Abstract
Nonsense-mediated mRNA decay (NMD), also called mRNA surveillance, is an important pathway used by all organisms that have been tested to degrade mRNAs that prematurely terminate translation and, as a consequence, eliminate the production of aberrant proteins that could be potentially harmful. In mammalian cells, NMD appears to involve splicing-dependent alterations to mRNA as well as ribosome-associated components of the translational apparatus. To date, human (h) Upf1 protein (p) (hUpf1p), a group 1 RNA helicase named after its Saccharomyces cerevisiae orthologue that functions in both translation termination and NMD, has been the only factor shown to be required for NMD in mammalian cells. Here, we describe human orthologues to S. cerevisiae Upf2p and S. cerevisiae Upf3p (Caenorhabditis elegans SMG-4) based on limited amino acid similarities. The existence of these orthologues provides evidence for a higher degree of evolutionary conservation of NMD than previously appreciated. Interestingly, human orthologues to S. cerevisiae Upf3p (C. elegans SMG-4) derive from two genes, one of which is X-linked and both of which generate multiple isoforms due to alternative pre-mRNA splicing. We demonstrate using immunoprecipitations of epitope-tagged proteins transiently produced in HeLa cells that hUpf2p interacts with hUpf1p, hUpf3p-X, and hUpf3p, and we define the domains required for the interactions. Furthermore, we find by using indirect immunofluorescence that hUpf1p is detected only in the cytoplasm, hUpf2p is detected primarily in the cytoplasm, and hUpf3p-X localizes primarily to nuclei. The finding that hUpf3p-X is a shuttling protein provides additional indication that NMD has both nuclear and cytoplasmic components.
The biogenesis of functionally mature mRNAs in mammalian cells is remarkably involved and inherently subject to inefficiencies and inaccuracies that result in the generation of abnormal translational reading frames. Mammalian mRNAs are transcribed initially as precursors, most of which contain multiple introns that must be removed by the process of pre-mRNA splicing. If transcription initiates incorrectly or an intron either fails to be removed or is removed using one or more abnormal splice sites, then product mRNA has the potential to harbor a premature termination codon (PTC) that could derive from an upstream reading frame, a retained intron, or a shift in the reading frame.
In order to cope with the generation of PTCs and their potential to result in deleterious proteins that function in new or dominant-negative ways, mammalian cells have evolved a pathway called nonsense-mediated mRNA decay (NMD) or mRNA surveillance (reviewed in references 20, 28, 30, 31, and 32). This pathway surveys all translated mRNAs, whether they be normal or defective, in order to degrade those that prematurely terminate translation more than 50 to 55 nucleotides (nt) upstream of the final exon-exon junction (7, 8, 41, 43, 44, 48, 49)—a feature of most PTCs but not most normal termination codons (34). These and other data indicate that NMD is mechanistically linked to nuclear pre-mRNA splicing.
Depending on the particular mRNA and its conditions of expression, NMD can take place in association with nuclei or after export to the cytoplasm. One unresolved issue of NMD pertains to the precise cellular site of nucleus-associated NMD that, like cytoplasmic NMD, requires a process that is experimentally indistinguishable from cytoplasmic translation. In theory, nucleus-associated NMD could take place either during mRNA transport from the nucleus to the cytoplasm and depend on cytoplasmic ribosomes or within the nucleoplasm and depend on nuclear ribosomes (8, 28, 44, 48, 49). The link between splicing and NMD exists for both nucleus-associated (7, 8, 44, 48, 49) and cytoplasmic (41) NMD. We have proposed that the link involves proteins that are deposited by the process of splicing at or near exon-exon junctions of product mRNA and remain bound to mRNA long enough to interact with translational factors. Recent studies using HeLa cell nuclear extracts and cross-linking in vitro have identified several proteins, including the nuclear matrix-associated splicing coactivator SRm160, that form a tight complex at or near exon-exon junctions as a direct consequence of splicing and remain associated with mRNA after its release from the spliceosome (26). Thus, evidence now exists that pre-mRNA splicing can influence mRNp structure. Another unresolved issue of NMD pertains to how the translational apparatus interacts with splicing-marked mRNA in a way that elicits NMD.
NMD typifies not only mammalian cells but all cells that have been examined. trans-acting factors known to be required for NMD are best understood for Saccharomyces cerevisiae and Caenorhabditis elegans, which are readily amenable to genetic analyses. Loss-of-function mutations affecting the S. cerevisiae Upf1 protein (p) (also known as Nam7p, Sal1p, Ifs2p, or Mof4p), Upf2p (also known as Nmd2p, Isf1p, or Sua1p), Upf3p (also known as Sua6p), or any one of SMG-1 through SMG-7 of C. elegans eliminate NMD without general effects on the decay of mRNAs lacking PTCs (4, 9, 17, 23, 24, 25, 35, 37). In yeast, polysome-associated mRNAs are substrates for NMD (50). Consistent with this, all three Upf proteins associate with ribosomes (3, 4) and Upf1p binds release factors (RFs) 1 and 3 to enhance translation termination and elicit NMD (11). Upf1p also forms a complex with Dcp2p (also known as Nmd1p [11]), a protein required for the mRNA decapping step of NMD (13). In fact, all three Upf proteins appear to function in translation termination and monitor translational fidelity since they interact (18, 19, 46), deleting any single UPF gene results in a nonsense suppression phenotype (9, 11, 25), and the mof4-1 allele of the UPF1 gene as well as an upf3-Δ strain demonstrate increased programmed −1 frameshifting (10, 39). The C. elegans orthologue to S. cerevisiae Upf1p is the phosphoprotein SMG-2 (35). SMG-4 appears to be the C. elegans orthologue to S. cerevisiae Upf3p and derives from alternatively spliced RNA (R. Aronoff, R. Baran, and J. Hodgkin, unpublished data). SMG-3 appears to be the C. elegans orthologue to S. cerevisiae Upf2p (S. Kuchma and P. Anderson, personal communication).
Until now, the only Upf-like or SMG-like factor identified for mammalian cells has been human (h) Upf1p (hUpf1p) (1, 36). hUpf1p is required for NMD in mammalian cells, as evidenced by the finding that a cysteine in place of an arginine at position 844 (R844C) within the RNA helicase domain has a dominant-negative effect on the pathway (42). Despite sequence and, by extrapolation, functional similarities among S. cerevisiae Upf1p, C. elegans SMG-2, and hUpf1p indicating that NMD evolved before most eukaryotes (1, 25, 35, 36), there may be significant differences among the three organisms in the factors that elicit NMD. First, yeast Upf1p has never been reported to be phosphorylated, in contrast to both SMG-2 (35) and hUpf1p (M. Pal, Y. Ishigaki, E. Nagy, and L. E. Maquat, unpublished data), although data demonstrating that epitope-tagged Upf1p can migrate as a doublet in acrylamide (4) suggest that it may be posttranslationally modified. Second, expression of either yeast Upf1p in SMG-2 mutant worms or hUpf1p in upf1 mutant yeast fails to restore NMD (35, 36). Third, even a hybrid hUpf1p flanked by the extreme N and C termini of yeast Upf1p, which is capable of binding RFs 1 and 3 and functioning in nonsense suppression in yeast, fails to function in NMD in yeast (11, 36). Finally, four of the seven SMG factors are without known orthologues in either yeast or humans.
Here, we identify and describe human orthologues to S. cerevisae Upf2p and S. cerevisae Upf3p (C. elegans SMG-4). Using comparative genomics and rapid amplification of cDNA ends (RACE), the results of cDNA analyses indicate that there is a single human orthologue to S. cerevisae Upf2p, which we have called hUpf2p. In contrast, there are multiple human orthologues to S. cerevisae Upf3p (C. elegans SMG-4) that derive from two separate genes, one of which is X-linked and both of which produce alternatively spliced RNAs. The full-length versions are called hUpf3p-X and hUpf3p. Immunoprecipitations of epitope-tagged proteins transiently produced in HeLa cells demonstrate that hUpf1p, hUpf2p, hUpf3p-X, and hUpf3p copurify, providing evidence for a role in NMD. Indirect immunofluorescence assays of protein localization in HeLa cells reveal that hUpf1p is detected exclusively in the cytoplasm, hUpf2p is detected primarily in the cytoplasm, and hUpf3p-X is mostly nuclear. Results of protein shuttling in interspecies heterokaryons indicate that hUpf3p-X shuttles rapidly between nuclei and the cytoplasm. Apparent similarities and differences of these proteins in S. cerevisiae, C. elegans, and humans are discussed.
MATERIALS AND METHODS
Isolation and sequence analysis of full-length cDNAs.
Two expressed sequence tags (ESTs; accession no. AA 8120190 and AA 447286) that appeared to encode different portions of a human orthologue to S. cerevisiae Upf2p (17) and one EST (accession no. NA 442937) and genomic sequence (accession no. DJ 327A19) that appeared to encode different human orthologues of S. cerevisiae Upf3p (C. elegans SMG-4) were identified by using the BLAST algorithm (http://www.ncbi.nlm.nih.gov/BLAST/) and dbest (nonredundant GenBank plus EMBL plus DDJB plus PDB EST) database. Primers were generated from these sequences and used to amplify a HeLa-cell Marathon cDNA library (Clontech) by using the Advantage cDNA polymerase mix (Clontech) and RACE-PCR. In order to ensure that the resulting cDNAs harbored an unmutagenized coding region, subsequent studies were confined to those cDNAs that harbored a coding region identical to (i) at least two out of three independently amplified coding regions and (ii) available database sequences.
Epitope-tagged expression plasmids.
pCI-Neo-FLAG-hUPF1 WT, which encodes wild-type (WT) hUpf1p, has been described (35a).
