Abstract
The pandemic caused by the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has impacted public health and the world economy and fueled a worldwide race to approve therapeutic and prophylactic agents, but so far there are no specific antiviral drugs. Understanding the biology of the virus is the first step in structuring strategies to combat it, and in this context several studies have been conducted with the aim of understanding the replication mechanism of SARS-CoV-2 in vitro systems. In this work, studies using transmission and scanning electron microscopy and 3D electron microscopy modeling were performed with the goal of characterizing the morphogenesis of SARS-CoV-2 in Vero-E6 cells. Several ultrastructural changes were observed—such as syncytia formation, cytoplasmic membrane projections, lipid droplets accumulation, proliferation of double-membrane vesicles derived from the rough endoplasmic reticulum, and alteration of mitochondria. The entry of the virus into cells occurred through endocytosis. Viral particles were observed attached to the cell membrane and in various cellular compartments, and extrusion of viral progeny took place by exocytosis. These findings allow us to infer that Vero-E6 cells are highly susceptible to SARS-CoV-2 infection as described in the literature and their replication cycle is similar to that described with SARS-CoV and MERS-CoV in vitro models.
Keywords: SARS-CoV-2, Vero-E6 cells, ultrastructural studies, morphogenesis, transmission electron microscopy, scanning electron microscopy, 3D electron microscopy modeling
1. Introduction
The severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), that is the causative agent of the coronavirus disease 2019 (COVID-19) pandemic [1], belongs to the Coronaviridae family (Coronavirinae subfamily) which includes four genera: Alphacoronavirus, Betacoronavirus, Gammacoronavirus, and Deltacoronavirus. While Alphacoronavirus and Betacoronavirus exclusively infect mammalian species, Gammacoronavirus and Deltacoronavirus have a wider host range that includes avian species [1,2]. The SARS-CoV-2 is grouped within the Betacoronavirus genus together with Middle East Respiratory Syndrome Coronavirus (MERS-CoV) and SARS-CoV, which are both also highly pathogenic, and HCoV-HKU1, HCoV-OC43, which causes seasonal and usually mild respiratory tract infections.
Coronaviruses are enveloped with positive-sense single-stranded RNA genome of 26–32 kb and have the largest genomes among RNA viruses. Phylogenetic analysis of SARS-CoV-2 revealed that is closely related (88–89% similarity) to SARS-like coronaviruses from bats—such as bat-SL-CoVZC45 (GenBank no. MG772933.1) and bat-SL-CoVZXC21 (GenBank no. MG772934.1)—and shares lower identity to SARS-CoV (~79% similarity) and MERS-CoV (~50% similarity) [3,4,5].
In vitro studies on the morphogenesis of coronaviruses demonstrated that the attached of these viruses to the host cell occurs from the interaction of the virus Spike protein (S protein) with the cellular ACE-2 (angiotensin converting enzyme 2) receptor [6,7,8]. Internalization of the particle occurs by endocytosis or through fusion between virus envelope and cellular membrane [9]. In SARS-CoV and MERS-CoV, the first moment of replication occurs on virus-induced double-membrane structures derived from the rough endoplasmic reticulum (RER), commonly referred to as ‘double-membrane vesicles’ (DMVs), located mostly in perinuclear areas [7,10,11]. These structures are also generated by other RNA viruses, such as Arteriviruses, Picornaviruses, and the Flaviviruses of genus Hepacivirus [12,13]. Viral RNA filaments inside DMVs were observed in studies in vitro with SARS-CoV-2, associating these compartments with viral replication [14].
Viruses with positive-sense single-stranded RNA genome replicate in host cell cytoplasm and induce intracellular membranous compartments harboring the sites of viral RNA synthesis. These replication factories are supposed to concentrate the components of the replicase and to shield replication intermediates from the host cell innate immune defense [15].
Given the fact that there is no therapeutic treatment for SARS-CoV-2 to date, studies in vitro are needed to understand the mechanism of infection and the virus–cell interaction, as well as to design effective strategies to inhibit virus morphogenesis. In this case, electron microscopy becomes an indispensable tool of analysis. The development of effective intervention strategies relies on the knowledge of molecular and cellular mechanisms of coronavirus infections, which highlights the significance of studying virus–host interactions at the molecular level to identify targets for antiviral intervention and to elucidate critical viral and host determinants that are decisive for the development of severe disease [16].
Vero cells, which are widely used in virus studies and in the production of human vaccines, have been shown to be a relevant model for studies of coronaviruses including SARS-CoV, SARS-CoV-2, and MERS-CoV [17,18,19,20,21,22,23].
In this work, we characterize the replicative cycle of SARS-CoV-2 from Vero-E6 lineage cells infected with a human clinical sample, using transmission electron microscopy and helium ion microscopy.
2. Materials and Methods
2.1. Virus Sample
A SARS-CoV-2, lineage B.1.1, was isolated from the nasopharyngeal swab from Brazilian patient by the National Influenza Centre (NIC) at Fiocruz, Rio de Janeiro. Total RNA was extracted from clinical sample using the QIAmp Viral RNA mini kit (Qiagen North Rhine-Westphalia, Hilden, Germany). Viral detection was done by real time reverse transcription polymerase chain reaction (RT-PCR) with TaqMan primers and probes (IDT) specific for the genes encoding the Envelope protein (E) and the viral RNA-dependent RNA Polymerase (RdRp), as described previously [2]. Reactions were performed with the Qiagen One Step RT-PCR kit (Qiagen, Germantown, MD, USA). Synthetic RNA sequences corresponding to E and RdRP targets [2] were used as positive controls. Putative coinfections in this sample were discarded by differential diagnosis analysis for other 17 respiratory viral pathogens and mycoplasm pneumonieae, as described previously [24]. Whole-genome sequences of isolate evaluated in this study are available in the Global initiative on sharing all influenza data (GISAID) under the accession numbers EPI_ISL_415105. For the experiments, the virus was amplified in Vero-E6 cells (African green monkey kidney). Virus titers were calculated by the tissue culture infectious dose at 50% (TCID50/mL) and the virus stocks kept in −80 °C freezer. All procedures were performed in a biosafety level 3 laboratory, according to WHO guidelines. The access to the genetic patrimony of the virus isolated is registered in Sistema Nacional de Gestão do Patrimônio Genético (SisGen ACCF49F). This research is approved by the Ethics Committee of Instituto Oswaldo Cruz (protocol number 2453470).
2.2. Cells and Virus Infection
Prior to infection, Vero-E6 cultures were maintained in DMEM supplemented with 10% fetal bovine serum (FBS) and 100 U/mL of penicillin-streptomycin (1× Pen-Strep) and cultured at 37 °C and 5% CO2 [25]. All cell culture reagents were acquired from Gibco (Waltham, MA, USA). For infection, monolayers were washed twice with phosphate buffered saline (PBS), inoculated with multiplicity of infection of 0.01 diluted in non-supplemented DMEM and incubated for 1 h at 37 °C for virus adsorption. After adsorption time, the inoculum was removed and cells were kept at 37 °C in DMEM supplemented with 2% FBS and 1× PenStrep. Non-infected control cultures (mock) were prepared using pure non-supplemented DMEM as infected cells. Monolayers were inspected daily under light microscope for observation of cytopathic effect (CPE), until 72 h post infection (pi). All procedures were performed in a biosafety level 3 laboratory, according to WHO guidelines.
2.3. Transmission Electron Microscopy (TEM) and Helium Ion Microscopy (HIM)
For transmission electron microscopy analysis, the infected and non-infected control (mock) monolayers of cells were tripsinized at 48 and 72 h pi. Cell suspensions were fixated in 2.5% glutaraldehyde in sodium cacodylate buffer (0.2 M, pH 7.2), post-fixated in 1% buffered osmium tetroxide, dehydrated in acetone, embedded in epoxy resin, and polymerized at 60 °C over the course of three days [26,27]. Ultrathin sections (50–70 nm) were obtained from the resin blocks. The sections were picked up using copper grids (300 mesh and no coating) and observed using Cs Corrected FEI Titan 80–300 (ThermoFisher Scientific, Waltham, MA, USA) and Hitachi HT 7800 (Hitachi, Tokyo, Japan) transmission electron microscopes. In the FEI Titan images were collected in scanning transmission electron microscopy (STEM) mode using a high-angle annular-dark field detector (HAADF) working at 80 kV high tension, 19 mrad convergence angle and probe size around 0.1 nm, 100 pA beam current, 2 up to 8 μs dwell time, and 2048 × 2048 pixels per image. For analysis in helium ion microscopy, infected and non-infected control (mock) monolayers were grown on sterile glass coverslips and fixed at 48 and 72 h pi, in 2.5% glutaraldehyde in sodium cacodylate buffer (0.2 M, pH 7.2), dehydrated in ethanol and submitted to critical-point-drying. The cells were analyzed in an Orion NanoFab Helium Ion microscope (Zeiss, Baden-Württemberg, Oberkochen, Germany) equipped with a flood gun and an Everhart–Thornley secondary electrons detector, flood gun is used to allow analyzing the cells without any conductive coating. Images were collected at 30 kV high tension, around 0.8 pA beam current, 32 up to 256-line average, 1 µs dwell time, 100 µs flood gun time, and 2048 × 2048 pixels per image.
2.4. Focused Ion Beam Scanning Electron Microscopy (FIB-SEM) and 3D Modeling
FIB-SEM image acquisition was carried out with a Zeiss Auriga crossbeam electron microscope (Zeiss, Baden-Württemberg, Oberkochen, Germany). The epoxide-embedded sample (block) of Vero-E6 cells 48h pi with SARS-CoV-2 prepared for TEM as described above was mounted on a support stub, coated with a 10 nm layer of gold and transferred to the microscope chamber. The sample was tilted 54° towards the ion column in order for the block surface to be oriented perpendicularly to the Ga+ ion beam. The area of interest was exposed by FIB milling of a U-shaped trench. Milling conditions were 30 kV acceleration voltage and a beam current of 16 nA (for coarse milling) and 2 nA for polishing. FIB slicing was performed at 20 nm thickness with an ion beam current of 1 nA. Imaging of the block surface was carried out at 1.8 kV using a backscattered electron detector and 5.4 nm pixel size.
