Abstract
In anautogenous mosquitoes, vitellogenesis, the key event in egg maturation, requires a blood meal. Consequently, mosquitoes are vectors of many devastating human diseases. An important adaptation for anautogenicity is the previtellogenic arrest (the state of arrest) preventing the activation of the yolk protein precursor (YPP) genes Vg and VCP prior to blood feeding. A novel GATA factor (AaGATAr) that recognizes GATA binding motifs (WGATAR) in the upstream region of the YPP genes serves as a transcriptional repressor at the state of arrest. Importantly, AaGATAr can override the 20-hydroxyecdysone transactivation of YPP genes, and its transcriptional repression involves the recruitment of CtBP, one of the universal corepressors. AaGATAr transcript is present only in the adult female fat body. Furthermore, in nuclear extracts of previtellogenic fat bodies with transcriptionally repressed YPP genes, there is a GATA binding protein forming a band with mobility similar to that of AaGATAr. The specific repression of YPP genes by AaGATAr in the fat body of the female mosquito during the state of arrest represents an important molecular adaptation for anautogenicity.
The recent resurgence of several of the most threatening mosquito-borne diseases is due to the failure to generate effective vaccines and to the rise of resistance in mosquitoes and in pathogens to insecticides and preventative drugs, respectively. Malaria is a particularly devastating mosquito-borne disease, taking a heavy toll on the human population in many parts of the world (4).
Mosquitoes serve as disease vectors because they require blood feeding for their egg development. In anautogenous mosquitoes, vitellogenesis, the cornerstone of egg maturation, is initiated only after a female mosquito ingests vertebrate blood. This blood meal triggers a hormonal cascade with 20-hydroxyecdysone (20E) as the terminal signal, which activates yolk protein precursor (YPP) genes in the metabolic tissue, called the fat body (11, 15, 29). An important adaptation for anautogenicity is the developmental arrest (the state of arrest), which prevents the activation of YPP genes in previtellogenic females prior to blood feeding. Understanding the molecular nature of the state of arrest and the mechanisms underlying blood meal activation of YPP genes is of paramount importance for current efforts that utilize molecular genetics to develop novel strategies for controlling mosquito-mediated disease transmission.
In the anautogenous mosquito Aedes aegypti, a key model vector, the fat body produces several YPPs: vitellogenin, vitellogenic carboxypeptidase, and vitellogenic cathepsin B (5, 6, 8). Genes encoding vitellogenic carboxypeptidase and vitellogenin have been cloned (10, 31). Analysis of the regulatory regions of Vg and VCP genes by binding and expression assays have suggested that both of these genes are under the synergistic control of the hormone- and tissue-specific gene-regulatory hierarchies (D. Martín, V. Kokoza, S. F. Wang, and A. S. Raikhel, unpublished data).
At the state of arrest, the ecdysteroid receptor is the target of the 20E signaling modification in the mosquito fat body. The functional ecdysteroid receptor is a heterodimer of the ecdysone receptor (EcR) and the retinoid X receptor homolog, Ultraspiracle (37, 38). In A. aegypti, two EcR isoforms and two Ultraspiracle isoforms have been cloned (7, 19, 36; Wang, C. Li, and Raikhel, unpublished data). We have found that at the state of arrest, AHR38, the mosquito homolog of DHR38 and vertebrate NGFI-B/Nurr1 orphan receptors, is the key factor inhibiting the ecdysone response in the A. aegypti fat body. At this stage, AHR38 interacts strongly with the AaUSP protein, preventing the formation of a functional ecdysteroid receptor (40).
In addition to the hormone-specific gene-regulatory hierarchy, transcriptional activation of Vg and VCP genes is under the control of a tissue-specific GATA factor (Martín and Raikhel, unpublished data). In this paper, we report a mosquito homolog of the GATA family of transcription factors (AaGATAr) that recognizes GATA binding motifs in the upstream region of the YPP genes, Vg and VCP. Significantly, AaGATAr acts as a repressor of YPP genes in cell transfection assays. The transcriptional repression by AaGATAr involves the corepressor CtBP (23, 27). AaGATAr mRNA is only present in adult female fat bodies and the binding activity corresponding to AaGATAr in the nuclei of previtellogenic fat bodies.
Thus, we have identified a novel transcriptional repressor, belonging to the GATA family of transcription factors, which recruits CtBP, one of the universal corepressors. Our data further suggest the involvement of AaGATAr in the specific repression of YPP genes in the fat body of the A. aegypti female at the state of arrest.
MATERIALS AND METHODS
Animals.
Mosquitoes, A. aegypti, were reared according to the method described by Hays and Raikhel (16). Vitellogenesis was initiated by allowing females 3 to 5 days after eclosion to feed on an anesthetized white rat.
Cloning and sequencing of cDNA.
Two degenerated oligonucleotides derived from highly conserved amino acid sequences of the GATA finger region (from human, mouse, chicken, and fruit fly GATA proteins) were designed: GATAS (a sense strand composed of 29 nucleotides corresponding to the amino acid residues -ECVNCG-) and GATAR (an antisense 27-nucleotide strand corresponding to amino acid sequence -GCANCV-). The sequences of the oligonucleotides were as follows: GATAS, 5′-AGATCTAGAGARTGYGTNAAYTGYGGNGC-3′; and GATAR, 5′-AGAGAATTCNCCRCANGCRTTRCANAC-3′, where R is A or G; Y is C or T; and N is A, T, C, or G.
To facilitate cloning of the amplified product, anchor sequences containing EcoRI and XbaI restriction sites were added 5′ of the forward and the reverse primers.
Total RNA prepared from previtellogenic female fat bodies was used to generate randomly primed double-stranded cDNA using reverse transcription (RT) (Gibco BRL). The initial PCRs were performed in a final volume of 50 μl containing 0.5 μg of each of the two degenerate primers, 10 ng of cDNA, and 0.25 U of AmpliTaq gold polymerase (Perkin-Elmer). After a denaturation step of 2 min at 94°C, amplifications were carried out for 10 cycles at 94°C for 30 s, 45°C for 1 min, and 72°C for 2 min, followed by 30 cycles at 94°C for 30 s, 55°C for 30 s, and 72°C for 1 min. The amplified fragment was subcloned into vector pGEMT-easy (Promega) for sequencing. After confirmation of its sequence, the 243-bp PCR fragment was used as a probe to screen a λZAPII cDNA library generated from fat bodies of vitellogenic female mosquitoes from 6 to 48 h post-blood meal (PBM) (9). After four rounds of screening, one clone with an insert size of around 3.8 kb was isolated and sequenced from both strands. The clone did not contain the initiation sequence. The missing part of the 5′ end sequence was obtained by 5′ rapid amplification of cDNA ends-PCR (Gibco BRL) using an antisense primer located 397 bp downstream of the AaGATAr cDNA start (5′-ACAAACGAGGAGCATGTAAACACT-3′). Total RNA purified from previtellogenic fat bodies was used as a template for RT. An 800-bp PCR fragment was subcloned into pGEMT-easy for sequencing and contained the initiation sequence, completing then the entire open reading frame (ORF).
Alignments of nucleotide and amino acid sequences.
The software package of the Genetics Computer Group (GCG, version 9.1) of the University of Wisconsin was used for sequence alignments, which were carried out with Pileup and were not further hand refined; alignment was displayed with the Box option. Phylogenetic analyses were carried out using amino acid sequences, with the Phylogeny Inference Package (Phylip, version 3.57c). We followed the method of neighbor joining, and the distances between different sequences were estimated with Kimura's formula and the application Protdist. Bootstrap analyses were carried out with the application Seqboot in the Phylip package, and the procedure was repeated 100 times.
