Abstract
Nuclear matrix attachment regions (MARs), which flank the immunoglobulin μ heavy-chain enhancer on either side, are required for the activation of the distal variable-region (VH) promoter in transgenic mice. Previously, we have shown that the MARs extend a local domain of chromatin accessibility at the μ enhancer to more distal sites. In this report, we examine the influence of MARs on the formation of a nucleoprotein complex at the enhancer and on the acetylation of histones, which have both been implicated in contributing to chromatin accessibility. By in vivo footprint analysis of transgenic μ gene constructs, we show that the occupancy of factor-binding sites at the μ enhancer is similar in transcriptionally active wild-type and transcriptionally inactive ΔMAR genes. Chromatin immunoprecipitation experiments indicate, however, that the acetylation of histones at enhancer-distal nucleosomes is enhanced 10-fold in the presence of MARs, whereas the levels of histone acetylation at enhancer-proximal nucleosomes are similar for wild-type and ΔMAR genes. Taken together, these data indicate that the function of MARs in mediating long-range chromatin accessibility and transcriptional activation of the VH promoter involves the generation of an extended domain of histone acetylation, independent of changes in the occupancy of the μ enhancer.
Transcriptional enhancers are thought to augment gene expression by inducing changes in chromatin accessibility and by facilitating the recruitment of RNA polymerases to linked promoters (6, 21, 61). Enhancer-induced changes in chromatin accessibility involve multiple mechanisms. Specific proteins, termed pioneer proteins, are able to bind directly to nucleosomal DNA (13). Moreover, various enhancer-binding proteins have been shown to interact with components of chromatin remodeling complexes that increase chromatin accessibility in an ATP-dependent manner (73, 77). Finally, some proteins that are bound at enhancers and/or promoters can associate with histone acetyltransferase complexes (HATs) that mediate acetylation of H3 and H4 core histones (9, 70). This type of histone modification is targeted locally to enhancers and/or promoters and is associated with the activation of promoters. In addition, histone acetylation has been correlated with the generation of an extended domain of general DNase I sensitivity and chromatin accessibility (8, 28, 67). However, the molecular mechanisms underlying the generation of long-range chromatin accessibility are still obscure.
The murine immunoglobulin heavy chain gene, which has been studied extensively as a model for tissue-specific gene expression, contains an intronic locus control region (LCR), located 1.5 kb downstream of the variable region (VH) promoter in the rearranged μ gene (20). Similar to other LCRs, this intronic regulatory region of the μ gene confers proper regulation and high-level expression upon transgenes irrespective of the site of chromosomal integration (18, 23, 37). Three regulatory elements contribute to the function of the intronic μ LCR. First, the Eμ enhancer spans a region of 220 bp and contains multiple transcription factor-binding sites, termed μE1 through μE5, μA, μB, and Octa (an octamer) (20). Studies addressing the identity of proteins that interact with these sites have shown that both ubiquitous proteins, such as the basic helix-loop-helix factors E47 and TFE3, and tissue-specific proteins, such as Ets1 and Pu.1, assemble into a stable and cell-type-specific nucleoprotein complex in vitro (3, 50, 53, 68). Second, a promoter for noncoding germ line Iμ transcripts is located at the 3′ boundary of the μ enhancer (42, 71). Finally, the Eμ enhancer is flanked on either side by nuclear matrix attachment regions (MARs) (14). Specific sequences in the MARs have been shown to interact with a B-cell-specific protein, Bright, which appears to antagonize the binding of a widely expressed protein, NF-μNR (30, 78).
In tissue culture transfection assays, the activation of the VH promoter requires only the μ enhancer, whereas in germ line transformation assays, the activation of the VH promoter requires both the enhancer and the flanking MARs (23). A function of the MARs in the regulation of chromatin structure was inferred from multiple experiments. First, the chromatin of transgenes lacking the MARs shows a pattern of DNase I digestion characteristic of inactive genes (23). Second, we found that the μ enhancer in combination with a flanking MAR can confer accessibility upon a distal site in nuclear chromatin, whereas the enhancer alone mediates only local chromatin accessibility (33). In these experiments, we replaced the VH promoter with a promoter for a bacteriophage RNA polymerase, which allowed an assessment of chromatin accessibility in the absence of endogenous transcription and in the absence of interactions between enhancer- and promoter-bound factors. Finally, the immunoglobulin MARs were also shown to antagonize methylation-dependent repression of long-range enhancer function (22). Taken together, these data suggest that the MARs are important components of the μ LCR that allow enhancer function over large distances.
MARs, also known as scaffold-associated regions, are short AT-rich DNA sequences that are widespread throughout the eukaryotic genome and associate with a proteinaceous matrix obtained after histone depletion of the nucleus (41, 57). MARs may have a role in organizing chromatin loops and in the functional insulation of chromatin domains from transcriptional silencing caused by adjacent heterochromatin regions (16, 27). In addition, MARs are frequently colocalized with enhancers or with the boundaries of genes. In association with enhancers, MARs have been shown to stimulate the expression of linked genes (39, 58, 60). However, MARs can also interfere with the interactions between promoters and enhancers when placed between these elements (27). Thus, the function of MARs appears to be dependent upon the context of regulatory elements.
In principle, MARs could confer long-range function upon the μ enhancer by altering the nucleoprotein complex that is assembled at the enhancer. Alternatively, the MARs could be involved in propagating histone modification and chromatin accessibility. Here, we describe experiments that are aimed at addressing these possible mechanisms of MAR function. By genomic footprinting of transgenes containing various parts of the μ LCR, we showed that the enhancer alone is fully occupied by DNA-binding proteins, despite its inability to activate a linked VH promoter. We also showed that the MARs allow the generation of an extended domain of histone acetylation, which could account for the long-range function of the μ enhancer in combination with MARs.
MATERIALS AND METHODS
Cell lines and cell culture.
All pre-B-cell lines were grown at 37°C and 5% CO2 in RPMI medium containing 10% fetal bovine serum and 50 μM β-mercaptoethanol. The pre-B-cell lines derived from transgenic mice used in this study have been described (31–33) except for the μΔMAR cell lines. The new μΔMAR pre-B-cell lines are identical to those described previously (23) except that they contain polylinker sequences (XbaI, NotI, and HinfI) flanking the 220-bp enhancer (Enh 220) fragment at both sides, allowing the specific amplification of the transgenic ΔMAR enhancer by ligation-mediated PCR (LM-PCR). The μΔMAR transgenic animals were obtained by microinjection of a XhoI/SalI DNA fragment containing the μΔMAR gene of plasmid pμΔ2 + 1 (23). The Abelson murine leukemia virus was used to immortalize the pre-B cells from the fetal livers of these animals (31, 63).
RNA analysis.
Total RNA from B cells was isolated using the acid-phenol–guanidinium method (12). S1 probes for the transgenic μ and the β-actin genes were obtained by linear PCR amplification (30 cycles) using Taq DNA polymerase (Boehringer Mannheim) from 10 pmol of specific 5′-end-, 32P-labeled oligonucleotides as primers and plasmids containing μ (p1–27 digested with XhoI) and β-actin (pmβactin digested with EcoRI) sequences as template DNA. The oligonucleotides used as primers to obtain the S1 probes were 5′-GGC CAT CTC CTG CTC GAA GTC-3′ for β-actin and 5′-CCC AGC TGC ATT TCA TTG TAA GG-3′ for the VH probe. Hybridization of the probes (∼10,000 cpm) to 3 to 5 μg of total RNA and digestion with S1 nuclease (Pharmacia) were carried out at 37°C using standard methods (1). The protected fragments were ethanol precipitated in the presence of 20 μg of yeast tRNA and electrophoresed through 8% acrylamide–7 M urea gels in 0.5× Tris-borate-EDTA.
In vivo footprinting.
In vivo footprinting was performed by incubation of pre-B cells with dimethyl sulfate (DMS; Aldrich) followed by LM-PCR amplification of the transgenic enhancer from the genomic DNA isolated from these cells, essentially as described by Ausubel et al. and by Garrity and Wold (1, 24). For in vivo DMS treatment, 50-ml aliquots of cell cultures containing ∼5 × 107 cells were collected by centrifugation (200 × g, 5 min), resuspended in 1 ml of prewarmed culture medium, and incubated for 5 min at 37°C. At this point, 2 to 10 μl of freshly prepared DMS (10% [vol/vol] in ethanol) was added and the samples were further incubated at 37°C for 1 min. To stop methylation, the mixtures were quickly transferred to 49 ml of ice-cold phosphate-buffered saline (PBS) with 0.2% (vol/vol) β-mercaptoethanol (PBS–β-ME), and cells were collected immediately by centrifugation. The cells were washed in 50 ml of ice-cold PBS–β-ME and finally resuspended in 300 μl of the same buffer. Genomic DNA was isolated after sodium dodecyl sulfate (SDS)-proteinase K digestion by phenol-chloroform extractions and ethanol precipitation. Control genomic DNA (50 to 100 μg) in 175 μl of TE buffer (10 mM Tris-HCl [pH 8.0], 1 mM EDTA), isolated from cells not incubated with DMS, was methylated in vitro with 25 μl of DMS (0.25 to 1% [vol/vol] in water) for 2 min at 22 to 37°C. Reactions were stopped by adding 50 μl of DMS stop buffer (1.5 M sodium acetate, pH 7.0; 1 M β-mercaptoethanol; 100 μg of tRNA/ml), and DNA was recovered by ethanol precipitation. The DNA pellets were resuspended in 200 μl of 1 M piperidine (Aldrich) and heated at 90°C (30 min). Next, samples were frozen on dry ice, and the piperidine was evaporated in a Speed-Vac centrifuge (Sorvall). The single-stranded DNAs were resuspended in TE buffer, extracted with phenol-chloroform, and ethanol precipitated twice before being used as templates in LM-PCRs (0.5 μg). A set of three nested oligonucleotides with increasing melting temperatures (Tm) were utilized in LM-PCR as primers for Vent DNA polymerase (New England Biolabs) (26). Typically, ∼1 pmol of oligonucleotide 1 was used for first-strand DNA synthesis (95°C for 7 min, 30 min at the Tm of oligonucleotide 1, and 76°C for 10 min). The products of this reaction were ligated overnight at 17°C using T4 DNA ligase (Promega) to the annealed LM-PCR1 and -2 primers. The ligated products were amplified (18 to 20 cycles) with 10 pmol of oligonucleotide 2 and LM-PCR1 (95°C for 1 min, 2 min at the Tm of oligonucleotide 2, and 76°C for 3.5 min). For the final extension, ∼2 pmol of 5′-end-, 32P-labeled oligonucleotide 3 was added to the reaction mixture during 2 cycles (95°C for 1 min, 2 min at the Tm of oligonucleotide 3, and 76°C for 10 min). The products of each reaction were phenol-chloroform extracted, ethanol precipitated, and resuspended in 25 μl of 95% formamide loading buffer. The samples were heated at 95°C (3 min), and ∼3 to 5 μl was electrophoresed through 6% polyacrylamide–7 M urea gels in 0.5× Tris-borate-EDTA.
