Abstract
Two hundred and forty-four ustilaginomycetous yeast or yeast-like strains were isolated from the soil, skin of animals or humans and plant materials during the past 20 years. Among them, 203 strains represent 39 known species, whereas 41 strains represent several novel species based on the sequence analyses of the rDNA genes [18S rDNA, Internal Transcribed Spacer (ITS) regions, 26S rDNA D1/D2 domain] and three protein genes (RPB1, RPB2, and TEF1). In this study, one new order, one new family, four new genera, twenty new species, and two new combinations were proposed. They are Franziozymales ord. nov., Franziozymaceae fam. nov., Baueromyces gen. nov., Franziozyma gen. nov., Guomyces gen. nov., Yunzhangomyces gen. nov., Baueromyces planticola sp. nov., Franziozyma bambusicola sp. nov., Gjaerumia cyclobalanopsidis sp. nov., Gjaerumia pseudominor sp. nov., Jamesdicksonia aceris sp. nov., Jaminaea lantanae sp. nov., Kalmanozyma hebeiensis sp. nov., Langdonia ligulariae sp. nov., Meira hainanensis sp. nov., Meira pileae sp. nov., Meira plantarum sp. nov., Phragmotaenium parafulvescens sp. nov., Sporisorium cylindricum sp. nov., Sympodiomycopsis europaea sp. nov., Tilletiopsis lunata sp. nov., Tilletiopsis pinicola sp. nov., Yunzhangomyces clavatus sp. nov., Yunzhangomyces cylindricus sp. nov., Yunzhangomyces qinlingensis sp. nov., Yunzhangomyces orchidis sp. nov., Guomyces nicotianae comb. nov., and Yunzhangomces scirpi comb. nov.
Keywords: Ustilaginomycotina, yeasts, smuts, 28 new taxa, molecular phylogeny
Introduction
The subphylum Ustilaginomycotina (Basidiomycota, Fungi) comprises a variety of lifestyles. The majority of species are biotrophic pathogens known as smuts, whereas some anamorphic yeast lineages are saprotrophs or, possibly, mycoparasites (Begerow et al., 2000, 2014, 2017; Bauer et al., 2001; Wang et al., 2014, 2015). The asexual yeast species were placed in nine genera, namely, Acaromyces (Boekhout et al., 2003), Farysizyma (Inácio et al., 2008), Jaminaea (Sipiczki and Kajdacsi, 2009), Malassezia (Boekhout et al., 2003), Meira (Boekhout et al., 2003), Moniliella (Stolk and Dakin, 1966), Pseudozyma (Boekhout, 1995), Sympodiomycopsis (Sugiyama et al., 1991), and Tilletiopsis (Gokhale, 1972; Boekhout, 1991; Boekhout et al., 1995), and reclassified in four classes in the Ustilaginomycotina, namely, Exobasidiomycetes, Malasseziomycetes, Moniliellomycetes, and Ustilaginomycetes (Bauer et al., 2001; Hibbett et al., 2007; Begerow et al., 2014; Wang et al., 2014). Most of these yeast genera are monophyletic, however, the genera Pseudozyma and Tilletiopsis were polyphyletic (Begerow et al., 2000, 2006, 2014; Fell et al., 2000; Stoll et al., 2003, 2005; Boekhout et al., 2011; McTaggart et al., 2012a,b). Recently, those two anamorphic polyphyletic genera were revised by Wang et al. (2015) based on multigene phylogenetic analyses. Except Pseudozyma alboarmeniaca pro tem., P. thailandica pro tem., P. tsukubaensis pro tem., P. hubeiensis pro tem., and P. pruni pro tem., the other species of Pseudozyma were transferred to Ustilago, Moesziomyces, Triodiomyces, Sporisorium, Langdonia, Kalmanozyma, and Dirkmeia, and the genus Pseudozyma was treated as a synonym of Ustilago (Wang et al., 2015). Three species were kept in the emended genus, Tilletiopsis, while the others were transferred to the genera Phragmotaenium, Gjaerumia, Robbauera, and Golubevia. Rhodotorula bacarum was treated as a synonym of Microstroma album, while Rhodotorula hinnulea and Rhodotorula phylloplana were synonymized and transferred to Microstroma as one single species Microstroma phylloplanum (Wang et al., 2015). Then, it was transferred to a newly described anamorphic genus Pseudomicrostroma by Kijpornyongpan and Aime (2017). All species in Farysizyma have been transferred to the teleomorphic genus, Farysia (Wang et al., 2015).
Benefiting from the molecular phylogenetic analyses, a large number of asexual fungi, especially at genus and higher ranks in Ustilaginomycotina were discovered (Begerow et al., 2014). Recently, three monotypic asexual genera in Ustilaginomycotina were proposed (Nasr et al., 2014; Albu et al., 2015; Sun et al., 2018). The genus Fereydounia represents the first yeast species in Urocystidales (Nasr et al., 2014). Violaceomyces is a yeast-like fungus in Violaceomycetales (Albu et al., 2015) and Capitulocladosporium is a Cladosporium-like fungus, but phylogenetically related to Violaceomycetales and Uleiellales in Ustilaginomycetes (Sun et al., 2018). Six Tilletiopsis-like yeast novel species in Exobasidiomycetes were described based on the phylogenetic analyses of multi-loci and LSU rDNA by Richter et al. (2019).
Over the past 22 years, more than 1,500 basidiomycetous yeast strains isolated from the soil, skin of animals or humans, and plant materials have been identified by analyzing the D1/D2 domain of the ribosomal large subunit DNA (D1/D2) and the ITS sequences in the State Key Laboratory of Mycology, China. Most of them belonging to Agaricomycotina and Pucciniomycotina. In addition, eight new genera, three families, and two orders have been documented in the article published by Li et al. (2020). In this study, a similar approach undertaken by Li et al. (2020) was used to propose one new order, one new family, four new genera, twenty new species, and two new combinations in the Ustilaginomycotina.
Materials and Methods
Strain Sampling and Phenotype Analyses
The yeast or yeast-like strains studied are listed in Table 1. Strains were isolated from plant leaves by using the improved ballistoconidia-fall method proposed by Nakase and Takashima (1993) and from the soil, tree bark, and rotten wood by an enrichment method described by Li et al. (2020). Yeasts were isolated from the skin of humans and animals by using the following protocol. The samples from human faces and heads and the skin of animals were collected with sterile swabs. Swabs were gently rolled back-and-forth 2–4 times across the skin and were then streaked onto Leeming and Notman agar plates (Leeming and Notman, 1987). The phenotypic and biochemotaxonomic characters were examined according to the methods introduced by Kurtzman et al. (2011). The sexual test and the ballistoconidium-forming activity of all the new species were investigated as described by Li et al. (2020).
TABLE 1.
List of yeast or yeast-like species employed and GenBank numbers determined in this study.
| Species | Holotype | CBS collection | Laboratory strain |
18S + ITS + D1/D2 | RPB1 | RPB2 | EF1 | ||
| Ustilaginaceae (Ustilaginales, Ustilaginomycetes) | |||||||||
| Sporisorium cylindricum sp. nov. | CGMCC 2.3756 | CBS 15755 | WZS28.2B | Wuzhishan Montain, Hainan province, China, Octomber 2007 | Leaf of unidentified plant | MN901699 | MN901756 | MN901781 | MN901669 |
| CGMCC 2.3576 | YX3.6 | Kunming country, Yunnan province, China, May 2007 | leaf of unidentified plant | MN901698 | / | / | MN901666 | ||
| Kalmanozyma hebeiensis sp. nov. | CGMCC 2.3457 | CBS 15483 | H8A4 | Hebei province, China, April 2007 | leaf of unidentified plant | MN901700 | / | MN901775 | MN901662 |
| Langdonia walkerae | CGMCC 2.4680 | CBS 144911 | JX1243 | China, Sptember 2012 | leaf of unidentified plant | MN901702 | / | MN901787 | MN901675 |
| Langdonia ligulariae sp. nov. | CGMCC 2.6313 | CBS 15581 | XZ146B3 | Lulang county, Tibet, China, Sptember 2014 | leaf of Ligularia tsangchanensis | MN901697 | MN901766 | / | MN901684 |
| Brachybasidiaceae (Exobasidiales, Exobasidiomycetes) | |||||||||
| Meira plantarum sp. nov. | CGMCC 2.4430 | CBS 12491 | FJYZ8-3 | Fuzhou county, Fujian province, China, August 2011 | Leaf of unidentified plant | MN901704 | MN901757 | MN901782 | MN901670 |
| CGMCC 2.4432 | FJYZ11-7 | Fuzhou county, Fujian province, China, August 2011 | Leaf of unidentified plant | MT896142 | / | / | / | ||
| CGMCC 2.4431 | FJYZ5-8 | Fuzhou county, Fujian province, China, August 2011 | Leaf of unidentified plant | MT896141 | / | / | / | ||
| CGMCC 2.6306 | CBS 144914 | XZ123E33 | Beibengxiang, Motuo county, Tibet, China, Sptember 2014 | Leaf of unidentified plant | MT896139 | MN901764 | / | MN901682 | |
| Meira pileae sp. nov. | CGMCC 2.6305 | CBS 144915 | XZ123B4 | Beibengxiang, Motuo county, Tibet, China, Sptember 2014 | Leaf of Pilea sp. | MT896138 | / | MN901791 | MN901681 |
| Meira hainanensis sp. nov. | CGMCC 2.3537 | CBS 15497 | WZS12.12 | Wuzhishan Montain, Hainan province, China, May 2007 | Leaf of unidentified plant | MN901703 | MN901753 | MN901777 | MN901664 |
| Yunzhangomyces orchidis sp. nov. | CGMCC 2.3451 | CBS 15753 | WZS24.28 | Wuzhishan Montain, Hainan province, China, Apir 2007 | Leaf of Orchidaceae | MN901726 | / | / | MN901661 |
| Yunzhangomyces clavatus sp. nov. | CGMCC 2.4433 | CBS 144908 | FJYZ8-4 | Fuzhou country, Fujian province, China, August 2011 | Leaf of unidentified plant | MN901724 | MN901758 | MN901783 | MN901671 |
| CBS 144917 | XZ128D | Heilongxiang, Motuo county, Tibet, China, Sptember 2014 | Leaf of Impatiens sp. | MN901725 | MN901765 | MN901792 | MN901683 | ||
| Yunzhangomyces qinlingensis sp. nov. | CGMCC 2.4533 | CBS 144910 | ZHH5D15 | Qinling, Heihe, Shaanxi province, China March 2012 | Leaf of unidentified plant | MN901729 | MN901759 | MN901785 | MN901673 |
| Yunzhangomyces cylindricus sp. nov. | CGMCC 2.6304 | CBS 15585 | HLJ17B1 | Daliangzi river national forest park, Heilongjiang province, China, August 2015 | Leaf of unidentified plant | MN901728 | / | MN901790 | MN901680 |
| Yunzhangomyces sp. | HLJ9.21 | Daliangzi river national forest park, Heilongjiang province, China, August 2015 | Leaf of unidentified plant | MN901727 | / | / | / | ||
| Georgefischeriales (Exobasidiomycetes) | |||||||||
| Phragmotaenium parafulvescens sp. nov. | CGMCC 2.3573 | CBS 15754 | SY9.2 | Sanya country, Hainan province, China, May 2007 | Leaf of unidentified plant | MN901716 | MN901754 | MN901778 | MN901665 |
| Gjaerumia pseudominor sp. nov. | CGMCC 2.5616 | CBS 144912 | TY1AS | Heilongjiang province, China, August 2015 | Leaf of unidentified plant | MN901705 | MN901761 | MN901789 | MN901677 |
| Gjaerumia cyclobalanopsidis sp. nov. | CGMCC 2.6419 | CBS 144918 | GT31C4 | Gutianshan, Zhejiang province, China, June 2011 | Leaf of Cyclobalanopsis sp. | MT896140 | / | / | / |
| Jamesdicksonia aceris sp. nov. | CGMCC 2.5679 | CBS 144913 | HLJ11A4A | Heilongjiang province, China, August 2015 | Leaf of unidentified plant | MN901731 | / | MN901796 | MN901687 |
| CGMCC 2.2370 | CBS 144907 | CB297 | Changbai mountain, Jilin province, October 1998 | Leaf of unidentified plant | MN901732 | / | MN901795 | MN901686 | |
| CGMCC 2.5602 | CBS 144916 | XZ155C4 | Bomi, Tibet, China, Sptember 2014 | Leaf of Acer pectinatum | MN901734 | / | MN901793 | MN901685 | |
| XZ156C4 | Bomi, Tibet, China, Sptember 2014 | Leaf of unidentified plant | MN901735/MN901736 | / | / | / | |||
| Entylomatales (Exobasidiomycetes) | |||||||||
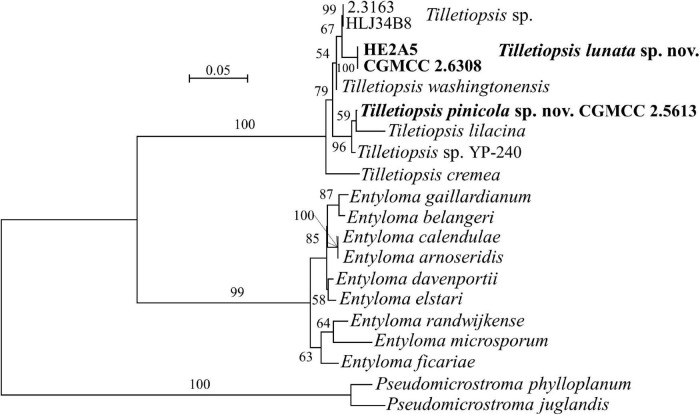
| Tilletiopsis pinicola sp. nov. | CGMCC 2.5613 | CBS 15775 | CBS 15775 | Heilongjiang province, China, August 2015 | A leaf of Pinus sp. | MN901708 | MN901760 | MN901788 | MN901676 |
| Tilletiopsis lunata sp. nov. | CGMCC 2.6308 | DSM 111865 | HE6AB1 | Huzhong, Heilongjiang Province, China, August 2015 | Leaf of unidentified plant | MN901706 | MN901763 | / | MN901679 |
| CGMCC 2.6307 | DSM 111865 | HE2A5 | Huzhong, Heilongjiang Province, China, August 2015 | Leaf of unidentified plant | MN901707 | MN901762 | / | MN901678 | |
| Microstromatales (Exobasidiomycetes) | |||||||||
| Jaminaea lantanae sp. nov. | CGMCC 2.3529 | HE2A5 | HK13.4 | Haikou country, Hainan province, China, May 2007 | Leaf of Lantana camara | MN901709 | / | MN901776 | MN901663 |
| CGMCC 2.3622 | HK13.4-2 | Haikou country, Hainan province, China, May 2007 | Leaf of Lantana camara | MN901710 | / | MN901779 | MN901667 | ||
| Sympodiomycopsis europaea sp. nov. | CGMCC 2.3119 | CBS 15470 | G1.1 | Germany, March 2006 | Leaf of unidentified plant, | MN901717 | MN901748 | MN901771 | MN901657 |
| CGMCC 2.3181 | G7.21 | Germany, March 2006 | Leaf of unidentified plant, | MN901718 | MN901752 | MN901774 | MN901660 | ||
| CGMCC 2.3120 | G4.3 | Germany, March 2006 | Leaf of unidentified plant, | MN901719 | / | / | / | ||
| CGMCC 2.3121 | G7.1-3 | Germany, March 2006 | Leaf of unidentified plant, | MN901720 | MN901749 | MN901772 | MN901658 | ||
| CGMCC 2.3122 | G7.2-2 | Germany, March 2006 | Leaf of unidentified plant, | MN901721 | / | / | / | ||
| CGMCC 2.3123 | G11.2 | Germany, March 2006 | Leaf of unidentified plant, | MN901722 | / | / | / | ||
| CGMCC 2.3124 | G16 | Germany, March 2006 | Leaf of unidentified plant, | MN901723 | / | / | / | ||
| Baueromyces planticola sp. nov. | CGMCC 2.4532 | CBS 144909 | XS9B4 | Xingshan country, Hubei province, China, March 2012 | Leaf of unidentified plant | MN901712 | / | MN901784 | MN901672 |
| CGMCC 2.4534 | GZMT1C2 | Maotai county, Guizhou province | Leaf of unidentified plant | MN901713 | / | MN901786 | MN901674 | ||
| CGMCC 2.4535 | FJS8A1 | Fanjingshan, Guizhou province | Leaf of unidentified plant | MN901714 | / | / | / | ||
| CGMCC 2.4536 | FJS8A1B | Fanjingshan, Guizhou province | Leaf of unidentified plant | MN901715 | / | / | / | ||
| Franziozymales (Exobasidiomycetes) | |||||||||
| Franziozyma bambusicola sp. nov. | CGMCC 2.2620 | CBS 15774 | XZ4C4 | Bomi county, Tibet, China, Sptember 2004 | A leaf of bamboo | MK415411 | MK415413 | MK415414 | MK415412 |
| XZ4A1 | Bomi county, Tibet, China, Sptember 2004 | A leaf of bamboo | MZ045837 | / | / | / | |||
PCR and DNA Sequencing
Deoxyribonucleic acid (DNA) was extracted following the method proposed by Wang and Bai (2008). The 18S (SSU) rDNA sequences were amplified according to Wang et al. (2003). The ITS (including the 5.8S rDNA) and 26S (LSU) rDNA D1/D2 regions were sequenced using the methods described previously (Wang and Bai, 2004). Amplification reactions and sequencing of the three protein genes, namely, two RNA polymerase II subunits (RPB1 and RPB2) and the translation elongation factor 1-α (TEF1), were performed as described in Wang et al. (2014). GenBank sequence accession numbers determined during this study are listed in Table 1.
Molecular Phylogenetic Analyses
Sequence alignments were performed with the MAFFT algorithm (Katoh and Standley, 2013) using the G-INS-i algorithm. The model GTR + I + G, the best nucleotide substitution model determined in MEGA 7.0 (Kumar et al., 2016), was selected for Bayesian inference (BI) and Maximum likelihood (ML) analyses. BI analysis was carried using MrBayes 3.1.2 (Ronquist et al., 2012) with the parameter settings proposed by Wang et al. (2015). ML phylogenetic reconstruction was performed using RAxML-HPC 7.2.8 (Stamatakis, 2006) with 500 bootstrap replicates. A Bayesian posterior probability (PP) of ≥0.9 or a bootstrap percentage (BP) of ≥70% was set as significantly supported in the constructed trees. The new alignments and trees in this study were deposited in TreeBASE (Nos. S28175).
Results and Discussion
Diversity and Ecology
Two hundred and forty-four ustilaginomycetous yeast or yeast-like strains isolated from soil (20%, 49/244), the skin of animals or humans (11%, 27/244), and plant materials (69%, 168/244), including leaves, tree bark, and rotten wood, were identified as 39 known species distributed in 15 genera, i.e., Entyloma, Exobasidium, Gjaerumia, Golubevia, Langdonia, Meira, Moesziomyces, Mycosarcoma, Phragmotaenium, Pseudozyma pro. tem, Quambalaria, Robbauera, Sporisorium, Tilletiopsis, and Ustilago, and 20 undescribed species (Table 1 and Supplementary Table 1) based on the ITS and D1/D2 sequence analyses.
