to our knowledge, there have been few publications addressing neurological manifestations in vivo. We hope to generate and test hypotheses through RNA-Sequencing (RNA-Seq) to gain mechanistic insights into the modes of action of cobalt.
2. Materials and Methods
2.1. Experimental Animals and Research Design
Experiments were performed in adult male Sprague Dawley (SD) rats obtained from Charles River (UK). The body weight range at the start of the experiments was 210–280 g. Food and water were provided ad libitum. Their body weight and general aspects of health were monitored daily.
Freshly-made cobalt chloride hexahydrate (CoCl2.6H2O; Sigma-Aldrich, Dorset, UK) solutions dissolved in distilled water (dH2O) and dH2O were sterilised through a 0.22 μm syringe-driven filter (Merck Millipore, Watford, UK).
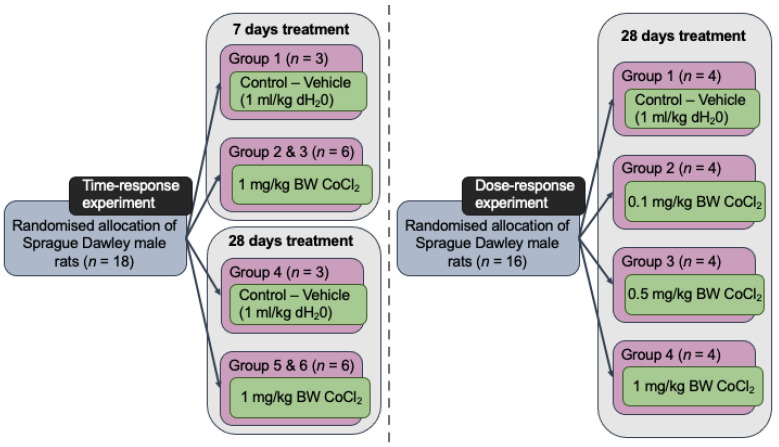
Two separate in vivo experiments were performed successively. The first experiment was a time-response experiment in which a 7-day cobalt treatment was compared against a 28-day treatment at a fixed cobalt dose. Animals were treated daily with either 1 mL/kg body weight dH2O (controls) or 1 mg/kg body weight (BW) CoCl2 doses injected i.p. (treated groups). The second in vivo experiment was a dose-response experiment. The animals were given 1 mL/kg dH2O i.p. in the case of the control group, or a range of cobalt solutions- 0.1, 0.5, or 1 mg/kg BW i.p. injections for 28 days. Figure 1 depicts the sample size and design information diagrammatically.
Figure 1.
Design of in vivo time- and dose-response experiments showing group distribution and sample size. All injections, both control and Co-treated rats, were carried out intraperitonially.
2.2. Sacrifice of Animals and Tissue Harvest
At the end of the exposure time, animals were killed by carbon dioxide (CO2) asphyxiation in a CO2 chamber. Each organ or brain part was dissected separately, weighed, and sections of tissue were stored appropriately to preserve its metal content and molecular RNAs. Blood samples were collected through cardiac puncture immediately before death and mixed with 200 µL of heparin (1000 IU/mL diluted 1:10; Sigma-Aldrich, Dorset, UK) to avoid clotting.
2.3. Tissue Cobalt Content Measured by ICP-MS
Quantification of cobalt content in all organs was obtained via inductively coupled plasma mass spectrometry (ICP-MS) analysis. For this, 100 mg of tissue from each organ/area of interest was taken and stored at −80 °C until further sample digestion. To obtain a liquid solution of the samples suitable for metal detection, 0.5 mL concentrated nitric acid was used per sample (HNO3; TraceSELECTTM Ultra, Sigma-Aldrich (Fluka), Dorset, UK) followed by 0.25 mL 30% hydrogen peroxide (w/w) (H2O2; Sigma-Aldrich, Dorset, UK). Each reagent was to left act for 20 min at 100 °C in a hot block to ensure the decomposition of the matrix. A quantity of 0.25 mL was transferred together with 9.75 mL ultrapure water into acid-washed tubes to avoid trace metal contamination. Standard dilutions were prepared from Cobalt Standard for atomic absorption spectrometry stock (TraceCERT®, Sigma-Aldrich (Fluka), Dorset, UK). The 1:40 dilution samples were serially introduced to the Agilent 7700× octopole collision ICP-MS system (Agilent Technologies, Cheadle, UK). Scandium or indium was used as the internal standard and data were obtained from the maximum signal with the ICP-MS operating in helium mode.
2.4. RNA Extraction
2.4.1. Isolation of RNA
To reduce RNA degradation, 50 or 100 mg from specific segments of brain tissue were dissected and placed in RNase-free tubes with 0.5 mL of RNAlater (Ambion, Life Technologies, Paisley, UK). RNase-free aerosol-resistant filter tips were used for all pipetting steps. Tissues were further cut into thin pieces and completely submerged in the stabilisation solution for overnight storage at 4 °C. RNAlater reagent was removed the next day and the tissues were frozen at −80 °C until RNA extraction.
Tissue was then resuspended in 1 mL QIAzol lysis reagent (Qiagen, Crawley, UK) and a cone ball steel bead was placed inside the tube. The tissue was disrupted with a horizontal Retsch MM200 Mixer Mill (Retsch GmbH, Haan, Germany) set at 30 Hz for 1 min intervals (×3). The lysate was transferred to another tube with 4 mL of QIAzol lysis reagent and the contents vortexed. The pre-processed samples were further prepared according to the protocol of the RNeasy Plus Universal Midi Kit (Qiagen, Crawley, UK).
2.4.2. Quality Check of RNA Samples
RNA purity characterised by the ratio of the absorbances 260/280 nm and 260/230 nm as well as nucleic acid concentration were quantified with a Nanodrop-2000c spectrophotometer (Labtech International, Heathfield, UK). Sample integrity was assessed through microfluidic chip-based analysis (Experion RNA StdSensChip; Bio-Rad, Watford, UK) in the Experion Automated Electrophoresis System (Bio-Rad, Watford, UK) and evaluated with the RNA Quality Indicator (RQI) number [29]. The results of these analyses for pref. cortex, cerebellum, and hippocampus are displayed in Tables S1–S3, respectively, in the Supplementary Materials.
2.5. RNA Sequencing (RNA-Seq)
2.5.1. Sample Pooling for RNA-Sequencing
To reduce the RNA-Seq costs, four biological samples for each comparison group were pooled into a single sample. The necessary volume from each sample was adjusted to obtain an RNA quantity of 5–20 μg depending on the brain area yield. The individual sample volumes were pooled into the corresponding RNase-free tubes for each group and the final contents were briefly vortexed.
2.5.2. RNA-Sequencing Analysis by BGI
RNA-sequencing of the pooled samples was performed by BGI Tech Solutions (Hong Kong, China) Co. Ltd. using a BGISEQ-500 sequencing platform with depth of 20 million base pairs (Mb) clean reads per sample and a 50 single-end bases (50SE) read length. Filtering of clean reads and their mapping to the UCSC rn6 rat reference genome were carried out by BGI, in addition to the quantification of gene fold-change.
2.5.3. Gene Ontology (GO) and KEGG Pathway Enrichment Analysis
Further bioinformatic analyses such as hierarchical clustering or generation of Venn diagrams were performed in MATLAB software (The MathWorks Inc. Natick, MA, USA. Release R2021a Update 5, 9.10.0.1739362). The software tool Cytoscape and its plugin ClueGO were used to conduct the Gene Ontology analysis of the DEGs [30,31]. ClueGO allows for a comparison to different reference ontology sets such as Molecular Function (GO MF; 8 April 2016), Biological Process (GO BP; 8 April 2016), and Cellular Component (GO CC; 8 April 2016), which describe specific gene function and cellular location aspects of gene activity as well as the Kyoto Encyclopaedia of Genes and Genomes (KEGG; 14 June 2016) for the specific enrichment of known pathways. Additionally, other online software was used to create protein–protein interaction (PPI) networks of gene-protein products: STRING (https://string-db.org/) [32] (last accessed: 24 December 2020).
2.6. Quantitative Real-Time Polymerase Chain Reaction (RT-qPCR)
Real-time quantitative PCR (RT-qPCR) was used to obtain the expression of genes of interest in individual samples in order to validate the RNA-Seq results from the pooled samples. All RT-qPCR experiments were performed in accordance with the minimum information for the publication of quantitative real-time PCR experiments (MIQE) guidelines [33]. The checklist for RT-qPCR assays performed with samples from the in vivo experiments is fully displayed in Table S4 in the Supplementary Materials.
The LunaScript RT SuperMix Kit (New England Biolabs, Hitchin, UK) was utilised to generate cDNA for the dose-response experiment samples. To each RT+ or RT- reaction, enough volume of each sample was added to attain 1 μg RNA in each tube. The thermal cycler (Model 480, Perkin Elmer, Warrington, UK) was pre-heated at 25 °C, and primers were allowed to anneal for 2 min at this temperature, continuing at 55 °C for cDNA synthesis during 10 min, and the final RT inactivation at 95 °C for 1 min. After cooling down the samples, the cDNA samples were stored at −20 °C until further use for RT-qPCR reactions.
The primer sequences were designed to span an intron or exon–exon junction through the NCBI Primer-BLAST tool (https://blast.ncbi.nlm.nih.gov/Blast.cgi) to selectively amplify cDNA (last accessed: 3 March 2020). The primer design criteria followed is displayed in Table S5 in the Supplementary Materials. Oligos were synthesised and purchased from Integrated DNA Technologies (IDT, Leuven, Belgium). The genes with their accession number and primer sequence, amplicon length, and melting temperature are presented in Table S6.
