Abstract
Zika virus (ZIKV), a re-emerging virus, causes congenital brain abnormalities and Guillain–Barré syndrome. It is mainly transmitted by Aedes mosquitoes, but infections are also linked to sexual transmissions. Infectious ZIKV has been isolated, and viral RNA has been detected in semen over a year after the onset of initial symptoms, but the mode of long-term persistence is not yet understood. ZIKV can proliferate in human Sertoli cells (HSerC) for several weeks in vitro, suggesting that it might be a reservoir for persistent ZIKV infection. This study determined proteomic changes in HSerC during ZIKV infections by TMT-mass spectrometry analysis. Levels of 4416 unique Sertoli cell proteins were significantly altered at 3, 5, and 7 days after ZIKV infection. The significantly altered proteins include enzymes, transcription regulators, transporters, kinases, peptidases, transmembrane receptors, cytokines, ion channels, and growth factors. Many of these proteins are involved in pathways associated with antiviral response, antigen presentation, and immune cell activation. Several immune response pathway proteins were significantly activated during infection, e.g., interferon signaling, T cell receptor signaling, IL-8 signaling, and Th1 signaling. The altered protein levels were linked to predicted activation of immune response in HSerC, which was predicted to suppress ZIKV infection. ZIKV infection also affected the levels of critical regulators of gluconeogenesis and glycolysis pathways such as phosphoglycerate mutase, phosphoglycerate kinase, and enolase. Interestingly, many significantly altered proteins were associated with cardiac hypertrophy, which may induce heart failure in infected patients. In summary, our research contributes to a better understanding of ZIKV replication dynamics and infection in Sertoli cells.
Keywords: Zika virus, Sertoli cells, sexual transmission, persistence in semen, immune response, glycolysis, carbohydrate metabolism, cardiovascular disease
1. Introduction
Zika virus (ZIKV) belongs to the family Flaviviridae, and is a single-stranded positive-sense RNA virus. Other members of the family include Japanese encephalitis virus (JEV), Yellow fever virus (YFV), West Nile virus (WNV) and Dengue virus (DENV). ZIKV was first discovered in Rhesus monkeys in Uganda’s Zika rainforest in 1947 [1].
However, the virus remained undiagnosed for a long time due to the disease’s non-specific flu-like symptoms and a lack of diagnostic screening [2,3]. In 2007, ZIKV re-emerged in the Pacific islands, spreading to over 80 countries/territories, including Latin America, the United States, and Southeast Asia [4,5,6,7,8]. The virus has been linked to microcephaly in babies [9] and Guillain–Barré syndrome (GBS) in adults [10,11]. Therefore, the World Health Organization (WHO) considers ZIKV to be a threat to global health, and declared a global health emergency in 2016 [12].
In endemic regions, ZIKV is mainly transmitted by mosquito bites (Aedes aegypti and A. albopictus) [13]. However, the virus can persist in the male reproductive tract and contribute to sexual transmission [14,15,16]. After the initial onset of symptoms, ZIKV RNA and infectious virus particles were detected in semen from infected males after 414 days [17] and 69 days [18], respectively. Sexual transmission of ZIKV has been reported in 14 countries so far [19]. ZIKV was found in murine testis and monkey models after inoculation, causing severe testicular damage [20,21,22,23]. Unlike the animal models, ZIKV infection does not cause any noticeable impact on human testis morphology. However, persistent ZIKV infection in the male genital tract may impact testicular hormone levels and sperm quality [24,25,26]. These findings indicate that, like many other viruses [27], ZIKV exploits the immune-privileged environment of the testis, which lacks an adaptive immune system [28], and hides for a prolonged time. However, the cellular reservoir of ZIKV testicular persistence is not clearly understood.
Sertoli, Leydig, and germ cells are the most prevalent cells found in human testes [29]. Sertoli cells constitute the major part of the testicular environment and play a critical role in the development of spermatogonial stem cells into mature sperm [30,31]. The testis–blood barrier, which protects the male reproductive system from pathogens, is also established by Sertoli cells [32]. Another major cell type in the testes is the Leydig cell, which secretes the male sex hormone testosterone and is essential for the development of male reproductive organs and characteristics [33]. In mouse testis, Sertoli cells are the most susceptible to ZIKV infection [34]. A previous study has demonstrated that ZIKV can cross the blood–testis barrier and efficiently infect Sertoli cells [35]. Moreover, the virus not only can infect Sertoli cells, but can persist for several days/weeks without any observable cytopathic impact [34,36,37].
We previously used an aptamer-based assay and demonstrated that ZIKV infection caused alterations in the levels of proteins involved in spermatogenesis in HSerC [36]. In order to better understand the mechanism(s) of viral persistence and its impact on the male genital tract, we extended the previous study by using a complementary tandem mass tag (TMT)-based 2D LC/MS/MS mass spectrometry-based approach to investigate Sertoli cell proteomic alterations after ZIKV infection. We measured ~8000 proteins across three time points and identified approximately 4400 Sertoli cell proteins significantly affected by ZIKV. The possible significance of these proteins in cellular processes, signaling pathways, and disease pathogenesis were investigated through bioinformatics analyses.
2. Materials and Methods
2.1. Cells
Primary Sertoli cells (HSerC) isolated from human testis were purchased from ScienCell Research Laboratories, CA, USA (Catalog #4520). The cells were cultured at 37 °C in 5% CO2 in poly-l-lysine-(Sigma−Aldrich, ON, Canada, Cat.# P4707) coated culture plates in Sertoli Cell Medium (ScienCell Research Laboratories, CA, USA, Cat. #4521). Following the company’s recommendation, Sertoli cells were trypsinized to detach them from the culture surface and passaged every 2–3 days for maintenance. All experiments were conducted using the cells that had reached passage 7.
2.2. Virus
The Zika virus strain used in this study (ZIKV/Homo sapiens/PRI/PRVABC59/2015) was donated by Dr. David Safronetz, Chief of Special Pathogens, National Microbiology Laboratory, Public Health Agency of Canada. For future use, the ZIKV strain was expanded in Vero cells (ATCC, Manassas, VA, USA, Cat. #CCL-81) and kept at −80 °C in 10% FBS (Thermo Fisher Scientific, Waltham, CA, USA, Cat #A4766801). The stock virus was titered in Vero cells by plaque assay.
2.3. Infection
HSerC were grown in 75 cm2 cell culture flasks treated with poly-l-lysine. At 70% confluency, cells were infected with ZIKV at a multiplicity of infection (MOI) of 3 plaque-forming units (PFU) per cell. Culture plates were rocked every 10–15 min for 2 h in a 37 °C incubator (Thermo electron corporation, Waltham, MA, USA) to allow the virus to attach to cell surfaces, and cells were overlaid with Sertoli cell media containing 2.5% FBS. At 3, 5, and 7 days after infection, ZIKV-infected and mock, non-infected cells were collected. All experiments were done in three biological replicates.
2.4. Protein Extraction and Quantification
The Sertoli cells infected with ZIKV and time-matched mock-treated cells were scraped from the culture plates after 3, 5 and 7 days of infection. Centrifugation at 600× g for 8 min pelleted the cells, which were then washed three times with sterile ice-cold PBS(Thermo Fisher Scientific, Waltham, CA, USA). The pelleted cells were then lysed in 4% SDS in 100 mM HEPES buffer pH 8.5 by sonication. Centrifugation at 14,000× g for 10 min at 4 °C was used to remove insoluble cellular components. Bradford Protein Assay was used to determine the protein concentrations in the supernatants (Bio-Rad, Hercules, CA, USA, Cat. 5000001).
2.5. Immunoblotting
The protein concentrations in the cell lysates were measured using the Bradford Protein Assay, and 20 µg of protein were resolved in 10% SDS-PAGE gels and transferred to 0.2 µm nitrocellulose membranes. Anti-PSMA2 (Cell Signaling, Danvers, MA, USA, Cat. 2455), anti-ZIKV NS1 (BioFront Technologies, Tallahassee, FL, USA, Cat. BF-1225-06), anti-ZIKV NS3 (Genetex, Irvine, CA, USA, Cat No. GTX133309), anti-ZIKV Env (Genetex, Cat No. GTX133314), anti-STAT1 (Cell Signaling, Cat. 9176S), and anti-Beta-Actin (Cell Signaling, Cat. 3700S) antibodies were used to detect protein targets. Anti-rabbit (Cell Signaling, Cat. #7074) or anti-mouse (Cell Signaling, Cat. #7076) secondary antibody was used to identify the primary antibody conjugates to the targeted proteins. After overlaying with ECL reagents, protein bands were photographed with an Amersham Imager 680 (Gelifesciences, MA, USA). To quantify band intensities, Image J version. 1.53e (NIH, Bethesda, MD, USA) was used, and Graphpad Prism version 6.0. (La Jolla, CA, USA) was used to visualize them graphically.
2.6. Tandem Mass Tags (TMT) Mass Spectrometry Analyses and Protein Quantification
To determine the impact of ZIKV infection on the cellular proteome, a total of 18 protein samples were collected from ZIKV- and mock-infected Sertoli cells at 3, 5 and 7 days post-infection (dpi). Proteins were digested into peptides by the SP3 (single-pot solid-phase-enhanced sample preparation) methods as described elsewhere [38,39]. In summary, peptides were eluted after digestion of the proteins with trypsin for 14 h at 37 °C. Six-plex TMT labeling was performed for mock and infected samples of the same time points following the manufacturer’s (Thermo Fisher Scientific, Waltham, CA, USA) instructions. An equal amount of six TMT labeled samples were mixed together and 2D LC/MS/MS was performed using an Orbitrap Q Exactive HF-X instrument [40] (Thermo Fisher Scientific, Bremen, Germany). For identification of the peptide/proteins, ZIKV (Thai strain) and human (Uniprot 2016) databases were used as references. The intensities of TMT6 peptide level reporter tags were averaged over a ±0.1 Da window and corrected for isotopic overlap between channels using the batch-specific correction matrix provided. The sum of peptide level TMT6 reporter tag intensities for each protein was transformed into a log2 scale for easier differential analysis.
2.7. Statistical and Bioinformatics Analyses
Initially, alterations in the levels of any individual protein expression were determined by the differences between Log2 values (Delta log) of an infected and time-matched mock sample. Then, the delta Log2 values were converted to fold-change for each of the proteins. The p-value was determined by the Students t-test (2 tails) and Z-score analyses based on the protein expression difference of all three replicates. p-value < 0.05 and Z-score values of ≥.96 σ and ≤−1.96 σ were considered significant as described before [41]. The lists of significantly altered proteins were uploaded into Ingenuity Pathway Analysis (IPA) software, and core analysis was done with a cut-off value p-value < 0.05 and fold change above 1.5 or below −1.5. The IPA core analysis predicted the top affected canonical pathways, bio-functions, interconnecting networks and upstream molecules based on these protein level changes. The Western blot image band intensities were quantified using Image J version1.53e software (NIH, Bethesda, MD, USA), and statistical analyses were performed by one-way or two-way ANOVA (p-values < 0.05) in GraphPad Prism version 6.0. MORPHEUS (Broad Institute, Cambridge, MA, USA), a free internet software program, was used to generate the heatmaps.
3. Results
3.1. Infectivity of ZIKV and Its Cytopathic Effect in Primary HSerC
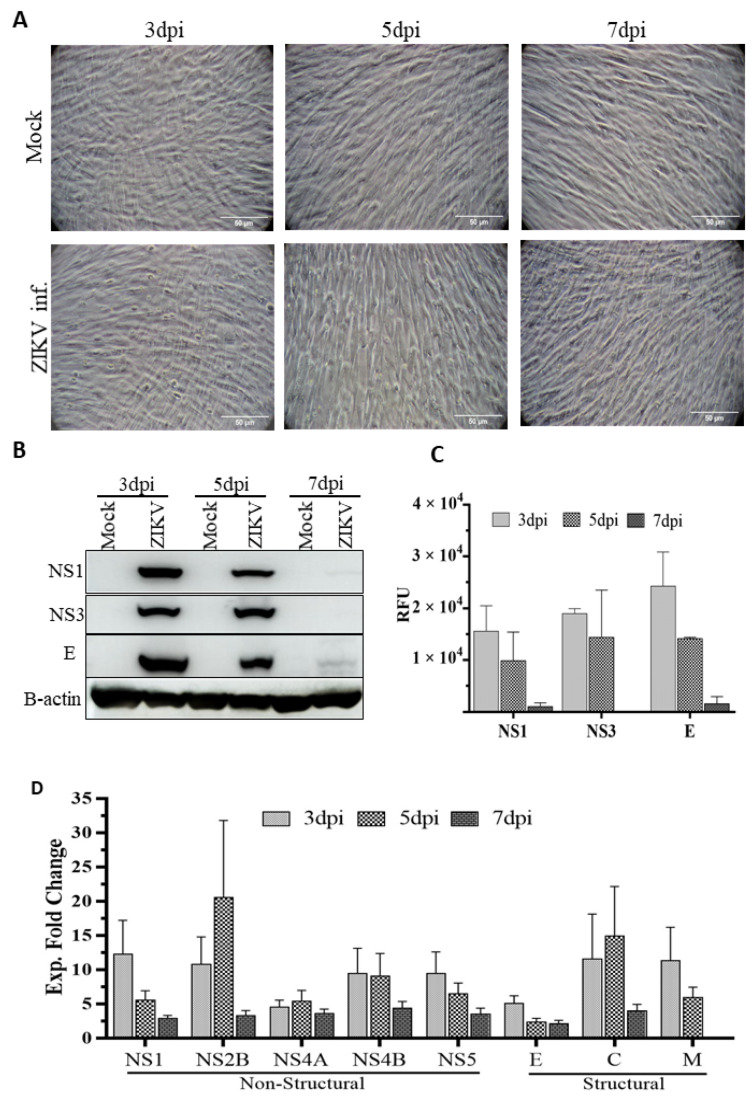
After infecting HSerC with ZIKV at MOI = 3, viral protein expression levels, which could result from increased expression, lower protein turnover, or a combination of both, and cytopathic effects were monitored at 3, 5 and 7 dpi, ZIKV infection did not induce any cytopathic effects (Figure 1A). However, three viral proteins (NS1, NS3, and E) were expressed at all time points in the infected cells, with maximal levels at 3 dpi; by 5 and 7 dpi, their expressions had dropped significantly (Figure 1B,C). Mass spectrometry analysis of Zika viral protein levels revealed that most of the proteins were at their highest levels at 3 dpi, with the exception of NS2B, NS4A, and C, which peaked at 5 dpi. All ZIKV proteins were expressed to the lowest level at 7 dpi (Figure 1D). In our previous study, we did not observe any significant protein level alterations at the early stage (1 dpi) of replication. However, virus titer peaks at 5 dpi and declines at 7 dpi [36]. Therefore, we selected day 3 (mid), day 5 (peak viral titer) and day 7 (late stage) for subsequent proteomic analyses based on these observations.
Figure 1.
