Abstract
Complex genetic and biochemical interactions between HOX proteins and members of the TALE (i.e., PBX and MEIS) family have been identified in embryonic development, and some of these interactions also appear to be important for leukemic transformation. We have previously shown that HOXA9 collaborates with MEIS1 in the induction of acute myeloid leukemia (AML). In this report, we demonstrate that HOXB3, which is highly divergent from HOXA9, also genetically interacts with MEIS1, but not with PBX1, in generating AML. In addition, we show that the HOXA9 and HOXB3 genes play key roles in establishing all the main characteristics of the leukemias, while MEIS1 functions only to accelerate the onset of the leukemic transformation. Contrasting the reported functional similarities between PREP1 and MEIS1, such as PBX nuclear retention, we also show that PREP1 overexpression is incapable of accelerating the HOXA9-induced AML, suggesting that MEIS1 function in transformation must entail more than PBX nuclear localization. Collectively, these data demonstrate that MEIS1 is a common leukemic collaborator with two structurally and functionally divergent HOX genes and that, in this collaboration, the HOX gene defines the identity of the leukemia.
The homeodomain-containing transcription factors of the HOX gene family, regulators of pattern formation and tissue identity during embryogenesis, have also been identified previously as regulators of hemopoietic cell proliferation and differentiation (40). In the hematopoietic system, HOX gene expression is largely confined to primitive cells (11, 32), and the enforced expression of HOX genes (i.e., HOXB4, HOXB3, and HOXA10) in mouse hemopoietic cells results in distinct phenotypes, affecting various hemopoietic lineages (33, 34, 39).
In agreement with their regulatory functions, aberrant expression of HOX genes is associated with leukemic transformation both in mice and in humans. In a subset of human myeloid leukemias, a recurrent translocation between the HOXA9 and NUP98 genes results in the expression of the fusion oncoprotein NUP98-HOXA9 (6, 24). Recently, the HOXA9 gene was also shown to be the single most highly correlated gene (out of 6,817 genes tested) for poor prognosis in human acute myeloid leukemia (AML) (12), thus suggesting a potential key role for this gene in human leukemia, beyond that caused by the HOXA9-NUP98 chromosomal translocation. By applying either retroviral insertional mutagenesis or retroviral overexpression, roles for the HOXA7, HOXA9, HOXA10, HOXB3, and HOXB8 genes in leukemic transformation in mice have also been established previously (17, 25, 28, 34, 38, 39). The lineage-specific effects produced by the overexpression of different HOX genes in mouse bone marrow cells, which often precede acute leukemic transformation (34, 39), raise the possibility that HOX genes may influence the typical phenotypic variations seen between subsets of acute leukemia.
A number of studies have demonstrated that HOX proteins collaborate in the in vitro DNA binding with members of the TALE (three-amino-acid loop extension) subclass of homeodomain-containing proteins comprising the PBC (mammalian PBX and Drosophila melanogaster EXD proteins) and MEIS (mammalian MEIS and PREP1 and Drosophila HTH proteins) families (20). This interaction shows moderate specificity, with HOX proteins from paralog groups 1 to 10 interacting with PBX proteins, whereas interaction with MEIS proteins is limited to HOX paralogs 9 to 13 (36). The cooperative interaction between PBX (or EXD) and HOX proteins has been shown elsewhere to enhance the DNA binding affinity and specificity of HOX proteins (20) and is essential for at least some of the HOX-dependent developmental programs (2, 29). In contrast, a functional role for a dimeric HOX-MEIS complex has not been established so far (31). Members of the MEIS family can, however, form a stable heterocomplex with PBX (or EXD) in both DNA-dependent and -independent manners (5, 8, 30), and interaction with MEIS induces nuclear localization of PBX proteins by preventing their nuclear export (1, 3, 15, 27). Recently, indirect interaction between HOX and MEIS proteins (or HTH) was established by the identification of HOX-PBX-MEIS heterotrimeric complexes (4, 37). Studies both with Drosophila and with mice have, furthermore, shown that formation of such a trimeric complex is essential for the execution of at least some HOX-dependent developmental programs (9, 14, 31).
Members of the PBX and MEIS families are also involved in human and mouse leukemias. PBX1 is part of the fusion protein E2A-PBX1 found in 10 to 20% of human pediatric pre-B acute lymphoblastic leukemia patients (16, 26). By applying retroviral co-overexpression, we have also previously demonstrated a strong collaboration between HOXA9 and E2A-PBX1 in the induction of AML (38). In addition, MEIS1 is frequently activated by retroviral integration in myeloid leukemias in BXH-2 mice and genetically interacts with HOXA7 and HOXA9 genes in AML (17, 22, 25). Thus, in leukemic transformation, as in the regulation of pattern formation and tissue identity during embryogenesis, an important genetic interaction has been established between HOX and TALE genes.
Based on the above results, we wanted to gain further insight into the nature of the collaboration between HOX proteins and members of the MEIS and PBC families in leukemic transformation. The results presented herein identify MEIS1 as a common collaborator with two divergent HOX genes, i.e., HOXA9 and HOXB3. The specificity of this collaboration was proven by the lack of genetic cooperativity between HOX and the two other TALE genes tested, i.e., PBX1 and PREP1. Using overexpression studies in bone marrow cells, we also demonstrate that each HOX gene studied predisposes to leukemias that are phenotypically distinct and that MEIS1 acts primarily to accelerate the occurrence of these leukemias without altering their phenotype.
MATERIALS AND METHODS
Animals.
All mice, both donors (C57BL/6Ly-Pep3b × C3H/HeJ)F1 [(PepC3)F1] and recipients (C57BL/6J × C3H/HeJ)F1 [(B6C3)F1], were bred and maintained as previously reported (38).
Generation of recombinant retroviruses.
The retroviral vectors used in this study, i.e., MSCV-HOXA9-pgk-neo (no. 412), MSCV-HOXB3-pgk-neo (no. 245), MSCV-PBX1b-pgk-puro (no. 448), and MSCV-MEIS1a-pgk-puro (no. 515), have all been described before (17, 34). The MSCV-PREP1-pgk-puro (no. 682) retrovirus was generated by subcloning the human PREP1 cDNA (5) into the HpaI site of the MSCV-pgk-puro retrovirus. The MSCV-pgk EGFP vector (generous gift from K. Humphries, Terry Fox Laboratory, Vancouver, British Columbia, Canada) served as a backbone to generate the MSCV-MEIS1a-pgk-EYFP (no. 722) retroviral vector (enhanced yellow fluorescent protein [EYFP] cDNA from Clontech) used in part of these studies. High-titer helper-free retrovirus producer cells were generated from GP+E-86 and BOSC-23 viral packaging cells and tested as reported previously (17).
Retroviral infection and transplantation of primary murine bone marrow cells.
Both double and single retroviral infections of primary murine bone marrow cells, followed by transplantation of infected cells, were done as previously described (38).
In vitro cultures and FACS analysis.
For myeloid clonogenic progenitor assays, cells were cultured in methylcellulose cultures as described previously (38). Bone marrow cells harvested from the cocultivation with virus-producing cells or recovered from reconstituted leukemic mice were plated at a concentration of 2 × 103 to 8 × 103 cells/ml or 3 × 104 cells/ml, respectively. In an effort to derive cell lines from the leukemic mice, their bone marrow and/or spleen cells were grown in liquid cultures of Iscove's medium containing 10% fetal calf serum, 10−5 M β-mercaptoethanol, 2 mM glutamine, and 200 mg of transferrin per ml, in the presence or absence of 5 ng of murine interleukin-3 (IL-3) per ml or 0.5 ng of granulocyte-macrophage colony-stimulating factor per ml. To analyze the effect of MEIS1 or HOXA9 on in vitro proliferation, EYFP+ or enhanced green fluorescent protein-positive (EGFP+) cells were purified, as previously described (39), from the bone marrow of the EGFP-control, HOXA9-EGFP, and MEIS1-EYFP mice and grown in Dulbecco's modified Eagle's medium containing 15% fetal calf serum, 10−5 M β-mercaptoethanol, 2 mM glutamine, 200 mg of transferrin per ml, 6 ng of murine IL-3 per ml, 10 ng of human IL-6 per ml, 50 ng of murine steel factor per ml, and 3 U of human urinary erythropoietin per ml. For fluorescence-activated cell sorting (FACS) analysis, cells from the bone marrow, spleen, and thymus of EGFP and MEIS1-EYFP mice were analyzed as previously described (17).