The image series were aligned and the structures of interest were manually segmented for 3D models production using the open source software IMOD [28].
2.5. Measurement of Virus Particle Size
The images were collected in a Zeiss Orion Nanofab helium ion microscope. The particle size statistical analysis was performed using ImageJ software. Each virus particle was selected manually and deleted from the images, after thresholding images they were converted into binary and then only virus particles were selected and counted using the command particle analysis from the menu analysis. About 600 virus particles were analyzed and the average size and standard deviation were extracted.
2.6. Immunofluorescence Staining
The cells were stained as previously described [29,30]. In short, Vero-E6 cells were seeded in coverslips and after 48 and 72 h of infection, were fixated using 3.7% formaldehyde. Cells were rinsed three times with PBS containing 0.1 mM CaCl2 and 1 mM MgCl2 (PBS/CM) and then permeabilized with 0.1% Triton X-100 plus 0.2% BSA in PBS/CM for 10 min (PBS/CM/TBSA). Cells were stained with rabbit polyclonal antibody anti-spike antibody (#56578—NOVUSBIO) at 1:250 dilution for overnight, followed by a rabbit anti-IgG Dylight 550 at 1:1000 dilution for 1 h. The coverslips were mounted in slides using an antifade mounting medium (VECTASHIELD®, Burlingame, CA, USA). Nuclear recognition was based on DAPI staining (1 μg/mL) for 5 min. Fluorescence was analyzed by fluorescence microscopy with an 100× objective lens (Olympus, Tokyo, Japan).
3. Results
Analysis of Vero-E6 cells infected with SARS-CoV-2 under inverted light microscopy demonstrated a CPE, that was mostly evident from 48 h pi. The CPE appeared as rounding and detaching of cells and formation of syncytia (data not shown).
3.1. Ultrastructural Cellular Change Caused by SARS-CoV-2 Infection
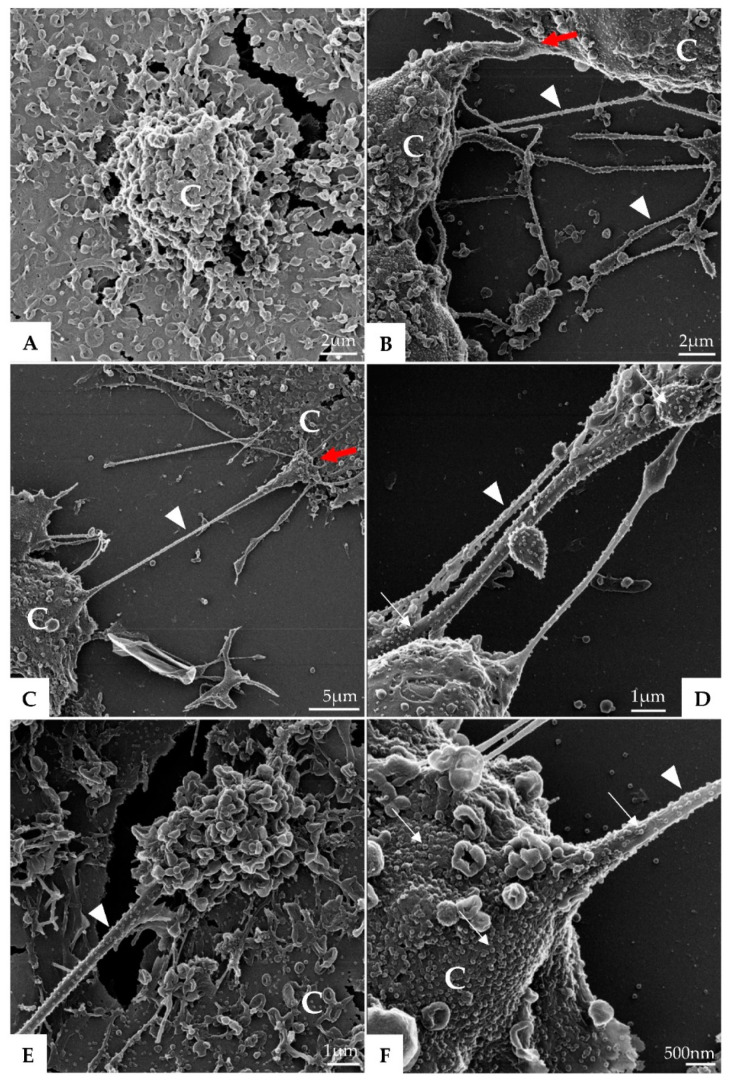
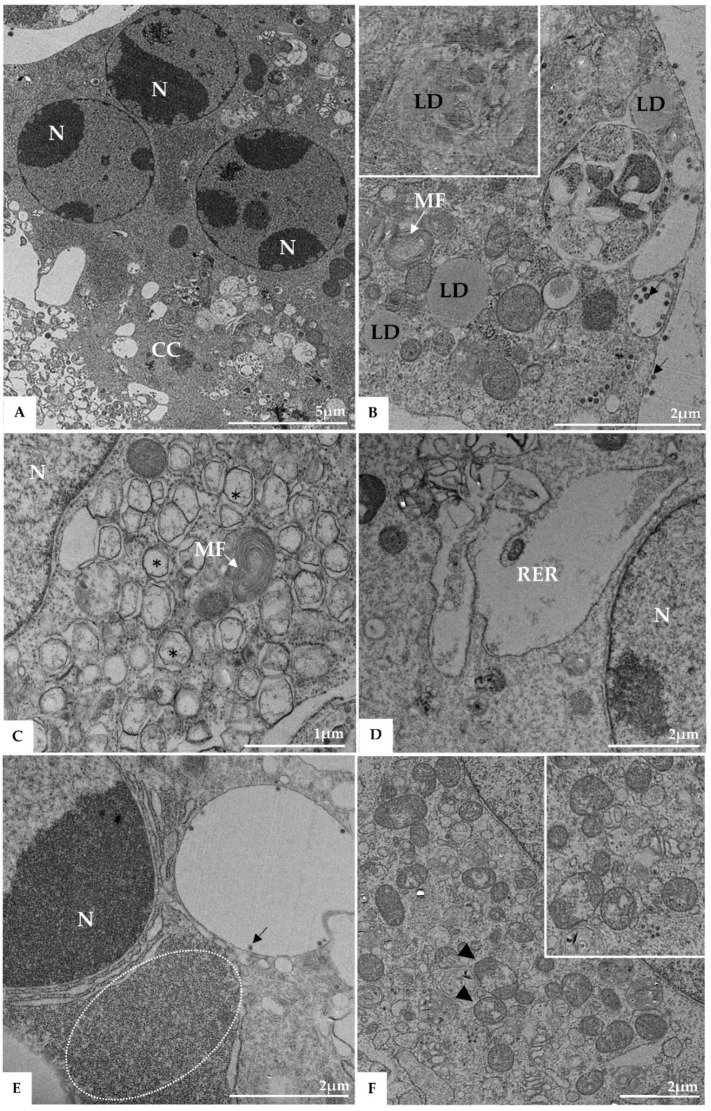
Morphological analysis of Vero-E6 cells at 48 and 72 h pi by using of MET and HIM showed changes associated to SARS-CoV-2 infection, including: (I) Cell activation—Cell activation was evidenced by a marked plasmatic membrane projection (filopodia and microvilli) (Figure 1B–F and Figure 2B–F), connection between adjacent cells mediated by these filopodia (Figure 1B–C, Figure 2B–D, and Figure 3B), viral particles associated with filopodia and microvilli (Figure 1B–F, Figure 2B–F, and Figure 3A) and to the cell membrane (Figure 1B–F and Figure 2B–F). Filopodia, as well as any changes in cell topography, were not observed in non-infected Vero-E6 cells at 48 (Figure 1A) or 72 h (Figure 2A) of cultivation. (II) Syncytia—Syncytial formation (multinucleated cells resulting from cell fusion) in Vero-E6 cells monolayer infected with SARS-CoV-2 were commonly observed as shown in the Figure 4A. (III) Lipid droplets (LD) accumulation (Figure 4B). (IV) Proliferation of double membrane vesicles (DMV). These vesicles were observed throughout the cytosol (Figure 4C). (V) Numerous myelin figures (Figure 4B,C). (VI) Thickening of the RER (Figure 4D), increased number of more electron-dense ribosomes (Figure 4E), and mitochondrial swelling and vacuolation (Figure 4F) were also observed.
Figure 1.
Vero-E6 cells 48 h post-infection with SARS-CoV-2 (HIM images). (A) Uninfected Vero-E6 cell at 48 h of cultivation (cell control). (B–F) Infected cells presenting filopodia (arrowhead); connection (red arrow) between adjacent cells mediated by filopodia was observed. Virus particles (thick arrow) attached to cell filopodia and with cell membrane were observed. Cell (C).
Figure 2.
Vero-E6 cells 72 h post-infection with SARS-CoV-2 (HIM images). (A) Uninfected Vero-E6 cell at 72 h of cultivation (cell control). (B–E) Infected Vero-E6 monolayer, connection (red arrow) between cells mediated by filopodia was observed (B–D). Virus particles (thick arrow, blue structures) was detected attached to cell filopodia (green, image (D)) and with cell membrane (green or red, image (D–F). Cell (C), filopodia (arrowhead). The images (D,E) were colored in Adobe photoshop.
