Abstract
The Mre11 complex has been implicated in diverse aspects of the cellular response to DNA damage. We used in situ fractionation of human fibroblasts to carry out cytologic analysis of Mre11 complex proteins in the double-strand break (DSB) response. In situ fractionation removes most nucleoplasmic protein, permitting immunofluorescent localization of proteins that become more avidly bound to nuclear structures after induction of DNA damage. We found that a fraction of the Mre11 complex was bound to promyelocyte leukemia protein bodies in undamaged cells. Within 10 min after gamma irradiation, nuclear retention of the Mre11 complex in small granular foci was observed and persisted until 2 h postirradiation. In light of the previous demonstration that the Mre11 complex associated with ionizing radiation (IR)-induced DSBs, we infer that the protein retained under these conditions was associated with DNA damage. We also observed increased retention of Rad51 following IR treatment, although IR induced Rad51 foci were distinct from Mre11 foci. The ATM kinase, which phosphorylates Nbs1 during activation of the S-phase checkpoint, was not required for the Mre11 complex to associate with DNA damage. These data suggest that the functions of the Mre11 complex in the DSB response are implicitly dependent upon its ability to detect DNA damage.
The human Mre11 complex, consisting of Mre11, Rad50, and Nbs1, functions in diverse aspects of the cellular response to DNA damage (43). Previous cytologic analyses have provided evidence that this complex associates with damaged DNA. This aspect of Mre11 complex function is diminished in human Mre11 complex mutants that exhibit S-phase checkpoint deficiency (44). We have therefore hypothesized that the complex influences the S-phase checkpoint either because it is itself a sensor of DNA damage or because it functions in close proximity to a sensor (8, 48).
In previous studies, we observed that the formation of ionizing radiation (IR)-induced foci (IRIF) of Mre11 complex proteins was strictly dependent upon the prior induction of double-strand breaks (DSBs). IRIF formation and IRIF multiplicity were influenced by the number of DSBs as well as by mutations affecting the DSB response. On that basis, the cytology of the Mre11 complex was inferred to reflect its role in the response to DSBs (35). Indeed, Mre11 complex IRIF formation has been used as an index of the DNA damage response in a variety of contexts (8, 12, 17, 19, 24, 33, 48, 49, 53, 54).
The interpretation that DSB-induced formation of Mre11 complex IRIF reflected an interaction with DNA damage was supported by the technique of partial-volume irradiation. This methodology permits the induction of DSBs in discrete subnuclear volumes and was used to demonstrate that the complex associates with DSBs while DSB repair is ongoing (39). In contrast, under the conditions previously employed, IRIF were detectable only at time points when the vast majority of DSB repair was complete. Thus, we have proposed that these structures represent Mre11 complex accumulation at irreparable or slowly repaired lesions (35). As such, IRIF observed at later time points provide indirect information regarding the DSB response and likely do not occur at sites of DNA damage or ongoing DSB repair.
Having used partial-volume irradiation to establish that Mre11 complex proteins bind damaged DNA during the time course of DSB repair, a method of in situ fractionation was adapted to analyze damaged cells at earlier time points. We reasoned that after IR, only a subset of the total Mre11 complex would be associated with DNA damage and the remaining nucleoplasmic pool would obscure the damage-associated protein from immunofluorescent detection.
We found that after detergent extraction, relocalization of Mre11 complex proteins was readily observed as early as 10 min post IR. The kinetics of IR-induced Mre11 complex relocalization in extracted cells were similar to those observed in partial-volume irradiation (39). Thus, we interpret this relocalization to reflect association of the complex with DNA damage. Surprisingly, Ku70, a protein suggested to bind DNA ends from in vitro analyses (18, 50), did not relocalize in response to IR, nor did it colocalize with the Mre11 complex. The ATM kinase phosphorylates Nbs1 in response to DNA damage, and this event is required for activation of the S-phase checkpoint (17, 32, 51, 53). In this report, we show that ATM does not influence the complex's ability to relocalize in response to DSB induction. This result suggests that ATM phosphorylation of Nbs1 during S-phase checkpoint activation occurs after the Mre11 complex has associated with damaged DNA.
MATERIALS AND METHODS
Cell lines.
Human 37Lu and IMR90 primary diploid fibroblasts were obtained from the American Type Culture Collection and were used at passages 9 to 15 and 12 to 17, respectively. 180BR primary fibroblasts were obtained from C. Arlett (MRC Cell Mutation Unit, Brighton, United Kingdom) and were used at passages 14 to 20. Ataxia-telangiectasia (A-T) primary fibroblasts (AT3BI) were obtained from J. Murnane (University of California San Francisco) and were used at passages 16 to 20. All cells were grown at 37°C in 5% CO2 in Dulbecco modified Eagle medium with 10% fetal calf serum (FCS) and were negative in monthly mycoplasma tests.
Antibodies.
All of the primary immunologic reagents used were directed against human proteins. Affinity purification of Mre11 and Rad50 rabbit antisera were previously described (11). Nbs1 rabbit antiserum and Mre11 monoclonal antibody (MAb) 8F3 were also previously described (8), as was Nbs1 monoclonal antibody (48). Mouse anti-PML (promyelocytic leukemia protein) MAb was from Santa Cruz. Rabbit Rad51 antiserum was a kind gift from A. Shinohara (Osaka University, Osaka, Japan), mouse anti-Ku70 MAb N3H10 and anti-Ku86 MAb N9C1 were obtained from R. Burgess, University of Wisconsin-Madison. All secondary antibodies were from Jackson Immunoresearch Laboratories.
Immunofluorescence and in situ cell fractionation.