Chromatin extracts.
Approximately 2 × 107 pre-B cells were fixed by adding 1/10 of the culture volume of fixing buffer (11% [vol/vol] formaldehyde, 100 mM NaCl, 0.5 mM EGTA, 50 mM Tris-HCl [pH 8.0]). After incubation at 37°C (10 min) and 4°C (50 min), the formaldehyde was quenched by adding 1/20 of the original volume of 2.5 M glycine. Subsequent steps were performed at 4°C. Fixed cells were harvested by centrifugation (200 × g for 5 min), washed in PBS, and resuspended in 15 ml of Triton buffer (10 mM Tris-HCl [pH 8.0], 10 mM EDTA, 0.5 mM EGTA, 0.25% [vol/vol] Triton X-100). After a 15-min incubation, Triton-washed cells were centrifuged (200 × g, 5 min), resuspended in 15 ml of NaCl buffer (10 mM Tris-HCl [pH 8.0], 1 mM EDTA, 0.5 mM EGTA, 200 mM NaCl), and incubated for an additional 15 min. Finally, the samples were centrifuged, resuspended in 1 ml of sonication buffer (10 mM Tris-HCl [pH 8.0], 1 mM EDTA, 0.5 mM EGTA, 1% [wt/vol] SDS) containing a cocktail of protease inhibitors (1 mM phenylmethylsulfonyl fluoride, 10 μg of aprotinin/ml, 0.8 μg of pepstatin/ml, 0.6 μg of leupeptin/ml), sonicated (Branson 450, using a microtip, setting 5, and 100% duty) for 10 to 15 bursts of 10 s, and separated by cooling on ice. This treatment yielded DNA fragments with an average size of 0.5 kb. Cell debris was removed by centrifugation (14,000 × g, 5 min), and the supernatant was stored at −80°C as bulk chromatin extracts. The concentrations of protein and DNA in these extracts were estimated by their absorbances at 260 nm (reported as the optical density at 260 nm [OD260]).
Chromatin immunoprecipitations.
Chromatin extracts were diluted to 6 OD260 units/ml in immunoprecipitation buffer (IP buffer; 140 mM NaCl, 1% Triton X-100, 0.1% sodium deoxycholate, 1 mM phenylmethylsulfonyl fluoride, 100 μg of yeast tRNA/ml, 100 μg of bovine serum albumin/ml) and preincubated for 1 h at 4°C with 10 μl of a protein A-Sepharose suspension/ml. A protein A-Sepharose suspension (50% [vol/vol]) was reconstituted in PBS and washed several times in IP buffer. After a short centrifugation (1,000 × g, 2 min), to remove the protein A-Sepharose beads, the chromatin extracts were divided into 600-μl aliquots. One of these aliquots was kept for isolation of input DNA. Specific serum recognizing acetylated H4 or acetylated H3 histones (α-AcH4 or α-AcH3; Upstate Biotechnology) and serum recognizing the B220 surface B cell as a negative control marker (α-B220; Calbiochem) were added to the other aliquots at a 1:100 dilution (6 μl). After incubation at 4°C (5 h), 40 μl of the protein A-Sepharose suspension was added to each tube, and the samples were incubated for an additional 2 h. Protein A beads were harvested by centrifugation (1,000 × g; 2 min) in Spin-X microcentrifuge tubes (Costar-Corning). Eluates were kept as unbound samples. The protein A beads were washed twice with 0.5 ml of IP buffer, once in 0.5 ml of IP buffer containing 500 mM NaCl, once in 0.5 ml of wash buffer (10 mM Tris [pH 8.0], 250 mM LiCl, 1 mM EDTA, 0.25% sodium deoxycholate), and finally twice with 0.5 ml of TE buffer. The chromatin bound to the protein A-Sepharose beads was eluted in 300 μl of elution buffer (50 mM Tris-HCl [pH 8.0], 10 mM EDTA, 1% [wt/vol] SDS) after heating at 65°C for 15 min. Bound and input chromatin samples were adjusted to 0.5% (wt/vol) SDS in 600 μl. All samples were incubated overnight at 65°C to reverse formaldehyde cross-links. Next, RNA was digested at 37°C (30 min) with 3 μl of DNase-free RNase A (10 mg/ml). Then, the samples were deproteinized for 2 to 3 h at 37°C by adding 10 μl of proteinase K (12 mg/ml). After phenol-chloroform extractions, the DNA was ethanol precipitated using glycogen (10 μg; Sigma) as a carrier. Precipitated DNA was resuspended in 100 μl of TE, and the concentration was estimated as the OD260.
Semiquantitative PCR.
A succession of fourfold dilutions of template DNA were made to quantify the amount of specific DNA fragments in the immunoprecipitation samples by PCR. The amount of template DNA in the starting tube of the dilution series was ∼20 ng. An initial amplification was performed using 20 cycles (94°C, 1 min; 55 to 60°C, 1 min; 72°C, 1 min) in 50 μl of PCR buffer (10 mM Tris-HCl [pH 8.3], 1.5 to 3 mM MgCl2, 50 mM KCl, 250 μM concentrations of each deoxynucleoside triphosphate, 0.001% [wt/vol] gelatin, 0.5 μM concentrations of each oligonucleotide, and 1 U of Taq polymerase). After this PCR, 15 μl was transferred to a new tube containing 50 μl of fresh PCR buffer and cycled 20 times under identical conditions. Ten microliters of each amplification product was analyzed in 2 to 3% Tris-acetate-EDTA–agarose gels. For amplification of the single-copy endogenous sequences we used two PCRs of 22 cycles each. The pairs of oligonucleotides used for PCR were as follows: VDJ-1 and VDJ-2 for the rearranged transgenic VDJ region, MB-1A and MB-1B for the endogenous mb-1 promoter, T7–1 and T7–2 for the distal T7 region, and T3–1 and T3–2 for the proximal T3-large T region.
Oligonucleotides.
All the oligonucleotides used in the LM-PCR experiments were purified from 20% acrylamide–7 M urea gels. The annealed oligonucleotides LM-PCR1 (5′-GCG GTG ACC CGG GAG ATC TGA ATT C-3′) and LM-PCR2 (5′-GAA TTC AGA TC-3′) were used as linker primers for ligation to the first-strand extension products of the LM-PCR. The sets of three nested oligonucleotides were as follows: SPR-1 (5′-CTA AAT ACA TTT TAG AAG TCG ATA AAC-3′), SPR-2 (5′-GTC GAT AAA CTT AAG TTT GGG GAA ACT AG-3′), and SPR-3 (5′-CTT AAG TTT GGG GAA ACT AGA ACT ACT CAA GC-3′) for the top strand in the wild-type μ transgene; 5′MAR-1 (5′-GAC ATT ACT TAA AGT TTA ACC GAG G-3′), 5′MAR-2 (5′-TTA ACC GAG GAA TGG GAG TGA GGC T-3′), and 5′MAR-3 (5′-CGA GGA ATG GGA GTG AGG CTC TCT CAT AC-3′) for the bottom strand in the wild-type μ transgene; 3J-1 (5′-AAC CAC CAA CCA GCA TGT TCA A-3′), 3J-2.2 (5′-GCA TGT TCA ACC GAA ATA AGT CTA GAG C-3′), and 3J-3.3 (5′-GTT CAA CCG AAA TAA GTC TAG AGC GGC CGG AAT-3′) for the top strand in the ΔMAR μ transgene; 5J-1 (5′-CAT CTA GCC TCG GTC TCA AAA GG-3′), 5J-2.2 (5′-TCT CAA AAG GGG TAG TTG CTG TCT AGA GC-3′), and 5J-3.3 (5′-AAG GGG TAG TTG CTG TCT AGA GCG GCC GCT GA-3′) for the bottom strand in the ΔMAR μ transgene; and LT-1.2 (5′-CAA AAG ATC ATT AAA TCT GTT TGT TG-3′), LT-2 (5′-CTG TTT GTT GGG GAT CCT CTA GAG TCT TC-3′), and LT-3 (5′-TGG GGA TCC TCT AGA GTC TTC CCT TTA GTG-3′) for the bottom strand in all the T3T7 transgenes. The oligonucleotides SPR1, SPR-2, and SPR-3 were used for amplification of the top strand of MPE-T3T7 and PE-T3T7. LM-PCRs using these primers indicated that >95% of the signal detected came from the high-copy-number T3T7 transgenic DNA and not from endogenous μ enhancer. In the case of E-T3T7 and ME-T3T7, the primers used for the top strand were CμLM-1 (5′-CAG TGT TGG GAA GGT TCT GAT AC-3′), CμLM-2 (5′-GTT CTG ATA CCC TGG ATG ACT TCA G-3′), and CμLM-3 (5′-CTG GAT GAC TTC AGT GTT GTT CTG GTA GTT CC-3′). These CμLM primers hybridized in the Cμ “stuffer” DNA fragment adjacent to Enh 90 in E-T3T7 and ME-T3T7 (36). For amplification of DNA from the chromatin immunoprecipitation experiments, the new oligonucleotides used were VDJ-1 (5′- GCC TCA GTC AAG TTG TCC TGC-3′), VDJ-2 (5′-GTA GTC CAT AGC ATA GTA AG-3′), MB-1A (5′-AGG GAT CCA TGG TGA TGA AC-3′), MB-1B (5′-CAA ACA GGC GTA TGA CAA GA-3′), T7–1 (5′-GGG AGA AGA ACA TGG AAG ACT CAG-3′), T7–2 (5′-TGC AAG TTT AAC ATA GCA GTT ACC-3′), T3–1 (5′-GGA TAG GAT GGA TAT AAT GTT TGG), and T3–2 (5′ -GGG CAA ATT AAC ATT TAA AGC TAG-3′).
