Abstract
Transglutaminase 2 (TGase 2), or tissue transglutaminase, catalyzes either ɛ-(γ-glutamyl)lysine or N1,N8-(γ-glutamyl)spermidine isopeptide bonds. TGase 2 expression has been associated with apoptosis, and it has been proposed that its activation should lead to the irreversible assembly of a cross-linked protein scaffold in dead cells. Thus, TGase 2-catalyzed protein polymerization contributes to the ultrastructural changes typical of dying apoptotic cells; it stabilizes the integrity of the apoptotic cells, preventing the release of harmful intracellular components into the extracellular space and, consequently, inflammation and scar formation. In order to perform a targeted disruption of the enzyme, we prepared a construct deleting part of exons 5 and 6, containing the active site, and intron 5. Complete absence of TGase 2 was demonstrated by reverse transcription-PCR and Western blot analysis. TGase activity measured on liver and thymus extracts showed, however, a minimal residual activity in TGase 2−/− mice. PCR analysis of mRNA extracted from the same tissues demonstrated that at least TGase 1 (normally present in the skin) is also expressed in these tissues and contributes to this residual activity. TGase 2−/− mice showed no major developmental abnormalities, and histological examination of the major organs appeared normal. Induction of apoptosis ex vivo in TGase 2−/− thymocytes (by CD95, dexamethasone, etoposide, and H2O2) and in vitro on TGase 2−/− mouse embryonal fibroblasts (by retinoids, UV, and H2O2) showed no significant differences. A reduction in cross-linked apoptotic bodies with a modestly increased release of lactate dehydrogenase has been detected in some cases. Together our results show that TGase 2 is not a crucial component of the main pathway of the apoptotic program. It is possible that the residual enzymatic activity, due to TGase 1 or redundancy of other still-unidentified TGases, can compensate for the lack of TGase 2.
Transglutaminase 2 (TGase 2; also called tissue transglutaminase or TG C) belongs to the transglutaminase (EC 2.3.2.13) family, which includes intracellular and extracellular enzymes catalyzing Ca2+-dependent reactions resulting in the formation of ɛ-(γ-glutamyl)lysine cross-links and/or in the covalent incorporation of di- and polyamines and histamine (25, 26). The establishment of these covalent cross-links leads to the posttranslational modification and, in many instances, the oligomerization of substrate proteins. The resulting protein polymers are resistant to breakage and chemical attack and can release polypeptides only through the proteolytic degradation of protein chains. At least seven distinct types of TGases in mammals have been characterized: TGase 1 (or TG K), TGase 2, TGase 3 (or TG E), TGase X, coagulation factor XIII, band 4.2., and prostate TGase. At least four transglutaminases (TGases 1, 2, 3, and X) are expressed and synthesized during terminal differentiation and death of human epidermal keratinocytes (44, 45), where they contribute to the formation of the cornified envelope.
The TGase 2 gene is constitutively expressed both during development (29, 48) and in adult tissues (for a review, see reference 36). In both cases a tight correlation between TGase 2 expression and occurrence of apoptosis has been found. This includes, for example, interdigital web formation (29), implantation of the embryo in utero (35), and mammary gland regression (31, 46). In addition, the presence and activity of the enzyme have been shown to increase in cells undergoing apoptosis in several models (2, 9, 10, 21, 23–25, 32–34, 37, 40). Indeed, during apoptosis de novo transcription of the TGase 2 gene is induced by several factors (e.g., retinoic acid [RA], prostaglandin E2, interleukin 6, and tumor growth factor β). Moreover, in addition to transcriptional regulation (24, 28, 41), TGase 2 can also be modulated posttranscriptionally (1, 23, 50) during apoptosis. TGase 2 activation leads to the assembly of intracellular cross-linked protein polymers, which irreversibly modifies cell organization, contributing to the wide ultrastructural changes occurring in cells undergoing apoptosis (9, 10, 39). This extensive TGase 2-dependent protein polymerization stabilizes apoptotic cells before their clearance by phagocytosis, thus contributing to the prevention of inflammation in the surrounding tissues (39).
In addition to its cross-linking activity, TGase 2 acts as the Gαh subunit, associated with the 50-kDa β subunit (Gβh), of the GTP-binding protein (Gh) in a ternary complex associated with the rat liver α1-adrenergic receptor (30). Thus, TGase 2-Gαh is a multifunctional protein, which by binding GTP in a Gαh-GTP complex can modulate receptor-stimulated phospholipase C activation.
In order to clarify the role of TGase 2 in apoptosis we have generated mice lacking TGase 2 by homologous recombination techniques. Our results, however, show that the disruption of TGase 2 does not produce a major phenotype and that apoptosis still occurs normally in the absence of TGase 2.
MATERIALS AND METHODS
Reagents.
Ham's F-12 and minimal essential media were from Gibco (Berlin, Germany), and fetal calf serum was from HyClone (Oud-Beijerland, The Netherlands). HEPES, bovine serum albumin (BSA), RNase A, propidium iodide (PI), Triton X-100, RA, N,N′-dimethylcasein, and putrescine were obtained from Sigma Chemical (St. Louis, Mo.). The mouse monoclonal anti-TGase 2 antibodies (clone CUB 7402 and clone CUB 7402+TG100) were purchased from Neo-Markers (Union City, Calif.). All electrophoresis reagents and secondary antibodies were from Bio-Rad (Richmond, Calif.) [3H]putrescine was obtained from Amersham (Arlington Heights, Ill.).
Generation of TGase 2-deficient mice.
A genomic clone containing the genomic sequence 3′ of exon 5 was isolated by screening a 129/SvJ mouse genomic library (Stratagene, La Jolla, Calif.). The targeting vector (Fig. 1A) was constructed by cloning an ∼4-kb EcoRV/BamHI fragment of this clone, containing the sequence from intron 6 to exon 9, into the pPNT vector (49) 3′ of the neomycin resistance gene between the XbaI and BamHI unique sites. An ∼2-kb fragment containing intron 3 was generated by PCR using primers designed on the basis of the sequence of exons 4 and 5. This fragment was cloned into the XhoI site of the pPNT vector 5′ of the neomycin resistance gene. This construct deletes 1.2 kb containing part of exon 5, intron 5, exon 6, and a small piece of intron 6 up to the EcoRV site.
