Abstract
The cyclin-dependent kinase (CDK)-activating kinase (CAK) is involved in cell cycle control, transcription, and DNA repair (E. A. Nigg, Curr. Opin. Cell. Biol. 8:312–317, 1996). However, the mechanisms of how CAK is integrated into these signaling pathways remain unknown. We previously demonstrated that abrogation of MAT1 (ménage à trois 1), an assembly factor and targeting subunit of CAK, induces G1 arrest (L. Wu, P. Chen, J. J. Hwang, L. W. Barsky, K. I. Weinberg, A. Jong, and V. A. Starnes, J. Biol. Chem. 274:5564–5572, 1999). This result led us to investigate how deregulation of CAK by MAT1 abrogation affects the cell cycle G1 exit, a process that is regulated most closely by phosphorylation of retinoblastoma tumor suppressor protein (pRb). Using mammalian cellular models that undergo G1 arrest evoked by antisense MAT1 abrogation, we found that deregulation of CAK inhibits pRb phosphorylation and cyclin E expression, CAK phosphorylation of pRb is MAT1 dose dependent but cyclin D1/CDK4 independent, and MAT1 interacts with pRb. These results suggest that CAK is involved in the regulation of cell cycle G1 exit while MAT1-modulated CAK formation and CAK phosphorylation of pRb may determine the cell cycle specificity of CAK in G1 progression.
The cyclin-dependent kinase (CDK)-activating kinase (CAK), an enzyme consisting of CDK7 (29, 60), cyclin H (17), and MAT1 (18, 55, 65), was originally implicated in cell cycle control by virtue of its ability to catalyze T-loop phosphorylation of CDKs in most eukaryotic cells (8, 16, 27, 37, 41, 43, 54). Subsequently, CAK was found to exist either in a free form or in association with the general transcription factor TFIIH (1, 15, 45, 47, 50, 53). Since TFIIH is required for both initiation of RNA polymerase II-catalyzed transcription and nucleotide excision repair (5, 10, 20, 57), identification of CAK as a component of TFIIH indicates that CAK has potential roles in these two processes. Given that CAK functions in the regulation of the cell cycle, transcription, and DNA repair, the challenging question of how CAK activity is modulated and integrated into these signaling pathways remains largely unanswered.
MAT1 (for ménage à trois 1) is an assembly factor and targeting subunit of CAK. To date, CAK without MAT1 has not been isolated from cells and the known functions of MAT1 always have been associated with CAK. MAT1 forms an active ternary CAK by assembling and stabilizing the association of CDK7 with cyclin H. This occurs in the absence of activating phosphorylation of the T loop of CDK7, which is an alternate mechanism different from the in vitro binary association of CDK7-cyclin H that requires T-loop phosphorylation in order for CDK7 to bind cyclin H (18, 33). Thus, the discovery of MAT1 as the third subunit of CAK (7, 18, 55, 65, 66) yields new insight into the regulation of CDK7-cyclin H. More interestingly, recent studies have found that the MAT1 protein, previously shown to function as an assembly factor for CDK7-cyclin H, also modulates CAK substrate specificity. For instance, addition of MAT1 to recombinant binary CAK (CDK7-cyclin H) switches its substrate preference to favor the RNA polymerase II large-subunit C-terminal domain over CDK2 (64); MAT1 enhances the ability of CDK7-cyclin H to phosphorylate isolated POU domains of octamer transcription factors (Oct factors) (25); and efficient phosphorylation of tumor suppressor p53 protein (p53) by CDK7-cyclin H is MAT1 dependent (28). Since the addition of MAT1 does not alter CDK7-cyclin H phosphorylation of CDK2 but does enhance CDK7-cyclin H phosphorylation of the C-terminal domain, p53, and Oct factors (25, 28, 64), MAT1 probably acts as a targeting subunit of CAK rather than an enhancer of CDK7-cyclin H phosphorylation of CDK. Although it is still unclear how MAT1 shifts CAK substrate specificity from its originally defined CDKs to other substrates, a growing number of studies have suggested that in addition to the role of MAT1 in assembling and stabilizing an active CAK, MAT1-mediated protein-protein interactions may play an important role in determining CAK substrate specificity. This prediction is supported by the following evidence: (i) MAT1 interacts with p53 and is required for CDK7-cyclin H to phosphorylate p53 efficiently (28); (ii) the interaction between POU domains and MAT1 can target CAK to Oct factors and result in their phosphorylation (25); and (iii) TFIIH lacking the CAK subcomplex will recover its transcriptional activity completely in the presence of free ternary CAK, while MAT1 interacts with both XPB (ERCC3) and XPD (ERCC2), two helicase subunits of TFIIH that mediate the association of CAK with core TFIIH (11, 45, 46). Initially, the discovery that CAK activity remains constant throughout the cell cycle (3, 44, 56) was a surprise. However, the above data reveal that CAK actually may be regulated either via MAT1-mediated CAK formation or via MAT1-dependent protein-protein interactions that target CAK to its different substrates at a precise time and in a defined order during cell cycle progression.
Cells exiting from the G1 phase of the cell cycle commit themselves to traversing the remainder of the growth cycle. It is well known that the cell cycle is regulated by the catalytic activities of CDKs (13, 24, 40, 41, 51). In turn, CDK1, CDK2, and CDK4 are activated by CAK phosphorylation (8, 16, 27, 37, 43, 54). In mammalian cells, the cyclin D-CDK4 complex is linked most closely to the regulation of G1 exit because it phosphorylates and inactivates retinoblastoma tumor suppressor protein (pRb) at G1 exit (24, 41, 42, 51, 58). Although CAK activation of cyclin D1-CDK4 has been assumed to be an important step for D1-CDK4 phosphorylation of pRb in vitro (8, 27, 37), there is no direct evidence yet to prove the existence of a CAK-cyclin D1-CDK4-pRb pathway. Recently, two new regulatory mechanisms of G1 exit have been revealed. These studies show that accumulation of hypophosphorylated (active) pRb and inhibition of E2F is not sufficient to arrest cells (68) and that the release of E2F-mediated transrepression of cell cycle genes but not transactivation by E2F triggers the G1/S transition (21). Thus, these additional G1 regulatory mechanisms that differ from well-accepted cyclin D-CDK4-pRb- or E2F-mediated transactivation pathways provide valuable information to study G1 progression.
In our previous studies, deregulation of CAK function via abrogation of MAT1 was found to induce G1 arrest (59). This result raises the intriguing possibility that CAK may regulate G1 exit. Since phosphorylation of pRb is a crucial event in driving cells through G1 into S (24, 31, 42, 51, 58), we investigated whether MAT1-modulated CAK activity enables cells to exit G1 and how MAT1 targets CAK activity to its putative G1 substrates, including cyclin D-CDK4 and pRb, to initiate G1 exit.
MATERIALS AND METHODS
Cell culture, transduction, and transfection.
Human osteosarcoma MG-63 and U-2 OS cells (American Type Culture Collection) were cultured in RPMI 1640 containing 10% fetal bovine serum (FBS). A 462-bp antisense MAT1 fragment cloned into a retroviral pG1xSvNa vector was transduced into cells using a retrovirus-mediated gene transfer system (59). The G418 concentration for selection of stable clones was determined by a 7-day lethal-dose test. After 48 h of posttransduction incubation, U-2 OS and MG-63 cells next were selected for 7 days with 0.2 mg of G418 per ml. About 20 single stable clones were picked up from each G1AsMatSvNa MAT1 antisense-transduced cell line and further expanded for detection of the MAT1 expression phenotype. G418-resistant colonies from G1xSvNa vector-transduced cells were pooled as a control. The same dose of G418 used for selection was supplemented in medium for maintenance of the stable clones. The G1AsMatSvNa (MAT1-AS)-transduced cells were used as experimental samples, while G1xSvNa (vector)-transduced cells and nontransduced (blank) cells served as controls. The retroviral G1nBgSvNa vector bearing a nucleus-targeted β-galactosidase was used for testing of gene transfer efficiency. Gene transfer efficiency was measured by determining the percentage of β-galactosidase-positive cells upon exposure to 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal) under a phase-contrast microscope as described previously (59). The gene transfer efficiency of the retroviral vector was about 31% under our experimental conditions.
To examine the specificity of antisense MAT1 in the above pRb-positive cellular models, we used the pRb-negative human osteosarcoma Saos-2 cell line (American Type Culture Collection) as a control. Saos-2 cells were cultured in RPMI 1640 containing 15% FBS. pcDNA3/AsMat (MAT1-AS), a pcDNA3 vector (Invitrogen) containing the same 462-bp antisense MAT1 fragment as above, was transfected into Saos-2 cells using Effectene (Qiagen) as described by the manufacturer. The G418 concentration for selection of stable clones was determined by a 7-day lethal-dose test. After 48 h of posttransfection incubation, Saos-2 cells next were selected for 7 days with 0.6 mg of G418 per ml. About 20 single stable clones were picked up from pcDNA3/AsMat (MAT1-AS)-transfected Saos-2 cells and expanded for detection of the MAT1 expression phenotype. G418-resistant colonies from pcDNA3 (vector)-transfected Saos-2 cells were pooled as a control. The same dose of G418 used for selection was added to the medium for maintenance of the stable clones. Both pcDNA3 vector-transfected and nontransfected blank cells were used in parallel as controls.