RESULTS
Formation of an enhancer complex in wild-type and ΔMAR μ transgenes.
In previous experiments, we observed marked differences in the levels of expression of wild-type and ΔMAR μ transgenes, and we found that gene constructs containing MARs generate an extended chromatin accessibility relative to gene constructs lacking MARs (23, 32, 33) (summarized in Fig. 1A and B). These observations raised the question of whether transcription factors are bound to the μ enhancer in the absence of flanking MARs. To analyze the nucleoprotein complex at the μ enhancer (Eμ), we performed in vivo footprinting experiments that detect the occupancy of individual factor-binding sites (19, 24). Toward this end, we incubated μ wild-type and ΔMAR transgenic pre-B cells with DMS and analyzed the methylation pattern of G residues in the μ enhancer region by an LM-PCR (24). To avoid amplification of endogenous μ enhancer sequences, we generated a new set of ΔMAR transgenic pre-B-cell lines in which polylinker sequences are inserted immediately adjacent to the enhancer. These polylinker sequences allow specific amplification of the transgenic μ enhancer by LM-PCR and prevent any background amplification from the endogenous locus, which would obscure the detection of incomplete factor occupancy at the transgenic enhancer. Using these sequences as anchors for transgene-specific oligonucleotides in the LM-PCR assay, no amplification of the endogenous locus could be detected in a nontransgenic mouse line (data not shown). Consistent with previous experiments (23), S1 nuclease protection assays of transgenic pre-B-cell RNA indicated that the levels of expression of the modified ΔMAR μ transgene in multiple lines are reduced by a factor of 30 to 1,000 relative to those of the wild-type μ transgene (Fig. 1C).
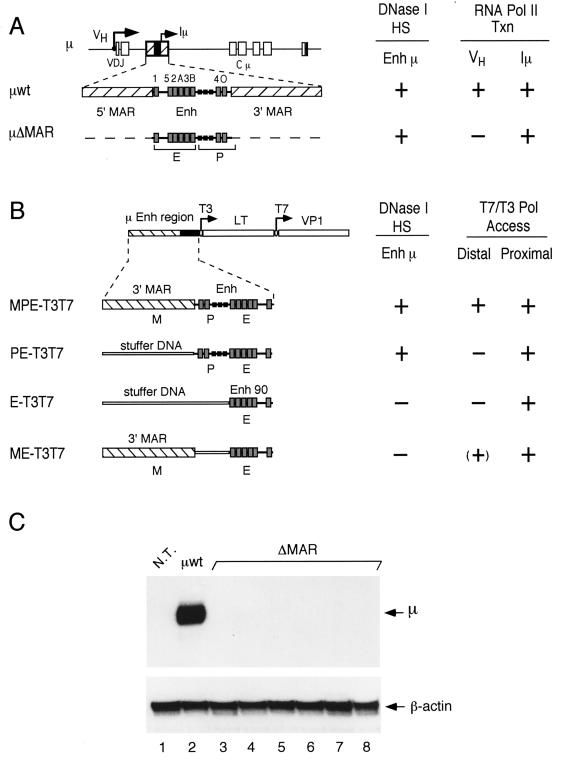
FIG. 1.
Schematic diagram of transgenes containing μ enhancer sequences. (A) Structure of the rearranged μ wild-type and ΔMAR transgenes containing the intragenic enhancer between the rearranged VDJ and Cμ exons. The positions of the VH and Iμ promoters are shown by arrows. The intragenic μ enhancer (Enh), including the Iμ promoter and binding sites for transcription factors (gray boxes) (20), is flanked by MARs (hatched boxes). (B) Structure of T3T7 transgenes in which μ enhancer sequences are linked to bacteriophage T3 and T7 promoters (arrows) at enhancer-proximal and -distal (1 kb) positions (33). The MPE-T3T7 gene construct contains the entire μ enhancer fragment (Enh), which includes the Iμ promoter (P) and enhancer core (E), and a single flanking MAR (M). The PE-T3T7 gene contains the Enh fragment without MAR sequences. The construct E-T3T7 contains only Enh 90, and the construct ME-T3T7 contains the Enh 90 fragment and a single MAR. DNA fragments (1 kb) from the large T (LT) and VP1 genes of simian virus 40 acted as reporters of transcription from the T3 and T7 promoters (33). “Stuffer” sequences replace μ LCR fragments in order to maintain the spatial relationship of the transgene components. To the right of each transgene (A and B), the data obtained by Forrester et al. (23) and Jenuwein et al. (33) are summarized. The formation of DNase I-hypersensitive sites (HS) at the μ enhancer and the generation of transcripts (VH and Iμ) by endogenous RNA polymerase II (Pol II Txn) or transcripts (T7, distal; T3, proximal) by exogenous bacteriophage RNA polymerases (T7/T3 Pol Access) are indicated by a plus sign. (C) Total RNA samples isolated from the B-cell cultures used in the in vivo footprinting experiments were probed for the presence of VH-initiated transcripts by S1 nuclease protection assay. Detection of the β-actin mRNA was used as an internal control. As expected from previous data (23), the transgenic enhancer in all μ ΔMAR cell lines did not activate the distal VH promoter at a detectable level (lanes 3 through 8). The total RNA from nontransgenic pre-B cells (N.T.; lane 1) and from μ-transgenic pre-B cells (μwt; lane 2) were used as controls.
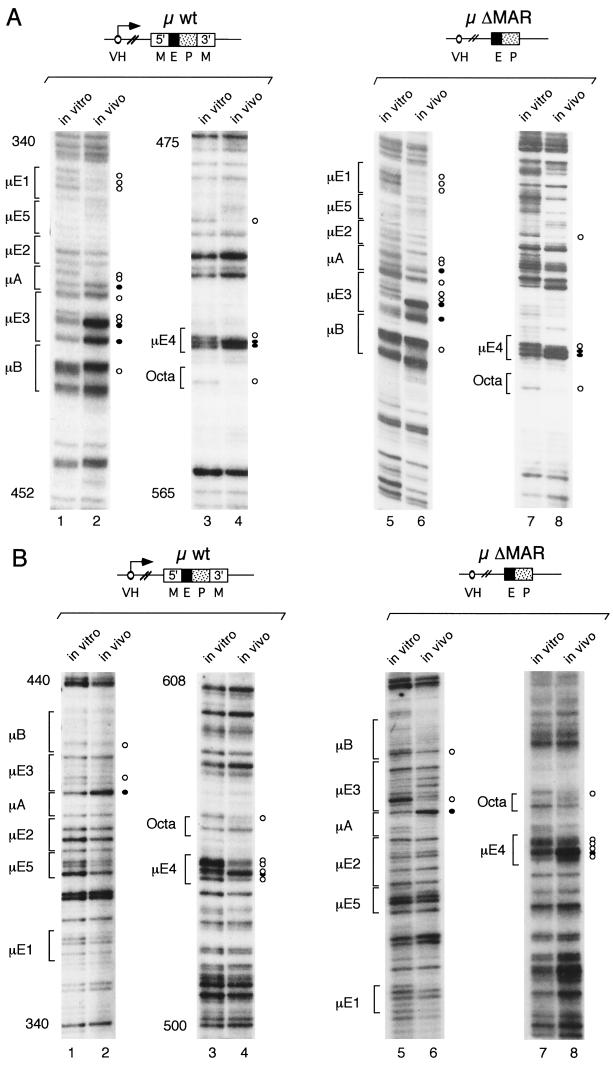
In vivo methylation of DNA in the transgenic pre-B-cell line 19–1-4, carrying a wild-type μ transgene (∼20 gene copies), and in vitro methylation of the corresponding purified genomic DNA revealed a specific pattern of protected and hyperreactive guanosine residues at the transgenic μ enhancer (Fig. 2). This pattern is similar to that previously reported for the endogenous μ locus in mature B-cell lines (19). The genomic footprints, observed on both DNA strands of the wild-type μ transgene, coincide with the known transcription factor-binding sites μE1 to Octa and reveal the presence of a fully assembled nucleoprotein complex at the μ enhancer in vivo (Fig. 2, lanes 1 through 4). Analysis of in vivo-methylated genomic DNA from the ΔMAR transgenic line 61 (∼20 gene copies) revealed a pattern of protections and enhancements of methylated guanosines similar to that found with the wild-type μ transgene (Fig. 2, lanes 5 through 8). This DMS modification pattern of the ΔMAR μ transgene was confirmed for four additional ΔMAR μ transgenic lines (data not shown). A similar pattern of DMS modifications was also observed with the PE-T3T7 transgene, which contains the 220-bp μ enhancer fragment but lacks both MAR sequences and the VH promoter (Fig. 3, lanes 1 through 4). Although some subtle differences were detected in the patterns of in vivo footprints of the wild-type and the ΔMAR transgenic lines, this analysis indicates that a nucleoprotein enhancer complex is stably assembled in both the transcriptionally active wild-type and the transcriptionally inactive ΔMAR μ transgenes (summarized in Fig. 4).
FIG. 2.