DNA, RNA, and protein analyses.
The probes used for RNA and DNA analysis were a XhoI/SalI fragment of pMC1neo (neo), a HindIII/ClaI fragment of MSCV-pgk-puro, or the full-length 1.4-kb HOXA9, 1.6-kb HOXB3, 1.5-kb MEIS1, 1.8-kb PBX1, and 1.8-kb PREP1 cDNAs, labeled with 32P by random primer extension. For Western blot analysis, total-cell lysates from HOXA9 or PREP1 viral producer cells (GP+E-86) were prepared as previously described (18). A polyclonal antibody to PREP1 was used as described previously (5).
RESULTS
Generation of bone marrow transplantation chimeras.
To determine whether the leukemic transformation induced by the previously reported genetic interaction between HOXA9 and MEIS1 was specific for these two genes or whether similar interactions could be detected with other HOX-TALE pairs, transplantation chimeras were generated using bone marrow cells engineered to retrovirally overexpress HOXB3 or HOXA9 together with either PBX1, MEIS1, or PREP1. In addition, various control mice were also generated (all transplantation chimeras that were part of these studies are outlined in Table 1).
TABLE 1.
Absolute numbers of untransduced and transduced myeloid colony-forming cellsa transplanted per mouse
| Expt no. and mouse group (n) | No. of CFC injected/mouseb
|
|||
|---|---|---|---|---|
| Untransduced | G418r | Puror | G418r and Puror | |
| Expt 1 | ||||
| Neo (6) | 2,200 | 2,100 | ||
| Puro (4) | 4,200 | 2,300 | ||
| PBX1 (6) | 3,300 | 1,400 | ||
| MEIS1 (6) | 3,300 | 1,600 | ||
| HOXA9 (7) | 950 | 2,200 | ||
| HOXB3 (6) | 3,400 | 2,300 | ||
| HOXA9-MEIS1 (6) | 2,100 | 1,800 | 1,000 | 250 |
| HOXB3-PBX1 (6) | 2,500 | 1,200 | 500 | 250 |
| HOXB3-MEIS1 (6) | 2,800 | 1,500 | 1,000 | 400 |
| Expt 2 | ||||
| HOXA9 (6) | 1,200 | 700 | ||
| HOXA9-MEIS1 (6) | 450 | 300 | 40 | 15 |
| HOXA9-PREP1 (6) | 1,600 | 650 | 500 | 300 |
The number of transduced long-term repopulating cells (LTRC) injected per mouse can be estimated based on our previous results which determined the frequency of LTRC at 1 per 100 colony-forming cells (CFC) (33) and estimation of gene transfer to LTRC equal to that of CFC (33).
The number of transduced CFC injected per mouse was determined as follows: (number of bone marrow cells injected per mouse) × (CFC frequency in the injected bone marrow inoculum) × (percentage of CFC resistant to puromycin and/or G418).
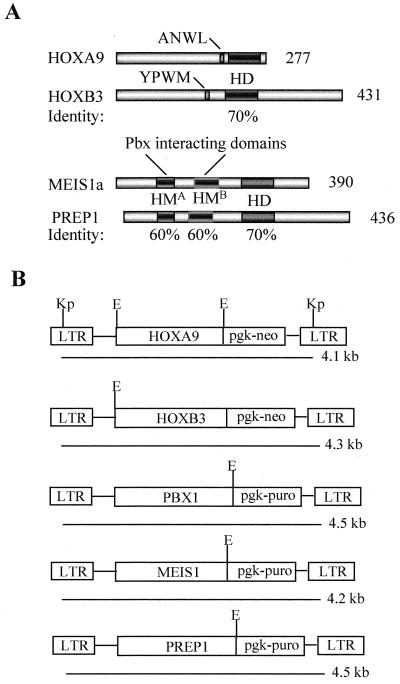
The decision to use HOXB3 and HOXA9 genes for these studies was based on the premises that the products of both of these genes have the capacity to induce AML when overexpressed (34, 38) and on sequence comparison studies which showed that the proteins encoded by these genes represent two of the most divergent (clustered) HOX proteins, which bear similarity only in their homeodomains (i.e., they are highly divergent in their N- and C-terminal regions [Fig. 1A]).
FIG. 1.
Diagrammatic representation of the HOXA9, HOXB3, MEIS1a, and PREP1 proteins and the retroviral constructs used in this study. (A) Sequence comparison of the HOXA9 and HOXB3 proteins and the MEIS1a and the PREP1 proteins used in this study. Both HOXA9 and HOXB3 proteins have a motif (ANWL in HOXA9 and YPWM in HOXB3) N-terminal to the homeodomain that is essential for their interaction with PBX proteins. Apart from their homeodomains, which are 70% identical, these proteins do not display significant sequence similarity. The MEIS1a and the PREP1 proteins share sequence similarity only in their homeodomains (70%) and in the N-terminal HMA (60%) and HMB (60%) domains that mediate interactions with PBX proteins. (B) Diagrammatic representation of the integrated MSCV-HOXA9, MSCV-HOXB3, MSCV-PBX1, MSCV-MEIS1, and MSCV-PREP1 proviruses. The expected sizes of the full-length long terminal repeat (LTR)-driven viral transcripts are shown. Restriction sites indicated are KpnI (Kp) (shown only for the HOXA9 virus but present in all constructs) and EcoRI (E). HD, homeodomain; HM, Homothorax-Meis domain.
The choice of the TALE genes was based on previous studies which demonstrated their involvement in leukemic transformation (i.e., PBX1 as part of E2A-PBX1 or MEIS1 as a genetic collaborator with HOXA9) (17, 21) or as a functional homolog to MEIS1 (PREP1). PREP1 was preferred over MEIS2 or MEIS3 because it is the most divergent member of the family (Fig. 1A), which still retains most of the functional capabilities of MEIS1, including its ability to regulate nuclear trafficking of PBX (1, 3, 15) and to bind identical DNA regulatory sequences (9, 14).
The bone marrow transplantation chimeras were generated by injecting bone marrow cells, immediately following their retroviral infection, into lethally irradiated mice. As the number of transduced cells transplanted per mouse can affect the time frame in which the leukemia develops (U. Thorsteinsdottir and G. Sauvageau, unpublished data), a proportion of the infected bone marrow cells was used to determine the number of transduced hemopoietic progenitors (resistant to G418 [Neor] and/or puromycin [Puror]) injected per mouse in each experimental group (Table 1). No preselection was performed prior to transplantation, thus rendering recipients of doubly infected cells (e.g., MEIS1 plus HOXB3) chimeras consisting of a mixture of non-, single-, and double-transduced cells. The exact composition of each chimera at the time of bone marrow transplantation is detailed in Table 1.
HOXB3 collaborates with MEIS1, but not with PBX1, to induce AML.