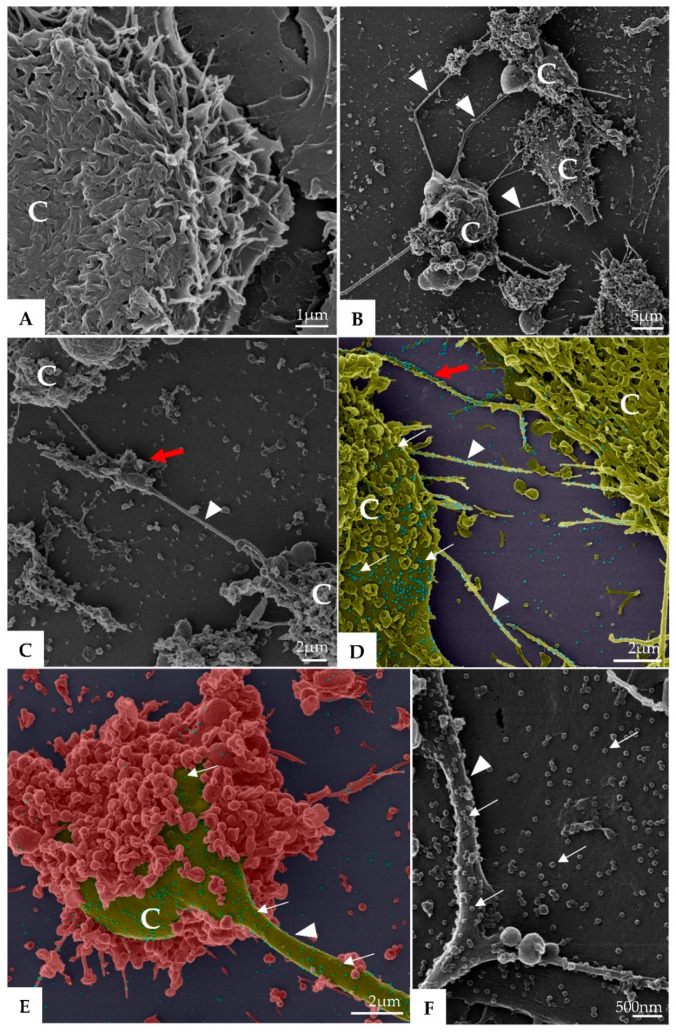
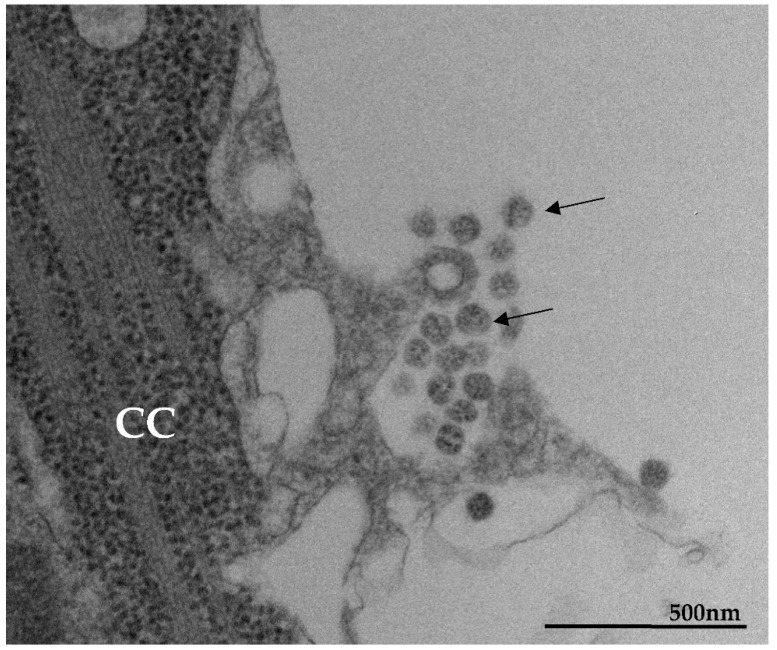
Figure 3.
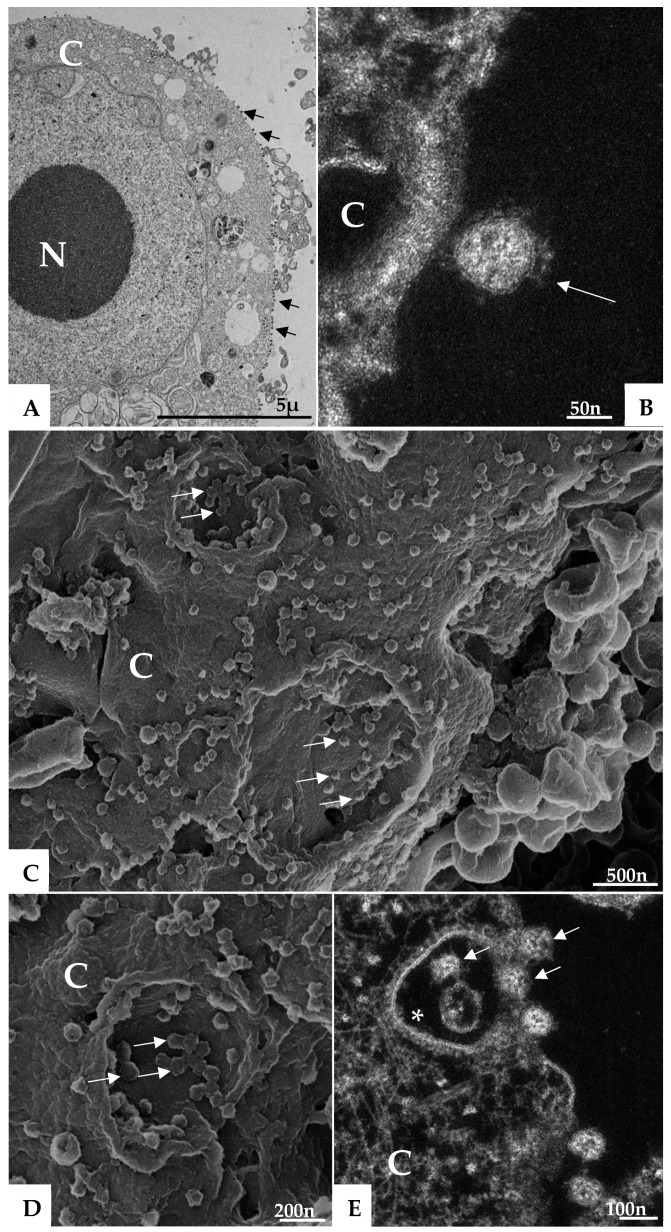
Microvilli in Vero-E6 cells 48 h post-infection with SARS-CoV-2 (TEM images). Microvilli (arrowhead) presenting adsorbed SARS-CoV-2 particles (thick arrow) (A). Interaction between adjacent cells mediated by Microvilli (B). Microvilli (arrowhead), cell cytoplasm (CC), nucleus (N).
Figure 4.
Syncytia formation (A), proliferation of lipid droplets (LD) (B), and double membrane vesicle (*) (C), myelin figures (MF) (B,C), thickening of the rough endoplasmic reticulum (RER) (D), electron-dense ribosomes (marked area) (E), and degeneration of mitochondria (arrowhead) (F) in Vero-E6 cells 48 h post-infection with SARS-CoV-2 (TEM images). Virus particles (arrow), cell cytoplasm (CC), nucleus (N).
3.2. SARS-CoV-2 Particles Diameter and Morphology
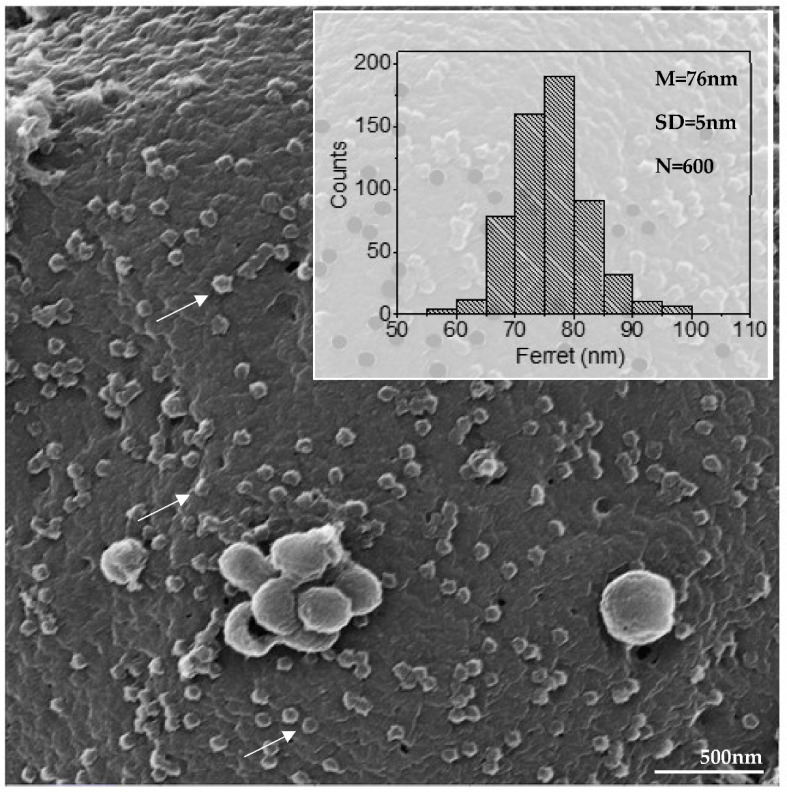
Morphometric analysis by helium ion microscopy of SARS-CoV-2 particles attached on the Vero-E6 cell membrane showed that their mean diameter was 76 nanometers (Figure 5). MET showed particles with a spherical morphology exhibiting spikes on their envelopes (Figure 6B).
Figure 5.
Diameter of SARS-CoV-2 particles attached in cell membrane (arrow) (HIM image). The average diameter of the SARS-CoV-2 particles was around 76 nanometers. Mean (M), standard deviation (SD), number of measured particles (N), virus particles (arrow).
Figure 6.
