Abstract
Extracellular signal-regulated kinase 1 (ERK1) and ERK2 are important components in signal transduction pathways involved in many cellular processes, including cell differentiation and proliferation. These proteins consist of a central kinase domain flanked by short N- and C-terminal noncatalytic domains. While the regulation of ERK2 by sequences within the kinase domain has been extensively studied, little is known about the small regions outside of the kinase domain. We performed mutational analysis on the N-terminal, noncatalytic domain of ERK2 in an attempt to determine its role in ERK2 function and regulation. Deleting or mutating amino acids 19 to 25 (ERK2-Δ19-25) created an ERK2 molecule that could be phosphorylated in response to growth factor and serum stimulation in a MEK (mitogen-activated protein kinase kinase or ERK kinase)-dependent manner but had little kinase activity and was unable to bind to MEK in vivo. Since MEK acts as a cytoplasmic anchor for the ERKs, the lack of a MEK interaction resulted in the aberrant nuclear localization of ERK2-Δ19-25 mutants in serum-starved cells. Assaying these mutants for their ability to affect ERK signaling revealed that ERK2-Δ19-25 mutants acted in a dominant-negative manner to inhibit transcriptional signaling through endogenous ERKs to an Elk1-responsive promoter in transfected COS-1 cells. However, ERK2-Δ19-25 had no effect on the phosphorylation of RSK2, an ERK2 cytoplasmic substrate, whereas a nonactivatable ERK (T183A) that retained these sequences could inhibit RSK2 phosphorylation. These results suggest that the N-terminal domain of ERK2 profoundly affects ERK2 localization, MEK binding, kinase activity, and signaling and identify a novel dominant-negative mutant of ERK2 that can dissociate at least some transcriptional responses from cytoplasmic responses.
The extracellular signal-regulated kinases (ERKs), or mitogen-activated protein (MAP) kinases, are ubiquitous serine/threonine protein kinases that lie in a signaling pathway downstream of the ras oncogene. These kinases are involved in signaling cascades that regulate a number of cellular pathways, including the control of both cell proliferation and differentiation (29). The two best-characterized ERKs, ERK1 and ERK2, are activated by phosphorylation on threonine and tyrosine in a TEY sequence within the kinase domain (19, 31) by the dual-specificity kinases MEK1 and MEK2 (MAP kinase kinase kinase or ERK kinase). Activation of various MAP kinase family members by MEKs in mammalian cells has recently been shown to be facilitated by scaffold proteins that selectively bind and bring together specific components of the signaling pathway (35, 41).
In cultured cells deprived of serum and growth factors, ERK1 and ERK2 are predominantly inactive and reside in the cytoplasm (10, 18, 27, 34). Ectopic expression of ERKs causes their localization in the nucleus and in the cytoplasm of serum-starved as well as proliferating cells (13, 18). Cytoplasmic retention of the ERKs under serum-starved conditions can be controlled by association with the MEKs and by MAP kinase phosphatase 3 (MKP-3), which act as cytoplasmic anchors for the ERKs due to the presence of nuclear export signals in these anchoring proteins (5, 13, 15, 28). Immunofluorescence studies have demonstrated that MEKs appear to be predominantly cytoplasmic in both quiescent and proliferating cells (27, 48); however, nuclear localization of MEK has been observed under certain conditions (22, 40).
Upon mitogen stimulation of the cell, the ERKs are quickly phosphorylated by the MEKs and a portion of the active ERK population translocates to the nucleus (10, 18, 27). Phosphorylation of the ERKs results in their dimerization, which facilitates their nuclear localization (6, 25). Nuclear localization appears to occur by both active transport and passive diffusion (1). In addition, neosynthesis of unstable proteins appears to be required to facilitate sustained ERK nuclear localization, suggesting that these unidentified labile proteins may serve as nuclear anchors for the ERKs (26). ERK activation and nuclear translocation have been demonstrated to be required for cellular proliferation and S-phase entry in fibroblasts (5, 30).
The ERK proteins consist of a central kinase domain flanked by short N- and C-terminal extensions. Crystallographic studies of ERK2 have demonstrated that the phosphorylation of the activating sites causes major structural changes within the kinase domain involving rotation of the phosphorylation lip that allows the active kinase to bind substrate (6, 47). The N- and C-terminal extensions outside of the kinase domain lie on the surface of the molecule and undergo only small alterations in position upon phosphorylation of ERK2 (6, 47).
While the activation and regulation of the ERKs has been extensively studied, little is known about the short N-terminal region that resides outside of the kinase domain. A portion of the C-terminal domain was shown to be required for ERK dimerization and nuclear translocation (6). Recent reports have focused on the C-terminal domain's role as a docking site for interactions between ERK2 and its regulators, as well as its role in the subcellular localization of ERK2 (33, 39).
We used deletion and substitution mutagenesis of short sequences within the N-terminal noncatalytic domain of ERK2 in an effort to determine what function, if any, it has in regulation of the protein's activation and function. ERK2 mutants containing mutations in amino acids 19 to 25 did not associate with MEK in cotransfected cells and had little kinase activity but did retain the ability to be phosphorylated on their activating sites in a MEK-dependent manner in response to mitogens. The failure of these mutants to bind to MEK was associated with their nuclear localization even in serum-starved cells. Screening these mutants for their ability to affect signaling to ERK substrates revealed that they acted as dominant-negatives in signaling to nuclear, but not to cytoplasmic, ERK substrates. These results identify the N-terminal domain as being important in regulating the activity, MEK association, substrate targeting, and localization of ERK2.
MATERIALS AND METHODS
Cell culture, transfections, and plasmids.
COS-1 cells were grown in Dulbecco's modified Eagle medium (DMEM) (Life Technologies, Rockville, Md.) supplemented with 10% fetal calf serum (FCS) at 37°C with 5% CO2. All transfections were performed using Lipofectamine (Life Technologies). FLAG-ERK2 has been described previously (37). ERK2 mutants were generated using the Transformer mutagenesis kit (Clontech, Palo Alto, Calif.). ERK2-Δ3-7 and ERK2-Δ10-19 also contain a C-terminal nonfunctional KT3 tag sequence, which accounts for the increase in the size of the protein product. Hemagglutinin (HA)-MEK constructs have been described previously (8). 5X GAL4-luciferase and GAL4-Elk1 were provided by Richard Maurer. HA-tagged Ras V12 was provided by Channing Der. HA-RSK2 was provided by Thomas Sturgill.
Luciferase assays.
COS-1 cells were cotransfected in triplicate with 1 μg of 5X GAL4-luciferase, 50 ng of GAL4-Elk1, 100 ng of HA-Ras V12 (or 100 ng of HA-MEK1 S218/222D), and a FLAG-ERK2 plasmid as indicated. Differing amounts of ERK2 plasmid were required to obtain equal protein expression. Wild-type (WT) ERK2 (WT-ERK2) plasmid amounts were 5, 10, 50, and 100 ng. ERK2-Δ10-19 amounts were 10, 20, 100, and 200 ng. ERK2-Δ10-25 and ERK2-Δ19-25 plasmid amounts were 40, 80, 400, and 800 ng. All transfection amounts were brought up to a total of 1.95 μg of DNA per transfection with the appropriate empty vector. After transfection the cells were placed in serum-free DMEM overnight and harvested at 24 h. Luciferase activity was determined on a Monolight 2010 Luminometer (Analytical Luminescence, Ann Arbor, Mich.). Western blot analyses for protein expression were performed on an equal amount of lysate from each sample. For the MEK1-versus-MEK2 experiment, 100 ng of either one was cotransfected with 100 ng of WT-ERK2, 200 ng of ERK2-Δ10-19, or 800 ng of an ERK2-Δ19-25 mutant. For the competition experiment, 100 ng of HA-MEK1 S218/222D was cotransfected with 800 ng of ERK2-Δ19-25 or empty vector. WT-ERK2 was titrated in with 5, 10, 25, 50, and 100 ng of plasmid.
Coimmunoprecipitations.
COS-1 cells were cotransfected with 2 μg of HA-MEK2 plasmid and a total of 4 μg of FLAG-ERK2 plasmid and empty vector in order to obtain equal ERK2 protein expression. After transfection the cells were incubated overnight in DMEM supplemented with 10% FCS. The following day the cells were washed twice and placed in serum-free DMEM for 4 h to inactivate the ERK pathway. The cells were harvested in hypotonic buffer, and immunoprecipitations were performed as described previously (7, 13).