Cells were seeded onto 18-mm glass coverslips. At 48 h later, cells were gamma irradiated at 12 Gy in a Mark I 137Cs source at 2.5 Gy/min or mock irradiated. After various recovery times, the cells were processed for immunofluorescent staining. Before fixation, the in situ cell fractionation procedure was performed as previously described (40). Briefly, the coverslips were washed in phosphate-buffered saline (PBS), incubated in cytoskeleton buffer [10 mM piperazine-N,N′-bis(2-ethanesulfonic acid) (PIPES; pH 6.8), 100 mM NaCl, 300 mM sucrose, 3 mM MgCl2, 1 mM EGTA, 0.5% Triton X-100] for 5 min on ice, followed by incubation in cytoskeleton stripping buffer (10 mM Tris-HCl [pH 7.4], 10 mM NaCl, 3 mM MgCl2, 1% [vol/vol] Tween 40, 0.5% [vol/vol] sodium deoxycholate) for 5 min on ice. After several washes with ice-cold PBS, the cells were fixed in modified Streck tissue fixative for 30 min and permeabilized in 0.5% Triton X-100 solution for 15 min at room temperature as previously described (29). Cells were blocked with 10% FCS in PBS and incubated with primary antibody for 1 h and with secondary antibody for 30 min at room temperature. All antibodies were diluted in 5% FCS–PBS. Cells were then washed, counterstained with 4′,6′-diamidino-2-phenylindole (DAPI), and mounted as previously described (35). Primary antibody dilutions were as follows: Mre11, Nbs1, and Rad50 antisera, 1:300, 1:1,000, and 1:200 respectively; mouse anti-Mre11 and anti-Nbs1, both 1:200; mouse anti-PML, 1:400; Rad51 antiserum, 1:200; mouse anti-Ku70 and anti-Ku86, both 1:400. All secondary antibodies were used at 1:150, except goat anti-rabbit immunoglobulin G-Texas Red, which was used at 1:1,000. Controls for double labeling were performed in which one of the primary antibodies was omitted or replaced with preimmune serum.
Most of the results presented here were obtained by immunostaining with Mre11 antiserum. However, similar results were obtained with Mre11 MAb, both Nbs1 MAb and polyclonal antibody, and Rad50 antiserum.
Images were captured by using a charge-coupled device camera (Princeton Instruments, Trenton, N.J.), and grayscale images were processed by using IP Labs software (Signal Analytics Corporation, Vienna, Va.) and Photoshop 5.5 (Adobe, San Jose, Calif.).
RESULTS
We adapted an in situ fractionation procedure to examine functional relationships between the Mre11 complex and other proteins involved in the response to DSBs. These experiments provide novel insight because they compare the intranuclear dispositions of DSB response proteins while DNA damage is present and DSB repair is ongoing.
Three distinct localization patterns of the Mre11 complex in response to DNA damage.
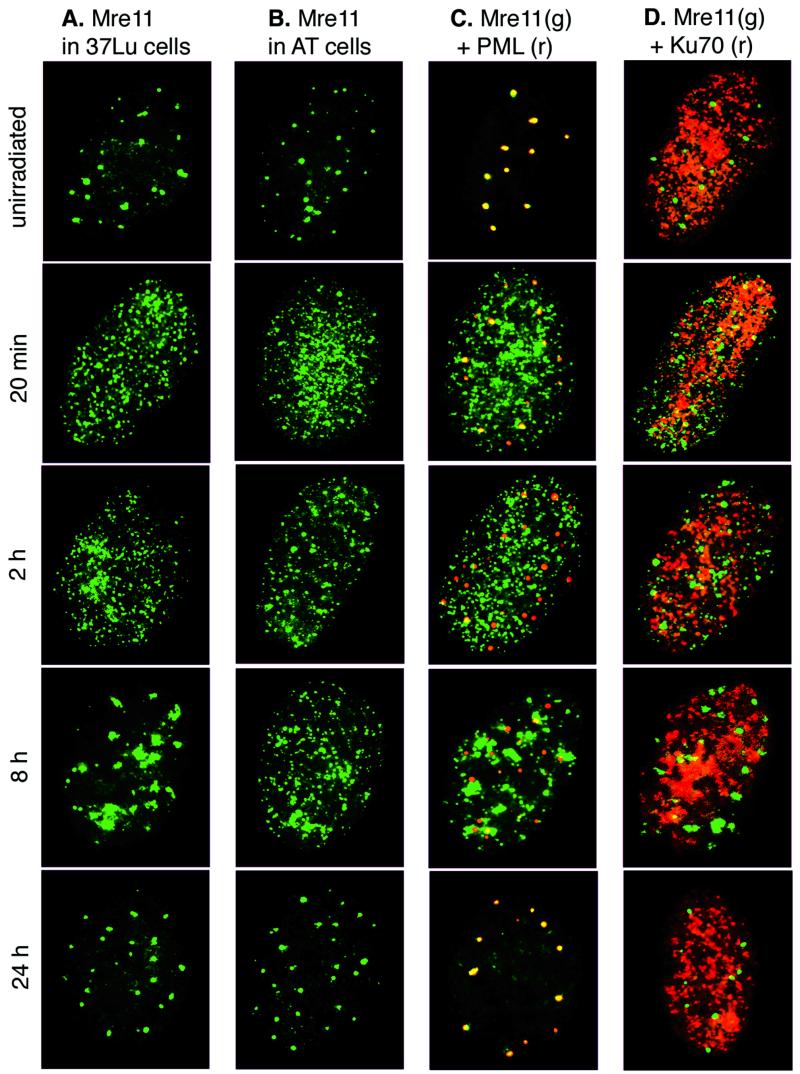
We used indirect immunofluorescence to observe the Mre11 complex following in situ fractionation of primary human fibroblasts. 37Lu and IMR90 normal human diploid fibroblasts were gamma irradiated and detergent extracted prior to fixation. Subsequent indirect immunofluorescence analysis with Mre11 and Nbs1 antisera revealed three distinct localization patterns for the proteins of the Mre11 complex.