Among 39 known yeast or yeast-like species isolated from the environment in this study, nine species were frequently isolated, while the other 29 species seem to be rare (Supplementary Table 1). Eighty strains of Tilletiopsis washingtonensis were obtained from eight provinces in China, which occupy 32.8% isolate frequency (80 strains/244 total isolated strains). The other frequently isolated species are Mycosarcoma maydis (Ustilago maydis) (9.4%), Pseudozyma hubeiensis pro. tem (6.6%), Moesziomyces aphidis (6.1%), Golubevia pallescens (5.7%), Phragmotaenium oryzicola (5.3%), Moesziomyces antarcticus (4.1%), Gjaerumia minor (3.7%), and Meira geulakonigii (2.9%) (Supplementary Table 1). T. washingtonensis commonly occurred on leaves in agreement with the observation of Boekhout (1991, 2011). However, it was also isolated from the soil, rotten wood, and tree bark (Supplementary Table 1). Although My. maydis is an important plant pathogen on corn, it was not isolated from other plant materials and soils, but from the skin of cows, sheep (lambs), and shepherds (Supplementary Table 1). The cases of human My. maydis infection have been reported (Patel et al., 1995; Teo and Tay, 2006; McNeil and Palazzi, 2012; Peraica et al., 2014; Agata and Marta, 2018). The research from Crotzer and Levetin (1996) indicated that the dispersal of smut spores was intervened by human activity, especially by plant harvesting. The smut spores seem to be transferred from plants to humans or animals by air currents (Crotzer and Levetin, 1996). Mo. aphidis was isolated both from plants (leaves), animals (cows), and in the soil. Ex. reticulatum, Go. pallescens, Ph. oryzicola, Ps. fusiformata, Ps. tsukubaensis, and T. lilacina were also isolated from the soil and from plant materials (Supplementary Table 1).
The below analyses illustrate the undescribed diversity of yeasts in Ustilaginomycotina, most of which represent rare taxa. A few not included potentially conspecific strains were not available for the study, inactive, or lost. These descriptions were made on a limited number of isolates because more strains could not be obtained despite of extensive sampling and analysis of more than 200 isolates.
The two most frequently used for identification of yeast genetic markers, ribosomal ITS, and D1/D2 domains of LSU proved their utility for identification and delimitation of species in Ustilaginomycotina. While nucleotide sequences of D1/D2 domains are often too conservative to distinguish closely related species, this region is useful for phylogenetic analyses. In contrast, the variability of ITS is often sufficient to identify new species from pair-wise similarity comparisons (see below).
New Taxon Delineation and Phylogenetic Placement
The Ustilaginomycotina includes mainly parasitic fungi and few of saprobic yeast or yeast-like members (Begerow et al., 2014). Traditionally, the phenotypic and ecological species concept with species identification based on the combination of host plants and morphological characteristics was applied for the plant pathogenetic fungi (Vánky, 2012; Begerow et al., 2014; Boekhout et al., 2021), but the integrative species concept with the incorporation of phenotypic and ecological characteristics and molecular data (e.g., rDNA and protein genes) have also been used (Stoll et al., 2003, 2005; Begerow et al., 2006, 2014; Sipiczki and Kajdacsi, 2009; McTaggart et al., 2012a,b; Kijpornyongpan and Aime, 2017; Richter et al., 2019; Boekhout et al., 2021). For the yeast or yeast-like fungi, the molecular data, combined with morphological and physiological characters, was mainly used to identify species and diagnose genus (Kurtzman et al., 2011; Begerow et al., 2014, 2017; Richter et al., 2019; Boekhout et al., 2021). Nearly 100 anamorphic yeast or yeast-like species in Ustilaginomycotina have been reported (Boekhout et al., 2003; Kurtzman et al., 2011; Begerow et al., 2014; Nasr et al., 2014; Albu et al., 2015; Wang et al., 2015; Sun et al., 2018; Richter et al., 2019), but only few of them have been connected to the sexual taxa or sexual stage, such as Pseudozyma prolifica (teleomorph My. maydis), Pseudozyma tsukubaensis (teleomorph Macalpinomyces spermophorus), Mo. aphidis, Mo. antarcticus, and Mo. rugulosus (Wang et al., 2015; Kruse et al., 2017; Tanaka and Honda, 2017; Li et al., 2019; Tanaka et al., 2019). However, most of them were dispersed in separated phylogenetic clades from the teleomorphic genera (Wang et al., 2015). Therefore, the phylogenetic and phenotypic species concept for anamorphic genera was proposed in this study. We tried to compare phylogenetic distances from available data for sexual species and undertaken them as a reference on newly described species in the genera, including both sexual and asexual species.Thus, the phylogenetic species concept was used here for the new asexual species delimitation in the teleomorphic genera because those yeast members have no the host data and sexual stage. This concept also applied in smut and yeasts communities (Kijpornyongpan and Aime, 2017; Richter et al., 2019). The yeast species identification benchmarks suggested by Fell et al. (2000), Scorzetti et al. (2002), Kurtzman and Fell (2006), Kurtzman (2014, 2015), Kurtzman et al. (2015), Vu et al. (2016, 2019), and Li et al. (2020) were also considered, but not followed strictly in this study.
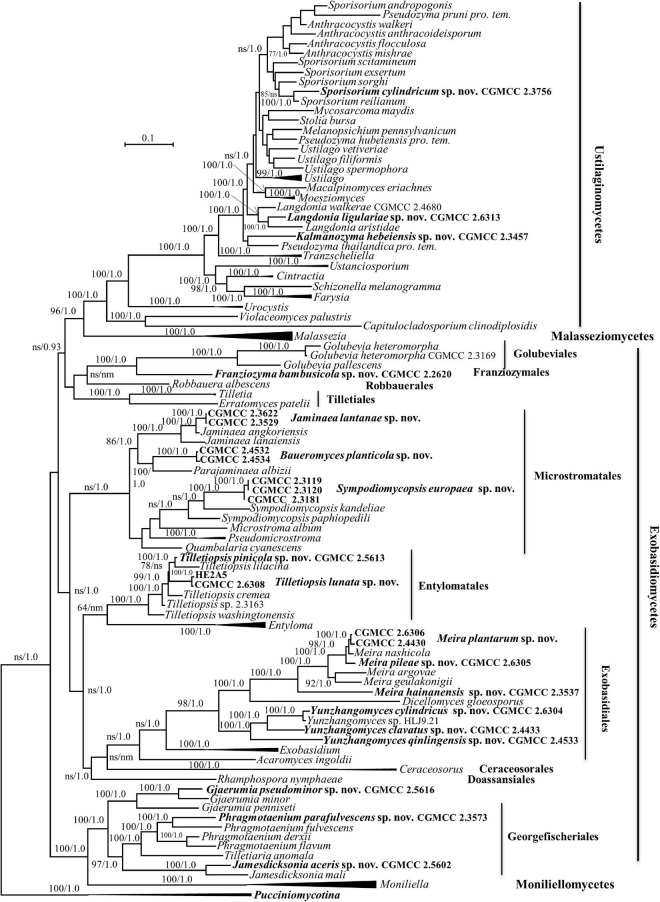
Forty-one strains (Table 1) were grouped into 20 novel species based on the phylogenetic and physiological comparison. Thirty of these strains represent 14 new species that are distributed in the genera Gjaerumia, Jamesdicksonia, Jaminaea, Kalmanozyma, Langdonia, Meira, Phragmotaenium, Sporisorium, Sympodiomycopsis, and Tilletiopsis. However, the 11 additional strains, representing six unknown taxa, occur in three unique phylogenetic positions in the phylogenetic trees (Figures 1–6 and Supplementary Figures 1–3) and cannot be assigned to any existing genera. Therefore, three new genera, namely, Baueromyces gen. nov., Franziozyma gen. nov., and Yunzhangomyces gen. nov, are proposed to accommodate these six novel species (see Taxonomy section).
FIGURE 1.
Phylogenetic tree inferred using the combined sequences of SSU rDNA, LSU rDNA D1/D2 domains, Internal Transcribed Spacer (ITS; including 5.8S rDNA), RPB1, RPB2, and TEF1, depicting the phylogenetic positions of new taxa (in bold) within Ustilaginomycotina. The tree backbone was constructed using maximum likelihood analysis. Bootstrap percentages of maximum likelihood analysis over 50% from 1,000 bootstrap replicates and posterior probabilities of Bayesian inference above 0.9 are shown respectively from left to right on the deep and major branches. Bar = 0.1 substitutions per nucleotide position. ns, not supported (BP < 50% or PP < 0.9); nm, not monophyletic; the compressed genera are monophyletic, the species in those clades were listed in Table 1 and Supplementary Table S2.
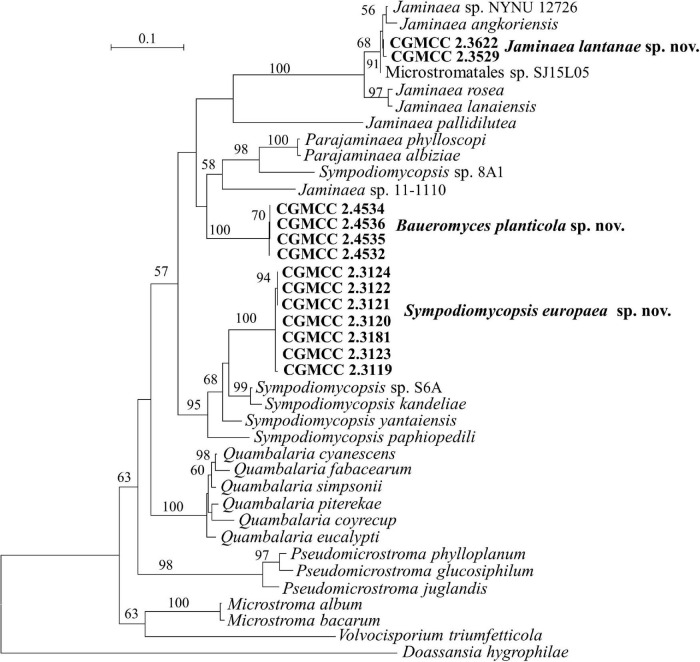
FIGURE 6.
Phylogeny of new yeast or yeast-like species in the Microstromatales inferred from the sequences of the LSU rDNA D1/D2 domains and ITS region (including 5.8S rDNA) by maximum likelihood analysis and over 50% from 1,000 bootstrap replicates is shown. Bar = 0.1 substitutions per nucleotide position.
Note that the ex-type strains (or reference strains) of known species were used for sequence similarity analyses for novel species comparisons, and that the GenBank and strain numbers can be found in Supplementary Table 2.
New Species Identification in the Ustilaginaceae (Ustilaginales, Ustilaginomycetes)
Five strains, CGMCC 2.3576, CGMCC 2.3756, CGMCC 2.3457, CGMCC 2.4680, and CGMCC 2.6313, belong to the family Ustilaginaceae (Table 1 and Figures 1, 2).
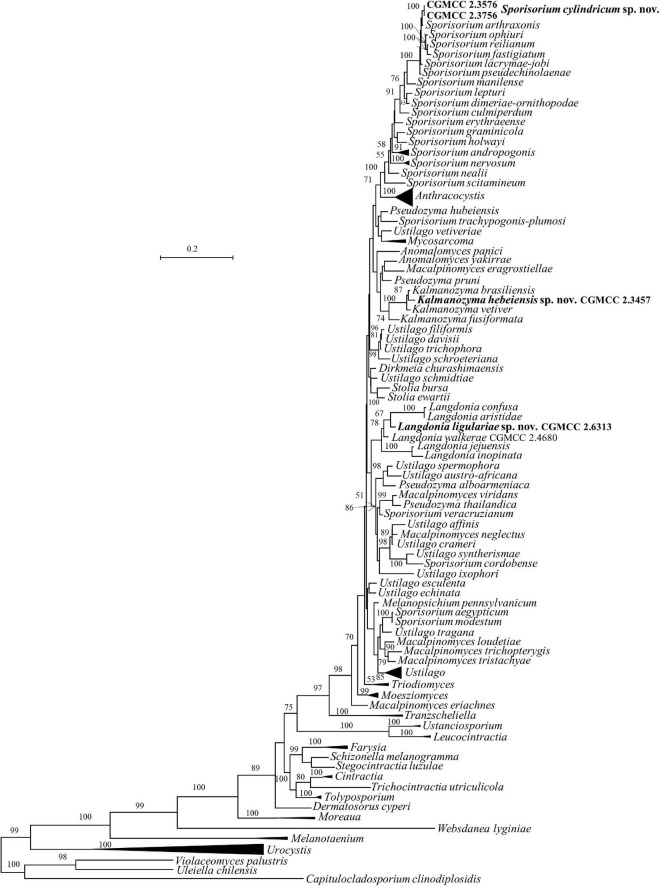
FIGURE 2.
Phylogeny of new yeast or yeast-like species in the Ustilaginomycetes inferred from the sequences of the LSU rDNA D1/D2 domains and ITS region (including 5.8S rDNA) by maximum likelihood analysis and over 50% from 1000 bootstrap replicates is shown. Bar = 0.2 substitutions per nucleotide position. the compressed genera are monophyletic, the species in those clades were listed in Table 1 and Supplementary Table S2.
As taxa within other smut genera, the species concept of Sporisorium was traditionally delimited based on the host and morphological characters (Vánky, 2012; Begerow et al., 2014). McTaggart et al. (2012a,b) revised the generic concept of Sporisorium based on multi-gene phylogenetic analysis, the morphological characters, and host specificity. In their study, 81 species of Sporisorium were used to perform the phylogenetic analysis and 34 species were kept in the revised Sporisorium (McTaggart et al., 2012a,b), while other members were transferred to Anthracocystis, Langdonia, Stollia, etc. The asexual species Pseudozyma graminicola was transferred to Sporisorium based on the phylogenetic analysis (Wang et al., 2015). Our two new isolates, CGMCC 2.3576 and CGMCC 2.3756, were phylogenetically related to S. arthraxonis, S. ophiuri, S. fastigiatum, S. reilianum, S. lacrymae-jobi, and S. pseudechinolaenae and separated from them in the ITS+LSU and multi-gene trees (Figures 1, 2). The six parasitic species differ from each other by 1–6 nt (0.16–1%) and 6–29 nt (0.9–3.9%) in the D1/D2 and the ITS regions, respectively. Strains CGMCC 2.3576 and CGMCC 2.3756 with identical D1/D2 and ITS sequences differ from those six species by more than 22 nucleotide (nt) (3%) mismatches (including substitutions and deletions) and 3–7 nt (0.5–1.1%) in the ITS and the D1/D2 regions, respectively.
The genus Langdonia includes ten species, namely, L. aristidae, L. aristidaria, L. aristidicola, L. clandestina, L. confusa, L. fraseriana, L. goniospora, L. inopinata, L. jejuensis, and L. walkerae (Begerow et al., 2014; Wang et al., 2015; Alqurashi et al., 2021), among which, nine species have a sexual stage except L. jejuensis. Seven of them have rDNA sequences (Supplementary Table 2) and differ from each other by 1–22 nt (0.16–3.6%) and 7–82 nt (1–12%) in the D1/D2 and ITS regions, respectively. Strain CGMCC 2.6313 are placed in the Langdonia clade and phylogenetically distinct from other known species (Figures 1, 3). The newly published species Langdonia walkerae was described based on sexual characters and molecular data with ten specimens collected from Aristida stricta and Aristida beyrichiana (Poaceae) in the southeastern United States in 2018 (Alqurashi et al., 2021). CGMCC 2.4680, with the asexual yeast stage, was isolated from the leaf of an unidentified plant in China in September 2012. It has two nt differences from L. walkerae in the ITS region, which indicated that CGMCC 2.4680 belong to L. walkerae (data not shown). This is the first case in the genus Langdonia for the connection between the sexual and asexual states. CGMCC 2.6313 has affinity with L. aristidae and L. confusa, and differs from them by more than 9 nt and 72–75 nt (10%) in the D1/D2 and ITS regions, respectively.
FIGURE 3.
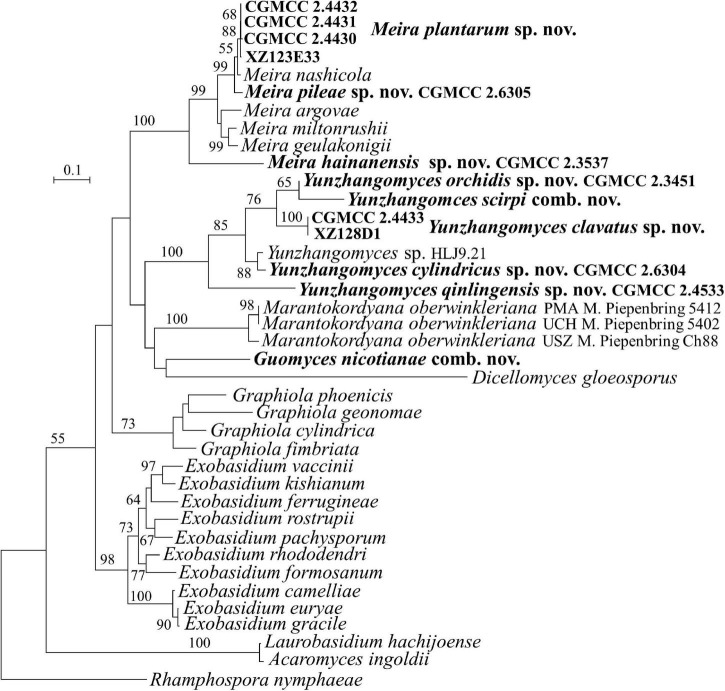
Phylogeny of new yeast or yeast-like species in the Exobasidiales inferred from the sequences of the LSU rDNA D1/D2 domains and ITS region (including 5.8S rDNA) by maximum likelihood analysis and over 50% from 1,000 bootstrap replicates is shown. Bar = 0.1 substitutions per nucleotide position.
Strain CGMCC 2.3457 locates in the anamorphic genus Kalmanozyma clade (Figures 1, 2). It differs from K. brasiliensis, K. vetiver, and K. fusiformata by 5–7 nt in the D1/D2 domain and by 23–71 nt (3–8%) in the ITS region.
New Taxon Identification in the Brachybasidiaceae (Exobasidiales, Exobasidiomycetes)
The family Brachybasidiaceae contains Brachybasidium, Dicellomyces, Exobasidiellum, Kordyana, Meira, and Proliferobasidium (Begerow et al., 2014). Eleven strains representing seven new species are placed in Brachybasidiaceae (Table 1, Figures 1, 3 and Supplementary Figures 1, 3). The anamorphic genus Meira comprises six species, namely, Me. argovae, Me. geulakonigii, Me. miltonrushii, Me. nicotianae, Me. nashicola, and Me. siamensis (Yasuda et al., 2006; Tanaka et al., 2008; Rush and Aime, 2013; Limtong et al., 2017; Cao et al., 2018). Strains CGMCC 2.4430, CGMCC 2.4431, CGMCC 2.4432, CGMCC 2.6305, CGMCC 2.6306, and CGMCC 2.3537 are clustered in the Meira clade (Figures 1, 3 and Supplementary Figures 1, 3). The former five strains form two groups represented by strains CGMCC 2.4430 and CGMCC 2.6305, respectively, and are closely related to Meira nashicola (Figures 1, 3). The CGMCC 2.4430 group, including four strains, has identical D1/D2 sequences and differ from one another by 0–7 nt (0–0.9%) in the ITS region, which indicates that they are conspecific. The CGMCC 2.6305 group, represented by a single strain, differs from the CGMCC 2.4430 group by 3 nt in the D1/D2 domain and 42 nt (5.4%) in the ITS region. These two groups differ from Me. nashicola by 1–7 nt in the D1/D2 domain, and by 24–27 nt (4–5%) in the ITS region, indicating that they are different species. Strain CGMCC 2.3537 was located in a basal branch in the Meira clade, and differs from Meira sp. 07F1061 (JX575187) and Meira sp. 08F0291 (JX575186) by 6–7 nt and from other know Meira species by more than 51 nt (8%) in the D1/D2 domain.