For RSK2 and GAL4-ELK experiments, the cells were starved for 5 h and either harvested or stimulated for 10 min with 10 ng of epidermal growth factor (EGF) per ml. Cells were harvested in hypotonic buffer (20 mM HEPES [pH 7.4], 2 mM EDTA, 2 mM EGTA) and lysed by centrifugation. For the GAL4-Elk1 experiment, the cells were sonicated prior to centrifugation. Coimmunoprecipitations were performed as described above.
Immunofluorescence.
COS-1 cells were cotransfected with HA-MEK2 and FLAG-ERK2 plasmids as indicated. The following day the cells were replated onto coverslips and later serum starved for 4 h before fixing in 4% paraformaldehyde and permeabilizing with 0.2% Triton X-100. The fixed cells were blocked in 20% goat serum and then probed with monoclonal M2 anti-FLAG antibody (Sigma) and polyclonal anti-HA antibody (Babco, Richmond, Calif.) in 5% goat serum. After washing in 0.05% Tween 20 in phosphate-buffered saline, the cells were probed with fluorescein isothiocyanate-conjugated anti-mouse and Texas red-conjugated anti-rabbit antibodies (Jackson Immunoresearch, West Grove, Pa.). Cells were DAPI stained (Sigma), dried, and mounted with Vectashield mounting media (Vector Laboratories, Burlingame, Calif.). Indirect immunofluorescence examination was performed on a Leica microscope.
Kinase assays.
COS-1 cells were transfected with ERK2 plasmids to obtain equal protein expression. All transfections were brought up to 2.0 μg of total DNA with pCDNA3 vector. The following day, the cells were serum starved for 4 h and either harvested or stimulated for 5 min with either 10% FCS or 10 ng of EGF (Upstate Biotechnology, Lake Placid, N.Y.) per ml. For the PD098059 experiment, either 50 μM PD098059 (BIOMOL, Plymouth Meeting, Pa.) or the dimethyl sulfoxide (DMSO) vehicle was added for the last hour of serum starvation and during the stimulation. The cells were harvested, and the kinase assays were performed on immunoprecipitated FLAG-ERKs as described previously (35). Phosphorylated myelin basic protein (MBP) was cut from the membrane and quantitated by Cerenkov counting. FLAG-ERKs were visualized by Western blotting using M5 anti-FLAG antibody (Sigma). Anti-phospho-ERK blotting was performed using a polyclonal antibody that specifically recognizes the dually phosphorylated (T183 and Y185), active forms of the ERKs (46).
Cell fractionation.
COS-1 cells were transfected with 1 μg of HA-MEK2 and ERK expression plasmids and empty vector to obtain equal protein expression. Cells were serum starved for the last 4 h before harvest at 24 h posttransfection. Cells were harvested in RSB (10 mM Tris-HCl [pH 7.4], 2 mM MgCl2, 2 mM phenylmethylsulfonyl fluoride, 5 μg of aprotinin per ml, 1 μg of leupeptin per ml, 200 μM Na orthovanadate) and left on ice for 5 min. Triton X-100 was added from a 10% stock to a final concentration of 0.5%, and the cells were left for another 5 min on ice. The cells were sheared by passage through a 21-gauge needle three times and then spun for 5 min at 1,000 × g through a 1 M sucrose pad to pellet the nuclei. The supernatant (cytoplasm and membranes) was removed, and the pellets (nuclei) were washed once in 3 ml of RSB and centrifuged as before. Equal cell equivalents were run on sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) gels and immunoblotted.
RSK phosphorylation.
COS-1 cells were transfected with 0.1 μg of HA-RSK2, 0.1 μg of HA-RasV12, and either 0.875 μg of WT-ERK2, 2 μg of ERK2 T183A, or 7 μg of ERK2-Δ19-25-7A to obtain equal ERK expression. Cells were put in 10% serum overnight and serum starved for 5 h before harvest at 24 h in FLAG buffer (35). HA RSK2 was immunoprecipitated with 12CA5 antibody preconjugated to protein A-agarose (Roche), and the pellets were washed three times in lysis buffer and run on a 10 to 15% acrylamide–SDS gel. Phospho-RSK2 was determined by immunoblotting with an antibody to phospho-Ser 380 of RSK1 (Upstate Biotechnology), a site of autophosphorylation in response to ERK phosphorylation (11).
RESULTS
N-terminal ERK2 mutants.
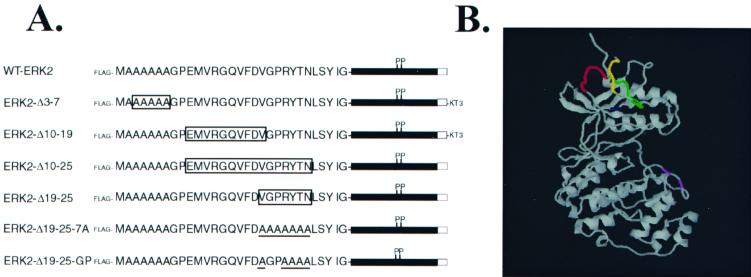
ERK2 consists of a central Ser/Thr kinase domain that is flanked by short N- and C-terminal domains that lie on the surface of the protein (47). To examine roles that the N-terminal region may play in the function of ERK2, a series of deletion and substitution mutants was constructed. These constructs were placed into the pCDNA3 vector containing a sequence coding for an N-terminal FLAG epitope (Fig. 1A). Figure 1B shows the crystal structure of unphosphorylated ERK2 (47), with the sequences affected by the mutations highlighted.
FIG. 1.
Diagram of ERK2 mutants and crystal structure of ERK2, with mutated regions highlighted. (A) The amino acid sequence of the N terminus of murine ERK2 up to the beginning of the kinase domain is shown, with the open boxes indicating sequences that were deleted from the various mutants and the underlining indicating sequences that were mutated to Ala. The shaded box indicates the kinase domain, and the open box indicates the C terminus. All constructs encode an N-terminal FLAG epitope tag, while mutants ERK2-Δ3-7 and ERK2-Δ10-19 contain an additional nonfunctional KT3 tag at the C terminus, which accounts for their decreased mobility in SDS-PAGE compared to wild-type ERK2. (B) The crystal structure of unphosphorylated ERK2, solved by Zhang and colleagues (47), is shown using the RasMol program. Mutated sequences are colored as follows: ERK2-Δ3-7 is yellow, ERK2-Δ10-19 is green, ERK2-Δ10-25 is green and red, and ERK2-Δ19-25 is red. Lys 52 is shown in blue; Thr 183 and Tyr 185 are shown in purple.
Mutants lacking residues 19 to 25 are phosphorylated in response to mitogens but have little activity.
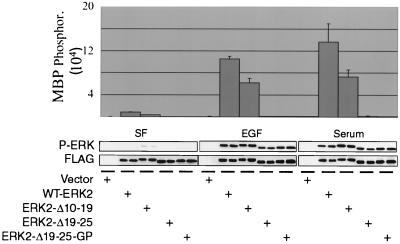
As an initial means to characterize these N-terminal ERK2 mutants, we determined their ability to be phosphorylated and activated in response to mitogens. WT-ERK2 and the ERK2 mutants were transiently transfected into COS-1 cells, and the cells were serum starved and then stimulated with EGF or serum for 10 min. Immunoprecipitated FLAG-ERKs were assayed for their ability to phosphorylate MBP (Fig. 2), an ERK2 substrate that does not have an ERK2 docking site (39). As expected, the activity of WT-ERK2 was greatly stimulated by both serum and by EGF, with kinase activity being induced 15.6-fold and 12-fold, respectively, over that seen in starved cells. The activity of ERK2-Δ10-19 was also stimulated in response to mitogens (20-fold by serum and 17.7-fold by EGF), although the overall level of activity was not as high as that of WT-ERK2. This suggests that deletion of a portion of the N terminus in ERK2-Δ10-19 caused this mutant to lose activity compared to WT-ERK2, although the fold stimulations in activity compared to WT-ERK2 were similar.
FIG. 2.
ERK2-Δ19-25 mutants are phosphorylated in response to mitogens but have little kinase activity. COS-1 cells were transfected with either empty vector, WT-ERK2, or an ERK2 mutant. The following day the cells were serum starved for 4 h before stimulation for 5 min with either 10 ng of EGF per ml or 10% serum. FLAG-ERK immunoprecipitates were used for an in vitro kinase assay using MBP as the substrate. The reaction mixtures were immunoblotted with antibodies for FLAG and phospho-ERK to determine protein expression and phosphorylation. SF, serum-free; P-ERK, phospho-ERK.