Northern blot analysis.
Total RNA from mosquitoes of different stages, taken from tissues as well as from whole males, was prepared using the RNeasy Qiagen kit. Fifteen micrograms of total RNA was separated in a 1.2% agarose–formaldehyde gel, blotted to a nylon membrane (N+-Hybond; Amersham), hybridized, and washed under stringent conditions. The probe used was a 2.5-kb EcoRI fragment of the full-length AaGATAr cDNA clone containing the DNA-binding domain, which was labeled by random priming (32) with [32P]dATP (3,000 Ci/mmol) (New England Nuclear). Northern blots were also hybridized with a labeled 0.8-kb EcoRI fragment from the VCP gene (10) and with a 0.85-kb EcoRI fragment from the A. aegypti actin gene (11).
In vitro transcription and translation.
The entire AaGATAr cDNA was cloned into pBluescript SK(+). A coupled in vitro transcription-translation TNT system (Promega), utilizing the T7 promoter, was used for expression of the AaGATAr cDNA. To monitor the in vitro reaction, the synthesized protein was labeled with [35S]methionine (1,200 Ci/mmol) from ICN Radiochemicals, and the radiolabeled product was visualized by electrophoresis and autoradiography.
EMSA.
Preparation of nuclear extracts from A. aegypti fat bodies and the electrophoretic mobility shift assay (EMSA) using the nuclear extracts were carried out according to the method described by Miura et al. (22). DNA probes for EMSA were made by annealing together complementary oligonucleotides. The GATA binding site (box A) in the Drosophila mulleri alcohol dehydrogenase gene (Adh) (1) was used as a positive control for binding. The oligonucleotides (only sense strands are shown) used to generate the different probes were D. mulleri box A, 5′-AGTGGTATTGATAAGAC-3′; AaVgGATAa, 5′-TTAATGCTTATCATCGCG-3′; AaVgGATAb, 5′-TTTTGCTTATCTTACTATCTTCA-3′; AaVgGATAc, 5′-GAATTTCAACAATGATAGCCTTTCA-3′. (Boldface letters correspond to the core GATA motif within the primers.)
Plasmid construction and cell transient transfection.
The pPac-ABF and Adh-1-(BoxA)6/CAT plasmids were provided by T. Abel (1). The pAc5-AaGATAr expression plasmid was constructed by subcloning the entire AaGATAr cDNA into the expression vector pAc5/V5/His(C) (Invitrogen), under the control of the actin 5C promoter. The reporter plasmid (AaVgGATAb)4/CAT was constructed by ligating four copies of the AaVgGATAb oligonucleotide 5′-AGCTTTTTGCTTATCTTACTATCTTCAATT-3′ (only one strand is shown; the HindIII site is underlined) into the HindIII site of Adh-1/CAT (21) just upstream of the Drosophila Adh promoter controlling the expression of CAT. The 0.6Vg-Luc reporter plasmid was constructed by cloning a 0.6-kb KpnI-SmaI fragment of the 5′ regulatory region of the A. aegypti Vg gene (31) into the promoterless plasmid pGL3basic (Promega). This construct include sequences from −600 to +115 bp of the Vg gene. The pAc5-dCtBP expression vector was constructed by excising the entire dCtBP cDNA from pGEX-3X-dCtBP (provided by S. Parkhurst) with BamHI, blunt ending the recessed termini with Klenow, and cloning the cDNA into pAc5/V5/His(C) digested with EcoRV. pAc5-AaGATArΔCtBP was obtained by PCR using two primers flanking the CtBP binding site within the AaGATAr protein. Expression vectors pAc5-AaEcR and pAc5-AaUSPb are described in reference 36. All constructs were confirmed by restriction analysis and sequencing.
Transfections were carried out using the Schneider Drosophila cell line (S2; Invitrogen), as described by Wang et al. (36) with minor modifications. Transfection was conducted with LipofectACE (Gibco BRL) with a DNA-to-lipid ratio of 1:10 (wt/wt).
GST pulldown assay.
AaGATAr, AaGATArΔCtBP, and Drosophila ABF were 35S labeled with a coupled in vitro transcription translation protocol (TNT; Promega). GST-dCtBP was provided by S. Parkhurst and was prepared as described previously (27). Binding assays were performed as described previously (40). Generation of the mutated AaGATArΔCtBP protein was carried out by PCR using Pfu polymerase (Promega) and a pair of primers flanking the PCDLSYK sequence. Sequencing was performed to verify the deletion.
Nucleotide sequence accession number.
The AaGATAr sequence has been submitted to the DDBJ/EMBL/GenBank database under accession no. AJ400338.
RESULTS
Cloning and characterization of AaGATAr cDNA.
Two motifs were selected from a highly conserved GATA DNA-binding domain for the design and synthesis of degenerated primers, which were then used for the initial PCR isolation of the cDNA encoding a mosquito GATAr (see Materials and Methods). RT-PCR of the fat body total RNA from previtellogenic females yielded a cDNA fragment of 256 bp. The sequence of this fragment exhibited a high degree of identity with the GATA DNA-binding domain. This cDNA fragment was then used to screen the λZAP II vitellogenic fat body cDNA library. The clone with the longest insert (3.8 kb) was sequenced from both strands. The missing 5′ portion of the cDNA was obtained by 5′ rapid amplification of cDNA ends-PCR using a specific antisense primer designed from the isolated clone.
The entire sequence of the putative AaGATAr cDNA was 4,275 bp long and encoded a protein of 866 amino acids with a predicted molecular mass of 93.83 kDa (Fig. 1). The deduced amino acid sequence started with a methionine at nucleotide 412 and ended with a stop codon at nucleotide 3016. The initiation codon conformed to the Kozak consensus sequence (20) (Fig. 1, double underlined). This putative methionine start codon was preceded by several in-frame stop codons, indicating that the AaGATAr sequence represented a full-length ORF. At the 3′ end, the AaGATAr polyadenylation signal AATAAA was followed by an additional 9-bp sequence and lacked a poly(A) tail, indicating that this cDNA was not a full-length transcript. To verify that the cloned cDNA contained a translatable ORF, it was expressed in a coupled TNT system under the control of the T7 promoter. Sodium dodecyl sulfate-polyacrylamide gel electrophoresis and fluorography showed that the in vitro-synthesized protein closely corresponded to its expected molecular size (data not shown).
FIG. 1.
Nucleotide and deduced amino acid sequence of AaGATAr. Boldface, stop codons; double underline, Kozak sequence; single underline, polyadenylation signal; boldfaced italics, phosphorylation signal; dotted underline, putative glycosylation sequences; striped box, putative nuclear localization signal; grey box, CtBP interaction motif. The DNA-binding domain is boxed.