PCR experiments were performed with triplicate RT+ per sample, one no-reverse transcriptase control (RT-) per sample, and one no-template control (NTC) per gene and with the corresponding reference gene controls. The SYBR Green detection method was used for the detection of amplification with the kit PowerUpTM SYBRTM Green Master Mix (Applied Biosystems, Thermo Fisher Scientific, Paisley, UK). The forward and reverse primers (10 pM/μL) and molecular-grade water were mixed with Master Mix (2×). Samples consisting of 1 μL cDNA were pipetted into MicroAmp Fast Optical 96-well reaction plates (Applied Biosystems, Paisley, UK) with the previous mix to a final volume of 20 μL per well. The specific thermal cycling parameters (Table S7) were set according to the optimised PowerUpTM SYBRTM Green Master Mix for fast cycling mode in the StepOnePlus Real-Time PCR system (Applied Biosystems, Paisley, UK). At the end of the cycling process, a melt curve was produced and inspected for the occurrence of primer dimers, misprimes, and possible contamination of genomic DNA.
The method used to calculate the relative fold-change was the comparative CT method [34], with CT being the threshold cycle detected over the 40 run cycles. For gene normalisation, the expression of typical brain reference genes Ywhaz, Tbp, and Pes1 was studied. Primer sequences are shown in Table S8. After quantification, the RefFinder web-based tool: https://www.heartcure.com.au/reffinder/ [35] was used to define the most stable reference gene for each tissue (last accessed: 2 March 2020). Full results of RefFinder analyses for each tissue are shown in Figures S1 and S2 in the Supplementary Materials.
2.7. Statistics
Shapiro–Wilk and Levene’s tests were used as preliminary methods to evaluate data normality and homogeneity of variances, respectively. Independent samples t-test was performed to establish statistical comparisons between the two groups (i.e., control and treatment groups). For statistical comparisons between more groups, the tests selected were one-way analysis of variance (ANOVA) together with the Dunnett’s post-hoc test. Statistical significance was declared when p-value < 0.05. The statistical software used was IBM SPSS Statistics 25 (IBM Corp. Armonk, NY, USA. Released 2017. IBM SPSS Statistics for Macintosh, version 25.0).
3. Results
3.1. Cobalt Accumulates in Organs and in Different Brain Structures
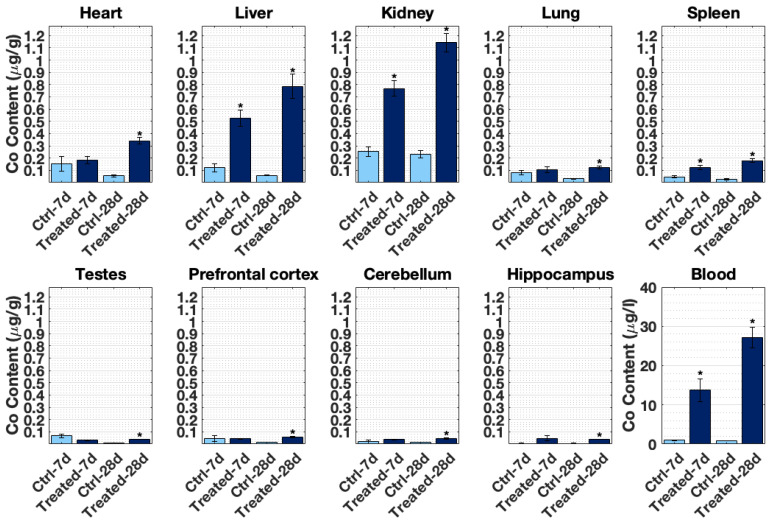
Figure 2 displays cobalt accumulation in all tissues tested and blood after 7- and 28-day treatments. ICP-MS results reveal that kidney, liver, and heart incorporated the highest Co content among the rats’ organs in that order. After 28 days, pref. cortex, cerebellum, and hippocampus assimilated significant amounts of cobalt (p < 0.01, compared with the control rats). This was not the case for the 7-day treatment. Co levels detected in blood from the treated rats by ICP-MS were within the range found in MoM patients. After 28 days, Co detected in rat blood was 27.14 ± 2.70 µg/L compared with the average levels of 1–2 µg/L reported in MoM hip resurfacing patients [36]. The maximum cobalt concentration found in a patient with a THR prosthesis was 6521 µg/L [37].
Figure 2.
Cobalt content in SD male rats’ tissues (ng/g) and blood (μg/L) at 7- and 28-days of daily i.p. CoCl2 injection treatment as assessed by ICP-MS analysis. Control groups were instead injected with distilled water following the same procedures. Figure presents mean ± SEM calculated from n = 3 samples in control groups (dH2O) and n = 6 in treatment groups (1 mg/kg B.W. CoCl2). * significantly different between control group and treatment group at a given time-point as assessed by two sample t-test (p < 0.05).
Several tissues in Figure 2 presented increased cobalt accumulation from 7- to 28-days, indicating possible cobalt time-dependent accumulation in the heart, liver, kidney, spleen, pref. cortex, and blood. However, many of the control samples in the 7-day treatment indicated high levels of cobalt (heart, liver, kidney, lung, spleen, testes, pref. cortex, and cerebellum). This augmentation is likely an artefact of ICP-MS measurements. For values closer to the Co detection limit, it could be that cobalt content is pushed to higher values.
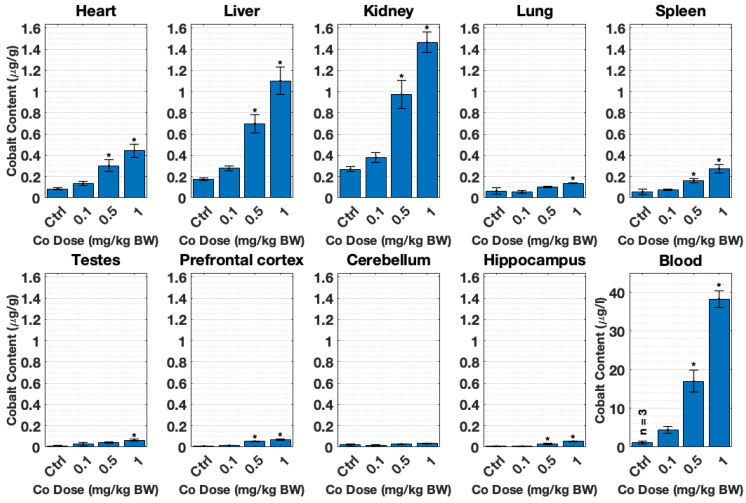
Figure 3 shows the cobalt content detected by ICP-MS in all organs after 28 days of cobalt dose-treatment: 0.1, 0.5, and 1 mg/kg B.W. CoCl2. This metal content analysis revealed an incremental accumulation of cobalt concentrations in tissues with increased doses. Thus, there is a dose-response accumulation of cobalt, which was significant after 0.5 mg/kg B.W. CoCl2 in most tissues. Kidney, liver, and heart in that order accumulated most cobalt in line with the time-response experiment trend (Figure 2). The pref. cortex and hippocampus also accumulated significant levels of cobalt (p < 0.01). The same issue was observed with regard to the offset in the control samples, which were closer to the detection limit of the ICP-MS due to their low cobalt content. This effect became obvious in the case of the cerebellum control group.
Figure 3.
Organ cobalt content (µg/g, tissue; µg/L, blood) obtained by ICP-MS after tissue and blood collection. SD male rats were treated with dH2O (control group) or different doses of CoCl2: 0.1, 0.5, and 1 mg/kg B.W. Animals were dosed daily with i.p. injections for 28 days. Each group presents mean ± SEM from n = 4 rats, and * significant differences in control and treatment means as tested by one-way ANOVA (p < 0.05).
3.2. The Transcriptional Response to the Cobalt Doses Selected Is Non-Proportional
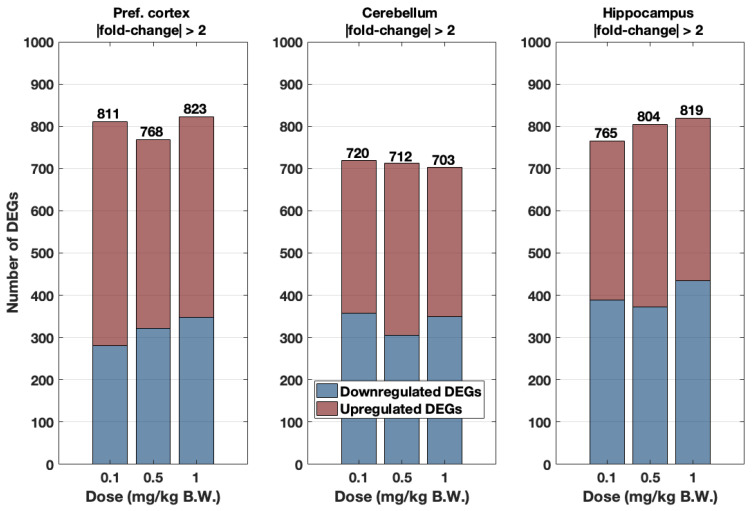
The cobalt tissue content measured through ICP-MS determined that the pref. cortex and hippocampus of 0.5 and 1 mg/kg BW CoCl2-treated groups had significantly greater accumulated cobalt compared to their control groups (Figure 3). However, to evaluate whether the incremental dose cobalt treatment resulted in a progressive transcriptomic response, the number of genes was plotted for all brain areas and the three cobalt treatments: 0.1, 0.5, and 1 mg/kg B.W. CoCl2 in Figure 4 (threshold = |fold change| > 2). Although the number of DEGs progressively increased in the hippocampus, there was no clear dose-response in terms of the number of DEGs in the pref. cortex and cerebellum.
Figure 4.
Number of upregulated (red) and downregulated (blue) DEGs (cutoff |fold change| > 2 only) in the pref. cortex, cerebellum, and hippocampus according to cobalt dose treatment: 0.1, 0.5, and 1 mg/kg B.W. CoCl2. Animals were dosed i.p. daily for 28 days with those doses or dH2O. Data were extracted from RNA-Seq experiments in which n = 4 samples were pooled to obtain n’ = 1, except in the case of the hippocampus treatment group 0.5 mg/kg B.W. CoCl2, where n’ = n = 3 as well as for 1 mg/kg B.W. CoCl2 where n’ = n = 1.