Cytopathic impact of ZIKV infection on HSerC and viral protein expression. HSerC were infected with ZIKV an MOI of 3. (A) Cytopathic impact of ZIKV infection was observed at 3, 5 and 7 days post infection (dpi) under bright−field microscopy at 200× magnification. Scale bar is 50 μm. (B) Viral protein expression was determined by Western blot using ZIKV-NS1, NS3 and ENV monoclonal antibodies at 3, 5 and 7 dpi. (C) Quantitative expression of ZIKV viral proteins determined by densitometry analysis of Western blot images using Image J and normalized to B-actin expression. (D) ZIKV proteins expression detected after 3, 5 and 7 dpi by mass spectrometry. Log2 expressions of ZIKV proteins were compared with mock-treated cells and converted to Fold Change (FC) Abbreviations. dpi = Days post infection. Exp = Expression. RFU = Relative fluorescence units.
3.2. The Impact of ZIKV Infection on the HSerC Cellular Proteome
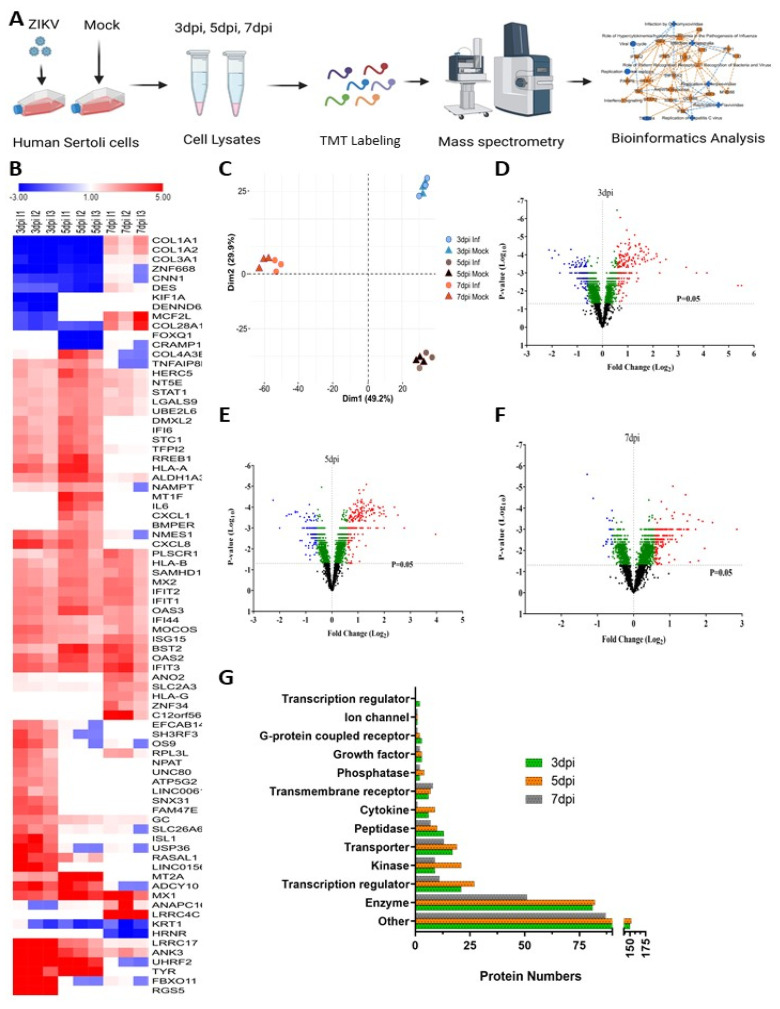
TMT-based mass spectrometry was used to determine the proteomic alterations in protein levels caused by ZIKV infection in HSerC, which detected more than 6000 proteins from each sample (Figure 2A). Among them, 4423 unique protein levels were significantly affected considering all three time points. A total of 2367 (853 up-regulated and 1514 down-regulated), 2363 (1651 up-regulated and 712 down-regulated), and 1782 (1209 up-regulated and 573 down-regulated) proteins were significantly (p value < 0.05) affected at 3, 5 and 7 dpi, respectively (Table 1).
Figure 2.
Detection of cellular protein level alterations in ZIKV-infected Sertoli cells by mass spectrometry. (A) Schematic flow diagram of the study design. (B) Heatmap of most affected proteins (Fold change ≥ 2.5 or ≤−2.5; p value < 0.05) by ZIKV infection in Sertoli cells. Red and blue colors indicate up-regulation and down-regulation, respectively. (C) PCA plot of proteomic data from mock- and ZIKV-infected cells, from all three replicates. Volcano plots displaying the protein level alterations after ZIKV infection at 3 dpi (D), 5 dpi (E), and 7 dpi (F). Red = significantly up-regulated (FC > 1.5; p-value < 0.05), blue = significantly down-regulated (FC < −1.5; p-value < 0.05), green = significantly affected (p-value < 0.05, but FC < ±1.5), black = not significantly affected (p-value > 0.05). (G) Classification of significantly altered (Fold change ≥ 1.5 or ≤ −1.5; p value < 0.05) protein types at different time points after ZIKV infection. Abbreviations. FC = Fold change; dpi = Days post infection; inf = Infected.
Table 1.
Numbers of significantly affected ZIKV-infected Sertoli proteins.
| Number That Are Significant | Total Unique | 3 dpi | 5 dpi | 7 dpi |
|---|---|---|---|---|
| and F.C. > 1.00 | 4423 | 853 | 1651 | 1209 |
| and F.C. < 1.00 | 1514 | 712 | 573 | |
| and F.C. > 1.10 | 4361 | 832 | 1632 | 1197 |
| and F.C. < −1.10 | 1495 | 700 | 559 | |
| and F.C. > 1.25 | 2234 | 393 | 744 | 647 |
| and F.C. < −1.25 | 755 | 329 | 193 | |
| and F.C. > 1.33 | 1342 | 286 | 468 | 382 |
| and F.C. < −1.33 | 395 | 197 | 90 | |
| and F.C. > 1.50 | 619 | 180 | 246 | 176 |
| and F.C. < −1.50 | 140 | 97 | 22 | |
| and F.C. > 1.66 | 351 | 119 | 161 | 94 |
| and F.C. < −1.66 | 71 | 45 | 7 | |
| and F.C. > 2.00 | 173 | 70 | 95 | 39 |
| and F.C. < −2.00 | 24 | 14 | 3 | |
| and F.C. > 2.50 | 79 | 36 | 38 | 19 |
| and F.C. < −2.50 | 6 | 8 | 1 |
Significance was determined by t-test and Z-score as detailed in Materials and Methods from three biological replicates. The 173 specific proteins whose levels were altered > 2.0-fold in either direction are listed in Table 2.
Heatmaps of the most affected proteins (Fold change ≥ 2.5 or ≤ −2.5; p-value < 0.05) revealed that certain protein levels were affected at specific time periods while other proteins were significantly affected at all times (Figure 2B). The topmost up-regulated proteins were Regulator of G-protein signaling 5 (RGS5, Fold change (FC): 45.4), F-box only protein 11 (FBXO11, FC: 41.1), E3 ubiquitin-protein ligase UHRF2 (UHRF2, FC: 17.5), Tyrosinase (TYR, FC: 15.6), Ankyrin-3 (ANK3, FC: 13.2) and the down-regulated were Forkhead box protein Q1 (FOXQ1, FC: −4.74), Kinesin-like protein (KIF1A, FC: −4.37), Collagen alpha-1(I) chain (COL1A1, FC: −3.95), Protein cramped-like (CRAMP1, FC: −4.02), and Hornerin (HRNR, FC: −3.93) (Figure 2B, Table 2). Principal component analysis (PCA) also showed that the protein samples were clustered by time point (Figure 2C).
Table 2.
List of protein levels altered at least 2-fold and significant by t-test (p-value < 0.05) or Z-score (≥1.96σ or ≤−1.96σ).
| Swprot | Gene | Protein | 3 dpi | 5 dpi | 7 dpi | |||
|---|---|---|---|---|---|---|---|---|
| Inf/Mock F.C. | p-Value | Inf/Mock F.C. | p-Value | Inf/Mock F.C. | p-Value | |||
| Up-regulated proteins | ||||||||
| O15539 | RGS5 | Regulator of G-protein signaling 5 | 45.46 | 5.5 × 10−3 | ||||
| Q86XK2 | FBXO11 | F-box only protein 11 | 41.16 | 4.9 × 10−3 | −1.01 | 7.9 × 10−1 | 1.11 | 1.9 × 10−1 |
| Q96PU4 | UHRF2 | E3 ubiquitin-protein ligase UHRF2 | 17.55 | 1.1 × 10−3 | 5.74 | 2.4 × 10−4 | −1.10 | 2.5 × 10−1 |
| Q12955 | ANK3 | Ankyrin-3 | 13.21 | 8.1 × 10−4 | 3.36 | 6.5 × 10−4 | 2.29 | 3.8 × 10−3 |
| Q8N6Y2 | LRRC17 | Leucine-rich repeat-containing protein 17 | 9.94 | 4.9 × 10−4 | 2.82 | 1.4 × 10−4 | 1.31 | 9.5 × 10−3 |
| O15544 | LINC01565 | Protein GR6 | 5.71 | 1.8 × 10−4 | ||||
| P14679 | TYR | Tyrosinase | 4.85 | 2.6 × 10−4 | 15.63 | 1.8 × 10−3 | ||
| O95294 | RASAL1 | RasGAP-activating-like protein 1 | 4.80 | 8.5 × 10−4 | 2.08 | 4.0 × 10−2 | ||
| Q9P275 | USP36 | Ubiquitin carboxyl-terminal hydrolase 36 | 4.44 | 1.2 × 10−4 | 1.03 | 7.0 × 10−1 | 1.06 | 3.5 × 10−1 |
| Q96PN6 | ADCY10 | Adenylate cyclase type 10 | 4.39 | 8.8 × 10−5 | 4.19 | 2.8 × 10−4 | 1.00 | 9.1 × 10−1 |
| P61371 | ISL1 | Insulin gene enhancer protein ISL-1 | 4.11 | 1.6 × 10−4 | ||||
| P10145 | CXCL8 | Interleukin-8 | 3.83 | 1.8 × 10−4 | 3.07 | 4.5 × 10−4 | ||
| P20591 | MX1 | Interferon-induced GTP-binding protein Mx1 | 3.53 | 8.1 × 10−5 | 5.24 | 1.0 × 10−4 | 4.54 | 4.8 × 10−4 |
| Q6ZV65 | FAM47E | Protein FAM47E | 3.53 | 4.9 × 10−5 | ||||
| Q13438 | OS9 | Protein | 3.42 | 2.1 × 10−4 | 0.99 | 8.0 × 10−1 | 1.04 | 5.7 × 10−1 |
| Q8N9S9 | SNX31 | Sorting nexin-31 | 3.31 | 1.0 × 10−4 | ||||
| O14879 | IFIT3 | Interferon-induced protein with tetratricopeptide repeats 3 | 3.30 | 3.5 × 10−5 | 3.41 | 9.5 × 10−5 | 3.75 | 4.2 × 10−4 |
| Q8TEJ3 | SH3RF3 | SH3 domain-containing RING finger protein 3 | 3.22 | 2.4 × 10−4 | 1.01 | 8.1 × 10−1 | ||
| P30447 | HLA-A | HLA class I histocompatibility antigen, A-23 alpha chain | 3.17 | 1.8 × 10−4 | 3.96 | 1.5 × 10−4 | ||
| Q96EN8 | MOCOS | Molybdenum cofactor sulfurase | 3.12 | 1.4 × 10−4 | 2.20 | 6.7 × 10−4 | 2.23 | 3.4 × 10−3 |
| Q9BXS9 | SLC26A6 | Solute carrier family 26 member 6 | 3.10 | 7.0 × 10−5 | 1.21 | 1.1 × 10−2 | 1.13 | 1.5 × 10−1 |
| P29728 | OAS2 | 2’-5’-oligoadenylate synthase 2 | 2.98 | 1.9 × 10−4 | 3.54 | 1.8 × 10−4 | 3.28 | 9.1 × 10−4 |
| Q9C002 | NMES1 | Normal mucosa of esophagus-specific gene 1 protein | 2.96 | 1.7 × 10−4 | 2.91 | 1.4 × 10−4 | 1.18 | 1.3 × 10−1 |
| P05161 | ISG15 | Ubiquitin-like protein ISG15 | 2.92 | 5.9 × 10−5 | 2.20 | 4.3 × 10−5 | 2.93 | 2.1 × 10−4 |
| P02774 | GC | Vitamin D-binding protein | 2.87 | 4.5 × 10−5 | 1.86 | 3.8 × 10−4 | 1.34 | 4.1 × 10−5 |
| Q9Y6K5 | OAS3 | 2’-5’-oligoadenylate synthase 3 | 2.85 | 4.3 × 10−5 | 3.60 | 1.1 × 10−4 | 2.30 | 1.4 × 10−3 |
| P09913 | IFIT2 | Interferon-induced protein with tetratricopeptide repeats 2 | 2.82 | 6.5 × 10−5 | 2.83 | 5.7 × 10−5 | 2.83 | 8.4 × 10−4 |
| Q14207 | NPAT | Protein NPAT | 2.78 | 3.3 × 10−4 | ||||
| P09914 | IFIT1 | Interferon-induced protein with tetratricopeptide repeats 1 | 2.76 | 2.4 × 10−5 | 3.05 | 8.4 × 10−5 | 2.76 | 9.0 × 10−4 |
| Q06055 | ATP5G2 | ATP synthase F(0) complex subunit C2, mitochondrial | 2.73 | 2.8 × 10−5 | ||||
| Q8TCB0 | IFI44 | Interferon-induced protein 44 | 2.68 | 1.1 × 10−4 | 2.07 | 4.5 × 10−4 | 2.37 | 7.5 × 10−4 |
| O75071 | EFCAB14 | EF-hand calcium-binding domain-containing protein 14 | 2.67 | 1.7 × 10−3 | 1.06 | 2.9 × 10−1 | ||
| Q8N2C7 | UNC80 | Protein unc-80 homolog | 2.66 | 1.6 × 10−4 | ||||
| Q92901 | RPL3L | 60S ribosomal protein L3-like | 2.65 | 2.4 × 10−3 | 1.97 | 1.6 × 10−2 | ||
| Q92766 | RREB1 | Ras-responsive element-binding protein 1 | 2.56 | 7.7 × 10−4 | 3.72 | 2.1 × 10−4 | ||
| Q96LM1 | LINC00615 | Putative uncharacterized protein encoded by LINC00615 | 2.56 | 3.0 × 10−3 | ||||
| P47895 | ALDH1A3 | Aldehyde dehydrogenase family 1 member A3 | 2.50 | 3.4 × 10−5 | 3.86 | 9.7 × 10−5 | 1.38 | 2.9 × 10−2 |
| P20592 | MX2 | Interferon-induced GTP-binding protein Mx2 | 2.48 | 9.3 × 10−5 | 2.98 | 1.2 × 10−4 | 2.80 | 4.1 × 10−4 |
| Q96AZ6 | ISG20 | Interferon-stimulated gene 20 kDa protein | 2.47 | 3.0 × 10−5 | 2.46 | 3.0 × 10−4 | ||
| Q9BXU1 | STK31 | Serine/threonine-protein kinase 31 | 2.47 | 3.2 × 10−4 | ||||