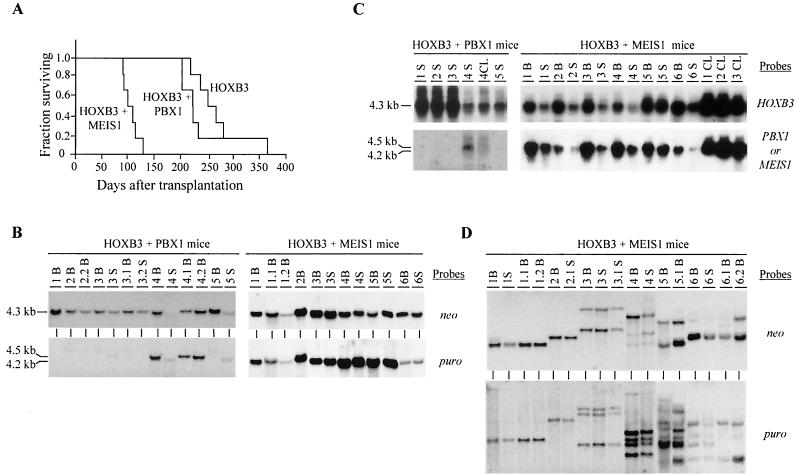
All recipients of HOXB3-transduced bone marrow cells (either alone or in combination with PBX1 or MEIS1) eventually developed AML but with different latencies (Fig. 2A). MEIS1, but not PBX1, could significantly accelerate the occurrence of AML in the HOXB3 chimeras, thus indicating a genetic collaboration between HOXB3 and MEIS1 in the induction of AML (Fig. 2A). The initial mixed nature of our chimeras (Table 1) can be exploited to further support these conclusions. Although doubly transduced cells represented only 21% of the HOXB3-transduced cells initially injected to generate the HOXB3-MEIS1 chimeras (Table 1), the AML that developed in all of the HOXB3-MEIS1 mice contained both intact MEIS1 and HOXB3 proviruses (Fig. 2B, right panel). In contrast, the presence of both the PBX1 and HOXB3 proviruses was detected in only two of the five HOXB3-PBX1 chimeras analyzed (see mouse 4 and its secondary recipients 4.1 and 4.2 and mouse 5 spleen in Fig. 2B, left panel). This is consistent with the bone marrow transplantation inoculum in which approximately one-fifth of the HOXB3-transduced cells were also infected with the PBX1 retrovirus, thus demonstrating the absence of oncogenic interaction between these two genes. This establishes that HOXB3 collaborates with MEIS1, but not PBX1, in leukemic transformation.
FIG. 2.
Demonstration of collaboration between HOXB3 and MEIS1, but not HOXB3 and PBX1, in leukemogenesis. (A) Survival graph demonstrating the collaboration between HOXB3 and MEIS1, but not PBX1, in the development of AML. The survival of the HOXB3-MEIS1 mice was significantly shorter than that of the HOXB3 mice (P < 0.001, two-tailed Student's t test) and the HOXB3-PBX1 mice (P < 0.007). The survival of the HOXB3-PBX1 mice was not significantly different from that of the HOXB3 mice. (B) Southern blot analyses of genomic DNA isolated from the bone marrow and/or spleen of the HOXB3-PBX1 and HOXB3-MEIS1 chimeras. DNA was digested with KpnI to release the integrated HOXB3 (4.3-kb), MEIS1 (4.2-kb), or PBX1 (4.5-kb) proviral fragments. The membranes were hybridized with a neo-specific probe to detect the HOXB3 provirus and a puro-specific probe to detect the MEIS1 or PBX1 provirus. (C) Northern blot analysis of total RNA (10 μg) isolated from bone marrow or spleen cells of the HOXB3-PBX1 and HOXB3-MEIS1 mice. The membranes were hybridized with full-length HOXB3, MEIS1, or PBX1 cDNA probes. (D) Southern blot analysis of DNA isolated from bone marrow of primary and secondary HOXB3-MEIS1 mice. The DNA was digested with EcoRI, which cuts the integrated provirus once, thus generating a unique fragment for each proviral integration site. The membranes were hybridized first with a neo-specific probe for detection of the HOXB3 proviral fragment(s) (top panel) and then subsequently with a puro-specific probe to detect the MEIS1 proviral fragment(s) (bottom panel). In panels B, C, and D, each primary recipient is identified with a specific number and its secondary recipients or cell lines generated from each primary recipient, with a derivative thereof (e.g., 1.1 and 1.2 and CL1, CL2, etc.). B, bone marrow; S, spleen; CL, cell lines.
Clonal analysis of proviral integration sites demonstrated that the HOXB3- and MEIS1-induced AMLs were mono- or biclonal (Fig. 2D). Furthermore, the numbers of clones detected with a probe (neo) that detects HOXB3 proviral integration sites and with a probe (puro) that detects MEIS1 proviral integrations were the same, strongly suggesting that all of the leukemic clones detected in the HOXB3-MEIS1 mice contained both the HOXB3 and MEIS1 proviruses (Fig. 2D). Northern blot analysis of total RNA isolated from the leukemic cells confirmed that these clones expressed both retrovirally derived mRNAs (Fig. 2C).
The AML induced by co-overexpression of HOXB3 and MEIS1 was readily transplanted to secondary recipients that developed AML in 41 ± 6 days (data not shown). The leukemias in the secondary mice (labeled as a derivative of a number, e.g., 1.1 or 3.1, etc.) contained both the HOXB3 and MEIS1 proviruses with the same clonal composition as that detected in the primary mice (Fig. 2D). Growth factor-independent cell lines were as easily generated from the HOXB3- and MEIS1-induced leukemias (n = 6) as from the control HOXA9-MEIS1-induced leukemias (see below), and high expression of both the HOXB3 and MEIS1 retrovirally derived messages could be detected in these cell lines (CL in Fig. 2C). Collectively, these results, together with our previous demonstration of collaboration between HOXA9 and MEIS1 in AML induction (17), demonstrate that MEIS1 can act as a common collaborator with highly structurally and functionally diverse HOX genes in leukemic transformation.
Overexpression of MEIS1 alone does not predispose to leukemia.
Although it has been previously demonstrated both for fibroblasts and for mouse bone marrow cells that PBX1 lacks an inherent transformation ability (17, 18, 21), the oncogenic potential of MEIS1 when activated alone has not been thoroughly evaluated. As outlined in Table 1, a number of control chimeras, overexpressing only a single HOX or TALE gene, were generated for the experiments described above. These chimeras were thus used to compare the leukemogenic potential of the MEIS1 gene with that of PBX1, HOXA9, or HOXB3. The number of transduced cells transplanted per mouse was high for each of the four groups of chimeras, with MEIS1 mice receiving numbers that were ∼70% of those received by the HOX mice (Table 1).