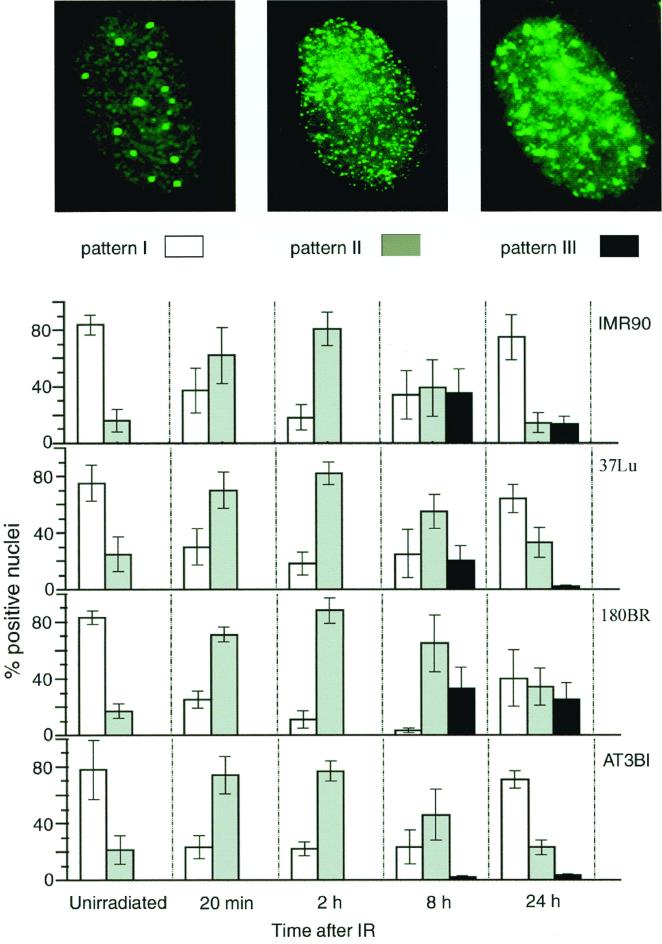
The majority (60 to 90%) of unirradiated cells contained large aggregates of Mre11, Rad50, and Nbs1. These structures also stain with antibody to PML, in agreement with previous studies (34) (Fig. 1 and 2A and C); Nbs1 and Rad50 data not shown). In contrast, from 20 to 120 min after IR, a punctate pattern of evenly distributed IRIF was observed (pattern II, gray bars in Fig. 1). The increase in pattern II IRIF was associated with a concomitant decrease in Mre11 staining of PML bodies (i.e., the type I pattern) (Fig. 1 and 2C). Increased numbers of cells exhibiting pattern II were detectable as early as 10 min after IR and by 20 min post IR, 50 to 80% of cells exhibited this pattern, with a maximum at 2 h post IR (70 to 95% of cells). At 2 h, the type II pattern began to change and Mre11 foci often looked larger and more sparse than at 20 min post IR (Fig. 2A). Based on previous assessments of DSB repair kinetics in IMR90 and 37Lu (35, 39), the formation of pattern II coincides temporally with DSB repair. Mre11 complex association with PML bodies decreased substantially following DNA damage, and by 2 h post IR, most PML bodies did not stain with Mre11 antiserum (Fig. 2C). We did not detect any changes in multiplicity or size of PML bodies following response to IR.
FIG. 1.
Kinetics of Mre11 complex relocalization in response to DNA damage. 37Lu, IMR90 (normal diploid fibroblasts), 180BR (DNA ligase IV-deficient fibroblasts), or AT3BI (ATM-deficient fibroblasts) cells were either irradiated at 12 Gy or mock irradiated and processed for immunofluorescent staining with Mre11 antiserum at the indicated time points following IR. The percentages of cells displaying patterns I (white bars), II (gray bars), and III (black bars) were calculated after scoring at least 200 nuclei for each time point. Data are the mean ± the standard deviation of three to six independent experiments.
FIG. 2.
Mre11 complex relocalization in normal and mutant cells. Cells were irradiated or mock treated and processed for single immunofluorescent staining with Mre11 antiserum (A and B) or for double staining with Mre11 antiserum (green) and PML (red) (C) or Ku70 (red) (D) MAb at the indicated time points after IR in normal human 37Lu fibroblasts (A, C, and D) or AT3BI A-T fibroblasts (B). Grayscale images were pseudocolored using IP Labs software and superimposed in Adobe Photoshop 5.5.
At 8 h post IR, the type II staining pattern decreased and the pattern III of Mre11 localization appeared (black bars in Fig. 1). The large, irregularly shaped foci that define this time point were identical to IRIF (35). IRIF appear to result from aggregation of type II pattern foci or by accretion of Mre11 complex proteins to a subset of the type II IRIF present at early time points (≤2 h). IRIF observed at 8 h did not coincide with PML bodies, and at 24 h post IR, Mre11 complex proteins resumed their association with PML bodies (Fig. 2C).
In summary, we observed a cyclical progression of Mre11 complex localization that began with the type I PML-associated pattern in unirradiated cells, followed by the formation of type II IRIF within 10 to 20 min after IR treatment (Fig. 1 and 2A and C). By 8 h, these structures appeared to coalesce into type III IRIF, which ultimately disappear so that at 24 h post IR, the Mre11 complex was again seen associated with PML bodies.
The Mre11 complex is cytologically distinct from Ku70, Ku86, and Rad51.
Genetic analysis of Saccharomyces cerevisiae has clearly demonstrated that the Mre11 complex functions in recombinational DNA repair, affecting both homologous recombination and nonhomologous end joining (NHEJ) (23, 43). In contrast, the Ku heterodimer appears to function specifically in NHEJ (9), whereas Rad51 is uniquely required for DSB repair by homologous recombination (27). We asked whether these disparate functions would manifest as distinct cytologic behaviors in IR-treated cells.
In unirradiated cells, in situ cell fractionation revealed a punctate staining of Ku70 and Ku86 evenly distributed throughout the nucleus. Ku70 (Fig. 2D) and Ku86 (data not shown) remained in essentially the same dispersed punctate pattern following irradiation, in contrast to Mre11 complex proteins. Similarly, the Ku complex in nonextracted cells was uniformly distributed in the nucleus irrespective of prior IR treatment (data not shown).
Under the conditions used, IR-induced alterations in the disposition of the Ku complex were not detected. Nevertheless, the role of the Mre11 complex in NHEJ suggested by analysis of S. cerevisiae mre11 mutants prompted us to assess whether IR-dependent Mre11 complex foci would colocalize with Ku70 or Ku86. Irradiated cells were fractionated, fixed, and doubly stained with Ku70 MAb and Mre11 antiserum. Ku70 and Mre11 did not colocalize at PML bodies in unirradiated cells or at other nuclear structures at any time points following irradiation (Fig. 2D; Ku86 data not shown).