| Q9Y3Z3 | SAMHD1 | Deoxynucleoside triphosphate triphosphohydrolase SAMHD1 | 2.46 | 1.0 × 10−4 | 2.82 | 4.5 × 10−5 | 2.49 | 1.1 × 10−3 |
| P52823 | STC1 | Stanniocalcin-1 | 2.43 | 2.6 × 10−4 | 2.98 | 1.4 × 10−4 | ||
| P02795 | MT2A | Metallothionein-2 | 2.42 | 8.0 × 10−6 | 6.81 | 6.8 × 10−4 | ||
| P30464 | HLA-B | HLA class I histocompatibility antigen, B-15 alpha chain | 2.41 | 2.3 × 10−4 | 2.49 | 1.6 × 10−4 | 2.11 | 1.5 × 10−3 |
| A6ND36 | FAM83G | Protein FAM83G | 2.33 | 1.6 × 10−4 | 1.77 | 1.9 × 10−4 | 1.14 | 7.0 × 10−4 |
| P48307 | TFPI2 | Tissue factor pathway inhibitor 2 | 2.32 | 1.4 × 10−4 | 3.11 | 1.9 × 10−4 | 1.18 | 4.1 × 10−2 |
| O95760 | IL33 | Interleukin-33 | 2.30 | 4.4 × 10−4 | 2.35 | 1.1 × 10−4 | ||
| Q6UXH9 | PAMR1 | Inactive serine protease PAMR1 | 2.26 | 2.1 × 10−4 | 1.12 | 7.5 × 10−2 | ||
| P09912 | IFI6 | Interferon alpha-inducible protein 6 | 2.23 | 5.0 × 10−4 | 2.87 | 2.2 × 10−4 | ||
| Q71F56 | MED13L | Mediator of RNA polymerase II transcription subunit 13-like | 2.23 | 3.2 × 10−4 | 1.04 | 8.1 × 10−1 | ||
| Q8TDJ6 | DMXL2 | DmX-like protein 2 | 2.21 | 1.1 × 10−3 | 3.02 | 1.9 × 10−4 | ||
| Q6P589 | TNFAIP8L2 | Tumor necrosis factor alpha-induced protein 8-like protein 2 | 2.20 | 1.9 × 10−4 | 2.59 | 6.9 × 10−4 | −1.13 | 2.2 × 10−1 |
| P18464 | HLA-B | HLA class I histocompatibility antigen, B-51 alpha chain | 2.18 | 7.6 × 10−4 | 2.78 | 1.5 × 10−4 | 2.62 | 9.1 × 10−4 |
| Q10589 | BST2 | Bone marrow stromal antigen 2 | 2.14 | 3.7 × 10−5 | 3.95 | 2.1 × 10−4 | 3.47 | 3.5 × 10−4 |
| P01033 | TIMP1 | Metalloproteinase inhibitor 1 | 2.11 | 4.2 × 10−5 | 2.02 | 4.5 × 10−4 | 1.11 | 1.3 × 10−1 |
| Q96J88 | EPSTI1 | Epithelial-stromal interaction protein 1 | 2.10 | 8.8 × 10−4 | 1.57 | 7.8 × 10−3 | ||
| O14933 | UBE2L6 | Ubiquitin/ISG15-conjugating enzyme E2 L6 | 2.09 | 2.2 × 10−5 | 2.71 | 2.5 × 10−4 | 1.73 | 7.4 × 10−3 |
| P30685 | HLA-B | HLA class I histocompatibility antigen, B-35 alpha chain | 2.09 | 1.3 × 10−4 | 2.28 | 2.8 × 10−4 | 2.00 | 2.0 × 10−3 |
| Q86UQ4 | ABCA13 | ATP-binding cassette sub-family A member 13 | 2.09 | 1.9 × 10−3 | ||||
| P42224 | STAT1 | Signal transducer and activator of transcription 1-alpha/beta | 2.08 | 1.4 × 10−5 | 2.74 | 5.8 × 10−5 | 1.71 | 3.3 × 10−3 |
| P15407 | FOSL1 | Fos-related antigen 1 | 2.08 | 1.5 × 10−4 | 2.18 | 3.2 × 10−4 | ||
| O14791 | APOL1 | Apolipoprotein L1 | 2.07 | 5.5 × 10−4 | ||||
| Q9UII4 | HERC5 | E3 ISG15--protein ligase HERC5 | 2.07 | 5.5 × 10−4 | 3.23 | 5.5 × 10−4 | 1.75 | 3.4 × 10−3 |
| O00182 | LGALS9 | Galectin-9 | 2.07 | 1.8 × 10−4 | 2.69 | 1.2 × 10−4 | 1.67 | 3.9 × 10−3 |
| Q14520 | HABP2 | Hyaluronan-binding protein 2 | 2.06 | 6.7 × 10−4 | 1.65 | 2.4 × 10−4 | 1.38 | 6.3 × 10−3 |
| Q8IXQ6 | PARP9 | Poly [ADP-ribose] polymerase 9 | 2.05 | 2.8 × 10−5 | 2.11 | 3.3 × 10−4 | 1.77 | 1.6 × 10−3 |
| Q03405 | PLAUR | Urokinase plasminogen activator surface receptor | 2.05 | 5.0 × 10−5 | 1.53 | 2.0 × 10−3 | 1.41 | 7.8 × 10−4 |
| Q96L93 | KIF16B | Kinesin-like protein KIF16B | 2.04 | 2.1 × 10−3 | 1.13 | 1.4 × 10−1 | ||
| Q29960 | HLA-C | HLA class I histocompatibility antigen, Cw-16 alpha chain | 2.04 | 2.1 × 10−4 | 2.37 | 4.4 × 10−4 | ||
| P25774 | CTSS | Cathepsin S | 2.02 | 4.8 × 10−4 | 1.79 | 3.3 × 10−4 | ||
| P04733 | MT1F | Metallothionein-1F | 3.99 | 5.9 × 10−4 | ||||
| Q9Y5P4 | COL4A3BP | Collagen type IV alpha-3-binding protein | 1.08 | 1.3 × 10−1 | 3.52 | 2.6 × 10−4 | −1.03 | 5.5 × 10−1 |
| P05231 | IL6 | Interleukin-6 | 3.51 | 1.1 × 10−4 | ||||
| P21589 | NT5E | 5’-nucleotidase | 1.98 | 8.2 × 10−5 | 2.87 | 1.2 × 10−4 | 1.88 | 1.6 × 10−3 |
| P09341 | CXCL1 | Growth-regulated alpha protein | 2.76 | 6.6 × 10−4 | ||||
| Q8N8U9 | BMPER | BMP-binding endothelial regulator protein | 2.62 | 7.6 × 10−5 | ||||
| O15162 | PLSCR1 | Phospholipid scramblase 1 | 1.84 | 9.3 × 10−4 | 2.61 | 5.0 × 10−4 | 2.81 | 6.2 × 10−4 |
| P43490 | NAMPT | Nicotinamide phosphoribosyltransferase | 1.55 | 3.6 × 10−5 | 2.60 | 1.5 × 10−4 | −1.01 | 8.9 × 10−1 |
| P02549 | SPTA1 | Spectrin alpha chain, erythrocytic 1 | 1.16 | 2.8 × 10−2 | 2.49 | 8.1 × 10−6 | ||
| Q9H5V8 | CDCP1 | CUB domain-containing protein 1 | 1.87 | 2.7 × 10−4 | 2.47 | 1.9 × 10−4 | 1.44 | 7.9 × 10−4 |
| P04179 | SOD2 | Superoxide dismutase [Mn], mitochondrial | 1.54 | 1.2 × 10−3 | 2.47 | 8.6 × 10−5 | −1.27 | 2.2 × 10−2 |
| O43581 | SYT7 | Synaptotagmin-7 | 2.45 | 7.5 × 10−3 | ||||
| Q8NAP3 | ZBTB38 | Zinc finger and BTB domain-containing protein 38 | 1.15 | 6.6 × 10−3 | 2.44 | 4.6 × 10−4 | 1.06 | 3.6 × 10−1 |
| Q53G44 | IFI44L | Interferon-induced protein 44-like | 1.83 | 3.3 × 10−4 | 2.34 | 1.5 × 10−4 | 2.38 | 5.3 × 10−4 |
| Q14457 | BECN1 | Beclin-1 | 1.77 | 1.1 × 10−4 | 2.32 | 6.7 × 10−5 | 1.03 | 5.9 × 10−1 |
| P35354 | PTGS2 | Prostaglandin G/H synthase 2 | 1.99 | 3.7 × 10−4 | 2.31 | 1.5 × 10−5 | −1.13 | 2.9 × 10−1 |
| Q8WZ74 | CTTNBP2 | Cortactin-binding protein 2 | 2.27 | 5.4 × 10−4 | 1.24 | 1.9 × 10−2 | ||
| P26022 | PTX3 | Pentraxin-related protein PTX3 | 1.54 | 3.3 × 10−4 | 2.26 | 2.5 × 10−3 | −1.05 | 5.1 × 10−1 |
| Q5EBM0 | CMPK2 | UMP-CMP kinase 2, mitochondrial | 1.91 | 1.1 × 10−4 | 2.25 | 9.5 × 10−5 | 1.84 | 1.3 × 10−3 |
| P28845 | HSD11B1 | Corticosteroid 11-beta-dehydrogenase isozyme 1 | 1.56 | 1.6 × 10−3 | 2.23 | 2.6 × 10−4 | 1.19 | 3.7 × 10−2 |
| Q8WWZ7 | ABCA5 | ATP-binding cassette sub-family A member 5 | 2.23 | 5.5 × 10−5 | 1.13 | 1.2 × 10−1 | ||
| O95786 | DDX58 | Probable ATP-dependent RNA helicase DDX58 | 1.79 | 1.2 × 10−4 | 2.23 | 2.1 × 10−4 | 2.11 | 1.3 × 10−3 |
| P04732 | MT1E | Metallothionein-1E | 1.26 | 3.8 × 10−3 | 2.22 | 1.5 × 10−4 | 1.91 | 2.3 × 10−3 |
| Q96RQ9 | IL4I1 | l-amino-acid oxidase | 1.74 | 3.3 × 10−4 | 2.22 | 1.6 × 10−4 | 1.28 | 2.8 × 10−2 |
| P01892 | HLA-A | HLA class I histocompatibility antigen, A-2 alpha chain | 1.78 | 2.8 × 10−4 | 2.22 | 4.1 × 10−4 | 1.60 | 4.9 × 10−3 |
| Q4VCS5 | AMOT | Angiomotin | 1.92 | 4.8 × 10−4 | 2.20 | 1.1 × 10−4 | ||
| P00973 | OAS1 | 2’-5’-oligoadenylate synthase 1 | 1.99 | 1.0 × 10−4 | 2.20 | 1.3 × 10−3 | 2.02 | 3.5 × 10−3 |
| Q5TEJ8 | THEMIS2 | Protein THEMIS2 | 2.18 | 6.1 × 10−5 | ||||
| Q96C10 | DHX58 | Probable ATP-dependent RNA helicase DHX58 | 1.35 | 1.6 × 10−2 | 2.18 | 1.8 × 10−4 | 1.08 | 2.6 × 10−1 |
| P13500 | CCL2 | C-C motif chemokine 2 | 2.17 | 7.9 × 10−4 | ||||
| Q9BY76 | ANGPTL4 | Angiopoietin-related protein 4 | 1.58 | 4.7 × 10−4 | 2.17 | 8.3 × 10−4 | 1.36 | 1.0 × 10−2 |
| Q9Y6I4 | USP3 | Ubiquitin carboxyl-terminal hydrolase 3 | 1.01 | 7.1 × 10−1 | 2.16 | 1.3 × 10−4 | ||
| O94808 | GFPT2 | Glutamine--fructose-6-phosphate aminotransferase [isomerizing] 2 | 1.65 | 6.4 × 10−5 | 2.15 | 2.6 × 10−4 | 1.38 | 6.8 × 10−3 |
| P15153 | RAC2 | Ras-related C3 botulinum toxin substrate 2 | 1.58 | 2.7 × 10−4 | 2.14 | 5.2 × 10−5 | 1.04 | 3.8 × 10−1 |
| P19525 | EIF2AK2 | Interferon-induced, double-stranded RNA-activated protein kinase | 1.59 | 1.1 × 10−4 | 2.13 | 1.1 × 10−4 | 1.91 | 1.6 × 10−3 |
| Q9HB58 | SP110 | Sp110 nuclear body protein | 1.80 | 1.7 × 10−3 | 2.11 | 9.3 × 10−4 | 1.81 | 1.2 × 10−3 |
| P05534 | HLA-A | HLA class I histocompatibility antigen, A-24 alpha chain | 1.58 | 4.0 × 10−4 | 2.09 | 1.9 × 10−4 | 1.98 | 8.2 × 10−4 |
| P52926 | HMGA2 | High mobility group protein HMGI-C | 1.28 | 2.5 × 10−2 | 2.09 | 2.2 × 10−5 | −1.16 | 5.7 × 10−2 |
| O75508 | CLDN11 | Claudin-11 | 1.05 | 5.9 × 10−1 | 2.09 | 3.9 × 10−4 | 1.27 | 2.1 × 10−3 |
| P07148 | FABP1 | Fatty acid-binding protein, liver | 2.09 | 4.4 × 10−4 | −1.09 | 4.6 × 10−1 | ||
| Q7Z402 | TMC7 | Transmembrane channel-like protein 7 | 2.08 | 2.3 × 10−4 | ||||
| Q01973 | ROR1 | Inactive tyrosine-protein kinase transmembrane receptor ROR1 | 2.08 | 4.6 × 10−4 | 1.41 | 2.1 × 10−3 | ||
| Q8IVT2 | MISP | Mitotic interactor and substrate of PLK1 | 1.75 | 3.8 × 10−4 | 2.07 | 4.0 × 10−4 | ||
| Q3MIT2 | PUS10 | Putative tRNA pseudouridine synthase Pus10 | 2.06 | 4.3 × 10−3 | −1.31 | 5.5 × 10−3 | ||
| P27701 | CD82 | CD82 antigen | 1.34 | 1.6 × 10−2 | 2.05 | 1.5 × 10−5 | 1.22 | 4.3 × 10−2 |
| Q15646 | OASL | 2’-5’-oligoadenylate synthase-like protein | 1.82 | 1.3 × 10−4 | 2.05 | 2.8 × 10−4 | 1.99 | 1.8 × 10−3 |
| P01584 | IL1B | Interleukin-1 beta | 1.88 | 9.0 × 10−5 | 2.04 | 2.1 × 10−4 | ||
| Q7Z3S9 | NOTCH2NL | Notch homolog 2 N-terminal-like protein | 1.13 | 1.0 × 10−1 | 2.02 | 1.7 × 10−4 | 1.03 | 6.1 × 10−1 |
| P68871 | HBB | Hemoglobin subunit beta | 1.54 | 3.6 × 10−3 | 2.02 | 2.4 × 10−4 | 1.31 | 1.3 × 10−2 |
| Q9BYX4 | IFIH1 | Interferon-induced helicase C domain-containing protein 1 | 1.82 | 1.8 × 10−4 | 2.01 | 2.5 × 10−4 | 2.09 | 2.7 × 10−3 |
| P16070 | CD44 | CD44 antigen | 1.42 | 8.9 × 10−4 | 2.01 | 2.2 × 10−4 | 1.32 | 3.6 × 10−3 |
| P28838 | LAP3 | Cytosol aminopeptidase | 1.66 | 2.3 × 10−5 | 2.00 | 1.4 × 10−4 | 1.42 | 6.2 × 10−3 |
| Q92597 | NDRG1 | Protein NDRG1 | 1.21 | 1.6 × 10−3 | 2.00 | 1.4 × 10−4 | 1.75 | 7.5 × 10−4 |