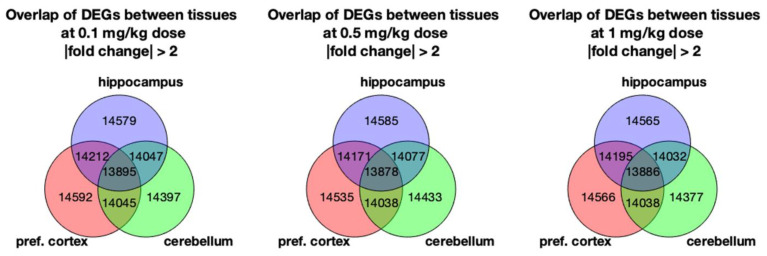
Figure 5 shows the Venn diagram of DEGs found in the pref. cortex, cerebellum, and hippocampus. The diagram intersections display common DEGs between groups, with the pref. cortex and hippocampus demonstrating a higher number of common genes than the cerebellum for every dose. Although the number of overlapping genes between brain areas indicate that the hippocampus, cerebellum, and pref. cortex are all part of the same tissue (i.e., the brain), the different DEGs also point towards regionalised areas with specific functions. It can also be appreciated that there were few differences in the number of common genes between CoCl2 dose regimes. Moreover, it was not possible to identify sets of DEGs that followed a dose-response fold change after hierarchical clustering of the common genes (Figure S3). Thus, we can conclude that the number and range of the doses used does not prompt a dose-response. This does not mean that the transcriptional response elicited is not relevant.
Figure 5.
Venn diagrams showing the number of overlapping DEGs between the pref. cortex, cerebellum, and hippocampus at the different cobalt treatment doses: 0.1, 0.5, and 1 mg/kg B.W. CoCl2. Rats were treated by daily i.p. injection for 28 days. DEGs were obtained through RNA-Seq by comparing the brain parts’ mRNA abundance of the treatment groups against the controls (dH2O-treated).
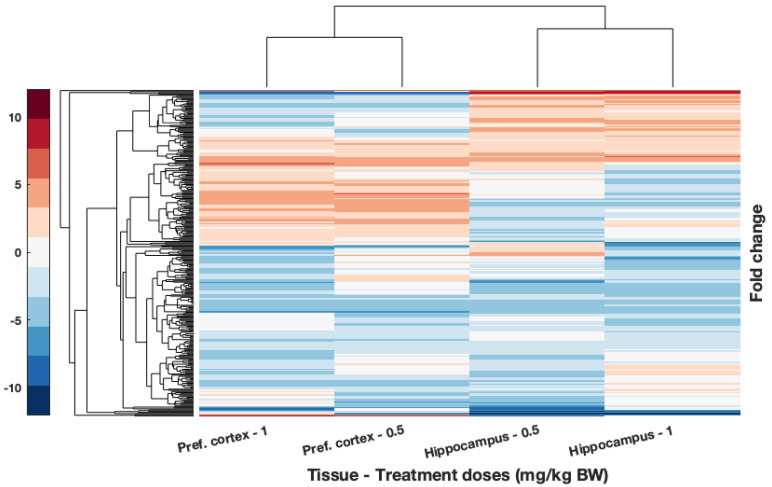
3.3. Global Transcriptional Response in the Pref. Cortex and Hippocampus
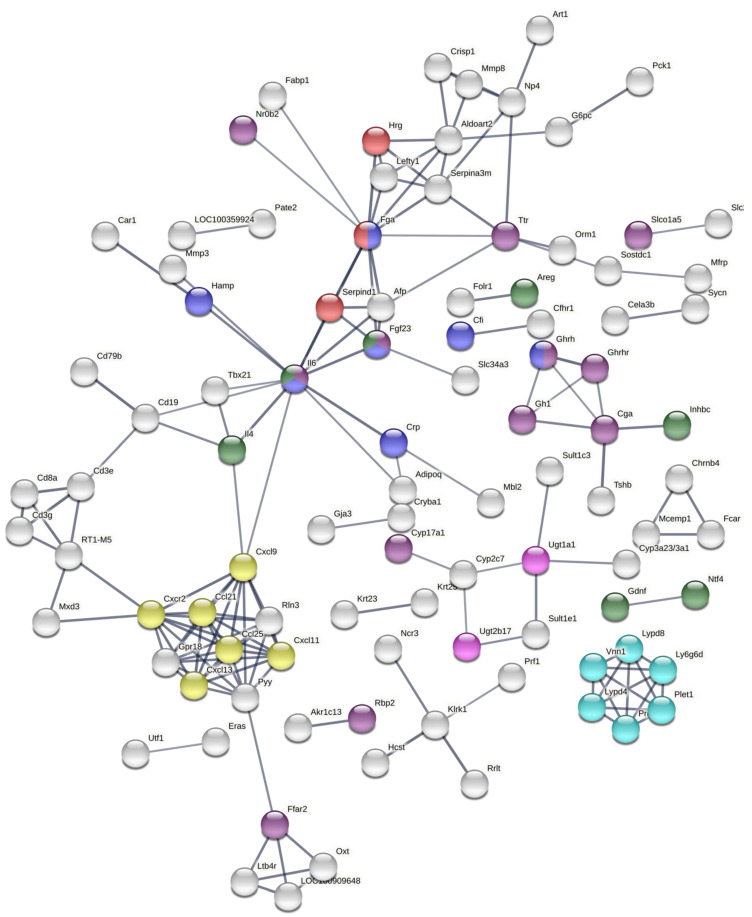
To further explore the effects on gene expression according to the dosage, this study focused on common DEGs for tissues with significant metal content accumulation. According to the results from the ICP-MS analyses, these tissues are the pref. cortex and hippocampus of rats dosed with 0.5 and 1 mg/kg B.W. CoCl2 (Figure 3). The result of this hierarchical clustering is displayed in Figure 6. The gene enrichment analysis of these genes generated with Cytoscape is displayed in Table 1. Several GO terms of importance involved in immunity and hormone activity could be observed. In addition, the protein–protein interaction (PPI) of DEGs–protein products was created to observe the possible links between the overlapped DEGs (Figure 7). The immune axis centred around interleukin-6 (IL6) was clearly separated from other clusters related to growth factors and hormone activity as well as some UDP-glucuronosyltransferases, specifically UGT enzymes that are involved in glucuronidation. Another cluster of interest reflected on the PPI is the glycosylphosphatidylinositol (GPI) anchor biosynthesis.
Figure 6.
Hierarchical clustering of DEGs from brain tissues with significant accumulation of cobalt: the pref. cortex and hippocampus from rats treated with 0.5 and 1 mg/kg B.W. CoCl2. DEGs were obtained from RNA-Seq comparing the RNA isolated from those tissues with those of controls treated with dH2O. Condition applied is for fold change to be over 2. Upregulated genes are shown in red while downregulated are displayed in blue. Hierarchical clustering and resulting dendrogram were generated with Euclidian distance. Samples analysed through RNA-Seq were pooled (n’ = 1) from n = 4 pref. cortex samples, n = 3 in hippocampus from 0.5 mg/kg B.W. CoCl2 treatment group, and n = 1 from 1 mg/kg B.W. CoCl2 treatment group.
Table 1.
Enriched GO terms obtained from DEGs of the pref. cortex and hippocampus in response to cobalt treatment with 0.5 and 1 mg/kg B.W. CoCl2 compared to control animals (dH2O). Rats were dosed for 28 days with i.p. injections. GO terms annotated were significantly enriched with p < 0.05. GO terms enriched belong to the Molecular Function (MF; 8 April 2016), Biological Process (BP; 8 April 2016), Cellular Component (CC; 8 April 2016), and KEGG (14 June 2016) GO databases.
| Gene Ontologies (GO) and GO Terms | Number of Genes with Annotations in the Ontology (%) | Number of Genes Represented in GO Terms (%) |
|---|---|---|
| Biological Process (BP) GO terms (8 April 2016) | 194 (72.12%) | 93 (34.57%) |
| Regulation of hormone levels | Afp, Bco1, Bik, Cga, Cyp11a1, Cyp17a1, Fam3b, Ffar2, Fga, Fgf23, Gh1, Ghrh, Ghrhr, Il6, Nr0b2, Nt5c1b, Pax8, Slc30a8, Slco1a5, Sult1e1 | |
| Response to interleukin-6 | Crp, Fga, Fgf23, Ghrh, Hamp, Il6, Pck1 | |
| T-helper 17 cell lineage commitment | Batf, Il6, Ly9 | |
| Response to vitamin | Cyp11a1, Fgf23, Folr1, Hamp, Orm1, Otc, Sult2a1, Tshb | |
| Response to pH | Acer1, Gh1, Gja3, Kcnk18, Pck1 | |
| Response to interleukin-1 | Ccl21, Ccl25, Cyp11a1, Il6, Mmp3, Pck1, Slc30a8 | |
| Cell chemotaxis | Ccl21, Ccl25, Ccr6, Cxcl13, Cxcl9, Cxcr2, Ffar2, Hrg, Stap1 | |
| Organ formation | Folr1, Foxh1, Gdnf, Ntf4, Pax8 | |
| Cell fate commitment | Batf, Elf5, Gata5, Gsx1, Gsx2, Il6, Ly9, Myl2, Ntf4, Olig3, Sostdc1 | |
| Molecular Function (MF) GO terms (8 April 2016) | 185 (68.77%) | 60 (22.3%) |
| Cytokine receptor binding | Bmp10, Ccl21, Ccl25, Cxcl13, Cxcl9, Gh1, Il6, Inhbc, Ntf4, Stap1 | |
| Heparin binding | Ang2, Comp, Cxcl13, Hrg, Mcpt4, Serpind1, Wisp3 | |
| Growth factor activity | Areg, Bmp10, Fgf23, Gdnf, Il6, Inhbc, Ntf4 | |
| Steroid binding | Comp, Crp, Cyp11a1, Fabp1, Sult1e1, Ugt1a1 | |
| Hormone activity | Bmp10, Cga, Gh1, Ghrh, Gpha2, Hamp, Inhbc, Pyy, Rln3, Tshb | |
| KEGG terms (14 June 2016) | 97 (36.06%) | 29 (10.78%) |
| Steroid hormone biosynthesis | Cyp11a1, Cyp17a1, Cyp2c7, Cyp3a23/3a1, Sult1e1, Ugt1a1, Ugt2b17 | |
| Hematopoietic cell lineage | Cd19, Cd3g, Cd8a, Fcer2, Il1r2, Il6 | |
| Cellular Component terms (8 April 2016) | 216 (80.3%) | 18 (6.69%) |
| External side of plasma membrane | Cd19, Cd8a, Cxcl9, Fcer2, Fga, Folr1, Hyal5, Il6, Itgad, Trpm8 | |
Figure 7.