Meira nicotianae was described by Cao et al. (2018), which occurred at a basal branch of the Meira clade in the LSU and ITS+LSU trees. However, Piepenbring et al. (2020) argued that the genus Meira was polyphyletic and that Me. nicotianae was separated from the Meira clade and was more closely related to Dicellomyces scirpi in the ITS + LSU tree. Our analyses (Figure 3 and Supplementary Figure 1) agreed with the result of Piepenbring et al. (2020). The conflicting placement of Me. nicotianae might be caused by incomplete taxon sampling. In the analyses of Piepenbring et al. (2020) and our study, more taxa, particularly in D. scirpi, were added for the phylogenetic tree construction, which resulted in the separation of Me. nicotianae from the genus Meira. Therefore, a new genus is proposed for Me. nicotianae in the following taxonomy section.
The genus Dicellomyces includes four species that parasitize on monocot plants (Begerow et al., 2014), D. gloeosporus, the type species, on Poaceae, D. calami on Arecaceae, D. scirpi on Cyperaceae, and D. bombacis on Bombacaceae. The latter species has been transferred to the genus Ceraceosorus (Ceraceosoraceae, Ceraceosorales) as C. bombacis (Cunningham et al., 1976). Piepenbring et al. (2020) indicated that D. scirpi probably represented a new genus based on the ITS + LSU sequence analysis and the comparison of host and morphological characters, including the shape of the sori, the presence of paraphyses, probasidial swellings, and the shape of conidia formed by germinating basidiospores. The molecular analyses from Piepenbring et al. (2020) showed that D. scirpi and D. gloeosporus were located in different phylogenetic clades, which was in agreement with Nasr et al. (2019) but differs from Tanaka et al. (2008). The phylogenetic incongruence might be caused by insufficient taxon sampling in the study of Tanaka et al. (2008).
Strains CGMCC 2.3451, CGMCC 2.4433, CGMCC 2.4533, CGMCC 2.6304, and XZ128D1 represent four undescribed species (Figures 1, 3) which differ from each other by more than 3% in the D1/D2 domain. They are all closely related to D. scirpi but differ from it by 4.4–11.5% in the D1/D2 domain. Our combined ITS, D1/D2 and the combined six-gene phylogenetic analyses confirm that D. scirpi and the five new strains are different from D. gloeosporus (Figures 1, 3 and Supplementary Figures 1, 3). Therefore, the D. scirpi clade was proposed as a new genus in the Brachybasidiaceae family (see Taxonomy section).
New Species Identification in the Georgefischeriales (Exobasidiomycetes)
Strains CGMCC 2.3573, CGMCC 2.5616, CGMCC 2.6419, CGMCC 2.5679, CGMCC 2.2370, CGMCC 2.5602, and XZ156C4 are placed in Georgefischeriales (Figures 1, 4 and Supplementary Figures 1, 2). The genus, Phragmotaenium, includes four yeast species and one plant infecting taxon (Begerow et al., 2014; Wang et al., 2015). Strain CGMCC 2.3537 is placed in the Phragmotaenium clade and differs from Ph. indicum, Ph. oryzicola, Ph. derxii, Ph. fulvescens, and Ph. flavum by 6–21 nt (1–3%) in the D1/D2 domain. More than 6% diversity between those taxa was found in the ITS region.
FIGURE 4.
Phylogeny of new yeast or yeast-like species in the Georgefischeriales inferred from the sequences of the LSU rDNA D1/D2 domains and ITS region (including 5.8S rDNA) by maximum likelihood analysis and over 50% from 1,000 bootstrap replicates is shown. Bar = 0.1 substitutions per nucleotide position.
The genus Gjaerumia comprises three parasitic smut fungi infecting the Asparagaceae, Melanthiaceae, and Xanthorrhoeaceae plant and two yeast species (Begerow et al., 2014; Wang et al., 2015). Gjaerumia marneyi, isolated from the phylloplane of Hibiscus tiliaceus (Malvaceae), was recently described and known as an asexual culturable yeast based on phylogenetic analysis (Tan et al., 2021). Strains CGMCC 2.5616 isolated from a leaf of an unidentified plant and CGMCC 2.6419 isolated from the leaf of Cyclobalanopsis sp. (Fagaceae) belong to the Gjaerumia clade and differ from the closely related species G. minor by 15 (2.5%) and 74 nt (11%) in the D1/D2 and ITS regions, respectively. These two novel strains differ from each other by 8 nt in the D1/D2 domain and 72 nt in the ITS region, which indicates that they belong to different species.
Five species of Jamesdicksonia, i.e., J. dactylidis, J. ischaemiana, J. irregularis, and J. mali, have available D1/D2 sequence (Supplementary Table 2). They differ from each other by 3–11 nt in this region. Strains CGMCC 2.5679, CGMCC 2.2370, and XZ156C4, isolated from the leaf of an unidentified plant, and CGMCC 2.5602, isolated the leaf of Acer pectinatum (Sapindaceae), cluster with the Jamesdicksonia species and are closely related to the asexual species J. mali isolated from apple (Malus, Rosaceae) recently described by Richter et al. (2019; Figures 1, 4). The four new strains have 0–3 and 3–5 nt differences in the D1/D2 domain and ITS region, respectively, which indicate that they are conspecific. They differ from J. mali CBS 111628 and CBS 111625 by 0–1 nt in the D1/D2 domain. However, there are 27–33 nt (6–7%) differences in the ITS region. They also have more divergence in the assimilation of carbon and nitrogen (see “Taxonomy” section, Table 2). The above phylogenetic and physiological comparisons indicate that they belong to different species. The other species of Jamesdicksonia differ from the four new strains by 6–8 nt in the D1/D2 region. Jamesdicksonia brizae, without the D1/D2 sequence, differs from the four new strains by more than 53 nt (8%) in the ITS region.
TABLE 2.
Physiological and biochemical characteristics of new species and their closest relatives.
| Characteristic a | Fermentation of glucose | Glucose | Galactose | L-Sorbose | Sucrose | Maltose | Cellobiose | Trehalose | Lactose | Melibiose | Raffinose | Melezitose | Inulin | Solube starch | D-Xylose | L-Arabinose | D-Arabinose | D-Ribose | L-Rhamnose | D-Glucosamine | N-Acetyl-D-glucosmine | Methanol | Ethanol | Glycerol | Erythritol | Ribitol | Galactitol | D-Mannitol | D-Glucitol | α-Methyl-D-glucoside | Salicin | D-glueonale | DL-Lactic acid | Succinic acid | Citric acid | Inositol | Hexadecane | Ammonium sulfate | Potassium nitrate | Sodium nitrite | L-lysine | Ethylamine | Cadaverine | Growth in vitamin-free medium | Starch like compounds formation | Growth with 50% glucose | Diazonium Blue B reaction | Hydrolysis of urea | The major ubiquinone | Growth at 17°C | Growth at 25°C | Growth at 30°C | Growth at 37°C |
| Sporisorium cylindricum sp. nov. | - | + | + | – | + | + | + | + | – | + | + | + | – | + | + | – | – | + | + | – | – | – | – | – | – | – | – | – | – | + | + | n | – | + | – | + | – | + | – | – | + | – | – | + | – | – | + | + | n | + | + | n | n |
| Kalmanozyma hebeiensis sp. nov. | – | + | + | – | + | + | + | + | lw | – | w | + | – | – | + | + | w | w | – | + | n | – | + | + | – | + | – | + | + | + | w | n | – | + | + | – | – | + | + | + | w | lw | – | + | – | – | + | + | n | + | + | n | n |
| Kalmanozyma vetiver | – | + | – | l | + | + | w | + | – | – | + | + | – | + | + | – | – | – | – | n | n | – | – | – | – | – | – | l | + | w | w | – | + | + | + | l | n | + | + | + | + | – | w | + | – | + | + | + | 10 | + | + | + | – |
| Kalmanozyma brasiliensis | – | + | + | + | + | + | + | + | – | – | + | + | + | – | + | + | – | + | – | l | + | – | + | + | + | + | – | + | + | n | + | + | – | + | – | l | – | n | + | + | + | n | l | + | – | – | + | + | n | + | + | + | – |
| Langdonia walkerae | – | + | – | – | + | – | w | w | w | – | w | w | w | w | + | + | – | – | – | – | – | – | – | – | – | – | – | w | – | – | – | n | – | w | – | – | – | + | + | + | + | + | + | – | – | – | + | + | n | + | + | n | n |
| Langdonia ligulariae sp. nov. | – | + | – | – | + | + | + | + | – | – | + | + | w | w | + | + | – | – | – | – | – | – | + | + | – | + | – | + | – | + | – | n | – | – | – | w | – | + | + | w | + | + | + | + | – | – | + | + | n | + | + | n | n |
| Langdonia jejuensis | – | + | l | l | + | + | l | + | + | – | + | + | – | – | + | + | – | + | – | – | l | – | – | – | l | – | – | + | + | – | l | l | – | – | + | – | + | n | + | + | + | – | + | + | – | – | + | + | 10 | + | + | + | + |
| Meira plantarum sp. nov. | – | + | + | – | + | + | + | + | – | + | + | + | – | – | + | + | – | + | – | – | n | – | – | – | + | – | v | + | + | – | lw | n | – | +, lw | – | – | – | + | + | + | lw, – | + | l | + | – | – | + | + | n | + | + | n | n |
| Meira pileae sp. nov. | – | + | + | – | + | + | + | + | – | + | + | + | + | – | + | + | w | w | – | – | n | – | w | w | w | w | – | w | w | – | w | – | – | lw | – | – | – | + | + | + | + | + | + | + | – | – | + | + | n | + | + | n | n |
| Meira nashicola | – | + | + | – | + | + | – | w | + | + | + | – | – | w | – | – | – | – | – | – | – | + | + | + | + | + | – | + | + | n | + | + | + | n | |||||||||||||||||||
| Meira hainanensis sp. nov. | – | + | + | – | + | + | + | + | – | + | + | + | + | + | + | + | + | + | – | – | n | – | + | w | + | lw | + | + | + | – | – | n | – | w | – | – | – | + | w | + | – | – | + | + | – | – | + | + | n | + | + | n | n |
| Meira argovae | – | + | + | – | + | + | + | + | v | v | + | + | + | + | + | + | + | + | – | – | n | – | + | + | + | + | + | + | + | – | + | – | + | + | + | – | n | + | + | n | n | n | v | – | – | + | + | n | + | + | + | – | |
| Yunzhangomyces orchidis sp. nov. | – | + | + | + | + | + | + | + | – | l | + | + | + | – | + | + | + | + | – | – | n | – | + | + | + | + | – | + | + | – | + | n | – | lw | lw | – | – | + | + | + | + | + | + | + | – | – | + | + | n | + | + | n | n |
| Yunzhangomyces clavatus sp. nov. | – | + | v | +, lw | + | + | v | + | – | + | + | + | – | – | + | + | – | v | – | – | n | – | +, lw | – | v | v | – | + | + | – | lw, – | n | – | w, – | – | – | – | + | + | + | v | + | + | + | – | – | + | + | n | + | + | n | n |
| Yunzhangomyces qinlingensis sp. nov. | – | + | + | + | + | + | – | + | – | – | + | + | – | – | + | + | lw | + | – | – | – | – | + | – | + | lw | – | + | + | lw | l | n | – | + | + | – | – | + | + | – | lw | lw | lw | + | – | – | + | + | n | + | + | n | n |
| Yunzhangomyces cylindricus sp. nov. | – | + | + | + | + | + | + | + | – | – | + | – | + | – | + | + | + | + | – | – | – | – | + | – | + | + | – | + | + | – | – | n | – | – | – | – | – | + | + | – | + | + | + | + | – | – | + | + | n | + | + | n | n |
| Phragmotaenium parafulvescens sp. nov. | – | + | lw | lw | + | + | – | + | lw | lw | + | + | + | + | + | + | + | – | – | – | – | – | – | + | – | lw | – | + | + | – | – | + | – | + | – | – | – | + | + | – | lw | – | – | + | – | – | + | + | n | + | + | n | n |
| Phragmotaenium fulvescens | – | + | + | v | + | + | + | + | + | + | + | + | + | + | + | + | + | + | – | – | n | – | v | + | + | + | – | + | + | – | – | v | v | + | + | – | n | n | + | + | n | n | n | – | – | – | + | + | 10 | + | + | + | – |
| Gjaerumia pseudominor sp. nov. | – | + | – | – | + | + | + | + | – | – | – | + | – | + | + | + | – | – | – | – | – | – | – | + | – | – | – | + | + | – | – | n | + | + | + | – | – | + | + | – | + | + | + | w | – | – | + | + | n | + | + | n | n |
| Gjaerumia cyclobalanopsidis sp. nov. | – | + | + | w | + | + | + | + | + | + | + | + | + | – | + | + | + | – | – | – | – | – | – | + | – | w | – | w | w | – | – | n | – | – | – | – | – | + | w | + | w | – | lw | – | – | – | + | + | n | + | + | n | n |
| Gjaerumia minor | – | + | + | v | + | + | + | + | + | + | + | + | + | + | + | + | + | + | – | – | n | – | v | + | v | + | – | + | + | v | – | v | + | + | + | – | n | + | v | n | n | n | – | – | – | + | + | 10 | + | + | v | – | |
| Jamesdicksonia aceri sp. nov. | – | + | v | – | + | + | + | + | v | – | +, w | +, w | v | v | v | + | + | + | + | – | n | – | – | + | + | + | – | + | + | – | – | n | – | v | – | – | – | + | + | – | + | + | + | + | – | – | + | + | n | + | + | n | n |
| Jamesdicksonia mali | – | + | + | – | + | + | + | + | + | + | + | + | + | + | l | + | – | + | w, – | + | – | – | w | w | – | – | + | + | – | – | + | – | + | – | – | + | + | + | w, – | – | – | + | + | n | + | + | w, – | n | |||||
| Tilletiopsis pinicola sp. nov. | – | + | + | – | + | + | – | + | – | w | + | + | – | – | + | + | + | + | – | – | – | – | + | + | + | – | – | – | w | – | – | n | – | w | w | – | – | + | + | – | + | + | + | + | – | – | + | + | n | + | + | n | n |
| Tilletiopsis lilacina | – | + | + | – | + | + | + | + | – | + | + | + | – | + | + | + | + | + | – | – | n | – | – | + | + | + | – | + | + | + | – | + | + | + | + | – | n | + | + | n | n | n | – | – | – | + | + | 10 | + | + | v | – | |
| Tilletiopsis lunata sp. nov. | – | + | + | – | + | + | – | + | – | n | + | + | + | + | + | + | – | + | – | – | – | – | + | + | + | v | – | + | + | – | – | – | – | + | + | – | – | + | + | v | + | – | + | + | – | – | + | + | n | + | + | n | n |
| Tilletiopsis washingtonensis | – | + | v | – | + | + | v | + | – | v | + | + | – | + | + | + | + | + | – | – | n | – | v | + | + | v | – | + | + | v | V | + | + | + | + | – | n | n | + | + | n | n | n | – | – | – | + | + | 10 | + | + | v | – |
| Jaminaea lantanae sp. nov. | – | + | – | + | l | lw | lw | – | – | – | – | lw | – | – | l | + | – | – | – | l | + | – | + | + | + | – | – | l | lw | lw | lw | – | – | – | – | – | – | + | – | – | + | + | + | + | – | – | + | + | n | + | + | n | n |
| Baueromyces planticola sp. nov. | – | + | + | – | + | +, w | + | +, lw | w | v | + | v | v | – | w | + | – | v | v | – | v | – | w, – | + | +, lw | – | – | + | v | w, – | lw, – | n | – | w, – | – | lw, – | – | + | +, w | – | w | w | w | + | – | – | + | + | n | + | + | n | n |
| Sympodiomycopsis europaea sp. nov. | – | + | + | + | + | + | – | lw | + | + | + | + | – | +, w | + | +, lw | – | + | – | – | n | – | + | + | + | – | – | + | +, lw | +, lw | – | n | – | w, – | lw, – | w, – | – | + | v | – | + | – | + | + | – | – | + | + | n | + | + | n | n |
| Sympodiomycopsis kandeliae | – | + | +, l | + | + | + | +, l | + | +, l | v | + | + | w, – | w, – | +, l | + | +, l | +, l | – | – | – | +, l | + | + | v | w | + | + | + | w | l, w | – | +, w | w | w, – | +, w | v | +, w | +, w | l, w | + | – | + | + | + | n | + | + | + | n | |||
| Sympodiomycopsis paphiopedili | – | + | + | + | + | + | + | + | + | + | + | + | – | – | + | + | + | + | – | – | n | – | + | + | + | + | – | + | + | + | – | + | – | + | + | + | n | + | + | – | + | – | + | – | + | n | + | 10 | + | + | + | – | |
| Franziozyma bambusicola sp. nov. | – | + | – | – | + | + | + | + | – | – | + | – | + | + | – | – | – | – | – | – | – | – | + | + | – | – | + | + | – | – | n | – | – | – | – | – | w | – | – | + | + | n | + | – | – | – | |||||||
| – | |||||||||||||||||||||||||||||||||||||||||||||||||||||
a+, positive; –, negative; l, latent; w, weak; lw, latent and weak; v, variable; n, not available.
Note: Strain CGMCC 2.6419 differs from the two Japanese strains, NIP003 (AB726595) and NIP007 (AB726598), by 2–3 nt in the D1/D2 domain, which indicate that they may be conspecific.
New Species Identification in the Entylomatales (Exobasidiomycetes)
Three species, namely, Tilletiopsis cremea, T. lilacina, and T. washingtonensis, were included in the revised genus Tilletiopsis (Wang et al., 2015). Strains HE6AB1, HE2A5, and CGMCC 2.5613 are clustered in the Tilletiopsis clade (Figures 1, 5 and Supplementary Figures 1, 3). The former two strains have 2 nt differences in both the D1/D2 and ITS regions, indicating their conspecificity. These two strains differ from strain CGMCC 2.5613 by 12 nt in the D1/D2 domain and by 23–24 nt in the ITS region. Strains HE6AB1 and HE2A5 are closely related to T. washingtonensis and differ from it by 3 and 10–12 nt (∼2%) in the D1/D2 domain and ITS region, respectively. Strain CGMCC 2.5613 has identical D1/D2 sequences with Tilletiopsis lilacina. However, they differ from each other by 11 nt (∼2%) in the ITS region. Physiological profiles of HE6AB1, HE2A5, and CGMCC 2.5613 differed from their closely related species T. washingtonensis and T. lilacina (see “Taxonomy” section, Table 2), and the phylogenetic analysis both indicated that they belong to two new species in Tilletiopsis.
FIGURE 5.
Phylogeny of new yeast or yeast-like species in the Entylomatales inferred from the sequences of the LSU rDNA D1/D2 domains and ITS region (including 5.8S rDNA) by maximum likelihood analysis and over 50% from 1,000 bootstrap replicates is shown. Bar = 0.05 substitutions per nucleotide position.
Note: Strain CGMCC 2.5613, Exobasidiomycetes sp. isolate CK927 (MH483605/MH474509) from lichen biocrust soil in Utah, USA and Tilletiopsis sp. isolate YP-240 (KU702544/KU702557) from Duke pine Forest soil in North Carolina, United States have identical D1/D2 sequences, but they differ from each other by 9–10 nt in the ITS region. A multigene approach is needed to determine whether or not they may represent different species.
New Species Identification in the Microstromatales (Exobasidiomycetes)
The Microstromales comprises Jaminaea, Parajaminaea, Pseudomicrostroma, Microstroma, Quambalaria, Sympodiomycopsis, and Volvocisporium. Parajaminaea, Pseudomicrostroma, and Microstroma are teleomorphic genera and contain both sexual and asexual species. The other genera in Microstromales belong to strictly anamorphic fungi. Our 14 isolates (Table 1) in Microstromales were all placed outside the known sexual genera (Figures 1, 6 and Supplementary Figures 1, 3).