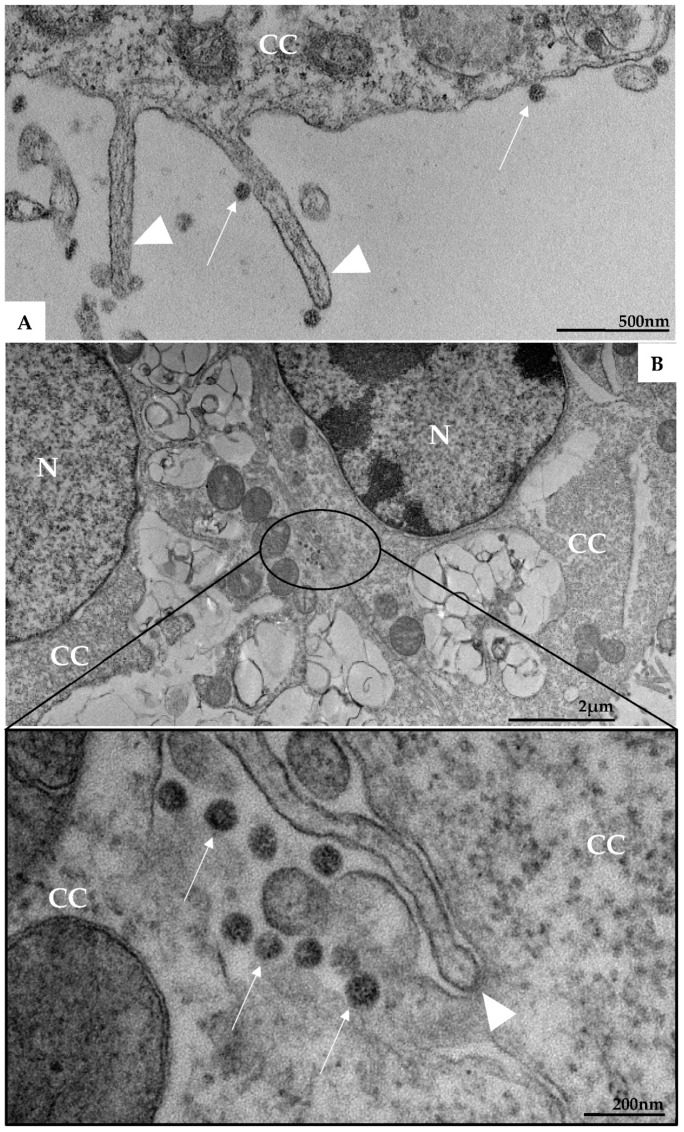
Attachment and endocytosis of SARS-CoV-2 particles (TEM (A), HAADF-STEM (B,E) and HIM (C,D) images). SARS-CoV-2 particles (arrows) attached to the cell membrane (A–D) and being internalized by endocytosis (arrow) (E). Endocytic vesicles coated in clathrin (*) (E). Cell cytoplasm (CC), cell membrane (CM), nucleus (N).
3.3. Morphogenesis of SARS-CoV-2 in Vero-E6 Cell
(I) Virus particle entry—Many virus particles attached to the cell surface (adsorption) (Figure 6A–E) was observed. The entry of the SARS-CoV-2 virus particles into the cells was most commonly realized by clathrin coated endocytic vesicles (Figure 6E), although the fusion of the virus envelope with the cell membrane has been observed also (data not shown).
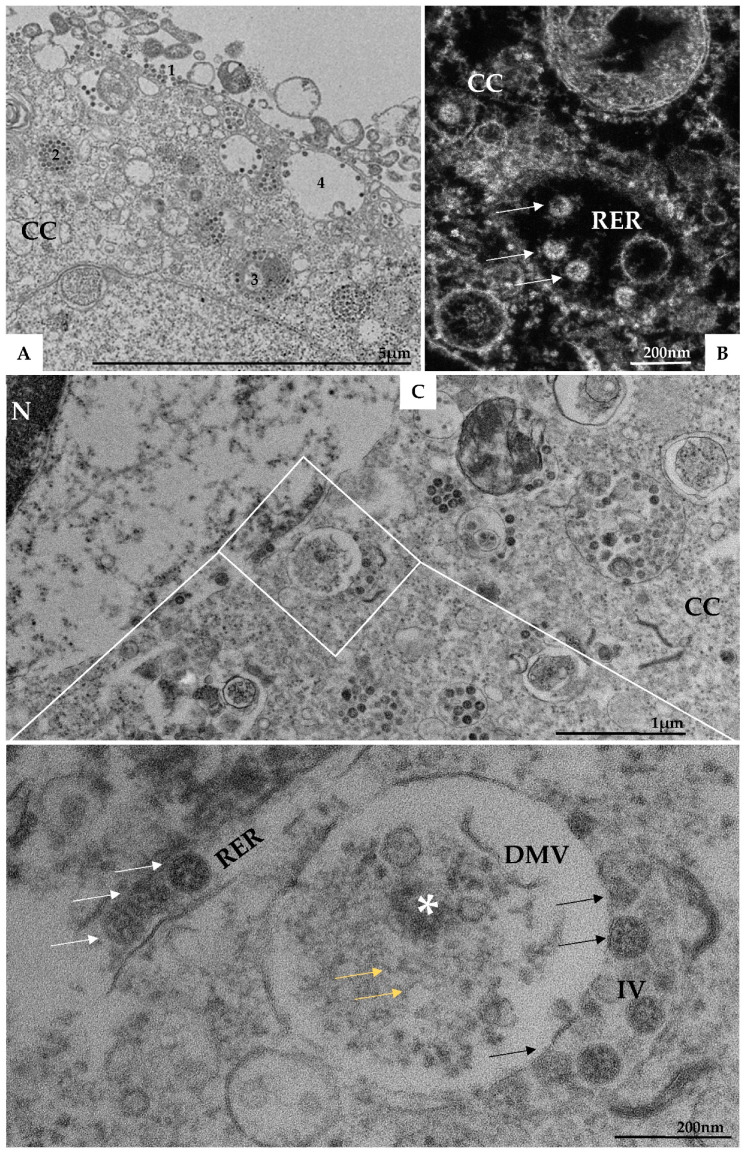
(II) Assembly of viral particles—Virus particles have been observed inside several cytoplasmic compartments including endosomes, RER, DMVs, intermediate vesicles (IV), and exosomes (Figure 7A,B). FIB-SEM images and three-dimensional (3D) modeling of the Vero-E6 cell, 48h pi, showed the interaction between different cellular compartments (Figure 8A–E see also Video S1).
Figure 7.
SARS-CoV-2 particles in lumen of several cellular compartments. (A) SARS-CoV-2 particles attached to the cell membrane (1), on endosomes (2), on the rough endoplasmic reticulum (3 and image (B) (arrow)), and being exocytosed via vesicles (4). (C) Presence of double membrane vesicle (DMV) containing in their lumen electron-dense material (*) and RNA-like filaments (yellow arrow). Budding of viral particles (black arrow) from these vesicles to into intermediate vesicles (IV) was observed. Virions were observed in rough endoplasmic reticulum cisterns (white arrow). Cell cytoplasm (CC), nucleus (N), and rough endoplasmic reticulum cisterns (RER). Transmission electron microscopy.
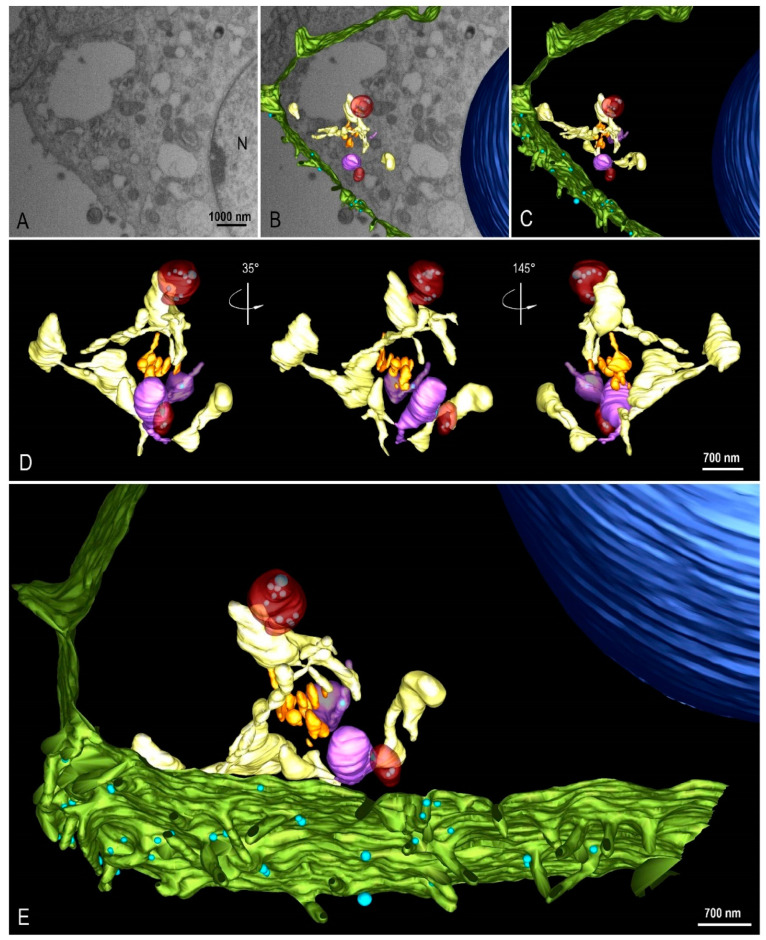
Figure 8.
3D modeling of Vero-E6 48 h post-infection with SARS-CoV-2. (A) FIB-SEM image of the block surface showing an infected cell. (B,C) Combination of a block surface image and the 3D model of structures of interest. (D) Different angle views of a region of interest showing the organization and interaction between different cellular compartments. (E) 3D model showing a global view of the model. Green (cell membrane), light blue (virus particle), dark blue (nucleus), yellow (rough endoplasmic reticulum), orange (Golgi complex), red (double membrane vesicle), purple (intermediate vesicle). See video in Supplementary Material (Video S1).