Protein–protein interaction (PPI) network obtained from the STRING web tool by analysing DEGs as their protein products. DEGs were obtained from RNA-Seq analyses of the pref. cortex and hippocampus from rats treated with 0.5 and 1 mg/kg B.W. CoCl2 against controls treated with dH2O for 28 days of i.p. injections. The following terms/keywords have been highlighted: cellular response to interleukin-6 (blue), regulation of hormone levels (purple), chemokine receptors bind chemokines (yellow), blood coagulation (red), UDP-glucuronosyltransferase activity (pink), growth factor activity (green), and post-translational modification: synthesis of GPI-anchored protein (cyan). The thickness of links between nodes represent the confidence in the interaction, only nodes connected with high confidence (0.7) are displayed.
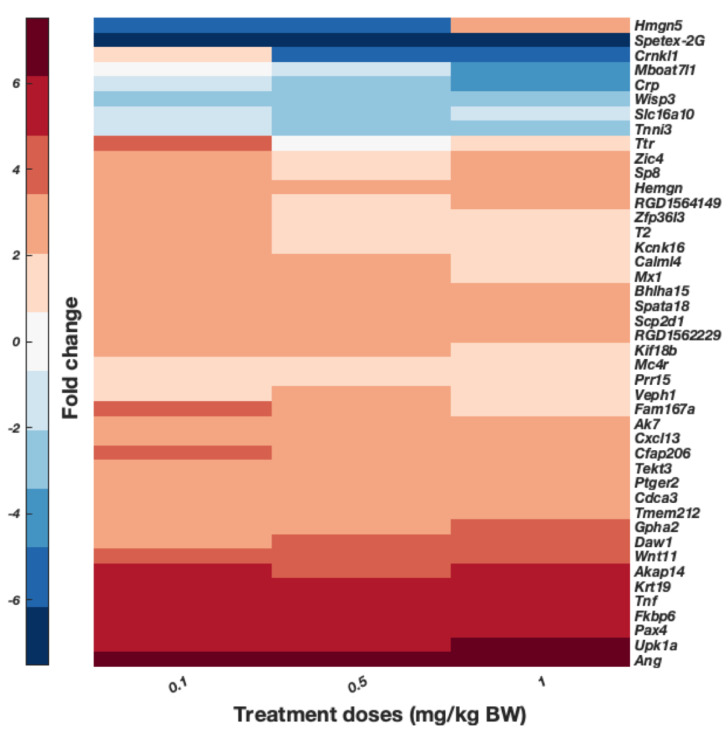
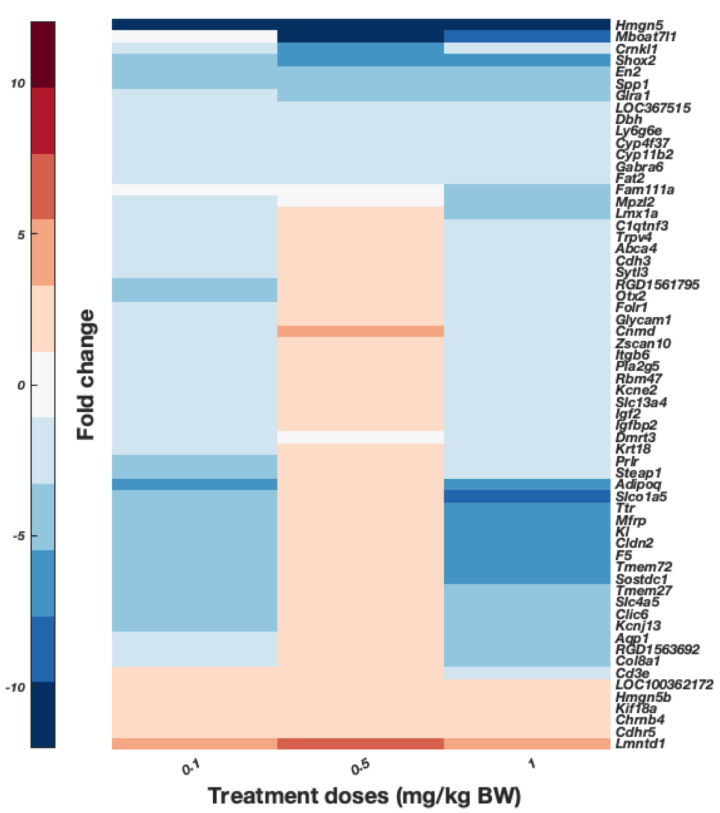
To better observe genes possibly involved in cobalt toxic mechanisms, the DEGs expressed over 2-fold change from the pref. cortex and hippocampus were plotted, as shown in Figure 8 and Figure 9, respectively. For display purposes only, DEGs with a significance threshold <0.05 obtained from the Poisson distribution and provided in the original RNA-Seq data are shown. These are displayed as arranged by hierarchical clustering analyses of the DEGs (dendrogram not shown). Figure 8 displays the common genes of the pref. cortex across the three doses, and presents several genes whose proteins have a role in inflammation and immunity such as Crp, Tnf, and Cxcl13. Figure 9 shows significant DEGs of the hippocampus over 2-fold change. Surprisingly, there are several markers attributed to the choroid plexus such as Clic6, Ttr, Kl, Col8a1, and others [38,39,40].
Figure 8.
DEGs obtained from the comparison of pref. cortex from rats dosed with 0.1, 0.5, and 1 mg/kg B.W. CoCl2 against the control group (dH2O). Animals were treated for 28 days with daily i.p. injections. DEGs displayed were obtained from the RNA-Seq analysis of pooled samples (n’ = 1 from n = 4 samples per group). Fold-change gene expression is indicated by colour, as described by the bar in the right side, upregulated genes are displayed in red while downregulated are blue. Genes are displayed as determined by the hierarchical clustering of DEGs over 2-fold-change (p < 0.05 from RNA-Seq Poisson distribution), dendrogram not shown (Euclidian distance).
Figure 9.
DEGs expressed in hippocampus of rats treated via i.p. with daily injections of 0.1, 0.5, and 1 mg/kg B.W. CoCl2 or dH2O (control groups) for 28 days. Pooled samples (n’ = 1 from n = 4 samples in group 0.1 mg/kg B.W. CoCl2, n = 3 from 0.5 mg/kg B.W. CoCl2, and n = 1 from 1 mg/kg B.W. CoCl2 group) were analysed through RNA-Seq and data are presented as the result of hierarchical clustering. DEGs in the graph are only those with fold-change >2 and p < 0.05 from RNA-Seq Poisson distribution, dendrogram is not shown. Colour bar presents fold-change: upregulated genes in red and downregulated genes in blue.
A number of genes were evaluated via RT-qPCR according to their high expression level in RNA-Seq data as well as their function. The genes selected were Tnf, Spata18, Ttr, and Akap14 in the pref. cortex and Kl in the hippocampus (see Figures S4 and S5 in the Supplementary Materials). In general, the RT-qPCR gene expression results agreed with the RNA-Seq data, and the fold expression of evaluated DEGs from RNA-Seq and RT-qPCR in this study did correlate. This is consistent with previous high-throughput comparisons between the two technologies [41] as well as research with pooled samples [42]. Thus, it was considered that the RT-qPCR results validate the RNA-Seq results.
4. Discussion
4.1. Metal Distribution Pattern Is Consistent with Previous Research and Cobalt Accumulates Significantly in the Brain
Figure 2 shows that there is a time-dependent cobalt accumulation trend in most tissues. This time-dependent trend might indicate that the longer cobalt levels remain elevated in blood, with a higher deposition of cobalt in nearly all organs tested, particularly in the heart, liver, and kidney. The accumulation of cobalt in most organs also appears to be proportional to the dose used, as displayed in Figure 3. In addition, cobalt levels were significantly increased in the pref. cortex and hippocampus of rats dosed with 0.5 and 1 mg/kg B.W. CoCl2 at 28 days.
The liver, kidney, and heart accumulated more metal than other organs (Figure 2 and Figure 3), which correlates with cobalt organ concentration in humans following cobalt radioisotope distribution [13]. This cobalt distribution follows the same pattern as previous experiments conducted by this research group [43,44] and other teams investigating cobaltism [45]. Most cobalt is excreted through urine [13,46], and given that both kidney and liver are involved in toxin and waste detoxification, it is not surprising that cobalt is predominantly concentrated in these tissues.
With regard to the cobalt in blood after the 28-day treatment with 1 mg/kg B.W., the level was 27.15 ± 2.70 µg/L in the time-response experiment while it was 38.24 ± 2.14 µg/L in the dose-response experiment. Given that the blood cobalt levels were under 100 µg/L, only subtle or moderate neurotoxicity was expected [1].