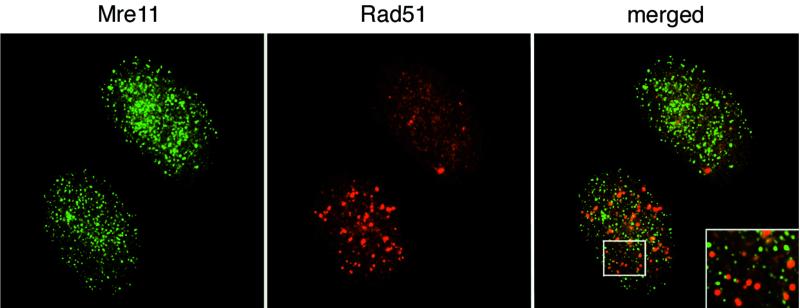
We previously showed that Rad51 and Mre11 IRIF did not colocalize nor were they coincident within the nucleus of a given cell at 8 h post IR (35). This result was somewhat surprising in light of the Mre11 complex's role in homologous recombination (6, 23). We asked whether Mre11 and Rad51 foci would colocalize in detergent-extracted cells at time points coinciding with ongoing DSB repair. Rad51 staining was not altered at 20 min post IR. However, at 2 and 8 h after IR, about 20% of cells contained Rad51 nuclear foci, as shown previously in nonextracted cells (22) (Fig. 3 and Table 1). Whereas the majority of cells (70 to 95%) showed relocalization of Mre11 in type II IRIF between 20 and 120 min post IR, the reorganization of Rad51 into foci occurred only in a subset (about 20%) of these cells over this time course (Table 1). We did not observe significant colocalization of Mre11 and Rad51 IRIF (Fig. 3).
FIG. 3.
Cytologic behavior of Mre11 and Rad51 proteins. 37Lu cells were irradiated and processed for double immunofluorescent staining with Rad51 antiserum and Mre11 MAb 8F3 2 h post IR. Note that although both cell types display pattern II of Mre11 localization, only one of them is positive for Rad51 foci. An enlarged fragment of this cell is shown as the insert to demonstrate the lack of colocalization of Mre11 and Rad51 foci.
TABLE 1.
Results of double immunostaining of 37Lu cells for Mre11 and Rad51 following DNA damagea
| Nuclear foci | Untreated | Time after IR
|
|||
|---|---|---|---|---|---|
| 20 min | 2 h | 8 h | 24 h | ||
| Mre11 type II pattern | 17 ± 9 | 60 ± 8 | 73 ± 5 | 52 ± 3 | 30 ± 15 |
| Rad51 | 3 ± 3 | 3 ± 3 | 28 ± 14 | 20 ± 8 | 4 ± 2 |
Data are expressed as percentages of cells displaying the indicated pattern. At least 200 nuclei were scored for each experiment to obtain percentage values. Data are the mean of four independent experiments ± the standard deviation.
When comparing the Rad51 and Mre11 complex responses, it is important to note that Rad51 levels are very low in G1 cells and are not induced by IR treatment (4, 15, 52). In contrast, the levels of Mre11 and Rad50 are relatively high and constant irrespective of cell cycle phase or the presence of DNA damage (11, 35). Hence, many fewer cells in an asynchronous culture are competent to form Rad51 as opposed to Mre11 foci. Consistent with previous data obtained with nonextracted cells (35, 39), Mre11 and Rad51 foci did not colocalize after detergent pretreatment of cells (Fig. 3). Coincident formation of Mre11 and Rad51 foci in irradiated cells was not observed in previous studies of nonextracted cells (35). The reduction in background staining afforded by pre-extraction of nucleoplasmic protein may increase the sensitivity of immunofluorescent detection and permit detection of Mre11 and Rad51 foci. Nevertheless, the general lack of colocalization is consistent with the distinct roles of the Mre11 complex and Rad51 in the DSB response.
Genetic determinants of cytologic behavior.
The Mre11 complex influences diverse endpoints in the cellular DNA damage response, including DNA recombination and cell cycle regulation (6, 23, 44). In the latter functions, the Mre11 complex and the ATM kinase collaborate to effect cell cycle checkpoint functions in S phase (44). To assess whether Mre11 complex relocalization was dependent upon ATM, we undertook cytologic analysis of cells with defects in DNA ligase IV and in the ATM kinase following detergent extraction.
The DNA ligase IV mutant cell line 180BR is profoundly DSB repair deficient (1, 2, 45). We previously showed that the half-life of IR-induced DSBs and associated relocalization of Mre11 were markedly increased in this cell line (35, 39). Consistent with the observation that the formation of type II IRIF is concomitant with DSB repair, we found that the progression from type II foci into PML-associated pattern I occurred more slowly in 180BR than in normal cells. At 8 h postirradiation, 15 to 30% of the normal cells returned to their unirradiated pattern of Mre11 localization, PML-associated pattern I. In contrast, PML-associated pattern I was not observed in 180BR cells at this time point and at 24 h, a substantial fraction of the IR-treated 180BR cells remained type III IRIF positive (Fig. 1). Hence, the persistence of IR-induced DSBs is correlated with the persistence of IRIF.
To test whether ATM activity is required for the complex to bind DNA damage, we examined the kinetics of DNA damage-dependent Mre11 complex relocalization in AT3BI primary A-T fibroblasts. With regard to the formation of IRIF that appear over the time course of DNA repair (type II), Mre11 and Nbs1 relocalization in these A-T cells was indistinguishable from that in the wild type (Fig. 1 and 2A and B). However, we did not detect IRIF-positive AT3BI cells following detergent extraction, whereas IRIF were present in irradiated cultures of the same cells without extraction (data not shown). In previous studies of nonextracted cells, we found that IRIF formation was severely reduced in simian virus 40-transformed A-T cells whereas primary A-T cells were only minimally affected (35, 48). Therefore, we conclude that IRIF did form in AT3BI cells but were removed by detergent extraction during in situ fractionation. Given the lack of an effect on the formation of type II IRIF, ATM-mediated Nbs1 phosphorylation does not appear to influence DNA damage recognition by the Mre11 complex.
DISCUSSION
In situ fractionation was carried out on gamma-irradiated cells to assess alterations of the intranuclear disposition of proteins that mediate recombinational DNA repair. The Mre11 complex and the Ku heterodimer exhibit distinct cytologic behaviors during the cellular response to DSBs. The data further indicate that association of the Mre11 complex with DNA damage does not require the ATM kinase.
Early versus late IRIF.