To generate pCI-Neo-hUPF3-X WT, the entire coding region of hUPF3-X cDNA together with 19 nt of 5′ untranslated region (UTR) and the two consecutive termination codons of the 3′ UTR were PCR amplified as two fragments that harbored an overlapping EcoRI site by using the Advantage cDNA polymerase mix (Clontech) and HeLa-cell Marathon-Ready cDNA (Clontech). Primers for the 5′ fragment of cDNA consisted of 5′CCCCGCTCGAGTTCAGCGGGGGACGTAGCCATGAAGGAAGAGAAGGAGCACAGGCC 3′ (sense; underlined nucleotides constitute the XhoI site) and 5′ CGCGGATCCTTATCAATCCTTTAATTTGTCCCTTTCTGG 3′ (no. 4 antisense). Primers for the 3′ fragment consisted of 5′ AGCTAAAGAAGATAGACAGAATTCCAG 3′ (sense) and 5′ TTTTCCTTTTGCGGCCGCTTATCACTCCTCTCCTCCTTCTTTTCTATGGC 3′ (antisense; underlined nucleotides constitute the NotI site). The 5′ fragment was cleaved with XhoI and EcoRI, the 3′ fragment was cleaved with EcoRI and NotI, and both were inserted simultaneously into the XhoI and NotI sites of pCI-Neo DNA. All PCR-generated clones were sequenced in their entirety. Regions harboring one or more mutated nucleotides were replaced with regions harboring the corresponding WT nucleotides by subcloning. Sequencing was used to confirm that the final construct was WT.
To generate pCI-Neo-hUPF2 WT, the entire coding region of hUPF2 cDNA together with 19 nt of 5′ UTR and the 3′ UTR termination codon were similarly inserted into pCI-Neo after PCR amplification using primers 5′ CCCCGCTCGAGGCTGATTGTCCTGGGTCACATAATGCCAG 3′ (sense) and 5′ CAAATAACATAGTCTTTCACTTCTTGGTCC 3′ (no. 9 antisense) to generate the 5′ fragment, primers 5′ CCCCGCTCGAGGCATGCACCCTGCTGGAGACATGTGGACCG 3′ (sense) and 5′ GGGAAGATCTTAGCGGCCGCTCATTACCTCAGATTCTCTTCATCGG 3′ (antisense) to generate the middle fragment, and primers 5′ GGCAGGTACAAGACTTGGAACGAG 3′ (sense) and 5′ ACTAAAGGGAAGCGGCCGCTCAACGTCTCCTCCCACCAGTC 3′ (antisense) to generate the 3′ fragment.
The resulting expression vectors were modified so as to produce amino-terminal-tagged HA-hUpf3p-X and T7-hUpf2p. Sequences encoding the hemagglutinin (HA) epitope were inserted as an XhoI- and EcoRI-cleaved PCR product that was synthesized using primers 5′ CCCCCGCTCGAGTTCAGCGGGGGACGTAGCCATGTACCCATACGACGTAAAAGACTACGCTAAGGAAGAGAAGGAGCACAGGCC 3′ (HA sense) and no. 4 antisense. Sequences encoding the T7 epitope were inserted as an XhoI- and EcoRV-cleaved PCR product that was synthesized using primers 5′ CCCCGACTCGAGGCTGATTGTCCTGGGTCACATAATGGCTAGCATGACTGGTGGACAGCAAATGGGTCCAGCTGAGCGTAAAAAGCCAGC 3′ (T7 sense) and no. 9 antisense.
pCI-Neo-hUPF3-X NES 3A, in which amino acids 54 to 58 were changed from VVIRRL to AVARRA, and pCI-Neo-hUPF3-X YVF→DVD, in which amino acids 117 to 119 were changed from YVF to DVD, were generated using overlap-extension PCR (21). For NES 3A, pCI-Neo-hUPF3-X WT was amplified using overlapping primers 5′ AGCAAGGCGGTAGCTCGAAGAGCACCTCCCACTTTGACCAAGGAGCAGCTTCAGG 3′ (sense; in which mutagenic nucleotides are italicized) and 5′ GGGAGGTGCTCTTCGAGCTACCGCCTTGCTCAGCGCTTCTTTCTTCTCCTTGTTGCG 3′ (antisense) and, as flanking primers, HA sense and no. 4 antisense. The resulting PCR product was inserted into the XhoI and EcoRI sites of pCI-Neo-hUPF3-X WT. For YVF→DVD, pCI-Neo-hUPF3-X WT was amplified using overlapping primers 5′ GATGGTGATGTAGACCTTGACAATAAAGGTCAGG 3′ (sense) and 5′ GTCAAGGTCTACATCACCATCAAAGCGATCCCTGAACAA 3′ (antisense) and the HA sense and no. 4 antisense flanking primers. pCI-Neo-hUPF3-X Δ(30-255) was generated from pCI-Neo-hUPF-X WT by digestion with PpuMI and EcoRI, Klenow filling the resulting 5′ overhangs, and circularization so as to create an in-frame deletion. pCI-Neo-hUPF3-X Δ(257-483) was similarly generated except PpuMI was omitted so that a nonsense codon was created at the filled EcoRI site.
To generate pCI-Neo-hUPF3 and pCI-Neo-hUPF3Δ, the entire coding region plus the termination codon of hUPF3 or hUPF3Δ cDNA was PCR amplified so as to contain 6 nt of 5′ UTR that promote optimal translation initiation efficiency (reviewed in reference 22) and a herpes simplex virus (HSV) epitope tag (Novagen) using the Advantage cDNA polymerase mix and HeLa-cell Marathon-Ready cDNA. Primers consisted of 5′ CCCCGCTCGAGGCCACCATGCAGCCTGAACTCGCTCCAGAGGATCCGGAAGATCTGTCGGCCCTAGAAGTGCAGTTCCACC 3′ (sense) and 5′ TTTTCCTTTTGCGGCCGCTCACTCTGCCTCTTCCCTCTTCTCAGGACC 3′ (antisense). The resulting PCR product was cleaved with XhoI and NotI and inserted into the corresponding sites of pCI-Neo.
pCI-Neo-hUPF2 Δ(94-133) was generated by overlap-extension PCR using overlapping primers 5′ TCAAAGAAAAAAGAAGAGGAAGAAGCTTGGGAACGAGACGACTTAAG 3′ (sense) and 5′ CTTAAATGATGTCGTTCCGAAGCTTCTTCCTCTTCTTTTTTCTTTGATTC 3′ (antisense). pCI-Neo-hUPF2 Δ(526-722) was generated by digestion with XbaI and Bsu36I followed by Klenow filling in the resulting 5′ overhangs and circularization so as to create an in-frame deletion. pCI-Neo-hUpf2p Δ(711-928) was generated by digestion with AflIII followed by circularization to create an in-frame deletion. pCI-Neo-hUPF2 Δ(709-1272) and pCI-Neo-hUPF2 Δ(787-1272) were derived by digestion with, respectively, BstEII and EcoRI or AflIII and EcoRI, Klenow filling in the resulting 5′ overhangs, and circularization to create a nonsense codon. All constructs were sequenced to ensure their composition.
HeLa-CCL2 cell transfections.
HeLa-CCL2 cells were propagated in minimal essential medium (MEM) containing 10% fetal bovine serum (FBS). One day prior to transfection, cells (∼3 × 105 to 4 × 105 per 60-mm-diameter dish) were cultured in antibiotic-free medium containing 10% FBS and, after reaching 80 or 100% confluency, were transfected using Lipofectamine PLUS Reagent or Lipofectamine 2000 (Life Technologies, Inc.), respectively, by following the manufacturer's directions.
RNA purification, Northern blot analysis, and reverse transcriptase (RT) PCR.
Total-cell RNA was isolated using Trizol (Life Technologies, Inc.). Poly(A)+ RNA was generated for Northern blot analysis (49) using the mRNA Isolation Kit (Dynal, Inc.). Uniformly 32P-labeled probes were synthesized from pCI-Neo-hUPF2, pCI-Neo-hUPF3-X, and pCI-Neo-hUPF3 using the Prime-a Gene kit (Promega) after cleavage with XhoI and NotI, which cleave to either side of each cDNA insert.
Total RNA Panels III and IV (2.5 μg; Clontech) or total HeLa-cell RNA (2.5 μg) were reverse transcribed for 1 h at 37°C using 500 ng of random hexamer (Promega) and Superscript II RT (200 U; Life Technologies) by following the Life Technologies protocol. The resulting cDNA was amplified using one-tenth of the RT reaction mixture, 0.2 mM concentrations of each deoxynucleotide, 5 μCi of [α-32P]dATP (3,000 Ci/mmol; Amersham), 20 pmol (0.4 μM) of each primer, and 5 U of Taq DNA polymerase (Life Technologies). To amplify hUPF3-X cDNA, the primers consisted of 5′ AGCACACGACTACTTCGAGTTCTTCG 3′ (sense) and 5′ CGCGGATCTTATCACAGTGTCTCTGGAGTAGATGTCATTTTCTC 3′ (antisense). To amplify hUPF3 cDNA, the primers consisted of 5′ CGCGGATCCTCATTACAGAGTCTCAGGGTTGGCACTGGTCTTCTC 3′ (sense) and 5′ TGCCAGAGCATACATCAACTTTAAAAACCAAGAGG 3′ (antisense). Primers for the amplification of G3PDH (glyceraldehyde-3-phosphate dehydrogenase) cDNA were commercially available (Clontech). For every PCR, each cycle consisted of denaturation for 10 s at 95°C, annealing for 1 min at 60°C, and extension for 1 min at 72°C for a total of 23 cycles. One-tenth of each PCR mixture was electrophoresed in a 10% polyacrylamide gel, and RT-PCR products were quantitated by PhosphorImaging (Molecular Dynamics).
Protein purification, immunoprecipitations, and Western blot analyses.
HeLa cells (one 60-mm-diameter dish) that had been mock transfected or transiently transfected with one or more epitope-tagged expression plasmids were rinsed with ice-cold phosphate-buffered saline (pH 7.4) and subsequently incubated in 600 μl of lysis buffer (16) (150 mM NaCl, 50 mM Tris-HCl [pH 7.4], and 0.4% NP-40 [Boehringer]) for 30 min at 4°C. The efficiency of tagged protein production was analyzed by Western blotting using either 1 μg of anti-FLAG (α-FLAG) antibody (M5; Sigma)/ml; 1 μg of α-T7 antibody (Novagen)/ml, 0.5 μg of α-HA antibody (high-affinity rat antibody; Boehringer)/ml, or 1 μg of α-HSV antibody (Novagen)/ml. Immunoprecipitations were performed at 4°C using 200 μl of lysate and 10 μg of α-T7 antibody (Novagen), 10 μg of α-HA antibody, 10 μg of α-HSV antibody, or 20 μg of purified α-hUpf1p antibody (35a). After 3 h, 30 μl of protein A-Sepharose (Boehringer; for α-T7 or α-HSV antibodies) or protein G-Sepharose (Boehringer; for α-FLAG and α-HA antibodies) that had been washed with lysis buffer were added for 2 h. Immunoprecipitates were then collected by centrifugation, pellets were washed twice with 1 ml of lysis buffer without NP-40 to eliminate unbound proteins, and bound proteins were analyzed by Western blotting using antibody to the appropriate epitope tag.