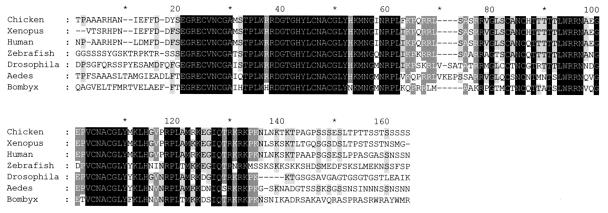
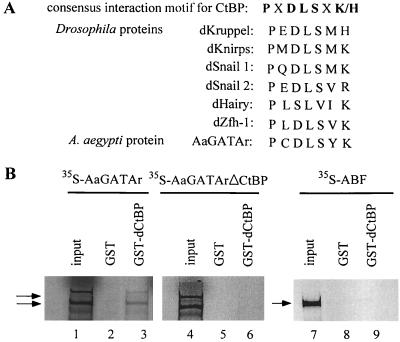
Analysis of the AaGATAr protein sequence revealed that it contained a double C.X2.C.X17.C.X2.C. zinc finger (Fig. 1), characteristic for the GATA DNA-binding transcription factors (25). Its DNA-binding domain showed a high level of conservation of the residues involved in direct contact with the DNA (24) (Fig. 2). Outside the DNA-binding domain, however, the level of conservation between AaGATAr and other GATA family members was very low. Characteristic features of AaGATAr included a putative nuclear localization signal located downstream of the second zinc finger (RKRKPK), several potential N-linked glycosylation sites (NXS/T), and a putative PKA phosphorylation site (RRPS) (Fig. 1). Surprisingly, AaGATAr also contained a motif (PCDLSYK) which was similar to the consensus binding motif PXDLSXK/H for the corepressor protein CtBP (Fig. 1) (23).
FIG. 2.
Similarities of AaGATAr to other members of the GATA family. Shown is an alignment of different GATA amino acid sequences belonging to different animal orders, displaying a high degree of similarity in the area corresponding to the DNA-binding domain. The alignment was made using a GCG PILEUP option, and results are displayed by using the BOX option. Sequence accession numbers are chicken (GATA4), P43691; Xenopus (GATA4), Q91677; human (GATA4), P43694; zebra fish (GATA3), Q91428; Drosophila (dGATAa), P52168; Aedes (AaGATAr), AJ400338; Bombyx (bGATAβ3), P52167.
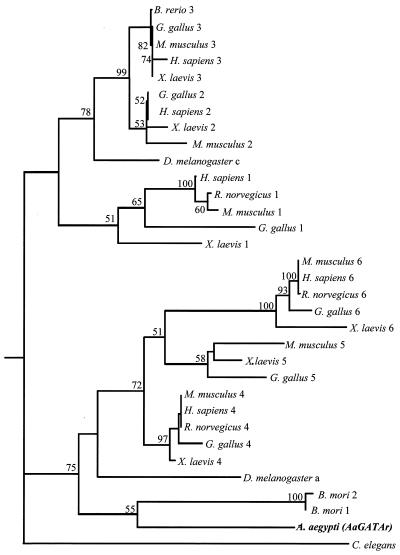
Next, we carried out phylogenetic analyses of amino acid sequences from proteins of the GATA family using the neighbor-joining and Kimura distance methods. Two zinc finger GATA proteins were clustered into two separated branches. The first included GATA factors of types 1, 2, and 3, while the second included types 4, 5, and 6 of vertebrate as well as insect and worm GATAs (Fig. 3). AaGATAr was classified as a member of the second group, being phylogenetically closest to Bombyx mori GATA 1 and 2 (Fig. 3).
FIG. 3.
Dendrogram generated from amino acid sequences corresponding to DNA-binding domains of GATA transcription factors. The method of neighbor joining, using the distance formula of Kimura, was applied to generate the tree. Numbers at the nodes correspond to bootstrap values in 100 replicates. The accession numbers for the GATA sequences used are A. aegypti, AJ400338; B. mori 1, P52167; B. mori 2, P52167; Brachydanio rerio, Q91428; Caenorhabditis elegans 1, P28515; Drosophila melanogaster a, P52168; D. melanogaster c, D50542; Gallus gallus 1, P17678; G. gallus 2, P23824; G. gallus 3, P23825; G. gallus 4, P43691; G. gallus 5, P43692; G. gallus 6, P43693; Homo sapiens 1, P15976; H. sapiens 2, P23769; H. sapiens 3, P23771; H. sapiens 4, P43694; H. sapiens 6, Q92908; Mus musculus 1, P17679; M. musculus 2, AB000096; M. musculus 3, P23772; M. musculus 4, Q08369; M. musculus 5, P97489; M. musculus 6, S82462; Rattus norvegicus 1, P43429; R. norvegicus 4, P46152; R. norvegicus 6, P46153; Xenopus laevis 1, P23767; X. laevis 2, P23770; X. laevis 3, P23773; X. laevis 4, Q91677; X. laevis 5, P43695; X. laevis 6, Q91678.
Tissue specificity and temporal pattern of AaGATAr mRNA expression.
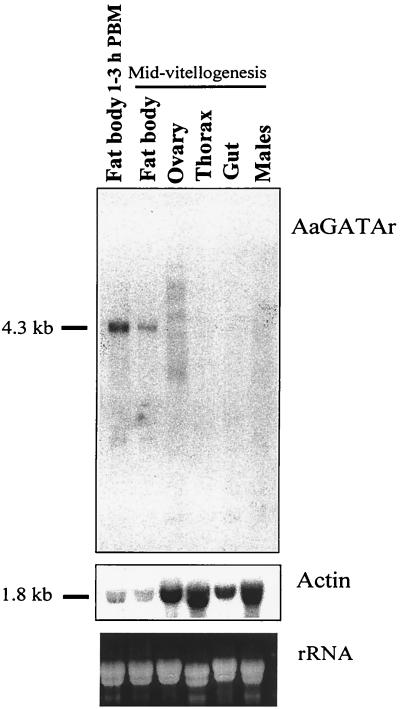
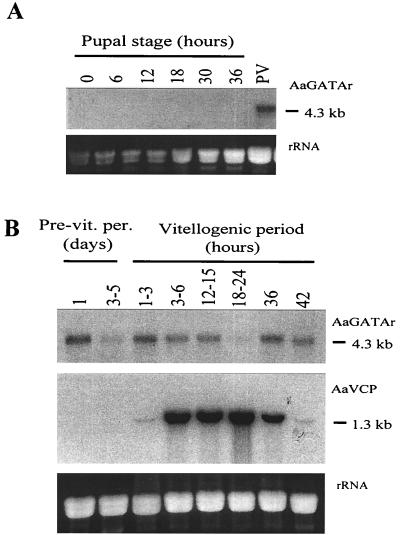
To determine the distribution of the AaGATAr mRNA in the adult mosquito, samples of total RNA from whole body of males as well as fat body, ovary, gut, and thorax of vitellogenic females were subjected to Northern blot analyses, using a 2.5-kb EcoRI AaGATAr cDNA fragment containing the DNA-binding domain as a probe. A single RNA transcript of 4.3 kb, matching the size of the AaGATAr cDNA, was found only in the female fat body (Fig. 4). No hybridization was detected in total RNA collected from pupae at different developmental time points. In contrast, the AaGATAr transcript was present in the fat bodies of female mosquitoes after eclosion (Fig. 5A). The same transcript size was detected when using a 1.7-kb cDNA fragment of the unique region of AaGATAr as a probe (data not shown).
FIG. 4.
Northern blot analysis of AaGATAr mRNA levels in males and different tissues of female mosquitoes. Total RNA was extracted from different female tissues as well as from whole males (15 μg per lane). Northern blots were hybridized with a 32P-labeled 2.5-kb fragment from the protein-coding region (including the DNA-binding domain) of the AaGATAr cDNA clone (upper panel). To control the integrity of the isolated RNA, the same blot was hybridized to A. aegypti actin cDNA (middle panel). Portions of the gel containing rRNA were stained with ethidium bromide to control for equivalent sample loading (lower panel).
FIG. 5.