Protein expression plasmids.
Recombinant protein expression vectors were constructed by using established cloning methods as previously described (60, 61). The coding cDNA sequences of CDK7, cyclin H, MAT1, cyclin A, cyclin D1, CDK2, and CDK4 were generated by reverse transcription-PCR methods. The PCR-amplified cDNA fragments were first cloned into TA cloning vectors (Invitrogen). These cDNA fragments, confirmed by nucleotide sequencing, were subcloned into the pET protein expression vector (Novagen). The expression capabilities of these constructs were tested by protein induction in BL21(DE3) cells as described previously (60).
GST fusion protein expression.
Glutathione S-transferase (GST) fusion proteins containing human pRb A, B, and C pocket sequences (GSTpRb-A/B/C) were provided by Y. K. Fung (USC/CHLA). GSTpRb-A/B/C was transformed into the BL21 strain, and a high level of expression was induced in the presence of 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG) for 4 h after the cultures reached an optical density at 600 nm of 1.0. The collected cell pellet was suspended in lysis buffer (20 mM Tris base [pH 8], 100 mM NaCl, 1 mM EDTA, 5 μg of leupeptin per ml, 0.1 mM phenylmethylsulfonyl fluoride [PMSF], 0.5% NP-40, 0.1 mg of lysozyme), and the cells were disrupted by sonication. The supernatant containing soluble fusion proteins was incubated with glutathione-Sepharose (Pharmacia Biotech) at 4°C for 1 h with gentle shaking. The expressed GSTpRb-A/B/C proteins bound to the resin were washed and collected by centrifugation at 4°C. The amount of purified GSTpRb-A/B/C protein was determined by Coomassie blue staining using bovine serum albumin (Sigma) as a standard.
In vitro translation.
In vitro transcription-translation of proteins cloned in pET or pGEM2 (Promega) systems was performed using the TNT-coupled reticulocyte lysate system (Promega) as specified by the manufacturer. CDK7, cyclin H, and MAT1 were individually translated first, and then the binary CAK (CDK7-cyclin H) and ternary CAK (CDK7-cyclin H-MAT1) complexes were assembled at 30°C for 1 h. These in vitro-assembled CAK complexes were used as enzymes for CAK assays or as protein complexes for pRb-MAT1 interaction analysis. To test whether CAK phosphorylation of pRb is MAT1 dependent, equal and constant amounts of CDK and cyclin H proteins translated from 1 μg of cDNA were mixed with MAT1 proteins translated from increasing amounts (0.25, 0.5, 1.0, and 1.5 μg) of cDNA. After incubation of the mixture at 30°C for 1 h, these assembled CAK complexes containing different amounts of MAT1 were immunoprecipitated using anti-CDK7 antibodies (Santa Cruz) and then used as enzymes for the CAK-pRb kinase assay. Either cyclin D1-CDK4 or cyclin A-CDK2 was cotranslated in the reticulocyte lysate as a substrate. The activities of these enzymes and substrates produced from the reticulocyte lysates were tested in parallel kinase assays. For MAT1-pRB interaction analysis, the recombinant proteins were simultaneously translated and labeled in the presence of [35S]methionine.
Kinase assay.
Kinase activity was measured in an immune-complex kinase assay in the presence of 5 μCi of [γ-32P]ATP; approximately 500 ng each of substrates and enzymes were used per reaction. Kinase buffer contained 50 mM Tris-HCl (pH 7.5), 10 mM MgCl2, 1 mM dithiothreitol, 50 μM ATP, 1 μg of aprotinin per ml, 1 μg of leupeptin per ml, and 1 μg of pepstatin per ml. The kinase reaction mixtures were incubated at 30°C for 30 min and then terminated by the addition of sodium dodecyl sulfate (SDS) loading buffer. The reaction mixtures were resolved by SDS-polyacrylamide gel electrophoresis (PAGE) and then transferred to a polyvinylidene fluoride membrane (Millipore). The radioactive signal was quantitated on a Molecular Dynamics PhosphorImager. GST-pRb containing the C pocket sequence (GSTpRb-C) was purchased from Santa Cruz Biotechnology and used as a substrate in the kinase assays.
Immunoprecipitation and Western blotting.
Subconfluent cells were harvested by trypsinization and lysed using universal lysis/immunoprecipitation buffer (50 mM Tris-HCl, 150 mM NaCl, 2 mM EDTA, 2 mM EGTA, 25 mM NaFl, 25 mM β-glycerophosphate [pH 7.5], 0.1 mM sodium orthovanadate, 0.1 mM PMSF, 5 μg of leupeptin per ml, 0.2% [vol/vol] Triton X-100, 0.5% [vol/vol] Nonidet P-40) as described previously (59). The protein concentration was measured by the Bradford method (Bio-Rad). Aliquots (500 μg) of cellular proteins were precleaned by adding 1.0 μg of the appropriate normal immunoglobulin G together with 20 μl of appropriate protein A+G-agarose conjugate (Santa Cruz) for 1 h at 4°C. The immunoprecipitations were performed with the appropriate antibody for 2 to 16 h at 4°C. Complexes bound to the protein A+G-agarose conjugate were washed five times with universal lysis/immunoprecipitation buffer and separated by SDS-PAGE. For immunoprecipitation of putative MAT1-pRb complexes, aliquots (2,000 μg) of cellular proteins were incubated with either MAT1 or pRb polyclonal antibodies. The Western blotting was performed as described previously (59). For Western blotting depiction of pRb phosphorylation status, the cell lysates were separated overnight at 4°C. For detection of CDK phosphorylation status, the cell lysates were separated for ≥5 h. All antibodies used in immunoprecipitations and Western blotting were purchased from Santa Cruz Biotechnology.
In vitro protein binding assay.
In vitro GST-tagged protein binding assays were performed as previously described (25) with minor modifications. GSTpRb-A/B/C proteins or GST fragments bound to glutathione-Sepharose (4 μg) were mixed with 15 μl of in vitro-translated, [35S]methionine-labeled proteins from the following list: MAT1, pGEM2-luciferase, cyclin D1, in vitro-assembled binary CAK, and in vitro-assembled ternary CAK (see “In vitro translation” above). These different mixtures were diluted with 200 μl of protein binding buffer (PBB) (20 mM Tris-HCl [pH 7.8], 20% [vol/vol] glycerol, 0.2 mM EDTA, 100 mM KCl, 0.25 mM PMSF, 0.3% [vol/vol] NP-40, 0.2 mg of BSA per ml) and incubated at 4°C for 1 h to initiate protein binding. After the binding mixtures were washed six times with PBB, the [35S]methionine-labeled proteins retained on the resin were eluted with SDS loading buffer, resolved by SDS-PAGE (10% polyacrylamide), and detected by autoradiography.
Cell cycle analysis.
Cell cycle analysis was described previously (59). Cells were prefixed with 1% paraformaldehyde and postfixed with 70% ethanol. The washed pellet was stained using a propidium iodide-RNase solution. The cell cycle status was analyzed with a FACScalibur flow cytometer using ModFit LT software.
Proliferation activation analyses.
Cell proliferation activation from a contact-inhibited state in an in vitro wound tissue model was performed as described previously (59). Confluent cells exhibiting contact inhibition were scraped with a pipette tip to create 1-mm wound tracks. Retrovirus-mediated β-galactosidase transduction was performed 12 h after the wound tracks were created. Proliferation activation was measured by counting the number of β-galactosidase-positive cells upon exposure to X-Gal. The time for closure of the wound tracks was recorded in parallel.
Cell proliferation analyses.
Equal numbers of cells were seeded in 24-well plates. At 24 h after seeding, the rate of cell duplication was determined by counting the cells for three consecutive days before the cells reached confluence. To determine the number of living cells in culture, the same number of cells were grown in 96-well plates for 72 h and then incubated with MTS tetrazolium compound (Promega) for the proliferation assay as described previously (59). The amount of formazan product, quantified by measuring the absorbance at 490 nm using a Kinetic Microplate Reader (Molecular Devices), is directly proportional to the number of living cells in culture.
RESULTS
pRb phosphorylation is inhibited in MAT1-AS-transduced cells.