Sipiczki and Kajdacsi (2009) proposed the genus Jaminaea based on rDNA phylogenetic analysis. The genus currently comprises four species, namely, J. angkorensis, J. lanaiensis, J. pallidilutea, and J. rosea (Sipiczki and Kajdacsi, 2009; Kijpornyongpan and Aime, 2017; Nasr et al., 2017). Strains CGMCC 2.3529, CGMCC 2.3622, and CGMCC 3662 have identical ITS and D1/D2 sequences. They are closely related to Jaminaea angkorensis (Figures 1, 6) and differ from it by 3 and 19 nt (3%) in the D1/D2 and ITS region, respectively.
Three species, namely, Sympodiomycopsis paphiopedili, S. kandeliae, and S. yantaiensis, were placed in the genus Sympodiomycopsis (Sugiyama et al., 1991; Wei et al., 2011; Chen et al., 2013). Strains CGMCC 2.3119, CGMCC 2.3120, CGMCC 2.3121, CGMCC 2.3122, CGMCC 2.3123, CGMCC 2.3124, and CGMCC 2.3181 differ from one another by 2 nt in the D1/D2 domain and by 3 nt in the ITS region, indicating that they are the same species. They differ from Sympodiomycopsis yantaiensis, S. paphiopedili, and S. kandeliae by 19–23 nt (4%) in the D1/D2 domain and by 60 nt (9%) in the ITS region.
Strains CGMCC 2.4532, CGMCC 2.4534, CGMCC 2.4535, and CGMCC 2.4536 form a separate branch with 100% BP support and are closely related to the genera Parajaminaea and Jaminaea in the Microstromatales (Figures 1, 6). These four strains have identical ITS sequences and 1 nt difference in the D1/D2 domain, indicating that they are conspecific. The D1/D2 and ITS sequence blast results showed that the four strains differ from the known species of Jaminaea, Parajaminaea, Pseudomicrostroma, Microstroma, Quambalaria, Sympodiomycopsis, and Volvocisporium by more than 20 nt (3%) and 90 nt (13%), respectively. The above data indicate that strains CGMCC 2.4532, CGMCC 2.4534, CGMCC 2.4535, and CGMCC 2.4536 represent a new genus in the Microstromatales because they cannot be placed in the existing genera in Microstromatales.
Note: The CGMCC 2.4532 group has identical ITS sequences with two strains 5CL1 (KJ460375) and 4FL2 (KJ460376) from Brazil, and identical D1/D2 sequences with strain BMA 85 (MH908976) from Brazil, which indicates they are conspecific.
New Species Identification in the Exobasidiomycetes Without Affiliation to a Known Order
Two strains, XZ4C4 and XZ4A1, have the same sequences in both the ITS and D1/D2 regions. A BLASTn search using the D1/D2 sequence of XZ4C4 showed that the top matched sequences were that of species in Microstromales, such as Ps. phylloplanum and Quambalaria cyanescens, with 94% similarity. However, the best match is with Golubevia pallescens and ‘Entyloma dahliae’ with 57–79% coverage and 79–83% similarity using ITS sequences as the query. To confirm the phylogenetic position of these two strains, a multiple gene phylogenetic tree was constructed (Figure 1). Strain XZ4C4, G. pallescens, and Golubevia heteromorpha, which was recently reported by Richter et al. (2019), form a clade with strong support (92% BP and 1.0 PP), but the former is clearly different from the latter since they are located in separate branches (Figure 1 and Supplementary Figures 1, 3). The sequence similarities in the D1/D2 and ITS regions between strain XZ4C4 and G. pallescens are 90.8 (535/589) and 71% (449/632), respectively, which are too distant to place XZ4C4 in the genus Golubevia and order Golubeviales. The above analyses indicated that strain XZ4C4 could represent a new order, distinct from Golubeviales. Therefore, Franziozyma bambusoicola gen. et sp. nov., Franziozymaceae fam. nov., and Franziozymales ord. nov. are proposed for strains XZ4C4 and XZ4A1.
Taxonomy
New Taxa in Ustilaginaceae (Ustilaginales, Ustilaginomycetes)
Sporisorium cylindricum Q.M. Wang, Y.Y. Li, M. Groenew. and M.M. Wang sp. nov. — MycoBank 839570
Etymology: the specific epithet cylindricum refers to the cell morphology of the type strain.
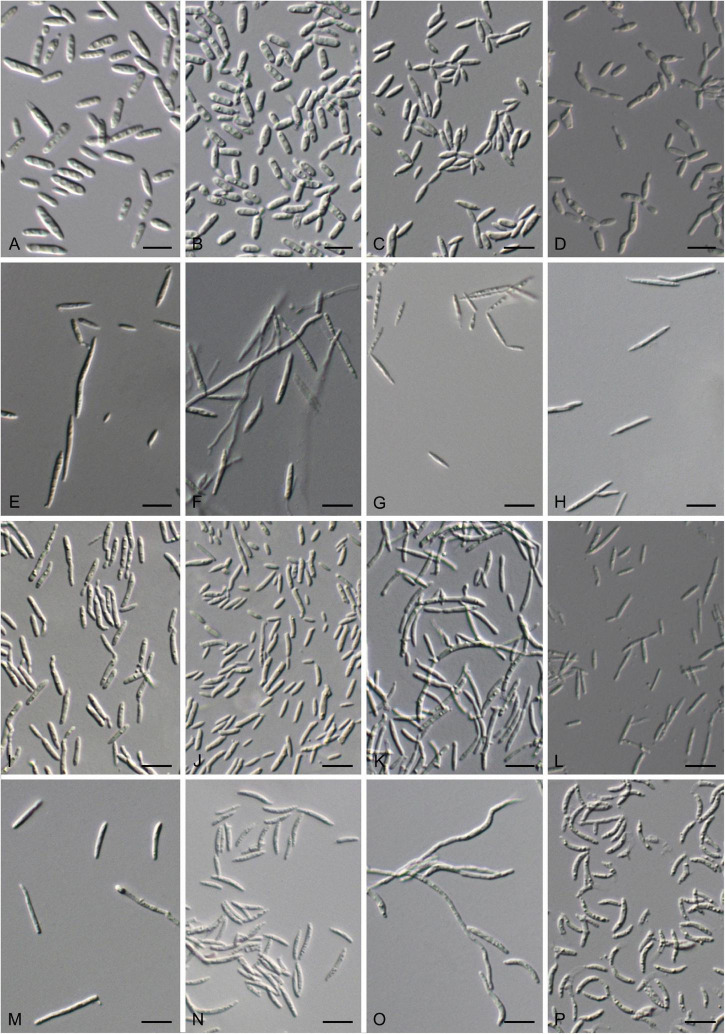
After 7 days at 17°C in YM broth, cells are cylindrical, 1.5–3.5 × 6.5–14.0 μm and single, a sediment is produced, budding is polar (Figure 7A). After 1 month at 17°C, a sediment and a film are formed. The streak culture is smooth, dull, whitish-cream, butyrous, and has an entire margin after 1 month at 17°C on YM agar. Pseudohyphae are absent in Dalmau plate culture on corn meal agar. Sexual structures are not produced on Potato Dextrose Agar (PDA), Yeast Mold (YM), CM, and V8 agars. Ballistoconidia are not observed.
FIGURE 7.
Vegetative cells grown in Yeast Mold (YM) broth for 7 days at 17°C and ballistoconidia produced on corn meal agar after 7 days at 17°C. (A) S. cylindricum CGMCC 2. 3756T; (B) K. hebeiensis CGMCC 2.3457T; (C) L. walkerae CGMCC 2. 4680; (D) L. ligulariae CGMCC 2. 6313; (E) M. plantarum CGMCC2.4430T; (F,G) M. pileae CGMCC 2.6305T; (H) M. hainanensis CGMCC 2.3537T; (I) Y. orchidis CGMCC 2.3451T; (J) Y. clavatus CGMCC 2.4433T; (K) Y. qinlingensis CGMCC 2.4533T; (L) Y. cylindricus CGMCC 2.6304T; (M,N) P. parafulvescens CGMCC 2.3573T; (O,P) G. pseudominor CGMCC 2.5616T. Bars = 10 μm. (G,N,P) are ballistoconidia.
Glucose is not fermented. Glucose, galactose, sucrose, maltose, cellobiose, trehalose, melibiose, raffinose, melezitose, soluble starch, D-ribose, D-xylose, L-rhamnose, α-Methyl-D-glucoside, salicin, succinic acid, and inositol are assimilated. L-sorbose, lactose, inulin, D-arabinose, L-arabinose, D-glucosamine, N-Acetyl-D-glucosmine, ethanol, glycerol, D-mannitol, D-glucitol, methanol, erythritol, ribitol, galactitol, D-glueonale, DL-lactic acid, citric acid, and hexadecane are not assimilated. Ammonium sulfate and L-lysine are assimilated. Potassium nitrate, sodium nitrite, ethylamine, and cadaverine are not assimilated. Maximum growth temperature is 30°C. Growth does occur in a vitamin-free medium. No starch-like substrate is produced. Growth does not occur on 50% (w/w) glucose-yeast extract agar. Urease and Diazonium blue B reactions are positive.
Typus: China, Tibet, obtained from a leaf of an unidentified plant, Oct. 2007, Qi-Ming Wang, holotype CGMCC 2. 3756T preserved in a metabolically inactive state in the China General Microbiological Culture Collection Center (CGMCC), Beijing, China. Ex-type CBS 15755 is deposited at the CBS collection of the Westerdijk Fungal Biodiversity Institute, Utrecht, Netherlands. Kunming county, Yunnan province, obtained from a leaf of an unidentified plant, May. 2007, Qi-Ming Wang, paratype CGMCC 2.3576.
Note: S. arthraxonis, S. ophiuri, S. fastigiatum, S. reilianum, S. lacrymae-jobi, and S. pseudechinolaenae are all parasitized on Poaceae. The two yeasts, CGMCC 2.3576 and CGMCC 2.3756, also isolated from leaves of the plant. Unfortunately, those plants were not identified. Except the worldwide distributed S. reilianum, the above five parasitic species mostly occur in the tropic region including southern China and Southeast Asia (Vánky, 2012). The two strains of S. cylindricum, CGMCC 2.3576 and CGMCC 2.3756, were isolated from the plant leaf collected in Yunan province (southern China) and Hanan province (southern China), respectively, which indicated that they have similar ecological and biogeographical characters to those parasitic species.
Kalmanozyma hebeiensis Q.M. Wang, Y.Y. Li, M. Groenew. and M.M. Wang sp. nov. — MycoBank 839571
Etymology: the specific epithet hebeiensis refers to the geography from which the type strain was isolated.
After 7 days at 17°C in YM broth, cells are cylindrical, 1.7–3.0 × 5.8–10.0 μm, and single or in pairs, a sediment is produced, budding is polar (Figure 7B). After 1 month at 17°C, a sediment and an incomplete ring are produced. The streak culture is smooth, gloomy, yellowish-cream, butyrous, and has an entire margin after 1 month at 17°C on YM agar. Pseudohyphae are formed in Dalmau plate culture on cornmeal agar. Sexual structures are not produced on PDA, YM, CM, and V8 agars. Ballistoconidia are not observed.
Glucose is not fermented. Glucose, galactose, sucrose, maltose, cellobiose, trehalose, lactose (latent and weak), raffinose (weak), melezitose, D-xylose, L-arabinose, D-arabinose (weak), D-ribose (weak), D-glucosamine, ethanol, glycerol, ribitol, D-mannitol, D-glucitol, α-Methyl-D-glucoside, salicin (weak), succinic acid and citric acid are assimilated. L-sorbose, melibiose, inulin, soluble starch, L-rhamnose, methanol, erythritol, galactitol, DL-lactic acid, inositol, and hexadecane are not assimilated. Ammonium sulfate, potassium nitrate, sodium nitrite, L-lysine (weak), and ethylamine (latent and weak) are assimilated. Cadaverine is not assimilated. Optimal growth is at 17–25°C. Growth does occur in a vitamin-free medium. No starch-like substrate is produced. Growth does not occur on 50% (w/w) glucose-yeast extract agar. Urease and Diazonium blue B reactions are positive.
Physiologically, K. hebeiensis differs from its closely related species, K. brasiliensis and K. vetiver, in its inability to assimilate L-sorbose and inositol (Table 2).
Typus: China, Hebei province, obtained from a leaf of an unidentified plant, October 2007, Qi-Ming Wang, holotype CGMCC 2.3457T preserved in a metabolically inactive state in the CGMCC, Beijing, China. Ex-type CBS 15483 is deposited at the CBS collection of the Westerdijk Fungal Biodiversity Institute, Utrecht, Netherlands.
Note: K. brasiliensis was isolated from the intestinal tract of chrysomelid larva associated with roots of Saccharum in Brazil (Oliveira et al., 2014). K. fusiformata was obtained from a barley leaf and from cauliflower (Boekhout, 2011). K. vetiveriae was isolated from phylloplane of Vetiveria zizanioides in Thailand. CGMCC 2.3457 was isolated from the plant. The host-substratum data indicated that species of Kalmanozyma are associated with the plant.
Yeast Stage Description of Langdonia walkerae
Langdonia walkerae was described by Alqurashi et al. (2021) based on a sexual stage. A yeast strain, CGMCC 2. 4680, isolated from a leaf of an unidentified plant, collected in September 2012 in China has 99.6% sequence similarity to L. walkerae in the ITS region, which indicated they belong to the same species. Here we described the yeast morphological and physiological characters as the asexual state of L. walkerae.
After 7 days at 17°C in YM broth, cells are fusiformis, long ovoid, and cylindrical, 1.1–2.5 × 3.5–11.8 μm and single, a sediment is produced, and budding is polar (Figure 7C). After 1 month at 17°C, a sediment and a film are produced. The streak culture is pale, yellowish-brown, flat, butyrous, glossy, slightly granulate, and has an entire margin after 1 month at 17°C on YM agar. Pseudohyphae are formed in Dalmau plate culture on cornmeal agar. Sexual structures are not produced on PDA, YM, CM, and V8 agars. Ballistoconidia are not observed.
Glucose is not fermented. Glucose, sucrose, cellobiose (weak), trehalose (weak), lactose (weak), raffinose (weak), melezitose (weak), inulin (weak), soluble starch (weak), D-xylose, L-arabinose, D-mannitol (weak), and succinic acid (weak) are assimilated. Galactose, L-sorbose, maltose, melibiose, D-arabinose, D-ribose, L-rhamnose, D-glucosamine, N-Acetyl-D-glucosmine, methanol, ethanol, glycerol, erythritol, ribitol, galactitol, D-glucitol, α-Methyl-D-glucoside, salicin, DL-lactic acid, citric acid, myo-inositol, and hexadecane are not assimilated. Ammonium sulfate, potassium nitrate, sodium nitrite, L-lysine, ethylamine, and cadaverine are assimilated. Optimal growth is at 17–25°C. Growth does not occur in vitamin-free medium. No starch-like substrate is produced. Growth does not occur on 50% (w/w) glucose-yeast extract agar Urease and Diazonium blue B reactions are positive.
Physiologically, L. walkerae differs from its closely related species, L. jejuensis, and L. ligulariae, in its inability to assimilate maltose and its ability to assimilate succinic acid (Table 2).
Langdonia ligulariae Q.M. Wang, Y.Y. Li, M. Groenew. and M.M. Wang sp. nov. — MycoBank 839578
Etymology: the specific epithet, ligulariae, refers to Ligularia, the plant genus from which the type strain was isolated.
After 7 days at 17°C in YM broth, cells are cylindrical to elongate, 1.5–2.7 × 4.0–12.0 μm and single, a sediment is produced, and budding is polar (Figure 7D). After 1 month at 17°C, a sediment and a ring are produced. The streak culture is yellowish-brown, flat, butyrous, arachnoid, and has an entire margin after 1 month at 17°C on YM agar. Pseudohyphae or hyphae are formed in Dalmau plate culture on cornmeal agar. Sexual structures are not produced on PDA, YM, CM, and V8 agars. Ballistoconidia are not observed.
Glucose is not fermented. Glucose, sucrose, maltose, cellobiose, trehalose, raffinose, melezitose, inulin (weak), soluble starch (weak), D-xylose, L-arabinose, ethanol, glycerol, ribitol, D-mannitol, α-Methyl-D-glucoside, and inositol (weak) are assimilated. Galactose, L-sorbose, lactose, melibiose, D-arabinose, D-ribose, L-rhamnose, D-glucosamine, N-Acetyl-D-glucosmine, methanol, erythritol, galactitol, D-glucitol, salicin, DL-lactic acid, succinic acid, citric acid, and hexadecane are not assimilated. Ammonium sulfate, potassium nitrate, sodium nitrite (weak), L-lysine, ethylamine, and cadaverine are assimilated. Optimal growth is at 17–25°C. Growth does occur in the vitamin-free medium. No starch-like substrate is produced. Growth does not occur on 50% (w/w) glucose-yeast extract agar Urease and Diazonium blue B reactions are positive.
Physiologically, L. ligulariae differs from its closely related species, L. walkerae and L. jejuensis, in its inability to assimilate lactose and its ability to use ethanol, glycerol, ribitol, and α-Methyl-D-glucoside (Table 2).
Typus: China, Tibet, obtained from a leaf of Ligularia tsangchanensis, September 2012, Qi-Ming Wang, holotype CGMCC 2. 6313T preserved in a metabolically inactive state in the CGMCC, Beijing, China. Ex-type CBS 15581 is deposited at the CBS collection of the Westerdijk Fungal Biodiversity Institute, Utrecht, Netherlands.
Note: All sexual Langdonia species infected the plant of Aristida (Poaceae). The asexual yeast species L. jejuensis was isolated from a leaf of Citrus unshiu (Rutaceae) in South Korea. CGMCC 2.6313 was isolated from a leaf of Ligularia tsangchanensis (Asteraceae) in Tibet, China. Although CGMCC 2.6313 and L. jejuensis were all isolated from the leaf of plant (Asteraceae and Rutaceae), they differ from the parasitic species of Langdonia whose host is the Poaceae grass, which indicated that the asexual yeast stage and the sexual stage of Langdonia may have different ecological inches in nature.
Brachybasidiaceae (Exobasidiales, Exobasidiomycetes)
Meira plantarum Q.M. Wang, Y.Y. Li, M. Groenew. and M.M. Wang sp. nov. — MycoBank 839580
Etymology: the specific epithet plantarum refers to the substrates from which the type strain was isolated.
After 7 days at 17°C in YM broth, cells are fusiform and cylindrical to elongate, 1.7–2.1 × 4.2–28.3 μm and single, a sediment is produced, and budding is polar (Figure 7E). After 1 month at 17°C, a sediment a film and are produced. On YM agar, after 1 month at 17°C, the colonies are firm to tough, whitish at first before becoming pale yellowish-brown with a velvety to the pruinose surface, and the margin is eroded. Hyphae are formed in Dalmau plate culture on cornmeal agar. Sexual structures are not produced on PDA, YM, CM, and V8 agars. Ballistoconidia are not observed.
Glucose is not fermented. Glucose, galactose, sucrose, maltose, cellobiose, trehalose, melibiose, raffinose, melezitose, D-xylose, L-arabinose, D-ribose, erythritol, galactitol (variable), D-mannitol, D-glucitol, salicin (latent and weak), and succinic acid (latent and weak) are assimilated. L-sorbose, lactose, inulin, soluble starch, D-arabinose, L-rhamnose, D-glucosamine, methanol, ethanol, glycerol, ribitol, α-Methyl-D-glucoside, DL-lactic acid, citric acid, inositol, and hexadecane are not assimilated. Ammonium sulfate, potassium nitrate, sodium nitrite, ethylamine, and cadaverine (latent) are assimilated. L-lysine (or latent and weak) is not assimilated. Optimal growth is at 17–25°C. Growth does occur in the vitamin-free medium. No starch-like substrate is produced. Growth does not occur on 50% (w/w) glucose-yeast extract agar Urease and Diazonium blue B reactions are positive.