The cobalt content of two hearts from MoM patients severely affected by systemic cobalt toxicity has been reported in the literature as 4.75 [47] and 8.32 µg/g [48] (reference values for heart cobalt content: 0.06 µg/g) [49]. In addition, other post-mortem analyses on patients with metal-on-polyethylene hip prostheses also revealed significantly elevated cobalt content averaging 0.12 µg/g (range: 0.006–6.299 µg/g) [49]. Table 2 shows a compact presentation of these figures and results for comparison. The higher values of 0.34 ± 0.03 µg/g and 0.44 ± 0.06 µg/g in the rats’ hearts at 28 days with 1 mg/kg B.W. CoCl2 treatment were also close to the average content in asymptomatic metal-on-polyethylene (MoP) patients (0.12 µg/g) [49]. Hence, the gradual dosing model used by this study and in previous studies [43] is comparable to the long-term systemic exposure of cobalt through circulating blood in MoM patients.
Table 2.
Cobalt concentrations of whole blood (WB), brain and heart tissues in the dose and time-response experiments and other studies that mimic gradual cobalt release through daily treatment [45], cobalt tissue analyses in the cardiac tissue, and serum of MoM patients with cobaltism [47,48] as well as post-mortem heart tissue from metal-on-polyethylene (MoP) patients [49]. Cobalt content values in unexposed human brain (<0.025 µg/g), heart (0.060 µg/g) and blood (<1 µg/L) were obtained from [36,49,50] in that order. The abbreviations are: i.p., intraperitoneal injections; WB, whole blood; avg., average; C, control; T, treatment; THA, total hip arthroplasty. * significantly different control and treatment groups as assessed by one-way ANOVA with Dunnett’s multiple comparison.
| Studies | Time-Response Study | Dose-Response Study | [45] | [47] | [48] | [49] | |
|---|---|---|---|---|---|---|---|
| Study outline | CoCl2 | CoCl2 | CoCl2 | MoP Patients | |||
| SD rats | SD rats | rabbits | MoM patient | MoM patient | |||
| 28 days | 28 days | 18 days | |||||
| 1 mg/kg BW | 1 mg/kg BW | 1354 µg/mL | |||||
| i.p. | i.p. | Intravenous infusion | |||||
| Tissues | n = 3 (C) and 6 (T) | n = 4 (C) and 4 (T) | n = 2 (C) and 4 (T) | case report | case report | n = 73 (C), 75 (THA) | |
| WB or serum (µg/L) | Cobalt exposed | 27.15 ± 2.70 * (WB) |
38.24 ± 2.14 * (WB) |
420.9 ± 154.5 (WB) |
192 (serum) |
287.6 (serum) |
Unknown |
| Controls/unexposed | 0.87 ± 0.00 | 1.17 ± 0.32 | 11.7 ± 2.7 | <1 | |||
| Brain (µg/g) |
Cobalt exposed | 0.06 ± 0.00 * (pref. cortex) | 0.07 ± 0.01 * (pref. cortex) | 0.2 ± 0.2 | Unknown | ||
| Controls | 0.02 ± 0.00 | 0.01 ± 0.00 | 0.06 ± 0.04 | <0.025 | |||
| Heart (µg/g) |
Cobalt exposed | 0.34 ± 0.03 * | 0.44 ± 0.06 * | 0.7 ± 0.5 | 4.75 | 8.32 | 0.12 (avg.); range: 0.006–6.299 |
| Controls/unexposed | 0.06 ± 0.01 | 0.09 ± 0.01 | 0.07 ± 0.1 | 0.06 | |||
To our knowledge, there is no information on the cobalt concentrations of MoM patients in the brain, probably due to the difficulty of obtaining samples from patients. In this study, the pref. cortex, and hippocampus had significant cobalt accumulated at 28 days (Figure 2 and Figure 3). A study by Apostoli et al. with rabbits dosed with cobalt for 18 days intravenously reported elevated brain levels, 0.2 ± 0.2 µg/g, from the average control levels, 0.06 ± 0.04 µg/g dry weight, and cobalt whole blood, 420.9 ± 154.5 µg/L (see Table 2 for reference [45]). The histology in several organs only reported damage to the eyes and the auditory systems. The model used for this study had much lower cobalt blood concentration and animals remained in good health, while Apostoli et al. described balance disturbance in rabbits due to vestibular damage. Given the literature and observations of this study, cobalt appeared to induce only subtle or moderate neurotoxicity in this study even when cobalt had significantly accumulated in the rats’ brains.
4.2. The Low Range Cobalt Dosage Used Does Not Lead to a Dose-Response
No dose-response was demonstrated either in the number or in the average fold change of DEGs elicited by cobalt treatment (Figure 4 and Figure S3). Previous studies suggest that the fraction of ionic cobalt remains constant throughout a wide range of cobalt concentrations in the blood due to its albumin binding capacity [13]. If cobalt is being sequestered by albumin, it is likely that the response to any administered cobalt treatment will be dampened. However, it is also possible for cobalt toxic effects to follow a dose response, but the few concentrations used here did not cover such a range of toxicity.
4.3. Overall Transcriptional Effects of Cobalt and the Choroid Plexus as a Target of Cobalt Toxicity
This study found some GO terms of interest that could be associated with cobalt toxicity in Table 1. This is the case of ‘steroid hormone biosynthesis’, which is comprised of a few genes of the Cytochrome P450 (Cyp prefix) family as well as by a couple UDP-glucuronosyltransferases (Ugt prefix) and a sulfotransferase (Sult prefix). The protein products of these gene families form part of the drug metabolism and detoxification pathways. CYPs are also involved in the biosynthesis of serotonin, dopamine [51], and steroid hormones [52]. The findings suggest that CYPs could be a target of cobalt since they normally bind to the heme metal substrate as some metal ions have been observed to inactivate members of the CYP family [53]. Nevertheless, there are other large GO terms linked with hormone homeostasis such as ‘regulation of hormone levels’, ‘steroid binding’, and ‘hormone activity’ in Table 1. Some of these are not directly related to steroid hormones e.g., essential thyroid related genes (Tshb), whose deletion leads to hypothyroidism, and other GO terms such as ‘response to retinoic acid’. Thus, nuclear receptors could possibly be regulated by cobalt. Nuclear receptors may bind steroids, retinoic acid, or thyroid hormones, and this binding depends on the nuclear receptor zinc finger domain. The CYP family synthesises some of the ligands that the nuclear receptors bind to, and further experiments could be performed to ascertain whether cobalt binds to CYPs or to nuclear receptors [54].
The PPI analysis (Figure 7) displayed a regulation of hormone levels consistently with the GO enrichment analysis as well as a network of drug-metabolising enzymes consisting of a few Cyp, Ugt, and Sult genes. The immune and haematopoietic axis centred around Il6 with important chemokine presence can also be observed. This immune response was also reflected in the GO enrichment from Table 1. Some of the immune-related GO terms are ‘response to interleukin-6′, ’T-helper 17 cell lineage commitment’, ‘response to interleukin-1′, ‘cell chemotaxis’, and ‘cytokine receptor binding’, which suggest immune cell differentiation, activity, and migration. Other factors related to blood coagulation such as Hrg, Serpind1, and Fga are displayed in Table 1. Activation of the immune system and dysregulation of haematopoietic transcriptional programmes could lead to several autoimmune and blood disease syndromes. Cobalt was a historical treatment of anaemia, and polycythaemia and skin rashes have been documented as a sporadic result of cobalt treatment [13]. Finally, the PPI also reported the presence of ‘GPI-anchored protein’ related transcripts. This is a post-transcriptional modification that mainly occurs in the endoplasmic reticulum, where most glycosylphosphatidylinositol (GPI) synthesis proteins function [55]. GPI-anchored protein synthesis depends on phospholipids, while the synthesis of steroid hormones is determined by cholesterol, hence, it is suggested here that cobalt modulates lipid metabolism. It is possible that cobalt could modulate lipid metabolism directly as cobalt has been seen to affect the rigidity of lipid membranes such as liposomes [56]. Lipid droplets were present in the spleen of a patient with a CoCr prosthesis [57]. Moreover, intracytoplasmic lipid and lipofuscin accumulation were found in a recent heart biopsy of a patient with arthroprosthetic cobaltism [48].
This study also found genetic markers in the pref. cortex (Figure 8) and hippocampus (Figure 9) almost exclusively attributable to the choroid plexus (e.g., Clic6, Klotho (Kl), transthyretin (Ttr), Veph1, some cilia markers, and Scl transporters). The molecular characterisation of the choroid plexus has only been achieved recently [38], and unfortunately the GO ontologies have not been adequately updated to report its presence. The choroid plexus is anatomically attached to the hippocampus and its joint dissection can go unnoticed when doing a fast isolation, as reported by specialists in the choroid plexus [39,58]. In fact, several studies investigating the effect of drugs or other interventions in the hippocampus have knowingly or unwittingly reported choroid plexus markers [58,59,60]. There are also markers of the choroid plexus such as transthyretin (Ttr) in the pref. cortex, and it is possible that part of the choroid plexus has also been included in brain samples other than the hippocampus, since the choroid plexus is distributed through all brain ventricles [58]. Different studies have revealed that heavy metals preferentially accumulate early on in the choroid plexus, which appears to retain them, thus protecting the brain [61,62,63,64,65,66]. Harrison-Brown et al. discovered that the penetration of cobalt in the cerebrospinal fluid (CSF) of MoM patients was limited to 15% of the cobalt in plasma [67]. They also found a nonlinear trend with a ceiling effect in the CSF cobalt accumulation in relation to Co plasma levels in blood. Thus, the choroid plexus could function as an absorptive barrier, and early cobalt accumulation and damage in the brain might occur in the choroid plexus.