In contrast, ERK2-Δ19-25 and ERK2-Δ19-25-GP (Fig. 2), as well as ERK2-Δ10-25 and ERK2-Δ19-25-7A (data not shown) had less than 1% of the kinase activity towards MBP compared to WT-ERK2 when cells were stimulated by either mitogen, suggesting that deletion or mutation of this region destroys the kinase activity of ERK2. This reduction of kinase activity was in agreement with experiments in which a large portion of the N terminus of ERK2 was replaced with that of p38 (43). However, ERK2-Δ19-25 and ERK2-Δ19-25-GP were nonetheless phosphorylated on the regulatory sites by endogenous MEKs in response to mitogens, as shown by a phospho-ERK blot of the immunoprecipitates from the kinase assay using an antibody that recognizes only the dually phosphorylated forms of the ERKs (46). So while ERK2-Δ19-25 and ERK2-Δ19-25-GP had little kinase activity, they were phosphorylated in response to EGF and serum to the same level as WT-ERK2 and ERK2-Δ10-19. Metabolic labeling of transfected cells demonstrated that ERK2-Δ19-25 was phosphorylated on Thr and Tyr to the same extent as WT-ERK2 in response to EGF stimulation (data not shown).
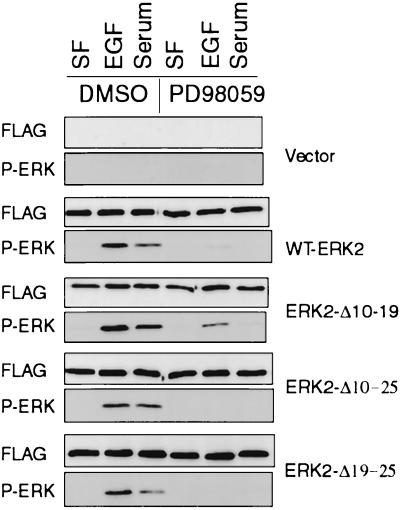
To confirm that the mutations had not changed the specificity of ERK2 as an in vivo substrate for MEK, the experiment was repeated with parallel dishes of cells treated with either DMSO or the MEK inhibitor PD098059 (Fig. 3). This drug specifically inhibits the activation of MEK1 and MEK2 (2). As expected, treatment of cells with PD098059 inhibited the phosphorylation of WT-ERK2 and ERK2-Δ10-19 by EGF and serum. PD098059 also inhibited the phosphorylation of ERK2-Δ10-25 and ERK2-Δ19-25 in response to mitogens, indicating that the observed phosphorylation of these mutants, as with WT-ERK2, was dependent on endogenous MEKs. Similar results were obtained with the MEK inhibitor U0126 (12), and both inhibitors were effective towards phosphorylation of ERK2-Δ19-25-7A and ERK2-Δ19-25-GP (data not shown). These mutants can therefore be phosphorylated in a MEK-dependent manner, but they have almost no kinase activity. The fact that endogenous MEKs still phosphorylate them to the same level as WT-ERK2 in vivo suggests that the mutations do not result in gross distortions of ERK2 structure, since MEK will not phosphorylate denatured ERKs (19, 36).
FIG. 3.
Phosphorylation of ERK2 mutants in response to EGF is MEK dependent. COS-1 cells were transfected and serum starved as described for Fig. 2. For the last hour of serum starvation the cells were treated with either DMSO or 50 μM PD098059 to inhibit MEK activation. The cells were then stimulated for 10 min with either EGF or serum in the presence of either DMSO or PD098059. The cells were harvested, and FLAG-ERK2 immunoprecipitates were immunoblotted with anti-FLAG and anti-phospho-ERK antibodies. SF, serum-free. P-ERK, phospho-ERK.
ERK proteins with mutations at residues 19 to 25 do not coimmunoprecipitate with MEK.
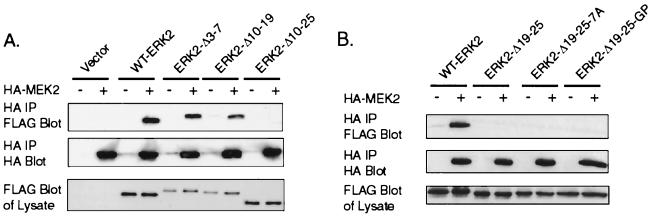
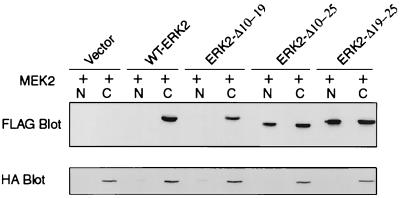
MEK binds to ERK through the N-terminal domain of MEK (3, 15). We next determined if the N-terminal domain of ERK2 was required for association with MEK. WT-ERK2 or an ERK2 deletion mutant was cotransfected with HA-MEK2 into COS-1 cells and harvested after serum starvation (Fig. 4). FLAG-ERK2–HA-MEK2 complexes were immunoprecipitated with antibodies to the HA epitope. The results indicate that WT-ERK2, ERK2-Δ3-7, and ERK2-Δ10-19 were all bound equally to MEK2 in serum-starved COS-1 cells (Fig. 4A). However, no ERK2 mutant that contained either a deletion or substitution mutation in residues 19 to 25 could be found to associate with MEK2 in coimmunoprecipitations (Fig. 4), suggesting that residues 19 to 25 of ERK2 were required for MEK2 association. Identical results were obtained in experiments using HA-MEK1 (data not shown).
FIG. 4.
Mutation of residues 19 to 25 of ERK2 inhibits binding to MEK. COS-1 cells were transfected with plasmids for HA-MEK2 and FLAG-ERKs as indicated. (A and B) The following day the cells were serum starved for 4 h before harvest at 24 h posttransfection. The cells were lysed in hypotonic buffer, and the proteins were immunoprecipitated with antibodies to the HA epitope. The immunoprecipitations were immunoblotted with antibodies to FLAG and HA. For the lysate blots, an equal amount of soluble cell lysate was run on SDS-PAGE gels and immunoblotted for FLAG-ERK. IP, immunoprecipitate.
ERK2 mutants that do not bind to MEK localize to the nucleus and cytoplasm of serum-starved cells.
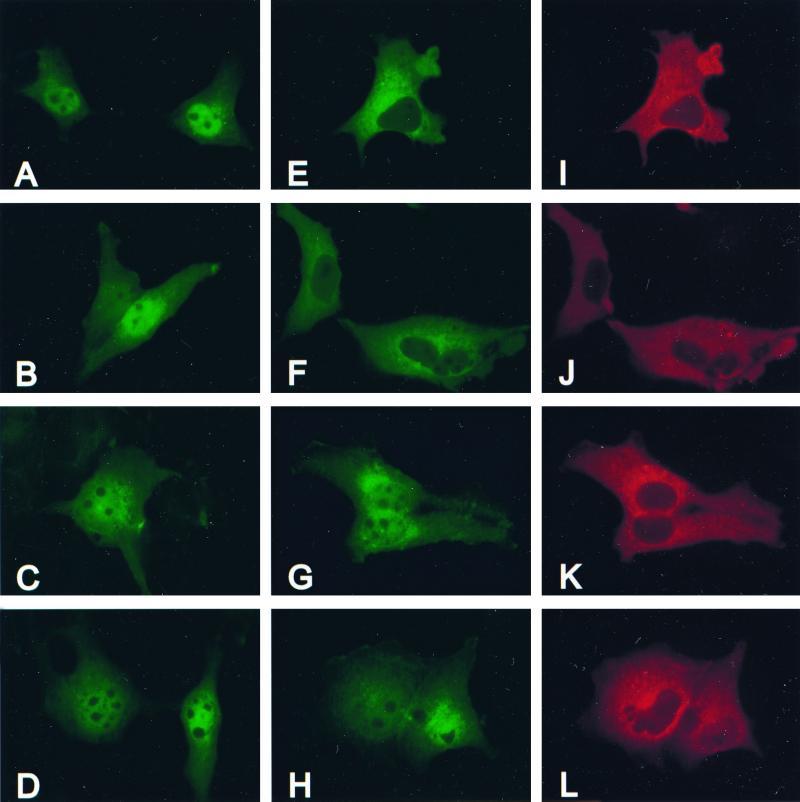
In cycling COS-1 cells grown in culture, ERK1 and ERK2 exist in the nucleus and cytoplasm. Four hours of serum withdrawal is sufficient to inactivate the ERK pathway, causing the ERKs to be retained in the cytoplasm (data not shown). Recent reports have identified the MEKs as cytoplasmic anchors for the ERKs, retaining the ERKs in the cytoplasm under conditions of serum starvation (15). To determine if the localization of ERK2-Δ19-25 and ERK2-Δ19-25-GP in serum-starved cells was altered compared to WT-ERK2 by their inability to bind to MEK, COS-1 cells were transfected with wild-type and mutant ERKs, with or without cotransfected HA-MEK2. The cells were serum-starved before fixation and processed for immunofluorescence. In the absence of cotransfected MEK2, WT-ERK (Fig. 5A), ERK2-Δ10-19 (Fig. 5B), ERK2-Δ19-25 (Fig. 5C), and ERK2-Δ19-25-GP (Fig. 5D) localized in the nucleus and the cytoplasm. The nuclear localization of exogenous ERK was most likely due to the inability of the endogenous ERK anchoring proteins (e.g., MEKs) to hold the excess FLAG-ERKs in the cytoplasm, causing the ERKs to enter the nucleus (15). Cotransfection of HA-MEK2 resulted in the redistribution of WT-ERK2 to the cytoplasm in serum-starved cells (Fig. 5E). In addition, ERK2-Δ3-7 (data not shown) and ERK2-Δ10-19 (Fig. 5F) behaved in a manner similar to WT-ERK2, with mostly cytoplasmic staining and little nuclear staining in starved cells cotransfected with MEK2. This supported the coimmunoprecipitation data in Fig. 4, demonstrating that the ERKs that bind to MEK2 colocalize with it in the cytoplasm of serum-starved cells. In contrast, ERK2-Δ19-25 (Fig. 5G), ERK2-Δ19-25-GP (Fig. 5H), ERK2-Δ19-25-7A (data not shown), and ERK2-Δ10-25 (data not shown) had prominent staining in both the nucleus and the cytoplasm of cells cotransfected with MEK2. The overexpression of MEK2 within the cell did not appear to alter the localization of these ERK-Δ19-25 mutants. Therefore, it appeared that the mutation of the 7-amino-acid sequence in the N terminus of ERK2 prevented the colocalization of these mutants with MEK2 in vivo, allowing these mutant ERKs to enter the nucleus under conditions of serum starvation, a time when ERK2 is normally retained in the cytoplasm. In all of the cotransfected cells HA-MEK2 was localized in the cytoplasm (Fig. 5I to L), consistent with previous reports that defined a nuclear export signal present in the N terminus of MEK (14). Localization of HA-MEK2 did not appear to be modified by the presence of exogenous ERK (data not shown).