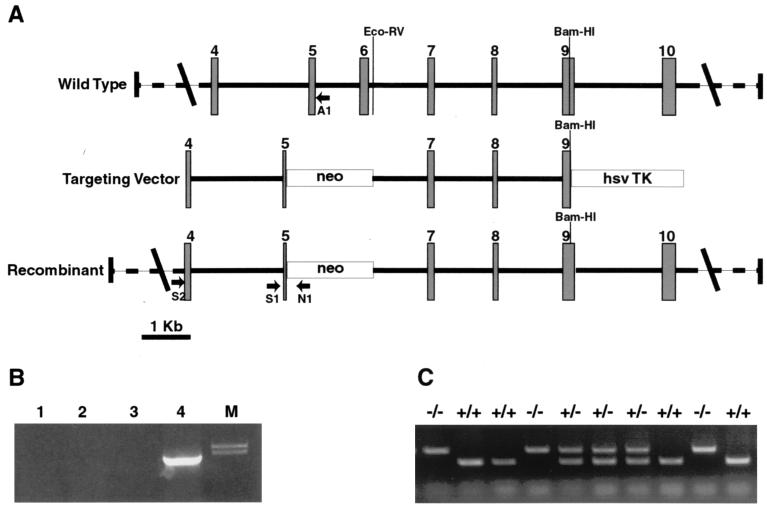
FIG. 1.
(A) Schematic representation of the targeting construct used to generate TGase 2 knockout mice. An EcoRV/BamHI genomic fragment (from intron 6 to exon 9) was cloned at the 5′ end of the neomycin resistance gene into the pPNT vector (49). Subsequently, a PCR fragment from exon 4 to exon 5 was cloned into this vector at the 5′ end of the neomycin resistance gene, using an XhoI site. As a result, part of exon 5 and the totality of exon 6 (containing the active site) and intron 5 were deleted. The arrows show the positions of the primers used for the screening of the recombinant clones (N1 and S2) and the genotyping of the mice (A1, N1, and S1). Intron positions were obtained by direct sequencing or PCR analysis with primers located in the flanking exons. The position of exon 7 was not determined, and therefore its position in the scheme is arbitrary. hsv TK, herpes simplex virus thymidine kinase. (B) PCR screening for the recombinant clone using a primer in the neomycin resistance gene (N1) and a primer on the TGase sequence (S2) in a region upstream of the fragment used to generate the targeting vector. Lanes 1 to 3 contain wild-type clones, and lane 4 contains the recombinant clone used to generate chimeras. (C) PCR screening for the genotyping of the mice. Three different primers (see Materials and Methods) were used at the same time: an antisense primer on the neomycin resistance gene (N1), an antisense primer in intron 5 (A1) deleted in the targeted alleles, and a sense primer in intron 4 (S1) present in both wild-type and targeted alleles.
The targeting vector was linearized by NotI and electroporated into embryonic stem cells. Cells were selected with G418 and ganciclovir. Surviving clones were screened by PCR using the following primers: S2 (5′AGCCGATGATGTGTACCTAGAC3′) and N1 (5′ACGAGACTAGTGAGACGTGC3′) (Fig. 1B). The following PCR program was used: 95°C for 5 min, followed by 40 cycles of 94°C for 1 min, 58°C for 1 min, and 72°C for 1 min. PCR products were resolved on a 0.8% agarose gel and stained with ethidium bromide. The positive clone was then confirmed by Southern blotting.
Cells from the positive clone were microinjected into C57BL/6 blastocysts and transferred into pseudopregnant recipients by Genome Systems, St. Louis, Mo. The six chimeric animals obtained were bred with C57BL/6 mice. The genotype of the subsequent offspring was determined by PCR using the following primers: S1 (5′TACTCCAGCTTCCTGTTCTG3′), A1 (5′TCCTGACCTGAGTCCTCGTC3′), and N1 (Fig. 1). The following PCR program was used: 95°C for 5 min, followed by 30 cycles of 94°C for 1 min, 56°C for 45 s, and 72°C for 1 min. PCR products were resolved on a 1% agarose gel and stained with ethidium bromide. Each mouse was individually genotyped before every experiment.
Cell cultures.
Mouse embryonic fibroblasts (MEFs) and thymocytes were grown in a 1:1 mixture of minimal essential medium and Ham's F-12 medium supplemented with 10% heat-inactivated fetal calf serum, 1.2 g of sodium bicarbonate per liter, and 15 mM HEPES at 37°C with 5% CO2 in a humidified atmosphere.
Western blotting.
Livers were homogenized in 3 ml of cold lysis buffer containing 100 mM Tris-HCl (pH 7.4), 10 mM KCl, 2 mM MgCl2, 0.1% Triton X-100, 1 mM EDTA, and 1 mM phenylmethylsulfonyl fluoride. They were then centrifuged, and the protein content of the supernatants was determined using the Bradford method (Bio-Rad). Proteins were normalized to 30 μg/lane, separated on sodium dodecyl sulfate (SDS)–12% polyacrylamide gels, and blotted onto nitrocellulose sheets. Filters were washed twice with phosphate-buffered saline (PBS) containing 0.1% Tween 20 before blocking nonspecific binding overnight with 10% nonfat milk and 5% BSA dissolved in PBS–0.1% Tween 20. The TGase 2 antigen was detected by incubation for 2 h with a 1:1 mixture of mouse monoclonal anti-TGase 2 antibodies (1:300 in PBS–0.1% Tween 20). Nitrocellulose filters were washed five times, and detection was performed by horseradish peroxidase-conjugated goat anti-mouse monoclonal antibody (1:2,500 in PBS–0.1% Tween 20 with 10% milk and 5% BSA) for 1 h at room temperature, using the ECL method (Amersham).
Enzyme assay.
TGase activity was determined by measuring the incorporation of [3H]putrescine into N,N′-dimethylcasein (22, 26). The reaction mixture contained 150 mM Tris-HCl buffer (pH 8.3), 90 mM NaCl, 10 mM dithiothreitol, 15 mM CaCl2, 12.5 mg of N,N′-dimethylcasein/ml, and 0.2 mM putrescine containing 1 μCi of [3H]putrescine. Proteins from different tissue and cellular extracts (0.1 to 0.3 mg) were incubated with the reaction mixture in a final volume of 150 μl at 37°C. After 20 min of incubation, the reaction was stopped by spotting 100-μl quadruplicate aliquots onto Whatman 3MM filter paper. Unbound [3H]putrescine was removed by washing with large volumes of 15, 10, and 5% trichloroacetic acid and absolute ethanol. Filters were then air dried and the radioactivity was measured by liquid scintillation counting.
PCR analysis of TGases 2 and 1.