Immunofluorescence microscopy.
HeLa cells were grown in MEM-α supplemented with 10% FBS, and 5 × 105 to 7 × 105 cells were transiently transfected with 1 to 2 μg or 10 μg of pCI-Neo-FLAG-hUPF1, pCI-Neo-T7-hUPF2, or pCI-Neo-HA-hUpf3-X using either Lipofectamine PLUS Reagent or Lipofectamine 2000. After either 24 or 40 h, the cells were seeded on a 60-mm-diameter dish holding three coverslips and were fixed (5). Epitope-tagged proteins were localized by indirect immunofluorescence using α-FLAG, α-T7, or α-HA antibody and rhodamine-conjugated secondary antibody (Sigma) raised against either mouse (for α-FLAG or α-T7 antibody) or rat (for α-HA antibody). For heterokaryon analyses, HeLa cells were transfected and seeded on coverslips as described above. After 24 h, the medium was replaced, and the cells were incubated 4 h later in the presence of 1 × 106 to 2 × 106 mouse NIH 3T3 cells and 50 μg of cycloheximide/ml. After 3 h, the concentration of cycloheximide was increased to 100 μg/ml. After 30 min, the cells were fused (37) using polyethylene glycol 1500 (Roche Molecular Biochemicals), washed extensively with phosphate-buffered saline, and incubated for an additional 1 to 2 h in the presence of 100 μg of cycloheximide/ml. Epitope-tagged proteins were localized as described above. Cells were simultaneously stained with 5 μg of Hoechst 33258 (Sigma)/ml in order to distinguish mouse and human nuclei.
Nucleotide sequence accession numbers.
GenBank accession numbers for nucleotide sequences are as follows: for hUPF2 cDNA, AF318574; hUPF3 cDNA, AF318575; and hUPF3X cDNA, AF318576.
RESULTS
Evidence for human orthologues to S. cerevisiae Upf2p and S. cerevisiae Upf3p (C. elegans SMG-4).
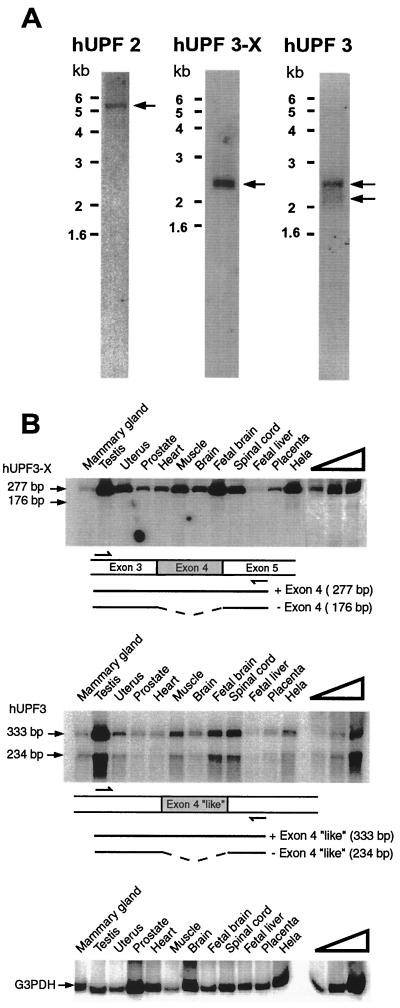
Two ESTs (accession no. AA 8120190 and AA 447286) that appeared to encode portions of hUpf2p were obtained by comparing the coding potential in all frames and both directions of cDNA sequences in the dbest database to S. cerevisiae Upf2p (17, 18) by using the BLAST algorithm. Primers designed from each EST, RACE-PCR product, and HeLa-cell Marathon-Ready cDNA (Clontech) were then used to obtain sequences from hUPF2 5′ and 3′ UTRs and the complete coding region. The derived cDNA harbored a total of 75 nt of 5′ UTR, 3,816 nt of coding region, and 1,810 nt of 3′ UTR up to the site of polyadenylation (Fig. 1; data not shown). Consistent with the derived sum of 5,701 nt, the analysis of HeLa-cell poly(A)+ RNA by Northern blotting using cDNA sequences from the coding region indicated that hUPF2 mRNA is ∼5.4 kb (Fig. 2A).
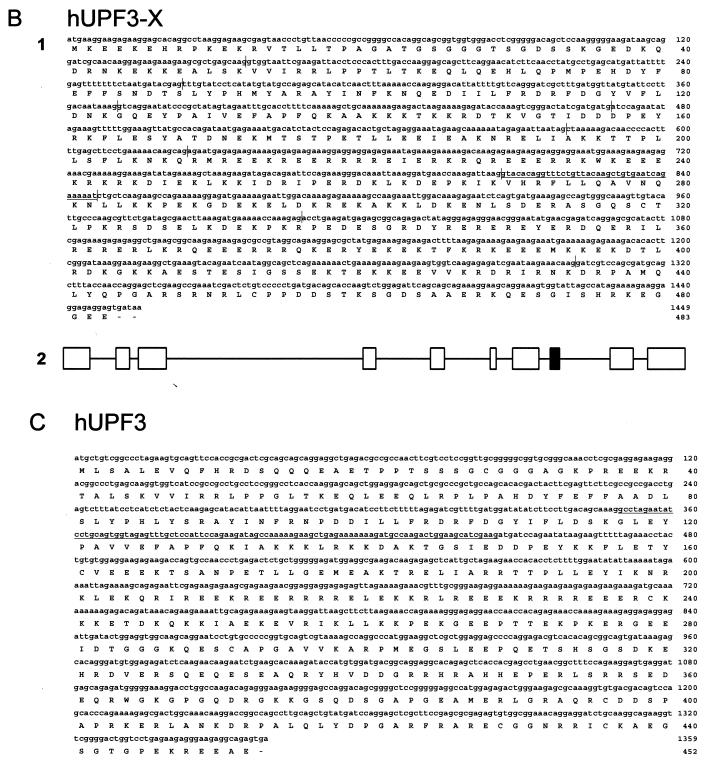
FIG. 1.
Characterization of hUPF2 (A), hUPF3-X (B1), and hUPF3 (C) cDNAs. Nucleotide and deduced amino acid sequences of hUPF2, hUPF3-X, and hUPF3 cDNAs are numbered at the right. (B1) Vertical lines in the hUPF3-X nucleotide sequence correspond to exon-exon junctions deduced from the X-chromosome sequence of PAC clone DJ 327A19. Underlined sequences correspond to an exon absent in the fibroblast-derived EST AA071043, suggesting that it is alternatively spliced. (B2) Exon-intron organization of the hUPF3-X gene. Introns are represented at one-tenth the scale of the exons. The black box corresponds to the exon absent from EST AA071043. (C) Underlined sequences in the hUPF3 nucleotide sequence correspond to the alternatively spliced exon evident from the analysis of HeLa cell RNA (see Results).
FIG. 2.
Analysis of hUPF2, hUPF3-X, and hUPF3 transcripts. (A) Poly(A)+ RNA from 75 μg of total HeLa-cell RNA was subject to Northern blotting and probed with coding region sequences from hUPF2, hUPF3-X, or hUPF3 cDNAs. hUPF2 mRNA migrates at ∼5.4 kb, hUPF3-X mRNA migrates at ∼2.4 kb, and hUPF3 mRNAs migrate at ∼2.1 and ∼2.4 kb. (B) cDNA was generated using total RNA (2.5 μg) from either the specified human tissue (Clontech) or HeLa cells. hUPF3-X, hUPF3, and, as a control, G3PDH cDNA were PCR amplified. In order to assay for exon skipping, hUPF3-X cDNA was amplified from exon 3 to exon 5, and hUPF3 cDNA was amplified from sequences corresponding to hUPF3-X exon 3 to sequences corresponding to hUPF3-X exon 5. Partial arrows specify the positions of PCR primer annealing. The right-most four lanes contain twofold (hUPF3-X) or threefold (hUPF3 and G3PDH) serial dilutions of HeLa-cell RNA in order to demonstrate a linear relationship between the amounts of input cDNA and RT-PCR products. Results are representative of two independently performed experiments.
hUPF2 cDNA encodes a 1,272-amino-acid protein having a predicted molecular mass of 148 kDa and a PI of 5.5. Relative to Upf2p from S. cerevisiae and Saccharomyces pombe, the latter of which was deduced from a single intron-less genomic sequence (accession no. Z98974) available in the nr (nonredundant GenBank plus EMBL plus DDJB plus PDB without EST) database, hUpf2p is, respectively, 22 and 21% identical and 39 and 35% similar (Fig. 3). hUpf2p harbors two distinctly large regions that are missing from S. cerevisiae Upf2p. One consists of 135 amino acids at the extreme N terminus, a part of which (amino acids 94 to 133; Fig. 3) contains sequences similar to those of one of the two domains in S. cerevisiae Upf2p required for binding to Upf1p (18, 19; see below). These N-terminal amino acids are also notable for their multiple acidic and basic repeats, which, the PROSITE algorithm revealed, contain numerous regions similar to nuclear localization sequences (NLS). The second region of hUpf2p that is missing from the S. cerevisiae orthologue consists of amino acids 502 to 567 and is localized in the middle of the protein. hUpf2p amino acids 164 to 179 are the most conserved among the three species, and the corresponding sequences in S. cerevisiae Upf2p have been proposed to constitute part of an NLS (17). Also notable is the absence of approximately 30 amino acids from the C terminus of hUpf2p that correspond to approximately half of the acidic repeat of S. cerevisiae Upf2p. No functional property or role has been ascribed to acidic repeats, which are shared with numerous nucleolar proteins, including nucleolin (14). Another potentially important feature of hUpf2p is the FIGEL motif (amino acids 659 to 663) and surrounding amino acids extending from positions 657 to 713, which are 32 and 30% identical and 55 and 53% similar to amino acids contained in the domain necessary for the binding of human eIF4A to eIF4GI, eIF4GII, and related factors, respectively (27). Recently, the corresponding region in S. cerevisiae Upf2p(Nmd2p), eIF4G, and cap binding protein 80 (CBP80) has been named NIC, and the NIC domain of Upf2p has been proposed to have a regulatory role by interacting with the translation initiation complex similarly to eIF4G (2).
FIG. 3.