FIG. 5.
ERK2-Δ19-25 mutants localize to nucleus and cytoplasm of serum starved cells cotransfected with MEK2. COS-1 cells were cotransfected with plasmids for FLAG-ERKs and empty vector (A to D) or FLAG-ERKs (E to H) and HA-MEK2 (I to L). The cells were serum starved before fixing in 4% paraformaldehyde at 48 h posttransfection. Fixed cells were probed with monoclonal M2 anti-FLAG and polyclonal anti-HA antibodies. Empty vector was cotransfected with plasmids for WT-ERK2 (A), ERK2-Δ10-19 (B), ERK2-Δ19-25 (C), or ERK2-Δ19-25-GP (D). Parallel cultures were cotransfected with WT-ERK2 (E) and MEK2 (I), ERK2-Δ10-19 (F) and MEK2 (J), ERK2-Δ19-25 (G) and MEK2 (K), or ERK2-Δ19-25-GP (H), and MEK2 (L).
To confirm the intracellular localization of WT-ERK2 and the ERK2 mutants under conditions of serum starvation, cell fractionation was performed on serum-starved COS-1 cells cotransfected with HA-MEK2 and FLAG-ERKs. As expected, transfected MEK2 was localized in the cytoplasm of serum-starved COS-1 cells (Fig. 6). WT-ERK2 and ERK2-Δ10-19 were also found exclusively in the cytoplasm of serum-starved cells, presumably being anchored there by the cotransfected MEK2. However, in agreement with the immunofluorescence data (Fig. 5), ERK2-Δ10-25 and ERK2-Δ19-25 were localized both in the cytoplasm and in the nucleus (Fig. 6). These mutants appeared to be equally distributed in the nucleus and cytoplasm regardless of the presence of cotransfected MEK2.
FIG. 6.
Cellular distribution of FLAG-ERK proteins by fractionation. COS-1 cells were cotransfected with plasmids for HA-MEK2 and either a FLAG-ERK2 or empty vector. The cells were washed and serum starved for 4 h before harvest at 48 h posttransfection. Nuclear and cytoplasmic fractions were prepared, and equal cell equivalents of each lysate were subjected to SDS-PAGE. Proteins were transferred to nitrocellulose and blotted with antibodies to FLAG and HA. N, nucleus; C, cytoplasm.
ERK2-Δ19-25-GP is deficient in association with ERK substrates in vivo.
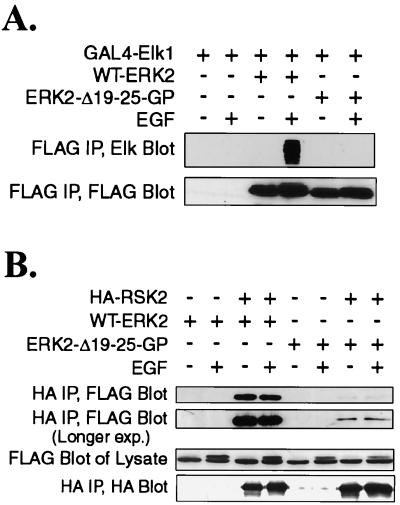
To determine if the mutation affected the ability of ERK2-Δ19-25-GP to associate with known ERK2 substrates in vivo, we performed coimmunoprecipitation experiments with Elk1, a member of the ets transcription factor family and a nuclear ERK target, and RSK2, an ERK cytoplasmic target. The D domain of Elk1 associates with an unknown region of ERK2 (45). COS-1 cells were cotransfected with plasmids for a GAL4-Elk1 fusion protein and either WT-ERK or ERK2-Δ19-25-GP. The cells were later serum-starved and stimulated with EGF. GAL4-Elk1 was coimmunoprecipitated with WT-ERK2 only from stimulated cells, but not from serum-starved cells, in agreement with a previous report that only phosphorylated ERK2 was able to associate with Elk1 (45). GAL4-Elk1 could not be coimmunoprecipitated with ERK2-Δ19-25-GP when the cells were starved or stimulated with EGF, demonstrating that ERK2-Δ19-25-GP was deficient in Elk1 binding even when it was phosphorylated.
ERK2 has been reported to bind to the cytoplasmic substrate RSK through aspartic acid residues in the C terminus of ERK2 interacting with lysine residues in RSK (39). We tested the ability of WT-ERK2 and ERK2-Δ19-25-GP to associate with RSK2 in coimmunoprecipitation experiments in cotransfected COS-1 cells that were starved or stimulated with EGF (Fig. 7B). In agreement with results reported by Smith et al. (38), WT-ERK2 was associated with RSK2 in both starved and stimulated cells. However, ERK2-Δ19-25-GP was only weakly associated with RSK2 under either condition. These data demonstrate that ERK2-Δ19-25-GP was deficient in binding to ERK substrates.
FIG. 7.
ERK2-Δ19-25-GP has reduced association with ERK2 substrates. (A) COS-1 cells were cotransfected with plasmids for GAL4-Elk1 and either WT-ERK2 or ERK2-Δ19-25-GP. The cells were serum starved for 5 h and stimulated with EGF for 15 min as indicated. FLAG-ERK immunoprecipitates were run on a gel and immunoblotted with antibodies for Elk1 and FLAG. (B) Cells were cotransfected with plasmids for HA-RSK2 and either WT-ERK2 or ERK2-Δ19-25-GP. The cells were starved and stimulated with EGF as described for panel A. Anti-HA immunoprecipitates were run on a gel and immunoblotted with antibodies for FLAG and HA. IP, immunoprecipitate; exp., exposure.
ERK2-Δ19-25 mutants inhibit ERK nuclear signaling to Elk1.