Total RNA was extracted from mouse livers, using the RNeasy minikit from Qiagen (Crawley, United Kingdom). Reverse transcription (RT)-PCRs were performed with the RT-PCR One Step System (Life Technologies, Paisley, United Kingdom), using 100 ng of total RNA, according to the manufacturer's instructions. The primers used for the amplification of TGase 2 were TG30 (5′GACAACAACTATGGGGATGGT3′) and TG9B (5′ATCATCTCGCTCTTGTTCGTC3′). The primers used for the amplification of TGase 1 were MTG1F (5′ACCACCACAGTGCTCCGATG3′) and MTG1R (CCACACGTGGAAGTTCCAAAC3′). The following PCR program was used in all cases: 42°C for 30 min and 94°C for 2 min, followed by 35 cycles of 94°C for 30 s, 57°C for 30 s, and 70°C for 30 s. PCR products were resolved on a 1.6% agarose gel and stained with ethidium bromide.
Determination of cell death.
To estimate DNA fragmentation, a mixture of floating cells and cells mechanically recovered from flasks, which had been subjected to different treatments, were collected at 800 × g for 10 min and fixed with a 1:1 solution of PBS and methanol-acetone (4:1, vol/vol) at −20°C. The cell cycle was evaluated by flow cytometry using PI staining (40 mg/ml) (22) in the presence of 13 kU of RNase A per ml (20 min of incubation at 37°C) on a FACS-Calibur flow cytometer (Becton Dickinson, San Jose, Calif.). Cells were excited at 488 nm using a 15-mW argon laser, and the fluorescence was monitored at 578 nm at a rate of 150 to 300 events/s. Ten thousand events were evaluated using the Cell Quest program (Becton Dickinson). Electronic gating (FSC-a/vs/FSC-h) was used, when appropriate, to eliminate cell aggregates.
LDH release.
For measurement of lactate dehydrogenase (LDH) levels, a kit was used according to the manufacturer's instructions (Sigma Chemical). Briefly, the cell culture supernatant was incubated with pyruvate and NADH, and the LDH activity was determined photometrically at 340 nm.
Quantification of cross-linked apoptotic bodies.
Cross-linked apoptotic bodies were estimated on cells cultured in 175-cm2 flasks, as previously described (26). Cells floating in the culture medium were collected by centrifugation at 800 × g for 10 min and pooled with the cells mechanically recovered from flasks. After being washed in PBS, cells were suspended in 1 ml of lysis buffer (10 mM KCl, 2 mM MgCl2, and 0.5% Triton X-100 in 10 mM Tris-HCl, pH 7.4) containing 1 mM phenylmethylsulfonyl fluoride and 2 mM iodoacetamide. After centrifugation the pellet was washed in lysis buffer, suspended in a 2% sodium dodecyl sulfate solution containing 5% β-mercaptoethanol, and boiled, and the number of detergent-insoluble apoptotic bodies was scored using a phase-contrast microscope (Diaphot; Nikon) and normalized to milligrams of protein.
RESULTS
Generation of TGase 2-deficient mice.
The TGase 2 gene was disrupted by homologous recombination. The targeting vector deletes 1,200 bp of the TGase gene from exon 5 to intron 6. This deletion includes exon 6, which contains the active site. The loss of the catalytic site abolishes the protein cross-linking activity of TGase 2, consequently removing its presumed role in the formation of the apoptotic body. Figure 1 shows the targeting vector and the screening strategy.
TGase 2−/− mice show no clear phenotypic abnormality (macroscopic or microscopic); they develop normally and are capable of reproducing at the expected frequency.
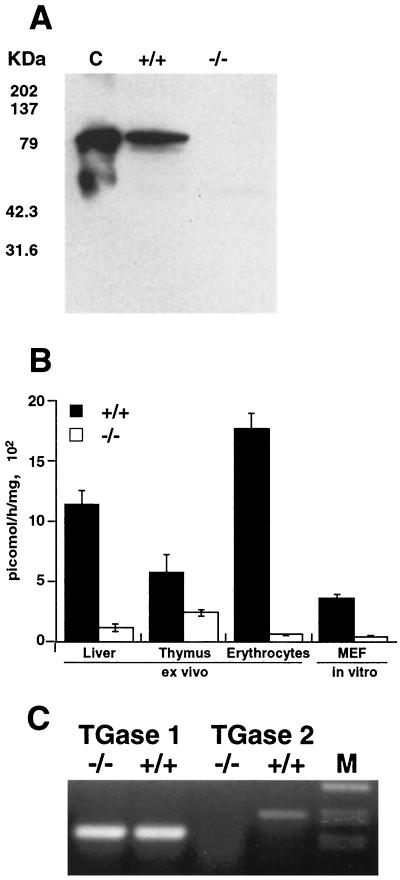
In order to confirm that no TGase 2 protein is produced in TGase 2-deficient mice, we performed Western blot analysis on liver, thymus, brain, and erythrocyte extracts from wild-type animals (TGase 2+/+) and from TGase 2−/− mice. Our results show the absence of TGase 2 protein in the −/− animals (Fig. 2A shows liver extracts). However, measurement of TGase activity (Fig. 2B) in different tissues extracted from TGase 2−/− animals showed that some residual TGase enzyme activity was still present. Indeed, while erythrocytes showed a reduction in activity close to 100%, thymocytes showed the highest level of residual TGase activity, which is quite significant compared with the activity of TGase 2+/+ animals. RT-PCR of RNA extracted from thymus tissues of both +/+ and −/− animals showed that no transcript for TGase 2 was present in −/− animals (Fig. 2C).
FIG. 2.
(A) Western blotting performed on liver extracts from wild-type and TGase-knockout mice with anti-TGase 2 antibody. Thirty micrograms of total protein was loaded in each lane. Recombinant guinea pig TGase 2 (0.1 μg) was used as a positive control (C). This blot is representative of experiments performed on different tissues from four different animals per group. (B) TGase enzymatic activity measured in extracts from different mouse tissues and cultured MEFs in both wild-type and TGase-knockout mice. Activity was measured as incorporation of [3H]putrescine into casein. Standard deviations for 10 different evaluations are shown. (C) RT-PCR performed on RNA extracted from thymus tissues of wild-type and TGase-knockout mice using primers for TGase 2 and TGase 1.
In order to detect and identify TGases different from type 2, we performed RT-PCRs using both specific and degenerate primers for the catalytic site region of TGases. Figure 2C shows that similar amounts of TGase 1 were expressed in −/− and +/+ animals. This should account for the residual TGase enzymatic activity.