| P11166 | SLC2A1 | Solute carrier family 2, facilitated glucose transporter member 1 | 1.65 | 6.2 × 10−4 | 1.91 | 7.9 × 10−4 | 2.12 | 9.2 × 10−6 |
| P09871 | C1S | Complement C1s subcomponent | 1.08 | 2.3 × 10−1 | 1.25 | 5.7 × 10−3 | 2.10 | 1.3 × 10−3 |
| P11169 | SLC2A3 | Solute carrier family 2, facilitated glucose transporter member 3 | 1.28 | 3.2 × 10−3 | 1.21 | 2.2 × 10−2 | 2.71 | 2.3 × 10−5 |
| Q96DE5 | ANAPC16 | Anaphase-promoting complex subunit 16 | −1.12 | 2.0 × 10−1 | 1.13 | 8.3 × 10−3 | 3.57 | 4.4E−10 |
| P00966 | ASS1 | Argininosuccinate synthase | −1.12 | 1.1 × 10−1 | −1.13 | 2.1 × 10−2 | 2.08 | 1.6 × 10−3 |
| Q2UY09 | COL28A1 | Collagen alpha-1(XXVIII) chain | −1.93 | 1.2 × 10−3 | −1.71 | 3.1 × 10−3 | 2.95 | 3.6 × 10−2 |
| P04264 | KRT1 | Keratin, type II cytoskeletal 1 | −1.40 | 1.9 × 10−1 | −1.98 | 1.9 × 10−2 | −2.41 | 2.5 × 10−6 |
| Q9HCJ2 | LRRC4C | Leucine-rich repeat-containing protein 4C | 7.23 | 1.3 × 10−3 | ||||
| Q8IXR9 | C12orf56 | Uncharacterized protein C12orf56 | 3.90 | 8.3 × 10−3 | ||||
| O15068 | MCF2L | Guanine nucleotide exchange factor DBS | −1.93 | 1.6 × 10−3 | 3.47 | 3.1 × 10−2 | ||
| Q9NQ90 | ANO2 | Anoctamin-2 | 1.23 | 2.1 × 10−2 | 2.86 | 1.2 × 10−3 | ||
| P17693 | HLA-G | HLA class I histocompatibility antigen, alpha chain G | 2.59 | 1.9 × 10−4 | ||||
| Q8IZ26 | ZNF34 | Zinc finger protein 34 | 2.51 | 2.2 × 10−3 | ||||
| Q9Y225 | RNF24 | RING finger protein 24 | 1.13 | 5.7 × 10−2 | 2.42 | 8.5 × 10−4 | ||
| P04222 | HLA-C | HLA class I histocompatibility antigen, Cw-3 alpha chain | 2.18 | 1.6 × 10−4 | ||||
| Q13794 | PMAIP1 | Phorbol-12-myristate-13-acetate-induced protein 1 | 2.12 | 3.1 × 10−4 | ||||
| Q96B67 | ARRDC3 | Arrestin domain-containing protein 3 | 2.09 | 2.3 × 10−4 | ||||
| Q13772 | NCOA4 | Nuclear receptor coactivator 4 | −1.04 | 6.0 × 10−1 | 2.08 | 1.3 × 10−3 | ||
| Q13137 | CALCOCO2 | Calcium-binding and coiled-coil domain-containing protein 2 | 1.19 | 3.4 × 10−2 | −1.04 | 4.1 × 10−1 | 2.03 | 7.2 × 10−4 |
| Down-regulated proteins | ||||||||
| Q12756 | KIF1A | Kinesin-like protein KIF1A | −4.38 | 5.4 × 10−5 | ||||
| P02452 | COL1A1 | Collagen alpha-1(I) chain | −3.95 | 1.5 × 10−4 | −3.39 | 2.6 × 10−4 | 2.19 | 2.7 × 10−2 |
| Q96K58 | ZNF668 | Zinc finger protein 668 | −3.66 | 6.0 × 10−5 | −2.63 | 1.6 × 10−4 | 1.03 | 3.6 × 10−1 |
| P08123 | COL1A2 | Collagen alpha-2(I) chain | −3.39 | 1.0 × 10−4 | −3.15 | 2.2 × 10−4 | 1.97 | 3.7 × 10−2 |
| Q8IWF6 | DENND6A | Protein DENND6A | −3.20 | 6.3 × 10−4 | ||||
| P02461 | COL3A1 | Collagen alpha-1(III) chain | −2.87 | 3.2 × 10−4 | −2.97 | 2.2 × 10−4 | 1.33 | 1.1 × 10−2 |
| Q9H1P3 | OSBPL2 | Oxysterol-binding protein-related protein 2 | −2.47 | 3.1 × 10−4 | 1.41 | 4.9 × 10−3 | ||
| Q8IZX4 | TAF1L | Transcription initiation factor TFIID subunit 1-like | −2.46 | 5.1 × 10−4 | −1.69 | 3.8 × 10−3 | ||
| Q69YL0 | NCBP2-AS2 | Uncharacterized protein NCBP2-AS2 | −2.42 | 6.9 × 10−4 | −1.51 | 3.6 × 10−3 | 1.66 | 2.3 × 10−2 |
| Q96JG9 | ZNF469 | Zinc finger protein 469 | −2.41 | 5.0 × 10−5 | 1.03 | 7.2 × 10−1 | ||
| P52732 | KIF11 | Kinesin-like protein KIF11 | −2.34 | 6.7 × 10−4 | ||||
| P09486 | SPARC | SPARC | −2.32 | 3.7 × 10−4 | −2.08 | 9.4 × 10−4 | 1.41 | 9.8 × 10−3 |
| P20908 | COL5A1 | Collagen alpha-1(V) chain | −2.29 | 2.4 × 10−4 | −1.99 | 4.9 × 10−4 | 1.40 | 4.5 × 10−2 |
| Q96PQ7 | KLHL5 | Kelch-like protein 5 | −2.20 | 3.6 × 10−4 | −1.11 | 1.2 × 10−1 | ||
| P51911 | CNN1 | Calponin-1 | −2.15 | 7.1 × 10−4 | −2.51 | 1.7 × 10−4 | 1.12 | 2.0 × 10−1 |
| Q8N7X1 | RBMXL3 | RNA-binding motif protein, X-linked-like-3 | −2.13 | 5.0 × 10−3 | −2.22 | 4.0 × 10−3 | ||
| O00767 | SCD | Acyl-CoA desaturase | −2.13 | 4.9 × 10−4 | −1.69 | 2.1 × 10−3 | 1.31 | 5.7 × 10−3 |
| P61916 | NPC2 | Epididymal secretory protein E1 | −2.10 | 4.3 × 10−4 | −2.09 | 3.1 × 10−4 | −1.52 | 4.1 × 10−3 |
| Q05682 | CALD1 | Caldesmon | −2.08 | 2.7 × 10−4 | −1.95 | 6.8 × 10−4 | −1.15 | 1.1 × 10−1 |
| Q8N806 | UBR7 | Putative E3 ubiquitin-protein ligase UBR7 | −2.06 | 1.3 × 10−3 | 1.47 | 2.6 × 10−3 | 1.03 | 6.3 × 10−1 |
| Q15113 | PCOLCE | Procollagen C-endopeptidase enhancer 1 | −2.06 | 4.6 × 10−4 | −1.65 | 7.8 × 10−4 | 1.39 | 1.7 × 10−2 |
| O75094 | SLIT3 | Slit homolog 3 protein | −2.04 | 3.5 × 10−4 | ||||
| Q07352 | ZFP36L1 | Zinc finger protein 36, C3H1 type-like 1 | −2.00 | 1.4 × 10−4 | −1.03 | 5.6 × 10−1 | 1.42 | 5.6 × 10−4 |
| P52736 | ZNF133 | Zinc finger protein 133 | −2.00 | 1.7 × 10−3 | ||||
| Q9C009 | FOXQ1 | Forkhead box protein Q1 | −4.75 | 4.6 × 10−5 | ||||
| Q96RY5 | CRAMP1 | Protein cramped-like | −4.03 | 1.1 × 10−3 | 1.01 | 7.7 × 10−1 | ||
| P17661 | DES | Desmin | −1.49 | 5.7 × 10−3 | −2.63 | 1.8 × 10−4 | 1.39 | 4.5 × 10−2 |
| P35527 | KRT9 | Keratin, type I cytoskeletal 9 | −1.55 | 1.3 × 10−1 | −2.35 | 4.0 × 10−3 | −2.17 | 3.4 × 10−5 |
| Q9NRM1 | ENAM | Enamelin | −1.59 | 3.2 × 10−4 | −2.31 | 2.8 × 10−4 | 1.51 | 3.1 × 10−3 |
| O43854 | EDIL3 | EGF-like repeat and discoidin I-like domain-containing protein 3 | −1.70 | 1.1 × 10−3 | −2.16 | 8.2 × 10−4 | −1.33 | 2.6 × 10−3 |
| Q86YZ3 | HRNR | Hornerin | −3.94 | 1.8 × 10−13 | ||||
dpi = days post infection. FC = fold change, red = significantly up-regulated; green = significantly down-regulated, blue = p-value < 0.05. Table is sorted first by significantly up-regulated and downregulated proteins at 3 dpi, then by those significantly down-regulated at 5 dpi; then by significantly up- and down-regulated at 7 dpi.
The volcano plot analysis of the proteins revealed that the majority of the highly affected proteins were down-regulated at 3 dpi (Figure 2D). In contrast, the most significantly up-regulated were at 5 and 7 dpi (Figure 2E,F). The altered proteins belong to different classes of proteins, including enzymes, transcription regulators, kinases, transporters, peptides, cytokines, transmembrane receptors, phosphatases, G protein-coupled receptors, growth factors, ion channels, transcription regulators and others (Figure 2G). We applied +/−1.5-fold fold-change cut-off with p < 0.05 for subsequent more complete bioinformatics analysis.
3.3. Impact of ZIKV Infection on Cellular Signaling Pathways and Function in HSerC
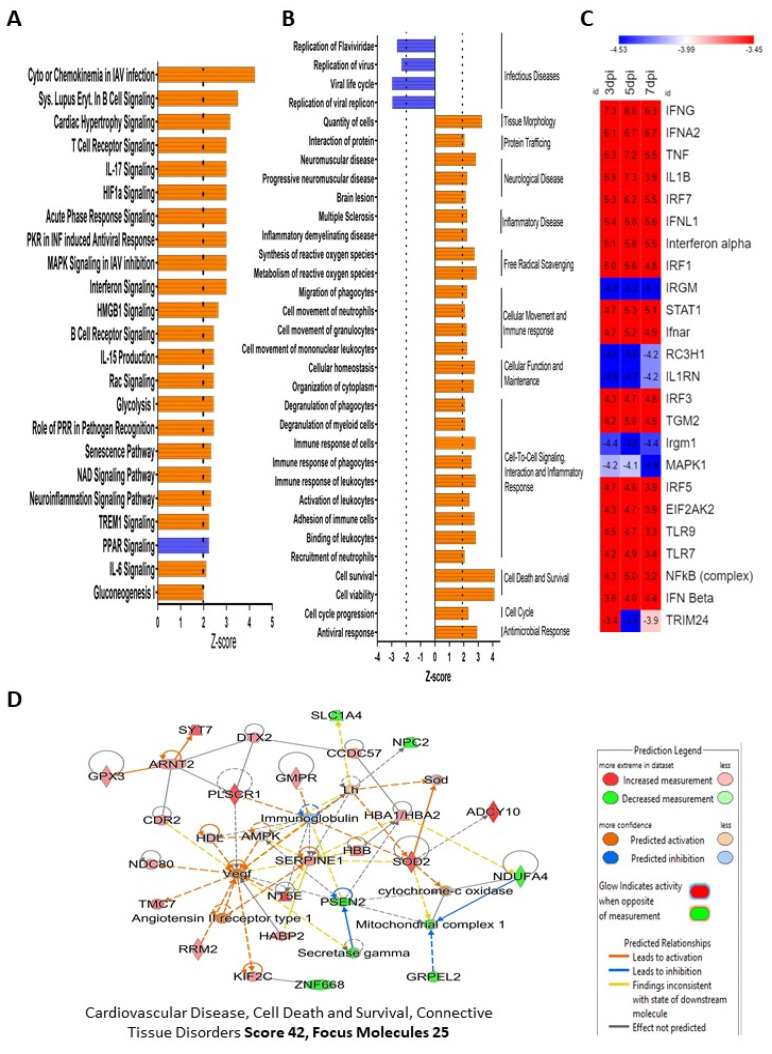
All proteins and their measured quantities at 3, 5 and 7 dpi were analyzed by Ingenuity Pathway Analysis (IPA) software to understand predicted impacts of ZIKV infection on cellular signaling pathways, bio-functions, and protein–protein networks. A total of 15, 31 and 13 Sertoli cell canonical pathways were predicted to be significantly affected at 3, 5 and 7 dpi, respectively, after ZIKV infection (Figure 3A, Figure S1A). GP6 signaling pathway was inhibited at 3 dpi, but was activated significantly at 7 dpi. Natural killer cell signaling was significantly down-regulated only at 7 dpi. Seven signaling pathways were predicted to be significantly activated at all three-time points; they are hypercytokinemia/hyperchemokinemia in the pathogenesis of influenza, interferon signaling, Systemic Lupus Erythematosus in B cell signaling pathway, role of PKR in interferon induction and antiviral response, neuroinflammation signaling pathway, role of pattern recognition receptors in recognition of bacteria and viruses, and NAD signaling pathway (Figure 3, Figure S1A). Interestingly, a few canonical pathways were only affected at 5 dpi, e.g., IL-17 signaling, IL-6 signaling, Cardiac hypertrophy signaling (enhanced), HIF1α signaling, Glycolysis I, Gluconeogenesis I, PPAR signaling, HMGB1 signaling, Rac signaling, etc. IPA predicted the altered proteins were significantly (Z score >1.96 or <−1.96) associated with a total of 156 diseases and functions, which were divided into 55 for 3 dpi, 95 for 5 dpi and 63 for 7 dpi (Table S1 Pathways associated with infectious disease were predicted to be significantly inhibited, whereas those associated with antimicrobial response, inflammatory disease, neurological disease, tissue morphology, cell death and survival, and cellular movement were the major disease and cellular function categories predicted to be activated at all three time points (Figure 3B; Figure S2). IPA also predicted a total of 801 unique upstream regulators to be activated/inhibited by ZIKV infection in Sertoli cells at 3 dpi (n = 400, activated, n = 279; inhibited 121), 5 dpi (n = 616, activated, n = 442; inhibited 174), and 7 dpi (n = 299, activated, n = 206; inhibited 93), which includes cytokines, enzymes, G-protein coupled receptors, growth factors, kinases, microRNAs, peptidases, phosphatase, transcription regulators, transmembrane receptor and transporters (Table S2). The top ten predicted upstream regulators are IFNG, IFNA2, TNF, IL1B, IRF7, IFNL1, Interferon-alpha, IFR1, IRGM and STAT1 (Figure 3C). The functional analysis of the top 15 upstream regulators showed that they are associated with activation of T-lymphocytes, leukocytes, cells, recruitment of leukocytes, antiviral response, and RNA virus replication (Figure S1B).