In vivo footprints in the μ enhancer of μ wild-type and ΔMAR transgenes. LM-PCR was performed using in vitro and in vivo DMS-methylated DNAs and specific oligonucleotides to amplify the top (A) and bottom (B) strands of the transgenic μ enhancer (see Materials and Methods). The products from these LM-PCRs, from both in vitro and in vivo samples, were separated in denaturing 6% polyacrylamide–urea gels and exposed to X-ray film to reveal the G ladders. The nucleotides are numbered as described by Ephrussi et al. (19). The DNA-binding sites of transcription factors (20) in the μ enhancer are labeled on the left of each panel. The G residues that are protected from DMS methylation in vivo are marked with an open circle. The G residues whose reactivity to DMS is enhanced in vivo are marked with a closed circle.
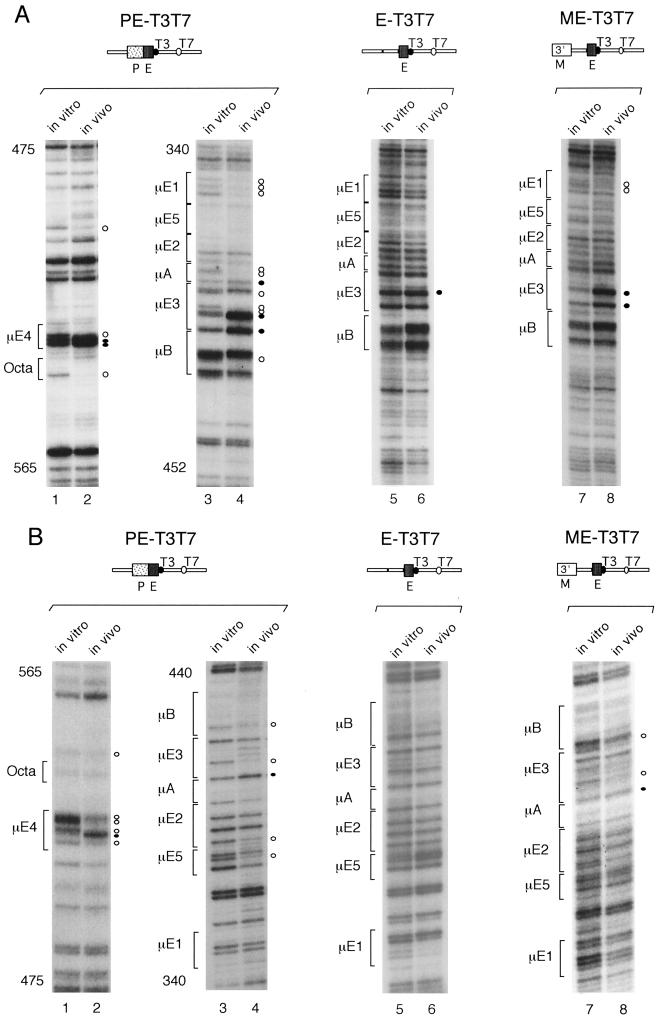
FIG. 3.
In vivo DMS footprints in the μ enhancer fragment of T3T7 transgenes. As described for Fig. 2, the genomic DMS-modified DNA from transgenic pre-B cells was isolated and subjected to LM-PCR to amplify the top (A) and bottom (B) strands of the transgenic μ enhancer. The pre-B-cell lines used had the transgenes integrated in a transcriptionally inactive chromatin (33). In vitro and in vivo DMS methylation patterns were visualized by autoradiography. The nucleotides are numbered as described by Ephrussi et al. (19). The protected and hyperreactive guanosine residues are marked with open and closed circles, respectively.
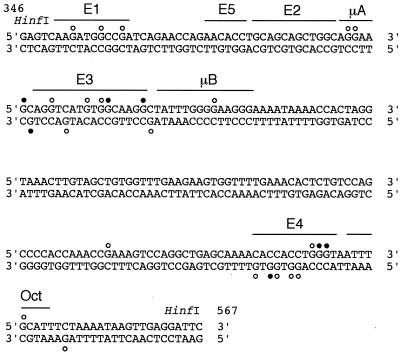
FIG. 4.
Summary of the in vivo footprints detected by DMS treatment in the μ enhancer of wild-type and ΔMAR μ transgenes. Guanosine bases with protection from or enhanced reactivities to DMS are indicated with open and closed circles, respectively. The binding sites for transcription factors in the μ enhancer are indicated (20). The DNA sequences represented are from the top and bottom strands of the 220-bp-long HinfI fragment of the μ LCR. The nucleotides are numbered as described by Ephrussi et al. (19).
Enhancer occupancy and DNase I-hypersensitive site formation.
In previous studies, we examined the ability of parts of the μ enhancer, alone or in combination with a MAR, to induce changes in nuclear chromatin in the absence of a linked VH promoter (32, 33). This analysis indicated that a minimal 90-bp core element of the μ enhancer (Enh 90), which includes sequences from the μE1 to the μB-binding site but lacks the Iμ promoter, confers local factor access in nuclear chromatin, independently of an active transcriptional state (32). This minimal enhancer core element, however, does not generate a DNase I-hypersensitive site in nuclear chromatin, whereas Enh 220, including the core element of the enhancer and the Iμ promoter, forms a DNase I-hypersensitive site (23, 32, 33).
To relate the different abilities of various μ enhancer fragments to generate DNase I-hypersensitive sites with the formation of specific nucleoprotein complexes, we performed in vivo DMS footprinting on transgenes containing Enh 90 or Enh 220 alone or in combination with a single MAR. The PE-T3T7 transgene, which contains the Enh 220 fragment in the absence of a MAR, has a T7 promoter at a distal position, 1 kb away from the μ enhancer, and a T3 promoter immediately adjacent to the enhancer (33). The PE-T3T7 transgene and the MPE-T3T7 transgene, which contains a single MAR, both lack the VH promoter, and they are not transcribed by endogenous RNA polymerases in the transgenic lines 1–29 (five gene copies) and 3–2 (three gene copies), respectively (33). As mentioned above, the PE-T3T7 transgene generated footprints similar to those detected with the wild-type and ΔMAR μ transgenes (Fig. 3, lanes 1 through 4), indicating that the formation of a stable nucleoprotein complex at the μ enhancer is dependent neither on a MAR nor on a linked VH promoter. However, the Iμ promoter contributes to the stability of the μ enhancer complex because the E-T3T7 transgene, containing the Enh 90 fragment alone, did not show any detectable footprints and the ME-T3T7 transgene, containing the Enh 90 fragment and a MAR, generated only weak footprints (Fig. 3, lanes 5 through 8). Together, these data suggest that the minimal μ enhancer core element requires the adjacent Iμ promoter region to form a stable nucleoprotein complex in nuclear chromatin, which correlates with the generation of a DNase I-hypersensitive site.
Histone H3 and H4 acetylation at enhancer-distal nucleosomes in μ transgenes.
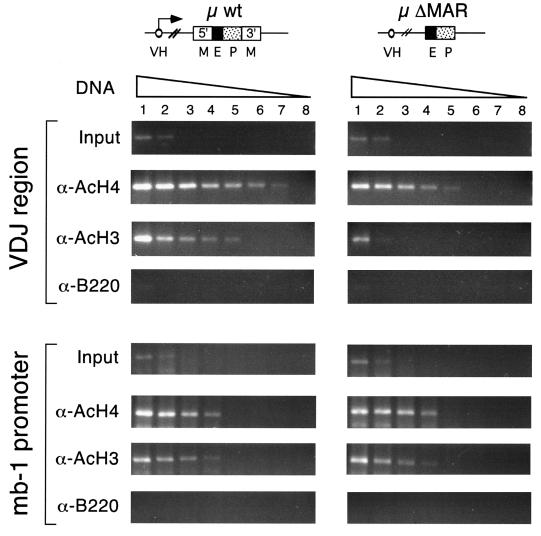
Acetylation of the basic N-terminal tails of core histones has a positive effect on transcription by generating a domain of accessible chromatin that facilitates the binding of activators and RNA polymerase to their target sites (44, 70, 76). Since the MARs of the μ gene extend chromatin accessibility to distal sites in vivo, we examined the possibility that MARs augment the level of H3 and H4 acetylation at the distal VH promoter. Toward this end, we prepared sonicated extracts from formaldehyde-cross-linked transgenic pre-B cells (see Materials and Methods), immunoprecipitated chromatin fragments with antiserum directed against acetylated histones H3 and H4 (15, 55), and detected transgene-specific VH promoter DNA by semiquantitative PCR amplification. To estimate the relative enrichment of a specific genomic DNA sequence in the immunoprecipitated material, we used a serial dilution of the starting amount of template DNA to ensure that the different samples could be compared within their linear range of amplification. Parallel amplification reactions, using purified genomic DNA from the bulk chromatin extracts (input DNA), allowed us to assess the enrichment of the transgene-specific VDJ sequences in the immunoprecipitated samples (Fig. 5, top). Amplification of the mb-1 promoter, which is active specifically in the B-cell lineage (35), was used as an internal control for the immunoprecipitation (Fig. 5, bottom).
FIG. 5.
Acetylation of H3 and H4 histones in the VDJ region of wild-type and ΔMAR μ transgenes. Chromatin extracts from formaldehyde-cross-linked wild-type and ΔMAR μ transgenic pre-B cells were immunoprecipitated with α-AcH4, α-AcH3, or α-B220 (negative control) antibodies. The DNAs of the bound fractions were isolated, and the transgenic VDJ sequences in the immunoprecipitated DNA were quantified by PCR. The amplification of the promoter region of the endogenous mb-1 gene was done as an internal control. The levels of enrichment in the immunoprecipitations were estimated by comparison with the amplification products of DNA isolated from the bulk chromatin extracts (input DNA). Eight serial fourfold dilutions of template DNA were done to allow a quantitative determination in the PCR assays. The amounts of template DNA (in nanograms) were 20 (lane 1), 5 (lane 2), 1.25 (lane 3), 0.31 (lane 4), 0.078 (lane 5), 0.019 (lane 6), 0.009 (lane 7), and 0.002 (lane 8). The VDJ region of the wild-type μ transgenes was immunoprecipitated with the α-AcH4 and α-AcH3 antibodies more efficiently than the VDJ region of the ΔMAR μ transgenes, indicating a preferential acetylation of H4 and H3 in the VDJ region of MAR-containing transgenes. These differences are not observed in the endogenous mb-1 promoter control.