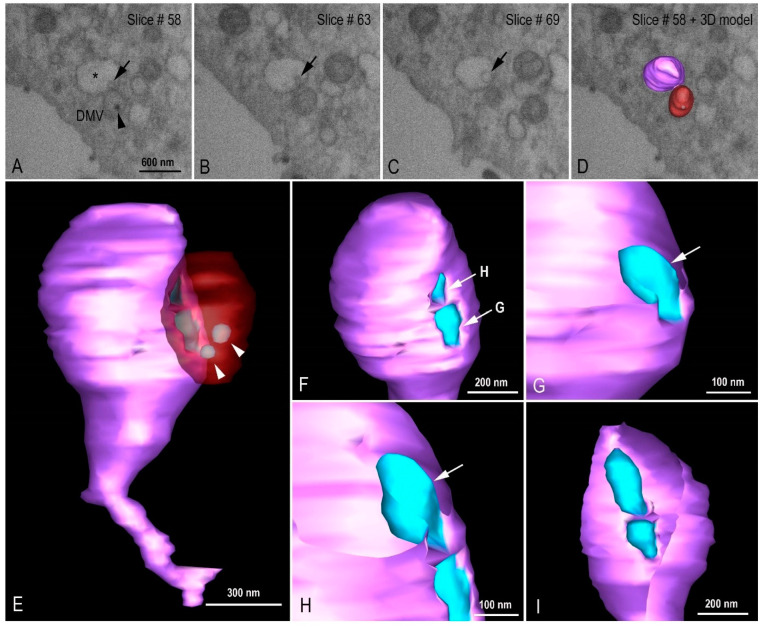
Using transmission electron microscopy (Figure 7C) and three-dimensional reconstruction (Figure 9A–E) the DMVs were observed located close to the RER, suggesting that DMVs result from remodeling of the RER membranes. Inside the DMVs, accumulation of electron-dense material, RNA-like filaments were observed (Figure 7C). IVs associated with these DMVs were observed as well as the presence of virus particles budding from the DMVs membranes into IVs lumen, suggesting that part of the assembly of viral particles can occur in this location (Figure 7C). The budding of the viral particles from the DMV into IV was also demonstrated in three-dimension reconstruction (Figure 10A–I, see also Video S2).
Figure 9.
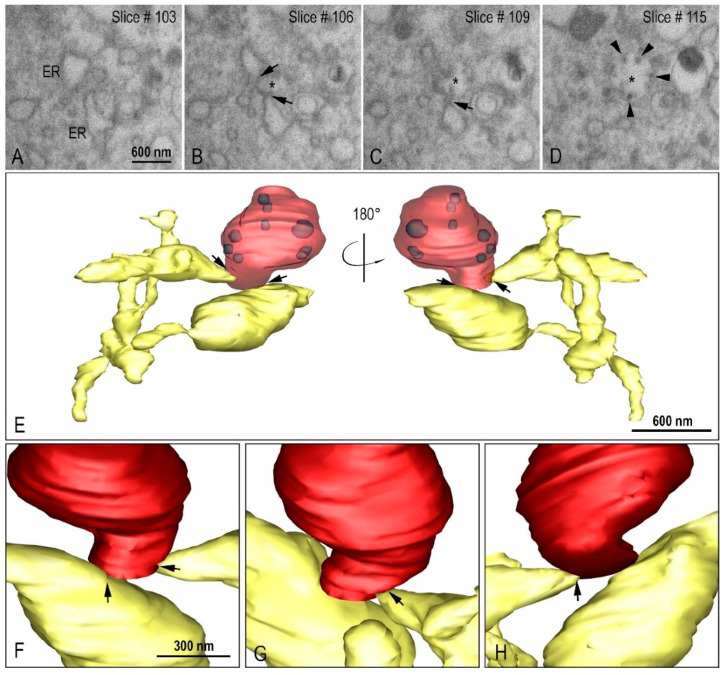
Interaction between the rough endoplasmic reticulum (ER) and double membrane vesicle (DMV). (A–D) FIB-SEM image sequence showing the ER and its connection (arrows) to a DMV (asterisk). Some virus particles can be seen inside the DMV (arrow heads). (E–H) 3D model of the DMV (red), virus particles (purple) inside the DMV and the ER (yellow). The regions of connection of the ER to the DMV are indicated with arrows. Interconnections within the ER network can be observed.
Figure 10.
Virion budding events at intermediate vesicle (IV). (A–D) FIB-SEM image sequence exhibiting different virus budding stages (arrows) at IV (asterisk) and the proximity with a double membrane vesicle (DMV). Virus particles can be observed inside the DMV (arrow head). (E–I) 3D model showing the DMV (red with transparence), virus particles (light blue, arrow heads), and the IV (purple). (F) Extra vesicular view of two virus particle budding regions. The arrows indicate the cutting plane showed in G and H. (G) Initial stage of virus particle budding. (H) Final stage of virus particle budding. (I) Intra vesicular view of the virus budding. See video in Supplementary Material (Video S2).
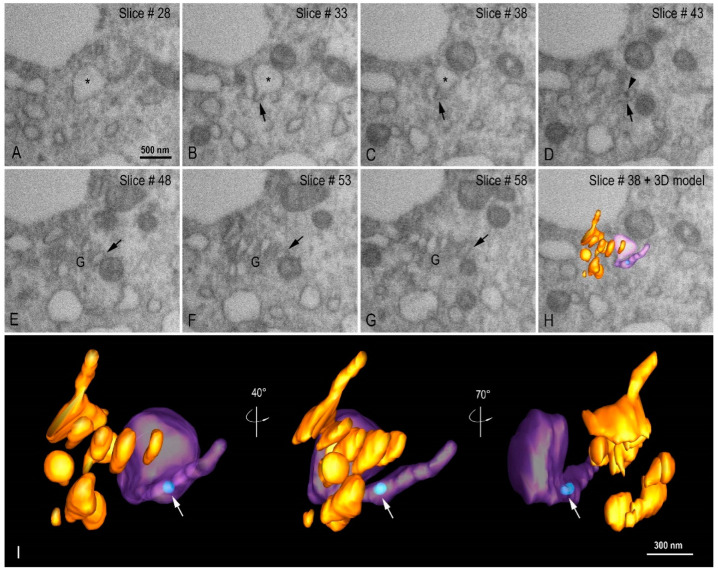
Golgi complex was observed by FIB-SEM image sequence and 3D modeling (Figure 11A–I, see also Video S3) close to the IVs containing virus particles, a fact that allows us to infer a possible relationship between both.
Figure 11.
Organization of the Golgi complex (G) and intermediate vesicle (IV). (A–H) FIB-SEM image sequence showing an IV (asterisk) with a tubular region (arrow) in close proximity with to the Golgi complex. A virus particle can be seen inside the tubular region of the IV (arrow head). (I) 3D model of the G (orange), IV (purple in transparence), and a virus particle (light blue, arrow) at different angle views. See video in Supplementary Material (Video S3).
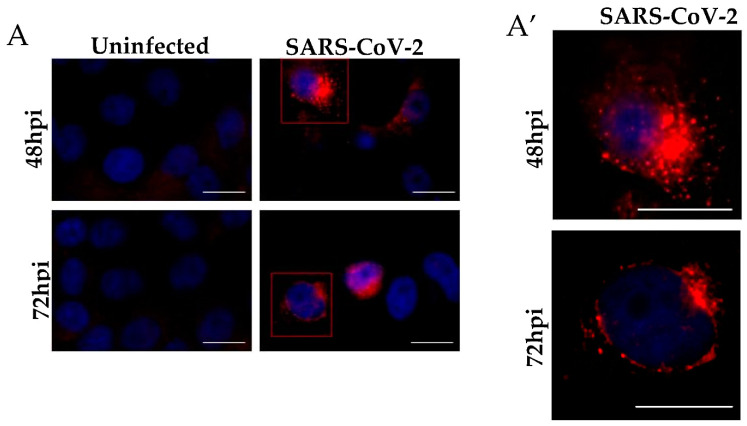
Immunolabeling tests using the immunofluorescence technique of Vero-E6 cells demonstrated the detection of SARS-CoV-2 Spike protein diffused throughout the cytosol both at 48 (Figure 12B,F) and 72 h pi (Figure 12D,H), suggesting viral replication and spreading infection. Detection of Spike protein of SARS-CoV-2 were not observed in the uninfected cells with 48 (Figure 12A,E) and 72 (Figure 12C,G) h of cultivation.
Figure 12.
Immunofluorescence analyses of Vero-E6 cells after SARS-CoV-2 infection for 48 and 72 h. (A) Uninfected cells with 48 and 72 h of cultivation. (A′) Representative zoom images. The virus was detected by the presence of the spike protein (red) performed by immunofluorescence, and cell nuclei stained with DAPI (blue). Hours post-infection (h pi). Scale bar 20 μm.
(III) Extrusion of virus particles. Virions were detected in multiple vesicles at the cell periphery and the release of the particles occurred by exocytosis (Figure 13). Fusion of the exocytic vesicle membrane with the cell cytoplasmic membrane was observed.
Figure 13.
Extrusion of SARS-CoV-2 particles (MET). The particles released by exocytosis. Cell cytoplasm (CC), virus particles (arrow).
4. Discussion
TEM, STEM, HIM, and 3D modeling were used in the present study aiming to characterize the main virus-induced cellular changes as well as the replicative cycle of SARS-CoV-2 in Vero-E6 cells infected with a Brazilian clinical sample.
We observed topographic changes in Vero-E6 cells at 48 and 72 h pi by SARS-CoV-2 evidenced by a marked cellular activation by the presence of numerous filopodia with associated viral particles. Connection between adjacent cells mediated by these filopodia was also observed. We believe that the role of filopodia in this in vitro model is associated with infection maximization. In vitro studies with Vero cells with SARS-CoV and SARS-CoV-2 corroborate our findings [23,31,32]. Enveloped viruses can spread by two different routes: the cell-free aqueous environment or by cell-to-cell contact [33,34,35]. Cell-to-cell contact spreading has unique advantages. The transfer speed is greater because the replication cycle of release, transmission, and entry can proceed in a faster manner. Another advantage of cell-to-cell contact is immune evasion because the limited exposure in extracellular space avoids interaction with neutralizing antibodies. Lastly, by exploiting cell-to-cell communication, the physical and immunological barriers can be overcome to spread the infection [34,35]. Viruses can hijack the filopodia system for their own use during the life cycle under the following strategies: viral surfing (the binding of virus to filopodia induced a rapid lateral movement toward the cell body), filopodial retraction (the filopodia on virally infected cells pull the virus inward), and filopodial bridges to assist virus transport between the cells [35,36].