The formation of Mre11 complex late IRIF occurs in the normal cellular response to DSBs. Conversely, this process is affected by mutations that alter the cellular DSB response (8, 35, 48). These observations support the utility of IRIF formation as an index of the cellular DSB response. An important caveat to this interpretation is that late IRIF formation is not readily detectable until the bulk of DSB repair is complete (4 to 8 h). By using ultrasoft X rays to induce localized DNA damage, the Mre11 complex has been directly linked to sites of DNA damage within the time course of DSB repair (30 to 90 min) (39). However, this methodology is limiting because it precludes analysis prior to 30 min post IR, precludes manipulations such as cell synchrony or microinjection, and requires a synchrotron to generate ultrasoft X rays.
Based on the anticipation that Mre11 complex members and other proteins that function in DSB detection or repair would become more avidly associated with chromatin in the presence of DNA damage, we adapted a method of detergent extraction to examine gamma-irradiated cells. Relocalization of the Mre11 complex occurred within 10 min of DSB induction and persisted until approximately 2 h post IR. The type II pattern is also seen in unirradiated cells within S phase, suggesting that reorganization of the complex is induced by naturally occurring breaks during DNA replication (R. S. Maser et al., unpublished data). Interestingly, the formation of early (i.e., type II) IRIF is accompanied by a decrease in Mre11 complex staining of PML bodies (Fig. 2C). The function(s) of PML bodies is not clearly established, nor is it clear that their composition is uniform in all cell types and under all cellular conditions. A number of enzymes with protein modification activities reside in PML bodies (30, 36). Therefore, it is conceivable that the departure of Mre11 complex members from PML bodies is attributable to DNA damage-induced modification of Mre11 complex or other PML-associated proteins.
Prior to extraction, the majority of the Mre11 complex was present in the nucleoplasm based on analysis of extracts prepared under conditions identical to those employed for in situ fractionation (data not shown). Hence, Mre11 complex proteins that ultimately associate with DNA damage most likely originate in the nucleoplasmic pool. In this regard, the ideas that PML bodies serve as reservoirs for Mre11 complex proteins and that Mre11 proteins leave PML bodies to associate with DNA damage seem less favorable, although the data do not rigorously exclude these possibilities.
By 8 h, a subset of the early IRIF appeared to coalesce into large, irregularly shaped aggregates. This observation is consistent with the interpretation that IRIF seen in nonextracted cells at late time points represent irreparable or slowly repaired lesions that nucleate multimerization of the Mre11 complex (35). In this regard, it is interesting that the time course over which chromosomal aberrations accumulate in irradiated cells corresponds well to the 6- to 12-h time course over which IRIF appear in nonextracted cells (7, 35).
In DSB repair-deficient 180BR cells, the half-life of IR-induced DSBs is markedly increased (2) and the transition from type II IRIF to the PML-associated pattern is delayed in those cells (Fig. 1). This validates the interpretation that type II IRIF correspond to sites of DNA damage and repair and suggests that the formation of IRIF is dependent upon the process of DSB repair.
Independent functions of the Mre11 and Ku complexes.
The role of the Ku complex in NHEJ is firmly established by in vitro and in vivo analyses (3, 28, 37). The observation that Ku heterodimers bind DNA ends in vitro and activate the DNA-dependent protein kinase catalytic subunit strongly suggests the mechanism by which these proteins function in intact cells to facilitate NHEJ (13, 14, 25, 47). In this context, our inability to detect IR-induced relocalization of the Ku complex is unexpected. It is conceivable that the presumptive Ku-DNA complex is not stable under the extraction conditions employed. However, a substantial amount Ku was retained after extraction, irrespective of prior irradiation. This associated fraction may represent Ku complex functions not directly related to DNA damage processing (10, 14, 16, 20, 21, 42).
A recent report presented evidence that hamster Mre11 and Ku colocalized at radiation-induced foci and that the mouse Mre11 and Ku proteins physically interacted (19). Given that we failed to detect relocalization of Ku complex proteins by in situ fractionation in human cells, it is perhaps not surprising that we failed to observe colocalization of Mre11 at Ku foci in these experiments. This may reflect a difference in the reagents used or possibly in the properties of rodent and human Ku complex proteins. It is clear from genetic analyses of S. cerevisiae that the yeast Mre11 and Ku complexes function in distinct pathways (5, 6, 31, 41). Crosses between Ku-deficient mice and murine Mre11 complex mutants will be important to address the functional interactions between these complexes in mammals.
Mre11 complex association with DNA damage is ATM independent.
Mutations in MRE11 and NBS1 comprise the genetic basis for two human syndromes that bear phenotypic similarity to A-T. The A-T-like disorder (A-TLD) and Nijmegen breakage syndrome (NBS) are caused by mutations in the MRE11 and NBS1 genes, respectively (8, 48). The mechanistic basis for the phenotypic similarities of A-T and NBS and A-TLD cells is unclear. It has been established that ATM phosphorylates Nbs1 (17, 32, 51, 53) and that this event is necessary for activation of the S-phase checkpoint (32, 53). In NBS and A-TLD cells, the formation of Mre11, as well as Nbs1 IRIF, is virtually abrogated (8, 48). Similarly, we were unable to detect the formation of early (type II) IRIF of either Mre11 or Nbs1 in these cells (data not shown). In NBS and A-TLD cells, the Mre11 complex is predominantly cytoplasmic; thus, the failure to form foci when breaks are present may be due in part to the complex's aberrant subcellular localization (44).
In contrast, the subcellular localization of the Mre11 complex is normal in A-T cells (R. S. Maser, unpublished data). The formation of Mre11 (Fig. 3) and Nbs1 (data not shown) type II IRIF in that context was indistinguishable from that of wild-type cells. This result argues that ATM-mediated Nbs1 phosphorylation is not required for the Mre11 complex to associate with DNA damage. These data support the hypothesis that the influence of the Mre11 complex on S-phase checkpoint activation is dependent upon DNA damage detection, as well as Nbs1 phosphorylation. This raises the possibility that ATM phosphorylates Nbs1 that is bound to DSBs, as opposed to nucleoplasmic Nbs1. Finally, because DSB repair is not grossly affected in A-T cells, phosphorylation of Nbs1 does not appear relevant to the DSB repair functions of the complex (26, 38, 46).
Conclusion.