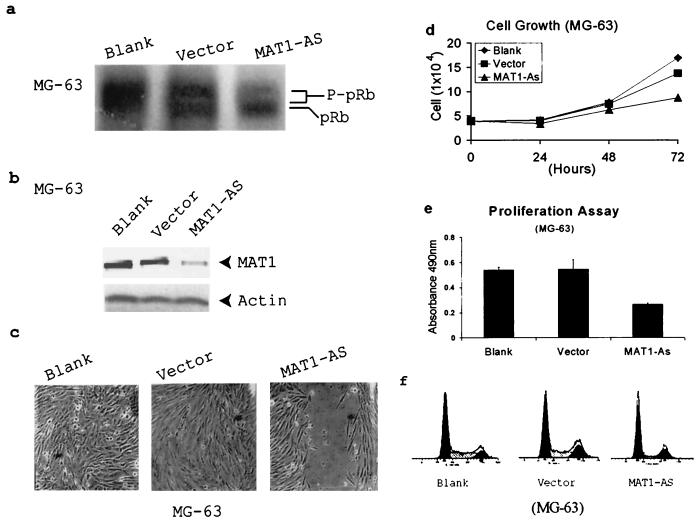
It is known that pRb function is regulated by its phosphorylation status. Hypophosphorylation of pRb in vivo will repress E2F transcription activity and arrest cells in the G1 phase (12, 22, 23, 67). To test whether the G1 arrest induced by abrogation of MAT1 is due to insufficient pRb phosphorylation, we demonstrated the in vivo pRb phosphorylation status in MAT1-AS-transduced MG-63 cells by Western blotting. Equal amounts of cellular proteins from G1AsMatSvNa (MAT1-AS)-transduced, G1xSvNa (vector)-transduced, and nontransduced (blank) cultures were separated by SDS-PAGE (6% polyacrylamide) overnight at 4°C. The blot was incubated with human pRb polyclonal antibodies. The results demonstrate that in vivo phosphorylation of pRb indeed is inhibited in these MAT1-AS-transduced MG-63 cells (Fig. 1a); these cells also showed a corresponding decrease in MAT1 protein expression, inhibited cell proliferation, and cell cycle G1 arrest (Fig. 1b to f).
FIG. 1.
Deregulation of CAK leads to inhibition of pRb phosphorylation and G1 arrest in MAT1-AS-transduced MG-63 cells. (a) Western blotting depicts pRb phosphorylation. P-pRb, hyperphosphorylated form of pRb. (b) Western blotting detects MAT1 protein expression. Actin detection was performed on the same blot as the protein loading control. (c) Analyses of cell proliferation activation in an in vitro injured tissue. Transduced and nontransduced confluent MG-63 cells were scraped to release cells from contact inhibition. The time for closure of the wound track was assessed under a phase-contrast microscope. Blank and Vector cells show closure of the wound track at 72 h, while the wound track in MAT1-AS-transduced cells still was not closed at that time. The time for closure of the wound track in MAT1-AS-transduced cells was 192 h (data not shown). (d) Cell growth analysis. The same numbers of transduced and nontransduced MG-63 cells were plated; cell duplication was monitored by counting the cells up to 3 days before they reached confluence. The growth curves represent the mean and standard deviation from the cells of triplicate wells. (e) Cell proliferation assay. Transduced and nontransduced subconfluent MG-63 cells were incubated with MTS tetrazolium compound and quantified by measurement of the absorbance at 490 nm to determine the proportion of living cells in culture. The data represent the mean and standard deviation of triplicate wells. (f) Cell cycle analysis. The cell cycle profile in MAT1-AS-transduced cells showed 67% cells in the G0/G1 phase and 12% in the S phase, which is 34% more cells in the G0/G1 phase and 54% fewer cells in the S phase compared with controls.
cCAK immunoprecipitated from MAT1-AS-transduced cells phosphorylate pRb much less efficiently.
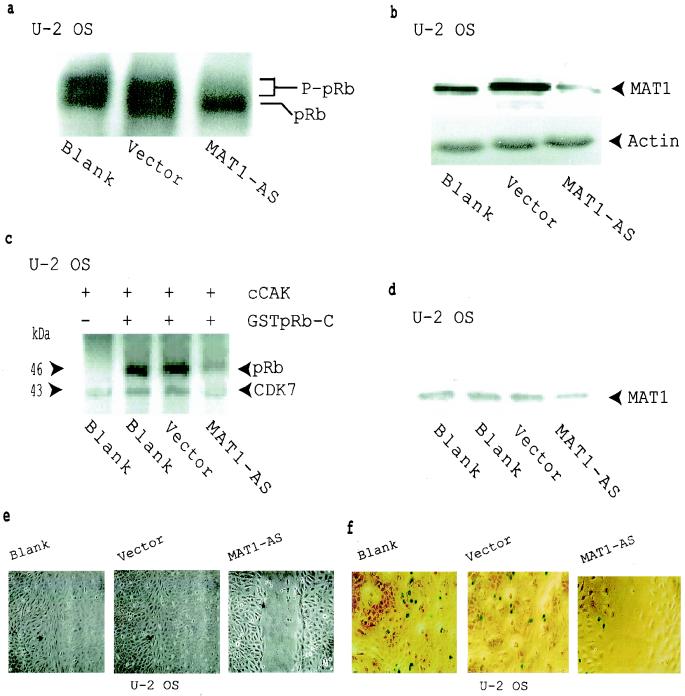
To determine whether the inhibited pRb phosphorylation in MAT1-AS-transduced cells is due directly to the deregulation of CAK function and not due to some other unknown cellular event, we used human CDK7 antibodies to immunoprecipitate cellular CAK (cCAK) complexes from G1AsMatSvNa (MAT1-AS)-transduced, G1xSvNa (vector)-transduced, and nontransduced (blank) U-2 OS cells. These cCAK complexes then were used as enzymes while GSTpRb-C fragments were used as substrates for kinase assays. cCAK complexes immunoprecipitated from MAT1-AS-transduced cells were much less able to phosphorylate pRb (Fig. 2c, lane 4). At the same time, in vivo pRb phosphorylation, MAT1 protein expression, cell proliferation, and cell proliferation activation were significantly inhibited in these MAT1-abrogated U-2 OS cells (Fig. 2a, b, e, and f).
FIG. 2.
Inhibition of pRb phosphorylation and cell proliferation in MAT1-AS-transduced U-2 OS cells. (a) Western blotting shows that pRb phosphorylation was inhibited in MAT1-AS-transduced cells. P-pRB, hyperphosphorylated form of pRB. (b) Western blotting analysis of MAT1 expression. Actin detection was performed on the same blot as the protein loading control. (c) cCAK phosphorylation of pRb. cCAKs immunoprecipitated from transduced and nontransduced U-2-OS cells were used as enzymes for the kinase assay; GSTpRb-C was used as a substrate. CDK7 was autophosphorylated in these reactions. (d) Equal amounts of CAK immunoprecipitates used in panel c were resolved by SDS-PAGE for Western blotting detection of the MAT1 level. (e) Analyses of cell proliferation activation in U-2 OS cells as described in the legend to Fig. 1c. The time for closure of the wound track in vector-transduced and nontransduced blank cells was 72 h, while the wound track in MAT1-AS-transduced cells was still open at that time. The time for closure of the wound track in MAT1-AS-transduced cells was 120 h (data not shown). (f) Cell proliferation was tested by retrovirus-mediated β-galactosidase transduction. Transduced and nontransduced confluent U-2 OS cells were scraped to release cells from contact inhibition. At 24 h after the wound track was created, proliferation activation was tested by transduction of the retroviral G1nBgSvNa vector into the wound track. At 48 h posttransduction, activated U-2 OS cell proliferation was measured at the margin of the wound track by counting the blue β-galactosidase-positive cells upon exposure to X-Gal under a phase-contrast microscope. The percentage of β-galactosidase-positive cells was 9% in MAT1-AS-transduced cells, 32% in nontransduced blank cells, and 30% in vector-transduced cells.
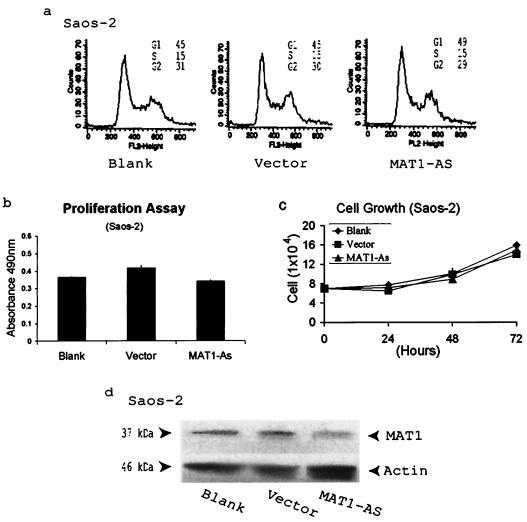
Our data suggest that antisense abrogation of MAT1 expression inhibits CAK phosphorylation of pRb. To ensure the target specificity of antisense MAT1 and to preclude any irrelevant effect of antisense MAT1 in our pRb-positive models, we examined the specificity of antisense MAT1 abrogation by transfection of MAT1-AS into pRb-negative Saos-2 cells. By analyses of the cell cycle profile and cell proliferation of these transfected Saos-2 cells, we found that lowering the MAT1 expression in these transfectants neither arrested cell cycle progression nor inhibited cell proliferation compared with the situation in vector-transfected and nontransfected cells (Fig. 3).
FIG. 3.
Antisense MAT1 abrogation has a negative impact on both cell cycle progression and cell proliferation in pRb-negative Saos-2 cells. (a) No cell cycle arrest is induced in MAT1-AS-transfected cells compared with vector-transfected and nontransfected blank cells. (b) Cell proliferation analysis of transfected and nontransfected Saos-2 cells as described in the legend to Fig. 1e. (c) Cell growth analysis of transfected and nontransfected Saos-2 cells as described in the legend to Fig. 1d. (d) Western blotting analysis of MAT1 expression. Protein loading was monitored by detection of actin on the same blot as that used for Western blotting.