The choroid plexus is also a place for steroid hormone biosynthesis [68] and it hosts metabolising enzymes to deal with and metabolise xenobiotics [69]. Many immune cells are resident in the choroid plexus, which works as the site for immune trafficking with the brain [70]. B and T lymphocytes can infiltrate the choroid plexus and affect its function under certain challenges, thus leading to inflammation [71,72,73]. In particular, the ‘cell chemotaxis’ GO term includes Cxcl13, a chemokine that recruits B lymphocytes, and it has been involved in lymphoid infiltration in the choroid plexus in a mouse model of neuropsychiatric lupus [73]. Moreover, very recently, modulation of Otx2 expression in the choroid plexus has been seen to regulate anxiogenic behaviour in mice [74], and in two studies, the choroid plexus transcriptome quickly responded to stress tests in mice [39,58]. Given that the choroid plexus has been implicated in depression disorders [75] and that some patients with elevated cobalt levels in their blood showed signs of neuropsychiatric symptoms such as depression [8,10], one might speculate that cobalt toxicity in the choroid plexus could impair its function, and contribute towards mood dysregulation.
5. Conclusions
In summary, although the rat pref. cortex and hippocampus accumulated lower amounts of cobalt than other tissues, these accumulations were still significant at similar elevated circulating cobalt levels to those found in some patients with MoM implants. We found that the common transcriptional response to cobalt in the brain areas analysed involved hormone and drug-metabolising activity, in addition to also describing a powerful immune response, perhaps mediated by inteleukin-6 (IL-6). An underlying dysfunction in lipid metabolism is also likely. We suggest that these mechanisms could be instigated as a consequence of cobalt ion binding and substitution of native metal ions of CYPs or nuclear receptors. In the future, researchers should consider evaluating the markers of inflammation and lymphoid cell activation as well as the steroidogenic activity in the choroid plexus in response to cobalt. Thus, we have generated a mechanistic hypothesis for cobalt neurotoxicity that could be explored further and have relevant implications for patients with MoM implants who develop neurological health issues.
Acknowledgments
Katie Henderson, Sarunya Laovitthayanggoon, Laura Beattie, and Ibrahim Alanazi provided invaluable technical and human support during the experiments. We are also grateful for the assistance and technical support of the Biological Procedures Unit (BPU) from the University of Strathclyde.
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/toxics10020059/s1, Table S1. Quality check of RNA samples from the pref. cortex tissue obtained from the dose-response in vivo experiments, Table S2. Quality check of RNA samples from the cerebellum tissues obtained from the in vivo dose-response experiments, Table S3. Quality check of RNA samples from the hippocampus tissues obtained from the in vivo dose-response experiments, Table S4. MIQE checklist from the MIQE guidelines for reproducibility and assessment of experimental RT-qPCR conditions, Table S5. Criteria for the design of the primers selected through NCBI Primer-BLAST tool, Table S6. Primer sequences of targeted genes designed for the in vivo dose-response experiment, Table S7. Fast PCR thermal cycling steps based on PowerUpTM SYBRTM Green Master Mix instructions for StepOnePlus Real-Time PCR system, Table S8: Primer sequences of control genes, Figure S1. RefFinder ranking of three reference genes Ct values in pref. cortex samples from the in vivo dose-response experiment, Figure S2. RefFinder ranking of three reference genes Ct values in hippocampus samples from the in vivo dose-response experiment, Figure S3. Hierarchical clustering of DEGs over 2-fold-change from RNA-Seq data obtained from the comparison of the pref. cortex, cerebellum, and hippocampus of rats treated with three concentrations of cobalt against those of controls treated with dH2O, Figure S4: Fold change mRNA gene expression levels obtained from RNA-Seq and RT-qPCR, Figure S5: Quantification of Tnf, Spata18, Ttr, and Akap14 delta (CT) values (CT target gene–CT internal control) in the pref. cortex and Kl in the hippocampus through RT-qPCR.
Author Contributions
Conceptualization, S.G.-A., R.J.T. and M.H.G.; Methodology, S.G.-A., R.J.T. and M.H.G.; Software, S.G.-A.; Validation, S.G.-A.; Formal analysis, S.G.-A.; Investigation, S.G.-A.; Resources, R.J.T. and M.H.G.; Data curation, S.G.-A.; Writing—original draft preparation, S.G.-A.; Writing—review and editing, S.G.-A., R.J.T. and M.H.G.; Visualization, S.G.-A.; Supervision, R.J.T. and M.H.G.; Project administration, M.H.G.; Funding acquisition, M.H.G. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by an Engineering the Future (ETF) studentship awarded by the University of Strathclyde (internal funding, Faculty of Engineering, University of Strathclyde).
Institutional Review Board Statement
All animal procedures were conducted under the UK Home Office project licenses, 60/4341 and PDE5626B67. Ethic Committee Name: UK Home Office and AWERB. Approval Code: 60/4341 and PDE5626B67 (UK Home Office licence numbers). Approval Date: 25th April 2017.
Informed Consent Statement
Not applicable.
Data Availability Statement
All data underpinning this publication are openly available from the University of Strathclyde KnowledgeBase at https://doi.org/10.15129/cbbe122f-4ee1-4bfb-9ec7-46c664bf10f2 (accessed on 29 November 2021).
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Kovochich M., Finley B.L., Novick R., Monnot A.D., Donovan E., Unice K.M., Fung E.S., Fung D., Paustenbach D.J. Understanding outcomes and toxicological aspects of second generation metal-on-metal hip implants: A state-of-the-art review. Crit. Rev. Toxicol. 2018;48:853–901. doi: 10.1080/10408444.2018.1563048. [DOI] [PubMed] [Google Scholar]
- 2.Langton D.J., Jameson S.S., Joyce T.J., Hallab N.J., Natu S., Nargol A.V.F. Early failure of metal-on-metal bearings in hip resurfacing and large-diameter total hip replacement. J. Bone Jt. Surg. Br. 2010;92:38–46. doi: 10.1302/0301-620X.92B1.22770. [DOI] [PubMed] [Google Scholar]
- 3.Sidaginamale R.P., Joyce T.J., Bowsher J.G., Lord J.K., Avery P.J., Natu S., Nargol A.V.F., Langton D.J. The clinical implications of metal debris release from the taper junctions and bearing surfaces of metal-on-metal hip arthroplasty. Bone Jt. J. 2016;98:925–933. doi: 10.1302/0301-620X.98B7.37029. [DOI] [PubMed] [Google Scholar]
- 4.Goode A.E., Perkins J.M., Sandison A., Karunakaran C., Cheng H., Wall D., Skinner J.A., Hart A.J., Porter A.E., McComb D.W., et al. Chemical speciation of nanoparticles surrounding metal-on-metal hips. Chem. Commun. 2012;48:8335. doi: 10.1039/c2cc33016d. [DOI] [PubMed] [Google Scholar]
- 5.MDA/2017/018. Medical Device Alert. All Metal-On-Metal (MoM) Hip Replacements: Updated Advice For Follow-Up of Patients. [(accessed on 29 November 2021)]; Available online: http://www.mhra.gov.uk/
- 6.Epstein M., Emri I., Hartemann P., Hoet P., Leitgeb N., Martínez Martínez L., Proykova A., Rizzo L., Rodriguez-Farré E., Rushton L., et al. Final Opinion on the Safety of Metal-on-Metal Joint Replacements with a Particular Focus on Hip Implants. Sci. Comm. Emerg. New. Identified Heal. Risks, no. September, 2014. [(accessed on 29 November 2021)]. Available online: https://ec.europa.eu/health/sites/default/files/scientific_committees/emerging/docs/scenihr_o_042.pdf.