Figure 3.
IPA-predicted activation and inhibition of bio-functions, canonical pathways, protein–protein networks, and upstream molecules after ZIKV infection in HSerC. (A) Top-most canonical pathways predicted to be significantly activated or inhibited by IPA at 5 days post ZIKV infection. (B) Top bio-functions and predicted activation or inhibition Z-scores indicated at 5 dpi. Activation is indicated by a positive Z-score and the inhibited bio-functions are indicated by negative Z-scores. (C) The top 25 upstream regulators, predicted to be affected at 3, 5 and 7 dpi after ZIKV infection in HSerC. Red indicates up-regulation, blue indicates down-regulation. Numbers in boxes show the significance of alteration measured by Z-score. (D) The most affected protein networks at 5 dpi by ZIKV infection in HSerC. Red and green represent up-regulation and down-regulation, respectively; gray proteins denote that they were recognized in the present study, but not significantly regulated; colorless proteins interact with molecules in the network, but were not identified in our study. Abbreviations. dpi = Days post infection.
IPA also built protein–protein networks of the significantly affected proteins based on their direct or indirect interactions. There were 12, 9, and 10 protein–protein interaction networks (Score > 20, Molecules > 13) predicted by IPA analysis of the altered proteins at 3, 5 and 7 dpi, respectively. The predicted networks were associated with different cellular functions and diseases, including cell morphology, cellular assembly and organization, neurological disease, lipid metabolism, molecular transport, cardiovascular disease, connective tissue disorders, antimicrobial response, cell cycle, and endocrine system development (Table S3). Cancer, connective tissue disorders, organismal injury and abnormalities (Score 38, Focus Molecules 23) were the most affected predicted protein–protein networks at 3 dpi, cardiovascular disease, cell death and survival, and connective tissue disorders (Score 42, Focus Molecules 25) were the most affected at 5 dpi, and antimicrobial response, and infectious diseases, inflammatory response (Score 41, Focus Molecules 22) were predicted to be the most affected at 7 dpi (Figure 3D; Figure S2C,D).
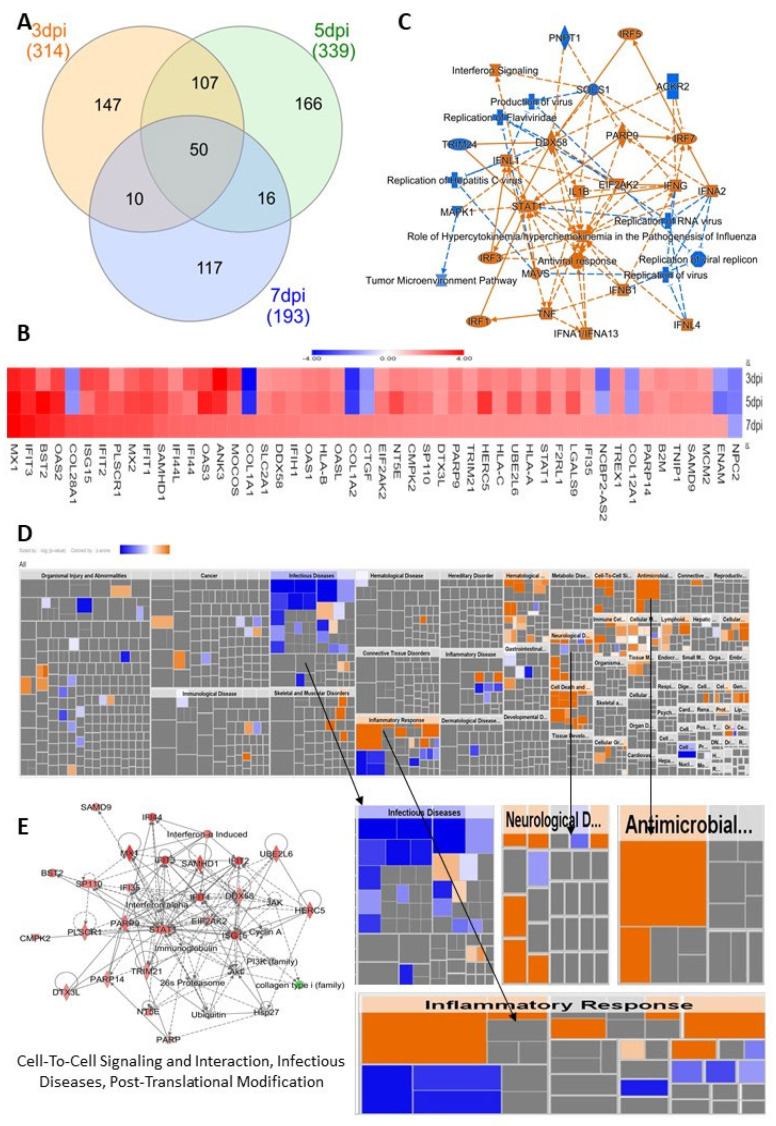
3.4. HSerC Activates Immune Response against ZIKV Infection
To understand the impact of ZIKV infections across the three time points, we selected the list of 50 commonly affected proteins (Figure 4A,B) and performed IPA core analysis. This analysis showed that the most affected proteins were predicted to be associated with canonical pathways involving replication of flavivirus, replication of RNA virus and interferon signaling pathways (Figure 4C). The infectious disease, inflammatory response, neurological disease, and antimicrobial disease responses were the most affected disease and functions predicted by heatmap analysis of IPA (Figure 4D). The protein–protein network analysis predicted their links with cell−to−cell signaling and interaction, infectious diseases, and post−translational modification (Figure 4E).
Figure 4.
Impact of commonly affected proteins in HSerC by ZIKV on cellular functions and diseases. (A) Venn diagram of altered proteins at 3, 5 and 7 dpi. (B) Summary of IPA analysis showing the most affected bio-functions, canonical pathways and affected protein networks and their relationship by the commonly affected proteins. (C) Heatmap of the 50 commonly affected proteins at 3, 5 and 7 dpi. Red/orange and blue represent up-regulation and down-regulation, respectively. (D) Heat map of the disease and biofunctions affected by the commonly affected proteins. (E) The protein–protein interaction networks affected by the commonly altered proteins. Red and green represent up-regulation and down-regulation, respectively; gray proteins denote that they were recognized in the present study, but not significantly regulated; colorless proteins interact with molecules in the network, but were not identified in our study. Abbreviations. dpi = Days post infection.
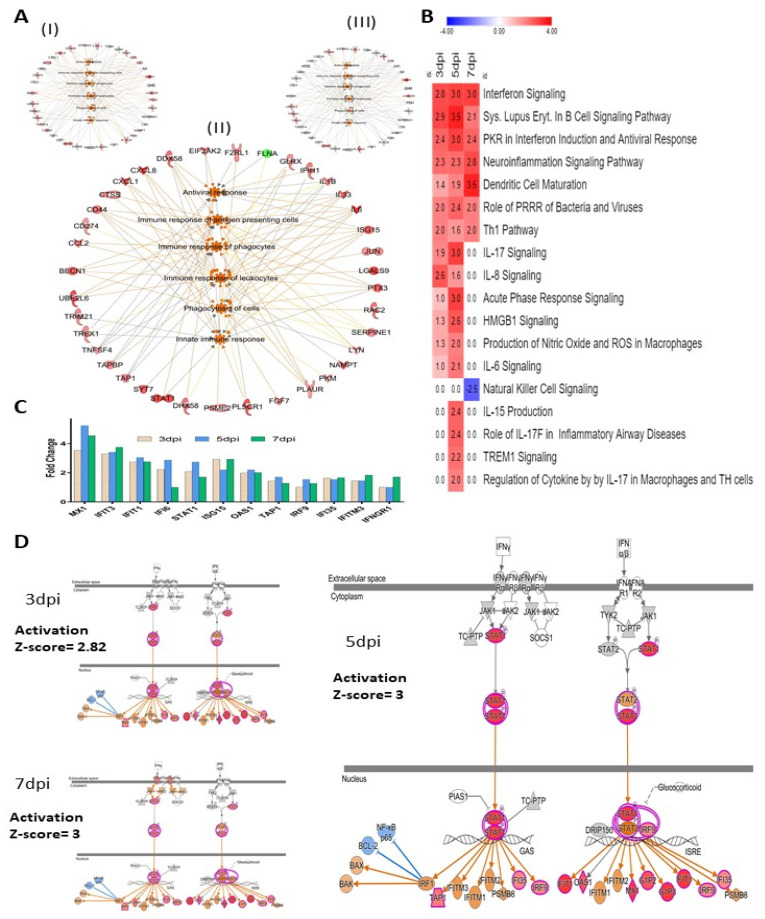
The immune response of antigen-presenting cells, immune response of phagocytes and leukocytes, activation of antiviral response, phagocytosis of cells and innate immune response also were predicted to be activated at 3, 5 and 7 days post-ZIKV infection (Figure 5A). Several proteins involved in immune response regulating signaling pathways were also significantly stimulated by ZIKV infection, which includes interferon signaling, PKR interferon induction and antiviral response, role of pattern recognition receptor in recognition (PRRR) of bacteria and virus, Th1 pathway, HMGB1, TREM1, IL-17, IL-8, IL-6, IL-15 signaling, etc. (Figure 5B). Across the three time points after ZIKV infection, interferon signaling was one of the most affected signaling pathways in Sertoli cells. ZIKV caused significant alteration of 12 protein levels that are predicted to regulate interferon signaling pathways (Figure 5C). Based on the expression values of these proteins, IPA predicted a significant activation of the pathways at 3 dpi (Z-score 2.82), 5 dpi (Z-score 3.0), and 7 dpi (Z-score 3.0) (Figure 5D).
Figure 5.
Immune response in HSerC after ZIKV infection. (A) Bio-functions related to immune response pathways affected at (I) 3 dpi, (II) 5 dpi, and (III) 7 dpi. (B) Canonical pathways associated with immune response pathways affected by ZIKV infection in Sertoli cells. Red and blue represent up-regulation and down-regulation, respectively. The number in each box shows the activation Z-score predicted by IPA. (C) Proteins in Interferon signaling pathways affected by ZIKV infection. (D) Activation of Interferon signaling pathway by ZIKV infection. Red and green represent up-regulation and down-regulation, respectively; gray proteins denote that they were recognized in the present study, but not significantly regulated; colorless proteins interact with molecules in the network, but were not identified in our study. Abbreviations. dpi = Days post infection.
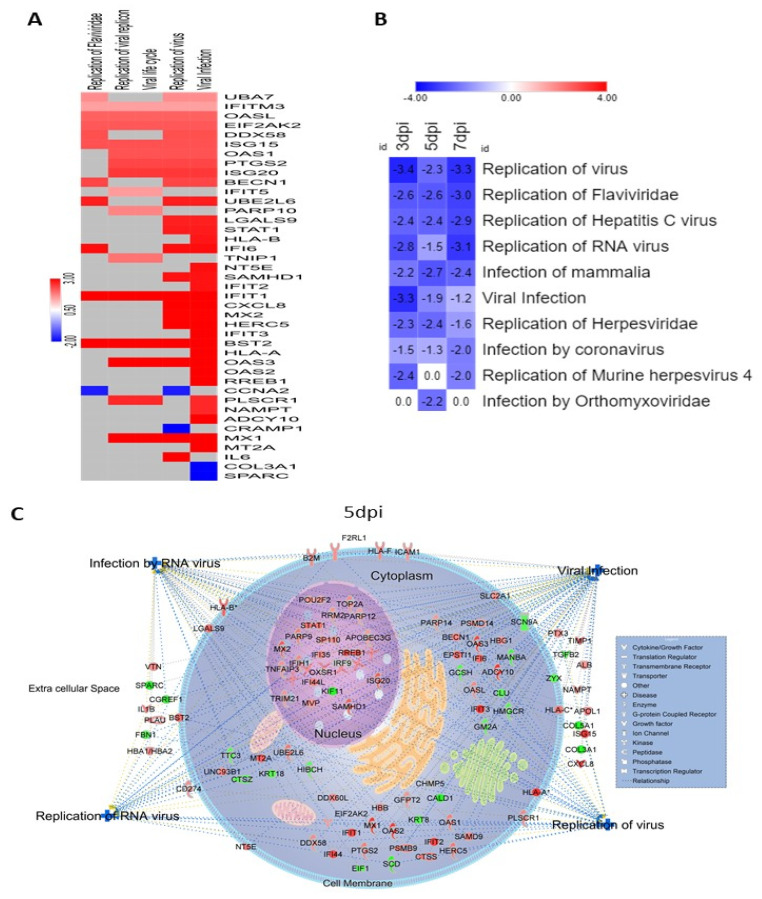
We identified several proteins associated with establishment of viral infection and replication that were significantly affected by ZIKV infection in HSerC (Figure 6A). Based on their expressions, IPA predicted that proteins associated with Flaviviridae infection and replication were significantly inhibited (Figure 6B). Interestingly, these analyses also predicted that proteins associated with replication of some other viruses such as Hepatitis C virus, Herpesviridae, murine herpesvirus 4, Orthomyxoviridae, and coronavirus were affected (Figure 6B). These proteins associated with viral infection and replication are predicted to be found in the cytoplasm, nucleus, cell membrane, and extracellular space of Sertoli cells (Figure 6C).
Figure 6.