Abrogation of MAT1 inhibits cyclin E expression but does not alter cyclin D1-CDK phosphorylation of pRb.
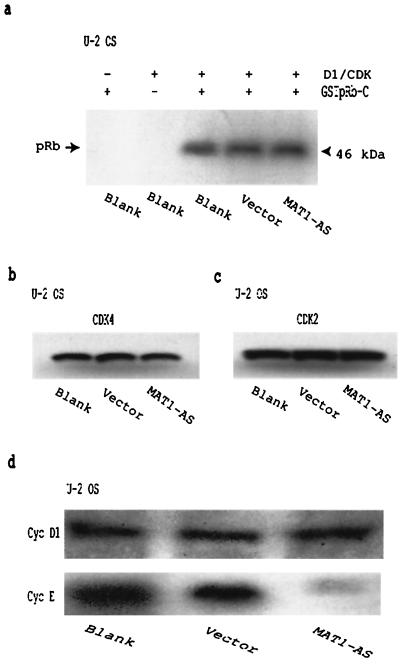
Because CAK activates cyclin D1-CDK4 in vitro, it has been assumed that this activation is an immediate upstream event that initiates cyclin D1-CDK4 phosphorylation of pRb, although there is no direct evidence yet to prove the existence of this connection. One may hypothesize that if CAK activation of cyclin D1-CDK4 is directly connected to subsequent cyclin D1-CDK4 phosphorylation of pRb, then abrogation of MAT1 should affect the activation of cyclin D1-CDK4 as well as the ability of cyclin D1-CDK4 to phosphorylate pRb. To test this hypothesis, we needed to perform on CDK4 the same experiments described above for analyzing pRb phosphorylation (Fig. 1a and 2a and c). That is, we needed to determine the in vivo CDK4 phosphorylation status in MAT1-AS-transduced cells and to examine whether decreased MAT1 altered cellular cyclin D1-CDK4 phosphorylation of pRb. Since cyclin D2 and D3 also associate with CDK4 in vivo and cyclin D1 associates with both CDK4 and CDK6, it was impossible for us to immunoprecipitate in vivo cyclin D1-CDK4 complexes but it was possible to isolate cyclin D1-CDK complexes by using cyclin D1 antibodies. Thus, we analyzed the cellular cyclin D1-CDK phosphorylation of pRb. We used cyclin D1 antibodies to immunoprecipitate cellular cyclin D1-CDK complexes from G1AsMatSvNa (MAT1-AS)-transduced, G1xSvNa (vector)-transduced, and nontransduced (blank) U-2 OS cells as enzymes for kinase assays using GSTpRb-C as a substrate. The results show that abrogation of MAT1 does not affect the cellular cyclin D1-CDK phosphorylation of pRb (Fig. 4a). Since MAT1 is more likely to act as a targeting subunit of CAK than as an enhancer of CDK7-cyclin H phosphorylation of CDK (25, 28, 64), the phosphorylation status of CDK2 and CDK4 should not be affected by MAT1 abrogation. To test this hypothesis, we also examined the CDK2 and CDK4 phosphorylation status in MAT1-AS-transduced cells by Western blotting using CDK2 and CDK4 antibodies. The phosphorylation status of CDK2 and CDK4 showed no change in MAT1-AS-transduced cells compared with vector-transduced and nontransduced cells (Fig. 4b and c). These experiments suggest that deregulation of CAK through MAT1 abrogation does not affect cyclin D1-CDK4 activation and phosphorylation.
FIG. 4.
Abrogation of MAT1 inhibits cyclin E expression in U-2 OS cells but has no effect on CDK phosphorylation. (a) Cellular cyclin D1-CDK phosphorylation of pRb. Cellular cyclin D1-CDK complexes immunoprecipitated from transduced and nontransduced U-2 OS cells were used as enzymes for the kinase assay, and GSTpRb-C was used as a substrate. (b and c) Western blotting analysis of CDK4 (b) and CDK2 (c) phosphorylation status. (d) Western blotting analysis of cyclin D1 and cyclin E expression.
While cellular CDK levels tend to remain constant, the levels of cyclins vary in abundance with the periodicity of the cell cycle, and the association of cyclins with CDKs regulates critical transition points (39, 62). Cyclin D1-CDK phosphorylates pRb at mid-G1 phase, while cyclin E-CDK2 phosphorylates pRb in the late G1 phase (9, 24, 52). Therefore, determination of cyclin D1 and cyclin E expression in MAT1-AS-transduced cells could answer two questions: (i) whether deregulated CAK function resulting from MAT1 abrogation alters the expression of G1 cyclins involved in pRb phosphorylation and (ii) the specific stage of the G1 phase at which MAT1 modulates CAK phosphorylation of pRb (mid-G1 phase or late G1). Cellular proteins from subconfluent MAT1-AS-transduced, vector-transduced, and nontransduced blank U-2 OS cells were subjected to SDS-PAGE, and the expression of cyclin D1 and cyclin E was examined by Western blotting. We found that MAT1 abrogation significantly inhibited cyclin E expression but had no affect on cyclin D1 expression (Fig. 4d). The results suggest that CAK functions in G1 phase and that MAT1-modulated CAK phosphorylation may occur before late G1 but after the mid-G1 stage of the cell cycle.
CAK phosphorylation of pRb is cyclin D1-CDK4 independent.
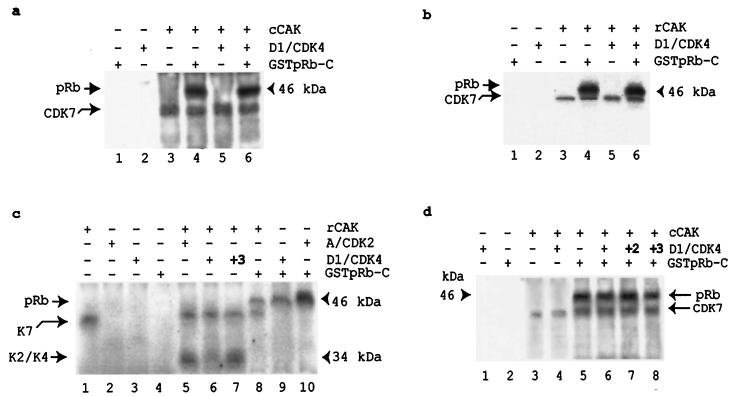
The activation and phosphorylation of CDK2 and CDK4 are MAT1 independent (25, 28, 64) (Fig. 4a to c), but MAT1 is required to target CAK activity to its non-CDK substrates (25, 28, 64) (Fig. 1a and 2a and c). Thus, we wished to determine the relationship between CAK phosphorylation of cyclin D1-CDK4 and CAK phosphorylation of pRb. We used human CDK7 polyclonal antibodies to immunoprecipitate cellular CAK complexes from G1AsMatSvNa (MAT1-AS)-transduced, G1xSvNa (vector)-transduced, and nontransduced (blank) U-2 OS cells. These immunoprecipitated complexes were used as enzymes for CAK assays, while in vitro-translated cyclin D1-CDK4 complexes were used as substrates. The results showed that cyclin D1-CDK4 phosphorylation was not detectable (data not shown). Next, we used immunoprecipitated cellular CAK from nontransduced (blank) U-2 OS cells as enzymes for the kinase assay with cyclin D1-CDK4, GSTpRb-C, or a mixture of the two were used as substrates. We found that cellular CAK readily phosphorylated pRb, but we were still unable to detect cellular CAK phosphorylation of cyclin D1-CDK4 (Fig. 5a). We were concerned that a cellular inhibitor that blocks CAK phosphorylation of cyclin D1-CDK4 might have been coimmunoprecipitated and thus was preventing CAK phosphorylation of cyclin D1-CDK4. To avoid this possible contamination, we used recombinant proteins as both enzymes and substrates. Under the same experimental conditions, our results showed that recombinant CAK (rCAK) complexes readily phosphorylated pRb but rCAK phosphorylation of cyclin D1-CDK4 still was not detected (Fig. 5b). Another possibility is that cyclin D1-CDK4 might be poorly translated or that the cyclin D1-CDK4 heterodimer might form less efficiently in reticulocyte lysates. Thus, we used cyclin A-CDK2 as a control in parallel with cyclin D1-CDK4. Also, we tested the activities of each preparation produced from in vitro translation in a parallel kinase assay (Fig. 5c, lanes 8 to 10). The results showed that CAK still readily phosphorylated both cyclin A-CDK2 and pRb (lanes 5 and 8) but phosphorylated cyclin D1-CDK4 very weakly (lane 6) unless three times the amount of cyclin D1-CDK4 was used (lane 7). Because we finally observed that CAK phosphorylated cyclin D1-CDK4 when three times the amount of cyclin D1-CDK4 was used in the kinase reaction, we next tested whether CAK enhanced cyclin D1-CDK4 phosphorylation of pRb by adding CAK to cyclin D1-CDK4-pRb reaction mixtures. The results showed that although CAK readily phosphorylated pRb (Fig. 5d, lane 5), CAK did not enhance the cyclin D1-CDK4 phosphorylation of pRb (lanes 6 to 8). Interestingly, CAK also did not phosphorylate cyclin D1-CDK4 in the presence of pRb even after the amount of cyclin D1-CDK4 was increased threefold (lane 8), despite being able to phosphorylate cyclin D1-CDK4 in the absence of pRb (Fig. 5c, lane 7). These results indicate that CAK phosphorylation of pRb is cyclin D1-CDK4 independent and that CAK favors pRb as its substrate in vitro.