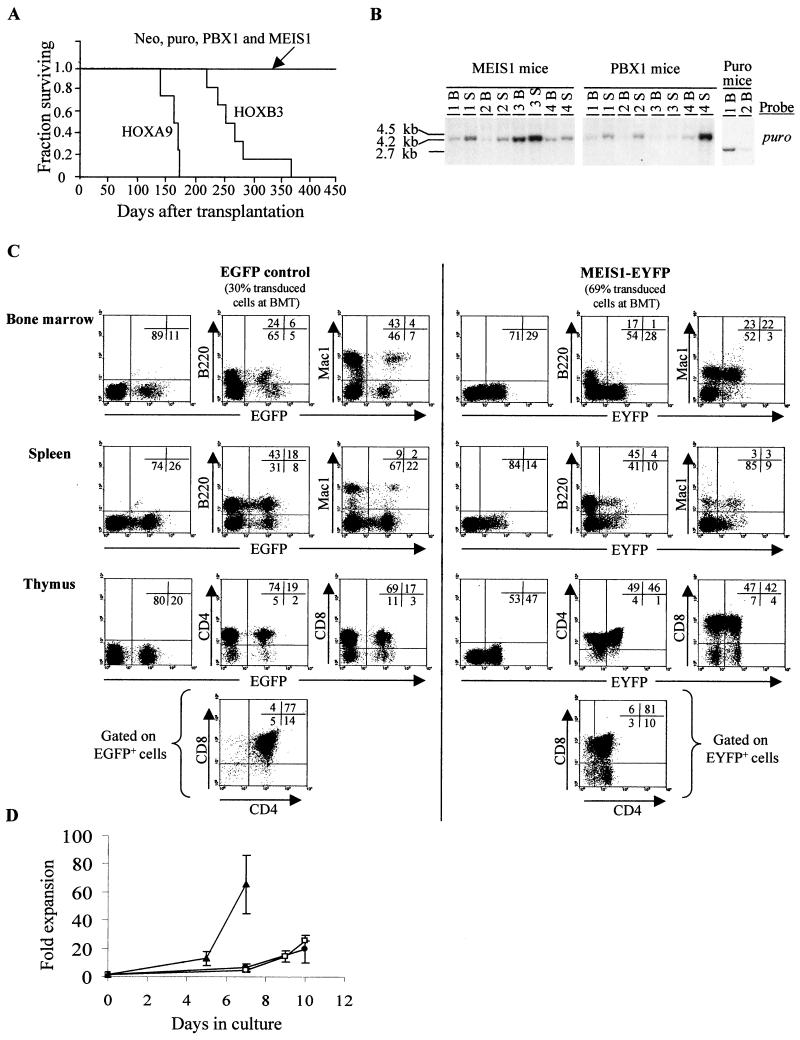
At 13 months posttransplantation, all mice in both the HOXA9 and HOXB3 groups had developed AML (all leukemias were mono- or biclonal [data not shown]), whereas the MEIS1 mice, like the PBX1 mice, appeared to thrive normally for the observation period of 15 months (Fig. 3A). At that time, four MEIS1 and PBX1 mice were sacrificed for more detailed analysis. By FACS and morphological analyses together with in vitro progenitor assays, the only hematological abnormality detected in both groups of mice was a slight enlargement of their spleen (MEIS1 mice, 0.23 ± 0.15 g, and PBX1 mice, 0.24 ± 0.20 g, versus untransplanted control, 0.1 g), which, however, was also frequently detected in the neo and puro control mice (0.22 ± 0.20 g) analyzed at a similar time point. Southern blot analysis of DNA isolated from bone marrow and spleen demonstrated the presence of the intact MEIS1 or PBX1 provirus in these organs, indicating that these mice had indeed been repopulated by MEIS1- or PBX1-transduced hematopoietic stem cells capable of long-term repopulation (Fig. 3B). Furthermore, Northern blot analysis of total RNA isolated from these same tissues revealed the expected expression of the retrovirally derived MEIS1 message (n = 3 mice [data not shown]). The low intensity of the proviral signal detected in hematopoietic tissues of most of the mice is an indicator of low-level repopulation by transduced cells, thus underscoring the fact that neither MEIS1 or PBX1 gave a proliferative advantage to hematopoietic cells.
FIG. 3.
Overexpression of MEIS1 is not permissive for B-lymphoid development but neither induces proliferation of bone marrow cells nor predisposes recipients to lymphoid or myeloid leukemias. (A) Survival graph of chimeras reconstituted with HOXA9-, HOXB3-, MEIS1-, or PBX1-transduced bone marrow cells, demonstrating, for the observation period of 450 days, that only the chimeras engineered to overexpress HOXB3 or HOXA9, but not MEIS1 or PBX1, developed leukemia. (B) Southern blot analyses of genomic DNA isolated from the bone marrow and spleen of puro-control, PBX1, and MEIS1 mice. DNA was digested with KpnI to release the integrated puro (2.7-kb), MEIS1 (4.2-kb), or PBX1 (4.5-kb) proviral fragments. The membranes were hybridized with a puro-specific probe to detect the control, MEIS1, and PBX1 proviruses. (C) Flow cytometric analysis of hematopoietic cells from bone marrow, spleen, and thymus of EGFP control and MEIS1-EYFP mice transplanted 60 days earlier with EGFP- or MEIS1-EYFP-transduced bone marrow cells, respectively. Numbers in the inset quadrant represent the percentages of live cells in the corresponding quadrant. (D) In vitro proliferation of HOXA9-EGFP (▴)-, EGFP-control (□)-, and MEIS1-EYFP (●)-positive bone marrow cells isolated from corresponding mouse chimeras at 60 days after transplantation. B, bone marrow; S, spleen; BMT, bone marrow transplantation.
In order to assess in greater detail the effect of overexpression of MEIS1 on the regeneration of the various hemopoietic lineages, another set of transplantation chimeras were generated as described above, but this time bone marrow cells were engineered to overexpress MEIS1 through the MSCV-MEIS-pgk-EYFP retroviral vector. These mice (n = 4 control mice, and n = 4 MEIS1 mice) were then sacrificed at 60 days posttransplantation, and the contribution of transduced cells to the myeloid and T- and B-lymphoid lineages was analyzed by FACS (Fig. 3C). In the bone marrow and spleen of the MEIS1 mice, the proportion and absolute numbers of myeloid cells (Mac1+ [Fig. 3C and data not shown, respectively]) were within the normal range, whereas the B-lymphoid cells (B220+) were slightly reduced. The contribution of transduced cells (EYFP+) to the myeloid lineage was within the expected range considering the initial gene transfer (69%), thus suggesting that MEIS1 had little effect on the proliferation or differentiation of myeloid cells in vivo. However, the contribution of MEIS1-transduced cells to the regeneration of B-lymphoid cells in both the bone marrow and the spleen was very low for all of the four mice analyzed (Fig. 3C), indicating that high levels of MEIS1 are incompatible with B-cell development. In contrast, overexpression of MEIS1 had no detectable effect on T-lymphoid development, as evidenced by a relatively high proportion of transduced cells in the thymus and their normal distribution in the thymic CD4 and CD8 subpopulations (Fig. 3C). As none of the MEIS1 chimeras that have been generated in our laboratory have developed any hematological malignancies (n = 20, of which n = 13 were ≥14 months posttransplantation when analyzed), this effect of MEIS1 on the B-lymphoid lineage does not appear to predispose such cells to leukemia (Fig. 3A and data not shown).
To determine the proliferative capacity of MEIS1-overexpressing cells, EYFP+ bone marrow cells from the MEIS1 chimeras were grown in vitro under conditions that stimulate the proliferation of primitive myeloid cells. In agreement with the finding of the effect of MEIS1 in vivo, the proliferative capacity of MEIS1-overexpressing bone marrow cells in vitro was similar to that of control (EGFP+) cells (Fig. 3D), thus supporting the conclusion that MEIS1 does not confer a proliferative advantage to bone marrow cells. In contrast, HOXA9-transduced bone marrow cells derived from HOXA9-EGFP chimeras (n = 4 mice) showed ∼10-fold-greater expansion than that of control bone marrow cells for a 7-day culture period (Fig. 3D).
Taken together, these data demonstrate that MEIS1 displays a very low leukemogenic potential when overexpressed alone in hematopoietic cells, in contrast to its clear leukemogenic effect when co-overexpressed with HOXA9 or HOXB3 (Fig. 2A and 4A).
FIG. 4.