Viral replication was suppressed by the cellular response in HSerC after ZIKV infection. (A). Heat map of the proteins involved in virus infection, lifecycle and replication at 5 dpi. Red and blue represent up-regulation and down-regulation of protein expression, respectively. (B) Replication of different family of viruses were predicted to be inhibited by the significantly affected proteins at 3, 5 and 7 dpi. The number in the box indicates the Z-score of the representative function or disease. (C) Cellular localization of the altered proteins predicted to be involved in the regulation of viral infection and in infection by, and replication of, RNA viruses. Red and green represent up-regulation and down-regulation of protein expression, respectively. Abbreviations. dpi = Days post infection.
3.5. ZIKV Infection Impacts Carbohydrate Metabolism in Sertoli Cells
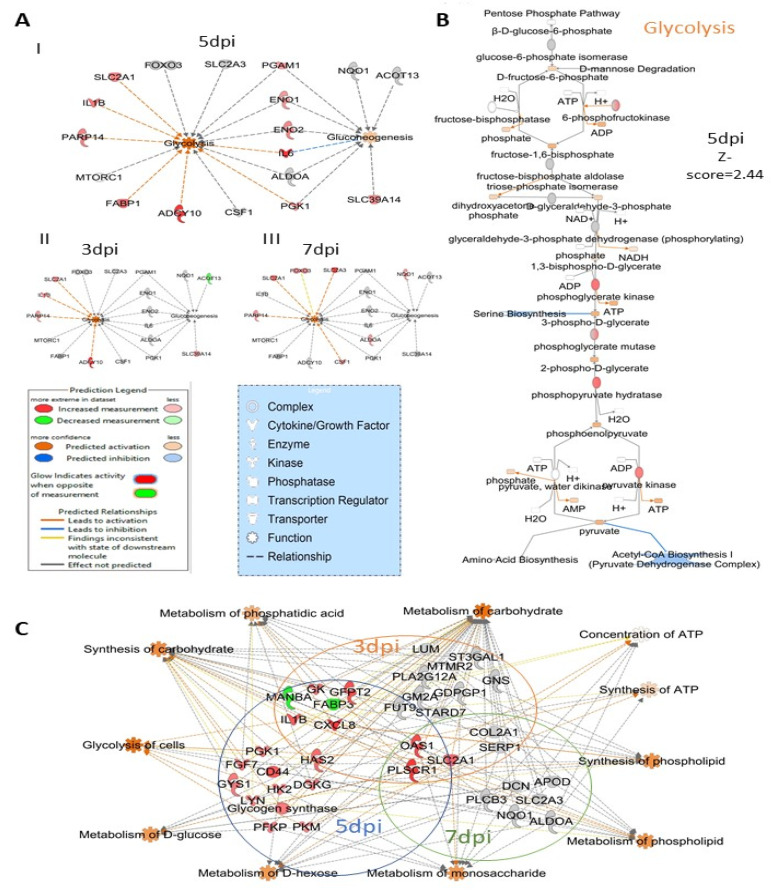
At 5 dpi ZIKV caused significant activation of glycolysis (Z-score = 2.44) and gluconeogenesis (Z-score = 2.0) pathways (Figure 3A). There were 18 Sertoli cell proteins associated with the glycolysis and gluconeogenesis pathway that were significantly affected by ZIKV infection. Eleven of their levels (SLC2A1, IL1B, PRP14, FABP1, ADCY10, PGK1, IL6, ENO2, ENO1, PGAM1 and SL39A14) were altered (Figure 7A) at 5 dpi and predicted to significantly activate (Z score = 2.44) the glycolysis pathway (Figure 7B). ZIKV infection significantly affected 20, 19 and 11 proteins related to carbohydrate metabolism and cellular energy production pathways at 3, 5 and 7 dpi, respectively (Figure 7C; Figure S3). These proteins are associated with the synthesis of ATP and its concentration in cells, metabolism of carbohydrate, phosphatidic acid, d-glucose, d-hexose, monosaccharide, phospholipids and synthesis of carbohydrate and phospholipid.
Figure 7.
ZIKV infection impacts carbohydrate metabolism in HSerC. (A) Proteins and enzymes involved in glycolysis and gluconeogenesis pathways affected by ZIKV infection at I. 5 dpi, II. 3 dpi and III. 7 dpi. IPA predicted the impact of ZIKV infection on (B) glycolysis pathway at 5 dpi in Sertoli cells. (C) Proteins involved in carbohydrate metabolism and energy production in cells affected by ZIKV infection. Red and green represent up-regulation and down-regulation of protein levels, respectively. Abbreviations. Dpi = Days post infection.
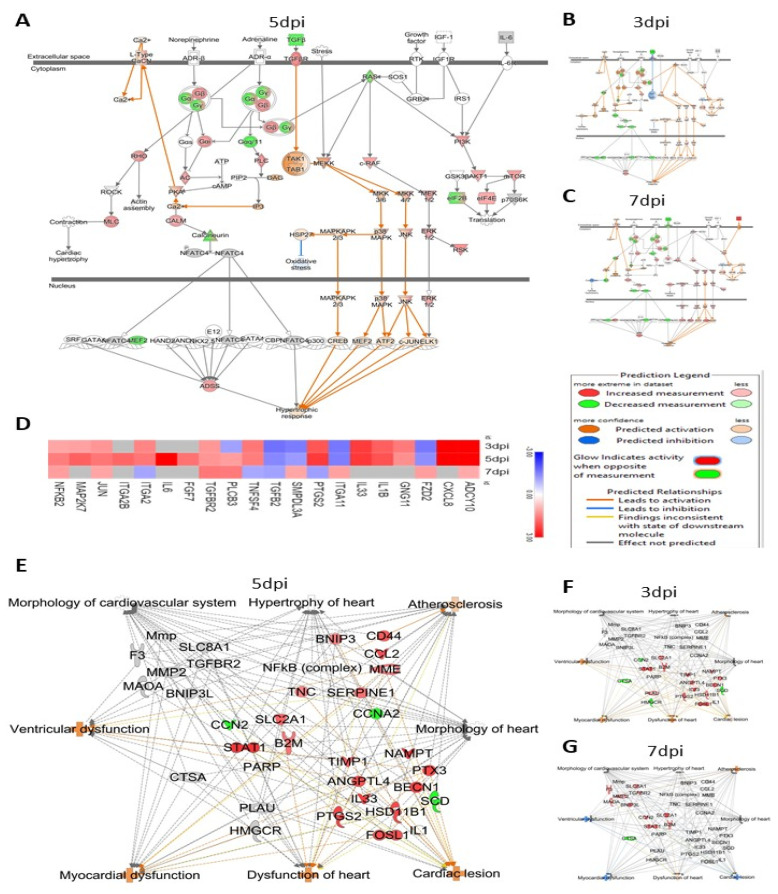
3.6. ZIKV Infection Significantly Affects Proteins That May Increase Cardiovascular Disease
ZIKV infection significantly affected 20 proteins (Figure 8D) involved in the cardiac hypertrophy pathway and significantly activated (Z-score = 3.1) proteins within the pathway at 5 dpi (Figure 8A). However, at 3 dpi and 7 dpi, the proteins in this pathway were not significantly impacted by ZIKV infection (Figure 8B,C). IPA has a database of 617 proteins associated with the increase of cardiovascular disease. Among these, 23, 28 and 14 proteins were significantly affected by 3, 5 and 7 days post-ZIKV infection, respectively. The significantly altered proteins are associated with atherosclerosis, ventricular dysfunction, myocardial dysfunction, cardiac lesion, hypertrophy of heart, morphology of heart and cardiovascular system and dysfunction of heart (Figure 8E–G).
Figure 8.
ZIKV infection significantly affects the levels of proteins involved in the cardiac hypertrophy pathway. IPA predicted activation of the cardiac hypertrophy pathway by Zika virus infection by (A) 3 dpi, (B) 7 dpi and (C) 5 dpi. (D). Heatmap of the proteins involved in cardiac hypertrophy pathway altered by ZIKV infection in HSerC. In the heatmap, red and blue represent up-regulation and down-regulation, respectively. Gray indicates the proteins were not detected at the respective time point. HSerC proteins associated with pathways that increase cardiovascular disease, that are affected by ZIKV infection at (E) 5 dpi (F) 3 dpi and (G) 7 dpi. Red and green represent up-regulation and down-regulation of protein expression, respectively. Abbreviations. dpi = Days post infection.
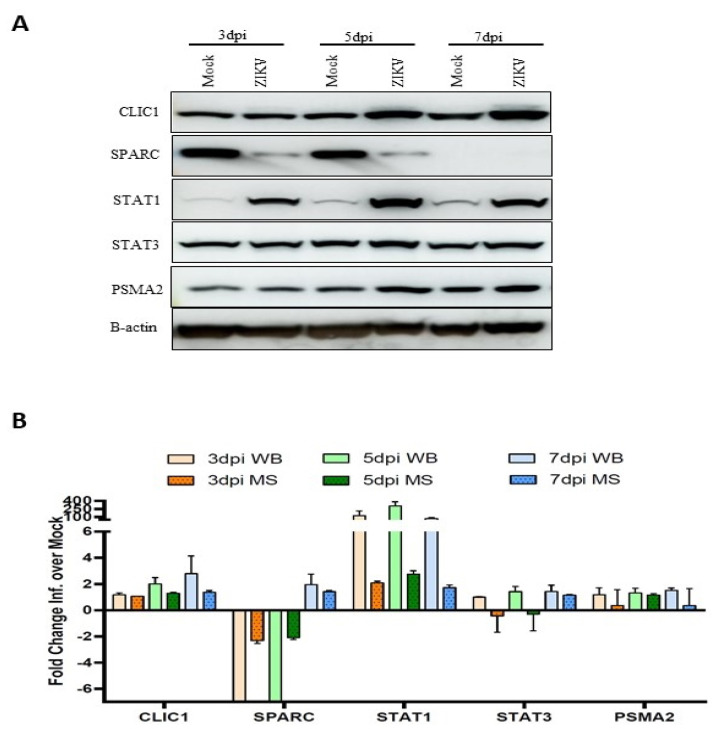
3.7. Validation of Mass Spec Data by Western Blot
Five proteins (CLIC1, SPARC, STAT1, STAT3 and PSMA2) were selected for validation based on their expression fold-change and availability of commercial antibodies. Western blot was done (Figure 9A), and the intensities of protein bands were measured to determine the fold difference between the infected and mock conditions. The fold change of each protein measured by Western blot at three different time points was compared side-by-side to the expression values determined by mass spectrometry (Figure 9B). All five proteins followed the same trend of expression by Western blot as measured by TMT-mass spectrometry.
Figure 9.
Western blot validation of proteomic changes detected by mass spectrometry (MS) analysis. Sertoli cell lysates were collected after ZIKV infection (MOI:3) at 3, 5 and 7 dpi. In total, 20 µg of proteins were separated by SDS-PAGE gel electrophoresis. (A) Expression of CLIC1, SPARC, STAT1, STAT3 and PSMA2 were detected by Western blot using specific antibodies. (B) Expression values of the proteins from Western-blot were quantified by ImageJ V1.8.0 from three replicates and plotted side-by-side to MS expression values. Expression of protein by Western blot was normalized to B-actin expression. Abbreviations: dpi = Days post infection; WB = Western blot; MS = Mass spectrometry; ZIKV = Zika virus.
4. Discussion
The mechanism(s) underlining the long-term persistence of ZIKV in male semen is not yet clearly understood. The sexual transmission of the virus raises the risk of initiating outbreaks in non-endemic regions, even in the absence of a mosquito vector. Furthermore, the pathologic consequences of viral persistence in the male genital tract are unclear. In our previous study, to investigate ZIKV infection-induced HSerC proteome alterations, we applied SOMAscan, a multi-plexed targeted technology that can identify over 1300 proteins from each sample [36]. Although SOMAscan, as a targeted approach, can identify many non-abundant proteins, it is still limited in its detection limit to the specified 1305 proteins [42]. Therefore, in this study, we used a six-plex TMT-based mass spectrometry approach to determine additional HSerC proteome alterations to provide a broader insight on the impact of ZIKV infection. However, unlike the previous study, we excluded the 1 dpi time point due to a small number of significantly affected proteins [36] and included 7 dpi to analyze the impact of ZIKV on Sertoli cells at a later stage. In addition, as previously reported [36,37,43], Zika virus infection had no apparent cytopathic impact on Sertoli cells after infection (Figure 1A). However, ZIKV replication [36] and viral protein expression (Figure 1B,C) were significantly reduced at 7 dpi in Sertoli cells, but infectious virus has been detected for at least 6 weeks in these cells [37]. This indicates that ZIKV initially undergoes a robust infection in Sertoli cells, but it is subsequently controlled, likely by cellular immune responses and later it maintains a low level of persistence. In this study, we have seen that most of the significantly affected proteins were down−regulated at 3 dpi, and up−regulated at 5 dpi as observed before [36]. This confirms a consistent time−dependent switch of proteomic alteration caused by ZIKV infection in Sertoli cells. Among the most affected proteins, FBXO11 has been associated with Neurodevelopmental disorders, mental retardation, and autism [44,45]. However, KIF1A expression anomaly was linked with neurological disorders in children [46]. Therefore, the alteration of FBXO11 and KIF1A could be associated with microcephaly and Guillain–Barré syndrome that develop during and/or after ZIKV infection. UHRF2 is one of the central regulators of cell cycle machinery [47]. TYR is responsible for initiating the conversion of tyrosine to melanin, which is responsible for pigmentation of eye, skin and hair [48]. Abnormalities in TYR expression may cause albinism [49,50]. FOXQ1 is a transcription regulator that controls the cell cycle, cell proliferation and is associated with cancers [51,52,53,54], and that inhibits macrophage recruitment [55], and activates Wnt signaling [56].
4.1. HSerC Activates Immune Response against ZIKV Infection
The human testis maintains an immune-privileged environment, separated from the body cavity by the blood–testis barrier (BTB), made with Sertoli cells [57]. However, to combat invading microbial pathogens, the testis possesses a local immune defense system [28]. In response to ZIKV infection, HSerC induced changes in the levels of proteins predicted to activate a strong immune response at 3, 5 and 7 dpi. According to the analysis of the 50 predominantly affected proteins, the inflammatory and antimicrobial response pathways were significantly triggered after ZIKV infection of HSerC (Figure 4D). The protein–protein interaction analysis also predicted antimicrobial response, infectious diseases, and inflammatory response as activated across the three-time points (Table S3). In addition, at least 17 cellular signaling pathways associated with immune response were predicted to be significantly activated in HSerC after ZIKV infection (Figure 5B). In agreement with our findings, a previous study based on transcriptomic analysis [35] also determined that interferon signaling, IL-15, role of PKR interferon induction and antiviral response, HMGB1 and role of pattern recognition receptor in recognition of bacteria and virus were significantly affected by ZIKV infection of Sertoli cells. Further analysis of the significantly affected proteins (Figure 5A) and upstream molecules (Figure S1B) indicated that proteins involved in immune response of antigen-presenting cell pathways (APCs), phagocytes, leukocytes and T lymphocytes were significantly activated. Sertoli cells’ potent immune response may have inhibited viral replication, as anticipated by IPA (Figure 6B), also seen by the decreased quantity of viral proteins at 5 and 7 dpi (Figure 1B,C). Interestingly, proteins associated with antiviral response against Flaviviridae may significantly suppress other human viruses such as Hepatitis C virus, Herpesvirus, and Coronavirus (Figure 6A,B). This suggests that targeting commonly used host proteins or pathways might lead to the development of a broad-spectrum antiviral therapy in the future.