We used in situ fractionation and immunofluorescence to analyze functional relationships among proteins with demonstrated roles in the cellular response to DNA damage. The primary advantage of this approach is that it facilitates cytologic examination of DSB repair using conventional irradiation while DSBs are present. Beyond providing further support for the hypothesis that the Mre11 complex detects DNA damage, the data define distinct functional roles in this regard for the Mre11 complex and the Ku complex. Finally, the data demonstrate that the ability of the Mre11 complex to bind DNA damage does not depend upon the ATM kinase.
ACKNOWLEDGMENTS
We are grateful to the members of the lab for insights throughout the course of this study and to Mark Kaplan and Mary Ellen Perry for comments and suggestions on the manuscript.
This work was supported by the Milwaukee Foundation, the National Institutes of Health (GM56888 and GM59413), and the Department of Energy (ER62859).
Footnotes
Report 3564 from the University of Wisconsin-Madison Laboratory of Genetics.
REFERENCES
- 1.Badie C, Iliakis G, Foray N, Alsbeih G, Cedervall B, Chavaudra N, Pantelias G, Arlett C, Malaise E P. Induction and rejoining of DNA double-strand breaks and interphase chromosome breaks after exposure to X rays in one normal and two hypersensitive human fibroblast cell lines. Radiat Res. 1995;144:26–35. [PubMed] [Google Scholar]
- 2.Badie C, Iliakis G, Foray N, Alsbeih G, Pantellias G E, Okayasu R, Cheong N, Russell N S, Begg A C, Arlett C F, et al. Defective repair of DNA double-strand breaks and chromosome damage in fibroblasts from a radiosensitive leukemia patient. Cancer Res. 1995;55:1232–1234. [PubMed] [Google Scholar]
- 3.Baumann P, West S C. DNA end-joining catalyzed by human cell-free extracts. Proc Natl Acad Sci USA. 1998;95:14066–14070. doi: 10.1073/pnas.95.24.14066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bishop D K, Ear U, Bhattacharyya A, Calderone C, Beckett M, Weichselbaum R R, Shinohara A. Xrcc3 is required for assembly of Rad51 complexes in vivo. J Biol Chem. 1998;273:21482–21488. doi: 10.1074/jbc.273.34.21482. [DOI] [PubMed] [Google Scholar]
- 5.Boulton S J, Jackson S P. Saccharomyces cerevisiae Ku70 potentiates illegitimate DNA double-strand break repair and serves as a barrier to error-prone DNA repair pathways. EMBO J. 1996;15:5093–5103. [PMC free article] [PubMed] [Google Scholar]
- 6.Bravo R, Macdonald-Bravo H. Existence of two populations of cyclin/proliferating cell nuclear antigen during the cell cycle: association with DNA replication sites. J Cell Biol. 1987;105:1549–1554. doi: 10.1083/jcb.105.4.1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bressan D A, Baxter B K, Petrini J H J. The Mre11-Rad50-Xrs2 protein complex facilitates homologous recombination-based double-strand break repair in Saccharomyces cerevisiae. Mol Cell Biol. 1999;19:7681–7687. doi: 10.1128/mcb.19.11.7681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brown J M, Evans J W, Kovacs M S. Mechanism of chromosome exchange formation in human fibroblasts: insights from “chromosome painting.”. Environ Mol Mutagen. 1993;22:218–224. doi: 10.1002/em.2850220407. [DOI] [PubMed] [Google Scholar]
- 9.Carney J P, Maser R S, Olivares H, Davis E M, Le Beau M, Yates J R, 3rd, Hays L, Morgan W F, Petrini J H J. The hMre11/hRad50 protein complex and Nijmegen breakage syndrome: linkage of double-strand break repair to the cellular DNA damage response. Cell. 1998;93:477–486. doi: 10.1016/s0092-8674(00)81175-7. [DOI] [PubMed] [Google Scholar]
- 10.Critchlow S E, Jackson S P. DNA end-joining: from yeast to man. Trends Biochem Sci. 1998;23:394–398. doi: 10.1016/s0968-0004(98)01284-5. [DOI] [PubMed] [Google Scholar]
- 11.Difilippantonio M J, Zhu J, Chen H T, Meffre E, Nussenzweig M C, Max E E, Ried T, Nussenzweig A. DNA repair protein Ku80 suppresses chromosomal aberrations and malignant transformation. Nature. 2000;404:510–514. doi: 10.1038/35006670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dimitrova D S, Todorov I T, Melendy T, Gilbert D M. Mcm2, but not RPA, is a component of the mammalian early G1-phase prereplication complex. J Cell Biol. 1999;146:709–722. doi: 10.1083/jcb.146.4.709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dolganov G M, Maser R S, Novikov A, Tosto L, Chong S, Bressan D A, Petrini J H J. Human Rad50 is physically associated with hMre11: identification of a conserved multiprotein complex implicated in recombinational DNA repair. Mol Cell Biol. 1996;16:4832–4841. doi: 10.1128/mcb.16.9.4832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dong Z, Zhong Q, Chen P L. The Nijmegen breakage syndrome protein is essential for Mre11 phosphorylation upon DNA damage. J Biol Chem. 1999;274:19513–19516. doi: 10.1074/jbc.274.28.19513. [DOI] [PubMed] [Google Scholar]
- 15.Featherstone C, Jackson S P. DNA double-strand break repair. Curr Biol. 1999;9:R759–R761. doi: 10.1016/S0960-9822(00)80005-6. [DOI] [PubMed] [Google Scholar]
- 16.Featherstone C, Jackson S P. Ku, a DNA repair protein with multiple cellular functions? Mutat Res. 1999;434:3–15. doi: 10.1016/s0921-8777(99)00006-3. [DOI] [PubMed] [Google Scholar]