FIG. 5.
CAK phosphorylation of pRb is cyclin D1-CDK4 independent. (a) cCAK phosphorylates pRb in the presence or absence of cyclin D1-CDK4. (b) rCAK readily phosphorylates pRb, while rCAK phosphorylation of cyclin D1-CDK4 is not detectable. (c) rCAK phosphorylation of cyclin-CDK complexes. rCAK readily phosphorylates cyclin A-CDK2, while rCAK phosphorylation of cyclin D1-CDK4 is not detectable (lanes 5 and 6) unless three times the amount of cyclin D1-CDK4 is used as a substrate (lane 7). The kinase activities of all enzyme preparations produced from the in vitro translation system were tested in parallel using GSTpRb-C as substrates (lanes 8 to 10). The migration positions of phosphorylated CDK2 and CDK4 have been confirmed by Western blotting in parallel (data not shown). (d) CAK does not enhance the cyclin D1-CDK4 phosphorylation of pRb.
MAT1 enhances CDK7-cyclin H phosphorylation of pRb.
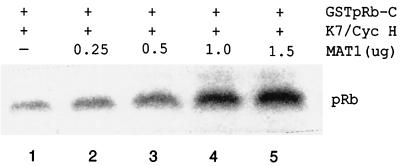
Since our data show that CAK phosphorylates pRb and that pRb phosphorylation is inhibited in MAT1-AS-transduced cells, we wanted to determine whether CAK phosphorylation of pRb truly is modulated by MAT1. Because MAT1 is an assembly factor of CAK, we first investigated whether MAT1 determines the CAK substrate specificity for pRb by assembling and stabilizing the association of the CDK7-cyclin H complex. We assembled binary CAK and ternary CAK in vitro by using equal and constant amounts of CDK7 and cyclin H while increasing the amount of MAT1 (see Materials and Methods). These assembled CAK complexes containing different amounts of MAT1 were used as enzymes for kinase assays with GSTpRb-C substrates. We found that ternary CAK phosphorylated pRb more efficiently than binary CAK did (Fig. 6, lanes 1 and 4) and that CAK phosphorylation of pRb was MAT1 dose dependent (Fig. 6). The results suggest that MAT1 enhances the CAK phosphorylation of pRb through MAT1 assembly and stabilization of the CDK7-cyclin H complex.
FIG. 6.
CAK phosphorylation of pRb is MAT1 dose dependent. Equal and constant amounts of CDK7 and cyclin H proteins were mixed with increasing amounts of MAT1 protein. Both CDK7 and cyclin H proteins were translated separately from 1 μg of cDNA; MAT1 protein was translated from 0.25, 0.5, 1.0, and 1.5 μg of cDNA.
MAT1 interacts with pRb.
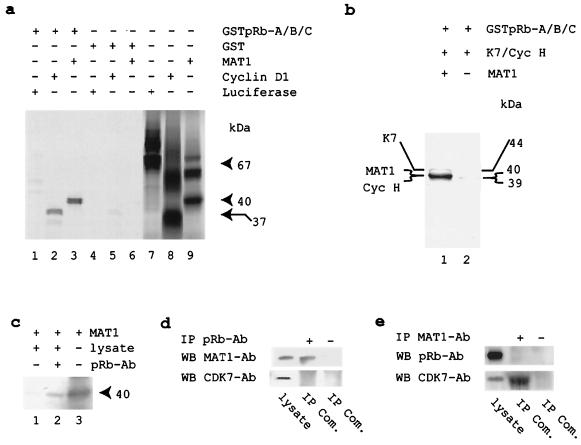
The above data show that CAK phosphorylation of pRb is MAT1 dose dependent. To date, CAK has not been found in pRb complexes, nor has pRb been copurified with CAK or CAK-associated complexes, e.g., TFIIH. Thus, the challenging question arises of how CAK is targeted to pRb phosphorylation. We note that CAK phosphorylates p53 and MAT1 interacts with p53 (28) and that the interactions between the MAT1 and POU domains of Oct factors can target CAK to Oct factors and result in their phosphorylation (25). We wondered if the altered CAK substrate specificity was determined by MAT1-mediated protein-protein interactions in the fashion of MAT1 targeting CAK to p53 or Oct factors. Therefore, we investigated whether MAT1 interacts with pRb. To test for a MAT1-pRb interaction in vitro, we used a modified GST domain-mediated protein binding assay as described previously (25). As controls, pGEM2-luciferase (negative binding of pRb) and His-cyclin D1 (positive binding of pRb) were used in parallel. The results show that MAT1 interacts with pRb in vitro (Fig. 7a) and that pRb interacts with ternary CAK (CDK7-cyclin H-MAT1) but not binary CAK (CDK7-cyclin H) (Fig. 7b). Further, we incubated in vitro-translated and [35S]methionine-labeled His-MAT1 with U-2 OS cellular proteins to generate putative MAT1-pRb binding complexes and then immunoprecipitated these complexes using pRb antibodies. The SDS-PAGE analysis showed that MAT1 also interacts with cellular pRb (Fig. 7c). To determine whether MAT1 interacts with pRb under physiological conditions that do not involve any overexpression, we immunoprecipitated putative MAT1-pRb binding complexes from cellular proteins extracted from subconfluent U-2 OS cells by using either pRb or MAT1 antibodies, while immunoprecipitation without antibody was used as a negative control in parallel. We analyzed these cellular complexes for speculative binding partners by using the corresponding antibodies on Western blots; Ewing's sarcoma cell lysate was used as a positive control of Western blotting in parallel. We found that MAT1 associated with pRb (Fig. 7d) but that pRb was not detectable in complexes immunoprecipitated by MAT1 antibodies (Fig. 7e) and CDK7 was not found in pRb complexes (Fig. 7d). Thus, these initial results indicate that (i) MAT1 interacts with pRb in a cellular context and (ii) the majority of cellular MAT1 exists as a subunit of active CAK complexes. These results suggest that the interaction between MAT1 and pRb may target CAK to pRb and result in pRb phosphorylation.
FIG. 7.
MAT1 interacts with pRb. (a) Monomeric MAT1 interacts with recombinant pRb. The migration positions of in vitro-translated and [35S]methionine-labeled recombinant proteins in SDS-PAGE showed 67-kDa pGEM2-luciferase, 37-kDa His-cyclin D1, and 40-kDa His-MAT1. (b) Ternary CAK, but not binary CAK, interacts with recombinant pRb. The migration positions of in vitro-translated and [35S]methionine-labeled recombinant proteins in SDS-PAGE showed 44-kDa CDK7 and 39-kDa cyclin H. The migration position of cyclin H was confirmed by Western blotting (data not shown). (c) MAT1 interacts with cellular pRb. In vitro-translated and [35S]methionine-labeled MAT1 was incubated with U-2 OS cellular proteins and then immunoprecipitated by pRb antibodies. (d) MAT1, but not CDK7, was coimmunoprecipitated by pRb antibodies. Ewing's sarcoma cellular proteins (15 μg) were used as positive control in Western blotting. IP Com., immunoprecipitated complexes. (e) MAT1 antibodies coimmunoprecipitate CDK7 but not pRb.
DISCUSSION
The restriction point of the G1 phase controls the transition between serum-dependent, extracellular signal regulation and the serum-independent, autonomous program. This restriction point currently is represented by pRb phosphorylation connecting the cell cycle clock with transcription. The molecular basis for these pRb-imposed checkpoints during G1 progression, as well as their potential relationship to one another, remains unclear. One widely accepted pathway that regulates this transition and enables cells to exit G1 involves cyclin D-CDK4 phosphorylation of pRb and concomitant E2F transactivation (14, 24, 26, 41, 42, 48, 51, 58). Recently, a novel G1 exit pathway has revealed an additional avenue for regulating E2F-mediated transrepression of cell cycle genes, in contrast to E2F-mediated transactivation (21). We present evidence showing that MAT1 integrates CAK activity into pRb phosphorylation to regulate G1 exit. Considering that CAK is involved in both cell cycle control and transcription, it is not surprising that CAK phosphorylation of pRb connects the cell cycle clock with transcription and activates the expression of late-G1-phase genes to enable G1 exit.
CAK regulates G1 exit.