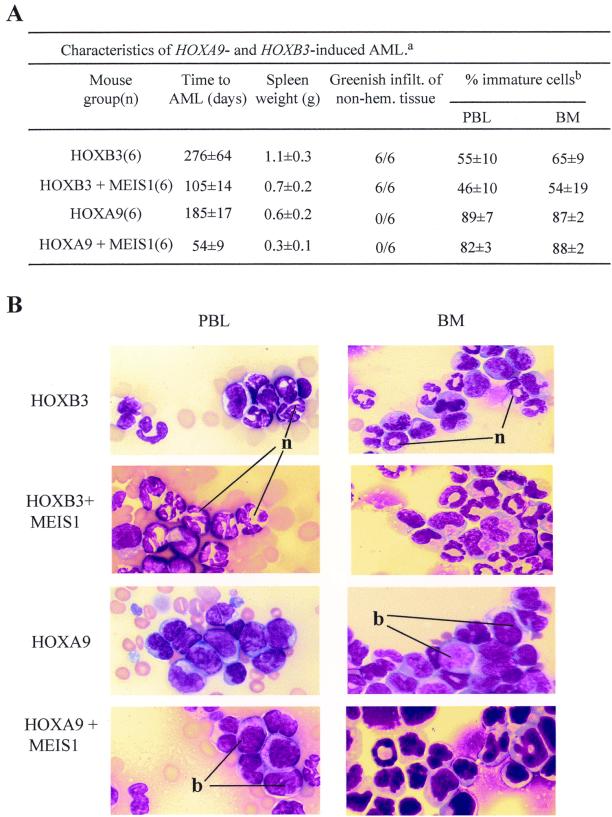
Differences between HOXB3- and HOXA9-induced AMLs. (A) Main characteristics of the AMLs that developed in HOXB3, HOXB3-MEIS1, HOXA9, and HOXA9-MEIS1 bone marrow chimeras. a, Results are expressed as the means ± standard deviations for the indicated number of mice. b, Determination of the proportion of immature and mature cells in hematopoietic tissue of the leukemic mice was based on morphological criteria, i.e., mature cells with segmented nuclei and immature cells, blast-like. For each tissue sample, n = 200 cells were counted from n = 3 representative mice in each group. infilt., infiltration; non-hem., nonhematopoietic. (B) Wright staining of peripheral blood smears (PBL) and bone marrow (BM) cytospins from representative leukemic HOXB3, HOXB3-MEIS1, HOXA9, and HOXA9-MEIS1 mice. Magnification, ×100 for all. n, neutrophil; b, blast.
The phenotypes of the AMLs that developed in transplanted mice are HOX gene dependent.
As discussed above, enforced expression of either HOXB3 or HOXA9 in mouse bone marrow cells induced mono- or biclonal transplantable AML in the recipients. However, despite transplantation of each HOXB3 and HOXA9 mouse with a similar dose of transduced cells (Table 1), the AMLs that developed in these two groups of primary recipients differed with respect to the latency (for HOXB3 chimeras, two times longer than for HOXA9 chimeras [Fig. 3A and 4A]), the differentiation status (much higher proportion of mature cells in the HOXB3-induced AML [Fig. 4]), and the tissue infiltration (much more pronounced in the HOXB3-induced AML [Fig. 4A]).
To determine the effects of MEIS1 co-overexpression on the phenotype of these two different HOX-induced AMLs, the HOXB3-MEIS1-induced leukemias were compared to those that developed in chimeras transplanted with bone marrow cells overexpressing HOXA9 or HOXB3 alone or co-overexpressing HOXA9 plus MEIS1 (Table 1). Although MEIS1 coexpression accelerated the occurrence of both the HOXB3- and HOXA9-induced AMLs by approximately threefold (Fig. 2A and 5A), it had no detectable effect on the phenotypic characteristics of their AML (Fig. 4 and data not shown). Thus, for example, the AML that developed in the HOXB3-MEIS1 chimeras had all the main characteristics of the AML that developed in the HOXB3 chimeras, such as the high proportion of mature myeloid cells and the massive greenish infiltration in nonhematopoietic tissue (Fig. 4). In contrast, the AMLs which occurred in the HOXA9-MEIS1 chimeras, like those of the HOXA9 chimeras, were characterized by only moderate infiltration into nonhematopoietic tissues and the presence of mostly immature (i.e., blast) cells in their hematopoietic organs (Fig. 4). Thus, although MEIS1 accelerates the occurrence of the HOXB3- and HOXA9-induced leukemias, the HOX gene involved ultimately sets the limit for this acceleration and the phenotype of the leukemia. These data, together with the finding of the lack of leukemogenic effect by MEIS1 when overexpressed alone in hematopoietic cells, thus strongly suggest that HOX genes determine the identity of the HOX-MEIS1-induced leukemias, with MEIS1 acting mainly to heighten their leukemogenic potential.
FIG. 5.
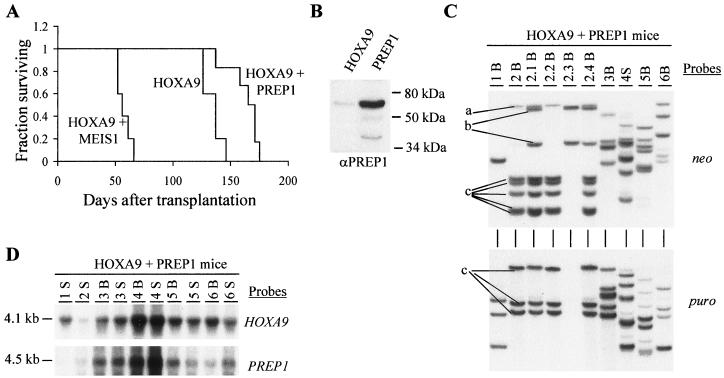
Demonstration of lack of collaboration between HOXA9 and PREP1 in leukemogenesis. (A) Survival graph demonstrating that co-overexpression of PREP1 with HOXA9, in contrast to that with MEIS1, does not accelerate the occurrence of the HOXA9-induced AML. The survival of the HOXA9-MEIS1 mice was significantly shorter than that of the HOXA9 mice (P < 0.001, two-tailed Student's t test) and the HOXA9-PREP1 mice (P < 0.001). (B) Western blot analysis of total-cell lysates from the HOXA9 and PREP1 viral producer cells. The membrane was probed with rabbit anti-human PREP1 polyclonal antibody. The position of the full-length 64-kDa PREP1 protein is indicated. Two minor products, as previously described (5), are also detected (one generated by an internal ATG site). (C) Southern blot analysis of DNA isolated from bone marrow of primary and secondary HOXA9-PREP1 mice. The DNA was digested with EcoRI, which cuts the integrated provirus once, thus generating a unique fragment for each proviral integration site. The membranes were hybridized first with a neo-specific probe for the detection of the HOXA9 proviral fragment(s) (top panel) and subsequently with a puro-specific probe to detect the PREP1 proviral fragment(s) (bottom panel). For clarity, the three different clones detected in the primary and secondary recipients of mouse 2 are labeled a, b, and c. (D) Northern blot analysis of total RNA (10 μg) isolated from bone marrow and spleen cells of the HOXA9-PREP1 mice. The membranes were hybridized with full-length HOXA9 and PREP1 cDNA probes. In panels C and D, each primary recipient is identified with a specific number, and its secondary recipients are identified with a derivative thereof (e.g., 1.1, 1.2, etc.). B, bone marrow; S, spleen.
PREP1, in contrast to MEIS1, does not accelerate the onset of HOXA9-induced AML.
To determine whether other members of the MEIS family could also accelerate the HOX-induced AML, HOXA9 and PREP1 were co-overexpressed in mouse bone marrow cells (see experiment 2, Table 1). PREP1 was selected for its reported functional similarities with MEIS1 (15) and its maximal divergence from MEIS1 in regions that exclude the conserved homeodomain and in the amino-terminal HMA and HMB motifs which mediate interaction with PBX proteins (5) (Fig. 1B).
Despite initial transplantation of the HOXA9-PREP1 mice with an ∼20-fold-higher number of doubly transduced cells than that for the HOXA9-MEIS1 mice (Table 1, experiment 2), the HOXA9-PREP1 mice developed AML with a latency similar to (or even longer than) that of the HOXA9 mice (Fig. 5A). In contrast, the HOXA9-MEIS1 mice, as previously reported (17), developed AML with an approximately three-times-shorter latency period (Fig. 5A). The leukemias that developed in the HOXA9-PREP1 mice were all AML and were morphologically similar to those that developed with HOXA9 (Fig. 4B). Thus, in contrast to other reported functional similarities with MEIS1, PREP1 cannot accelerate the occurrence of HOXA9-induced leukemias.
To exclude the possibility that the lack of collaboration between HOXA9 and PREP1 was caused by a failure to generate PREP1 protein from the PREP1 provirus, Western blot analysis was performed on total cellular lysates from the PREP1 and HOXA9 viral producer cells. As opposed to low levels of endogenous PREP1 present in the HOXA9 viral producer cells, high levels of PREP1 protein were detected in the PREP1 producer cells (Fig. 5B).