- 7.Rizzetti M.C., Liberini P., Zarattini G., Catalani S., Pazzaglia U., Apostoli P., Padovani A. Loss of sight and sound. Could it be the hip? Lancet. 2009;373:1052. doi: 10.1016/S0140-6736(09)60490-6. [DOI] [PubMed] [Google Scholar]
- 8.Tower S.S. Arthroprosthetic Cobaltism: Neurological and Cardiac Manifestations in Two Patients with Metal-on-Metal Arthroplasty: A Case Report. J. Bone Jt. Surg. Am. 2010;92:2847–2851. doi: 10.2106/JBJS.J.00125. [DOI] [PubMed] [Google Scholar]
- 9.Mao X., Wong A.A., Crawford R.W. Cobalt toxicity--an emerging clinical problem in patients with metal-on-metal hip prostheses? Med. J. Aust. 2011;194:649–651. doi: 10.5694/j.1326-5377.2011.tb03151.x. [DOI] [PubMed] [Google Scholar]
- 10.Green B., Griffiths E., Almond S. Neuropsychiatric symptoms following metal-on-metal implant failure with cobalt and chromium toxicity. BMC Psychiatry. 2017;17:33. doi: 10.1186/s12888-016-1174-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Catalani S., Rizzetti M.C., Padovani A., Apostoli P. Neurotoxicity of cobalt. Hum. Exp. Toxicol. 2012;31:421–437. doi: 10.1177/0960327111414280. [DOI] [PubMed] [Google Scholar]
- 12.Machado C., Appelbe A., Wood R. Arthroprosthetic Cobaltism and Cardiomyopathy. Hear. Lung. Circ. 2012;21:759–760. doi: 10.1016/j.hlc.2012.03.013. [DOI] [PubMed] [Google Scholar]
- 13.Paustenbach D.J., Tvermoes B.E., Unice K.M., Finley B.L., Kerger B.D. A review of the health hazards posed by cobalt. Crit. Rev. Toxicol. 2013;43:316–362. doi: 10.3109/10408444.2013.779633. [DOI] [PubMed] [Google Scholar]
- 14.Ho J.H., Leikin J.B., Dargan P.I., Archer J.R., Wood D.M., Brent J. Metal-on-Metal Hip Joint Prostheses: A Retrospective Case Series Investigating the Association of Systemic Toxicity with Serum Cobalt and Chromium Concentrations. J. Med. Toxicol. 2017;13:321–328. doi: 10.1007/s13181-017-0629-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Karovic O., Tonazzini I., Rebola N., Edström E., Lövdahl C., Fredholm B.B., Dare E. Toxic effects of cobalt in primary cultures of mouse astrocytes. Similarities with hypoxia and role of HIF-1α. Biochem. Pharmacol. 2007;73:694–708. doi: 10.1016/j.bcp.2006.11.008. [DOI] [PubMed] [Google Scholar]
- 16.Wang P., Li L., Zhang Z., Kan Q., Chen S., Gao F. Time-dependent homeostasis between glucose uptake and consumption in astrocytes exposed to CoCl2 treatment. Mol. Med. Rep. 2016;13:2909–2917. doi: 10.3892/mmr.2016.4873. [DOI] [PubMed] [Google Scholar]
- 17.Gómez-Arnaiz S., Tate R.J., Grant M.H. Cytotoxicity of cobalt chloride in brain cell lines-a comparison between astrocytoma and neuroblastoma cells. Toxicol. Vitr. 2020;68:104958. doi: 10.1016/j.tiv.2020.104958. [DOI] [PubMed] [Google Scholar]
- 18.Li P., Ding D., Salvi R., Roth J.A. Cobalt-Induced Ototoxicity in Rat Postnatal Cochlear Organotypic Cultures. Neurotox. Res. 2015;28:209–221. doi: 10.1007/s12640-015-9538-8. [DOI] [PubMed] [Google Scholar]
- 19.Kikuchi S., Ninomiya T., Kohno T., Kojima T., Tatsumi H. Cobalt inhibits motility of axonal mitochondria and induces axonal degeneration in cultured dorsal root ganglion cells of rat. Cell Biol. Toxicol. 2018;34:93–107. doi: 10.1007/s10565-017-9402-0. [DOI] [PubMed] [Google Scholar]
- 20.Chimeh U., Zimmerman M.A., Gilyazova N., Li P.A. B355252, a novel small molecule, confers neuroprotection against cobalt chloride toxicity in mouse hippocampal cells through altering mitochondrial dynamics and limiting autophagy induction. Int. J. Med. Sci. 2018;15:1384–1396. doi: 10.7150/ijms.24702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Naves T., Jawhari S., Jauberteau M.O., Ratinaud M.H., Verdier M. Autophagy takes place in mutated p53 neuroblastoma cells in response to hypoxia mimetic CoCl2. Biochem. Pharmacol. 2013;85:1153–1161. doi: 10.1016/j.bcp.2013.01.022. [DOI] [PubMed] [Google Scholar]
- 22.Fung F.K., Law B.Y., Lo A.C. Lutein Attenuates Both Apoptosis and Autophagy upon Cobalt (II) Chloride-Induced Hypoxia in Rat Műller Cells. PLoS ONE. 2016;11:e0167828. doi: 10.1371/journal.pone.0167828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mou Y.H., Yang J.Y., Cui N., Wang J.M., Hou Y., Song S., Wu C.F. Effects of cobalt chloride on nitric oxide and cytokines/chemokines production in microglia. Int. Immunopharmacol. 2012;13:120–125. doi: 10.1016/j.intimp.2012.03.017. [DOI] [PubMed] [Google Scholar]
- 24.McMullen P.D., Pendse S.N., Black M.B., Mansouri K., Haider S., Andersen M.E., Clewell R.A. Addressing systematic inconsistencies between in vitro and in vivo transcriptomic mode of action signatures. Toxicol. Vitr. 2019;58:1–12. doi: 10.1016/j.tiv.2019.02.014. [DOI] [PubMed] [Google Scholar]
- 25.Joseph P. Transcriptomics in toxicology. Food Chem. Toxicol. 2017;109:650–662. doi: 10.1016/j.fct.2017.07.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Garoui E., Amara I.B., Driss D., Elwej A., Chaabouni S.E., Boudawara T., Zeghal N. Effects of Cobalt on Membrane ATPases, Oxidant, and Antioxidant Values in the Cerebrum and Cerebellum of Suckling Rats. Biol. Trace Elem. Res. 2013;154:387–395. doi: 10.1007/s12011-013-9746-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Caltana L., Merelli A., Lazarowski A., Brusco A. Neuronal and Glial Alterations Due to Focal Cortical Hypoxia Induced by Direct Cobalt Chloride (CoCl2) Brain Injection. Neurotox. Res. 2009;15:348–358. doi: 10.1007/s12640-009-9038-9. [DOI] [PubMed] [Google Scholar]
- 28.Kajiwara K., Sunaga K., Tsuda T., Sugaya A., Sugaya E., Kimura M. Peony root extract upregulates transthyretin and phosphoglycerate mutase in mouse cobalt focus seizure. Biochem. Biophys. Res. Commun. 2008;371:375–379. doi: 10.1016/j.bbrc.2008.04.094. [DOI] [PubMed] [Google Scholar]
- 29.Denisov V., Strong W., Walder M., Gingrich J., Wintz H. Development and Validation of RQI: An RNA Quality Indicator for the Experion Automated Electrophoresis System. [(accessed on 29 November 2021)];Bio-Rad Bull. 2008 :5761. Available online: http://www.gene-quantification.org/Bio-Rad-bulletin-5761.pdf. [Google Scholar]
- 30.Shannon P., Markiel A., Ozier O., Baliga N.S., Wang J.T., Ramage D., Amin N., Schwikowski B., Ideker T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bindea G., Mlecnik B., Hackl H., Charoentong P., Tosolini M., Kirilovsky A., Fridman W.-H., Pagès F., Trajanoski Z., Galon J. ClueGO: A Cytoscape plug-in to decipher functionally grouped gene ontology and pathway annotation networks. Bioinformatics. 2009;25:1091–1093. doi: 10.1093/bioinformatics/btp101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Szklarczyk D., Gable A.L., Lyon D., Junge A., Wyder S., Huerta-Cepas J., Simonovic M., Doncheva N.T., Morris J.H., Bork P., et al. STRING v11: Protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019;47:D607–D613. doi: 10.1093/nar/gky1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bustin S.A., Benes V., Garson J.A., Hellemans J., Huggett J., Kubista M., Mueller R., Nolan T., Pfaffl M.W., Shipley G.L. The MIQE Guidelines: Minimum Information for Publication of Quantitative Real-Time PCR Experiments. Clin. Chem. 2009;55:611–622. doi: 10.1373/clinchem.2008.112797. [DOI] [PubMed] [Google Scholar]
- 34.Schmittgen T.D., Livak K.J. Analyzing real-time PCR data by the comparative CT method. Nat. Protoc. 2008;3:1101–1108. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
- 35.Xie F., Xiao P., Chen D., Xu L., Zhang B. miRDeepFinder: A miRNA analysis tool for deep sequencing of plant small RNAs. Plant Mol. Biol. 2012;80:75–84. doi: 10.1007/s11103-012-9885-2. [DOI] [PubMed] [Google Scholar]
- 36.Sidaginamale R.P., Joyce T.J., Lord J.K., Jefferson R., Blain P.G., Nargol A.V.F., Langton D.J. Blood metal ion testing is an effective screening tool to identify poorly performing metal-on-metal bearing surfaces. Bone Jt. Res. 2013;2:84–95. doi: 10.1302/2046-3758.25.2000148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zywiel M.G., Brandt J.M., Overgaard C.B., Cheung A.C., Turgeon T.R., Syed K.A. Fatal cardiomyopathy after revision total hip replacement for fracture of a ceramic liner. Bone Jt. J. 2013;95:31–37. doi: 10.1302/0301-620X.95B1.30060. [DOI] [PubMed] [Google Scholar]
- 38.Lun M.P., Johnson M.B., Broadbelt K.G., Watanabe M., Kang Y.J., Chau K.F., Springel M.W., Malesz A., Sousa A.M.M., Pletikos M., et al. Spatially Heterogeneous Choroid Plexus Transcriptomes Encode Positional Identity and Contribute to Regional CSF Production. J. Neurosci. 2015;35:4903–4916. doi: 10.1523/JNEUROSCI.3081-14.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mathew R.S., Mullan H., Blusztajn J.K., Lehtinen M.K. Comment on ‘Multiple repressive mechanisms in the hippocampus during memory formation. ’ Science. 2016;353:453. doi: 10.1126/science.aaf1288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sathyanesan M., Girgenti M.J., Banasr M., Stone K., Bruce C., Guilchicek E., Wilczak-Havill K., Nairn A., Williams K., Sass S., et al. A molecular characterization of the choroid plexus and stress-induced gene regulation. Transl. Psychiatry. 2012;2:e139. doi: 10.1038/tp.2012.64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Everaert C., Luypaert M., Maag J.L., Cheng Q.X., Dinger M.E., Hellemans J., Mestdagh P. Benchmarking of RNA-sequencing analysis workflows using whole-transcriptome RT-qPCR expression data. Sci. Rep. 2017;7:1559. doi: 10.1038/s41598-017-01617-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Assefa A.T., Vandesompele J., Thas O. On the utility of RNA sample pooling to optimize cost and statistical power in RNA sequencing experiments. BMC Genom. 2020;21:312. doi: 10.1186/s12864-020-6754-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Laovitthayanggoon S., Henderson C.J., McCluskey C., MacDonald M., Tate R.J., Grant M.H., Currie S. Cobalt Administration Causes Reduced Contractility with Parallel Increases in TRPC6 and TRPM7 Transporter Protein Expression in Adult Rat Hearts. Cardiovasc. Toxicol. 2019;19:276–286. doi: 10.1007/s12012-018-9498-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Afolaranmi G.A., Akbar M., Brewer J., Grant M.H. Distribution of metal released from cobalt–chromium alloy orthopaedic wear particles implanted into air pouches in mice. J. Biomed. Mater. Res. A. 2012;100:1529–1538. doi: 10.1002/jbm.a.34091. [DOI] [PubMed] [Google Scholar]