Using these chromatin immunoprecipitation assays, we estimated that the transgene-specific VDJ region was enriched in the α-AcH4-immunoprecipitated DNA from the wild-type μ transgene approximately 16-fold (two serial 1:4 dilutions) relative to the corresponding VDJ region from the ΔMAR μ transgene samples (Fig. 5, top). An even more pronounced difference was observed in the samples that were immunoprecipitated with α-AcH3 antibodies. In these samples, the VDJ region of the wild-type μ transgene was enriched approximately 64-fold (three serial 1:4 dilutions) relative to the corresponding region of the ΔMAR μ transgene (Fig. 5, top). As a control, no significant difference in the efficiency of immunoprecipitation of the endogenous mb-1 promoter sequences was detected in wild-type and ΔMAR samples (Fig. 5, bottom). In addition, the specificity of the immunoprecipitations was confirmed by the use of an unrelated α-B220 antibody. Together, these data indicate that the overall level of histone acetylation in the distal VH promoter region of the ΔMAR transgene is significantly reduced relative to that of the wild-type μ transgene.
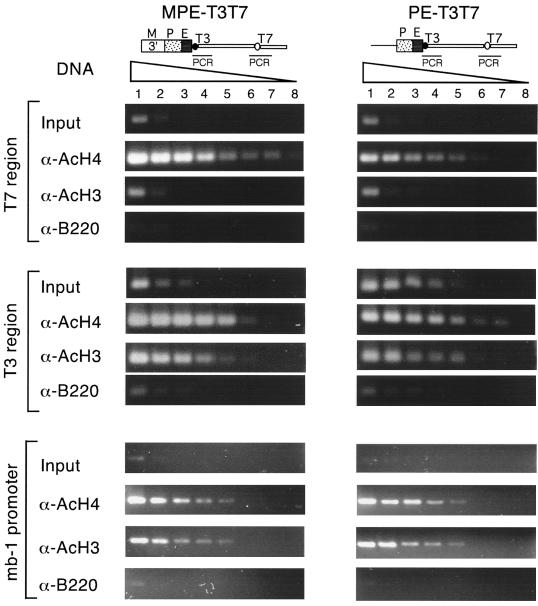
The different transcriptional states of the wild-type and ΔMAR μ transgenes raise the question of whether the enhanced levels of histone acetylation in the wild-type gene are an indirect consequence of an active transcriptional state or a direct MAR-dependent effect. To address this question, we examined the acetylation status of histones H3 and H4 in the MPE-T3T7 and PE-T3T7 transgenes, neither of which is transcribed by endogenous RNA polymerases (36). In the α-AcH4-immunoprecipitated samples, the enhancer-distal T7 promoter region of the MPE-T3T7 transgene was enriched approximately 16-fold relative to that of the PE-T3T7 transgene (Fig. 6, top). In contrast, the level of H4 acetylation in the enhancer-proximal T3 promoter region was similarly high in both MPE-T3T7 and PE-T3T7 transgenes (Fig. 6, middle). Analysis of the acetylation status of histone H3 also revealed high levels of acetylation in the T3 promoter region of both transgenes, whereas no significant H3 acetylation was detected in the distal T7 promoter region of either the MPE-T3T7 or the PE-T3T7 transgene. As a control, no significant difference in the efficiencies of immunoprecipitation of mb-1 promoter sequences was detected in the MPE-T3T7 and PE-T3T7 samples (Fig. 6, bottom). Thus, the level of histone H4 acetylation at an enhancer-distal site is higher in the MPE-T3T7 transgene than in the PE-T3T7 transgene. However, no augmented H3 acetylation was detected at the enhancer-distal region of the MPE-T3T7 transgene.
FIG. 6.
Acetylation of H3 and H4 histones at μ enhancer-distal and -proximal positions in T3T7 transgenes. Chromatin extracts from formaldehyde-cross-linked MPE-T3T7 and PE-T3T7 transgenic pre-B cells were immunoprecipitated as described for Fig. 5. In the pre-B-cell lines used in these experiments, the transgenes are integrated in transcriptionally inactive chromatin (33). Transgene-specific sequences in the immunoprecipitated chromatin and input DNA were quantitated by PCR assays. Specific primers were used for amplification of ∼350-bp DNA fragments that include either the distal T7 region, the proximal T3 region, or the endogenous mb-1 promoter as an internal control. Eight serial fourfold dilutions of the template DNA were used to allow a semiquantitative PCR. The distal T7 region of the MPE-T3T7 transgene was immunoprecipitated with α-AcH4 antiserum with a higher efficiency than that of the PE-T3T7 transgene.
Taken together, the chromatin immunoprecipitation experiments indicate that the chromatin of the MAR-containing μ transgenes contains approximately 10- to 60-fold-higher levels of acetylated histones H4 and H3 than the chromatin of the ΔMAR μ transgenes. Moreover, the acetylation of histone H4 appears to be independent of transcription by endogenous RNA polymerases, whereas the acetylation of H3 correlates with an active transcriptional state.
DISCUSSION
Gene activation in eukaryotes has been proposed to consist of a multistep process that includes changes in chromatin structure, modifications of histones, and transcriptional activation of promoters (6, 21, 25, 70). The causal relationship and sequence of these events are still obscure. Using the immunoglobulin μ gene locus as a model, we have previously shown that the intronic LCR can induce changes in chromatin structure (33). Notably, the enhancer is necessary and sufficient to mediate local chromatin accessibility independently of transcriptional activation. However, the enhancer requires a flanking MAR to confer accessibility at distal sites. In this study, we show that MARs are not involved in assembling a stable nucleoprotein complex at the enhancer but are involved in generating an extended domain of histone acetylation. This histone hyperacetylation is independent of changes in the occupancy of enhancer sequences by DNA-binding proteins and, at least for H4, can be detected even in MAR-containing transgenes that are transcriptionally silent (i.e., MPE-T3T7). Thus, the MARs act independently of the assembly of an enhancer complex and may function prior to transcriptional activation.
MAR-independent assembly of an enhancer complex in nuclear chromatin.
Our in vivo footprinting data obtained with the intronic μ enhancer suggest that a nucleoprotein complex is assembled at the transcriptionally inactive ΔMAR μ transgene. Although the footprinting data reveal similar patterns of DMS enhancement and protection in wild-type and ΔMAR transgenes, the experiments do not indicate whether the nucleoprotein complexes are identical. Small differences in the DMS pattern are observed, which could be interpreted as an indication of an alteration in binding of proteins to the enhancer by the association either with non-DNA-binding proteins or with MAR-binding proteins. Some μ enhancer-binding proteins, such as Pu.1 and Oct-1, have been shown to associate with cofactors. In particular, the association of Oct-1 with its cofactor Oca-B alters the DMS interference pattern of Oct-1 (64), and Pu.1 can interact with the HAT CREB-binding protein (81). MAR-binding proteins may affect the nucleoprotein complex at the enhancer either directly, by interactions with μ enhancer-binding proteins, or indirectly, by recruiting chromatin-modifying complexes. To date, however, no data are available on the interaction of the MAR-binding proteins Bright and NF-μNR, which augment or repress enhancer function, with components of the μ enhancer complex or with chromatin-modifying complexes (30, 78). Finally, we cannot rule out the possibility that different proteins are bound at the μ enhancer in the presence and absence of MARs. We consider this possibility unlikely because the μ enhancer alone displays the same cell type specificity as the enhancer in combination with a MAR (32, 50, 53). MARs actually repress intronic enhancer function in non-B cells (65, 79).
The full occupancy of the enhancer in the ΔMAR transgene contrasts with the lack of detectable transcription from the distal VH promoter. However, the enhancer occupancy is consistent with the previous observation that the enhancer-proximal Iμ promoter is functional in the ΔMAR μ transgene (23). In addition, Nikolajczyk and colleagues have shown that a small enhancer fragment, including the μE5 and μB elements, is sufficient to confer accessibility upon an adjacent site in the context of in vitro-assembled chromatin (52). Thus, the assembly of transcription factors at the μ enhancer can direct local promoter activation, but it is insufficient to activate distal promoters in nuclear chromatin.
The MAR-independent formation of a stable nucleoprotein complex at the μ enhancer correlates with the previously observed appearance of a DNase I-hypersensitive site in ΔMAR transgenes (23). Moreover, the full occupancy of the μ enhancer and the formation of a DNase I-hypersensitive site in ΔMAR transgenes are both dependent upon the presence of the Iμ promoter. The partial occupancy observed at the enhancer core (Enh 90) lacking the Iμ promoter could reflect a partial occupancy in a multicopy gene array or, alternatively, a full occupancy in a subset of cells. The requirement of the Iμ promoter for full enhancer occupancy is reminiscent of the requirement of a linked promoter for the formation of a DNase I-hypersensitive site at a transgenic β-globin enhancer (62). However, the activity of the Iμ promoter does not seem to be necessary for the generation of a stable nucleoprotein complex, because we also observed full occupancy of the enhancer in the transcriptionally inactive PE-T3T7 transgene. Instead, the role of Iμ promoter sequences in augmenting factor occupancy at the μ enhancer could involve reciprocal stabilization of enhancer- and promoter-bound proteins, which may be necessary for the formation of a stable nucleoprotein complex in higher-order chromatin (70). The dependence of the enhancer complex on the presence of the Iμ promoter is in contrast to experiments showing that the 90-bp μ enhancer core can induce accessibility, albeit at a reduced level, at an enhancer-proximal bacteriophage promoter (32, 33). The accessibility assay using a bacteriophage RNA polymerase allows a positive readout of factor access in subsets of gene copies or cells, whereas full occupancy of the enhancer is required to detect clear genomic footprints. Thus, these differences in the assays may account for the apparent difference in the contribution of the Iμ promoter to enhancer complex formation and chromatin accessibility.