- 17.Flygare J, Benson F, Hellgren D. Expression of the human RAD51 gene during the cell cycle in primary human peripheral blood lymphocytes. Biochim Biophys Acta. 1996;1312:231–236. doi: 10.1016/0167-4889(96)00040-7. [DOI] [PubMed] [Google Scholar]
- 18.Gao Y, Sun Y, Frank K M, Dikkes P, Fujiwara Y, Seidl K J, Sekiguchi J M, Rathbun G A, Swat W, Wang J, Bronson R T, Malynn B A, Bryans M, Zhu C, Chaudhuri J, Davidson L, Ferrini R, Stamato T, Orkin S H, Greenberg M E, Alt F W. A critical role for DNA end-joining proteins in both lymphogenesis and neurogenesis. Cell. 1998;95:891–902. doi: 10.1016/s0092-8674(00)81714-6. [DOI] [PubMed] [Google Scholar]
- 19.Gatei M, Young D, Cerosaletti K M, Desai-Mehta A, Spring K, Kozlov S, Lavin M F, Gatti R A, Concannon P, Khanna K. ATM-dependent phosphorylation of nibrin in response to radiation exposure. Nat Genet. 2000;25:115–119. doi: 10.1038/75508. [DOI] [PubMed] [Google Scholar]
- 20.Getts R C, Stamato T D. Absence of a Ku-like DNA end binding activity in the xrs double-strand DNA repair-deficient mutant. J Biol Chem. 1994;269:15981–15984. [PubMed] [Google Scholar]
- 21.Goedecke W, Eijpe M, Offenberg H H, van Aalderen M, Heyting C. Mre11 and Ku70 interact in somatic cells, but are differentially expressed in early meiosis. Nat Genet. 1999;23:194–198. doi: 10.1038/13821. [DOI] [PubMed] [Google Scholar]
- 22.Gu Y, Seidl K J, Rathbun G A, Zhu C, Manis J P, van der Stoep N, Davidson L, Cheng H L, Sekiguchi J M, Frank K, Stanhope-Baker P, Schlissel M S, Roth D B, Alt F W. Growth retardation and leaky SCID phenotype of Ku70-deficient mice. Immunity. 1997;7:653–665. doi: 10.1016/s1074-7613(00)80386-6. [DOI] [PubMed] [Google Scholar]
- 23.Gu Y, Sekiguchi J, Gao Y, Dikkes P, Frank K, Ferguson D, Hasty P, Chun J, Alt F W. Defective embryonic neurogenesis in Ku-deficient but not DNA-dependent protein kinase catalytic subunit-deficient mice. Proc Natl Acad Sci USA. 2000;97:2668–2673. doi: 10.1073/pnas.97.6.2668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Haaf T, Golub E I, Reddy G, Radding C M, Ward D C. Nuclear foci of mammalian Rad51 recombination protein in somatic cells after DNA damage and its localization in synaptonemal complexes. Proc Natl Acad Sci USA. 1995;92:2298–2302. doi: 10.1073/pnas.92.6.2298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Haber J E. The many interfaces of Mre11. Cell. 1998;95:583–586. doi: 10.1016/s0092-8674(00)81626-8. [DOI] [PubMed] [Google Scholar]
- 26.Ito A, Tauchi H, Kobayashi J, Morishima K, Nakamura A, Hirokawa Y, Matsuura S, Ito K, Komatsu K. Expression of full-length NBS1 protein restores normal radiation responses in cells from Nijmegen breakage syndrome patients. Biochem Biophys Res Commun. 1999;265:716–721. doi: 10.1006/bbrc.1999.1737. [DOI] [PubMed] [Google Scholar]
- 27.Jin S, Weaver D T. Double-strand break repair by Ku70 requires heterodimerization with Ku80 and DNA binding functions. EMBO J. 1997;16:6874–6885. doi: 10.1093/emboj/16.22.6874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jorgensen T J, Shiloh Y. The ATM gene and the radiobiology of ataxia-telangiectasia. Int J Radiat Biol. 1996;69:527–537. doi: 10.1080/095530096145535. [DOI] [PubMed] [Google Scholar]
- 29.Kanaar R, Hoeijmakers J H, van Gent D C. Molecular mechanisms of DNA double strand break repair. Trends Cell Biol. 1998;8:483–489. doi: 10.1016/s0962-8924(98)01383-x. [DOI] [PubMed] [Google Scholar]
- 30.Kanaar R, Hoeijmakers J H J. Recombination and joining: different means to the same ends. Genes Funct. 1997;1:165–174. doi: 10.1046/j.1365-4624.1997.00016.x. [DOI] [PubMed] [Google Scholar]
- 31.Kodym R, Hurth E. Determination of radiation-induced DNA strand breaks in individual cells by non-radioactive labelling of 3′ OH ends. Int J Radiat Biol. 1995;68:133–139. doi: 10.1080/09553009514551031. [DOI] [PubMed] [Google Scholar]
- 32.Kretz-Remy C, Tanguay R M. SUMO/sentrin: protein modifiers regulating important cellular functions. Biochem Cell Biol. 1999;77:299–309. [PubMed] [Google Scholar]
- 33.Lee S E, Moore J K, Holmes A, Umezu K, Kolodner R D, Haber J E. Saccharomyces Ku70, Mre11/Rad50, and RPA proteins regulate adaptation to G2/M arrest after DNA damage. Cell. 1998;94:399–409. doi: 10.1016/s0092-8674(00)81482-8. [DOI] [PubMed] [Google Scholar]
- 34.Lim D S, Kim S T, Xu B, Maser R S, Lin J, Petrini J H J, Kastan M B. ATM phosphorylates p95/Nbs1 in an S-phase checkpoint pathway. Nature. 2000;404:613–617. doi: 10.1038/35007091. [DOI] [PubMed] [Google Scholar]
- 35.Liu Y, Li M, Lee E Y, Maizels N. Localization and dynamic relocalization of mammalian Rad52 during the cell cycle and in response to DNA damage. Curr Biol. 1999;9:975–978. doi: 10.1016/s0960-9822(99)80427-8. [DOI] [PubMed] [Google Scholar]
- 36.Lombard D B, Guarente L. Nijmegen breakage syndrome disease protein and MRE11 at PML nuclear bodies and meiotic telomeres. Cancer Res. 2000;60:2331–2334. [PubMed] [Google Scholar]
- 37.Maser R S, Monsen K J, Nelms B E, Petrini J H J. hMre11 and hRad50 nuclear foci are induced during the normal cellular response to DNA double-strand breaks. Mol Cell Biol. 1997;17:6087–6096. doi: 10.1128/mcb.17.10.6087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Maul G G, Negorev D, Bell P, Ishov A M. Review: properties and assembly mechanisms of ND10, PML bodies, or PODs. J Struct Biol. 2000;129:278–287. doi: 10.1006/jsbi.2000.4239. [DOI] [PubMed] [Google Scholar]