Interestingly, all leukemias that developed in the primary HOXA9-PREP1 mice contained and expressed both the HOXA9 and PREP1 proviruses (Fig. 5C and 5D). This does not indicate genetic collaboration but rather reflects the very high double gene transfer for the HOXA9 and PREP1 retroviruses, as ∼50% of HOXA9-transduced myeloid progenitors that were transplanted initially also contained the PREP1 provirus (Table 1). Definitive proof for the absence of genetic interaction between HOXA9 and PREP1 was provided by the clonal analysis and transplantation of the leukemias that developed in these mice. For example, of three leukemic clones (i.e., Fig. 5C, a, b, and c) detected in primary recipient 2, only one clone (clone c) contained both the HOXA9 and PREP1 proviruses (HOXA9 at five integration sites and PREP1 at three), while the two other clones (a and b) contained only the HOXA9 provirus (in clone a at one integration site and in clone b at two integration sites). When the leukemic cells from this primary mouse (mouse 2) were transplanted to secondary recipients, the PREP1-containing clone c could be outcompeted by clone b lacking PREP1 (Fig. 5C, compare 2B with 2.3B). This demonstrates that PREP1 was not essential for the maintenance of the HOXA9-induced leukemia.
These data demonstrate a lack of collaboration between HOXA9 and PREP1 in leukemic transformation, thus underscoring the specificity of the collaboration between HOX genes and MEIS1.
DISCUSSION
Previous studies favored the possibility that HOX and TALE genes would collaborate in specific pairs, with the pentapeptide-containing HOX proteins (e.g., HOXB3) collaborating with PBX and HOX proteins from paralogous groups 9 to 13 (e.g., HOXA9) collaborating with MEIS (17, 18). The studies reported in this paper clearly indicate that MEIS1 is a common leukemogenic collaborator with the two highly divergent HOX genes HOXB3 and HOXA9. These data would thus argue against the concept of specific collaborating pairs but would rather support a common mechanism in leukemias induced by HOX genes and MEIS1. The specificity of the MEIS1-HOX collaboration for leukemic transformation was evidenced by the inability of another MEIS family member, PREP1, to substitute for MEIS1 in accelerating the HOXA9-induced AML. Evidence presented herein also establishes the lack of oncogenicity of MEIS1 when overexpressed alone in primitive bone marrow cells and shows that the leukemogenic potential and phenotypes of the leukemias induced by the HOX-MEIS1 pair are largely dependent on the HOX gene involved, with MEIS1 acting mainly to accelerate the onset of these leukemias.
The nature of the collaboration between MEIS1 and HOX genes in the induction of AML.
Biochemical and genetic studies have demonstrated the importance of HOX-PBX (2, 29) and, most recently, HOX-PBX-MEIS heterocomplex formation for the execution of some HOX-dependent developmental programs (9, 14, 31). Previously, we showed that HOXB3- or HOXB4-induced transformation of Rat-1 fibroblasts is dependent on endogenous PBX1 levels and is enhanced by co-overexpression of PBX1, underscoring a role for a complex containing HOX and PBX in transformation (18). However, with respect to transformation of hemopoietic cells, no such collaboration can be detected between HOXB3 and PBX1 but can be detected rather between HOXB3 and MEIS1. These findings were most surprising, considering that HOXB3-induced transformation of Rat-1 fibroblasts was not enhanced by the coexpression of MEIS1 (J. Krosl and G. Sauvageau, unpublished observation). This emphasizes the importance of the cell type used to study HOX-induced transformation (i.e., primitive bone marrow cells for leukemias). In both Drosophila and mammalian development, the nuclear localization of EXD or PBX is dependent on the presence of HTH or MEIS, whereas a MEIS-independent mechanism appears to operate to maintain PBX nuclear localization in fibroblast cell lines (1, 3, 15, 27). Although it has not been determined for primitive hemopoietic cells, the inability of PBX1 to accelerate the HOX (-A9 or -B3)-induced leukemias when overexpressed could be explained by its cytoplasmic, rather than nuclear, localization in the absence of MEIS proteins. In support of a role for PBX proteins in HOX-induced leukemias, the tryptophan motif of HOXA9 (essential for HOXA9-PBX interaction) was recently demonstrated to be necessary for HOXA9-induced in vitro immortalization of myeloid progenitor cells (35), although another study suggests that it might be dispensable (7). The ability of MEIS1 to induce AML in collaboration with HOX proteins must, however, entail more than retaining endogenous PBX protein in the nucleus. This is evident by our demonstration here that the PREP1 protein, which is capable of inducing nuclear localization of EXD and PBX, in both Drosophila and mammalian cells (3, 15), lacks the ability to accelerate the HOXA9-induced leukemias. Together, these data indicate that in the HOX-induced leukemias the MEIS1 protein must have another role, in addition to one potentially involving PBX, which cannot be accomplished by PREP1.
It was recently demonstrated in two hematopoietic cell lines (i.e., U-937 and KG1) that the HOXA9 protein is part of a trimeric complex with both PBX2 and MEIS1 (37). This suggests that at least some HOX gene functions in hematopoietic cells could be dependent on such a trimeric complex formation. However, definitive proof of whether a similar trimeric complex is the foundation for the collaboration between HOX and MEIS1 proteins in leukemic transformation can be accomplished only with the use of appropriate HOX and TALE mutants, or by the identification of transforming targets which would require HOX-PBX and MEIS interactions for their full activation.
HOX genes determine the identity of the HOX- and MEIS1-induced AML.
Although co-overexpression of MEIS1 accelerated the occurrence of both the HOXB3- and HOXA9-induced AML, their phenotypes remained HOX gene dependent. This observation is not restricted to HOXB3 and HOXA9, as the occurrence of the AML induced by expression of the human fusion protein NUP98-HOXA9 is also accelerated by MEIS1, without affecting its phenotype (E. Kroon et al., unpublished data). The underlying mechanism responsible for the differences between the HOXA9- and HOXB3-induced leukemias is currently unknown. Previous and ongoing studies by our group have demonstrated that, when overexpressed in mouse bone marrow cells, the four HOX genes tested thus far generate distinct hematopoietic phenotypes (33, 34, 39; U. Thorsteinsdottir et al., unpublished data). This suggests that a subset of target genes, possibly responsible for cellular identity, is differentially regulated by each HOX gene product, thereby predisposing target cells to leukemias with different characteristics.
We show here that MEIS1, in contrast to most clustered and nonclustered HOX genes (e.g., TCL-3 or HOX11), does not predispose target cells to leukemia when overexpressed in mouse bone marrow cells (13, 34, 39). This difference might be attributed to the inability of MEIS1, as shown here both in vivo and in vitro, to confer any proliferative advantage on primitive hematopoietic cells. In contrast, we and others have shown previously that overexpression of all of the HOX genes tested so far, as well as the nonclustered HOX11 gene, enhances the proliferative potential of primitive hematopoietic cells (34, 38, 39). The ability of MEIS1 to possess leukemogenic potential when co-overexpressed with HOX genes raises the possibility that it could engage in similar collaboration with other oncogenes that enhance cellular proliferation. This hypothesis is currently being evaluated in our laboratory.
Functional differences between the MEIS family members MEIS1 and PREP1.