TGase 2−/− thymocytes and MEFs show normal induction of apoptosis.
Since a large number of reports suggest a role for TGase 2 in apoptosis, including in vivo in the thymus (47), we studied apoptosis induced ex vivo in mouse thymocytes and in vitro in MEFs.
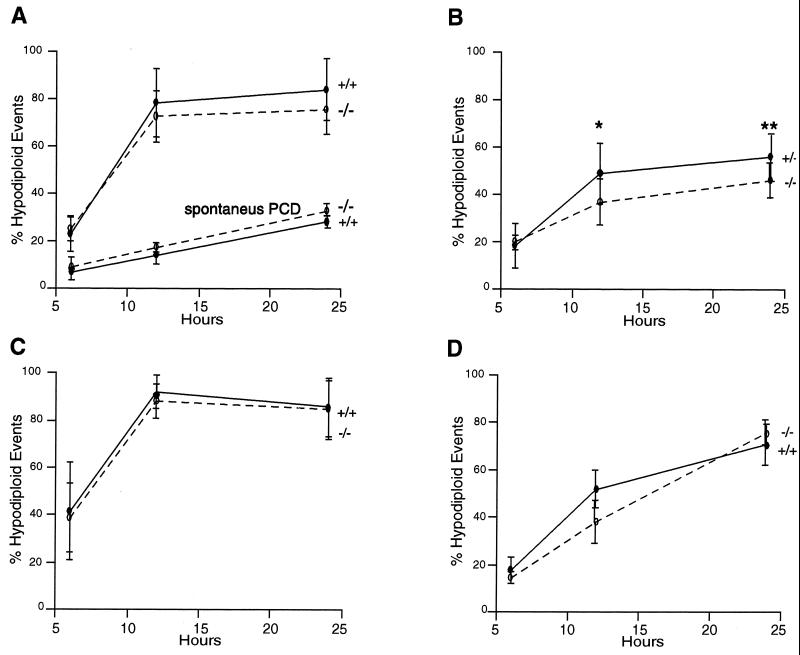
Apoptosis was induced with different stimuli, namely, etoposide (Fig. 3A), CD95 ligation (Fig. 3B), dexamethasone (Fig. 3C), and H2O2 (Fig. 3D). Figure 3A shows also the spontaneous level of apoptosis of thymocytes ex vivo. No relevant difference in the number of cells undergoing apoptosis was observed between +/+ and −/− animals with any of the treatments.
FIG. 3.
Hypodiploid events in wild-type and knockout mouse thymocytes, either left untreated (spontaneous programmed cell death [PCD]) (bottom curves in panel A) or treated with etoposide (25 μM) (top curves in panel A), anti-CD95 agonist antibody (1 μg/ml) (B), dexamethasone (10 μM) (C), and H2O2 (30 μM) (D). Cells were treated for 6, 12, and 24 h, then fixed with a 1:1 solution of PBS and methanol-acetone (4:1, vol/vol) at −20°C and stained with PI. The number of events (10,000 collected) with hypodiploid DNA was measured by flow cytometry. The data reported are averages of four independent experiments performed on different mice. ∗, P = 0.0352 for +/+ versus −/− mice; ∗∗, P = 0.0451 for +/+ versus −/− mice (according to the Student t test). Differences not indicated are not statistically significant.
The evaluation of apoptosis ex vivo was performed on cells presenting a significant TGase enzymatic activity (Fig. 2B), at least in part due to TGase 1. We therefore performed similar experiments on cultured MEFs, which show the lowest residual enzymatic activity (Fig. 2B).
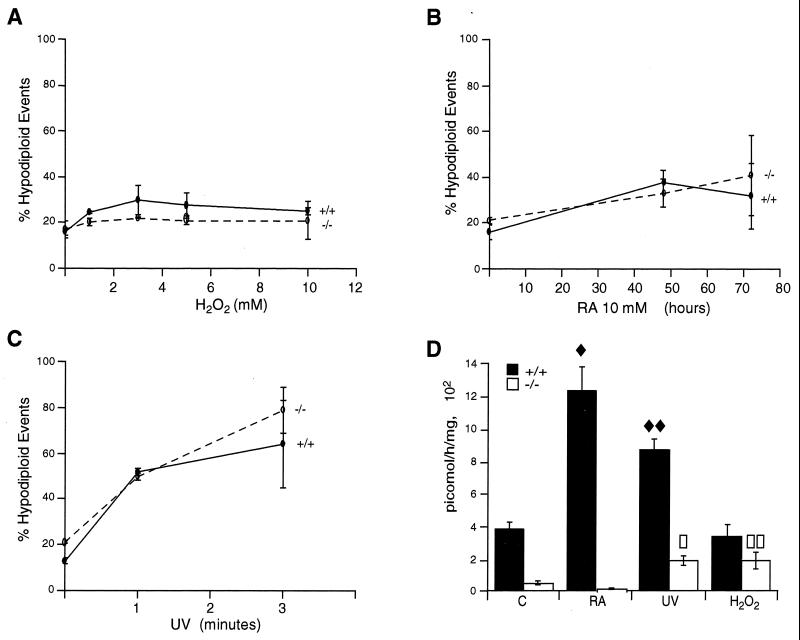
As in the ex vivo studies, no difference was observed when +/+ and −/− MEFs were treated in vitro with H2O2 (Fig. 4A), RA (Fig. 4B), or UV (Fig. 4C). Treatment of +/+ MEFs with RA and UV resulted in an increase in TGase activity; −/− MEFs had a much lower basal activity that increased with UV and H2O2 treatment, while RA treatment resulted in a decrease of TGase activity (Fig. 4D). Since TGase 1 is negatively regulated by retinoids, while TGase 2 is upregulated (50), these results are in keeping with the evidence that the minimal residual activity observed in MEFs is due to TGase 1 (Fig. 4D). Indeed, as for thymocytes, mRNA for TGase 1 was also detected in MEFs by RT-PCR (data not shown).
FIG. 4.