Northern blot analysis of AaGATAr mRNA levels during pupal development and in the female mosquito fat body during the first vitellogenic cycle. (A) Total RNA from various stages in A. aegypti development (15 μg per lane). Northern blots were hybridized with a 32P-labeled 2.5-kb fragment from the protein-coding region (including the DNA-binding domain) of the AaGATAr cDNA clone (upper panel). (B) Total RNA was extracted from fat bodies dissected from previtellogenic mosquitoes on indicated days (Pre-vit.per.) after eclosion and from vitellogenic fat bodies at indicated hours PBM (15 μg per lane). Northern blots were hybridized with the same 32P-labeled 2.5-kb fragment from the AaGATAr cDNA clone (upper panel) and with a 32P-labeled 0.8-kb fragment from the AaVCP gene (middle panel). Portions of the gel containing rRNA were stained with ethidium bromide to control for equivalent sample loading (lower gels in panels A and B).
Detailed analysis of the AaGATAr expression in the female fat bodies showed that its transcript was present throughout the previtellogenic, vitellogenic, and postvitellogenic periods (Fig. 5B). The AaGATAr transcript was more abundant in newly eclosed females and newly fed females (1 to 3 h PBM) as well as in those at the postvitellogenic stage (36 and 42 h PBM) (Fig. 5B). Thus, the AaGATAr gene appears to be a fat body-specific gene expressed only in adult females.
AaGATAr protein recognizes GATA binding sites and binds to putative GATA elements in 5′ regions of mosquito YPP genes.
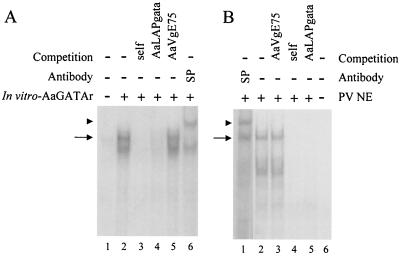
Next, we elucidated whether the AaGATAr protein recognized the DNA-binding site characteristic for the GATA family of transcription factors. The TNT-expressed AaGATAr was tested in EMSAs using the box A sequence from the D. mulleri alcohol dehydrogenase (Adh) gene as a probe, which had been shown to bind a truncated version of the Drosophila GATAb protein, ABF (1). In our EMSAs, a binding complex was observed with AaGATAr and the box A response element (Fig. 6A, lane 2). The specificity of the complex was confirmed by its competition with a 50-fold molar excess of either the box A DNA (Fig. 6A, lane 3) or a putative GATA binding site from the A. aegypti lysosomal aspartic protease (LAP) gene (12) (Fig. 6A, lane 4) but was not confirmed by nonspecific competitors like a putative E75 binding site from the A. aegypti Vg gene (Fig. 6A, lane 5). Furthermore, the specificity of the complex was also established by a supershift, with the antibodies recognizing the first zinc finger domain of B. mori GATA (13) (Fig. 6A, lane 6). Taken together, these results indicated that AaGATAr was specifically bound to GATA DNA motifs in vitro.
FIG. 6.
AaGATAr protein binds to a GATA binding site, and identification is performed for a functional GATA protein in fat body nuclear extracts of previtellogenic fat bodies. −, absence of substance; +, presence of substance. (A) EMSA using in vitro-transcribed and -translated AaGATAr and 32P-labeled Drosophila box A element as a probe. The GATA-specific complex is indicated by an arrow (lane 2). Fiftyfold molar excesses of cold probe (lane 3) or the AaLAP GATA binding site (lane 4) were included as specific competitors. The same excess of cold double-stranded oligonucleotide of an unrelated sequence (AaVgE75 binding site) was included in lane 5. The DNA-AaGATAr complex was supershifted by a B. mori anti-GATA serum (lane 6). An arrowhead indicates the position of the supershifted complex. In vitro-expressed full-length AaGATAr showed a small product due to an internal initiation site of transcription. (B) Nuclear extracts from previtellogenic fat bodies were examined by EMSA with AaVgGATAb element as a probe. The GATA complex was also supershifted by a B. mori anti-GATA serum (lane 1). An arrowhead indicates the position of the supershifted complex. The GATA-specific complex is indicated by an arrow. For competition analysis of this complex, 50-fold molar excesses of cold probe (lane 4) or AaLAP GATA oligonucleotide (lane 5) were included as specific competitors. The same amount of cold double-stranded oligonucleotide of an unrelated sequence was included in lane 3.
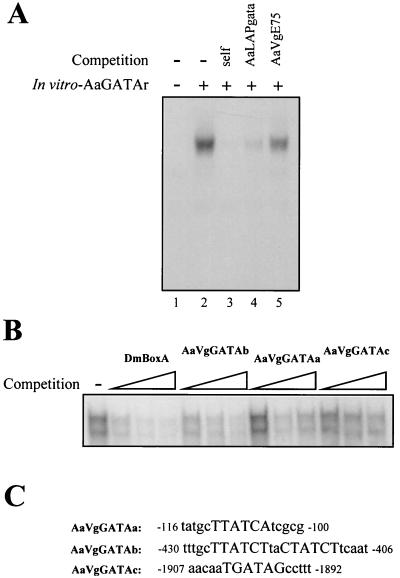
Analysis of the 5′ regulatory region of Vg and VCP genes showed the presence of several GATA binding sites. As an example, AaGATAr specifically bound to a putative binding site from the Vg gene designated AaVgGATAb (Fig. 7A, lanes 2 to 5).
FIG. 7.
In vitro-synthesized AaGATAr protein binds to upstream elements of the A. aegypti YPP genes. (A) AaGATAr protein binds to a sequence in the Vg gene. An EMSA was done with in vitro-produced AaGATAr and AaVgGATAb as probes. A complex due to specific interaction between DNA and protein is clearly detected (lane 2). Fiftyfold molar excesses of cold probe (lane 3) and AaLAP GATA binding site (lane 4) were included as specific competition. The same excess of a cold, unrelated oligonucleotide (AaVgE75) was included in lane 5. (B) Relative affinity of different AaVg GATA binding sites for AaGATAr protein. Binding of AaGATAr to Drosophila box A element in the absence (−) or in the presence of increasing amounts of cold competing probes (self-competition [DmBoxA.], AaVgGATAa, AaVgGATAb, and AaVgGATAc) was performed to compare the binding affinity of the different AaVg GATA elements. (C) GATA motifs in the upstream regulatory regions of vitellogenin used in the competition analysis.
To demonstrate the occurrence of different relative affinities of AaGATAr binding to different GATA binding sites within the 5′ regulatory region of the Vg gene, we carried out an EMSA competition analysis. The binding complex formed by the AaGATAr protein and the box A response element was competed with increasing concentrations of one of three different AaVg GATA response elements (designated as AaVgGATAa, AaVgGATAb, and AaVgGATAc) (Fig. 7C) as well as with the Drosophila box A element as a positive control. The competition levels indicated that the DNA-binding affinity of AaGATAr towards the different AaVg GATA binding sites followed the order AaVgGATAb→AaVgGATAa→AaVgGATAc (Fig. 7B).
Previtellogenic fat body nuclear extracts contain a GATAr-like factor bound to GATA elements in the YPP genes.
To identify the occurrence of nuclear factors that bind specifically to these GATA sequences in the Vg gene at the state of previtellogenic arrest, we carried out EMSAs with nuclear extracts prepared from fat bodies of previtellogenic female mosquitoes. A retardation complex was observed when the radiolabeled AaVgGATAb response element was incubated with the nuclear extract (Fig. 6B, lane 2). To determine the specificity of this interaction, competition experiments were performed using excess cold-specific or nonspecific oligonucleotides (Fig. 6B, lanes 3 to 5). As done before, the specificity of the complex was also established by a supershift with the anti-GATA serum (Fig. 6B, lane 1). Developmental changes of this GATA binding complex in the fat body nuclei showed that it was present during the stage of arrest (3 to 5 days posteclosion), as well as at 60 h PBM, when YPP synthesis was completely halted (not shown).