MAR-dependent acetylation differs for H3 and H4 core histones.
Previous analyses of transcriptionally active and inactive gene loci have implicated the acetylation of core histones in the opening of the higher-order chromatin structure and in gene activation (28, 60, 75). However, only recently has substantial progress been made in the recognition of histone acetylation as a general mechanism in transcriptional regulation (26, 70). An increasing number of transcription factors have either intrinsic HAT and/or histone deacetylase activities or associate with cofactors that contain HAT activities (2, 8–11, 40). In addition, the basic amino-terminal tails of core histones that are targets for acetylation are essential for cell viability in vivo and for higher-order chromatin structure in vitro (43). Finally, acetylation of the amino-terminal tails of H4 and H3 has been shown to enhance the binding of transcription factors to nucleosomal templates (74, 76).
Enhancers have been shown to function, in part, by recruiting HATs (69) and by targeting acetylation of H3 and H4 histones to linked promoters by DNA looping and protein-protein interactions (56). Most reported examples of histone hyperacetylations concern H3 and/or H4 modifications that are detected in the immediate vicinity of enhancers or promoters. However, two recent studies have linked long-range histone acetylation to the regulation of chromatin accessibility. The intronic enhancer region of the T-cell receptor α/δ locus has been shown to impart long-range H3 hyperacetylation in transgenic mice, which has been correlated with active transcription and V(D)J recombination (46). Likewise, long-range histone acetylation has also been found in the human β-globin locus, which is independent of both transcription and the presence of the LCR (66). Our analysis now identifies the MAR of the μ locus as the first regulatory element that appears to be specifically involved in the generation of long-range histone acetylation.
Our analysis of histone acetylation in the chromatin of μ transgenes revealed a notable difference between histones H3 and H4. Acetylation of H4 was detected at enhancer-distal positions in both the transcriptionally active wild-type μ gene and the transcriptionally inactive MAR-containing MPE-T3T7 transgene. In contrast, acetylation of H3 at the enhancer-distal position was detected only in the VDJ region of the transcriptionally active μ gene, although enhancer-proximal H3 acetylation was found in both μ and MPE-T3T7 transgenes. HATs have been shown to differ in their histone preference, and the recruitment of H3-restricted HATs may be associated with the assembly of active transcription complexes (26, 40, 70). Although the level of H4 acetylation at enhancer-distal positions is augmented by transcription, the effect of transcription is smaller than that of the presence of MARs. Specifically, the presence of MARs increases the level of long-range H4 acetylation ∼10-fold in the transcriptionally inactive MPE-T3T7 transgene and in the transcriptionally active μ gene.
Acetylation of the amino-terminal tail of H4 has been previously implicated in the long-range regulation of chromatin structure. For example, H4 acetylation has been found to colocalize with the coding regions of transcribed genes (54). In addition, the silencing of inactive X chromosomes in females is also accompanied by underacetylated histone H4 (36). Moreover, the amino-terminal tail of H4 was shown to be essential for the formation of the RAP1-SIR3-H4 complex and for silencing of chromatin domains in yeast (26). A model that could explain the pivotal role of H4 acetylation in the long-range regulation of chromatin structure was inferred from the high-resolution three-dimensional structure of the nucleosome (45). This structural analysis revealed an electrostatic interaction between two adjacent nucleosomes that is mediated by the basic N tail of H4 and a conserved acidic region in H2A and H2B. Disruption of these interactions by acetylation could lead to a disorganization of the compact higher-order structure of the chromatin fiber (72). Therefore, MAR-mediated long-range hyperacetylation of H4 could result in an extended disorganization of the chromatin fiber.
How do MARs confer long-range enhancer function and histone acetylation?
Several mechanisms could account for the regulation of long-range enhancer action and histone H4 acetylation. First, MARs could directly facilitate the recruitment of HATs to the enhancer region. Several HATs have been found to associate with enhancer-binding proteins, and additional HATs that preferentially acetylate H4 may be recruited by interaction with MAR-binding proteins. Secondly, MARs could affect histone acetylation indirectly by targeting the transgene to specific subnuclear regions. The model of the eukaryotic interphase nucleus as a structurally and functionally organized compartment has gained experimental support in recent years (16, 47). The putative existence of an underlying structure directly involved in maintaining a specific nuclear organization is an appealing possibility (4, 41, 57). The association of biologically relevant processes (like RNA transcription, RNA processing, and DNA replication) with the nuclear matrix (57) suggests the possibility that nuclear processes take place in functional domains within the nucleus. Since most of the cellular HAT and HDAC activities are retained within the nuclear matrix preparations (17), it is tempting to speculate that MARs could enhance the reversible acetylation of a genetic locus by its anchoring to the nuclear matrix.
Thirdly, MARs could affect chromatin structure and histone acetylation indirectly via changes in the CpG modification of DNA. Recently, it was shown that MARs can overcome a CpG methylation-dependent repression of transcription (22). In that study, it was found that μ genes that have been methylated in vitro at all CpG dinucleotides, prior to their stable transfection to B-cell lines, required MARs to efficiently initiate transcription from the distal VH promoter. Moreover, the DNA of the CpG-methylated ΔMAR genes was kept in a DNase I-resistant chromatin structure in which H3 and H4 were underacetylated near the VH promoter (22). Thus, CpG methylation of transfected DNA reproduced, at least in part, the MAR dependence observed in transgenic animals. Importantly, a link between CpG methylation and histone deacetylation has been discovered through the recruitment of HDAC activities by MeCP2 (34, 49), a protein that binds to methylated CpG dinucleotides and that also recognizes MAR sequences (48, 80). Other proteins that bind methylated CpG and repress transcription at a distance, like MBD1, also seem to act by recruiting HDAC activities (51). Indeed, MARs from different origins had been implicated in DNA demethylation at CpG residues (38), and it has also been shown that loss of the transcriptional activity of a transgene is accompanied by DNA methylation and histone deacetylation (59). According to this scheme, recruitment of DNA demethylases may be regulated by MARs (5). Finally, the binding of HMG-I(Y) to MAR sequences has been shown to antagonize the binding of histone H1 in vitro (82). Notably, histone H1 has been recently shown to act as a repressor of core histone acetylation (29).
How do MARs mediate the generation of an extended domain of histone acetylation? The model of MAR function to target DNA regions to a specific subnuclear compartment does not require the propagation of histone modification from the enhancer to distal sites. Instead, subnuclear targeting of DNA could result in a domain-wide histone acetylation that is independent of enhancer-promoter interactions by looping. Alternatively, MARs could extend enhancer-induced local chromatin accessibility to distal sites by changes in DNA topology. Consistent with this possibility, MARs are recognized by topoisomerases that could alter chromatin structure (7).
In conclusion, our analysis suggests a multistep model of gene activation. First, the assembly of a stable enhancer complex can be governed by the enhancer core and the Iμ promoter alone. This enhancer complex allows localized H3 and H4 acetylation, but it does not result in distal histone acetylation and promoter activation. In combination with flanking MARs, the enhancer mediates extended H4 acetylation and chromatin accessibility, even in the absence of detectable transcription. This transcription-competent state can be converted into a fully active state by the addition of a VH promoter, which results in transcription and distal H3 acetylation.
ACKNOWLEDGMENTS
We thank W. C. Forrester for valuable discussions. L.A.F. especially thanks Juan Galceran and Mikael Sigvardsson for their continued help and support.
L.A.F. and M.W. were holders of postdoctoral fellowships from M.E.C. of Spain and from DFG of Germany, respectively. This work was funded by a grant from the National Institutes of Health to Rudolf Grosschedl.