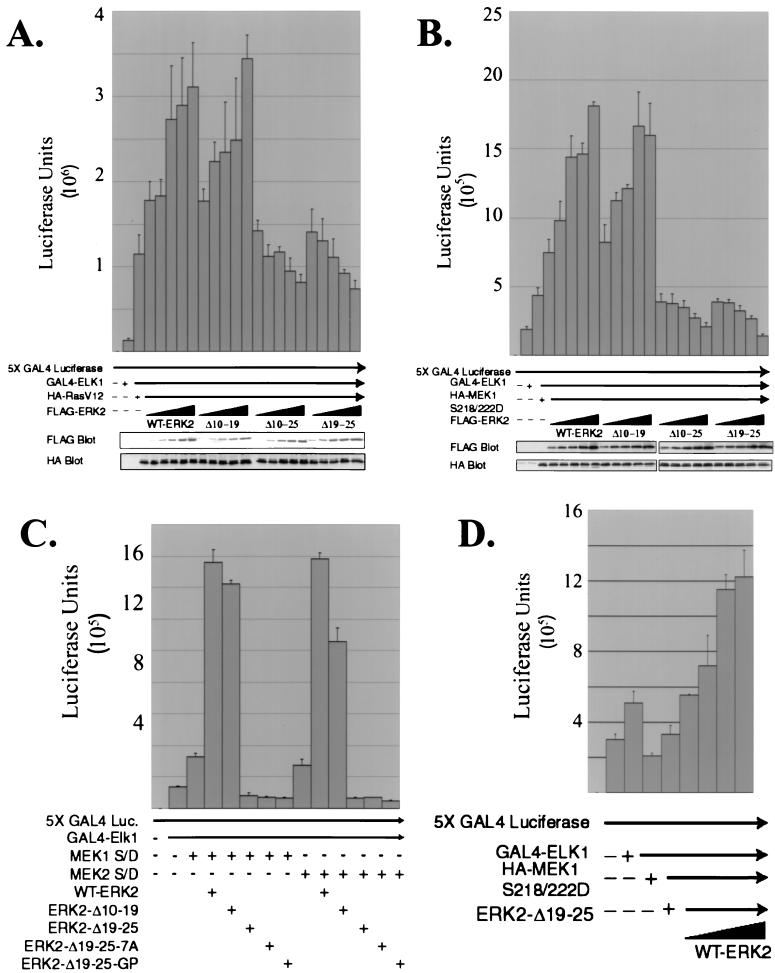
To screen these mutants for their effect on ERK nuclear signaling, we determined their ability to stimulate transcriptional activation of Elk1. MAP kinases including the ERKs, JNKs, and p38 proteins are able to phosphorylate and activate Elk1, phosphorylating residues in the C-terminal transcriptional activation domain (9, 16, 17, 24, 42). The ability of WT-ERK2 and the ERK2 mutants to signal to a fusion protein consisting of the DNA binding region of GAL4 fused to the transcriptional activation domain of Elk1 was assayed by use of a luciferase reporter construct containing five GAL4 consensus binding sites in the promoter (32). A constitutively activated form of Ras, RasV12, was also cotransfected to activate the ERK pathway under serum-free conditions. The addition of RasV12 was able to stimulate Elk1-specific transcription sixfold over the basal activity seen by addition of GAL4-Elk1 alone (Fig. 8A). The RasV12 was presumably activating endogenous MAP kinase pathways, including the ERKs and stress-activated protein kinases, to phosphorylate and activate GAL4-Elk1. Cotransfection with increasing amounts of WT-ERK2 further stimulated GAL4-Elk1 transcriptional activity in the presence of RasV12 in a dose-dependent manner. Addition of increasing amounts of ERK2-Δ3-7 or ERK2-Δ10-19 also stimulated Elk1 transcriptional activity to levels similar to those stimulated by WT-ERK2 (Fig. 8A and data not shown). However, expression of ERK2-Δ10-25 or ERK2-Δ19-25 inhibited signaling to Elk1 in a dose-dependent manner compared to cells that received RasV12 and empty vector (Fig. 8A), with both mutants decreasing luciferase activity by about 30%. This data suggested that ERK2 molecules lacking amino acids 19 to 25 were capable of acting in a dominant-negative manner to inhibit signaling of RasV12 through endogenous MAP kinase pathways to Elk1. Similar results were obtained with ERK2-Δ19-25-7A and ERK2-Δ19-25-GP (data not shown). Blotting for expression of the transfected constructs shows that equal amounts of RasV12 were present in each sample and that expression of WT-ERK2 and of the deletion mutants was comparable.
FIG. 8.
ERK2-Δ19-25 mutants act as dominant negatives in signaling to an Elk1 responsive reporter. (A) COS-1 cells were cotransfected in triplicate with plasmids for 5X GAL4-luciferase, GAL4-ELK1, HA-RasV12, and increasing amounts of the indicated FLAG-ERK2 construct. The cells were incubated in serum-free DMEM overnight before harvesting at 24 h and determination of luciferase activity. An equal amount of protein from one lysate of each triplicate closest to the mean was immunoblotted with anti-FLAG and anti-HA antibodies to determine the expression of HA-RasV12 and FLAG-ERK2 in each sample. (B) The same experiment as described for panel A was performed, except MEK1 S218/222D was transfected in place of HA-RasV12. (C) 5X GAL4-luciferase, GAL4-ELK1, ERK2 plasmids, and either mutationally activated MEK1 or MEK2 were transfected into COS-1 cells in triplicate and luciferase assays were performed on cell lysates as described for panel B. (D) COS-1 cells were cotransfected in triplicate with 5X GAL4-luciferase, GAL4-ELK1, HA-MEK1 S218/222D, ERK2-Δ19-25 and increasing amounts of WT-ERK2, as indicated. Luciferase activity was determined 24 h after transfection. S/D, S218/222D.
The ability of ERK2-Δ10-25 and ERK2-Δ19-25 to only partially inhibit signaling stimulated by RasV12 could mean that additional signaling to GAL4-Elk1 occurs through stimulation of the JNKs and p38 stress-activated kinases as well as the ERKs. To specifically target signaling through the ERKs to GAL4-Elk1, we performed the same experiment using a mutationally activated form of MEK1, MEK1 S218/222D, instead of RasV12, so that the only signaling to GAL4-Elk1 would be through the ERK pathway. Cotransfection of MEK1 S218/222D with the GAL4-Elk1 reporter system increased luciferase activity twofold over that seen in the absence of MEK1 S218/222D, suggesting that indeed only a portion of the signaling by RasV12 to GAL4-Elk1 may have occurred through the ERKs. As was seen in Fig. 8A, titration of increasing amounts of WT-ERK2, ERK2-Δ3-7, or ERK2-Δ10-19 increased luciferase activity in a dose-dependent manner over that seen with MEK1 S218/222D and empty vector (Fig. 8B and data not shown). However, expression of ERK2-Δ10-25 or ERK2-Δ19-25 effectively inhibited the luciferase expression activated by MEK1 S218/222D in a dose-dependent fashion. While approximately 30% of signaling of RasV12 to GAL4-Elk1 was inhibited by ERK2-Δ10-25 or ERK2-Δ19-25 (Fig. 8A), there was complete inhibition of signaling by ERK2-Δ10-25 or ERK2-Δ19-25 when MEK1 S218/222D was the upstream stimulus (Fig. 8B). Thus, these ERK mutants were able to completely inhibit the ability of MEK1 S218/222D to signal through endogenous ERKs to GAL4-Elk1. To determine if this inhibition was specific for a MEK isoform, we repeated the experiment using mutationally activated MEK1 or mutationally activated MEK2 in the presence of a single amount of WT-ERK2 or ERK2 mutant (Fig. 8C). The data demonstrate that signaling by both isoforms of MEK through endogenous ERKs to Elk1 was inhibited by ERK2-Δ19-25, ERK2-Δ19-25-7A, or ERK2-Δ19-25-GP.
To determine if increased expression of WT-ERK2 can rescue the dominant-negative effect of ERK2-Δ19-25 on signaling to GAL4-Elk1, the luciferase assays were performed using a constant amount of ERK2-Δ19-25 cotransfected with MEK1 S218/222D (Fig. 8D). The expression of increasing amounts of WT-ERK2 was able to overcome the dominant-negative affect of ERK2-Δ19-25 and restore signaling to GAL4-Elk1 in a dose-dependent manner.
ERK2-Δ19-25-7A does not inhibit phosphorylation of the ERK substrate RSK2.