AaGATAr protein acts as a transcriptional repressor.
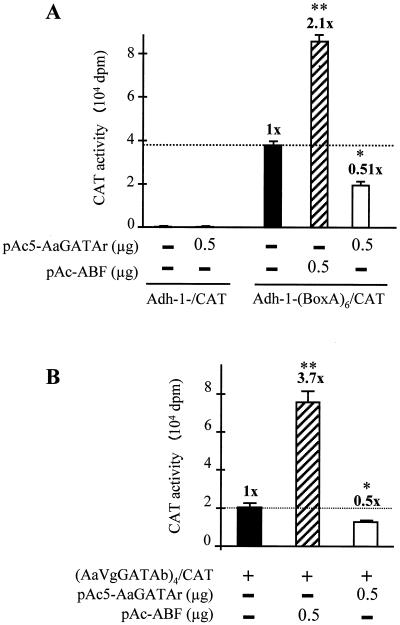
To establish whether AaGATAr was able to regulate transcription upon binding to GATA binding sites, we used the transient transfection assay in a Drosophila Schneider (S2) cell line. We employed an adh-1-(BoxA)6-CAT reporter construct that contained six copies of the box A site upstream of the adh-1 promoter with the CAT gene as a reporter. The pAc5-AaGATAr expression vector contained AaGATAr cDNA under the control of the constitutive actin 5C promoter. As a positive control, the pPac5-ABF vector (Drosophila ABF controlled by the actin 5C promoter) (1) was used. As shown in Fig. 8A, while ABF was able to activate the reporter gene (up to 2.1-fold), AaGATAr repressed the reporter activity (51% inhibition).
FIG. 8.
AaGATAr represses gene transcription via GATA-specific recognition sites. The Drosophila Schneider cell line was transfected with the indicated plasmids by the lipid-mediated method. −, absence of substance; +, presence of substance. (A) Chloramphenicol acetyltransferase (CAT) activity from cells transfected with 100 μg of a reporter plasmid containing six copies of box A upstream of the Adh promoter driving the expression of CAT, with either 0.5 μg of pPac5-ABF (Drosophila GATAb isoform controlled by the actin promoter) or pAc5-AaGATAr (AaGATAr controlled by the actin promoter). As a control, the reporter plasmid Adh-1/CAT (lacking any GATA sequences) was used. (B) CAT activity from cells transfected with 100 ng of a reporter plasmid containing four copies of the AaVgGATAb binding site upstream of the Adh promoter driving the expression of CAT, with 0.5 μg of pPAc5-ABF or pAc5-AaGATAr. In all cases, cells were transfected for 12 h and incubated for 48 h at 22°C. The plasmid pAc5-LacZ was used in all assays for transfection efficiency. Transfection was performed in two independent experiments in triplicate. Significant differences (t test) between treated and respective controls are represented by asterisks (∗, P ≤ 0.05; ∗∗, P ≤ 0.005). Each value represents the mean ± standard deviation.
To verify that AaGATAr could exert similar repression via the GATA binding sites present in the Vg gene, we modified the adh-1-(BoxA)6-CAT reporter plasmid, replacing six copies of box A with four copies of the AaVgGATAb binding site. This construct was cotransfected with either the ABF or AaGATAr expression vectors; ABF was able to activate the reporter up to 3.7-fold, whereas AaGATAr repressed the CAT activity about 50% (Fig. 8B). It is worth noting that in the absence of pPac-ABF or pAc5-AaGATAr, the reporter construct showed a high basal activity consistent with the presence of Drosophila GATAb in S2 cells (1).
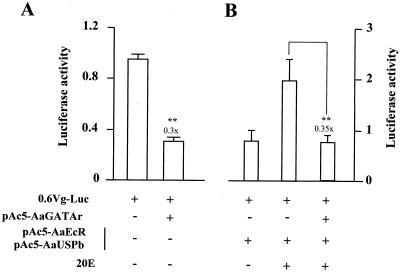
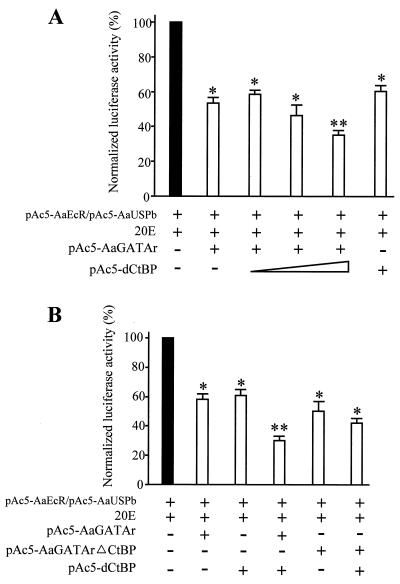
Next, we carried out cell transfection experiments utilizing fragments of the 5′ regulatory regions of the mosquito Vg gene containing GATA binding sites in the context of their natural flanking sequences. The luciferase reporter was driven by the Vg promoter containing 0.6 kb of the 5′ untranscribed region (−600 to +115 bp) with both AaGATAb and AaGATAa binding sites (0.6Vg-Luc) (Fig. 9). Cotransfection of the 0.6Vg-Luc construct with the AaGATAr expression plasmid resulted in a noteworthy 70% reduction of the basal reporter activity (Fig. 9A). In addition to GATA binding sites, this region contained the ecdysone response element sufficient for a two- to threefold ecdysone-dependent activation after cotransfection with AaEcR and AaUSP expression plasmids (Martín, Wang, and Raikhel, unpublished data). Because of that, cotransfection of the 0.6Vg-Luc construct with the AaEcR and AaUSP expression plasmids resulted in about a 2.5-fold induction of transcriptional activity in the presence of 20E (Fig. 9B). Importantly, this ecdysone-dependent transactivation was abolished, a 65% total reduction, as a result of the cotransfection of AaGATAr (Fig. 9B). To ensure that the AaGATAr-dependent repression was not due to a nonspecific squelching of the general transcription machinery, cell transfections were carried out with Adh-1/CAT and pAc5-LacZ (reporters without GATA binding sites) as well as with 0.6Vg-Luc (reporter with GATA binding sites), along with pAc5-AaGATAr at the same time. Whereas AaGATAr inhibited the activity of 0.6Vg-Luc, it did not affect the expression of the other reporter plasmids which were driven by promoters independent of GATA (data not shown).
FIG. 9.
AaGATAr inhibits basal and 20E-dependent activation of the A. aegypti vitellogenin gene. Luciferase activity is measured in arbitrary units. (A) Luciferase enzyme activity in Drosophila Schneider cells transfected with 100 ng of a reporter gene containing 0.6 kb of the 5′ regulatory region of the Vg gene, which controls the expression of luciferase and pAc5-AaGATAr (500 ng). (B) The Vg gene-luciferase reporter and pAc5-AaGATAr were cotransfected along with the expression vectors pAc5-AaEcR (12.5 ng) and pAc5-AaUSPb (12.5 ng). Cells were treated, where indicated, with 1 μM 20E for 36 h after transfection and incubated at 22°C. The plasmid pAc5-LacZ was used in all assays for transfection efficiency. Transfection was performed in two independent experiments in triplicate. Significant differences (t test) between treated and respective controls are represented by asterisks (∗∗, P ≤ 0.005). Each value represents the mean ± standard deviation.