Syncytial formation in Vero-E6 cells monolayer 48 and 72 h pi with SARS-CoV-2 were commonly observed in our analysis. In ultrastructural studies of Vero cells infected with SARS-CoV multinucleated syncytial cells were occasionally seen [37]. Studies with patients with COVID-19 showed lungs contain infected pneumocytes with abnormal morphology and frequent multinucleation [38]. The authors infer that generation of these syncytia results from activation of the SARS-CoV-2 spike protein at the cell plasma membrane level.
Our analysis demonstrated accumulation of LDs in infected cells, which is corroborated by a previous study conducted by our group [29]. In the above-mentioned study, in vitro SARS-CoV-2 infection were seen to modulate pathways of lipid synthesis and uptake as monitored by testing for CD36, SREBP-1, PPARγ, and DGAT-1 expression in monocytes and triggered LD formation in different human cell lines. Ultrastructural analysis by transmission electron microscopy of Vero-E6 cells infected with SARS-CoV-2 showed viral particles colocalizing with LDs, suggesting that LDs might serve as an assembly platform. Indeed, inhibition of LD formation in Vero-E6 lead to decreased SARS-CoV-2 replication [29]. Our data are also corroborated with the findings of Nardacci et al. (2021) that demonstrated that SARS-CoV-2 infection induce the accumulation of LDs, both in cultured cells and in type II pneumocytes of lung from infected patients. In addition, LDs were often observed in close contact with swollen mitochondria and may participate in mechanisms of cytopathogenesis [39].
SARS-CoV-2 particles attached to the cell surfaces with an average diameter of 76 nanometers and with spherical morphology exhibiting spikes on their envelope, which is characteristic of viruses belonging to the Coronaviridae family. This data is very consistent with previous in vitro studies of SARS-CoV [9] and SARS-CoV-2 in which the virion diameter ranged from 60 to 140 nm [14,38,40,41,42].
We observed that the entry of the SARS-CoV-2 into the host cell occurs most commonly by endocytosis, although the fusion of the virus envelope with the cell membrane has also been observed. This is consistent with our previous ultrastructural studies with SARS-CoV-2 [23] and others from literature with SARS-CoV and MERS-CoV which support both mechanisms [9,43,44].
In the present study, DMVs located close to the RER were observed, suggesting that these vesicles result from remodeling of the RER membranes. Presence of RNA-like filaments and electron-dense material inside the lumen of DMVs was observed. DMVs are induced during the replication of a variety of RNA viruses [45,46,47,48] and were identified as the sole compartment where viral RNA transcription occurs for coronaviruses [49]. Morphological analysis of different cell lines infected with SARS-CoV, MERS-CoV, and SARS-CoV-2—as well as in our studies—has demonstrated that the DMVs are derived from the RER. In the studies concerning SARS-CoV and MERS-CoV, the nonstructural proteins 3 and 4 were predominantly responsible for orchestrating this process [7,50,51,52]. In some of these models, the RER lumen constitutes the space between the DMV’s inner and outer membrane, while the enclosed space is of cytoplasmic origin and enriched in double-stranded RNA (dsRNA) [14].
Our data corroborates the findings from the SARS-CoV and MERS-CoV studies, as well as the analysis with Vero-E6 and human pulmonary cell lines (Calu3) infected with SARS-CoV-2, in which DMVs containing viral RNA inside were found [14,23,32]. Wolff et al. [47], using cellular cryoelectron microscopy, identified a molecular pore complex spanning both membranes of the DMVs, which interconnects the interior of the vesicles with the cytoplasm and would allow the release of newly synthesized RNA from the DMV interior into the cytoplasm.
We observed IVs associated with DMVs, as well as the presence of virus particles budding from the DMVs membranes into IVs lumen, suggesting that part of the assembly of particles can occur in this localization. Close proximity of IVs with the Golgi complex was observed, a fact that allows us to infer a possible relationship between IVs and the Golgi complex. Knoops et al. [7] and Siu et al. [53] observed large assemblies of convoluted membranes, often in close proximity to DMV clusters in Vero-E6 cells infected with SARS-CoV. The authors suggest that these structures are probably identical to the “reticular inclusions” that were firstly observed in cells infected with mouse hepatitis coronavirus [54] and were later referred to as ‘clusters of tubular cisternal elements’, which may have a connection to the endoplasmic reticulum–Golgi intermediate compartment (ERGIC) [55]. The structural proteins membrane (M), nucleocapsid (N), envelope (E), and S—together with the genomic RNA—drive the assembly of new virus particles, which in the case of other coronaviruses bud into ERGIC [56].
We observed virions in multiple smooth vesicles at the cell periphery, close to the cell membrane. Fusion of membrane of the smooth vesicles with the cell membrane resulting in the release of the particles was also observed, too. This mechanism of viral progeny release was detected in our previous studies with SARS-CoV-2 [24], and corroborates data presented by Qian et al. [57] and Qinfen et al. [9] from in vitro studies with alveolar type II cells and Vero-E6 cells infected with SARS-CoV.
In our immunolabeling tests by the immunofluorescence technique, the robust presence of Spike protein was observed in all Vero-E6 cell cytoplasms at both 48 and 72 h pi, proving the high permissiveness of this cell line to SARS-CoV-2 infection, as demonstrated in our ultrastructural studies.
Our analyses demonstrated how essential electron microscopy is when it comes to the analysis of a causative agent of an unknown disease, a situation already experienced in 2003 with SARS-CoV [58], Ebola [59,60,61], Hendra [62], Nipah [63], and monkeypox [64].
This study showed that the Vero-E6 cell is a good in vitro model for studies of SARS-CoV-2 biogenesis and that it can be used for pre-clinical studies.
Acknowledgments
The authors would like to thank IOC for providing us with its technology and facilities during the course of the research, the Plataforma de Microscopia Eletrônica Rudolph Barth, IOC, Fiocruz, the NB3 Platform (IOC, Fiocruz), the CENABIO (Federal University of Rio de Janeiro) and the Willian Ferreira (INMETRO, Rio de Janeiro).
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/v14020201/s1, Video S1. Three-dimensional reconstruction of Vero-E6 48 h post-infection with SARS-CoV-2. Combination of a FIB-SEM image sequence and the 3D model presenting different cellular compartments engaged in SARS-CoV-2 replication. Green (cell membrane), light blue (virus particle), dark blue (nucleus), yellow (rough endoplasmic reticulum), orange (Golgi complex), red (double membrane vesicle), purple (intermediate vesicle). Video S2. Three-dimensional reconstruction of SARS-CoV-2 particles budding events at intermediate vesicle. 3D model showing different virus budding stages and the proximity with a double membrane vesicle. Red with transparence (double membrane vesicle), light blue (virus particles), purple (intermediate vesicle). Video S3. Three-dimensional reconstruction of organization of the Golgi complex and intermediate vesicle. Combination FIB-SEM image sequence and the 3D model showing an intermediate vesicle with a tubular region in close proximity with the Golgi complex. A virus particle can be seen inside the tubular region of the intermediate vesicle. Orange (Golgi complex), purple in transparence (intermediate vesicle), light blue (virus particle).
Author Contributions
Conceptualization, formal analysis, funding acquisition, investigation, methodology, project administration, resources, writing—original draft, writing—review and editing, D.F.B.-V.; Formal analysis, investigation, methodology, writing—review and editing, M.A.N.d.S., A.L.T.d.A., A.d.C.R., W.G.-D., B.S.A., P.T.B., S.S.G.D. and V.C.S.; Funding acquisition, formal analysis, investigation, methodology, writing—review and editing, M.E.M., M.D.M., F.C.M. and M.M.S.; Funding acquisition, writing—review and editing, O.M.B. and T.M.L.S. All authors have read and agreed to the published version of the manuscript.
Funding
This research was supported by Laboratório de Morfologia e Morfogênese Viral, Instituto Oswaldo Cruz (IOC), Fiocruz, FIOTEC (grant number IOC-023-FIO-18-2-58), Fundação de Amparo à Pesquisa do Estado do Rio de Janeiro (FAPERJ) (grant number: E-27/2021/269465) to D.F.B.-V., Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq) (grant number: 301992/2017-7) to O.M.B., Laboratório de Vírus Respiratório e do Sarampo, IOC, Fiocruz, FAPERJ (grant number: E-26/210.151/2020), Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES) and CNPq for the student fellowships to M.D.M. T.M.L.S.’s group was supported by CNPq (441019/2020-0, 307162/2017-6), FAPERJ (E-26/210.182/2020, E-26/201.067/2021, E-26/210.112/2020). CAPES, CNPq, and FAPERJ also supported the National Institutes of Science and Technology Program (INCT) on Diseases of Neglected populations (INCT-IDPN, Finance Code 465313/2014-0). Thanks are due to Fiocruz under the auspices of the Inova Program (B3-Bovespa funding, Finance Code VGPDI-032-ARVC-20). The funders had no role in the study design, data collection, analysis, and preparation of or decision to publish the manuscript.