Comparison of Upf2 proteins from H. sapiens, S. pombe, and S. cerevisiae. Amino acid sequences are numbered at the right. White-letter amino acids in black or grey boxes correspond to S. pombe and S. cerevisiae amino acids that are, respectively, identical or similar to those of H. sapiens. Black-letter amino acids in grey boxes correspond only to those that are identical or similar between S. cerevisiae and S. pombe (rather than to those that are identical or similar to those of H. sapiens). Amino acids underlined with a thin or a thick line represent S. cerevisiae sequences known to interact with S. cerevisiae Upf3p or Upf1p, respectively (18, 19). Notably, hUpf2p sequences that constitute the two putative hUpf1p binding sites derive from the N terminus (broken-line box; amino acids 94 to 133) and the C terminus (thick underline; amino acids 1085 to 1124 and 1167 to 1194). hUpf2p sequences that constitute the putative hUpf3p binding site are specified by the thin line, which signifies the major binding determinant, and the broken line, which signifies sequences that contribute to binding. Amino acids underlined with a double line correspond to the putative NLS in S. cerevisiae (17).
Using similar methodologies and the nr database, an X-chromosome-derived genomic sequence (accession no. DJ 327A19) that potentially encodes portions of the human orthologue to S. cerevisiae Upf3p (C. elegans SMG-4) (25; R. Aronoff, R. Baran, and J. Hodgkin, unpublished data) was obtained. Considering that another hUPF3 gene also exists (see below), the X-linked DNA was designated hUPF3-X. Primers designed from the genomic hUPF3-X sequence, RACE-PCR product, and HeLa-cell Marathon-Ready cDNA were used to obtain a more complete hUPF3-X cDNA sequence, including 20 nt of 5′ UTR, all 1,449 nt of the coding region, and 850 nt of 3′ UTR up to the site of polyadenylation. The cDNA encodes a 483-amino-acid protein having a predicted molecular mass of 58 kDa and a PI of 9.5. Consistent with the derived sum of 2,319 nt, hUPF3-X mRNA is ∼2.4 kb (Fig. 2). The hUPF3-X gene consists of 11 exons spanning 18.9 kbp (Fig. 1). The existence of an hUPF3-X EST (accession no. AA 071043) from fibroblasts lacking exon 8 suggests that hUPF3-X pre-mRNA could be alternatively spliced to generate two distinct mRNAs: one containing exon 8 and the other lacking exon 8 and encoding a protein that lacks amino acids 270 to 282. In fact, the analysis of RT-PCR products that extend from hUPF3-X exon 7 to exon 9 indicate that exon 8 is alternatively spliced in HeLa-cell RNA so as to generate approximately equal amounts of exon 8-containing and exon 8-lacking RNA (data not shown). As would be expected, the difference of 39 nt was not detectable by Northern blot analysis (Fig. 2A).
Another EST (accession no. NA 442937) appeared to derive from a gene different from the hUpf3p-X gene. RACE-PCR product and HeLa-cell Marathon-Ready cDNA were used to obtain two hUPF3 cDNAs that harbored 2,296 and 2,197 nt upstream of the site of polyadenylation. The sole difference between the two cDNAs was the presence or absence of 99 bp, indicating that the cDNAs derived from alternatively spliced versions of a common pre-mRNA. Notably, amino acids 117 to 149 encoded by these 99 bp are similar to all of those encoded by exon 4 of the hUPF3-X gene. The isoform containing these 33 amino acids was designated hUpf3p, and the isoform lacking these 33 amino acids was designated hUpf3pΔ. Consistent with the existence of hUPF3 transcript alternative splicing, the analysis of HeLa-cell poly(A)+ RNA by Northern blotting using sequences from the coding region of the 2,296-bp cDNA revealed ∼2.1- and ∼2.4-kb RNA sequences (Fig. 2A). Since the difference in size between the two RNAs is larger than the alternatively spliced exon, the difference may reflect additional differences in, e.g., the transcription start site or poly(A) tail length. Currently, there is no evidence for additional alternative splicing of hUPF3 transcripts. Proof for alternative splicing of the 99-nt exon of hUPF3 RNA that resembles exon 4 of hUPF3-X transcripts was obtained by analyzing HeLa cell RNA by using RT-PCR (Fig. 2B; sequencing data not shown). In fact, data obtained using RT-PCR indicate that this exon is alternatively spliced in every human tissue examined, albeit with different efficiencies, depending on the tissue (Fig. 2B). As would be predicted from the Northern blot analysis of hUPF3-X RNA (Fig. 2A), and consistent with the sequence analysis of hUPF3-X cDNA, the skipping of hUPF3-X exon 4 was barely detected in HeLa cells (Fig. 2B). Likewise, hUPF3-X exon 4 skipping was either barely detected or undetected in each of the human tissues that was examined (Fig. 2B). The failure to detect hUPF3 sequences similar to the alternatively spliced exon 8 of hUPF3-X by either cDNA sequencing or searching available EST databases (Fig. 4) exemplifies an additional difference in the alternative splicing of hUPF3 and hUPF3-X transcripts.
FIG. 4.
Comparison of Upf3-X and hUpf3 proteins from H. sapiens, C. elegans, S. pombe, and S. cerevisiae. (A) Amino acid alignment of hUpf3p-X and hUpf3p according to the coding potential of hUPF3-X gene exons. Amino acid sequences are numbered at the left. White-letter amino acids in black or grey boxes correspond to amino acids that are, respectively, identical or similar between hUpf3p-X and hUpf3p. STOP, end of the coding region. Italicized amino acids specify alternatively spliced exons. (B) Amino acid alignment is provided only for regions that show significant similarity. White-letter amino acids in black or grey boxes correspond to amino acids that are, respectively, identical and similar to those of H. sapiens, and black-letter amino acids in grey boxes correspond only to amino acids that are identical or similar among the three other species. Amino acids underlined with a thin line represent S. cerevisiae sequences shown be required for the interaction with S. cerevisiae Ufp2p (18). The region between arrowheads specifies the S. cerevisiae NES that spans amino acids 88 to 97 (40). (C) Regions corresponding to putative NES, NLS, or acidic-basic domains are specified with black, grey, or lined boxes, respectively. The S. cerevisiae Upf2p interacting domain is underlined, and the broken-line box sets off the region shown in panel B.
hUPF3 cDNA encodes a 452-amino-acid protein having an estimated molecular mass of 52 kDa and a PI of 8.9, while hUPF3Δ cDNA encodes a 420-amino-acid protein having an estimated molecular mass of 49 kDa and a PI of 8.6. Identity and similarity between hUpf3p and hUpf3p-X are 42 and 60%, respectively. N-terminal amino acids 38 to 236 are the most conserved and manifest 86% similarity. C-terminal amino acids 202 to 453 are considerably more divergent even though some sequences, e.g., those that could comprise one of several NLS, appear to be conserved. Remarkably, the C termini of both proteins are rich in acidic amino acids (25% for both) and basic amino acids (27% for hUpf3p and 31% for hUpf3-X).
A search of the nr database uncovered the UPF3 gene of S. pombe (YD33), which consists of three exons and two intervening introns (data not shown). It was not possible to align the entire amino acid sequence of hUpf3p-X and hUpf3p to the sequence of Upf3p(SMG-4) of C. elegans, S. pombe, or S. cerevisiae due to the considerable degree of divergence at the C termini. However, conserved domains, some of which have been shown to be functional for S. cerevisiae Upf3p, do exist in hUpf3p-X and hUpf3p (Fig. 4) and include a nuclear export signal (NES; 40) in addition to sequences required for an interaction with Upf2p (19). Using the PROSITE algorithm, other motifs common to hUpf3p-X, hUpf3p, and putative orthologues in other species were found to consist of one or more putative NLS in all but the S. pombe orthologue (Fig. 4). Also, an acidic-basic region was shared by the N termini of hUpf2p, hUpf3p, and C. elegans SMG-4 but not S. cerevisiae Upf3p (Fig. 4). As was found for hUpf2p, however, a comparison of hUpf3p-X or hUpf3 and its orthologues in other species reveals that the relative positions of comparable domains can vary.
By using the two-hybrid analysis to assay genetically for interactions between full-length and deletion-bearing S. cerevisiae proteins, amino acids 1 to 181 of Upf1p and 947 to 1061 of Upf2p have been shown to be required for the Upf1p-Upf2p interaction (17, 18, 19, 45), and amino acids 564 to 933 of Upf2p and 78 to 278 of Upf3p have been shown to be required for the Upf2p-Upf3p interaction (19). Notably, because two-hybrid analysis is performed in the presence of S. cerevisiae proteins, either interaction may be direct or involve bridging by one or more cellular proteins. Sequence comparisons of yeast and human orthologues indicate that all Upf protein interactions could be conserved in mammalian cells.
Mutations located towards the C-terminal region of hUpf2p and towards the N-terminal region of hUpf3p-X inhibit the hUpf2p–hUpf3p-X interaction.