REFERENCES
- 1.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K. Current protocols in molecular biology. New York, N.Y: Greene and Wiley-Interscience; 1993. [Google Scholar]
- 2.Bannister A J, Kouzarides T. The CBP co-activator is a histone acetyltransferase. Nature. 1996;384:641–643. doi: 10.1038/384641a0. [DOI] [PubMed] [Google Scholar]
- 3.Beckmann H, Su L K, Kadesch T. TFE3: a helix-loop-helix protein that activates transcription through the immunoglobulin enhancer muE3 motif. Genes Dev. 1990;4:167–179. doi: 10.1101/gad.4.2.167. [DOI] [PubMed] [Google Scholar]
- 4.Belmont A S, Dietzel S, Nye A C, Strukov Y G, Tumbar T. Large-scale chromatin structure and function. Curr Opin Cell Biol. 1999;11:307–311. doi: 10.1016/S0955-0674(99)80041-6. [DOI] [PubMed] [Google Scholar]
- 5.Bhattacharya S K, Ramchandani S, Cervoni N, Szyf M. A mammalian protein with specific demethylase activity for mCpG DNA. Nature. 1999;397:579–583. doi: 10.1038/17533. [DOI] [PubMed] [Google Scholar]
- 6.Blackwood E M, Kadonaga J T. Going the distance: a current view of enhancer action. Science. 1998;281:60–63. doi: 10.1126/science.281.5373.60. [DOI] [PubMed] [Google Scholar]
- 7.Bode J, Kohwi Y, Dickinson L, John T, Klehr D, Mielke C, Kohwi-Shigematsu T. Biological significance of unwinding capability of nuclear matrix-associating DNAs. Science. 1992;255:195–197. doi: 10.1126/science.1553545. [DOI] [PubMed] [Google Scholar]
- 8.Brehm A, Miska E A, McCance D J, Reid J L, Bannister A J, Kouzarides T. Retinoblastoma protein recruits histone deacetylase to repress transcription. Nature. 1998;391:597–601. doi: 10.1038/35404. [DOI] [PubMed] [Google Scholar]
- 9.Brown C E, Lechner T, Howe L, Workman J L. The many HATs of transcription coactivators. Trends Biochem Sci. 2000;25:15–19. doi: 10.1016/s0968-0004(99)01516-9. [DOI] [PubMed] [Google Scholar]
- 10.Brownell J E, Zhou J, Ranalli T, Kobayashi R, Edmondson D G, Roth S Y, Allis C D. Tetrahymena histone acetyltransferase A: a homolog to yeast Gcn5p linking histone acetylation to gene activation. Cell. 1996;84:843–851. doi: 10.1016/s0092-8674(00)81063-6. [DOI] [PubMed] [Google Scholar]
- 11.Chen H, Lin R J, Schiltz R L, Chakravarti D, Nash A, Nagy L, Privalsky L M, Nakatani Y, Evans R M. Nuclear receptor coactivator ACTR is a novel histone acetyltransferase and forms a multimeric activation complex with P/CAF and CBP/p300. Cell. 1997;90:569–580. doi: 10.1016/s0092-8674(00)80516-4. [DOI] [PubMed] [Google Scholar]
- 12.Chomczynski P, Sacchi N. Method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- 13.Cirillo L A, Zaret K S. An early developmental transcription factor complex that is more stable on nucleosome core particles than on free DNA. Mol Cell. 1999;4:961–969. doi: 10.1016/s1097-2765(00)80225-7. [DOI] [PubMed] [Google Scholar]
- 14.Cockerill P N, Yuen M-H, Garrard W T. The enhancer of the immunoglobulin heavy chain locus is flanked by presumptive chromosomal loop anchorage elements. J Biol Chem. 1987;262:5394–5397. [PubMed] [Google Scholar]
- 15.Crane-Robinson C, Wolffe A P. Immunological analysis of chromatin: FIS and CHIPS. Trends Genet. 1998;14:477–480. doi: 10.1016/s0168-9525(98)01621-7. [DOI] [PubMed] [Google Scholar]
- 16.Davie J R. The nuclear matrix and the regulation of chromatin organization and function. Int Rev Cytol. 1995;162A:191–250. doi: 10.1016/s0074-7696(08)61232-2. [DOI] [PubMed] [Google Scholar]
- 17.Davie J R. Nuclear matrix, dynamic histone acetylation and transcriptionally active chromatin. Mol Biol Rep. 1997;24:197–207. doi: 10.1023/a:1006811817247. [DOI] [PubMed] [Google Scholar]
- 18.Dillon N, Grosveld F. Chromatin domains as potential units of eukaryotic gene function. Curr Opin Genet Dev. 1994;4:260–264. doi: 10.1016/s0959-437x(05)80053-x. [DOI] [PubMed] [Google Scholar]
- 19.Ephrussi A, Church G, Tonegawa S, Gilbert W. B lineage-specific interactions of an immunoglobulin enhancer with cellular factors in vivo. Science. 1985;227:134–140. doi: 10.1126/science.3917574. [DOI] [PubMed] [Google Scholar]
- 20.Ernst P, Smale S T. Combinatorial regulation of transcription II: the immunoglobulin μ heavy chain gene. Immunity. 1995;2:427–438. doi: 10.1016/1074-7613(95)90024-1. [DOI] [PubMed] [Google Scholar]
- 21.Felsenfeld G, Boyes J, Chung J, Clark D, Studitsky V. Chromatin structure and gene expression. Proc Natl Acad Sci USA. 1996;93:9384–9388. doi: 10.1073/pnas.93.18.9384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Forrester W C, Fernández L A, Grosschedl R. Nuclear matrix attachment regions antagonize methylation-dependent repression of long-range enhancer-promoter interactions. Genes Dev. 1999;13:3003–3014. doi: 10.1101/gad.13.22.3003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Forrester W C, van Genderen C, Jenuwein T, Grosschedl R. Dependence of enhancer-mediated transcription of the immunoglobulin μ gene on nuclear matrix attachment regions. Science. 1994;265:1221–1225. doi: 10.1126/science.8066460. [DOI] [PubMed] [Google Scholar]
- 24.Garrity P A, Wold B. Effects of different DNA polymerases in ligation-mediated PCR: enhanced genomic sequencing and in vivo footprinting. Proc Natl Acad Sci USA. 1992;89:1021–1025. doi: 10.1073/pnas.89.3.1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Grosveld F. Activation by locus control regions? Curr Opin Genet Dev. 1999;9:152–157. doi: 10.1016/S0959-437X(99)80023-9. [DOI] [PubMed] [Google Scholar]
- 26.Grunstein M. Histone acetylation in chromatin structure and transcription. Nature. 1997;389:349–352. doi: 10.1038/38664. [DOI] [PubMed] [Google Scholar]
- 27.Hart C M, Laemmli U K. Facilitation of chromatin dynamics by SARs. Curr Opin Genet Dev. 1998;8:519–525. doi: 10.1016/s0959-437x(98)80005-1. [DOI] [PubMed] [Google Scholar]
- 28.Hebbes T R, Clayton A L, Thorne A W, Crane-Robinson C. Core histone hyperacetylation co-maps with generalized DNase I sensitivity in the chicken β-globin chromosomal domain. EMBO J. 1994;13:1823–1830. doi: 10.1002/j.1460-2075.1994.tb06451.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Herrera J E, West K L, Schiltz R L, Nakatani Y, Bustin M. Histone H1 is a specific repressor of core histone acetylation in chromatin. Mol Cell Biol. 2000;20:523–529. doi: 10.1128/mcb.20.2.523-529.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Herrscher R F, Kaplan M H, Lelsz D L, Das C, Scheuermann R, Tucker P W. The immunoglobulin heavy-chain matrix-associating regions are bound by Bright: a B cell-specific trans-activator that describes a new DNA-binding protein family. Genes Dev. 1995;9:3067–3082. doi: 10.1101/gad.9.24.3067. [DOI] [PubMed] [Google Scholar]
- 31.Jenuwein T, Grosschedl R. Complex pattern of immunoglobulin μ gene expression in normal and transgenic mice: nonoverlapping regulatory sequences govern distinct tissue specificities. Genes Dev. 1991;5:932–943. doi: 10.1101/gad.5.6.932. [DOI] [PubMed] [Google Scholar]
- 32.Jenuwein T, Forrester W C, Qiu R G, Grosschedl R. The immunoglobulin μ enhancer core establishes local factor access in nuclear chromatin independent of transcriptional stimulation. Genes Dev. 1993;7:2016–2032. doi: 10.1101/gad.7.10.2016. [DOI] [PubMed] [Google Scholar]
- 33.Jenuwein T, Forrester W C, Fernandez-Herrero L A, Laible G, Dull M, Grosschedl R. Extension of chromatin accessibility by nuclear matrix attachment regions. Nature. 1997;385:269–272. doi: 10.1038/385269a0. [DOI] [PubMed] [Google Scholar]
- 34.Jones P L, Veenstra G J C, Wade P A, Vermaak D, Kass S U, Landsberger N, Strouboulis J, Wolffe A P. Methylated DNA and MeCP2 recruit histone deacetylase to repress transcription. Nat Genet. 1998;19:187–191. doi: 10.1038/561. [DOI] [PubMed] [Google Scholar]
- 35.Kashiwamura S, Koyama T, Matsuo T, Steinmetz M. Structure of the murine mb-1 gene encoding a putative sIgM-associated molecule. J Immunol. 1990;145:337–343. [PubMed] [Google Scholar]
- 36.Keohane A M, Lavender J S, O'Neill L P, Turner B M. Histone acetylation and X inactivation. Dev Genet. 1998;22:65–73. doi: 10.1002/(SICI)1520-6408(1998)22:1<65::AID-DVG7>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- 37.Kioussis D, Festenstein R. Locus control regions: overcoming heterochromatin-induced gene inactivation in mammals. Curr Opin Genet Dev. 1997;7:614–619. doi: 10.1016/s0959-437x(97)80008-1. [DOI] [PubMed] [Google Scholar]
- 38.Kirillov A, Kistler B, Mostoslavsky R, Cedar H, Wirth T, Bergman Y. A role for nuclear NF-κB in B-cell-specific demethylation of the Igκ locus. Nat Genet. 1996;13:435–441. doi: 10.1038/ng0895-435. [DOI] [PubMed] [Google Scholar]
- 39.Klehr D, Maas K, Bode J. Scaffold-attached regions from the human interferon β domain can be used to enhance the stable expression of genes under the control of various promoters. Biochemistry. 1991;30:1264–1270. doi: 10.1021/bi00219a015. [DOI] [PubMed] [Google Scholar]
- 40.Kuo M-H, Zhou J, Jambeck P, Churchill E A M, Allis C D. Targeted histone acetyltransferase activity of yeast Gcn5p is required for the activation of downstream genes in vivo. Genes Dev. 1998;12:627–639. doi: 10.1101/gad.12.5.627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Laemmli U K, Käs E, Poljak L, Adachi Y. Scaffold-associated regions: cis-acting determinants of chromatin structural loops and functional domains. Curr Opin Genet Dev. 1992;2:275–285. doi: 10.1016/s0959-437x(05)80285-0. [DOI] [PubMed] [Google Scholar]
- 42.Lennon G G, Perry R P. Cμ-containing transcripts initiate heterogeneously within the IgH enhancer region and contain a novel 5′-nontranslatable exon. Nature. 1985;318:475–478. doi: 10.1038/318475a0. [DOI] [PubMed] [Google Scholar]