Physiologically, M. plantarum differs from its closely related species, M. nashicola and M. pileae, in its inability to assimilate D-arabinose (Table 2).
Typus: China, Fuzhou county, Fujian province, obtained from a leaf of an unidentified plant, Oct. 2011, Qi-Ming Wang, holotype CGMCC 2.4430T preserved in a metabolically inactive state in the CGMCC, Beijing, China. Ex-type CBS 12491 is deposited at the CBS collection of the Westerdijk Fungal Biodiversity Institute, Utrecht, Netherlands. Fuzhou county, Fujian province, obtained from a leaf of an unidentified plant, October 2011, Qi-Ming Wang, paratypes CGMCC 2.4431 and CGMCC 2.4432. Beibengxiang, Motuo county, Tibet, obtained from a leaf of an unidentified plant, September 2014, Qi-Ming Wang, paratype CGMCC 2.6306.
Meira pileae Q.M. Wang, Y.Y. Li, M. Groenew. and M.M. Wang sp. nov. — MycoBank 839582
Etymology: the specific epithet pileae refers to Pilea, the plant genus from which the type strain was isolated.
After 7 days at 17°C in YM broth, cells are cylindrical to elongate, 1.3–2.3 × 5.0–25.0 μm and single, a sediment is produced, budding is polar, hyphae are narrow, and 1.2–2.5 μm (Figure 7F). After 1 month at 17°C, a film and a sediment are produced. On YM agar, after 1 month at 17°C, the colonies are firm to tough with a whitish velvety to the pruinose surface, dull, and the margin is eroded. Hyphae are formed in Dalmau plate culture on cornmeal agar. Sexual structures are not produced on PDA, YM, CM, and V8 agars. Ballistoconidia are fusiform or cylindrical (1.0–2.1 × 6.7–12.5 μm; Figure 7G).
Glucose is not fermented. Glucose, galactose, sucrose, maltose, cellobiose, trehalose, melibiose, inulin, raffinose, melezitose, D-xylose, L-arabinose, D-arabinose (weak), ethanol (weak), glycerol (weak), ribose (weak), erythritol (weak), D-mannitol (weak), D-glucitol (weak), salicin, and succinic acid (latent and weak) are assimilated. L-sorbose, lactose, galactitol, soluble starch, L-rhamnose, D-glucosamine, methanol, α-Methyl-D-glucoside, D-glueonale, DL-lactic acid, citric acid, inositol, and hexadecane are not assimilated. Ammonium sulfate, potassium nitrate, sodium nitrite, L-lysine, ethylamine, and cadaverine are assimilated. Optimal growth is at 17–25°C. Growth does occur in the vitamin-free medium. No starch-like substrate is produced. Growth does not occur on 50% (w/w) glucose-yeast extract agar Urease and Diazonium blue B reactions are positive.
Physiologically, Me. pileae differs from its closely related species, Me. nashicola and Me. Plantarum, in its ability to assimilate inulin (Table 2).
Typus: China, Beibengxiang, Motuo county, Tibet, obtained from a leaf of Pilea sp., September 2014, Qi-Ming Wang, holotype CGMCC 2.6305T preserved in a metabolically inactive state in the CGMCC, Beijing, China. Ex-type CBS 144915 is deposited at the CBS collection of the Westerdijk Fungal Biodiversity Institute, Utrecht, Netherlands.
Meira hainanensis Q.M. Wang, Y.Y. Li, M. Groenew. and M.M. Wang sp. nov. — MycoBank 839584
Etymology: the specific epithet hainanensis refers to the geographic origin of the type strain, Hainan province, China.
After 7 days at 17°C in YM broth, cells are fusiform and cylindrical to elongate, 1.0–1.7 × 6.7–20.0 μm and single, a sediment is produced, budding is polar, hyphae are narrow (1.2–2.5 μm; Figure 7H). After 1 month at 17°C, a thick film and a sediment are produced. The colonies are firm to tough, whitish at first, before becoming pale yellowish-brown with the farinose surface, and the margin is eroded after 1 month at 17°C on YM agar. Hyphae are formed in Dalmau plate culture on cornmeal agar. Sexual structures are not produced on PDA, YM, CM, and V8 agars. Ballistoconidia are not observed.
Glucose is not fermented. Glucose, galactose, sucrose, maltose, cellobiose, trehalose, melibiose, raffinose, melezitose, inulin, soluble starch, D-xylose, L-arabinose, D-arabinose, D-ribose, ethanol, glycerol (weak), erythritol, ribitol (latent and weak), galactitol, D-mannitol, D-glucitol, and succinic acid (weak) are assimilated. L-sorbose, lactose, L-rhamnose, D-glucosamine, methanol, α-Methyl-D-glucoside, salicin, DL-lactic acid, citric acid, inositol, and hexadecane are not assimilated. Ammonium sulfate, potassium nitrate (weak), sodium nitrite, and cadaverine are assimilated. L-lysine and ethylamine are not assimilated. Optimal growth is at 17–25°C. Growth does occur in the vitamin-free medium. No starch-like substrate is produced. Growth does not occur on 50% (w/w) glucose-yeast extract agar Urease and Diazonium blue B reactions are positive.
Physiologically, Me. hainanensis differs from its closely related species Me. argovae in its inability to assimilate salicin and citric acid (Table 2).
Typus: China, Wuzhishan Montain, Hainan province, obtained from a leaf of an unidentified plant, May 2007, Qi-Ming Wang, holotype CGMCC 2.3537T preserved in a metabolically inactive state in the CGMCC, Beijing, China. Ex-type CBS 15497 is deposited at the CBS collection of the Westerdijk Fungal Biodiversity Institute, Utrecht, Netherlands.
Note: Limtong et al. (2017) indicated that species of Meira seem to be relate to plants and organisms associated with plants. Me. argovae and Me. geulakonigii were isolated from mite cadavers (Tetranychus cinnabarinus and Phyllocoptruta oleivira) from citrus leaves (Ricinus communis and Citrus paradisi) in Israel (Boekhout et al., 2003). Me. nashicola, Me. miltonrushii, and Me. siamensis were obtained from the leaf or fruit of plant (Yasuda et al., 2006; Tanaka et al., 2008; Rush and Aime, 2013; Limtong et al., 2017). Our three described Meira species were all isolated from the leaf of plant (Table 1). Some strains of Meira had been reported as endophytes of plant species (Paz et al., 2007a; Rush and Aime, 2013). Me. argovae, Me. geulakonigii, and Me. nashicola were proposed to use as a biological control agent against mites and powdery mildew fungi (Boekhout et al., 2003; Sztejnberg et al., 2004; Paz et al., 2007b; Gerson et al., 2008). The biological control property of our described Meira species, Me. miltonrushii and Me. siamensis need to be tested in the future.
Yunzhangomyces Q.M. Wang, E. Tanaka, M. Groenew., and D. Begerow gen. nov. — MycoBank 839581
Etymology: the genus is named in honor of Yun-Zhang Wang for his pioneering work on the taxonomy of smuts.
This genus is proposed for the branch represented by Dicellomyces scirpi, which formed a separate branch from the genera in the Brachybasidiaceae family (Exobasidiales, Exobasidiomycetes). The genus is mainly circumscribed by the description of Dicellomyces scirpi and the phylogenetic analysis of the six-genes sequences (Figure 1).
This genus includes sexual and asexual species. Sexual member infecting Scirpus sylvaticus (Cyperaceae); basidia developing in gelatinous basidiocarps breaking through epidermis, swollen, not persistent probasidia, with paraphyses, sterigmata 2; producing allantoid or coiled conidia (Parmasto, 1968; Reid, 1976; Ingold, 1985; Piepenbring et al., 2020). Asexual species present butyrous, yellow or brown colonies, smooth or eroded margin with budding cells present.
Type species: Yunzhangomyces scirpi (Raitv.) Q.M. Wang, E. Tanaka, M. Groenew. and D. Begerow.
Yunzhangomyces scirpi (Raitv.) Q.M. Wang, E. Tanaka, M. Groenew. and D. Begerow, comb. nov. — MycoBank 839585
Basionym: Dicellomyces scirpi Raitv., in Parmasto, Eesti NSV Tead. Akad. Toim. 17(2): 223 (1968).
Yunzhangomyces orchidis Q.M. Wang, E. Tanaka, M. Groenew. and D. Begerow sp. nov. — MycoBank 839587
Etymology: the specific epithet orchidis refers to plant host, Orchidaceae sp., from which the type strain was isolated.
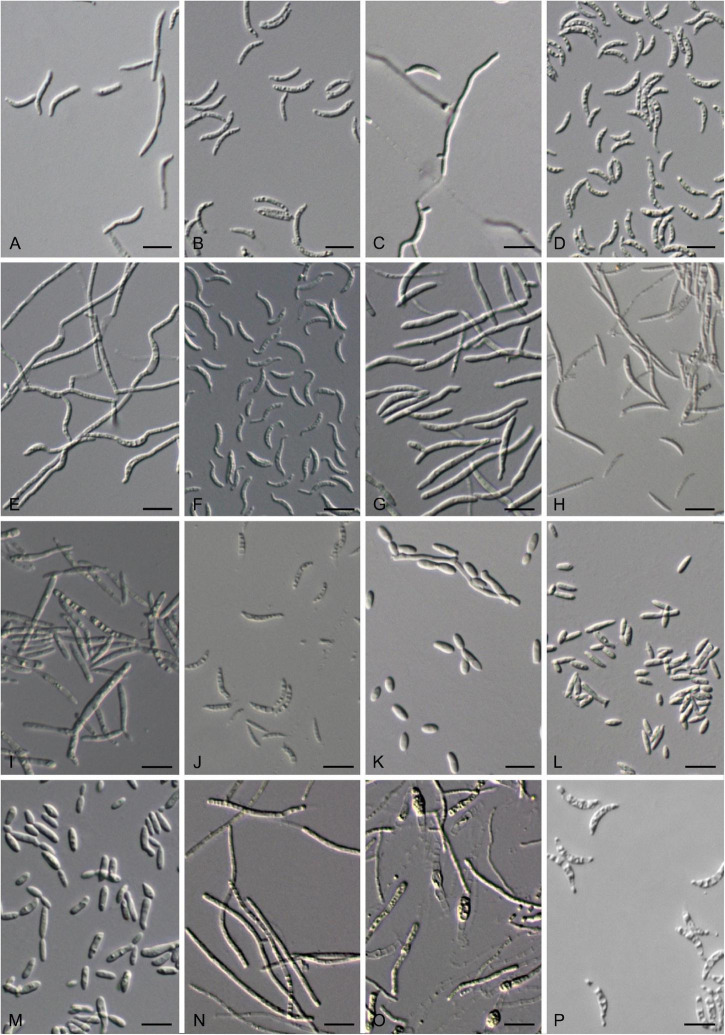
After 7 days at 17°C in YM broth, cells are cylindrical to elongate, .08–1.7 × 2.8–16.7 μm and single, a sediment is produced, budding is polar, and hyphae are narrow (1.2-2.5 μm; Figure 7I). After 1 month at 17°C, a thick film and a sediment are produced. The streak culture is pale yellowish to brown with smooth and glistening surface, butyrous, and has an entire margin after 1 month at 17°C on YM agar. Pseudohyphae are formed in Dalmau plate culture on cornmeal agar. Sexual structures are not produced on PDA, YM, CM, and V8 agars. Ballistoconidia are not observed.
Glucose is not fermented. Glucose, galactose, L-sorbose, sucrose, maltose, cellobiose, trehalose, melibiose (latent), raffinose, melezitose, inulin, D-xylose, L-arabinose, D-arabinose, D-ribose, ethanol, glycerol, erythritol, ribitol, D-mannitol, D-glucitol, salicin, succinic acid (latent and weak), and citric acid (latent and weak) are assimilated. Lactose, soluble starch, L-rhamnose, D-glucosamine, methanol, galactitol, α-Methyl-D-glucoside, DL-lactic acid, inositol, and hexadecane are not assimilated. Ammonium sulfate, potassium nitrate, sodium nitrite, L-lysine, ethylamine, and cadaverine are assimilated Optimal growth is at 17–25°C. Growth does occur in the vitamin-free medium. No starch-like substrate is produced. Growth does not occur on 50% (w/w) glucose-yeast extract agar Urease and Diazonium blue B reactions are positive.
Physiologically, Y. orchidis differs from its closely related culturable species Y. clavatus in its ability to assimilate inulin, D-arabinose, and glycerol (Table 2).
Typus: China, Wuzhishan Montain, Hainan province, obtained from a leaf of Orchidaceae sp., Apr. 2007, Qi-Ming Wang, holotype CGMCC 2.3451T preserved in a metabolically inactive state in the CGMCC, Beijing, China. Ex-type CBS 15753 is deposited at the CBS collection of the Westerdijk Fungal Biodiversity Institute, Utrecht, Netherlands.
Yunzhangomyces clavatus Q.M. Wang, E. Tanaka, M. Groenew. and D. Begerow sp. nov. — MycoBank 839589
Etymology: the specific epithet clavatus refers to the vegetative cell morphology of the type strain.
After 7 days at 17°C in YM broth, cells are ovoid to elongate, cylindrical, club-shaped, 1.3–1.7 × 5.8–16.7 μm and single, a sediment is produced, budding is polar, and hyphae are narrow (1.2–2.5 μm; Figure 7J). After 1 month at 17°C, a ring and a sediment are produced. The streak culture is pale brown, butyrous, and the surface is glistening with slight wrinkle and has an entire margin after 1 month at 17°C on YM agar. Pseudohyphae are formed in Dalmau plate culture on cornmeal agar. Sexual structures are not produced on PDA, YM, CM, and V8 agars. Ballistoconidia are not observed.
Glucose is not fermented. Glucose, galactose (variable), L-sorbose (or latent and weak), sucrose, maltose, cellobiose (variable), trehalose, melibiose, raffinose, melezitose, D-xylose, L-arabinose, D-ribose (variable), ethanol (or latent and weak), erythritol (variable), ribitol (variable), D-mannitol, and D-glucitol are assimilated. Lactose, inulin, soluble starch, D-arabinose, L-rhamnose, D-glucosamine, methanol, glycerol, galactitol, α-Methyl-D-glucoside, salicin (or latent and weak), succinic acid (or weak), DL-lactic acid, citric acid, inositol, and hexadecane are not assimilated. Ammonium sulfate, potassium nitrate, sodium nitrite, L-lysine (variable), ethylamine, and cadaverine are assimilated. Optimal growth is at 17–25°C. Growth does occur in the vitamin-free medium. No starch-like substrate is produced. Growth does not occur on 50% (w/w) glucose-yeast extract agar Urease and Diazonium blue B reactions are positive.
Physiologically, Y. clavatus differs from its closely related species, Y. orchis, in its inability to assimilate inulin, D-arabinose, and glycerol (Table 2).
Typus: China, Fuzhou county, Fujian province, obtained from a leaf of the unidentified plant, August 2011, Qi-Ming Wang, holotype CGMCC 2.4433T preserved in a metabolically inactive state in the CGMCC, Beijing, China. Ex-type CBS 144908 is deposited at the CBS collection of the Westerdijk Fungal Biodiversity Institute, Utrecht, Netherlands; Heilongxiang, Motuo county, Tibet, obtained from a leaf of Impatiens sp., September 2014, Qi-Ming Wang, paratype CBS 144917.
Yunzhangomyces qinlingensis Q.M. Wang, E. Tanaka, M. Groenew. and D. Begerow sp. nov. — MycoBank 839593
Etymology: the specific epithet qinlingensis refers to the geographic origin of the type strain, Qinling mountain, China.
After 7 days at 17°C in YM broth, cells are cylindrical to elongate, club-shaped, 1.2–1.8 × 6.0–20.8 μm and single, a sediment is produced, budding is polar, and hyphae are narrow, 1.2–2.5 μm (Figure 7K). After 1 month at 17°C, a ring and a sediment are produced. The streak culture is pale yellow, butyrous, the surface is smooth and glistening and has an entire margin after 1 month at 17°C on YM agar. Pseudohyphae and hyphae are absent in Dalmau plate culture on cornmeal agar. Sexual structures are not produced on PDA, YM, CM, and V8 agars. Ballistoconidia are not observed.
Glucose is not fermented. Glucose, galactose, L-sorbose, sucrose, maltose, trehalose, raffinose, melezitose, D-xylose, L-arabinose, D-arabinose (latent and weak), D-ribose, ethanol, erythritol, ribitol (latent and weak), D-mannitol, D-glucitol, α-Methyl-D-glucoside (latent and weak), salicin (latent), succinic acid and citric acid are assimilated. Cellobiose, lactose, melibiose, inulin, soluble starch, L-rhamnose, D-glucosamine, N-Acetyl-D-glucosmine, methanol, glycerol, galactitol, DL-lactic acid, inositol, and hexadecane are not assimilated. Ammonium sulfate, potassium nitrate, L-lysine (latent and weak), ethylamine (latent and weak), and cadaverine (latent and weak) are assimilated. Sodium nitrite is not assimilated. Optimal growth is at 17–25°C. Growth does occur in the vitamin-free medium. No starch-like substrate is produced. Growth does not occur on 50% (w/w) glucose-yeast extract agar Urease and Diazonium blue B reactions are positive.
Physiologically, Y. qinlingensis differs from its closely related species Y. cylindricus in its inability to assimilate cellobiose and inulin and its ability to use melezitose, succinic acid, and citric acid (Table 2).
Typus: China, Qinling, Shaanxi province, obtained from a leaf of the unidentified plant, March 2012, Qi-Ming Wang, holotype CGMCC 2.4533T preserved in a metabolically inactive state in the China General Microbiological Culture Collection Center (CGMCC), Beijing, China. Ex-type CBS 144910 is deposited at the CBS collection of the Westerdijk Fungal Biodiversity Institute, Utrecht, Netherlands.
Yunzhangomyces cylindricus Q.M. Wang, E. Tanaka, M. Groenew. and D. Begerow sp. nov. — MycoBank 839595
Etymology: the specific epithet cylindricus refers to the vegetative cell morphology of the type strain.
After 7 days at 17°C in YM broth, cells are cylindrical, club-shaped, 1.2–1.8 × 6.0–20.8 μm and single, a sediment is produced, budding is polar, and hyphae are narrow (1.2–2.5 μm; Figure 7L). After 1 month at 17°C, a ring and a sediment are produced. The streak culture is pale yellow, butyrous, and the surface is smooth and glistening and has an entire margin after 1 month at 17°C on YM agar. Pseudohyphae and hyphae are absent in Dalmau plate culture on cornmeal agar. Sexual structures are not produced on PDA, YM, CM, and V8 agars. Ballistoconidia are not observed.
Glucose is not fermented. Glucose, galactose, L-sorbose, sucrose, maltose, cellobiose, trehalose, raffinose, inulin, D-xylose, L-arabinose, D-arabinose, D-ribose, ethanol, erythritol, ribitol, D-mannitol, and D-glucitol are assimilated. Lactose, melibiose, melezitose, soluble starch, L-rhamnose, D-glucosamine, N-Acetyl-D-glucosmine, methanol, glycerol, galactitol, α-Methyl-D-glucoside, salicin, DL-lactic acid, succinic acid, citric acid, inositol, and hexadecane are not assimilated. Ammonium sulfate, potassium nitrate, L-lysine, ethylamine, and cadaverine are assimilated. Sodium nitrite is not assimilated. Maximum Optimal growth is at 17–25°C. Growth does occur in the vitamin-free medium. No starch-like substrate is produced. Growth does not occur on 50% (w/w) glucose-yeast extract agar Urease and Diazonium blue B reactions are positive.