Institutional Review Board Statement
The study was conducted according to the guidelines of the Declaration of Helsinki, and approved by the Human Ethics Committee of Fiocruz, protocol code 2453470.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest exists.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Gorbalenya A.E., Baker S.C., Baric R.S., Groot R.J., Drosten C., Gulyaeva A.A., Haagmans B.L., Lauber C., Leontovich A.M., Neuman B.W., et al. The species severe acute respiratory syndrome-related coronavirus: Classifying 2019-nCoV and naming it SARS-CoV-2. Nat. Microbiol. 2020;5:536–544. doi: 10.1038/s41564-020-0695-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Corman V.M., Landt O., Kaiser M., Molenkamp R., Meijer A., Chu D.K.W., Bleicker T., Brünink S., Schneider J., Schmidt M.L., et al. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Eurosurveillance. 2020;25:2000045. doi: 10.2807/1560-7917.ES.2020.25.3.2000045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jiang S., Du L., Shi Z. An emerging coronavirus causing pneumonia outbreak in Wuhan, China: Calling for developing therapeutic and prophylactic strategies. Emerg. Microbes Infect. 2020;9:275–277. doi: 10.1080/22221751.2020.1723441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lu H., Stratton C.W., Tang Y.W. Outbreak of pneumonia of unknown etiology in Wuhan China: The mystery and the miracle. J. Med. Virol. 2020;92:401–402. doi: 10.1002/jmv.25678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ren L.-L., Wang Y.-M., Wu Z.-Q., Xiang Z.-C., Guo L., Xu T., Jiang Y.-Z., Xiong Y., Li Y.-J., Li X.-W., et al. Identification of a novel coronavirus causing severe pneumonia in human: A descriptive study. Chin. Med. J. 2020;133:1015–1024. doi: 10.1097/CM9.0000000000000722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Letko M., Marzi A., Munster V. Functional assessment of cell entry and receptor usage for SARS- CoV-2 and other lineage B betacoronaviruses. First functional assessment of the interaction of the SARS-CoV-2 Spike protein receptor binding domain with the cellular receptor ACE2. Nat. Microbiol. 2020;5:562–569. doi: 10.1038/s41564-020-0688-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Knoops K., Kikkert M., Worm S.H.E.V.D., Zevenhoven-Dobbe J.C., Van Der Meer Y., Koster A.J., Mommaas A.M., Snijder E.J. SARS-Coronavirus Replication Is Supported by a Reticulovesicular Network of Modified Endoplasmic Reticulum. PLoS Biol. 2008;6:e226. doi: 10.1371/journal.pbio.0060226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Walls A.C., Park Y.J., Tortorici M.A., Wall A., McGuire A.T., Veesler D. Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Recommended resource describing key aspects of SARS-CoV-2 Spike features. Cell. 2020;181:281–292.e6. doi: 10.1016/j.cell.2020.02.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang Q., Cui J., Huang X., Zheng H., Huang J., Fang L., Li K., Zhang J. The life cycle of SARS coronavirus in Vero E6 cells. J. Med. Virol. 2004;73:332–337. doi: 10.1002/jmv.20095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Goldsmith C.S., Tatti K.M., Ksiazek T.G., Rollin P., Comer J.A., Lee W.W., Rota P.A., Bankamp B., Bellini W.J., Zaki S.R. Ultrastructural Characterization of SARS Coronavirus. Emerg. Infect. Dis. 2004;10:320–326. doi: 10.3201/eid1002.030913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Snijder E.J., van der Meer Y., Zevenhoven-Dobbe J., Onderwater J.J., van der Meulen J., Koerten H.K., Mommaas A.M. Ultrastructure and origin of membrane vesicles associated with the severe acute respiratory syndrome coronavirus replication complex. J. Virol. 2006;80:5927–5940. doi: 10.1128/JVI.02501-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Netherton C.L., Wileman T. Virus factories, double membrane vesicles and viroplasm generated in animal cells. Curr. Opin. Virol. 2011;1:381–387. doi: 10.1016/j.coviro.2011.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Blanchard E., Roingeard P. Virus-induced double-membrane vesicles. Cell. Microbiol. 2015;17:45–50. doi: 10.1111/cmi.12372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Klein S., Cortese M., Winter S.L., Wachsmuth-Melm M., Neufeldt C.J., Cerikan B., Stanifer M.L., Boulant S., Barten-schlager R., Chlanda P. SARS-CoV-2 structure and replication characterized by in situ cryo-electron tomography. Nat. Commun. 2020;11:5885. doi: 10.1038/s41467-020-19619-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Harak C., Lohmann V. Ultrastructure of the replication sites of positive-strand RNA viruses. Virology. 2015;479:418–433. doi: 10.1016/j.virol.2015.02.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.V’kovski P., Kratzel A., Steiner S., Stalder H., Thiel T. Coronavirus biology and replication: Implications for SARS-CoV-2. Nat. Rev. Microbiol. 2021;19:155–170. doi: 10.1038/s41579-020-00468-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Matsuyama S., Nao N., Shirato K., Kawase M., Saito S., Takayama I., Nagata N., Sekizuka T., Katoh H., Kato F. Enhanced isolation of SARS-CoV-2 by TMPRSS2-expressing cells. Proc. Natl. Acad. Sci. USA. 2020;117:7001–7003. doi: 10.1073/pnas.2002589117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Banerjee A., Nasir J.A., Budylowski P., Yip L., Aftanas P., Christie N., Ghalami A., Baid K., Raphenya A.R., Hirota J.A. Isolation, sequence, infectivity and replication kinetics of SARS-CoV-2. Emerg. Infect. Dis. 2020;26:2054–2063. doi: 10.3201/eid2609.201495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tseng C.-T.K., Tseng J., Perrone L., Worthy M., Popov V., Peters C.J. Apical Entry and Release of Severe Acute Respiratory Syndrome-Associated Coronavirus in Polarized Calu-3 Lung Epithelial Cells. J. Virol. 2005;79:9470–9479. doi: 10.1128/JVI.79.15.9470-9479.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mossel E.C., Huang C., Narayanan K., Makino S., Tesh R.B., Peters C.J. Exogenous ACE2 Expression Allows Refractory Cell Lines to Support Severe Acute Respiratory Syndrome Coronavirus Replication. J. Virol. 2005;79:3846–3850. doi: 10.1128/JVI.79.6.3846-3850.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kaye M., Druce J., Tran T., Kostecki R., Chibo D., Morris J., Catton M., Birch C. SARS–associated Coronavirus Replication in Cell Lines. Emerg. Infect. Dis. 2006;12:128–133. doi: 10.3201/eid1201.050496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ogando N.S., Dalebout T.J., Zevenhoven-Dobbe J.C., Limpens R.W., van der Meer Y., Caly L., Druce J., de Vries J.J.C., Kikkert M., Bárcena M., et al. SARS-coronavirus-2 replication in Vero E6 cells: Replication kinetics, rapid adaptation and cytopathology. J. Gen. Virol. 2020;101:925–940. doi: 10.1099/jgv.0.001453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Barreto-Vieira D.F., da Silva M.A.N., Garcia C.C., Miranda M.D., Matos A.D.R., Caetano B.C., Resende P.C., Motta F.C., Siqueira M.M., Wendell Girard D., et al. Morphology and morphogenesis of SARS-CoV-2 in Vero-E6 cells. Mem. Inst. Oswaldo Cruz. 2021;116:e200443. doi: 10.1590/0074-02760200443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Matos A.D.R., Motta F.C., Caetano B.C., Ogrzewalska M., Garcia C.C., Lopes J.C.O., Miranda M., Livorati M.T.F.P., Abreu A., Brown D., et al. Identification of SARS-CoV-2 and additional respiratory pathogens cases under the investigation of COVID-19 initial phase in a Brazilian reference laboratory. Mem. Inst. Oswaldo Cruz. 2020;115:e200232. doi: 10.1590/0074-02760200232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Szretter K.J., Balish A.L., Katz J.M. Influenza: Propagation, quantification, and storage. Curr. Protoc. Microbiol. 2006;3:15G.1. doi: 10.1002/0471729256.mc15g01s3. [DOI] [PubMed] [Google Scholar]
- 26.Barreto-Vieira D.F., Barth-Schatzmayr O.M., Schatzmayr H.G. Manual de Técnicas. Interciência; Rio de Janeiro, Brasil: 2010. Modelo animal experimental para o estudo da patogênese dos vírus dengue sorotipos 1 e 2. [Google Scholar]
- 27.Barth O.M., da Silva M.A.N., Barreto-Vieira D.F. Low impact to fixed cell processing aiming transmission electron microscopy. Mem. Inst. Oswaldo Cruz. 2016;111:411–413. doi: 10.1590/0074-02760150433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kremer J.R., Mastronarde D.N., McIntosh J.R. Computer Visualization of Three-Dimensional Image Data Using IMOD. J. Struct. Biol. 1996;116:71–76. doi: 10.1006/jsbi.1996.0013. [DOI] [PubMed] [Google Scholar]