Hypodiploid events in wild-type and knockout MEFs treated with 1, 3, 5, and 10 mM H2O2 for 15 min in PBS, washed, and then cultured in medium for 24 h (A); treated with 10 μM RA for 48 and 72 h (B); or treated with UV irradiation for 1 or 3 min in PBS and then cultured in medium for 24 h (C). Cells were fixed with a 1:1 solution of PBS and methanol-acetone (4:1, vol/vol) at −20°C and stained with PI. The number of events with hypodiploid DNA was measured by flow cytometry (see Materials and Methods). The data reported are averages of four independent experiments. (D) Corresponding transglutaminase activity in wild-type and knockout MEFs, untreated (C) or treated with 10 μM RA for 48 h, UV irradiation for 3 min followed by 24 h of culture, and 5 mM H2O2 followed by 24 h of culture. The data reported are averages of four independent experiments. Statistical differences between treated cells and the corresponding controls were evaluated. In detail, differences were statistically significant as follows: ⧫, P = 0.0189 for control and RA-treated +/+ cells; ⧫⧫, P = 0.0013 for control and UV-treated +/+ cells; □, P = 0.0294 for control and UV-treated −/− cells; and □□, P = 0.0475 for control and H2O2-treated −/− cells. Differences not indicated were not statistically significant. All differences between +/+ and −/− cells are statistically significant.
Cross-linked apoptotic body formation is present in TGase 2−/− mice.
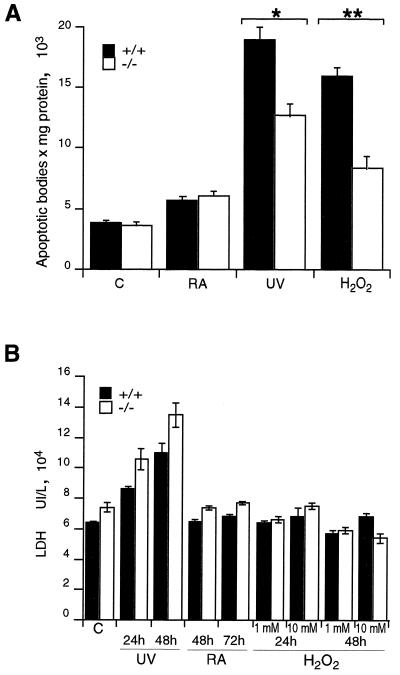
It has been consistently suggested that TGase 2 induction during apoptosis results in the formation of cross-linked insoluble apoptotic bodies. In order to investigate the possibility that the formation of cross-linked apoptotic bodies is impaired in TGase 2−/− mice, we measured the number of insoluble apoptotic bodies in control or RA-, UV-, and H2O2-treated MEFs. Figure 5A shows that apoptotic bodies also formed in TGase 2−/− cells, even though the number of cross-linked apoptotic bodies was significantly reduced in UV- and H2O2-treated −/− MEFs.
FIG. 5.
(A) Cross-linked apoptotic body formation in wild-type and knockout MEFs. Cells were treated with 10 μM RA for 48 h, UV irradiation for 3 min followed by culture for 24 h, or 3 mM H2O2 for 15 min followed by culture for 24 h. See Materials and Methods for details. (B) LDH release in wild-type and knockout MEFs. Cells were left untreated (C) or treated with UV irradiation for 3 min followed by culture for 24 and 48 h, 10 μM RA for 48 and 72 h, or 1 and 10 mM H2O2 for 15 min followed by culture for 24 and 48 h. The data reported are averages of four independent experiments. Statistical analysis was performed according to the Student t test to compare +/+ and −/− cells: ∗, P = 0.0014; ∗∗, P = 0.0003. Differences not indicated are not statistically significant.
The reduction of cross-linked apoptotic bodies could be accompanied by an increased release of cytoplasmic material from cells undergoing apoptosis. In order to evaluate this aspect, we measured the release of LDH from +/+ and −/− MEFs after induction of apoptosis with UV, RA, and H2O2. Figure 5B shows that LDH release was only moderately increased (not statistically significant) in +/+ versus −/− MEFs. Treatment with UV, with which some necrosis was expected, showed a significant increase in LDH release in both +/+ and −/− cells. Therefore, our results are consistent with an essentially normal induction of apoptosis.
DISCUSSION
We have generated TGase 2-deficient mice through homologous recombination techniques. No TGase 2 was detectable in these mice by RT-PCR or Western blotting. The mice were viable and fertile and showed no developmental abnormalities. Apoptosis induced with different agents in both fibroblasts and thymocytes is normal. The reduction in enzymatic activity in −/− mice showed only a minor effect on both cross-linked apoptotic body formation (Fig. 5A) and LDH release (Fig. 5B), with no major consequences for the mice. Although we cannot exclude the possibility that TGase 2 deficiency may play a role in pathological situations (11, 38), aged mice (up to 20 months of age) do not show abnormalities such as cancer development and generation of autoimmunity (data not shown). It might be necessary to cross the mice into a more permissive genetic background in order to reveal an overt phenotype.
Deletion of various genes involved in apoptosis does not always produce an evident phenotype. Mice with deleted caspases 1, 2, 6, and 11 do not show evident developmental abnormalities (51), while in other cases a very specific or minimal phenotype is observed. Disruptions of other genes produce a phenotype only when animals are stressed with specific inducers requiring that protein, namely, radiation on p53−/− (8) or liposaccharides on caspase 1−/− (16, 18) cells. Even though the accredited model for apoptosis indicates the requirement for the apoptosome and in particular for apaf-1 and caspases 3 and 9 (for a review, see references 6 and 17), the knockout of the genes for these proteins shows that thymocytes are still able to undergo apoptosis (for a review, see references 5 and 51). This has elicited various explanations, including the existence of additional unknown pathways or compensation mechanisms.
Similarly, there are different possible interpretations for the lack of phenotype in TGase 2 animals: (i) TGase 2 is not involved in apoptosis; (ii) it is not involved in the central, essential apoptotic machinery, but it is part of a regulatory or side pathway elicited only by specific inducers or only in specific tissues; or (iii) there is redundancy in the system.