Of the four mammalian MEIS family members, the MEIS1, MEIS2, and MEIS3 proteins share a high sequence similarity over the entire protein sequence (e.g., MEIS1 versus MEIS2, 77.2%, and MEIS1 versus MEIS3, 69.9%), which is highest in their homeodomain and their PBX interaction domain, HM (23). In contrast, apart from the homeodomain and the HM domain, the PREP1 protein does not share high sequence similarity with other members of the MEIS family. Despite this difference, PREP1 can substitute for MEIS1 or HTH in directing PBX or EXD nuclear localization (3, 15) and, like MEIS1, can form a heterotrimer with HOXB1 and PBX1 on the HOXB2 enhancer element (9, 14). In addition, recent studies using transgenic flies have shown functional conservation between HTH and PREP1 (15). The inability of PREP1 to accelerate the HOXA9-induced leukemias described here represents direct evidence for a functional difference between the PREP1 and MEIS1 proteins. On the basis of these studies, this difference is likely mediated through parts of the MEIS1 and PREP1 proteins other than the homeodomain or the HM domain and thus may involve functions other than DNA binding and interaction with PBC proteins.
The finding that PREP1 is incapable of accelerating the HOX-induced leukemias and the low overall sequence similarity between PREP1 and the three other MEIS family members also raise the possibility that, in vertebrates, PREP1 could have evolved to perform functions (perhaps antagonistic) distinct from those of other family members. This difference could thus allow an additional level of regulation within HOX- and TALE-dependent pathways. Interestingly, the PREP1 and MEIS1 protein levels are differentially regulated upon retinoic acid treatment of embryonic carcinoma P19 cells, with PREP1 protein levels dominating in untreated cells and MEIS1 dominating after retinoic acid treatment (9). Furthermore, in adult mouse tissues PREP1 is expressed ubiquitously (10), whereas MEIS1 expression appears to be more specific (10). These studies, together with the data presented here, are thus suggestive of dissimilar regulatory roles for MEIS1 and PREP1 proteins.
In summary, the results of the present study are highly suggestive that genetic interaction with MEIS1 is part of a common mechanism in HOX-induced leukemias. These studies also establish that each of the two HOX genes tested has the capacity to determine the phenotype of the leukemias, independently of MEIS1 co-overexpression. The inability of PREP1 to substitute for MEIS1 indicates that MEIS1 function in this collaboration must involve more than PBX nuclear retention. The existence of such a common mechanism, together with the growing evidence that HOX genes and their cofactors are causal oncogenes for human leukemia, reinforces the importance of defining the (common) molecular basis underlying HOX-induced transformation.
ACKNOWLEDGMENTS
We acknowledge Nadine Mayotte for expert technical assistance and Marie-Eve Leroux and Stephane Matte for their expertise and help regarding the maintenance and manipulation of the animals kept at the specific-pathogen-free facility. The support of Nathalie Tessier is also acknowledged for FACS analyses. Robert G. Hawley is acknowledged for his MSCV vectors.
This work was supported by a grant from the National Cancer Institute of Canada (NCI-C). U.T. is the recipient of a Leukemia Research Fund of Canada Fellowship, E.K. is the recipient of a Leukemia and Lymphoma Society of America Fellowship, L.J. is the recipient of a Medical Research Council (MRC) of Canada Fellowship, and Guy Sauvageau is an MRC Clinician-Scientist Scholar.
REFERENCES
- 1.Abu-Shaar M, Ryoo H D, Mann R S. Control of the nuclear localization of extradenticle by competing nuclear import and export signals. Genes Dev. 1999;13:935–945. doi: 10.1101/gad.13.8.935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Azpiazu N, Morata G. Functional and regulatory interactions between Hox and extradenticle genes. Genes Dev. 1998;12:261–273. doi: 10.1101/gad.12.2.261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Berthelsen J, Kilstrup-Nielsen C, Blasi F, Mavilio F, Zappavigna V. The subcellular localization of PBX1 and EXD proteins depends on nuclear import and export signals and is modulated by association with PREP1 and HTH. Genes Dev. 1999;13:946–953. doi: 10.1101/gad.13.8.946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Berthelsen J, Zappavigna V, Feretti E, Mavilio F, Blasi F. The novel homeoprotein Prep1 modulates Pbx-Hox protein cooperativity. EMBO J. 1998;17:1434–1445. doi: 10.1093/emboj/17.5.1434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Berthelsen J, Zappavigna V, Mavilio F, Blasi F. Prep1, a novel functional partner of Pbx proteins. EMBO J. 1998;17:1423–1433. doi: 10.1093/emboj/17.5.1423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Borrow J, Shearman A M, Stanton V P, Becher R, Collins T, Williams A J, Dubé I D, Katz F, Kwong Y L, Morris C, Ohyashiki K, Toyama K, Rowley J, Housman D E. The t(7;11)(p15;p15) translocation in acute myeloid leukemia fuses the genes for nucleoporin NUP98 and class I homeoprotein HOXA9. Nat Genet. 1996;12:159–167. doi: 10.1038/ng0296-159. [DOI] [PubMed] [Google Scholar]
- 7.Calvo K R, Sykes D B, Pasillas M, Kamps M P. Hoxa9 immortalizes a granulocyte-macrophage colony-stimulating factor-dependent promyelocyte capable of biphenotypic differentiation to neutrophils or macrophages, independent of enforced Meis expression. Mol Cell Biol. 2000;20:3274–3285. doi: 10.1128/mcb.20.9.3274-3285.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chang C P, Jacobs Y, Nakamura T, Jenkins N A, Copeland N G, Cleary M L. Meis proteins are major in vivo DNA binding partners for wild-type but not chimeric Pbx proteins. Mol Cell Biol. 1997;17:5679–5687. doi: 10.1128/mcb.17.10.5679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ferreti E, Marshall H, Pöpperl H, Maconochie H, Krumlauf R, Blasi F, Berthelsen J. Segmental expression of Hoxb2 in r4 requires two separate sites that integrate cooperative interactions between Prep1, Pbx and Hox proteins. Development. 2000;127:155–166. doi: 10.1242/dev.127.1.155. [DOI] [PubMed] [Google Scholar]
- 10.Ferreti E, Schulz H, Talarico D, Blasi F. The PBX-regulating protein PREP1 is present in different PBX-complexed forms in mouse. Mech Dev. 1999;83:53–64. doi: 10.1016/s0925-4773(99)00031-3. [DOI] [PubMed] [Google Scholar]
- 11.Giampaolo A, Sterpetti P, Bulocrini D, Samoggia P, Pelosi P, Valtieri F, Peschle C. Key functional role and lineage-specific expression of HOXB cluster genes in purified hematopoietic progenitor differentiation. Blood. 1994;84:3637–3647. [PubMed] [Google Scholar]
- 12.Golub T R, Slonim D K, Tamayo P, Huard C, Gaasenbeek M, Mesirov J P, Coller H, Loh M L, Downing J R, Caligiuri M A, Bloomfield C D, Lander E S. Molecular classification of cancer: class discovery and class prediction by gene expression monitoring. Science. 1999;286:531–537. doi: 10.1126/science.286.5439.531. [DOI] [PubMed] [Google Scholar]
- 13.Hawley R G, Fong A Z, Reis M D, Zhang N, Lu M, Hawley T S. Transforming function of the HOX11/TCL3 homeobox gene. Cancer Res. 1997;57:337–345. [PubMed] [Google Scholar]