- 29.Dias S.S.G., Soares V.C., Ferreira A.C., Sacramento C.Q., Fintelman-Rodrigues N., Temerozo J.R., Teixeira L., Nunes da Silva M.A., Barreto E., Mattos M., et al. Lipid droplets fuel SARS-CoV-2 replication and production of inflammatory mediators. PLoS Pathog. 2020;16:e1009127. doi: 10.1371/journal.ppat.1009127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Souza-Moreira L., Soares V.C., Dias S.D.S.G., Bozza P.T. Adipose-derived Mesenchymal Stromal Cells Modulate Lipid Metabolism and Lipid Droplet Biogenesis via AKT/mTOR –PPARγ Signalling in Macrophages. Sci. Rep. 2019;9:1–11. doi: 10.1038/s41598-019-56835-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ng M.L., Lee J.W.M., Leong M.L.N., Ling A.E., Tan H.C., Ooi E.E. Topographic changes in SARS coronavirus-infected cells at late stages of infection. Emerg. Infect. Dis. 2004;10:1907–1914. doi: 10.3201/eid1011.040195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Caldas L.A., Carneiro F.A., Higa L.M., Monteiro F.L., da Silva G.P., da Costa L.J., Durigon E.L., Tanuri A., de Souza W. Ultrastructural analysis of SARS-CoV-2 interactions with the host cell via high resolution scanning electron microscopy. Sci. Rep. 2020;10:16099. doi: 10.1038/s41598-020-73162-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sherer N.M., Lehmann M.J., Jimenez-Soto L.F., Horensavitz C., Pypaert M., Mothers W. Retroviruses can establish filopodial bridges for efficient cell-to-cell transmission. Nat. Cell Biol. 2007;9:310–315. doi: 10.1038/ncb1544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mothes W., Sherer N.M., Jin J., Zhong P. Virus Cell-to-Cell Transmission. J. Virol. 2010;84:8360–8368. doi: 10.1128/JVI.00443-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chang K., Baginski J., Hassan S.F., Volin M., Shukla D., Tiwari V. Filopodia and Viruses: An Analysis of Membrane Processes in Entry Mechanisms. Front. Microbiol. 2016;7:300. doi: 10.3389/fmicb.2016.00300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tiwari V., Tarbutton M.S., Shukla D. Diversity of heparan sulfate and HSV entry: Basic understanding and treatment strategies. Molecules. 2015;20:2707–2727. doi: 10.3390/molecules20022707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Braga L., Ali H., Secco I., Chiavacci E., Neves G., Goldhill D., Penn R., Jimenez-Guardeño J.M., Ortega-Prieto A.M., Bussani R., et al. Drugs that inhibit TMEM16 proteins block SARS-CoV-2 spike-induced syncytia. Nature. 2021;594:88–93. doi: 10.1038/s41586-021-03491-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhu N., Dingyu Z., Wenling W., Xingwang L., Bo Y., Song J., Zhao X., Huang B., Shi W., Lu R., et al. A novel coronavirus from patients with pneumonia in China, 2019. N. Engl. J. Med. 2020;382:723–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nardacci R., Colavita F., Castilletti C., Lapa D., Matusali G., Meschi S., Del Nonno F., Colombo D., Capobianchi M.R., Zumla A., et al. Evidences for lipid involvement in SARS-CoV-2 cytopathogenesis. Cell Death Dis. 2021;12:1–12. doi: 10.1038/s41419-021-03527-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Laue M., Kauter A., Hoffmann T., Möller L., Michel J., Nitsche A. Morphometry of SARS-CoV and SARS-CoV-2 particles in ultrathin plastic sections of infected Vero cell cultures. Sci. Rep. 2021;11:3515. doi: 10.1038/s41598-021-82852-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Werion A., Belkhir L., Perrot M., Schmit G., Aydin S., Chen Z., Penaloza A., De Greef J., Yildiz H., Pothen L., et al. SARS-CoV-2 causes a specific dysfunction of the kidney proximal tubule. Kidney Int. 2020;98:1296–1307. doi: 10.1016/j.kint.2020.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Park W.B., Kwon N.J., Choi S.J., Kang C.K., Choe P.G., Kim J.Y., Yun J., Lee G.W., Seong M.W., Kim N.J., et al. Virus isolation from the first patient with SARS-CoV-2 in Korea. J. Korean Med. Sci. 2020;35:e84. doi: 10.3346/jkms.2020.35.e84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Song Z., Xu Y., Bao L., Zhang L., Yu P., Qu Y., Zhu H., Zhao W., Han Y., Qin C. From SARS to MERS, thrusting coronaviruses into the spotlight. Viruses. 2019;11:59. doi: 10.3390/v11010059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ng M., Tan S., See E., Ooi E., Ling A. Early events of SARS coronavirus infection in Vero cells. J. Med Virol. 2003;71:323–331. doi: 10.1002/jmv.10499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Paul D., Hoppe S., Saher G., Krijnse-Locker J., Bartenschlager R. Morphological and Biochemical Characterization of the Membranous Hepatitis C Virus Replication Compartment. J. Virol. 2013;87:10612–10627. doi: 10.1128/JVI.01370-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.De Wilde A.H., Raj V.S., Oudshoorn D., Bestebroer T.M., Van Nieuwkoop S., Limpens R.W.A.L., Posthuma C.C., Van Der Meer Y., Barcena M., Haagmans B.L., et al. MERS-coronavirus replication induces severe in vitro cytopathology and is strongly inhibited by cyclosporin A or interferon-α treatment. J. Gen. Virol. 2013;94:1749–1760. doi: 10.1099/vir.0.052910-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wolff G., Melia C.E., Snijder E.J., Bárcena M. Double-Membrane Vesicles as Platforms for Viral Replication. Trends Microbiol. 2020;28:1022–1033. doi: 10.1016/j.tim.2020.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhou X., Cong Y., Veenendaal T., Klumperman J., Shi D., Mari M., Reggiori F. Ultrastructural Characterization of Membrane Rearrangements Induced by Porcine Epidemic Diarrhea Virus Infection. Viruses. 2017;9:251. doi: 10.3390/v9090251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Snijder E.J., Limpens R.W.A.L., de Wilde A.H., de Jong A.W.M., Zevenhoven-Dobbe J.C., Maier H.J., Faas F.F.G.A., Koster A.J., Bárcena M. A unifying structural and functional model of the coronavirus replication organelle: Tracking down RNA synthesis. PLOS Biol. 2020;18:e3000715. doi: 10.1371/journal.pbio.3000715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Oudshoorn D., Rijs K., Limpens R.W.A.L., Groen K., Koster A.J., Snijder E.R., Kikkert M., Bárcena M. Expression and cleavage of middle east respiratory syndrome coronavirus nsp3-4 polyprotein induce the formation of double-membrane vesicles that mimic those associated with coronaviral RNA replication. mBio. 2017;8:e01658-17. doi: 10.1128/mBio.01658-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mendonça L., Howe A., Gilchrist J.B., Sun D., Knight M.L., Zanetti-Domingues L.C., Bateman B., Krebs A.S., Chen L., Radecke J., et al. SARS-CoV-2 assembly and egress pathway revealed by correlative multi-modal multi-scale cryo-imaging. Nat Commun. 2021;30:4629. doi: 10.1038/s41467-021-24887-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cortese M., Lee J.Y., Cerikan B., Neufeldt C.J., Oorschot V.M.J., Köhrer S., Hennies J., Schieber N.L., Ronchi P., Mizzon G., et al. Integrative imaging reveals SARS-CoV-2-induced reshaping of subcellular morphologies. Cell Host Microbe. 2020;28:853–866.e5. doi: 10.1016/j.chom.2020.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Siu Y.L., Teoh K.T., Chan C.M., Kien F., Escriou N., Tsao S.W., Nicholls J.M., Altmeyer R., Peiris J.S.M., Bruzzone R., et al. The M, E, and N structural proteins of the severe acute respiratory syndrome coronavirus are required for efficient assembly, trafficking, and release of virus-like particles. J. Virol. 2008;82:11318–11330. doi: 10.1128/JVI.01052-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.David-Ferreira J.F., Manaker R.A. An electron microscope study of the development of a mouse hepatitis virus in tissue culture cells. J. Cell Biol. 1965;24:57–78. doi: 10.1083/jcb.24.1.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Krijnse-Locker J., Ericsson M., Rottier P.J.M., Griffiths G. Characterization of the budding compartment of mouse hepatitis virus: Evidence that transport from the RER to the Golgi complex requires only one vesicular transport step. J. Cell Biol. 1994;124:55–70. doi: 10.1083/jcb.124.1.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Perlman S., Masters P.S. Coronaviridae: The Viruses and Their Replication. 7th ed. Volume 1 LWW; New York, NY, USA: 2020. [Google Scholar]
- 57.Qian Z., Travanty E.A., Oko L., Edeen K., Berglund A., Wang J., Ito Y., Holmes K.V., Mason R.J. Innate immune response of human alveolar type II cells infected with severe acute respiratory syndrome-coronavirus. Am. J. Respir. Cell Mol. Biol. 2013;48:742–748. doi: 10.1165/rcmb.2012-0339OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ksiazek T.G., Erdman D., Goldsmith C.S., Zaki S.R., Peret T., Emery S., Tong S., Urbani C., Comer J.A., Lim W., et al. A Novel Coronavirus Associated with Severe Acute Respiratory Syndrome. N. Engl. J. Med. 2003;348:1953–1966. doi: 10.1056/NEJMoa030781. [DOI] [PubMed] [Google Scholar]
- 59.Johnson K.M., Lange J.V., Webb P.A., Murphy F.A. Isolation and partial characterization of a new virus causing acute hæmorrhagic fever in zaire. Lancet. 1977;309:569–571. doi: 10.1016/S0140-6736(77)92000-1. [DOI] [PubMed] [Google Scholar]
- 60.Bowen E.T., Lloyd G., Harris W.J., Platt G.S., Baskerville A., Vella E.E. Viral haemorrhagic fever in southern Sudan and northern Zaire. Preliminary studies on the etiological agent. Lancet. 1977;1:571–573. doi: 10.1016/S0140-6736(77)92001-3. [DOI] [PubMed] [Google Scholar]
- 61.Pattyn S., Groen G., Jacob W., Piot P., Courteille G. Isolation of marburg-like virus from a case of hæmorrhagic fever in zaire. Lancet. 1977;309:573–574. doi: 10.1016/S0140-6736(77)92002-5. [DOI] [PubMed] [Google Scholar]
- 62.Murray K., Selleck P., Hooper P., Hyatt A., Gould A., Gleeson L., Westbury H., Hiley L., Selvey L., Rodwell B. A morbillivirus that caused fatal disease in horses and humans. Science. 1995;268:94–97. doi: 10.1126/science.7701348. [DOI] [PubMed] [Google Scholar]
- 63.Chua K.B., Bellini W.J., Rota P.A., Harcourt B.H., Tamin A., Lam S.K., Ksiazek T.G., Rollin P.E., Zaki S.R., Shieh W.-J., et al. Nipah Virus: A Recently Emergent Deadly Paramyxovirus. Science. 2000;288:1432–1435. doi: 10.1126/science.288.5470.1432. [DOI] [PubMed] [Google Scholar]
- 64.Centers for Disease Control and Prevention Multistate outbreak of monkeypox—Illinois, Indiana, and Wisconsin, 2003. MMWR Morb. Mortal Wkly. Rep. 2003;52:537–540. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Not applicable.