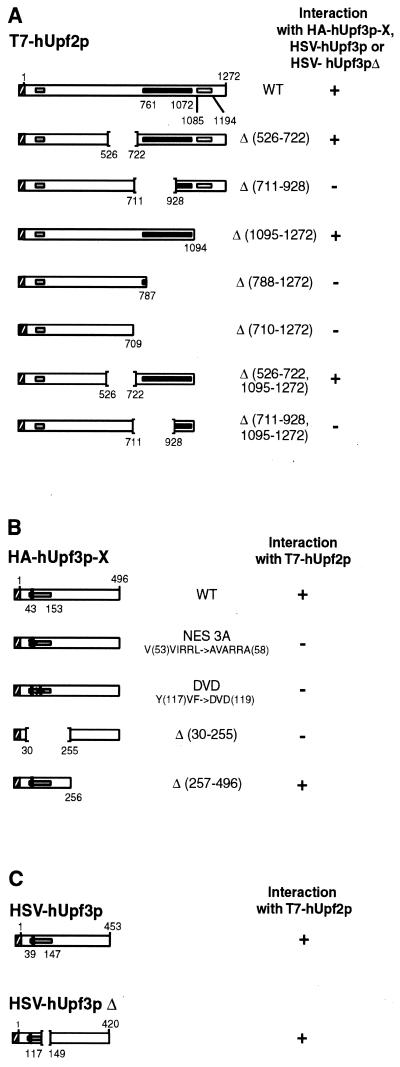
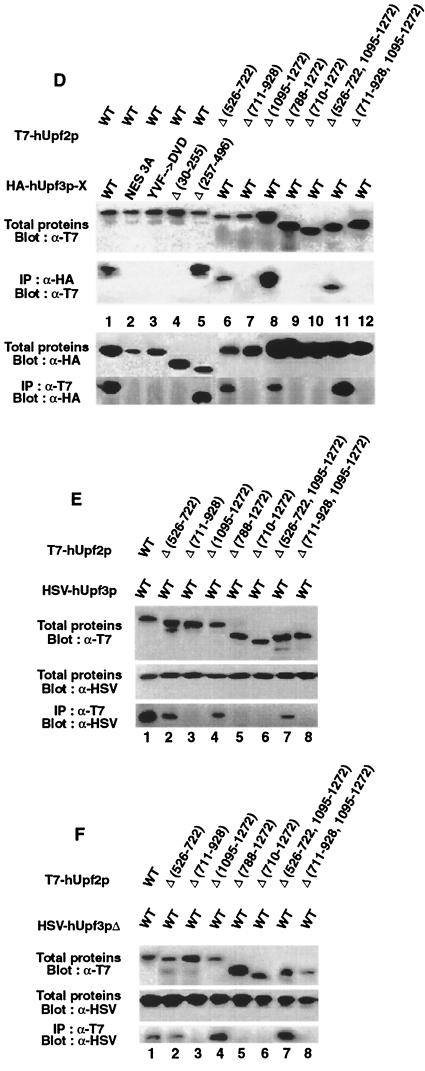
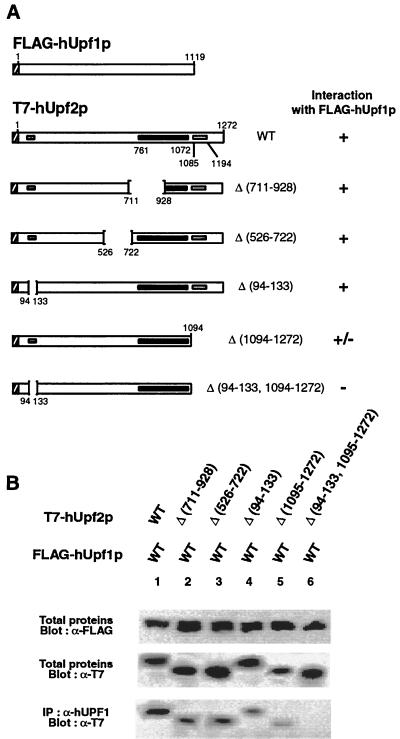
In order to explore the possibility that the human orthologues interact, WT hUpf2p and WT hUpf3p-X were transiently coproduced in HeLa cells from cDNA expression vectors as T7- and HA-tagged proteins, respectively (Fig. 5A and B). Western blot analysis of total protein demonstrated that each protein was expressed (Fig. 5D, lane 1). The findings that T7-hUpf2p was immunoprecipitated with α-HA antibody and HA–hUpf3p-X was immunoprecipitated with α-T7 antibody (Fig. 5D, lane 1) indicated that the two proteins interact either directly or indirectly. In order to define sequences within each protein required for the interaction, proteins harboring a deletion were produced and assayed. Results for T7-hUpf2p demonstrated that amino acids 526 to 722 or 1095 to 1272 could be deleted without consequence to the interaction with HA–hUpf3p-X, whereas deletion of amino acids 711 to 928 eliminated the interaction (Fig. 5D, lanes 6 to 8, 11, and 12). These results are consistent with predictions made based on the degree of conservation between yeast and human proteins, considering that hUpf2p amino acids 761 to 1072 correspond to the domain of S. cerevisiae Upf2p that interacts with S. cerevisiae hUpf3p (Fig. 3). Results for HA–hUpf3p-X demonstrated that mutation of either the putative NES (NES 3A; amino acids 53 to 58) or amino acids 117-119 (YVF→DVD) or deletion of amino acids 30 to 255 eliminated the interaction with T7-hUpf2p, whereas deletion of amino acids 257 to 483 was of no consequence to the interaction (Fig. 5D, lanes 2 to 5).
FIG. 5.
Characterization of the interaction between hUpf2p and hUpf3p-X or hUpf3p. (A) Diagram of WT and mutated T7-hUpf2p. Striped, black, and grey boxes specify, respectively, the T7 epitope tag, putative hUpf3p binding site, and putative hUpf1p binding sites. Δ, amino acid deletion. (B) Diagram of WT and mutated HA–hUpf3p-X. NES, putative NES, the counterpart of which has function in S. cerevisiae Upf2p (40). Striped, black, and grey boxes specify, respectively, the HA epitope tag, putative NES, and putative hUpf2p binding site. (C) Diagram of WT HSV-hUpf3p and WT HSV-hUpf3pΔ. Striped, black, and grey boxes specify, respectively, the HSV epitope tag, putative NES, and putative hUpf2p binding site. (D) Total proteins (10 μl of lysate) from 104 HeLa cells that had been transiently transfected with the specified T7-hUPF2 and HA–hUPF3-X expression vectors were subjected to Western blot analysis using α-T7 or α-HA antibody either before or after immunoprecipitation (IP) with α-T7 or α-HA antibody as specified. (E and F) Total proteins (10 μl of lysate) from 104 HeLa cells that had been transiently transfected with the specified T7-hUPF2 and HSV-hUPF3 or HSV-hUPF3Δ expression vectors were subjected to Western blot analysis using α-T7 or α-HSV antibody either before or after immunoprecipitation with α-T7 antibody as specified. Results typify three independently performed experiments that, taken as a whole, rule out the possibility that the absence of an interaction is attributable to a low expression level of any particular epitope-tagged protein.
Mutations located towards the C-terminal region of hUpf2p also inhibit the interaction with hUpf3p and hUpf3pΔ.
In order to determine if hUpf2p also interacts with hUpf3p or hUpf3pΔ, an isoform hUpf3p generated by alternative splicing, T7-tagged WT hUpf2p, and HSV-tagged hUpf3p or hUpf3Δ were transiently coproduced in HeLa cells (Fig. 5E and F). A priori, while an interaction between hUpf2p and hUpf3p was predicted, an interaction between hUpf2p and hUpf3pΔ was not predicted given that the 33 codons removed by alternative splicing encode part of the putative Upf2p interaction domain (Fig. 5C). The results of immunoprecipitations with α-T7 antibody followed by Western blotting with α-HSV antibody demonstrated that T7-hUpf2p interacted with both HSV-hUpf3p and HSV-hUpf3pΔ (Fig. 5E and F, lanes 1). The finding that T7-hUpf2p interacted with HSV-hUpf3pΔ indicates that hUpf3p amino acids 117 to 149 are not required for the interaction. This finding is compatible with the data indicating that deletion of amino acids 151 to 204 of S. cerevisiae Upf3p eliminates the interaction with Upf2p (19) provided that elimination does not depend on the deletion of amino acids 175 to 205, which correspond to hUpf2p amino acids 117 to 147 (Fig. 4). Deletion of hUpf2p amino acids 711 to 928, 788 to 1272, 710 to 1272, or 711 to 928 together with 1095 to 1272, which were shown to preclude the interaction with hUpf3p-X (Fig. 5D, lanes 7, 9, 10, and 12), also precluded an interaction with hUpf3p and hUpf3pΔ (Fig. 5E and F, lanes 3, 5, 6, and 8). However, deletion of hUpf2p amino acids 526 to 722 or amino acids 1095 to 1272 did not preclude the interaction with hUpf3p and hUpf3pΔ (compare Fig. 5D, lanes 6, 8, and 11, and Fig. 5E and F, lanes 2, 4, and 7), in agreement with results obtained for the interaction with hUpf3p-X.
Evidence that hUpf1p interacts with hUpf2p.
Considering that the interaction between Upf2p and Upf3p is conserved between S. cerevisiae and Homo sapiens, it is reasonable to think that the same would be true of the interaction between Upf2p and Upf1p. First, the interaction between Upf2p and Upf1p in S. cerevisiae is required for NMD, as analyses of deletion and point mutations within one or the other of the two proteins have demonstrated (17, 18, 19, 45). Second, the bipartite nature of the Upf1p binding site of S. cerevisiae Upf2p, consisting of amino acids 947 to 985 and 1034 to 1061, appears to be conserved in hUpf2p as C-terminal amino acids 1085 to 1124 and 1167 to 1194 (Fig. 2). Interestingly, an additional conservation of only the first part of the bipartite site is present in N-terminal hUpf2p amino acids 94 to 133, which is rich in acidic and basic amino acids (Fig. 2). Relative to the corresponding S. cerevisiae Upf2p sequences that bind Upf1p, the N-terminal hUpf2 sequence is 46% similar, while the C-terminal hUpf2p sequences are 28% similar for the first part and 46% similar for the second part.
In order to test for an interaction between hUpf1p and hUpf2p, FLAG-tagged WT hUpf1p and T7-tagged WT hUpf2p were transiently coproduced in HeLa cells. Previous studies have demonstrated that FLAG-hUpf1p is functional in NMD, binds RNA, and associates with ribosomes, as does its yeast counterpart (35a, 40). α-hUpf1p antibody immunoprecipitated T7-hUpf2p only in cells that had been transfected with both FLAG-hUpf1p and T7-hUpf2p expression vectors (Fig. 6B, lane 1; data not shown). Therefore, hUpf1p and hUpf2p interact, consistent with a role for hUpf2p in NMD. Since α-hUpf1p antibody reacts with both FLAG-hUpf1p and endogenous HeLa-cell hUpf1p, the failure to detect the interaction between hUpf1p and hUpf2p in cells lacking FLAG-hUpf1p (data not shown) probably indicates that the level of endogenous HeLa-cell hUpf1p is too low to allow a detectable interaction with T7-hUpf2p under the conditions employed. We resorted to using α-hUpf1p antibody since, for reasons that may reflect epitope accessibility, α-FLAG antibody failed to immunoprecipitate T7-hUpf2p (data not shown). Similarly, α-T7 antibody failed to immunoprecipitate FLAG-hUpf1p (data not shown). Deletion of T7-hUpf2p amino acids 94 to 133 resulted in a reproducibly slight weakening of the interaction with FLAG-hUpf1p (Fig. 6B, lane 4; data not shown), deletion of T7-hUpf2p amino acids 1095 to 1272 resulted in a reproducibly significant weakening of the interaction with FLAG-hUpf1p (Fig. 6, lane 5; data not shown), and deletion of amino acids 94 to 133 together with 1095 to 1272 precluded the interaction with FLAG-hUpf1p (Fig. 6B, lane 6). These findings corroborate the importance of T7-hUpf2p amino acids 94 to 133 for hUpf1p binding but indicate that these amino acids are less important for binding than amino acids 1095 to 1272. Deletion of T7-hUpf2p amino acids 711 to 928, a region important for hUpf3p-X, hUpf3p, and hUpf3pΔ binding (Fig. 5D, lane 7; Fig. 5E and F, lanes 3), was of no consequence to the interaction with FLAG-hUpf1p (Fig. 6B, lane 2). This result indicates that the interaction between hUpf2p and hUpf1p is independent of the interaction between hUpf2p and hUpf3p. Similarly, deletion of T7-hUpf2p amino acids 526 to 722 was inconsequential to the interaction with hUpf1p (Fig. 6B, lane 3).