Physiologically, Y. cylindricus differs from its closely related species Y. qinlingensis in its inability to assimilate melezitose, succinic acid, and citric acid and its ability to use cellobiose and inulin (Table 2).
Typus: China, Daliangzi river national forest park, Heilongjiang province, obtained from a leaf of the unidentified plant, August 2015, Qi-Ming Wang, holotype CGMCC 2.6304T preserved in a metabolically inactive state in the CGMCC, Beijing, China. Ex-type CBS 15585 is deposited at the CBS collection of the Westerdijk Fungal Biodiversity Institute, Utrecht, Netherlands.
Note: All the members of the genus Yunzhangomyces were isolated from the leaf of plant, which indicated that they are plant associates.
Guomyces Q.M. Wang, E. Tanaka, M. Groenew., and D. Begerow gen. nov. — MycoBank 839579
Etymology: The genus is named in honor of Lin Guo for her pioneering contributions to the taxonomy of smuts.
This genus is proposed for the branch represented by Meira nicotianae, which formed a separate branch from the genera in the Brachybasidiaceae family (Exobasidiales, Exobasidiomycetes). The genus is mainly circumscribed by the phylogenetic analysis of the ITS+LSU sequences (Figure 3 and Supplementary Figure 3).
Sexual reproduction is not known. Colonies were butyrous, yellow with eroded margin. Budding cells present or not. Hyphae are formed. Ballistoconidia are not produced.
Type species: Guomyces nicotianae (H.K. Wang and F.C. Lin) Q.M. Wang, E. Tanaka, M. Groenew., and D. Begerow.
Guomyces nicotianae (H.K. Wang and F.C. Lin) Q.M. Wang, E. Tanaka, M. Groenew., and D. Begerow, comb. nov. — MycoBank 839598
Basionym: Meira nicotianae H.K. Wang and F.C. Lin, in Cao et al. Phytotaxa 365 (2): 176 (2018).
Georgefischeriales (Exobasidiomycetes)
Phragmotaenium parafulvescens Q.M. Wang, Y.Y. Li, M. Groenew., and M.M. Wang sp. nov. — MycoBank 839586
Etymology: the specific epithet, parafulvescens, refers to a similar colony morphology to that of Phragmotaenium fulvescens.
After 7 days at 17°C in YM broth, cells are cylindrical, 1.0–1.7 × 10.8–25.0 μm and single, a sediment is produced, budding is polar, and hyphae are narrow (1.2–2.5 μm; Figure 7M). After 1 month at 17°C, a ring and a sediment are produced. The streak culture is cream to pale yellow, butyrous, and the surface is glistening with slight wrinkle and has an entire margin after 1 month at 17°C on YM agar. Hyphae are formed in Dalmau plate culture on cornmeal agar. Sexual structures are not produced on PDA, YM, CM, and V8 agars. Ballistoconidia are allantoid or falcate, 1.0–1.6 × 7.5–16.7 μm (Figure 7N).
Glucose is not fermented. Glucose, galactose (latent and weak), L-sorbose (latent and weak), sucrose, maltose, trehalose, Lactose (latent and weak), melibiose (latent and weak), raffinose, melezitose, inulin, soluble starch, D-xylose, L-arabinose, D-arabinose, glycerol, ribitol (latent and weak), D-mannitol, D-glucitol, D-gluenoale, and succinic acid are assimilated. Cellobiose, D-ribose, L-rhamnose, D-glucosamine, N-Acetyl-D-glucosmine, methanol, ethanol, erythritol, galactitol, α-Methyl-D-glucoside, salicin, DL-lactic acid, citric acid, inositol, and hexadecane are not assimilated. Ammonium sulfate, potassium nitrate, and L-lysine (latent and weak) are assimilated. Sodium nitrite, ethylamine, and cadaverine are not assimilated. Optimal growth is at 17–25°C. Growth does occur in the vitamin-free medium. No starch-like substrate is produced. Growth does not occur on 50% (w/w) glucose-yeast extract agar Urease and Diazonium blue B reactions are positive.
Physiologically, P. parafulvescens differs from its closely related species, P. fulvescens, in its inability to assimilate cellobiose, D-ribose, erythritol, citric acid, and sodium nitrite and its positive growth in vitamin-free medium (Table 2).
Typus: China, Sanya county, Hainan province, obtained from a leaf of the unidentified plant, May 2007, Qi-Ming Wang, holotype CGMCC 2.3573T preserved in a metabolically inactive state in the CGMCC, Beijing, China. Ex-type CBS 15754 is deposited at the CBS collection of the Westerdijk Fungal Biodiversity Institute, Utrecht, Netherlands.
Gjaerumia pseudominor Q.M. Wang, Y.Y. Li, M. Groenew., and M.M. Wang sp. nov. — MycoBank 839588
Etymology: the specific epithet pseudominor refers to the similar colony morphology to that of Gjaerumia minor.
After 7 days at 17°C in YM broth, cells are cylindrical or allantoid, 1.2–1.8 × 6.7–22.5 μm and single, a sediment is produced, budding is polar, and hyphae are narrow (1.2–2.5 μm; Figure 7O). After 1 month at 17°C, a sediment is produced. The streak culture is cream to light-yellowish, butyrous, and the surface is glistening with smooth and has an entire margin after 1 month at 17°C on YM agar. Hyphae are formed, narrow and cylindrical in Dalmau plate culture on cornmeal agar. Sexual structures are not produced on PDA, YM, CM, and V8 agars. Ballistoconidia are allantoid or falcate, 1.0–1.6 × 6.7–12.5 μm (Figure 7P).
Glucose is not fermented. Glucose, sucrose, maltose, cellobiose, trehalose, melezitose, solube starch, D-xylose, L-arabinose, glycerol, D-mannitol, D-glucitol, DL-lactic acid, succinic acid, and citric acid are assimilated. Galactose, L-sorbose, lactose, melibiose, raffinose, inulin, D-arabinose, D-ribose, L-rhamnose, D-glucosamine, N-Acetyl-D-glucosmine, methanol, ethanol, erythritol, ribitol, galactitol, α-Methyl-D-glucoside, salicin, inositol, and hexadecane are not assimilated. Ammonium sulfate, potassium nitrate, L-lysine, ethylamine, and cadaverine are assimilated. Sodium nitrite is not assimilated. Optimal growth is at 17–25°C. Growth does occur in a vitamin-free medium (weak). No starch-like substrate is produced. Growth does not occur on 50% (w/w) glucose-yeast extract agar Urease and Diazonium blue B reactions are positive.
Physiologically, G. pseudominor differs from its closely related species G. minor and G. cyclobalanopsidis in its inability to assimilate galactose, lactose, melibiose, raffinose, inulin, D-arabinose, and D-ribose (Table 2).
Typus: China, Heilongjiang province, obtained from a leaf of the unidentified plant, August 2015, Qi-Ming Wang, holotype CGMCC 2.5616T preserved in a metabolically inactive state in the CGMCC, Beijing, China. Ex-type CBS 144912 is deposited at the CBS collection of the Westerdijk Fungal Biodiversity Institute, Utrecht, Netherlands.
Gjaerumia cyclobalanopsidis Q.M. Wang, Y.Y. Li, M. Groenew. and M.M. Wang sp. nov. — MycoBank 839591
Etymology: the specific epithet cyclobalanopsidis refers to Cyclobalanopsis, a plant host from which the type strain was isolated.
After 7 days at 17°C in YM broth, cells are cylindrical or allantoid, 1.0–1.6 × 7.3–23.6 μm and single, a sediment is produced, budding is polar, and hyphae are narrow (1.2–2.5 μm; Figure 8A). After 1 month at 17°C, a film and sediment are produced. The streak culture is cream to yellowish, butyrous, the surface is wrinkled with gloomy, and has an entire margin after 1 month at 17°C on YM agar. Hyphae are formed, narrow and cylindrical in Dalmau plate culture on cornmeal agar. Sexual structures are not produced on PDA, YM, CM, and V8 agars. Ballistoconidia are allantoid or falcate (1.0–1.7 × 8.3–14.2 μm; Figure 8B).
FIGURE 8.
Vegetative cells grown in YM broth for 7 days at 17°C and ballistoconidia produced on cornmeal agar after 7 days at 17°C. (A,B) G. cyclobalanopsidis CGMCC 2.6419T; (C–F) J. aceris CGMCC 2.5602T; (G,H) T. pinicola CGMCC 2.5613T; (I,J) T. lunata CGMCC 2.6308T; (K) J. lantanae CGMCC 2.3529T; (L) S. europaea CGMCC 2.3119T; (M) B. planticola CGMCC 2.4532T; (N–P) F. bambusicola CGMCC 2.2620T. Bars = 10 μm. (B,D,F,H,P) are ballistoconidia.
Glucose is not fermented. Glucose, galactose, L-sorbose (weak), sucrose, maltose, cellobiose, trehalose, lactose, melibiose, raffinose, melezitose, inulin, D-xylose, L-arabinose, D-arabinose, glycerol, ribitol (weak), D-mannitol (weak), and D-glucitol (weak) are assimilated. Solube starch, D-ribose, L-rhamnose, D-glucosamine, N-Acetyl-D-glucosmine, methanol, ethanol, erythritol, galactitol, α-Methyl-D-glucoside, salicin, DL-lactic acid, succinic acid, citric acid, inositol, and hexadecane are not assimilated. Ammonium sulfate, potassium nitrate (weak), sodium nitrite, L-lysine (weak), and cadaverine (latent and weak) are assimilated. Ethylamine is not assimilated. Optimal growth is at 17–25°C. Growth does not occur in vitamin-free medium. No starch-like substrate is produced. Growth does not occur on 50% (w/w) glucose-yeast extract agar Urease and Diazonium blue B reactions are positive.
Physiologically, G. cyclobalanopsidis differs from its closely related species G. pseudominor and G. minor in its inability to assimilate soluble starch, DL-lactic acid, and succinic acid (Table 2).
Typus: China, Gutianshan, Zhejiang province, obtained from a leaf of Cyclobalanopsis sp., June 2011, Qi-Ming Wang, holotype CGMCC 2.6419T preserved in a metabolically inactive state in the CGMCC, Beijing, China. Ex-type CBS 144918 is deposited at the CBS collection of the Westerdijk Fungal Biodiversity Institute, Utrecht, Netherlands.
Jamesdicksonia aceris Q.M. Wang, Y.Y. Li, M. Groenew. and M.M. Wang sp. nov. — MycoBank MB839779
Etymology: the specific epithet aceris refers to Acer, a plant host from which the type strain was isolated.
After 7 days at 17°C in YM broth, cells are allantoid to elongate, 0.8–1.8 × 7.0–11.8 μm and single, a sediment is produced, budding is polar, hyphae are narrow (1.2–2.5 μm; Figures 8C,E). After 1 month at 17°C, a film and sediment are produced. The streak culture is cream to cream to pink, butyrous, the surface is smooth or slightly wrinkled with glistening, and has an entire margin after 1 month at 17°C on YM agar. Hyphae are formed in Dalmau plate culture on cornmeal agar. Sexual structures are not produced on PDA, YM, CM, and V8 agars. Ballistoconidia are allantoid, falcate, 0.8–1.7 × 7.5–15.8 μm (Figures 8D,F).
Glucose is not fermented. Glucose, galactose (variable), sucrose, maltose, cellobiose, trehalose, lactose (variable), raffinose (or weak), melezitose (or weak), inulin (variable), soluble starch (variable), D-xylose (variable), L-arabinose, D-arabinose, D-ribose, L-rhamnose, glycerol, erythritol, ribitol, D-mannitol, D-glucitol, and succinic acid (variable) are assimilated. L-sorbose, melibiose, D-glucosamine, methanol, ethanol, galactitol, α-Methyl-D-glucoside, salicin, DL-lactic acid, citric acid, inositol, and hexadecane are not assimilated. Ammonium sulfate, potassium nitrate, L-lysine, ethylamine, and cadaverine are assimilated. Sodium nitrite is not assimilated. Optimal growth is at 17–25°C. Growth does occur in the vitamin-free medium. No starch-like substrate is produced. Growth does not occur on 50% (w/w) glucose-yeast extract agar Urease and Diazonium blue B reactions are positive.
Physiologically, J. aceris differs from its closely related species J. mali in its inability to assimilate melibiose, D-glucosamine, and sodium nitrite and its ability to use D-arabinose, ribitol, and cadaverine (Table 2).
Typus: China, Bomi, Tibet, obtained from a leaf of Acer pectinatum, September 2014, Qi-Ming Wang, holotype CGMCC 2.5602T preserved in a metabolically inactive state in the CGMCC, Beijing, China. Ex-type CBS 144916 is deposited at the CBS collection of the Westerdijk Fungal Biodiversity Institute, Utrecht, Netherlands. Bomi, Tibet, obtained from a leaf of the unidentified plant, September 2014, Qi-Ming Wang, paratype XZ156C4. Heilongjiang province, obtained from a leaf of an unidentified plant, Aug. 2015, Qi-Ming Wang, paratype CGMCC 2.5679. Jilin province, Changbai mountain, obtained from a leaf of the unidentified plant, Oct. 1998, Feng-Yan Bai, paratype CGMCC 2.2370.
Entylomatales (Exobasidiomycetes)
Tilletiopsis pinicola Q.M. Wang, Y.Y. Li, M. Groenew. and M.M. Wang sp. nov. — MycoBank 839596
Etymology: the specific epithet pinicola refers to Pinus plant host from which the type strain was isolated.
After 7 days at 17°C in YM broth, cells are cylindrical, 1.3–2.7 × 6.7–28.3 μm and single, a sediment is produced, budding is polar, and hyphae are narrow (1.2–2.5 μm; Figure 8G). After 1 month at 17°C, a film and sediment are produced. The streak culture is pale yellow, butyrous, the surface is glistening with slightly granular, and has an entire margin after 1 month at 17°C on YM agar. Pseudohyphae are formed in Dalmau plate culture on cornmeal agar. Sexual structures are not produced on PDA, YM, CM, and V8 agars. Ballistoconidia are lunate to sickle-shaped or cylindrical (0.8–1.3 × 8.3–19.1 μm; Figure 8H).
Glucose is not fermented. Glucose, galactose, sucrose, maltose, trehalose, melibiose (weak), raffinose, melezitose, D-xylose, L-arabinose, D-arabinose, D-ribose, ethanol, glycerol, erythritol, D-glucitol (weak), succinic acid (weak), and citric acid (weak) are assimilated. L-sorbose, cellobiose, lactose, inulin, soluble starch, L-rhamnose, D-glucosamine, N-Acetyl-D-glucosmine, methanol, ribitol, galactitol, D-mannitol, α-Methyl-D-glucoside, salicin, DL-lactic acid, inositol, and hexadecane are not assimilated. Ammonium sulfate, potassium nitrate, L-lysine, ethylamine, and cadaverine are assimilated. Sodium nitrite is not assimilated. Optimal growth is at 17–25°C. Growth does occur in the vitamin-free medium. No starch-like substrate is produced. Growth does not occur on 50% (w/w) glucose-yeast extract agar Urease and Diazonium blue B reactions are positive.
Physiologically, T. pinicola differs from its closely related species, T. lilacina, in its inability to assimilate cellobiose, soluble starch, ribitol, D-mannitol, α-Methyl-D-glucoside, DL-lactic acid, and sodium nitrite and its ability to assimilate ethanol and grow in the vitamin-free medium (Table 2).
Typus: China, Heilongjiang province, obtained from a leaf of Pinus sp., August 2015, Qi-Ming Wang, holotype CGMCC 2.5613T preserved in a metabolically inactive state in the CGMCC, Beijing, China. Ex-type CBS 15775 is deposited at the CBS collection of the Westerdijk Fungal Biodiversity Institute, Utrecht, Netherlands.
Tilletiopsis lunata Q.M. Wang, Y.Y. Li, M. Groenew. and M.M. Wang sp. nov. — MycoBank 839594
Etymology: the specific epithet, lunata, refers to the vegetative cell morphology of the type strain.
After 7 days at 17°C in YM broth, cells are cylindrical, 1.0–2.5 × 10.0–25.2 μm and single, a sediment is produced, budding is polar, and hyphae are narrow (1.2–2.5 μm; Figure 8I). After 1 month at 17°C, an easy-dispersed film and sediment are produced. The streak culture is pale-yellowish, butyrous, flat, and the surface is venous and has an entire margin after 1 month at 17°C on YM agar. Hyphae are formed in Dalmau plate culture on cornmeal agar. Sexual structures are not produced on PDA, YM, CM, and V8 agars. Ballistoconidia are lunate to sickle-shaped (0.8–2.1 × 8.3–21.6 μm; Figure 8J).
Glucose is not fermented. Glucose, galactose, sucrose, maltose, trehalose, raffinose, melezitose, inulin, soluble starch, D-xylose, L-arabinose, D-ribose, ethanol, glycerol, erythritol, ribitol (variable), D-mannitol, D-glucitol, succinic acid, and citric acid are assimilated. L-sorbose, cellobiose, lactose, D-arabinose, L-rhamnose, D-glucosamine, N-Acetyl-D-glucosmine, methanol, galactitol, α-Methyl-D-glucoside, salicin, D-glueonale, DL-lactic acid, inositol, and hexadecane are not assimilated. Ammonium sulfate, potassium nitrate, sodium nitrite (variable), L-lysine, and cadaverine are assimilated. Ethylamine is not assimilated. Optimal growth is at 17–25°C. Growth does occur in the vitamin-free medium. No starch-like substrate is produced. Growth does not occur on 50% (w/w) glucose-yeast extract agar Urease and Diazonium blue B reactions are positive.
Physiologically, T. lunata differs from its closely related species, T. washingtonensis, in its inability to assimilate D-arabinose, D-glueonale, and DL-lactic acid and its ability to use inulin, and its positive growth in the vitamin-free medium (Table 2).
Typus: China, Huzhong, Heilongjiang Province, obtained from a leaf of the unidentified plant, August 2015, Qi-Ming Wang, holotype CGMCC 2.6308T preserved in a metabolically inactive state in the CGMCC, Beijing, China. Ex-type DSM 111865 is deposited at the German Collection of Microorganisms and Cell Cultures, Braunschweig, Germany. Huzhong, Heilongjiang Province, obtained from a leaf of the unidentified plant, August 2015, Qi-Ming Wang, paratype CGMCC 2.6307.
Note: The two new species of Tilletiopsis and the known species, T. cremea and T. lilacina, are all isolated from plant leaves (Boekhout et al., 2011). T. washingtonensis seems to be a common inhabitant of the phyllosphere, but it also occurs in the soil, rotten wood, and tree bark (Supplementary Table 1).
Microstromatales (Exobasidiomycetes)
Jaminaea lantanae Q.M. Wang, Y.Y. Li, M. Groenew. and M.M. Wang sp. nov. — MycoBank 839590
Etymology: the specific epithet lantanae refers to Lantana plant host, from which the type strain was isolated.
After 7 days at 17°C in YM broth, cells are ovoid to elongate, ellipsoidal, 1.7–2.9 × 3.3–10.0 μm and single, a sediment is produced, budding is polar, and hyphae are narrow (1.2–2.5 μm; Figure 8K). After 1 month at 17°C, an easy-dispersed film and sediment are produced. The streak culture is pink, butyrous, flat, the surface is wrinkled and moist with glistening, and has an entire margin after 1 month at 17°C on YM agar. Hyphae are absent in Dalmau plate culture on cornmeal agar. Sexual structures are not produced on PDA, YM, CM, and V8 agars. Ballistoconidia are not observed.