- 45.Apostoli P., Catalani S., Zaghini A., Mariotti A., Poliani P.L., Vielmi V., Semeraro F., Duse S., Porzionato A., Macchi V., et al. High doses of cobalt induce optic and auditory neuropathy. Exp. Toxicol. Pathol. 2013;65:719–727. doi: 10.1016/j.etp.2012.09.006. [DOI] [PubMed] [Google Scholar]
- 46.Daniel J., Ziaee H., Pradhan C., Pynsent P.B., McMinn D.J. Renal Clearance of Cobalt in Relation to the Use of Metal-on-Metal Bearings in Hip Arthroplasty. J. Bone Jt. Surg. Am. 2010;92:840–845. doi: 10.2106/JBJS.H.01821. [DOI] [PubMed] [Google Scholar]
- 47.Martin J.R., Spencer-Gardner L., Camp C.L., Stulak J.M., Sierra R.J. Cardiac cobaltism: A rare complication after bilateral metal-on-metal total hip arthroplasty. Arthroplast. Today. 2015;1:99–102. doi: 10.1016/j.artd.2015.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Allen L.A., Ambardekar A.V., Devaraj K.M., Maleszewski J.J., Wolfel E.E. Missing elements of the history. N. Engl. J. Med. 2014;370:559–566. doi: 10.1056/NEJMcps1213196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wyles C.C., Wright T.C., Bois M.C., Amin S., Fayyaz A., Jenkins S.M., Wyles S.P., Day P.L., Murray D.L., Trousdale R.T., et al. Myocardial cobalt levels are elevated in the setting of total hip arthroplasty. J. Bone Jt. Surg. Am. 2017;99:e118. doi: 10.2106/JBJS.17.00159. [DOI] [PubMed] [Google Scholar]
- 50.Garcia F., Ortega A., Domingo J.L., Corbella J. Accumulation of metals in autopsy tissues of subjects living in Tarragona County, Spain. J. Environ. Sci. Heal. Part. A. 2001;36:1767–1786. doi: 10.1081/ESE-100106258. [DOI] [PubMed] [Google Scholar]
- 51.Ferguson C.S., Tyndale R.F. Cytochrome P450 enzymes in the brain: Emerging evidence of biological significance. Trends Pharmacol. Sci. 2011;32:708–714. doi: 10.1016/j.tips.2011.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pikuleva I.A., Waterman M.R. Cytochromes P450: Roles in Diseases. J. Biol. Chem. 2013;288:17091–17098. doi: 10.1074/jbc.R112.431916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Dixit V.A., Warwicker J., De Visser S.P. How Do Metal Ions Modulate the Rate-Determining Electron-Transfer Step in Cytochrome P450 Reactions? Chem. A Eur. J. 2020;26:15270–15281. doi: 10.1002/chem.202003024. [DOI] [PubMed] [Google Scholar]
- 54.Honkakoski P., Negishi M. Regulation of cytochrome P450 (CYP) genes by nuclear receptors. Biochem. J. 2000;347:321–337. doi: 10.1042/bj3470321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kinoshita T. Biosynthesis and biology of mammalian GPI-anchored proteins. Open Biol. 2020;10:190290. doi: 10.1098/rsob.190290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Umbsaar J., Kerek E., Prenner E.J. Cobalt and nickel affect the fluidity of negatively-charged biomimetic membranes. Chem. Phys. Lipids. 2018;210:28–37. doi: 10.1016/j.chemphyslip.2017.11.016. [DOI] [PubMed] [Google Scholar]
- 57.Urban R.M., Tomlinson M.J., Hall D.J., Jacobs J.J. Accumulation in liver and spleen of metal particles generated at nonbearing surfaces in hip arthroplasty. J. Arthroplast. 2004;19:94–101. doi: 10.1016/j.arth.2004.09.013. [DOI] [PubMed] [Google Scholar]
- 58.Stankiewicz A.M., Goscik J., Majewska A., Swiergiel A.H., Juszczak G.R. The Effect of Acute and Chronic Social Stress on the Hippocampal Transcriptome in Mice. PLoS ONE. 2015;10:e0142195. doi: 10.1371/journal.pone.0142195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Cho J., Yu N.K., Choi J.H., Sim S.E., Kang S.J., Kwak C., Lee S.-W., Kim J.-I., Choi D.I., Kim V.N., et al. Multiple repressive mechanisms in the hippocampus during memory formation. Science. 2015;350:82–87. doi: 10.1126/science.aac7368. [DOI] [PubMed] [Google Scholar]
- 60.Schneider J.S., Anderson D.W., Sonnenahalli H., Vadigepalli R. Sex-based differences in gene expression in hippocampus following postnatal lead exposure. Toxicol. Appl. Pharmacol. 2011;256:179–190. doi: 10.1016/j.taap.2011.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Watanabe T., Natt O., Boretius S., Frahm J., Michaelis T. In vivo 3D MRI staining of mouse brain after subcutaneous application of MnCl2. Magn. Reson. Med. 2002;48:852–859. doi: 10.1002/mrm.10276. [DOI] [PubMed] [Google Scholar]
- 62.Sudarshana D.M., Nair G., Dwyer J.T., Dewey B., Steele S.U., Suto D.J., Wu T., Berkowitz B.A., Koretsky A.P., Cortese I.C.M., et al. Manganese-enhanced MRI of the brain in healthy volunteers. Am. J. Neuroradiol. 2019;40:1309–1316. doi: 10.3174/ajnr.A6152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Steuerwald A.J., Blaisdell F.S., Geraghty C.M., Parsons P.J. Regional Distribution and Accumulation of Lead in Caprine Brain Tissues Following a Long-Term Oral Dosing Regimen. J. Toxicol. Environ. Heal. Part. A. 2014;77:663–678. doi: 10.1080/15287394.2014.880328. [DOI] [PubMed] [Google Scholar]
- 64.Manton W.I., Kirkpatrick J.B., Cook J.D. Does the choroid plexus really protect the brain from lead? Lancet. 1984;324:351. doi: 10.1016/S0140-6736(84)92719-3. [DOI] [PubMed] [Google Scholar]
- 65.Takeda A., Takefuta S., Ijiro H., Okada S., Oku N. 109Cd transport in rat brain. Brain Res. Bull. 1999;49:453–457. doi: 10.1016/S0361-9230(99)00080-5. [DOI] [PubMed] [Google Scholar]
- 66.Nakamura M., Yasutake A., Fujimura M., Hachiya N., Marumoto M. Effect of methylmercury administration on choroid plexus function in rats. Arch. Toxicol. 2011;85:911–918. doi: 10.1007/s00204-010-0623-8. [DOI] [PubMed] [Google Scholar]
- 67.Harrison-Brown M., Scholes C., Field C., McQuilty R., Farah S.B., Nizam I., Kerr D., Kohan L. Limited penetration of cobalt and chromium ions into the cerebrospinal fluid following metal on metal arthroplasty: A cross-sectional analysis. Clin. Toxicol. 2020;58:233–240. doi: 10.1080/15563650.2019.1636993. [DOI] [PubMed] [Google Scholar]
- 68.Quintela T., Goncalves I., Carreto L.C., Santos M.A.S., Marcelino H., Patriarca F.M., Santos C.R.A. Analysis of the Effects of Sex Hormone Background on the Rat Choroid Plexus Transcriptome by cDNA Microarrays. PLoS ONE. 2013;8:e60199. doi: 10.1371/journal.pone.0060199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Gradinaru D., Minn A.L., Artur Y., Minn A., Heydel J.M. Drug metabolizing enzyme expression in rat choroid plexus: Effects of in vivo xenobiotics treatment. Arch. Toxicol. 2009;83:581–586. doi: 10.1007/s00204-008-0386-7. [DOI] [PubMed] [Google Scholar]
- 70.Ghersi-Egea J.F., Strazielle N., Catala M., Silva-Vargas V., Doetsch F., Engelhardt B. Molecular anatomy and functions of the choroidal blood-cerebrospinal fluid barrier in health and disease. Acta Neuropathol. 2018;135:337–361. doi: 10.1007/s00401-018-1807-1. [DOI] [PubMed] [Google Scholar]
- 71.Zhu L., Stein L.R., Kim D., Ho K., Yu G.Q., Zhan L., Larsson T.E., Mucke L. Klotho controls the brain–immune system interface in the choroid plexus. Proc. Natl. Acad. Sci. USA. 2018;115:E11388–E11396. doi: 10.1073/pnas.1808609115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Baruch K., Ron-Harel N., Gal H., Deczkowska A., Shifrut E., Ndifon W., Mirlas-Neisberg N., Cardon M., Vaknin I., Cahalon L., et al. CNS-specific immunity at the choroid plexus shifts toward destructive Th2 inflammation in brain aging. Proc. Natl. Acad. Sci. USA. 2013;110:2264–2269. doi: 10.1073/pnas.1211270110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Stock A.D., Der E., Gelb S., Huang M., Weidenheim K., Ben-Zvi A., Putterman C. Tertiary lymphoid structures in the choroid plexus in neuropsychiatric lupus. JCI Insight. 2019;4:e124203. doi: 10.1172/jci.insight.124203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Vincent C., Gilabert-Juan J., Gibel-Russo R., Alvarez-Fischer D., Krebs M.O., Le Pen G., Prochiantz A., Di Nardo A.A. Non-cell-autonomous OTX2 transcription factor regulates anxiety-related behavior in the mouse. Mol. Psychiatry. 2021;26:1–12. doi: 10.1038/s41380-021-01132-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Turner C.A., Thompson R.C., Bunney W.E., Schatzberg A.F., Barchas J.D., Myers R.M., Akil H., Watson S.J. Altered choroid plexus gene expression in major depressive disorder. Front. Hum. Neurosci. 2014. 8:238. doi: 10.3389/fnhum.2014.00238. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data underpinning this publication are openly available from the University of Strathclyde KnowledgeBase at https://doi.org/10.15129/cbbe122f-4ee1-4bfb-9ec7-46c664bf10f2 (accessed on 29 November 2021).