FIG. 6.
hUpf1p and hUpf2p coimmunoprecipitate. (A) Diagrams of epitope-tagged proteins (here, open inset boxes indicate hUpf1p binding sites), as well as FLAG-hUpf1p. (B) Total proteins (10 μl of lysate) from 104 HeLa cells that had been transiently transfected with the specified combination of FLAG-hUpf1p and either WT or mutated T7-hUpf2p expression vectors were subjected to Western blot analysis using α-FLAG or α-T7 antibody. Immunoprecipitations (IP) with α-hUpf1p antibody were analyzed by Western blotting using α-T7 antibody.
hUpf1p is detected exclusively in the cytoplasm, hUpf2p is detected primarily in the cytoplasm, and hUpf3p-X is detected primarily in nuclei.
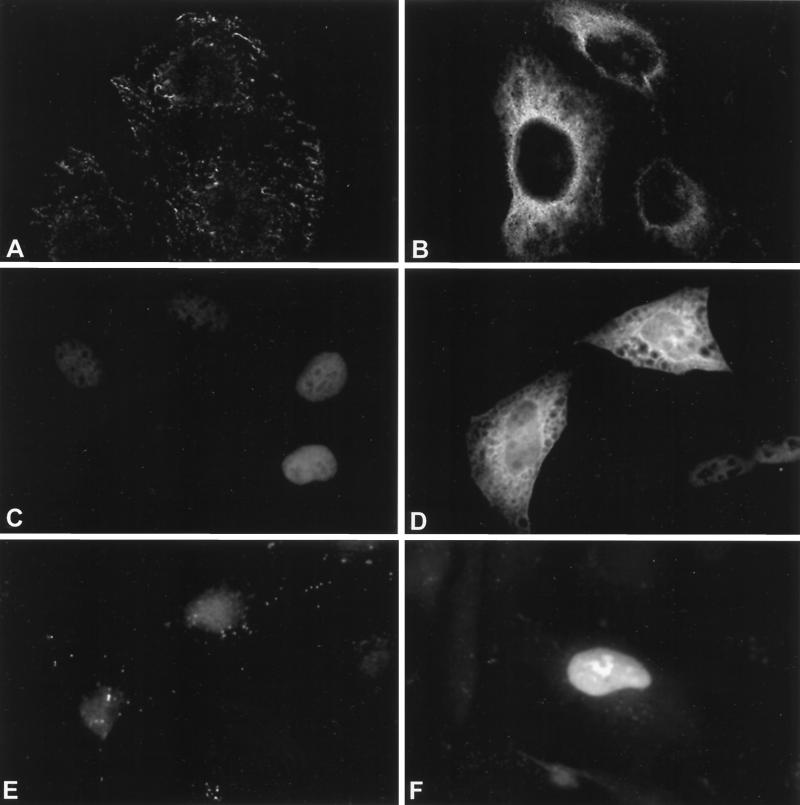
Insight into protein function often derives from information on where in the cell the protein resides. To determine the intracellular location of hUpf1p, hUpf2p, and hUpf3p-X, HeLa cells were transiently transfected with expression plasmids that produce FLAG-hUpf1p, T7-hUpf2p, HA–hUpf3p-X, or, as a control, T7-hnRNP A1. The location of each transiently produced protein was then determined by indirect immunofluorescence using antibody against the appropriate epitope tag. None of the antibodies reacted with mock-transfected cells except for antibody to the T7 epitope tag, which reacted slightly with nuclei (Fig. 7A, C, and E). T7-hnRNP A1 was detected exclusively in nuclei (data not shown) as described previously (37). FLAG-hUpf1p was detected exclusively in the cytoplasm (Fig. 7B). Endogenous hUpf1p was also found to be cytoplasmic by Western analysis using an α-hUpf1p antibody that reacts specifically with hUpf1p (data not shown). Localization of hUpf1p to the cytoplasm is consistent with its presence in postnuclear extracts of human Raji and U937 cells (1), its purification with polysomes and ribosomal subunits (35a), as well as indirect immunofluorescence studies of human Raji and U937 cells that employed peptide antibodies (1). Furthermore, S. cerevisiae Upf1p localizes to the cytoplasm but not the nucleus (3, 4). T7-hUpf2p was detected primarily in cytoplasm (Fig. 7D). The background of nuclear reactivity evident in mock-transfected cells (Fig. 7C) precluded our determining if a smaller fraction of T7-hUpf2p localizes to nuclei. While the intracellular distribution of S. cerevisiae Upf2p has never been reported, Upf2p is known to associate with polyribosomes (4). Moreover, the finding that targeting a dominant-negative Upf2p variant to nuclei alleviated the inhibition of NMD suggests that function in NMD is cytoplasmic (17). Notably, cytoplasmic FLAG-hUpf1p and T7-hUpf2p occasionally appeared concentrated in the vicinity of the nuclear envelope (e.g., Fig. 7B and D). HA–hUpf3p-X localized predominantly to nuclei (Fig. 7F). This result was unexpected given that Upf3p in S. cerevisiae localizes primarily to the cytoplasm even though it is exported from nuclei to the cytoplasm by a NES (40).
FIG. 7.
hUpf1p and hUpf2p are primarily cytoplasmic, while hUpf3p-X is primarily nuclear. HeLa cells were mock transfected (left) or transiently transfected with FLAG-hUpf1p, T7-hUpf2p, or HA–hUpf3p-X expression vectors (right) and fixed. The subcellular location of each protein was determined by indirect immunofluorescence using antibody against each epitope tag (FLAG [A and B], T7 [C and D], and HA [E and F]) and an appropriate rhodamine-conjugated secondary antibody.
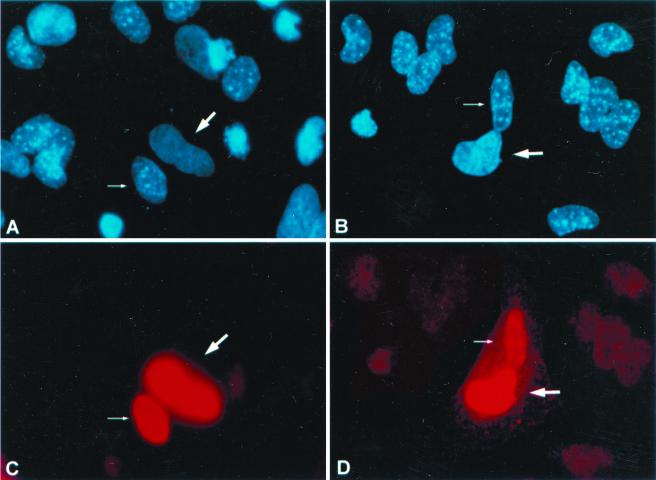
To confirm the observations that hUpf3p-X is primarily nuclear and able to function in association with cytoplasmic hUpf1p and hUpf2p, HeLa cells were transiently transfected with the HA–hUpf3p-X expression vector or, as a control, the T7-hnRNP A1 expression vector, and the cells subsequently fused to mouse NIH 3T3 cells to form heterokaryons (33). Before and during fusion, the cells were treated with cycloheximide to prevent additional protein synthesis. At 2 h postfusion, the cells were fixed and HA–hUpf3p-X or T7-hnRNP A1 was localized using antibody against the appropriate epitope tag. Human and mouse nuclei were distinguished using the dye Hoechst 33458, which stains intranuclear bodies only in mouse nuclei (Fig. 8A and B). T7-hnRNP A1 produced in HeLa cells prior to heterokaryon formation was detected within mouse nuclei of the heterokaryons and, thus, shuttles as has been reported previously (Fig. 8C; 37). Significantly, HA–hUpf3p-X was also found to shuttle (Fig. 8D).
FIG. 8.
hUpf3p-X shuttles between nuclei and cytoplasm. HeLa cells were transfected with either the T7-hnRNP A1 (A and C) or HA-hUpf3p-X (B and D) expression vector. At 24 h posttransfection, cells were incubated with cycloheximide, subsequently fused with mouse NIH 3T3 cells using polyethylene glycol to form heterokaryons, incubated further with cycloheximide, and fixed. Expressed proteins were localized by indirect immunofluorescence (bottom). Cells were simultaneously incubated with Hoechst 33258 for differential staining of human and mouse nuclei (top).
DISCUSSION
Until this report, mammalian orthologues to the three S. cerevisiae Upf and seven C. elegans SMG factors known to be required for NMD consisted solely of hUpf1p (1, 36, 42), which is orthologous to S. cerevisiae Upf1p (23) and C. elegans SMG-2 (35). Therefore, our finding of human orthologues to S. cerevisiae Upf2p and S. cerevisiae Upf3p (C. elegans SMG-4) provides evidence for a higher degree of conservation of the NMD pathways in yeast, worms, and mammals than was previously appreciated. Nevertheless, differences between humans and lower eukaryotes became apparent with the discovery of four human isoforms related to the single isoform of S. cerevisiae Upf3p and the two isoforms of C. elegans SMG-4: (i) hUpf3p-X, which derives from an X-linked gene, (ii) a version of hUpf3p-X that is a product of exon skipping, (iii) hUpf3p, which derives from a second gene, and (iv) a version of hUpf3p, called hUpf3pΔ, that is a product of exon skipping (Fig. 1 to 4). Notably, C. elegans SMG-4 also derives from alternatively spliced RNA, but the two resulting isoforms have different C termini (R. Aronoff, R. Baran, and J. Hodgkin, unpublished data) in contrast to the isoforms generated by alternative splicing of hUPF3-X or hUPF3 transcripts. Isoforms of hUpf3p-X, which derive from a gene containing 10 exons, differ by the presence or absence of exon 8 (Fig. 1; data not shown). While the exonic organization of the hUPF3 gene remains to be determined, hUpf3pΔ is the result of skipping the hUPF3 equivalent of hUPF3-X exon 4 (Fig. 1, 2, and 4). Results of database analyses and RT-PCR (Fig. 2) indicate that there is no hUPF3 mRNA equivalent to the exon-skipped hUPF3-X mRNA, just as there is no hUPF3-X mRNA equivalent to the exon-skipped hUPF3 mRNA. Differences in exon skipping together with differences in amino acid sequence are likely to confer differences in function.