Glucose is not fermented. Glucose, L-sorbose, sucrose (latent), maltose (latent and weak), cellobiose (latent and weak), melezitose (latent and weak), D-xylose (latent), L-arabinose, D-glucosamine (latent), N-Acetyl-D-glucosmine, ethanol, glycerol, erythritol, D-mannitol (latent), D-glucitol (latent and weak), α-Methyl-D-glucoside (latent and weak), and salicin (latent and weak) are assimilated. Galactose, trehalose, melibiose, raffinose, lactose, inulin, soluble starch, D-arabinose, D-Ribose, L-rhamnose, methanol, ribitol, galactitol, D-glueonale, DL-lactic acid, succinic acid, citric acid, inositol, and hexadecane are not assimilated. Ammonium sulfate, L-lysine, ethylamine, and cadaverine are assimilated. Potassium nitrate and sodium nitrite are not assimilated. Optimal growth is at 17–25°C. Growth does occur in the vitamin-free medium. No starch-like substrate is produced. Growth does not occur on 50% (w/w) glucose-yeast extract agar Urease and Diazonium blue B reactions are positive.
Typus: China, Haikou county, Hainan province, obtained from a leaf of Lantana camara, Aug. 2007, Qi-Ming Wang, holotype CGMCC 2.3529T preserved in a metabolically inactive state in the CGMCC, Beijing, China. Ex-type CBS 15493 is deposited at the CBS collection of the Westerdijk Fungal Biodiversity Institute, Utrecht, Netherlands. Haikou county, Hainan province, obtained from a leaf of Lantana camara, Aug. 2007, Qi-Ming Wang, paratype CGMCC 2.3622.
Note: J. angkorensis was isolated from fallen decaying leaves in Cambodia, J. lanaiensis from marine driftwood in Hawaiian, J. pallidilutea from plant material from mangrove in Iran, J. rosea from phylloplane of Plumeria in Florida (Sipiczki and Kajdacsi, 2009; Wei et al., 2011; Kijpornyongpan and Aime, 2017; Nasr et al., 2017). J. lantanae was isolated from the leaf of Lantana camara. The above data indicated that the members of Jaminaea are widespread along with plant associates.
Sympodiomycopsis europaea Q.M. Wang, Y.Y. Li, M. Groenew. and F.Y. Bai sp. nov. — MycoBank 839583
Etymology: the specific epithet, europaea, refers to the geographic origin of the type strain, Europe.
After 7 days at 17°C in YM broth, cells are or ellipsoidal to elongate, cylindrical, 1.5–1.8 × 4.2–12.5 μm and single, a sediment is produced, budding is polar, hyphae are narrow (1.2–2.5 μm; Figure 8L). After 1 month at 17°C, a ring and a sediment are produced. The streak culture is pink colored, butyrous, flat, and the surface is rachnoid with dull and has an entire margin after 1 month at 17°C on YM agar. Pseudohyphae are formed in Dalmau plate culture on cornmeal agar. Sexual structures are not produced on PDA, YM, CM, and V8 agars. Ballistoconidia are not observed.
Glucose is not fermented. Glucose, galactose, L-sorbose, sucrose, maltose, trehalose (latent and weak), lactose, melibiose, raffinose, melezitose, soluble starch (weak), D-xylose, L-arabinose (or latent and weak), D-ribose, ethanol, glycerol, erythritol, D-mannitol, D-glucitol (or latent and weak), and α-Methyl-D-glucoside (or latent and weak) are assimilated. Cellobiose, inulin, D-arabinose, L-rhamnose, D-glucosamine, methanol, ribitol, galactitol, salicin, DL-lactic acid, succinic acid (or weak), citric acid (or latent and weak), inositol (or weak), and hexadecane are not assimilated. Ammonium sulfate, potassium nitrate (variable), L-lysine (weak), and cadaverine are assimilated. Sodium nitrite and ethylamine are not assimilated. Optimal growth is at 17–25°C. Growth does occur in the vitamin-free medium. No starch-like substrate is produced. Growth does not occur on 50% (w/w) glucose-yeast extract agar Urease and Diazonium blue B reactions are positive.
Physiologically, S. europaea differs from its closely related species, S. kandeliae and S. paphiopedili, in its inability to assimilate cellobiose, D-arabinose, and grow in the 50% glucose medium (Table 2).
Typus: Germany, obtained from a leaf of the unidentified plant, March 2006, Feng-Yan Bai, holotype CGMCC 2.3119T preserved in a metabolically inactive state in the CGMCC, Beijing, China. Ex-type CBS 15470 is deposited at the CBS collection of the Westerdijk Fungal Biodiversity Institute, Utrecht, Netherlands; Obtained from a leaf of the unidentified plant, Mar. 2006, Feng-Yan Bai, paratypes CGMCC 2.3181, CGMCC 2.3120, CGMCC 2.3121, CGMCC 2.3122, CGMCC 2.3123 and CGMCC 2.3124.
Note: The species of Sympodiomycopsis seem to associate with plant substrate. S. paphiopedili was isolated from the nectar of Paphiopedilum primulinum, S. kandeliae from flowers of Kandelia candel in the mangrove forest, S. yantaiensis from insect frass from the trunk of Sophora japonica, and the newly described species S. europaea also from the leaf of plant.
Baueromyces Q.M. Wang, D. Begerow, and M. Groenew. gen. nov. — MycoBank 839577
Etymology: the genus is named in honor of Robert Bauer for his pioneering work on the taxonomy of smuts.
This genus is proposed for the branch represented by strain CGMCC 2.4532, which formed a separate branch from the genera in the Microstromatales (Exobasidiomycetes). The genus is mainly circumscribed by the phylogenetic analysis of the six-gene sequences (Figure 1).
Sexual reproduction not known. Colonies are butyrous, pink, and have smooth margins. Budding cells are present. Ballistoconidia are not produced. Hyphae are not formed.
Type species: Baueromyces planticola Q.M. Wang, D. Begerow and M. Groenew.
Baueromyces planticola Q.M. Wang, D. Begerow, and M. Groenew. sp. nov. — MycoBank 839597
Etymology: the specific epithet, planticola, refers to the substrate origin of the type strain, plant.
After 7 days at 17°C in YM broth, cells are cylindrical, 1.7–2.5 × 4.2–10.0 μm and single, a sediment is produced, budding is polar, hyphae are narrow (1.2–2.5 μm; Figure 8M). After 1 month at 17°C, a film or ring and a sediment are produced. The streak culture is cream to pink, butyrous, flat, and the surface is smooth or slightly wrinkled with glistening and has an entire margin after 1 month at 17°C on YM agar. Pseudohyphae and Hyphae are absent in Dalmau plate culture on cornmeal agar. Sexual structures are not produced on PDA, YM, CM, and V8 agars. Ballistoconidia are not observed.
Glucose is not fermented. Glucose, galactose, sucrose, maltose (week), cellobiose, trehalose (latent and weak), lactose (weak), melibiose (variable), raffinose, melezitose (variable), inulin, D-xylose (weak), L-arabinose, D-ribose (variable), L-rhamnose (variable), N-Acetyl-D-glucosmine (variable), glycerol, erythritol (latent and weak), D-mannitol, and D-glucitol are assimilated. L-sorbose, soluble starch, D-arabinose, D-glucosamine, methanol, ethanol (or weak), ribitol, galactitol, α-Methyl-D-glucoside (weak), salicin (or latent and weak), DL-lactic acid, succinic acid (or weak), citric acid, inositol (or latent and weak), and hexadecane are not assimilated. Ammonium sulfate, potassium nitrate (or weak), L-lysine (or weak), ethylamine (or weak), and cadaverine (or weak) are assimilated. Sodium nitrite (or latent and weak) is not assimilated. The maximum growth temperature is 30–32°C. Growth does occur in the vitamin-free medium. No starch-like substrate is produced. Growth does not occur on 50% (w/w) glucose-yeast extract agar Urease and Diazonium blue B reactions are positive.
Typus: China, Xingshan County, Hubei province, obtained from a leaf of the unidentified plant, March 2012, Qi-Ming Wang, holotype CGMCC 2.4532T preserved in a metabolically inactive state in the CGMCC, Beijing, China. Ex-type CBS 144909 is deposited at the CBS collection of the Westerdijk Fungal Biodiversity Institute, Utrecht, Netherlands. Maotai county, Guizhou province, obtained from a leaf of the unidentified plant, March 2012, Qi-Ming Wang, CGMCC 2.4534. Fanjingshan, Guizhou province, obtained from a leaf of the unidentified plant, March 2012, Qi-Ming Wang, paratypes CGMCC 2.4535 and CGMCC 2.4536.
Franziozymales Q.M. Wang, D. Begerow, and M. Groenew. ord. nov. — MycoBank 839576
Member of Exobasidiomycetes. The diagnosis of the order Franziozymales is based on the description of the genus Franziozyma. The nomenclature of the order is based on the genus Franziozyma.
Type family: Franziozymaceae Q.M. Wang, D. Begerow, M. Groenew.
Franziozymaceae Q.M. Wang, D. Begerow and M. Groenew. fam. nov. — MycoBank 839575
Member of Franziozymales (Exobasidiomycetes). The diagnosis of the family Franziozymaceae is based on the description of the genus, Franziozyma. The nomenclature of the family is based on the genus.
Type genus: Franziozyma Q.M. Wang, D. Begerow and M. Groenew.
Genus accepted: Franziozyma Q.M. Wang, D. Begerow and M. Groenew.
Franziozyma Q.M. Wang, D. Begerow, and M. Groenew. gen. nov. — MycoBank 839574
Etymology: the genus is named in honor of Franz Oberwinkler for his pioneering work on the taxonomy of smuts.
This genus is proposed for the branch represented by strain XZ4C4T, which formed a separate branch from Golubeviales and other orders in Exobasidiomycetes. The genus is mainly circumscribed by the phylogenetic analysis of the six loci dataset (Figure 1).
Sexual reproduction is not known. Colonies are butyrous, white, margin, or eroded. Budding cells present or not. Hyphae are formed. Ballistoconidia are produced.
Type species: Franziozyma bambusicola Q.M. Wang, D. Begerow and M. Groenew.
Franziozyma bambusicola Q.M. Wang, D. Begerow and M. Groenew. sp. nov. — MycoBank 839573
Etymology: the specific epithet bambusicola refers to the origin of the substrate of the type strain of this species.
After 10 days at 17°C on 5% malt extract agar, colonies are cream, soft or tough, usually glabrous, or sometimes pubescent, shiny or dull, ridged, and with an eroded margin. Hyphae are narrow and cylindrical, usually 1.0–1.3 × 30–60 μm, sometimes regularly branched (Figure 8N). Chlamydospores occur intercalarily or terminally and are single, subglobose or ellipsoidal, 4.0–6.0 × 6–10 μm (Figure 8O). Sexual structures are not produced on YM, YPD, V8, and CM agars. Ballistoconidia are ellipsoidal orallantoid, 2.3–2.7 × 13.6–18.2 μm (Figure 8P).
Glucose is not fermented. Glucose, sucrose, maltose, cellobiose, trehalose, raffinose, inulin, soluble starch, glycerol, erythritol, D-mannitol, and D-glucitol are assimilated. Galactose, L-sorbose, lactose, melibiose, melezitose, D-xylose, L-arabinose, D-arabinose, D-ribose, L-rhamnose, D-glucosamine, methanol, ethanol, ribitol, galactitol, α-Methyl-D-glucoside, salicin, DL-lactate, and succinate, citrate, myo-inositol, and hexadecane are not assimilated. The maximum growth temperature is 22°C. Growth in the vitamin-free medium is weak. No starch-like substrate is produced. Growth does not occur on 50% (w/w) glucose-yeast extract agar Urease and Diazonium blue B reactions are positive.
Typus: China, Bomi County, Tibet, obtained from a leaf of bamboo, Sep. 2004, Qi-Ming Wang, holotype CGMCC 2.2620T preserved in a metabolically inactive state in the CGMCC, Beijing, China. Ex-type CBS 15774 is deposited at the CBS collection of the Westerdijk Fungal Biodiversity Institute, Utrecht, Netherlands. Bomi county, Tibet, obtained from a leaf of bamboo, September 2004, Qi-Ming Wang, paratype XZ4A1.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author Contributions
Q-MW conceived and designed the project. Q-MW, Y-YL, Y-TG, and FW performed sampling and yeast isolation. Y-YL, A-HL, M-MW, and Q-MW performed phenotypic characterization and analyzed the molecular data. B-QZ registered the taxa in MycoBank submitted the sequence data in TreeBASE. Q-MW, M-MW, MG, and DB wrote the manuscript. ET and F-YB revised the manuscript. ET supported the sequences generated in his laboratory. All authors contributed to the article and approved the submitted version.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We thank Prof. Long Wang for his advice on nomenclatural matters and help to improve the English language. We also thank the reviewers for their invaluable comments and suggestions.
Funding
This study was supported by grants Nos. 31961133020 and 31570016 from the National Natural Science Foundation of China (NSFC), No. 2021FY100905 from the Ministry of Science and Technology of China, No. BE 2201/28-1 from the Deutsche Forschungsgemeinschaft of Germany (DFG), and No. G-2019-1-012 from Institute for Fermentation, Osaka (IFO).
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2021.777338/full#supplementary-material
Phylogeny of new yeast or yeast-like species in the Exobasidiomycetes inferred from the sequences of the LSU rDNA D1/D2 domains and ITS region (including 5.8S rDNA) by maximum likelihood analysis and over 50% from 1,000 bootstrap replicates is shown. Bar = 0.1 substitutions per nucleotide position. The compressed genera are monophyletic, the species in those clades were listed in Table 1 and Supplementary Table 2.
Phylogeny of new yeast or yeast-like species in the Exobasidiomycetes inferred from the sequences of the LSU rDNA D1/D2 domains by maximum likelihood analysis and over 50% from 1,000 bootstrap replicates is shown. Bar = 0.05 and 0.02 substitutions per nucleotide position. The compressed genera are monophyletic, the species in those clades were listed in Table 1 and Supplementary Table 2.
Phylogeny of new yeast or yeast-like species in the Exobasidiomycetes inferred from the sequences of the LSU rDNA D1/D2 domains by maximum-likelihood analysis and over 50% from 1000 bootstrap replicates is shown. Bar = 0.05 and 0.02 substitutions per nucleotide position. The compressed genera are monophyletic, the species in those clades were listed in Table 1 and Supplementary Table 2.