The lack of effect on apoptosis of targeted disruption of the TGase 2 gene is in apparent contrast to previous evidence in favor of a role for TGase 2 in the apoptotic program. Indeed, it has been shown previously that transfection of an antisense TGase 2 construct into cell lines confers resistance to apoptosis induction, while sense transfectants show enhanced spontaneous apoptosis (22). There are several reasons for this disparity. First, not all models of apoptosis require TGase 2. For example, CD95 ligation elicits apoptosis independently of the steady-state levels of TGase 2 protein (3); correspondingly, there is no change in TGase enzymatic activity during CD95-induced apoptosis (3). Second, other TGases may assume the protein cross-linking role of TGase 2. Indeed, the TGase 2−/− mice showed different degrees of TGase enzymatic activity in different tissues. Our data show the presence of TGase 1 in both +/+ and −/− mice. Recently, TGase 1 has been shown to exist in tissues different from the skin, namely, in the central nervous system (15). Furthermore, the TGase activity levels in −/− thymocytes is inhibited by GTP, a property of TGase X (E. Candi, G. Melino, et al., unpublished observation), suggesting its expression in lymphoid tissue. Therefore, the possibility that other TGases, and particularly TGase 1, can compensate for TGase 2 loss cannot be excluded. TGases show very different biochemical properties, such as kcat/Km ratio, residue preference, and yield (26). Therefore, it is unlikely that there is a perfect compensation among distinct TGases. In fact, TGase 1 knockout animals show a lethal phenotype, despite the presence of four distinct TGases in the skin (20). However, point mutations for TGase 1 in humans, with complete loss of TGase enzymatic activity (4), are compatible with life but cause a skin disease known as lamellar ichthyosis (12, 42), suggesting a different degree of compensation in humans.
Despite the large body of data suggesting an involvement of TGase 2 in apoptosis, its precise role in this process is not evident from the present gene disruption study. While the TGase 2−/− animals could be used to study other functions of the enzyme, particularly by further crossbreeding to evaluate its contribution in pathologies such as celiac (7, 27) and Huntington (13, 14, 19, 43) diseases, clarification of the importance of TGase 2 in apoptosis may well require the generation of animals deficient in multiple TGases.
ACKNOWLEDGMENTS
We thank Mauro Piacentini, Gennaro Ciliberto, and Richard A. Knight for generous support, critical discussions, and helpful suggestions. This work could not have been completed without the generous help of Francesca Bernassola, Eleonora Candi, Marco Corazzari, Daniela Barcaroli, and Marco Ranalli. We thank Giuseppe Bertini, Giancarlo Cortese, and Pierino Piccoli (S.S.D. SAFU, Instituto Fisioterapici Ospedalieri, Rome, Italy) for technical assistance and mouse husbandry.
The work was partially supported by grants from MURST, MinSan, Associazione Neuroblastoma, AIRC, Telethon (E 872 and E 1257), and EU (QLG1-1999-00739).
REFERENCES
- 1.Achyuthan K E, Greenberg C S. Identification of a guanosine triphosphate-binding site on guinea pig liver transglutaminase. Role of GTP and calcium ions in modulating activity. J Biol Chem. 1987;262:1901–1906. [PubMed] [Google Scholar]
- 2.Amendola A, Gougeon M L, Poccia F, Bondurand A, Fesus L, Piacentini M. Induction of “tissue” transglutaminase in HIV-pathogenesis: evidence for high rate of apoptosis of CD4+ T lymphocytes and accessory cells in lymphoid tissues. Proc Natl Acad Sci USA. 1996;93:11057–11062. doi: 10.1073/pnas.93.20.11057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bernassola F, Scheurpflug C, Herr I, Krammer P H, Debatin K M, Melino G. Induction of apoptosis by IFNgamma in human neuroblastoma cell lines through the CD95/CD95L autocrine circuit. Cell Death Differ. 1999;6:652–660. doi: 10.1038/sj.cdd.4400537. [DOI] [PubMed] [Google Scholar]
- 4.Candi E, Melino G, Lahm A, Ceci R, Rossi A, Kim I-G, Ciani B, Steinert P M. Transglutaminase 1 mutations in lamellar ichthyosis. J Biol Chem. 1998;273:13693–13702. doi: 10.1074/jbc.273.22.13693. [DOI] [PubMed] [Google Scholar]
- 5.Cecconi F. Apaf1 and the apoptotic machinery. Cell Death Differ. 1999;6:1087–1098. doi: 10.1038/sj.cdd.4400602. [DOI] [PubMed] [Google Scholar]
- 6.De Laurenzi V, Melino G. Apoptosis. The little devil of death. Nature. 2000;406:135–136. doi: 10.1038/35018190. [DOI] [PubMed] [Google Scholar]
- 7.Dieterich W, Ehnis T, Bauer M, Donner P, Volta U, Riecken E O, Schuppan D. Identification of tissue transglutaminase as the autoantigen of celiac disease. Nat Med. 1997;3:797–801. doi: 10.1038/nm0797-797. [DOI] [PubMed] [Google Scholar]
- 8.Donehower L A, Harvey M, Slagle B L, McArthur M J, Montgomery C A, Jr, Butel J S, Bradley A. Mice deficient for p53 are developmentally normal but susceptible to spontaneous tumours. Nature. 1992;356:215–221. doi: 10.1038/356215a0. [DOI] [PubMed] [Google Scholar]
- 9.Fesus L, Thomazy V, Autuori F, Ceru' M P, Tarcsa E, Piacentini M. Apoptotic hepatocytes become insoluble in detergents and chaotropic agents as a result of transglutaminase action. FEBS Lett. 1989;245:150–154. doi: 10.1016/0014-5793(89)80210-8. [DOI] [PubMed] [Google Scholar]
- 10.Fesus L, Davies P J A, Piacentini M. Apoptosis: molecular mechanisms in programmed cell death. Eur J Cell Biol. 1991;56:170–177. [PubMed] [Google Scholar]
- 11.Hettasch J M, Bandarenko N, Burchette J L, Lai T S, Marks J R, Haroon Z A, Peters K, Dewhirst M W, Iglehart J D, Greenberg C S. Tissue transglutaminase expression in human breast cancer. Lab Investig. 1996;75:637–645. [PubMed] [Google Scholar]