AaGATAr interacts with the corepressor CtBP.
We identified a putative recognition site (PCDLSYK) for CtBP in AaGATAr (Fig. 1 and 10A). We used the glutathione S-transferase (GST) pulldown in vitro binding assay to examine the interaction between Drosophila CtBP (dCtBP) and AaGATAr. Drosophila CtBP, fused to GST (GST-dCtBP), was immobilized on glutathione-Sepharose beads and incubated with in vitro-expressed 35S-labeled AaGATAr. This assay showed that GST-dCtBP specifically interacted with AaGATAr (Fig. 10B, lanes 1 to 3). The mutant AaGATArΔCtBP with a deleted putative CtBP binding site was expressed and radiolabeled in vitro (Fig. 10B, lane 4); however, it failed to bind GST-dCtBP (Fig. 10B, lane 6). As a negative control, we tested in the same assay the Drosophila ABF protein, a transcriptional GATA activator. Drosophila ABF was prepared in a way similar to that for AaGATAr (Fig. 10B, lane 7) and subjected to binding with GST-dCtBP. No interaction between GST-dCtBP and ABF was detected (Fig. 10B, lane 9).
FIG. 10.
AaGATAr interacts with the corepressor dCtBP in vitro. (A) Consensus interaction motif for CtBP. Shown are Drosophila and mosquito repressors that have been demonstrated to bind to the CtBP corepressor protein. (B) 35S-labeled full-length AaGATAr protein (lane 1) interacts with GST-dCtBP (lane 3) but not with GST alone (lane 2). Mutated AaGATAr protein lacking the CtBP recognition site (AaGATArΔCtBP) (lane 4) is not able to retain GST-dCtBP (lane 6) or GST alone (lane 5). As a control, 35S-labeled ABF protein (lane 7) shows no interaction with GST-dCtBP (lane 9) or GST alone (lane 8). In vitro-expressed full-length AaGATAr showed a smaller product, due to an internal initiation site of transcription.
We then cotransfected Drosophila S2 cells with both pAc5-AaGATAr and pAc5-dCtBP, along with the 0.6Vg-Luc plasmid as a reporter. As shown, whereas AaGATAr was able to repress the ecdysone-dependent stimulation of the reporter by 53%, addition of increasing amounts of dCtBP resulted in an increase of the repression in a dose-dependent manner (up to a 70% inhibition with a 1:1 AaGATAr/dCtBP ratio) (Fig. 11A). When we transfected Drosophila cells with the mutated AaGATArΔCtBP protein, it displayed the same degree of repression that AaGATAr did alone, but this repression was not increased by dCtBP cotransfection (Fig. 11B).
FIG. 11.
dCtBP cooperates with AaGATAr to repress transcription. (A) Luciferase enzyme activity in Drosophila Schneider cells transfected with 100 ng of the 0.6Vg-Luc reporter gene along with the expression vectors pAc5-AaEcR (12.5 ng), pAc5-USPb (12.5 ng), pAc5-AaGATAr (500 ng), and pAc5-dCtBP (10, 100, and 500 ng). (B) Luciferase activity of Drosophila Schneider cells transfected with 100 ng of the 0.6Vg-Luc reporter gene along with expression vectors pAc5-AaEcR (12.5 ng), pAc5-USPb (12.5 ng), pAc5-AaGATAr (250 ng), pAc5-dCtBP (250 ng), and pAc5-AaGATArΔCtBP (250 ng). Cells were treated with 1 μM 20E for 36 h after transfection. The plasmid pAc5-LacZ was used in each assay for transfection efficiency. Transfections were performed in two independent experiments in triplicate. Significant differences (t test) between treated and respective controls are represented by asterisks (∗, P ≤ 0.05; ∗∗, P ≤ 0.005). Each value represents the mean ± standard deviation.
DISCUSSION
AaGATAr is a novel member of the GATA family of transcription factors that serves as a repressor.
In this work, we report the characterization of a mosquito transcription factor belonging to the GATA family. Analysis of the amino acid sequence of the AaGATAr protein revealed that it contained a double C.X2.C.X17.C.X2.C. zinc finger, a signature motif of the GATA DNA-binding transcription factors (25). Moreover, the Pileup analysis based on the alignment of the DNA-binding domains of GATA family members confirmed that AaGATAr belongs to the GATA family and is related to the subfamily that includes isoforms 4 through 6 (Fig. 3). Furthermore, AaGATAr protein recognizes the DNA-binding sites characteristic for the GATA family of transcription factors and specifically binds to several putative GATA elements in 5′ regulatory regions of the mosquito YPP genes Vg and VCP.
Previously, the large family of GATA-related transcription factors was known for its transactivating properties (3, 26). The novel aspect of our study is the finding that AaGATAr acts as a transcriptional repressor. AaGATAr was able to repress the adh-1-(BoxA)6-CAT reporter construct that contained six copies of box A GATA elements. Moreover, in the transient assay, AaGATAr could exert similar repression via the GATA binding sites present in the Vg gene. Even more significant is the ability of AaGATAr to overcome and repress ecdysone-dependent activation of the 5′ regulatory regions of the mosquito Vg gene containing GATA binding sites in the context of their natural flanking sequences, as well as the ecdysone response element. Recently, it has been shown that transcriptional cofactors of the FOG (Friend of GATA) family can repress GATA-mediated activation (14, 18). However, to our knowledge, this is the first report of a GATA transcription factor that serves as a specific transcriptional repressor.
AaGATAr directly interacts with the corepressor CtBP.
AaGATAr is involved in transcriptional repression by recruiting the corepressor protein CtBP. CtBP has been shown to interact with a highly conserved PXDLSXK sequence within several zinc finger repressors which are involved in the restricted patterns of gene expression during Drosophila embryogenesis (such as krüppel, snail, knirps, hairy, and zfh-1) (23, 27, 39) with the vertebrate proteins CtIP, ZEB, and BKLF (28, 33, 35) and the viral protein Ad2/5E1A (34). As in the case of these repressors, the interaction with CtBP depends on the integrity of a highly conserved motif, PCDLSYK, in AaGATAr. The CtBP recognition motif has not been found in any other member of the GATA family of transcription factors, which is consistent with the previously reported role of GATA proteins as transcriptional activators (3, 26).
Cotransfection experiments validated the importance of CtBP in the AaGATAr-mediated repression of the Vg gene. Cotransfection with increasing amounts of dCtBP resulted in an elevation of the repression levels of the Vg gene in a dose-dependent manner. Although CtBP significantly enhanced AaGATAr repression, AaGATAr alone or the mutated AaGATArΔCtBP was still able to repress up to 50% of the ecdysone-dependent stimulation of the reporter, suggesting the presence of an additional mode of repression associated with the AaGATAr action. The Drosophila developmental transcriptional repressor hairy provides an example of such a dual repression mechanism. The action of this repressor is independently mediated by two corepressors, Groucho and dCtBP (23, 27).
Previously, GATA activators have been shown to interact with cofactors, such as FOG in vertebretes and u-shaped in Drosophila, which can repress GATA-mediated activation. Interestingly, FOG and u-shaped contain the CtBP recognition motifs and subsequently recruit CtBP to display repression activity (14, 17, 18, 35). Thus, CtBP appears to play a critical role in either the repression of GATA-mediated activation through binding to a corepressor or a direct interaction with the GATA repressor, as reported in this paper.
AaGATAr serves the unique role of a specific repressor at the state of vitellogenic arrest in the mosquito female.