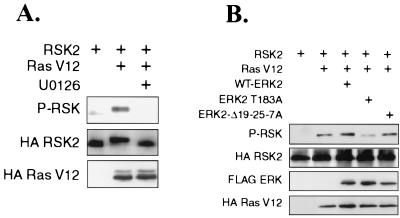
To determine if ERK2-Δ19-25 mutants inhibited signaling not only to a nuclear target but to a cytoplasmic ERK substrate as well, we assessed the ability of ERK2-Δ19-25-7A to affect phosphorylation of the ERK substrate RSK2. RSK isoforms are phosphorylated and activated upon ERK activation and translocate to the nucleus (10). ERKs phosphorylate Ser 363 of RSK1, stimulating kinase activity and autophosphorylation of RSK1 at Ser 380 (11). Analogous phosphorylation sites exist in RSK2 (11). To determine if RSK2 activation was dependent on MEK activity in response to activated Ras, cells cotransfected with RasV12 were treated with DMSO or the MEK inhibitor U0126 (12). Immunoblotting HA-RSK immunoprecipitates with antibodies against phosphorylated serine 380 of RSK1 showed that RSK2 was only weakly phosphorylated under serum-starved conditions but that its phosphorylation could be stimulated by cotransfection of RasV12 (Fig. 9A). This increase in RSK2 phosphorylation was MEK dependent, as addition of the MEK inhibitor U0126 inhibited RSK2 phosphorylation. To determine the effect of ERK cotransfection on RSK phosphorylation, WT-ERK2, ERK2 T183A, or ERK2-Δ19-25-7A was cotransfected with RasV12 (Fig. 9B). Cotransfection of WT-ERK2 increased the amount of phospho-HA-RSK2, while cotransfection of ERK2 T183A, a phosphorylation site mutant and a dominant-negative ERK2 that can bind to MEK, inhibited RSK2 phosphorylation. However, ERK2-Δ19-25-7A had no effect on the phosphorylation of RSK2. These data demonstrate that ERK2-Δ19-25-7A did not act as a dominant negative in signaling to a cytoplasmic ERK substrate.
FIG. 9.
ERK2-Δ19-25-7A does not inhibit phosphorylation of cytoplasmic ERK substrate RSK2. (A) COS-1 cells were transfected with plasmids for HA-RSK2 and HA-RasV12 and serum starved for 5 h before harvest at 24 h. The MEK inhibitor U0126 was added for the last 2 h before harvest. HA immunoprecipitates were immunoblotted for phospho-RSK and HA-RSK2. Lysates were immunoblotted for HA-RasV12. (B) COS-1 cells were transfected as described for panel A with the addition of cotransfection with the indicated ERK plasmid. HA-RSK2 was immunoprecipitated and immunoblotted with antibodies for HA-phosphorylated RSK. Lysates were immunoblotted for FLAG-ERK and HA-RasV12. P-RSK, phospho-RSK.
DISCUSSION
In this paper we characterize the role of the short N-terminal, noncatalytic domain of p42 ERK2 in ERK2 regulation. Mutation of amino acids 19 to 25 (VGPRYTN) in the N terminus caused profound effects in ERK2 activity, MEK binding, localization, and signaling. Mutation of these residues to alanine gave identical results, as did leaving the glycine and proline intact and replacing the other five residues with alanine. This region lies in a random coil structure at one end of the molecule and appears to be necessary for kinase activity of ERK2, as mutations in this region generated a mutant protein with little activity (Fig. 2). This is in agreement with a previously described chimera of ERK2 and p38, in which the N terminus of ERK2 through subdomain II was replaced with that of p38. As with ERK2-Δ19-25, this p38-ERK2 chimera suffered a major loss in kinase activity (43).
While the binding site for the ERKs on MEKs has been localized to a small region in the N terminus of MEK, the region(s) of the ERKs required for MEK binding are as yet unclear. Experiments by Brunet and Pouyssegur (4) suggest that MAP kinases receive upstream signals in a region between kinase subdomains III and IV of the MAP kinases. Other investigators have proposed that there are several sites on the ERKs that are required for recognition by the MEKs (43). In agreement with this, recent reports identify acidic residues in the C terminus (39) and a broad region in the N terminus (44) of ERK2 as being necessary for ERK phosphorylation and MEK binding. The crystal structure of unphosphorylated ERK2 (47) shown in Fig. 1B shows that the N and C termini of ERK2 lie on the surface of the molecule. The observation that both N- and C-terminal mutations affect binding to MEKs suggests that the tertiary structure formed by these regions is important in MEK binding. In addition, it has been observed that ERK2-Δ10-25 and ERK2-Δ19-25 cannot bind to MKP-3 (S. Eblen, unpublished observation), a cytoplasmic anchor and phosphatase for the ERKs (5, 28), in agreement with Tanoue et al. (39) who suggest that MEK and MKP-3 share a common binding site. We propose that the N- and C-terminal regions collectively form a protein-protein interaction domain, with both regions being necessary and neither sufficient to form a MEK/MKP3 binding site. This sequence is identical in ERK1 except for the last residue, which is a glutamine, suggesting that this conserved region may also be required for ERK1 activity and interactions. In addition, recent work from our laboratory has identified a protein called MP1 that appears to stabilize the interaction between MEK1 and ERK1 specifically, demonstrating that other proteins may also be involved in the MEK/ERK interaction (35).
Besides being activators of ERK2, the MEKs bind to and retain ERK1 and ERK2 in the cytoplasm of unstimulated cells, preventing premature nuclear entry of the ERKs (15). In serum-starved cells cotransfected with plasmids for WT-ERK2 and MEK2, WT-ERK2 was retained in the cytoplasm (Fig. 5 and 6). However, under the same conditions, ERK2-Δ19-25 mutants were localized in both the nucleus and the cytoplasm, supporting the coimmunoprecipitation experiments by demonstrating that these mutants do not colocalize with MEK in serum-starved cells. This inability to bind to MEK resulted in the aberrant nuclear localization of these mutants under conditions of serum starvation.
One of the best-characterized nuclear targets of the ERKs and the stress-activated protein kinases is the nuclear transcription factor Elk1, which contains several MAP kinase phosphorylation sites in the C-terminal transactivation domain (20). Phosphorylation of these sites by MAP kinase family members increases transcriptional activation by Elk1 (9, 16, 17, 24, 42). Using Elk1 transcriptional activity as a readout of ERK activation in a luciferase reporter system (32), cotransfection of WT-ERK2 or ERK2 mutants that bind to MEK with either a mutationally activated form of MEK1, MEK2, or Ras increased transcriptional activation of Elk1 in a dose-dependent manner (Fig. 8). However, cotransfection with ERK2-Δ10-25 or ERK2-Δ19-25 partially inhibited the ability of activated Ras to signal through endogenous ERKs to Elk1. Inhibition may be only partial due to the ability of Ras to activate other MAP kinase pathways to activate Elk1. This concept was supported by the observation that ERK2-Δ10-25 and ERK2-Δ19-25 were able to fully inhibit the Elk1 activation by activated MEK1 or MEK2, which can only signal through ERK1 and ERK2. Thus, these ERK mutants act as dominant-negatives in inhibiting signaling through endogenous ERKs. Interestingly, no inhibition of phosphorylation of the ERK2 cytoplasmic substrate RSK2 was observed in the presence of ERK2-Δ19-25-7A, suggesting that ERK2-Δ19-25 mutants may only inhibit signaling to nuclear substrates, while not affecting cytoplasmic substrates. The differences in the effects on RSK and ELK activation are most likely not due to differences in the ability of ERK2-Δ19-25 mutants to bind to these substrates, as ERK2-Δ19-25 mutants showed no binding to GAL4-Elk1 and had a greatly reduced ability to bind to RSK compared to WT-ERK2 in coimmunoprecipitation experiments (Fig. 7).
A recent report describes the creation of dominant-negative ERK mutants by the addition of a CAAX box to the C terminus, causing membrane localization (21). The authors propose that these proteins inhibit ERK nuclear signaling by dimerizing with endogenous ERKs, tethering them to the membrane and preventing nuclear localization. We believe that the dominant-negative mutants that we have described here, which were created by mutating a natural part of ERK2, also inhibit ERK nuclear signaling through their localization, albeit in a different manner. Our hypothesis is that the aberrant constitutive nuclear localization of a pool of ERK2-Δ19-25 mutants inhibits endogenous ERK nuclear signaling. This unregulated, inappropriate nuclear localization of a kinase-dead ERK could interfere with the ability of endogenous ERKs to signal to the nucleus by affecting the nuclear translocation or retention of endogenous ERKs. ERKs require neosynthesis of labile nuclear anchor proteins in order to be retained in the nucleus for extended periods of time after initial translocation (26). ERK2-Δ19-25 could act as a dominant negative by either binding to proteins that facilitate nuclear import or by taking up nuclear binding sites for ERKs. In either scenario, these mutants would decrease the amount of active endogenous ERKs in the nucleus at any given time. While the coimmunoprecipitation experiments discussed above suggest that these mutants have a decreased ability to bind substrate, these interactions may have a stronger affinity in vivo. Elk1 contains a MAP kinase binding domain, called the D domain, responsible for binding to ERKs and stress-activated protein kinases (45), while other ERK substrates contain an FXFP motif that ERKs bind (23). Binding of ERKs through these motifs is often required for ERK to phosphorylate their substrates (23, 45). If ERK2-Δ19-25, which has little kinase activity even when phosphorylated, could bind to ERK nuclear substrates and keep endogenous ERKs from binding and phosphorylating them, transcriptional activation by endogenous ERKs could be inhibited. Other methods of determining in vivo interactions, such as the immunofluorescence and fractionation experiments (Fig. 5 and 6) used in conjunction with the MEK coimmunoprecipitations, would be required to determine the extent of these interactions.