- 14.Jacobs Y, Schnabel C A, Cleary M L. Trimeric association of Hox and TALE homeodomain proteins mediates Hoxb2 hindbrain enhancer activity. Mol Cell Biol. 1999;19:5134–5142. doi: 10.1128/mcb.19.7.5134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jaw T J, You L-R, Knoepfler P S, Yao L-C, Pai C-Y, Tang C-Y, Chang L-P, Berthelsen J, Blasi F, Kamps M P, Sun Y H. Direct interaction of two homeoproteins, Homothorax and Extradenticle, is essential for EXD nuclear localization and function. Mech Dev. 2000;91:279–291. doi: 10.1016/s0925-4773(99)00316-0. [DOI] [PubMed] [Google Scholar]
- 16.Kamps M P, Murre C, Sun X H, Baltimore D. A new homeobox gene contributes the DNA binding domain of the t(1;19) translocation protein in pre-B ALL. Cell. 1990;60:547–555. doi: 10.1016/0092-8674(90)90658-2. [DOI] [PubMed] [Google Scholar]
- 17.Kroon E, Krosl J, Thorsteinsdottir U, Baban S, Buchberg A M, Sauvageau G. Hoxa9 transforms primary bone marrow cells through specific collaboration with Meis1a but not Pbx1b. EMBO J. 1998;17:3714–3725. doi: 10.1093/emboj/17.13.3714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Krosl J, Baban S, Krosl G, Rozenfeld S, Largman C, Sauvageau G. Cellular proliferation and transformation induced by HOXB4 and HOXB3 proteins involves cooperation with PBX1. Oncogene. 1998;16:3403–3412. doi: 10.1038/sj.onc.1201883. [DOI] [PubMed] [Google Scholar]
- 19.Look A T. Oncogenic transcription factors in the human acute leukemias. Science. 1997;278:1059–1064. doi: 10.1126/science.278.5340.1059. [DOI] [PubMed] [Google Scholar]
- 20.Mann R S. The specificity of homeotic gene function. Bioessays. 1995;17:855–863. doi: 10.1002/bies.950171007. [DOI] [PubMed] [Google Scholar]
- 21.Monica K, LeBrun D P, Dedera D A, Brown R, Cleary M L. Transformation properties of the E2a-Pbx1 chimeric oncoprotein: fusion with E2a is essential, but the Pbx1 homeodomain is dispensable. Mol Cell Biol. 1994;14:8304–8314. doi: 10.1128/mcb.14.12.8304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Moskow J J, Bullrich F, Huebner K, Daar I O, Buchberg A M. Meis1, a PBX1-related homeobox gene involved in myeloid leukemia in BXH-2 mice. Mol Cell Biol. 1995;15:5434–5443. doi: 10.1128/mcb.15.10.5434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nakamura T, Jenkins N A, Copeland N G. Identification of a new family of Pbx-related homeobox genes. Oncogene. 1996;15:2235–2242. [PubMed] [Google Scholar]
- 24.Nakamura T, Largaespada D A, Lee M P, Johnson L A, Ohyashiki K, Toyama K, Chen S J, Willman C L, Chen I M, Feinberg A P, Jenkins N A, Copeland N G, Shaughnessy J D. Fusion of the nucleoporin gene NUP98 to HOXA9 by the chromosome translocation t(7;11)(p15; p15) in human myeloid leukaemia. Nat Genet. 1996;12:154–158. doi: 10.1038/ng0296-154. [DOI] [PubMed] [Google Scholar]
- 25.Nakamura T, Largaespada D A, Shaughnessy J D, Jenkins N A, Copeland N G. Cooperative activation of Hoxa and Pbx1-related genes in murine myeloid leukaemias. Nat Genet. 1996;12:149–153. doi: 10.1038/ng0296-149. [DOI] [PubMed] [Google Scholar]
- 26.Nourse J, Mellentin J D, Galili N, Wilkinson J, Stanbridge E, Smith S D, Cleary M C. Chromosomal translocation t(1;19) results in synthesis of a homeobox fusion mRNA that codes for a potential chimeric transcription factor. Cell. 1990;60:535–546. doi: 10.1016/0092-8674(90)90657-z. [DOI] [PubMed] [Google Scholar]
- 27.Pai C Y, Kuo T S, Jaw T J, Kurant E, Chen C T, Bessarab D A, Salzberg A, Sun Y H. The Homothorax homeoprotein activates the nuclear localization of another homeoprotein, Extradenticle, and suppresses eye development in Drosophila. Genes Dev. 1998;12:446. doi: 10.1101/gad.12.3.435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Perkins A C, Cory S. Conditional immortalization of mouse myelomonocytic, megakaryocytic and mast cell progenitors by the Hox-2.4 homeobox gene. EMBO J. 1993;12:3835–3846. doi: 10.1002/j.1460-2075.1993.tb06062.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pöpperl H, Bienz M, Studer M, Chan S-K, Aparicio S, Mann R S, Krumlauf R. Segmental expression of Hoxb-1 is controlled by a highly conserved autoregulatory loop dependent upon exd/pbx. Cell. 1995;81:1031–1042. doi: 10.1016/s0092-8674(05)80008-x. [DOI] [PubMed] [Google Scholar]
- 30.Rieckhof G E, Casares F, Ryoo H D, Abu-Shaar M, Mann R S. Nuclear translocation of extradenticle requires homothorax, which encodes an extradenticle-related homeodomain protein. Cell. 1997;91:171–183. doi: 10.1016/s0092-8674(00)80400-6. [DOI] [PubMed] [Google Scholar]
- 31.Ryoo H D, Marty T, Casares F, Affolter M, Mann R S. Regulation of Hox target genes by a DNA bound Homothorax/Hox/Extradenticle complex. Development. 1999;126:5137–5148. doi: 10.1242/dev.126.22.5137. [DOI] [PubMed] [Google Scholar]
- 32.Sauvageau G, Lansdorp P M, Eaves C J, Hogge D E, Dragowska W H, Reid D S, Largman C, Lawrence H J, Humphries R K. Differential expression of homeobox genes in functionally distinct CD34+ subpopulations of human bone marrow cells. Proc Natl Acad Sci USA. 1994;91:12223–12227. doi: 10.1073/pnas.91.25.12223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sauvageau G, Thorsteinsdottir U, Eaves C J, Lawrence H J, Largman C, Lansdorp P M, Humphries R K. Overexpression of HOXB4 in hematopoietic cells causes the selective expansion of more primitive populations in vitro and in vivo. Genes Dev. 1995;9:1753–1765. doi: 10.1101/gad.9.14.1753. [DOI] [PubMed] [Google Scholar]
- 34.Sauvageau G, Thorsteinsdottir U, Hough M R, Hugo P, Lawrence H J, Largman C, Humphries R K. Over-expression of HOXB3 in hematopoietic cells causes defective lymphoid development and progressive myeloproliferation. Immunity. 1997;6:13–22. doi: 10.1016/s1074-7613(00)80238-1. [DOI] [PubMed] [Google Scholar]
- 35.Schnabel C A, Jacobs Y, Cleary M L. HoxA9-mediated immortalization of myeloid progenitors requires functional interactions with TALE cofactors Pbx and Meis. Oncogene. 2000;19:608–616. doi: 10.1038/sj.onc.1203371. [DOI] [PubMed] [Google Scholar]
- 36.Shen W F, Montgomery J C, Rozenfeld S, Moskow J J, Lawrence H J, Buchberg A M, Largman C. The AbdB-like proteins stabilize DNA binding by the Meis1 homeodomain proteins. Mol Cell Biol. 1997;17:6448–6458. doi: 10.1128/mcb.17.11.6448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shen W-F, Rozenfeld S, Kwong A, Kömüves L G, Lawrence H J, Largman C. HOXA9 forms triple complexes with PBX2 and MEIS1 in myeloid cells. Mol Cell Biol. 1999;19:3051–3061. doi: 10.1128/mcb.19.4.3051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Thorsteinsdottir U, Krosl J, Kroon E, Haman A, Hoang T, Sauvageau G. The oncogene E2A-Pbx1a collaborates with Hoxa9 to acutely transform primary bone marrow cells. Mol Cell Biol. 1999;19:6355–6366. doi: 10.1128/mcb.19.9.6355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Thorsteinsdottir U, Sauvageau G, Hough M R, Lawrence H J, Largman C, Humphries R K. Overexpression of HOXA10 in murine hematopoietic cells perturbs both myeloid and lymphoid differentiation and leads to acute myeloid leukemia. Mol Cell Biol. 1997;17:495–505. doi: 10.1128/mcb.17.1.495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Thorsteinsdottir U, Sauvageau G, Humphries R K. Hox homeobox genes as regulators of normal and leukemic hematopoiesis. Hematol Oncol Clin N Am. 1997;11:1221–1237. doi: 10.1016/s0889-8588(05)70491-3. [DOI] [PubMed] [Google Scholar]