- 12.Huber M, Rettler I, Bernasconi K, Frenk E, Lavrijsen S P, Ponec M, Bon A, Lautenschlager S, Schorderet D F, Hohl D. Mutations of keratinocyte transglutaminase in lamellar ichthyosis. Science. 1995;267:525–528. doi: 10.1126/science.7824952. [DOI] [PubMed] [Google Scholar]
- 13.Igarashi S, Koide R, Shimohata T, Yamada M, Hayashi Y, Takano H, Date H, Oyake M, Sato T, Sato A, Egawa S, Ikeuchi T, Tanaka H, Nakano R, Tanaka K, Hozumi I, Inuzuka T, Takahashi H, Tsuji S. Suppression of aggregate formation and apoptosis by transglutaminase inhibitors in cells expressing truncated DRPLA protein with an expanded polyglutamine stretch. Nat Genet. 1998;18:111–117. doi: 10.1038/ng0298-111. [DOI] [PubMed] [Google Scholar]
- 14.Kahlem P, Green H, Djian P. Transglutaminase action imitates Huntington's disease: selective polymerization of Huntingtin containing expanded polyglutamine. Mol Cell. 1998;1:595–601. doi: 10.1016/s1097-2765(00)80059-3. [DOI] [PubMed] [Google Scholar]
- 15.Kim S Y, Grant P, Lee J H, Pant H C, Steinert P M. Differential expression of multiple transglutaminases in human brain. Increased expression and cross-linking by transglutaminases 1 and 2 in Alzheimer's disease. J Biol Chem. 1999;274:30715–30721. doi: 10.1074/jbc.274.43.30715. [DOI] [PubMed] [Google Scholar]
- 16.Kuida K, Lippke J A, Ku G, Harding M W, Livingstone D J, Su M S-S, Flavell R A. Altered cytokine export and apoptosis in mice deficient in interleukin-1 beta converting enzyme. Science. 1995;281:2000–2002. doi: 10.1126/science.7535475. [DOI] [PubMed] [Google Scholar]
- 17.Kumar S. Mechanisms mediating caspase activation in cell death. Cell Death Differ. 1999;6:1060–1066. doi: 10.1038/sj.cdd.4400600. [DOI] [PubMed] [Google Scholar]
- 18.Li P, Allen H, Banerjee S, Franklin S, Herzog L, Johnstone C, McDowell J, Paskind M, Rodman L, Salfeld J, Townes E, Tracey D, Wardwell S, Wei F-Y, Wong W W, Kamen R, Seshadri T. Mice deficient in IL-1 beta-converting enzyme are defective in production of mature IL beta and resistant to endotoxic shock. Cell. 1995;80:401–411. doi: 10.1016/0092-8674(95)90490-5. [DOI] [PubMed] [Google Scholar]
- 19.Lorand L. DRPLA aggregation and transglutaminase, revisited. Nat Genet. 1998;20:231. doi: 10.1038/3033. [DOI] [PubMed] [Google Scholar]
- 20.Matsuki M, Yamashita F, Ishida-Yamamoto A, Yamada K, Kinoshita C, Fushiki S, Ueda E, Morishima Y, Tabata K, Yasuno H, Hashida M, Iizuka H, Ikawa M, Okabe M, Kondoh G, Kinoshita T, Takeda J, Yamanishi K. Defective stratum corneum and early neonatal death in mice lacking the gene for transglutaminase 1 (keratinocyte transglutaminase) Proc Natl Acad Sci USA. 1998;95:1044–1049. doi: 10.1073/pnas.95.3.1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Melino G, Farrace M G, Ceru' M P, Piacentini M. Correlation between transglutaminase activity and polyamine levels in human neuroblastoma cells. Effect of retinoic acid and alpha-difluoromethylornithine. Exp Cell Res. 1988;179:429–445. doi: 10.1016/0014-4827(88)90281-9. [DOI] [PubMed] [Google Scholar]
- 22.Melino G, Annicchiarico-Petruzzelli M, Piredda L, Candi E, Gentile V, Davies P J A, Piacentini M. Tissue transglutaminase and apoptosis: sense and antisense transfection studies with human neuroblastoma cells. Mol Cell Biol. 1994;14:6584–6596. doi: 10.1128/mcb.14.10.6584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Melino G, Bernassola F, Corasaniti M T, Knight R A, Nistico G, Finazzi-Agro A. S-nitrosylation regulates apoptosis. Nature. 1997;388:432–433. doi: 10.1038/41237. [DOI] [PubMed] [Google Scholar]
- 24.Melino G, Draoui M, Piacentini M, Bellincampi L, Bernassola F, Reichert U, Cohen P. Retinoic acid receptors a and g mediate tissue-transglutaminase induction in human neuroblastoma cells undergoing apoptosis. Exp Cell Res. 1997;255:55–61. doi: 10.1006/excr.1997.3656. [DOI] [PubMed] [Google Scholar]
- 25.Melino G, Piacentini M. Tissue transglutaminase in apoptosis: a downstream or a multifunctional upstream effector? FEBS Lett. 1998;430:59–63. doi: 10.1016/s0014-5793(98)00521-3. [DOI] [PubMed] [Google Scholar]
- 26.Melino G, Candi E, Steinert P M. Assay for transglutaminases in cell death. Methods Enzymol. 2000;322:433–472. doi: 10.1016/s0076-6879(00)22042-9. [DOI] [PubMed] [Google Scholar]
- 27.Molberg O, Mcadam S N, Korner R, Quarsten H, Kristiansen C, Madsen L, Fugger L, Scott H, Noren O, Roepstorff P, Lundin K E, Sjostrom H, Sollid L M. Tissue transglutaminase selectively modifies gliadin peptides that are recognized by gut-derived T cells in celiac disease. Nat Med. 1998;4:713–717. doi: 10.1038/nm0698-713. [DOI] [PubMed] [Google Scholar]
- 28.Nagy L, Saydak M, Shipley N, Lu S, Basilion J P, Yan Z H, Syka P, Chandraratna R, Heyman R, Davies P J A. Identification and characterization of a versatile retinoid response element (RARE/RXRE) in the promoter of the mouse tissue transglutaminase gene. J Biol Chem. 1996;271:4355–4365. doi: 10.1074/jbc.271.8.4355. [DOI] [PubMed] [Google Scholar]
- 29.Nagy L, Thomazy V A, Saydak M M, Stein J P, Davies P J A. The promoter of mouse tissue transglutaminase gene directs tissue-specific, retinoid-regulated and apoptosis-linked expression. Cell Death Differ. 1997;4:534–547. doi: 10.1038/sj.cdd.4400290. [DOI] [PubMed] [Google Scholar]
- 30.Nakaoka H, Perez D M, Baek K J, Das T, Husain A, Misono K, Im M, Graham R M. Gh: a GTP-binding protein with transglutaminase activity and receptor signaling function. Science. 1994;264:1593–1596. doi: 10.1126/science.7911253. [DOI] [PubMed] [Google Scholar]