- 39.Mittnacht S, Weinberg R A. G1/S phosphorylation of the retinoblastoma protein is associated with an altered affinity for the nuclear compartment. Cell. 1991;65:381–393. doi: 10.1016/0092-8674(91)90456-9. [DOI] [PubMed] [Google Scholar]
- 40.Moore J K, Haber J E. Cell cycle and genetic requirements of two pathways of nonhomologous end-joining repair of double-strand breaks in Saccharomyces cerevisiae. Mol Cell Biol. 1996;16:2164–2173. doi: 10.1128/mcb.16.5.2164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Murnane J P. Cell cycle regulation in response to DNA damage in mammalian cells: a historical perspective. Cancer Metastasis Rev. 1995;14:17–29. doi: 10.1007/BF00690208. [DOI] [PubMed] [Google Scholar]
- 42.Nelms B E, Maser R S, MacKay J F, Lagally M G, Petrini J H J. In situ visualization of DNA double-strand break repair in human fibroblasts. Science. 1998;280:590–592. doi: 10.1126/science.280.5363.590. [DOI] [PubMed] [Google Scholar]
- 43.Nickerson J A, Krockmalnic G, He D C, Penman S. Immunolocalization in three dimensions: immunogold staining of cytoskeletal and nuclear matrix proteins in resinless electron microscopy sections. Proc Natl Acad Sci USA. 1990;87:2259–2263. doi: 10.1073/pnas.87.6.2259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nugent C I, Bosco G, Ross L O, Evans S K, Salinger A P, Moore J K, Haber J E, Lundblad V. Telomere maintenance is dependent on activities required for end repair of double-strand breaks. Curr Biol. 1998;8:657–660. doi: 10.1016/s0960-9822(98)70253-2. [DOI] [PubMed] [Google Scholar]
- 45.Nussenzweig A, Chen C, da Costa Soares V, Sanchez M, Sokol K, Nussenzweig M C, Li G C. Requirement for Ku80 in growth and immunoglobulin V(D)J recombination. Nature. 1996;382:551–555. doi: 10.1038/382551a0. [DOI] [PubMed] [Google Scholar]
- 46.Petrini J H. The mammalian Mre11-Rad50-nbs1 protein complex: integration of functions in the cellular DNA-damage response. Am J Hum Genet. 1999;64:1264–1269. doi: 10.1086/302391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Petrini J H. The mre11 complex and ATM: collaborating to navigate S phase. Curr Opin Cell Biol. 2000;12:293–296. doi: 10.1016/s0955-0674(00)00091-0. [DOI] [PubMed] [Google Scholar]
- 48.Riballo E, Critchlow S E, Teo S H, Doherty A J, Priestley A, Broughton B, Kysela B, Beamish H, Plowman N, Arlett C F, Lehmann A R, Jackson S P, Jeggo P A. Identification of a defect in DNA ligase IV in a radiosensitive leukaemia patient. Curr Biol. 1999;9:699–702. doi: 10.1016/s0960-9822(99)80311-x. [DOI] [PubMed] [Google Scholar]
- 49.Shiloh Y. Ataxia-telangiectasia and the Nijmegen breakage syndrome: related disorders but genes apart. Annu Rev Genet. 1997;31:635–662. doi: 10.1146/annurev.genet.31.1.635. [DOI] [PubMed] [Google Scholar]
- 50.Smith G C, Jackson S P. The DNA-dependent protein kinase. Genes Dev. 1999;13:916–934. doi: 10.1101/gad.13.8.916. [DOI] [PubMed] [Google Scholar]
- 51.Stewart G, Maser R S, Stankovic T, Bressan D A, Kaplan M I, Jaspers N G J, Byrd P J, Petrini J H J, Taylor A M R. The DNA double strand break repair gene hMre11 is mutated in individuals with a new ataxia telangiectasia like disorder (ATLD) Cell. 1999;99:577–587. doi: 10.1016/s0092-8674(00)81547-0. [DOI] [PubMed] [Google Scholar]
- 52.Wang Y, Cortez D, Yazdi P, Neff N, Elledge S J, Qin J. BASC, a super complex of BRCA1-associated proteins involved in the recognition and repair of aberrant DNA structures. Genes Dev. 2000;14:927–939. [PMC free article] [PubMed] [Google Scholar]
- 53.Weaver D T. What to do at an end: DNA double-strand break repair. Trends Genet. 1995;11:388–392. doi: 10.1016/s0168-9525(00)89121-0. [DOI] [PubMed] [Google Scholar]
- 54.Wu X, Ranganathan V, Weisman D S, Heine W F, Ciccone D N, O'Neill T B, Crick K E, Pierce K A, Lane W S, Rathbun G, Livingston D M, Weaver D T. ATM phosphorylation of Nijmegen breakage syndrome protein is required in a DNA damage response. Nature. 2000;405:477–482. doi: 10.1038/35013089. [DOI] [PubMed] [Google Scholar]
- 55.Yamamoto A, Taki T, Yagi H, Habu T, Yoshida K, Yoshimura Y, Yamamoto K, Matsushiro A, Nishimune Y, Morita T. Cell cycle-dependent expression of the mouse Rad51 gene in proliferating cells. Mol Gen Genet. 1996;251:1–12. doi: 10.1007/BF02174338. [DOI] [PubMed] [Google Scholar]
- 56.Zhao S, Weng Y C, Yuan S S, Lin Y T, Hsu H C, Lin S C, Gerbino E, Song M H, Zdzienicka M Z, Gatti R A, Shay J W, Ziv Y, Shiloh Y, Lee E Y. Functional link between ataxia-telangiectasia and Nijmegen breakage syndrome gene products. Nature. 2000;405:473–477. doi: 10.1038/35013083. [DOI] [PubMed] [Google Scholar]
- 57.Zhong Q, Chen C F, Li S, Chen Y, Wang C C, Xiao J, Chen P L, Sharp Z D, Lee W H. Association of BRCA1 with the hRad50-hMre11–p95 complex and the DNA damage response. Science. 1999;285:747–750. doi: 10.1126/science.285.5428.747. [DOI] [PubMed] [Google Scholar]
- 58.Zhu X-D, Kuster B, Mann M, Petrini J H J, de Lange T. Cell cycle regulated association of Rad50/Mre11/Nbs1 with TRF2 and human telomeres. Nat Genet. 2000;25:347–352. doi: 10.1038/77139. [DOI] [PubMed] [Google Scholar]