In S. cerevisiae, Upf1p, Upf2p, and Upf3p form a complex (4, 19, 45, 46). Results of immunoprecipitations using extracts from HeLa cells that transiently produced different combinations of epitope-tagged hUpf proteins indicate that hUpf1p interacts with hUpf2p and hUpf2p interacts with hUpf1p, hUpf3p-X, hUpf3p, and hUpf3pΔ (Fig. 5 and 6). In general, hUpf2p amino acids required for the interaction with hUpf1p and hUpf3p-X and hUpf3p-X and hUpf3p amino acids required for the interaction with hUpf2p correspond to those predicted from the corresponding interaction in S. cerevisiae. However, two observations are particularly worthy of comment. First, results generated from the analysis of hUpf3pΔ indicate that amino acids 117 to 149 of hUpf3p are not required for the interaction with hUpf2p (Fig. 5F). Therefore, our results narrow the interaction site relative to what would be predicted from comparable studies of S. cerevisiae Upf3p (19). Second, of the two hUpf2p sites that interact with hUpf1p, amino acids 94 to 133, which comprise the N-terminal site, lack the bipartite nature that characterizes both amino acids 1085 to 1194, which comprise the C-terminal site, as well as the corresponding site in S. cerevisiae (Fig. 2). More specifically, the Upf1p-Upf2p interaction in S. cerevisiae requires Upf2p amino acids 947 to 985 and 1034 to 1061 (19). hUpf2p amino acids 94 to 133, which strikingly resemble only the first portion of the bipartite domain in Upf2p, contribute less significantly to the interaction with hUpf2p than amino acids 1085 to 1124 and 1167 to 1194, which respectively resemble the first and second portions of the bipartite domain in Upf2p (Fig. 6B).
Genetic and biochemical analyses of C. elegans SMG-2, SMG-3, and SMG-4, which are orthologous to S. cerevisiae Upf1p, Upf2p, and Upf3p, respectively (35; S. Kuchma and P. Anderson, personal communication), offer considerable insight into function considering that smg-3 and smg-4 mutants are defective in SMG-2 phosphorylation (35). Extrapolating from these findings, hUpf2p and each of or some combination of hUpf3p-X, hUpf3p, or hUpf3pΔ would be expected to affect the phosphorylation of hUpf1p. Additional evidence for this possibility derives from the finding that hUpf1p is a phosphoprotein that is subject to serum-induced phosphorylation by a phosphotidylinositol-3-kinase-related kinase (35a).
Insight into protein function can often be obtained be information on intracellular localization. hUpf1p and hUpf2p produced in HeLa cells as epitope-tagged proteins from transiently introduced expression vectors were detected primarily if not exclusively in the cytoplasm, occasionally concentrated near the nuclear envelope (Fig. 7). A cytoplasmic location is consistent with their putative roles in translation termination and NMD as well as with the ribosomal association of HeLa-cell hUpf1p (35a) and S. cerevisiae Upf1p, Upf2p, and Upf3p (4). In contrast, Upf3p-X produced under similar conditions was found primarily in nuclei (Fig. 7F). hUpf3p-X was also found to shuttle rapidly between nuclei and cytoplasm (Fig. 8D), making it possible to complex with cytoplasmic hUpf1p and hUpf2p as results from immunoprecipitations would predict. Considering that hUpf3p-X, hUpf3p, and hUpf3pΔ each harbor sequences corresponding to the NES known to be functional in S. cerevisiae (40) in addition to multiple putative NLS, hUpf3 and hUpf3Δ may also shuttle between nuclei and cytoplasm. S. cerevisiae Upf3p also shuttles between nuclei and cytoplasm but localizes primarily to the cytoplasm (40). Notably, the cytoplasmic distribution that typifies Upf3p in S. cerevisiae when expressed from a centromeric plasmid was found to extend to nuclei when Upf3p was expressed at an eightfold higher level from a 2μ plasmid (40), indicating that our observed nuclear location of hUpf3p-X could theoretically reflect expression at abnormal intracellular levels. Countering this idea, hUpf3p-X was undetectable in the cytoplasm. Furthermore, the cellular location of hUpf1p is cytoplasmic regardless of whether the protein derives from a transiently introduced plasmid or the HeLa-cell genome (Fig. 7; data not shown).
Issues that remain to be resolved include the precise role of each hUpf protein in translation termination and NMD. It will be of interest to determine if functional differences exist between the four alternatively spliced products of the hUPF3-X and hUPF3 genes. It will also be important to determine if similarity between the FIGEL-containing domain of hUpf2p and the eIF4A and eIF4AIII binding site of the eIF4G family of translation initiation factors indicates that there is a physical connection between translation termination and mRNA degradation at the 5′ end, which appears to be the first nucleolytic step of NMD (29). However, attempts to coimmunoprecipitate T7-hUpf2p and HA-eIF4A or HA-eIF4AIII have failed to detect an interaction (data not shown). Recent studies of S. cerevisiae have demonstrated that Upf1p interacts with Hrp1p, which has been proposed to mark transcripts at so-called downstream sequence elements (15) much as splicing-dependent proteins have been proposed to mark the exon-exon junctions of mammalian mRNAs (7, 8, 26, 41, 43, 44, 48, 49). Therefore, another issue to be resolved is the relationship between those hUpf proteins that shuttle between nuclei and cytoplasm and the splicing-dependent mark, at least some component(s) of which must also shuttle, considering its role in not only nucleus-associated NMD but cytoplasmic NMD (41). It will also be of great interest to resolve if mammalian cells have orthologues to C. elegans SMG-1, SMG-5, SMG-6, and SMG-7, the first of which appears to be a phosphotidylinositol-3-kinase-related kinase required for SMG-2 phosphorylation and the rest of which are required for SMG-2 dephosphorylation (35).
ACKNOWLEDGMENTS
We thank Xiaolei Sun and Mahadeb Pal for reagents, Xiaojie Li, Saikat Pal, and Deborah Ogden for technical assistance, Javier Cáceres for helpful advice regarding the heterokaryon assays, Javier Cáceres and Adrian Krainer for the T7-hnRNP A1 expression vector, and Nahum Sonenberg for helpful conversations.
This work was supported by Public Health Service Research grants DK 33933 and GM 59614 (L.E.M.), a fellowship from the Association pour la Recherche sur le Cancer (G.S.), and NCI core grant CA 16056 for support of the Roswell Park Cell Analysis Facility (J.B.).
REFERENCES
- 1.Applequist S E, Selg M, Raman C, Jäck H M. Cloning and characterization of HUPF1, a human homolog of the Saccharomyces cerevisiae nonsense mRNA-reducing UPF1 protein. Nucleic Acids Res. 1997;25:814–821. doi: 10.1093/nar/25.4.814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aravind L, Koonin E V. Eukaryote-specific domains in translation initiation factors: implications for translational regulation and evolution of the translational system. Genome Res. 2000;10:1172–1184. doi: 10.1101/gr.10.8.1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Atkin A L, Altamura N, Leeds P, Culbertson M R. The majority of yeast UPF1 co-localizes with polyribosomes in the cytoplasm. Mol Biol Cell. 1995;6:611–625. doi: 10.1091/mbc.6.5.611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Atkin A L, Schenkman L R, Eastham M, Dahlseid J N, Lelivelt M J, Culbertson M R. Relationship between yeast polyribosomes and Upf proteins required for nonsense mRNA decay. J Biol Chem. 1997;272:22163–22172. doi: 10.1074/jbc.272.35.22163. [DOI] [PubMed] [Google Scholar]
- 5.Cáceres J F, Screaton G R, Krainer A R. A specific subset of SR proteins shuttles continuously between the nucleus and the cytoplasm. Genes Dev. 1998;12:55–66. doi: 10.1101/gad.12.1.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cali B M, Kuchma S L, Latham J, Anderson P. smg-7 is required for mRNA surveillance in Caenorhabditis elegans. Genetics. 1999;151:605–616. doi: 10.1093/genetics/151.2.605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carter M S, Li S, Wilkinson M F. A splicing-dependent regulatory mechanism that detects translation signals. EMBO J. 1996;15:5965–5975. [PMC free article] [PubMed] [Google Scholar]
- 8.Cheng J, Belgrader P, Zhou X, Maquat L E. Introns are cis effectors of the nonsense-codon-mediated reduction in nuclear mRNA abundance. Mol Cell Biol. 1994;14:6317–6325. doi: 10.1128/mcb.14.9.6317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cui Y, Hagan K W, Zhang S, Peltz S W. Identification and characterization of genes that are required for the accelerated degradation of mRNAs containing a premature translational termination codon. Genes Dev. 1995;9:423–436. doi: 10.1101/gad.9.4.423. [DOI] [PubMed] [Google Scholar]
- 10.Cui Y, Dinman J D, Peltz S W. Mof4–1 is an allele of the UPF1/IFS2 gene which affects both mRNA turnover and −1 ribosomal frameshifting efficiency. EMBO J. 1996;15:5726–5736. doi: 10.1002/j.1460-2075.1996.tb00956.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Czaplinski K, Ruiz-Echevarria M J, Paushkin S V, Han X, Weng Y, Perlick H A, Dietz H C, Ter Avanesyan M D, Peltz S W. The surveillance complex interacts with the translation release factors to enhance termination and degrade aberrant mRNAs. Genes Dev. 1998;12:1665–1677. doi: 10.1101/gad.12.11.1665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Czaplinski K, Weng Y, Hagan K W, Peltz S W. Purification and characterization of the Upf1 protein: a factor involved in translation and mRNA degradation. RNA. 1995;1:610–623. [PMC free article] [PubMed] [Google Scholar]