Cells exiting G1 and entering S must pass through early G1, mid-G1, late G1, and the G1/S transition. Phosphorylation of pRb at mid-G1 by cyclin D-CDK4 and at late G1 by cyclin E-CDK2 is required for cells to exit G1; during these periods the pRb phosphorylation status undergoes cyclical changes. Previous data showing that abrogation of MAT1 induces G1 arrest (59) imply that CAK regulates G1 progression. We present data here to show a correlation between MAT1 abrogation and CAK phosphorylation of pRb in the regulation of G1 exit. If MAT1-modulated CAK phosphorylation of pRb is necessary for G1 exit, then MAT1-AS-transduced cells that show proliferation inhibition and G1 arrest (Fig. 1c to f and 2e and f) also should show inhibited pRb phosphorylation (Fig. 1a and 2a and c). This lowering of MAT1 expression in pRb-positive cells should specifically deregulate CAK-pRb signaling (Fig. 1 and 2) without altering the CDK phosphorylation status (Fig. 4a to c), while the lowering of MAT1 expression in pRb-negative cells (Fig. 3d) should not arrest cell cycle progression and inhibit cell proliferation (Fig. 3a to c). Furthermore, we present additional evidence to support our hypothesis that MAT1-modulated CAK phosphorylation of pRb regulates G1 exit. We find that MAT1 abrogation does not alter the level of cyclin D1 but significantly inhibits cyclin E expression (Fig. 4d). Also, cellular CAK isolated from MAT1-AS cells phosphorylates pRb much less efficiently than does CAK isolated from control cells (Fig. 2c), but cellular cyclin D1-CDK isolated from MAT1-AS cells shows no change in pRb phosphorylation compared with controls (Fig. 4a). Importantly, in vitro CAK phosphorylation of pRb is MAT1 dose dependent (Fig. 6), supporting the data showing that abrogation of MAT1 inhibits the CAK phosphorylation of pRb (Fig. 1a and 2a and c). These findings suggest that (i) MAT1 modulates CAK phosphorylation of pRb to regulate G1 exit; (ii) CAK phosphorylation of pRb may occur after the mid-G1 phase, when cyclin D-CDK4 phosphorylates pRb, but before the late G1 phase, when cyclin E-CDK2 phosphorylates pRb; and (iii) CAK phosphorylation of pRb may initiate or enhance cyclin E expression.
CAK phosphorylation of pRb is cyclin D1-CDK4 independent.
Cyclins D1, D2, and D3 are induced in G1 and form complexes with CDK2, CDK4, CDK5, or CDK6 (2, 24, 34–36, 38, 63). The cyclin D1-CDK4 complex phosphorylates pRb in the middle of the G1 phase, releasing E2F-DP proteins from the pRb complex and resulting in G1 exit and S-phase entry (22, 23, 26, 30, 48). Since CAK activates cyclin D1-CDK4 in vitro (8, 27, 37) and MAT1 determines CAK substrate specificity (25, 28, 64), we previously hypothesized that CAK activation of cyclin D1-CDK may have a direct connection for cyclin D1-CDK4 phosphorylation of pRb or that CAK may directly phosphorylate pRb to regulate G1 progression (59). Our data suggest that CAK regulates G1 exit through direct phosphorylation of pRb in the late G1 phase because of the following evidence. First, in MAT1-AS-transduced cells showing proliferation inhibition and G1 arrest (Fig. 1c to f and 2e and f), pRb phosphorylation was inhibited (Fig. 1a and 2a and c) but there was no effect on the phosphorylation of cyclin D-CDK4 or cyclin A-CDK2 (Fig. 4a to c). Second, both cellular and recombinant CAK readily phosphorylate pRb and CAK phosphorylates pRb in the presence or absence of cyclin D1-CDK4 (Fig. 2c and 5). Third, CAK favors pRb as its substrate over cyclin D1-CDK4 when both targets are present (Fig. 5a and b and 5d, lanes 6 to 8). Finally, CAK phosphorylation of pRb is MAT1 dose dependent (Fig. 6) and cyclin D1-CDK4 independent (Fig. 5). It is important to note that our experiments do not preclude the possibility of a CAK-cyclin D1-CDK4-pRb signal transduction; rather, we present a new CAK-pRb pathway in G1 progression. Distinguishing the difference and establishing the links between the CAK-pRb and cyclin D1-CDK4-pRb pathways will be pursued in the future.
MAT1 modulates CAK substrate specificity in the regulation of G1 exit.
Although we know that MAT1 shifts the substrate preference of the CDK7-cyclin H complex from the originally defined CDK2 to a different set of substrates (25, 28, 46, 64), the question that arises is how MAT1 targets CAK activity to its various substrates. Does this occur because MAT1 assembles and stabilizes the association of CDK7-cyclin H, or does MAT1 alter CAK substrate specificity via a MAT1-mediated protein-protein interaction? Our results here suggest that MAT1 targets CAK activity to pRb phosphorylation both by assembling and stabilizing the association of CDK7-cyclin H and through a MAT1-pRb interaction. Indeed, when increasing amounts of MAT1 are added to CDK7 and cyclin H, CAK phosphorylation of pRb is stoichiometrically enhanced (Fig. 6). Also, in vitro-translated MAT1 is able to interact either with recombinant pRb or cellular pRb (Fig. 7a and c) and pRb can bind to ternary CAK but not to binary CAK (Fig. 7b). These data not only support the hypothesis that the novel domains of MAT1 are involved in gene regulation and protein-protein interactions (4, 6, 19, 32, 49; P. S. Freemont, I. M. Hanson, and J. Trowsdale, Letter, Cell 64:483–484, 1991) but also suggest that MAT1 determines CAK substrate specificity through MAT1-mediated CAK formation and MAT1-modulated protein-protein interaction. The present study also addresses whether the MAT1-pRb interaction occurs in vivo. We observed that MAT1 existed in the complexes immunoprecipitated by pRb antibodies while CDK7 was not detectable (Fig. 7d) whereas CDK7 was found in the complexes immunoprecipitated by MAT1 antibodies but pRb was not detectable (Fig. 7e). These results indicate that in a cellular context, MAT1 is responsible for interacting with both CDK7 and pRb whereas there is no detectable association between pRb and CKD7. Since we detected both MAT1 (Fig. 7d) and E2F-1 (data not shown) in the same complexes immunoprecipitated by pRb antibodies, it suggests that MAT1 interacts with pRb and exists in pRb-E2F complexes. However, a challenging question also arises: by existing in pRb complexes, does MAT1 serve as a bridge to recruit CDK7-cyclin H to phosphorylate pRb at a precise time and in a defined order during the cell cycle progression? To address this issue in the future, we need to perform a comprehensive analysis of G1 progression to determine the precise stage of the G1 phase at which MAT1-mediated CAK phosphorylation of pRb occurs. We also need to identify the domain(s) of MAT1 that interacts with pRb and regulates CAK-pRb phosphorylation by using a recombinant baculovirus expression system as described recently by Busso et al. (4), in which distinct regions of MAT1 that regulate CDK7 kinase and TFIIH transcription activities have been identified.
Here and in the past (59), the physiological role of MAT1 and CAK in specific cell cycle phases has been assessed by removing MAT1 protein from mammalian cells. These data not only reveal that MAT1-modulated CAK phosphorylation of pRb regulates cell cycle G1 exit but also support the results by other groups that the presence of MAT1 assembles an active CAK and determines the substrate specificity of CAK (25, 28, 64). The discovery of MAT1-modulated CAK phosphorylation of pRb suggests that transcription may be altered following cell cycle events at G1 exit. Taken together with other studies uncovering novel mechanisms involved in G1 progression (21, 68), our work will bring us closer to a comprehensive understanding of G1 regulation. To define the precise physiological function of CAK-pRb signal transduction and to understand how MAT1 itself is regulated, we first need to focus on (i) identifying the CAK phosphorylation sites on pRb and determining how phosphorylation of these sites by CAK is needed to promote G1 exit and (ii) delineating a CAK-pRb pathway by using microarrays to detect biologically relevant gene clusters induced by MAT1 abrogation.
ACKNOWLEDGMENTS
We thank Yuen Kai Fung for providing the GSTpRb-A/B/C construct and W. French Anderson for providing the retroviral pG1xSvNa vector.