- 31.Nemes Z, Jr, Friis R R, Aeschlimann D, Saurer S, Paulsson M, Fesus L. Expression and activation of tissue transglutaminase in apoptotic cells of involuting rodent mammary tissue. Eur J Cell Biol. 1996;70:125–133. [PubMed] [Google Scholar]
- 32.Piacentini M, Fesus L, Farrace M G, Ghibelli L, Piredda L, Melino G. The expression of “tissue” transglutaminase in two human cancer cell lines is related with the programmed cell death (apoptosis) Eur J Cell Biol. 1991;54:246–254. [PubMed] [Google Scholar]
- 33.Piacentini M, Annicchiarico-Petruzzelli M, Oliverio S, Piredda L, Biedler J L, Melino G. Phenotype-specific “tissue” transglutaminase regulation in human neuroblastoma cells in response to retinoic acid: correlation with cell death by apoptosis. Int J Cancer. 1992;52:271–278. doi: 10.1002/ijc.2910520220. [DOI] [PubMed] [Google Scholar]
- 34.Piacentini M, Fesus L, Melino G. Multiple cell cycle access to the apoptotic death programme in human neuroblastoma cells. FEBS Lett. 1993;320:150–154. doi: 10.1016/0014-5793(93)80081-5. [DOI] [PubMed] [Google Scholar]
- 35.Piacentini M, Autuori F. Immunohistochemical localization of tissue transglutaminase and Bcl-2 in rat uterine tissues during embryo implantation and post-partum involution. Differentiation. 1994;57:51–61. doi: 10.1046/j.1432-0436.1994.5710051.x. [DOI] [PubMed] [Google Scholar]
- 36.Piacentini M. Tissue transglutaminase: a candidate effector element of physiological cell death. Curr Top Microbiol Immunol. 1995;200:163–176. doi: 10.1007/978-3-642-79437-7_12. [DOI] [PubMed] [Google Scholar]
- 37.Piacentini M, Piredda L, Starace D, Annicchiarico-Petruzzelli M, Mattel M, Oliverio S, Farrace M G, Melino G. Differential growth properties of S- and N-type human neuroblastoma cell variants transplanted into SCID mice: correlation with apoptosis and effect of ethanol. J Pathol. 1996;180:415–422. doi: 10.1002/(SICI)1096-9896(199612)180:4<415::AID-PATH684>3.0.CO;2-A. [DOI] [PubMed] [Google Scholar]
- 38.Piacentini M, Colizzi V. Tissue transglutaminase: apoptosis versus autoimmunity. Immunol Today. 1999;20:130–134. doi: 10.1016/s0167-5699(98)01416-9. [DOI] [PubMed] [Google Scholar]
- 39.Piredda L, Amendola A, Colizzi V, Davies P J A, Farrace M G, Fraziano M, Gentile V, Uray I, Piacentini M, Fesus L. Lack of “tissue” transglutaminase protein cross-linking leads to leakage of macromolecules from dying cells: relationship to development of autoimmunity in MRLIpr/Ipr mice. Cell Death Differ. 1997;4:463–472. doi: 10.1038/sj.cdd.4400267. [DOI] [PubMed] [Google Scholar]
- 40.Piredda L, Farrace M G, Lo Bello M, Malorni W, Melino G, Petruzzelli R, Piacentini M. Identification of 'tissue' transglutaminase binding proteins in neural cells committed to apoptosis. FASEB J. 1999;13:355–364. doi: 10.1096/fasebj.13.2.355. [DOI] [PubMed] [Google Scholar]
- 41.Ritter S J, Davies P J. Identification of a transforming growth factor-beta1/bone morphogenetic protein 4 (TGF-beta1/BMP4) response element within the mouse tissue transglutaminase gene promoter. J Biol Chem. 1998;273:12798–12806. doi: 10.1074/jbc.273.21.12798. [DOI] [PubMed] [Google Scholar]
- 42.Russell L J, DiGiovanna J J, Rogers G R, Steinert P M, Hashem N, Compton J G, Bale S J. Mutations in the gene for transglutaminase 1 in autosomal recessive lamellar ichthyosis. Nat Genet. 1995;9:279–283. doi: 10.1038/ng0395-279. [DOI] [PubMed] [Google Scholar]
- 43.Saudou F, Finkbeiner S, Devys D, Greenberg M E. Huntingtin acts in the nucleus to induce apoptosis but death does not correlate with the formation of intranuclear inclusions. Cell. 1998;95:55–66. doi: 10.1016/s0092-8674(00)81782-1. [DOI] [PubMed] [Google Scholar]
- 44.Steinert P M. A model for the hierarchical structure of the human epidermal cornified cell envelope. Cell Death Differ. 1995;2:33–40. [PubMed] [Google Scholar]
- 45.Steinert P M, Candi E, Tarcsa E, Marekov L N, Sette M, Paci M, Ciani B, Guerrieri P, Melino G. Transglutaminase crosslinking and structural studies of the human small proline rich 3 protein. Cell Death Differ. 1999;6:916–930. doi: 10.1038/sj.cdd.4400568. [DOI] [PubMed] [Google Scholar]
- 46.Strange R, Li F, Saurer S, Bukhard A, Friis R R. Apoptotic cell death and tissue remodelling during mouse mammary gland involution. Development. 1992;115:49–58. doi: 10.1242/dev.115.1.49. [DOI] [PubMed] [Google Scholar]
- 47.Szondy Z, Molnar P, Nemes Z, Boyiardzis M, Kedei N, Toth R, Fesus L. Differential expression of transglutaminase during in vivo apoptosis of thymocytes induced via distinct signalling pathways. FEBS Lett. 1997;404:307–313. doi: 10.1016/s0014-5793(97)00140-3. [DOI] [PubMed] [Google Scholar]
- 48.Thomazy V A, Davies P J. Expression of tissue transglutaminase in the developing chicken limb is associated both with apoptosis and endochondral ossification. Cell Death Differ. 1999;6:146–154. doi: 10.1038/sj.cdd.4400464. [DOI] [PubMed] [Google Scholar]
- 49.Tybulewicz V L, Crawford C E, Jackson P K, Bronson R T, Mulligan R C. Neonatal lethality and lymphopenia in mice with a homozygous disruption of the c-abl proto-oncogene. Cell. 1991;65:1153–1163. doi: 10.1016/0092-8674(91)90011-m. [DOI] [PubMed] [Google Scholar]
- 50.Zhang L X, Mills K J, Dawson M I, Collins S J, Jetten A M. Evidence for the involvement of retinoic acid receptor RAR alpha-dependent signaling pathway in the induction of tissue transglutaminase and apoptosis by retinoids. J Biol Chem. 1995;270:6022–6029. doi: 10.1074/jbc.270.11.6022. [DOI] [PubMed] [Google Scholar]
- 51.Zheng T S, Hunot S, Kuida K, Flavell R A. Caspase knockouts: matters of life and death. Cell Death Differ. 1999;6:1043–1053. doi: 10.1038/sj.cdd.4400593. [DOI] [PubMed] [Google Scholar]