4.2. ZIKV Affects Proteins Involved in Carbohydrate Metabolism
Viruses use biomolecules from the host and induce anabolism to create the macromolecules needed for virion replication and assembly. As a result, it is no surprise that viral infection forces host cells to modify their metabolism in order to support effective virus replication [58]. Many oncogenic viruses use the glycolysis pathway, such as human papillomavirus (HPV), hepatitis C virus (HCV), hepatitis B virus (HBV), Kaposi’s sarcoma-associated herpesvirus (KSHV), Epstein–Barr virus (EBV), Merkel cell polyomavirus (MCPyV), and Adenovirus [59,60]. However, several non-oncogenic viruses were also found to activate the glycolysis pathways, including Herpes simplex virus 1 and 2 [61], and Human cytomegalovirus [58]. Dengue virus (DENV), another relative of ZIKV, depends on the glycolysis pathways for its replication [62]. High concentrations of glucose can restrict the growth of ZIKV in human kidney cells [63]. ZIKV infection increases glucose incorporation into the TCA cycle in mosquito cells, but the mechanism is not clear [64]. In HSerC, proteins involved in glycolysis and gluconeogenesis pathways were significantly activated by ZIKV infection (Figure 3A and Figure 7B). In addition, the metabolism of d-fructose, monosaccharides, phosphatidic acid, phospholipids were also predicted to be significantly activated by the significantly altered proteins (Figure 7C, Figure S3). Previously, we detected PGAM1, a key regulator of glycolysis and gluconeogenesis pathways, as significantly up-regulated by ZIKV infection in HSerC [36]. However, proteins involved in energy generation in Sertoli cells by lactate and lipid oxidization through the Peroxisome Proliferator-Activated Receptor (PPAR) signaling pathway [65] were significantly down-regulated by ZIKV infection (Figure 3A), as reported previously [36]. These results suggest that ZIKV hijacks carbohydrate metabolism to usurp cellular energy generation in HSerC. The glycolysis pathway could be a potential target against ZIKV and needs further investigation in the future.
4.3. ZIKV Infection Affects Proteins Associated with an Increase in Cardiovascular Disease
Surprisingly, we found many proteins involved in cardiac disease that were significantly affected (Figure 8E–G) during ZIKV infection of HSerC. These proteins are associated with significant activation of atherosclerosis, ventricular dysfunction, myocardial dysfunction, and cardiac lesion (Figure 8E–G). RGS5 is one of the topmost affected proteins detected in HSerC after ZIKV infection. It is a GTPase activator that protects against cardiac hypertrophy and cardiac fibrosis [66]. Cardiovascular disease was a frequently detected protein–protein network predicted by IPA at all three−time points after ZIKV infection (Table S3). In addition, levels of at least 20 proteins involved in the cardiac hypertrophy signaling pathway were significantly altered by ZIKV, causing significant predicted activation of the pathway (Figure 3A and Figure 8A) at 5 dpi in HSerC. Thus far, cardiovascular complications have been associated with ZIKV infection by a case report [67], an observational study [68], and in an animal study [69]. Although identified by us in HSerC, the cardiac hypertrophy signaling pathway could be a possible mechanism of cardiac complications in ZIKV-infected patients. However, the current study has limitations, as it was performed in a non-cardiac cell line. To fully comprehend the link between ZIKV and cardiovascular abnormalities and its mechanism, extensive investigation is necessary.
5. Conclusions
Understanding the mechanisms of ZIKV persistence in the male genital tract is critical for treatment of the disease, antiviral/vaccine development and overall control of viral transmission. Multiple studies have suggested that Sertoli cells could be a potential reservoir for this viral persistence. Sertoli cells build the blood–testis barrier that maintains an isolated environment for optimal development of the germ cells and provides sufficient nutrients and signals for sequential differentiation into sperm by spermatogenesis [30]. ZIKV infection of Sertoli cells may impact sperm development and male fertility. Moreover, a clear understanding of the replication dynamics of the virus in an immune−privileged environment is necessary for antiviral or vaccine development. Previous studies of ZIKV−infected Sertoli cells have reported the roles of a particular protein [70] or its impact on the BTB [71]. However, this is the first study that uses mass spectrometry to understand global proteomic alterations induced by ZIKV infection in HSerC at 3, 5 and 7 dpi. Longitudinal proteome analysis facilitates the identification of critical proteins and pathways for the successful ZIKV infection cycle and provides detailed knowledge that may be used in therapeutic intervention. Many host proteins and signaling pathways crucial for ZIKV replication were found in this study, and these could be targeted for antiviral or diagnostic tool development in the future.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/v14020377/s1, Supplementary Table S1. Disease and functions affected by ZIKV infection in HSerC. Supplementary Table S2. IPA predicted the upstream regulators affected by ZIKV infection in Sertoli cells. Supplementary Table S3. The protein–protein networks affected by ZIKV infection at 3,5 and 7 dpi in HSerC. Supplementary Figure S1. (A) Comparison of canonical pathways affected at 3, 5 and 7 dpi during ZIKV infection in HSerC. Red indicates up-regulation, blue indicates down-regulation. Numbers in the boxes show the significance of alteration measured by Z-score. (B) Top 15 upstream regulators associated with cellular function and disease. Supplementary Figure S2. IPA-predicted activation and inhibition of bio-functions, and protein–protein network at 3 and 7 dpi in HSerC. Top bio-functions and their predicted activation or inhibition Z-scores indicated at (A) 3 and (B) 7 dpi. Activation is indicated by positive Z-Score and the inhibited bio-functions are indicated by negative Z-scores. The most effected protein–protein networks at (C) 3 dpi and (D) 7 dpi by ZIKV infection in HSerC. Red and green represent up-regulation and down-regulation, respectively; gray proteins denote that they were recognized in the present study, but not significantly regulated; colorless proteins interact with molecules in the network, but were not identified in our study. Abbreviations: dpi = Days post infection. Supplementary Figure S3: Proteins involved in the metabolism of carbohydrate and energy production in cells affected by ZIKV infection at 3 dpi and 7 dpi. Red and green represent up-regulation and down-regulation, respectively; gray proteins denote that they were recognized in the present study, but not significantly regulated; colorless proteins interact with molecules in the network, but were not identified in our study. Abbreviations: dpi = Days post infection.
Author Contributions
Conceptualization, M.-u.R. and K.M.C.; methodology, Y.L.; software, M.-u.R.; validation, V.S., formal analysis, M.-u.R. and K.M.C.; investigation, M.-u.R. and K.M.C.; writing—original draft preparation, M.-u.R.; writing—review and editing, M.-u.R. and K.M.C., visualization, M.-u.R. and K.M.C.; supervision, K.M.C.; project administration, K.M.C.; funding acquisition, K.M.C. All authors have read and agreed to the published version of the manuscript.
Funding
This study was funded by grant MOP-106713 from the Canadian Institutes of Health Research to KMC. MuR was supported by a Research Manitoba Studentship.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Spectra (in MGF format) and an overall log2 protein expression matrix are available at the University of California, San Diego’s MassIVE archive (massive.ucsd.edu) under the accession MSV000088634.
Conflicts of Interest
The authors declare they have no conflicts of interest with this study.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Dick G.W.A., Kitchen S.F., Haddow A.J. Zika Virus (I). Isolations and serological specificity. Trans. R. Soc. Trop. Med. Hyg. 1952;46:509–520. doi: 10.1016/0035-9203(52)90042-4. [DOI] [PubMed] [Google Scholar]
- 2.Shuaib W., Stanazai H., Abazid A.G., Mattar A.A. Re-Emergence of Zika Virus: A Review on Pathogenesis, Clinical Manifestations, Diagnosis, Treatment, and Prevention. Am. J. Med. 2016;129:879.e7–879.e12. doi: 10.1016/j.amjmed.2016.02.027. [DOI] [PubMed] [Google Scholar]
- 3.Petersen L.R., Jamieson D.J., Powers A.M., Honein M.A. Zika Virus. N. Eng. J. Med. 2016;374:1552–1563. doi: 10.1056/NEJMra1602113. [DOI] [PubMed] [Google Scholar]
- 4.Ferraris P., Yssel H., Missé D. Zika virus infection: An update. Microbes Infect. 2019;21:353–360. doi: 10.1016/j.micinf.2019.04.005. [DOI] [PubMed] [Google Scholar]
- 5.Lessler J., Chaisson L.H., Kucirka L.M., Bi Q., Grantz K., Salje H., Carcelen A.C., Ott C.T., Sheffield J.S., Ferguson N.M., et al. Assessing the global threat from Zika virus. Science. 2016;353:6300. doi: 10.1126/science.aaf8160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ribeiro L.S., Marques R.E., de Jesus A.M.R., de Almeida R.P., Teixeira M.M. Zika crisis in Brazil: Challenges in research and development. Curr. Opin. Virol. 2016;18:76–81. doi: 10.1016/j.coviro.2016.04.002. [DOI] [PubMed] [Google Scholar]
- 7.Baud D., Gubler D.J., Schaub B., Lanteri M.C., Musso D. An update on Zika virus infection. Lancet. 2017;390:2099–2109. doi: 10.1016/S0140-6736(17)31450-2. [DOI] [PubMed] [Google Scholar]
- 8.WHO Zika Epidemiology Update. 2019. [(accessed on 24 December 2021)]. Available online: https://www.who.int/emergencies/diseases/zika/zika-epidemiology-update-july-2019.pdf.
- 9.Faizan M.I., Abdullah M., Ali S., Naqvi I.H., Ahmed A., Parveen S. Zika Virus-Induced Microcephaly and Its Possible Molecular Mechanism. Intervirology. 2016;59:152–158. doi: 10.1159/000452950. [DOI] [PubMed] [Google Scholar]
- 10.Shang P., Zhu M., Wang Y., Zheng X., Wu X., Zhu J., Feng J., Zhang H.L. Axonal variants of Guillain-Barré syndrome: An update. J. Neurol. 2021;268:2402–2419. doi: 10.1007/s00415-020-09742-2. [DOI] [PubMed] [Google Scholar]
- 11.Brasil P., Sequeira P.C., Freitas A.D.A., Zogbi H.E., Calvet G.A., de Souza R.V., Siqueira A.M., de Mendonca M.C.L., Nogueira R.M.R., de Filippis A.M.B., et al. Guillain-Barré syndrome associated with Zika virus infection. Lancet. 2016;387:1482. doi: 10.1016/S0140-6736(16)30058-7. [DOI] [PubMed] [Google Scholar]
- 12.WHO Zika Situation Report. 2016. [(accessed on 25 August 2017)]. Available online: http://www.who.int/emergencies/zika-virus/situation-report/25-august-2016/en.