If the dominant-negative effects of the ERK2-Δ19-25 are due to saturation of nuclear targets, one might expect cytoplasmic substrates to be unaffected by expression of the mutant. This is what we observed: ERK2-Δ19-25-7A did not inhibit RSK2 phosphorylation, while treatment with U0126 (12), a MEK inhibitor, and ERK T183A, a dominant-negative phosphorylation site mutant of ERK2 that can still bind to MEK, both inhibited the phosphorylation of RSK2 induced by RasV12. Whether this scenario will be observed with other ERK substrates requires a further analysis of many more ERK targets. The data suggest that this mutant ERK is able to differentially inhibit signaling by specific pools of endogenous ERK proteins. Thus, in addition to elucidating regulatory functions of the ERK N terminus, this study reports the construction of a novel dominant-negative ERK mutant that should be useful in dissecting the diverse role of MAP kinases in cell regulation.
ACKNOWLEDGMENTS
This work was supported by Public Health Service Grants GM47332 and CA40042 from the National Institute of Health. S. Eblen was supported by National Research Service Award 5F32 GM18672-02.
We thank the members of the Weber lab, PWP, and Jesse Kwiek for helpful discussions. We also thank Scott Weed for pCDNA3-FLAG, Channing Der for RasV12, Tom Sturgill for RSK2, Richard Maurer for GAL4-Elk1 and 5X GAL4 luciferase, and Alexis Rahal for technical assistance.
REFERENCES
- 1.Adachi M, Fukuda M, Nishida E. Two co-existing mechanisms for nuclear import of MAP kinase: passive diffusion of a monomer and active transport of a dimer. EMBO J. 1999;18:5347–5358. doi: 10.1093/emboj/18.19.5347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alessi D R, Cuenda A, Cohen P, Dudley D T, Saltiel A R. PD 098059 is a specific inhibitor of the activation of mitogen-activated protein kinase kinase in vitro and in vivo. J Biol Chem. 1995;270:27489–27494. doi: 10.1074/jbc.270.46.27489. [DOI] [PubMed] [Google Scholar]
- 3.Bardwell L, Cook J G, Chang E C, Cairns B R, Thorner J. Signaling in the yeast pheromone response pathway: specific and high-affinity interaction of the mitogen-activated protein (MAP) kinases Kss1 and Fus3 with the upstream MAP kinase kinase Ste7. Mol Cell Biol. 1996;16:3637–3650. doi: 10.1128/mcb.16.7.3637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brunet A, Pouyssegur J. Identification of MAP kinase domains by redirecting stress signals into growth factor responses. Science. 1996;272:1652–1655. doi: 10.1126/science.272.5268.1652. [DOI] [PubMed] [Google Scholar]
- 5.Brunet A, Roux D, Lenormand P, Dowd S, Keyse S, Pouyssegur J. Nuclear translocation of p42/p44 mitogen-activated protein kinase is required for growth factor-induced gene expression and cell cycle entry. EMBO J. 1999;18:664–674. doi: 10.1093/emboj/18.3.664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Canagarajah B J, Khokhlatchev A, Cobb M H, Goldsmith E J. Activation mechanism of the MAP kinase ERK2 by dual phosphorylation. Cell. 1997;90:859–869. doi: 10.1016/s0092-8674(00)80351-7. [DOI] [PubMed] [Google Scholar]
- 7.Catling, A. D., S. T. Eblen, H. J. Schaeffer, and M. J. Weber. Scaffold protein regulation of the MAP kinase cascade. Methods Enzymol., in press. [DOI] [PubMed]
- 8.Catling A D, Schaeffer H J, Reuter C W, Reddy G R, Weber M J. A proline-rich sequence unique to MEK1 and MEK2 is required for raf binding and regulates MEK function. Mol Cell Biol. 1995;15:5214–5225. doi: 10.1128/mcb.15.10.5214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cavigelli M, Dolfi F, Claret F X, Karin M. Induction of c-fos expression through JNK-mediated TCF/Elk-1 phosphorylation. EMBO J. 1995;14:5957–5964. doi: 10.1002/j.1460-2075.1995.tb00284.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen R H, Sarnecki C, Blenis J. Nuclear localization and regulation of erk- and rsk-encoded protein kinases. Mol Cell Biol. 1992;12:915–927. doi: 10.1128/mcb.12.3.915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dalby K N, Morrice N, Caudwell F B, Avruch J, Cohen P. Identification of regulatory phosphorylation sites in mitogen-activated protein kinase (MAPK)-activated protein kinase-1a/p90rsk that are inducible by MAPK. J Biol Chem. 1998;273:1496–1505. doi: 10.1074/jbc.273.3.1496. [DOI] [PubMed] [Google Scholar]
- 12.Favata M F, Horiuchi K Y, Manos E J, Daulerio A J, Stradley D A, Feeser W S, Van Dyk D E, Pitts W J, Earl R A, Hobbs F, Copeland R A, Magolda R L, Scherle P A, Trzaskos J M. Identification of a novel inhibitor of mitogen-activated protein kinase kinase. J Biol Chem. 1998;273:18623–18632. doi: 10.1074/jbc.273.29.18623. [DOI] [PubMed] [Google Scholar]
- 13.Fukuda M, Gotoh I, Adachi M, Gotoh Y, Nishida E. A novel regulatory mechanism in the mitogen-activated protein (MAP) kinase cascade. Role of nuclear export signal of MAP kinase kinase. J Biol Chem. 1997;272:32642–32648. doi: 10.1074/jbc.272.51.32642. [DOI] [PubMed] [Google Scholar]
- 14.Fukuda M, Gotoh I, Gotoh Y, Nishida E. Cytoplasmic localization of mitogen-activated protein kinase kinase directed by its NH2-terminal, leucine-rich short amino acid sequence, which acts as a nuclear export signal. J Biol Chem. 1996;271:20024–20028. doi: 10.1074/jbc.271.33.20024. [DOI] [PubMed] [Google Scholar]
- 15.Fukuda M, Gotoh Y, Nishida E. Interaction of MAP kinase with MAP kinase kinase: its possible role in the control of nucleocytoplasmic transport of MAP kinase. EMBO J. 1997;16:1901–1908. doi: 10.1093/emboj/16.8.1901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gille H, Kortenjann M, Thomae O, Moomaw C, Slaughter C, Cobb M H, Shaw P E. ERK phosphorylation potentiates Elk-1-mediated ternary complex formation and transactivation. EMBO J. 1995;14:951–962. doi: 10.1002/j.1460-2075.1995.tb07076.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gille H, Strahl T, Shaw P E. Activation of ternary complex factor Elk-1 by stress-activated protein kinases. Curr Biol. 1995;5:1191–1200. doi: 10.1016/s0960-9822(95)00235-1. [DOI] [PubMed] [Google Scholar]
- 18.Gonzalez F A, Seth A, Raden D L, Bowman D S, Fay F S, Davis R J. Serum-induced translocation of mitogen-activated protein kinase to the cell surface ruffling membrane and the nucleus. J Cell Biol. 1993;122:1089–1101. doi: 10.1083/jcb.122.5.1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Her J H, Lakhani S, Zu K, Vila J, Dent P, Sturgill T W, Weber M J. Dual phosphorylation and autophosphorylation in mitogen-activated protein (MAP) kinase activation. Biochem J. 1993;296:25–31. doi: 10.1042/bj2960025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hill C S, Marais R, John S, Wynne J, Dalton S, Treisman R. Functional analysis of a growth factor-responsive transcription factor complex. Cell. 1993;73:395–406. doi: 10.1016/0092-8674(93)90238-l. [DOI] [PubMed] [Google Scholar]