The stage specificity of gene expression can be regulated through different mechanisms, which may include a permanent shutdown of temporally expressed genes or the use of alternative promoters in order to shift the regulation of a particular gene from one stage-specific regulatory hierarchy to another. In Drosophila, transcriptional repressors such as krüppel, snail, hairy, knirps, or zhf-1 are responsible for the correct timing of the expression of genes involved in development of the embryo through their sequential shutdown (23, 27, 28, 39). The stage-specific expression of the Drosophila alcohol dehydrogenase gene is achieved through the alternative activation of two tandem promoters (30). The proximal promoter is active during embryonic development and early larval stages, while the distal promoter is active in late larval stages and adulthood. The switch to the late distal promoter is accomplished through a permanent shutdown of the early proximal promoter by a zinc finger repressor, AEF-1 (30). Interestingly, AEF-1 has also been implicated in the permanent repression of Drosophila fat body-specific YPP genes in other female tissues (2).
Cyclic egg production in insects is activated either by environmental factors such as food and mating or by internal developmental and endocrine signals. Unlike other patterns of temporal regulation of gene expression which involve permanent repression of stage-specific genes, expression of YPP genes in insects with cyclic egg production suggests the need for a temporal alteration of transcriptional repression and activation of the same genes. However, the precise mechanism of such a cyclic regulation of YPP genes has not been elucidated.
Analyses of tissue and temporal distribution of the AaGATAr transcript have demonstrated that it is expressed only in the fat body of adult female mosquitoes. This specificity suggests that AaGATAr acts as a fat body-specific repressor regulating the transcriptional activity of genes expressed during the reproductive phase of female life. Furthermore, EMSAs of female fat body nuclear extracts have shown that a retardation band with mobility and specificity of binding similar to those of AaGATAr exists at the state of arrest, as well as during postvitellogenic stages when YPP genes are shut off. Taken together, our results from transcript abundance and binding activity suggest that AaGATAr represents a specific factor involved in temporal transcriptional repression of YPP genes before and between vitellogenic cycles.
In anautogenous mosquitoes, such as A. aegypti, this temporal alteration of transcriptional repression and activation involved in the regulation of YPP genes is under the strict control of a hormonal cascade, initiation of which requires a blood meal. In this work, we have uncovered a significant element of the underlying molecular mechanism of the state of arrest, which involves a unique GATA repressor. Importantly, we showed that AaGATAr can not only inhibit the basal activity presented by the natural Vg promoter but can also overcome its ecdysone-dependent activation. Impairing ecdysone-dependent activation by AaGATAr during previtellogenesis is essential because at that period, detectable amounts of 20E as well as the components of the EcR, AaEcR and AaUSP are present (15, 40). Without comparable studies on other insects with cyclic egg production, it is difficult to speculate whether in evolution, anautogenous mosquitoes have developed a unique repression mechanism utilizing GATAr. Alternatively, they may have adapted a more general mechanism for the cyclic regulation of YPP genes by developing its strict control via a blood meal-activated hormonal cascade.
In summary, the characterization of AaGATAr and its response elements in the Vg and VCP genes allows us to better understand the complex regulatory program controlling the expression of YPP genes in anautogenous mosquitoes, which serve as vectors of pathogens responsible for some major infectious diseases in humans.
ACKNOWLEDGMENTS
We thank T. Abel for the kind gift of (BoxA)6/CAT plasmid and pPac-ABF clones, S. Parkhurst for the GST-dCtBP clone, and K. Iatrou for antibodies to B. mori GATA. We also thank A. Hays for his invaluable help in maintaining mosquitoes, M. Trail for editing the manuscript, and D. Arnosti for helpful discussions of the manuscript.
This work was supported by grant AI-24716 from the National Institutes of Health to A. S. Raikhel. D. Martín was a recipient of a postdoctoral research grant from the Spanish Ministry of Education and Culture. M.-D. Piulachs was supported in part by a travel grant from the Spanish Ministry of Education and Culture.
REFERENCES
- 1.Abel T, Michelson A M, Maniatis T. A Drosophila GATA family member that binds to Adh regulatory sequences is expressed in the developing fat body. Development. 1993;119:623–633. doi: 10.1242/dev.119.3.623. [DOI] [PubMed] [Google Scholar]
- 2.An W, Wensink P. Integrating sex- and tissue-specific regulation within a single Drosophila enhancer. Genes Dev. 1995;9:256–266. doi: 10.1101/gad.9.2.256. [DOI] [PubMed] [Google Scholar]
- 3.Bossard P, Zaret K. GATA transcription factors as potentiators of gut endoderm differentiation. Development. 1998;125:4909–4917. doi: 10.1242/dev.125.24.4909. [DOI] [PubMed] [Google Scholar]
- 4.Butler D. Time to put malaria control on the global agenda. Nature. 1997;386:535–536. doi: 10.1038/386535a0. [DOI] [PubMed] [Google Scholar]
- 5.Chen J-S, Cho W-L, Raikhel A S. Analysis of mosquito vitellogenin cDNA. Similarity with vertebrate phosvitins and arthropod serum proteins. J Mol Biol. 1994;237:641–647. doi: 10.1006/jmbi.1994.1261. [DOI] [PubMed] [Google Scholar]
- 6.Cho W-L, Deitsch K W, Raikhel A S. An extraovarian protein accumulated in mosquito oocytes is a carboxypeptidase activated in embryos. Proc Natl Acad Sci USA. 1991;88:10821–10824. doi: 10.1073/pnas.88.23.10821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cho W-L, Kapitskaya M, Raikhel A S. Mosquito ecdysteroid receptor: analysis of the cDNA and expression during vitellogenesis. Insect Biochem Mol Biol. 1995;25:19–27. doi: 10.1016/0965-1748(94)00045-j. [DOI] [PubMed] [Google Scholar]
- 8.Cho W-L, Tsao S M, Hays A R, Walter R, Chen J S, Snigirevskaya E S, Raikhel A S. Mosquito cathepsin B-like protease involved in embryonic degradation of vitellin is produced as a latent extraovarian precursor. J Biol Chem. 1999;274:13311–13321. doi: 10.1074/jbc.274.19.13311. [DOI] [PubMed] [Google Scholar]
- 9.Cho W-L, Raikhel A S. Cloning of cDNA for mosquito lysosomal aspartic protease. J Biol Chem. 1992;267:21823–21829. [PubMed] [Google Scholar]
- 10.Deitsch K W, Raikhel A S. Cloning and analysis of the locus for mosquito vitellogenic carboxypeptidase. Insect Mol Biol. 1993;2:205–213. doi: 10.1111/j.1365-2583.1994.tb00140.x. [DOI] [PubMed] [Google Scholar]
- 11.Deitsch K W, Chen J S, Raikhel A S. Indirect control of yolk protein genes by 20-hydroxyecdysone in the fat body of the mosquito, Aedes aegypti. Insect Biochem Mol Biol. 1995;25:449–454. doi: 10.1016/0965-1748(94)00082-a. [DOI] [PubMed] [Google Scholar]