References
- Agata W., Marta P. S. (2018). Insight into Mycosarcoma maydis (Ustilago maydis) – benefits and harmful effects of the phytopathogenic fungus for humans. Biomed. J. Sci. Technol. Res. 4 3746–3748. 10.26717/BJSTR.2018.04.001005 [DOI] [Google Scholar]
- Albu S., Toome M., Aime M. C. (2015). Violaceomyces palustris gen. et sp. nov. and a new monotypic lineage, Violaceomycetales ord. nov. in Ustilaginomycetes. Mycologia 107 1193–1204. 10.3852/14-260 [DOI] [PubMed] [Google Scholar]
- Alqurashi A. S., Kerrigan J., Savchenko K. G. (2021). Morphological and molecular characterization of Langdonia walkerae sp. nov. infecting Aristida stricta and A. beyrichiana in long leaf pine-grass land ecosystems in the southeastern USA. Fungal Syst. Evol. 8 39–47. 10.3114/fuse.2021.08.04 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauer R., Begerow D., Oberwinkler F., Piepenbring M., Berbee M. L. (2001). “Ustilaginomycetes,” in The Mycota, VII, Part B: Systematics and Evolution, eds McLaughlin D. J., McLaughlin E. G., Lemke P. A. (Berlin: Springer-Verlag; ), 57–83. 10.12664/mycobiota.2013.01.06 [DOI] [Google Scholar]
- Begerow D., Bauer R., Boekhout T. (2000). Phylogenetic placements of ustilaginomycetous anamorphs as deduced from nuclear LSU rDNA sequences. Mycol. Res. 104 53–60. 10.1017/S0953756299001161 [DOI] [Google Scholar]
- Begerow D., Kemler M., Feige A., Yurkov A. (2017). “Parasitism in yeasts,” in Yeasts in Natural Ecosystems: Ecology, eds Buzzini P., Lachance M. A., Yurkov A. (Cham: Springer; ), 179–210. 10.1007/978-3-319-61575-2_7 [DOI] [Google Scholar]
- Begerow D., Schäfer A. M., Kellner R., Yurkov A., Kemler M., Oberwinkler F., et al. (2014). “Ustilaginomycotina,” in The Mycota, Vol. VII, Part A: Systematics and Evolution, 2nd Edn, eds McLaughlin D. J., Spatafora J. W. (Berlin: Springer-Verlag; ), 295–329. 10.1007/978-3-642-55318-9_11 [DOI] [Google Scholar]
- Begerow D., Stoll M., Bauer R. (2006). A phylogenetic hypothesis of Ustilaginomycotina based on multiple gene analyses and morphological data. Mycologia 98 906–916. 10.1080/15572536.2006.11832620 [DOI] [PubMed] [Google Scholar]
- Boekhout T. (1991). A revision of ballistoconidia-forming yeasts and fungi. Stud. Mycol. 33 1–194. 10.1016/j.simyco.2015.10.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boekhout T. (1995). Pseudozyma Bandoni emend. Boekhout, a genus for yeast-like anamorphs of Ustilaginales. J. Gen. Appl. Microbiol. 41 359–366. 10.2323/jgam.41.359 [DOI] [Google Scholar]
- Boekhout T. (2011). “Tilletiopsis Derx ex Derx (1930),” in The Yeasts, A Taxonomic Study, 5th Edn. eds Kurtzman C. P., Fell J. W., Boekhout T. (Amsterdam: Elsevier; ), 2003–2014. 10.1016/B978-0-444-52149-1.00160-9 [DOI] [Google Scholar]
- Boekhout T., Aime M. C., Begerow D., Gabaldón T., Heitman J., Kemler M., et al. (2021). The evolving species concepts used for yeasts: from phenotypes and genomes to speciation networks. Fungal Diver. 109 27–55. 10.1007/s13225-021-00475-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boekhout T., Fell J. W., O’Donnell K. (1995). Molecular systematics of some yeast-like anamorphs belonging to the Ustilaginales and Tilletiales. Stud. Mycol. 38 175–183. [Google Scholar]
- Boekhout T., Fonseca I., Sampaio J. P., Bandoni R. J., Fell J. W., Kwon-Chung K. J. (2011). “Discussion of teleomorphic and anamorphic basidiomycetous yeasts,” in The Yeasts, A Taxonomic Study, 5th Edn. eds Kurtzman C. P., Fell J. W., Boekhout T. (Amsterdam: Elsevier; ), 1339–1372. 10.1016/B978-0-444-52149-1.00100-2 [DOI] [Google Scholar]
- Boekhout T., Theelen B., Houbraken J., Robert V., Scorzetti G., Gafni A., et al. (2003). Novel anamorphic mite-associated fungi belonging to the Ustilaginomycetes: Meira geulakonigii gen. nov., sp. nov., Meira argovae sp. nov. and Acaromyces ingoldii gen. nov., sp. nov. Int. J. Syst. Evol. Microbiol. 53 1655–1664. 10.1099/ijs.0.02434-0 [DOI] [PubMed] [Google Scholar]
- Cao Y., Pu-Dong L. I., Zhao J., Wang H. K., Wang Q. (2018). Morph-molecular characterization of Meira nicotianae sp. nov. a novel basidiomycetous, anamorphic yeast-like fungus associated with growth improvement in tobacco plant. Phytotaxa 365 169–181. 10.11646/phytotaxa.365.2.4 [DOI] [Google Scholar]
- Chen L., Zhang L., Li Z. H., Hui F. L. (2013). Sympodiomycopsis yantaiensis sp. nov., a basidiomycetous yeast isolated from insect frass. Int. J. Syst. Evol. Microbiol. 63 3501–3505. 10.1099/ijs.0.053686-0 [DOI] [PubMed] [Google Scholar]
- Crotzer V., Levetin E. (1996). The aerobiological significance of smut spores in Tulsa, Oklahoma. Aerobiologia 12 177–184. 10.1007/BF02248147 [DOI] [Google Scholar]
- Cunningham J. L., Bakshi B. K., Lentz P. L., Gilliam M. S. (1976). Two new genera of leaf parasitic fungi (Basidiomycetidae: Brachybasidiaceae). Mycologia 68 640–654. 10.1080/00275514.1976.12019948 [DOI] [Google Scholar]
- Fell J. W., Boekhout T., Fonseca Á, Scorzetti G., Statzell-Tallman A. (2000). Biodiversity and systematics of basidiomycetous yeasts as determined by large-subunit rDNA D1/D2 domain sequence analysis. Int. J. Syst. Evol. Microbiol. 50 1351–1372. 10.1099/00207713-50-3-1351 [DOI] [PubMed] [Google Scholar]
- Gerson U., Gafni A., Paz Z., Sztejnberg A. (2008). A tale of three acaropathogenic fungi in Israel: Hirsutella, Meira and Acaromyces. Exp. Appl. Acarol. 46 183–194. 10.1007/s10493-008-9202-6 [DOI] [PubMed] [Google Scholar]
- Gokhale A. A. (1972). Studies on the genus Tilletiopsis. Nova Hedwigia 23 795–809. [Google Scholar]
- Hibbett D. S., Binder M., Bischoff J. F., Blackwell M., Cannon P. F., Eriksson O. E., et al. (2007). A higher-level phylogenetic classification of the fungi. Mycol. Res. 111 509–547. 10.1016/j.mycres.2007.03.004 [DOI] [PubMed] [Google Scholar]
- Inácio J., Landell M. F., Valente P., Wang P. H., Wang Y. T., Yang S. H., et al. (2008). Farysizyma gen. nov., an anamorphic genus in the Ustilaginales to accommodate three novel epiphytic basidiomycetous yeast species from America, Europe and Asia. FEMS Yeast Res. 8 499–508. 10.1111/j.1567-1364.2008.00377.x [DOI] [PubMed] [Google Scholar]
- Ingold C. T. (1985). Dicellomyces scirpi: its conidial stage and taxonomic position. Trans. Br. Mycol. Soc. 84 542–545. 10.1016/S0007-1536(85)80021-8 [DOI] [Google Scholar]
- Katoh K., Standley D. M. (2013). MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 30 772–780. 10.1093/molbev/mst010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kijpornyongpan T., Aime M. C. (2017). Taxonomic revisions in the Microstromatales: two new yeast species, two new genera, and validation of Jaminaea and two Sympodiomycopsis species. Mycol. Prog. 16 495–505. 10.1007/s11557-017-1276-2 [DOI] [Google Scholar]
- Kruse J., Doehlemann G., Kemen E., Thines M. (2017). Asexual and sexual morphs of Moesziomyces revisited. IMA Fungus 8 117–129. 10.5598/imafungus [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S., Stecher G., Tamura K. (2016). MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33 1870–1874. 10.1093/molbev/msw054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurtzman C. P. (2014). Use of gene sequence analyses and genome comparisons for yeast systematics. Int. J. Syst. Evol. Microbiol. 64 325–332. 10.1099/ijs.0.054197-0 [DOI] [PubMed] [Google Scholar]
- Kurtzman C. P. (2015). Identification of food and beverage spoilage yeasts from DNA sequence analyses. Int. J. Food Microbiol. 213 71–78. 10.1016/j.ijfoodmicro.2015.05.023 [DOI] [PubMed] [Google Scholar]
- Kurtzman C. P., Fell J. W. (2006). “Yeast systematics and phylogeny–implications of molecular identification methods for studies in ecology,” in Biodiversity and Ecophysiology of Yeasts, eds Lachance M. A., Kurtzman C. P., Fell J. W. (Berlin: Springer; ), 11–30. 10.1007/3-540-30985-3_2 [DOI] [Google Scholar]
- Kurtzman C. P., Fell J. W., Boekhout T., Robert V. (2011). “Methods for isolation, phenotypic characterization and maintenance of yeasts,” in The Yeasts, A Taxonomic Study, 5th Edn. eds Kurtzman C. P., Fell J. W., Boekhout T. (Amsterdam: Elsevier; ), 87–110. 10.1016/b978-0-444-52149-1.00007-0 [DOI] [Google Scholar]
- Kurtzman C. P., Mateo R. Q., Kolecka A., Theelen B., Robert V., Boekhout T. (2015). Advances in yeast systematics and phylogeny and their use as predictors of biotechnologically important metabolic pathways. FEMS Yeast Res. 15:fov050. 10.1093/femsyr/fov050 [DOI] [PubMed] [Google Scholar]
- Leeming J. P., Notman F. H. (1987). Improved methods for isolation and enumeration of Malassezia furfur from human skin. J. Clin. Microbiol. 25 2017–2019. 10.1128/jcm.25.10.2017-2019.1987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li A. H., Yuan F. X., Groenewald M., Bensch K., Yurkov A. M., Li K., et al. (2020). Diversity and phylogeny of basidiomycetous yeasts from plant leaves and soil: proposal of two new orders, three new families, eight new genera and one hundred and seven new species. Stud. Mycol. 96 17–140. 10.1016/j.simyco.2020.01.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y. M., Shivas R. G., Li B. J., Cai L. (2019). Diversity of Moesziomyces (Ustilaginales, Ustilaginomycotina) on Echinochloa and Leersia (Poaceae). Mycokeys 52 1–16. 10.3897/mycokeys.52.30461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Limtong S., Polburee P., Chamnanpa T., Khunnamwong P., Limtong P. (2017). Meira siamensis sp. nov., a novel anamorphic ustilaginomycetous yeast species isolated from the vetiver grass phylloplane. Int. J. Syst. Evol. 67 2418–2422. 10.1099/ijsem.0.001969 [DOI] [PubMed] [Google Scholar]
- McNeil J. C., Palazzi D. L. (2012). Ustilago as a cause of fungal peritonitis: case report and review of the literature. J. Pediatr. Infect. Dis. Soc. 1 337–339. 10.1093/jpids/pis043 [DOI] [PubMed] [Google Scholar]
- McTaggart A. R., Shivas R. G., Geering A. D. W., Callaghan B., Vánky K., Scharaschkin T. (2012a). Soral synapomorphies are significant for the systematics of the Ustilago-Sporisorium-Macalpinomyces complex (Ustilaginaceae). Persoonia 29 63–77. 10.3767/003158512X660562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McTaggart A. R., Shivas R. G., Geering A. D. W., Vánky K., Scharaschkin T. (2012b). Taxonomic revision of Ustilago, Sporisorium and Macalpinomyces. Persoonia 29 116–132. 10.3767/003158512X661462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakase T., Takashima M. (1993). A simple procedure for the high frequency isolation of new taxa of ballistosporous yeasts living on the surfaces of plants. RIKEN Rev. 3 33–34. [Google Scholar]
- Nasr S., Lutz M., Amoozegar M. A., Eparvier V., Stien D., Fazeli S. A. S., et al. (2019). Graphiola fimbriata: the first species of Graphiolaceae (Exobasidiales, Basidiomycota) described only based on its yeast stage. Mycol. Prog. 18 359–368. 10.1007/s11557-018-1450-1 [DOI] [Google Scholar]
- Nasr S., Mohammadimehr M., Geranpayeh V. M., Amoozegar M. A., Shahzadeh F. S. A., Yurkov A. (2017). Jaminaea pallidilutea sp. nov. (Microstromatales), a basidiomycetous yeast isolated from plant material of mangrove forests in Iran. Int. J. Syst. Evol. Microbiol. 67 4405–4408. 10.1099/ijsem.0.002302 [DOI] [PubMed] [Google Scholar]
- Nasr S., Soudi M. R., Fazeli S. A. S., Nguyen H. D. T., Lutz M., Piątek M. (2014). Expanding evolutionary diversity in the Ustilaginomycotina: Fereydouniaceae fam. nov. and Fereydounia gen. nov., the first urocystidalean yeast lineage. Mycol. Prog. 13 1217–1226. 10.1007/s11557-014-1012-0 [DOI] [Google Scholar]
- Oliveira J. V. C., Borges T. A., Corrêa D. S. R. A., Freitas L. F. D., Rosa C. A., Goldman G. H., et al. (2014). Pseudozyma brasiliensis sp. nov., a xylanolytic, ustilaginomycetous yeast species isolated from an insect pest of sugarcane roots. Int. J. Syst. Evol. Microbiol. 64 2159–2168. 10.1099/ijs.0.060103-0 [DOI] [PubMed] [Google Scholar]
- Parmasto E. (1968). A new species and a new family of the Aphyllophorales. Eesti NSV Tead. Akad. Toim. 17 223–228. [Google Scholar]
- Patel R., Roberts G. D., Kelly D. G., Walker R. C. (1995). Central venous catheter infection due to Ustilago species. Clin. Infect. Dis. 21 1043–1044. 10.1093/clinids/21.4.1043 [DOI] [PubMed] [Google Scholar]
- Paz Z., Burdman S., Gerson U., Sztejnberg A. (2007a). Antagonistic effects of the endophytic fungus Meira geulakonigii on the citrus rust mite Phyllocoptruta oleivora. J. Appl. Microbiol. 103 2570–2579. 10.1111/j.1365-2672.2007.03512.x [DOI] [PubMed] [Google Scholar]
- Paz Z., Gerson U., Sztejnberg A. (2007b). Assaying three new fungi against citrus mites in the laboratory, and a field trial. Biocontrol 52 855–862. 10.1007/s10526-006-9060-2 [DOI] [Google Scholar]
- Peraica M., Richter D., Rašić D. (2014). Mycotoxicoses in children. Arh. Hig. Rada Toksikol. 65 347–363. 10.2478/10004-1254-65-2014-2557 [DOI] [PubMed] [Google Scholar]
- Piepenbring M., Hartmann M., Hofmann T. A., Lutz M. (2020). Two new species in a new genus and a critical revision of Brachybasidiaceae (Exobasidiales, Basidiomycota) in honor of Franz Oberwinkler. Mycol. Prog. 19 351–365. 10.1007/s11557-020-01564-w [DOI] [Google Scholar]
- Reid D. A. (1976). Dicellomyces scirpi (Basidiomycetes) new to Britain. Trans. Br. Mycol. Soc. 66 536–538. 10.1016/S0007-1536(76)80232-X [DOI] [Google Scholar]
- Richter C., Yurkov A. M., Boekhout T., Stadler M. (2019). Diversity of Tilletiopsis-like fungi in Exobasidiomycetes (Ustilaginomycotina) and description of six novel species. Front. Microbiol. 10:2544. 10.3389/fmicb.2019.02544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronquist F., Teslenko M., van der Mark P., Ayres D. L., Darling A., Höhna S., et al. (2012). MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 61 539–542. 10.1093/sysbio/sys029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rush T. A., Aime M. C. (2013). The genus Meira: phylogenetic placement and description of a new species. Antonie van Leeuwenhoek 103 1097–1106. 10.1007/s10482-013-9889-1 [DOI] [PubMed] [Google Scholar]
- Scorzetti G., Fell J. W., Fonseca A., Statzell-Tallman A. (2002). Systematics of basidiomycetous yeasts: a comparison of large subunit D1/D2 and internal transcribed spacer rDNA regions. FEMS Yeast Res. 2 495–517. 10.1111/j.1567-1364.2002.tb00117 [DOI] [PubMed] [Google Scholar]
- Sipiczki M., Kajdacsi E. (2009). Jaminaea angkorensis gen. nov., sp. nov., a novel anamorphic fungus containing an S943 nuclear small–subunit rRNA group IB intron represents a basal branch of Microstromatales. Int. J. Syst. Evol. Microbiol. 59 914–920. 10.1099/ijs.0.003939-0 [DOI] [PubMed] [Google Scholar]
- Stamatakis A. (2006). RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22 2688–2690. 10.1093/bioinformatics/btl446 [DOI] [PubMed] [Google Scholar]
- Stolk A. C., Dakin J. C. (1966). Moniliella, a new genus of Moniliales. Antonie Van Leeuwenhoek 32 399–409. 10.1007/BF02097491 [DOI] [PubMed] [Google Scholar]
- Stoll M., Begerow D., Oberwinkler F. (2005). Molecular phylogeny of Ustilago, Sporisorium, and related taxa based on combined analyses of rDNA sequences. Mycol. Res. 109 342–356. 10.1017/S0953756204002229 [DOI] [PubMed] [Google Scholar]
- Stoll M., Piepenbring M., Begerow D., Oberwinkler F. (2003). Molecular phylogeny of Ustilago and Sporisorium species (Basidiomycota, Ustilaginales) based on internal transcribed spacer (ITS) sequences. Can. J. Bot. 81 976–984. 10.1556/AVet.48.2000.1.10 [DOI] [PubMed] [Google Scholar]
- Sugiyama J., Tokuoka K., Suh S. O., Hirata A., Komagata K. (1991). Sympodiomycopsis: a new yeast-like anamorph genus with basidiomycetous nature from orchid nectar. Antonie Van Leeuwenhoek 59 95–108. 10.1007/BF00445653 [DOI] [PubMed] [Google Scholar]
- Sun L. Y., Sun X., Guo L. D. (2018). Capitulocladosporium clinodiplosidis gen. et sp. nov., a hyphomyceteous ustilaginomycete from midge. Mycol. Prog. 17 307–318. 10.1007/s11557-017-1352-7 [DOI] [Google Scholar]
- Sztejnberg A., Paz Z., Boekhout T., Gafni A., Gerson U. (2004). A new fungus with dual biocontrol capabilities: reducing the numbers of phytophagous mites and powdery mildew disease damage. Crop Prot. 23 1125–1129. 10.1016/j.cropro.2004.04.004 [DOI] [Google Scholar]
- Tan Y. P., Bishop-Hurley, Shivas R. G. (2021). Gjaerumia marneyi Y.P. Tan, Bishop-Hurley and R.G. Shivas, sp. nov. Index Fungorum 495:2. [Google Scholar]
- Tanaka E., Honda Y. (2017). Teleomorph–anamorph connection of Macalpinomyces spermophorus with Pseudozyma tsukubaensis and corresponding erythritol production. Mycoscience 58 445–451. 10.1016/j.myc.2017.06.006 [DOI] [Google Scholar]
- Tanaka E., Koitabashi M., Kitamoto H. (2019). A teleomorph of the ustilaginalean yeast Moesziomyces antarcticus on barnyard grass in Japan provides bioresources that degrade biodegradable plastics. Antonie Van Leeuwenhoek 112 599–614. 10.1007/s10482-018-1190-x [DOI] [PubMed] [Google Scholar]
- Tanaka E., Shimizu K., Imanishi Y., Yasuda F., Tanaka C. (2008). Isolation of basidiomycetous anamorphic yeast-like fungus Meira argovae found on Japanese bamboo. Mycoscience 49 329–333. 10.1007/s10267-008-0429-1 [DOI] [Google Scholar]
- Teo L. H., Tay Y. K. (2006). Ustilago species infection in humans. Br. J. Dermatol. 155 1096–1097. 10.1111/j.1365-2133.2006.07515.x [DOI] [PubMed] [Google Scholar]
- Vánky K. (2012). Smut Fungi of the World. St. Paul, MN: APS. [Google Scholar]
- Vu D., Groenewald M., de Vries M., Gehrmann T., Stielow B., Eberhardt U., et al. (2019). Large-scale generation and analysis of filamentous fungal DNA barcodes boosts coverage for kingdom fungi and reveals thresholds for fungal species and higher taxon delimitation. Stud. Mycol. 92 135–154. 10.1016/j.simyco.2018.05.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vu D., Groenewald M., Szöke S., Cardinali G., Eberhardt U., Stielow B., et al. (2016). DNA barcoding analysis of more than 9000 yeast isolates contributes to quantitative thresholds for yeast species and genera delimitation. Stud. Mycol. 85 91–105. 10.1016/j.simyco.2016.11.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q. M., Bai F. Y. (2004). Four new yeast species of the genus Sporobolomyces from plant leaves. FEMS Yeast Res. 4 579–586. 10.1016/j.femsyr.2003.11.002 [DOI] [PubMed] [Google Scholar]
- Wang Q. M., Bai F. Y. (2008). Molecular phylogeny of basidiomycetous yeasts in the Cryptococcus luteolus lineage (Tremellales) based on nuclear rDNA and mitochondrial cytochrome b gene sequence analyses: proposal of Derxomyces gen. nov. and Hannaella gen. nov., and description of eight novel Derxomyces species. FEMS Yeast Res. 8 799–814. 10.1111/j.1567-1364.2008.00403.x [DOI] [PubMed] [Google Scholar]
- Wang Q. M., Bai F. Y., Zhao J. H., Jia J. H. (2003). Bensingtonia changbaiensis sp. nov. and Bensingtonia sorbi sp. nov., novel ballistoconidium-forming yeast species from plant leaves. Int. J. Syst. Evol. Microbiol. 53 2085–2089. 10.1099/ijs.0.02736-0 [DOI] [PubMed] [Google Scholar]
- Wang Q. M., Begerow D., Groenewald M., Liu X. Z., Theelen B., Bai F. Y., et al. (2015). Multigene phylogeny and taxonomic revision of yeasts and related fungi in the Ustilaginomycotina. Stud. Mycol. 81 55–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q. M., Theelen B., Groenewald M., Bai F. Y., Boekhout T. (2014). Moniliellomycetes and Malasseziomycetes, two new classes in Ustilaginomycotina. Persoonia 33 41–47. 10.3767/003158514X682313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei Y. H., Liou G. Y., Liu H. Y., Lee F. L. (2011). Sympodiomycopsis kandeliae sp. nov., a basidiomycetous anamorphic fungus from mangroves, and reclassification of Sympodiomycopsis lanaiensis as Jaminaea lanaiensis comb. nov. Int. J. Syst. Evol. Microbiol. 61 469–473. 10.1099/ijs.0.021865-0 [DOI] [PubMed] [Google Scholar]
- Yasuda F., Izawa H., Yamagishi D., Akamatsu H., Kodama M., Otani H. (2006). Meira nashicola sp. nov., a novel basidiomycetous, anamorphic yeastlike fungus isolated from Japanese pear fruit with reddish stain. Mycoscience 47 36–40. 10.1007/s10267-005-0266-4 [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Phylogeny of new yeast or yeast-like species in the Exobasidiomycetes inferred from the sequences of the LSU rDNA D1/D2 domains and ITS region (including 5.8S rDNA) by maximum likelihood analysis and over 50% from 1,000 bootstrap replicates is shown. Bar = 0.1 substitutions per nucleotide position. The compressed genera are monophyletic, the species in those clades were listed in Table 1 and Supplementary Table 2.
Phylogeny of new yeast or yeast-like species in the Exobasidiomycetes inferred from the sequences of the LSU rDNA D1/D2 domains by maximum likelihood analysis and over 50% from 1,000 bootstrap replicates is shown. Bar = 0.05 and 0.02 substitutions per nucleotide position. The compressed genera are monophyletic, the species in those clades were listed in Table 1 and Supplementary Table 2.
Phylogeny of new yeast or yeast-like species in the Exobasidiomycetes inferred from the sequences of the LSU rDNA D1/D2 domains by maximum-likelihood analysis and over 50% from 1000 bootstrap replicates is shown. Bar = 0.05 and 0.02 substitutions per nucleotide position. The compressed genera are monophyletic, the species in those clades were listed in Table 1 and Supplementary Table 2.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.