- 13.Dunckley T, Parker R. The DCP2 protein is required for mRNA decapping in Saccharomyces cerevisiae and contains a functional MutT motif. EMBO J. 1999;18:5411–5422. doi: 10.1093/emboj/18.19.5411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ginisty H, Sicard H, Roger B, Bouvet P. Structure and functions of nucleolin. J Cell Sci. 1999;112:761–772. doi: 10.1242/jcs.112.6.761. [DOI] [PubMed] [Google Scholar]
- 15.González C I, Ruiz-Echevarría M J, Vasudevan S, Henry M F, Peltz S W. The yeast hnRNP-like protein Hrp1/Nab4 marks a transcript for nonsense-mediated mRNA decay. Mol Cell. 2000;5:489–499. doi: 10.1016/s1097-2765(00)80443-8. [DOI] [PubMed] [Google Scholar]
- 16.Harlow E, Lane D. Using antibodies: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1999. [Google Scholar]
- 17.He F, Jacobson A. Identification of a novel component of the nonsense-mediated mRNA decay pathway by use of an interacting protein screen. Genes Dev. 1995;9:437–454. doi: 10.1101/gad.9.4.437. [DOI] [PubMed] [Google Scholar]
- 18.He F, Brown A H, Jacobson A. Interaction between Nmd2p and Upf1p is required for activity but not for dominant-negative inhibition of the nonsense-mediated mRNA decay pathway in yeast. RNA. 1996;2:153–170. [PMC free article] [PubMed] [Google Scholar]
- 19.He F, Brown A H, Jacobson A. Upf1p, Nmd2p, and Upf3p are interacting components of the yeast nonsense-mediated mRNA decay pathway. Mol Cell Biol. 1997;17:1580–1594. doi: 10.1128/mcb.17.3.1580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hentze M W, Kulozik A E. A perfect message: RNA surveillance and nonsense-mediated decay. Cell. 1999;96:307–310. doi: 10.1016/s0092-8674(00)80542-5. [DOI] [PubMed] [Google Scholar]
- 21.Ho S N, Hunt H D, Horton R M, Pullen J K, Pease L R. Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene. 1989;77:51–59. doi: 10.1016/0378-1119(89)90358-2. [DOI] [PubMed] [Google Scholar]
- 22.Kozak M. Initiation of translation in prokaryotes and eukaryotes. Gene. 1999;234:187–208. doi: 10.1016/s0378-1119(99)00210-3. [DOI] [PubMed] [Google Scholar]
- 23.Lee B S, Culbertson M R. Identification of an additional gene required for eukaryotic nonsense mRNA turnover. Proc Natl Acad Sci USA. 1995;92:10354–10358. doi: 10.1073/pnas.92.22.10354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Leeds P, Peltz S W, Jacobson A, Culbertson M R. The product of the yeast UPF1 gene is required for rapid turnover of mRNAs containing a premature translational termination codon. Genes Dev. 1991;5:2303–2314. doi: 10.1101/gad.5.12a.2303. [DOI] [PubMed] [Google Scholar]
- 25.Leeds P, Wood J M, Lee B S, Culbertson M R. Gene products that promote mRNA turnover in Saccharomyces cerevisiae. Mol Cell Biol. 1992;12:2165–2177. doi: 10.1128/mcb.12.5.2165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Le Hir H, Moore M J, Maquat L E. Pre-mRNA splicing alters mRNP composition: evidence for a stable association of proteins at exon-exon junctions. Genes Dev. 2000;14:1098–1108. [PMC free article] [PubMed] [Google Scholar]
- 27.Li Q, Imataka H, Morino S, Rogers G W, Jr, Richter-Cook N J, Merrick W C, Sonenberg N. Eukaryotic translation initiation factor 4AIII(eIF4AIII) is functionally distinct from eIF4I and eIF4AII. Mol Cell Biol. 1999;19:7336–7346. doi: 10.1128/mcb.19.11.7336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li S, Wilkinson M R. Nonsense surveillance in lymphocytes? Immunity. 1998;8:135–141. doi: 10.1016/s1074-7613(00)80466-5. [DOI] [PubMed] [Google Scholar]
- 29.Lim S-K, Maquat L E. Human β-globin mRNAs that harbor a nonsense codon are degraded in murine erythroid tissues to intermediates that have a 5′ cap-like structure. EMBO J. 1992;11:3271–3278. doi: 10.1002/j.1460-2075.1992.tb05405.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Maquat L E. When cells stop making sense: effects of nonsense codons on RNA metabolism in vertebrate cells. RNA. 1995;1:453–465. [PMC free article] [PubMed] [Google Scholar]
- 31.Maquat L E. Defects in RNA splicing and the consequence of shortened translational reading frames. Am J Hum Genet. 1996;59:279–286. [PMC free article] [PubMed] [Google Scholar]
- 32.Maquat L E. Nonsense-mediated RNA decay in mammalian cells: a splicing-dependent means to down-regulate the levels of mRNAs that prematurely terminate translation. In: Sonenberg N, Hershey J W B, Mathews M B, editors. Translational control of gene expression. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 2000. pp. 849–868. [Google Scholar]
- 33.Michael W M, Eder P S, Dreyfuss G. The K nuclear shuttling domain: a novel signal for nuclear import and nuclear export in the hnRNP K protein. EMBO J. 1997;16:3587–3598. doi: 10.1093/emboj/16.12.3587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nagy E, Maquat L E. A rule for termination-codon position within intron-containing genes: when nonsense affects RNA abundance. Trends Biochem Sci. 1998;23:198–199. doi: 10.1016/s0968-0004(98)01208-0. [DOI] [PubMed] [Google Scholar]
- 35.Page M F, Carr B, Anders K R, Grimson A, Anderson P. SMG-2 is a phosphorylated protein required for mRNA surveillance in Caenorhabditis elegans and related to Upf1p of yeast. Mol Cell Biol. 1999;19:5943–5951. doi: 10.1128/mcb.19.9.5943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35a.Pal, M., Y. Ishigaki, E. Nagy, and L. E. Maquat. Evidence that phosphorylation of human Upf1 protein varies with intracellular location and is mediated by a wortmannin-sensitive and rapamycin-sensitive PI 3-kinase-related kinase signaling pathway. RNA, in press. [DOI] [PMC free article] [PubMed]
- 36.Perlick H A, Medghalchi S M, Spencer F A, Kendzior R J, Jr, Dietz H C. Mammalian orthologues of a yeast regulator of nonsense transcript stability. Proc Natl Acad Sci USA. 1996;93:10928–10932. doi: 10.1073/pnas.93.20.10928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Piñol-Roma S, Dreyfuss G. Shuttling of pre-mRNA binding proteins between nucleus and cytoplasm. Nature. 1992;355:730–732. doi: 10.1038/355730a0. [DOI] [PubMed] [Google Scholar]
- 38.Pulak R, Anderson P. mRNA surveillance by the Caenorhabditis elegans smg genes. Genes Dev. 1993;7:1885–1897. doi: 10.1101/gad.7.10.1885. [DOI] [PubMed] [Google Scholar]
- 39.Ruiz-Echevarria M J, Yasenchak J M, Han X, Dinman J D, Peltz S W. The Upf3 protein is a component of the surveillance complex that monitors both translation and mRNA turnover and affects viral propagation. Proc Natl Acad Sci USA. 1998;95:8721–8726. doi: 10.1073/pnas.95.15.8721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shirley R L, Lelivelt M J, Schenkman L R, Dahlseid J N, Culbertson M R. A factor required for nonsense-mediated mRNA decay in yeast is exported from the nucleus to the cytoplasm by a nuclear export signal sequence. J Cell Sci. 1998;111:3129–3143. doi: 10.1242/jcs.111.21.3129. [DOI] [PubMed] [Google Scholar]
- 41.Sun X, Moriarty P M, Maquat L E. Nonsense-mediated decay of glutathione peroxidase 1 mRNA in the cytoplasm is dependent on intron position and not restricted to newly synthesized mRNA. EMBO J. 2000;19:4734–4744. doi: 10.1093/emboj/19.17.4734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sun X, Perlick H A, Dietz H C, Maquat L E. A mutated human homologue to yeast Upf1 protein has a dominant-negative effect on the decay of nonsense-containing mRNAs in mammalian cells. Proc Natl Acad Sci USA. 1998;95:10009–10014. doi: 10.1073/pnas.95.17.10009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sun X, Maquat L E. mRNA surveillance in mammalian cells: the relationship between introns and translation termination. RNA. 2000;6:1–8. doi: 10.1017/s1355838200991660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Thermann R, Neu-Yilik G, Deters A, Frede U, Wehr K, Hagemeier C, Hentze M W, Kulozik A E. Binary specification of nonsense codons by splicing and cytoplasmic translation. EMBO J. 1998;17:3484–3494. doi: 10.1093/emboj/17.12.3484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Weng Y, Czaplinski K, Peltz S W. Genetic and biochemical characterization of the mutations in the ATPase and helicase regions of Upf1 protein. Mol Cell Biol. 1996;16:5477–5490. doi: 10.1128/mcb.16.10.5477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Weng Y, Czaplinski K, Peltz S W. Identification and characterization of mutations in the UPF1 gene that affect nonsense suppression and the formation of the Upf protein complex but not mRNA turnover. Mol Cell Biol. 1996;16:5491–5506. doi: 10.1128/mcb.16.10.5491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Weng Y, Czaplinski K, Peltz S W. ATP is a cofactor of the Upf1 protein that modulates its translation termination and RNA binding activities. RNA. 1998;4:205–214. [PMC free article] [PubMed] [Google Scholar]
- 48.Zhang J, Sun X, Qian Y, LaDuca J P, Maquat L E. At least one intron is required for the nonsense-mediated decay of triosephosphate isomerase mRNA: a possible link between nuclear splicing and cytoplasmic translation. Mol Cell Biol. 1998;18:5272–5283. doi: 10.1128/mcb.18.9.5272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhang J, Sun X, Qian Y, Maquat L E. Intron function in the nonsense-mediated decay of β-globin mRNA: indications that pre-mRNA splicing in the nucleus can influence mRNA translation in the cytoplasm. RNA. 1998;4:801–815. doi: 10.1017/s1355838298971849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhang S, Welch E M, Hogan K, Brown A H, Peltz S W, Jacobson A. Polysome-associated mRNAs are substrates for the nonsense-mediated mRNA decay pathway in Saccharomyces cerevisiae. RNA. 1997;3:234–244. [PMC free article] [PubMed] [Google Scholar]