- 12.Dittmer N T, Raikhel A S. Analysis of the mosquito lysosomal aspartic protease gene: an insect housekeeping gene with fat body-enhanced expression. Insect Biochem Mol Biol. 1997;27:323–335. doi: 10.1016/s0965-1748(97)00007-6. [DOI] [PubMed] [Google Scholar]
- 13.Drevet J R, Skeiky Y A W, Iatrou K. GATA-type zinc finger motif-containing sequences and chorion gene transcription factors of the silkworm Bombyx mori. J Biol Chem. 1994;269:10660–10667. [PubMed] [Google Scholar]
- 14.Fox A H, Liew C, Holmes M, Kowalski K, Mackay J, Crossley M. Transcriptional cofactors of the FOG family interact with GATA proteins by means of multiple zinc fingers. EMBO J. 1999;18:2812–2822. doi: 10.1093/emboj/18.10.2812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hagedorn H H. The role of ecdysteroids in the adult insect. In: Downer R G H, Laufer H, editors. Endocrinology of insects. New York, N.Y: Alan R. Liss; 1983. pp. 271–304. [Google Scholar]
- 16.Hays A R, Raikhel A S. A novel protein produced by the vitellogenic fat body and accumulated in mosquito oocytes. Roux's Arch Dev Biol. 1990;199:114–121. doi: 10.1007/BF02029559. [DOI] [PubMed] [Google Scholar]
- 17.Heanlin M, Cubadda Y, Blondeau F, Heitzler P, Lutz Y, Simpson P, Ramain P. Transcriptional activity of Pannier is regulated negatively by heterodimerization of the GATA DNA-binding domain with cofactor encoded by the U-shaped gene of Drosophila. Genes Dev. 1997;11:3096–3108. doi: 10.1101/gad.11.22.3096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Holmes M, Turner J, Fox A, Chisholm O, Crossley M, Chong B. hFOG-2, a novel zinc finger protein, binds the co-repressor mCtBP2 and modulates GATA-mediated activation. J Biol Chem. 1999;274:23491–23498. doi: 10.1074/jbc.274.33.23491. [DOI] [PubMed] [Google Scholar]
- 19.Kapitskaya M, Wang S-F, Cress D E, Dhadialla T S, Raikhel A S. The mosquito ultraspiracle homologue, a partner of ecdysteroid receptor heterodimer: cloning and characterization of isoforms expressed during vitellogenesis. Mol Cell Endocrinol. 1996;121:119–132. doi: 10.1016/0303-7207(96)03847-6. [DOI] [PubMed] [Google Scholar]
- 20.Kozak M. Point mutations define a sequence flanking the AUG initiator codon that modulates translation by eukaryotic ribosomes. Cell. 1986;44:283–292. doi: 10.1016/0092-8674(86)90762-2. [DOI] [PubMed] [Google Scholar]
- 21.Krasnow M A, Saffman E E, Kornfeld K, Hogness D S. Transcriptional activation and repression by Ultrabithorax proteins in cultured Drosophila cells. Cell. 1989;57:1031–1043. doi: 10.1016/0092-8674(89)90341-3. [DOI] [PubMed] [Google Scholar]
- 22.Miura K, Wang S F, Raikhel A S. Two distinct subpopulations of ecdysone receptor complex in the female mosquito during vitellogenesis. Mol Cell Endocrinol. 1999;156:111–120. doi: 10.1016/s0303-7207(99)00136-7. [DOI] [PubMed] [Google Scholar]
- 23.Nibu Y, Zhang H, Bajor E, Barolo S, Small S, Levine M. dCtBP mediates transcriptional repression by Knirps, Knuppel and Snail in the Drosophila embryo. EMBO J. 1998;17:7009–7020. doi: 10.1093/emboj/17.23.7009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Omichinski J G, Clore G M, Schadd O, Felsenfeld G, Trainor C, Appella E, Stahl S J, Gronenborn A M. NMR structure of a specific DNA complex of Zn-containing DNA binding domain of GATA-1. Science. 1993;261:438–446. doi: 10.1126/science.8332909. [DOI] [PubMed] [Google Scholar]
- 25.Orkin S H. GATA binding transcription factors in hematopoietic cells. Blood. 1992;80:575–581. [PubMed] [Google Scholar]
- 26.Orkin S H. Transcriptional factors and hematopoietic development. J Biol Chem. 1995;270:4955–4958. doi: 10.1074/jbc.270.10.4955. [DOI] [PubMed] [Google Scholar]
- 27.Poortinga G, Watanabe M, Parkhurst S M. Drosophila CtBP: a Hairy-interacting protein required for embryonic segmentation and Hairy-mediated transcriptional repression. EMBO J. 1998;17:2067–2078. doi: 10.1093/emboj/17.7.2067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Postigo A A, Dean D C. ZEB represses transcription through interaction with the corepressor CtBP. Proc Natl Acad Sci USA. 1999;96:6683–6688. doi: 10.1073/pnas.96.12.6683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Raikhel A S. Vitellogenesis in mosquitoes. Adv Dis Vector Res. 1992;9:1–39. [Google Scholar]
- 30.Ren B, Maniatis T. Regulation of Drosophila Adh promoter switching by an initiator-targeted repression mechanism. EMBO J. 1998;17:1076–1086. doi: 10.1093/emboj/17.4.1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Romans P, Tu Z, Ke Z, Hagedorn H. Analysis of a vitellogenin gene of the mosquito, Aedes aegypti and comparisons to vitellogenins from other organisms. Insect Biochem Mol Biol. 1995;25:939–958. doi: 10.1016/0965-1748(95)00037-v. [DOI] [PubMed] [Google Scholar]
- 32.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 33.Schaeper U, Subramanian T, Lim L, Boyd J M, Chinnadurai G. Interaction between a cellular protein that binds to the C-terminal region of adenovirus E1A (CtBP) and a novel cellular protein is disrupted by E1A through a conserved PLDLS motif. J Biol Chem. 1998;273:8549–8552. doi: 10.1074/jbc.273.15.8549. [DOI] [PubMed] [Google Scholar]
- 34.Swirnoff A H, Apel E D, Svaren J, Sevetson B R, Zimonjic D B, Popescu N C, Milbrandt J. Nab1, a corepressor of NGFI-A (Egr-1), contains an active repression domain. Mol Cell Biol. 1998;18:512–524. doi: 10.1128/mcb.18.1.512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Turner J, Crossley M. Cloning and characterization of mCtBP2, a co-repressor that associates with basic Kruppel-like factor and other mammalian transcriptional regulators. EMBO J. 1998;17:5129–5140. doi: 10.1093/emboj/17.17.5129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang S F, Li C, Zhu J, Miura K, Miksicek R J, Raikhel A S. Differential expression and regulation by 20-hydroxyecdysone of mosquito ultraspiracle isoforms. Dev Biol. 2000;218:99–113. doi: 10.1006/dbio.1999.9575. [DOI] [PubMed] [Google Scholar]
- 37.Yao T P, Segraves W A, Oro A E, McKeown M, Evans R M. Drosophila ultraspiracle modulates ecdysone receptor function via heterodimer formation. Cell. 1992;71:63–72. doi: 10.1016/0092-8674(92)90266-f. [DOI] [PubMed] [Google Scholar]
- 38.Yao T P, Forman B M, Jiang Z, Cherbas L, Chen J D, McKeown M, Cherbas P, Evans R M. Functional ecdysone receptor is the product of EcR and Ultraspiracle genes. Nature. 1993;366:476–479. doi: 10.1038/366476a0. [DOI] [PubMed] [Google Scholar]
- 39.Zhang H, Levine M. Groucho and dCtBP mediate separate pathways of transcriptional repression in the Drosophila embryo. Proc Natl Acad Sci USA. 1999;96:535–540. doi: 10.1073/pnas.96.2.535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhu J, Miura K, Chen L, Raikhel A S. AHR38, a homolog of NGFI-B, inhibits formation of the functional ecdysteroid receptor in the mosquito Aedes aegypti. EMBO J. 2000;19:253–262. doi: 10.1093/emboj/19.2.253. [DOI] [PMC free article] [PubMed] [Google Scholar]