REFERENCES
- 1.Adamczewski J P, Rossignol M, Tassan J P, Nigg E A, Moncollin V, Egly J M. MAT1, cdk7 and cyclin H form a kinase complex which is UV light-sensitive upon association with TFIIH. EMBO J. 1996;15:1877–1884. [PMC free article] [PubMed] [Google Scholar]
- 2.Bates S, Bonetta L, MacAllan D, Parry D, Holder A, Dickson C, Peters G. CDK6 (PLSTIRE) and CDK4 (PSK-J3) are a distinct subset of the cyclin-dependent kinases that associate with cyclin D1. Oncogene. 1994;9:71–79. [PubMed] [Google Scholar]
- 3.Brown A J, Jones T, Shuttleworth J. Expression and activity of p40MO15, the catalytic subunit of cdk- activating kinase, during Xenopus oogenesis and embryogenesis. Mol Biol Cell. 1994;5:921–932. doi: 10.1091/mbc.5.8.921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Busso D, Keriel A, Sandrock B, Poterszman A, Gileadi O, Egly J M. Distinct regions of MAT1 regulate cdk7 kinase and TFIIH transcription activities. J Biol Chem. 2000;275:22815–22823. doi: 10.1074/jbc.M002578200. [DOI] [PubMed] [Google Scholar]
- 5.Chalut C, Moncollin V, Egly J M. Transcription by RNA polymerase II: a process linked to DNA repair. Bioessays. 1994;16:651–655. doi: 10.1002/bies.950160910. [DOI] [PubMed] [Google Scholar]
- 6.Cohen C, Parry D A. Alpha-helical coiled coils and bundles: how to design an alpha-helical protein. Proteins. 1990;7:1–15. doi: 10.1002/prot.340070102. [DOI] [PubMed] [Google Scholar]
- 7.Devault A, Martinez A M, Fesquet D, Labbe J C, Morin N, Tassan J P, Nigg E A, Cavadore J C, Doree M. MAT1 (‘menage a trois’), a new RING finger protein subunit stabilizing cyclin H-cdk7 complexes in starfish and Xenopus CAK. EMBO J. 1995;14:5027–5036. doi: 10.1002/j.1460-2075.1995.tb00185.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Diehl J A, Sherr C J. A dominant-negative cyclin D1 mutant prevents nuclear import of cyclin-dependent kinase 4 (CDK4) and its phosphorylation by CDK-activating kinase. Mol Cell Biol. 1997;17:7362–7374. doi: 10.1128/mcb.17.12.7362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dou Q P, An B. RB and apoptotic cell death. Front Biosci. 1998;3:d419–d430. doi: 10.2741/a288. [DOI] [PubMed] [Google Scholar]
- 10.Drapkin R, Reinberg D. The multifunctional TFIIH complex and transcriptional control, Trends Biochem. Sci. 1994;19:504–508. doi: 10.1016/0968-0004(94)90139-2. [DOI] [PubMed] [Google Scholar]
- 11.Drapkin R, Le Roy G, Cho H, Akoulitchev S, Reinberg D. Human cyclin-dependent kinase-activating kinase exists in three distinct complexes. Proc Natl Acad Sci USA. 1996;93:6488–6493. doi: 10.1073/pnas.93.13.6488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dyson N. pRB, p107 and the regulation of the E2F transcription factor. J Cell Sci Suppl. 1994;18:81–87. doi: 10.1242/jcs.1994.supplement_18.12. [DOI] [PubMed] [Google Scholar]
- 13.Elledge S J. Cell cycle checkpoints: preventing an identity crisis. Science. 1996;274:1664–1672. doi: 10.1126/science.274.5293.1664. [DOI] [PubMed] [Google Scholar]
- 14.Ewen M E, Sluss H K, Sherr C J, Matsushime H, Kato J, Livingston D M. Functional interactions of the retinoblastoma protein with mammalian D-type cyclins. Cell. 1993;73:487–497. doi: 10.1016/0092-8674(93)90136-e. [DOI] [PubMed] [Google Scholar]
- 15.Feaver W J, Svejstrup J Q, Henry N L, Kornberg R D. Relationship of CDK-activating kinase and RNA polymerase II CTD kinase TFIIH/TFIIK. Cell. 1994;79:1103–1109. doi: 10.1016/0092-8674(94)90040-x. [DOI] [PubMed] [Google Scholar]
- 16.Fesquet D, Labbe J C, Derancourt J, Capony J P, Galas S, Girard F, Lorca T, Shuttleworth J, Doree M, Cavadore J C. The MO15 gene encodes the catalytic subunit of a protein kinase that activates cdc2 and other cyclin-dependent kinases (CDKs) through phosphorylation of Thr161 and its homologues. EMBO J. 1993;12:3111–3121. doi: 10.1002/j.1460-2075.1993.tb05980.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fisher R P, Morgan D O. A novel cyclin associates with MO15/CDK7 to form the CDK-activating kinase. Cell. 1994;78:713–724. doi: 10.1016/0092-8674(94)90535-5. [DOI] [PubMed] [Google Scholar]
- 18.Fisher R P, Jin P, Chamberlin H M, Morgan D O. Alternative mechanisms of CAK assembly require an assembly factor or an activating kinase. Cell. 1995;83:47–57. doi: 10.1016/0092-8674(95)90233-3. [DOI] [PubMed] [Google Scholar]
- 19.Freemont P S. The RING finger. A novel protein sequence motif related to the zinc finger. Ann N Y Acad Sci. 1993;684:174–192. doi: 10.1111/j.1749-6632.1993.tb32280.x. [DOI] [PubMed] [Google Scholar]
- 20.Goodrich J A, Tjian R. Transcription factors IIE and IIH and ATP hydrolysis direct promoter clearance by RNA polymerase II. Cell. 1994;77:145–156. doi: 10.1016/0092-8674(94)90242-9. [DOI] [PubMed] [Google Scholar]
- 21.He S, Cook B L, Deverman B E, Weihe U, Zhang F, Prachand V, Zheng J, Weintraub S J. E2F is required to prevent inappropriate S-phase entry of mammalian cells. Mol Cell Biol. 2000;20:363–371. doi: 10.1128/mcb.20.1.363-371.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Helin K, Harlow E, Fattaey A. Inhibition of E2F-1 transactivation by direct binding of the retinoblastoma protein. Mol Cell Biol. 1993;13:6501–6508. doi: 10.1128/mcb.13.10.6501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hiebert S W, Chellappan S P, Horowitz J M, Nevins J R. The interaction of RB with E2F coincides with an inhibition of the transcriptional activity of E2F. Genes Dev. 1992;6:177–185. doi: 10.1101/gad.6.2.177. [DOI] [PubMed] [Google Scholar]
- 24.Hunter T, Pines J. Cyclins and cancer. II. Cyclin D and CDK inhibitors come of age. Cell. 1994;79:573–582. doi: 10.1016/0092-8674(94)90543-6. [DOI] [PubMed] [Google Scholar]
- 25.Inamoto S, Segil N, Pan Z Q, Kimura M, Roeder R G. The cyclin-dependent kinase-activating kinase (CAK) assembly factor, MAT1, targets and enhances CAK activity on the POU domains of octamer transcription factors. J Biol Chem. 1997;272:29852–29858. doi: 10.1074/jbc.272.47.29852. [DOI] [PubMed] [Google Scholar]
- 26.Kato J, Matsushime H, Hiebert S W, Ewen M E, Sherr C J. Direct binding of cyclin D to the retinoblastoma gene product (pRb) and pRb phosphorylation by the cyclin D-dependent kinase CDK4. Genes Dev. 1993;7:331–342. doi: 10.1101/gad.7.3.331. [DOI] [PubMed] [Google Scholar]
- 27.Kato J Y, Matsuoka M, Polyak K, Massague J, Sherr C J. Cyclic AMP-induced G1 phase arrest mediated by an inhibitor (p27Kip1) of cyclin-dependent kinase 4 activation. Cell. 1994;79:487–496. doi: 10.1016/0092-8674(94)90257-7. [DOI] [PubMed] [Google Scholar]
- 28.Ko L J, Shieh S Y, Chen X, Jayaraman L, Tamai K, Taya Y, Prives C, Pan Z Q. p53 is phosphorylated by CDK7-cyclin H in a p36MAT1-dependent manner. Mol Cell Biol. 1997;17:7220–7229. doi: 10.1128/mcb.17.12.7220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Levedakou E N, He M, Baptist E W, Craven R J, Cance W G, Welcsh P L, Simmons A, Naylor S L, Leach R J, Lewis T B, et al. Two novel human serine/threonine kinases with homologies to the cell cycle regulating Xenopus MO15, and NIMA kinases: cloning and characterization of their expression pattern. Oncogene. 1994;9:1977–1988. [PubMed] [Google Scholar]
- 30.Levine A J. p53, the cellular gatekeeper for growth and division. Cell. 1997;88:323–331. doi: 10.1016/s0092-8674(00)81871-1. [DOI] [PubMed] [Google Scholar]
- 31.Lundberg A S, Weinberg R A. Control of the cell cycle and apoptosis. Eur J Cancer. 1999;35:531–539. [PubMed] [Google Scholar]
- 32.Lupas A, Van Dyke M, Stock J. Predicting coiled coils from protein sequences. Science. 1991;252:1162–1164. doi: 10.1126/science.252.5009.1162. [DOI] [PubMed] [Google Scholar]
- 33.Martinez A M, Afshar M, Martin F, Cavadore J C, Labbe J C, Doree M. Dual phosphorylation of the T-loop in cdk7: its role in controlling cyclin H binding and CAK activity. EMBO J. 1997;16:343–354. doi: 10.1093/emboj/16.2.343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Matsushime H, Roussel M F, Sherr C J. Novel mammalian cyclins (CYL genes) expressed during G1. Cold Spring Harbor Symp Quant Biol. 1991;56:69–74. doi: 10.1101/sqb.1991.056.01.010. [DOI] [PubMed] [Google Scholar]