- 13.Faye O., Freire C.C., Iamarino A., Faye O., de Oliveira J.V.C., Diallo M., Zanotto P.M., Sall A.A. Molecular Evolution of Zika Virus during Its Emergence in the 20th Century. PLoS Negl. Trop. Dis. 2014;8:36. doi: 10.1371/journal.pntd.0002636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Turmel J.M., Abgueguen P., Hubert B., Vandamme Y.M., Maquart M., Le Guillou-Guillemette H., Leparc-Goffart I. Late sexual transmission of Zika virus related to persistence in the semen. Lancet. 2016;387:2501. doi: 10.1016/S0140-6736(16)30775-9. [DOI] [PubMed] [Google Scholar]
- 15.Hastings A.K., Fikrig E. Focus: Infectious Diseases: Zika Virus and Sexual Transmission: A New Route of Transmission for Mosquito-borne Flaviviruses. [(accessed on 1 January 2022)];Yale J. Biol. Med. 2017 90:325. Available online: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC5482308/ [PMC free article] [PubMed] [Google Scholar]
- 16.Moreira J., Peixoto T.M., Siqueira A.M., Lamas C.C. Sexually acquired Zika virus: A systematic review. Clin. Microbiol. Infect. 2017;23:296–305. doi: 10.1016/j.cmi.2016.12.027. [DOI] [PubMed] [Google Scholar]
- 17.Bujan L., Mansuy J.M., Hamdi S., Pasquier C., Joguet G. 1 year after acute Zika virus infection in men. Lancet Infect. Dis. 2020;20:25–26. doi: 10.1016/S1473-3099(19)30678-4. [DOI] [PubMed] [Google Scholar]
- 18.Arsuaga M., Bujalance S.G., Díaz-Menéndez M., Vázquez A., Arribas J.R. Probable sexual transmission of Zika virus from a vasectomised man. Lancet Infect. Dis. 2016;16:1107. doi: 10.1016/S1473-3099(16)30320-6. [DOI] [PubMed] [Google Scholar]
- 19.Le Tortorec A., Matusali G., Mahé D., Aubry F., Mazaud-Guittot S., Houzet L., Dejucq-Rainsford N. From ancient to emerging infections: The odyssey of viruses in the male genital tract. Physiol. Rev. 2020;100:1349–1414. doi: 10.1152/physrev.00021.2019. [DOI] [PubMed] [Google Scholar]
- 20.Govero J., Esakky P., Scheaffer S.M., Fernandez E., Drury A., Platt D.J., Gorman M.J., Richner J.M., Caine E.A., Salazar V., et al. Zika virus infection damages the testes in mice. Nature. 2016;540:438–442. doi: 10.1038/nature20556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ma W., Li S., Ma S., Jia L., Zhang F., Zhang Y., Zhang J., Wong G., Zhang S., Lu X., et al. Zika Virus Causes Testis Damage and Leads to Male Infertility in Mice. Cell. 2016;167:1511–1524.e10. doi: 10.1016/j.cell.2016.11.016. [DOI] [PubMed] [Google Scholar]
- 22.Osuna C.E., Lim S.Y., Deleage C., Griffin B.D., Stein D., Schroeder L.T., Omange R., Best K., Luo M., Hraber P.T., et al. Zika viral dynamics and shedding in rhesus and cynomolgus macaques. Nat. Med. 2016;22:1448–1455. doi: 10.1038/nm.4206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hirsch A.J., Smith J.L., Haese N.N., Broeckel R.M., Parkins C.J., Kreklywich C., DeFilippis V.R., Denton M., Smith P.P., Messer W.B., et al. Zika Virus infection of rhesus macaques leads to viral persistence in multiple tissues. PLoS Pathog. 2017;13:e1006219. doi: 10.1371/journal.ppat.1006219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mansuy J.M., Suberbielle E., Chapuy-Regaud S., Mengelle C., Bujan L., Marchou B., Delobel P., Gonzalez-Dunia D., Malnou C.E., Izopet J., et al. Zika virus in semen and spermatozoa. Lancet Infect. Dis. 2016;16:1106–1107. doi: 10.1016/S1473-3099(16)30336-X. [DOI] [PubMed] [Google Scholar]
- 25.Joguet G., Mansuy J.M., Matusali G., Hamdi S., Walschaerts M., Pavili L., Guyomard S., Prisant N., Lamarre P., Dejucq-Rainsford N., et al. Effect of acute Zika virus infection on sperm and virus clearance in body fluids: A prospective observational study. Lancet Infect. Dis. 2017;17:1200–1208. doi: 10.1016/S1473-3099(17)30444-9. [DOI] [PubMed] [Google Scholar]
- 26.Paz-Bailey G., Rosenberg E.S., Doyle K. Persistence of Zika Virus in Body Fluids—Preliminary Report. N. Engl. J. Med. 2018;379:1234. doi: 10.1056/NEJMoa1613108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Salam A.P., Horby P.W. The Breadth of Viruses in Human Semen. Emerg. Infect. Dis. 2017;23:1922–1924. doi: 10.3201/eid2311.171049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhao S., Zhu W., Xue S., Han D. Testicular defense systems: Immune privilege and innate immunity. Cell. Mol. Immunol. 2014;11:428–437. doi: 10.1038/cmi.2014.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Griswold M.D. The central role of Sertoli cells in spermatogenesis. Semin. Cell Dev. Biol. 1998;9:411–416. doi: 10.1006/scdb.1998.0203. [DOI] [PubMed] [Google Scholar]
- 30.Griswold M.D. 50 years of spermatogenesis: Sertoli cells and their interactions with germ cells. Biol. Reprod. 2018;99:87–100. doi: 10.1093/biolre/ioy027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.França L.R., Hess R.A., Dufour J.M., Hofmann M.C., Griswold M.D. The Sertoli cell: One hundred fifty years of beauty and plasticity. Andrology. 2016;4:189–212. doi: 10.1111/andr.12165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kaur G., Thompson L.A., Dufour J.M. Seminars in Cell & Developmental Biology. Academic Press; Cambridge, MS, USA: 2014. Sertoli cells—Immunological sentinels of spermatogenesis; p. 36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen H., Ge R.S., Zirkin B.R. Leydig cells: From stem cells to aging. Mol. Cell. Endocrinol. 2009;306:9–16. doi: 10.1016/j.mce.2009.01.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sheng Z.Y., Gao N., Wang Z.Y., Cui X.Y., Zhou D.S., Fan D.Y., Chen H., Wang P.G., An J. Sertoli cells are susceptible to ZIKV infection in mouse testis. Front. Cell. Infect. Microbiol. 2017;7:272. doi: 10.3389/fcimb.2017.00272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Strange D.P., Green R., Siemann D.N., Gale M., Verma S. Immunoprofiles of human Sertoli cells infected with Zika virus reveals unique insights into host-pathogen crosstalk. Sci. Rep. 2018;8:1–15. doi: 10.1038/s41598-018-27027-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rashid M.U., Zahedi-Amiri A., Glover K.K., Gao A., Nickol M.E., Kindrachuk J., Wilkins J.A., Coombs K.M. Zika virus dysregulates human sertoli cell proteins involved in spermatogenesis with little effect on tight junctions. PLoS Negl. Trop. Dis. 2020;14:1–23. doi: 10.1371/journal.pntd.0008335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kumar A., Jovel J., Lopez-Orozco J., Limonta D., Airo A.M., Hou S., Stryapunina I., Fibke C., Moore R.B., Hobman T.C. Human sertoli cells support high levels of zika virus replication and persistence. Sci. Rep. 2018;8:1–11. doi: 10.1038/s41598-018-23899-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sielaff M., Kuharev J., Bohn T., Hahlbrock J., Bopp T., Tenzer S., Distler U. Evaluation of FASP, SP3, and iST Protocols for Proteomic Sample Preparation in the Low Microgram Range. J. Proteome Res. 2017;16:4060–4072. doi: 10.1021/acs.jproteome.7b00433. [DOI] [PubMed] [Google Scholar]
- 39.Glover K.K.M., Zahedi-Amiri A., Lao Y., Spicer V., Klonisch T., Coombs K.M. Zika Infection Disrupts Proteins Involved in the Neurosensory System. Front. Cell Dev. Biol. 2020;8:571. doi: 10.3389/fcell.2020.00571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kelstrup C.D., Bekker-Jensen D.B., Arrey T.N., Hogrebe A., Harder A., Olsen J.V. Performance Evaluation of the Q Exactive HF-X for Shotgun Proteomics. J. Proteome Res. 2018;17:727–738. doi: 10.1021/acs.jproteome.7b00602. [DOI] [PubMed] [Google Scholar]
- 41.Coombs K.M., Berard A., Xu W., Krokhin O., Meng X., Cortens J.P., Kobasa D., Wilkins J., Brown E.G. Quantitative Proteomic Analyses of Influenza virus-Infected Cultured Human Lung Cells. J. Virol. 2010;84:10888–10906. doi: 10.1128/JVI.00431-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Candia J., Cheung F., Kotliarov Y., Fantoni G., Sellers B., Griesman T., Huang J., Stuccio S., Zingone A., Ryan B.M., et al. Assessment of Variability in the SOMAscan Assay. Sci. Rep. 2017;7:1–13. doi: 10.1038/s41598-017-14755-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Siemann D.N., Strange D.P., Maharaj P.N., Shi P.-Y., Verma S. Zika Virus Infects Human Sertoli Cells and Modulates the Integrity of the In Vitro Blood-Testis Barrier Model. J. Virol. 2017;91:22. doi: 10.1128/JVI.00623-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jansen S., van der Werf I.M., Innes A.M., Afenjar A., Agrawal P.B., Anderson I.J., Atwal P.S., van Binsbergen E., van den Boogaard M.J., Castiglia L., et al. De novo variants in FBXO11 cause a syndromic form of intellectual disability with behavioral problems and dysmorphisms. Eur. J. Hum. Genet. 2019;27:20. doi: 10.1038/s41431-018-0292-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gregor A., Sadleir L.G., Asadollahi R., Azzarello-Burri S., Battaglia A., Ousager L.B., Boonsawat P., Bruel A.L., Buchert R., Calpena E., et al. De Novo Variants in the F-Box Protein FBXO11 in 20 Individuals with a Variable Neurodevelopmental Disorder. Am. J. Hum. Genet. 2018;103:305–316. doi: 10.1016/j.ajhg.2018.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nemani T., Steel D., Kaliakatsos M., DeVile C., Ververi A., Scott R., Getov S., Sudhakar S., Male A., Mankad K., et al. KIF1A-related disorders in children: A wide spectrum of central and peripheral nervous system involvement. J. Peripher. Nerv. Syst. 2020;25:117–124. doi: 10.1111/jns.12368. [DOI] [PubMed] [Google Scholar]
- 47.Mori T., Ikeda D.D., Yamaguchi Y., Unoki M. NIRF/UHRF2 occupies a central position in the cell cycle network and allows coupling with the epigenetic landscape. FEBS Lett. 2012;586:1570–1583. doi: 10.1016/j.febslet.2012.04.038. [DOI] [PubMed] [Google Scholar]
- 48.Wilde S., Timpson A., Kirsanow K., Kaiser E., Kayser M., Unterländer M., Hollfelder N., Potekhina I.D., Schier W., Thomas M.G., et al. Direct evidence for positive selection of skin, hair, and eye pigmentation in Europeans during the last 5000 y. Proc. Natl. Acad. Sci. USA. 2014;111:4832–4837. doi: 10.1073/pnas.1316513111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ghodsinejad Kalahroudi V., Kamalidehghan B., Arasteh Kani A., Aryani O., Tondar M., Ahmadipour F., Chung L.Y., Houshmand M. Two Novel Tyrosinase (TYR) Gene Mutations with Pathogenic Impact on Oculocutaneous Albinism Type 1 (OCA1) PLoS ONE. 2014;9:106656. doi: 10.1371/journal.pone.0106656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Spritz R.A., Oh J., Fukai K., Holmes S.A., Ho L., Chitayat D., France T.D., Musarella M.A., Orlow S.J., Schnur R.E., et al. Novel Mutations of the Tyrosinase (TYR) Gene in Type I Oculocutaneous Albinism (OCA1) [(accessed on 1 January 2022)];Hum. Mutat. 1997 10:171–174. doi: 10.1002/(SICI)1098-1004(1997)10:2<171::AID-HUMU11>3.0.CO;2-X. Available online: https://www.proquest.com/openview/0754b19d664ffd0592b6b7b688100ca7/1?pq-origsite=gscholar&cbl=30498. [DOI] [PubMed] [Google Scholar]
- 51.Kaneda H., Arao T., Tanaka K., Tamura D., Aomatsu K., Kudo K., Sakai K., De Velasco M.A., Matsumoto K., Fujita Y., et al. FOXQ1 is overexpressed in colorectal cancer and enhances tumorigenicity and tumor growth. Cancer Res. 2010;70:2053–2063. doi: 10.1158/0008-5472.CAN-09-2161. [DOI] [PubMed] [Google Scholar]
- 52.Gao M., Shih I.M., Wang T.L. The role of forkhead box Q1 transcription factor in ovarian epithelial carcinomas. Int. J. Mol. Sci. 2012;13:13881–13893. doi: 10.3390/ijms131113881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sun H.T., Cheng S.X., Tu Y., Li X.H., Zhang S. FoxQ1 Promotes Glioma Cells Proliferation and Migration by Regulating NRXN3 Expression. PLoS ONE. 2013;8:e55693. doi: 10.1371/journal.pone.0055693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Xia L., Huang W., Tian D., Zhang L., Qi X., Chen Z., Shang X., Nie Y., Wu K. Forkhead box Q1 promotes hepatocellular carcinoma metastasis by transactivating ZEB2 and VersicanV1 expression. Hepatology. 2014;59:958–973. doi: 10.1002/hep.26735. [DOI] [PubMed] [Google Scholar]
- 55.Tang H., Zheng J., Bai X., Yue K.L., Liang J.H., Li D.Y., Wang L.P., Wang J.L., Guo Q. Forkhead Box Q1 Is Critical to Angiogenesis and Macrophage Recruitment of Colorectal Cancer. Front. Oncol. 2020;10:2561. doi: 10.3389/fonc.2020.564298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Christensen J., Bentz S., Sengstag T., Shastri V.P., Anderle P. FOXQ1, a novel target of the Wnt pathway and a new marker for activation of Wnt signaling in solid tumors. PLoS ONE. 2013;8:3. doi: 10.1371/journal.pone.0060051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mruk D.D., Cheng C.Y. The Mammalian Blood-Testis Barrier: Its Biology and Regulation. Endocr. Rev. 2015;36:564–591. doi: 10.1210/er.2014-1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Vastag L., Koyuncu E., Grady S.L., Shenk T.E., Rabinowitz J.D. Divergent effects of human cytomegalovirus and herpes simplex virus-1 on cellular metabolism. PLoS Pathog. 2011;7:e1002124. doi: 10.1371/journal.ppat.1002124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Thaker S.K., Ch’ng J., Christofk H.R. Viral hijacking of cellular metabolism. BMC Biol. 2019;17:1–15. doi: 10.1186/s12915-019-0678-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Fisher T.N., Ginsberg H.S. Accumulation of organic acids by HeLa cells infected with type 4 adenovirus. Proc. Soc. Exp. Biol. Med. 1957;95:47–51. doi: 10.3181/00379727-95-23113. [DOI] [PubMed] [Google Scholar]
- 61.Lewis V.J., Scott L.V. Nutritional requirements for the production of herpes simplex virus. I. Influence of glucose and glutamine of herpes simplex virus production by HeLa cells. J. Bacteriol. 1962;83:475–482. doi: 10.1128/jb.83.3.475-482.1962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Fontaine K.A., Sanchez E.L., Camarda R., Lagunoff M. Dengue virus induces and requires glycolysis for optimal replication. J. Virol. 2015;89:2358–2366. doi: 10.1128/JVI.02309-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Reslan A., Haddad J.G., Koundi L.M., Desprès P., Bascands J.L., Gadea G. Zika Virus Growth in Human Kidney Cells Is Restricted by an Elevated Glucose Level. Int. J. Mol. Sci. 2021;22:1–13. doi: 10.3390/ijms22052495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Thaker S.K., Chapa T., Garcia G., Jr., Gong D., Schmid E.W., Arumugaswami V., Sun R., Christofk H.R. Differential Metabolic Reprogramming by Zika Virus Promotes Cell Death in Human versus Mosquito Cells. Cell Metab. 2019;29:1206–1216. doi: 10.1016/j.cmet.2019.01.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Jutte N.H.P.M., Eikvar L., Levy F.O., Hansson V. Metabolism of palmitate in cultured rat Sertoli cells. Reproduction. 1985;73:497–503. doi: 10.1530/jrf.0.0730497. [DOI] [PubMed] [Google Scholar]
- 66.Li H., He C., Feng J., Zhang Y., Tang Q., Bian Z., Bai X., Zhou H., Jiang H., Heximer S.P., et al. Regulator of G protein signaling 5 protects against cardiac hypertrophy and fibrosis during biomechanical stress of pressure overload. Proc. Natl. Acad. Sci. USA. 2010;107:13818–13823. doi: 10.1073/pnas.1008397107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Aletti M., Lecoules S., Kanczuga V., Soler C., Maquart M., Simon F., Leparc-Goffart I. Transient myocarditis associated with acute Zika virus infection. Clin. Infect. Dis. 2017;64:678–679. doi: 10.1093/cid/ciw802. [DOI] [PubMed] [Google Scholar]
- 68.Minhas A.M., Nayab A., Iyer S., Narmeen M., Fatima K., Khan M.S., Constantin J. Association of Zika Virus with Myocarditis, Heart Failure, and Arrhythmias: A Literature Review. Cureus. 2017;9 doi: 10.7759/cureus.1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Li X.F., Dong H.L., Huang X.Y., Qiu Y.F., Wang H.J., Deng Y.Q., Zhang N.N., Ye Q., Zhao H., Liu Z.Y., et al. Characterization of a 2016 Clinical Isolate of Zika Virus in Non-human Primates. EBioMedicine. 2016;12:170. doi: 10.1016/j.ebiom.2016.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Strange D.P., Jiyarom B., Sadri-Ardekani H., Cazares L.H., Kenny T.A., Ward M.D., Verma S. Paracrine IFN Response Limits ZIKV Infection in Human Sertoli Cells. Front. Microbiol. 2021;12:1044. doi: 10.3389/fmicb.2021.667146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Nie Y., Hui L., Guo M., Yang W., Huang R., Chen J., Wen X., Zhao M., Wu Y. Rearrangement of Actin Cytoskeleton by Zika Virus Infection Facilitates Blood–Testis Barrier Hyperpermeability. Virol. Sin. 2021;36:692–705. doi: 10.1007/s12250-020-00343-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Spectra (in MGF format) and an overall log2 protein expression matrix are available at the University of California, San Diego’s MassIVE archive (massive.ucsd.edu) under the accession MSV000088634.