- 43.Ling X, Harkness T A, Schultz C M, Adams G F, Grunstein M. Yeast histone H3 and H4 amino termini are important for nucleosome assembly in vivo and in vitro: redundant and position-independent functions in assembly but not in gene regulation. Genes Dev. 1996;10:686–699. doi: 10.1101/gad.10.6.686. [DOI] [PubMed] [Google Scholar]
- 44.Luger K, Richmond T J. The histone tails of the nucleosome. Curr Opin Genet Dev. 1998;8:140–146. doi: 10.1016/s0959-437x(98)80134-2. [DOI] [PubMed] [Google Scholar]
- 45.Luger K, Mäder A W, Richmond R K, Sargent D F, Richmond T J. Crystal structure of the nucleosome core particle at 2.8 Å resolution. Nature. 1997;389:251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- 46.McMurry M T, Krangel M S. A role for histone acetylation in the developmental regulation of V(D)J recombination. Science. 2000;287:495–498. [PubMed] [Google Scholar]
- 47.Misteli T, Spector D. The cellular organization of gene expression. Curr Opin Cell Biol. 1998;10:323–331. doi: 10.1016/s0955-0674(98)80007-0. [DOI] [PubMed] [Google Scholar]
- 48.Nan X, Campoy F J, Bird A. MeCP2 is a transcriptional repressor with abundant binding sites in genomic chromatin. Cell. 1997;88:471–481. doi: 10.1016/s0092-8674(00)81887-5. [DOI] [PubMed] [Google Scholar]
- 49.Nan X, Ng H-H, Johnson C A, Laherty C D, Turner B M, Eisenman R N, Bird A. Transcriptional repression by the methyl-CpG-binding protein MeCP2 involves a histone deacetylase complex. Nature. 1998;393:386–389. doi: 10.1038/30764. [DOI] [PubMed] [Google Scholar]
- 50.Nelsen B, Tian G, Erman B, Gregoire J, Maki R, Graves B, Sen R. Regulation of lymphoid-specific immunoglobulin mu heavy chain gene enhancer by ETS-domain proteins. Science. 1993;261:82–86. doi: 10.1126/science.8316859. [DOI] [PubMed] [Google Scholar]
- 51.Ng H H, Jeppesen P, Bird A. Active repression of methylated genes by the chromosomal protein MBD1. Mol Cell Biol. 2000;20:1394–1406. doi: 10.1128/mcb.20.4.1394-1406.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nikolajczyk B S, Sanchez J A, Sen R. ETS protein-dependent accessibility changes at the immunoglobulin mu heavy chain enhancer. Immunity. 1999;11:11–20. doi: 10.1016/s1074-7613(00)80077-1. [DOI] [PubMed] [Google Scholar]
- 53.Nikolajczyk B S, Cortes M, Feinman R, Sen R. Combinatorial determinants of tissue-specific transcription in B cells and macrophages. Mol Cell Biol. 1997;17:3527–3535. doi: 10.1128/mcb.17.7.3527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.O'Neill L P, Turner B M. Histone H4 acetylation distinguishes coding regions of the human genome from heterochromatin in a differentiation-dependent but transcription-independent manner. EMBO J. 1995;14:3946–3957. doi: 10.1002/j.1460-2075.1995.tb00066.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Orlando V V, Strutt H, Paro R. Analysis of chromatin structure by in vivo formaldehyde cross-linking. Methods. 1997;11:205–214. doi: 10.1006/meth.1996.0407. [DOI] [PubMed] [Google Scholar]
- 56.Parekh B S, Maniatis T. Virus infection leads to localized hyperacetylation of histone H3 and H4 at the IFN-β promoter. Mol Cell. 1999;3:125–129. doi: 10.1016/s1097-2765(00)80181-1. [DOI] [PubMed] [Google Scholar]
- 57.Pederson T. Thinking about a nuclear matrix. J Mol Biol. 1998;277:147–159. doi: 10.1006/jmbi.1997.1618. [DOI] [PubMed] [Google Scholar]
- 58.Phi-Van L, von Kries J P, Ostertag W, Strätling W H. The chicken lysozyme 5′ matrix attachment region increases transcription from a heterologous promoter in heterologous cells and dampens position effects on the expression of transfected genes. Mol Cell Biol. 1990;10:2302–2307. doi: 10.1128/mcb.10.5.2302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Pikaart J M, Recillas-Targa F, Felsenfeld G. Loss of transcriptional activity of a transgene is accompanied by DNA methylation and histone deacetylation and is prevented by insulators. Genes Dev. 1998;12:2852–2862. doi: 10.1101/gad.12.18.2852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Pogo B G T, Allfrey V G, Mirsky A E. RNA synthesis and histone acetylation during the course of gene activation in lymphocytes. Proc Natl Acad Sci USA. 1966;55:805–812. doi: 10.1073/pnas.55.4.805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ptashne M, Gann A. Transcriptional activation by recruitment. Nature. 1997;386:569–577. doi: 10.1038/386569a0. [DOI] [PubMed] [Google Scholar]
- 62.Reitman M, Lee E, Westphal H, Felsenfeld G. An enhancer/locus control region is not sufficient to open chromatin. Mol Cell Biol. 1993;13:3990–3998. doi: 10.1128/mcb.13.7.3990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Rosenberg N, Baltimore D. A quantitative assay for transformation of bone marrow cells by Abelson murine leukemia virus. J Exp Med. 1976;143:1453–1463. doi: 10.1084/jem.143.6.1453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sauter P, Matthias P. Coactivator OBF-1 makes selective contacts with both the POU-specific domain and the POU homeodomain and acts as a molecular clamp on DNA. Mol Cell Biol. 1998;18:7397–7409. doi: 10.1128/mcb.18.12.7397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Scheuermann R, Chen U. A developmental-specific factor binds to suppressor sites flanking the immunoglobulin heavy chain enhancer. Genes Dev. 1989;3:1255–1266. doi: 10.1101/gad.3.8.1255. [DOI] [PubMed] [Google Scholar]
- 66.Schübeler D, Francanstel C, Cimbora D M, Reik A, Martin D I K, Groudine M. Nuclear localization and histone acetylation: a pathway for chromatin opening and transcriptional activation of the human β-globin locus. Genes Dev. 2000;14:940–950. [PMC free article] [PubMed] [Google Scholar]
- 67.Sealy L, Chalkley R. DNA associated with hyperacetylated histone is preferentially digested by DNase I. Nucleic Acids Res. 1978;5:1863–1876. doi: 10.1093/nar/5.6.1863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sen R, Baltimore D. Multiple nuclear factors interact with the immunoglobulin enhancer sequences. Cell. 1986;46:705–716. [PubMed] [Google Scholar]
- 69.Sheridan P L, Mayall T P, Verdin E, Jones K A. Histone acetyltransferases regulate the HIV-1 enhancer activity in vitro. Genes Dev. 1997;11:3327–3340. doi: 10.1101/gad.11.24.3327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Struhl K. Histone acetylation and transcriptional regulatory mechanisms. Genes Dev. 1998;12:599–606. doi: 10.1101/gad.12.5.599. [DOI] [PubMed] [Google Scholar]
- 71.Su L, Kadesch T. The immunoglobulin heavy-chain enhancer functions as the promoter for Iμ sterile transcription. Mol Cell Biol. 1990;10:2619–2624. doi: 10.1128/mcb.10.6.2619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.van Holde K, Zlatanova J. What determines the folding of the chromatin fiber? Proc Natl Acad Sci USA. 1996;93:10548–10555. doi: 10.1073/pnas.93.20.10548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Varga-Weisz P D, Becker P B. Chromatin-remodeling factors: machines that regulate? Curr Opin Cell Biol. 1998;10:346–353. doi: 10.1016/s0955-0674(98)80010-0. [DOI] [PubMed] [Google Scholar]
- 74.Vettese-Dadey M, Grant P A, Hebbes T R, Crane-Robinson C, Allis C D, Workman J L. Acetylation of histone H4 plays a primary role in enhancing transcription factor binding to nucleosomal DNA in vitro. EMBO J. 1996;15:2508–2518. [PMC free article] [PubMed] [Google Scholar]
- 75.Vidali G, Boffa L C, Bradbury E M, Allfrey V G. Butyrate suppression of histone deacetylation leads to accumulation of multiacetylated forms of H3 and H4 and increased DNase I sensitivity of the associated DNA sequences. Proc Natl Acad Sci USA. 1978;75:2239–2243. doi: 10.1073/pnas.75.5.2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Vitolo J M, Thiriet C, Hayes J J. The H3–H4 N-terminal tail domains are the primary mediators of transcription factor IIIA access to 5S DNA within a nucleosome. Mol Cell Biol. 2000;20:2167–2175. doi: 10.1128/mcb.20.6.2167-2175.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wade P A, Wolffe A P. Transcriptional regulation: SWItching circuitry. Curr Biol. 1999;9:221–224. doi: 10.1016/s0960-9822(99)80134-1. [DOI] [PubMed] [Google Scholar]
- 78.Wang Z, Goldstein A, Zong R-T, Lin D, Neufeld E J, Scheuermann R H, Tucker P W. Cux/CDP homeoprotein is a component of NF-μNR and represses the immunoglobulin heavy chain intronic enhancer by antagonizing the Bright transcription activator. Mol Cell Biol. 1999;19:284–295. doi: 10.1128/mcb.19.1.284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Wasylyk C, Wasylyk B. The immunoglobulin heavy-chain B-lymphocyte enhancer efficiently stimulates transcription in non-lymphoid cells. EMBO J. 1986;5:553–560. doi: 10.1002/j.1460-2075.1986.tb04246.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Weitzel J M, Buhrmester H, Stratling W H. Chicken MAR-binding protein ARBP is homologous to rat methyl-CpG-binding protein MeCP2. Mol Cell Biol. 1997;17:5656–5666. doi: 10.1128/mcb.17.9.5656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Yamamoto F, Kihara-Negishi H, Yamada T, Hashimoto Y, Oikawa T. Physical and functional interactions between the transcription factor PU.1 and the coactivator CBP. Oncogene. 1999;18:1495–1501. doi: 10.1038/sj.onc.1202427. [DOI] [PubMed] [Google Scholar]
- 82.Zhao K, Kas E, Gonzalez E, Laemmli U K. SAR-dependent mobilization of histone H1 by HMG-I/Y in vitro: HMG-I/Y is enriched in H1-depleted chromatin. EMBO J. 1993;12:3237–3247. doi: 10.1002/j.1460-2075.1993.tb05993.x. [DOI] [PMC free article] [PubMed] [Google Scholar]