- 21.Hochholdinger F, Baier G, Nogalo A, Bauer B, Grunicke H H, Überall F. Novel membrane-targeted ERK1 and ERK2 chimeras which act as dominant negative, isotype-specific mitogen-activated protein kinase inhibitors of Ras-Raf-mediated transcriptional activation of c-fos in NIH 3T3 cells. Mol Cell Biol. 1999;19:8052–8065. doi: 10.1128/mcb.19.12.8052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jaaro H, Rubinfeld H, Hanoch T, Seger R. Nuclear translocation of mitogen-activated protein kinase kinase (MEK1) in response to mitogenic stimulation. Proc Natl Acad Sci USA. 1997;94:3742–3747. doi: 10.1073/pnas.94.8.3742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jacobs D, Glossip D, Xing H, Muslin A J, Kornfeld K. Multiple docking sites on substrate proteins form a modular system that mediates recognition by ERK MAP kinase. Genes Dev. 1999;13:163–175. [PMC free article] [PubMed] [Google Scholar]
- 24.Janknecht R, Ernst W H, Pingoud V, Nordheim A. Activation of ternary complex factor Elk-1 by MAP kinases. EMBO J. 1993;12:5097–5104. doi: 10.1002/j.1460-2075.1993.tb06204.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Khokhlatchev A V, Canagarajah B, Wilsbacher J, Robinson M, Atkinson M, Goldsmith E, Cobb M H. Phosphorylation of the MAP kinase ERK2 promotes its homodimerization and nuclear translocation. Cell. 1998;93:605–615. doi: 10.1016/s0092-8674(00)81189-7. [DOI] [PubMed] [Google Scholar]
- 26.Lenormand P, Brondello J M, Brunet A, Pouyssegur J. Growth factor-induced p42/p44 MAPK nuclear translocation and retention requires both MAPK activation and neosynthesis of nuclear anchoring proteins. J Cell Biol. 1998;142:625–633. doi: 10.1083/jcb.142.3.625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lenormand P, Sardet C, Pages G, L'Allemain G, Brunet A, Pouyssegur J. Growth factors induce nuclear translocation of MAP kinases (p42mapk and p44mapk) but not of their activator MAP kinase kinase (p45mapkk) in fibroblasts. J Cell Biol. 1993;122:1079–1088. doi: 10.1083/jcb.122.5.1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Muda M, Boschert U, Dickinson R, Martinou J C, Martinou I, Camps M, Schlegel W, Arkinstall S. MKP-3, a novel cytosolic protein-tyrosine phosphatase that exemplifies a new class of mitogen-activated protein kinase phosphatase. J Biol Chem. 1996;271:4319–4326. doi: 10.1074/jbc.271.8.4319. [DOI] [PubMed] [Google Scholar]
- 29.Nishida E, Gotoh Y. The MAP kinase cascade is essential for diverse signal transduction pathways. Trends Biochem Sci. 1993;18:128–131. doi: 10.1016/0968-0004(93)90019-j. [DOI] [PubMed] [Google Scholar]
- 30.Pages G, Lenormand P, L'Allemain G, Chambard J C, Meloche S, Pouyssegur J. Mitogen-activated protein kinases p42mapk and p44mapk are required for fibroblast proliferation. Proc Natl Acad Sci USA. 1993;90:8319–8323. doi: 10.1073/pnas.90.18.8319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Payne D M, Rossomando A J, Martino P, Erickson A K, Her J H, Shabanowitz J, Hunt D F, Weber M J, Sturgill T W. Identification of the regulatory phosphorylation sites in pp42/mitogen-activated protein kinase (MAP kinase) EMBO J. 1991;10:885–892. doi: 10.1002/j.1460-2075.1991.tb08021.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Roberson M S, Misra-Press A, Laurance M E, Stork P J, Maurer R A. A role for mitogen-activated protein kinase in mediating activation of the glycoprotein hormone α-subunit promoter by gonadotropin-releasing hormone. Mol Cell Biol. 1995;15:3531–3539. doi: 10.1128/mcb.15.7.3531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rubinfeld H, Hanoch T, Seger R. Identification of a cytoplasmic-retention sequence in ERK2. J Biol Chem. 1999;274:30349–30352. doi: 10.1074/jbc.274.43.30349. [DOI] [PubMed] [Google Scholar]
- 34.Sanghera J S, Peter M, Nigg E A, Pelech S L. Immunological characterization of avian MAP kinases: evidence for nuclear localization. Mol Biol Cell. 1992;3:775–787. doi: 10.1091/mbc.3.7.775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schaeffer H J, Catling A D, Eblen S T, Collier L S, Krauss A, Weber M J. MP1: a MEK binding partner that enhances enzymatic activation of the MAP kinase cascade. Science. 1998;281:1668–1671. doi: 10.1126/science.281.5383.1668. [DOI] [PubMed] [Google Scholar]
- 36.Seger R, Ahn N G, Posada J, Munar E S, Jensen A M, Cooper J A, Cobb M H, Krebs E G. Purification and characterization of mitogen-activated protein kinase activator(s) from epidermal growth factor-stimulated A431 cells. J Biol Chem. 1992;267:14373–14381. [PubMed] [Google Scholar]
- 37.Slack J K, Catling A D, Eblen S T, Weber M J, Parsons J T. c-Raf-mediated inhibition of epidermal growth factor-stimulated cell migration. J Biol Chem. 1999;274:27177–27184. doi: 10.1074/jbc.274.38.27177. [DOI] [PubMed] [Google Scholar]
- 38.Smith J A, Poteet-Smith C E, Malarkey K, Sturgill T W. Identification of an extracellular signal-regulated kinase (ERK) docking site in ribosomal S6 kinase, a sequence critical for activation by ERK in vivo. J Biol Chem. 1999;274:2893–2898. doi: 10.1074/jbc.274.5.2893. [DOI] [PubMed] [Google Scholar]
- 39.Tanoue T, Adachi M, Moriguchi T, Nishida E. A conserved docking motif in MAP kinases common to substrates, activators and regulators. Nat Cell Biol. 2000;2:110–116. doi: 10.1038/35000065. [DOI] [PubMed] [Google Scholar]
- 40.Tolwinski N S, Shapiro P S, Goueli S, Ahn N G. Nuclear localization of mitogen-activated protein kinase kinase 1 (MKK1) is promoted by serum stimulation and G2-M progression. Requirement for phosphorylation at the activation lip and signaling downstream of MKK. J Biol Chem. 1999;274:6168–6174. doi: 10.1074/jbc.274.10.6168. [DOI] [PubMed] [Google Scholar]
- 41.Whitmarsh A J, Cavanagh J, Tournier C, Yasuda J, Davis R J. A mammalian scaffold complex that selectively mediates MAP kinase activation. Science. 1998;281:1671–1674. doi: 10.1126/science.281.5383.1671. [DOI] [PubMed] [Google Scholar]
- 42.Whitmarsh A J, Shore P, Sharrocks A D, Davis R J. Integration of MAP kinase signal transduction pathways at the serum response element. Science. 1995;269:403–407. doi: 10.1126/science.7618106. [DOI] [PubMed] [Google Scholar]
- 43.Wilsbacher J L, Goldsmith E J, Cobb M H. Phosphorylation of MAP kinases by MAP/ERK involves multiple regions of MAP kinases. J Biol Chem. 1999;274:16988–16994. doi: 10.1074/jbc.274.24.16988. [DOI] [PubMed] [Google Scholar]
- 44.Xu B, Wilsbacher J L, Collisson T, Cobb M H. The N-terminal ERK-binding site of MEK1 is required for efficient feedback phosphorylation by ERK2 in vitro and ERK activation in vivo. J Biol Chem. 1999;274:34029–34035. doi: 10.1074/jbc.274.48.34029. [DOI] [PubMed] [Google Scholar]
- 45.Yang S H, Yates P R, Whitmarsh A J, Davis R J, Sharrocks A D. The Elk-1 ETS-domain transcription factor contains a mitogen-activated protein kinase targeting motif. Mol Cell Biol. 1998;18:710–720. doi: 10.1128/mcb.18.2.710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zecevic M, Catling A D, Eblen S T, Renzi L, Hittle J C, Yen T J, Gorbsky G J, Weber M J. Active MAP kinase in mitosis: localization at kinetochores and association with the motor protein CENP-E. J Cell Biol. 1998;142:1547–1558. doi: 10.1083/jcb.142.6.1547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhang F, Strand A, Robbins D, Cobb M H, Goldsmith E J. Atomic structure of the MAP kinase ERK2 at 2.3 A resolution. Nature. 1994;367:704–711. doi: 10.1038/367704a0. [DOI] [PubMed] [Google Scholar]
- 48.Zheng C F, Guan K L. Cytoplasmic localization of the mitogen-activated protein kinase activator MEK. J Biol Chem. 1994;269:19947–19952. [PubMed] [Google Scholar]