- 35.Matsushime H, Ewen M E, Strom D K, Kato J Y, Hanks S K, Roussel M F, Sherr C J. Identification and properties of an atypical catalytic subunit (p34PSK-J3/cdk4) for mammalian D type G1 cyclins. Cell. 1992;71:323–334. doi: 10.1016/0092-8674(92)90360-o. [DOI] [PubMed] [Google Scholar]
- 36.Matsushime H, Quelle D E, Shurtleff S A, Shibuya M, Sherr C J, Kato J Y. D-type cyclin-dependent kinase activity in mammalian cells. Mol Cell Biol. 1994;14:2066–2076. doi: 10.1128/mcb.14.3.2066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Matsuoka M, Kato J Y, Fisher R P, Morgan D O, Sherr C J. Activation of cyclin-dependent kinase 4 (cdk4) by mouse MO15-associated kinase. Mol Cell Biol. 1994;14:7265–7275. doi: 10.1128/mcb.14.11.7265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Meyerson M, Harlow E. Identification of G1 kinase activity for cdk6, a novel cyclin D partner. Mol Cell Biol. 1994;14:2077–2086. doi: 10.1128/mcb.14.3.2077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Morgan D O. Principles of CDK regulation. Nature. 1995;374:131–134. doi: 10.1038/374131a0. [DOI] [PubMed] [Google Scholar]
- 40.Nasmyth K. Viewpoint: putting the cell cycle in order. Science. 1996;274:1643–1645. doi: 10.1126/science.274.5293.1643. [DOI] [PubMed] [Google Scholar]
- 41.Nigg E A. Cyclin-dependent kinase 7: at the cross-roads of transcription, DNA repair and cell cycle control? Curr Opin Cell Biol. 1996;8:312–317. doi: 10.1016/s0955-0674(96)80003-2. [DOI] [PubMed] [Google Scholar]
- 42.Nurse P. Ordering S phase and M phase in the cell cycle. Cell. 1994;79:547–550. doi: 10.1016/0092-8674(94)90539-8. [DOI] [PubMed] [Google Scholar]
- 43.Poon R Y, Yamashita K, Adamczewski J P, Hunt T, Shuttleworth J. The cdc2-related protein p40MO15 is the catalytic subunit of a protein kinase that can activate p33cdk2 and p34cdc2. EMBO J. 1993;12:3123–3132. doi: 10.1002/j.1460-2075.1993.tb05981.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Poon R Y, Yamashita K, Howell M, Ershler M A, Belyavsky A, Hunt T. Cell cycle regulation of the p34cdc2/p33cdk2-activating kinase p40MO15. J Cell Sci. 1994;107:2789–2799. doi: 10.1242/jcs.107.10.2789. [DOI] [PubMed] [Google Scholar]
- 45.Reardon J T, Ge H, Gibbs E, Sancar A, Hurwitz J, Pan Z Q. Isolation and characterization of two human transcription factor IIH (TFIIH)-related complexes: ERCC2/CAK and TFIIH. Proc Natl Acad Sci USA. 1996;93:6482–6487. doi: 10.1073/pnas.93.13.6482. . (Erratum, 93:10538.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rossignol M, Kolb-Cheynel I, Egly J M. Substrate specificity of the cdk-activating kinase (CAK) is altered upon association with TFIIH. EMBO J. 1997;16:1628–1637. doi: 10.1093/emboj/16.7.1628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Roy R, Adamczewski J P, Seroz T, Vermeulen W, Tassan J P, Schaeffer L, Nigg E A, Hoeijmakers J H, Egly J M. The MO15 cell cycle kinase is associated with the TFIIH transcription-DNA repair factor. Cell. 1994;79:1093–1101. doi: 10.1016/0092-8674(94)90039-6. [DOI] [PubMed] [Google Scholar]
- 48.Sanchez I, Dynlacht B D. Transcriptional control of the cell cycle. Curr Opin Cell Biol. 1996;8:318–324. doi: 10.1016/s0955-0674(96)80004-4. [DOI] [PubMed] [Google Scholar]
- 49.Schwabe J W, Klug A. Zinc mining for protein domains. Nat Struct Biol. 1994;1:345–349. doi: 10.1038/nsb0694-345. [DOI] [PubMed] [Google Scholar]
- 50.Serizawa H, Makela T P, Conaway J W, Conaway R C, Weinberg R A, Young R A. Association of Cdk-activating kinase subunits with transcription factor TFIIH. Nature. 1995;374:280–282. doi: 10.1038/374280a0. [DOI] [PubMed] [Google Scholar]
- 51.Sherr C J. Cancer cell cycles. Science. 1996;274:1672–1677. doi: 10.1126/science.274.5293.1672. [DOI] [PubMed] [Google Scholar]
- 52.Sherr C J, Roberts J M. Inhibitors of mammalian G1 cyclin-dependent kinases. Genes Dev. 1995;9:1149–1163. doi: 10.1101/gad.9.10.1149. [DOI] [PubMed] [Google Scholar]
- 53.Shiekhattar R, Mermelstein F, Fisher R P, Drapkin R, Dynlacht B, Wessling H C, Morgan D O, Reinberg D. Cdk-activating kinase complex is a component of human transcription factor TFIIH. Nature. 1995;374:283–287. doi: 10.1038/374283a0. [DOI] [PubMed] [Google Scholar]
- 54.Solomon M J, Harper J W, Shuttleworth J. CAK, the p34cdc2 activating kinase, contains a protein identical or closely related to p40MO15. EMBO J. 1993;12:3133–3142. doi: 10.1002/j.1460-2075.1993.tb05982.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tassan J P, Jaquenoud M, Fry A M, Frutiger S, Hughes G J, Nigg E A. In vitro assembly of a functional human CDK7-cyclin H complex requires MAT1, a novel 36 kDa RING finger protein. EMBO J. 1995;14:5608–5617. doi: 10.1002/j.1460-2075.1995.tb00248.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tassan J P, Schultz S J, Bartek J, Nigg E A. Cell cycle analysis of the activity, subcellular localization, and subunit composition of human CAK (CDK-activating kinase), J. Cell Biol. 1994;127:467–478. doi: 10.1083/jcb.127.2.467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wang Z, Svejstrup J Q, Feaver W J, Wu X, Kornberg R D, Friedberg E C. Transcription factor b (TFIIH) is required during nucleotide-excision repair in yeast. Nature. 1994;368:74–76. doi: 10.1038/368074a0. [DOI] [PubMed] [Google Scholar]
- 58.Weinberg R A. The retinoblastoma protein and cell cycle control. Cell. 1995;81:323–330. doi: 10.1016/0092-8674(95)90385-2. [DOI] [PubMed] [Google Scholar]
- 59.Wu L, Chen P, Hwang J J, Barsky L W, Weinberg K I, Jong A, Starnes V A. RNA antisense abrogation of MAT1 induces G1 phase arrest and triggers apoptosis in aortic smooth muscle cells. J Biol Chem. 1999;274:5564–5572. doi: 10.1074/jbc.274.9.5564. [DOI] [PubMed] [Google Scholar]
- 60.Wu L, Yee A, Liu L, Carbonaro-Hall D, Venkatesan N, Tolo V T, Hall F L. Molecular cloning of the human CAK1 gene encoding a cyclin-dependent kinase-activating kinase. Oncogene. 1994;9:2089–2096. [PubMed] [Google Scholar]
- 61.Wu L, Liu L, Yee A, Carbonaro-Hall D, Tolo V, Hall F. Molecular cloning of the human CYCG1 gene encoding a G-type cyclin: overexpression in human osteosarcoma cells. Oncol Rep. 1994;1:705–711. doi: 10.3892/or.1.4.705. [DOI] [PubMed] [Google Scholar]
- 62.Wu L, Williams R, Carbonaro-Hall D, Liu L, Tolo V, Hall F. Sequential and progressive cyclin expression in human osteosarcoma cells: diagnostic and therapeutic implications. Int J Oncol. 1993;3:859–867. doi: 10.3892/ijo.3.5.859. [DOI] [PubMed] [Google Scholar]
- 63.Xiong Y, Zhang H, Beach D. D type cyclins associate with multiple protein kinases and the DNA replication and repair factor PCNA. Cell. 1992;71:505–514. doi: 10.1016/0092-8674(92)90518-h. [DOI] [PubMed] [Google Scholar]
- 64.Yankulov K Y, Bentley D L. Regulation of CDK7 substrate specificity by MAT1 and TFIIH. EMBO J. 1997;16:1638–1646. doi: 10.1093/emboj/16.7.1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yee A, Nichols M A, Wu L, Hall F L, Kobayashi R, Xiong Y. Molecular cloning of CDK7-associated human MAT1, a cyclin-dependent kinase-activating kinase (CAK) assembly factor. Cancer Res. 1995;55:6058–6062. [PubMed] [Google Scholar]
- 66.Yee A, Wu L, Liu L, Kobayashi R, Xiong Y, Hall F L. Biochemical characterization of the human cyclin-dependent protein kinase activating kinase. Identification of p35 as a novel regulatory subunit. J Biol Chem. 1996;271:471–477. doi: 10.1074/jbc.271.1.471. [DOI] [PubMed] [Google Scholar]
- 67.Zamanian M, La Thangue N B. Adenovirus E1a prevents the retinoblastoma gene product from repressing the activity of a cellular transcription factor. EMBO J. 1992;11:2603–2610. doi: 10.1002/j.1460-2075.1992.tb05325.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zhang H S, Gavin M, Dahiya A, Postigo A A, Ma D, Luo R X, Harbour J W, Dean D C. Exit from G1 and S phase of the cell cycle is regulated by repressor complexes containing HDAC-Rb-hSWI/SNF and Rb–hSWI/SNF. Cell. 2000;101:79–89. doi: 10.1016/S0092-8674(00)80625-X. [DOI] [PubMed] [Google Scholar]